Abstract
Background
Human immunodeficiency virus type 1 (HIV-1) sequence diversity and the presence of archived epitope mutations in antibody binding sites are a major obstacle for the clinical application of broadly neutralizing antibodies (bNAbs) against HIV-1. Specifically, it is unclear to what degree the viral reservoir is compartmentalized and if virus susceptibility to antibody neutralization differs across tissues.
Methods
The Last Gift cohort enrolled 7 people with HIV diagnosed with a terminal illness and collected antemortem blood and postmortem tissues across 33 anatomical compartments for near full-length env HIV genome sequencing. Using these data, we applied a Bayesian machine-learning model (Markov chain Monte Carlo–support vector machine) that uses HIV-1 envelope sequences and approximated glycan-occupancy information to quantitatively predict the half-maximal inhibitory concentrations (IC50) of bNAbs, allowing us to map neutralization resistance pattern across tissue reservoirs.
Results
Predicted mean susceptibilities across tissues within participants were relatively homogenous, and the susceptibility pattern observed in blood often matched what was predicted for tissues. However, selected tissues, such as the brain, showed evidence of compartmentalized viral populations with distinct neutralization susceptibilities in some participants. Additionally, we found substantial heterogeneity in the range of neutralization susceptibilities across tissues within and between individuals, and between bNAbs within individuals (standard deviation of log2(IC50) >3.4).
Conclusions
Blood-based screening methods to determine viral susceptibility to bNAbs might underestimate the presence of resistant viral variants in tissues. The extent to which these resistant viruses are clinically relevant, that is, lead to bNAb therapeutic failure, needs to be further explored.
Keywords: HIV, neutralization susceptibilities, bNAbs, tissues, prediction modeling
We report on the landscape of predicted human immunodeficiency virus type 1 (HIV-1) neutralization susceptibilities to broadly neutralizing antibodies across multiple anatomical compartments by applying a novel machine-learning model to tissue HIV-1 Envelope sequences collected postmortem from 7 participants in the Last Gift cohort.
Highly potent and broadly neutralizing antibodies (bNAbs) against human immunodeficiency virus type 1 (HIV-1) are being developed as therapeutic and prophylactic tools (reviewed in [1]). bNAbs have demonstrated that they can temporarily reduce plasma viral loads in viremic persons with HIV-1 (PWH) [2–4] or delay viral rebound once antiretroviral therapy (ART) is stopped. The rapid emergence of neutralization-resistant viral variants, however, frequently results in virological failure of bNAb treatment, creating a significant challenge for clinical development [5–7]. Neutralization-resistant breakthrough viruses are thought to emerge either from viral populations with preexisting bNAb epitope escape variants or due to viral recombination (as discussed by Cohen et al [8]). Furthermore, bulk peripheral blood mononuclear cell (PBMC) viral outgrowth cultures used for screening study participants have failed to detect preexisting bNAb resistance in the reservoir in some individuals [7]. Indeed, HIV-1 is integrated into the genome of various cells and various tissue compartments such as in lymph nodes and gut-associated lymphoid tissues, creating important sanctuaries for persistent HIV infection [9–11]. In addition, compartmentalization of viral quasispecies has been described for multiple tissues such as the genital tract [12] and the central nervous system (CNS) [13].
The fragmented knowledge about the HIV reservoir is partly due to restricted access to human tissues. To address tissue availability, the Last Gift cohort, a perimortem observational research study is enrolling PWHs diagnosed with a terminal illness from a non-HIV condition for collection of antemortem blood and postmortem tissues across multiple anatomical compartments. Sequence analysis of near full-length (FL) env HIV single genomes showed that viral diversity of HIV-RNA populations in blood and HIV-DNA populations in tissues varied between tissues across participants; however, evidence of identical intact FL env proviruses within and across tissues was found [14]. These findings raised the question of how much viral compartmentalization impacts overall neutralization susceptibility.
We recently reported on a novel computational model, the Bayesian Markov chain Monte Carlo–support vector regression (MCMC–SVR), for the prediction of viral neutralization sensitivity by incorporating both HIV env sequence and approximated glycan occupancy as variables to quantitatively predict the half maximal inhibitory concentrations (IC50) of multiple bNAbs [15]. When combined with single-genome sequencing of blood- or tissue-derived HIV-1 env, it allows the rapid and accurate mapping of the HIV-1 reservoir for predicted resistance to anti-HIV antibodies. Here, we applied this model to 791 near FL env single genome sequences generated from reservoir viruses in 33 tissues obtained from 7 participants in the Last Gift cohort. We were primarily interested in determining if we would find evidence of bNAb susceptibility compartmentalization in tissues.
METHODS
Study Cohort Data
For our analyses, we included 791 near FL HIV-1 env sequences from 7 Last Gift cohort participants across 33 tissues. A subset of the sequences has been previously described by Chaillon et al [14]. For details on the cohort, blood and tissue sampling, and the sequencing approach, see the Supplementary Methods. Additionally, 404 full-length HIV-1 envelope sequences from paired cerebrospinal fluid (CSF) and blood plasma samples from 9 participants in a previously published study [15] were included in our analysis.
bNAb Neutralization Sensitivity Prediction
As previously described [16], the Bayesian MCMC–SVR was used. Unlike our previous strategy to perform point estimation of neutralization susceptibility using SVR, a least-square LS–SVR model [17] was used as a supervised learning model to predict neutralization sensitivity with predicted intervals using the selected optimal features, providing additional information of predicted antibody sensitivity for robust evaluation. More specifically, we assumed that the variance is constant and independent from the input feature set, called homoscedasticity. LS–support vector machine (SVM) with Radial Basis Function (RBF) kernel in the LS–SVMlab matlab package was used to fit the model for further evaluation (see the Supplementary Methods).
Additive Effect Model for Antibody Combination
In order to evaluate the combination effect of bNAbs, an additive-effect model [16] was applied to calculate the combined IC50 using the following formula: combined_IC50 = 1/ sum_i(1/IC50_i), where combined_IC50 represents the combined IC50 and i denotes the selected single bNAbs that recognize distinct epitopes. Details on predicting bNAb combinations that match blood and tissue sequences are provided in the Supplementary Methods.
Statistical Analyses
Two-sided Wilcoxon rank sum tests, 1-way analysis of variance (ANOVA), and linear and logistic regression were used for pairwise and across group comparisons as well as for comparison of the proportion of resistant viruses across participants and tissues. A detailed statistical plan is described in the Supplementary Methods.
RESULTS
IC50 Prediction-Model Optimization
Our previously reported computational framework [16] for quantitatively estimating log-scaled IC50 values (log2[IC50]) included 2 main steps: env sequence position selection using MCMC and single IC50 value prediction model by SVR. To further determine the range of predicted neutralization sensitivities, we upgraded the second step by estimating the pointwise neutralization sensitivity with approximated prediction intervals using LS–SVM regression [17]. We focused on 9 bNAbs that are currently in clinical development, specifically the CD4bs antibodies VRC01, 3BNC117, N6, and VRC07-523; the V3 loop glycan antibodies PGT121 and 10-1074; the V2 apex glycan antibodies PGDM1400 and PG9; and the membrane-proximal external region (MPER) antibody 10E8 [1]. As previously reported [16], the model performed robustly across all 9 bNAbs with exceptional correlation (R2 values ranging from 0.86 to 0.93; Supplementary Figure 1) between predicted and measured IC50 values when tested with paired env sequences and measured IC50 value information from the Los Alamos CATNAP database [18].
Neutralization Susceptibility Across Tissues
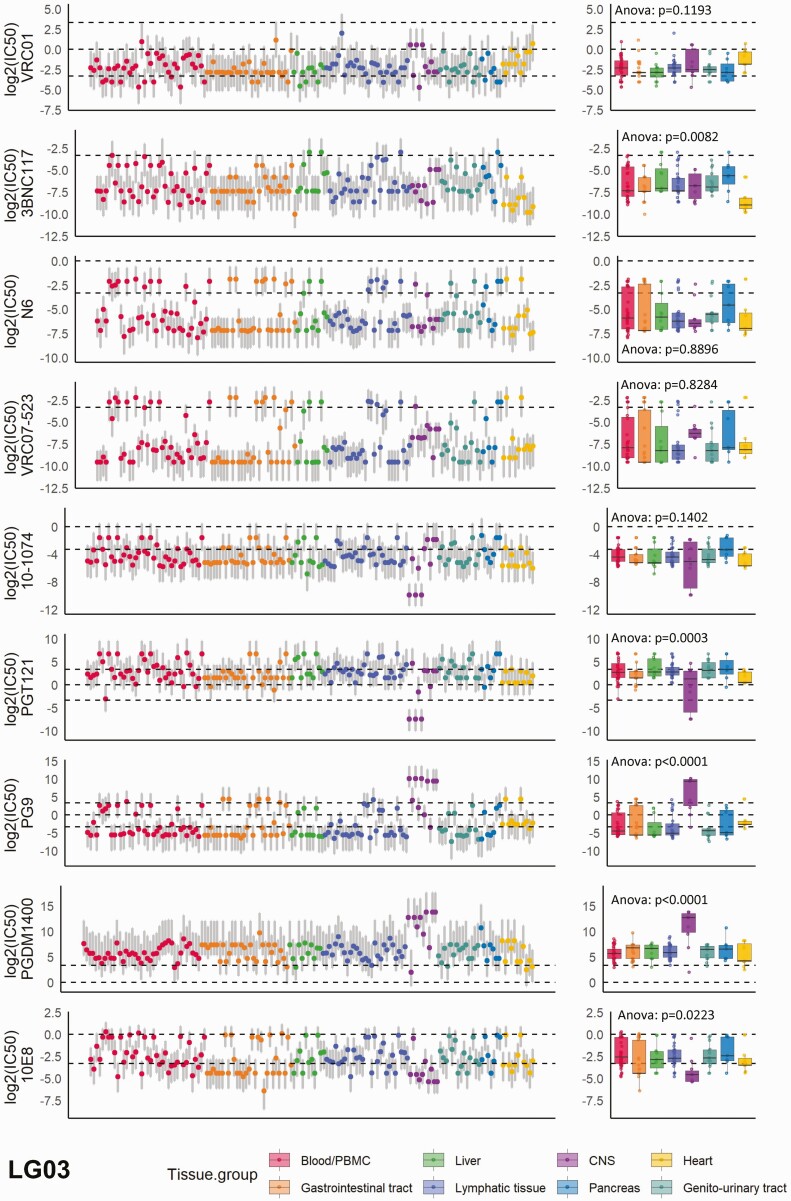
A total of 791 near FL env sequences across 33 tissues that had been derived from 7 participants in the Last Gift cohort were available for the analysis (Supplementary Figure 2). Two participants (LG01 and LG04) had stopped ART prior to autopsy, leading to plasma viremia of 13 500 and 48 000 copies/mL, respectively, at the time of their death. The remaining 5 participants had remained virologically suppressed on ART. Overall, 29 to 176 sequences (median, 123) from a median of 17 tissues per participant were available (Supplementary Figure 2A). To improve comparability, we grouped tissues based on organ system association, such as gastrointestinal tract or genitourinary tract, resulting in 8 tissue groups including blood/PBMCs (Supplementary Figure 2B). As expected, we observed substantial heterogeneity of the predicted neutralization sensitivities, as determined by IC50 values in micrograms per milliliter, across bNAbs (mean standard deviation [SD] of log2[IC50] = 3.44), across sequences to each single bNAb (mean SD of log2[IC50] = 2.76), across participants (mean SD of log2[IC50] = 3.30), and across tissues (mean SD of log2[IC50] = 3.57), as shown to be exemplary for participant LG03 (Figure 1) but also across participants (Supplementary Table 1 and Supplementary Figure 3).
Figure 1.
The distribution of neutralization susceptibilities to the 9 selected broadly neutralizing antibodies (bNAbs) across all tissues; example is shown for participant LG03 in the Last Gift cohort. Each dot represents the predicted neutralization susceptibility with the respective prediction range (gray vertical lines) as calculated by our model. The dashed lines from bottom to top denote half-maximal inhibitory concentrations (IC50) = 0.1 µg/mL, IC50 = 1 µg/mL, and IC50 = 10 µg/mL. The box plots show the median values with the first and third quartiles across all tissue compartments for each bNAb. One-way ANOVA was used for across-group comparison. See also Supplementary Figure 3 for data on the remaining participants. Abbreviations: ANOVA, analysis of variance; CNS, central nervous system; PBMC, peripheral blood mononuclear cell.
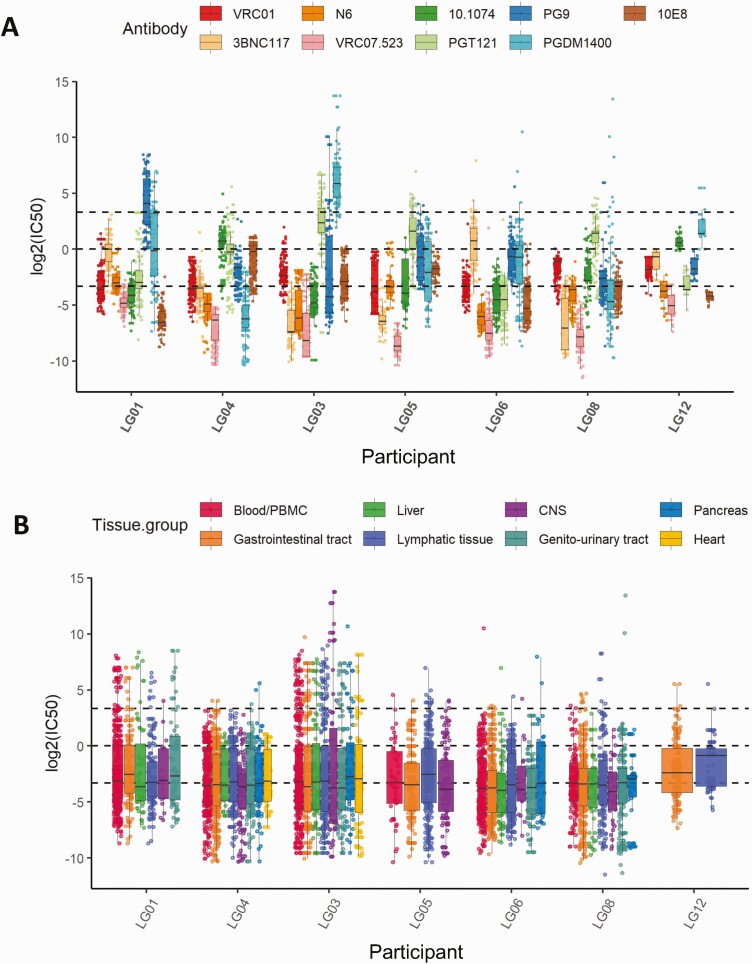
As a first analysis, we compared the distribution of predicted neutralization susceptibilities to all 9 bNAbs and across all tissues between each of the 7 participants. A wide bNAb-dependent variability of IC50 value distributions was observed (Figure 2A), consistent with the diversity of susceptibilities that we have previously reported in PBMCs [16]. While the geometric mean IC50 values for all sequences across all bNAbs and tissues significantly differed between individuals (1-way ANOVA test, P < 2e-16), they nominally only ranged from .076 to .204 µg/mL (Supplementary Figures3 and 4A) when adjusting for tissue group and antibody. More importantly, the susceptibility pattern per bNAb was also distinctively different between participants. Despite these interparticipant differences, we observed a clear hierarchy of predicted neuralization activity between the bNAbs. Overall and reflecting their known superior neutralization breadth, the CD4bs antibodies performed best followed by MPER-specific, V2 apex–specific, and V3 glycan–specific antibodies. Indeed, the predicted IC50 values for VRC07-523 were the lowest across all tissues/participants (2-sided Wilcoxon test with Benjamini–Hochberg correction, P < .001) and ranged from 0.003 to 0.035 µg/mL (Supplementary Figure 4B). The CD4bs antibodies N6 and 3BNC117 followed closely. For some bNAbs, such as the V2 apex glycan antibody PGDM1400, we observed a wide spread of predicted IC50 values that ranged from 0.013 to 58.83 µg/mL.
Figure 2.
A, The heterogeneity of neutralization susceptibilities for individual broadly neutralizing antibodies (bNAbs) across all tissues per participant. B, The homogeneous distribution of predicted neutralization in different tissue compartments for each participant across all bNAbs. The box plots show the median values with the first and third quartiles across the bNAbs and tissues. The dashed lines from bottom to top denote half-maximal inhibitory concentrations (IC50) = 0.1 µg/mL, IC50 = 1 µg/mL, and IC50 = 10 µg/mL. Abbreviations: CNS, central nervous system; PBMC, peripheral blood mononuclear cell.
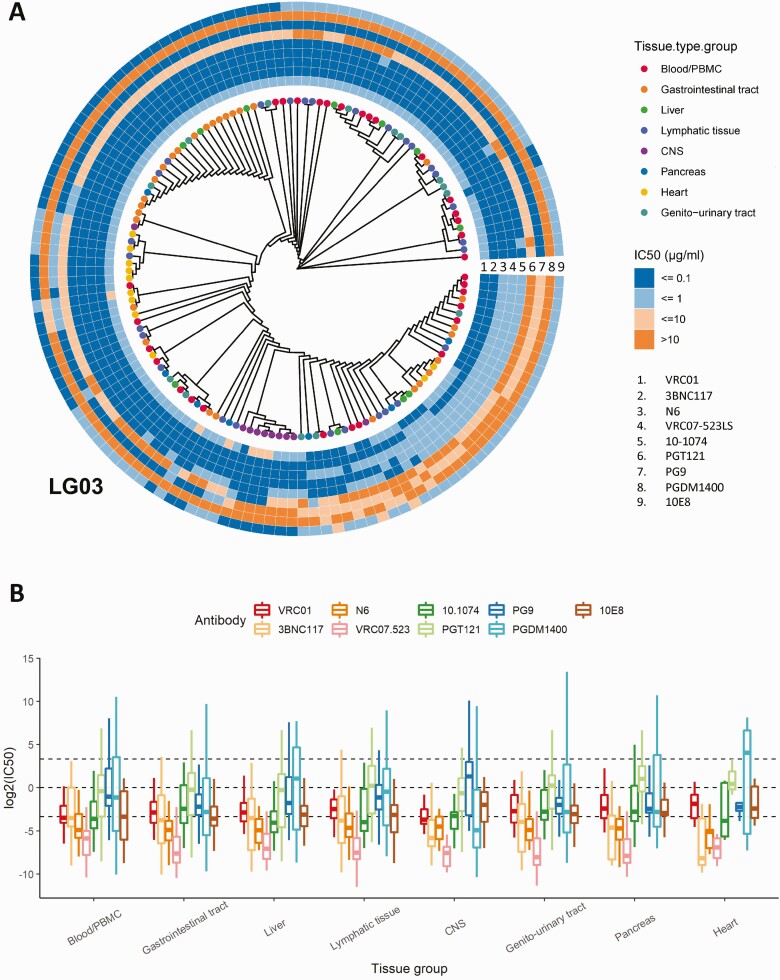
Figure 3.
A, Maximum likelihood phylogeny for participant LG03. Tips are colored by tissue compartment as in the legend, and each sequence is paired with the predicted half-maximal inhibitory concentration (IC50) value range information for the 9 tested broadly neutralizing antibodies (bNAbs). The tree has been modified to allow for better visualization by displaying tree topology without branch length information. IC50 values are color coded in 4 groups (≤0.1 µg/mL to >10 µg/mL), and each ring represents a separate bNAb (1 through 9) as per the legend. See also Supplementary Figure 5 for data on the remaining participants. B, The heterogenous distribution of predicted neutralization in different tissue compartments for the selected 9 bNAbs. The box plots show the median values with the first and third quartiles across the bNAbs. Abbreviations: CNS, central nervous system; PBMC, peripheral blood mononuclear cell.
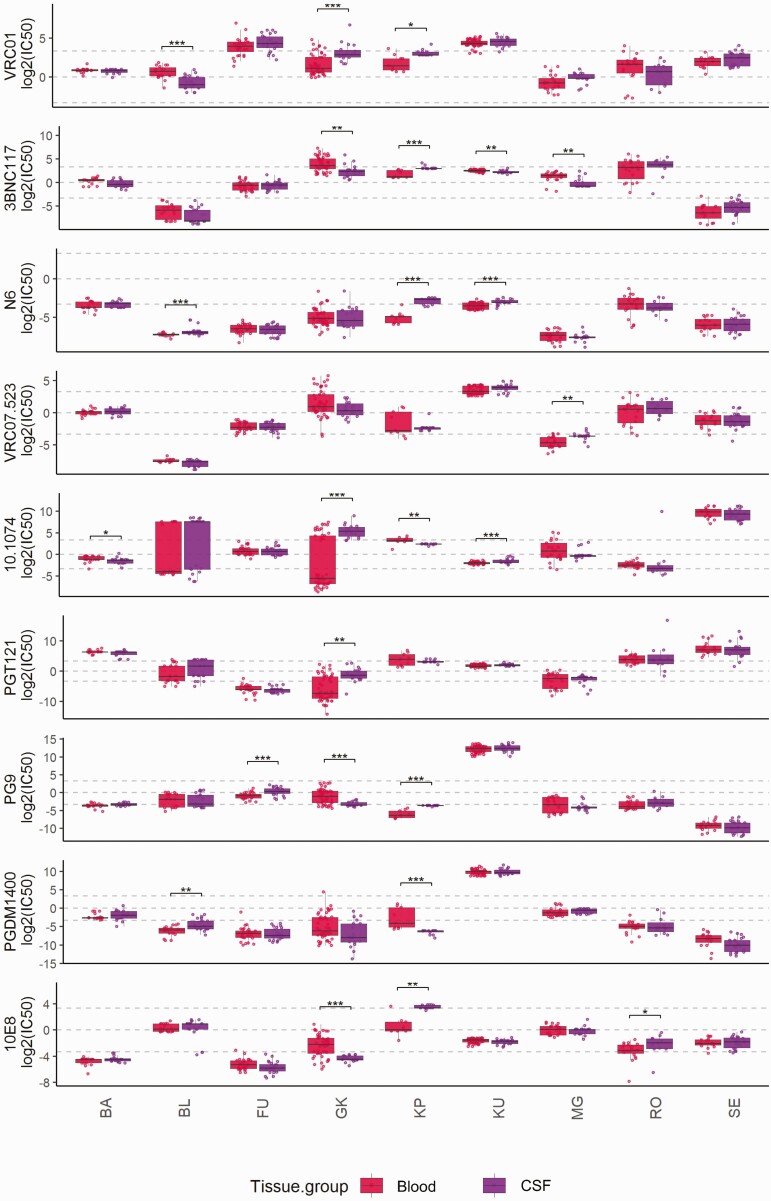
Figure 4.
The distribution of neutralization susceptibilities to the 9 selected broadly neutralizing antibodies comparing blood with CSF derived env sequences from Stefic et al [15]. Each box represents the median (central line) and IQR (25% and 75% percentiles) and the two whiskers represent 1.5 * IQR. The two-sided wilcoxon test is used to test the mean difference followed by multiple test correction using the Benjamini-Hochberg procedure across all the patients. (* represents an adjusted p-value < 0.05 . ** represents an adjusted p-value <0.01 . ***represents an adjusted p-value <0.001). Abbreviation: CSF, cerebrospinal fluid.
To determine if we would observe differences in susceptibilities across tissues, we first compared tissues within each participant and across all bNAbs (Figure 2B and Supplementary Figure3). Overall, there was little heterogeneity between predicted mean IC50 values across tissues in each individual. However, a substantial range in IC50 values was observed in each individual’s viruses, with predicted IC50 values considered to be likely resistant (mean, 15.2% of all sequences per participant; min/max, 10%/21.1%) or resistant (mean, 4.3% of all sequences per participant; min/max, 0.3%/15.2%). When we compared tissue groups across all participants and bNAbs, we found the expected span of IC50 values (min, 0.0003 µg/mL; max, >100 µg/mL). The overall means ranged from 0.077 µg/mL to 0.133 µg/mL and did not generally differ between tissues, except for CNS-derived sequences for which the predicted mean IC50s differed significantly compared with blood/PBMCs, lymphatic tissue, and gastrointestinal tissue (Wilcox test with Benjamini–Hochberg correction; P = .002; P = .0008; and P = .03, respectively; Supplementary Figure 4C). In this dataset, we did not find evidence of general tissue-specific compartmentalization of viruses with a divergent neutralization susceptibility pattern. Specifically, susceptibilities in lymphatic tissues that have previously been considered sanctuary viral reservoir sites due to reduced immune pressure [19] did not differ overall from other tissues. Furthermore, phylogenetic analyses with paired predicted IC50 value information, as demonstrated for participant LG03 (see Figure 3A and Supplementary Figure 5 for the remaining participants), also revealed the presence of a nearly identical FL env population across different tissues with respective identical neutralization susceptibility pattern. Clear differences, however, became apparent again when we focused specifically on individual bNAb susceptibilities (Figure 3B). As described above, across all participants, VRC07-523 demonstrated the lowest predicted IC50 values in all tissues (mean, 0.0245 µg/mL); for other bNAbs, such as the V2 apex glycan antibodies, frequent resistant sequences across most tissues were predicted (mean, 30.9% of all sequences per participant; min/max, 2.8%/99%). Taken together, these data across tissues highlight that bNAb cocktails for therapeutic approaches should ideally be personalized by identifying combinations with the most promising complementary breadth.
bNAb Susceptibilities in Blood Match Susceptibilities in Deep Tissue Reservoirs
Given the diversity of bNAb-specific susceptibilities within each individual (Figure 2), pretherapy screening to determine the most effective bNAb combination has been suggested. There are multiple approaches including quantitative viral outgrowth assays and proviral DNA env sequence–based infectious molecular clone testing (Monogram Biosciences) [1]. These strategies, however, are based on the viral quasispecies that are found in blood/PBMCs at the time of the assay, potentially failing to quantify the real diversity of viral escape variants present in PWH. We, therefore, determined if the susceptibilities observed in the PBMC sequences would be representative of all other tissues and compared the frequency of sequences with predicted IC50 values >1 µg/mL per bNAb in blood/PBMCs with the different tissues. In the majority of PWH, we saw consistency between the proportion of resistant strains in blood/PBMCs and tissues. However, in some individuals and tissues, we observed a difference in the proportion of sequences predicted to be resistant to single bNAbs (Table 1). Specifically, we found sequences with a resistance pattern in the CNS, the genitourinary tract, and the gastrointestinal tissues that were not observed in blood/PBMCs within the same individuals (Table 1). These data suggest that while virus susceptibilities in blood/PBMCs seem to serve as unexpectedly reliable predictors of susceptibilities in the tissues, the exceptions that we found also suggest that blood/PBMCs as the sole source for assessing viral reservoir susceptibility might miss potentially resistant strains that could result in viral rebound and bNAb therapeutic failure.
Table 1.
Differences in Frequency of Sequences With Predicted Half-Maximal Inhibitory Concentrations >1 µg/mL for Broadly Neutralizing Antibodies Comparing Blood/Peripheral Blood Mononuclear Cells With Other Tissues for Each Participant
| Participant | Tissue Group | Broadly Neutralizing Antibody | P Value | Proportion_differences |
|---|---|---|---|---|
| LG01 | Liver | 3BNC117 | .006 | –0.5434 |
| Lymphatic tissue | 3BNC117 | <.001 | –0.4757 | |
| CNS | 3BNC117 | .004 | –0.6862 | |
| LG03 | CNS | PG9 | .004 | 0.5179 |
| CNS | PGT121 | .008 | –0.4230 | |
| LG04 | CNS | 10-1074 | <.001 | –0.7103 |
| Gastrointestinal tract | 1E + 09 | <.001 | –0.5562 | |
| Gastrointestinal tract | PGT121 | <.001 | 0.4348 | |
| Lymphatic tissue | PGT121 | <.001 | 0.4108 | |
| LG05 | Gastrointestinal tract | PG9 | <.001 | –0.68 |
| LG06 | Genitourinary tract | 3BNC117 | .002 | –0.4666 |
| Gastrointestinal tract | PG9 | <.001 | –0.5422 | |
| Genitourinary tract | PG9 | <.001 | –0.5666 |
Abbreviation: CNS, central nervous system.
For participant LG08, no statistically significant differences were observed.
For participant LG12, no blood/ peripheral blood mononuclear cell sequences were available. Shown are only differences that reached statistical significance (P < .05).
Isolated Compartments With Resistant Strains: CNS
Overall, we saw limited variation in the tissue-specific predicted susceptibility pattern, although CNS-derived sequences seemed to differ the most from other tissues with regard to the mean IC50 values across all bNAbs but also the presence of resistant viral variants that were not found in blood. For example, in participant LG03, we observed distinct populations of env sequences derived from the CNS that showed high levels of resistance against selected bNAbs, such as PG9 and PGDM1400 (Figure 1). Indeed, it has been suggested that the CNS, due to the blood–brain barrier, might harbor viruses with distinct and potentially resistant susceptibility profiles and could be the source of viral rebound and/or breakthrough viremia in bNAb-treated individuals [15, 20]. To further explore this specific compartment, we analyzed the paired blood and CSF env sequence data from a study by Stefic et al [15]. In their study, 9 individuals provided 241 blood and 163 cerebrospinal fluid (CSF) sequences (mean, 27 and 18 per individual; Figure 4). Overall, the pattern of neutralization susceptibility was similar between blood and CSF in most individuals, that is, participants BA, RO, and SE showed none or only minimal differences for individual bNAbs (Figure 4). In contrast, in participants KP and GK, CSF sequences were predicted to be significantly more resistant to neutralization compared with blood for selected bNAbs such as 10E8 and 10-1074 (adjusted P values, .003 and .0007, respectively), while blood-derived sequences were predictably more resistant to PGDM1400 and 10E8 (adjusted P values, .0004 and .0002, respectively; Figure 4 and Supplementary Figure 6A and 6B). Nevertheless, despite these isolated differences, in this second dataset we confirmed general consistency of neutralizing susceptibility patterns between viruses in blood and those in the CNS compartment.
Combination bNAb Regimens Based on Blood Sequences to Overcome Tissue Resistance
Despite the similarities between neutralization susceptibility patterns in blood and tissues, the findings of isolated tissue viruses with predicted neutralization resistance could pose a risk for bNAb therapy that would have been chosen based on blood-based susceptibility testing. We were, therefore, interested in determining if bNAb combination regimens, selected based on PBMC viral sequence susceptibility predictions, would likely also cover resistant viruses that were present in tissues but not visible in blood at the time of sampling. Using CD4bs bNAbs as the basis due to their superior breadth and combining with different other bNAb classes to maximally cover epitope regions, we applied an additive-effect model to calculate the combined IC50 values for different bNAb combinations [16, 21, 22]. We found that in order to cover all viruses in each participant at IC50 values <0.1 µg/mL, only the triple combination of VRC07-523LS + 10-1074 and 10E8 achieved this goal (Supplementary Table 2). However, the CD4bs antibody 3BNC117 or VRC01 and the V2-apex antibodies PGDM1400 or PG9 when combined with 10-0174 and 10E8 in a quadruple combination also covered all viruses at the respective IC50 level. Interestingly, for 3 participants (LG04, LG05, and LG06), VRC07-523 alone covered all viruses at IC50 values <0.1 µg/mL. In addition, bNAb combinations predicted to cover all viruses in blood at IC50 values <0.1 µg/mL also covered tissue viruses in more than 98% across participants (mean, 98%; min/max, 88%/100%). These data, therefore, suggest that despite isolated resistant viral variants in tissues, blood/PBMC-derived sequence analyses to identify bNAb combinations successfully identified bNAb cocktails with presumably sufficient breadth to neutralize all reservoir viruses in the respective individual.
DISCUSSION
The extent to which neutralization-resistant strains are present in different tissue compartments has not been comprehensively determined. Our analysis of 7 participants in the Last Gift cohort now suggests that a wide distribution of viral neutralization susceptibilities can be observed within an individual consistent with the known within-host viral diversity (reviewed in [23]). Specifically, for each participant, we discovered individual neutralization susceptibility signatures across the tested bNAbs, suggesting that the selection of bNAb regimens will likely require a personalized approach, similar to what is done for ART regimens that are selected based on the viral genotypic resistance pattern [24]. Nevertheless, despite these inter- and within-participant differences in susceptibility, we observed a clear hierarchy of predicted neuralization activity between he bNAbs. Reflecting their exquisite breadth, the CD4bs antibodies performed best in covering viral sequences across individuals and tissues, suggesting that they serve as a cornerstone for bNAb cocktails.
Overall, little heterogeneity was observed between predicted mean IC50 values across tissues in each individual, arguing against a general tissue-specific compartmentalization of viruses with divergent neutralization susceptibility patterns. Indeed, we (and others [14, 25, 26]) have previously demonstrated that there is an active exchange of viruses between tissues and that the reservoir viruses across tissues are rather similar during ART and in the absence of detectable plasma viremia. Blood/PBMCs served as an unexpectedly reliable predictor of the tissue distribution of virus susceptibilities. This is an observation that might support the further development of blood-based assays for geno- and phenotypic assessment of viral neutralization resistance prior to initiation of bNAb therapy.
Despite the overall homology of the predicted susceptibility pattern across tissues, some exceptions were observed. Specifically, compared with blood, the CNS seemed to harbor more resistant viruses in some rare cases. Whether these or the small number of other resistant strains indeed can fuel viremic rebound during bNAb therapy is unknown. One caveat to our data is that for our analysis we used the env sequences alone without further information if these envelopes belong to replication-competent or possibly replication-defective viruses. Characterizing each env sequence, therefore, might overestimate the level of resistance that is present in a specific viral reservoir. Nevertheless, despite isolated predicted resistant viral variants in tissues, blood/PBMC-derived sequence analyses to identify bNAb combinations suggested bNAb cocktails with presumably sufficient breadth to neutralize all reservoir viruses in the respective individual. Only 1 triple bNAb combination was modeled to cover all sequences across all participants at highly potent IC50 values of <0.1 µg/mL. However, several quadruple bNAb combinations also achieved this goal. These are, however, theoretical considerations, assuming that the bNAb levels and activity are similar and unaffected across all tissues. How well bNAbs penetrate into tissues once administered intravenously is currently still under investigation [27]. Specifically, the blood–brain barrier will prevent therapeutic levels of bNAbs in the CNS, and this compartment and the viral reservoir it harbors needs particular attention for future selections of bNAb therapies.
In summary, using a machine-learning prediction model, we were able to map the landscape of viral neutralization resistance across tissue reservoirs of several individuals. Our results suggest that given the significant viral diversity and the unique bNAb susceptibility profiles within each individual, concepts for personalized bNAb regimens should be considered. Alternatively, bNAb combinations with extraordinarily broad viral coverage might be necessary to overcome resistant strains. These concepts should be evaluated in future clinical trials.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. C. W., A. C., and B. J. conceptualized the study. C. W. and D. A. L. developed the model modifications. C. W., A. C., and T. E. S. analyzed the human immunodeficiency virus type 1 viral sequences. T. E. S., C. W., and A. C. performed statistical analyses. S. G. and D. M. S. developed and lead the Last Gift cohort and performed all rapid autopsy procedures. All authors contributed to the writing and editing of the manuscript and approved the final version.
Acknowledgments. We acknowledge the Last Gift cohort participants and their relatives, the Last Gift cohort study staff and community advisors, and all Last Gift cohort collaborators.
Financial support. This work was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (NIH, NIAID; P01 AI131385 to D. M. S., R01 AI106408 to B. J., National Institute on Drug Abuse (NIDA) R01 DA055491 to A. C., and NIDA DP2 DA051915 to S. G.). A. C., S. G., and D. M. S. are supported by the James B. Pendleton Charitable Trust and by the San Diego Center for AIDS Research, an NIH-funded program (P30 AI036214) that is supported by the following NIH institutes and centers: NIAID, NCI, NHLBI, NIA, NICHD, NIDA, NIDCR, NIDDK, NIGMS, NIMH, NIMHD, FIC, and OAR. A. C. is also supported by NIH grants R01 MH128153 and R01 DK131532. This project was also supported by funding from the Ragon Institute of MGH, MIT, and Harvard.
Contributor Information
Chuangqi Wang, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA.
Timothy E Schlub, University of Sydney, Faculty of Medicine and Health, Sydney School of Public Health, Sydney, New South Wales, Australia.
Wen Han Yu, Bill & Melinda Gates Medical Research Institute, Cambridge, Massachusetts, USA.
C Sabrina Tan, Division of Infectious Diseases, Center for Virology and Vaccine Research, Beth Israel Deaconess Medical Center, Boston, Massachusetts, USA.
Karl Stefic, Department of Virology, Tours University Hospital, Tours, France.
Sara Gianella, Division of Infectious Diseases and Global Public Health, University of California–San Diego, San Diego, California, USA.
Davey M Smith, Division of Infectious Diseases and Global Public Health, University of California–San Diego, San Diego, California, USA; VA San Diego Healthcare System, San Diego, California, USA.
Douglas A Lauffenburger, Department of Biological Engineering, Massachusetts Institute of Technology, Cambridge, Massachusetts, USA.
Antoine Chaillon, Division of Infectious Diseases and Global Public Health, University of California–San Diego, San Diego, California, USA.
Boris Julg, Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology, and Harvard University, Cambridge, Massachusetts, USA.
References
- 1. Julg B, Barouch D.. Broadly neutralizing antibodies for HIV-1 prevention and therapy. Semin Immunol 2021; 101475. [DOI] [PubMed] [Google Scholar]
- 2. Caskey M, Klein F, Lorenzi JC, et al. . Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 2015; 522:487–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lynch RM, Boritz E, Coates EE, et al. . Virologic effects of broadly neutralizing antibody VRC01 administration during chronic HIV-1 infection. Sci Transl Med 2015; 7:319ra–206. [DOI] [PubMed] [Google Scholar]
- 4. Caskey M, Schoofs T, Gruell H, et al. . Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat Med 2017; 23:185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bar KJ, Sneller MC, Harrison LJ, et al. . Effect of HIV antibody VRC01 on viral rebound after treatment interruption. N Engl J Med 2016; 375:2037–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Scheid JF, Horwitz JA, Bar-On Y, et al. . HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 2016; 535:556–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mendoza P, Gruell H, Nogueira L, et al. . Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 2018; 561:479–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cohen YZ, Lorenzi JCC, Krassnig L, et al. . Relationship between latent and rebound viruses in a clinical trial of anti-HIV-1 antibody 3BNC117. J Exp Med 2018; 215:2311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lorenzo-Redondo R, Fryer HR, Bedford T, et al. . Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature 2016; 530:51–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rothenberger MK, Keele BF, Wietgrefe SW, et al. . Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci U S A 2015; 112:E1126–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Banga R, Procopio FA, Noto A, et al. . PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med 2016; 22:754–61. [DOI] [PubMed] [Google Scholar]
- 12. Miller RL, Ponte R, Jones BR, et al. . HIV diversity and genetic compartmentalization in blood and testes during suppressive antiretroviral therapy. J Virol 2019; 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stam AJ, Nijhuis M, van den Bergh WM, Wensing AM.. Differential genotypic evolution of HIV-1 quasispecies in cerebrospinal fluid and plasma: a systematic review. AIDS Rev 2013; 15:152–61. [PubMed] [Google Scholar]
- 14. Chaillon A, Gianella S, Dellicour S, et al. . HIV persists throughout deep tissues with repopulation from multiple anatomical sources. J Clin Invest 2020; 130:1699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stefic K, Chaillon A, Bouvin-Pley M, et al. . Probing the compartmentalization of HIV-1 in the central nervous system through its neutralization properties. PLoS One 2017; 12:e0181680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu WH, Su D, Torabi J, et al. . Predicting the broadly neutralizing antibody susceptibility of the HIV reservoir. JCI Insight 2019; 4:e130153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. De Brabanter K, De Brabanter J, Suykens JA, De Moor B.. Approximate confidence and prediction intervals for least squares support vector regression. IEEE Trans Neural Netw 2011; 22:110–20. [DOI] [PubMed] [Google Scholar]
- 18. Yoon H, Macke J, West AP Jr., et al. . CATNAP: a tool to compile, analyze and tally neutralizing antibody panels. Nucleic Acids Res 2015; 43: W213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fukazawa Y, Lum R, Okoye AA, et al. . B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med 2015; 21:132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kapoor A, Tan CS.. Immunotherapeutics to treat HIV in the central nervous system. Curr HIV/AIDS Rep 2020; 17:499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wagh K, Bhattacharya T, Williamson C, et al. . Optimal combinations of broadly neutralizing antibodies for prevention and treatment of HIV-1 clade c infection. PLoS Pathog 2016; 12:e1005520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kong R, Louder MK, Wagh K, et al. . Improving neutralization potency and breadth by combining broadly reactive HIV-1 antibodies targeting major neutralization epitopes. J Virol 2015; 89:2659–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hemelaar J. Implications of HIV diversity for the HIV-1 pandemic. J Infect 2013; 66:391–400. [DOI] [PubMed] [Google Scholar]
- 24.Available at: https://clinicalinfo.hiv.gov/en/guidelines/adult-and-adolescent-arv/whats-new-guidelines.
- 25. De Scheerder MA, Vrancken B, Dellicour S, et al. . HIV rebound is predominantly fueled by genetically identical viral expansions from diverse reservoirs. Cell Host Microbe 2019; 26:347–358.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bozzi G, Simonetti FR, Watters SA, et al. . No evidence of ongoing HIV replication or compartmentalization in tissues during combination antiretroviral therapy: implications for HIV eradication. Sci Adv 2019; 5:eaav2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carias AM, Schneider JR, Madden P, et al. . Anatomic distribution of intravenously injected IgG takes approximately 1 week to achieve stratum corneum saturation in vaginal tissues. J Immunol 2021; 207:505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.