Abstract
Differences in methods for biospecimen collection, processing, and storage can yield considerable variability and error. Therefore, best practices for standard operating procedures are critical for successful discovery, development, and validation of disease biomarkers. Here, we describe standard operating procedures developed for biospecimen collection during the DREAM (Diabetes RElated to Acute pancreatitis and its Mechanisms) Study within the Type 1 Diabetes in Acute Pancreatitis Consortium (T1DAPC). Notably these protocols were developed using an integrative process based on prior consortium experience and with input from working groups with expertise in immunology, pancreatitis and diabetes. Publication and adoption consistent biospecimen protocols will inform future studies and allow for better comparisons across different metabolic research efforts.
Keywords: laboratory protocol, pancreas, immunology, type 1 diabetes, biosamples, biorepository
INTRODUCTION
Scientific breakthrough research would not be possible without the participation of individuals who selflessly donate biological specimens, which are used as a research bridge between basic and translational research. Best practices for specimen collection, processing and storage are key for consistency and validity of research findings emanating from biospecimen testing.1–3 Thus, the careful curation and storage of biospecimens is also part of both ethical and scientific importance.1,3,4
The type 1 diabetes in acute pancreatitis consortium (T1DAPC), supported by the National Institute for Diabetes and Digestive and Kidney Diseases (NIDDK), has established a biospecimen committee to develop standard operating procedures (SOP) for biospecimen collection during the DREAM (Diabetes RElated to Acute pancreatitis and its Mechanisms) Study to improve our understanding of the mechanistic underpinnings resulting in the development of diabetes after acute pancreatitis (AP). This SOP builds upon the document developed for the Chronic Pancreatitis Diabetes Pancreas Cancer Consortium (CPDPC), supported by the National Cancer Institute (NCI) and the NIDDK.2
There are several unique components of the T1DAPC biospecimen committee structure worth reporting. Notably several working groups were queried (the immunology, the pancreatitis and the diabetes working groups), and by an integrative process their efforts (reported elsewhere in this collection) were incorporated into the biorepository design as detailed below. The T1DAPC SOPs will serve as a valuable resource for investigators wishing to engage in performing research within and outside of the consortium with an interest in pancreatitis related diabetes.
OBJECTIVES
In addition to the previously outlined objectives of the DREAM Study, a major collaborative effort within the consortium will be the establishment of an annotated repository of biospecimens (serum, plasma, peripheral blood mononuclear monocytes [PBMC], DNA, RNA, and stool). The T1DAPC biorepository will allow the identification of biomarkers to achieve the following objectives: 1) risk stratification of pancreatitis participants, 2) early detection of type 1 diabetes, 3) narrowing knowledge gaps of interrelationships between the endocrine and exocrine pancreas, and 4) informing the development of future strategies to prevent or reverse diabetes after AP.
MEETINGS
The T1DAPC formed a Biospecimen Committee, including 2 Co-Chairs (C.W. and D.L.C.) and a data coordinating center (DCC) Biostatistician (A.M.D.), which met monthly for 18 months. The Biospecimen Committee was composed of representatives of all the clinical centers, members of the T1DAPC Working Groups (pancreatitis, diabetes, immunology) and the NIDDK. The committee co-chairs and DCC Biostatistician met at a minimum weekly to organize the committee and provided monthly reports to the T1DAPC Steering Committee as requested.
RESOURCES UTILIZED
CPDPC SOPs were used as a starting point along with T1DAPC clinical center SOPs. Blood and stool (as detailed in Table 1 and 2) were the initial T1DAPC SOPs to be developed for the DREAM Study. It was decided that if ancillary studies were developed that needed additional biospecimens, the CPDPC SOP would be consulted for initial guidance (pancreas juice, urine, saliva, tissue and other).
TABLE 1.
Detailed Specimens Collected in T1DAPC
| Specimen Type | Collection | Type of Preservative | Short-term Stabilization | Centrifuge | Processing Time | Aliquot | Long-term Storage |
|---|---|---|---|---|---|---|---|
| Plasma | Blood draw | EDTA tube* (purple top) |
4°C (refrigerated or on wet ice) | 1200 g at room temperature for 10 min with brake on | <4 h | 250 μL × 20 | −80°C |
| Serum | Blood draw | Silicone-coated (red top) | Room temperature for 30–60 min, then 4°C until centrifugation | 1200 g at room temperature for 10 min with brake on | <4 h | 250 μL × 20 | −80°C |
| DNA | Blood draw | PAXgene tube* | Room temperature | NA | NA | NA | −80°C |
| RNA | Blood draw | Tempus tube† | Room temperature, then −20°C for 24 h | NA | NA | NA | −80°C |
| Blood in cell stabilizers | Blood draw | Sodium heparin tube (green top), PROT1 proteomic stabilizer | Room temperature | NA | <12 h | 600 μL × 6 (250 μL blood added to 350 μL PROT1) | −80°C |
| Stool for fecal elastase | Home collection | NA | Room temperature | NA | NA | NA | −80°C |
| Stool for metabolome | Home collection | OMNImet-GUT‡ | Room temperature | Vortex each tube at medium setting for 60 s | NA | ~ 0.8 mL × 3 | −80°C |
| Stool for microbiome | Home collection | OMNIgene-GUT‡ | Room temperature | Vortex each tube at medium setting for 60 s | NA | ~ 0.8 mL × 3 | −80°C |
Mix gently by inverting 2 to 3 times
Shake vigorously or vortex for 10 seconds
For a minimum of 30 seconds, shake the sealed tube as hard and fast as possible in a back and forth motion.
NA indicates not applicable.
TABLE 2.
Detailed Metabolic Procedure Specimens Collected in T1DAPC
| Specimen Type | Collection | Metabolic Test | Type of Preservative | Short-term Stabilization | Centrifuge | Processing Time | Aliquot | Long-term Storage |
|---|---|---|---|---|---|---|---|---|
| Plasma | Blood draw | OGTT (6 time points) | 2 ml P800 tube* | 4°C (refrigerated or on wet ice) | 1200 g at room temperature for 10 min with brake on | <4 h | 250 μL × 4 per time point | −80°C |
| MMTT (8 time points) | 8.5 ml P800 tube* | 1200 g at room temperature for 20 min with brake on | 250 μL × 4 and 500 μL × 6 per time point | |||||
| Serum | Blood draw | OGTT (6 time points) | 6 ml Silicone-coated (red top) | Room temperature for 30–60 min, then 4°C until centrifugation | 1200g at room temperature for 10 minutes with brake on | <4 h | 250 μL × 12 per time point | −80°C |
| MMTT (8 time points) | 4 ml Silicone-coated (red top) | 250 μL × 8 per time point | ||||||
| FSIGTT (21 time points) | 3 or 5 ml Silicone-coated (red top) | 500 μL × 3 or 500 μL × 5 per time point |
Mix gently by slowly inverting 8 to 10 times
OGTT indicates oral glucose tolerance test; MMTT, mixed meal tolerance test; FSIGTT, frequently sampled intravenous glucose test.
PROCESS
Similarly to the aforementioned CPDPC effort, consensus was obtained for best practices and SOP developed and added to the manual of operations (MOP). These were relatively straightforward for standard serum, plasma and stool collections since these have been well characterized and utilized across many consortia.2,5,6 The consortium currently has several clinical centers, some with satellite sites, from which study participants are enrolled7 (Fig. 1). Most samples collected from these participants are processed according to the SOPs and aliquots stored locally until shipment to the study biorepository, Penn State University Institute for Personalized Medicine (PSU-IPM) and the NIDDK Central Repository (DKCR) (Figs. 1 and 2; Tables 1 and 2). However, the working groups specific to the T1DAPC brought unique insight to the biorepository. The rationale and mechanisms of obtaining these samples are described in detail elsewhere.7,8 Specifically, the immunology working group strongly supported the use of centralized and standardized processing of PBMC9 to minimize the variability introduced by having multiple laboratories process these samples and to standardize processing and storage. The heparinized (green top) blood samples have an aliquot taken with blood cell stabilizers added, frozen, stored and remaining green top samples sent same day with overnight shipping to a centralized PBMC processing facility. Cryopreserved PBMC are then periodically shipped to the central repository on liquid nitrogen until needed for functional assays (Fig. 1).
FIGURE 1.
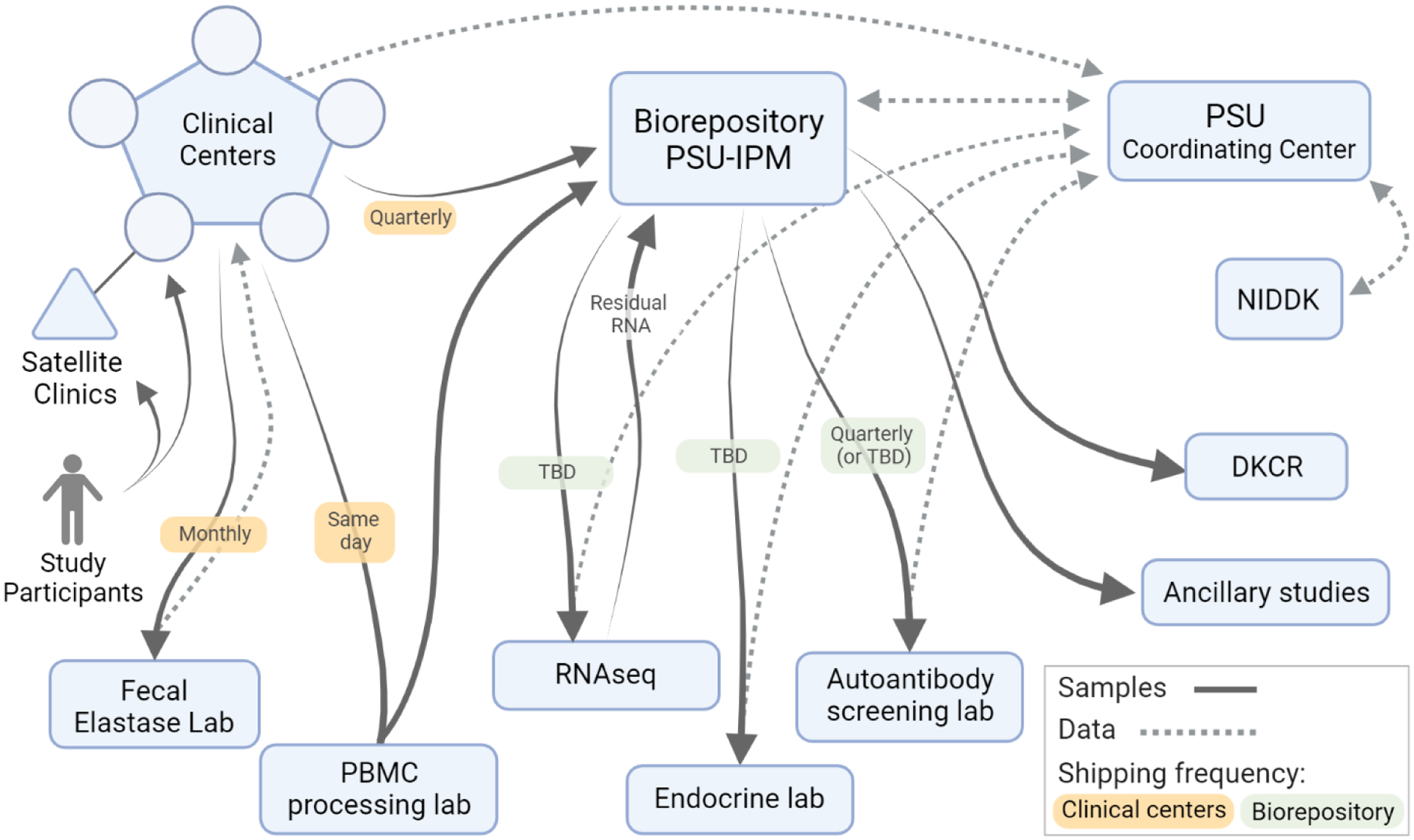
The overall flow of biospecimens and data within the T1DAPC. DKCR, NIDDK Central Repository; PBMC, peripheral blood mononuclear cells; PSU-IPM, Penn State University Institute for Personalized Medicine; TBD, to be determined.
FIGURE 2.
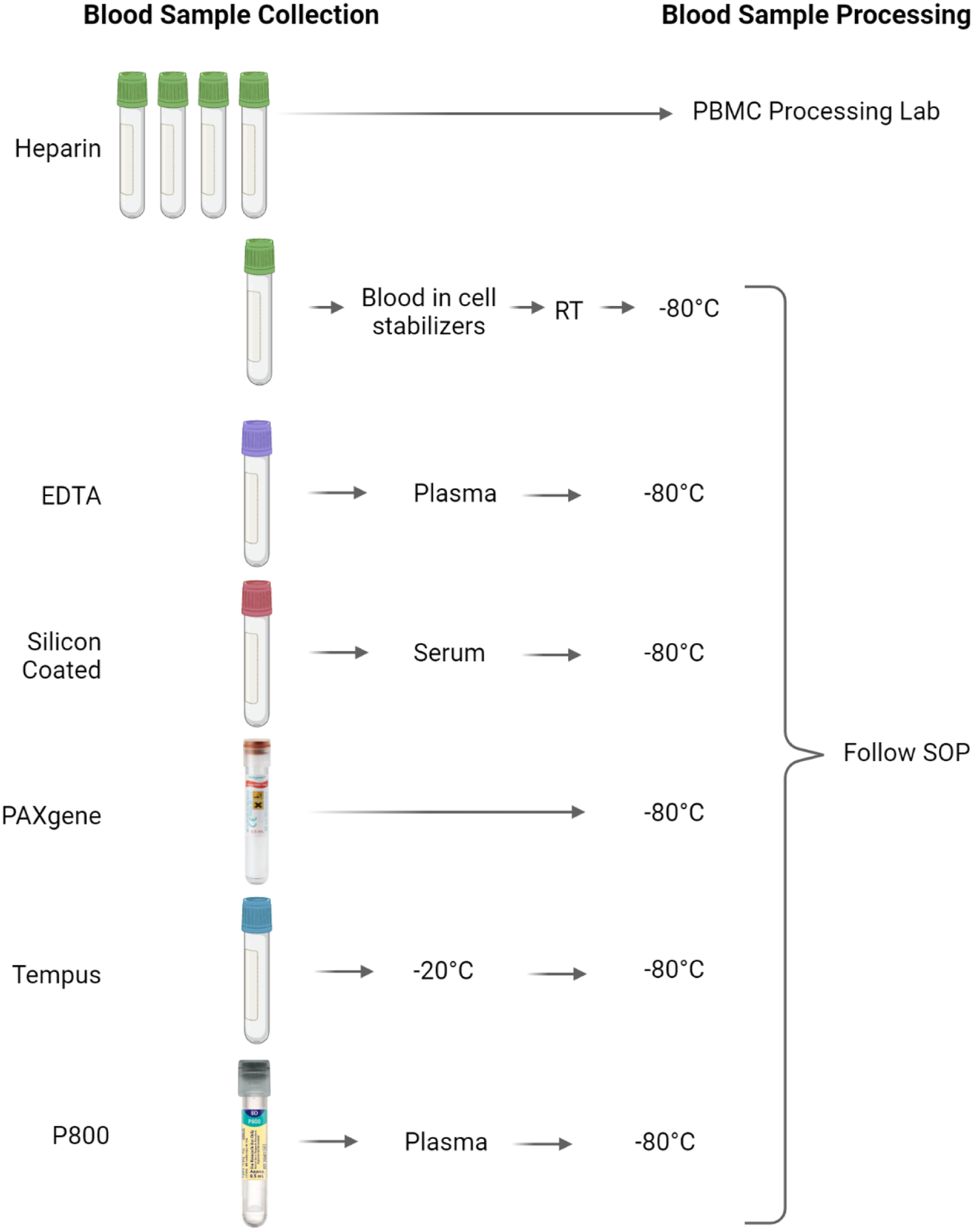
Blood sample collection.
The pancreatitis working group finalized and advised the biospecimen committee on collection of stool samples as outlined Table 1 and Figure 1. The clinical centers will send an aliquot of stool samples to JOLI Diagnostic, Inc. (Williamsviille, NY, http://jolidiagnostic.com) for fecal elastase measurement with data being sent back to the clinical site. The remainder of stool samples are initially collected in OMNIgene-GUT and OMNImet-GUT tubes (DNA Genotek, Ottawa, Canada, https://www.dnagenotek.com), and then aliquoted, for future microbiome and metabolome studies respectively (Table 1). These samples will be sent to the central repository periodically for storage and distribution as indicated (Fig. 1).
The diabetes working group focused on aspects related to monitoring progression to diabetes as described in the diabetes working group manuscript.10 A unique aspect of this is timed sampling of blood following a glucose or standardized meal challenge and the use of P800 plasma collection tubes (Becton Dickinson, Franklin Lakes, NJ) containing a cocktail of protease inhibitors specifically used to stabilize labile hormones (Fig. 2 and Table 2). The clinical sites are responsible for aliquoting and storing these samples and then periodically sending batched samples to the study biorepository (Fig. 1).
The biorepository is responsible for sending samples periodically to the designated endocrine laboratory (Indiana University Center for Diabetes and Metabolic Diseases Translation Core) for analysis of the samples from the glucose or meal challenge procedure. Also, periodically a serum aliquot from each participant visit will be sent to the autoantibody laboratory (Barbara Davis Center for Diabetes - University of Colorado) for analysis of traditional T1D associated autoantibodies.11 Finally, the Tempus tube (Applied Biosystems, Waltham, Mass) samples will be distributed for RNA transcriptome analysis as described in the immunology working group manuscript.8 A PAXgene tube (Qiagen, Germantown, Md) will be collected once from each participant in the study for future genomic studies (Fig. 1).
The biorepository will keep in reserve 20–30% of samples for eventual long-term storage at the DKCR. Ancillary studies are anticipated throughout the duration of the study and upon approval samples will be sent to investigators.
PREPARATION OF WRITTEN SOPs
Representatives from each clinical center with expertise in immunology, diabetes, pancreatitis and stool testing were solicited for their opinion on the initial versions of the SOPs and a final version was approved by the Steering Committee and included in the MOP.
TRAINING
Because sample processing was relatively straightforward, biospecimen collection and processing was incorporated into DREAM Coordinator training. Coordinators and lab technicians were required to undergo a separate Data Management System (DMS) training specific to sample management.
BIOINFORMATICS DATA MANAGEMENT SYSTEM
The Sample Tracking (ST) module of the DCC DMS12 serves as the bioinformatics data management system in T1DAPC. In ST, users can generate sample specific barcoded labels. After labeling samples, users will use ST to associate participant information (ID, visit, collection date) with the sample barcode, as well as sample volume. Sample tracking is also used to ship samples from the sites to labs and the biorepository, and for the labs and biorepository to mark samples received.
LONG-TERM STORAGE/PRESERVATION
The Penn State Institute for Personalized Medicine was selected to serve as the T1DAPC biorepository. Aliquoted samples are stored in 2 mL cryovials suitable for long-term storage. These will be stored in −80°C freezers at the clinical sites until shipment to the biorepository. Upon shipment to the biorepository, samples will be maintained in −80°C freezers, or liquid nitrogen (LN) tanks for PBMCs, located at two separate sites within the Penn State College of Medicine and supported by separate electric grids and backup generators. All −80°C freezers have direct CO2 backup systems, while the LN freezer is connected directly to an LN storage tank that provides automated backup. All −80°C and LN freezers are monitored continuously (24 h/d; 7 d/wk) by remote monitoring with output providing continuous temperature records of each unit as well as notification systems in case of temperature deviation.
Location and status of each sample are maintained in a FreezerPro database (version 7.5.1 Azenta Life Sciences, Chelmsford, Mass). Accuracy of the location information in FreezerPro is routinely verified by monthly spot checks and immediately following the upload of any large data sets.
DISCUSSION
We have successfully developed an SOP within the T1DAPC for the DREAM Study to facilitate the identification and validation of biomarkers for risk stratification, early detection and to improve our understanding of pancreatitis related diabetes. It has been well described in the literature that differences in specimen collection, processing, and storage methods can become a considerable source of error in studies that relate to the discovery, development, and validation of biomarkers.1–3 This trend is particularly true for biospecimens collected for pancreas research in which prior biomarker development is lacking. Thus, it is essential that the procedures for collection, handling, processing and storage of biospecimens be tested, standardized, and carefully documented to optimize biological sample use for pancreas research.
The DREAM biospecimens will be a rich source for biomarker investigations that will improve our understanding of pancreatitis related diabetes and will have far reaching impact for the present and future. Biospecimens will be available to the scientific community through ancillary study collaboration with a T1DAPC clinical center during the lifetime of the T1DAPC. In the future, approximately 20–30% of the DREAM biospecimens along with matching essential data elements will be stored at the DKCR and will be available for the wider scientific community at the end of the DREAM Study.
Grant Support:
Research reported in this publication was supported by funding from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) for the Type 1 Diabetes in Acute Pancreatitis Consortium (T1DAPC) under award numbers
U01 DK127367 (Minnesota),
U01 DK127377 (Pittsburgh),
U01 DK127378 (Illinois at Chicago),
U01 DK127382 (Indiana),
U01 DK127384 (DCC, Penn State),
U01 DK127388 (Ohio State),
U01 DK127392 (University of Florida - Advent Health),
U01 DK127395 (Stanford),
U01 DK127400 (Johns Hopkins), and
U01 DK127403 (Cedars-Sinai).
U01 DK 127404 (Benaroya - Seattle)
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflicts of interest/disclosures:
The following authors disclose potential conflicts of interest: D.K.A. advisory board for National Pancreas Foundation and American Pancreatic Association Foundation, royalties from McGraw-Hill; M.D.B research support from Viacyte and Dexcom, advisory board for Insulet; P.J.L. advisory board for AbbVie; R.E.P. consulting activities with Bayer AG, Corcept Therapeutics Incorporated, Dexcom, Gasherbrum Bio, Inc., Hanmi Pharmaceutical Co., Hengrui (USA) Ltd, Merck, Novo Nordisk, Pfizer, Rivus Pharmaceuticals, Inc., Sanofi, Scohia Pharma, Inc., and Sun Pharmaceutical Industries, research support from Hanmi Pharmaceutical Co., Janssen, Metavention, Novo Nordisk, Poxel SA, and Sanofi, and speaking fees from Novo Nordisk; E.K.S. speaking fees from MedScape; C.S. advisory board for Vertex Pharmaceuticals. The remaining authors declare no conflict of interest.
Abbreviations
- AP
Acute Pancreatitis
- CPDPC
Chronic Pancreatitis Diabetes Pancreas Cancer Consortium
- DCC
Data Coordinating Center
- DMS
Data Management System
- DKCR
NIDDK Central Repository
- DREAM
Diabetes RElated to Acute pancreatitis and its Mechanisms
- LN
Liquid Nitrogen
- MOP
Manual of Operations
- NCI
National Cancer Institute
- NIDDK
National Institute of Diabetes and Digestive and Kidney Diseases
- PBMC
Peripheral Blood Mononuclear Cells
- PSU-IPM
Penn State University Institute for Personalized Medicine
- SOP
Standard Operating Procedures
- ST
Sample Tracking Module
- T1DAPC
Type 1 Diabetes in Acute Pancreatitis Consortium
Contributor Information
Clive Wasserfall, Department of Pathology, Immunology and Laboratory Medicine, University of Florida Diabetes Institute, Gainesville, FL.
Anne-Marie Dyer, Department of Public Health Sciences, Penn State College of Medicine, Hershey, PA.
Cate Speake, Diabetes Clinical Research Program, Center for Interventional Immunology, Benaroya Research Institute at Virginia Mason, Seattle, WA.
Dana K. Andersen, Division of Digestive Disease and Nutrition, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD.
Kendall Thomas Baab, Department of Public Health Sciences, Penn State College of Medicine, Hershey, PA.
Melena D. Bellin, Departments of Pediatrics and Surgery, University of Minnesota Medical School, Minneapolis, MN.
James R. Broach, Department of Biochemistry and Molecular Biology, Penn State College of Medicine, Hershey, PA.
Martha Campbell-Thompson, Department of Pathology, Immunology and Laboratory Medicine, University of Florida Diabetes Institute, Gainesville, FL.
Vernon M. Chinchilli, Department of Public Health Sciences, Penn State College of Medicine, Hershey, PA.
Peter J. Lee, Division of Gastroenterology, Hepatology, and Nutrition, The Ohio State University Wexner Medical Center, Columbus, OH.
Walter G. Park, Division of Gastroenterology, Stanford University School of Medicine, Palo Alto, CA.
Richard E. Pratley, AdventHealth Translational Research Institute, Orlando, FL.
Jami L. Saloman, Division of Gastroenterology, Hepatology and Nutrition, University of Pittsburgh, Pittsburgh, PA.
Emily K. Sims, Department of Pediatrics, Indiana University School of Medicine, Indianapolis, IN.
Gong Tang, Department of Biostatistics, Graduate School of Public Health, University of Pittsburgh, Pittsburgh, Pennsylvania..
Dhiraj Yadav, Division of Gastroenterology, Hepatology and Nutrition, University of Pittsburgh, Pittsburgh, PA.
Cemal Yazici, Division of Gastroenterology and Hepatology, Department of Medicine, University of Illinois at Chicago, Chicago, IL.
Darwin L. Conwell, Division of Gastroenterology, Hepatology, and Nutrition, The Ohio State University Wexner Medical Center, Columbus, OH.
REFERENCES
- 1.Feng Z, Kagan J, Pepe M, et al. The Early Detection Research Network’s Specimen reference sets: paving the way for rapid evaluation of potential biomarkers. Clin Chem. 2013;59:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fisher WE, Cruz-Monserrate Z, McElhany AL, et al. Standard operating procedures for biospecimen collection, processing, and storage: From the Consortium for the Study of Chronic Pancreatitis, Diabetes, and Pancreatic Cancer. Pancreas. 2018;47:1213–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore HM, Kelly AB, Jewell SD, et al. Biospecimen reporting for improved study quality (BRISQ). Cancer Cytopathol. 2011;119:92–101. [DOI] [PubMed] [Google Scholar]
- 4.Biorepositories and Biospecimen Research Branch. NCI Best Practices for Biospecimen Resources. Bethesda, MD; National Cancer Institute, National Institutes of Health: 2016. Available at: https://biospecimens.cancer.gov/bestpractices/. Accessed April 2022. [Google Scholar]
- 5.Sanderson-November M, Silver S, Hooker V, et al. Biorepository best practices for research and clinical investigations. Contemp Clin Trials. 2021;116:106572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hullsiek KH, George M, Brown SK. Designing and managing a flexible and dynamic biorepository system: a 15-year perspective from the CPCRA, ESPRIT, and INSIGHT clinical trial networks. Curr Opin HIV AIDS. 2010;5:538–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hart PA, Papachristou GI, Park WG, et al. Rationale and design for the Diabetes RElated to Acute pancreatitis and its Mechanisms (DREAM) study: A prospective cohort study from the Type 1 Diabetes in Acute Pancreatitis Consortium (T1DAPC). Pancreas. 2022;51:XXX–XXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casu A, Grippo PJ, Wasserfall C, et al. Evaluating the Immunopathogenesis of Diabetes Following Acute Pancreatitis in the Diabetes RElated to Acute Pancreatitis and Its Mechanisms (DREAM) Study: From the Type 1 Diabetes in Acute Pancreatitis Consortium (T1DAPC). Pancreas. 2022;51:XXX–XXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmed S, Cerosaletti K, James E, et al. Standardizing T-Cell biomarkers in type 1 diabetes: challenges and recent advances. Diabetes. 2019;68:1366–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dungan KM, Hart PA, Andersen DK, et al. Assessing the pathophysiology of hyperglycemia in the Diabetes RElated to Acute Pancreatitis and Its Mechanisms Study (DREAM): From the Type 1 Diabetes in Acute Pancreatitis Consortium (T1DAPC). Pancreas. 2022;51:XXX–XXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.So M, O’Rourke C, Ylescupidez A, et al. Characterising the age-dependent effects of risk factors on type 1 diabetes progression. Diabetologia. 2022;65:684–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dyer AM, Baab K, Merchlinski A, et al. Rationale and development of a data coordinating center to support the Type 1 Diabetes in Acute Pancreatitis Consortium (T1DAPC). Pancreas. 2022;51:XXX–XXX. [DOI] [PMC free article] [PubMed] [Google Scholar]


