Esophageal cancer, with 544,076 deaths in 2020,1 includes esophageal adenocarcinoma and esophageal squamous cell carcinoma (ESCC). ESCC comprises 90% of esophageal cancers globally. The 5-year survival rate remains poor (15%–25% worldwide) because patients usually present with late disease.2 Although ESCC can be diagnosed by means of esophagogastroduodenoscopy (EGD) with biopsy, EGD is not widely available in low-income countries.2 Inexpensive, safe, accessible diagnostic alternatives will likely improve diagnosis and outcome.
DNA methylation often silences gene expression, inducing loss-of-function of tumor suppressor genes. Hypermethylation occurs at high levels and frequencies in ESCC. However, these potential biomarkers have not been tested extensively in minimally invasively–obtained cytologic samples from patients with ESCC, where they could increase detection of ESCC.
Less invasive, nonendoscopic sampling techniques have examined Barrett’s esophagus, a precursor of esophageal adenocarcinoma.3–6 A recent article described samples of mostly squamous esophageal dysplasia obtained via endoscopic biopsy, brushings, or older balloons in Iran and China.7 A study by Middleton et al8 was the first to use a sponge-based approach to screen for ESCC in Tanzania. Our approach uses quantitative methylation-specific polymerase chain reaction instead of histology, which requires pathologists to interpret findings of paraffin-embedded cell clots. Our study combines sponge-based sampling with a predictive biomarker panel to develop an accessible ESCC screening method in low-income countries, where EGD is not widely available. We used epigenetic changes (DNA methylation levels) as a basis for this strategy.
To discover and select epigenetic biomarkers, we first searched PubMed and The Cancer Genome Atlas for ESCC markers described previously (Supplementary Figure 1A). We then selected CpG sites using minimum and maximum γ-value (percent methylation) thresholds: high methylation levels in ESCC, but low levels in all types of normal tissues. Markers emerging from this step were tested in fully methylated DNA, unmethylated DNA, and finally paired normal–tumor tissues. Markers passing these steps were used to construct a multivariable model on a training set composed of EsophaCap sponge samples. Finally, this model was applied to a separate independent test set.
For the paired normal–tumor tissue step, the following 6 markers were analyzed: cg20655070, SALL1, SLC35F1, TAC1, ZNF132, and ZNF542. Only 5 of these—cg20655070, SLC35F1, TAC1, ZNF132, and ZNF542—were significantly hypermethylated in 48 tumor- vs normal-tissue pairs (P = .02, P = .0007, P =.03, P < .0001, and P = .009, respectively) (Supplementary Figure 1B).
For the sponge step, 35 patients with ESCC and 59 non-ESCC control subjects were analyzed; 5 other patients were excluded due to inability to swallow the sponge device. No hematochezia, chest pain, ulceration, perforation, or detached capsules occurred.
Characteristics of training- and test-set participants, including age, sex, tobacco use, alcohol use, race, and ESCC stage at time of sample collection, are shown in Supplementary Table 1. Median age (P = .01) and a history of smoking (P = .001) were significantly higher in patients with ESCC in the training set, although there was no difference among current smokers (18% vs 16%).
In the training set, cg20655070, SLC35F1, TAC1, ZNF132, and ZNF542 all exhibited significantly higher methylation levels in ESCCs than controls (Figure 1A). Receiver operating characteristic analyses yielded individual areas under the receiver operating characteristic curve of 0.73, 0.84, 0.78, 0.86, and 0.85, respectively (Figure 1B). Notably, the area under the receiver operating characteristic curve of age alone was 0.70 (Figure 1B).
Figure 1.
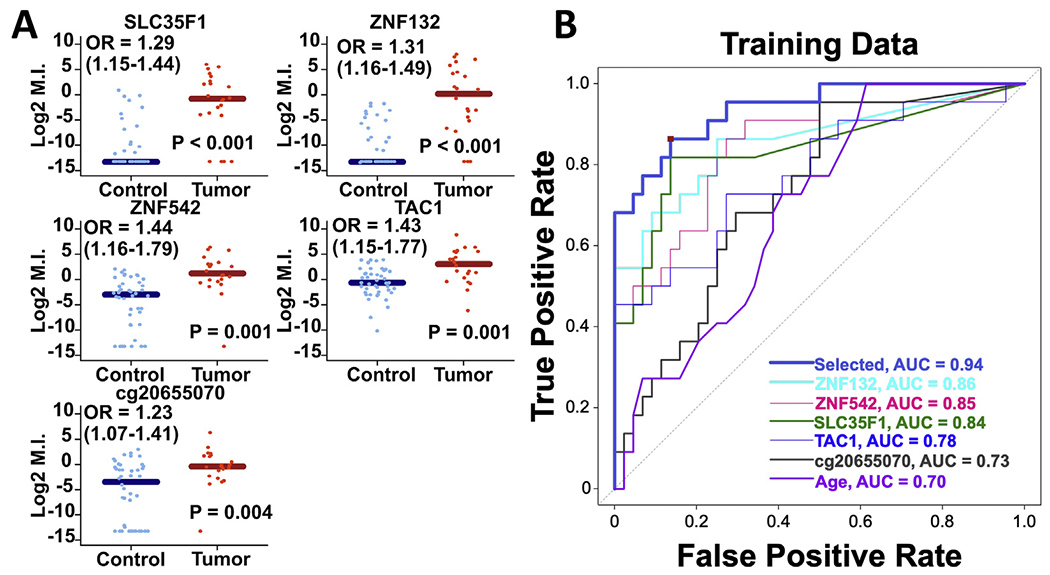
(A) Comparison of methylation levels between control and esophageal squamous cell carcinoma (ESCC) samples collected via the sponge device in the training set. Sample DNA was collected via the retrievable esophageal sponge device. Horizontal lines represent median methylation index values with 95% CI. cg20655070 (P = 0.004), SLC35F1 (P < 0.001), TAC1 (P = 0.001), ZNF132 (P < 0.001), and ZNF542 (P = 0.001) methylation levels were significantly higher in cytologic DNA of ESCC patients vs patients without neoplastic disease. (B) Univariate ROC analysis of diagnostic performance of cg20655070, SLC35F1, TAC1, ZNF132, and ZNF542 in DNA collected with the retrievable esophageal sponge capsule in the training set. The scores generated from the classification algorithm, as indicated by the dark blue plot labeled “Selected” (cg20655070 + SLC35F1 + ZNF132 + age), represent the weighted combination of biomarkers and clinical variables as identified by the LASSO approach with 5-fold cross-validation. The red square corresponds to Youden’s point and represents the optimum combination of sensitivity (0.86) and specificity (0.86) in the training set data.
A 3-marker classification algorithm (cg20655070, SLC35F1, and ZNF132) that included age as a fourth parameter was constructed on the basis of a score of the linear sum of weighted methylation levels for each individual. In the training set, this algorithm yielded an area under the receiver operating characteristic curve of 0.94 (Figure 1B). With the model index cutoff at 0.115, the formula yielded specificity = 0.86 and sensitivity = 0.86 in the training set. In an independent, subsequently collected, test set of 13 ESCC and 15 control sponges, the algorithm successfully classified 90% of patients, with specificity = 0.867 and sensitivity = 0.923; there were 2 false positives and 1 false negative in the test set, with positive predictive value = 0.86 and negative predictive value = 0.93.
Notably, the model showed 100% accuracy in 7 training-set patients with early-stage ESCC (4 with stage 1 and 3 with stage 2).
Thus, we present a systematic application of a stepwise approach to identify and validate discriminatory methylation-specific biomarkers for diagnosing ESCC in patient samples obtained using a minimally invasive, non-endoscopic strategy. This method and model, with potential generalizability to the at-risk population, meet a potential ESCC screening need because EGD is not widely available in ESCC-prone countries. Application of this screening strategy may ultimately improve early ESCC diagnosis, therapeutic intervention, and survival.
Candidate gene selection on the basis of both the published literature and The Cancer Genome Atlas data permitted identification of discriminant epigenetic biomarkers, some independently corroborated by others. Two biomarkers, ZNF542 and cg20655070, were found in the literature search. Notably, although Wang et al3 used The Cancer Genome Atlas data in their biomarker selection process, they did not apply their panels to nonendoscopic esophageal sponges.
No adverse events resulted from swallowing the sponge-capsule device, demonstrating its safety, convenience, ease, and efficacy. Previous studies in other diseases confirmed its simplicity in use and ability to sample the entire mucosa—of relevance to ESCC, which occurs in the proximal, middle, or distal esophagus. In Iyer et al,5 85% of participants expressed a willingness to swallow the device a second time.
In our training set, the ages of patients with ESCC were higher than the control subjects (P = .01); this difference was expected because most esophageal cancers occur in older individuals; the worldwide median age is 67.5 years. Similarly, tobacco consumption was higher in patients with ESCC than in control subjects, which is not surprising because this is the most significant risk factor for ESCC.9 In a recent Uganda study, smoking had the greatest population-attributable fraction for ESCC; most of our ESCC test-set patients (85%) and many ESCC training-set patients (43%) were from Uganda.10
In summary, we described a multimarker methylation biomarker panel that accurately discriminates between ESCC and nontumor patients in cytologic samples acquired via a minimally invasive sponge-capsule device. This strategy represents a low-risk, cost-effective esophageal sampling and diagnostic strategy that merits further study in larger prospective high-risk population screening trials.
Supplementary Material
Funding
This work was supported by grants from the National Institutes of Health (CA211457). Dr Meltzer is an American Cancer Society Clinical Research Professor and the Harry and Betty Myerberg/Thomas R. Hendrix Professor of Gastroenterology. This research was also supported by a generous gift from Peter T. Nicholl.
Conflicts of interests
These authors disclose the following: Stephen J. Meltzer is a founder of, and holds equity in, Capsulomics, Inc. Johns Hopkins University School of Medicine owns equity in Capsulomics, Inc. Ke Ma and Yulan Cheng are coinventors of technology licensed to Capsulomics, Inc. Ke Ma also owns equity in Capsulomics, Inc. Mouen A. Khashab is a consultant for Boston Scientific, Olympus America, Medtronic, Apollo Endosurgery, Pentax, and GI Supply, and receives royalties from UpToDate and Elsevier. Eun Ji Shin is a consultant for Boston Scientific. Vikesh K. Singh is a consultant to Abbvie and Ariel Precision Medicine and a scientific advisory board member to and stock option holder of Kyttaro, received grant funding from Abbvie, Orgenesis, and Theraly. Corey Nolet: Advisor to Capsulomics, Inc. Dennis Gong: Employed by Capsulomics, Inc. The remaining authors disclose no conflicts.
Abbreviations used in this paper:
- EGD
esophagogastroduodenoscopy
- ESCC
esophageal squamous cell carcinoma
Esophageal Squamous Cell Carcinoma Consortium
Boniface A. E. Lumori,1 Christopher K. Opio,2 Simran Jit,3 Ludmila Danilova,4 Zhe Wang,3,5 Cem Simsek,3 Saowanee Ngamruengphong,3 Eun Ji Shin,3 Mouen A. Khashab,3 Vikesh K. Singh,3 Alan H. Tieu,3 Corey Nolet,6 Dennis Gong,6,7 Kai-Hua Chang,8 Vishnu Prasath,8 Robert C. Bollinger,9 Tza-Huei Wang,7 Josephine Feliciano,10 Vincent K. Lam,10 Richard Battafarano,9 Michelle Turner,10 Peggy Lang,10 Kristen A. Marrone,10 and Hao Wang11; 1Department of Internal Medicine, Mbarara University of Science and Technology, Mbarara, Uganda; 2Aga Khan University Hospital, Nairobi, Kenya; 3Division of Gastroenterology and Hepatology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland; 4Division of Biostatistics, Department of Oncology, Johns Hopkins University School of Medicine, Baltimore, Maryland; 5Department of Gastroenterology, Tongji University School of Medicine, Shanghai, China; 6Capsulomics, Inc., Baltimore, Maryland; 7Department of Biomedical Engineering, Johns Hopkins University, Baltimore, Maryland; 8Department of Surgery, Johns Hopkins University School of Medicine, Baltimore, Maryland; 9Division of Infectious Diseases, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland; 10Department of Oncology, Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University School of Medicine, Baltimore, Maryland; and 11Department of Internal Medicine, Makerere University College of Health Sciences, Kampala, Uganda.
Footnotes
Supplementary Material
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2022.04.021.
CRediT Authorship Contributions
Ke Ma, MD (Conceptualization: Lead; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Writing – original draft: Lead; Writing – review & editing: Lead).
Andrew Kalra, BS (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Validation: Equal; Writing – original draft: Equal; Writing – review & editing: Equal).
Hua-Ling Tsai, ScM (Data curation: Equal; Formal analysis: Lead; Software: Lead; Visualization: Lead; Writing – review & editing: Supporting).
Samson Okello, MD (Resources: Supporting).
Boniface Elias Amee Lumori, MD (Resources: Supporting).
Christopher K. Opio, MD (Conceptualization: Supporting; Funding acquisition: Supporting; Supervision: Supporting).
Simran Jit, MBBS (Resources: Supporting).
Ludmila Danilova, PhD (Formal analysis: Supporting).
Zhe Wang, PhD (Formal analysis: Supporting; Investigation: Supporting).
Cem Simsek, MD (Resources: Supporting).
Saowanee Ngamruengphong, MD (Resources: Supporting).
Eun Ji Shin, MD (Resources: Supporting).
Mouen A. Khashab, MD (Resources: Supporting).
Vikesh K. Singh, MD (Resources: Supporting).
Alan H. Tieu, MD (Resources: Supporting).
Corey Nolet, PhD. (Formal analysis: Supporting; Software: Supporting).
Dennis Gong, BS (Formal analysis: Supporting).
Kai-Hua Chang, MD (Resources: Supporting).
Vishnu Prasath, BS (Resources: Supporting).
Robert C. Bollinger, MD, MPH (Conceptualization: Equal; Methodology: Supporting; Writing – review & editing: Supporting).
Tza-Huei Wang, PhD (Conceptualization: Supporting; Funding acquisition: Supporting; Methodology: Supporting; Supervision: Supporting).
Josephine Feliciano, MD (Resources: Supporting).
Vincent K. Lam, MD (Resources: Supporting).
Richard Battafarano, MD, PhD (Resources: Supporting).
Michelle Turner, MSN, CRNP (Resources: Supporting).
Peggy Lang, MSN, CRNP (Resources: Supporting).
Kristen A. Marrone, MD (Resources: Supporting).
Hao Wang, PhD (Formal analysis: Supporting; Funding acquisition: Supporting; Supervision: Supporting).
Yulan Cheng, MD (Conceptualization: Equal; Data curation: Supporting; Formal analysis: Supporting; Methodology: Equal; Validation: Equal).
Stephen J. Meltzer, MD (Conceptualization: Equal; Funding acquisition: Lead; Methodology: Equal; Supervision: Lead; Writing – original draft: Equal; Writing – review & editing: Equal).
Contributor Information
KE MA, Division of Gastroenterology and Hepatology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
ANDREW KALRA, Division of Gastroenterology and Hepatology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
HUA-LING TSAI, Division of Biostatistics, Department of Oncology, Johns Hopkins University School of Medicine, Baltimore, Maryland.
SAMSON OKELLO, Department of Internal Medicine, Mbarara University of Science and Technology, Mbarara, Uganda.
YULAN CHENG, Division of Gastroenterology and Hepatology, Department of Medicine, Johns Hopkins University School of Medicine, Baltimore, Maryland.
STEPHEN J. MELTZER, Division of Gastroenterology and Hepatology, Department of Medicine and Department of Oncology, Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University School of Medicine, Baltimore, Maryland
References
- 1.Sung H, et al. CA Cancer J Clin 2021;71:209–249. [DOI] [PubMed] [Google Scholar]
- 2.Couch G, et al. Cancer Prev Res 2016;9:558–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z, et al. Clin Cancer Res 2019;25:2127–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moinova HR, et al. Sci Transl Med 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iyer PG, et al. Am J Gastroenterol 2018;113:1156–1166. [DOI] [PubMed] [Google Scholar]
- 6.Iyer PG, et al. Gastrointest Endosc 2021;94:498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin Y, et al. Cancer Epidemiol Biomarkers Prev 2020;29:2642–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Middleton DRS, et al. Int J Cancer 2021;148:1208–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abnet CC, et al. Gastroenterology 2018;154:360–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okello S, et al. BMC Cancer 2016;16:446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


