Abstract
The Clostridium perfringens tetracycline resistance determinant from the 47-kb conjugative R-plasmid pCW3 is unique in that it consists of two overlapping genes, tetA(P) and tetB(P), which mediate resistance by different mechanisms. Detailed transcriptional analysis has shown that the inducible tetA(P) and tetB(P) genes comprise an operon that is transcribed from a single promoter, P3, located 529 bp upstream of the tetA(P) start codon. Deletion of P3 or alteration of the spacing between the −35 and −10 regions significantly reduced the level of transcription in a reporter construct. Induction was shown to be mediated at the level of transcription. Unexpectedly, a factor-independent terminator, T1, was detected downstream of P3 but before the start of the tetA(P) gene. Deletion or mutation of this terminator led to increased read-through transcription in the reporter construct. It is postulated that the T1 terminator is an intrinsic control element of the tet(P) operon and that it acts to prevent the overexpression of the TetA(P) transmembrane protein, even in the presence of tetracycline.
The Tet P determinant from the 47-kb conjugative R-plasmid pCW3 from Clostridium perfringens is unique among tetracycline resistance determinants in that it consists of two overlapping genes, tetA(P) and tetB(P), which mediate resistance by different mechanisms. The tetA(P) gene is 1,260 bp in length and encodes a 46-kDa protein, TetA(P), which is responsible for the active efflux of tetracycline from the cell (44). The TetA(P) protein is predicted to contain 12 transmembrane domains but is atypical because it does not have the typical structure or conserved motifs that are common to the other classes of tetracycline efflux proteins (7, 21). The tetB(P) gene, which overlaps the tetA(P) gene by 17 nucleotides (nt), is 1,956 bp in length and encodes a putative 72.6-kDa protein. The TetB(P) protein has significant amino acid sequence identity (37 to 39%) to Tet(M)-like cytoplasmic ribosomal protection proteins (44).
The tet(P) genes are the most widely distributed tetracycline resistance genes in C. perfringens, being found in both conjugative and nonconjugative tetracycline-resistant strains from diverse geographical locations and environmental sources (1, 24). Conjugative transfer of tetracycline resistance is invariably associated with plasmids that are either identical to or closely related to pCW3 (2, 3, 41). In these conjugative isolates, resistance is inducible. Inducible resistance is also observed when pCW3 is introduced into derivatives of strains CW234 and CW362, whereas in a strain 13 background tetracycline resistance is constitutively expressed, suggesting that induction requires an as-yet-unidentified host-encoded factor (19, 38). Resistance is also constitutively expressed in nonconjugative isolates.
Analysis of the approximately 1 kb of sequence data that are available upstream of tetA(P) (44) has revealed that this region is AT rich, having an overall G+C content of 22%, which is similar to the normal 24 to 27% G+C content of C. perfringens DNA (9, 20). There is a highly AT-rich region between bp 377 and 575, which has a G+C content of only 14% (44). Although several sequences with similarity to the consensus C. perfringens ς70-like promoter sequence (39) can be identified, the AT-rich nature of the upstream region has prevented the precise identification of the tet(P) promoter. No recognizable promoter appears to be present between the start codons of the tetA(P) and tetB(P) genes, and a potential factor-independent terminator (ΔG = −21.3 kcal mol−1) is present at the end of the tetB(P) gene, suggesting that these genes comprise an operon (44).
The objective of this study was to carry out detailed transcriptional analysis of the pCW3-encoded tet(P) genes. The results have shown that the tetA(P) and tetB(P) genes comprise an operon that is transcribed from a single promoter, P3, located 529 bp upstream of the tetA(P) start codon. Induction was shown to be at the level of transcription. A potential factor-independent terminator, T1, which is located some 390 bp downstream from the transcriptional start point but before the start of the tetA(P) gene, was also identified.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial plasmids used in this study are described in Table 1. Escherichia coli strains were derivatives of DH5α (Life Technologies) and were routinely grown at 37°C in 2× yeast extract-tryptone (YT) supplemented with ampicillin (100 μg/ml), chloramphenicol (20 μg/ml), erythromycin (150 μg/ml), or tetracycline (10 μg/ml). C. perfringens strains were derivatives of the chlorate- and streptomycin-resistant strain JIR33 (19) and were cultured under anaerobic conditions at 37°C in fluid thioglycolate medium (Difco), nutrient broth (38), or Trypticase peptone glucose broth (40). Solid media were prepared by the addition of 1.5% (wt/vol) bacteriological agar (Oxoid) prior to sterilization. C. perfringens strains grown on agar medium were incubated in an atmosphere of 80% N2–10% H2–10% CO2 in an anaerobic jar (Oxoid) or in an anaerobic chamber (COY Laboratory Products Inc.). Antibiotics were added where appropriate, unless otherwise stated, to the indicated concentrations: chloramphenicol (5 μg/ml), erythromycin (50 μg/ml), tetracycline (0.5 or 5 μg/ml), or streptomycin (1 mg/ml). Potassium chlorate-resistant (chlorate-resistant) strains were cultured on the appropriate solid medium containing 1% (vol/vol) saturated potassium chlorate solution.
TABLE 1.
Relevant characteristics of plasmids
| Plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| pCW3 | 47 kb; Tcr Tra+ | 2 |
| pJIR71 | pUC18Ω[PstI; pJIR39, 2.9 kb, contains tetA(P) and part of tetB(P)]; Tcr | 1 |
| pPSV | EmrlacZ′ catP′; pIP404 replication origin (C. perfringens); pUC18 replication origin (E. coli) | 29 |
| pJIR418 | Cmr EmrlacZ′; pIP404 replication origin (C. perfringens); pUC18 replication origin (E. coli) | 45 |
| pJIR1438 | pPSV HindIII/SphIΩ[HindIII/SphI; pJIR71, 839 bp, contains upstream region tetA(P)] | This study |
| pJIR1494 | pPSV HindIII/SphIΩ(ΔP3 SOE-PCR product; HindIII/SphI, 810 bp) | This study |
| pJIR1617 | pPSV HindIII/SphIΩ(ΔT1 SOE-PCR product; HindIII/SphI, 808 bp) | This study |
| pJIR1618 | pPSV HindIII/SphIΩ(ΔPE2 SOE-PCR product; HindIII/SphI, 810 bp) | This study |
| pJIR1644 | pPSV HindIII/SphIΩ(HindIII/SphI; pJIR1694, 838 bp, has ΔA at bp 525b) | This study |
| pJIR1645 | pPSV HindIII/SphIΩ(HindIII/SphI; pJIR1716, 839 bp, contains G-to-A change at bp 941b) | This study |
| pJIR1716 | Random mutant of pJIR71, shows increased tetracycline resistance, contains G-to-A change at bp 941b and A-to-G change at bp 2218b(T386A) | This study |
| pJIR1722 | pJIR1634 Asp718/T4 polymerase Ω(EcoRI/HindIII/T4 polymerase; pJIR1465, 0.25 kb, contains RK2 oriT) | This study |
Cmr, chloramphenicol resistance; Emr, erythromycin resistance; Tcr, tetracycline resistance; Tra+, transfer proficient.
The coordinates relate to the published nucleotide sequence (44) (GenBank accession no. L20800).
DNA isolation and molecular techniques.
Plasmid DNA was routinely isolated from E. coli strains using either the Magic Minipreps DNA Purification System (Promega), the High Pure Plasmid Isolation kit (Roche Molecular Biochemicals), or an alkaline lysis method (32). Plasmid DNA was prepared from C. perfringens cells as previously described (4, 27). Restriction endonuclease digestion and ligation of DNA were performed according to the manufacturer's instructions (Roche Molecular Biochemicals). Transformation of E. coli (42) and C. perfringens (43) cells was performed as described previously. PCR amplification was carried out with Taq DNA polymerase (Roche Molecular Biochemicals). PCR products for nucleotide sequencing and cloning were purified as described previously (26).
Nucleotide sequence analysis was performed either by the dideoxynucleotide chain termination method using a T7 Sequencing kit (Pharmacia) or the ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction kit (PE Applied Biosystems) and an ABI 373A automated fluorescent sequencing apparatus in accordance with the manufacturer's instructions. Sequence analysis was carried out with Sequencher 3.0 software (Gene Codes Corporation). Oligonucleotide primers (Table 2) used for the preparation of probes, reverse transcriptase PCR (RT-PCR), nucleotide sequencing, or primer extension were synthesized using a 392 DNA/RNA synthesizer (PE Applied Biosystems).
TABLE 2.
Oligonucleotide primers
| Primer no. | Sequence (5′-3′) | Position (template) | Usea |
|---|---|---|---|
| 212 | TCGGGGACTATTACTA | 2449–2464b | LP |
| 274 | AACCTGTGGTTATGTAT | 3683–3667b | LP |
| 1366 | CACAGATTGTATGGGGATTAGG | 1364–1385cd | PCR, LP |
| 1367 | CATTTATAGAAAGCACAGTAGC | 2128–2107cd | PCR, LP |
| 1369 | GCTACTGTGCTTTCTATAAATG | 2107–2128cd | PCR, RT-PCR, S |
| 1370 | ATGTGTCAAATAATATTCTTGT | 2657–2636cd | PCR, RT |
| 1947 | TTATTAACCATTAATCATCACC | 1073–1052c | PCR, PE, S |
| 2980 | AATAAGTAAACAGGTAACGTCT | 1657–1679e | LP |
| 2981 | GCTCCTTGGAAGCTGTCAGTAG | 2344–2322e | LP |
| 3333 | CTCTTCCAACTGATTTTATAGC | 2412–2395c | PE, S |
| 3334 | GCCTCATAATCATATAACCTAA | 933–912c | PE, S |
| 3534 | AGCATTGCAGGATATAGTTTC | 296–276g | SOE, S |
| 3644 | CAAGATTTTTCATATACT | 659–642c | PE, S, LP |
| 4392 | AATACCGTGTAAAGATGTAACATGCTCATTAT | 773–750cf | SOE |
| 4393 | AAAGCTGTACATATAACCTAAATATAAACATA | 923–901cf | SOE |
| 4400 | GGTTATATGTACAGCTTTTTTTGTTTAAGGAG | 955–977cf | SOE |
| 4401 | ACATCTTTACACGGTATTTTTATGTCTATTTT | 802–824cf | SOE |
| 4402 | ATAAAAATAGTAGTTATGCTAAAATATAAAC | 534–555cf | SOE, LP |
| 4403 | CATAACTACTATTTTTATATTTATATTTATCC | 504–482cf | SOE |
| 8034 | CACGGTATTTTTATGTCTATTTT | 802–824c | LP |
| 8133 | AACATAAAATATAGAACT | 906–889c | LP |
S, sequencing; LP, oligonucleotides used for preparing labeled probes; PE, primer extension.
Refers to relevant position within the Tn4451 sequence (6), GenBank accession no. U15027; see also the work of Lyras et al. (26).
Refers to relevant position within the Tet P determinant (44), GenBank accession no. L20800.
See the work of Lyras and Rood (25).
Refers to relevant position within the Erm B determinant of C. perfringens (8), GenBank accession no. U18931.
Oligonucleotide contains 9 bp at the 5′ end which are complementary to the corresponding primer, which was used in deletion of the putative promoter regions.
Refers to relevant position within the catP sequence (48), GenBank accession no. M74769.1.
Construction of the tetA(P)-catP transcriptional fusion, pJIR1438.
An 839-bp pJIR71-derived HindIII/SphI fragment, which carried the upstream region and the start of the tetA(P) gene (44), was cloned into the C. perfringens promoter probe shuttle vector pPSV (29) to construct pJIR1438.
Construction of the deletion derivatives pJIR1617, pJIR1618, and pJIR1494.
These deletion derivatives were constructed from a pJIR1438 template by splice overlap extension (SOE)-PCR (15, 16). Initially, for each deletion two separate PCRs were performed with pJIR1438 as a template. The first PCR, using the primers UP and either 4393, 4392, or 4403 (Table 2), amplified the DNA upstream of each of the respective primer extension endpoints. The second PCR utilized primers 3534, which binds downstream of the catP translational initiation codon, and either 4400, 4401, or 4402 (Table 2) to amplify the region downstream of each of these endpoints. The resultant PCR products contained complementary sequences and were purified from a 1.0% low-melting-point agarose (FMC BioProducts) gel with the Magic PCR Preps DNA Purification system (Promega). The two products specific for each deletion derivative were then mixed, and a third PCR was performed using primers UP and 3534. The PCR consisted of 30 cycles of 1 min of denaturation at 91°C, 1 min of annealing at 37°C, and 3 min of extension at 71°C. The resultant SOE-PCR products were excised and extracted as before. These products were digested with HindIII and SphI and ligated to HindIII/SphI-digested pPSV DNA. The resultant recombinant plasmids pJIR1617, pJIR1618, and pJIR1494 carried deletions in the T1, PE2, and P3 regions, respectively. The insert in each of the recombinant plasmids was sequenced to confirm that the precise deletion had occurred and that no other changes were present.
Determination of chloramphenicol MICs.
For each of the deletion mutants, chloramphenicol MICs were determined in both E. coli and C. perfringens at 37°C as described previously (21). Briefly, for E. coli strains, overnight broth cultures were diluted 1:25 into fresh 2× YT broth containing erythromycin and grown until the turbidity at 550 nm was 0.7 to 0.8. Cultures were then diluted 1:100 in fresh broth. Duplicate 10-μl aliquots were then placed onto 2× YT containing chloramphenicol at concentrations ranging from 0 to 200 μg/ml. The cultures were incubated for 18 to 20 h at 37°C, and the MIC was determined as the lowest concentration of chloramphenicol that completely inhibited growth. Assays were repeated three times. For C. perfringens, an essentially identical procedure was followed with the exceptions that brain heart infusion medium was substituted for 2× YT medium and chloramphenicol concentrations in the range of 0 to 80 μg/ml were used.
Preparation of C. perfringens RNA.
Total RNA was extracted from 20 or 100 ml of C. perfringens broth cultures using Trizol reagent (Gibco-BRL) as described previously (25) with the additional step of treatment with RNase-free DNase (Promega). For strain JIR33(pCW3), RNA was extracted from cultures that had been grown either in the presence or in the absence of tetracycline (5 μg/ml) from starter cultures grown in the presence or absence of medium containing subinhibitory levels of tetracycline (0.5 μg/ml). For strains harboring the tetA(P)-catP transcriptional fusion plasmid pJIR1438 and its derivatives, cells were grown in medium containing erythromycin (50 μg/ml). RNA either was used directly or was stored at −70°C following the addition of 10 μl of 2 M NaCl and 2 volumes of 100% ethanol. The concentration of RNA was determined by measuring the absorbance at 260 nm using a DMS 100 UV-visible spectrophotometer (Varian Technology). The RNA concentration was calculated based on the assumption that an absorbance reading of 1 was equivalent to an RNA concentration of 40 μg/ml (42).
RT-PCR.
RT reactions were performed on total RNA using a commercially available Reverse Transcription system (Promega) with slight modifications to the recommended protocol. RT reactions were performed in a final volume of 20 μl, which contained 5 mM MgCl2, 1× RT buffer, 1 mM (each) deoxynucleoside triphosphates, 1 U of RNasin, 15 U of avian myeloblastosis virus RT, 3 μM oligonucleotide primer, and 2.5 to 5 μg of substrate RNA. Reaction mixtures were incubated at 42°C for 1 h, and reactions were terminated by boiling the mixtures for 5 min, followed by incubation on ice for 5 min. cDNA products were amplified in 25-μl PCR mixtures using 5 μl of the RT reaction mixture as the template and a standard PCR protocol of 30 cycles of a 1-min denaturation step at 95°C, a 2-min annealing step at 50°C, and a 3-min extension step at 72°C. PCR products were analyzed on 1.5% agarose gels and were sequenced to confirm that they represented the correct nucleotide sequence.
Primer extension analysis.
Primer extension analysis was carried out using a commercially available Primer Extension system (Promega) as described before (25). Initial experiments involved primers 1947 (Table 2), which was complementary to the tetA(P) translational initiation codon, and 3333 (Table 2), which was complementary to a region 133 bp downstream of the initiation codon of tetB(P). Further primer extension analysis was performed using oligonucleotides 3334 and 3644. Oligonucleotides were 5′ end labeled with 40 μCi of [γ-32P]dATP (4,000 Ci [148 TBq]/mmol) (GeneWorks) using T4 polynucleotide kinase (Promega) and then annealed with approximately 50 μg of total C. perfringens RNA. Extension reactions were carried out using avian myeloblastosis virus RT according to the manufacturer's instructions (Promega). cDNA products were ethanol precipitated and separated on an 8% polyacrylamide gel containing 8 M urea.
RNA dot blots.
RNA samples for dot blotting were applied to a Hybond-N+ nylon membrane using a dot blot apparatus (Minifold SRC 96; Schleicher and Schuell). RNA (25 μg) was precipitated and resuspended in RNA dilution buffer (diethyl pyrocarbonate-treated distilled H2O, 20× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 37% [wt/vol] formaldehyde [5:3:2]), and aliquots (100 μl) were heated at 65°C for 15 min to denature the RNA before it was applied to the filter. The filters were then air dried and cross-linked under UV light (312 nm) for 3 to 5 min.
Radioactively labeled DNA probes specific for the tet(P) upstream region, and the catP and erm(B) genes, were derived by PCR with the following primer pairs: 4402 and 3644 for P3-PE2, 8034 and 8133 for PE2-T1, 274 and 212 for catP, and 2981 and 2980 for erm(B). Oligonucleotides that bound to the coding strand were 5′ end labeled with [γ-32P]dATP using T4 polynucleotide kinase (18). PCR was performed using pJIR71, pJIR45, and pJIR418 templates for the tet(P)-, catP-, and erm(B)-specific probes, respectively. The amplified labeled products were either isolated from an agarose gel using the BRESAclean DNA purification kit (GeneWorks), if nonspecific products were also present, or purified directly from the PCR with the BRESAclean nucleic acid purification kit. In each experiment, probes of equivalent specific activity, as determined in a scintillation counter, were utilized for each blot.
Prehybridization was carried out for at least 1 h at 60°C in hybridization solution, which contained 1% bovine serum albumin, 5× SSC, and 1% sodium dodecyl sulfate. Probe DNA in hybridization solution was boiled for 10 min and placed on ice prior to use. Hybridization was carried out at 60°C, followed by stringency washes (0.1× SSC, 0.1% sodium dodecyl sulfate) at 60°C. After washing, the membranes were exposed to a storage phosphor screen (Molecular Dynamics), which was analyzed using a STORM Phosphoimager system and ImageQuant image analysis software (Molecular Dynamics).
Northern hybridization.
Northern analysis was performed on either 20 μg of RNA for analysis of the pJIR1438 derivatives or 10 μg of RNA for the analysis of strains carrying pCW3. RNA samples were denatured in RNA sample buffer (150 μl of formamide, 52.5 μl of 37% [wt/vol] formaldehyde, 3 μl of 50× running buffer, 15 μl of 1-mg/ml ethidium bromide) at 65°C for 5 to 10 min and separated through 1.5% agarose gels containing formaldehyde. Standards included either unlabeled RNA markers (Promega) or [α-32P]ATP (3,000 Ci [111 TBq]/mmol) (GeneWorks)-labeled RNA markers prepared with the RiboMark labeling system (Promega) in accordance with the manufacturer's instructions. RNA was transferred overnight at 4°C to a Hybond-N+ nylon membrane by capillary transfer using 10× SSC. The nylon membrane was air dried and cross-linked under UV light (312 nm) for 3 to 5 min.
DNA probes specific for the tet(P) upstream region and the catP and erm(B) genes were derived by PCR as described for the dot blots. The DNA probe specific for the tetA(P) gene was derived in an analogous manner, with primers 1366 and 1367 (Table 2). Prehybridization, hybridization, and detection were carried out as described for dot blot hybridization analysis.
RESULTS
Transcriptional organization of the tet(P) genes.
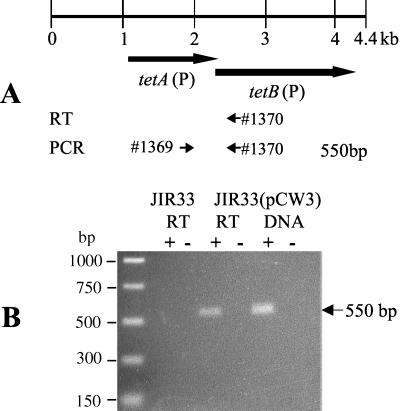
To determine if the tetA(P) and tetB(P) genes were arranged in an operon, Northern blots were initially performed with separate gene-specific probes on RNA that was isolated from C. perfringens cells that harbored pCW3. Unfortunately, a hybridizing smear was consistently observed for both the tetA(P) and tetB(P) probes (data not shown), suggesting that the RNA transcript was unstable. Therefore, the alternative method of RT-PCR analysis was performed using primer 1370, which was specific for the putative tetA(P)-tetB(P) transcript and bound 323 nt downstream of the tetB(P) start codon (Fig. 1), and RNA from JIR33(pCW3), which exhibits an inducible tetracycline resistance phenotype (19). The resultant cDNA molecules were then amplified using PCR with primers 1369 and 1370, which produce a product of 550 bp. The oligonucleotide 1369 binds within tetA(P), 179 bp upstream of the tetB(P) start codon. An RT-dependent product was observed, indicating that the tetA(P) and tetB(P) genes were transcriptionally coupled (Fig. 1). To determine whether a promoter was present upstream of the tetB(P) gene, primer extension analysis was carried out on RNA from JIR33(pCW3) using primer 3333, which was complementary to a region 133 nt downstream of the initiation codon of tetB(P). No cDNA products were detected (data not shown). Based on these results, it was concluded that the tetA(P) and tetB(P) genes formed an operon.
FIG. 1.
RT-PCR analysis of tet(P) RNA. (A) Schematic showing oligonucleotides utilized in RT-PCR experiments. The locations and extents of the tetA(P) and tetB(P) genes are shown. The locations of the oligonucleotide primers used in RT-PCR analysis are indicated by the numbered arrows. (B) Agarose gel electrophoresis of RT-PCR products. RT-PCR analysis was performed using total RNA that was isolated from JIR33(pCW3) cells grown in the presence of tetracycline (5 μg/ml) and from JIR33 grown without tetracycline. The positive controls, labeled DNA, used pCW3 templates extracted from JIR33(pCW3). The + and − labels refer to reactions performed in the presence and absence of RT, respectively. The + and − under the DNA label refer to PCRs performed in the presence and absence of DNA template, respectively.
Primer extension analysis of the tet(P) operon.
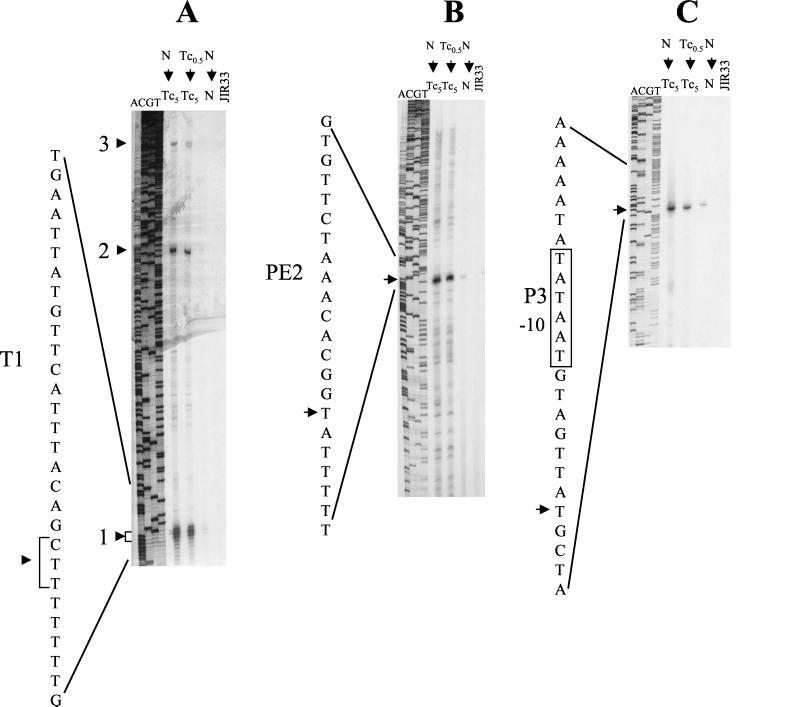
To identify promoters upstream of tetA(P), primer extension analysis was performed on RNA extracted from JIR33(pCW3) cells that had been grown in the presence and absence of tetracycline (5 μg/ml), from starter cultures grown in the presence and absence of subinhibitory but inducing levels of tetracycline (0.5 μg/ml). Total RNA extracted from JIR33 was used as the negative control. Primer extension reactions were carried out with primer 1947, which was complementary to the beginning of the tetA(P) coding region. Three major cDNA products were consistently obtained from RNA extracted from JIR33(pCW3) cells that had been exposed to tetracycline, whereas either smaller amounts of product or no products were observed for cells grown in the absence of tetracycline (Fig. 2A).
FIG. 2.
Primer extension analysis of tet(P) transcripts. cDNA products were produced by RT extension of tet(P) transcripts using the 32P-end-labeled oligonucleotides 1947 (A), 3334 (B), and 3644 (C) (Fig. 3). RNA was extracted from the inducible strain JIR33(pCW3), which had been grown in the media indicated. Primer extension reactions were performed on 50 μg of total RNA. RNA extracted from JIR33 was subjected to the same treatment and used as negative controls. The sequencing lanes labeled ACGT were produced using the same oligonucleotide on a pJIR71 template, which contains the tet(P) upstream region. The three major cDNA products detected are indicated by the arrows and are labeled 1, 2, and 3. The −10 region of the putative P3 promoter is indicated, as are the T1 and PE2 regions. N, nutrient broth; Tc0.5, nutrient broth containing subinhibitory tetracycline (0.5 μg/ml); Tc5, nutrient broth containing tetracycline (5 μg/ml).
Primer extension endpoints may represent transcriptional start points and thereby identify promoters. However, they may also result from mRNA processing or from the presence of secondary mRNA structures that lead to RT stalling. The smallest product observed in these experiments was located approximately 102 nt upstream of the tetA(P) translational initiation codon and appeared to be a result of the latter process (Fig. 3). It was subsequently shown to represent the site of a transcriptional terminator, designated T1. The second product, designated PE2, was mapped to a position 256 nt upstream of the tetA(P) initiation codon (Fig. 2B), and the third was mapped to a point 522 nt upstream of this ATG codon (Fig. 2C). The latter was subsequently shown to be the transcript originating from the tet promoter, P3.
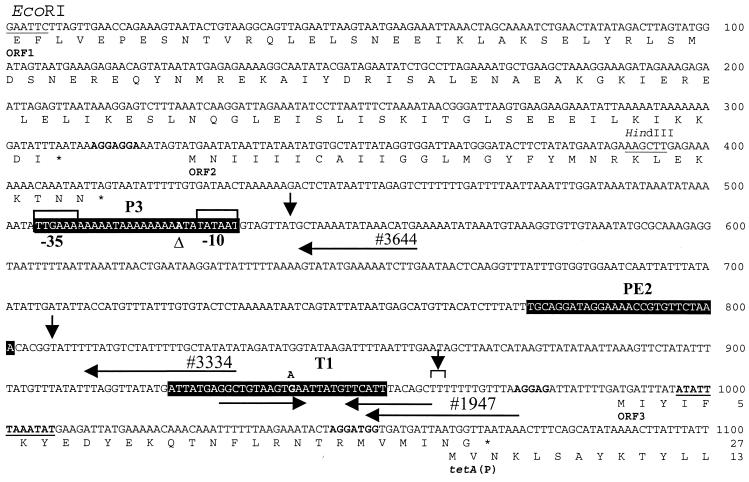
FIG. 3.
Location of putative promoters in the tetA(P) upstream region. The positions of the oligonucleotide primers 1947, 3334, and 3644, which were used for primer extension analyses, are indicated by the labeled arrows above the sequence. The primer extension endpoints are indicated by the downward-pointing arrows. The −10 and −35 regions of the P3 promoter are boxed. Also indicated are other features of the upstream region including ORF1, ORF2, ORF3, and the start of the tetA(P) gene (44). The deduced amino acid sequence is shown below the nucleotide sequence. Stop codons are indicated by the asterisks. Potential ribosome binding sites are shown in boldface. The inverted repeat that forms the factor-independent transcriptional terminator, T1, is indicated by the horizontal arrows. The 12-bp palindromic sequence previously identified is underlined and shown in boldface (44). The positions of the P3-525ΔA and T1-G941A mutations within the constructs pJIR1644 and pJIR1645, respectively, are also indicated above the sequence. The regions deleted in the construction of ΔP3, ΔPE2, and ΔT1 derivatives are indicated in reversed type. Coordinates refer to the published sequence (44), GenBank accession no. L20800.
Mutagenesis and analysis of T1, PE2, and P3.
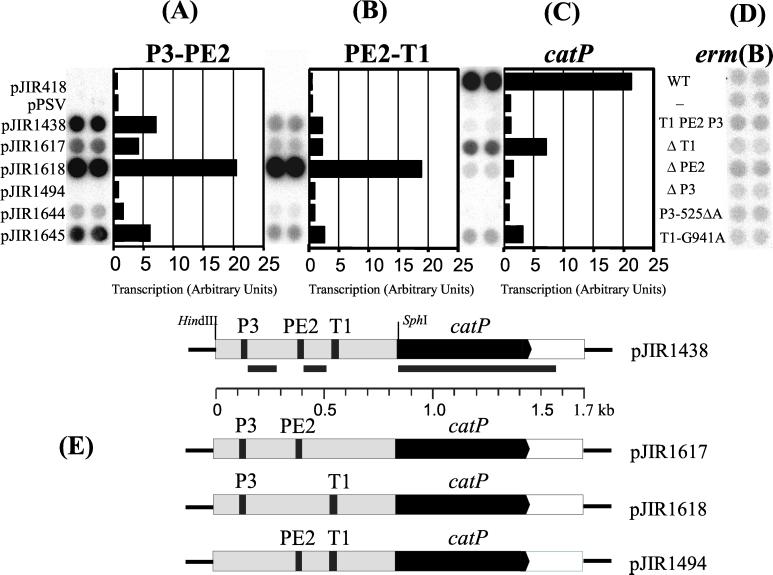
To examine the roles of T1, PE2, and P3 in gene expression, SOE-PCR (15, 16) was used to construct separate plasmids that each contained deletions in the regions upstream of the primer extension endpoints. Because of difficulties experienced with the genetic manipulation of shuttle vector constructs containing the tet(P) operon, it was necessary to use a reporter construct containing an alternative antibiotic resistance gene. An 839-bp HindIII/SphI pJIR71-derived fragment, which carried the relevant upstream region and the start of the tetA(P) gene (44), was cloned into the C. perfringens promoter probe shuttle vector pPSV (29) to construct pJIR1438 (Fig. 4). SOE-PCR was then used to delete each of the T1, PE2, and P3 regions, and each of the resultant PCR fusion products was cloned into pPSV to construct pJIR1617, pJIR1618, and pJIR1494, respectively (Fig. 4). pPSV contains a promoterless chloramphenicol acetyltransferase gene, catP, which confers chloramphenicol resistance (29).
FIG. 4.
Transcriptional analysis of promoter mutants by RNA dot blot hybridization. Total RNA was extracted from JIR33 cells harboring the constructs indicated, grown in the presence of erythromycin (50 μg/ml). RNA extracted from JIR33 was used as a negative control. RNA (25 μg) was spotted in duplicate onto each filter and probed with γ-32P-labeled probes specific for the P3-PE2, PE2-T1, catP, and erm(B) transcripts. A schematic of the tet(P)-catP transcriptional fusion showing the extent of the hybridizing probes, indicated by the black bars, is shown in panel E, together with a diagrammatic representation of each of the deletion mutants. RNA was quantified from each of the dot blots with a Storm Phosphoimager and ImageQuant software (Molecular Dynamics). For each blot, the average volume (the integrated intensity of the pixels) of each duplicate dot for each construct was calculated. This value was then standardized for the RNA levels of the erm(B) control. The levels of transcript are expressed as arbitrary units, and each value represents a ratio of average volume per dot/average volume per erm(B) control. WT, wild type.
In addition to these constructs, two other plasmids that contained mutations in the tetA(P) upstream region were analyzed. These plasmids were derived from pJIR1694 and pJIR1716, which had been detected during previous studies that involved random mutagenesis of the tetA(P) gene (7). The plasmid pJIR1694 (7) had a deletion of an A residue at position 525, between the −35 and −10 regions of the putative promoter P3 (Fig. 3). This deletion resulted in a reduction of the tetracycline MIC in E. coli from 30 to 15 μg/ml (7). The plasmid pJIR1716 had a mutation of a G residue to an A residue at bp 941, which was within the T1 region (Fig. 3). This plasmid also had a change from an A to a G residue at bp 2218 [TetA(P)-T386A]. The result of these two mutations was a hyper-tetracycline-resistance phenotype in E. coli, with a tetracycline MIC of 50 μg/ml (T. Bannam and J. Rood, unpublished data). The HindIII/SphI fragments, containing the upstream regions and the start of the tetA(P) genes from pJIR1694 and pJIR1716, were cloned into pPSV to construct pJIR1644 and pJIR1645, respectively. Note that the TetA(P)-T386A mutation encoded on pJIR1716 was not present in the upstream region used to construct pJIR1645. Therefore, any phenotypic changes observed with this plasmid must be solely the result of the G941A mutation in the T1 region.
All of the pPSV derivatives, as well as the recombinant plasmid pJIR418 (45), which carries the wild-type catP gene and its native promoter, were introduced into C. perfringens strain JIR33 for further analysis. The effect of each of the deletions and mutations on promoter activity was assayed in vivo by determining the MICs of chloramphenicol. The results (Table 3) showed that deleting (pJIR1617) or mutating (pJIR1645) the T1 region led to a sixfold increase in MIC. Similarly, deleting the PE2 region (pJIR1618) resulted in a greater-than-twofold increase in MIC. By contrast, deleting the putative promoter region P3 (pJIR1494), or mutating it by changing the spacing between the −35 and −10 sequences from 17 to 16 bp, resulted in a decrease in MIC, essentially to that observed for the vector control, pPSV. These results suggested that P3 was the promoter responsible for transcription of the tet(P) operon.
TABLE 3.
MICs for the deletion mutants of E. coli and C. perfringens
| Plasmid | Mutation | Chloramphenicol MIC (μg/ml)
|
|
|---|---|---|---|
| E. coli (DH5α) | C. perfringens (JIR33) | ||
| None | None | NDa | <0.5 |
| pJIR418 | Positive control | >200 | 60 |
| pPSV | Vector control | 15 | 1.5 |
| pJIR1438 | Wild type | 52.5 | 4 |
| pJIR1617 | ΔT1 | >200 | 25 |
| pJIR1618 | ΔPE2 | 67.5 | 10 |
| pJIR1494 | ΔP3 | 22.5 | 1.5 |
| pJIR1644 | P3-525ΔA | ND | 2.5 |
| pJIR1645 | T1-G941A | ND | 25 |
ND, not done.
Transcriptional analysis of the tetA(P)-catP fusions.
To determine the transcriptional status of the upstream regions, primer extension analysis was performed on each of the reporter constructs of the deletion derivatives and the random mutants. Analysis of RNA extracted from cells carrying either pJIR1617 (ΔT1) or pJIR1645 (T1-G941A) revealed cDNA products corresponding to products previously mapped downstream of T1, PE2, and P3 (data not shown). Analysis of pJIR1618 (ΔPE2) revealed cDNA products corresponding to those detected downstream of T1 and P3 but barely detectable levels of product 2. Finally, for pJIR1494 (ΔP3), no cDNA products corresponding to those products mapped downstream of PE2 and P3 were detected and only extremely low levels of product 1 were observed (data not shown). Only low levels of all three major cDNA products were observed for pJIR1644 (P3-525ΔA). Taken together, these results provided further evidence that P3 was the tet(P) promoter.
Quantitative RNA dot blot analysis of the transcriptional fusions.
Dot blot hybridization analysis was performed on RNA isolated from JIR33 derivatives harboring the shuttle vector constructs. The RNA was hybridized with DNA probes that were specific for the regions between P3 and PE2 (P3-PE2 probe), PE2 and T1 (PE2-T1 probe), and for the catP gene (catP probe) (Fig. 4E). As a quantitative control for RNA concentration, the RNA preparations were also probed using an erm(B)-specific probe. Since the erm(B) gene located on pPSV is constitutively expressed, the level of transcription of this gene should be equivalent for each of the derivatives studied (Fig. 4D).
When the wild-type upstream region was present (pJIR1438), the level of catP mRNA was slightly higher than that of background (Fig. 4C). Hybridization analysis with the other probes showed that in cells containing pJIR1438 there was more upstream transcript present than catP transcript (Fig. 4A and B). Changes in the T1 region, whether by deletion (pJIR1617) or point mutation (pJIR1645), resulted in an increase in the level of catP transcript. This increase was greater in the deletant than in the random mutant (Fig. 4C). In the upstream region, lower levels of transcript than that seen for the catP probe were observed. There was also a decreased amount of transcript within the P3-to-PE2 region compared to that observed for the wild type. Again, this effect was more distinct in the deletant (Fig. 4A). By contrast, a wild-type level of hybridization was observed for both the T1 deletant and the T1-G941A mutant with the PE2-T1 probe (Fig. 4B).
Deletion of the PE2 region (pJIR1618) resulted in a slight increase in the amount of catP transcript, to a level less than twofold higher than that observed for the wild type (Fig. 4C). The effect of this deletion on the upstream transcript(s) was more dramatic (Fig. 4A and B), with an increase of just under threefold and slightly more than eightfold compared to the wild type for the P3-PE2 and P2-T1 probes, respectively (Fig. 4A and B).
Finally, deletion of the P3 promoter (pJIR1494) resulted in a decrease in the level of all transcripts to that of background (pPSV) (Fig. 4A to C). Slightly higher levels of hybridization compared to background were observed for the P3-521ΔA mutant, pJIR1644. These results were in agreement with the primer extension analysis and MIC determinations. They confirmed that P3 was the tet(P) promoter. When this promoter was deleted or even changed slightly by altering the spacing between the −35 and −10 regions, reporter gene expression was reduced to background levels. This result implied that there were no significant promoters downstream of P3. The results also implied that sequences downstream of the P3 transcriptional start point were involved in modulating catP expression resulting from the P3 promoter.
Putative RNA secondary structures in the tet(P) upstream region.
Examination of the region upstream of the tetA(P) gene for RNA folding predictions using the programs Foldrna (52) and mfold (28, 53) revealed that the T1 and PE2 regions were associated with potential RNA stem-loop secondary structures with ΔG values of −17.1 and −13.3 kcal/mol, respectively. Modeling of the upstream regions of the ΔPE2 and ΔT1 transcripts revealed no significant changes from that observed for the wild type, apart from the deletion of the appropriate structures. The T1 structure consisted of a stem containing four G+C pairs followed by a string of T residues and resembled the characteristic structure of a factor-independent terminator (10, 36, 37). Analysis of this potential structure by use of an algorithm (10) designed to predict Rho-independent terminators suggested that it was a good terminator candidate. This algorithm calculates a value, d, which correlates with a predicted termination efficiency, expressed as a percentage. In general, positive d values are regarded as the necessary requirement to describe the structure of E. coli Rho-independent terminators (10). The T1 region was calculated as having a d value of 35.51, which correlated with an in vitro termination efficiency of ∼86%. The deletion of this region would inhibit the formation of this structure, and changes in this stem-loop structure caused by the G-to-A transition at position 941, which prevents the formation of a GC base pair, reduce the ΔG to −11.1 kcal/mol. This potentially creates a less efficient terminator with a calculated d value of 17.95, which correlates with a termination efficiency of ∼60%. Based on these data and in conjunction with the MIC and dot blot hybridization results, it was concluded that the structure located in the T1 region represented a transcriptional terminator.
Examination of the PE2 structure revealed that it was also followed by a string of T residues, with an A residue separating the T residues from the stem-loop structure. This structure also bears some resemblance to factor-independent terminators, but analysis similar to that performed for T1 revealed that the PE2 region did not meet the minimum conditions that are used to define a terminator. This result does not rule out the possibility that this region of RNA secondary structure is acting as a terminator in C. perfringens. Even in E. coli, a small proportion of terminators do not conform to the constraints of this algorithm and have negative d values (10).
Evidence for small RNA transcripts derived from the tet(P) upstream region.
If the predicted secondary structures were formed in vivo, then short 5′-proximal RNA transcripts representing terminated mRNA molecules would be present. To detect these molecules, Northern hybridization analysis was performed on purified RNA preparations extracted from the JIR33 derivatives used in the previous experiments, using radioactively labeled P3-PE2 and catP-specific probes (Fig. 4E).
Using the P3-PE2 probe, two small hybridizing bands, which were estimated to be approximately 250 and 410 nt in size, were observed for the wild-type construct, pJIR1438 (Fig. 5A, lane 2). If the 5′ ends of these bands mapped to the transcriptional start point downstream of P3, their 3′ ends would map near the PE2 and T1 structures, respectively. Further evidence that the 410-nt transcript represented a product terminating at T1 came from examination of the profile of the ΔT1 and G941A mutants. When probed with the P3-PE2 probe, only the 250-nt RNA band was observed for the T1 deletant (Fig. 5A, lane 3), whereas both the 250-nt band and a faintly hybridizing 410-nt band were observed for the T1-G941A mutant (Fig. 5A, lane 7). Two larger bands estimated as approximately 1.43 and 0.77 kb in size were also observed for both mutants (Fig. 5A). These bands also hybridized to the catP probe (Fig. 5B, lanes 3 and 7). Weakly hybridizing bands of approximately 1.43 and 0.77 kb in size were also observed for wild-type and ΔPE2 RNA when probed with catP (Fig. 5B, lane 4).
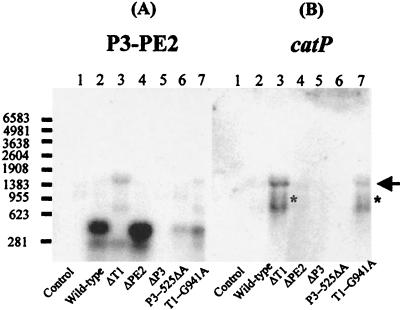
FIG. 5.
Northern hybridization analyses of RNA from each of the promoter mutants. Total RNA was isolated from cells of JIR33 derivatives grown in the presence of erythromycin (50 μg/ml). RNA (20 μg) was subjected to electrophoresis through 1.5% formaldehyde gels and transferred to nylon membranes, which were subsequently probed with the γ-32P-labeled P3-PE2- and catP-specific probes, respectively (Fig. 4). The arrow represents full-length tetA(P)-catP transcript. The asterisks mark the faintly hybridizing 1.0- to 1.1-kb transcripts observed in the blot probed with catP. The size of RNA markers is indicated at the left in bases. Lanes: 1, pPSV; 2, pJIR1438; 3, pJIR1617; 4, pJIR1618; 5, pJIR1494; 6, pJIR1644; 7, pJIR1645.
Promoter fusion constructs initiating at P3 and terminating at the catP transcriptional terminator would yield an ∼1,460-nt transcript. As a result, it was concluded that the 1.43-kb band (Fig. 5) represented a full-length mRNA transcript initiating at the transcriptional start point downstream of P3. The 0.77-kb and 1.0- to 1.1-kb hybridizing bands observed in the T1 deletant and T1 mutant most probably represent transcripts resulting from RNA processing or degradation.
Analysis of the PE2 deletant with the P3-PE2 probe revealed that the two smaller hybridizing bands were present but at greater intensity than that observed for the wild type (Fig. 5A, lane 4). The first of these bands was smaller, correlating with the 29-bp deletion of the PE2 region. The presence of the second band, slightly less than 250 nt, in this deletant indicated that the 3′ end of the smaller RNA transcript did not map to the PE2 stem-loop structure, as this sequence was not present in the deletant. The 3′ end of this transcript may map further downstream, or alternatively, it may represent a breakdown product obtained from RNA processing. In agreement with the results already obtained, no hybridization to any of the probes was observed with RNA from the P3 deletant (Fig. 5, lane 5), and only a very faintly hybridizing 410-nt P3-specific band was observed for RNA from the P3-525ΔA mutant (Fig. 5, lane 6).
The observation of the smaller RNA transcripts in the reporter system prompted two questions. Firstly, could the small transcripts be observed from the upstream region of the tet(P) operon on the native plasmid, pCW3? Secondly, did the regulation of the tet(P) genes involve a transcriptional attenuation mechanism similar to that proposed for the tet(M) gene of Tn916 (49)? If such a mechanism existed, then in the absence of tetracycline, tet(P) transcription would terminate in the upstream region, whereas when tetracycline was added, there would be read-through of transcription from the upstream region into the tet(P) coding region.
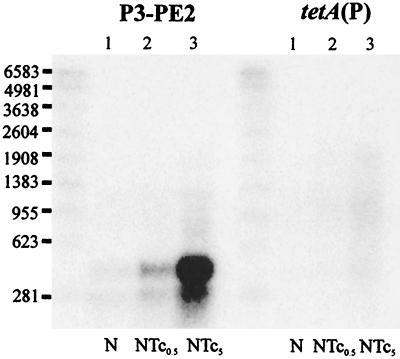
To answer these questions, Northern hybridization analysis was performed using total RNA extracted from JIR33(pCW3) cultures grown in the presence or absence of tetracycline and using probes specific for the P3-PE2 and tetA(P) regions. Northern analysis with the P3-PE2 probe showed that both of the small RNA bands observed with the reporter constructs were present in the pCW3-derived RNA preparations (Fig. 6). In addition, hybridizing smears that presumably represented a highly unstable full-length transcript were also observed from cells exposed to tetracycline. Interestingly, the intensity of hybridization of both the small RNA bands and the smeared RNA increased upon tetracycline induction. Although the signal intensity observed with the tetA(P)-specific probe was relatively low, a broadly smeared band, which commenced at the 23S rRNA band and increased in intensity upon exposure to tetracycline, was observed (Fig. 6). Similar profiles were observed when these experiments were repeated with different RNA preparations. Despite numerous attempts, it was not possible to obtain better resolution of the full-length tet(P) transcript. However, these results were consistent with the hypothesis that induction of tetracycline resistance was at the level of transcription from the P3 promoter and did not involve transcriptional attenuation at T1.
FIG. 6.
Northern hybridization analysis of tet(P) RNA. Total RNA was isolated from JIR33(pCW3) cells grown in the media indicated. RNA (10 μg) was subjected to electrophoresis through 1.5% formaldehyde gels and transferred to nylon membranes. Nylon membranes were subsequently probed with γ-32P-labeled DNA probes specific for either the P3-PE2 or tetA(P) transcripts as indicated. Lanes: 1, nutrient broth (N); 2, nutrient broth containing subinhibitory tetracycline (0.5 μg/ml) (NTc0.5); 3, nutrient broth containing tetracycline (5 μg/ml) (NTc5). Numbers at left are RNA sizes in bases.
DISCUSSION
In this study, we have shown that the tetA(P) and tetB(P) genes comprise an operon that is transcribed from a single promoter, P3, which is located 529 bp upstream of the translational initiation codon of the tetA(P) gene. This conclusion is based on the results obtained from RT-PCR, primer extension analysis, reporter gene expression, and Northern hybridization analysis.
Although several potential promoter sequences were present between P3 and the start of the tetA(P) gene, analysis of the ΔP3 and P3-525ΔA mutants provided strong evidence that there were no functional promoters located downstream of the P3 promoter. Analysis of this promoter revealed that it was identical to the consensus C. perfringens ς70 promoter (39) and contained only one mismatch to the E. coli ς70 consensus sequence (14). The spacing between the −35 and −10 regions, at 17 nt, is the optimal spacing observed for C. perfringens (39) and E. coli (13) promoter sequences. This spacing appears to be a stringent constraint on promoter function because it is required for recognition by the RNA polymerase holoenzyme (5, 14, 30, 33, 47, 51). Deviations from this spacing, as observed for the P3ΔA mutant with the reporter construct in this study, have a severe effect on expression. The close similarity of the promoter to the consensus ς70 sequence suggests that this promoter, if not subjected to any regulatory constraints, would act as a strong promoter in vivo (14, 35).
Transcription of the tet(P) operon appears to be under tight regulatory control, which is not surprising given that the TetA(P) protein is a transmembrane protein that is involved in the active efflux of tetracycline from the cell (44). In E. coli, the constitutive expression of tetracycline efflux proteins has been shown elsewhere to reduce the competitive fitness of the resultant strain (23, 34). High-level expression of the tetracycline resistance proteins TetA(B) and TetA(K) has also been shown previously to be highly toxic to the host cell (11, 12).
In gram-positive bacteria, tetracycline resistance genes may be regulated by transcriptional or translational attenuation (17, 22, 31, 49), translational coupling (46), or a tetracycline-responsive repressor protein (50). The results from this study are consistent with the hypothesis that in C. perfringens the induction of tetracycline resistance is at the level of initiation of transcription from the tet(P) promoter, P3. This postulate suggests the involvement of a regulatory protein that modulates P3 promoter activity. We have been unable to identify such a protein, although we have previously shown that inducible expression of the tet(P) operon requires an as-yet-unidentified host-encoded factor (19).
Once induced, transcription of the tet(P) operon is dependent upon read-through of the factor-independent transcriptional terminator, T1, which is located 390 bp downstream of the transcriptional start point. Northern blots using probes specific for the upstream region identified two small RNA transcripts, approximately 410 and 250 nt in size. The size of the larger fragment is consistent with a transcript originating from the P3 promoter and terminating at T1. Mutations that affected the formation or stability of this structure (ΔT1 and T1-G941A) resulted in high-level expression of the downstream gene. The presence of such a structure suggested the possibility that regulation was mediated by a transcriptional attenuation mechanism. However, the experimental data did not support this hypothesis (Fig. 6). Instead, it is postulated that T1 is an intrinsic control element that acts to prevent the overexpression of the TetA(P) protein, even in the presence of tetracycline.
The role of the PE2 region is more difficult to understand. Deletion of PE2 in the reporter construct leads to significantly increased levels of the mRNA transcripts detected by the P3-PE2 and PE2-T1 probes and slightly increased chloramphenicol resistance. However, the nature of these transcripts appears unaltered compared to that of the wild type (Fig. 5A), apart from the expected small reduction in size resulting specifically from the deletion. It is possible that, like T1, PE2 is also a terminator sequence but is less efficient and that its effects are modulated by the presence of the downstream T1 terminator. Alternatively, deletion of PE2 may act to stabilize the transcripts originating from the P3 promoter.
ACKNOWLEDGMENTS
This research was supported by grants from the Australian Research Council. P.A.J. was the recipient of an Australian Postgraduate Award.
REFERENCES
- 1.Abraham L J, Berryman D I, Rood J I. Hybridization analysis of the class P tetracycline resistance determinant from the Clostridium perfringens R-plasmid, pCW3. Plasmid. 1988;19:113–120. doi: 10.1016/0147-619x(88)90050-9. [DOI] [PubMed] [Google Scholar]
- 2.Abraham L J, Rood J I. Cloning and analysis of the Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid. 1985;13:155–162. doi: 10.1016/0147-619x(85)90038-1. [DOI] [PubMed] [Google Scholar]
- 3.Abraham L J, Rood J I. Molecular analysis of transferable tetracycline resistance plasmids from Clostridium perfringens. J Bacteriol. 1985;161:636–640. doi: 10.1128/jb.161.2.636-640.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abraham L J, Wales A J, Rood J I. Worldwide distribution of the conjugative Clostridium perfringens tetracycline resistance plasmid, pCW3. Plasmid. 1985;14:37–46. doi: 10.1016/0147-619x(85)90030-7. [DOI] [PubMed] [Google Scholar]
- 5.Ayers D G, Auble D T, deHaseth P L. Promoter recognition by Escherichia coli RNA polymerase. Role of the spacer DNA in functional complex formation. J Mol Biol. 1989;207:749–756. doi: 10.1016/0022-2836(89)90241-6. [DOI] [PubMed] [Google Scholar]
- 6.Bannam T L, Crellin P K, Rood J I. Molecular genetics of the chloramphenicol-resistance transposon Tn4451 from Clostridium perfringens: the TnpX site-specific recombinase excises a circular transposon molecule. Mol Microbiol. 1995;16:535–551. doi: 10.1111/j.1365-2958.1995.tb02417.x. [DOI] [PubMed] [Google Scholar]
- 7.Bannam T L, Rood J I. Identification of structural and functional domains of the tetracycline efflux protein TetA(P) from Clostridium perfringens. Microbiology. 1999;145:2947–2955. doi: 10.1099/00221287-145-10-2947. [DOI] [PubMed] [Google Scholar]
- 8.Berryman D I, Rood J I. The closely related ermB-ermAM genes from Clostridium perfringens, Enterococcus faecalis (pAMβ1), and Streptococcus agalactiae (pIP501) are flanked by variants of a directly repeated sequence. Antimicrob Agents Chemother. 1995;39:1830–1840. doi: 10.1128/aac.39.8.1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cummins C S, Johnson J L. Taxonomy of the clostridia: wall composition and DNA homologies in Clostridium butyricum and other butyric-acid producing clostridia. J Gen Microbiol. 1971;67:33–46. [Google Scholar]
- 10.d'Aubenton Carafa Y, Brody E, Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J Mol Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 11.Eckert B, Beck C F. Overproduction of transposon Tn10-encoded tetracycline resistance protein results in cell death and loss of membrane potential. J Bacteriol. 1989;171:3557–3559. doi: 10.1128/jb.171.6.3557-3559.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guay G G, Rothstein D M. Expression of the tetK gene from Staphylococcus aureus in Escherichia coli: comparison of substrate specificities of TetA(B), TetA(C), and TetK efflux proteins. Antimicrob Agents Chemother. 1993;37:191–198. doi: 10.1128/aac.37.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harley C B, Reynolds R P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horton R M, Cai Z L, Ho S N, Pease L R. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 16.Horton R M, Hunt H D, Ho S N, Pullen J K, Pease L R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 17.Hoshino T, Ikeda T, Tomizuka N, Furukawa K. Nucleotide sequence of the tetracycline resistance gene of pTHT15, a thermophilic Bacillus plasmid: comparison with staphylococcal TcR controls. Gene. 1985;37:131–138. doi: 10.1016/0378-1119(85)90265-3. [DOI] [PubMed] [Google Scholar]
- 18.Iannello R C. DNaseI footprinting using PCR-generated end-labeled DNA probes. In: Tymss M J, editor. In vitro transcription and translation. Totowa, N.J: Humana Press Inc.; 1995. pp. 379–391. [DOI] [PubMed] [Google Scholar]
- 19.Johanesen, P. A., D. Lyras, and J. I. Rood. Induction of pCW3-encoded tetracycline resistance in Clostridium perfringens involves a host-encoded factor. Plasmid, in press. [DOI] [PubMed]
- 20.Johnson J L, Francis B S. Taxonomy of the clostridia: ribosomal acid homologies among the species. J Gen Microbiol. 1975;88:229–244. doi: 10.1099/00221287-88-2-229. [DOI] [PubMed] [Google Scholar]
- 21.Kennan R M, McMurry L M, Levy S B, Rood J I. Glutamate residues located within putative transmembrane helices are essential for TetA(P)-mediated tetracycline efflux. J Bacteriol. 1997;179:7011–7015. doi: 10.1128/jb.179.22.7011-7015.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan S A, Novick R P. Complete nucleotide sequence of pT181, a tetracycline-resistance plasmid from Staphylococcus aureus. Plasmid. 1983;10:251–259. doi: 10.1016/0147-619x(83)90039-2. [DOI] [PubMed] [Google Scholar]
- 23.Lee S W, Edlin G. Expression of tetracycline resistance in pBR322 derivatives reduces the reproductive fitness of plasmid-containing Escherichia coli. Gene. 1985;39:173–180. doi: 10.1016/0378-1119(85)90311-7. [DOI] [PubMed] [Google Scholar]
- 24.Lyras D, Rood J I. Genetic organization and distribution of tetracycline resistance determinants in Clostridium perfringens. Antimicrob Agents Chemother. 1996;40:2500–2504. doi: 10.1128/aac.40.11.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyras D, Rood J I. Transposition of Tn4451 and Tn4453 involves a circular intermediate that forms a promoter for the large resolvase, TnpX. Mol Microbiol. 2000;38:588–601. doi: 10.1046/j.1365-2958.2000.02154.x. [DOI] [PubMed] [Google Scholar]
- 26.Lyras D, Storie C, Huggins A S, Crellin P K, Bannam T L, Rood J I. Chloramphenicol resistance in Clostridium difficile is encoded on Tn4453 transposons that are closely related to Tn4451 from Clostridium perfringens. Antimicrob Agents Chemother. 1998;42:1563–1567. doi: 10.1128/aac.42.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyristis M, Bryant A E, Sloan J, Awad M M, Nisbet I T, Stevens D L, Rood J I. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol Microbiol. 1994;12:761–777. doi: 10.1111/j.1365-2958.1994.tb01063.x. [DOI] [PubMed] [Google Scholar]
- 28.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 29.Matsushita C, Matsushita O, Koyama M, Okabe A. A Clostridium perfringens vector for the selection of promoters. Plasmid. 1994;31:317–319. doi: 10.1006/plas.1994.1035. [DOI] [PubMed] [Google Scholar]
- 30.McKane M, Gussin G N. Changes in the 17 bp spacer in the P(R) promoter of bacteriophage lambda affect steps in open complex formation that precede DNA strand separation. J Mol Biol. 2000;299:337–349. doi: 10.1006/jmbi.2000.3757. [DOI] [PubMed] [Google Scholar]
- 31.Mojumdar M, Khan S A. Characterization of the tetracycline resistance gene of plasmid pT181 of Staphylococcus aureus. J Bacteriol. 1988;170:5522–5528. doi: 10.1128/jb.170.12.5522-5528.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morelle G. A plasmid extraction procedure on a miniprep scale. Focus. 1989;11:7–8. [Google Scholar]
- 33.Mulligan M E, Brosius J, McClure W R. Characterization in vitro of the effect of spacer length on the activity of Escherichia coli RNA polymerase at the TAC promoter. J Biol Chem. 1985;260:3529–3538. [PubMed] [Google Scholar]
- 34.Nguyen T N, Phan Q G, Duong L P, Bertrand K P, Lenski R E. Effects of carriage and expression of the Tn10 tetracycline-resistance operon on the fitness of Escherichia coli K12. Mol Biol Evol. 1989;6:213–225. doi: 10.1093/oxfordjournals.molbev.a040545. [DOI] [PubMed] [Google Scholar]
- 35.Record M T J, Reznikoff W S, Craig M L, McQuade K L, Schalx P J. Escherichia coli RNA polymerase (Eς70), promoters, and the kinetics of the steps of transcription initiation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 792–820. [Google Scholar]
- 36.Reynolds R, Bermudez-Cruz R M, Chamberlin M J. Parameters affecting transcription termination by Escherichia coli RNA polymerase. I. Analysis of 13 rho-independent terminators. J Mol Biol. 1992;224:31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- 37.Richardson J P, Greenblatt J. Control of RNA chain elongation and termination. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 822–848. [Google Scholar]
- 38.Rood J I. Transferable tetracycline resistance in Clostridium perfringens strains of porcine origin. Can J Microbiol. 1983;29:1241–1246. doi: 10.1139/m83-193. [DOI] [PubMed] [Google Scholar]
- 39.Rood J I, Cole S T. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol Rev. 1991;55:621–648. doi: 10.1128/mr.55.4.621-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rood J I, Maher E A, Somers E B, Campos E, Duncan C L. Isolation and characterization of multiply antibiotic-resistant Clostridum perfringens strains from porcine feces. Antimicrob Agents Chemother. 1978;13:871–880. doi: 10.1128/aac.13.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rood J I, Scott V N, Duncan C L. Identification of a transferable tetracycline resistance plasmid (pCW3) from Clostridium perfringens. Plasmid. 1978;1:563–570. doi: 10.1016/0147-619x(78)90013-6. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 43.Scott P T, Rood J I. Electroporation-mediated transformation of lysostaphin-treated Clostridium perfringens. Gene. 1989;82:327–333. doi: 10.1016/0378-1119(89)90059-0. [DOI] [PubMed] [Google Scholar]
- 44.Sloan J, McMurry L M, Lyras D, Levy S B, Rood J I. The Clostridium perfringens Tet P determinant comprises two overlapping genes: tetA(P), which mediates active tetracycline efflux, and tetB(P), which is related to the ribosomal protection family of tetracycline-resistance determinants. Mol Microbiol. 1994;11:403–415. doi: 10.1111/j.1365-2958.1994.tb00320.x. [DOI] [PubMed] [Google Scholar]
- 45.Sloan J, Warner T A, Scott P T, Bannam T L, Berryman D I, Rood J I. Construction of a sequenced Clostridium perfringens-Escherichia coli shuttle plasmid. Plasmid. 1992;27:207–219. doi: 10.1016/0147-619x(92)90023-4. [DOI] [PubMed] [Google Scholar]
- 46.Stasinopoulos S J, Farr G A, Bechhofer D H. Bacillus subtilis tetA(L) gene expression: evidence for regulation by translational reinitiation. Mol Microbiol. 1998;30:923–932. doi: 10.1046/j.1365-2958.1998.01119.x. [DOI] [PubMed] [Google Scholar]
- 47.Stefano J E, Gralla J D. Spacer mutations in the lac ps promoter. Proc Natl Acad Sci USA. 1982;79:1069–1072. doi: 10.1073/pnas.79.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steffen C, Matzura H. Nucleotide sequence analysis and expression studies of a chloramphenicol-acetyltransferase-coding gene from Clostridium perfringens. Gene. 1989;75:349–354. doi: 10.1016/0378-1119(89)90282-5. [DOI] [PubMed] [Google Scholar]
- 49.Su Y A, He P, Clewell D B. Characterization of the tet(M) determinant of Tn916: evidence for regulation by transcription attenuation. Antimicrob Agents Chemother. 1992;36:769–778. doi: 10.1128/aac.36.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tauch A, Puhler A, Kalinowski J, Thierbach G. TetZ, a new tetracycline resistance determinant discovered in gram-positive bacteria, shows high homology to gram-negative regulated efflux systems. Plasmid. 2000;44:285–291. doi: 10.1006/plas.2000.1489. [DOI] [PubMed] [Google Scholar]
- 51.Warne S E, deHaseth P L. Promoter recognition by Escherichia coli RNA polymerase. Effects of single base pair deletions and insertions in the spacer DNA separating the −10 and −35 regions are dependent on spacer DNA sequence. Biochemistry. 1993;32:6134–6140. doi: 10.1021/bi00075a003. [DOI] [PubMed] [Google Scholar]
- 52.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–288. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]
- 53.Zuker M, Mathews D H, Turner D H. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J, Clark B F C, editors. RNA biochemistry and biotechnology. New York, N.Y: Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]