Abstract
Rhodobacter sphaeroides has multiple homologues of most of the Escherichia coli chemotaxis genes, organized in three major operons and other, unlinked, loci. These include cheA1 and cheR1 (che Op1) and cheA2, cheR2, and cheB1 (che Op2). In-frame deletions of these cheR and cheB homologues were constructed and the chemosensory behaviour of the resultant mutants examined on swarm plates and in tethered cell assays. Under the conditions tested, CheR2 and CheB1 were essential for normal chemotaxis, whereas CheR1 was not. cheR2 and cheB1, but not cheR1, were also able to complement the equivalent E. coli mutants. However, none of the proteins were required for the correct polar localization of the chemoreceptor McpG in R. sphaeroides. In E. coli, CheR binds to the NWETF motif on the high-abundance receptors, allowing methylation of both high- and low-abundance receptors. This motif is not contained on any R. sphaeroides chemoreceptors thus far identified, although 2 of the 13 putative chemoreceptors, McpA and TlpT, do have similar sequences. This suggests that CheR2 either interacts with the NWETF motif of E. coli methyl-accepting chemotaxis proteins (MCPs), even though its native motif may be slightly different, or with another conserved region of the MCPs. Methanol release measurements show that R. sphaeroides has an adaptation system that is different from that of Bacillus subtilis and E. coli, with methanol release measurable on the addition of attractant but not on its removal. Intriguingly, CheA2, but not CheA1, is able to phosphorylate CheB1, suggesting that signaling through CheA1 cannot initiate feedback receptor adaptation via CheB1-P.
Since bacteria are too small to sense a gradient along their length, they employ a system of temporal sensing. This allows them to detect concentration changes over distances equivalent to several body lengths. Many bacterial species possess a chemosensory system that allows the comparison of the current concentration of chemoeffectors in the environment to that encountered a few seconds previously. Bacterial chemotaxis has been studied most extensively in Escherichia coli and Salmonella enterica serovar Typhimurium (for reviews, see references 2, 8, and 46). In a homogeneous environment E. coli shows a random swimming pattern of “runs” (periods of smooth swimming) and “tumbles” (periods of reorientation caused by flagellum bundle disruption). When the concentration of an attractant is increased, the cells exhibit a greatly reduced frequency of tumbling, increasing the chance that they will swim toward the source of attractant. After a few seconds the cells adapt, reverting to their prestimulus tumbling bias, and are capable of responding to a subsequent change in chemoeffector concentration. Sensory adaptation is essential for temporal sensing and for the accumulation of bacteria in environments that are optimal for growth.
Methyl-accepting chemotaxis proteins (MCPs) are both the site of initial signal transduction and of adaptation in the chemosensory pathway (for reviews, see references 10 and 28). These proteins function as homodimers. They have two transmembrane helices flanking a periplasmic ligand-binding domain and a cytoplasmic signaling domain. The latter domain is highly conserved among the MCPs of eubacterial and archaeal species (27). It contains specific glutamate residues in two regions, K1 and R1, which are methylated during adaptation. Methyl groups are transferred to the specific glutamate residues of the MCPs from S-adenosylmethionine by a constitutively active methyltransferase, CheR (44). These groups are removed by the methylesterase, CheB (54), and released as methanol (18). CheB activity is increased on its phosphorylation by CheA-P (15). CheB has a regulatory N-terminal domain and a C-terminal catalytic, amidase-esterase domain (9). Inhibition of the catalytic domain is reduced upon phosphorylation of the regulatory domain by CheA-P (23). The ligand occupancy of the periplasmic domain of the MCP reflects the current chemoeffector concentration, whereas the methylation state of the cytoplasmic domain is a “record” of the concentration a few seconds previously. Methylated MCPs are more proficient at CheA activation than unmethylated MCPs (5). When a repellent binds or an attractant leaves the ligand-binding domain of the MCP, a signal is transmitted through a cytoplasmic linker protein CheW to CheA (6, 7, 29). CheA autophosphorylates and the phosphoryl group is transferred either to CheY or, at a slower rate, to CheB. CheY-P binds to FliM, effecting a switch in the direction of flagellar motor rotation and hence a tumble. CheB-P demethylates the MCPs, reducing the level of CheA activation by the MCPs. Conversely, when the attractant concentration increases, CheY-P and CheB-P levels decrease. In response to a persistent stimulus, the cells achieve a steady state of MCP methylation. This restores the prestimulus pattern of runs and tumbles, allowing a response to subsequent changes in concentration of chemoeffector.
By modulating the methylation state of the MCPs, CheR and CheB-P enable adaptation of the chemosensory system to a background level of chemoeffector. CheR and CheB have been extensively studied in E. coli (for recent reviews, see references 9 and 17). CheR is predominantly found associated with the extreme C-terminal pentapeptide, NWETF, of the high-abundance receptor proteins Tsr and Tar in a 1:1 molar ratio (53). The Trp at the second position and the Phe at the last position are critical (38). The NWETF motif is not present in the “low-abundance” receptors Tap and Trg. Cells expressing only low-abundance receptors exhibit poor swarming, impaired MCP methylation, and compromised adaptation (11, 51). Fusion of the NWETF motif to the C terminus of Trg greatly enhances methylation and chemotaxis in a strain expressing Trg as the sole receptor (12). This suggests that the binding of CheR to this motif is critical for methylation and thus adaptation.
Sensory adaptation in nonenteric bacteria shows some variations on the E. coli paradigm. For example, in the gram-positive bacterium Bacillus subtilis, CheY-P causes periods of smooth-swimming as opposed to tumbling. In addition, this bacterium shows CheB-dependent methanol release both upon addition and removal of attractant (19, 47). Remethylation of the MCPs appears to be dependent on phosphorylated CheY (20). B. subtilis has two further proteins, CheC and CheD, which are required for normal methylation (33, 34). CheC inhibits CheR activity and hence reduces MCP methylation. CheD is thought to facilitate CheA activation and is required for CheR activity (33). The archaeon Halobacterium salinarum also shows increased turnover of methyl groups in response to both positive and negative chemical and light stimuli (1, 43). Together, these differences suggest that chemotaxis and adaptation are more complex in many other species than they are in enteric bacteria.
The purple, nonsulfur, α-subgroup bacterium Rhodobacter sphaeroides provides an alternative model system for the study of chemotaxis. It can grow aerobically, photoheterotrophically, and anaerobically in the dark by using a variety of alternative electron acceptors. It exhibits taxis toward various chemoeffectors, light, and oxygen (4). Transport and partial metabolism are required for some chemosensory responses (16), and the extent of the responses can depend on the growth conditions. Unlike E. coli and B. subtilis, R. sphaeroides changes swimming direction by interrupting flagellar rotation (2, 3). Upon an increase in attractant concentration the frequency of stops decreases (32). R. sphaeroides possesses multiple homologues of the E. coli chemotaxis genes arranged in three operons and at other unlinked loci. che Op1 contains cheD, cheY1, cheA1, cheW1, cheR1, and cheY2, and che Op2 contains cheY3, cheA2, cheW2, cheW3, cheR2, cheB1, and tlpC (13, 37, 49). Thirteen chemoreceptors, including both membrane-spanning and cytoplasmic or transducer-like proteins (Tlps), have been identified to date. These are differentially expressed according to the environmental condition (14). It is not known whether the products of the che operons operate through independent, linear pathways or whether there is significant cross talk between the components of these operons. It is interesting that, while most of the receptors identified in R. sphaeroides have conserved glutamate residues which could be involved in adaptation, only two (McpA and TlpT) contain a motif with limited homology to the CheR-binding site (50; www.jgi.doe.gov/JGI_microbial/html/rhodobacter/rhodob_homepage.html). R. sphaeroides also possesses a homologue of the B. subtilis adaptation protein, CheD, but no CheC encoding gene has been identified.
The discovery of CheR and CheB homologues in R. sphaeroides and the identification of MCPs with conserved glutamyl residues suggest a role for methylation-dependent chemotaxis. In this study we set out to examine the function of CheR1, CheR2, and CheB1 in R. sphaeroides chemotaxis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids are shown in Tables 1 and 2. R. sphaeroides strains were grown in succinate medium (39) at 30°C, either aerobically or photoheterotrophically as described previously (26). E. coli strains were grown at 37°C in Luria-Bertani (LB) medium. The antibiotics kanamycin, nalidixic acid, and streptomycin were used at 25 μg/ml, and ampicillin was used at 100 μg/ml.
TABLE 1.
Strains used in this study
| Strain | Characteristicsa | Source or reference |
|---|---|---|
| R. sphaeroides | ||
| WS8N | Spontaneous nalidixic acid-resistant mutant of wild-type WS8 | 41 |
| JPA117 | Δche Op1 derivative of WS8N | 13 |
| JPA500 | WS8N containing an mcpG-egfp fusion in place of the wild-type mcpG in the chromosome | 48 |
| JPA516 | ΔcheB1 derivative of JPA500 | This study |
| JPA517 | ΔcheB1 derivative of WS8N | This study |
| JPA564 | ΔcheR2 derivative of JPA500 | This study |
| JPA565 | ΔcheR2 derivative of WS8N | This study |
| JPA568 | ΔcheR1 derivative of WS8N | This study |
| JPA569 | ΔcheR1 derivative of JPA500 | This study |
| E. coli | ||
| DH5α | Strain that supports blue-white screening of colonies for cloning experiments | Gibco-BRL |
| S17-1λpir | Strain capable of mobilizing the suicide vector pK18mobsacB into R. sphaeroides; Smr | 31 |
| RP437 | Wild type for chemotaxis | J. S. Parkinson |
| RP1254 | ΔcheR derivative of RP437 | J. S. Parkinson |
| RP4972 | ΔcheB derivative of RP437 | J. S. Parkinson |
Smr, streptomycin resistant.
TABLE 2.
Plasmids used in this study
| Plasmid | Characteristicsa | Source or reference |
|---|---|---|
| pUC19 | High-copy-number cloning vector; Ap | Pharmacia |
| pK18mobsacB | Allelic exchange suicide vector mobilized by E. coli S17-1λpir; allows blue-white screening for inserts; Kmr Sucs | 36 |
| pCHE3.1 | 3.1-kb EcoRI fragment from R. sphaeroides che Op1, in pUC18 | 49 |
| pJPA112 | 3.1-kb EcoRI fragment from R. sphaeroides che Op2, in pUC19 | 13 |
| pJPA113 | 4.6-kb BamHI fragment from R. sphaeroides che Op2, in pUC19 | 13 |
| pACM35 | 0.42-kb EcoRI/PstI fragment from pCHE3.1 in pUC19 | This study |
| pACM36 | Blunted 0.4-kb BglII/PstI fragment from pCHE3.1, ligated into the blunted PstI site of pACM35 so that the upstream and downstream flanking regions of cheR1 were joined in frame | This study |
| pAG1 | ΔcheR1 construct derived from pACM36 in pK18mobsacB. | This study |
| pHV3 | 0.54-kb EcoRI/BamHI PCR fragment that contained the upstream flanking sequence of cheR2 cloned into pK18mobsacB | This study |
| pHV4 | 0.54-kb BamHI/HindIII PCR fragment that contained the downstream flanking sequence of cheR2 cloned into appropriately cut pHV3 to generate a ΔcheR2 in-frame construct | This study |
| pTJC10 | 1.4-kb PstI/PvuII fragment from pJPA112 containing upstream flanking sequence of cheB1 and 1.4-kb EcoRI/BamHI fragment from pJPA113 containing downstream flanking sequence ligated together into pUC19 to generate a ΔcheB1 construct | This study |
| pTJC11 | ΔcheB1 construct derived from pTJC10 in pK18mobsacB | This study |
| pREP4 | Carries lacIq gene; reduces “leaky” expression from the tac promoter of pQE30; Kmr | Qiagen |
| pQE30 | Ptac-based expression vector; introduces RGS(H6) sequence at N termini of expressed proteins; Apr | Qiagen |
| pQEA1 | PCR fragment containing the coding sequences of cheA1 cloned into the KpnI/HindIII sites of pQE30 | 37 |
| pQEA2 | PCR fragment containing the coding sequences of cheA2 cloned into the KpnI/PstI sites of pQE30 | 37 |
| pQE30R1 | PCR fragment containing the coding sequences of cheR1 cloned into the BamHI/HindIII sites of pQE30 | This study |
| pQE30R2 | PCR fragment containing the coding sequences of cheR2 cloned into the BamHI/HindIII sites of pQE30 | This study |
| pQE30B1 | PCR fragment containing the coding sequences of cheB1 cloned into the BamHI/HindIII sites of pQE30 | This study |
| pEGFP-N1 | GFP protein fusion vector; Kmr | Clontech |
| pGW41 | mcpG-egfp fusion and downstream flanking sequence from mcpG in pK18mobsacB | 48 |
Kmr, Karamycin resistant; Sucs, sucrose sensitive; Apr, ampicillin resistant; GFP, green fluorescent protein.
Molecular genetic techniques
All cloning steps were carried out by standard methods (35). Sequencing-quality plasmid DNA was extracted by using the Plasmid Midi-Kit (Qiagen) or the WizardPlus kit (Promega), sequenced by the University of Oxford Biochemistry sequencing service, and analyzed with the GCG software package (University of Wisconsin). PCRs were carried out by using Pfu DNA polymerase (Stratagene and Promega) and purified as described previously (26). All primers were synthesized by Genosys Biotechnologies, Inc.
Construction of deletion strains
Regions upstream and downstream from the gene of interest were cloned together into pK18mobsacB, as detailed below, and sequenced to ensure that they were in frame and contained no PCR misincorporation errors. These constructs were introduced into the chromosome of R. sphaeroides by allelic exchange as described previously (13, 36).
cheR1
A 0.42-kb region immediately upstream of cheR1 was cut from pCHE3.1 with EcoRI and PstI and cloned into pUC19 to generate plasmid pACM35. A 0.4-kb fragment immediately downstream of cheR1 was cut from pCHE3.1 with BglII and PstI, pACM35 was linearized with PstI, the single-stranded ends of both fragments were filled in with T4 DNA polymerase, and the two were ligated together. The resultant plasmid with the upstream and downstream regions in the correct orientation was designated pACM36. The entire insert from this pUC19 derivative was excised with EcoRI and HindIII and ligated into similarly cut pK18mobsacB to generate the final construct pAG1.
cheR2
A 0.54-kb region immediately upstream of cheR2 was amplified by PCR by using primers that encompassed the start codon and that included 5′ EcoRI and 3′ BamHI sites. A 0.54-kb region that contained the downstream flanking sequence of cheR2, including the ribosome-binding site of cheB1, was amplified by PCR with primers that included 5′ BamHI and 3′ HindIII sites. The first PCR product was cloned into appropriately cut pK18mobsacB to produce plasmid pHV3. The second PCR product was ligated into pHV3 cut with BamHI and HindIII to generate pHV4.
cheB1
A 1.4-kb region immediately upstream of cheB1 was cut from pJPA112 with PstI and PvuII. A 1.4-kb region immediately downstream of cheB1 was cut from pJPA113 with EcoRI, the single-stranded ends filled in with T4 DNA polymerase and then cut again with BamHI. These were cloned together into pUC19 cut with PstI and BamHI to generate pTJC10. The entire insert from this plasmid was excised with PstI and BamHI and ligated into similarly cut pK18mobsacB to generate pTJC11.
R. sphaeroides behavioral assays
Swarm plates containing 0.25% agar (BiTek; Difco) and 100 μM attractant were inoculated and incubated as described elsewhere (26). Each experiment was performed in triplicate and repeated three times to generate nine data sets.
Aerobic and photoheterotrophic cells were analyzed by the tethered cell assay as described in detail previously (26). At least three data sets, which together included at least 10 cells, were collected for each strain.
Growth rates were measured as described in Martin et al. (26).
Construction of expression plasmids.
The coding sequences (excluding the ATG start codon) of cheR1, cheR2, and cheB1 were amplified by PCR with primers incorporating exogenous 5′ and 3′ restriction sites facilitating in-frame cloning into pQE30 (Qiagen). The fragments were cloned such that expression was under the control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible tac promoter and that the protein products would have an N-terminal tag of six-histidine residues to facilitate purification. The cheR1, cheR2, and cheB1 fragments had 5′ BamHI and 3′HindIII sites and, once cloned, generated expression plasmids pQE30R1, pQE30R2, and pQE30B1, respectively. The cloning of cheA1 and cheA2 in pQE30 has been described previously (37). Sequence analysis of the clones revealed no PCR misincorporation errors.
E. coli behavioral assay.
E. coli swarming assays were carried out by a modification of the method of Wolfe et al.(52). RP437, RP1254, and RP4972 were each transformed with pREP4 prior to transformation with the cheR1, cheR2, or cheB1 expression plasmids. pREP4, which expresses the LacIq repressor protein, minimizes “leaky” expression from Ptac. The strains carrying the appropriate expression plasmids were grown overnight in LB broth with ampicillin and kanamycin at 30°C. LB swarm plates containing 0.25% agar, antibiotics as appropriate, and IPTG at 0, 1, 10, 100, or 1,000 μM were inoculated in triplicate with 5 μl of the stationary-phase culture and incubated at 30°C. RP437 swarm plates were photographed after 8 h, and swarm plates of the mutant strains were photographed after 15 h of incubation.
Growth rates were measured as described by Shah et al. (37).
Methanol release experiments
A modified version of the methanol release assay developed for R. sphaeroides was used (21). All experiments were carried out under photoheterotrophic conditions. Cells were grown to an optical density at 660 nm (OD660) of between 0.8 and 1.2, and a volume adjusted for cell number was harvested by centrifugation. The pellet was washed once in 1 ml of HEPES-Cm buffer (10 mM Na-HEPES [pH 7.2]–50 μg of chloramphenicol ml−1, sparged with nitrogen), resuspended in 1 ml of HEPES-Cm, and incubated for 40 min with illumination at 50 μM m−2 s−1 with 40 μl of l-[methyl-3H]methionine (0.06 μmol ml−1; specific activity, 185 mCi mmol−1; Amersham). The cells were harvested, washed in HEPES-Cm buffer, and loaded onto a sterile 0.45-μm (pore size) syringe filter (Nalgene). HEPES-Cm buffer was passed through the filter for 30 min to remove the excess radiolabel. The flow rate was adjusted to 1 ml min−1. Each experiment included monitoring of (i) prestimulus behavior (HEPES-Cm buffer for 10 min), (ii) the response to 1 mM sodium propionate in HEPES-Cm (10 min), and (iii) the response to its replacement by HEPES-Cm without attractant (10 min). Next, 0.5-ml samples were collected and subjected to vapor-phase transfer into 7 ml of Optiphase Hi-Safe scintillation fluid (Fisher) for 16 h, and the radioactivity was measured in a Beckman 5000TD scintillation counter. At least three data sets were collected for each experiment.
Purification of CheA1, CheA2, and CheB1
Cells containing the appropriate expression plasmids were grown in 2YT medium with antibiotics as appropriate to an OD600 of 0.8 at 37°C. Expression was induced by the addition of 0.1 mM IPTG for 20 h at 18°C. After induction, cells were harvested by centrifugation (6,000 × g, 15 min) and resuspended in lysis buffer (10% [vol/vol] glycerol; 50 mM Tris-HCl, pH 8.0; 150 mM NaCl; 10 mM imidazole; 1 mM dithiothreitol [DTT]) in a volume of 60 ml per liter of the original culture. Cells were lysed by sonication on ice for six 20-s intervals (Vibracell; Sonics and Materials, Inc.). Lysates were cleared by centrifugation (35,000 × g, 15 min) and filtration of the supernatant through a 0.45-μm (pore-size) syringe filter. The filtered supernatant was applied to a Ni-nitrilotriacetic acid agarose column (Qiagen) equilibrated with lysis buffer. The column was washed with 60 column volumes of lysis buffer prior to elution of the protein in lysis buffer supplemented with 250 mM imidazole.
Phosphotransfer assays.
All reactions were performed in TGMNKD buffer (50 mM Tris-HCl, pH 8.0; 10% [vol/vol] glycerol; 5 mM MgCl2; 150 mM NaCl; 50 mM KCl; 1 mM DTT) at 20°C. Then, a 5 μM concentration of either CheA1 or CheA2 was preincubated with 0.5 mM [γ-32P]ATP (14 GBq mmol−1; Amersham) for 15 min before the addition of CheB1 in a final reaction volume of 100 μl. A 10-μl sample was taken prior to addition of CheB1 (T = 0). After the addition of CheB1, 10-μl samples were taken at the intervals shown and quenched in 5 μl of 3× sodium dodecyl sulfate (SDS)-EDTA loading dye (7.5% [wt/vol] SDS; 90 mM EDTA; 37.5 mM Tris-HCl, pH 6.8; 37.5% [vol/vol] glycerol; 3% [vol/vol] β-mercaptoethanol). Samples were heated to 65°C for 30 s prior to SDS-polyacrylamide gel electrophoresis (PAGE) on 15% gels according to the method of Laemmli (22). Gels were dried and exposed to phosphor screens (Kodak), and the radioactive bands were detected by using an SF-PhosphorImager with ImageQuant version 5.0 software (Molecular Dynamics).
Construction of egfp fusions and fluorescence microscopy.
A pK18mobsacB derivative, pGW41, that contained mcpG fused in-frame to egfp from pEGFP-N1 (Clontech), together with downstream flanking sequences, was used to generate strain JPA500 in a recent study (48). This strain contained mcpG-egfp in place of the wild-type gene in the chromosome. The pK18mobsacB derivatives for the deletion of cheR1, cheR2, and cheB1 (pAG1, pHV4, and pTJC11, respectively) were introduced into JPA500 to generate deletion mutants in the mcpG-egfp fusion background (JPA569, JPA564, and JPA516, respectively).
Fluorescence microscopy was performed as described previously (48).
RESULTS
Deletion analysis of cheR1, cheR2, and cheB1.
The predicted amino acid sequences of CheR1 (encoded in che Op1, accession no. X80205) and of CheR2 and CheB1 (encoded in che Op2, accession no. AJ000977) were compared with the equivalent proteins from E. coli and B. subtilis. The CheR1, CheR2, and CheB1 of R. sphaeroides have 43, 37, and 47% identity, respectively, with the corresponding E. coli proteins and were therefore designated homologues of these adaptation enzymes.
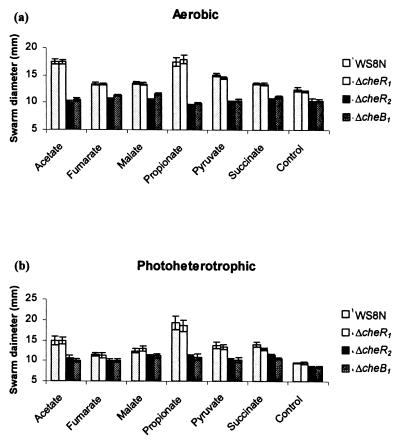
Unmarked strains were constructed with cheR1 (JPA568), cheR2 (JPA565), or cheB1 (JPA517) deleted in frame. The responses of these strains to gradients of chemoattractants (see Materials and Methods) was compared to the wild-type strain WS8N on swarm plates, under both aerobic and photoheterotrophic conditions (Fig. 1). Deletion of cheR1 had no significant effect on swarming ability in comparison to the wild-type strain under both environmental conditions (P > 0.05). However, deletion of either cheR2 or cheB1 resulted in a similar, significant decrease in swarm size to most attractants tested under both environmental conditions (P < 0.05). Only the swarms toward fumarate and malate under photoheterotrophic conditions showed no statistical difference; however, these attractants also cause very limited swarming with the wild type under photoheterotrophic conditions. All of the mutants showed normal growth rates (data not shown).
FIG. 1.
Comparison of the swarm diameters of strains JPA568 (ΔcheR1), JPA565 (ΔcheR2), and JPA517 (ΔcheB1) with WS8N (wild type) under aerobic (a) and photoheterotrophic (b) conditions to 100 μM concentrations of attractants (control is no added attractant). The swarm diameters of strains WS8N (wild type), JPA568 (ΔcheR1), JPA565 (ΔcheR2), and JPA517 (ΔcheB1) are shown from left to right for each attractant. The error bars represent 1 standard error of the mean (SEM) from nine experiments. Strains JPA565 and JPA517 have significantly reduced swarm diameters to most attractants compared to strains WS8N and JPA568 under both growth conditions.
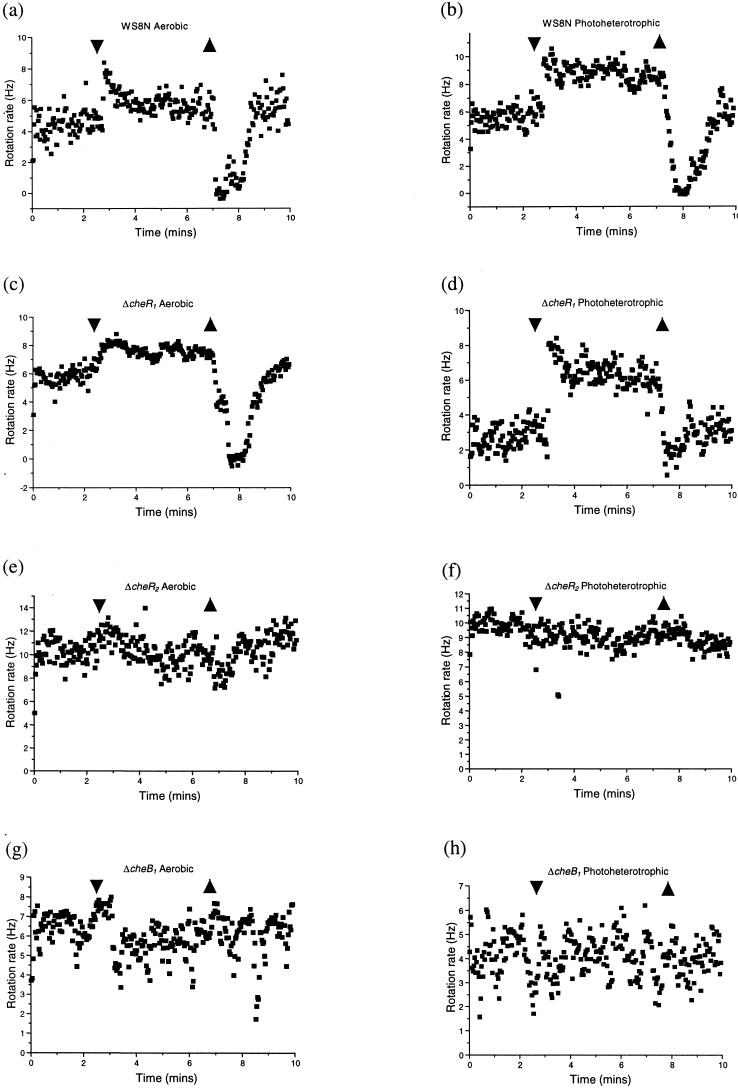
The tethering of cells by their flagella in a flow chamber allows a direct assessment of changes in the flagellar stopping frequency of individual cells in response to the addition and removal of a chemoeffector. The responses of the cheR1, cheR2, and cheB1 mutants to the addition and removal of 1 mM propionate were compared to those of the wild type (Fig. 2). The addition of 1 mM propionate caused the flagellar rotation rate of both aerobically and photoheterotrophically grown WS8N to increase, possibly as a result of a decrease in the number of short stops. Upon attractant removal flagellar rotation stopped, followed by adaptation to prestimulus behavior over a period of 1 to 2 min (Fig. 2a and b). The ΔcheR1 strain displayed wild-type responses to the addition and removal of propionate under both environmental conditions (Fig. 2c and d). The rotation rate of unstimulated cells showed some variability, but this was not strain dependent. In contrast, responses were lost to both the addition and the removal of propionate under both environmental conditions in both the ΔcheR2 and ΔcheB1 strains (Fig. 2e, f, g, and h). Thus, under the conditions tested, CheR2 and CheB1 were essential for chemotaxis in both swarm plate and tethered cell assays, whereas CheR1 was dispensable. However, strains deleted for either cheR2 or cheB1 retained a stopping frequency similar to unstimulated wild-type cells, unlike the corresponding E. coli mutants that are either exclusively smooth swimming or tumbly, respectively (data not shown; see references 42 and 54).
FIG. 2.
Behavior of wild-type (26) and mutant strains in the tethered cell assay. The strains and the environmental conditions are indicated above each graph in panels a to h. Addition and removal of 1 mM propionate is shown by arrows. The wild-type (WS8N) and ΔcheR1 (JPA568) strains showed normal responses, whereas the ΔcheR2 (JPA565) and ΔcheB1 (JPA517) strains failed to respond. Each strain was assayed at least three times. Graphs show the average response of a field of view that included at least three cells. Individual cells showed significant variability in their unstimulated rotation rates. However, this result was not strain dependent.
Expression of CheR1, CheR2, and CheB1 of R. sphaeroides in wild-type and mutant strains of E. coli
CheR1, CheR2, and CheB1 of R. sphaeroides were expressed as N-terminal His6 fusions from plasmid pQE30 in E. coli strains RP437 (wild type), RP1254 (ΔcheR), and RP4972 (ΔcheB). In each case, upon induction with IPTG, bands of the correct size were detected by SDS-PAGE (data not shown). The effects of this expression on swarming behavior are shown in Fig. 3. CheR1 did not restore swarming to RP1254 at any level of induction (data not shown). CheR2 complemented RP1254 when induced at low levels (up to 10 μM IPTG [Fig. 3a]), restoring the ability of RP1254 to form chemotaxis rings in swarm plates. CheR2 inhibited swarming of RP437 at IPTG concentrations of ≥100 μM, but there was a severe inhibition of growth at these levels of induction (data not shown). At lower levels of induction there was no effect on swarming (Fig. 3b). CheB1 complemented RP4972 only in the absence of IPTG (Fig. 3c). IPTG concentrations of 1, 10, and 100 μM abolished complementation by CheB1 but did not affect growth rate (data not shown). CheB1 inhibited swarming of RP437 in the absence of IPTG without affecting the growth rate (Fig. 3b). Thus, there is some expression from the noninduced Ptac promoter even in the presence of pREP4. Complementation of both the E. coli mutant strains required an incubation period of 15 h to achieve swarm sizes comparable to those obtained with the wild-type strain, RP437, after 8 h. However, swarm rings were observed in all cases where partial complementation was observed, indicating restoration of some chemotaxis. Microscopic examination revealed that the presence of R. sphaeroides CheB1 reduced the high tumbling bias of RP4972 and CheR2 reduced the smooth bias of RP1254 (data not shown). Together, these data suggest that CheR2 and CheB1 from R. sphaeroides function as adaptation enzymes and are able, at least in part, to complement the equivalent E. coli mutations.
FIG. 3.
Effect of expression of pQE30, pQE30R2, and pQE30B1 in E. coli strains RP1254 (ΔcheR) (a), RP437 (wild type) (b), and RP4972 (ΔcheB) (c). The plasmids in RP1254 were induced with 10 μM IPTG but were not induced in RP437 and RP4972. The swarm plates were incubated for 15 h at 30°C.
Methanol release during the chemotactic response.
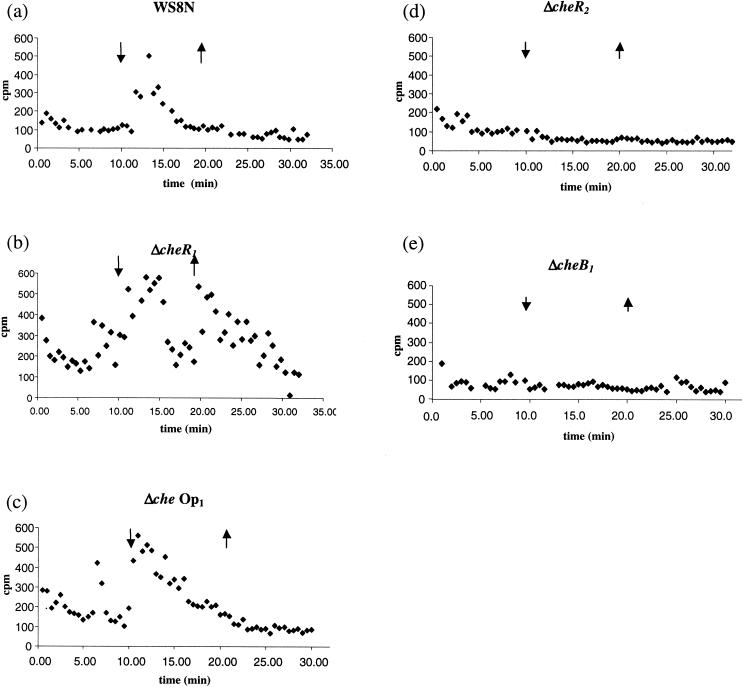
To investigate the methylation of R. sphaeroides MCPs, the pattern of methanol release from the wild-type strain, WS8N, was assessed. WS8N cells showed a release of methanol upon the addition of 1 mM propionate but not upon its removal (Fig. 4a). This is different from the pattern of methanol release seen both in E. coli and in B. subtilis. To determine the roles of CheR1, CheR2, and CheB1 in this methanol production, the three deletion strains JPA568, JPA565, and JPA517 were investigated. In cells deleted for cheR1, methanol was detected upon both the addition and the removal of 1 mM propionate (Fig. 4b). Interestingly, a mutant lacking all of che Op1 (JPA117) showed a wild-type response to the addition of propionate with methanol release upon addition but not upon its removal (Fig. 4c). At least three data sets were collected for each strain. Thus, the methanol released upon the removal of propionate in the ΔcheR1 strain must depend on one or more components encoded by che Op1 other than cheR1.
FIG. 4.
Methanol release from wild-type and mutant strains upon the addition and removal of 1 mM propionate (shown by arrows). (a) WS8N (wild-type) showed release upon the addition of attractant. (b) ΔcheR1 (JPA568) showed release upon attractant removal as well as upon addition. (c) Δche Op1 showed a wild-type methanol release profile. (d and e) Methanol release was not detected in ΔcheR2 (JPA565) (d) or in ΔcheB1 (JPA517) (e).
Strains deleted of either cheR2 or cheB1 showed no methanol release upon either the addition or the removal of propionate (Fig. 4d and e). This indicates that CheR2 and CheB1 are required for the methanol production observed in the wild type.
Phosphorylation of CheB1 by CheA1 and CheA2.
The transfer of phosphoryl groups from purified CheA1 and CheA2 to purified CheB1 in the presence of ATP was measured in vitro. CheA1 and CheA2 both autophosphorylated when preincubated with [γ-32P]ATP. CheB1 was added to each of the phosphorylated CheA preparations, and the transfer of label was monitored (Fig. 5). CheA2-32P was able to transfer phosphoryl groups to CheB1, but there was no detectable phosphotransfer from CheA1-32P. Control reactions showed that there was no change in the level of label on the CheAs when CheB1 was not added. Furthermore, CheB1 was not phosphorylated in reaction mixtures containing labeled ATP but lacking either of the CheAs (data not shown).
FIG. 5.
CheA2-dependent phosphorylation of CheB1 in the presence of [γ-32P]ATP. Phosphorimages after separation of reaction products by SDS–15% PAGE are shown for CheA1 and CheB1 (a) and for CheA2 and CheB1 (b). Both of the CheA preparations contain phosphorylatable proteolytic fragments of CheA, which are similar in size to CheB1 (visible in the T = 0 samples). Control experiments in which CheB1 was omitted showed that these bands did not change in intensity throughout the experiment (data not shown).
Effect of deletions on MCP localization.
In E. coli MCPs are localized to the poles of the cell, and this localization is dependent on CheW and CheA but not on CheR and CheB (24, 25, 40). A chemoreceptor of R. sphaeroides, McpG, also forms clusters at a single pole of the cell. Correct localization of this MCP is dependent upon components of che Op2 (26, 48). We investigated the effect of CheR1, CheR2, and CheB1 on the localization of McpG in R. sphaeroides cells by generating strains containing the mcpG-egfp fusion in mutants deleted of cheR1 (JPA569), cheR2 (JPA564), and cheB1 (JPA516). The pattern of fluorescence of the McpG-GFP fusion strain in a WS8N background (JPA500) was compared to that shown by the fusion in the deletion backgrounds (Table 3). The localization of the fusion protein was scored as “one pole” (as seen in 93.7% of cells with a wild-type background [JPA500])or “aberrant.” Aberrant localization included cells showing similar levels of fluorescence at both poles and fluorescence not associated with the cell pole. Wild-type cells that did not contain the mcpG-egfp fusion did not fluoresce. The McpG-GFP fusion localized to one pole in 95.4% of the ΔcheR1 cells, 94.9% of the ΔcheR2 cells, and 97.0% of the ΔcheB1 cells (Table 3). A similar localization profile was observed with photoheterotrophically grown cells (data not shown). Therefore, localization of McpG does not depend upon CheR1, CheR2, or CheB1. This indicates that the adaptation proteins of R. sphaeroides are not essential for normal McpG clustering at a pole.
TABLE 3.
Distribution of McpG-GFP fluorescence in aerobically grown cellsa
| Strain | Total no. of fluorescing cells | No. of cells with normal fluorescence | No. of cells with aberrant fluorescence | % Aberrant fluorescence ± SEM |
|---|---|---|---|---|
| mcpG-egfp | 527 | 494 | 33 | 6.3 ± 1.4 |
| ΔcheR1 mcpG-egfp | 527 | 503 | 24 | 4.6 ± 1.3 |
| ΔcheR2 mcpG-egfp | 602 | 571 | 31 | 5.1 ± .95 |
| ΔcheB1 mcpG-egfp | 866 | 840 | 26 | 3.0 ± .75 |
All percentages are given with respect to the total number of fluorescing cells from at least five independent experiments. None of the mutant strains showed a greater degree of aberrant fluorescence than in the wild-type background.
DISCUSSION
Several observations have led to the suggestion that MCPs and methylation-dependent adaptation may have a role in R. sphaeroides chemotaxis. In this study we examined the R. sphaeroides adaptation enzyme homologues CheR1, CheR2, and CheB1 by genetic and biochemical approaches.
Deletion of the che Op1 encoded cheR1 does not affect the ability of R. sphaeroides to respond to the compounds tested either on swarm plates or in tethered cell assays under aerobic or photoheterotrophic conditions, a finding consistent with the report of Hamblin et al. (13). The inability of CheR1 to complement a ΔcheR mutant of E. coli indicates that CheR1 is unable to substitute for the CheR of E. coli. This is of interest since CheR1 shares greater sequence identity with the E. coli protein than does CheR2 (43 and 37%, respectively). The methanol release data do, however, suggest that CheR1 has a role, as yet unidentified, in R. sphaeroides chemotaxis.
The deletion of either cheR2 or cheB1, which both lie within che Op2, resulted in a nonchemotactic phenotype under aerobic and photoheterotrophic conditions, both on swarm plates and in tethered cell assays. Both CheR2 and CheB1 can substitute for the corresponding homologues in E. coli. These findings suggest that CheR2 is the principal methyltransferase under laboratory conditions and that CheB1 functions as a methylesterase. The formation of swarm rings by the complemented strains demonstrates that CheR2 and CheB1 support chemotaxis in E. coli and are not simply restoring the tumble bias. CheR2 and CheB1, unlike CheR1, must therefore allow adaptation of these E. coli MCPs. The CheR of E. coli binds to the conserved NWETF motif on the high-abundance receptors, allowing methylation of both high- and low-abundance receptors (12). This motif has not been identified on any R. sphaeroides chemoreceptors, including the highly expressed McpG, although McpA and TlpT have similar motifs (GWEDF and GFEDF, respectively). However, McpA is expressed at low levels under the conditions tested, and its deletion has no effects on chemosensory behavior (unpublished data), whereas transmembrane prediction analysis shows that TlpT is probably a cytoplasmic protein. The ability of CheR2 to function in the E. coli chemosensory pathway is interesting, since it suggests that CheR2 is either able to interact with this motif, even though its native binding motif may be different, or CheR2 may interact with a different, conserved region of the MCPs.
In E. coli, null mutants of cheR and cheB cause smooth-swimming and tumbly phenotypes, respectively (42, 54). However, these strains retain the ability to respond (although not adapt) to changes in chemoattractant concentration (45, 54). Thus, while the strains appear nonchemotactic on swarm plates, changes in flagellar switching frequency are observed on the addition or the removal of attractant in tethered cell and free-swimming assays. In R. sphaeroides, deletion of either cheR2 or cheB1 results in a strain that is nonchemotactic on swarm plates. However, unlike the equivalent E. coli mutants, these strains are incapable of responding to the addition or the removal of attractant in the tethered cell assay. In addition, these strains retain a stopping frequency similar to that of unstimulated wild-type cells. Therefore, while R. sphaeroides contains homologues of the E. coli adaptation enzymes that will complement the equivalent E. coli mutations, the phenotypes observed on their deletion are significantly different. Further investigations are ongoing to determine whether these differences are the result of the multiple homologues of the adaptation enzymes present in R. sphaeroides or reflect a more substantial difference in the adaptation pathway between E. coli and R. sphaeroides.
Modified experimental conditions allowed us to demonstrate for the first time methanol release upon challenge with chemoeffector. Methanol production was observed upon the addition of 1 mM propionate but not upon its removal. This is in contrast to the pattern of methanol release in E. coli, where release is associated with the increased methylesterase activity of CheB-P, resulting in reduced activation of CheA by the MCPs. In R. sphaeroides methanol release upon the addition of propionate was abolished in both a ΔcheB1 strain and in a ΔcheR2 strain. Taken together with the complementation data, this suggests that CheR2 is the principal methyltransferase and that CheB1 is the principal methylesterase.
Unexpectedly, methanol release was observed upon both the addition and the removal of propionate in the ΔcheR1 strain. However, despite this difference in the pattern of methanol release, the ΔcheR1 strain was capable of exhibiting normal chemotaxis both on swarm plates and in the tethered cell assay. This is a similar pattern to that observed in wild-type B. subtilis and H. salinarum cells (19, 30, 47). In B. subtilis the additional proteins CheC and CheD are required for chemoreceptor methylation. orf1 in che Op1 (49) has 29% similarity with cheD from B. subtilis. The methanol release profile in the Δche Op1 strain was similar to that of wild-type cells. Therefore, the methanol produced upon the removal of propionate in the ΔcheR1 strain depends on components of che Op1, possibly the cheD homologue. This would suggest that there is an interaction between the CheD and CheR1 of R. sphaeroides. In B. subtilis, CheD has been shown to interact with the protein CheC (34), although the mechanism by which CheD regulates chemotaxis in B. subtilis remains unclear. However, despite the presence of CheD, R. sphaeroides has no apparent CheC homologue. Investigations are under way to determine whether any CheR1-CheD interaction exists and also the role of these proteins in adaptation in R. sphaeroides. It is clear that both CheR2 and CheB1 share some functional similarity with the equivalent adaptation enzymes of E. coli, as demonstrated by the complementation and methanol release data. However, adaptation to chemical stimuli in R. sphaeroides is more complex than in E. coli and also shows some similarity with the system of B. subtilis.
CheB1 has an important role in methylation-dependent chemotaxis. It is essential for chemotaxis in swarm plates and in tethered cell assays. CheB1 is also required for a normal pattern of methanol production. Complementation data show that it shares functional similarity with its E. coli counterpart and hence may be phosphorylated by E. coli CheA. In vitro phosphotransfer assays demonstrated that in R. sphaeroides CheB1 is phosphorylated by CheA2 but not by CheA1 (although both CheA1 and CheA2 can autophosphorylate in vitro). This finding is surprising since it suggests that any signal mediated by CheA1 cannot be subject to feedback adaptation via CheB1-P. The intriguing disparity between the ability of the CheAs to phosphorylate CheB1 is not readily identifiable from a comparison of the sequences of the two histidine protein kinases.
In agreement with a previous study in which it was shown that components of che Op1were not required for McpG-GFP clustering (48), CheR1 is not required for correct McpG-GFP localization. Furthermore, neither of the che Op2-encoded proteins, CheR2 and CheB1, was required for the aggregation of McpG in a chemoreceptor complex. Similarly, in E. coli CheR and CheB are not required for receptor clustering (24). The methylation state of the deletion mutants is likely to be altered with respect to the wild-type and therefore, as in E. coli, the methylation state of the chemoreceptor complex does not affect receptor localization. In this regard at least, the CheR1, CheR2, and CheB1 proteins of R. sphaeroides have properties similar to those of their counterparts in E. coli.
We have investigated the roles of CheR1, CheR2, and CheB1 from R. sphaeroides. It is clear that adaptation to chemoeffectors in this bacterium will not conform to the E. coli paradigm. The mechanism of adaptation may share some similarity with the gram-positive organism B. subtilis and the archeon H. salinarum, but there are also crucial differences between the species. The data presented here illustrate selectivity and discrimination within the histidine protein kinase and the response regulator components of the R. sphaeroides che pathways. The completion of the R. sphaeroides genome indicates additional CheR and CheB homologues, and investigations into the role of the multiple proteins in behavior and adaptation are under way to derive an alternative model by which bacteria may adapt to chemotactic stimuli.
ACKNOWLEDGMENTS
We thank J. S. Parkinson for the E. coli ΔcheR and ΔcheB mutants.
This work was supported by the BBSRC, and T.J.C. was the recipient of an undergraduate bursary from the Nuffield Foundation.
REFERENCES
- 1.Alam M, Lebert M, Oesterhelt D, Hazelbauer G L. Methyl-accepting taxis proteins in Halobacterium halobium. EMBO J. 1989;8:631–639. doi: 10.1002/j.1460-2075.1989.tb03418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armitage J P. Bacterial tactic responses. Adv Microb Physiol. 1999;41:229–289. doi: 10.1016/s0065-2911(08)60168-x. [DOI] [PubMed] [Google Scholar]
- 3.Armitage J P, Macnab R M. Unidirectional intermittent rotation of the flagellum of Rhodobacter sphaeroides. J Bacteriol. 1987;169:514–518. doi: 10.1128/jb.169.2.514-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armitage J P, Schmitt R. Bacterial chemotaxis: Rhodobacter sphaeroides and Sinorhizobium meliloti—variations on a theme? Microbiology. 1997;143:3671–3682. doi: 10.1099/00221287-143-12-3671. [DOI] [PubMed] [Google Scholar]
- 5.Borkovich K A, Alex L A, Simon M I. Attenuation of sensory receptor signaling by covalent modification. Proc Natl Acad Sci USA. 1992;89:6756–6760. doi: 10.1073/pnas.89.15.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borkovich K A, Kaplan N, Hess J F, Simon M I. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc Natl Acad Sci USA. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borkovich K A, Simon M I. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell. 1990;63:1339–1348. doi: 10.1016/0092-8674(90)90429-i. [DOI] [PubMed] [Google Scholar]
- 8.Bren A, Eisenbach M. How signals are heard during bacterial chemotaxis: protein-protein interactions in sensory signal propagation. J Bacteriol. 2000;182:6865–6873. doi: 10.1128/jb.182.24.6865-6873.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djordjevic S, Stock A M. Structural analysis of bacterial chemotaxis proteins: components of a dynamic signaling system. J Struct Biol. 1998;124:189–200. doi: 10.1006/jsbi.1998.4034. [DOI] [PubMed] [Google Scholar]
- 10.Falke J J, Bass R B, Butler S L, Chervitz S A, Danielson M A. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol. 1997;13:457–512. doi: 10.1146/annurev.cellbio.13.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng X, Baumgartner J W, Hazelbauer G L. High- and low-abundance chemoreceptors in Escherichia coli: differential activities associated with closely related cytoplasmic domains. J Bacteriol. 1997;179:6714–6720. doi: 10.1128/jb.179.21.6714-6720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng X, Lilly A A, Hazelbauer G L. Enhanced function conferred on low-abundance chemoreceptor Trg by a methyltransferase-docking site. J Bacteriol. 1999;181:3164–3171. doi: 10.1128/jb.181.10.3164-3171.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamblin P A, Maguire B A, Grishanin R N, Armitage J P. Evidence for two chemosensory pathways in Rhodobacter sphaeroides. Mol Microbiol. 1997;26:1083–1096. doi: 10.1046/j.1365-2958.1997.6502022.x. [DOI] [PubMed] [Google Scholar]
- 14.Harrison D M, Skidmore J, Armitage J P, Maddock J R. Localization and environmental regulation of MCP-like proteins in Rhodobacter sphaeroides. Mol Microbiol. 1999;31:885–892. doi: 10.1046/j.1365-2958.1999.01226.x. [DOI] [PubMed] [Google Scholar]
- 15.Hess J F, Oosawa K, Kaplan N, Simon M I. Phosphorylation of three proteins in the signalling pathway of bacterial chemotaxis. Cell. 1988;53:79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- 16.Jeziore-Sassoon Y, Hamblin P A, Bootle W C, Poole P S, Armitage J P. Metabolism is required for chemotaxis to sugars in Rhodobacter sphaeroides. Microbiology. 1998;144:229–239. doi: 10.1099/00221287-144-1-229. [DOI] [PubMed] [Google Scholar]
- 17.Jurica M S, Stoddard B L. Mind your B's and R's: bacterial chemotaxis, signal transduction and protein recognition. Structure. 1998;6:809–813. doi: 10.1016/s0969-2126(98)00082-3. [DOI] [PubMed] [Google Scholar]
- 18.Kehry M R, Doak T G, Dahlquist F W. Stimulus-induced changes in methylesterase activity during chemotaxis in Escherichia coli. J Biol Chem. 1984;259:11828–11835. [PubMed] [Google Scholar]
- 19.Kirby J R, Kristich C J, Feinberg S L, Ordal G W. Methanol production during chemotaxis to amino acids in Bacillus subtilis. Mol Microbiol. 1997;24:869–878. doi: 10.1046/j.1365-2958.1997.3941759.x. [DOI] [PubMed] [Google Scholar]
- 20.Kirby J R, Saulmon M M, Kristich C J, Ordal G W. CheY-dependent methylation of the asparagine receptor, McpB, during chemotaxis in Bacillus subtilis. J Biol Chem. 1999;274:11092–11100. doi: 10.1074/jbc.274.16.11092. [DOI] [PubMed] [Google Scholar]
- 21.Kort R, Crielaard W, Spudich J L, Hellingwerf K J. Color-sensitive motility and methanol release responses in Rhodobacter sphaeroides. J Bacteriol. 2000;182:3017–3021. doi: 10.1128/jb.182.11.3017-3021.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lupas A, Stock J. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J Biol Chem. 1989;264:17337–17342. [PubMed] [Google Scholar]
- 24.Lybarger S R, Maddock J R. Clustering of the chemoreceptor complex in Escherichia coli is independent of the methyltransferase CheR and the methylesterase CheB. J Bacteriol. 1999;181:5527–5529. doi: 10.1128/jb.181.17.5527-5529.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maddock J R, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 26.Martin A C, Wadhams G H, Armitage J P. The roles of the multiple CheW and CheA homologues in chemotaxis and in chemoreceptor localization in Rhodobacter sphaeroides. Mol Microbiol. 2001;40:1261–1272. doi: 10.1046/j.1365-2958.2001.02468.x. [DOI] [PubMed] [Google Scholar]
- 27.Morgan D G, Baumgartner J B, Hazelbauer G L. Proteins antigenically related to methyl-accepting chemotaxis proteins of Escherichia coli detected in a wide range of bacterial species. J Bacteriol. 1993;175:133–140. doi: 10.1128/jb.175.1.133-140.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mowbray S L, Sandgren M O. Chemotaxis receptors: a progress report on structure and function. J Struct Biol. 1998;124:257–275. doi: 10.1006/jsbi.1998.4043. [DOI] [PubMed] [Google Scholar]
- 29.Ninfa E G, Stock A, Mowbray S, Stock J. Reconstruction of the bacterial chemotaxis signal transduction system from purified components. J Biol Chem. 1991;266:9764–9770. [PubMed] [Google Scholar]
- 30.Nordmann B, Lebert M R, Alam M, Nitz S, Kollmannsberger H, Oesterhelt D, Hazelbauer G L. Identification of volatile forms of methyl groups released by Halobacterium salinarium. J Biol Chem. 1994;269:16449–16454. [PubMed] [Google Scholar]
- 31.Penfold R J, Pemberton J M. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene. 1992;118:145–146. doi: 10.1016/0378-1119(92)90263-o. [DOI] [PubMed] [Google Scholar]
- 32.Poole P S, Armitage J P. Motility response of Rhodobacter sphaeroides to chemotactic stimulation. J Bacteriol. 1988;170:5673–5679. doi: 10.1128/jb.170.12.5673-5679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosario M M, Kirby J R, Bochar D A, Ordal G W. Chemotactic methylation and behavior in Bacillus subtilis: role of two unique proteins, CheC and CheD. Biochemistry. 1995;34:3823–3831. doi: 10.1021/bi00011a040. [DOI] [PubMed] [Google Scholar]
- 34.Rosario M M, Ordal G W. CheC and CheD interact to regulate methylation of Bacillus subtilis methyl-accepting chemotaxis proteins. Mol Microbiol. 1996;21:511–518. doi: 10.1111/j.1365-2958.1996.tb02560.x. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multipurpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 37.Shah D S, Porter S L, Harris D C, Wadhams G H, Hamblin P A, Armitage J P. Identification of a fourth cheY gene in Rhodobacter sphaeroides and interspecies interaction within the bacterial chemotaxis signal transduction pathway. Mol Microbiol. 2000;35:101–112. doi: 10.1046/j.1365-2958.2000.01680.x. [DOI] [PubMed] [Google Scholar]
- 38.Shiomi D, Okumura H, Homma M, Kawagishi I. The aspartate chemoreceptor Tar is effectively methylated by binding to the methyltransferase mainly through hydrophobic interaction. Mol Microbiol. 2000;36:132–140. doi: 10.1046/j.1365-2958.2000.01834.x. [DOI] [PubMed] [Google Scholar]
- 39.Sistrom W R. A requirement for sodium in the growth of Rhodopseudomonas sphaeroides. J Gen Microbiol. 1960;22:778–785. doi: 10.1099/00221287-22-3-778. [DOI] [PubMed] [Google Scholar]
- 40.Skidmore J M, Ellefson D D, McNamara B P, Couto M M P, Wolfe A J, Maddock J R. Polar clustering of the chemoreceptor complex in Escherichia coli occurs in the absence of complete CheA function. J Bacteriol. 2000;182:967–973. doi: 10.1128/jb.182.4.967-973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sockett R E, Foster J C A, Armitage J P. Molecular biology of the Rhodobacter sphaeroides flagellum. FEMS Symp. 1990;53:473–479. [Google Scholar]
- 42.Springer W R, Koshland D E., Jr Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc Natl Acad Sci USA. 1977;74:533–537. doi: 10.1073/pnas.74.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spudich E N, Takahashi T, Spudich J L. Sensory rhodopsin I and II modulate a methylation/demethylation system in Halobacterium halobium. Proc Natl Acad Sci USA. 1989;86:7746–7750. doi: 10.1073/pnas.86.20.7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stock J B, Clarke S, Koshland D E., Jr The protein carboxymethyltransferase involved in Escherichia coli and Salmonella typhimurium chemotaxis. Meth Enzymol. 1984;106:310–321. doi: 10.1016/0076-6879(84)06031-6. [DOI] [PubMed] [Google Scholar]
- 45.Stock J B, Maderis A M, Koshland D E., Jr Bacterial chemotaxis in the absence of receptor carboxyl methylation. Cell. 1981;27:37–44. doi: 10.1016/0092-8674(81)90358-5. [DOI] [PubMed] [Google Scholar]
- 46.Stock J B, Surette M G. Chemotaxis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1103–1129. [Google Scholar]
- 47.Thoelke M S, Kirby J R, Ordal G W. Novel methyl transfer during chemotaxis in Bacillus subtilis. Biochemistry. 1989;28:5585–5589. doi: 10.1021/bi00439a037. [DOI] [PubMed] [Google Scholar]
- 48.Wadhams G H, Martin A C, Armitage J P. Identification and localization of a methyl-accepting chemotaxis protein in Rhodobacter sphaeroides. Mol Microbiol. 2000;36:1222–1233. doi: 10.1046/j.1365-2958.2000.01936.x. [DOI] [PubMed] [Google Scholar]
- 49.Ward M J, Bell A W, Hamblin P A, Packer H L, Armitage J P. Identification of a chemotaxis operon with 2 cheY genes in Rhodobacter sphaeroides. Mol Microbiol. 1995;17:357–366. doi: 10.1111/j.1365-2958.1995.mmi_17020357.x. [DOI] [PubMed] [Google Scholar]
- 50.Ward M J, Harrison D M, Ebner M J, Armitage J P. Identification of a methyl-accepting chemotaxis protein in Rhodobacter sphaeroides. Mol Microbiol. 1995;18:115–121. doi: 10.1111/j.1365-2958.1995.mmi_18010115.x. [DOI] [PubMed] [Google Scholar]
- 51.Weerasuriya S, Schneider B M, Manson M D. Chimeric chemoreceptors in Escherichia coli: signaling properties of Tar-Tap and Tap-Tar hybrids. J Bacteriol. 1998;180:914–920. doi: 10.1128/jb.180.4.914-920.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolfe A J, Conley M P, Kramer T J, Berg H C. Reconstitution of signalling in bacterial chemotaxis. J Bacteriol. 1987;169:1878–1885. doi: 10.1128/jb.169.5.1878-1885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu J G, Li J Y, Li G Y, Long D G, Weis R M. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- 54.Yonekawa H, Hayashi H, Parkinson J S. Requirement of the cheB function for sensory adaptation in Escherichia coli. J Bacteriol. 1983;156:1228–1235. doi: 10.1128/jb.156.3.1228-1235.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]