Abstract
Phosphofructokinase (PFK) is a key enzyme of the glycolytic pathway in all domains of life. Two related PFKs, ATP-dependent and PPi-dependent PFK, have been distinguished in bacteria and eucarya, as well as in some archaea. Hyperthermophilic archaea of the order Thermococcales, including Pyrococcus and Thermococcus spp., have recently been demonstrated to possess a unique ADP-dependent PFK (ADP-PFK) that appears to be phylogenetically distinct. Here, we report the presence of ADP-PFKs in glycogen-producing members of the orders Methanococcales and Methanosarcinales, including both mesophilic and thermophilic representatives. To verify the substrate specificities of the methanogenic kinases, the gene encoding the ADP-PFK from Methanococcus jannaschii was functionally expressed in Escherichia coli, and the produced enzyme was purified and characterized in detail. Compared to its counterparts from the two members of the order Thermococcales, the M. jannaschii ADP-PFK has an extremely low Km for fructose 6-phosphate (9.6 μM), and it accepts both ADP and acetyl-phosphate as phosphoryl donors. Phylogenetic analysis of the ADP-PFK reveals it to be a key enzyme of the modified Embden-Meyerhof pathway of heterotrophic and chemolithoautotrophic archaea. Interestingly, uncharacterized homologs of this unusual kinase are present in several eucarya.
The Embden-Meyerhof pathway is the most common route for the degradation of glucose. While several small variations in this glycolytic pathway are known, major modifications have been demonstrated in Pyrococcus furiosus and other hyperthermophilic archaea (4, 23). A combination of metabolic, biochemical, and genetic approaches have established that the pyrococcal glycolysis differs from the Embden-Meyerhof pathway by incorporating new conversions, novel enzymes, and unique control (9, 11, 15, 27, 28). First, the single-step conversion of glyceraldehyde-3-phosphate to 3-phospho (3P)-glycerate is catalyzed by a uniquely controlled glyceraldehyde-3-phosphate ferredoxin oxidoreductase (GAPOR) instead of the two-step reaction catalyzed by the conventional couple glyceraldehyde-3-phosphate dehydrogenase and phosphoglycerate kinase (15, 28). Second, instead of the classical ATP-dependent hexokinase, the pyrococcal pathway involves a novel ADP-dependent glucokinase (ADP-GLK) (11, 13). Third, a novel nonallosteric ADP-dependent phosphofructokinase (ADP-PFK) replaces the more common ATP-PFK (27).
The gene encoding an ADP-PFK was identified in the genome of P. furiosus and functionally expressed in Escherichia coli, and the encoded protein was thoroughly characterized (27). Primary structure comparison revealed the ADP-PFK to be a member of a novel enzyme family that did not show homology to known PFKs, which are monophyletic and include both ATP- and pyrophosphate (PPi)-dependent enzymes. However, the ADP-PFK appeared to have significant similarity to the ADP-GLK from P. furiosus, suggesting that they belong to the same novel family of kinases. Recently, the crystal structure of the ADP-GLK from Thermococcus litoralis was solved. Unexpected structural similarity was recognized with members of the ribokinase family (7).
Initial analysis of the first sequenced archaeal genome, that of the hyperthermophilic archaeon Methanococcus (Methanocaldococcus) jannaschii (29), suggested the presence of several glycolytic-enzyme-encoding genes but indicated the absence of a gene encoding a classical PFK (2, 24). Hence, it was suggested that the PFK from M. jannaschii could be ADP dependent and therefore undetectable in the sequence data (24). Indeed, an ortholog (MJ1604) with 48% identity (on the amino acid level) to the P. furiosus ADP-PFK was found to be encoded by the M. jannaschii genome (27). The presence of this hypothetical ADP-PFK in M. jannaschii suggests the presence of a modified Embden-Meyerhof pathway in methanogenic archaea as well. Previous studies on the genomic and enzyme levels indicated the presence of classical Embden-Meyerhof enzymes in bacteria, eucarya, and archaea (3). However, no attention was given to enzymes involved in the modified Embden-Meyerhof pathway.
To obtain insight into the presence and function of ADP-PFKs in representatives of different phylogenetic lineages, we investigated their distribution on both the genomic and functional levels. Moreover, the gene encoding the ADP-PFK from M. jannaschii was overexpressed in E. coli, and the purified enzyme was thoroughly characterized. The results provided evidence for the presence of ADP-PFKs in both mesophilic and thermophilic archaea and led us to propose an evolutionary model.
MATERIALS AND METHODS
Organisms and growth conditions.
All microorganisms were grown under an H2-CO2 atmosphere in 50 and 250 ml of medium, except that Methanosaeta concilii and P. furiosus were grown under an N2-CO2 atmosphere. P. furiosus (100°C) (DSM 3638) Methanococcus igneus (80°C) (DSM 5666), M. jannaschii (80°C) (DSM 2661), Methanococcus maripaludis (37°C) (DSM 2067), Methanococcus thermolithotrophicus (65°C) (DSM 2095), Methanopyrus kandleri (95°C) (DSM 6324), and Methanobacterium thermoautotrophicum Z-245 (65°C) (DSM 3720) (10) and Methanosarcina mazei (37°C) (DSM 2053) and M. concilii (37°C) (DSM 3671) (26) were grown as described previously (optimum temperatures given in parentheses). M. mazei and M. concilii were supplemented with 50 mM methanol and 30 mM acetate, respectively. Nina Brunner (University of Essen, Essen, Germany) kindly provided cells of Methanothermus fervidus (DSM 2088), grown at 80°C. Cells of Methanospirillum hungatei (DSM 864), grown at 37°C, were kindly provided by Frank de Bok (Wageningen University, Wageningen, The Netherlands).
Preparation of cell extracts from methanogens.
Cells were harvested from exponentially growing cultures by centrifugation (27,500 × g for 20 min) and suspended in 20 mM Tris-HCl (pH 7.5). The suspension was sonicated three times for 30 s each time. Cell debris was removed by centrifugation (16,000 × g for 15 min), and the resulting supernatant was used for enzyme measurements and protein determination. All manipulations in the preparation of cell extracts were performed anaerobically.
Enzyme activity assays in cell extracts.
All enzyme assays were performed anaerobically in N2-flushed stoppered 1-ml quartz cuvettes. Enzymatic activities were determined at temperatures close to the optimal growth temperature of the respective microorganism, with a maximum of 80°C. However, when auxiliary enzymes from mesophilic sources were included in assays, the measurements were performed at 37 (M. maripaludis, M. mazei, M. concilii, and M. hungatei) or 50°C (P. furiosus, M. igneus, M. jannaschii, M. thermolithotrophicus, M. kandleri, M. thermoautotrophicum, and M. fervidus), at which temperatures the heat-labile enzymes remained active and the activities of the thermostable enzymes could still be detected. The pH of buffers was adjusted at assay temperatures. Specific activities were calculated from linear rates and expressed in units per milligram of protein. One unit was defined as the amount of enzyme required to convert 1 μmol of substrate per min.
PFK activity was measured by following the oxidation of NADH in a coupled assay with fructose-1,6-bisphosphate aldolase, triosephosphate isomerase, and glycerol-3-phosphate dehydrogenase. The assay mixture contained 100 mM morpholineethanesulfonic acid (MES) (pH 6.5); 10 mM MgCl2; 0.2 mM NADH; 10 mM fructose-6-phosphate; 2.5 mM ATP, ADP, or acetyl-phosphate; 0.23 U of aldolase (from rabbit muscle); 11 U of triosephosphate isomerase (from rabbit muscle); 3.9 U of glycerol-3-phosphate dehydrogenase (from rabbit muscle); and 0.13 to 0.88 mg of protein. The absorbance of NADH was followed at 340 nm (ɛ = 6.18 mM−1 cm−1).
Hydrogenase activity was measured in hydrogen-flushed cuvettes using methyl viologen as an artificial electron acceptor. The assay mixture contained 100 mM Tris-HCl (pH 7.8), 0.1 mM dithiothreitol, and 1 mM methyl viologen. To measure maximal hydrogenase activity, the buffer was slightly prereduced by adding ca. 1 μl of a freshly prepared 100 mM sodium dithionite solution to the cuvette until an optical density at 578 nm of ca. 0.2 was reached. The reaction was started by adding 0.035 to 0.13 mg of protein. The reduction of methyl viologen was followed at 578 nm (ɛ = 9.7 mM−1 cm−1).
Protein concentrations were determined with Coomassie brilliant blue G250 as previously described (1).
Genome analysis.
Genome databases were screened for orthologs of modified and classical types of GLK and PFK, as well as of glyceraldehyde 3-phosphate dehydrogenase and glyceraldehyde 3-phosphate ferredoxin oxidoreductase genes, at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/ [update, August 2001]), using blastP (expect value [E] < 2e-04); at Genomes On Line Databases (http://igweb.integratedgenomics.com/GOLD/ [update, July 2001]); and at Kyoto Encyclopedia of Genes and Genomes (http://www.genome.ad.jp/kegg/ [update July 2001]). The Sulfolobus solfataricus genome was screened at Multipurpose Automated Genome Project Investigation (http://niji.imb.nrc.ca/sulfolobus/private/sulfolobus.html [update, January 2001]).
Multiple sequence alignment and tree construction.
Sets of amino acid sequences were aligned with the ClustalX program (version 1.64b) and edited manually. A neighbor-joining tree was constructed with complete 16S and 18S rRNA sequences using the ARB software package (http://www.mikro.biologie.tu-muenchen.de/pub/ARB/documentation/arb.ps) and modified based on the model of Woese et al. (30).
Cloning of the pfkC gene from M. jannaschii.
An ortholog of the recently characterized pfkC (formerly called pfkA) gene from P. furiosus (27) was detected in the M. jannaschii database (MJ1604). The following primer set was designed to amplify this M. jannaschii pfkC gene open reading frame by PCR: BG445 (5′-GCGCGCCATGGCATGTGATATTATGGAAATAA; sense) and BG446 (5′-GCGCGGATCCTTAGCTCATTCTTTTTTTATTTAA; antisense); NcoI and BamHI restriction sites are indicated in boldface. The 100-μl polymerase reaction mixture contained 100 ng of chromosomal M. jannaschii DNA, isolated as described before (22); 100 ng each of primers BG445 and BG446; 0.2 mM deoxynucleoside triphosphates; Pfu polymerase buffer; and 5 U of Pfu DNA polymerase and was subjected to 35 cycles of amplification (1 min at 94°C, 35 s at 50°C, and 3 min at 72°C) on a DNA thermal cycler (Perkin-Elmer Cetus). The PCR product was digested (NcoI/BamHI) and cloned into an NcoI/BamHI-digested pET9d vector, resulting in pLUW575, which was transformed into BL21(DE3). Sequence analysis of pLUW575 was done by the dideoxynucleotide chain termination method with a Li-Cor automatic sequencing system (model 4000L). Sequence data were analyzed using the computer program DNASTAR.
Purification and characterization of the E. coli-produced ADP-PFK from M. jannaschii.
An overnight culture of E. coli BL21(DE3) harboring pLUW575 was inoculated (1%) into 1 liter of Luria-Bertani medium with 50 μg of kanamycin/ml. After growth for 16 h at 37°C, the cells were harvested by centrifugation (2,200 × g for 20 min) and resuspended in 10 ml of a 20 mM Tris-HCl buffer (pH 7.8). The suspension was passed twice through a French press (100 MPa), and cell debris was removed by centrifugation (10,000 × g for 20 min). The resulting supernatant was heat treated for 30 min at 80°C, and precipitated proteins were removed by centrifugation.
The cell extract was filtered through a 0.45-μm-pore-size filter and applied to a Q-Sepharose fast-flow column (Amersham Pharmacia Biotech) that was equilibrated with a 20 mM Tris-HCl buffer (pH 7.8) containing 1 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate}. ADP-PFK activity eluted at 0.3 M NaCl in a 125-ml gradient from 0 to 1 M NaCl. Active fractions were pooled and desalted by ultrafiltration using a 10 mM potassium phosphate buffer (pH 7.0). The desalted pool was applied to a hydroxyapatite CHT5-1 column (Bio-Rad) that was equilibrated with 10 mM potassium phosphate buffer. The enzyme eluted in a 75-ml linear gradient (10 to 500 mM potassium phosphate) at 250 mM potassium phosphate. Active fractions were pooled, the buffer was changed for a 25 mM Tris-HCl buffer (pH 7.8) containing 1 mM CHAPS by ultrafiltration, and the pool was loaded onto a mono-Q HR 5/5 column (Amersham Pharmacia Biotech) that was equilibrated in the same buffer. The enzyme eluted from the column at 0.3 M NaCl in a 20-ml linear gradient from 0 to 1 M NaCl. Fractions showing ADP-PFK activity were pooled and concentrated 16-fold to a final volume of 460 μl. This concentrated pool was applied to a Superdex 200 HR 10/30 gel filtration column (Amersham Pharmacia Biotech) that was equilibrated with a 100 mM Tris-HCl buffer (pH 7.8) containing 100 mM NaCl, from which the protein eluted after 15 ml. The purity of the ADP-PFK was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE).
The purified enzyme was characterized by determining its specific activity, molecular mass, pH optimum, substrate specificity, kinetic parameters, and allosteric effectors as described before (27).
RESULTS
Genome analysis.
To investigate the presence of genes that potentially encode enzymes involved in the modified Embden-Meyerhof pathway, we screened recent releases of complete and incomplete genome sequences of archaea and thermophilic bacteria (Table 1).
TABLE 1.
Enzymes of the classical and modified Embden-Meyerhof pathways encoded in the different genomes of archaea and (hyper)thermophilic bacteria
| Genomea | Accession
no.
|
||||||
|---|---|---|---|---|---|---|---|
| ATP-GLK | ADP-GLK | ATP-PFK | ADP-PFK | PPi-PFK | GAPDHb | GAPOR | |
| Pfu | AF127910 | AF127909 | PF1729232 | AAC70892 | |||
| Pho | PH0589 | PH1645 | PH1830 | PH0457 | |||
| Pab | PAB0967 | PAB0213 | PAB0257 | PA1315 | |||
| Mja | MJ1604 | MJ1146 | MJ1185 | ||||
| Mma | Presentc | Present | |||||
| Afu | AF1732 | ||||||
| Mth | MT1009 | ||||||
| Hal | AAG20664 | AAG18725 | |||||
| Tac | TA0825 | TA1103 | |||||
| Sso | SSO0528 | ||||||
| Ape | APE2091 | APE0012d | APE0171 | ||||
| Tma | TM1469 | TM0209 | TM0289 | TM0688 | |||
| Aae | AQ1496 | AQ1708 | AQ1065 | ||||
| Tth | Present | Present | Present | ||||
Genome analyses were performed on the following organisms euryarchaea, Pfu, P. furiosus; Pho, P. horikoshii; Pab, P. abyssi; Mja, M. jannaschii; Mma, M. mazei; Afu, A. fulgidus; Mth, M. thermoautotrophicum; Hal, Halobacterium sp. strain NRC-1; Tac, T. acidophilum. crenarchaea, Sso, S. solfataricus; Ape, A. pernix. Bacteria, Tma, T. maritima; Aae, A. aeolicus; Tth, T. thermophilus.
GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Present, present in the genome but not yet annotated.
Orthologs of the novel GAPOR were identified in the genomes of all three Pyrococcus species (P. furiosus,Pyrococcus horikoshii and Pyrococcus abyssi) and in that of M. jannaschii. A classical NAD-dependent glyceraldehyde-3-phosphate dehydrogenase appeared to be present in all screened genomes. In P. furiosus, this glyceraldehyde-3-phosphate dehydrogenase is involved in gluconeogenesis, whereas GAPOR functions in the glycolytic direction (28).
Orthologs of the ADP-GLK were identified only in the genomes of all three Pyrococcus species. Genes encoding classical ATP-dependent hexokinases were identified in Halobacterium sp. strain NRC-1, Thermoplasma acidophilum, Aeropyrum pernix, and the hyperthermophilic bacteria Thermotoga maritima, Aquifex aeolicus, and Thermus thermophilus.
Finally, ADP-PFK orthologs were identified in the three Pyrococcus genomes, the M. jannaschii genome, and the genome of M. mazei Gö1. Remarkably, both an ATP-PFK and a PPi-PFK ortholog were identified in the genome of T. maritima, whereas in the closely related A. aeolicus, only an ATP-PFK ortholog was identified.
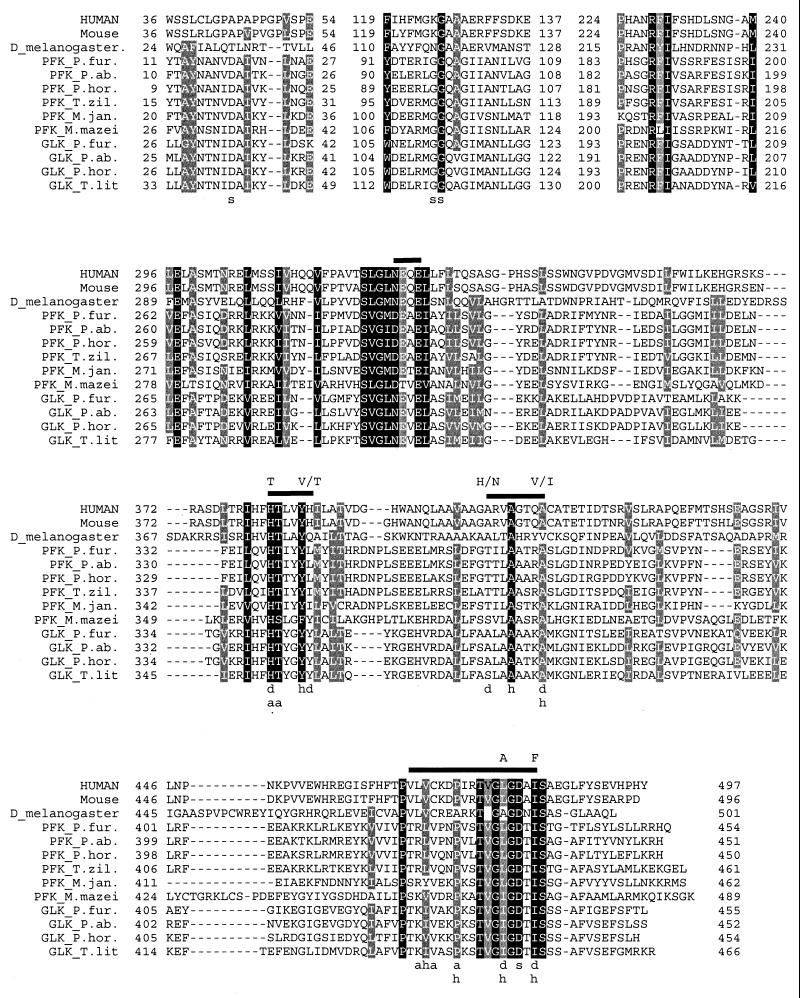
Interestingly, homologs of ADP-dependent sugar kinases (12 to 17% identity to the archaeal kinases) were identified in several eukaryotic genome sequences, i.e., those of Drosophila melanogaster (AAF49769), Caenorhabditis elegans (T32780), Mus musculus (BAB27619), and Homo sapiens (AAH06112) (query, ADP-GLK [AF127910]; blastP, E < 1e-07); no homologous sequence was identified in yeast. Based on the recently described ADP-GLK structure of T. litoralis (7), a multiple sequence alignment of the identified homologs was created showing the conservation of several structurally important active-site residues in the ADP-dependent sugar kinases and their eukaryal homologs (Fig. 1). The C. elegans amino acid sequence was omitted from the alignment because the C-terminal part of its sequence was fused with a putative transacylase, and therefore, important residues conserved at the C-terminus might have been lost.
FIG. 1.
Multiple alignment of ADP-dependent kinase homologs. Gaps introduced for optimal alignment are marked by hyphens. Conserved residues are shaded black. Residues that are present in at least one eukaryal sequence and in the majority of the archaeal sequences are shaded grey. Nucleotide binding loops are indicated by bars above the alignment. a, residues that form hydrogen bonds with ADP; d, residues involved in the discrimination between ADP and ATP (the residues in these positions specific for ATP-dependent ribokinase and adenosine kinase are indicated above the alignment); h, hydrophobic residues that form a pocket with the approximate shape of the adenosine moiety; s; sugar binding and/or activation residues (7). The sequences have the following accession numbers: Human, hypothetical H. sapiens protein, AAH06112; Mouse, hypothetical M. musculus protein, BAB27619; D_melanogaster, hypothetical protein, AAF49769; PFK_P.fur, P. furiosus ADP-PFK, AF127909; PFK_P.ab., putative P. abyssi ADP-PFK PAB0213, D75170; PFK_P.hor., putative P. horikoshii ADP-PFK PH1645, E71044; PFK_T.zil., T. zilligii ADP-PFK, AAF97356; PFK_M.jan., M. jannaschii ADP-PFK MJ1604, C64500; PFK_M.mazei, putative M. mazei ADP-PFK (A. Johan, personal communication); GLK_P.fur., P. furiosus ADP-GLK, AF127910; GLK_P.ab., putative P. abyssi ADP-GLK PAB0967, B75058; GLK_P.hor., putative P. horikoshii ADP-GLK PH0589, A71174; GLK_T.lit., T. litoralis ADP-GLK.
Distribution of ADP-PFK activity in methanogens.
The identification of orthologs of the ADP-PFK in M. jannaschii and M. mazei (Table 1) suggested that the modified Embden-Meyerhof pathway, as first described in the fermentative hyperthermophilic archaeon P. furiosus, may also be present in methanogens. This prompted us to determine PFK activity using ADP and ATP as the phosphoryl group donors in a wide range of methanogenic species belonging to various taxonomic groups. As a reference enzyme, hydrogenase activity was measured as well.
ADP-PFK activity was detected in cell extracts of the thermophilic methanogens M. jannaschii, M. igneus, and M. thermolithotrophicus and in the mesophilic methanogens M. maripaludis and M. mazei (Table 2). The presence of an ATP-PFK activity (5.6 mU/mg) in M. maripaludis had been reported previously (31). However, we could detect only activity of an ADP-PFK (6.1 mU/mg) but not that of an ATP-PFK in M. maripaludis. An ADP-PFK activity was also detected in the mesophilic M. concilii. In that organism, a high ATP-PFK activity was also detected. Since M. concilii is known to possess high adenylate kinase activity (8), the detected ADP-PFK activity might be the result of the combined action of the adenylate kinase and the ATP-PFK activity. In M. thermolithotrophicus, both ADP-PFK and ATP-PFK activities were detected.
TABLE 2.
ADP-PFK activities in archaeaa
| Microorganism | Growth temp (°C) | Hydrogenase activity (U/mg) | ADP-PFK activity (mU/mg) | ATP-PFK activity (mU/mg) |
|---|---|---|---|---|
| P. furiosus | 100 | NMb | 150 | 2.6 |
| T. litoralisd | 85 | NM | 200 | NM |
| M. jannaschii | 80 | 28 | 21 | ND |
| M. igneus | 80 | 67 | 16 | ND |
| M. thermolithotrophicus | 65 | 130 | 18 | 3.8 |
| M. maripaludis | 37 | 23 | 2.7 | ND |
| M. mazei | 37 | 140 | 6.1 | ND |
| M. concilii | 37 | NDc | 29 | 38 |
Enzyme activities were determined as described in Materials and Methods. No PFK activity could be detected in M. thermoautotrophicum, M. kandleri,M. fervidus, or M. hungatei extracts.
NM, not measured.
ND, not detectable.
Selig et al. 1997 (23).
Overexpression of the M. jannaschii pfkC gene in E. coli.
To gain insight into the substrate specificity of the methanogic ADP-PFK orthologs, we compared the properties of the enzyme of M. jannaschii to those of P. furiosus ADP-PFK. For this purpose, the M. jannaschii pfkC gene (MJ1604) was PCR amplified and cloned into pET9d, resulting in plasmid pLUW575. DNA sequence analysis of pLUW575 confirmed that the cloned pfkC gene showed the expected sequence. Sodium dodecyl sulfate-PAGE analysis of a cell extract of E. coli BL21(DE3) harboring pLUW575 revealed an additional band of 51.5 kDa, which corresponded to the calculated molecular mass (53.4 kDa) of the gene product (not shown). This band was absent in cell extracts of E. coli BL21(DE3) carrying the pET9d vector that also showed no ADP-PFK activity. In a cell extract of E. coli BL21(DE3) harboring pLUW575, an ADP-PFK activity of 0.8 U/mg was measured at 50°C, confirming that the cloned M. jannaschii pfkC gene indeed encoded an ADP-PFK. The enzyme could be produced up to 10% of total soluble cell protein after 16 h of cultivation at 37°C without inducing gene expression by adding isopropyl-1-thio-β-d-galactopyranoside.
Characteristics of the M. jannaschii ADP-PFK.
The E. coli-produced M. jannaschii ADP-PFK was purified to homogeneity. The native molecular mass of the enzyme, as determined by native PAGE at various acrylamide percentages was approximately 50.1 kDa, indicating that the M. jannaschii ADP-PFK is a monomer (not shown).
The purified enzyme had a specific activity of 8.2 U/mg at 50°C at the optimum pH of 6.5, only in the direction of phosphorylation. Apart from ADP, acetyl-phosphate could serve as an efficient phosphoryl group donor to the enzyme (Table 3). Divalent cations were required for activity, as indicated by complete lack of activity in the presence of EDTA. ADP-PFK activity was highest in the presence of CaCl2, followed by MgCl2 (Table 3). Both KCl and NaCl had negative effects on the ADP-PFK activity (84 and 88% activity in 500 mM KCl and NaCl, respectively). Furthermore, the enzyme activity was negatively affected by the addition of ATP or AMP to the assay mixture (53 and 24% activity in 10 mM ATP and AMP, respectively). However, the addition of fructose 2,6-bisphosphate, pyruvate, glucose, phosphoenolpyruvate, or citrate to the assay mixture had no effect on the activity. The enzyme showed Michaelis-Menten kinetics at 50°C, with the following constants, which were determined according to a computer-aided direct fit using the Michaelis-Menten equation: apparent Km values of 0.0096 ± 0.0007 and 0.49 ± 0.13 mM for fructose 6-phosphate and ADP, respectively, and apparent Vmax values of 11.2 ± 0.3 and 9.59 ± 0.74 U/mg for fructose 6-phosphate and ADP, respectively. For acetyl-phosphate as a phosphoryl group donor, an apparent Km value of 11.9 ± 1.8 mM and an apparent Vmax of 14.4 ± 1.0 U/mg at 50°C were determined. In a Hill plot, the kinetic data of fructose 6-phosphate, ADP, and acetyl-phosphate showed noncooperative binding of the substrates (not shown).
TABLE 3.
Phosphoryl group donor and cation dependence of the ADP-PFK from M. jannaschiia
| Phosphoryl group donor | Divalent cation | Sp act (mU/mg) | Relative activity (%) |
|---|---|---|---|
| ADP | Mg2+ | 8,200 | 100 |
| GDP | Mg2+ | 115 | 1.4 |
| ATP | Mg2+ | 24.6 | 0.3 |
| GTP | Mg2+ | 664 | 8.1 |
| Acetyl-phosphate | Mg2+ | 6,806 | 83 |
| Polyphosphate | Mg2+ | NDb | ND |
| Phosphoenolpyruvate | Mg2+ | ND | ND |
| Pyrophosphate | Mg2+ | ND | ND |
| ADP | Ca2+ | 9,840 | 120 |
| ADP | Co2+ | 6,396 | 78 |
| ADP | Mn2+ | 4,428 | 54 |
| ADP | Zn2+ | ND | ND |
Standard enzyme assays were done at 50°C, except that phosphoryl group donors and cations were varied as described in Materials and Methods.
ND, not detectable, i.e., the activity was less than 0.3% of the activity under optimal conditions.
DISCUSSION
Following the discovery of ADP-PFK activity in P. furiosus (9) and characterization of this novel enzyme (27), ADP-PFK activity has been detected in various members of the order Thermococcales (18, 23). The presence of a glycolytic pathway in methanogens has recently been proposed based on (i) enzyme analyses of M. maripaludis (31) and (ii) analysis of the genome sequence of M. jannaschii, which revealed several glycolytic orthologs (2). However, no PFK gene was detected (24). Here, we demonstrate the functional presence of ADP-PFKs in methanogenic archaea, analyze their distribution, and describe the unique catalytic properties of the purified enzyme from M. jannaschii.
The recent characterization of the amino acid sequence of the P. furiosus and Thermococcus zilligii ADP-PFK (20, 27) resulted in the identification of orthologs in the genomes of both chemolithoautotrophic (M. jannaschii and M. mazei) and heterotrophic (P. abyssi and P. horikoshii) archaea (Table 1). These data already suggested that a modified Embden-Meyerhof pathway, present in P. furiosus, might also be operational in methanogens. In addition, we detected PFK activity in all methanogens investigated, i.e., M. jannaschii, M. thermolithotrophicus, M. igneus, and M. maripaludis, of the order Methanococcales, and M. mazei, of the order Methanosarcinales (Table 2). Although ADP-PFK activity could be detected in M. concilii extracts, this activity was probably the result of the concerted action of ATP-PFK activity and high adenylate kinase activity (8). The high ADP-PFK activity (18 mU/mg) and relatively low ATP-PFK activity (3.8 mU/mg) detected in M. thermolithotrophicus might indicate that two types of PFKs, i.e., ADP and ATP dependent, are present in that organism. On the other hand, the P. furiosus PFK was shown to be active with ADP and to some extent with ATP (27). Therefore, the ADP-PFK and ATP-PFK activities detected in M. thermolithotrophicus might also be the result of a single enzyme with affinity for both ATP and ADP. In all other methanogens investigated, neither ADP-PFK nor ATP-PFK activity was detected.
Apart from the analysis of an ADP-PFK, the modified Embden-Meyerhof pathway in P. furiosus also differs from the conventional Embden-Meyerhof pathway at the level of glyceraldehyde 3-phosphate. The conversion of glyceraldehyde 3-phosphate by GAPOR has also been detected in Thermococcus spp. (23) and M. jannaschii (28; G. Schut and J. van der Oost, personal communication). This observation confirms predictions from the genome analyses (Table 1) that the glycolytic pathway in M. jannaschii resembles that in P. furiosus, not only in the ADP dependency of the PFK but also in regard to the conversion of glyceraldehyde 3-phosphate by GAPOR.
The activities of ADP-PFK and GAPOR found in cell extracts of M. jannaschii grown under standard autotrophic conditions (H2 and CO2), are an order of magnitude lower (PFK, 21 mU/mg; GAPOR, 100 mU/mg) than those in cell extracts of P. furiosus grown on saccharides (PFK, 150 mU/mg; GAPOR, 1,800 mU/mg) (11, 28). This suggests that, at least under the investigated conditions, the modified Embden-Meyerhof pathway is less prominent in M. jannaschii than in P. furiosus. Since the pathway in methanogens is assumed to be used for glycogen degradation during starvation, the enzyme activities might be induced in starved cells. Under starvation, the mesophilic archaeon M. maripaludis has been reported to degrade its glycogen storage, resulting in the release of glucose 1-phosphate, which is converted to glucose 6-phosphate to enter the Embden-Meyerhof pathway (31). Moreover, glycogen has been reported to be a reserve polysaccharide in Methanosarcina spp. and Methanosaeta spp. (16, 17). Likewise, the detected ATP-PFK activity in M. concilii and the ADP-PFK activity in Methanococcus spp. and M. mazei presumably play a role in the degradation of glycogen. All methanogens in which no PFK activity could be detected, such as M. thermoautotrophicum, M. kandleri, M. fervidus, and M. hungatei, appear to lack the capacity to synthesize and degrade glycogen.
Because of the different physiologies of heterotrophic and chemolithoautotrophic organisms, a comparison of the properties of the ADP-PFKs from both types of organisms was of interest. Therefore, the gene encoding the ADP-PFK from M. jannaschii (MJ1604) was expressed in E. coli, and the protein was purified and characterized in detail. The biochemical characteristics of this protein were compared to those of the ADP-dependent enzymes from P. furiosus (27) and T. zilligii (18) (Table 4). The M. jannaschii enzyme is the only ADP-PFK studied that was able to use acetyl-phosphate as a phosphoryl group donor. It remains unclear whether the specificity for acetyl-phosphate is a specific physiological adaptation of methanogens or a nonspecific result of a minor deviation in the substrate binding pocket of the M. jannaschii enzyme. The latter explanation appears to be the more likely, since no acetyl-phosphate-dependent PFK activity was detected in the other methanogens used in this study, including the acetoclastic ones (data not shown).
TABLE 4.
Biochemical comparison of ADP-PFKsa
| Parameter | Substrate | Value
|
||
|---|---|---|---|---|
| P. furiosusb | T. zilligiic | M. jannaschii | ||
| Molecular mass of native enzyme (kDa) | 180 | 200 | 51.5 | |
| Molecular mass of subunit (kDa) | 52 | 53.9 | 53.4 | |
| Allosteric regulation | No | No | No | |
| Apparent Km (mM) | Fructose 6-phosphate | 2.3 | 3.77 | 0.0096 |
| ADP | 0.11 | 0.11 | 0.49 | |
| Acetyl-phosphate | NDd | ND | 11.9 | |
| Fructose 1,6-bisphosphate | ND | 12.5 | ND | |
| AMP | ND | 0.56 | ND | |
| Apparent Vmax (U/mg) | Fructose 6-phosphate | 194 | 197 | 11.2 |
| ADP | 150 | 243 | 9.59 | |
| Acetyl-phosphate | ND | ND | 14.4 | |
The affinity of the M. jannaschii enzyme for fructose 6-phosphate is extremely high, and its catalytic efficiency (kcat/Km) of 1,038 mM−1 s−1 is the highest reported to date. Normally, the apparent Km for the phosphoryl group donor is lower than that for the cosubstrate fructose 6-phosphate, presumably a reflection of the average intracellular concentrations of both substrates (18). This contrasts with the properties of the M. jannaschii enzyme, which shows a 50-fold-lower apparent Km for fructose 6-phosphate than for ADP. This might reflect different intracellular concentrations of fructose 6-phosphate in the glycogen-degrading M. jannaschii and the sugar-fermenting P. furiosus and T. zilligii.
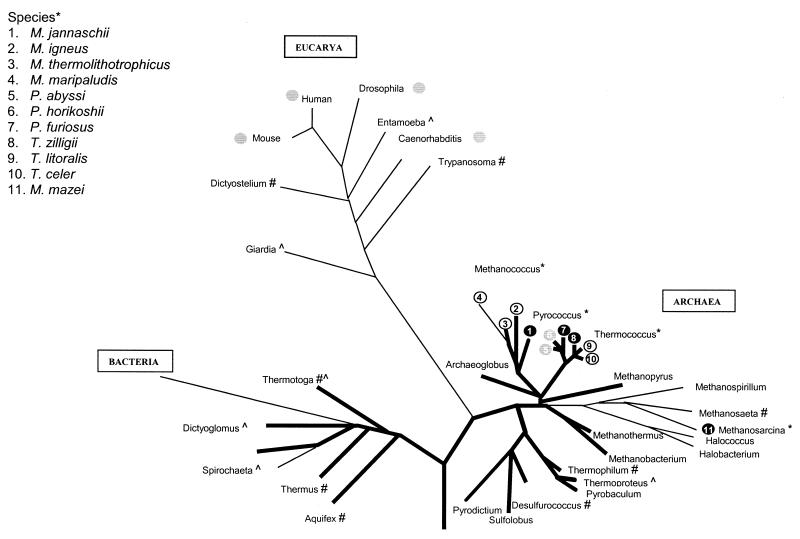
Our data extend the previously noted presence of ADP-PFKs in members of the order Thermococcales to their abundant presence in members of the closely related order Methanococcales. Furthermore, we show that PFKs are present in all tested glycogen-consuming methanogens of the genera Methanococcus, Methanosarcina, and Methanosaeta. The first two genera harbor ADP-PFKs, whereas M. concilii most likely contains an ATP-PFK rather than ADP-PFK activity. The presence of activity of an ADP-PFK in mesophilic archaea and that of ATP-PFK and PPi-PFK in hyperthermophilic archaea (6, 19, 25) indicates that ADP dependency is not essential for this glycolytic conversion at higher temperatures (Fig. 2).
FIG. 2.
Phylogenetic tree based on 16S and 18S rRNA sequences. The tree was constructed based on complete 16S and 18S rRNA sequences using the ARB software package. The thick lines represent the (hyper)thermophilic lineages, and the thin lines represent the mesophilic lineages. PFK activity or related sequences identified in the different species on the genomic and/or functional level are marked as follows: ∗, ADP-PFK; #, ATP-PFK; ^, PPi-PFK; ○, ADP-PFK activity present; , ADP-dependent (sugar) kinase homolog present; ●, both ADP-PFK activity and homologous gene identified.
Recently, the crystal structure of the ADP-dependent glucokinase from T. litoralis has been solved (7). Because of its distinct primary structure, it came as a surprise that the ADP-GLK structure had significant structural similarity to the ATP-dependent ribokinase family. Cocrystalization with ADP elucidated the position of the nucleotide binding site. Remarkably, minor modifications in amino acid composition appear to result in a clear preference for either ADP or ATP in the different kinases (7). The majority of the residues that were found to interact directly with ADP in the T. litoralis ADP-GLK are conserved in all ADP-dependent sugar kinase homologs, including the eukaryal sequences (Fig. 1). The size of the side chain of residue 353 (M. jannaschii ADP-PFK numbering) seems to be important for the ADP-dependent kinases to lose their ATP-dependent activity, as it excludes a nucleotide to adopt the proper position for donating the γ-phosphate (7). However, ATP can be phosphorylated to some extent by several ADP-PFKs (P. furiosus [27], M. jannaschii, and T. zilligii [18]). Interestingly, in the last two sequences a Leu or Ile residue has replaced a Tyr residue. ATP-dependent kinases of the ribokinase family in general have residues with small side chains (Val or Thr) at this position (Fig. 1) (7). The eukaryal sequences have more bulky His or Gln residues at the same position. Moreover, adjacent to Ile353 (M. jannaschii ADP-PFK numbering), there is a well-conserved Tyr residue (Tyr352) (except for the Phe residue in M. mazei) (Fig. 1). In all known members of the ATP-dependent ribokinase family, an extra residue is located between Tyr352 and Ile353 (not shown). This elongation of one residue may result in a rearrangement of the main chain to create an appropriate shape for the recognition of the α-phosphate of ATP in these ATP-dependent enzymes (7). This elongation is absent in the ADP-dependent sugar kinases and their eukaryal homologs. Ile444, close to the catalytic base Asp442 (M. jannaschii ADP-PFK numbering) seems to be one of the key residues that determine ADP specificity. These two residues are conserved in all ADP-PFK homologs, as well as in the eukaryal homologs (Fig. 1). ATP-dependent ribokinases show a Phe residue at the corresponding position (Fig. 1) (7). Hydrophobic amino acids forming a pocket for the right shape of the adenosine moiety are conserved in the eukaryal sequences as well (Fig. 1), suggesting that the identified homologs in eucarya are most likely kinases, with a possible preference for ADP as a phosphoryl donor. Unfortunately, no ADP-GLK/glucose-6-phosphate cocrystals are available at the moment that would allow identification of the sugar binding site of the ADP-dependent sugar kinases. Nevertheless, important residues involved in the binding of sugars in members of the ribokinase family (7) seem to be conserved in the ADP-dependent sugar kinases (Fig. 1). These putative crucial residues (Asp28 and Gly106-Gly107 of the M. jannaschii ADP-PFK) are conserved in all archaeal homologs but not in the eukaryal homologs. Hence, the structure-based sequence comparisons suggest that the eukaryal homologs are presumably (ADP-dependent) kinases but most likely do not phosphorylate similar sugar substrates.
A phylogenetic tree of the eukaryal and archaeal ADP-dependent (sugar) kinase homologs (not shown) is in good agreement with the dendrogram based on 16S and 18S rRNA sequences (Fig. 2), suggesting natural inheritance rather than horizontal gene transfer. This suggests that ADP-dependent kinases may have evolved before the separation of eucarya and archaea and, if present in the last common ancestor, were lost from bacteria. Such a loss may also have occurred in the crenarchaea and among the euryarchaea in Archaeoglobus (see Addendum), non-glycogen-consuming methanogens, and deeply rooted halophiles (Fig. 2). Probably because of their specific physiological characteristics (no Embden-Meyerhof pathway), the selective pressure to maintain these kinases was lost in these organisms. Only the heterotrophic order Thermococcales, as well as the glycogen-degrading orders Methanococcales and Methanosarcinales, are found to contain a functional variant of the Embden-Meyerhof pathway and, as such, benefit from harboring an ADP-PFK.
The observed presence of unique ADP-PFK activity and the corresponding genes in the phylogenetically closely related Pyrococcus spp. and Methanococcus spp., as well as the deeply branched M. mazei, suggests that the specific function of ADP-dependent sugar phosphorylation originated at least before the branching of Thermococcales and Methanococcales and that gene duplication in the order Thermococcales has led to two specific enzymes, i.e., ADP-PFK and ADP-GLK. The acquired sugar kinases and the gaining of specific glycosyl hydrolases and sugar transporters by Pyrococcus spp. (5) probably enabled these organisms to ferment sugars. The ADP-PFKs described here most likely function as a key step in a central metabolic pathway. Functional analysis of the eukaryal homologs will be the next step to gain more insight into the evolution of this enzyme family.
ACKNOWLEDGMENTS
This research was supported by the Earth and Life Sciences Foundation (ALW), which is subsidized by the Netherlands Organization for Scientific Research (NWO).
We thank Frank de Bok and Nina Brunner for providing cell extract methanogens. Erwin Zoetendal is gratefully acknowledged for constructing the phylogenetic tree. Finally, we thank Andre Johann (Goettingen Genomics Laboratory) for providing the M. mazei PFK sequence.
C.H.V. and J.E.T. contributed equally to this work.
ADDENDUM
While this report was being evaluated, Labes et al. described an ADP-GLK and an ADP-PFK activity in starch-degrading Archaeoglobus fulgidus strain 7324 (14). The corresponding genes have not yet been identified. In the genome sequence of type strain VC16, no genes encoding ADP-GLK or ADP-PFK could be identified (12).
REFERENCES
- 1.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 2.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 3.Dandekar T, Schuster S, Snel B, Huynen M, Bork P. Pathway alignment: application to the comparative analysis of glycolytic enzymes. Biochem J. 1999;343:115–124. [PMC free article] [PubMed] [Google Scholar]
- 4.de Vos W M, Kengen S W M, Voorhorst W G B, van der Oost J. Sugar utilization and its control in hyperthermophiles. Extremophiles. 1998;2:201–205. doi: 10.1007/s007920050061. [DOI] [PubMed] [Google Scholar]
- 5.Ettema T J G, van der Oost J, Huynen M. Modularity in the gain and loss of genes: applications for function prediction. Trends Genet. 2001;17:485–487. doi: 10.1016/s0168-9525(01)02384-8. [DOI] [PubMed] [Google Scholar]
- 6.Hansen T, Schönheit P. Purification and properties of the first-identified, archaeal, ATP-dependent 6-phosphofructokinase, an extremely thermophilic non-allosteric enzyme, from the hyperthermophile Desulfurococcus amylolyticus. Arch Microbiol. 2000;173:103–109. doi: 10.1007/s002039900114. [DOI] [PubMed] [Google Scholar]
- 7.Ito S, Fushinobu S, Yoshioka I, Koga S, Matsuzawa H, Wakagi T. Structural basis for the ADP-specificity of a novel glucokinase from a hyperthermophilic archaeon. Structure. 2001;9:205–214. doi: 10.1016/s0969-2126(01)00577-9. [DOI] [PubMed] [Google Scholar]
- 8.Jetten M S M, Stams A J M, Zehnder A J B. Isolation and characterization of acetyl-coenzyme A synthetase from Methanothrix soehngenii. J Bacteriol. 1989;171:5430–5435. doi: 10.1128/jb.171.10.5430-5435.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kengen S W M, de Bok F A M, van Loo N-D, Dijkema C, Stams A J M, de Vos W M. Evidence for the operation of a novel Embden-Meyerhof pathway that involves ADP-dependent kinases during sugar fermentation by Pyrococcus furiosus. J Biol Chem. 1994;269:17537–17541. [PubMed] [Google Scholar]
- 10.Kengen S W M, Luesink E J, Stams A J M, Zehnder A J B. Purification and characterization of an extremely thermostable beta-glucosidase from the hyperthermophilic archaeon Pyrococcus furiosus. Eur J Biochem. 1993;213:305–312. doi: 10.1111/j.1432-1033.1993.tb17763.x. [DOI] [PubMed] [Google Scholar]
- 11.Kengen S W M, Tuininga J E, de Bok F A M, Stams A J W, de Vos W M. Purification and characterization of a novel ADP-dependent glucokinase from the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1995;270:30453–30457. doi: 10.1074/jbc.270.51.30453. [DOI] [PubMed] [Google Scholar]
- 12.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Venter J C. The complete genome of the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 13.Koga S, Yoshioka I, Sakuraba H, Takahashi M, Sakasegawa S, Shimizu S, Ohshima T. Biochemical characterization, cloning, and sequencing of ADP-dependent (AMP-forming) glucokinase from two hyperthermophilic Archaea, Pyrococcus furiosus and Thermococcus litoralis. J Biochem. 2000;128:1079–1085. doi: 10.1093/oxfordjournals.jbchem.a022836. [DOI] [PubMed] [Google Scholar]
- 14.Labes, A., and P. Schönheit. 2001. Sugar utilization in the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus fulgidus strain 7324: starch degradation to acetate and CO2 via a modified Embden-Meyerhof pathway and acetyl-CoA synthetase (ADP-forming). Arch. Microbiol. 10.1007/s002030100330. [DOI] [PubMed]
- 15.Mukund S, Adams M W W. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase, a novel tungsten-containing enzyme with a potential glycolytic role in the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1995;270:8389–8392. doi: 10.1074/jbc.270.15.8389. [DOI] [PubMed] [Google Scholar]
- 16.Murray P A, Zinder S H. Nitrogen fixation by a methanogenic bacterium. Nature. 1984;312:284–286. doi: 10.1038/312286a0. [DOI] [PubMed] [Google Scholar]
- 17.Pellerin P, Gruson B, Prensier G, Albagnac G, Debiere P. Glycogen in Methanotrix. Arch Microbiol. 1987;146:377–381. [Google Scholar]
- 18.Ronimus R S, Koning J, Morgan H W. Purification and characterization of an ADP-dependent phosphofructokinase from Thermococcus zilligii. Extremophiles. 1999;3:121–129. doi: 10.1007/s007920050107. [DOI] [PubMed] [Google Scholar]
- 19.Ronimus R S, Morgan H W, Ding Y H R. Phosphofructokinase activities within the order Spirochaetales and the characterisation of the pyrophosphate-dependent phosphofructokinase from Spirochaeta thermophila. Arch Microbiol. 1999;172:401–406. doi: 10.1007/s002030050777. [DOI] [PubMed] [Google Scholar]
- 20.Ronimus R S, De Heus E, Morgan H W. Sequencing, expression, characterisation and phylogeny of the ADP-dependent phosphofructokinase from the hyperthermophilic euryarchaeal Thermococcus zilligii. Biochim Biophys Acta. 2001;1527:384–391. doi: 10.1016/s0167-4781(00)00301-8. [DOI] [PubMed] [Google Scholar]
- 21.Ronimus R S, Kawarabayasi Y, Kikuchi H, Morgan H W. Cloning, expression and characterisation of a family B ATP-dependent phosphofructokinase activity from the hyperthermophilic crenarchaeon Aeropyrum pernix. FEMS Microbiol Lett. 2001;202:85–90. doi: 10.1111/j.1574-6968.2001.tb10784.x. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 23.Selig M, Xavier K B, Santos H, Schönheit P. Comparative analysis of Embden-Meyerhof and Entner-Doudoroff glycolytic pathways in hyperthermophilic archaea and the bacterium Thermotoga. Arch Microbiol. 1997;167:217–232. doi: 10.1007/BF03356097. [DOI] [PubMed] [Google Scholar]
- 24.Selkov E, Maltsev N, Olsen G J, Overbeek R, Whitman W B. A reconstruction of the metabolism of Methanococcus jannaschiifrom sequence data. Gene. 1997;197:11–26. doi: 10.1016/s0378-1119(97)00307-7. [DOI] [PubMed] [Google Scholar]
- 25.Siebers B, Klenk H P, Hensel R. PPi-dependent phosphofructokinase from Thermoproteus tenax, an archaeal descendant of an ancient line in phosphofructokinase evolution. J Bacteriol. 1998;180:2137–2143. doi: 10.1128/jb.180.8.2137-2143.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stams A J M, van Dijk J B, Dijkema C, Plugge C M. Growth of syntrophic propionate-oxidizing bacteria with fumurate in the absence of methanogenic bacteria. Appl Environ Microbiol. 1993;59:1114–1119. doi: 10.1128/aem.59.4.1114-1119.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuininga J E, Verhees C H, van der Oost J, Kengen S W M, Stams A J M, de Vos W M. Molecular and biochemical characterization of the ADP-dependent phosphofructokinase from the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem. 1999;274:21023–21028. doi: 10.1074/jbc.274.30.21023. [DOI] [PubMed] [Google Scholar]
- 28.van der Oost J, Schut G, Kengen S W M, Hagen W R, Thomm M, de Vos W M. The ferredoxin-dependent conversion of glyceraldehyde-3-phosphate in the hyperthermophilic archaeon Pyrococcus furiosusrepresents a novel site of glycolytic regulation. J Biol Chem. 1998;273:28149–28154. doi: 10.1074/jbc.273.43.28149. [DOI] [PubMed] [Google Scholar]
- 29.Whitman, W. B., D. R. Boone, and Y. Koga. Bergey's manual of systematic bacteriology, 2nd ed. Springer, New York, N.Y.
- 30.Woese C R, Kandler O, Wheelis M L. Towards a natural system for organisms. Proposal for the domains Archaea, Bacteria and Eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J P, Ladapo J, Whitman W B. Pathway of glycogen metabolism in Methanococcus maripaludis. J Bacteriol. 1994;176:325–332. doi: 10.1128/jb.176.2.325-332.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]