Abstract
The shortening of the 3’ untranslated regions (3’UTRs) due to alternative polyadenylation (APA) has become an important characteristic of cancer. However, the function of APA-induced 3’UTR shortening in gastric cancer (GC) remains unclear. KHDRBS1 (sam68), as an RNA-binding protein (RBP), is significantly upregulated in GC. In this study, we found that the 3’UTR of KHDRBS1 is generally shortened in GC tissues compared to paracancer tissues. Moreover, KHDRBS1 mRNA with a shortened 3’UTR can escape the inhibitory effect of miRNAs, resulting in its increased expression in GC. Overexpression of KHDRBS1, especially KHDRBS1 with a shortened 3’UTR, promotes the growth and metastasis of GC in vivo and in vitro. In conclusion, the experimental results show that shortening of the KHDRBS1 mRNA 3’UTR can mediate the overexpression of KHDRBS1 in GC cells and promote the progression of GC.
Keywords: KHDRBS1, APA, gastric cancer, proliferation, metastasis
Introduction
Gastric cancer is one of the deadliest human malignant tumors in the world [1]. Although advances in modern medicine have improved the survival rate of GC patients, they still suffer from a poor prognosis [2]. Unfortunately, due to the lack of effective biomarkers, it is difficult to make accurate early diagnosis and prognosis prediction for GC patients. In addition, the treatment methods for GC are also relatively limited, mainly including surgery and adjuvant chemoradiotherapy [3]. Therefore, it is urgent to further explore the pathogenesis of GC to identify new diagnostic or prognostic biomarkers, which could promote the development of target therapy and improve the clinical prognosis for GC patients.
RBPs play important roles in the posttranscriptional regulation of RNA splicing, processing, transport, nuclear export, translation, localization, and stability [4-8]. Besides, RBPs are also closely associated with tumors and play a key role in malignant tumor progression and drug resistance [9-11]. Reports showed that the protein expression of KHDRBS1 is elevated in a variety of tumors [12]. Todaro et al. [13] showed that inhibiting KHDRBS1 and Rad51 suppresses breast cancer stem-like cells. Tian et al. [14] confirmed that KHDRBS1 can promote tumorigenesis in lung adenocarcinoma by regulating alternative splicing. Wan et al. [15] showed that KHDRBS1 is required for the growth and survival of nonmelanoma skin cancer. However, the function of KHDRBS1 and the underlying mechanism in GC are not clear.
Previous studies have observed that when T cells transition from a quiescent state to a proliferating state, 3’UTRs are globally shortened, showing that alternative polyadenylation (APA) is very important for T cell proliferation [16,17]. Overexpression or activation of oncogenes promotes the development of tumors, and studies have reported that 3’UTR shortening of oncogenes is sufficient to activate oncogenes to promote cancer transformation. This phenomenon mainly occurs because the shortened 3’UTRs evade the inhibitory effects of miRNAs [18-22], which process also plays important roles in cancers [23,24]. Since approximately half of human genes have multiple APA sites [25], the overexpression or activation of oncogenes caused by APA-mediated 3’UTR shortening may be ubiquitous in malignancies [26]. Clinical data also showed that in human breast, lung, colorectum and kidney cancer, the presence of short 3’UTRs in tumors predicts a poor survival prognosis for patients [26-29]. Begik et al. defined specific subsets of APA events to efficiently classify cancer types including gastric cancer [30]. Lai et al. provided an effective approach for genome-wide APA site profiling and reveals a link between APA modulation and gastric cancer metastasis [31]. Recently, Sun et al. [32] confirmed that CPSF6 is highly expressed in gastric cancer and inhibits the apoptosis of gastric cancer cells. However, the pathological function of APA-mediated 3’UTR shortening in gastric cancer and the underlying mechanism are still unclear.
Herein, we identified two novel short 3’UTR isoforms of KHDRBS1 generated by APA and found that KHDRBS1 with a short 3’UTR could escape the inhibitory effect of miRNAs, causing increased expression of KHDRBS1 in GC. Interestingly, GC cells with overexpression of the shorter 3’UTR KHDRBS1 isoform exhibited increased proliferation and migration capacities. These results show that 3’UTR shortening allows KHDRBS1 mRNA to circumvent miRNA-mediated repression and provides a mechanism for the increased expression of KHDRBS1 in GC.
Materials and methods
Cell lines and culture
Human GC cells (BGC-823, AGS, SNU5, HGC-27, MKN-45 and MGC-803) were acquired from Shanghai Cell Bank (Shanghai, China) or ATCC (the American Type Culture Collection). All of the cell lines were cultured in RPMI-1640 medium (E600028, Sangon Biotech, Shanghai, China) supplemented with 5% FBS (16010-159, Gibco, Thermo Fisher Scientific, USA); and incubated at 37°C in a humidified atmosphere with 5% CO2.
Clinical samples
Sixteen human gastric cancer tissues and adjacent non-tumor tissues were obtained at the first Affiliated Hospital of Anhui Medical University (Hefei, Anhui, China) in 2020. This study was carried out in accordance with the Declaration of Helsinki and approved by the Institutional Review Boards of Anhui Medical University (PJ2016-09-07). All the patients provided written informed consent form.
Immunohistochemistry
Clinical tissue samples were fixed with formalin and embedded in paraffin. The samples were sliced and used for immunohistochemical (IHC) staining. Staining was performed using antibodies against KHDRBS1 (1:100, 10222-1-AP, Proteintech Group, USA) and β-actin (1:200, 66009-1-Ig, Proteintech Group, USA) and the Rapid Immunohistochemical Kit (E-IR-R220, Elabscience, China) according to standard procedures.
Western blotting analysis
Western blotting (WB) analysis was conducted as described previously [33]. The antibodies against KHDRBS1 (1:1,000, 10222-1-AP, Proteintech Group, USA), β-actin (1:2,000, 66009-1-Ig, Proteintech Group, USA) and secondary antibodies (1:5,000, SA00001-2 and RP30009, Proteintech Group, USA) were used.
Quantitative RT-PCR
The mRNA levels of KHDRBS1 and GAPDH were detected using quantitative RT-PCR (RT-qPCR). The primers sequences were as follows: KHDRBS1 (F1: CAGAGGTGCCACTGTGACTC, R1: GAGGTGGAGGCAAAGGTATC; F2: GACTATGGACATGGGGAGG, R2: TGGGTGCTCTCTGTATGCTC; F3: ACACACAAACCTGTTAGTTTC, R3: CTTTACGGGATGCCTCAAATC; F4: CCCAAACTAGGCTACATTTC, R4: AGATTCAACCGCCATGTGC; F5: GAAGAGGTTGATGGTGGTG, R5: CAAGTGAACTTTTCATGGAG; F6: AGGGACACTGCAGCTGAATG, R6: GGCAGACTTACACATGTAGC) and GAPDH (F: CTGCCTCTACTGGCGCTG, R: GGTCAGGTCCACCACTGAC).
Plasmid construction and transfection
The KHDRBS1 ORF was amplified from AGS cDNA by PCR and cloned into the vector pSIN-GFP. In order to ensure expression of the long isoforms, the functional PAS signal motif (AATAAA or ATTAAA) was mutated to ACACAC. For the generation of stably transfected AGS and BGC-823 cells, pSIN-KHDRBS1-CDS, pSIN-KHDRBS1-CDS+3’UTRS, pSIN-KHDRBS1-CDS+3’UTRM, pSIN-KHDRBS1-CDS+3’UTRL or vector pSIN-GFP was transfected into 293T cells with lentivirus plasmids. Lentivirus particles were harvested and used to infect AGS and BGC-823 cells. The stably transfected cells were screened using 0.1 μg/mL puromycin.
Cell functional assays
For the MTT assay, cells were counted and plated in 96-well plates (1000 cells per well). Seventy-two hours later, 0.1 mg MTT was added to each well, and the absorbance at 570 nm was measured.
For the cell colony formation assay, cells were counted and seeded in 6-well plates (1000 cells per well). After 10-15 days, colony formation was examined.
For the Transwell assay, AGS and BGC-823 cells (5-10 × 104) were added to the top chamber with 8-µm pores. For invasion assays, Matrigel was added to chamber. The lower chambers contained medium containing 10% FBS. After 16-24 hours of incubation, the inserts were fixed with methanol and washed by PBS, and then they were stained with 0.1% crystal violet solution for 5-10 min. The cells were imaged with an Olympus IX-70 microscope and counted.
Xenograft assays
Five weeks old male nude mice were purchased from Slack Experimental Animal Company (Shanghai, China). Stably transfected AGS cells (5 × 106) were mixed with Matrigel and subcutaneously injected into the flanks of the animals. The tumor volume was measured every 3 days, and the tumors were harvested after 4-5 weeks.
Statistical analyses
We used SPSS 22.0 or GraphPad Prism 8.02 software to analyze the data. The data from cell functional assays were analyzed using unpaired two-tailed t-tests. P<0.05 was considered statistically significant.
Results
KHDRBS1 protein is elevated in GC
We detected the protein expression level of KHDRBS1 in 19 pairs of paraffin-embedded tissue samples with immunohistochemistry experiments. The results demonstrated that the protein level of KHDRBS1 in GC tissues was significantly increased versus paired paracancer tissues (Figure 1A). We also collected 16 paired fresh GC and paracancer tissue samples and detected the protein expression level of KHDRBS1 by Western blotting analysis. The experimental results showed that the protein level of KHDRBS1 was higher in 12 GC tissues than that in the paired paracancer tissues (Figure 1B). In addition, the protein level of KHDRBS1 was higher in most GC cell lines than that in the GSE-1 normal gastric epithelial cell line (Figure 1C).
Figure 1.
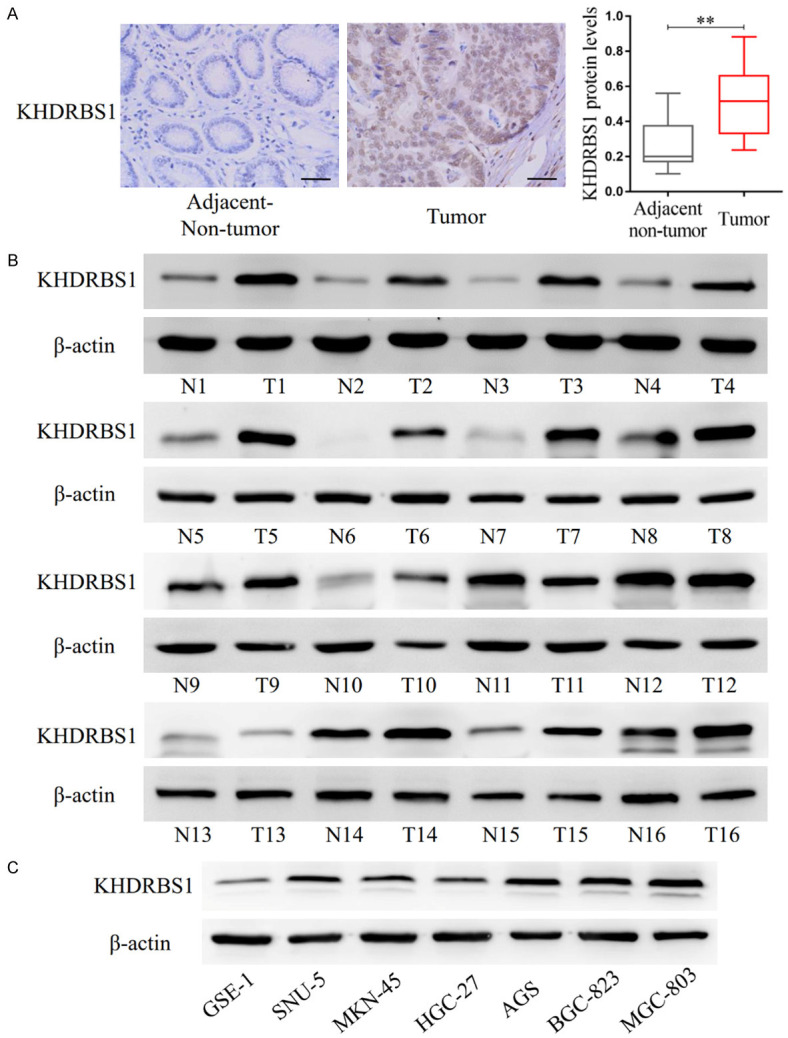
KHDRBS1 expression is increased in gastric cancer tissues. A. The protein expression levels of KHDRBS1 in 16 paired GC and paracancer tissues were detected by IHC staining. Scale bar, 50 μm. B. The protein expression levels of KHDRBS1 in 16 paired fresh GC and paracancer tissues were detected by WB. C. The protein expression levels of KHDRBS1 in normal gastric epithelial cell and cancer cells were detected by WB. (**P<0.01, Student’s t-test).
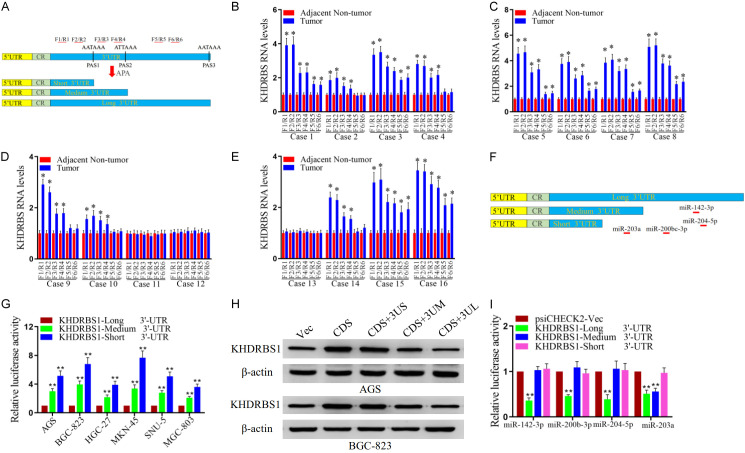
3’UTR shortening promotes the expression of KHDRBS1
By analyzing the characteristics of the KHDRBS1 sequence, it was found that there were two 5’ proximal APA sites (PAS1 and PAS2) in the 3’UTR. This suggested that KHDRBS1 has two short 3’UTR forms. For the upstream and downstream regions of PAS1 and PAS2, we designed 6 pairs of primers (F1/R1, F2/R2, F3/R3, F4/R4, F5/R5 and F6/R6) (Figure 2A). The real-time fluorescent quantitative PCR results are shown in Figure 2B-E. In the five samples (Cases 2, 4, 9, 10, and 14), only the expression of the KHDRBS1 transcript with a shortened 3’UTR was increased in the tumor tissues versus the paracancer tissues, while the expression level of the KHDRBS1 transcript with a long 3’UTR was not obviously different (Figure 2B, 2D and 2E). In the other 8 samples (Cases 1, 3, 5, 6, 7, 8, 15, and 16), the expression levels of KHDRBS1 transcripts containing long or short 3’UTRs were significantly increased, but the expression levels of KHDRBS1 transcripts containing short 3’UTRs were increased more than those of transcripts containing long 3’UTRs (Figure 2B, 2C and 2E). These experimental results indicate that the expression levels of KHDRBS1 transcripts containing short 3’UTRs were higher in GC tissues than in adjacent cancers. After comparing different transcripts, we found that the long 3’UTR contained more potential miRNA sites, which were predicted with TargetScan (Release 8.0) software (Figure 2F) [34]. Therefore, we cloned the 3’UTRs of different lengths into the psiCHECK2 plasmid, and an experiment with a fluorescent reporter vector confirmed that transcripts containing shorter 3’UTRs had higher translation activity (Figure 2G). Furthermore, we cloned KHDRBS1 with different 3’UTRs into gene expression plasmids. The experimental results showed that transcripts containing shorter 3’UTRs had higher expression efficiency than transcripts containing longer 3’UTRs. Invariably, the experimental results showed that the 3’UTR of the KHDRBS1 gene was shortened by APA, which led to its escape of miRNA-mediated inhibition and promoted the expression of KHDRBS1 (Figure 2H). In order to understand the impact of miRNAs including miR-142-3p, miR-200b-3p, miR-204-5p and miR-203a-3p on KHDRBS1, we detected the translation activity of KHDRBS1 3’UTRs using luciferase activity assay. The results showed that the miR-142-3p, miR-200b-3p and miR-204-5p significantly inhibited the translation activity of KHDRBS1 isoform with long 3’UTR, but did not affect the translation activity of the isoform with short or medium 3’UTR (Figure 2I). Furthermore, MiR-203a significantly inhibited the translation activity of the KHDRBS1 isoform with long and medium 3’UTR, but did not affect the translation activity of the isoform with short 3’UTR (Figure 2I). This luciferase activity assay partially explains why the mRNA with short 3’UTR has higher translation activity than the mRNA with long 3’UTR. Recent studies have confirmed that miR-200b-3p, miR-203a and miR-204-5p can directly inhibit the expression of KHDRBS1 through 3’UTR [35-37].
Figure 2.
The length of the 3’UTR affects KHDRBS1 expression. A. Schematic illustration of the KHDRBS1 isoform with a long or short 3’UTR. Green boxes show coding regions (CRs); blue boxes represent 3’UTRs. Positions of PAS1 and PAS2 are indicated by black vertical lines, and the sequences targeted by the primer pairs are indicated by red horizontal lines. B-E. qRT-PCR analyses of the mRNA expression of KHDRBS1 were performed. GAPDH was used as the control. The results are shown as the mean ± SD. F. Positions of the miRNA binding sites are indicated by red horizontal lines. G. The translation activity of the short 3’UTR, medium 3’UTR or long 3’UTR of KHDRBS1 in GC was examined by luciferase reporter assay. H. The protein level of KHDRBS1 was detected by WB. β-actin was used as the control. I. The translation activity of the short 3’UTR, medium 3’UTR or long 3’UTR of KHDRBS1 in GC cells after enforced expression of the specific miRNAs was assessed by luciferase reporter assay. (**P<0.01, Student’s t-test).
3’UTR shortening promotes cell proliferation in GC
To confirm whether ectopic expression of KHDRBS1 promotes the proliferation of GC, we first cloned KHDRBS1 with different 3’UTRs into a plasmid and established BGC-823 and AGS cells that stably overexpressed KHDRBS1. Then, we performed the cell counting, MTT, and colony formation assays. The results demonstrated that overexpression of KHDRBS1 promoted GC cells proliferation (Figure 3A, 3B) and colony formation rates in vitro (Figure 3C, 3D). Besides, we found that KHDRBS1 transcripts with longer or shorter 3’UTRs promoted GC cell proliferation, but KHDRBS1 transcripts with shorter 3’UTRs had a greater impact. In a word, these results indicated that the overexpression of KHDRBS1, especially 3’UTR shortening, can promote cell proliferation in GC.
Figure 3.
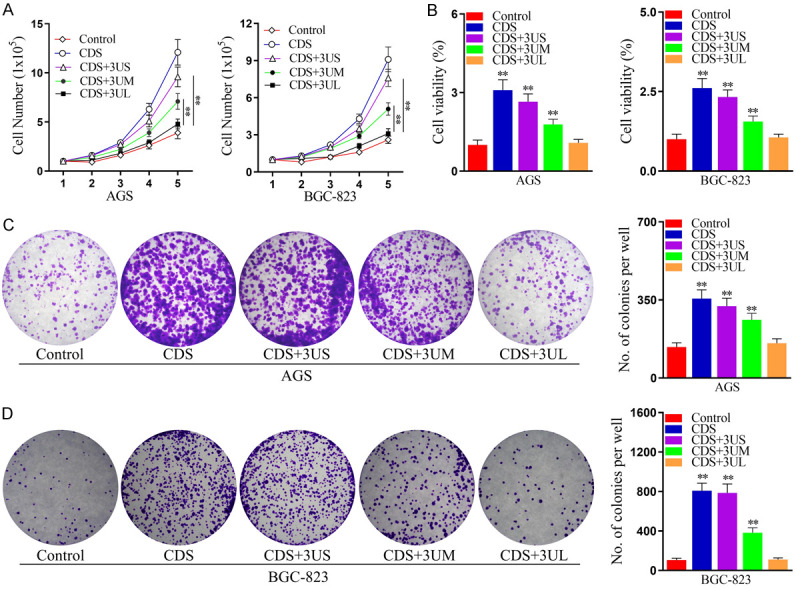
KHDRBS1, especially its shorter 3’UTR transcript, promotes the proliferation of human GC cells. A. Cell number was detected using the cell number assay in AGS and BGC-823 cells overexpressing KHDRBS1 or not (Control). B. Cell viability was measured by the MTT assay in AGS and BGC-823 cells overexpressing KHDRBS1 or not (Control). C, D. The colony formation ability of AGS and BGC-823 cells overexpressing KHDRBS1 or not (Control) was assessed using a colony formation assay. The data are shown as the mean ± SD. (**P<0.01, Student’s t-test).
3’UTR shortening promotes the migration and invasion in GC cells
In order to determine whether ectopic expression of KHDRBS1 promotes the migration and invasion in GC cells, we performed Transwell assays. The results indicated that overexpression of KHDRBS1 promoted GC cell migration (Figure 4A, 4B) and invasion (Figure 4C, 4D) in vitro. Moreover, we found that KHDRBS1 transcripts with longer or shorter 3’UTRs promoted GC cell migration and invasion, but KHDRBS1 transcripts with shorter 3’UTRs had a greater impact. Therefore, above results indicated that the overexpression of KHDRBS1, especially 3’UTR shortening, can promote the migration and invasion in GC cells.
Figure 4.
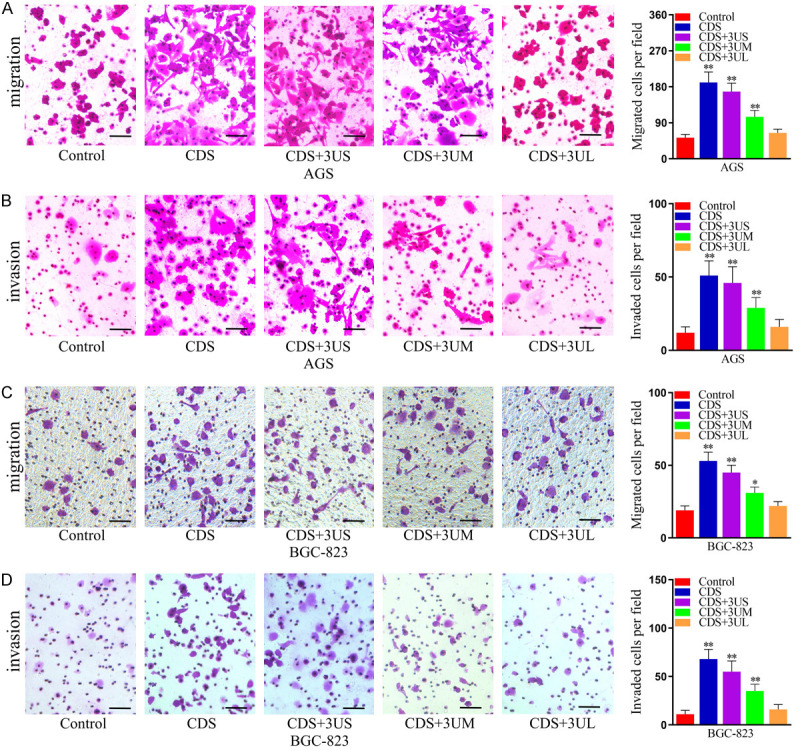
KHDRBS1, especially its shorter 3’UTR transcript, promotes the migration and invasion of human GC cells. A, B. Cell migration was measured by the Transwell migration assay in AGS and BGC-823 cells overexpressing KHDRBS1 or not (Control). C, D. Cell invasion was measured by the Transwell invasion assay in AGS and BGC-823 cells overexpressing KHDRBS1 or not (Control). Scale bars, 100 μm. The data are presented as the mean ± SD. (**P<0.01, Student’s t-test).
KHDRBS1, especially its shorter 3’UTR transcript, promotes GC growth and metastasis in vivo
To clarify the impact of overexpression of KHDRBS1 on GC growth and metastasis in vivo, we injected AGS cells stably transfected with the KHDRBS1 or control plasmid into the flanks of mice to construct subcutaneous tumorigenesis model. The results showed that the overexpression of KHDRBS1 significantly promoted GC growth and the transcript with a shorter 3’UTR had a more obvious impact (Figure 5A, 5B). Furthermore, we injected AGS cells into the tail vein to explore whether the abnormal expression of KHDRBS1 affects the metastatic ability of GC cells in vivo. The results showed that KHDRBS1 transcripts with 3’UTRs of diverse lengths could promote cell lung metastasis, but KHDRBS1 isoform with a shorter 3’UTR had a more significant impact (Figure 5C). Therefore, the above results confirmed that the overexpression of KHDRBS1, especially its transcript with a shorter 3’UTR, promotes cell growth and lung metastasis in GC.
Figure 5.
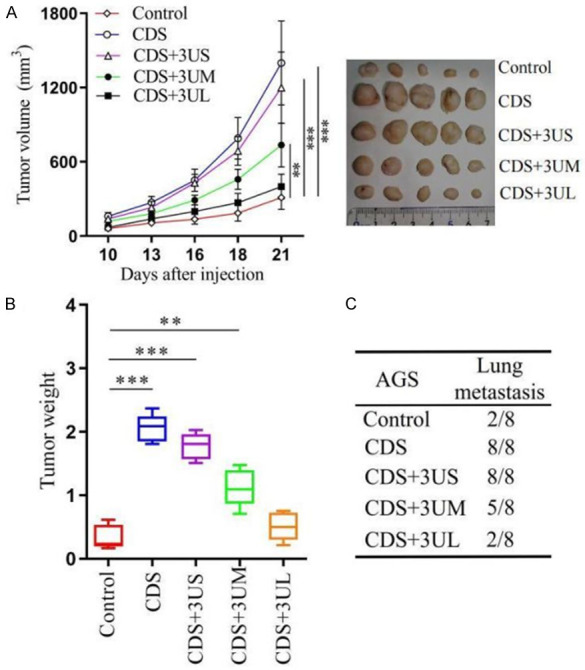
KHDRBS1 promotes the growth and metastasis of human GC cells in vivo. A. Xenograft assays: AGS cells were mixed with Matrigel and subcutaneously injected into the flanks of the mice. B. The weight of the AGS tumors at the end point is shown. C. Lung metastases model: the number of mice with lung metastases was determined one month after the tail vein injection. (**P<0.01, Student’s t-test).
Discussion
In eukaryotic cells, the polyadenylation (polyA) tails are ubiquitous in mRNAs. The 3’UTR of a particular gene has multiple different polyA sites, which is called alternative polyadenylation (APA) of the 3’UTR. At present, approximately half of human genes have been found to have APA sites [38]. In recent years, it has been discovered that APA regulatory factors, such as NUDT21, CSTF2, CPSF6 and PCF11, are closely related to tumor progression. The expression level of NUDT21 is significantly reduced in glioma and liver cancer, and NUDT21 can inhibit the progression of tumors by regulating the APA process of downstream genes [20,39,40]. CSTF2 promotes 3’UTR shortening of the downstream gene RAC1 through APA and promotes bladder cancer progression [41]. The expression of CPSF6 is elevated in liver cancer, and its overexpression promotes 3’UTR shortening of the downstream gene NQO1 and regulates the abnormal metabolism of liver cancer cells [42]. Low expression of PCF11 in neuroblastoma predicts a better survival prognosis for patients [43].
Increasing evidences have demonstrated that 3’UTR shortening is a common phenomenon in cancer and is closely associated with the poor prognosis of many malignancies [18,44,45]. 3’UTRs contain binding sites for a variety of cis-regulatory elements, such as miRNAs. The shortening of the 3’UTR may cause the loss of these regulatory elements and affect the expression level of genes, which in turn leads to many biological dysfunction [46]. The shortening of the IMP1 3’UTR due to APA resulted in the loss of multiple miRNA binding sites, including the let-7 binding site. Experimental results show that IMP1 transcripts with a shorter 3’UTR have a higher ability to promote cell transformation in fibroblasts [18]. IMP1 isoform with a shorter 3’UTR promotes colorectal cancer (CRC) metastasis [47]. The shorter 3’UTR of FNDC3B allows it to escape the inhibitory effects of miRNAs and leads to an increase in the expression of FNDC3B in nasopharyngeal carcinoma. In particular, the overexpression of FNDC3B transcripts with shorter 3’UTR promoted the development of nasopharyngeal carcinoma [48].
KHDRBS1 is an RNA-binding protein and plays an important role in the progression of various cancers [12]. Early studies have shown that acetylation may positively regulate the binding of KHDRBS1 to RNA substrates and enhance its function in promoting tumor cell proliferation [49]. So far, many studies have found the impacts of KHDRBS1 on cancers. A study demonstrated that knockdown of KHDRBS1 in prostate cancer can delay prostate cancer cell cycle progression and inhibit cell proliferation [50]. Another study found that KHDRBS1 are significantly upregulated in cervical cancer and the upregulation of KHDRBS1 indicates the poorer prognosis of patients. Moreover, KHDRBS1 promotes the migration and invasion of cervical cancer cells through the Akt/GSK-3β/Snail pathway [51]. Besides, elevated expression of KHDRBS1 and localization of the protein to the nucleus of CRC cells both predict disease progression and poor patient prognosis [52]. KHDRBS1 plays a key role in the progression of colon tumors by regulating NF-κB [53]. High expression of KHDRBS1 in lung cancer predicts poor survival prognosis, drives the splicing of the hnRNPA1-dependent oncogene PKM, and then promotes the progression of lung cancer [14,54]. KHDRBS1 inhibits the apoptosis of oral tongue squamous cell carcinoma cells through the antiapoptotic proteins caspase-9, caspase-3 and PARP [55]. KHDRBS1 is crucial for the growth and survival of nonmelanoma skin cancer by regulating DNA damage responses [15]. The upregulation of KHDRBS1 is associated with the poor prognosis in GC patients, and knockdown of KHDRBS1 can inhibit the ability of proliferation and metastasis of GC cells [56]. However, the function of KHDRBS1 in GC and the related mechanisms are still unclear.
In this study, the results showed that the level of KHDRBS1 expression is important for the progression of gastric cancer. APA-regulated KHDRBS1 3’UTR shortening could lead to its escape of miRNA-mediated gene repression and caused its high expression in GC. KHDRBS1, especially its isoform with a shorter 3’UTR, promoted GC progression. These results suggested that targeting KHDRBS1 may be useful for reducing tumor progression.
Acknowledgements
This work was supported by Postgraduate Innovation Special Foundation of Jiangxi Province (YC2020-B054).
Disclosure of conflict of interest
None.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–648. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 3.Joshi SS, Badgwell BD. Current treatment and recent progress in gastric cancer. CA Cancer J Clin. 2021;71:264–279. doi: 10.3322/caac.21657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosker KE, Fenstermacher SJ, Pazyra-Murphy MF, Elliott HL, Segal RA. The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability. Nat Neurosci. 2016;19:690–696. doi: 10.1038/nn.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murakawa Y, Hinz M, Mothes J, Schuetz A, Uhl M, Wyler E, Yasuda T, Mastrobuoni G, Friedel CC, Dolken L, Kempa S, Schmidt-Supprian M, Bluthgen N, Backofen R, Heinemann U, Wolf J, Scheidereit C, Landthaler M. RC3H1 post-transcriptionally regulates A20 mRNA and modulates the activity of the IKK/NF-kappaB pathway. Nat Commun. 2015;6:7367. doi: 10.1038/ncomms8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messina V, Meikar O, Paronetto MP, Calabretta S, Geremia R, Kotaja N, Sette C. The RNA binding protein SAM68 transiently localizes in the chromatoid body of male germ cells and influences expression of select microRNAs. PLoS One. 2012;7:e39729. doi: 10.1371/journal.pone.0039729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Jimenez F, Sanchez-Margalet V. Role of Sam68 in post-transcriptional gene regulation. Int J Mol Sci. 2013;14:23402–23419. doi: 10.3390/ijms141223402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pereira B, Billaud M, Almeida R. RNA-binding proteins in cancer: old players and new actors. Trends Cancer. 2017;3:506–528. doi: 10.1016/j.trecan.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Gebauer F, Schwarzl T, Valcarcel J, Hentze MW. RNA-binding proteins in human genetic disease. Nat Rev Genet. 2021;22:185–198. doi: 10.1038/s41576-020-00302-y. [DOI] [PubMed] [Google Scholar]
- 11.Qin H, Ni H, Liu Y, Yuan Y, Xi T, Li X, Zheng L. RNA-binding proteins in tumor progression. J Hematol Oncol. 2020;13:90. doi: 10.1186/s13045-020-00927-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bielli P, Busa R, Paronetto MP, Sette C. The RNA-binding protein Sam68 is a multifunctional player in human cancer. Endocr Relat Cancer. 2011;18:R91–R102. doi: 10.1530/ERC-11-0041. [DOI] [PubMed] [Google Scholar]
- 13.Turdo A, Gaggianesi M, Di Franco S, Veschi V, D’Accardo C, Porcelli G, Lo Iacono M, Pillitteri I, Verona F, Militello G, Zippo A, Poli V, Fagnocchi L, Beyes S, Stella S, Lattanzio R, Faldetta N, Lentini VL, Porcasi R, Pistone G, Bongiorno MR, Stassi G, De Maria R, Todaro M. Effective targeting of breast cancer stem cells by combined inhibition of Sam68 and Rad51. Oncogene. 2022;41:2196–2209. doi: 10.1038/s41388-022-02239-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu S, Chen W, Wang J, Qi L, Pan H, Feng Z, Tian D. SAM68 promotes tumorigenesis in lung adenocarcinoma by regulating metabolic conversion via PKM alternative splicing. Theranostics. 2021;11:3359–3375. doi: 10.7150/thno.51360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu K, Sun X, Xia X, Hobbs RP, Guo Y, Coulombe PA, Wan F. Sam68 is required for the growth and survival of nonmelanoma skin cancer. Cancer Med. 2019;8:6106–6113. doi: 10.1002/cam4.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3’ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elkon R, Drost J, van Haaften G, Jenal M, Schrier M, Oude Vrielink JA, Agami R. E2F mediates enhanced alternative polyadenylation in proliferation. Genome Biol. 2012;13:R59. doi: 10.1186/gb-2012-13-7-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayr C, Bartel DP. Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller S, Rycak L, Afonso-Grunz F, Winter P, Zawada AM, Damrath E, Scheider J, Schmah J, Koch I, Kahl G, Rotter B. APADB: a database for alternative polyadenylation and microRNA regulation events. Database (Oxford) 2014;2014:bau076. doi: 10.1093/database/bau076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun M, Ding J, Li D, Yang G, Cheng Z, Zhu Q. NUDT21 regulates 3’-UTR length and microRNA-mediated gene silencing in hepatocellular carcinoma. Cancer Lett. 2017;410:158–168. doi: 10.1016/j.canlet.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 21.Masamha CP, Xia Z, Peart N, Collum S, Li W, Wagner EJ, Shyu AB. CFIm25 regulates glutaminase alternative terminal exon definition to modulate miR-23 function. RNA. 2016;22:830–838. doi: 10.1261/rna.055939.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akman BH, Can T, Erson-Bensan AE. Estrogen-induced upregulation and 3’-UTR shortening of CDC6. Nucleic Acids Res. 2012;40:10679–10688. doi: 10.1093/nar/gks855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao F, Tian X, Zhang Z. The PCAT3/PCAT9-miR-203-SNAI2 axis functions as a key mediator for prostate tumor growth and progression. Oncotarget. 2018;9:12212–12225. doi: 10.18632/oncotarget.24198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian X, Zhang Z. miR-191/DAB2 axis regulates the tumorigenicity of estrogen receptor-positive breast cancer. IUBMB Life. 2018;70:71–80. doi: 10.1002/iub.1705. [DOI] [PubMed] [Google Scholar]
- 25.Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masamha CP, Wagner EJ. The contribution of alternative polyadenylation to the cancer phenotype. Carcinogenesis. 2018;39:2–10. doi: 10.1093/carcin/bgx096. [DOI] [PubMed] [Google Scholar]
- 27.Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, Li W. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3’-UTR landscape across seven tumour types. Nat Commun. 2014;5:5274. doi: 10.1038/ncomms6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gruber AJ, Zavolan M. Alternative cleavage and polyadenylation in health and disease. Nat Rev Genet. 2019;20:599–614. doi: 10.1038/s41576-019-0145-z. [DOI] [PubMed] [Google Scholar]
- 29.Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol. 2017;18:18–30. doi: 10.1038/nrm.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Begik O, Oyken M, Cinkilli Alican T, Can T, Erson-Bensan AE. Alternative polyadenylation patterns for novel gene discovery and classification in cancer. Neoplasia. 2017;19:574–582. doi: 10.1016/j.neo.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai DP, Tan S, Kang YN, Wu J, Ooi HS, Chen J, Shen TT, Qi Y, Zhang X, Guo Y, Zhu T, Liu B, Shao Z, Zhao X. Genome-wide profiling of polyadenylation sites reveals a link between selective polyadenylation and cancer metastasis. Hum Mol Genet. 2015;24:3410–3417. doi: 10.1093/hmg/ddv089. [DOI] [PubMed] [Google Scholar]
- 32.Shi X, Ding K, Zhao Q, Li P, Kang Y, Tan S, Sun J. Suppression of CPSF6 enhances apoptosis through alternative polyadenylation-mediated shortening of the VHL 3’UTR in gastric cancer cells. Front Genet. 2021;12:707644. doi: 10.3389/fgene.2021.707644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C, Zheng Q, Pan S, Chen W, Huang J, Cao Y, Tu Y, Li Z, Yu C, Jie Z. The RNA-binding protein NELFE promotes gastric cancer growth and metastasis through E2F2. Front Oncol. 2021;11:677111. doi: 10.3389/fonc.2021.677111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGeary SE, Lin KS, Shi CY, Pham TM, Bisaria N, Kelley GM, Bartel DP. The biochemical basis of microRNA targeting efficacy. Science. 2019;366:eaav1741. doi: 10.1126/science.aav1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng C, Lu T, Fan Z. miR-200b-3p alleviates TNF-alpha-induced apoptosis and inflammation of intestinal epithelial cells and ulcerative colitis progression in rats via negatively regulating KHDRBS1. Cytotechnology. 2021;73:727–743. doi: 10.1007/s10616-021-00490-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao D, Tian Y, Li P, Wang L, Xiao A, Zhang M, Shi T. MicroRNA-203 inhibits the malignant progression of neuroblastoma by targeting Sam68. Mol Med Rep. 2015;12:5554–5560. doi: 10.3892/mmr.2015.4013. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Tian H, Yuan J, Wu H, Wu J, Zhu X. CONSORT: Sam68 is directly regulated by miR-204 and promotes the self-renewal potential of breast cancer cells by activating the Wnt/Beta-catenin signaling pathway. Medicine (Baltimore) 2015;94:e2228. doi: 10.1097/MD.0000000000002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, Li W, Wagner EJ. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510:412–416. doi: 10.1038/nature13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan S, Li H, Zhang W, Shao Y, Liu Y, Guan H, Wu J, Kang Y, Zhao J, Yu Q, Gu Y, Ding K, Zhang M, Qian W, Zhu Y, Cai H, Chen C, Lobie PE, Zhao X, Sun J, Zhu T. NUDT21 negatively regulates PSMB2 and CXXC5 by alternative polyadenylation and contributes to hepatocellular carcinoma suppression. Oncogene. 2018;37:4887–4900. doi: 10.1038/s41388-018-0280-6. [DOI] [PubMed] [Google Scholar]
- 41.Chen X, Zhang JX, Luo JH, Wu S, Yuan GJ, Ma NF, Feng Y, Cai MY, Chen RX, Lu J, Jiang LJ, Chen JW, Jin XH, Liu HL, Chen W, Guan XY, Kang TB, Zhou FJ, Xie D. CSTF2-induced shortening of the RAC1 3’UTR promotes the pathogenesis of urothelial carcinoma of the bladder. Cancer Res. 2018;78:5848–5862. doi: 10.1158/0008-5472.CAN-18-0822. [DOI] [PubMed] [Google Scholar]
- 42.Tan S, Zhang M, Shi X, Ding K, Zhao Q, Guo Q, Wang H, Wu Z, Kang Y, Zhu T, Sun J, Zhao X. CPSF6 links alternative polyadenylation to metabolism adaption in hepatocellular carcinoma progression. J Exp Clin Cancer Res. 2021;40:85. doi: 10.1186/s13046-021-01884-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogorodnikov A, Levin M, Tattikota S, Tokalov S, Hoque M, Scherzinger D, Marini F, Poetsch A, Binder H, Macher-Goppinger S, Probst HC, Tian B, Schaefer M, Lackner KJ, Westermann F, Danckwardt S. Transcriptome 3’ end organization by PCF11 links alternative polyadenylation to formation and neuronal differentiation of neuroblastoma. Nat Commun. 2018;9:5331. doi: 10.1038/s41467-018-07580-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin Y, Li Z, Ozsolak F, Kim SW, Arango-Argoty G, Liu TT, Tenenbaum SA, Bailey T, Monaghan AP, Milos PM, John B. An in-depth map of polyadenylation sites in cancer. Nucleic Acids Res. 2012;40:8460–8471. doi: 10.1093/nar/gks637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lembo A, Di Cunto F, Provero P. Shortening of 3’UTRs correlates with poor prognosis in breast and lung cancer. PLoS One. 2012;7:e31129. doi: 10.1371/journal.pone.0031129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun Y, Fu Y, Li Y, Xu A. Genome-wide alternative polyadenylation in animals: insights from high-throughput technologies. J Mol Cell Biol. 2012;4:352–361. doi: 10.1093/jmcb/mjs041. [DOI] [PubMed] [Google Scholar]
- 47.Andres SF, Williams KN, Plesset JB, Headd JJ, Mizuno R, Chatterji P, Lento AA, Klein-Szanto AJ, Mick R, Hamilton KE, Rustgi AK. IMP1 3’UTR shortening enhances metastatic burden in colorectal cancer. Carcinogenesis. 2019;40:569–579. doi: 10.1093/carcin/bgy153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li YQ, Chen Y, Xu YF, He QM, Yang XJ, Li YQ, Hong XH, Huang SY, Tang LL, Liu N. FNDC3B 3’-UTR shortening escapes from microRNA-mediated gene repression and promotes nasopharyngeal carcinoma progression. Cancer Sci. 2020;111:1991–2003. doi: 10.1111/cas.14394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Busa R, Paronetto MP, Farini D, Pierantozzi E, Botti F, Angelini DF, Attisani F, Vespasiani G, Sette C. The RNA-binding protein Sam68 contributes to proliferation and survival of human prostate cancer cells. Oncogene. 2007;26:4372–4382. doi: 10.1038/sj.onc.1210224. [DOI] [PubMed] [Google Scholar]
- 50.Babic I, Jakymiw A, Fujita DJ. The RNA binding protein Sam68 is acetylated in tumor cell lines, and its acetylation correlates with enhanced RNA binding activity. Oncogene. 2004;23:3781–3789. doi: 10.1038/sj.onc.1207484. [DOI] [PubMed] [Google Scholar]
- 51.Li Z, Yu CP, Zhong Y, Liu TJ, Huang QD, Zhao XH, Huang H, Tu H, Jiang S, Zhang Y, Liu JH, Song LB. Sam68 expression and cytoplasmic localization is correlated with lymph node metastasis as well as prognosis in patients with early-stage cervical cancer. Ann Oncol. 2012;23:638–646. doi: 10.1093/annonc/mdr290. [DOI] [PubMed] [Google Scholar]
- 52.Liao WT, Liu JL, Wang ZG, Cui YM, Shi L, Li TT, Zhao XH, Chen XT, Ding YQ, Song LB. High expression level and nuclear localization of Sam68 are associated with progression and poor prognosis in colorectal cancer. BMC Gastroenterol. 2013;13:126. doi: 10.1186/1471-230X-13-126. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Fu K, Sun X, Wier EM, Hodgson A, Liu Y, Sears CL, Wan F. Sam68/KHDRBS1 is critical for colon tumorigenesis by regulating genotoxic stress-induced NF-kappaB activation. Elife. 2016;5:e15018. doi: 10.7554/eLife.15018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang Z, Xu Y, Sun N, Zhang M, Xie J, Jiang Z. High Sam68 expression predicts poor prognosis in non-small cell lung cancer. Clin Transl Oncol. 2014;16:886–891. doi: 10.1007/s12094-014-1160-3. [DOI] [PubMed] [Google Scholar]
- 55.Chen S, Li H, Zhuang S, Zhang J, Gao F, Wang X, Chen W, Song M. Sam68 reduces cisplatin-induced apoptosis in tongue carcinoma. J Exp Clin Cancer Res. 2016;35:123. doi: 10.1186/s13046-016-0390-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xiao J, Wang Q, Yang Q, Wang H, Qiang F, He S, Cai J, Yang L, Wang Y. Clinical significance and effect of Sam68 expression in gastric cancer. Oncol Lett. 2018;15:4745–4752. doi: 10.3892/ol.2018.7930. [DOI] [PMC free article] [PubMed] [Google Scholar]