Abstract
Hydrogen peroxide is generated during aerobic metabolism and is capable of damaging critical biomolecules. However, mutants of Escherichia coli that are devoid of catalase typically exhibit no adverse phenotypes during growth in aerobic media. We discovered that catalase mutants retain the ability to rapidly scavenge H2O2 whether it is formed internally or provided exogenously. Analysis of candidate genes revealed that the residual activity is due to alkyl hydroperoxide reductase (Ahp). Mutants that lack both Ahp and catalase could not scavenge H2O2. These mutants excreted substantial amounts of H2O2, and they grew poorly in air. Ahp is kinetically a more efficient scavenger of trace H2O2 than is catalase and therefore is likely to be the primary scavenger of endogenous H2O2. Accordingly, mutants that lack Ahp accumulated sufficient hydrogen peroxide to induce the OxyR regulon, whereas the OxyR regulon remained off in catalase mutants. Catalase still has an important role in wild-type cells, because the activity of Ahp is saturated at a low (10−5 M) concentration of H2O2. In contrast, catalase has a high Km, and it therefore becomes the predominant scavenger when H2O2 concentrations are high. This arrangement is reasonable because the cell cannot provide enough NADH for Ahp to rapidly degrade large amounts of H2O2. In sum, E. coli does indeed generate substantial H2O2, but damage is averted by the scavenging activity of Ahp.
Virtually all aerobic organisms contain enzymes that convert superoxide and hydrogen peroxide to innocuous products. The ubiquity of these scavenging enzymes suggests that exposure to O2− and H2O2 is an inevitable part of the aerobic lifestyle and that these species can damage cells. It follows that organisms that cannot scavenge them will fare poorly in an aerobic habitat, and in 1971 McCord et al. proposed that some obligate anaerobes may be unable to grow in air at least in part because they lack sufficient levels of scavenging enzymes (17). This reasoning also suggested that aerobes would be much less oxygen tolerant if they lacked superoxide dismutase (SOD) and catalase. In 1985 this prediction was partly affirmed by studies of sodA sodB mutants of Escherichia coli (3). These strains suffered elevated rates of DNA damage, could not catabolize nonfermentable carbon sources, and did not grow at all without extensive amino acid supplements. However, contrary to expectation, mutants that lacked catalase grew as well as their wild-type parents and exhibited no increase in mutation rate (14, 29).
The fitness of the catalase mutants did not reflect invulnerability to H2O2. Low concentrations of exogenous H2O2 (ca. 30 μM) are sufficient to inhibit the growth of E. coli. Although the growth-blocking cell lesions have not yet been identified, H2O2 can oxidize thiols, which may inactivate enzymes that have active-site sulfhydryl residues. Methionine sulfoxide adducts (21) and protein carbonyls (5) may also be products of enzyme oxidation. In addition, H2O2 can inactivate the exposed [4Fe-4S] clusters of aconitase B and fumarase B (S. M. Varghese, S. Korshunov, and J. A. Imlay, unpublished data). Finally, reactions between H2O2 and intracellular iron generate hydroxyl radicals, which in turn attack DNA. Micromolar concentrations of H2O2 are therefore mutagenic.
Thus, the robust performance of catalase mutants seemed to imply that these amounts of H2O2 are not normally generated during aerobic growth. There are presently no data that firmly establish the amount of H2O2 that is formed as a by-product of metabolism. No enzymes in E. coli, other than SOD, generate H2O2 as a deliberate, stoichiometric product. However, in vitro studies have shown that H2O2 can be formed by the adventitious oxidation of redox enzymes by molecular oxygen.
The main sources of H2O2 inside the cell are probably flavoenzymes, both because they are abundant and because flavins are amenable to the univalent electron transfer reaction that initiates the production of superoxide and H2O2 (6, 18). The rates at which some flavoenzymes generate these species have been measured in vitro, and extrapolations suggest that they may form about 10 μM H2O2 s−1 in vivo (18). Gonzalez-Flecha and Demple reported evidence of endogenous H2O2 production by E. coli (7). If these rates are correct, then the lack of phenotype of catalase mutants seems difficult to explain.
In this study, we discovered that mutants which are devoid of catalase still scavenge physiological concentrations of H2O2 as rapidly as do wild-type cells. By identifying and eliminating the remaining scavenging enzyme, alkyl hydroperoxide reductase, we were able to directly quantify endogenous H2O2 production and demonstrate that it is enough to poison scavengerless cells.
MATERIALS AND METHODS
Chemicals, enzymes, and media.
Cumene hydroperoxide, 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB), horseradish peroxidase (type II), hydrogen peroxide, o-dianisidine, o-nitrophenyl-β-d-galactopyranoside, plumbagin, and potassium cyanide were purchased from Sigma. Coomassie protein reagent was obtained from Pierce. Bovine liver catalase (20 mg/ml) was from Boehringer Mannheim, and Amplex red (AR) was from Molecular Probes. Dimethyl sulfoxide (DMSO) was purchased from Fisher. Water for buffers was purified with a Labconco Water Pro PS system using deionized water as the feedstock.
Luria broth (LB) contained (per liter) 10 g of Bactotryptone (Difco), 5 g of yeast extract (Difco), and 10 g of sodium chloride. To prevent the photochemical formation of hydrogen peroxide, LB medium was shielded from light and used within 24 h of its preparation. Glucose minimal medium consisted of minimal A salts (19) with 1 mM MgSO4 · 7H2O, 5 mg of thiamine, and 2 g of glucose per liter. However, to minimize the chemical production of hydrogen peroxide, the glucose medium used in some experiments (as noted) was prepared immediately before use and contained only 0.5 g of glucose per liter. Tetracycline, kanamycin, spectinomycin, and chloramphenicol were used at 12, 40, 120, and 20 μg/ml, respectively.
Growth conditions and strains.
Cultures were routinely grown at 37°C. Aerobic cultures were grown in shaking flasks; anaerobic cultures were grown in a Coy chamber (Coy Laboratory Products, Inc.) under 85% N2–10% H2–5% CO2. The optical densities (OD) of all cultures were measured at 600 nm.
The strains used in this study were derived from E. coli K-12 and are listed in Table 1; isogenic strains were used in all experiments. Mutations were introduced into strains via P1 transduction (19). To avoid the outgrowth of suppressed strains, the katG, katE, oxyR, and Tn10-linked ahp mutations were transduced and selected under anaerobic conditions. The presence of katG null mutations was confirmed by an o-dianisidine assay for hydroperoxidase I (HPI) activity (below). katE mutants failed to form bubbles on plates when colonies were overlaid with a drop of 30% H2O2. Mutants lacking oxyR were identified by their inability to induce HPI when anaerobic, exponentially growing cultures at 0.1 OD in LB medium were exposed to 60 μM H2O2 for 45 min. Mutations in gshA were confirmed by measurements of intracellular thiol levels using DTNB (13).
TABLE 1.
E. coli strains
| Strain | Genotype | Source or reference |
|---|---|---|
| UM1 | katE katG14 lacY rspL thi-1 | 14 |
| UM120 | katE12::Tn10 hfrH thi-1 | Peter Loewen |
| UM202 | katG17::Tn10 hfrH thi-1 | Peter Loewen |
| GK100 | ΔcydAB::cam Δ(cyoABCDE)456::kan | 12 |
| KM38 | As UM1 plus ΔcydAB::cam | P1(GK100) × UM1 |
| KM39 | As UMI plus Δ(cyoABCDE)456::kan | P1(GK100) × UM1 |
| SK2255 | zbe-279::Tn10 thyA6 rps120 decC1 | E. coli Genetic Stock Center |
| N9716 | As GC4468 plus ΔoxyR::spec | Gisela Storz |
| AS430 | ΔoxyR::spec ΔlacU169 rpsL | P1(N9716) × GC4468 |
| AB1157 | F−thr-1 leuB6 proA2 his-4 thi-1 argE2 lacY1 galK2 rspL supE44 ara-14 xyl-15 mtl-1 tsx-33 | 10 |
| MG1655 | F− wild-type | E. coli Genetic Stock Center |
| JI360 | As MG1655 plus katE12::Tn10 | P1(UM120) × MG1655 |
| JI361 | As MG1655 plus katG17::Tn10 | P1(UM202) × MG1655 |
| JI362 | As JI360 plus Δ(katE12::Tn10)1 (Tets) | Tets derivative of JI360 |
| JI364 | As JI361 plus Δ(katG17::Tn10)1 (Tets) | Tets derivative of JI361 |
| JI367 | As JI364 plus katE12::Tn10 | P1(UM120) × JI364 |
| JI370 | As MG1655 plus ΔahpCF′ kan::′ahpF | P1(MC4100ΔahpCF) × MG1655 |
| JI372 | As JI362 plus ΔahpCF′ kan::′ahpF | P1(MC4100ΔahpCF) × JI362 |
| JI374 | As JI364 plus ΔahpCF′ kan::′ahpF | P1(MC4100ΔahpCF) × JI364 |
| JI377 | As JI367 plus ΔahpCF′ kan::′ahpF | P1(MC4100ΔahpCF) × JI367 |
| MC4100 | araD139 Δ(argF-lac)169 λ−flhD5301 fruA25 relA1 rpsL150 rbsR22 deoC1 | E. coli Genetic Stock Center |
| MC4100ΔahpCF | As MC4100 plus ΔahpCF′ kan::′ahpF | Gisela Storz |
| GS022 | As MC4100 plus λRS45 Φ(katG::lacZ) | Gisela Storz |
| LC70 | As GS022 plus ΔahpCF′ kan::′ahpF | P1(MC4100ΔahpCF) × GS022 |
| LC74 | As LC70 plus ΔoxyR::spec | P1(AS430) × LC70 |
| LC80 | As GS022 plus katG17::Tn10 | P1(UM202) × GS022 |
| HDO3 | As GS022 plus katE12::Tn10 | P1(UM120) × GS022 |
| LC82 | As LC80 plus Δ(katG17::Tn10)2 (Tets) | Tets derivative of LC80 |
| LC84 | As LC82 plus katE12::Tn10 | P1(UM120) × LC82 |
| LC87 | As MC4100ΔahpCF plus zbe-279::Tn10 | P1(SK2255) × MC4100ΔahpCF |
| LC89 | As GS022 plus ΔahpCF′ kan::′ahpF zbe-279::Tn10 | P1(LC87) × GS022 |
Alkyl hydroperoxide reductase cannot easily be assayed in extracts, because its subunits dissociate (11, 26). However, ahpCF mutants formed a large zone of growth inhibition when 10 μl of 3% cumene hydroperoxide was spotted onto a filter disk and laid onto a mutant-seeded plate (32). The excision of Tn10 elements containing a tetracycline resistance marker was achieved at 42°C by standard methods (16).
H2O2 detection.
In the presence of H2O2, horseradish peroxidase (HRP) oxidizes AR to the fluorescent product resorufin. One milligram of AR was dissolved in 0.78 ml of DMSO, and 0.75 ml of this solution was then diluted into 18 ml of 50 mM potassium phosphate (KPi, pH 7.8) to generate a 200 μM stock solution. This solution was shielded from light. HRP was dissolved in 50 mM KPi (pH 7.8) to 0.02 mg/ml. To measure H2O2, 0.45 ml of sample was mixed with 0.25 ml of AR and 0.25 ml of HRP. Fluorescence was then measured in a Shimadzu RF Mini-150 fluorometer and converted to H2O2 concentration using a curve obtained from standard samples. Note that a small amount of H2O2 is generated by the dye/HRP detection system itself; this amount was accounted for by the standard curve.
H2O2 scavenging by whole cells.
Cultures were grown aerobically for at least four generations to 0.1 to 0.3 OD. Cells were pelleted in a microcentrifuge, washed twice, and resuspended in room temperature phosphate-buffered saline (PBS) at an OD of 0.1. H2O2 was added to the appropriate final concentration (see figure legends). At intervals, 0.45-ml aliquots were removed, diluted when necessary, and assayed immediately for H2O2 content by the AR/HRP method.
LB medium was used for most experiments. However, in experiments designed to measure the scavenging of trace H2O2, cultures were grown in minimal A glucose medium containing 0.5 mM each of the 20 l-amino acids. Cells were then washed and resuspended into fresh medium containing only 0.02% glucose and 0.05 mM amino acids, so that metabolism was active and could provide Ahp with reductants, yet the amount of H2O2 generated by the medium was minimal. When cyanide was included in the medium, fluorescence development was permitted to proceed for 10 min before a final reading was made, since the carryover of cyanide inhibits the activity of HRP.
Measurement of H2O2 accumulation in cell cultures.
To detect H2O2 formation by drug-treated cells, we exposed log-phase cultures (0.2 OD) in LB medium to 300 μM plumbagin, an amount that is sufficient to consume 8 μM oxygen per min in a cyanide-resistant (nonrespiratory) fashion. After 16 min, cells were removed by centrifugation, and the residual H2O2 in the medium was assayed by the HRP/o-dianisidine assay (18).
H2O2 can be detected with greater sensitivity in defined medium using the AR/HRP assay. Cells were grown anaerobically overnight in minimal A medium containing 0.2% glucose, diluted to 0.01 OD, and grown anaerobically for four generations. These log-phase cells were then subcultured to 0.05 OD in fresh aerobic minimal A medium containing 0.05% glucose. At 0.05 and 0.1 OD, the culture medium was assayed for H2O2. Hydrogen peroxide levels were also determined in sterile medium that was incubated at 37°C for an equivalent time. The low glucose concentration was used in order to minimize H2O2 production by salt-catalyzed glucose autooxidation (below).
H2O2 production rates.
In order to monitor continuously the intracellular formation of H2O2, extracellular H2O2 levels were measured. Cells were first grown overnight in minimal A 0.2% glucose medium containing 0.5 mM each of the 20 amino acids. Most cultures were then diluted to ≈0.001 OD and grown aerobically to an OD of ≈0.1; however, JI377 was grown only to an OD of ≈0.05 in order to measure H2O2 production before growth was significantly inhibited. Cells were then washed in fresh medium containing only 0.02% glucose and 0.05 mM amino acids, resuspended at an OD of 0.1 in the same medium, and incubated with shaking at either 25 or 37°C. Glucose was added to the minimal A salts immediately before use. Aliquots were removed at intervals, and their H2O2 content was measured. The rate of H2O2 production was normalized to the cytoplasmic volume of the suspended cells, using a standard ratio of 0.47 μl of internal volume per 1 ml of 1.0 OD of E. coli (10).
Total catalase activity.
Aerobic exponential-phase cells were harvested at 0.5 OD, washed in cold 50 mM KPi, resuspended in 1/4 the original volume, and lysed by passage through a French press. Extracts were centrifuged at 13,000 × g for 20 min to remove cell debris and then stored on ice. Catalase activity was measured in a 1-ml reaction mixture containing 50 μl of extract, 1 mM H2O2, and PBS (pH 7.3). At various time points, 10 μl was removed, diluted 1:500 in PBS, and assayed for H2O2 by the AR/HRP procedure.
HPI assay.
Cultures were grown aerobically for five to six generations in LB medium. Chloramphenicol was added, and cultures were incubated for 5 min. Cells were then washed in PBS containing chloramphenicol, and cell pellets were frozen on dry ice. Within an hour the cells were resuspended in 1/10 the culture volume of 10 mM KPi buffer (pH 6.4) and lysed by sonication. Debris was centrifuged out at 13,000 × g for 20 min. HPI activity was assayed by the o-dianisidine method (18).
β-Galactosidase assay.
Studies of katG::lacZ expression used a λRS45 φ(katG::lacZ) lysogen. Cultures were grown overnight under anaerobic conditions, diluted to an OD of 0.01 in LB, and grown anaerobically to an OD of 0.1. For measurement of anaerobic expression, chloramphenicol was added and cultures were harvested. For measurement of aerobic expression, cultures were shifted into air and grown with vigorous shaking to an OD of 0.3 to 0.4. Where indicated, 13,000 U of catalase was added every 15 min. Repeated additions were necessary because catalase rapidly loses activity in cell cultures.
At harvest, cultures were centrifuged. Pellets were washed in 50 mM cold KPi buffer (pH 7.0) with special concern to remove any residual exogenous catalase. Cells were resuspended in 50 mM KPi buffer (pH 7.0) at 1/10 the culture volume and lysed by French press. Cell debris was removed by centrifugation at 13,000 × g for 20 min. Extracts were incubated at 28°C for 5 min before being assayed. β-Galactosidase activity was assayed in a 1.2-ml reaction mixture consisting of 0.2 ml of ONPG (o-nitrophenyl-β-d-galactopyranoside, 4 mg/ml), extract, and Z-buffer (19) at 28°C. Change in absorption over time was monitored at 420 nm. Protein concentrations of all extracts were determined using a Coomassie dye-based assay by Pierce. All assays were performed on duplicate samples, and the values were then averaged.
Growth experiments.
Care was taken to determine whether the poor aerobic growth of the Ahp− Kat− strain (JI377) was due to endogenous H2O2 or H2O2 that was chemically generated by autooxidation of the medium. Low-peroxide medium, which contains ≈0.02 μM H2O2, was prepared by adding filter-sterilized glucose to anaerobic minimal A salts immediately before inoculation. During aerobic incubation at 37°C, this medium accumulates ≈0.04 μM H2O2 per h. JI377 was inoculated from an anaerobic overnight culture to 0.01 OD in anaerobic, low-peroxide medium and grown anaerobically to 0.1 OD. Cells were then subcultured into the same medium and shifted into air. Growth was monitored during the first 2 h, during which the vast majority of H2O2 present in the medium was generated by the cells.
In some experiments, anaerobic MG1655 was mixed with JI377 at a 9:1 ratio in LB, the mixed culture was diluted to 0.01 OD in aerobic LB, and the growth of both strains was monitored by intermittent dilution and plating on LB plates (to quantify total viable cells) and LB plus tetracycline (to quantify viable JI377). In other experiments JI377 was shifted into aerobic LB in pure culture, and 6,500 U of catalase was added every 15 min. JI377 growth was monitored by plating, because the absorbance of catalase interferes with measurement of biomass by optical density.
Disk diffusion.
Standing overnight cultures in LB medium were diluted to 0.005 OD in aerobic LB and then grown to an OD of ≈0.1. Cultures were diluted 1:10, and 100 μl was spread on LB plates. Round sterile filters (1-cm diameter) were placed in the center of the plates and spotted with 25 μl of 3% H2O2. Plates were incubated at 37°C overnight. The distance from the edge of the disk to the edge of the growth zone was measured. This experiment was performed in triplicate; mean values are reported.
RESULTS
Catalase is not the primary scavenger of low-level H2O2.
Inside bacteria, antibiotics such as paraquat, plumbagin, and juglone are cyclically reduced by redox enzymes and oxidized by molecular oxygen, thereby generating superoxide and, upon its dismutation, hydrogen peroxide. The rate at which H2O2 is made inside drug-treated E. coli can be determined by measurement of nonrespiratory oxygen consumption in the presence of cyanide. However, we were unable to detect H2O2 accumulating in the medium of plumbagin-treated wild-type cells, despite the fact that oxygen consumption measurements indicated that the amount of H2O2 should have been easily within our detection limits (data not shown).
E. coli contains two catalases, and we anticipated that these were responsible for scavenging the H2O2. However, when the experiments were repeated with strain UM1, which has point mutations in both of the catalase structural genes (katG and katE), H2O2 was again undetectable. By adding 1.5 μM H2O2 to the bacterial culture, we found that UM1 degraded H2O2 as rapidly as did its wild-type parent (data not shown).
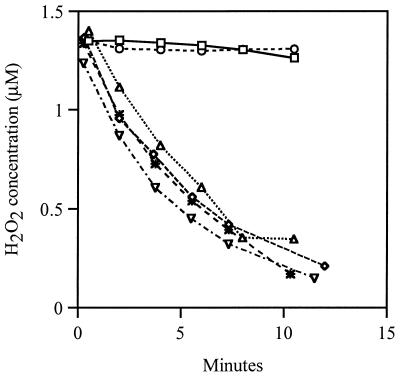
HPI, the catalase encoded by katG, exhibits weak NAD(P)H-dependent peroxidase activity in vitro, and the katG17 mutation that is present in UM1 eliminates catalase but not peroxidase activity. Therefore, we wondered whether the residual peroxidase activity of HPI was responsible for the scavenging activity of UM1. True null mutations of both katE and katG were transduced into the wild-type strain MG1655, and the H2O2 scavenging measurements were repeated. This catalase double mutant had no detectable catalase or HPI peroxidase activity in vitro, but it still scavenged 1.5 μM H2O2 approximately as rapidly as its wild-type parent (Fig. 1). Therefore, E. coli must have another means of efficiently scavenging H2O2.
FIG. 1.
Ahp scavenges H2O2 in a Kat− strain. Cultures of MG1655 (wild type, ▿), JI367 (katG katE, ◊), JI370 (ahpCF, *), JI377 (ahpCF katG katE, ○), JI374 (ahpCF katG, □), and JI372 (ahpCF katE, ▵) were grown aerobically in LB and resuspended in PBS at an OD of 0.1. H2O2 was added at a final concentration of 1.5 μM. At various time points after addition of H2O2, the H2O2 concentration was measured as described in Materials and Methods.
Ahp is the source of scavenging in a catalase null mutant.
Other mechanisms of scavenging were considered. Some α-ketoacids, such as pyruvate, can be excreted into the medium of glucose-fed cells, and these can chemically reduce H2O2 in an oxidative decarboxylation reaction (4). In separate work we have observed that both the respiratory cytochrome o and cytochrome d oxidases have weak peroxidase activities (A. Nguyen and J. A. Imlay, submitted for publication). Finally, alkyl hydroperoxide reductase (Ahp) has been shown to accept H2O2 as a substrate in vitro (22).
Spent medium was examined, but it did not have significant scavenging activity. Cyanide competitively inhibits H2O2 binding by cytochrome oxidases but did not diminish scavenging by a katG katE mutant; the same result was obtained when cyo and cyd mutations were placed in this background. Thus, neither growth medium nor cytochrome oxidases provided the catalase-independent scavenging activity.
Ahp is an NAD(P)H-dependent peroxidase that rapidly reduces organic hydroperoxides as diverse as cumene and t-butylhydroperoxide (11). Niimura et al. subsequently found some activity with hydrogen peroxide as the substrate, although we are not aware of any comparative study of turnover numbers (22). Ahp, like HPI, is a member of the OxyR regulon in diverse bacteria (15, 20, 24). OxyR is activated by organic hydroperoxides as well as by H2O2, and it has become accepted that the physiological role of Ahp is to scavenge organic hydroperoxides. Mutants that lack Ahp are indeed hypersensitive to growth inhibition by organic hydroperoxides, which are not substrates for catalase (32).
A null mutant lacking Ahp scavenged H2O2 as well as did its wild-type parent (Fig. 1). However, in contrast to the katG katE strain, an ahpCF katG katE triple mutant exhibited virtually no scavenging activity. An ahpCF katG mutant also failed to scavenge H2O2, although an ahpCF katE mutant did so (Fig. 1). Thus, Ahp provides the scavenging activity that persists in catalase null mutants.
HPI induction can compensate for loss of Ahp.
We wished to learn which enzyme, Ahp or HPI, was the predominant scavenger in wild-type cells. Because the ahpCF and katG single mutants each scavenged H2O2 at the same rate as did wild-type cells (Fig. 1), we inferred that one of the enzymes might be induced to compensate for the absence of the other. Both ahpCF and katG are positively regulated by the OxyR regulon, and a mutation in the constitutive scavenger might cause intracellular H2O2 to accumulate until OxyR activated the expression of the other, ultimately restoring wild-type levels of H2O2 scavenging.
We observed that an ahpCF mutant contained sevenfold more total catalase (including both HPI and HPII) than did wild-type cells. The o-dianisidine peroxidase activity, which specifically reflects the HPI titer, was increased 10-fold. Similarly, the β-galactosidase activity of an aerobic λRS45 katG::lacZ lysogen was increased 10-fold when the ahpCF null mutation was introduced, rising from 0.04 ± 0.01 to 0.39 ± 0.05 β-galactosidase U/mg. In contrast, the introduction of katG (LC80), katE (HDO3), or both katG and katE (LC84) mutations did not increase β-galactosidase activity at all (0.04 ± 0.01, 0.03 ± 0.01, and 0.04 ± 0.01 U/mg, respectively). These results agreed with the observations of Rosner and Storz (27) and Ritz et al. (25a).
Induction of katG in the ahpCF mutant was blocked by an oxyR mutation (Table 2). To ensure that induction of the OxyR regulon was not due to a suppressor mutation secondary to the ahpCF mutation, the ahpCF null allele was retransduced into the katG::lacZ lysogen under anaerobic conditions. Once again, the fusion was induced when the transductant was cultured in aerobic medium (data not shown). These data demonstrate that in wild-type cells Ahp rather than catalase is the primary scavenger of an endogenous inducer of the OxyR regulon.
TABLE 2.
Endogenous H2O2 activates the OxyR regulon in an ahpCF mutant
| Straina | Mean β-galactosidase activity (U/mg of protein) ± SD
|
||
|---|---|---|---|
| Anaerobic | Aerobic | Aerobic + catalaseb | |
| Ahp+ OxyR+ (GSO22) | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 |
| Ahp− OxyR+ (LC70) | 0.03 ± 0.01 | 0.35 ± 0.03 | 0.18 ± 0.02 |
| Ahp− OxyR− (LC74) | 0.03 ± 0.01 | 0.03 ± 0.01 | NDc |
Strains were isogenic and harbored λRS45 (katG::lacZ).
Exogenous catalase (30 μl) was added before the culture was aerated and then every 15 min until it was iced.
ND, not determined.
The induction of katG::lacZ in ahp cultures was partially averted by the addition to the growth medium of exogenous catalase (Table 2). Since catalase scavenges only H2O2, we infer that H2O2 is the species that accumulates in these cultures and activates the OxyR regulon. The extracellular catalase may have failed to completely block induction both because of its instability and because extracellular scavengers cannot fully eradicate intracellular H2O2 accumulation (30). Alternatively, it is possible that an additional inducer was present.
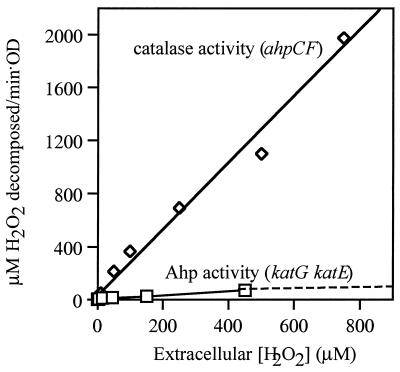
Ahp and HPI have discrete roles in scavenging H2O2.
Mutants that contain only one or the other scavenger exhibited very different dose-response curves (Fig. 2). Whole cells that contained only Ahp scavenged low concentrations of H2O2 very effectively, exhibiting a half-maximal rate when the extracellular concentration of H2O2 was about 5 μM. The activity became saturated when extracellular H2O2 exceeded 20 μM (Fig. 2). In contrast, HPI-expressing cells were not saturated by even millimolar concentrations of H2O2, consistent with its Km of 5.9 mM (9). The dose-response curves suggest that the primary role of HPI might be to scavenge higher concentrations of H2O2, against which Ahp is ineffective.
FIG. 2.
Dependence of scavenging rate on H2O2 concentration. Rates of H2O2 decomposition were measured in dilute suspensions of JI370 (ahpCF, ◊) and JI367 (katG katE, □). Rates were normalized to a value of 1.0 OD.
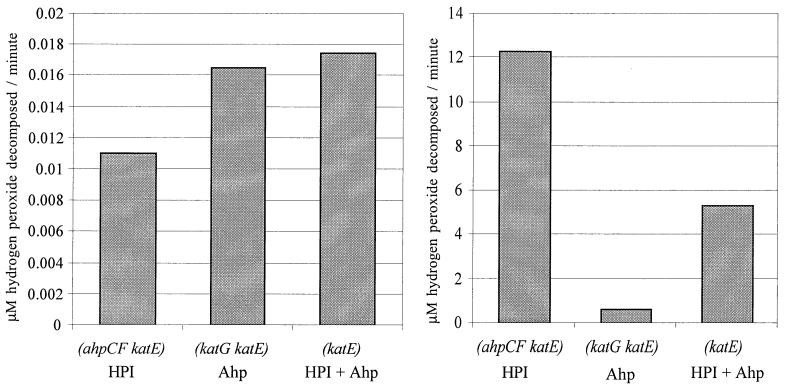
To directly contrast the kinetic behaviors of Ahp and HPI, we measured the rates at which cells decomposed low (0.1 μM) and high (150 μM) concentrations of H2O2. (To simplify interpretation, the strains used in these experiments lacked HPII. However, control experiments demonstrated that HPII provides significant scavenging activity only in stationary phase, when it is induced by RpoS [28].) An HPI− Ahp+ mutant scavenged 0.1 μM H2O2 slightly more rapidly than did an HPI+ Ahp− mutant, despite the fact that the latter strain has 10-fold-induced levels of HPI (Fig. 3, left panel). Thus, in wild-type E. coli most scavenging of low-dose H2O2 must be done by Ahp. Conversely, the HPI− Ahp+ strain was poor at degrading 150 μM H2O2, unlike the HPI+ Ahp− and HPI+ Ahp+ strains (Fig. 3, right panel). Although Ahp can be inactivated by H2O2 in vitro (11), its poor activity in vivo seemed not to stem from this problem, since normal scavenging activity was observed when the 150 μM H2O2 was washed away and the cells were exposed to 1.5 μM H2O2 (data not shown). The Ahp− mutant scavenged 150 μM H2O2 twice as rapidly as did the wild type, because HPI+ was induced (30).
FIG. 3.
Distinct efficiencies of Ahp and HPI at different H2O2 concentrations. H2O2 was added at a final concentration of 0.1 μM (right panel) and 150 μM (left panel) to cultures of JI372 (ahpCF katE), JI367 (katG katE), and JI362 (katE). Two minutes after addition of H2O2, the H2O2 concentration was measured as described in Materials and Methods.
These data indicate that Ahp and HPI have distinct roles: Ahp is more effective at scavenging very low concentrations of H2O2, while HPI is the more effective enzyme at higher concentrations. The dominance of HPI at supranormal levels of H2O2 was also apparent in disk diffusion assays, which test the ability of strains to degrade high concentrations of H2O2 to a level that permits growth. The katG mutant was hypersensitive, while the ahpCF mutant showed wild-type resistance (Table 3). The ahpCF mutation debilitated only strains that lacked HPI. As before, the addition of a katE mutation did not affect the sensitivity of any strain.
TABLE 3.
HPI (katG) is the primary scavenger of supranormal H2O2 concentrationsa
| Strain | Zone of inhibitionb (cm) |
|---|---|
| Wild type | 1.1 |
| katG | 1.6 |
| ahpCF | 1.1 |
| katG katE | 1.6 |
| ahpCF katG | 2.9 |
| ahpCF katG katE | 3.1 |
Assay was done by disk diffusion of 25 μl of 3% (880 mM) H2O2. Assay was done in triplicate, and trends were always consistent. One data set is shown. Strains used were MG1655 (wild type), JI364 (katG), JI370 (ahpCF), JI367 (katG katE), JI374 (ahpCF katG), and JI377 (ahpCF katG katE).
Distance of clearing from the edge of the disk.
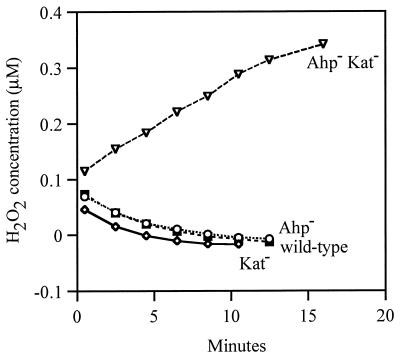
Calculation of endogenous H2O2 production during aerobic growth.
Substantial H2O2 accumulated in the medium of Ahp− Kat− cultures. The extracellular concentration of H2O2 rose to 1.8 μM during aerobic growth in minimal 0.05% glucose medium when cells were cultured for a single generation, from 0.05 to 0.10 OD600. In contrast, the H2O2 concentrations of wild-type (MG1655), Kat− (JI367), and Ahp− (JI370) cultures were below our detection limit of ≈0.04 μM H2O2. The fact that the H2O2 concentration was so low in Ahp− cultures was initially surprising, given that sufficient H2O2 was present to activate the OxyR protein. However, subsequent work indicated that the concentration of H2O2 inside these cells may have been substantially higher than that outside them (30).
Kat− Ahp− strains have <5% of the scavenging activity of wild-type cells, so that virtually all of the H2O2 that enters or is formed within these cells diffuses out without being scavenged (30). For this reason the measurement of excreted H2O2 is a valid proxy for measurement of endogenous H2O2 formation. Using this strain, we quantitated the rate at which E. coli generates H2O2 (Fig. 4). Measurements were made after exponentially growing cells were suspended in fresh glucose/amino acids medium at 37°C (and do not necessarily apply in other media). The rate of H2O2 formation, normalized to cell volume, was 14 μM H2O2/s at 37°C. This result is consistent with earlier predictions (Discussion).
FIG. 4.
H2O2 production by Ahp− Kat− cells. MG1655 (wild type), JI370 (ahpCF), JI367 (katG katE), and JI377 (ahpCF katG katE) were grown aerobically in LB and resuspended in 37°C minimal A salts containing 0.2% glucose. At various time points after resuspension, the H2O2 concentration of the medium was measured. (The H2O2 levels drop for the three scavenger-proficient strains because these strains degrade the 0.05 μM H2O2 that is present in the initial medium.)
Inability to scavenge endogenous H2O2 causes a growth defect.
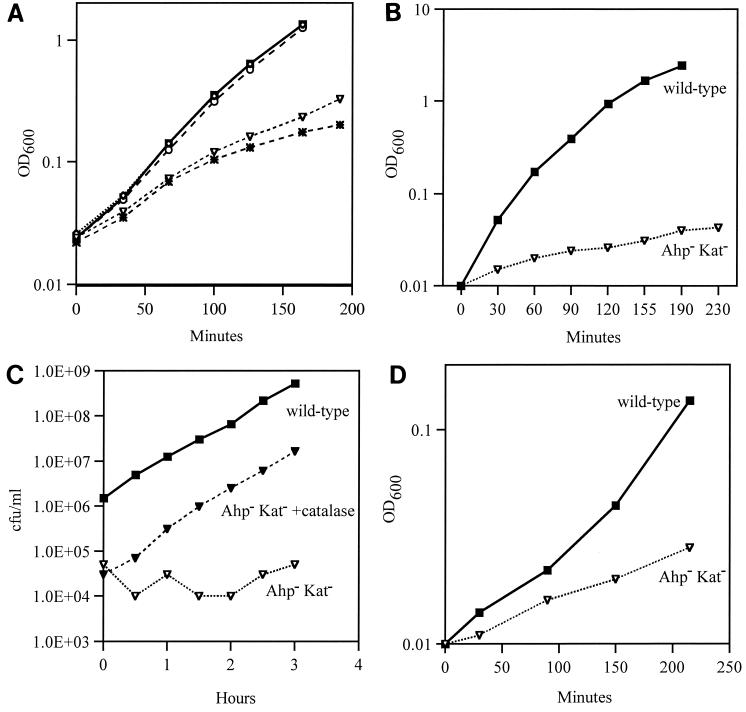
It has long been suspected that aerobic metabolism generates enough H2O2 to damage cells that cannot scavenge it. No growth defects were apparent in the ahpCF mutant; we presume this is because the cell compensates sufficiently for the loss of ahpCF by inducing katG. However, the Ahp− Kat− strain grew poorly in all aerobic media that were tested (Fig. 5). Growth slowed progressively over time, and when cells were repeatedly subcultured, growth often stopped entirely (Fig. 5B). This may reflect the continual accumulation of damage in the cell. Growth was particularly poor on defined media that lacked amino acids (data not shown). Wild-type growth was restored when catalase was added to the medium (Fig. 5C) or when the mutant was cultured anaerobically (not shown).
FIG. 5.
An Ahp− Kat− strain has an aerobic growth defect. (A) Growth of MG1655 (wild type, ▪), JI370 (ahpCF, ○), JI367 (katG katE, ◊), JI374 (katG ahpCF, *), and JI377 (katG katE ahpCF, ▿) in aerobic LB medium. (B) MG1655 and JI377 were grown aerobically in LB from 0.001 OD to mid-log phase, as for panel A. Cells were then subcultured into fresh LB at an OD of 0.01, and residual growth was observed. (C) Exogenous catalase protects against an aerobic growth defect in LB. MG1655 and JI377 were grown aerobically in fresh LB. Cultures were then subcultured to 0.01 OD in LB. Exogenous catalase was added to one culture of JI377 every 15 min to maintain catalase activity. At various time points, aliquots were removed from each culture and plated in selective top agar. Growth rate was determined the next day. (D) Endogenous H2O2 can be toxic to cells. Exponential anaerobic MG1655 and JI377 were subcultured into fresh aerobic peroxide-free minimal A glucose (0.2%) medium, and growth was monitored.
Special efforts were undertaken to confirm that the growth defect was due to endogenous H2O2 rather than H2O2 made by autooxidation of the glucose medium. Cells were cultured anaerobically in glucose medium to log phase and then diluted into fresh aerobic “peroxide-free” medium (Fig. 5D). Within the first half-hour the Ahp− Kat− strain grew more poorly than did its wild-type parent. Our measurements over this period confirmed that the H2O2 found in this culture was generated primarily by the cells rather than by glucose oxidation. Thus, aerobic E. coli generates enough H2O2 to debilitate the cell. In previous studies of catalase mutants, the toxicity of endogenous H2O2 was obscured by the scavenging action of Ahp.
The necessity for care in these experiments was underscored by the observation that medium that had been stored on the bench often contained micromolar amounts of H2O2, and dilution of even wild-type cells into stored medium was sufficient to transiently induce the OxyR regulon (data not shown).
DISCUSSION
H2O2 may be the sole physiological substrate of Ahp.
The results of this study indicate that in exponentially growing E. coli Ahp is responsible for the degradation of low concentrations of hydrogen peroxide. Previously, Ahp was primarily associated with the detoxification of organic hydroperoxides. However, the range of organic peroxides that are good substrates for the enzyme indicates that its active site can accommodate virtually any ROOH, so it is not surprising that it reacts with HOOH as well. Earlier genetic studies confirmed that Ahp provides cellular resistance to organic hydroperoxides but found little role in resistance to H2O2. This result is now understandable: because catalases cannot degrade organic hydroperoxides, Ahp is likely to be the only enzyme in E. coli with that catalytic ability. At the same time, the role of Ahp in H2O2 decomposition was obscured because the widely employed disk inhibition assays confront cells with high H2O2 concentrations that are more efficiently decomposed by catalases.
It is not clear whether E. coli must ever detoxify organic hydroperoxides in nature. Lipid hydroperoxides were suggested to be the physiological substrates for Ahp, but E. coli lacks the polyunsaturated fatty acids that appear to be necessary for lipid peroxidation (1), and we are not aware of any study that has documented the recovery of peroxidized lipids from this bacterium. It is possible that organic hydroperoxides are pseudosubstrates for an enzyme whose only role in nature is the decomposition of hydrogen peroxide.
Why does E. coli use multiple scavengers?
In general, all peroxidases will be inferior to catalases at scavenging high concentrations of H2O2 because peroxidases can turn over only as quickly as the cell can provide a reductant to them. Catalases escape this restriction. For example, the HPI catalase of Ahp− E. coli degraded 500 μM H2O2 at a rate of 7 × 107 molecules cell−1 s−1. To achieve the same rate using Ahp would require an equivalent amount of NADH, which substantially exceeds the rate at which metabolism generates NADH. (Glucose-saturated cells can generate enough NADH to respire 6 × 106 molecules of oxygen cell−1 s−1.) The disparity is even greater when one considers decomposition of higher concentrations of H2O2. Therefore, catalases allow cells to degrade high concentrations of H2O2 far more quickly than would peroxidases alone.
The preference for Ahp at low H2O2 concentrations may derive from its greater catalytic efficiency. Since normal intracellular concentrations of H2O2 are well below the Km of both enzymes, kcat/Km is the relevant kinetic parameter. The kcat/Km of the catalase activity of HPI is 9 × 105 M−1 s−1 (9); that of Ahp has not been reported, but the data of Niimura et al. (22) imply that it is at least 8 × 106 M−1 s−1. The implication is that at least 10-fold more molecules of HPI need to be synthesized to provide the scavenging activity provided by Ahp. A second disadvantage to catalase may stem from the fact that enzymes that require reactions with two molecules of substrate to complete a catalytic cycle can have difficulty at low substrate concentrations, when an intermediate state is long-lived. Compound I, the divalently oxidized catalase intermediate, can be unstable and in some circumstances may reversibly deactivate (9), which would further diminish the ability of catalase to scavenge trace amounts of H2O2. Peroxidases may comprise an evolutionary solution to this problem.
The kinetic efficiency of HPII (kcat/Km = 1.3 × 106 M−1 s−1) (23), the stationary-phase catalase, is similar to that of HPI. It seems reasonable that E. coli would increase its catalase titer in stationary phase, when there may not be sufficient NADH for Ahp to remain an effective scavenger. Gonzalez-Flecha and Demple reported that both catalases are induced as growth slows (8). It is not clear why scavenging efficiency could not be maintained by induction of HPI alone. For now, the benefit to E. coli of having two catalases instead of one remains obscure.
The compensatory interactions that we observed between catalase and Ahp synthesis have been observed previously in a wide range of bacteria. In Xanthomonas campestris, Bacteroides fragilis, and Pseudomonas aeruginosa, mutations in ahp cause catalase induction (20, 24, 26); this may be true in Bacillus subtilis as well (2). Conversely, katG mutations in Mycobacterium tuberculosis select for promoter upmutations in ahp (31). It seems likely that the division of labor found in E. coli, wherein Ahp scavenges low levels of H2O2 and catalase scavenges high levels, is widespread.
Mechanisms of H2O2 formation and cell damage.
The existence of a strain unable to scavenge the H2O2 it produces has enabled us to measure the rate at which H2O2 is formed. In earlier studies in vitro, we determined that the respiratory chain was likely to be the primary source of H2O2 during aerobic growth on glucose, largely because of the autooxidizability of the NADH dehydrogenase II (18). This enzyme adventitiously transfers electrons to oxygen from its reduced flavin in a direct second-order chemical reaction. Other flavoenzymes can do so as well, and in circumstances where these other enzymes are especially abundant, they may be the predominant sources of superoxide and H2O2.
The rate at which H2O2 is likely to be formed in vivo can be predicted from the in vitro data. We found that 9 molecules of H2O2 were formed per 10,000 electrons flowing through the NADH dehydrogenase II (18). We do not know the fraction of the respiratory flux that flows through this enzyme during growth on glucose, but given that exponentially growing cells consume about 3.2 mM O2/s, one extrapolates that the rate of H2O2 formation could be up to 12 μM/s. This is close to the rate determined in vivo in this study (14 μM/s). Efforts to test the sources of H2O2 production in vivo are under way.
Gonzalez-Flecha and Demple inferred rates of H2O2 production by intact, wild-type cells (ca. 1 to 2 μM/s) that are lower than those that we have reported (8). However, we observed rates similar to theirs when we suspended cells in room temperature buffer, as they did (3 μM/s). It is not surprising that H2O2 is most rapidly formed when metabolism is active and temperatures are high enough to overcome the activation energy (18). There was a procedural difference, however. Their calculation was based on measurements of steady-state concentrations of extracellular H2O2 in suspensions of cells: by assuming that catalase was the predominant scavenger of H2O2, they used measurements of its activity to calculate the rate of H2O2 production. In our experiments we found that H2O2 did not accumulate extracellularly due to the action of Ahp. These differences may reflect the different conditions under which the experiments were performed.
Inside growing cells, the steady-state concentration of H2O2 depends on the rates of its formation and of its dissipation. In this study we quantified the rate at which H2O2 is formed when E. coli is cultured under a particular set of growth conditions. In the accompanying work (30), we measured the processes that consume H2O2 and estimated the internal H2O2 concentration.
ACKNOWLEDGMENTS
We are grateful to Gigi Storz, Peter Loewen, Bruce Demple, Bob Gennis, and Jim Slauch for providing strains and discussion that helped us in this study, and we thank Holly Oliver for assistance with strain constructions.
This work was supported by grant GM49640 from the National Institutes of Health.
REFERENCES
- 1.Bielski B H J, Arudi R L, Sutherland M W. A study of the reactivity of HO2−/O2− with unsaturated fatty acids. J Biol Chem. 1983;258:4759–4761. [PubMed] [Google Scholar]
- 2.Bsat N, Chen L, Helmann J. Mutation of the Bacillus subtilis alkyl hydroperoxide reductase (ahpCF) operon reveals compensatory interactions among hydrogen peroxide stress genes. J Bacteriol. 1996;178:6579–6586. doi: 10.1128/jb.178.22.6579-6586.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlioz A, Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J. 1986;5:623–630. doi: 10.1002/j.1460-2075.1986.tb04256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Constantopoulos G, Barranger J. Nonenzymatic decarboxylation of pyruvate. Anal Biochem. 1984;139:353–358. doi: 10.1016/0003-2697(84)90016-2. [DOI] [PubMed] [Google Scholar]
- 5.Dukan S, Nyström T. Oxidative stress defense and deterioration of growth-arrested Escherichia coli cells. J Biol Chem. 1999;274:26027–26032. doi: 10.1074/jbc.274.37.26027. [DOI] [PubMed] [Google Scholar]
- 6.Gaudu P, Touati D, Niviere V, Fontecave M. The NAD(P)H-flavin oxidoreductase from Escherichia coli as a source of superoxide radicals. J Biol Chem. 1994;269:8182–8188. [PubMed] [Google Scholar]
- 7.Gonzalez-Flecha B, Demple B. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J Biol Chem. 1995;1995:13681–13687. doi: 10.1074/jbc.270.23.13681. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Flecha B, Demple B. Homeostatic regulation of intracellular hydrogen peroxide concentration in aerobically growing Escherichia coli. J Bacteriol. 1997;179:382–388. doi: 10.1128/jb.179.2.382-388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hillar A, Peters B, Pauls R, Loboda A, Zhang H, Mauk A G, Loewen P C. Modulation of the activities of catalase-peroxidase HPI of Escherichia coli by site-directed mutagenesis. Biochemistry. 2000;59:5868–5875. doi: 10.1021/bi0000059. [DOI] [PubMed] [Google Scholar]
- 10.Imlay J A, Fridovich I. Assay of metabolic superoxide production in Escherichia coli. J Biol Chem. 1991;266:6957–6965. [PubMed] [Google Scholar]
- 11.Jacobson F S, Morgan R W, Christman M F, Ames B N. An alkyl hydroperoxide reductase from Salmonella typhimurium involved in the defense of DNA against oxidative damage: purification and properties. J Biol Chem. 1989;264:1488–1496. [PubMed] [Google Scholar]
- 12.Kaysser T M, Ghaim J B, Georgiou C, Gennis R B. Methionine-393 is an axial ligand of the heme b558 component of the cytochrome bd ubiquinol oxidase from Escherichia coli. Biochemistry. 1995;34:13491–13501. doi: 10.1021/bi00041a029. [DOI] [PubMed] [Google Scholar]
- 13.Lawley P D, Thatcher C J. Methylation of deoxyribonucleic acid in cultured mammalian cells by N-methyl-N′-nitro-N-nitrosoguanidine: the influence of cellular thiol concentrations. Biochem J. 1970;116:693–707. doi: 10.1042/bj1160693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loewen P C. Isolation of catalase-deficient Escherichia coli mutants and genetic mapping of katE, a locus that affects catalase activity. J Bacteriol. 1984;157:622–626. doi: 10.1128/jb.157.2.622-626.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loprasert S, Fuangthong M, Whangsuk W, Atichartpongkul A, Mongkolsuk S. Molecular and physiological analysis of an OxyR-regulated ahpC promoter in Xanthomonas campestris pv. phaseoli. Mol Microbiol. 2000;37:1504–1514. doi: 10.1046/j.1365-2958.2000.02107.x. [DOI] [PubMed] [Google Scholar]
- 16.Maloy S R, Nunn W D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981;145:1110–1112. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCord J M, Keele B B, Jr, Fridovich I. An enzyme-based theory of obligate anaerobiosis: the physiological function of superoxide dismutase. Proc Natl Acad Sci USA. 1971;68:1024–1027. doi: 10.1073/pnas.68.5.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Messner K R, Imlay J A. The identification of primary sites of superoxide and hydrogen peroxide formation in the aerobic respiratory chain and sulfite reductase complex of Escherichia coli. J Biol Chem. 1999;274:10119–10128. doi: 10.1074/jbc.274.15.10119. [DOI] [PubMed] [Google Scholar]
- 19.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 20.Mongkolsuk S, Whangsuk W, Vattanaviboon P, Loprasert S, Fuangthong M. A Xanthomonas alkyl hydroperoxide reductase subunit C (ahpC) mutant showed an altered peroxide stress response and complex regulation of the compensatory response of peroxide detoxification enzymes. J Bacteriol. 2000;182:6845–6849. doi: 10.1128/jb.182.23.6845-6849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moskovitz J, Rahman M A, Strassman J, Yancey S O, Kushner S R, Brot N, Weissbach H. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J Bacteriol. 1995;177:502–507. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niimura Y, Poole L B, Massey V. Amphibacillus xylanus NADH oxidase and Salmonella typhimurium alkyl-hydroperoxide reductase flavoprotein components show extremely high scavenging activity for both alkyl hydroperoxide and hydrogen peroxide in the presence of S. typhimurium alkyl-hydroperoxide reductase 22-kDa protein component. J Biol Chem. 1995;270:25645–25650. doi: 10.1074/jbc.270.43.25645. [DOI] [PubMed] [Google Scholar]
- 23.Obinger C, Maj M, Nicholls P, Loewen P. Activity, peroxide compound formation, and heme d synthesis in Escherichia coli HPII catalase. Arch Biochem Biophys. 1997;342:58–67. doi: 10.1006/abbi.1997.9988. [DOI] [PubMed] [Google Scholar]
- 24.Ochsner U A, Vasil M L, Alsabbagh E, Parvatiyar K, Hassett D J. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of the katB-ankB. ahpB, and ahpC-ahpF. J Bacteriol. 2000;182:4533–4544. doi: 10.1128/jb.182.16.4533-4544.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds C M, Poole L B. Activity of one of two engineered heterodimers of AhpF, the NADH:peroxiredoxin oxioreductase from Salmonella typhimurium, reveals intrasubunit electron transfer between domains. Biochemistry. 2001;40:3912–3919. doi: 10.1021/bi002766h. [DOI] [PubMed] [Google Scholar]
- 25a.Ritz D, Patel H, Doan B, Zheng M, Aslund F, Storz G, Beckwith J. Thioredoxin 2 is involved in the oxidative stress response in Escherichia coli. J Biol Chem. 2000;275:2505–2512. doi: 10.1074/jbc.275.4.2505. [DOI] [PubMed] [Google Scholar]
- 26.Rocha E R, Smith C J. Role of the alkyl hydroperoxide reductase (ahpCF) gene in oxidative stress defense of the obligate anaerobe Bacteroides fragilis. J Bacteriol. 1999;181:5701–5710. doi: 10.1128/jb.181.18.5701-5710.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosner J L, Storz G. Effects of peroxides on susceptibilities of Escherichia coli and Mycobacterium smegmatis to isoniazid. Antimicrob Agents Chemother. 1994;38:1829–1833. doi: 10.1128/aac.38.8.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sak B D, Eisenstark A, Touati D. Exonuclease III and the catalase hydroperoxidase II in Escherichia coli are both regulated by the katF gene product. Proc Natl Acad Sci USA. 1989;86:3271–3275. doi: 10.1073/pnas.86.9.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schellhorn H E, Hassan H M. Response of hydroperoxidase and superoxide dismutase deficient mutants of Escherichia coli K-12 to oxidative stress. Can J Microbiol. 1988;34:1171–1176. doi: 10.1139/m88-206. [DOI] [PubMed] [Google Scholar]
- 30.Seaver L C, Imlay J A. Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J Bacteriol. 2001;183:7182–7189. doi: 10.1128/JB.183.24.7182-7189.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherman D R, Mdluli K, Hickey M J, Arain T M, Morris S L, Barry III C E, Stover C K. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science. 1996;272:1641–1643. doi: 10.1126/science.272.5268.1641. [DOI] [PubMed] [Google Scholar]
- 32.Storz G, Jacobson F S, Tartaglia L A, Morgan R W, Silveira L A, Ames B N. An alkyl hydroperoxide reductase induced by oxidative stress in Salmonella typhimurium and Escherichia coli: genetic characterization and cloning of ahp. J Bacteriol. 1989;171:2049–2055. doi: 10.1128/jb.171.4.2049-2055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]