Abstract
Influenza A(H3N2) virus exhibited complex seasonal patterns to evade pre-existing antibodies, resulting in changes in the antigenicity of the viron surface protein hemagglutinin (HA). To monitor the currently imported influenza viruses as well as to assess the capacity of health emergencies at the Shanghai port, we collected respiratory specimens of passengers from different countries and regions including some of Europe with influenza-like illness at the Shanghai port during 2016/2017, examined amino acid substitutions, and calculated the perfect-match vaccine efficacy using the p epitope model. Phylogenetic analysis of the HA genes revealed that influenza A(H3N2) viruses belonging to eight subclades were detected, and three amino acid substitutions in the subclade 3C.2a.4 were also added. Besides, two epidemic influenza virus strains were found in the 2016/2017 winter and 2016 summer. The results of lower predicted vaccine effectiveness in summer suggest that the imported A(H3N2) strains were not a good match for the A/Hong Kong/4801/2014 vaccine strain since the summer of 2017. Therefore, the Shanghai Port might stop the risk of the international spread of influenza for the first time, and curb the entry of A(H3N2) from overseas at the earliest stage of a probable influenza pandemic.
Subject terms: Viral genetics, Phylogenetics
Introduction
Influenza viruses in the Orthomyxoviridae family cause highly contagious respiratory diseases with potentially fatal outcomes. There are currently four types in this family, which are type A, B, C and D. Type D viruses have not been reported to infect human yet1. In contrast to type B and C viruses, type A viruses in humans evolve fast and spark a devastating pandemic2–4. In particular, H3N2 subtype viruses hold responsible for a major seasonal influenza epidemic. Additionally, H3N2 subtype viruses could escape host immunity through piecemeal recombination, antigen drift or antigen conversion, and finally induce a lethal new flu pandemic with a potential to kill millions5.
On the surface of H3N2 subtype viruses, hemagglutinin (HA) proteins are closely related to the antigen variation of the epidemic influenza virus, and the variants further trigger phylogenetic clade changes. Since the spring and the summer of 2009, there have been up to seven clades (clades 1 to 7) defined by phylogeny inference6. After 2011, the derivative of Clade 3C, 3C1, 3C2, and 3C3 were dominated in many regions7,8. Previous studies demonstrated similar rates for clades 3C.2a, 3C.3, and 3C.3a early in the season9. In 2014, more new genetic subclades with special HA mutation sites emerged, 3C.2a from 3C2, 3C.3a and 3C.3b from 3C310. Past research showed similar rates for clades 3C.2a, 3C.3, and 3C.3a early in the season, but 3C.2a dominated rapidly in the virus population for more than 70% by January 20157. The genetic subclade 3C.2a1 emerged at the end of the 2015/2016 season11 and has become predominant in the 2016/2017 season. Afterwards, A(H3N2) 3C.2a further divided into new genetic groups by genetic drift (3C.2a.2, 3C.2a.3 and 3C.2a.4)12–14.
Compared to former years, the influenza season 2016/2017 co-circulated earlier in China, particularly in the southern regions. The influenza cases continued to rise, and A(H3N2) viruses became the dominant strain. Hong Kong SAR government continues to report a significant number of serious influenza-related cases and deaths. By June 11st 2017, the Hong Kong Center for Health Protection had confirmed 223 cases, with 155 deaths15. A(H3N2) viruses were also dominant in Europe and North America. The laboratory experiments verified that the vaccine effect of influenza A was not ideal for people over 65 from Finland16 and Sweden17. There were higher mortality and hospitalization rate in the United States in the 2016/2017 flu season18. Therefore, it is extremely vital to evaluate whether the vaccine matches the strains in circulation.
Shanghai is one of the port cities of China to the world, from which H3N2 subtype data might indicate the worldwide trend of the viruses’ evolution to some extent. Here, we analyzed virological surveillance data at the Shanghai Port, described the phylogenetic evolution, inspected the antigen variation characteristics from the molecular level of the currently circulating viruses, and compared them with the vaccine and WHO reference viruses representing various genetic clades. Our findings highlighted the structural implications for the understanding of the phenotypic changes, evolution, and epidemiological monitoring of A(H3N2) viruses.
Results
Virological influenza surveillance during flu season 2016/2017
Virological influenza surveillance data in the Shanghai port were collected weekly. From February 2016 to September 2017, a total of 64 swab samples were collected from passengers of different countries, including 41 passed through Asia (25 from Hong Kong and 12 from Southeast Asia, especially), 16 passed through Europe, 7 passed through the America, and 7 passed through Oceania.
A(H3N2) virus activity increased from the 44th week of 2016, peaked in the 1st week of 2017 and decreased afterwards. The highest proportion of A(H3N2) was observed in summer (28/64, 43.7%), followed by winter (22/64, 34.3%) which outnumbered by that in spring and fall (11/64, 21.8%).
Phylogeny relationships of imported A(H3N2) viruses during the flu season 2016/2017
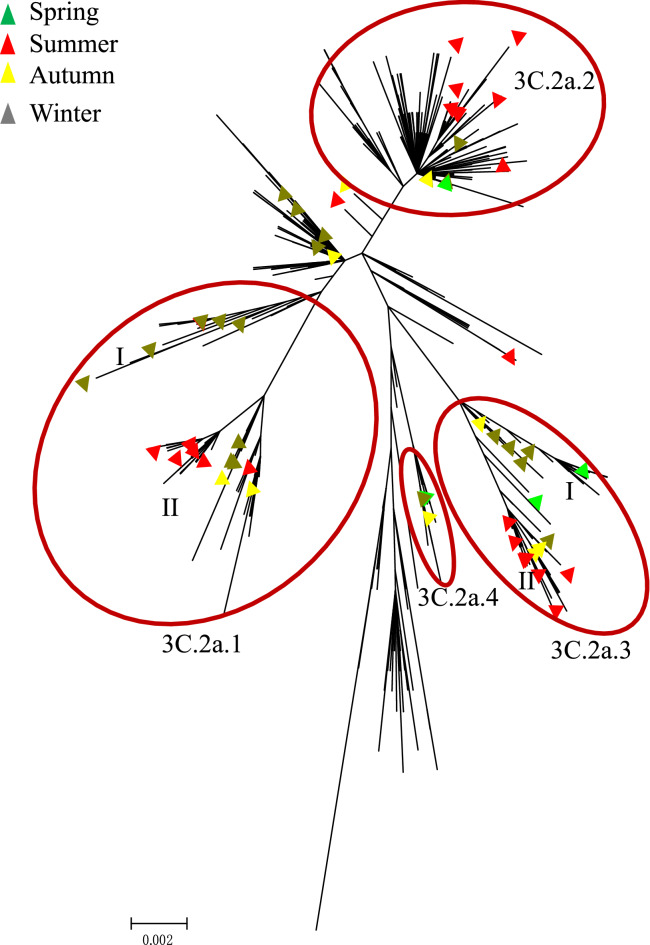
Of the 610 genetically characterized viruses, 546 were provided from GISAID EpiFlu databases. All 64 HA genes sequenced by the Shanghai Port belonged to the H3N2 3C.2a clade. This clade also included the vaccine strain A/HongKong/4801/2014, supporting the vaccine recommendation in the 2016–2018 northern hemisphere influenza season by WHO. Among the 64 viruses, the majority (n = 20, 31.2%) belonged to the subclade 3C.2a.1 represented by A/Singapore/INFIMH-16-0019/2016. The proportions for other subclades were 26.5% (3C.2a.2, n = 17), 25% (3C.2a.3, n = 16) and 6.2% (3C.2a.4, n = 4) (Fig. 1).
Figure 1.
Phylogenetic analysis of the HA segments circulating between 2016/2017.
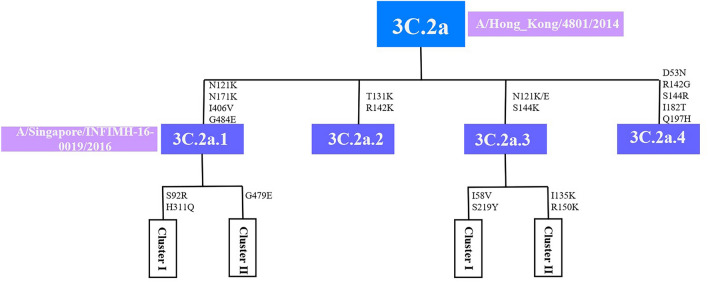
Individual clades of A(H3N2) are typically defined by amino acid substitutions that occur as they diversify from parental strains. Analysis of HA sequences indicated co-circulation of multiple variants in clade 3C.2a. All variants within subclade 3C.2a.1 shared four substitutions N121K, N171K, I406V and G484E. Three additional substitutions were observed in the 3C.2a.1 subcluster: S92R and H311Q in cluster I, G479E in cluster II. Variants 3C.2a.3 shared N121K/E and S144K (I58V and S219R in cluster I and T135K and R150K in cluster II), Variants 3C.2a.2 were characterized by T131K and R142K substitutions and variants 3C.2a.4 were characterized by D53N, R142G, S144R, I182T and Q197H (Fig. 2).
Figure 2.
Schematic diagram demonstrating the shared amino acid changes between clades 3C.2a, 3C.2a.1, 3C.2a.2, 3C.2a.3 and 3C.2a.4 on HA gene.
There were more 3C.2a.1 variants identified from samples collected in the 2017 summer (n = 11) than in the 2016/2017 winter (n = 7). This subclade was further divided into two homogenous sub-clusters (cluster I and II; Fig. 1). The strains from cluster I were concentrated in winter, and the cluster II strains were persisted more common in the summer months. Most viruses in the subgroup 3C.2a.3 happened in summer. And we also found that there was no prominent summer or winter trend of viruses clustered in 3C.2a.2.
The clade pattern of imported A(H3N2) influenza viruses, 2016/2017
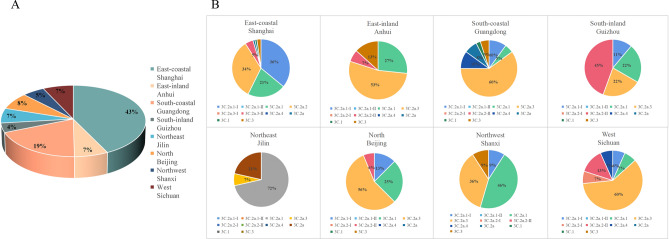
To analyze the geographical distribution of A(H3N2) in China, 31 provinces were classified into six regions based on geographic proximity: North (Beijing), East-coastal (Shanghai), East-inland (Anhui), South-coastal (Guangdong), South-inland (Guizhou), Northeast (Jilin), Northwest (Shanxi) and West (Sichuan). According to our phylogenetic analysis, the A(H3N2) number of the above six regions be counted (Fig. 3A). The Proportions for A(H3N2) in these regions were 5%, 43%, 7.2%, 19%, 4%, 7% ,5% and 7%, respectively. Interestingly, higher epidemic waves of A(H3N2) were observed in Eastern and Southern in China coastal areas, and we presumed that convenient transportation and dense population contributed to it19.
Figure 3.
The Proportions of Influenza A(H3N2) in China six regions (A) and H3N2 clade patterns in China eight provinces (B).
The genetic diversity results (Fig. 3B) indicated that the diversity increased in the East and South, especially coastal cities, Shanghai and Guangzhou. All clades and subclades of the current A(H3N2) were detected in both cities. 3C.2a.3 (60%) was dominant in Guangzhou, with a small proportion of 3C.2a.4 (10%), 3C.2a (3%), 3C.1 (2%), 3C.3 (5%), 3C.2a.1-I (10%) and 3C.2a.1 (5%). In contrast, 3C.2a.1, 3C.2a.1-I and 3C.2a.2 were the major subcluster in Shanghai, with proportions of 19.21%, 32.36% and 30.34%, respectively. 3C.2a.3-II (4%), 3C.1 (1%), 3C.3 (2%) and 3C.2a (1%) were also detected in this region. The diversity of the clade pattern and the dominant clade in these two coastal cities matched well with the trends of the current global A (H3N2), likely because of the higher density of migration and subtropical monsoon climate20.
Prediction of glycosylation sites in A(H3N2) viruses during flu season 2016/2017
There were two models of predicted glycosylation sites in the HA proteins of the A(H3N2) clade 3C.2a: 12 potential glycosylation sites (N8ST, N22GT, N38AT, N45SS, N63CT, N126WT, N133GT, N158YT, N165VT, N246ST, N285GS and N483GT) and 11 potential glycosylation sites (N8ST, N22GT, N38AT, N45SS, N63CT, N126WT, N133GT, N158YT, N165VT, N246ST and N285GS). All of virus strains detected at the Shanghai Port in the clade 3C.2a.1 had 11 potential glycosylation sites, and the rest in the other clades had 12 sites. Comparing to the vaccine strains 2016/2017 A/HongKong/4801/2014 (N8ST, N22GT, N38AT, N45SS, N63CT, N126WT, N133GT, N165VT, N246ST, N285GS and N483GT), the clade 3C.2a.1 virus did not have potential glycosylation site 483(N483GT), the viruses in the clade 3C.2a.2, the clade 3C.2a.3 and the clade 3C.2a.4 had the potential glycosylation site 158(N158YT).
Estimation of vaccine efficacy for A(H3N2)
To assess the effect of the accumulated mutations in the HA1 domain on predicted vaccine efficacy in a given year, the p epitope method was used to evaluate how closely the vaccine strain resembles the imported strain (Table 1). Theoretically, when p epitope in the dominant epitope is higher than 0.19, the vaccine efficacy becomes negative21,22. For the 2016/2017 season, the average p epitope for all A(H3N2) strains was 0.090, which indicated the vaccine efficacy (VE) against those strains was 52.96% (E = 24.89% of 47%, p epitope = 0) of that of a perfect match with the vaccine strain. However, from the 2016/2017 winter to the 2017 autumn, the VE value fluctuated first and then decreased, with the highest value in spring (VE = 58.51%), the lowest value in autumn (VE = 58.51%), and the inflection point in summer (VE = 49.29%). These results suggest that the A(H3N2) strains circulating in 2017 were separated from the vaccine strain and effectively reduced the VE starting in the summer.
Table 1.
Calculated vaccine efficacy using Pepitope model in dominant epitope A of influenza A(H3N2) circulating in Shanghai port during 2016/2017, winter, spring, summer and autumn.
| Influenzaseason | Vaccine strain | Dominantepitope | Pepitope | Vaccine efficacy 47% | Vaccine efficacy 100% |
|---|---|---|---|---|---|
| 2016/2017 | A/Hong_Kong/4801/2014 2016/2017 | A | 0.090 | 24.89% | 52.96% |
| Winter | 0.082 | 26.5% | 56.53% | ||
| Spring | 0.078 | 27.5% | 58.51% | ||
| Summer | 0.096 | 23.17% | 49.29% | ||
| Autumn | 0.105 | 21.00% | 44.68% |
Discussion
Human seasonal A(H3N2) virus epidemics in different zone have highly diverse patterns, especially in the northern hemisphere, where these patterns can exhibit semiannual or annual epidemic cycles. Moreover, HA segments of A(H3N2) viruses have undergone considerable genetic differentiation and evolved in seven genetic groups and multiple clades/subclades since 2009. It is the result of H3N2 viruses circulating via the network of temporally overlapping epidemics and high rates of migration, rather than local persistence. It has been suggested that the global persistence of A(H3N2) virus is the result of a migrating meta-population in which multiple different localities may seed seasonal epidemics in temperate regions in a given year23. Shanghai, the most developed and open city in China, attracts people around the world. So, the Shanghai Port has the characteristics of large passenger flow, high workload and wide international flight distribution. Around 44 million passengers enter and exit the Shanghai Port in 2017, accounting for 7.3 percent of the total number of people entering and exiting China's ports24. In addition, the specimens involved in this study were all from international travelers, who pass through cities with high population density or dense traffic, such as Europe, the United States, and Hong Kong, China. These cities or regions provide convenient channels for the mutation, transmission and spread of influenza25. Although surveillance for influenza at ports has been increasing, there have been few reports of interactions between ports and global epidemic trends. In this study, we performed genetic and evolutionary analyses for viruses obtained during 2016/2017 in order to investigate the evolution of the influenza virus during 2016/2017 at the Shanghai Port and predict the influenza A(H3N2) virus epidemic trends in the future.
The genetic and phylogenetic analyses in different countries and regions indicated that there has been a similar pattern existed among all of the evolutionary trends of A(H3N2) viruses discussed above. Previous research was reported by either ethnically homogenous (Taiwan26, Australian27 and Canada28) or the GISAID EpiFlu databases (isolated in Japan, Bangladesh, Australia, Thailand and the USA). In the current study, 31.2% viruses of the 3C.2a clade clustered in a subgroup carrying N121K, N171K and G484E. In Europe and Canada, the majority of A(H3N2) viruses also belonged to the subclade 3C.2a.1 during the same season28,29, whereas amino acid substitutions were used as clade markers just at N171K or N121K. Most of the rest viruses evolved away from the 3C.2a-A/Hong Kong/4801/2014-like clade by acquiring the genetic markers T131K, R142K and N121K/E, S144K which have been identified as being characteristic of clades 3C.2a.2 and 3C.2a.3. These markers were reported in most of other H3N2 influenza viruses isolated in the Denmark, Finland, Israel, Korean, Yokohama, Taiwan and the UK, in 2016/201730–36. Additionally, clade 3C.2a.3 was further grouped into cluster I (carrying I58V and S219Y) and cluster II (T135K and R150K). The substitutions in cluster I has appeared in report from the UK, and in cluster II emerged in the Taiwan putative clades 3C.2a.3a, which were isolated from severe patients. Members of the subclade 3C.2a.4 were characterized by two amino acid substitutions at R142G and S144R. Three other variants (D53N, I182T and Q197H) were not reported previously, which is likely to increases the odds of implications including alterations of the antigenic epitopes and immune escape. These reports, together with our results, suggested that subclusters within clade 3C.2a and eight subclades have emerged and expanded during this recent influenza epidemic. These changes were continuing to diversify worldwide with complicated and rapid dynamics.
During 2016/2017, the following two influenza epidemics occurred in this study: one epidemic between December 2016 and February 2017 and one epidemic between June 2017 and August 2017. These epidemics were consistent with the influenza epidemics observed in Hong Kong, Southeast Asia and Europe in 2017. According to the previous research on influenza seasons, Influenza cases usually appear between autumn and spring, with the influenza activity peaking after October37,38. However, the A(H3N2) virus seemed to frequently result in larger summer epidemics, such as those in 2010 and 201239,40. In our study, the H3N2-dominant summer wave occurred in 2017. The number of 2017 summer season was over than the 2016/2017 winter season. Research has shown that the I668V mutation in the PA subunit led to temperature-sensitive and attenuation in the 2016/2017 winter season virus strains and an adaptation to high temperatures in the 2017 summer viruses41. Adaptation to high temperatures may be the result of the natural evolution of the influenza virus and could explain the second epidemic that occurred during the summer of 2017 in this study.
Furthermore, we predicted the VE of the 2016/2017 season viruses by the p epitope model and found that predicted VE value to decline overall in 2016/2017. From 2016, A/Hong Kong/4801/2014 is regarded as the vaccine strain in the southern hemisphere, and was well-matched for the imported A(H3N2). Early and mid-season VE estimates for 2016/2017 showed that the A(H3N2) illness had observed antigenic drift and decreased predicted VE29. But in the 2016/2017, it was successively reported that the imported A(H3N2) strains had observed antigenic drift and decreasing predicted VE. Denmark and Finland’s studies indicated a drop in VE from the early period to the later period of the 2016/201742. Coincidentally, this phenomenon exists in 2017/2018 too. In USA analysis of 2017/2018 of A(H3N2) VE in prevention of hospitalization, VE was low across adult age groups and levels of frailty and chronic comorbidities43. We furthermore found that the main antigenic change is the substitution of amino acid 135, 150 and 131 in mid to late 2017 (Table S1). Amino acid 135 is located in a conserved region of the receptor-binding site in the antigenic epitope A and causes a loss of a glycosylation motif44. Changes in glycosylation motifs may be relevant to antigenicity, viral fitness and/or pathogenicity45. Amino acid 131 is located in the antigenic epitope A and conserved in 45% of all human H1, H2 and H3 viruses46. VE studies from Stockholm and Finland show that the proportion of samples containing T131K (36%) increased may be responsible for viral antigenic change and part of the observed VE drop33. Studies from northern Greece have provided evidence supporting indications that the specific T135K variant may be associated with vaccination failure. The T135K mutation was observed in 82% of the viruses originating from vaccinated patients44. Based on the Canadian report, the higher VE estimates may be due to the relatively infrequent (15%) circulation of the T135K variant in Canada47. T135K-R150K are also as genetic markers for clade 3C.2a.3, and viruses in this clade were isolated from patients with severe infections in Taiwan. This finding can be explained by the lower VE and shorter protection time for these clades compared with other circulating clades48. As a result, decreasing influenza vaccine protection with increasing time, A/Hong Kong/4801/2014 may not be as effective in eliciting immunity against future circulating A(H3N2) in the next influenza season. If the emerging A(H3N2) virus subpopulation continues to diversify from vaccine components, VE may decrease further by the end of the 2016/2017 season in terms of its antigenic properties or a broader cycle of variant strains.
In addition, compared to A/Hong Kong/4801/2014, the 3C.2a strains in our surveillance data had at least eleven potential N-glycosylation sites and had similar glycosylation patterns. Clade 3C.2a.1-like strains lost one potential glycosylation sites: 483(N483GT), Clade 3C.2a.2-like, 3C.2a.3-like, and 3C.2a.4-like strains gained one potential glycosylation site: 158(N158YT). Research shows that the mechanisms of glycosylation and deglycosylation in virus fall into three general classes: (i) mask antigenic epitopes and thus block binding to neutralizing antibodies49, (ii) adjust receptor-binding affinity50, or (iii) modulate the virulence of the Influenza viruses in mammals51. It’s could be concluded that the virus would make itself to elude the existed antibodies by changing herd immunity or increasing the viral fitness in an unknown mechanism.
In summary, our study indicates that new mutations and derivations of clade 3C.2a emerged continuously and rapidly during 2016/2017, which can reflect the trend of the current global influenza A(H3N2), and also has a paramount impact on the viral adaptation and transmission. So Shanghai might stop the risk of the international spread of influenza for the first time, and curbing the entry of A(H3N2) from overseas at the earliest stage of the influenza pandemic. Strengthening the entry monitoring of ports in coastal areas and improving the ability of ports health quarantine to deal with public health emergencis are required to investigate the antigenic effects and to avoid the next pandemic.
Methods
Ethics statements
This study was performed with the residual samples collected for the detection of influenza in the Shanghai Port. All the samples used for research have got the informed consent of passengers. The sample for the informed consent was attached as follows.
The study protocols were approved by the Institutional Review Board of Bio-X Life Science Research Center of Shanghai Jiao Tong University (IRB No. M202007). Informed consent was obtained from all subjects, and all methods were carried out in accordance with relevant guidelines and regulations.
Surveillance and data collection
The Shanghai port located on the south wing of the Yangtze River Delta Region. It is a key transportation hub combing water, land, and air transportation, facilitating the airborne survival and transmission of influenza viruses. During 2016/2017, a total of 64 throat swabs were collected from influenza-like illness cases in the Shanghai port. The basics epidemiological data including patients’ age, stopover sites and sampling dates were collected. The detailed list of viruses is provided in Table 2.
Table 2.
Influenza A(H3N2) virus strains sequenced in this study.
| Name | Patients’age | Nationality | Stopover sites | Sampling dates | Accession numbers |
|---|---|---|---|---|---|
| 201602161010.seq | 9 | China | Hong Kong | 2016.02.16 | OM956282 |
| 201608081702.seq | 29 | China | Australia, Hong Kong | 2016.08.08 | OM956281 |
| 201609171121.seq | 29 | China | Hong Kong, Australia | 2016.09.17 | OM956280 |
| 201610051316.seq | 31 | China | Hong Kong | 2016.10.05 | OM956279 |
| 201610241504.seq | 44 | China(Taiwan) | Japan, the USA, Hong Kong, Taiwan | 2016.10.24 | OM956257 |
| 201610251519.seq | 24 | China | Hong Kong | 2016.10.25 | OM956278 |
| 201611301787.seq | 25 | China | Hong Kong | 2016.11.30 | OM956277 |
| 201612141923.seq | 2 | China | Hong Kong | 2016.12.14 | OM956276 |
| 201612261085.seq | 20 | China | Hong Kong | 2016.12.26 | OM956275 |
| 201701051269.seq | 49 | Australia | Italy | 2017.01.05 | OM956256 |
| 201701071301.seq | 63 | The Netherlands | The Netherlands | 2017.01.07 | OM956255 |
| 201701111341.seq | 65 | China | Indonesia | 2017.01.11 | OM956228 |
| 201701121362.seq | 23 | China | The United Arab Emirates(UAE) | 2017.01.12 | OM956227 |
| 201701121363.seq | 62 | China | Vietnam | 2017.01.12 | OM956274 |
| 201701151395.seq | 38 | China | The USA | 2017.01.15 | OM956254 |
| 201701211465.seq | 13 | China | Britain | 2017.01.21 | OM956253 |
| 201701211466.seq | 53 | China | France | 2017.01.21 | OM956252 |
| 201701241505.seq | 3 | China | Italy | 2017.01.24 | OM956273 |
| 201701261555.seq | 36 | Germany | Germany | 2017.01.26 | OM956251 |
| 201701291611.seq | 30 | China | Macao | 2017.01.29 | OM956225 |
| 201702051704.seq | 10 | China | The USA, Hawaii | 2017.02.06 | OM956231 |
| 201702101743.seq | 58 | China | Iceland, Norway | 2017.02.10 | OM956250 |
| 201702121780.seq | 34 | China | Canada | 2017.02.12 | OM956249 |
| 201702131787.seq | 1 | The USA | The USA | 2017.02.13 | OM956248 |
| 201702211871.seq | 45 | China | Canada | 2017.02.22 | OM956247 |
| 201702211874.seq | 23 | Indonesia | Indonesia | 2017.02.21 | OM956258 |
| 201702221882.seq | 64 | Netherlands | Netherlands, France | 2017.02.22 | OM956246 |
| 201702261906.seq | 30 | China | The Netherlands | 2017.02.26 | OM956245 |
| 201703041965.seq | 30 | China | Germany, South Korea | 2017.03.06 | OM956244 |
| 201703081006.seq | 55 | Russia | Russia, Indonesia | 2017.03.08 | OM956243 |
| 201705111650.seq | 32 | Indonesia | Indonesia, Malaysia | 2017.05.11 | OM956224 |
| 201705191715.seq | 33 | China | Hong Kong | 2017.05.19 | OM956272 |
| 201706081022.seq | 67 | China | Turkey | 2017.06.08 | OM956242 |
| 201707231849.seq | 12 | China | Hong Kong | 2017.07.23 | OM956283 |
| 201707231857.seq | 59 | China | Hong Kong | 2017.07.23 | OM956271 |
| 201707241890.seq | 54 | China | Hong Kong, Thailand | 2017.07.24 | OM956270 |
| 201707261936.seq | 69 | China | Hong Kong | 2017.07.26 | OM956269 |
| 201707271994.seq | 21 | China | Macao, Hong Kong | 2017.07.27 | OM956268 |
| 201707271996.seq | 54 | China | Hong Kong | 2017.07.27 | OM956267 |
| 201707301041.seq | 8 | China | Indonesia | 2017.07.30 | OM956230 |
| 201707301044.seq | 72 | China | Russia | 2017.07.30 | OM956241 |
| 201708061243.seq | 21 | China | Japan | 2017.08.06 | OM956266 |
| 201708061245.seq | 26 | China | The USA, Canada | 2017.08.07 | OM956240 |
| 201708071293.seq | 38 | China | Hong Kong, Thailand | 2017.08.07 | OM956265 |
| 201708101387.seq | 20 | China | Hong Kong | 2017.08.10 | OM956264 |
| 201708111416.seq | 71 | China | France, Switzerland, Italy | 2017.08.11 | OM956239 |
| 201708111419.seq | 9 | China | Hong Kong, Macao | 2017.08.11 | OM956263 |
| 201708121441.seq | 5 | China | Hong Kong | 2017.08.12 | OM956262 |
| 201708131464.seq | 24 | China | South Korea | 2017.08.13 | OM956226 |
| 201708151516.seq | 76 | China | Russia | 2017.08.15 | OM956238 |
| 201708191682.seq | 16 | China | South Korea | 2017.08.19 | OM956223 |
| 201708221788.seq | 18 | China | Germany, France, Switzerland, Italy | 2017.08.22 | OM956237 |
| 201708241888.seq | 5 | China | Taiwan | 2017.08.24 | OM956221 |
| 201708251901.seq | 24 | China | Philippines, Hong Kong | 2017.08.25 | OM956220 |
| 201708251923.seq | 8 | China | France, Italy, Turkey | 2017.08.25 | OM956236 |
| 201708261954.seq | 22 | China | Hong Kong, Thailand | 2017.08.26 | OM956261 |
| 201708261955.seq | 19 | China | Hong Kong | 2017.08.26 | OM956260 |
| 201708281998.seq | 62 | China | Russia | 2017.08.28 | OM956235 |
| 201708301087.seq | 18 | China | Taiwan | 2017.08.30 | OM956229 |
| 201709041192.seq | 34 | China | Hong Kong, Australia | 2017.09.04 | OM956259 |
| 201709131430.seq | 46 | Poland | Poland | 2017.09.13 | OM956232 |
| 201709231691.seq | 30 | South Korea | Indonesia, South Korea | 2017.09.25 | OM956222 |
| 201709261774.seq | 33 | India | Hong Kong, India | 2017.09.26 | OM956234 |
| 201709301835.seq | 25 | China | Hong Kong | 2017.10.03 | OM956233 |
RNA purification and HA gene sequencing
Viral RNA was obtained from 140 μL of the sample by the QIAMP viral RNA extraction kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. RNA was eluted in 100 μl RNase/DNase elution buffer provided in the kit and stored at − 40 °C. Samples were subtyped based the Real-Time RT-PCR Diagnostic assay (Shanghai Biogerm Influenza H1/H3 typing assay) for detection of influenza virus in an ABI Prism 7500 thermocycler (Applied Biosystems, Foster City, California, USA). The HA genes were subsequently amplified by the samples with a Ct value of less than 30, and were subjected to reverse transcription and amplification using a PrimeScript One-step RT-PCR Kit Ver.2 (TaKaRa, Japan) as previously described52. RT-PCR products with the correct band size were selected by agarose gel electrophoresis and sequenced using ABI PRISM Dye Deoxy Terminator cycle sequencing kit (Life Technology, Foster City, CA, USA). These HA gene sequences were assembled using the SeqMan Pro software (DNASTAR, Madison, WI) and deposited in the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) with the accession numbers (OM956220-OM956283). Finally we obtained 64 full-length HA sequences.
Sequence alignment and phylogenetic analysis
Reference sequences of known clades and vaccine strains (the accession numbers in Supplementary Table S2) as recommended by WHO53 included in the evolutionary analysis were retrieved from GISAID EpiFlu databases. All HA gene sequences aligned by MAFFT program (MAFFT-7.220-WIN64 version). Protein translation were performed on the basis of nucleotide sequences using Molecular Evolutionary Genetics Analysis software (MEGA, version 6.0; http://www.megasoftware.net/). Phylogenetic tree was constructed using the maximum-likelihood method with a Hasegawa-Kishino-Yano (HKY) + gamma nucleotide substitution model and 1000 bootstrap replications. The ability to perform clade designations based on signature amino acids as compared to A/HongKong/4801/2014-likeA/H3N2-like clade 3C.2a viruses and A/Singapore/INFIMH-16-0019/2016-likeA/H3N2-like clade 3C.2a.1 viruses was confirmed with the 64 isolates depicted in the final tree, and extended to the other 546 isolates.
Prediction of glycosylation sites
The prediction of potential N-liked glycosylation sites was performed with an online server: NetNGlyc 1.054. This server considers the amino acid alignment N-X-S/T, where X can be any amino acid except Asp or Pro. A threshold value of > 0.5 suggests glycosylation.
Prediction of vaccine efficacy
The predicted VE of A(H3N2) was estimated using the Pepitope model, which is a measure of the antigenic distance between vaccine and circulating strains. Antigenic distance was calculated from the fraction of substituted amino acid residues in the dominant HA epitope21. The amino acid residues in the HA epitopes of A(H3N2) were pre-defined in the p epitope calculator to respectively possess 19, 21, 27, 41 and 22 amino acids55. The mathematical formula linking VE and the p epitope is given by VE = −2.47 × p epitope + 0.47 in which VE is 47% when p epitope = 0.
Supplementary Information
Acknowledgements
This study was supported by the National Science and Technology Major Project (2018ZX10101003-002-005), Shanghai Science and Technology Committee Project (18495810900) and Collaborative Research Grant KLMVI-OP-201903 of CAS Key Laboratory of Molecular Virology & Immunology, Institut Pasteur of Shanghai, Chinese Academy of Sciences.
Author contributions
Z.T conceived and designed the study; Z.Z. performed the experiments; S.L. analyzed data and discussed the results; X.Z. wrote the manuscript; J.H. and H.Z. collected the samples, B.Z. revised the manuscript.
Data availability
The datasets generated and analysed during the current study are available in the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/), ACCESSION NUMBER TO DATASETS: OM956220-OM956283.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zilong Zhang, Shenwei Li and Xiaolin Zhu
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-19228-y.
References
- 1.Hause BM, et al. Isolation of a novel Swine influenza virus from Oklahoma in 2011 which is distantly related to human influenza C viruses. PLoS Pathog. 2013;9:e1003176. doi: 10.1371/journal.ppat.1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mills CE, Robins JM, Lipsitch M. Transmissibility of 1918 pandemic influenza. Nature. 2004;432:904–906. doi: 10.1038/nature03063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stuart-Harris CH. Virus of the 1918 influenza pandemic. Nature. 1970;225:66. doi: 10.1038/225850a0. [DOI] [PubMed] [Google Scholar]
- 5.Laver WG, Air GM, Dopheide TA, Ward CW. Amino acid sequence changes in the haemagglutinin of A/Hong Kong (H3N2) influenza virus during the period 1968–77. Nature. 1980;283:66. doi: 10.1038/283454a0. [DOI] [PubMed] [Google Scholar]
- 6.European Centre for Disease Prevention and Control (ECDC). Influenza virus characterization, summary Europe, March 2012. http://ecdc.europa.eu/en/publications/Publications/1204-TED-CNRL-report.pdf. Accessed 1 August 2017.
- 7.Chambers BS, Parkhouse K, Ross TM, Alby K, Hensley SE. Identification of hemagglutinin residues responsible for H3N2 antigenic drift during the 2014–2015 influenza season. Cell Rep. 2015;12:1–6. doi: 10.1016/j.celrep.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.European Centre for Disease Prevention and Control. Influenza virus characterisation, summary Europe. Stockholm: ECDC; 2015. http://ecdc.europa.eu/en/publications/Publications/influenza-virus-characterisation-June-2015.pdf.
- 9.Stellrecht KA. Incidence of matrix genes mutations affecting PCR tests among influenza H3N2 clades circulating during the 2014/15 season. Diagn. Microbiol. Infect. Dis. 2018;91:239–244. doi: 10.1016/j.diagmicrobio.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 10.Review of the 2014 influenza season in the southern hemisphere. Wkly. Epidemiol. Rec.89, 529–541 (2014). [PubMed]
- 11.European Centre for Disease Prevention and Control. Influenza virus characterisation, Summary Europe, September 2016 2016 14 Feb 2017. http://ecdc.europa.eu/en/publications/Publications/influenza-virus-characterisation-september-2016.pdf
- 12.Yang J-R, Hsu S-Z, Kuo C-Y, Huang H-Y, Huang T-Y, Wang H-C. Liu M-T An epidemic surge of infuenza A(H3N2) virus at the end of the 2016–2017 season in Taiwan with an increased viral genetic heterogeneity. J. Clin. Virol. 2018;15:99–100. doi: 10.1016/j.jcv.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 13.Glatman-Freedman A, Drori Y, Beni SA, et al. Genetic divergence of influenza A (H3N2) amino acid substitutions mark the beginning of the 2016–2017 winter season in Israel. J. Clin. Virol. 2017;93:71–75. doi: 10.1016/j.jcv.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trebbien R, Fischer TK, Krause TG, et al. Changes in genetically drifted H3N2 influenza A viruses and vaccine effectiveness in adults 65 years and older during the 2016/17 season in Denmark. J. Clin. Virol. 2017;94:1–7. doi: 10.1016/j.jcv.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 15.Epidemiology and risk assessment of seasonal influenza during the summer of 2017 in China. Chin. J. Viral Dis.4, 8–15 2017.
- 16.National Institute for Health and Welfare. Finland. Influenssa leviää nyt – rokotuksen ehtii ottaa vielä. Helsinki: THL; 2017. Finnish. https://www.thl.fi/en/web/infektiotaudit/-/influenssa-leviaa-nyt-rokotuksen-ehtii-ottaa-viela.
- 17.Säsongsstatistik för influensa 2016/2017. Stockholm: Stockholm Smittskydd; 2017.http://www.vardgivarguiden.se/globalassets/behandlingsstod/smittskydd/statistik/influensa/influensasasongen-2016-2017.pdf.
- 18.Grohskopf LA, et al. Prevention and control of seasonal influenza with vaccines. MMWR Recommend. Rep. 2016;65:1–54. doi: 10.15585/mmwr.rr6505a1. [DOI] [PubMed] [Google Scholar]
- 19.Li TZ, Zheng R, Yv F, Zhang Q, Guo CP, Liang LC, et al. Clinical analysis of 979 cases of influenza A patients. Chin. J. Med. 2016;51:50–51. [Google Scholar]
- 20.Grais RF, Ellis JH, Glass GE. Assessing the impact of airline travel on geographic spread of pandemic influenza. Eur. J. Epidemiol. 2003;18:1065–1072. doi: 10.1023/A:1026140019146. [DOI] [PubMed] [Google Scholar]
- 21.Gupta V, Earl DJ, Deem MW. Quantifying influenza vaccine efficacy and antigenic distance. Vaccine. 2006;24:3881–3888. doi: 10.1016/j.vaccine.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tewawong N, et al. Assessing antigenic drift of seasonal influenza A(H3N2) and A(H1N1)pdm09 viruses. PLoS ONE. 2015;10:e0139958. doi: 10.1371/journal.pone.0139958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bahl J, Nelson MI, Chan KH, et al. Temporally structured metapopulation dynamics and persistence of influenza A H3N2 virus in humans. Proc. Natl. Acad. Sci. 2011;108:19359–19364. doi: 10.1073/pnas.1109314108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shanghai Municipai Office For Port Services. 5.25 million people enjoy 10-second self-service for customs clearance in Shanghai Port in 2017.2018. http://kab.sh.gov.cn/gzzt/002007/002007002/20180205/7813c833-abed-4dce-9dde-d024110da26a.html
- 25.Jiang ZY, Bai JJ, Cai J, Li RY, Jin ZY, Xu B. Analysis of temporal and spatial distribution characteristics of 2009 influenza A (H1N1) pandemic. J. GeoInf. Sci. 2012;14:794–798. [Google Scholar]
- 26.Tsou TP, Su CP, Huang WT, Yang JR, Liu MT. Influenza A(H3N2) virus variants and patient characteristics during a summer influenza epidemic in Taiwan, 2017. Euro Surveill. 2017;22:17–00767. doi: 10.2807/1560-7917.ES.2017.22.50.17-00767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan SG, Chilver MB, Carville KS, Deng YM, Grant KA, Higgins G, et al. Low interim influenza vaccine effectiveness, Australia, 1 May to 24 September 2017. Euro Surveill. 2017;22:66. doi: 10.2807/1560-7917.ES.2017.22.43.17-00707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skowronski DM, Chambers C, De Serres G, Dickinson JA, Winter AL, Hickman R, et al. Early season co-circulation of influenza A(H3N2) and B(Yamagata): Interim estimates of 2017/18 vaccine effectiveness, Canada, January 2018. Euro Surveill. 2018;23:66. doi: 10.2807/1560-7917.ES.2018.23.5.18-00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melidou A, Broberg E. European region influenza surveillance network. Predominance of influenza A(H3N2) virus genetic subclade 3C.2a1 during an early 2016/17 influenza season in Europe-Contribution of surveillance data from World Health Organization (WHO) European Region to the WHO vaccine composition consultation for northern hemisphere 2017/18. Vaccine. 35, 4828–35 (2017) [DOI] [PubMed]
- 30.Trebbien R, Fischer TK, Krause TG, Nielsen L, Nielsen XC, Weinreich LS, et al. Changes in genetically drifted H3N2 influenza A viruses and vaccine effectiveness in adults 65 years and older during the 2016/17 season in Denmark. J Clin. Virol. 2017;94:1–7. doi: 10.1016/j.jcv.2017.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Glatman-Freedman A, Drori Y, Beni SA, Friedman N, Pando R, Sefty H, et al. Genetic divergence of Influenza A(H3N2) amino acid substitutions mark the beginning of the 2016–2017 winter season in Israel. J. Clin. Virol. 2017;93:71–75. doi: 10.1016/j.jcv.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harvala H, Frampton D, Grant P, Raffle J, Ferns RB, Kozlakidis Z, et al. ICONIC Consortium. Emergence of a novel subclade of influenza A(H3N2) virus in London, December 2016 to January 2017. Euro Surveill. 2017;22:66. doi: 10.2807/1560-7917.ES.2017.22.8.30466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hergens MP, Baum U, Brytting M, et al. Mid-season real-time estimates of seasonal influenza vaccine effectiveness in persons 65 years and older in register-based surveillance, Stockholm County, Sweden, and Finland, January 2017. Eurosurveillance. 2017;22:66. doi: 10.2807/1560-7917.ES.2017.22.8.30469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawakami Chiharu,Yamayoshi Seiya,Akimoto Miki et al. Genetic and antigenic characterisation of influenza A(H3N2) viruses isolated in Yokohama during the 2016/17 and 2017/18 influenza seasons.Euro Surveill.24, (2019). [DOI] [PMC free article] [PubMed]
- 35.Il KJ, Sehee P, Hyuk C, et al. Distinct molecular evolution of influenza H3N2 strains in the 2016/17 season and its implications for vaccine effectiveness. Mol. Phylogenet. Evol. 2019;131:29–34. doi: 10.1016/j.ympev.2018.10.042. [DOI] [PubMed] [Google Scholar]
- 36.Yang J-R, et al. An epidemic surge of influenza A (H3N2) virus at the end of the 2016–2017 season in Taiwan with an increased viral genetic heterogeneity. J. Clin. Virol. 2018;99:15–2. doi: 10.1016/j.jcv.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 37.Saha S, Chadha M, Shu Y. Divergent seasonal patterns of influenza types A and B across latitude gradient in Tropical Asia. Influenza other respir. Virus. 2016;10:176–184. doi: 10.1111/irv.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu H, et al. Characterization of regional influenza seasonality patterns in China and implications for vaccination strategies: Spatio-temporal modeling of surveillance data. PLoS Med. 2013;10:e1001552. doi: 10.1371/journal.pmed.1001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Center for Disease Control and Prevention. Influenza Express Weekly, Influenza Season, 2009–2017 (2017) http://www.cdc.gov.tw/english/submenu.aspx?treeid=00ed75d6c887bb27&nowtreeid=8f1e239d0fd8877a
- 40.Yang JR, Cheng CY, Chen CY, Lin CH, Kuo CY, Huang HY, et al. A virus-like particle vaccination strategy expands its tolerance to H3N2 antigenic drift by enhancing neutralizing antibodies against hemagglutinin stalk. Antiviral Res. 2017;140:62–75. doi: 10.1016/j.antiviral.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 41.Wei D, Yu D-M, Wang M-J, et al. Genome-wide characterization of the seasonal H3N2 virus in Shanghai reveals natural temperature-sensitive strains conferred by the I668V mutation in the PA subunit. Emerg. Microbes Infect. 2018;171:66. doi: 10.1038/s41426-018-0172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maria-Pia H, Ulrike B, Mia B, et al. Mid-season real-time estimates of seasonal influenza vaccine effectiveness in persons 65 years and older in register-based surveillance, Stockholm county, Sweden, and Finland, January 2017. Eurosurveillance. 2017;6:66. doi: 10.2807/1560-7917.ES.2017.22.8.30469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin ET, Cheng C, Petrie JG, Elif A, Manjusha G, Middleton DB, et al. Low influenza vaccine effectiveness against a(h3n2)-associated hospitalizations in 2016–2017 and 2017–2018 of the hospitalized adult influenza vaccine effectiveness network (Haiven) J. Infect. Dis. 2020;223:2062–2071. doi: 10.1093/infdis/jiaa685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melidou A, Gioula G, Exindari M, Ioannou E, Gkolfinopoulou K, Georgakopoulou T, et al. Influenza A(H3N2) genetic variants in vaccinated patients in northern Greece. J. Clin Virol. 2017;94:29–32. doi: 10.1016/j.jcv.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 45.An Y, Mccullers JA, Alymova I, Parsons LM, Cipollo JF. Glycosylation analysis of engineered h3n2 influenza a virus hemagglutinins with sequentially added historically relevant glycosylation sites. J. Proteome Res. 2015;14:3957–3969. doi: 10.1021/acs.jproteome.5b00416. [DOI] [PubMed] [Google Scholar]
- 46.Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature. 2012;489:66. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skowronski DM, Chambers C, Sabaiduc S, Dickinson JA, Krajden M. Interim estimates of 2016/17 vaccine effectiveness against influenza a(h3n2), Canada, January 2017. Eurosurveillance. 2017;22:66. doi: 10.2807/1560-7917.ES.2017.22.6.30460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsou TP, Su CP, Huang WT, Yang JR, Liu MT. Influenza A(H3N2) virus variants and patient characteristics during a summer influenza epidemic in Taiwan, 2017. Euro Surveill. 2017;22:17–00767. doi: 10.2807/1560-7917.ES.2017.22.50.17-00767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Long J, Bushnell RV, Tobin JK, Pan K, Deem MW, Nara PL, Tobin GJ. Tobin GJ Evolution of H3N2 influenza virus in a guinea pig model. PLoS ONE. 2011;6:e20130. doi: 10.1371/journal.pone.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Owen RE, Yamada E, Thompson CI, Phillipson LJ, Thompson C, Taylor E, Zambon M, Osborn HM, Barclay WS. Borrow P Alterations in receptor binding properties of recent human influenza H3N2 viruses are associated with reduced natural killer cell lysis of infected cells. J. Virol. 2007;81:11170–11178. doi: 10.1128/JVI.01217-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spackman E, Senne DA, Myers TJ, Bulaga LL, Garber LP, Perdue ML, Lohman K, Daum LT. Suarez DL Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 2002;40:3256–3260. doi: 10.1128/JCM.40.9.3256-3260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 53.WHO. WHO Recommendations on the Composition of Influenza Virus Vaccines 2018. http://www.who.int/influenza/vaccines/virus/recommendations/en/
- 54.Prediction of N-glycosylation sites in human proteins. [Internet]. 2004. http://www.cbs.dtu.dk/services/NetNGlyc/.
- 55.Bonomo ME, Deem MW. Predicting influenza H3N2 vaccine efficacy from evolution of the dominant epitope. Clin. Infect. Dis. 2018;67:1129–1131. doi: 10.1093/cid/ciy323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed during the current study are available in the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/), ACCESSION NUMBER TO DATASETS: OM956220-OM956283.