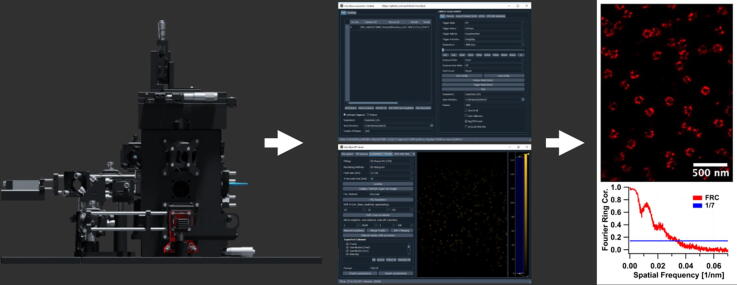
Graphical abstract
Keywords: Super-resolution, Fluorescence microscopy, Open-source, Single-molecule, TIRF, Industrial-grade CMOS
Abstract
Commercial super-resolution (SR) imaging systems require a high budget, while current more affordable open source microscopy systems lack modularity and sometimes are too complex or lack reliability. We present miEye – a cost-effective microscope designed for high-resolution wide-field fluorescence imaging. The build is constructed using a CNC milled aluminum microscope body and commercially available optomechanics, with open-source Python-based microscope control, data visualization, and analysis software integration. The data acquisition software works robustly with commonly used industrial-grade complementary metal oxide semiconductor (iCMOS) cameras, performs IR beam back-reflection-based automatic focus stabilization, and allows for laser control via an Arduino-based laser relay. The open-source nature of the design is aimed to facilitate adaptation by the community. The build can be constructed for a cost of roughly 50 k €. It contains SM-fiber and MM-fiber excitation paths that are easy to interchange and an adaptable emission path. Also, it ensures <5 nm/min stability of the sample on all axes, and allows achieving <30 nm lateral resolution for dSTORM and DNA-PAINT single-molecule localization microscopy (SMLM) experiments. Thus it serves as a cost-effective and adaptable addition to the open source microscopy community and potentially will allow high-quality SR imaging even for limited-budget research groups.
Specifications table.
| Hardware name | miEye |
| Subject area | Biological sciences (e.g., microbiology and biochemistry) |
| Hardware type | Imaging tools |
| Closest commercial analog | openFRAME by Cairn Research: https://www.cairn-research.co.uk/product/openframe-microscope/ |
| Open source license | CC BY-NC-SA 4.0 This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License |
| Cost of hardware | ∼50,000 € |
| Source file repository | OSF identifier: https://doi.org/10.17605/osf.io/j2fqy |
| OSHWA certification UID (OPTIONAL) |
Hardware in context
The SR microscopy enables biomedical researchers to obtain structural and functional information on biomolecules that were inaccessible by any other methods [1], [2]. This opens doors to ground-breaking discoveries both in living cells or organisms and in vitro [3]. However, SR microscopy is a high-end approach both hardware- and software-wise. There are commercial microscopy systems from major microscope manufacturers and also from smaller companies that can do both data acquisition and analysis but they are expensive, heavy-weighted, slow, lack adaptability, and still require skilled staff to operate them properly.
A common way to deal with the cost issues of commercial SR microscopy systems is to use shared equipment. However, often such an approach reduces flexibility in experimental design, puts constraints on the experimental schedule, and still applies usage fees [4]. In addition to that, the user must rely on specific light sources, filters, cameras, and usually dedicated software for microscope control offered by the provider of the commercial system. Therefore in many laboratories, SR microscopy set-ups are custom-made using commercially available microscope bodies [5]. Such solutions sometimes are cheaper, and allow for almost unlimited flexibility around hardware and software but require the involvement of dedicated highly skilled programmers and engineers which research labs typically do not invest into hiring due to limited funding or positions. There are a number of commercial software solutions (e.g. NIS Elements, Metamorph, etc.) for microscope control and data acquisition but it is expensive and hardly compatible with custom-microscopy set-ups.
To share the knowledge obtained when building their own, often innovative and advanced, microscopy set-ups researchers from various places opened up their designs by uploading them into publicly available repositories and describing how to build and operate them [6]. There are dozens of such innovative and advanced open-source microscopy approaches for various kinds of SR techniques already available and nicely summarized on the open microscopy website [7]. The notable SMLM approaches that work even at densely labeled samples from this list include: cellSTORM [8], [9], easySTORM [10], K2TIRF [11], lifeHack [12], liteTIRF [13], LSFM [14], miCube [15]. Not all of these examples include cost-effective solutions for light sources and detectors. General limitations of those systems are related to insufficient documentation, compatibility with open-source software, and the extremely time-consuming technical support which is usually not incentivized by the current funding and publication environment.
Data acquisition and analysis software for SR microscopy is developing rapidly. There are either Java-, Python-, or MATLAB-based open-source solutions for SR microscopy data acquisition and analysis that are summarized on the open microscopy website [7]. ImageJ and FiJi can be considered key open-source projects dedicated to image analysis [16], [17]. Now the Python-based Napari platform [18], [19] is gaining popularity as a tool for browsing, annotating, and analyzing large multi-dimensional datasets. Also, there is a highly advanced MATLAB-based data analysis package SMAP containing hundreds of plugins [20]. μManager is a project for microscope control and data acquisition that uses ImageJ as its base [21]. There are several Python-based developments for microscope control like ImSwitch [22], Microscope-Cockpit [23]. Some of them, like the Python microscope or PYME can perform both data acquisition and analysis or are close to that [22], [24], [25], [26]. Java-based projects like ImageJ, µManager, PycroManager [27], and others can be challenging as they have a higher entry barrier than Python for code adjustments and debugging. Python is becoming a more favorable option for scientists with its low entry barrier, and a large selection of maintained packages, especially with recent interests in machine learning.
The success of the famous ThunderSTORM plugin [28] for FiJi/ImageJ clearly demonstrates that simple and dedicated task projects are powerful and highly valuable although they might not be maintained anymore. Based on this and previously developed open-microscopy approaches one could tell that there is a demand in the SR microscopy community for bench-top SMLM microscopy systems that would be open-source, cost-effective, modular, utilize low-cost commercial products, achieve high-resolution, and have dedicated, preferably Python-based reliable software.
Hardware description
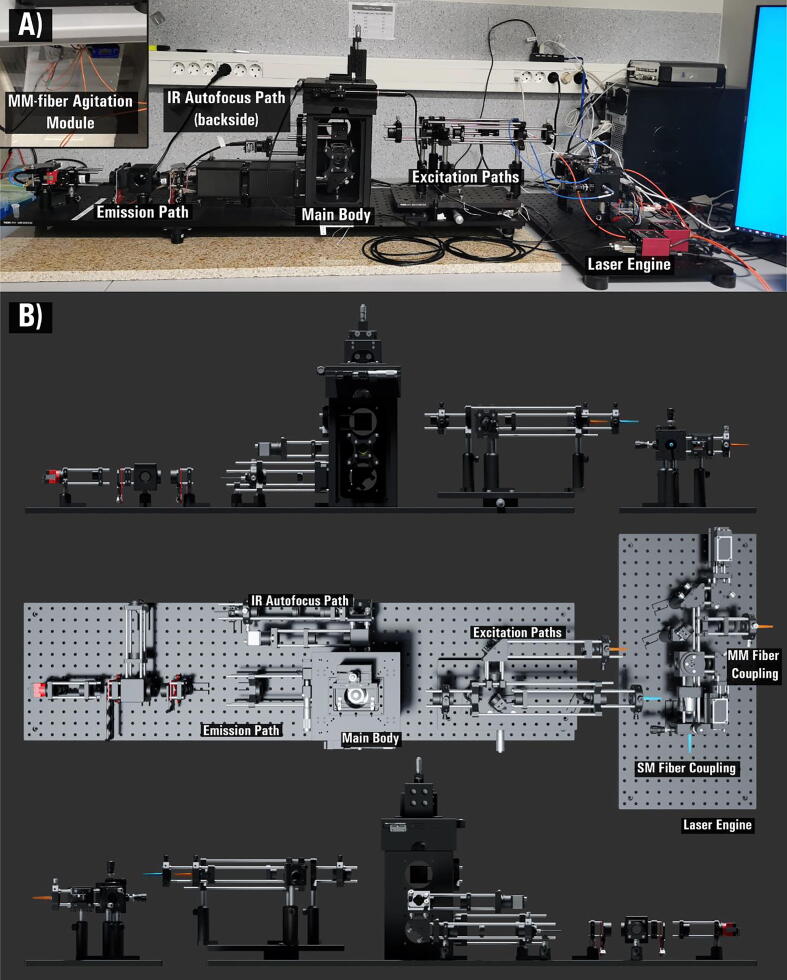
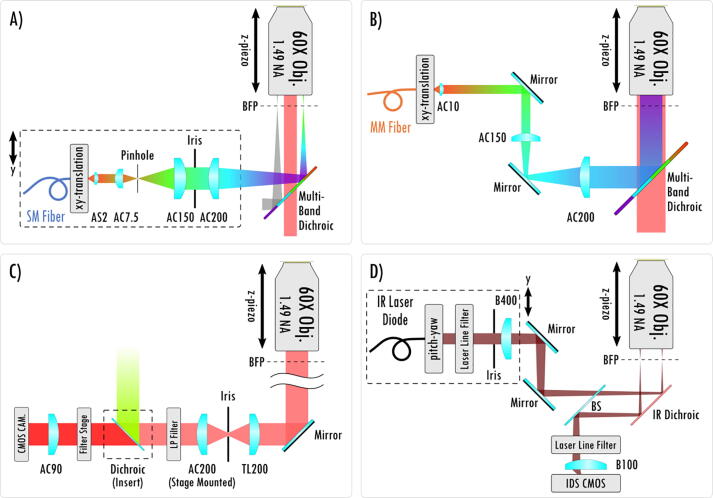
Here we present an open-source hardware and software framework entitled miEye. This is a bench-top cost-effective modular bespoke fluorescence microscopy system for SMLM studies of biomolecule structures and dynamics. We based the project on the recently developed open-source miCube platform [15]. In contrast to the original miCube, the miEye system has a three-layer microscope body adding a mid-layer for the IR path while preserving the recommended distance between the objective and its tube lens, alongside a cost-effective modular laser engine that can provide both single-mode fiber (SMF) and multi-mode fiber (MMF) outputs (Fig. 1A). It leverages two types of excitation schemes: 1) SMF objective-based TIRF (that also works for epi- and HILO), and 2) MMF output in widefield configuration on the sample plane for homogeneous illumination (Fig. 2A-B). Also, it contains a sample autofocus stabilization system and a modular emission path (Fig. 1B, Fig. 2C-D). Since miEye is an open-microscopy project, we maintain its GitHub repository [29], and our lab’s website [30]. The original miCube system has a two-layer microscope body with only SMF excitation for TIRF, HILO, or epi-mode excitation. Its emission path equipped with a single scientific complementary metal-oxide semiconductor (sCMOS) camera was less modular than ours. Also, it had no sample autofocusing system.
Fig. 1.
The open-source miEye single-molecule localization microscope. A) Photograph of the fully assembled miEye set-up with key features annotated. B) Render of the miEye with key features annotated. Top: front view, middle: top-side view, bottom: back-side view.
Fig. 2.
Schematic overview of the miEye instrument. A) The excitation path via single-mode fiber (SMF) for TIRF-, HILO-, and Epi-mode. B) The excitation path via multi-mode fiber (MMF) for Epi-mode when imaging MMF output on the sample plane. C) The fluorescence emission path. D) IR laser-based autofocusing path. AC: achromat lens, AS: aspheric lens, BFP: back-focal plane, TL: tube lens, B: B-coated N-BK7 optics, BS: beamsplitter.
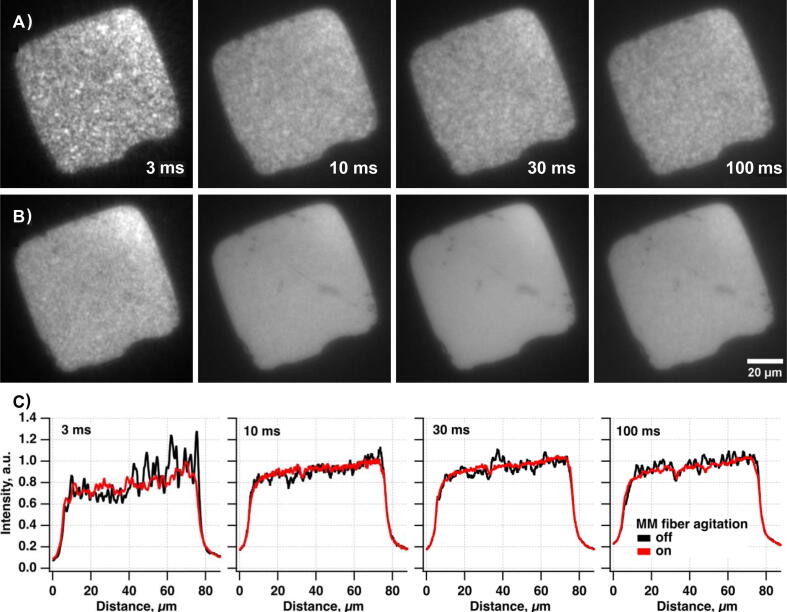
In our system, we employ inexpensive but highly capable industrial-grade CMOS (iCMOS) cameras. There are several SR set-ups published that employed the iCMOS as the main detectors [31], [32]. Recently a procedure for their characterization and calibration that helps to improve SR microscopy results was published [33]. We pair high-frame-rate iCMOS cameras, the possibility for simultaneous dual-channel imaging, and illumination via a 4-wavelength (405, 488, 520, 638 nm) laser diode combiner to maximize their utility (Fig. 2C). Also to improve the beam quality we pass the output of the free-space laser combiner through either MMF or SMF. The MMF agitator module (sometimes called mode-scrambler) is used in the case of the MMF excitation. It helps to minimize the speckles and ensures an even illumination profile. Our MMF agitator is built according to the previously published instructions [34]. Similar published alternatives like miCube [15] or liteTIRF [13] had no autofocusing capabilities. Also, they had either an SMF-based laser engine or a single laser source for TIRF-type excitation.
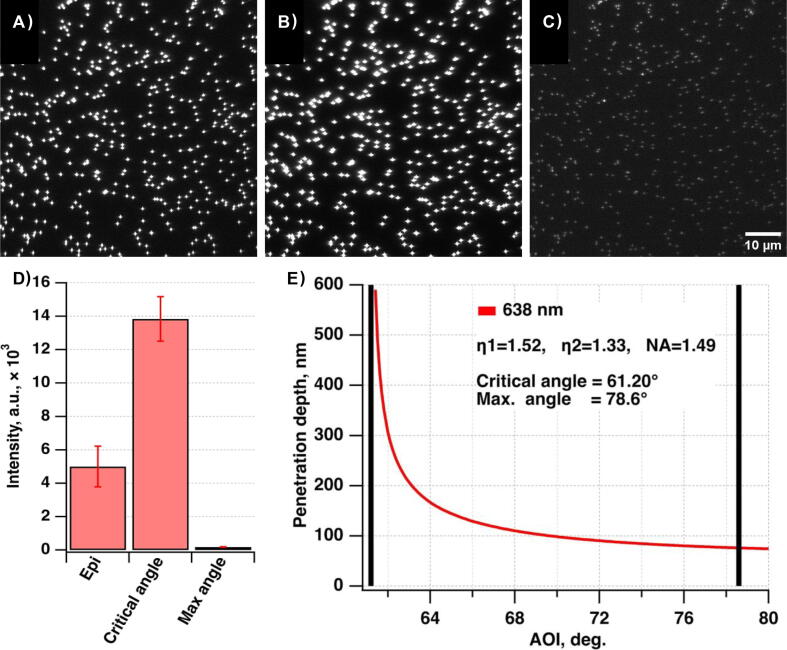
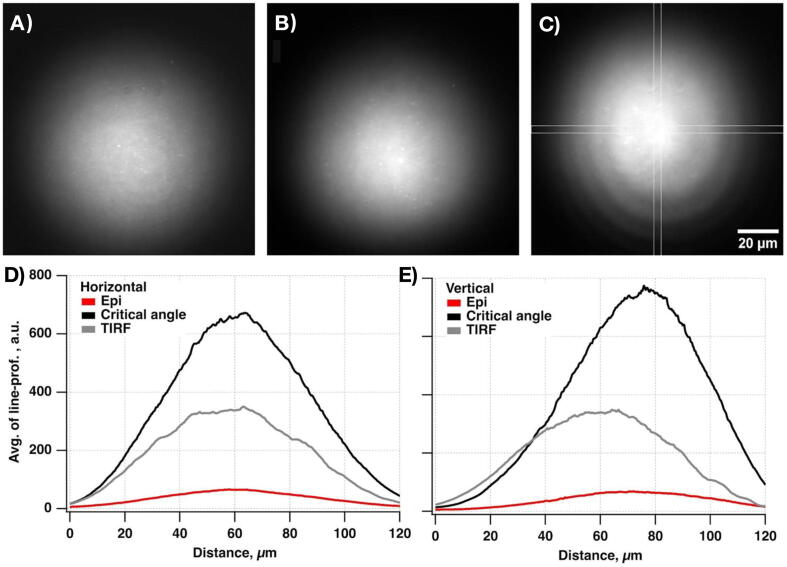
The TIRF system is well suited to imaging molecules near the coverslip surface in the presence of a fluorescent background from other fluorescent molecules diffusing in solution. It increases sensitivity and lowers noise by reducing background signals from fluorescently labeled molecules in the solution. The MMF output formation on the sample plane combined with a home-made speckle-reducer [34] creates a homogeneous flat illumination profile with a reduced risk of objective damage (the laser beam is not focused at the back focal plane of the objective in contrast with the TIRF configuration) and decreased laser power loss due to higher coupling efficiency than SMF. Expected laser coupling efficiencies for the SMF and MMF (Table 1.) demonstrate the advantages of introducing the MMF setting. Alternative settings of SMF combined with a refractive beam-shaper were not considered due to its higher cost and increased system complexity (e.g., output beam profile dependence on input beam diameter). Both excitation schemes work well for SMLM super-resolution imaging, whether in vitro or in cells. The only exception is that the TIRF scheme works better for DNA-PAINT [35] or similar methods [36], [37], [38] aiming to visualize reversibly binding fluorescent molecules while containing diffusing fluorescent molecules in solution.
Table 1.
Laser coupling efficiencies for the SMF and MMF.
| Wavelength [nm] | SMF coupling efficiency [%] | MMF coupling efficiency [%] |
|---|---|---|
| 638 | 46.3 | 71.0 |
| 520 | 37.1 | 69.0 |
| 488 | 16.6 | 67.1 |
| 405 | 8.6 | 60.0 |
The aforementioned attractive solutions that we introduced required us to develop Python-based microscope control, data acquisition, and data analysis modules. Note that μManager was discarded as it suffered from reoccurring crashes when combined with iCMOS cameras and adjusting or implementing any new features or hardware adapters (e.g., automatic focus stabilization functionality) posed an extremely time-consuming task. PYME did not offer drivers for the hardware we were implementing at the time and to avoid pipeline restrictions we did not adopt it. We added the modules into a single unified microEye Python package that is routinely maintained and updated [29], [39]. The microscope control module allows for controlling hardware components and performs sample focus stabilization (autofocusing) to counter sample drift in the Z-axis using a closed-feed-back loop between the IR laser position and the objective piezo stage. TiffViewer module offers interactive data processing and analysis, such as filtering of acquired images, cropping, and Fourier ring correlation (FRC) to estimate system resolution, thresholding, blob detection, and similar. The microEye is entirely Python-based and aims to be user-friendly. At the time of writing this manuscript an experimental adaptation of the C-written CPU/GPU-accelerated fit3Dcspline that is included in the Matlab-based superresolution microscopy analysis platform (SMAP) [40] was added purely in Python. μManager [21], Python microscope [24], or ImSwitch [22] are limited in terms of data analysis, ImageJ/FiJi [16], [17], and Napari [18], [19] have no data acquisition or microscope control possibilities. PYME [25] is the closest project to our microEye.
The miEye approach brings the possibility to perform high-end SR microscopy more simply with bench-top hardware and dedicated reliable software. A user is presented with a less complex interface, so they can focus on experimental design and obtaining data. A similar commercial system would cost >150 thousand Euro, while our system price is approximately 50 thousand Euro. This is even cheaper than what was published previously for other similar capabilities of SMLM open-source hardware set-ups. Thus, it opens doors for cutting-edge biomedical SR microscopy-related discoveries even with a limited budget.
Potential uses of the miEye project:
-
●
Use as a regular fluorescence microscope for various samples’ imaging.
-
●
Use for single-molecule fluorescence and super-resolution microscopy.
-
●
Use the software for controlling various bespoke microscopy systems.
Design files summary
| Design file name | File type | Open source license | Location of the file |
|---|---|---|---|
| miEye_V2.0.2 | FreeCAD Standard file format (.FCStd) | CC BY-NC-SA 4.0 | https://doi.org/10.17605/osf.io/j2fqy |
| M38_SM2_Tubelens_adapter_v2.1 | FreeCAD Standard file format (.FCStd) | CC BY-NC-SA 4.0 | https://doi.org/10.17605/osf.io/j2fqy |
| XYZ_stage_adapter | FreeCAD Standard file format (.FCStd) | CC BY-NC-SA 4.0 | https://doi.org/10.17605/osf.io/j2fqy |
| Setup_3D_Model | Blender file (.blend) in compressed archive (.zip) | CC BY-NC-SA 4.0 | https://doi.org/10.17605/osf.io/j2fqy |
| microEye | Compressed Python package (.zip) | GNU General Public License v3 (GPLv3) | https://doi.org/10.17605/osf.io/j2fqy |
| RelayBox | Compressed Arduino project (.zip) | GNU General Public License v3 (GPLv3) | https://doi.org/10.17605/osf.io/j2fqy |
miEye_V2.0.2: a CNC milled aluminum block finished with sanding and black anodization to be mounted onto the aluminum breadboard as the main miEye microscope body.
M38_SM2_Tubelens_adapter_v2.1: helps to mount the commonly used Nikon tube lens within “miEye_V2.0.2” closer to the objective to keep it within the recommended distance.
XYZ_stage_adapter: a CNC milled aluminum part finished with sanding and black anodization to mount the XYZ sample stage onto the main miEye microscope body.
Setup_3D_Model: 3D model of the miEye laser engine and the microscope.
microEye: a python toolkit for fluorescence microscopy that features iCMOS cameras. The Acquisition Module allows multi-cam image acquisition within one graphical user interface. The Control Module allows setting the laser excitation presets, manual focus, and automatic focus stabilization by monitoring the peak position of a totally internally reflected IR beam and moving the piezo stage accordingly. The Tiff Viewer Module allows accessing tiff images (single file and sequences) of order TYX (2D SMLM), and also allows for visualizing the filtering-localization process interactively.
Bill of materials summary
Bill of materials is uploaded as an editable spreadsheet to an open access online location: https://doi.org/10.17605/osf.io/j2fqy.
Please note that suppliers mentioned here are just suggestions. Material costs may vary over time and depend on location, and alternative sources may provide better prices than those listed here.
Build instructions
This guide will walk you through the process of assembling our fluorescence microscope build. This manuscript assumes general technical knowledge of Optics, Electronics, Lasers, and safety measures related to handling the used equipment.
General safety and assembly notice
The building and operation of the microscope involve using different electronic devices that include lasers, high-voltage piezo controllers, CMOS cameras, and Arduino among other devices. Thus, we strongly advise you to practice safety measures and read the safety instructions included within each device’s manual that are available at their respective websites by professionally trained personnel. Disclaimer: the authors provide an as-is guide and are not in any case held responsible for any accidents, damages, or injuries that might occur in any connection to using or misinterpreting this manuscript; the sole purpose of this manuscript is to provide aid in reproducing our scientific work. We recommend removing any jewelry and wearing gloves when handling optics along with safety goggles, and being grounded when handling electronic devices. Special care must be taken when translating the laser beam in between TIRF and HILO/EPI modes as the collimated laser beam exits the objective at a certain angle and depending on the microscope orientation/position it can point at eye level. Note that the IR laser is not visible to the human eye.
Disclaimer
The author provides an as-is guide and is not, in any case, held responsible for any accidents that might occur in any connection to using or misinterpreting this manuscript; the sole purpose of this manuscript is to provide aid in reproducing our scientific work.
The Laser Engine ( Fig. 3 )
Fig.3.
The miEye assembly visual guide – fully assembled laser engine.
The laser engine is assembled on a Thorlabs 30 × 60 mm2 breadboard (MB3060/M) standing on four Ø45.0 mm Sorbothane feet (AV6/M) (Fig. 3). Throughout this guide, if needed we will refer to breadboard mounting holes using XY coordinates as illustrated in Fig. 3. This part of the setup uses a mounting base (BA1S/M) along with post holders (PH75/M) and Ø12.7 mm optical posts (TR75/M) to install optomechanical elements. Note that attaching (BA1S/M) to (PH75/M) requires a 10 mm long (M6 × 1.0) screw. We will refer to such post assemblies as (PA.xxx) where xxx is the numerical identifier of each assembly element (e.g., BA1S/M.001, PH75/M.001 …). One can fix the post assemblies mounted on the breadboard using 16 mm long low-profile channel (M6 × 1.0) screws (SH6M16LP) or similar screws, we will omit to mention this part within the guide and the reader should secure all assemblies to avoid falling optics.
Single-Mode Fiber Coupling ( Fig. 4 )
-
1.
Attach one fiber end (polarization-maintaining (PM) SMF (P1-405BPM-FC-1)) to the SM1 threaded adapter plate (SM1FC.001) which enables us to mount the fiber onto the differential XY translator (ST1XY-D/M). Then, install the translator on the breadboard using post assembly (PA.001) around the mounting hole (5, 10).
-
2.
Screw in three 4-inch-long cage system rods (ER4.001-3) to the XY translator, and skip adding the fourth rod as illustrated to be able to replace the coupling objective as the 30 mm cage system prevents that.
-
3.
Add the 30 mm cage-compatible Z-Axis Translation Mount (SM1ZA) which allows for fine-tuning the coupling objective focal plane.
-
4.
Screw in a long working distance plan achromat objective (40X, NA 0.5, WD 3.0) to the Z-Axis Translation Mount (SM1ZA) using an adapter with external SM1 threads and internal RMS threads. Then, attach a 30 mm Cage Cube (C6WR.001) to the three rods (ER4.001-3) using its through holes, making sure to fix it tightly with the rods penetrating no more than the cube’s wall thickness.
-
5.
Later we will explain how to exploit the cube (C6WR.001) to couple an additional laser source using a dichroic mounted to a Ø1″ optic mount (B5CT2) that adapts to the kinematic cage cube platform (B4C/M). Mind that rods protruding into the cube (C6WR.001) through the top holes would obstruct adding and rotating the cube-compatible accessories including (B4C/M).
-
6.
As illustrated, attach two 4-inch-long cage system rods (ER4.004-5) to the opposite side of the cube (C6WR.001) and fix them tightly.
-
7.
Add a post assembly (PA.002) mounted right-angle kinematic mirror mount (KCB1C_M.001) with a dielectric coated mirror (BB1_E02.001), with the two previously added rods fixed into the kinematic mount cage rod bores. Then, add four 1-inch-long cage system rods (ER1.001-4) to the other side of the kinematic mirror mount (KCB1C_M.001).
-
8.
Mount a second right-angle kinematic mirror mount (KCB1C_M.002) with a dielectric coated mirror (BB1_E02.002) to the four previously assembled 1-inch-long rods.
-
9.
Similarly, add four 1-inch-long cage system rods (ER1.005-8) to the other side of the kinematic mirror mount (KCB1C_M.002).
-
10.
Mount a 30 mm to 16 mm cage adapter plate (SP05_M.001) to the four 1″ rods and fix them in place.
-
11.
Mount the four wavelengths laser diode combiner (IO_MatchBox_40A_48A_52A_64A_11_DM_CF) from “Integrated Optics” with its heatsink and mechanical shutter to the adapter plate (SP05_M.001) using four 1“ long compact cage assembly rods (SR1.001-4).
Fig. 4.
The miEye assembly visual guide – single-mode fiber (SMF) coupling.
Multi-Mode Fiber Coupling ( Fig. 5 )
-
1.
Start by mounting a 30 mm to 30 mm cage system Snap-On right-angle adapter (CP30Q) to the two 4-inch-long cage system rods (ER4.004–5). Then, mount four 4-inch-long cage system rods (ER4.006–9) to this right-angle adapter plate.
-
2.
Position an SM1-threaded 30 mm cage plate (CP33_M.001) along the four rods (ER4.006–9) and do not fix it until alignment. The coupling lens is the achromatic doublet (AC080_010_A) – a positive 10 mm lens of Ø8 mm – mounted within the Ø1″ adapter (AD8T) and fixed using a retaining ring. Then, fix the adapter containing the lens (AD8T) within a Ø1″ SM1 lens tube (SM1L05.001) using a retaining ring. Finally, attach the lens tube (SM1L05.001) to the cage plate (CP33_M.001).
-
3.
Attach one of the MMF (square-core MMF (150 µm × 150 µm core) that is an FC/PC patch cable (M103L05)) ends to the SM1 threaded adapter plate (SM1FC.002) which enables mounting the fiber to the XY translating lens mount (CXY1A.001). Then, fix the 30 mm cage system translating lens mount at the end of the four rods (ER4.006-9).
-
4.
Add a post assembly (PA.003) with the bottom plate (KBB25_M.001) of the KB25/M kinematic base mounted to the post using an M4 screw. Do not fix the position of this assembly until the alignment process, producing a 90-degree reflection towards the MMF coupling path.
-
5.
Attach a Ø1″ protected silver mirror (PF10_03_P01.001) within the mirror holder (MH25T.001) that is compatible with the compact kinematic mirror mount (KMSS_M.001). Afterward, fix the mirror mount (KMSS_M.001) to the top plate (KBT25_M.001) of the KB25/M kinematic base to produce a 45-degree angle-of-incidence (AOI) when attached to the bottom plate (KBB25_M.001) as illustrated.
Fig. 5.
The miEye assembly visual guide – multi-mode fiber (MMF) coupling.
Additional Laser Coupling into SMF ( Fig. 6 )
-
1.
Screw-in four 1-inch-long cage system rods (ER1.009-12) to the side of the cube (C6WR.001) as illustrated.
-
2.
Add a post assembly (PA.004) mounted right-angle kinematic mirror mount (KCB1C_M.003) with a dielectric coated mirror (BB1_E02.003), with the four previously added rods fixed into the kinematic mount cage rod bores. Then, add four 1-inch-long cage system rods (ER1.013-16) to the other side of the kinematic mirror mount (KCB1C_M.003).
-
3.
Mount a 30 mm to 16 mm cage adapter plate (SP05_M.002) to the four 1″ rods and fix them in place.
-
4.
Mount the three wavelengths laser combiner (IO_MatchBox_40A_48A_52A_XXY_11_DM_CF) from “Integrated Optics” with its heatsink and mechanical shutter to the adapter plate (SP05_M.002) using four 1“ long compact cage assembly rods (SR1.005-8).
-
5.
Insert the dichroic mounted on a kinematic cage cube platform (B4C_M, B5CT2) into the cube (C6WR.001).
Fig. 6.
The miEye assembly visual guide – additional laser coupling.
The Microscope Base ( Fig. 7 )
Fig. 7.
The miEye assembly visual guide – the microscope base.
The microscope is assembled on a Thorlabs 30 × 120 mm2 breadboard (MB30120/M) standing on 5 of Newport’s Mechanical Vibration Isolators (VIB100-0920) mounted onto the breadboard using Newport’s adapters (VIB100-BA-ADJ). For further mechanical vibration dampening, we place a 1-inch-thick marble plate below the breadboard on a regular table. Throughout this guide when needed we will refer to breadboard mounting holes using XY coordinates.
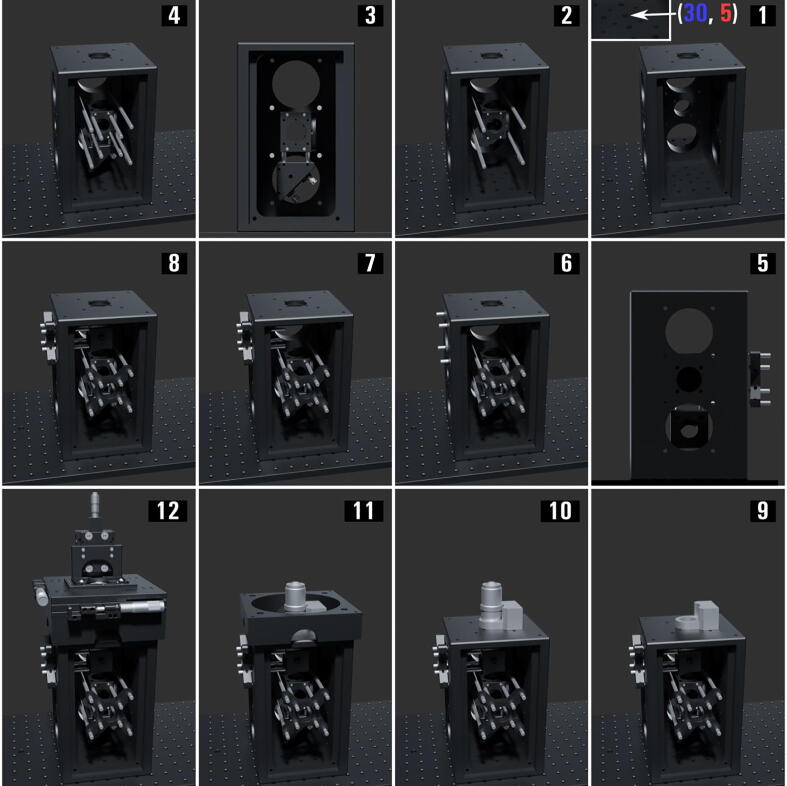
The Microscope Body ( Fig. 8 )
Fig. 8.
The miEye assembly visual guide – construction of the microscope body.
The microscope revolves around the body which is our iteration/design of miCube’s approach, a CNC milled and anodized steel modular skeleton. The design allows flexible adjustments of different microscopy configurations, but we will demonstrate our preferred one. The bottom level is dedicated to three different orientations of the emission path and uses the 60 mm cage system suitable for Ø 2” optics. We prefer to use the middle layer for the IR autofocus stabilization path. The top-level uses the wider 60 mm cage system to allow for various excitation settings. On the top and bottom faces, we have 30/60 mm cage system mounts allowing vertical element stacking within the microscope’s body. The cage system mounts are (M6 × 1.0) threaded and use Thorlabs thread adapters (AE4E6M) to attach the cage system rods to the body. Mounting holes on the bottom plate allow mounting the microscope body on optical breadboards. The top face has mounting threads (M36 × 0.75, M25 × 0.75) suitable for the piezo stage mounting adapter. An XYZ-Stage custom adapter mounts to the top face using four (M6 × 1.0) mounting holes.
-
1.
Position the microscope body at the breadboard hole (30, 5) so that the central mounting hole aligns with it as illustrated and fix it on the breadboard using 25 mm or longer M6 × 1.0 screws.
-
2.
Inside the cube, screw four 6-inch-long cage system rods (ER6.001-4) into the back plate’s middle 60 mm cage system mounting holes via four (AE4E6M) adapters. Prepare the cube (C6WR.002) with the IR dichroic mounted using a 30-mm-cage-compatible rectangular filter mount (FFM1) attached to a fixed cage cube platform (B3C_M). Rotate the platform (B3C_M) so that the dichroic is at 45-degree AOI with the IR beam coming from the back and reflected upwards toward the objective.
-
3.
Attach four 1-inch-long cage system rods (ER1.017-20) to the right-angle kinematic elliptical mirror mount (KCB1EC_M) equipped with a 1″ broadband dielectric elliptical mirror (BBE1-E02). Roughly attach the emission mirror mount to the cube (C6WR.002) as illustrated, pointing to the left.
-
4.
Attach four 3-inch-long cage system rods (ER3.001-4) to the cube (C6WR.002) side as shown in figure X.
-
5.
Add a 30 mm to 60 mm cage plate adapter (LCP33_M.001) so that the previous IR dichroic cube and mirror mount assembly attach to the 30 mm part using the 3-inch-long rods (ER3.001-4). Then attach the whole assembly to the 6-inch-long rods (ER6.001-4) allowing alignment along the y-axis.
-
6.
Screw four ½“ long cage system rods (ER05.001-4) to the left side top 60 mm cage system mounting holes via four (AE4E6M) adapters.
-
7.
Mount the 30 mm to 60 mm cage plate adapter (LCP33_M.002) to the cube via the ½“ long cage system rods (ER05.001-4). Add four 3-inch-long cage system rods (ER3.005-8) to the 30 mm cage system holes on the adapter (LCP33_M.002) without fixing them.
-
8.
Screw the 3-inch-long rods (ER3.005-8) into the side of the 30 mm cage cube with a dichroic filter mount (CM1_DCH_M) equipped with the multiband dichroic. Orient the cube (CM1_DCH_M) so that the excitation beam propagates through the right-side figure-wise.
-
9.
Screw the piezo stage adapter (Part NO.) to the cube’s top threaded hole (M36 × 0.75) then mount the Z-axis 100um piezo stage (FOC100) to the adapter.
-
10.
Screw the objective (Nikon APO TIRF X60 1.49NA) to the Z-axis piezo stage (FOC100). NOTE: This step is for illustrative purposes and to avoid any damage to the objective we recommend skipping this and adding the objective after completing all the main body assembly steps.
-
11.
Mount the custom-made XYZ Stage adapter (XYZ_Stage_adapter) to the top side of the cube using four M6 low-profile screws.
-
12.
To mount the XYZ manual micrometric stage (Piezo Concept MMS165 + MZA) reset the bottom micrometric handle to its minimum position revealing the two left mounting holes. After loosely screwing in two low-profile M6 screws flushed with the lower plate surface, push the spring-loaded bottom plate to reveal the other two holes on the right; then proceed with fixing the stage to the adapter. Note that we have replaced the manual micrometric handles with two 25 mm motorized actuators (Z825B).
-
13.
To add/remove the objective one must disassemble the l-shaped sample holder mounted to the manual Z stage by screwing off its four mounting screws.
The Excitation Path using SMF ( Fig. 9 )
-
1.
Mount the 25.0 mm translation stage (PT1/M) on the microscope base breadboard (MB30120/M) aligned with the hole (41, 4) as illustrated.
-
2.
Mounting the breadboard (MB1530F_M) to the translation stage (PT1/M) requires drilling the (MB1530F_M) threaded holes, then fixing it with three low-profile M6 screws.
-
3.
Mount two 30 mm to 60 mm cage plate adapters (LCP33_M.003-4) onto two post assemblies (PA.005-6), then connect the two adapters using three 10-inch-long cage system rods (ER10.001-3) using the 60 mm cage through holes; during the last step mount a 60 mm to 30 mm cage system right-angle adapter (LCP30) to two of the rods. Fix the post assemblies (PA.005-6) on the breadboard (MB1530F_M); note that the post assemblies (PA.005-6) mounted as illustrated do not align with the top opening of the main body at the translation stage (PT1/M) reset position, but that is intentional.
-
4.
Mount four 4-inch-long cage system rods (ER.010-13) to the cage plate adapter’s (LCP33_M.003) 30 mm cage system through holes as illustrated; similarly, add four 10-inch-long rods (ER10.004-7) to the adapter plate (LCP33_M.004) preferably having 16 cm of the rods available at the left of the plate for telescope adjustments.
-
5.
Mount the adjustable 2 mm aspheric collimator (CFC2_A) to an SM1-threaded collimator adapter (AD15F2), then pass the second SM patch cable (P1-405BPM-FC-1) end through an XY translating lens mount (CXY1A.002) then screw it in the collimator; follow that with screwing the collimator adapter (AD15F2) to the translating lens mount (CXY1A.002). Finally, fix the translating lens mount (CXY1A.002) on the 10-inch-long rods (ER10.004-7).
-
6.
Mount the first telescope Ø5 mm achromat (AC050_008_A_ML) to its SM1-threaded lens adapter (S1TM09), then screw the adapter within a Ø1″ SM1 lens tube (SM1L05.002). Attach the lens tube (SM1L05.002) to the vague system adapter (LCP33_M.004).
-
7.
Add an SM1-threaded 30 mm cage plate (CP33_M.002) along the four rods (ER10.004-7) and do not fix it till alignment, then attach the second telescope 150 mm achromat (AC254_150_A_ML.001) to the cage plate (CP33_M.002).
-
8.
Add the XY translating lens mount (CXY1A.003) with a 200 mm achromat (AC254_200_A_ML.001) attached to its moving side and an SM1 ring-actuated iris diaphragm (SM1D12D.001) to its fixed side.
Fig. 9.
The miEye assembly visual guide – assembly of the single-mode fiber (SMF) excitation path.
The Excitation Path using Multi-Mode Fiber ( Fig. 10 )
-
1.
Add a 150 mm achromat (AC254_150_A_ML.002) to the 60 mm to 30 mm cage system right-angle adapter (LCP30).
-
2.
Screw-in four 2-inch-long cage system rods (ER2.001-4) to the side of a right-angle kinematic mirror mount (KCB1_M.001) with a Ø1″ protected silver mirror (PF10_03_P01.002). Fix the mirror mount (KCB1_M.001) on post assembly (PA.007) and attach it to the right-angle adapter (LCP30) using the four rods.
-
3.
Screw in three 8-inch-long cage system rods (ER8.001-3) to the other side of the mirror mount (KCB1_M.001). Mount a coarse ±1 mm XY slip plate positioner (SPT1C_M.001) on the rods (ER8.001-3). Attach a 10 mm achromatic mounted doublet (AC080_010_A_ML) to the SM1-threaded adapter (SM1AD8) then mount the adapter to the slip plate positioner (SPT1C_M.001).
-
4.
Pass the second MM patch cable (M103L05) end through an XY translating lens mount (CXY1A.004) then screw it to the SM1 threaded adapter plate (SM1FC.003); follow that with screwing the fiber adapter (SM1FC.003) to the translating lens mount (CXY1A.004). Mount the translating lens mount (CXY1A.004) on the 8-inch-long rods (ER8.001-3) without fixing it. Add a 4-inch rod (ER4.014) to the available rod holes in plates (SPT1C_M.001, CXY1A.004) which allows fixing the distance between the fiber and lens while translating both on the 8-inch-long rods.
-
5.
Add a post assembly (PA.008) with the bottom plate (KBB25_M.002) of the KB25/M kinematic base mounted to the post using an M4 screw.
-
6.
Attach a Ø1″ protected silver mirror (PF10_03_P01.002) within the mirror holder (MH25T.002) that is compatible with the compact kinematic mirror mount (KMSS_M.002). Afterward, fix the mirror mount (KMSS_M.002) to the top plate (KBT25_M.002) of the KB25/M kinematic base to produce a 45-degree angle-of-incidence (AOI) when attached to the bottom plate (KBB25_M.002) as illustrated. Fix the position of the assembly (PA.008) to produce a 90-degree reflection towards the main body direction.
Fig. 10.
The miEye assembly visual guide – assembly of the multi-mode fiber (MMF) excitation path.
The IR Autofocus Path ( Fig. 11 )
Fig. 11.
miEye assembly visual guide – assembly of the IR autofocus path.
This part of the setup uses a mounting base (BA1S/M) along with post holders (PH20/M) and Ø12.7 mm optical posts (TR20/M) to install optomechanical elements. We recommend attaching (BA1S/M) to (PH20/M) using short (M6 × 1.0) screws not to obstruct the post-height adjustments. We will keep referring to such post assembly as (PA.xxx).
-
1.
Screw four ½“ long cage system rods (ER05.005-8) to the backside middle 30 mm cage system mounting holes via four (AE4E6M) adapters. Screw four rod adapters (ERSCB.001-4) to the cube-mounted non-polarizing beamsplitter (CCM1_BS014_M) as illustrated, then mount it to the main body on the ½” long rods.
-
2.
Screw four ½“ long cage system rods (ER05.009-12) to the side of the beamsplitter (CCM1_BS014_M). Screw four rod adapters (ERSCB.005-8) to a right-angle kinematic mirror mount (KCB1_M.002) equipped with a Ø1″ protected silver mirror (PF10_03_P01.003); then mount the mirror (KCB1_M.002) on the rods (ER05.009-12) and screw two rod-adapters (ERSCB.009-10) on its bottom side diagonally. Similarly, add two ½” long rods (ER05.013-14) diagonally to a right-angle kinematic mirror mount (KCB1_M.003) equipped with a Ø1″ protected silver mirror (PF10_03_P01.004); then mount it to the other mirror via rod adapters (ERSCB.009-10).
-
3.
Screw four ½“ long rods (ER05.015-18) to the side of the mirror mount (KCB1_M.003). Mount 30 mm translating cage segment plate (CPX1_M.001) on post assembly (PA.009) then fix the plate to the rods (ER05.015-18) using its boreholes.
-
4.
Screw four 10-inch-long rods (ER10.008-11) to the translating plate of (CPX1_M.001) then mount on it an SM1-threaded cage plate (CP33_M.003), an XY translating lens mount (CXY1A.005), and a post-assembly (PA.010) with a 30 mm cage mounting bracket (CP33B). This arrangement allows sliding the whole segment to adjust the TIRF angle.
-
5.
Mount the 400 mm B-coated N-BK7 mounted lens (LA1172_B_ML) on the cage plate (CP33_M.003), then attach to the lens mount an SM1 ring-actuated iris diaphragm (SM1D12D.002). Mount the laser line filter (FL830_10.001) within a lens tube (SM1L05.003) then attach it to the fixed part of the mount (CXY1A.005). Mount the collimated laser diode module (CPS830S) within its SM1-threaded kinematic pitch/yaw adapter (KAD8F) then screw the adapter to the translating part of the mount (CXY1A.005).
-
6.
Screw four 6-inch-long rods (ER6.005-8) to the side of the beamsplitter (CCM1_BS014_M), then mount on the same side another laser line filter (FL830_10.002) housed in the lens tube (SM1L05.004). Add on the rods (ER6.005-8) a cage plate (CP33_M.004) and mount on it a 100 mm B-coated N-BK7 mounted lens (LA1509_B_ML). Mount a 30 mm translating cage segment plate (CPX1_M.002) to the rods (ER6.005-8) using its boreholes.
-
7.
Add to the translating segment of the plate (CPX1_M.002) a set of one adapter with external SM1 threads and internal C-mount threads (SM1A10.001) and a 0.5″ long C-mount coupler (CMT2.001) with external threads; which enable attaching the autofocus tracking CMOS monochromatic camera (IDS_UI_3060CP_M_V2) to the SM1 mount and adjusting its orientation.
-
8.
Set both indicated translating segments at their center position, as it would help during the alignment process.
The Emission Path ( Fig. 12 )
Fig. 12.
The miEye assembly visual guide – assembly of the emission path.
This part of the setup uses different post mounts and post lengths to maintain the identifier count (PA.xxx).
-
1.
Screw four 8-inch-long cage system rods (ER8.004-7) to the left side bottom 60 mm cage system mounting holes via four (AE4E6M) adapters. Mount the Nikon 200 mm tube lens (MXA20696) to the CNC custom-made adapter (M38_SM2_Adapter), then attach the adapter (M38_SM2_Adapter) to an external SM2 coupler (SM2A20). The coupler (SM2A20) is in turn mounted on a coarse ± 2 mm slip plate positioner (SPT2_M) with the plate mounted on the rods (ER8.004-7).
-
2.
Add the post assembly (PA.011: BA1S_M, PH20_M, TR30_M) mounted 30 mm to 60 mm cage plate adapter (LCP33_M.005) with an SM1 ring-actuated iris diaphragm (SM1D12D.003) attach to the adapter plate. Roughly position it so that the iris diaphragm (SM1D12D.003) is at the focal plane of the tube lens (MXA20696).
-
3.
Add a post assembly (PA.012: BA1S_M, PH20_M, TR30_M) mounted SM1-threaded 30 mm cage plate (CP33_M.005), then screw four 1-inch-long rods (ER1.021-24) to the SM1-threaded dual-position piezo slider (ELL6K) and fix it to the cage plate (CP33_M.005). Add a 200 mm achromatic doublet (AC254_200_A_ML.002) to one of the slider’s (ELL6K) mounts, forming the 4f configuration with the tube lens (MXA20696). Note that one can replace rods (ER1.021-24) with 8-inch-long rods instead to avoid tilt alignment.
-
4.
Add a post assembly (PA.013: BA1_M, PH20_M, TR20_M) mounted right-turning kinematic fluorescence filter cube (DFM1B_M) along the emission path for two-channel imaging purposes which requires an application-specific dichroic (Semrock F38-640). From the main body side, mount a short-pass filter (Semrock FF01-775/SP-25) housed in a lens tube (SM1L03) to the cube (DFM1B_M). Screw four ½“ long rods (ER05.019-22) into the opposite side of the cube (DFM1B_M), and four ½” long rods (ER05.023-26) to the SM1-threaded four-position piezo slider (ELL9); using a thick cage plate (CP33T_M) mount the slider (ELL9) combining the rods (ER05.019-26) midway. Add the desired filters (Edmund Optics Ø25 mm OD 6: 697/75, 692/40, 575/35, 540/50) to the filter slider (ELL9).
-
5.
Add the post assembly (PA.014: BA1S_M, PH20_M, TR30_M) mounted coarse ±1 mm XY slip plate positioner (SPT1C_M.002), then attach to the positioner four 4-inch-long rods (ER4.015-18) and a set of one thermally insulating adapter with external SM1 threads and internal C-mount threads (SM1A10TS.001) and a 0.5″ long C-mount coupler (CMT2.002) with external threads; which enable attaching the CMOS monochrome camera (Alvium_1800_U_158m) to the SM1 mount of positioner (SPT1C_M.002) and adjusting its orientation. Add the cage plate (CP33_M.006) to the rods (ER4.015-18) and attach to it the achromatic doublet (89_Achromat_edmund_49_782) mounted in the lens tube (SM1L05.005).
-
6.
The second channel can be assembled using four 6-inch long rods (ER6.009-12) a cage plate (CP33_M.007) and a post assembly (PA.014: BA1S_M, PH20_M, TR30_M) mounted XY slip plate positioner (SPT1C_M.003); along with a set of one thermally insulating adapter with external SM1 threads and internal C-mount threads (SM1A10TS.002) and a 0.5″ long C-mount coupler (CMT2.003) with external threads to mount the second camera. (Optional)
Electronics ( Fig. 13 )
Fig. 13.
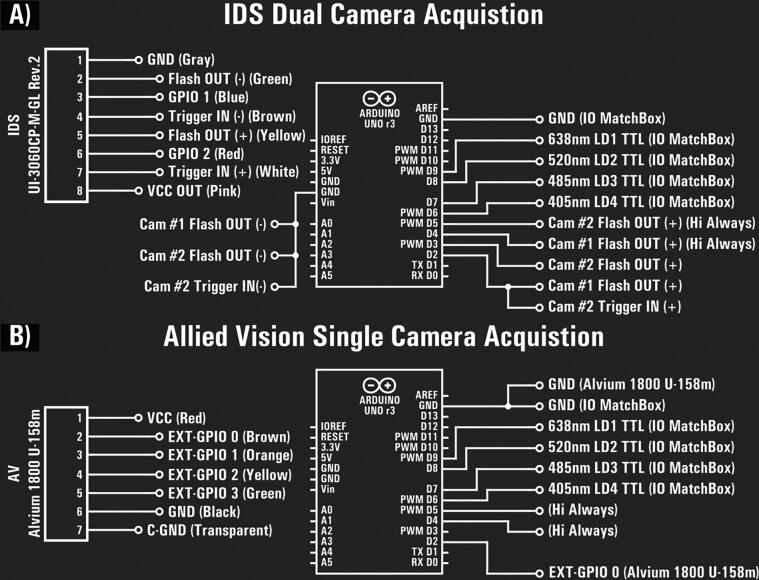
Circuit diagram of the Arduino Uno r3 laser relay. A) IDS dual camera laser triggering of IO MatchBox through the Arduino-registered camera flash signals. Common ground is used for all isolated GPIO optocouplers along with a high-always (Hi Always) digital output pin for each flash (+) wiring. B) AV camera GPIO and IO MatchBox wiring with the Arduino laser relay. The camera shares a non-isolated external ground with the Arduino.
The RelayBox handles the laser triggering by using the GPIO signals in a certain logic according to a configuration sent by the microEye’s control module.
Note that the user can decide how to wire up their triggering and define the logic within the Arduino. The wiring scheme provided in Fig. 13 is just a suggestion. The pins D4 and D5 are always high and pin D10 alternates between high and low for each loop cycle. Take good care during the wiring process, especially with non-isolated input/output.
-
1.
Connect the camera GPIOs in use to the Arduino board using a camera-specific GPIO cable; note to adapt the GPIO cable bare wires with male jumper wires we have used soldering and heat shrink wrap.
-
2.
Connect the triggering wires from the lasers to the Arduino board; use male-to-male jumper wires.
-
3.
Connect the USB cable from the Arduino board to the PC. (Disconnect when not in use)
Software Installation and setup
-
1.
Download and install the latest Python 3.9 stable release. (We tested with 3.9.4 and 3.9.7)
-
2.
Open a terminal and install the microEye package using pip.
“pip install microEye”
-
3.
Download the file requirements.txt and install the microEye required packages by executing the following line. (This may take a while)
“pip install -r /path/to/requirements.txt”
-
4.
Install the dependent hardware-specific drivers for the cameras if you intend to use the hardware control or acquisition modules. (microEye might not function as expected in case drivers are missing):
-
•
Integrated Optics Laser control software and drivers.
-
•
IDS uEye CMOS cameras: install IDS Software Suite 4.96.1 for Windows 32/64-bit. (We did not test it with Linux)
-
•
Allied Vision CMOS cameras: install the Vimba SDK 5.0 or 6.0 outside the Program Files directory to skip the run as admin requirement, then in a terminal navigate to directory […/Allied Vision/Vimba_5.0/VimbaPython/] where [setup.py] is located and execute:
“python -m pip install.”
-
•
Thorlabs CMOS cameras: install Thorcam in its default directory as it is initial to have uc480_64.dll in the path [C:\Program Files\Thorlabs\Scientific Imaging\ThorCam\uc480_64.dll]. (Note, Thorlabs cameras can get identified by Windows as IDS uEye cameras and run without this software)
-
5.
Other hardware used by the miEye microscope includes Thorlabs Kinesis® Software and Elliptec™ Software.
-
6.
Download and run examples to start using microEye.
Operation instructions
miEye test samples
-
1.
Glass coverslip with adsorbed fluorescently labeled protein. We used ATTO655 labeled streptavidin (Sigma-Aldrich, 02744) for coating the glass coverslip (170 µm thickness, Thermo Scientific, Menzel Coverslip 24x60mm #1.5 (0.16–0.19 mm)). A cleaned glass coverslip was assembled into the flow cell by attaching it to a 6 channel-plastic slide (80606, Ibidi) via a double-sided sticky tape (3 M, 9088–200). Streptavidin-ATTO655 was diluted in ultrapure DI water to the final concentration of 160 nM and loaded into the flow cell. After 5 min of incubation, the surface-unbound protein was washed away with 500 µL of ultrapure DI water.
-
2.
Dye sandwich. Before the assembly of the dye sandwich, a glass coverslip (170 µm thickness, Thermo Scientific, Menzel Coverslip 24x60mm #1.5 (0.16–0.19 mm)) was initially cleaned for 5 min using an air plasma generator (∼260 mTorr, highest power setting, ZEPTO-W6, Diener electronic). A 10 × 10 mm-sized hole was then cut out in a center of the double-sided sticky tape (3 M, 9088–200) piece (24 × 60 mm) using a scalpel and such tape was attached to the cleaned glass coverslip. Next, ATTO647N-NHS ester (AD 647 N, ATTO-TEC) was diluted to the final concentration of 2 µM in ultrapure DI water and ∼50 µL of this dye solution was added to the glass coverslip inside the cut area. The thick glass slide (26 × 76 × 1 mm, Heinz Herenz Hamburg, ref.: 1041010) was then put on top of the double-sided sticky tape spacer, thus forming a sandwich with a trapped ATTO647-NHS dye solution in between the two glass surfaces.
-
3.
Surface immobilized 200 nm diameter polystyrene Crimson beads (Invitrogen, F8806). The Crimson beads were diluted 4000 fold in ultrapure DI water and immobilized on the poly-l-Lysine (Cultrex, cat. no. 3438–100-01) coated glass coverslip (170 µm thickness, Thermo Scientific, Menzel Coverslip 24x60mm #1.5 (0.16–0.19 mm)). The excess Crimson beads were washed away by rinsing the coverslip with ultra-pure DI water. This coverslip was dried using nitrogen gas and attached to a thick glass slide (26 × 76 × 1 mm, Heinz Herenz Hamburg, ref.: 1041010) via a double-sided sticky tape (3 M, 9088–200) spacer. Inside this sandwich, we added a drop of ∼50 µL ultrapure DI water.
-
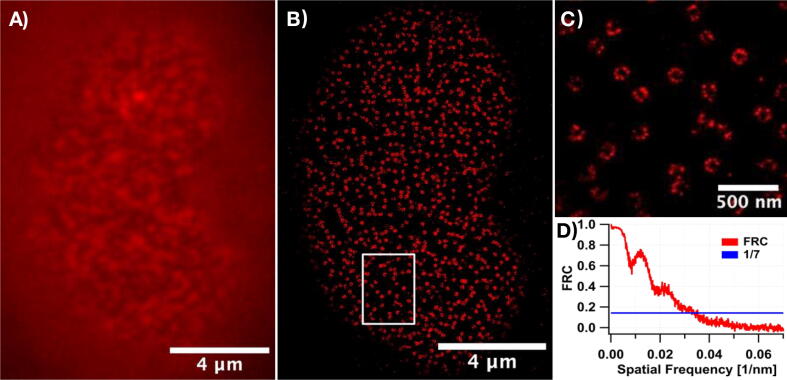
4.Preparation of the U-2 OS cells with labeled Nup96 complexes for dSTORM imaging.
-
a)U-2 OS-CRISPR-Nup96-SNAP clone #33 (300444, CLS GmbH) cell line was purchased from Cell Lines Service (Germany). Seeding, fixation, and staining of the cells were performed as described previously [42]. Two days before the fixation procedure, U2OS-Nup96-SNAP cells were seeded on 8 well-chambered glass coverslip (C8-1.5H-N, Cellvis) so that on the day of fixation the cells would reach the confluency of about 50–70 %. During this period, U2OS-Nup96-SNAP cells were grown in McCoy’s 5A medium (16600082, Thermo Fisher Scientific) supplemented with 100 µg/mL streptomycin (15070063, Thermo Fisher Scientific), 100 U/mL penicillin (15070063, Thermo Fisher Scientific), 1 mM sodium pyruvate (11360070, Thermo Fisher Scientific), 1 % MEM non-essential amino acids (11140035, Thermo Fisher Scientific) and 10 % fetal bovine serum (15595309, Fisher Scientific) at 37 °C and 5 % CO2.
-
b)On the imaging day, glass surface-attached U2OS-Nup96-SNAP cells were initially prefixed in phosphate-buffered saline (PBS; 10010023, Thermo Fisher Scientific) containing 2.4 % (w/v) formaldehyde (FA; 28906, Thermo Fisher Scientific) for 30 s. After the 3 min, permeabilization step using 0.4 % (v/v) Triton X-100 (3051.2, Carl Roth) in PBS, the complete fixation of the cells was performed for 30 min in 2.4 % (w/v) FA solution in PBS. The sample was then washed two times using PBS with 5 min incubation each time. The fixation of the cells was halted by submerging the sample in PBS containing 100 mM NH4Cl (5470.1, Carl Roth) for 5 min and then performing two 5 min-long washing steps in PBS. After quenching the remaining FA, a few drops of Image-iT FX Signal Enhancer (I36933, Thermo Fisher Scientific) were added onto the sample and incubated for 30 min. Staining of SNAP-tag labeled Nup96 was carried out at room temperature for 2 h using benzylguanine-conjugated AF647 (S9136S, New England Biolabs) diluted to the final concentration of 1 µM in PBS supplemented with 1 µM of dithiothreitol (R0862, Thermo Fisher Scientific) and 0.5 % (w/v) bovine serum albumin (T844.2, Carl Roth). The excess of the unbound dye was removed by washing the sample three times with 5 min incubations in PBS.
-
c)For dSTORM experiments, an imaging buffer (50 mM of Tris (2449.2, Carl Roth)-HCl (pH 8.0, at 25 °C), 10 mM of NaCl (3957.1, Carl Roth), 500 µg/mL of glucose oxidase (G2133-10KU, Sigma-Aldrich), 40 µg/mL of catalase (C100-50MG, Sigma-Aldrich), 10 % (w/v) d-Glucose (X997.2, Carl Roth) and 143 mM of 2-mercaptoethanol (4227.1, Carl Roth) in H2O) was added to the sample and then it was sealed with parafilm.
-
a)
Initial operation
Once the microscope hardware has been assembled, the code imported, and the software and its dependencies are installed correctly, the microscope is ready to be used. The initial use requires precise alignment of the optical paths and it involves some essential key steps described in the following subsections.
SMF laser coupling
-
1.
First, align the laser beam along the optical axis using an iris diaphragm (SM1D12D) or an alignment tool (DG10-1500-H1-MD). Adjust the kinematic mirror holder (KCB1C_M.002) so that the beam passes through an iris positioned between the mirror holder (KCB1C_M.001) and the cube (C6WR.001).
-
2.
Similarly, adjust the kinematic mirror holder (KCB1C_M.001) so that the beam passes through an iris positioned on the differential XY translator (ST1XY-D/M) instead of the adapter plate (SM1FC.001). Note, the objective should be removed during this alignment procedure.
-
3.
Iterate between (1) and (2) and consider whether you might need to adjust the XY position of the translator (ST1XY-D/M) to ensure beam alignment through the optical axis.
-
4.
Install the objective and SMF adapter back to their respective holders, and set the objective at one working distance from the SMF input.
-
5.
Use a laser power meter positioned in front of the SMF output to constantly monitor the SMF output power while performing fine alignment steps. Adjust the XY translator (ST1XY-D/M) along with the Z position of the translation mounted (SM1ZA) objective to reach the maximum of the laser power. Note, one needs to iterate again between (1) and (2) along with the Z position to achieve maximum coupling; this usually can take 1–2 h and a compromise between the four coupled wavelengths should be made. Expected coupling efficiencies for the SMF are provided in Table 1 and vary due to alignment variations within the laser combiner for each wavelength.
MMF laser coupling
-
1.
Position the post assembly (PA.003) and its mounted mirror on the compact kinematic mirror mount (KMSS_M.001) so that the beam is incident at the center of the mirror at a ∼45° angle. Then remove the coupling lens tube (SM1L05.001) temporarily.
-
2.
Iterate between adjusting the kinematic mirror mount (KMSS_M.001) and the MMF input XY position (CXY1A.001) to align the beam along the central optical axis.
-
3.
Position the coupling achromatic 10 mm lens one focal length away from the MMF end, then position a laser power meter in front of the MMF output to constantly monitor the MMF output power while performing fine alignment steps.
-
4.
Repeat step (2) until maximum laser power coupling is achieved. Expected coupling efficiencies for the MMF are provided in Table 1 and vary due to alignment variations within the laser combiner for each wavelength.
Additional laser coupling
-
1.
Similarly, adjust the kinematic mirror mount (KCB1C_M.003) and the kinematic cage cube platform (B4C_M) so that the additional laser beam propagates along the optical axis.
-
2.
Use a laser power meter positioned in front of the SMF output to constantly monitor the SMF output power while performing fine alignment steps.
-
3.
Repeat step (2) until maximum laser power coupling is achieved. Note, after this alignment procedure one might need to repeat the first SMF coupling procedure.
SMF excitation path alignment
-
1.
Remove both beam expander lenses (AC050_008_A_ML, AC254_150_A_ML.001), the TIRF lens (AC254_200_A_ML.001), the MMF mirror on the magnetic holder (KB25_M.002), and the objective.
-
2.
Move the 25.0 mm translation stage (PT1/M) at ∼20.8 mm position (that should be close to the setting required for Epi-mode).
-
3.
Insert lens tube with two frosted glass discs (aligning tool, see the parts list sheet available here: https://doi.org/10.17605/osf.io/j2fqy) instead of the objective.
-
4.
Adjust the collimator (CFC2_A) ring correctly to produce a collimated beam, simply point the output to a far away wall and try to minimize the beam size.
-
5.
Check out the off-axis centering of the beam in each of the cage system holders using an SM1 threaded iris diaphragm. If the beam is not passing through centers, adjust the SMF collimator’s holder (CXY1A.002). Note that we have replaced the translating lens mount (CXY1A.002) with a kinematic XY slip plate mount (KC1-S/M) as the collimator (CFC2_A) produces a beam that requires pitch and yaw adjustments.
-
6.
Install the first beam expander lens (AC050_008_A_ML) and realign the beam so it would not change direction after passing this element and diverge from the optical axis due to minor misalignment.
-
7.
Install the second beam expander lens (AC254_150_A_ML.001); again, as it is mounted on a fixed cage system plate the beam should not propagate off-axis. Position the cage plate (CP33_M.002) so that the distance between lenses (AC050_008_A_ML, AC254_150_A_ML.001) is equal to the sum of their focal lengths (∼157.5 mm). Then, check if the beam diameter after the beam expander is not varying drastically. Otherwise, adjust the position of the cage plate (CP33_M.002) so the second lens (AC254_150_A_ML.001) produces a beam with a roughly constant beam diameter.
-
8.
Install the TIRF lens (AC254_200_A_ML.001) in its XY translating lens mount (CXY1A.003). The distance from the objective’s mounting back focal plane to the TIRF lens should be roughly equal to its focal length of 200 mm. Check whether the beam is still on-axis after the TIRF lens. If not, adjust the TIRF lens position by translating it off-axis.
-
9.
Add an iris diaphragm on the multi-band dichroic cube (CM1_DCH_M) side facing the SMF excitation path, and another iris diaphragm on the plate adapter (LCP33_M.002). Perform adjustments to the post height, translation stage (PT1/M) position, and the post's XY positioning to allow the beam to propagate on-axis through the two irises.
-
10.
Then, adjust the multiband dichroic cube (CM1_DCH_M) position on-axis so the beam passes through the lower frosted glass alignment disk which is located close to the objective mount. Adjust the TIRF lens so the beam passes through the second top frosted glass disk of the alignment tool, you might need to revisit some of the steps (i) and (j).
-
11.
Install the objective instead of the alignment tool. Inspect the beam shape after the object (e.g. on the ceiling) for each of the excitation wavelengths. It should be making clear concentric rings with the center dot being the most intense. Also, the beam should be perpendicular to the sample plane. If it is not the case, adjust the TIRF lens off-axis to make it perpendicular to the sample plane, and translate it axially to make the concentric rings more clear. Note, if the beam expander was not configured correctly you might have to adjust the second lens' on-axis position by translating the plate (CP33_M.002).
-
12.
To adjust the excitation angle to the TIRF mode, add oil to the objective, and a glass coverslip covered with a fluorescently labeled protein. Start image acquisition, and turn off the auto-scaling of image intensity. Bring the sample into focus by first adjusting the manual Z-axis stage and then finely with the piezo stage. Once the surface view is crisp, start moving the 25.0 mm translation stage off-axis and monitor the intensity of the sample. Once you reach a reading close to 16.3 mm on the stage micrometer the intensity will increase which is characteristic of the critical angle and the evanescent field appearance. Then the stage can be translated even further to higher angles of incidence and thus the lower penetration depth of the evanescent field would be achieved.
MMF excitation path alignment
-
1.
Remove the two lenses (AC254_200_A_ML.001, AC254_150_A_ML.002, AC080_010_A_ML) along the MMF excitation path and install the mirror mounted on the magnetic holder (KB25_M.002).
-
2.
Install the lens (AC080_010_A_ML) back and position it roughly-one focal length away from the MMF by sliding the positioner (SPT1C_M.001) axially so that the beam after the lens would be almost collimated. Then check out whether the beam propagates along the optical axis of each cage system element. If not centered, adjust the off-axial position of the slip plate positioner (SPT1C_M.001) or the MMF off-axial position. If necessary, adjust the position and height of the post assembly (PA.008) mounted mirror and its kinematic holder (KMSS_M.002).
-
3.
Install the aforementioned alignment tool instead of the objective and check out whether the beam passes through the centers of the bottom and top plate of the aligning tool. If not centered, adjust the kinematic mirror holder (KMSS_M.002) and the 25.0 mm translation stage position (PT1/M).
-
4.
Install the lens (AC254_150_A_ML.002) back on the right-angle adapter (LCP30), just before the mirror on the magnetic holder. No adjustments should be needed, otherwise, revisit the previous steps.
-
5.
Install the TIRF lens (AC254_200_A_ML.001) back as aligned previously in the SMF excitation path, the TIRF lens and the lens (AC254_150_A_ML.002) should form a 4f configuration (we alter the 4f configuration to achieve the right magnification for us).
-
6.
The beam should be collimated after the TIRF lens, otherwise slide the plates (CXY1A.004, SPT1C_M.001) maintaining the distance between the plates by fixing the fourth rod (ER4.014). The beam after the TIRF lens also should pass through the centers of the bottom and top plate of the aligning tool. If not centered, adjust the kinematic mirror holder (KMSS_M.002) and the 25.0 mm translation stage position (PT1/M).
-
7.
Install the objective and add a sample of dye sandwich into the sample holder of the microscope. Bring the sample into focus.
-
8.
Start imaging with the appropriate laser. Then the size of the illuminated field of view is adjusted by changing the optical path length between the two lenses (AC080_010_A_ML, AC254_150_A_ML.002) keeping the distance between the MMF end and the first lens fixed; then by fine-tuning the position of the MMF end relative to the first lens we refocus the fiber output at the sample plane.
-
9.
The square-shaped illumination should become clear when properly refocused on the sample plane, which is observed very well when the agitation module is turned on.
IR autofocusing system alignment
-
1.
Reset the IR TIRF segment to the zero position on the translating plate (CPX1_M.001) and remove the laser line filter (FL830_10.001) and the lens (LA1172_B_ML).
-
2.
Adjust the pitch and yaw of the IR laser diode mount (KAD8F) to achieve alignment using two iris diaphragms along the segment optical axis. Use the aid of a Ø1″ IR Alignment Disk (VRC2D1) that can be easily inserted into Ø1″ diaphragms or any similar IR alignment tool to be able to see the IR beam.
-
3.
Adjust the two kinematic mirror holders (KCB1_M.002-3) so the beam passes through the center optical axis of the cube (C6WR.002) without the dichroic mirror.
-
4.
Orient the dichroic mirror within the cube (C6WR.002) for 45° AOI and move the cube along the rods so that the IR laser beam passes through the aligning tool installed in place of the objective. Revisit step (3) if necessary.
-
5.
Reinstall the lens (LA1172_B_ML) and roughly position it one focal length (∼400 mm) away from the BFP of the objective by observing the beam focused on the bottom frosted glass disk of the alignment tool.
-
6.
Remove the aligning tool and reinstall the objective, and the laser line filter (FL830_10.001).
-
7.
Place the sample and bring it into contact with the objective. Find the surface by fine focusing.
-
8.
Translate the IR segment off-axis to adjust the TIRF angle until the IR beam on the camera reaches TIR conditions (the spot will become more intense at the critical angle); one also might need to adjust the on-axis position of the lens (LA1509_B_ML).
-
9.
Fasten the IR TIRF segment.
Emission path alignment
-
1.
Remove all lenses along the emission path, the objective, the camera, and any filters.
-
2.
Attach a collimated laser source at the end of the alignment tool with a mount that allows for pitch/yaw and XY position adjustments, so the beam passes through the two frosted glass disk irises. Adjust the elliptical mirror kinematic mount (KCB1EC_M) knobs and its vertical position so the beam propagates on-axis along all apertures reaching the camera mount.
-
3.
Install the tube lens (MXA20696) back and adjust its off-axis position using the slip plate (SPT2_M) so the laser beam is on-axis.
-
4.
Slide the adapter plate (LCP33_M.005) with the iris diaphragm (SM1D12D.003) located at the focal plane of the tube lens (MXA20696) where the beam is focused; an alternative way is to position your phone camera instead of the alignment tool on the sample holder and slide till the iris diaphragm is sharply in focus with the camera focus set to infinity.
-
5.
Reinstall the second 4f configuration lens (AC254_200_A_ML.002) and adjust its height, position, and tilt so the beam stays on-axis. Note that one can replace rods (ER1.021-24) with 8-inch-long rods instead to avoid the tilt alignment. The beam should be collimated after the lens by translating it along the optical axis so the beam diameter on a far (>1 m) target/wall is minimized; an alternative is to place your phone camera instead of the detection camera and position the lens so the iris diaphragm is sharply in focus with the camera focus set to infinity.
-
6.
Adjust the filter cube (DFM1B_M) position, height, and tilt, so the beam alignment after it stays on-axis.
-
7.
Remove the alignment laser, and reinstall the camera (Alvium_1800_U_158m), its lens (89_Achromat_edmund_49_782), and any filters on the path. Adjust the lens on-axis position so the iris diaphragm is sharply in-focus, and shine some light on the iris diaphragm if needed. Adjust the XY position of the camera so the view is roughly at the center of the chip.
-
8.
Reinstall the objective, use a test sample and revisit the alignment if necessary.
Changing the modes of excitation
-
1.
In SMF excitation, adjust the position of the 25 mm translation stage so that the beam after the objective is perpendicular to the sample plane.
-
2.
To move from SMF excitation to MMF, install the magnetic base (KB25_M.001) mounted mirror at the laser engine, then install the magnetic base (KB25_M.002) mounted mirror at the excitation path. Note that changing from SMF to MMF mode does not require realignment as the used magnetic bases offer high repeatability.
-
3.
To move from the MMF excitation to the SMF, remove the two previously installed mirrors. By moving the 25.0 mm translation stage, one can move from Epi- to TIRF-mode.
Routine operation
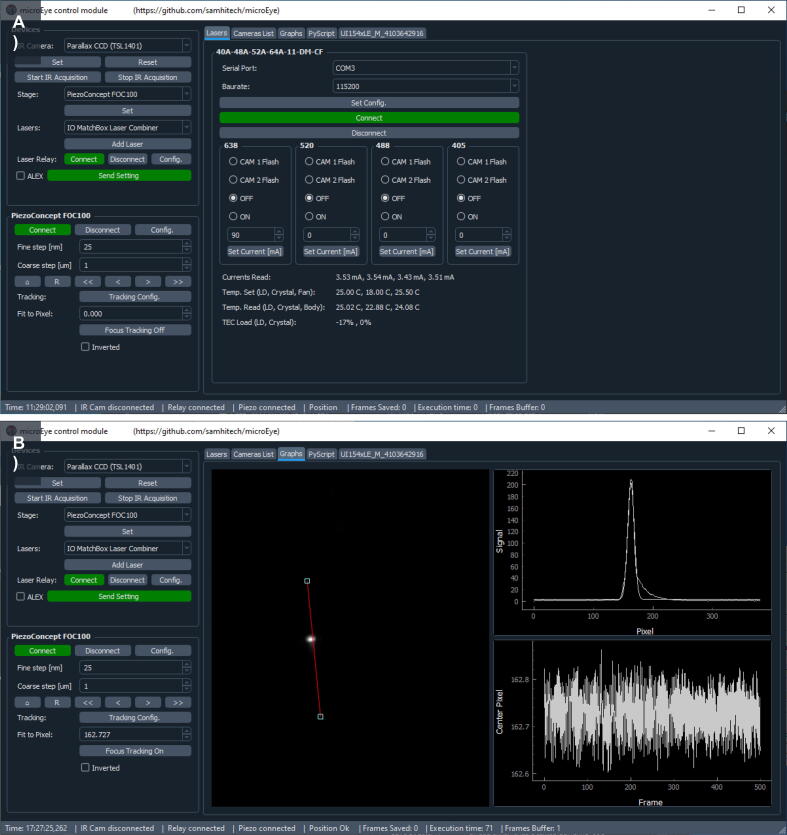
The microEye Control module GUI ( Fig. 14 )
-
1.
To start the hardware control GUI, run the example “control_module.py” script using Python. The GUI will appear (Fig. 14A), and you will be able to select a camera for IR beam recording, lasers, and a piezo controller for active focus stabilization.
-
2.For IR laser beam position acquisition, select the desired camera at the “Camera List” panel. A new tab on the right-hand side will appear (e.g. “UI154xLE_M_4103642916”).
-
a.Set parameters for this camera in this new panel: the pixel clock speed (we used 25 MHz), frame rate (∼14 frames/s), and frame exposure time (depends on the IR beam intensity and has to be set so that the beam would not oversaturate the detector).
-
b.Once settings are set, start IR acquisition by pushing the freerun button, then go to the ‘Graphs’ tab.
-
c.Move the sample into the approximate focus position using the manual coarse z-axis sample stage. The IR laser beam back-reflection will appear on the image as a linearly translating spot.
-
d.Position the line profile ROI so that it would follow the movement of the beam (Fig. 14B). Once that is done, the sample can be moved into focus more precisely using the piezo z-stage. Do this only after setting up the acquisition module and seeing the sample fluorescence on the camera.
-
a.
-
3.The miEye uses an “Integrated optics” 4-wavelengths laser diode combiner.
-
a.Plug the combiner power supply in a power socket and the USB cable in a USB-A 2.0 slot on the PC.
-
b.Make sure that the Arduino-based laser relay box is also connected to a USB-A 2.0 slot on the PC, and the trigger pins from the camera and laser combiner are connected to Arduino according to the build instructions.
-
c.Set the laser relay box COM port settings using the “config” button, then connect to it. (If successfully connected, the “connect” button turns from red to green.)
-
d.Select the combiner from the laser source pop-up menu and push the ‘Add Laser’ button. A new panel in the tab “Lasers” on the right-hand side will appear. There select the COM port, to which the combiner is connected, press the “set configuration” button, and then the “connect” button. (If successfully connected, the “connect” button turns from red to green.)
-
e.Four laser control panels will appear for each of the different lasers, and they allow for using the camera as a trigger, switching the laser continuously on or continuously off. After changing any of those triggering options, send the configuration to the laser relay box by pushing the “Send Setting” button.
-
a.
-
4.Connect the piezo controller to the PC via USB cable and switch its power button on.
-
a.In the “control module” GUI select the correct piezo controller and click the “Set” button. At the bottom left corner a piezo z-axis control panel will appear (e.g. “PiezoCoencept FOC100”).
-
b.Set the piezo controller COM port settings using the “config” button, then connect to it. (If successfully connected, the “connect” button turns from red to green.)
-
c.The panel then allows for moving the objective z-axis piezo stage in fine and coarse steps. Also, it performs active piezo stabilization based on detecting the central peak of the back-reflected IR beam and continuously driving the piezo Z-stage to keep the sample at the set z-position. To enable autofocusing press the “Autofocus off” button, and it will change into “Autofocus on”.
-
a.
Fig. 14.
The microEye GUI for the Control module contains a “Devices” selection panel (top left part of GUI), piezo control panel (bottom left of GUI), and specific tabs: “Lasers” (visible in panel A) for laser control (displayed in the top image of this figure), “Camera List” allows selecting camera for the back-reflected IR laser position imaging, “Camera title” for setting parameters of the selected camera, “Graphs” (visible in panel B) for displaying the camera output and visualizing the IR laser beam’s center position (displayed in the bottom image of this figure), and scripting (right side).
The microEye Acquisition module GUI (Fig. 15).
Fig. 15.
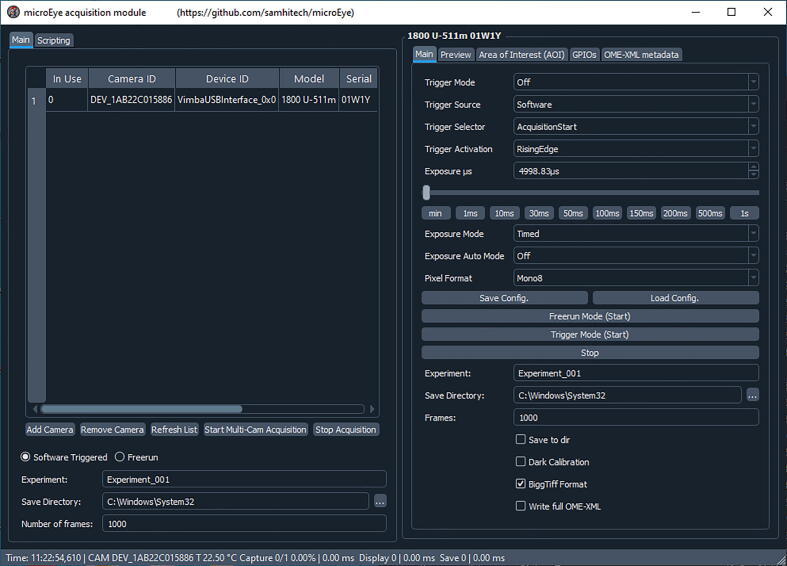
The microEye GUI for the Acquisition module contains the main panel (left side) for camera selection and specific panels for setting parameters for the selected camera (right side).
The microEye software GUI of the Acquisition module contains the main panel for camera selection and specific panels for setting parameters for each selected camera. The specific panel entitled “Main” is dedicated to setting triggering, exposure time and mode, pixel format, experiment filename, directory for storing acquired files, and a number of frames to acquire. Also, it contains options to enable: saving the acquired images, dark calibration mean and variance maps generation, storing the acquired images as BiggTiffFormat files and writing the full metadata when saving images. Specific panel “Preview” allows autoscaling of acquired images, setting the scale manually and displaying histograms of pixel intensity values. The panel “Area of Interest (AOI)” allows setting the area of interest for the camera. The panel “GPIOs” allows setting the triggering scheme of the camera. The panel “OME-XML metadata” allows storing parameters set in this acquisition module.
-
1.
The “Acquisition module” and “Control module” work as an independent process from each other, as the first control cameras settings and performs data acquisition, while the second manages the rest of the hardware used by the microscope; such as which laser will switch on during image series acquisition will depend on the configuration of the laser relay box that is sent to Arduino using the “Control module”.
-
2.
Before starting the module connect your cameras to PC.
-
3.
To start the “Acquisition module”, open the “acquisition_module.py” example using Python.
-
4.
The GUI will appear (Fig. 15), and there you will be able to add a camera. More than one camera can be added for dual-camera imaging, and the procedure is the same as adding a single camera.
-
5.
To add the camera, select a specific camera from the list of connected devices, then push the “Add Camera” button and a new panel will appear on the right-hand side.
-
6.
On the GPIOs tab, set the line 0 for triggering as output and to be active during acquisition or exposure for instance.
-
7.
Then set acquisition parameters: such as bit-depth “Pixel Format”, frame rate, and exposure time among any other camera-specific parameters.
-
8.
There is a possibility to set the area of interest to record only part of the sensor.
-
9.
At the bottom part of the panel several options are provided to enable storing data, along with saving images as bigTIFF format and including full metadata. Also, one can select the data storage directory where a folder with the experiment field is created with the data within.
-
10.
Once all of the aforementioned settings are selected, one can start acquiring image series by pushing the “Start freerun” button. If saving is enabled, the stack will be saved at the selected directory into a folder with the provided filename. Otherwise, the stack won’t be saved.
The microEye TiffViewer module GUI (Fig. 16, Fig. 17, Fig. 18).
-
1.
To start the Tiff Viewer module, open the “tiff_viewer.py” using Python.
-
2.
This GUI has three main panels: “File system” (Fig. 16), “Tiff options” (Fig. 17), and “Localization / Render” (Fig. 18).
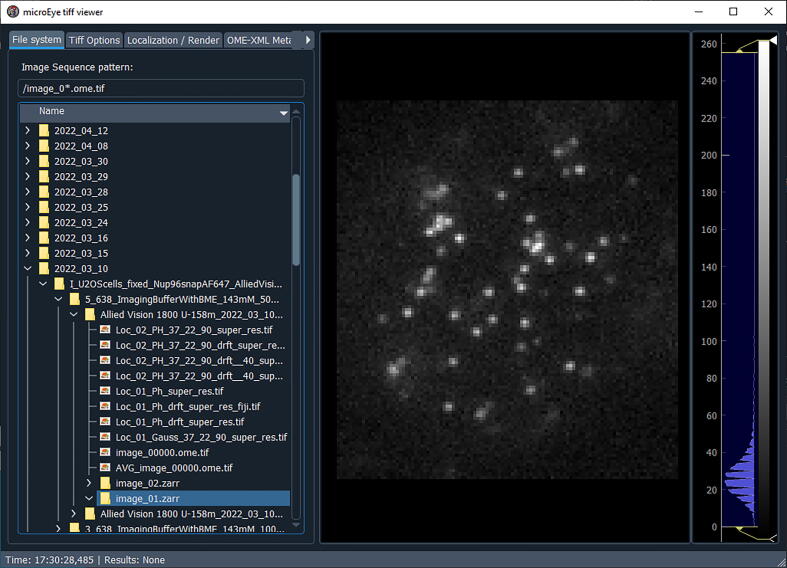
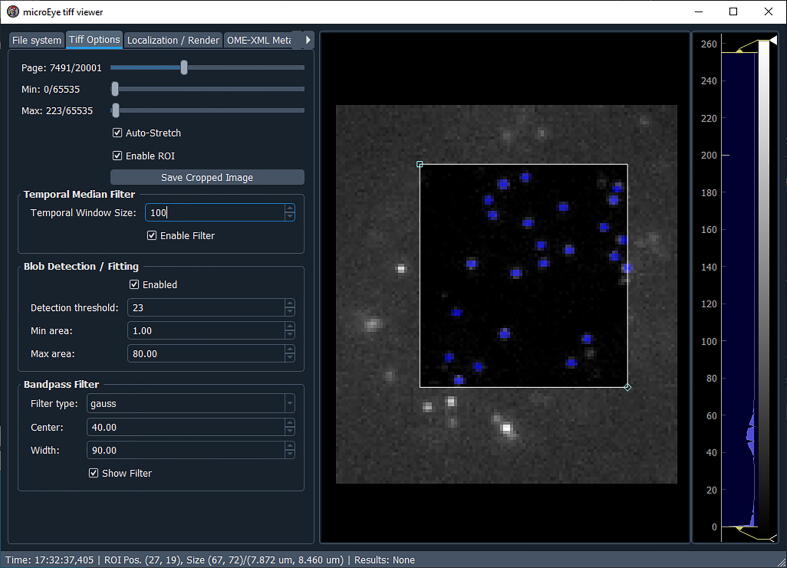
Fig. 16.
The microEye GUI for the Tiff viewer module’s “File system” panel.
Fig. 17.
The microEye GUI for the Tiff viewer module’s “Tiff options” panel.
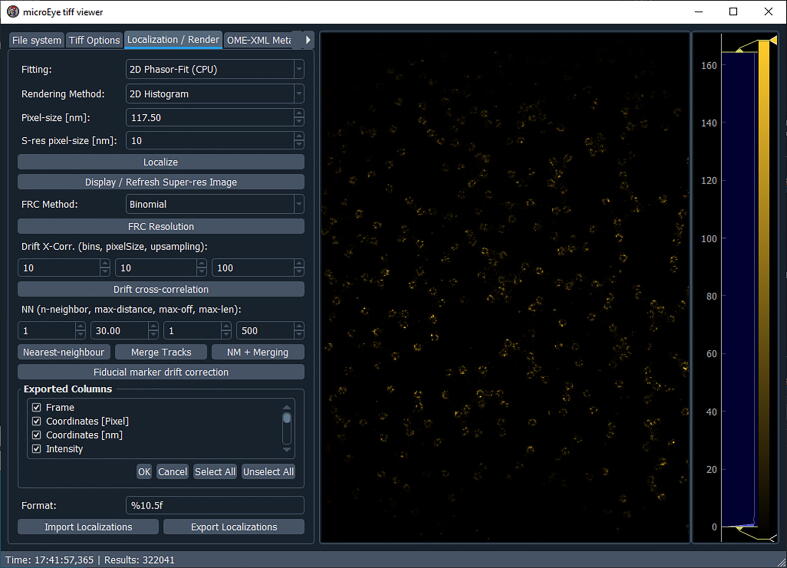
Fig. 18.
The microEye GUI for the Tiff viewer module’s “Localization / Render” panel.
Selecting the desired file for analysis
-
3.
First, in the “File system” panel select the folder with the image stack that you want to analyze by navigating to that file and double-clicking it (Fig. 16).
-
4.
The selected file will be displayed in the “Tiff Options” panel (Fig. 17).
Setting parameters for fluorescent spot detection ( Fig. 17 )
-
1.
In the “Tiff Options” panel, find the desired image by sliding through the image stack (use the “Page” slider).
-
2.
Set the lower and higher intensity levels of the initial look-up table (LUT) of the image (“Min” and “Max” sliders).
-
3.
There is a possibility to analyze only a part of the image that can be set using the “Enable ROI” check box. Once enabled, interactive markers for ROI on the right-hand side of the image will appear.
-
4.
Set the window size for “Temporal Median Filter” to remove non-blinking spots, and confirm its performance by sliding through the image stack.
-
5.
Set parameters for Blob Detection/Filtering. Set the detection sensitivity by adjusting the “Detection threshold” value, and set the minimal and maximal possible area of our fluorescent spots (“Min area” and “Max area”).
-
6.
Set image bandpass filtering parameters: “Filtering type”, center value, and width. If the “Show Filter” checkbox is enabled, an image displaying the filter shape will appear. Select the best performing filter type and then work the center/width parameters and adjust the detection threshold such that the desired spots would be detected (marked in blue).
Setting parameters for localization and rendering ( Fig. 18 )
-
1.
In the “Localization / Rendering” panel, select the fitting model (either Phasor or 2D Gaussian), rendering method (we recommend using 2D histogram rendering), projected pixel size of the camera, and the SR pixel size (recommendation is to use 10-fold lower value than the pixel size).
-
2.
Click the “Localize” button and the localization process will begin.
-
3.
Rendered images will be automatically displayed.
-
4.
To remove sample drift in X and Y, set “Drift X-Corr.” parameters: number bins (start by 10), pixel size (start with a similar value as the set SR pixel size), and upsampling (keep it around 100). Once parameters are set, click the “Drift cross-correlation” button.
-
5.
To group the localizations belonging to the same fluorescent spot, use Nearest-neighbor and merging. Set the “n-neighbor”, “max-distance”, “max-off”, “max-len” parameters, and group localizations by pressing: “Nearest-neighbor” -> “Merge Tracks” or just “NM + Merging”.
-
6.
The table with selected info (see the “Exported Columns” menu) including the SR image can be saved on the PC by clicking the “Export Localizations” button.
-
7.
Also, info from previously saved tables can be loaded for analysis.
-
8.
To estimate the resolution [41], select the FRC method and click the “FRC Resolution” button. If an error appears, try to lower the SR pixel size.
Routine maintenance
For cleaning of the objective lens, lenses, and filters follow procedures provided by the vendor of those items. Before an experiment (especially after changing the mode of excitation from SMF into MMF), it is a good practice to visually inspect the alignment of the whole path from the fiber end to the objective, otherwise, perform this routine weekly. Also use a glass coverslip covered with e.g. fluorescently labeled protein (no protein in solution, just on the surface) and check out whether the laser’s illumination profile is at the center of the image.
Validation and characterization
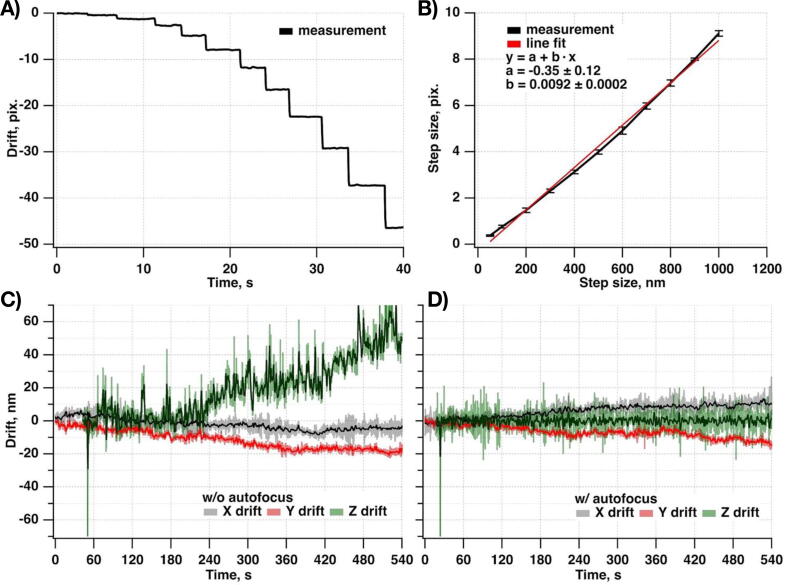
Stability of the system in X, Y, and Z axes
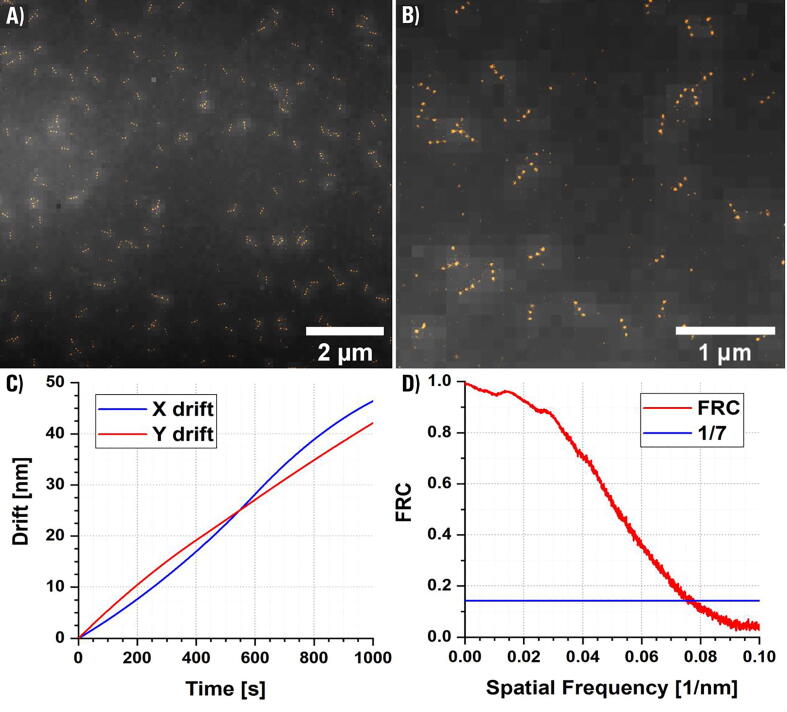
To estimate linear drift we performed SMF-based epi-mode imaging of surface-immobilized 200 nm diameter polystyrene Crimson beads (Invitrogen, F8806) under 638 nm wavelength excitation with 50 ms exposure time with and without closed-loop Z-axis drift-suppressions system in place for 10 min time period.
The calibration curve to convert the back-reflected IR beam displacement from pixels into sample drift along the Z-axis in nanometers was evaluated by performing individual piezo Z stage steps (11 steps: 50, 100, 200, 300, 400, 500, 600, 700, 800, 900, 1000 nm) while recording the IR beam position (Fig. 19A-B).
Fig. 19.
Typical drift experienced by the miEye. A-B) Calibration of the IR beam displacement from pixels into nm. Typical drift C) and D) in X (gray), Y (red), and Z (green) as experienced by the miEye. C) without, and D) with an active drift-suppressions system. Raw signals are represented in lighter colors, and filtered using 5-point box smoothing are represented by a darker color. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
During these experiments, we recorded the back-reflected IR laser beam position (estimated by a Gaussian function fit to the line profile taken along the movement axis of the IR beam on the camera) on the camera of the autofocusing system. We fitted fluorescent beads in the acquired image series using a 2D Gaussian function and extracted their X and Y center positions. Next, we estimated the average drift of both X, and Y center positions over time for all the detected beads. During this experiment, we introduced a 100 nm step in the Z direction using the objective piezo stage to mark the beginning of acquisition, and then the objective was moved back to the original focus location using autofocus which was later turned off immediately for the case without autofocus.
A typical drift measurement is shown in Fig. 19C-D. These curves indicate that the system experiences a drift of <7 nm/min in the lateral plane and in the axial plane without active drift-suppressions systems in place. The active Z-axis drift-suppression system allowed us to stabilize the Z-axis drift such that during the whole measurement it fluctuated 10 nm around the same stable value. Note that the IR beam position fluctuates due to mode jumps and therefore the apparent Z axis fluctuation is likely lower than reflected by this Z-drift curve. Future 3D SMLM implementation by introducing astigmatism into the emission path would allow us to investigate the source of the aforementioned Z-axis fluctuation.
Different modes of imaging
We tested image quality and the possibility to obtain different modes of excitation (Epi, TIRF: at the critical angle, and shallower TIRF: at a maximal angle that still gives a visible signal of all beads) for the SMF-based excitation scheme using the same aforementioned Crimson beads sample. Different angles of excitation were achieved by translating the whole excitation arm using the 25 mm translation stage.
First, we moved the stage to a position where the excitation beam was perpendicular to the sample surface (overlapping the beam and reflection at the objective back focal plane imaged on the camera), noted down the readings of the stage micrometer (i.e. 20.75 mm), and recorded 10 image-long series. Here we provide an average image over this series of images (Fig. 20A). Next, we adjusted the micrometer and observed the beads' fluorescence intensity (autoscaling of the grayscale was switched off) on the camera image. We translated the stage until we observed the maximal intensity of the beads, which indicated that we reached the critical angle (i.e. 16.28 mm), and again recorded 10 image-long series. An average image over this series of images showed an increased intensity of ∼3-fold compared to the Epi-mode (Fig. 20B and D). Finally, we translated the stage even more until we observed the minimal intensity of the beads (after that we saw a complete loss of the signal), which indicated that we reached the maximal possible angle (i.e. 15.78 mm), and again recorded 10 image-long series. An average image over this series of images showed a decrease of intensity of ∼25-fold compared to the Epi-mode, and 65-fold compared to the critical angle (Fig. 20C and D). Such change is expected since the penetration depth of the evanescent field, which is generated in TIRF excitation, is dependent on the angle of incidence and is comparable to the beads size (Fig. 20E).
Fig. 20.
Intensity of crimson beads at different modes (angles) of single-mode fiber (SMF) excitation. A)Epi (0–70 DU). B) Critical angle (0–160 DU). C) Maximal angle (0–40 DU). D) Barchart showing mean intensity (5003.7 ± 612.1 a.u.; 13843.8 ± 666.9 a.u.;179.7 ± 5.6 a.u.) of crimson beads. E) Angle of incidence and penetration depth dependence for 638 nm wavelength. According to calculations, the critical angle was at 61.20° for 638 nm wavelength excitation, and the maximal angle was estimated to be ∼78.6°.
According to calculations (e.g. “Calc TIRF” plugin of ImageJ [42]) critical angle of our imaging system (638 nm excitation and 1.49NA objective, ∼1.52 index of immersion oil) is ∼61.2° with the highest angle being ∼78.6°. The critical and maximal excitation angles of the images (Fig. 20B and C) were interpolated from Abbe sine condition [43] and are within 1.2 % and 2.7 % error, respectively. Note that the sample was kept in focus using our active drift-suppressions system during the whole experiment.
We measured the illumination profile with the SMF excitation under 638 nm wavelength excitation at Epi-, CA, and TIRF modes using the glass coverslip with adsorbed fluorescently labeled protein (Fig. 21A-C). We refer the reader to the “Samples for testing of the set-up” subsection of this work for this sample preparation instructions. By taking horizontal and vertical line scans we obtained Gaussian-like intensity profiles (Fig. 21D-E). For the Epi mode intensity at the center of illumination was ∼80 a.u. The intensity dropped from ∼750 a.u. to ∼345 a.u. when we moved from CA into the TIRF mode. Obtaining more homogeneous illumination with SMF excitation would require massively increasing the illuminated area and restricting the field-of-view to the center peak, sacrificing most of the laser intensity.
Fig. 21.
Illumination profile at different modes of excitation with single-mode fiber (SMF). A)Epi mode (0–80 DU). B) At a critical angle (0–660 DU). C) At the TIRF mode (0–345 DU). Lines indicate the position used for making the 30-pixels-wide line profiles. D) Horizontal, and E) vertical line profiles. The sample is a glass coverslip covered with sAv-ATTO655 molecules.
The MMF illumination (MMF output imaged on the sample plane) system with a fiber agitator is able to achieve the homogeneous illumination required for fast and high-resolution SMLM. To image illumination profiles, we prepared a “dye sandwich” in between two coverslips, and directly observed the illumination profile [44]. With fiber agitation, speckles are averaged out for exposure times >= 10 ms (Fig. 22A-B), with small residual speckles observed for the lowest 3 ms exposure time. The lateral profile with and without agitation allows comparing the amplitude of the speckles to the inhomogeneity of the illumination (Fig. 22C). Note that, for all lasers, the corners of the fiber are of slightly lower intensity.
Fig. 22.
Homogeneous illumination by speckle reduction and the output of the multi-mode fiber (MMF) imaging onto the objective’s sample plane. A) Images of the dye sandwich sample with the MMF agitation module switched off for the 638 nm diode laser at various exposure times. The actual exposure time is indicated at the bottom of each image. B) Same profiles with the MMF agitation module switched on at the speed of 1150 rpm. C) Lateral intensity profiles of (A) (black curve) and (B) (red curve). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Super-resolution imaging using a MMF
To demonstrate the SMLM imaging capabilities of our microscope system arranged in an MMF-based excitation scheme, we chose to perform dSTORM imaging of nuclear pore complexes (NPCs) composed of fluorescently labeled nucleoporin Nup96-SNAP-tag conjugates in U2OS cells. One of the main reasons behind this decision is that recently it has been shown that nuclear pores serve as a versatile and biologically relevant reference standard for benchmarking the SR imaging capabilities of various microscopes [45]. Since U2OS cell lines expressing Nup96 proteins endogenously tagged with different labels are available commercially, it was selected as indeed a convenient and informative sample for such a purpose.
dSTORM experiments were performed employing a MMF whose output was imaged onto the sample plane of an objective. To minimize the number of speckles generated in this type of excitation, the speed of the MMF agitator was maintained at 1150 rpm during the whole experiment. The sample was illuminated using a 638 nm laser with its power set to ∼3.6 kW/cm2 and focused on the lower flat envelope of the cell’s nucleus. Fluorescence emission was filtered through a 697/75 bandpass filter. The projected pixel size was estimated to be ∼127.9 nm in XY. The 20 000 frames-long movies were acquired at 20 Hz.
By analyzing and post-processing the dSTORM data acquired using a MMF-based excitation scheme, we were able to reconstruct a SR image of the fluorescently labeled NPCs distributed over the entire nucleus of a single U2OS cell (Fig. 23B). Individual NPCs with their characteristic ring-like structure are clearly resolved in the resulting SR image and in some of its parts even the eight corners, where the nucleoporin Nup96-SNAP-AF647 conjugates are located, of these NPCs can be seen (Fig. 23C). In comparison, the averaged image of the “raw” 20 000 frames-long stack imparts no details about the structural properties of these NPCs and it also barely provides any information about their arrangement across the lower surface of the cell’s nucleus since discrete NPCs are almost indistinguishable in such image (Fig. 23A). The resolution value of the aforementioned reconstructed SR image reported by the Fourier ring correlation was equal to 34.7 nm (Fig. 23D). By utilizing the Nup96-SNAP-containing NPCs as a quantitative reference structure and achieving such spatial resolution with this approach, we confirm the SR imaging capabilities of our microscope system set in a MMF-based configuration and demonstrate that the laser powers obtained in such excitation scheme are sufficient for a rapid and efficient dSTORM data acquisition when using low exposure times of the detection camera. Therefore, we highly anticipate that miEye will maintain the same or even better level of performance in other types of SMLM measurements of more sophisticated and less-characterized biological objects and systems.
Fig. 23.
dSTORM imaging of fluorescently labeled nuclear pore complexes (NPCs) of fixed U2OS-Nup96-SNAP cells. A) Averaged image of the unprocessed 20,000 frame-long image stack showing an individual nucleus of a U2OS cell. B) Reconstructed super-resolution image of Nup96-SNAP-AF647-containing NPCs located at the distinct positions of a single U2OS cell’s entire nucleus. C) Close-up view of a chosen nucleus area (indicated by a white box in panel B). Individual NPCs and their ring-like arrangement are visible. D) Fourier ring correlation (FRC) curve indicating the obtained spatial resolution of 34.7 nm.
Super-resolution imaging using SMF
To assay the achievable optical resolution of the miEye using the SMF excitation scheme, we employed a commercially available DNA-PAINT sample containing DNA nano-rulers with 80-nm spacing in between three docking sites. Using these DNA nano-rulers immobilized onto a coverslip surface, we performed DNA-PAINT with ATTO647N-labeled imager strands. The SR reconstruction and subsequent drift correction (Fig. 24C) show the resolved 80-nm-ruler-pattern (Fig. 24A-B). To quantify the achievable resolution we employed the FRC method that showed ∼14 nm localization precision (Fig. 24D). This demonstrated that high lateral resolution using DNA-PAINT with the miEye can be achieved.
Fig. 24.
TIRF microscopy image of sparsely distributed DNA nano-rulers with 80-nm spacing. Three independent imaging experiments under the 638 nm wavelength excitation were done with a similar outcome. A) Larger field-of-view (FOV) conventional TIRF microscopy average stack overlaid with reconstructed super-resolution image. B) Zoomed image of the larger FOV presented in panel A. C) Graph showing curves of X and Y drift used for drift correction using cross-correlation. D) FRC curve, which showed a resolution value of 14.3 nm.
Concluding remarks
The miEye bench-top microscopy system represents an addition to the current open source SMLM options available to researchers. It has proven ability in white light and fluorescent imaging in vitro and fixed cell culture, maintaining high resolution, stability, and reliability. The low-cost of the build compared to commercial options, modularity, and the open-source nature of the microscope facilitate its accessibility to a wide range of potential users. Our microEye Python package allows for reliable microscope control, easy data acquisition, visualization, and analysis. The microEye is an open-source project that targets the microscopy community and can be extended to support other imaging modalities or analysis methods.
Capabilities/limitations
-
•
TIRF microscopy imaging of single molecules.
-
•
dSTORM imaging.
-
•
DNA-PAINT imaging.
-
•
14 nm achievable resolution with calibration samples for DNA-PAINT.
-
•
Typical sample drift along X and Y axis is ∼3 nm/min.
-
•
Has an active drift-suppressions system to compensate for the Z-axis drift of the sample.
-
•
The dedicated Python package “microEye” enables reliable microscope control, data acquisition and analysis within a single environment.
-
•
The build uses a custom CNC milled three-layer cube as a core of the set-up thus any further changes are limited by this part.
-
•
Most of the other components are from Thorlabs catalog.
-
•
Currently it has no heating and therefore imaging at RT is only possible.
Ethics statements
Human and animal rights: none.
CRediT authorship contribution statement
Mohammad Nour Alsamsam: Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Aurimas Kopūstas: Investigation, Methodology, Writing – original draft. Meda Jurevičiūtė: Investigation, Methodology. Marijonas Tutkus: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This project has received funding from the European Social Fund (project No 09.3.3.-LMT-K-712-19-0113 for M.T.) under a grant agreement with the Research Council of Lithuania (LMTLT), Lithuanian Research Council (S-MIP-20-55 for M.T.), and also from Vilnius University (MSF-JM-10/2021).
References
- 1.Huang B., Bates M., Zhuang X. Super-resolution fluorescence microscopy. Annu. Rev. Biochem. 2009;78(1):993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu S., Hoess P., Ries J. Super-resolution microscopy for structural cell biology. Annu. Rev. Biophys. 2022;51(1):301–326. doi: 10.1146/annurev-biophys-102521-112912. [DOI] [PubMed] [Google Scholar]
- 3.Hell S.W., Sahl S.J., Bates M., Zhuang X., Heintzmann R., Booth M.J., Bewersdorf J., Shtengel G., Hess H., Tinnefeld P., Honigmann A., Jakobs S., Testa I., Cognet L., Lounis B., Ewers H., Davis S.J., Eggeling C., Klenerman D., Willig K.I., Vicidomini G., Castello M., Diaspro A., Cordes T. The 2015 super-resolution microscopy roadmap. J. Phys. D Appl. Phys. 2015;48 [Google Scholar]
- 4.Hohlbein J., Diederich B., Marsikova B., Reynaud E.G., Holden S., Jahr W., Haase R., Prakash K. Open microscopy in the life sciences: quo vadis? Nat. Methods. 2022;19(9):1020–1025. doi: 10.1038/s41592-022-01602-3. [DOI] [PubMed] [Google Scholar]
- 5.Single-Molecule Techniques: A Laboratory Manual, Q. Rev. Biol. 83 (2008) 406–406.
- 6.Pearce J.M. Laboratory equipment: cut costs with open-source hardware. Nature. 2014;505:618. doi: 10.1038/505618d. [DOI] [PubMed] [Google Scholar]
- 7.OM_Hardware.md at main HohlbeinLab/OpenMicroscopy, Github, n.d. https://github.com/HohlbeinLab/OpenMicroscopy (accessed July 7, 2022).
- 8.Diederich B., Then P., Jügler A., Förster R., Heintzmann R., Waigh T. cellSTORM-Cost-effective super-resolution on a cellphone using dSTORM. PLoS ONE. 2019;14(1):e0209827. doi: 10.1371/journal.pone.0209827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.10.1101/2020.09.04.283085.
- 10.Kwakwa K., Savell A., Davies T., Munro I., Parrinello S., Purbhoo M.A., Dunsby C., Neil M.A.A., French P.M.W. easySTORM: a robust, lower-cost approach to localisation and TIRF microscopy. J. Biophotonics. 2016;9:948–957. doi: 10.1002/jbio.201500324. [DOI] [PubMed] [Google Scholar]
- 11.G. Lab, K2 TIRF, (2020). https://ganzingerlab.github.io/K2TIRF/K2TIRF/component_table.html (accessed July 7, 2022).
- 12.J. Edwards, LifeHack Microscope: Home, (n.d.). https://holdenlab.github.io/LifeHackWebsite/home.html (accessed July 7, 2022).
- 13.Auer A., Schlichthaerle T., Woehrstein J.B., Schueder F., Strauss M.T., Grabmayr H., Jungmann R. Nanometer-scale multiplexed super-resolution imaging with an economic 3D-DNA-PAINT microscope. ChemPhysChem. 2018;19(22):3024–3034. doi: 10.1002/cphc.201800630. [DOI] [PubMed] [Google Scholar]
- 14.Prakash K. Laser-free super-resolution microscopy. Philos. Trans. A Math. Phys. Eng. Sci. 2021;379:20200144. doi: 10.1098/rsta.2020.0144. [DOI] [PubMed] [Google Scholar]
- 15.Martens K.J.A., van Beljouw S.P.B., van der Els S., Vink J.N.A., Baas S., Vogelaar G.A., Brouns S.J.J., van Baarlen P., Kleerebezem M., Hohlbein J. Visualisation of dCas9 target search in vivo using an open-microscopy framework. Nat. Commun. 2019;10:3552. doi: 10.1038/s41467-019-11514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sofroniew N., Lambert T., Evans K., Nunez-Iglesias J., Bokota G., Winston P., Peña-Castellanos G., Yamauchi K., Bussonnier M., Doncila Pop D., Can Solak A., Liu Z., Wadhwa P., Burt A., Buckley G., Sweet A., Migas L., Hilsenstein V., Gaifas L., Bragantini J., Rodríguez-Guerra J., Muñoz H., Freeman J., Boone P., Lowe A., Gohlke C., Royer L., Pierré A., Har-Gil H., McGovern A. napari: a multi-dimensional image viewer for Python. Zenodo. 2022 doi: 10.5281/ZENODO.3555620. [DOI] [Google Scholar]
- 19.napari — napari, (n.d.). https://napari.org/ (accessed July 7, 2022).
- 20.Ries J. SMAP: a modular super-resolution microscopy analysis platform for SMLM data. Nat. Methods. 2020;17:870–872. doi: 10.1038/s41592-020-0938-1. [DOI] [PubMed] [Google Scholar]
- 21.Edelstein A., Amodaj N., Hoover K., Vale R., Stuurman N. Computer Control of Microscopes Using µManager. Curr. Protocols Mol. Biol. 2010;92(1) doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreno X., Al-Kadhimi S., Alvelid J., Bodén A., Testa I. ImSwitch: Generalizing microscope control in Python. J. Open Source Softw. 2021;6:3394. [Google Scholar]
- 23.Phillips M.A., Susano Pinto D.M., Hall N., Mateos-Langerak J., Parton R.M., Titlow J., Stoychev D.V., Parks T., Susano Pinto T., Sedat J.W., Booth M.J., Davis I., Dobbie I.M. Microscope-Cockpit: Python-based bespoke microscopy for bio-medical science. Wellcome Open Res. 2021;6:76. doi: 10.12688/wellcomeopenres.16610.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D.M.S. Pinto, Python microscope, (n.d.). https://www.micron.ox.ac.uk/software/microscope/ (accessed July 7, 2022).
- 25.Marin Z., Graff M., Barentine A.E.S., Soeller C., Chung K.K.H., Fuentes L.A., Baddeley D. PYMEVisualize: an open-source tool for exploring 3D super-resolution data. Nat. Methods. 2021;18:582–584. doi: 10.1038/s41592-021-01165-9. [DOI] [PubMed] [Google Scholar]
- 26.python-microscopy, (2015). https://python-microscopy.org/ (accessed July 10, 2022).
- 27.Pinkard H., Stuurman N., Ivanov I.E., Anthony N.M., Ouyang W., Li B., Yang B., Tsuchida M.A., Chhun B., Zhang G., Mei R., Anderson M., Shepherd D.P., Hunt-Isaak I., Dunn R.L., Jahr W., Kato S., Royer L.A., Thiagarajah J.R., Eliceiri K.W., Lundberg E., Mehta S.B., Waller L. Pycro-Manager: open-source software for customized and reproducible microscope control. Nat. Methods. 2021;18:226–228. doi: 10.1038/s41592-021-01087-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ovesný M., Křížek P., Borkovec J., Svindrych Z., Hagen G.M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohammad Nour Alsamsam, microEye: A python toolkit for fluorescence microscopy, Github, n.d. https://github.com/samhitech/microEye (accessed July 21, 2022).
- 30.M. Tutkus, Tutkus lab, Tutkus Lab. (n.d.). https://tutkuslab.github.io/ (accessed July 21, 2022).
- 31.Diekmann R., Till K., Müller M., Simonis M., Schüttpelz M., Huser T. Characterization of an industry-grade CMOS camera well suited for single molecule localization microscopy – high performance super-resolution at low cost. Sci. Rep. 2017;7:14425. doi: 10.1038/s41598-017-14762-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van den Eynde R., Sandmeyer A., Vandenberg W., Duwé S., Hübner W., Huser T., Dedecker P., Müller M. Quantitative comparison of camera technologies for cost-effective super-resolution optical fluctuation imaging (SOFI) J. Phys. Photonics. 2019;1 [Google Scholar]
- 33.Diekmann R., Deschamps J., Li Y., Deguchi T., Tschanz A., Kahnwald M., Matti U., Ries J. Photon-free (s)CMOS camera characterization for artifact reduction in high- and super-resolution microscopy. Nat. Commun. 2022;13:3362. doi: 10.1038/s41467-022-30907-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schröder D., Deschamps J., Dasgupta A., Matti U., Ries J. Cost-efficient open source laser engine for microscopy. Biomed. Opt. Express. 2020;11:609–623. doi: 10.1364/BOE.380815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnitzbauer J., Strauss M.T., Schlichthaerle T., Schueder F., Jungmann R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 2017;12(6):1198–1228. doi: 10.1038/nprot.2017.024. [DOI] [PubMed] [Google Scholar]
- 36.Gao F., Mei E., Lim M., Hochstrasser R.M. Probing lipid vesicles by bimolecular association and dissociation trajectories of single molecules. J. Am. Chem. Soc. 2006;128(14):4814–4822. doi: 10.1021/ja058098a. [DOI] [PubMed] [Google Scholar]
- 37.Fazio T., Visnapuu M.-L., Wind S., Greene E.C. DNA curtains and nanoscale curtain rods: high-throughput tools for single molecule imaging. Langmuir. 2008;24:10524–10531. doi: 10.1021/la801762h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kopu̅stas A., Ivanovaitė Š., Rakickas T., Pocevičiu̅tė E., Paksaitė J., Karvelis T., Zaremba M., Manakova E., Tutkus M. Oriented soft DNA curtains for single-molecule imaging. Langmuir. 2021;37(11):3428–3437. doi: 10.1021/acs.langmuir.1c00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MicroEye, PyPI. (n.d.). https://pypi.org/project/microEye/ (accessed July 7, 2022).
- 40.Li Y., Mund M., Hoess P., Deschamps J., Matti U., Nijmeijer B., Sabinina V.J., Ellenberg J., Schoen I., Ries J. Real-time 3D single-molecule localization using experimental point spread functions. Nat. Methods. 2018;15(5):367–369. doi: 10.1038/nmeth.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nieuwenhuizen R.P.J., Lidke K.A., Bates M., Puig D.L., Grünwald D., Stallinga S., Rieger B. Measuring image resolution in optical nanoscopy. Nat. Methods. 2013;10(6):557–562. doi: 10.1038/nmeth.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.S. Rhode, Calculate TIRF, (n.d.). https://imagej.nih.gov/ij/plugins/tirf/index.html (accessed July 10, 2022).
- 43.Oheim M., Salomon A., Weissman A., Brunstein M., Becherer U. Calibrating evanescent-wave penetration depths for biological TIRF microscopy. Biophys. J. 2019;117(5):795–809. doi: 10.1016/j.bpj.2019.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deschamps J., Rowald A., Ries J. Efficient homogeneous illumination and optical sectioning for quantitative single-molecule localization microscopy. Opt. Express. 2016;24:28080–28090. doi: 10.1364/OE.24.028080. [DOI] [PubMed] [Google Scholar]
- 45.Thevathasan J.V., Kahnwald M., Cieśliński K., Hoess P., Peneti S.K., Reitberger M., Heid D., Kasuba K.C., Hoerner S.J., Li Y., Wu Y.-L., Mund M., Matti U., Pereira P.M., Henriques R., Nijmeijer B., Kueblbeck M., Sabinina V.J., Ellenberg J., Ries J. Nuclear pores as versatile reference standards for quantitative superresolution microscopy. Nat. Methods. 2019;16(10):1045–1053. doi: 10.1038/s41592-019-0574-9. [DOI] [PMC free article] [PubMed] [Google Scholar]