Abstract
Rumen microbes play an important role in ruminant energy supply and animal performance. Previous studies showed that yak (Bos grunniens) rumen microbiome and fermentation differ from other ruminants. However, little is understood about the features of the rumen microbiome that make yak adapted to their unique environmental and dietary conditions. This study was to investigate the rumen microbiome and metabolome to understand how yak adapt to the coarse forage and harsh environment in the Qinghai-Tibetan plateau. Nine female Qaidam yellow cattle (Bos taurus), 9 dzomo (hybrids of cattle and yak) and 9 female plateau yak (B. grunniens), about 5 to 6 years old, were used in this study. Rumen fermentation parameters, fibrolytic enzyme activities, and rumen metataxonomic were determined. Then 18 (6 samples per group) were selected for rumen metagenomic and metabolome analysis. Metataxonomic analysis revealed that the rumen microbiota was significantly different among plateau yak, Qaidam yellow cattle, and dzomo (P < 0.05). Metagenomic analysis displayed a larger gene pool encoding a richer repertoire of carbohydrate-active enzymes in the rumen microbiome of plateau yak and dzomo than Qaidam yellow cattle (P < 0.05). Some of the genes encoding glycoside hydrolases that mediate the digestion of cellulose and hemicellulose were significantly enriched in the rumen of plateau yak than Qaidam yellow cattle, but glycoside hydrolase 57 that primarily includes amylases was abundant in Qaidam yellow cattle (P < 0.05). The rumen fermentation profile differed also, Qaidam yellow cattle having a higher molar proportion of acetate but a lower molar proportion of propionate than dzomo and plateau yak (P < 0.05). Based on metabolomic analysis, rumen microbial metabolic pathways and metabolites were different. Differential metabolites are mainly amino acids, carboxylic acids, sugars, and bile acids. Changes in rumen microbial composition could explain the above results. The present study showed that the rumen microbiome of plateau yak helps its host to adapt to the Qinghai-Tibetan plateau. In particular, the plateau yak rumen microbiome has more enzymes genes involved in cellulase and hemicellulase than that of cattle, resulting higher fibrolytic enzyme activities in yak, further providing stronger fiber degradation function.
Keywords: High plateau ruminant, Rumen microbiome, Metagenome, Metabolome
1. Introduction
The rumen microbiota enables the digestion of feed and production of both the energy source (about 70% of the energy required by ruminants) in the form of volatile fatty acids (VFA, mainly acetate, propionate, and butyrate) and the protein source (about 70% of the total protein reaching the small intestines) in the form of microbial protein that the ruminants can directly utilize (Fan et al., 2020; Lin et al., 2019; Shabat et al., 2016; Stewart et al., 2018; Zebeli et al., 2015). Recent studies have revealed that rumen microbiota is associated with milk protein yield (Xue et al., 2020a), methane yield (Difford et al., 2018; Kamke et al., 2016), and feed efficiency (Li et al., 2019a). Besides the profound dietary effect (Li et al., 2019a; Spor et al., 2011), host genetics can also affect the rumen microbiota (Li et al., 2019a), and some rumen microbes may be “heritable” (Li et al., 2019b). Indeed, different species and breeds of ruminants can harbor their stable and heritable microbiota (Paz et al., 2016), probably a result of co-evolution and adaptation with the host.
The yak (Bos grunniens), the largest ruminant mammal living at the highest altitude in the world, has evolved to have some unique morphological and physiological mechanisms that allow them to adapt to the harsh environment (i.e., low oxygen, cold temperature, and limited availability of feed of low digestibility in the long winter) (Xin et al., 2019a). Compared to cattle, yak have shorter tongues and stronger and denser conical papillae, which allow for the consumption and digestion of the local forages that are poorly digestible (Shao et al., 2010). To adapt to the thin air and the low atmospheric oxygen concentration, yak have a larger heart and lung but smaller blood cells than cattle, all of which help improve the transport and supply of oxygen throughout their body (Durmowicz et al., 1993; Guan et al., 2017). Dzomo, the female hybrid of yak (B. grunniens) and taurine cattle (Bos taurus), adapt well to the local harsh environment and show hybridization advantage. Dzomo are more productive than yak in terms of both milk and meat yield, and possess better physiological adaptation to the high-altitude environment than cattle (Takase et al., 2002; Weir et al., 1974). A few studies have compared the rumen microbial composition between cattle and yak, but reports on rumen microbiota in dzomo are limited. Previous studies have shown differences in the rumen microbiota between yak and cattle, but the 2 ruminant species were kept at different altitudes (Xin et al., 2019b; Zhang et al., 2016). Christensenellaceae and Ruminococcaceae, which could be associated with feed efficiency (Myer et al., 2015; Perea et al., 2017), were enriched in yak compared to cattle (Xin et al., 2019b). Prevotella spp. increased in the rumen of yak compared to cattle (Zhang et al., 2016). Because alteration of rumen microbiota can impact rumen function and energy utilization in the host body, rumen microbiota may also contribute to host adaptation. We hypothesized that besides the adaptive evolution of the respiratory and the circulatory systems, the digestive system, especially the rumen microbiota, of yak had probably also undergone adaptive evolution to ingest and digest the available local feed. However, it remains to be determined if and how the yak rumen microbiota helps its host to adapt to its harsh environment.
Zhang et al. (2016) compared the rumen metagenome and rumen epithelial transcriptome of cattle and yak living at different altitudes. Their results showed that compared with cattle living at low altitudes, the rumen microbiome of the yak is enriched with VFA fermentation pathways and the yak rumen wall is more effective in absorbing VFA. That study suggests the contributions of the rumen microbiome to the adaptive evolution of ruminants living at high altitudes. However, their VFA results were obtained in vitro, and they only focused on VFA production and methanogenesis. Because diet, environment, and feeding can profoundly affect the rumen microbiome (Hugenholtz and de Vos, 2018; Woff et al., 2017), these confounding factors should be eliminated in comparative studies of different species or breeds of ruminants. The objective of this study was to elucidate the potential mechanism by which the rumen microbiota contributes to yak adaptation. To achieve this goal, we compared the rumen microbiota (composition and structure), fermentation, and function (both the metagenome and metabolome) among yak, cattle, and dzomo (designated as a species), all of which were kept at the same high altitude and fed the same diet. This study provided new knowledge of the yak rumen microbiome that might help understand its adaptation to high-altitude environments.
2. Materials and methods
2.1. Animal ethics statement
Animal care and experimental procedures were approved by the Institutional Animal Care and Use Committee (protocol number: NWAFAC1008) of the College of Animal Science and Technology of the Northwest A&F University (Yangling, Shaanxi, China).
2.2. Experiment design and sample collection
Nine female Qaidam yellow cattle (referred to as cattle hereafter), 9 female plateau yak (referred to as yak hereafter), and 9 female dzomo, each with similar body weight (about 200 kg) and age (5 to 6 years old), were used in this study. Because of the difficulty to weigh each of the grazing animals, each animal's bodyweight was visually estimated by an experienced herdsman. All study animals were raised in the Qinghai-Tibet plateau without any supplementary feed. Animals grazed at the same pasture each day and Kobresia myosuroides and Phragmites communis were the predominant pasture species. The ratio of K. myosuroides to P. communis was 9.1 ± 0.2. The sampling time was during winter, on December 31, 2017, after the animals had been grazing on this pasture for 62 days. Rumen fluid samples were collected before grazing in the morning from each animal using an oral stomach tube and a pump, both of which were thoroughly cleaned using fresh warm water between sample collections. The first 10 to 15 mL of the sample from each animal was discarded to avoid contamination from saliva. Ruminal pH was measured immediately after sampling using a pH meter. A subsample of 10 mL of ruminal fluid was filtered through 4 layers of cheesecloth. Five microliters were mixed with 1 mL of 25% metaphosphoric acid and stored at −40 °C until it was analysed for VFA. Another 5 mL of rumen fluid was centrifuged at 1,000 × g at 4 °C for 10 min, then the supernatant was immediately frozen in liquid nitrogen and then stored at −80 °C until plant cell-degrading enzyme activity analysis. The rest of the rumen fluid was immediately frozen in liquid nitrogen and then stored at −80 °C until analysis.
2.3. Analysis of VFA and plant cell-degrading enzyme activity
The rumen fluid samples were thawed at 4 °C, and the solid particles and protein were removed according to the procedures of Li et al. (2014a). The VFA concentrations were analysed using gas chromatography (Agilent Technologies 7820A GC system, Santa Clara, CA) equipped with a 30 m × 0.25 mm × 0.33 μm fused silica column (AE-FFAP, Atech Technologies Co. Ltd., Shanghai, China).
Centrifuged rumen fluid was thawed and immediately analyzed for the activities of carboxymethyl cellulase (CMCase), avicelase, xylanase, and acetylesterase using carboxymethyl cellulose, avicel, birchwood xylan, and p-nitrophenyl acetate, respectively, as the substrates. The enzyme assay reaction was incubated at 39 °C and pH 7.0 for 30 min except for the xylanase assay (for 15 min). The amounts of released reducing sugars were quantified using the dinitrosalicyclic acid colorimetry method at 540 nm (Cao et al., 2021; Miller, 1959), and the production of p-nitrophenol was determined at 415 nm (Yang and Yue, 2012; Yue and Yang, 2009). One milliunit of enzyme activity was defined as the amount of enzyme releasing 1 nmol of reducing sugar (e.g., xylose or glucose equivalent) or p-nitrophenol per min per microliter.
2.4. Metataxonomic analysis of rumen prokaryotes and fungi
Microbial DNA was extracted from each rumen sample using the E.Z.N.A. soil DNA Kit (Omega Bio-tek, Norcross, GA, U.S.) according to the manufacturer's protocols with additional bead-beating step. The final DNA concentration and purity were determined using a NanoDrop 2000 UV–vis spectrophotometer (Thermo Scientific, Wilmington, USA), followed by visual quality checking using agarose gel (1%) electrophoresis. Individual amplicon libraries were prepared for prokaryotes using PCR amplification of the V3–V4 hypervariable regions of the 16S rRNA gene with primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The ITS1 region of the fungal rRNA operon was amplified with primers 1737F (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and 2043R (5′-GCTGCGTTCTTCATCGATGC-3′) to prepare individual amplicon libraries for fungi. The PCR products were gel purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using QuantiFluor-ST (Promega, USA) according to the manufacturer's protocol. Purified amplicons were pooled at equimolar ratio and paired-end sequenced (2 × 300 bp) on an Illumina MiSeq platform (Illumina, San Diego, USA) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). The raw reads have been deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: PRJNA744001, PRJNA744022).
The raw sequence reads were demultiplexed, quality-filtered using Trimmomatic, and then merged using FLASH. Operational taxonomic units (OTUs) were clustered (de novo) with a 97% similarity cutoff using UPARSE (version 7.1 http://drive5.com/uparse/) and chimeric sequences were identified and removed using UCHIME. The representative 16S rRNA gene sequences of the OTUs were taxonomically classified using the RDP Classifier algorithm (http://rdp.cme.msu.edu/) against the Silva 128 database at a confidence threshold of 70%. The ITS sequences were taxonomically assigned using the UNITE 7.0 database (https://unite.ut.ee/).
2.5. Metagenomic sequencing and analysis
Each microbial DNA extract was fragmented to an average size of about 300 bp using Covaris M220 (Gene Company Limited, China). Individual sequencing libraries were prepared using the TruSeqTM DNA Sample Prep Kit (Illumina, San Diego, CA, USA). Paired-end sequencing was performed on Illumina NovaSeq (Illumina Inc., San Diego, CA, USA) at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China) using the NovaSeq Reagent Kit according to the manufacturer's instructions (www.illumina.com). The raw reads have been deposited into the NCBI Sequence Read Archive (SRA) database (Accession Number: PRJNA744415).
Adapter sequences were trimmed off from the paired-end reads using SeqPrep (https://github.com/jstjohn/SeqPrep). Low-quality reads (length < 50 bp, a quality value < 20, or having any N's) were removed using Sickle (https://github.com/najoshi/sickle). Host DNA was identified and removed after comparing all the reads with the genomes of cattle (https://www.ncbi.nlm.nih.gov/genome/?term=cattle) and yak (https://www.ncbi.nlm.nih.gov/genome/?term=yak) using BWA (http://bio-bwa.sourceforge.net). The cleaned metagenomic sequence reads were assembled using MEGAHIT (Li et al., 2014b) (https://github.com/voutcn/megahit). Contigs with a length of ≥300 bp were used for further analysis.
Open reading frames (ORFs) from each contig were predicted using MetaGene (Noguchi et al., 2006) (http://metagene.cb.k.u-tokyo.ac.jp/). All ORFs sharing ≥95% sequence identity over ≥90% of their length were clustered using CD-HIT (Fu et al., 2012) (http://www.bioinformatics.org/cd-hit/), and the longest sequence from each cluster was selected as its representative sequence to construct a non-redundant gene catalog. The quality-filtered sequence reads were mapped to the representative sequences with 95% identity using SOAPaligner (Li et al., 2008) (http://soap.genomics.org.cn/), and gene abundance in each sample was calculated as transcripts per million.
Representative sequences of the non-redundant gene catalog were compared to the NCBI NR database using BLASTP (Version 2.2.28+, http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Altschul et al., 1997) for taxonomic assignment, and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation was conducted also using BLASTP (Version 2.2.28+) against the KEGG database (Xie et al., 2011) (http://www.genome.jp/keeg/). Carbohydrate-active enzyme annotation was predicted using hmmscan (http://hmmer.janelia.org/search/hmmscan) against the CAZy database Version 5.0 (http://www.cazy.org/). The maximum e-value cutoff for all the annotations was set at 1e−5.
2.6. Metabolomic analysis of rumen fluid
One hundred microliters of each rumen fluid sample was subjected to metabolite extraction using 500 μL methanol: water (4:1, vol/vol) solution containing 2% L-2-chlorophenylalanine (as internal standard). Then the mixtures were vortexed for 10 s and centrifugation at 13,000 × g at 4 °C for 20 min. The supernatant was carefully transferred to a glass-derived bottle and vacuum-dried. After 80 μL methoxy amine hydrochloride (15 mg/mL in pyridine) was added, the samples were vortex-mixed for 2 min and incubated at 37 °C for 90 min to carry out the oximation reaction. Eighty microliters of trifluoroacetamide reagent containing 1% trimethylchlorosilane and 20 μL n-hexane was added to each sample, and then all samples were vortex-mixed for 2 min and incubated at 70 °C for 60 min. The samples were cooled to room temperature and analyzed using gas chromatography and mass spectrometry (GC–MS).
The rumen metabolome was analyzed using gas chromatography (Agilent 7890A, Agilent Technologies, Inc., Santa Clara, CA, USA) coupled to an Agilent 5975C mass selective detector (Agilent, USA) with an inert electron impact ionization (EI) source and ionization voltage at 70 eV. Briefly, metabolites were separated with an HP-5MS capillary column (30 m × 0.25 mm × 0.25 μm) using helium (99.999% purity) as the carrier gas at a constant flow rate (1 mL/min). The GC column temperature was programmed to hold at 60 °C for 0.5 min, rise to 310 °C at a rate of 8 °C/min, and then hold at 310 °C for 6 min. A quality control (QC) sample was prepared by pulling an equal volume of each sample, and the QC sample and rumen fluid samples were analyzed in the same manner. To assess the repeatability of the analysis, the QC sample was injected once every 10 rumen fluid samples. The GC–MS data were processed using the MassHunter workstation Quantitative Analysis package (version v10.0.707.0) to extract raw peaks, filter and calibrate data baselines, align peaks, deconvolute, identify peaks, and integrate peak areas. The resulting matrixes detected in at least 80% of the samples were retained. After filtering, the missing values of the raw data were filled up by half of the compound minimum. The peak area was normalized in the data analysis. The internal standard was used for data quality control (reproducibility), and the metabolic features whose relative standard deviation (RSD) exceeded that of the QC by >30% were discarded. Mass spectra of these metabolic features were identified using the Fiehn database (https://fiehnlab.ucdavis.edu/projects/fiehnlib).
The metabolomic data were analyzed using principal component analysis (PCA), and orthogonal partial least squares discriminate analysis (OPLS-DA) was used to determine the global metabolic differences among the 3 species. Statistical significance among species was declared at VIP value > 1 and P-value < 0.05. P-values were estimated using paired Student's t test for single-dimensional statistical analysis. A total of 27 differential metabolites among 2 of the 3 species were mapped into their biochemical pathways through metabolic enrichment and pathway analysis based on search against the KEGG database (http://www.genome.jp/kegg/) (Xie et al., 2011). These metabolites were classified according to the pathways into which they were mapped or the functions that they could perform. Differential metabolites were cross-listed with the pathways in the KEGG database, and the top differential pathways were identified (Xia et al., 2015). The relationship between different species and metabolites was visualized as a heat map using the “pheatmap” package in R (www.r-project.org).
2.7. Statistical analysis
Data are presented as mean ± SD and were analyzed by one way analysis of variance (ANOVA) or Kruskal–Wallis. Rumen fermentation parameters, fibrolytic enzyme activities, carbohydrate-active enzyme genes and alpha diversity were calculated using one way ANOVA followed by the Duncan test. Difference in microbial species was tested using the Kruskal–Wallis H test, with post hoc analysis done using Tukey–Kramer. Plots of principal coordinates analysis (PCoA) were based on the Bray–Curtis dissimilarity and statistical significance of difference was tested using analysis of similarities (ANOSIM) with 999 permutations. Differential KEGG pathways were analyzed using linear discriminant analysis effect size (LEfSe), and visualized using GraphPad Prism 7.0 software (GraphPad Software, Inc., La Jolla, CA, USA). Spearman's correlations between different rumen species and metabolites were analyzed by “psych” package in R and visualized as a heat map using the “pheatmap” package in R. Significance was set at P < 0.05. Statistical analyses were performed using GraphPad Prism 7.0 software (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS 19.0 for Windows (SPSS Inc., Chicago, IL, USA).
3. Results
3.1. Rumen fermentation parameters and fibrolytic enzyme activities
No significant difference was observed in rumen pH among the 3 species (Table 1), but the rumen concentration of total VFA in dzomo was lower (P < 0.05) than in cattle. The molar proportion of propionate in yak and dzomo was higher than in cattle (P < 0.05), while the molar proportion of acetate and valerate as well as acetate-to-propionate (A:P) ratio showed the opposite trend. Yak butyrate molar proportion was significantly higher than the other 2 species (P < 0.05). The rumen microbiome of yak had the highest (P < 0.05) activities of carboxymethyl cellulase and avicelase (Table 1).
Table 1.
Rumen fermentation parameters and fibrolytic enzyme activities among cattle, dzomo and yak.
| Item | Cattle | Dzomo | Yak | SEM | P-value1 |
|---|---|---|---|---|---|
| pH | 7.10 | 7.37 | 7.29 | 0.049 | 0.077 |
| Total VFA, mM | 74.00a | 58.07b | 70.08a,b | 2.651 | 0.032 |
| VFA proportion, mol/100 mol | |||||
| Acetate | 77.92a | 76.48b | 75.10c | 0.257 | <0.001 |
| Propionate | 13.23b | 14.10a | 14.38a | 0.166 | 0.008 |
| Butyrate | 7.11b | 7.61b | 8.56a | 0.179 | 0.001 |
| Isobutyrate | 0.58 | 0.70 | 0.70 | 0.025 | 0.092 |
| Isovalerate | 0.62 | 0.70 | 0.81 | 0.047 | 0.263 |
| Valerate | 0.53a | 0.42b | 0.45b | 0.019 | 0.040 |
| A:P ratio | 5.90a | 5.44b | 5.24b | 0.077 | <0.001 |
| Fibrolytic enzyme activities, mU2 | |||||
| Xylanase | 485.80 | 648.25 | 789.27 | 109.252 | 0.542 |
| Carboxymethyl cellulase | 158.02b | 182.63a,b | 220.69a | 9.578 | 0.021 |
| Avicelase | 187.76b | 204.60b | 248.14a | 6.736 | <0.001 |
| Acetylesterase | 149.76 | 154.75 | 146.54 | 3.571 | 0.065 |
VFA = volatile fatty acids.
a,b,cDifferent superscripts in a row designate a significant difference (P < 0.05).
P-values were calculated using one-way analysis of variance (ANOVA) (n = 9 per species).
mU: One milliunit of enzyme activity was defined as the amount of enzyme releasing 1 nmol of reducing sugar (e.g., xylose or glucose equivalent) or p-nitrophenol per min per microliter.
3.2. Rumen microbiota structure and composition
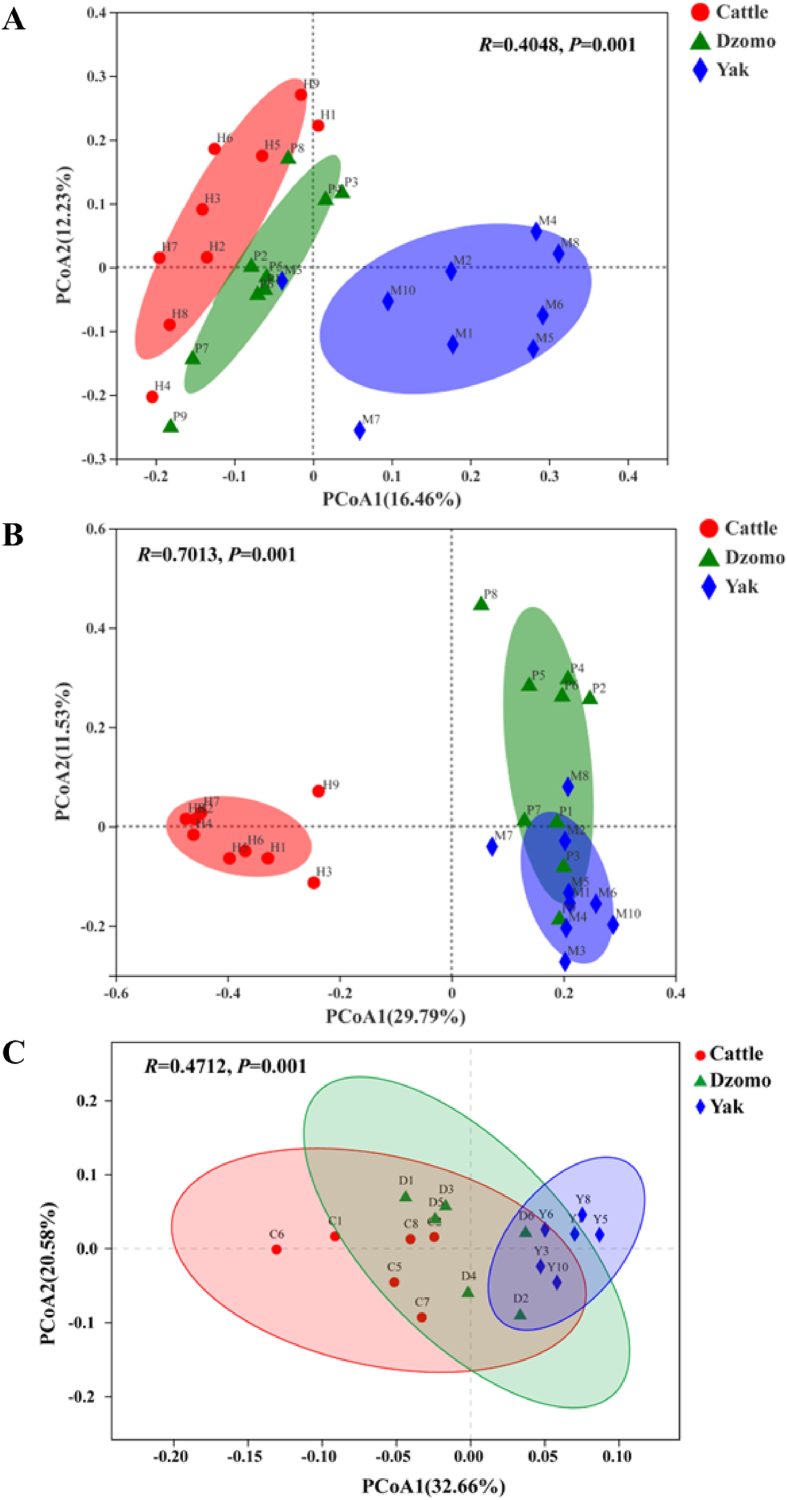
Using metataxonomic analysis, we compared the rumen microbiota among the 3 species. We obtained >1.38 million and 0.79 million clean sequences of 16S and ITS. To minimize the effects of sequencing depth on alpha and beta diversity measures, the number of reads was subsampled to a minimum number (35,784 and 32,060, respectively) of sequences across all of the samples. The sequencing depth coverage was >98% for bacteria and 99% for fungi for all the samples. The 3 species had different bacterial and fungal rumen microbiota. The ACE richness estimate of rumen bacteria was lower in yak (P < 0.05) than cattle and dzomo, but no significant difference was observed in Simpson's diversity index among the 3 species. Simpson evenness was higher (P < 0.05) for yak than for cattle and dzomo (Table S1). With respect to fungi, observed species richness (both ACE and Chao1 estimates) in cattle were lower (P < 0.05) than in dzomo and yak, but Simpson evenness for cattle was higher (P < 0.05) compared with yak and dzomo (Table S2). The β-diversity of the bacterial and fungal microbiotas was compared using PCoA based on Bray–Curtis dissimilarity, and ANOSIM analysis showed a difference (P = 0.001) in the overall rumen microbiota of both bacteria and fungi among the 3 species (Fig. 1A and B). Rumen bacterial microbiota of cattle and dzomo was more similar, while fungal microbiota of dzomo was more similar to yak compared to cattle. Overall, the rumen microbiota of bacteria and fungi of cattle clustered separately from that of yak, while that of dzomo fell in between.
Fig. 1.
Plots of principal coordinates analysis (PCoA) comparing the overall rumen microbiota among the 3 species. (A and B) Bacteria and fungi were based on Bray–Curtis dissimilarity determined by metataxonomics (n = 9 per species). (C) Rumen microorganism was based on Bray–Curtis dissimilarity determined by metagenomics (n = 6 per species). The ellipses represent the 95% of the samples belonging to each group. Statistical significance of difference was tested using analysis of similarities (ANOSIM) with 999 permutations.
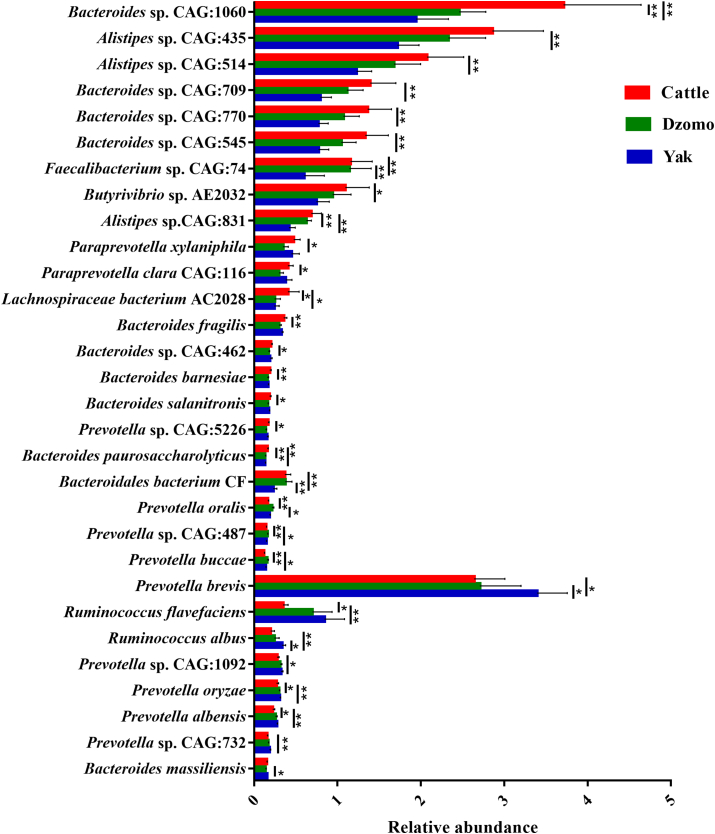
We obtained a total of >1.36 billion raw reads (>75.8 million reads per sample) totaling >206 GB of sequence data (>11.4 GB per sample). After the removal of host DNA and quality filtering, we obtained >1.35 billion reads in total and >75.1 million reads per sample. The average length of the ORFs was about 494 bp. PCoA analysis of the metagenomic data also revealed differences (P = 0.001) in the overall microbiota among the 3 species (Fig. 1C). Bacteroidetes, Firmicutes, Proteobacteria, and Euryarchaeota were the predominant phyla in all samples, with Bacteroidetes (53.71%) and Firmicutes (24.74%) being the most predominant. The relative abundance of Proteobacteria, Verrucomicrobia, Planctomycetes, Synergistetes, Cyanobacteria, Chloroflexi, and Acidobacteria was higher (P < 0.05) in yak than in cattle, while Chlamydiae was more predominant (P < 0.05) in cattle and Actinobacteria more predominant (P < 0.05) in dzomo (Fig. S1). At the genus level, Bacteroides, Alistipes, Butyrivibrio, Faecalibacterium, and Pseudobutyrivibrio were more predominant in cattle than in yak (P < 0.05), while the opposite was true (P < 0.05) for Ruminococcus, Selenomonas, Oscillabacter, Treponema, and Fibrobacter (Fig. S2). At the species level, nineteen species, most of which belonged to Bacteroides and Alistipes, were more predominant (P < 0.05) in cattle than in yak or dzomo; 4 species were more predominant (P < 0.05) in dzomo; and 8 species (most belonging to Prevotella) were more predominant (P < 0.05) in yak (Fig. 2).
Fig. 2.
Microbial species (identified in the rumen metagenome) that significantly differed in relative abundance among the 3 species. The difference was tested using the Kruskal–Wallis H test, with post hoc test done using the Tukey–Kramer test (n = 6 per species). Only the microbial species each with a relative abundance >0.1% in all samples were shown. Data present mean ± SD. ∗P < 0.05, ∗∗P < 0.01.
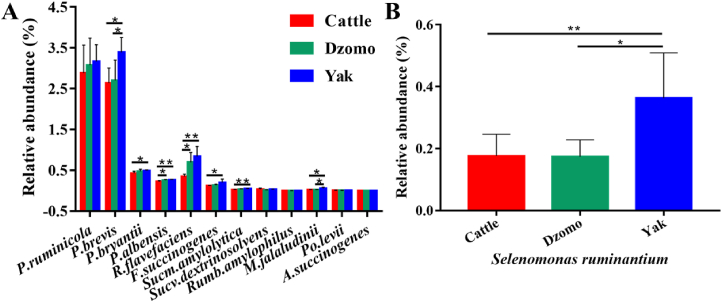
Of the 12 succinate-producing bacterial species, Prevotella brevis, Prevotella bryantii, Prevotella albensis, Fibrobacter succinogenes, Succinimonas amylolytica, and Mitsuokella jalaludinii were enriched (P < 0.05) in the yak rumen compared to the rumen microbiota of cattle or dzomo (Fig. 3A). So was the succinate-utilizing species identified, Selenomonas ruminantium (Fig. 3B). The bacterial genera likely using the acrylate pathway for propionate production did not differ (P > 0.05) in relative abundance among the 3 species (Fig. S3). The relative abundance of Ruminococcus flavefaciens, Ruminococcus albus, and F. succinogenes, the best-known species of cellulolytic bacteria in the rumen, was enriched (P < 0.05) in yak compared to cattle (Fig. S4).
Fig. 3.
Bacterial species likely involved in succinic acid pathway for the propionate production. (A) Identified bacterial species likely involved in succinate production. (B) Identified bacterial species likely involved in succinate utilization. The difference in relative abundance among the 3 species was tested using the Kruskal–Wallis H test, with post hoc test done using the Tukey–Kramer test (n = 6 per species). Data present mean ± SD. Abbreviations: P. = Prevotella; R. = Ruminococcus; F. = Fibrobacter; Sucm. = Succinimonas; Sucv. = Succinivibrio; Rumb. = Ruminobacter; M. = Mitsuokella; Po. = Porphyromonas; A. = Actinobacillus. ∗P < 0.05, ∗∗P < 0.01.
3.3. Carbohydrate-active enzymes and KEGG pathway
The carbohydrate-active enzymes (CAZymes) encoded by the rumen microbiome are of critical importance to feed digestion in ruminants (Hess et al., 2011; Moraïs and Mizrahi, 2019). The relative abundance of CAZymes in cattle was lower (P < 0.05) than in dzomo and yak (Table 2). The abundance of 6 types of CAZymes was quite different among the 3 species. The abundance of glycoside hydrolases (GHs) was higher (P < 0.05) in dzomo than cattle, whereas the abundance of glycosyl transferases (GTs) in yak was higher (P < 0.05) than cattle and dzomo (Table 2). Glycoside hydrolase and carbohydrate esterase (CE) families that are involved in polysaccharide degradation were further compared. Glycoside hydrolase 48, primarily exoglucanases (Moraïs and Mizrahi, 2019) and GH45, mostly endoglucanases (Moraïs and Mizrahi, 2019), were more abundant (P < 0.05) in yak than in cattle (Table 3). Among the GHs encoding hemicellulases, GH5 and GH44, both mostly xyloglucanases (Moraïs and Mizrahi, 2019), GH16 and GH17, (both mostly β-glucanases (Moraïs and Mizrahi, 2019), and GH11, primarily endoxylanases (Dai et al., 2015) were more (P < 0.05) abundant in yak than in cattle (Table 3). Similarly, polysaccharide lyase (PL) 11 (rhamnogalacturonan endolyase or exolyase) and PL4-4 (rhamnogalacturonan endolyase) were more abundant (P < 0.05) in yak than in cattle (Table S3). On the other hand, yak had the lowest (either significantly or numerically) abundance of the GH families encoding amylases (Table 3). Of the detected carbohydrate esterases, CE12 (mostly acetylesterase acting on xylan, pectin, or rhamnogalacturonan) was more abundant (P < 0.05) in yak than in cattle, whereas CE13 (pectin acetylesterase) was more abundant (P < 0.05) in cattle than in dzomo or yak (Table 3). Of the 15 most abundant GTs, GT2, GT4, and GT9 were more abundant (P < 0.05) in yak, while GT1 was more abundant (P < 0.05) in cattle (Table S4). Among the top 15 differential carbohydrate-binding modules (CBMs) detected (Table S5), CBM48, CBM72, CBM78, CBM12, CBM58, CBM5, and CBM41 were more abundant (P < 0.05) in cattle, whereas CBM67, CBM61, CBM40, CBM51, CBM57, CBM77, and CBM70 were enriched in yak compared to the other 2 ruminant species.
Table 2.
Abundance (in transcripts per million) of carbohydrate-active enzyme genes in the rumen metagenome of cattle, dzomo and yak.
| Item | Cattle | Dzomo | Yak | SEM | P-value1 |
|---|---|---|---|---|---|
| Sum | 44,914.81b | 48,529.87a | 48,156.05a | 528.905 | 0.002 |
| GH | 21,449.70b | 23,392.58a | 22,277.84a,b | 290.210 | 0.012 |
| GT | 9,081.16b | 9,665.29b | 10,837.93a | 216.862 | <0.001 |
| CE | 7,909.66 | 8,349.70 | 7,812.54 | 102.821 | 0.066 |
| CBM | 4,810.67 | 4,998.95 | 5,095.12 | 53.208 | 0.077 |
| PL | 826.66b | 1,068.62a | 1,084.87a | 32.479 | <0.001 |
| AA | 836.96b | 1,054.73a | 1,047.74a | 26.257 | <0.002 |
GH = glycoside hydrolases; GT = glycosyl transferases; CE = carbohydrate esterase; CBM = carbohydrate-binding modules; PL = polysaccharide lyases; AA = auxiliary activities.
a,bWithin a row, values with a different superscript designate a significant difference (P < 0.05).
P-values were calculated using one-way ANOVA (n = 6 per species).
Table 3.
Abundance (in transcripts per million) of cellulase, hemicellulose, acetylesterase and amylase genes in the rumen metagenome of cattle, dzomo, and yak.
| CAZymes | CAZymes family | Cattle | Dzomo | Yak | SEM | P-value1 |
|---|---|---|---|---|---|---|
| Cellulases | GH48 | 0.75b | 1.85a,b | 1.99a | 0.221 | 0.030 |
| GH6 | 0.00 | 0.16 | 0.00 | 0.038 | 0.137 | |
| GH5 | 111.47b | 156.61b | 212.09a | 12.010 | <0.001 | |
| GH8 | 130.50 | 133.56 | 122.58 | 2.804 | 0.270 | |
| GH9 | 363.70 | 390.48 | 371.90 | 7.236 | 0.320 | |
| GH45 | 0.28b | 0.45b | 2.24a | 0.330 | 0.016 | |
| Xylanases | GH10 | 481.40b | 536.92a | 505.02a,b | 9.039 | 0.030 |
| GH11 | 17.34b | 27.13a,b | 35.54a | 2.474 | 0.003 | |
| GH30 | 122.36 | 125.61 | 101.99 | 4.477 | 0.055 | |
| GH43 | 194.08 | 196.92 | 198.73 | 3.729 | 0.889 | |
| Mannase | GH5 | 111.47b | 156.61b | 212.09a | 12.01 | <0.001 |
| GH26 | 268.66a | 252.52a | 217.97b | 7.18 | 0.005 | |
| Xyloglucanases | GH5 | 111.47b | 156.61b | 212.09a | 12.010 | <0.001 |
| GH12 | 0.69 | 0.75 | 0.60 | 0.114 | 0.879 | |
| GH44 | 0.40c | 1.30b | 2.17a | 0.200 | <0.001 | |
| GH74 | 84.53 | 98.31 | 107.97 | 4.661 | 0.115 | |
| β-glucanases | GH5 | 111.47b | 156.61b | 212.09a | 12.010 | <0.001 |
| GH16 | 311.07b | 347.48a | 345.92a | 5.374 | 0.002 | |
| GH17 | 0.29b | 0.34b | 0.90a | 0.099 | 0.011 | |
| Acetylesterase | CE12 | 305.64b | 391.04a | 416.44a | 15.528 | 0.003 |
| CE13 | 3.98a | 1.16b | 0.53b | 0.445 | <0.001 | |
| CE16 | 0.00 | 0.13 | 0.00 | 0.033 | 0.160 | |
| Amylase | GH31 | 655.08a,b | 707.4a | 643.81b | 10.278 | 0.015 |
| GH13 | 425.55 | 454.01 | 399.72 | 11.603 | 0.163 | |
| GH57 | 219.73a | 185.77b | 186.45b | 4.857 | 0.001 | |
| GH77 | 270.10 | 270.45 | 263.52 | 3.459 | 0.681 | |
| GH15 | 3.20 | 3.33 | 1.89 | 0.325 | 0.133 | |
| GH19 | 4.55 | 1.87 | 4.07 | 0.568 | 0.118 |
CAZymes = carbohydrate-active enzymes; GH = glycoside hydrolases; CE = carbohydrate esterase.
a,b,cWithin a row, values with a different superscript designate a significant difference (P < 0.05).
P-values were calculated using one-way ANOVA (n = 6 per species).
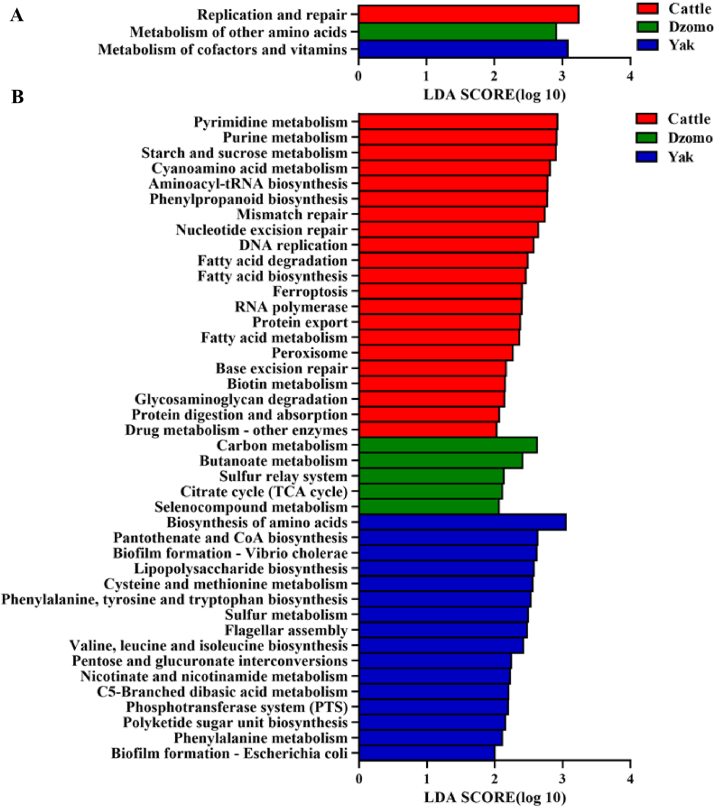
Replication and repair were enriched in cattle (P < 0.05); metabolic pathways of other amino acids were enriched in dzomo (P < 0.05); whereas metabolism of cofactors and vitamins was represented more abundantly (P < 0.05) in yak (Fig. 4A). At level 3 (Fig. 4B), 21, 16, and 5 metabolic pathways were enriched in cattle, yak, and dzomo (P < 0.05), respectively, compared with the other 2 ruminant species. The metabolism of pyrimidine, purine, starch and sucrose, cyanoamino acid, and aminoacyl-tRNA biosynthesis were the top 5 for cattle; amino acid biosynthesis, pantothenate and CoA biosynthesis, biofilm formation-Vibrio cholerae, lipopolysaccharide biosynthesis, cysteine and methionine metabolism were the top 5 for yak; and carbon metabolism, butanoate metabolism, citrate cycle, sulfur relay system, and selenocompound metabolism were enriched in dzomo. Differential modules based on LEfSe analysis are shown in Fig. S5.
Fig. 4.
Results of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. (A) Differential KEGG pathways among the 3 species at level 2. (B) Differential KEGG pathways among the 3 species at level 3. Differential KEGG pathways among the 3 species as determined using linear discriminant analysis effect size (LEfSe). Only the pathways that differed significantly (P < 0.05) between 2 of the 3 species with an LDA score >2 are shown. LDA = linear discriminant analysis.
3.4. Rumen metabolite profiles
A total of 185 individual metabolites were identified. Principal component analysis of all the samples and 3 QC samples showed that the analysis system was stable (Fig. S6A). As shown by OPLS-DA, rumen metabolite profiles were distinct among cattle, dzomo, and yak (Figs. S6B–D). After filtering using the student's t test (P < 0.05) and variable importance in projection (VIP > 1), 27 metabolites were found to differ between 2 ruminant species (Table 4). Notably, 16 metabolites differed (P < 0.05) between cattle and dzomo: Caproic acid, benzenepropanoic acid, 2-deoxyerythritol, lithocholic acid, dehydroascorbic acid, 4-(dimethylamino)azobenzene, phenylacetamide, uridine 5′-monophosphate, and z-phthalate were higher (P < 0.05) in cattle than dzomo, and lithocholic acid (FC = 25.65) and phenylacetamide (FC = 22.74) were the most upregulated metabolites in cattle. On the other hand, ethylene glycol, glycine, aminomalonate, 3-hydroxyphenylacetic acid, chenodeoxycholic acid, 2-picolinic acid, 3,6-anhydro-D-glucose were higher (P < 0.05) in dzomo than cattle, with glycine (FC = 17.86), chenodeoxycholic acid (FC = 12.82), 2-picolinic acid (FC = 22.22), 3,6-anhydro-D-glucose (FC = 15.39) being the most upregulated in dzomo. Compared to the rumen of cattle, the rumen of yak had a higher (P < 0.05) ethylene glycol, 2-hydroxypyrazinyl-2-propenoic acid, sebacic acid, 2-picolinic acid, 1,3-dihydroxypyridine, chenodeoxycholic acid, and 3,6-anhydro-D-glucose, but a lower (P < 0.05) serine, z-phthalate, 2-deoxyerythritol, undecanedioic acid, lithocholic acid, dehydroascorbic acid, glucose-6-phosphate, uridine 5′-monophosphate, and isochlorogenic acid. When compared to the rumen of dzomo, the yak rumen had higher valeric acid, boric acid, 1,3-dihydroxypyridine, benzenepropanoic acid, and sebacic acid, but a lower 5′-deoxy-5′-methylthioadenosine, glycine, aminomalonate, pimelic acid, 3-hydroxyphenylacetic acid, and isochlorogenic acid.
Table 4.
Rumen fluid metabolites that differed between 2 of the 3 ruminant species (n = 6 per species).
| Item | RT1, min | Cattle vs Dzomo |
Cattle vs Yak |
Dzomo vs Yak |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| VIP2 | P-value3 | FC4 | VIP | P-value | FC | VIP | P-value | FC | ||
| Ethylene glycol | 9.45 | 2.549 | 0.002 | 0.353 | 2.193 | 0.014 | 0.502 | 1.435 | 0.110 | 1.422 |
| Phenylacetamide | 9.47 | 2.229 | 0.025 | 22.741 | 1.678 | 0.186 | 2.422 | 1.148 | 0.095 | 0.106 |
| Valeric acid | 9.57 | 1.795 | 0.051 | 1.522 | 0.113 | 0.551 | 0.882 | 2.589 | 0.014 | 0.579 |
| Boric acid | 9.71 | 0.740 | 0.116 | 2.196 | 0.214 | 0.897 | 0.961 | 1.015 | 0.034 | 0.438 |
| 5′-Deoxy-5′-methylthioadenosine | 9.99 | 1.935 | 0.100 | 0.278 | 1.461 | 0.258 | 15.17 | 2.829 | 0.015 | 54.566 |
| 2-Picolinic acid | 10.5 | 1.736 | 0.001 | 0.045 | 2.415 | <0.001 | 0.041 | 0.287 | 0.602 | 0.900 |
| Caproic acid | 11.16 | 1.662 | 0.018 | 2.426 | 1.094 | 0.649 | 1.168 | 1.451 | 0.097 | 0.481 |
| Dehydroascorbic acid | 11.59 | 1.096 | <0.001 | 3.332 | 1.572 | <0.001 | 4.656 | 0.389 | <0.001 | 1.397 |
| 2-Deoxyerythritol | 12.32 | 1.843 | <0.001 | 7.787 | 2.457 | <0.001 | 9.590 | 0.238 | 0.055 | 1.231 |
| Glucose-6-Phosphate | 15.19 | 0.941 | 0.001 | 2.083 | 1.351 | <0.001 | 2.175 | 0.068 | 0.619 | 1.044 |
| Glycine | 15.5 | 4.329 | 0.001 | 0.056 | 1.584 | 0.071 | 0.351 | 4.127 | 0.002 | 6.274 |
| Serine | 16.32 | 0.733 | <0.001 | 3.044 | 1.035 | <0.001 | 4.202 | 0.236 | 0.050 | 1.381 |
| 1,3-Dihydroxypyridine | 16.71 | 0.343 | 0.180 | 2.424 | 1.080 | 0.035 | 0.275 | 1.149 | 0.010 | 0.113 |
| Benzenepropanoic acid | 17.34 | 6.440 | 0.030 | 1.666 | 3.032 | 0.739 | 1.065 | 7.269 | 0.003 | 0.639 |
| Aminomalonate | 18.17 | 1.386 | 0.026 | 0.716 | 0.413 | 0.816 | 1.031 | 1.461 | 0.030 | 1.441 |
| 3,6-Anhydro-D-glucose | 18.42 | 1.527 | <0.001 | 0.065 | 1.345 | 0.002 | 0.094 | 0.649 | 0.107 | 1.459 |
| Pimelic acid | 19.05 | 0.721 | 0.181 | 0.841 | 0.117 | 0.683 | 1.064 | 1.048 | 0.022 | 1.266 |
| 2-Hydroxypyrazinyl-2-propenoic acid | 19.07 | 1.236 | 0.665 | 0.823 | 6.336 | 0.048 | 0.505 | 3.694 | 0.168 | 0.613 |
| 3-Hydroxyphenylacetic acid | 20.25 | 1.452 | 0.004 | 0.515 | 0.132 | 0.799 | 0.948 | 1.396 | 0.007 | 1.839 |
| 4-(Dimethylamino)azobenzene | 21.56 | 1.149 | 0.002 | 17.479 | 0.689 | 0.249 | 1.752 | 0.713 | 0.090 | 0.100 |
| Isochlorogenic acid | 21.97 | 0.506 | 0.124 | 1.396 | 1.881 | <0.001 | 16.819 | 1.158 | 0.001 | 12.044 |
| Sebacic acid | 23.99 | 0.214 | 0.005 | 1.547 | 1.292 | 0.021 | 0.106 | 1.054 | 0.017 | 0.068 |
| Undecanedioic acid | 25.20 | 0.148 | 0.530 | 1.056 | 1.016 | 0.004 | 1.396 | 0.746 | <0.001 | 1.322 |
| Lithocholic acid | 25.60 | 1.376 | <0.001 | 25.649 | 1.228 | 0.021 | 3.142 | 0.420 | 0.268 | 0.123 |
| Chenodeoxycholic acid | 25.96 | 1.434 | 0.019 | 0.078 | 1.747 | 0.023 | 0.099 | 0.397 | 0.629 | 1.268 |
| Uridine 5′-monophosphate | 26.75 | 1.746 | 0.017 | 2.455 | 2.833 | <0.001 | 5.819 | 0.693 | 0.338 | 2.370 |
| Zinc phthalate | 31.29 | 2.933 | <0.001 | 6.861 | 4.010 | <0.001 | 8.566 | 0.396 | 0.110 | 1.248 |
RT = retention time.
VIP = variable importance in projection.
P-values were calculated using Student’ t-test (n = 6 per species).
FC = fold change. Take cattle vs dzomo for example, if a FC value is less than 1, it means that there is less metabolite in cattle than in dzomo.
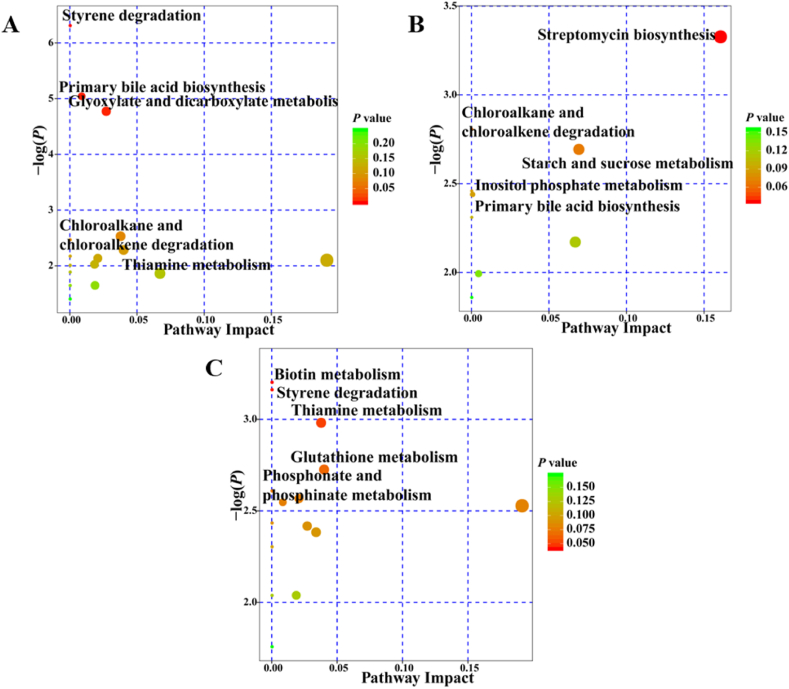
Metabolite pathway enrichment analysis based on the significantly different rumen metabolites revealed the pathways that potentially contributed to the observed difference in rumen metabolite profiles (Fig. 5). The pathways for styrene degradation, primary bile acid biosynthesis, glyoxylate and dicarboxylate metabolism, chloroalkane and chloroalkene degradation, and thiamine metabolism were different (P < 0.05) between cattle and dzomo (Fig. 5A). Between cattle and yak (Fig. 5B), the pathways involved in streptomycin biosynthesis, chloroalkane and chloroalkene degradation, starch and sucrose metabolism, inositol phosphate metabolism, and primary bile acid biosynthesis differed (P < 0.05). The pathways for biotin metabolism, styrene degradation, thiamine metabolism, glutathione metabolism, and phosphonate and phosphinate metabolism differed (P < 0.05) between dzomo and yak (Fig. 5C).
Fig. 5.
Metabolic pathways significantly differing in rumen metabolites among 3 species. (A) Significantly differing metabolic pathways between cattle and dzomo. (B) Significantly differing metabolic pathways between cattle and yak. (C) Significantly differing metabolic pathways between dzomo and yak. The X-axis and Y-axis represent the pathway impact and pathway enrichment, respectively. The redder the color indicate the smaller P-value from enrichment analysis, and the larger abscissa indicates the greater impact from the pathway topology analysis (n = 6 per species).
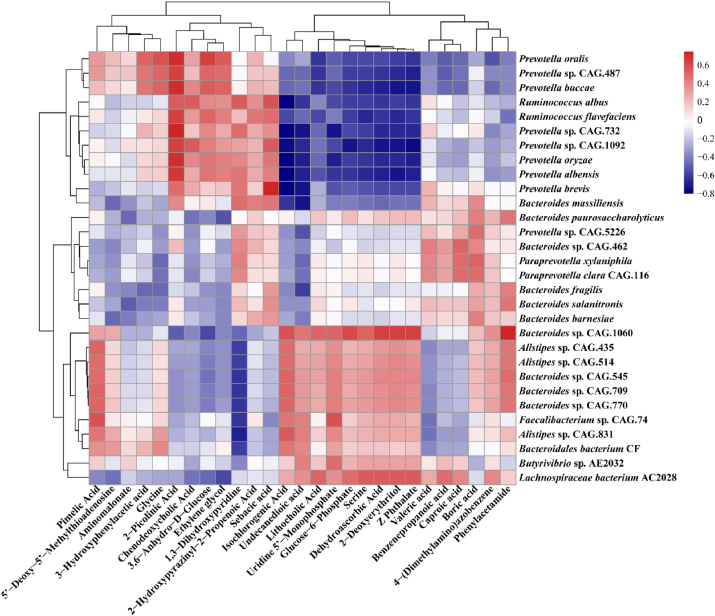
3.5. Correlation analysis of rumen microbiota and metabolites
Lithocholic acid and glucose-6-phosphate were positively correlated with Bacteroides sp. CAG.1060, Bacteroides sp. CAG.545, Bacteroides sp. CAG.709, Bacteroides sp. CAG.770, Alistipes sp. CAG.435, Alistipes sp. CAG.514, and Lachnospiraceae bacterium AC 2028 (Fig. 6). Chenodeoxycholic acid and 3,6-anhydro-D-glucose were positively correlated with 8 Prevotella species (P. oralis, Prevotella sp. CAG.487, P. buccae, P. oryzae, P. albensis, P. brevis, Prevotella sp. CAG.732, and Prevotella sp. CAG.1092) and 2 Ruminococcus sp. (R. flavefaciens and R. albus) (Fig. 6).
Fig. 6.
Pearson correlations between significantly different rumen microbial species and microbial metabolites. The color and the color intensity of the squares correspond to the direction and strength of the correlation based on the scale to the right.
4. Discussion
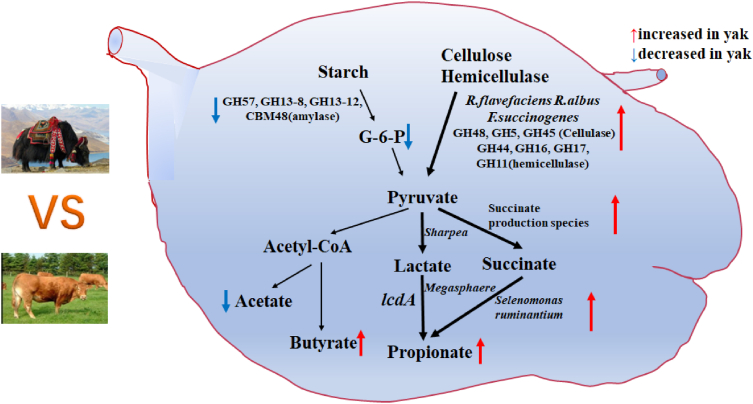
The yak is an indigenous livestock animal on the Qinghai Tibetan Plateau, and it is raised at altitudes between 3,000 and 5,000 m (Zhou et al., 2018). The high altitude and the environmental conditions are not suitable for domestic cattle. The cattle used in this experiment were Qaidam yellow cattle, which are well adapted to an altitude of 2,800 m, but their pulmonary artery pressure is still significantly higher than that of indigenous yak and Tibetan sheep (Zhou et al., 2018). Currently, little is known whether the rumen microbiome is co-evolved tighter with ruminants living in high-altitude environments. In this study, we integrated rumen fermentation profiles, metataxonomics, metagenomics, and metabolomics of the rumen microbiome comparatively to explore how the rumen microbiome might help yak to adapt to their living conditions, especially the poor dietary conditions unique to the Qinghai-Tibet high-altitude plateau. As summarized in Fig. 7, cellulase and hemicellulase, and PL families were enriched in the yak rumen microbiome compared to the rumen microbiome of cattle, which can help improve fiber degradation. On the other hand, yak had decreased amylase-containing GH families and CBM families, which might slow down starch degradation in the rumen and allow more starch to reach the small intestine. Additionally, the increased succinate-producing and utilizing bacterial species might promote propionate production. All the above are consistent with the ability of yak to live on the poor diet available on the high plateau.
Fig. 7.
A schematic depicting the microbial processes of carbohydrate utilization and volatile fatty acids formation in yak vs cattle.
Rumen microbiota is primarily responsible for the energy acquisition of ruminants, and many studies showed that differences in rumen microbiota may alter energy efficiency (Shi et al., 2014; Xue et al., 2020a). We first compared the rumen microbiota among the 3 species. Of the 21 phyla identified by metagenomics in this study, Bacteroidetes and Firmicutes, which are considered important microbes in providing the energy required by ruminant animals (Tremaroli and Backhed, 2012), were the most predominant. Studies have shown that with the increase in dietary energy level and concentrate proportion, the relative abundance of Firmicutes could increase (Ahmad et al., 2020; Liu et al., 2019). Similarly, the gut microbiota of obese people and mice cecum contained more Firmicutes and less Bacteroidetes (Ley et al., 2005, 2006), which agrees with the fact that the relative abundance of Firmicutes can be positively correlated with dietary energy concentration. Compared with the study of Ahmad et al. (2020), the relative abundance of Bacteroidetes in the 3 species was higher, but that of Firmicutes was lower. This discrepancy may be partially attributable to the concentrate proportions (30%, dry matter) in the diet of the cited study. The relative abundance of Proteobacteria and Verrucomicrobia was significantly higher in yak than in the other 2 species. Members of the phylum Proteobacteria are metabolically flexible in modifying their gene repertoire in response to changes in available substrate and energy sources (Moulin et al., 2001; Zhou et al., 2020), and their relative abundance was significantly higher in the rumen of cows with high milk yield and milk protein content, suggesting that Proteobacteria is positively correlated with feed efficiency (Xue et al., 2020a). Verrucomicrobia contains species that are highly specialized degraders of fucoidans and other complex polysaccharides (Sichert et al., 2020), and they may play an important role in polysaccharide degradation. In a previous transcriptomic study, Ruminococcus and Fibrobacter were shown to express most of the cellulase transcripts, and they, together with Prevotella, probably expressed the majority of hemicellulose transcripts in the rumen (Dai et al., 2015). The high abundance of Ruminococcus, Fibrobacter, and Prevotella and the activities of carboxymethyl cellulase and avicelase in the rumen of yak corroborate its greater ability to digest crude fiber. Xue et al. (2020a) compared the microbial composition between cows with high and low milk yield and milk protein content, and they found that the rumen species enriched in the high-yielding cows were mainly members of Prevotella, and P. buccae, P. albensis and R. flavefaciens were significantly higher in the high-yielding cows. We conclude that the above 4 bacterial species are positively correlated with feed efficiency. Interestingly, they were also significantly more predominant in the rumen of yak than in cattle. These species, probably among others, may help enable yak to adapt to their poor diet.
In the present study, yak were found to possess more hemicellulase (e.g., GH11, GH5, GH44, GH16, GH17) and rhamnogalacturonan endolyase (e.g., PL11and PL4-4) than cattle. Many CBMs associated with galactose binding (CBM61, CBM62, CBM51) and L-rhamnose binding (CBM67) were also enriched in yak. These results indicate that yak probably have a higher capacity to remove galactose from the main chain of hemicellulose. Rapid hemicellulose digestion can facilitate cellulose digestion in forage because the removal of hemicellulose can expose cellulose for microbial attachment and digestion. Although GH13, which contains well-known amylases, did not differ among the 3 species, its subfamilies GH13-8 (starch branching enzyme) and GH13-12 and GH13-14 (pullulanase) and their associated CBM (CBM48) were more abundant in cattle (Machovic and Janecek, 2008) (Table S6). Another starch binding CBM, CBM41 (Janeček et al., 2017; Zeng et al., 2019), was also enriched in cattle. The yak rumen microbiome likely increased its ability to degrade forage not only by enhancing cellulose degradation (as evidenced by enriched GHs involved in cellulose hydrolysis) but also by enhancing hemicellulose and pectin degradation. On the other hand, the rumen microbiome of cattle had a higher abundance of GHs and CBM involved in starch digestion and binding, respectively. Even though both the yak and cattle used in the present study were fed the same diet, the rumen microbiome of yak probably has evolved to be better adapted to the forage that yak consume, whereas the rumen microbiome of cattle, which are domesticated and have been fed diets containing starch, likely has evolved to better utilize starch. Yak produce milk of desirable characteristics (e.g., high milk fat, milk protein, milk total solids), but their milk yield is very low (about 1 kg per day) (Cui et al., 2016; Zhang et al., 2020). The lack of sufficient dietary energy is one limiting factor. It will be interesting to test if the rumen microbiome of yak can evolve to gradually adapt to starch-rich diets over the long-term.
It is well-known that forage typically increases acetate concentration in the rumen compared to a diet containing starch. In the present study, acetate molar proportion reached 75.1% to 77.9%, significantly higher than that reported in other studies (Yang et al., 2012; Yang and Beauchemin, 2006; Zebeli et al., 2006), and the acetate: propionate ratio was also much higher than that reported by others (Ahmad et al., 2020; Liu et al., 2019). Both were likely attributed to the forage-only diet used in the present study. One previous study showed that rumen fermentation to propionate could increase energy efficiency (Shabat et al., 2016). Indeed, glucose fermentation to acetate, propionate, and butyrate can provide 62%, 109%, and 78% of the energy stored in glucose, respectively (Ryle and Orskov, 1990). The gain of energy in propionate fermentation is because it incorporates hydrogen. This in turn reduces the amount of energy wasted via methane production from hydrogen. Two fermentation pathways are mainly responsible for the propionate production in the rumen: the acrylate pathway and succinic acid pathway (Xue et al., 2020b), with the former using lactate as the input substrate and acryloyl-CoA as an intermediate, while the latter uses oxaloacetate as the starting substrate and succinate as an intermediate. In this study, the increased relative abundance of succinate-producing and utilizing bacterial species seen in yak might be attributable to the higher propionate proportion detected in those species. On the other hand, because lactate-producing and utilizing bacterial genera did not differ in relative abundance among the 3 species, the acrylate pathway of propionate production might be similar across the 3 species. Isobutyrate, isovalerate, and 2-methyl-butanoic are considered essential growth factors for most fiber-degrading microorganisms (Allison et al., 1962; Yang, 2002; Zhang et al., 2013). The increased isobutyrate and isovalerate concentration (data was not shown) noted in the yak might facilitate the growth of some fiber-degrading bacteria, which corroborates the increased relative abundance of the 3 well-known cellulolytic bacteria: R. flavefaciens, R. albus, and F. succinogenes, in the yak. Branched-chain volatile fatty acids are used in synthesizing branched-chain amino acids (Allison et al., 1962). The higher branched-chain VFA concentration in the yak rumen may increase valine, leucine, and isoleucine biosynthesis. It should be noted that although all the 3 species had acetate as the major VFA, yak had the highest molar proportion of propionate but the lowest molar proportion of acetate. Thus, we speculated that in the absence of high-quality forage, the yak rumen microbiome is adapted to change rumen fermentation and enhance energy harvest.
Our metabolomic results showed that cattle enriched styrene degradation and streptomycin biosynthesis, while yak and dzomo enriched primary bile acid biosynthesis. Bile acids are important components of the bile, and they play an important role in fat metabolism, mainly in the intestinal and liver circulatory systems where they play a protective role through recirculation (Jia et al., 2018). The presence of primary and secondary bile acids in the rumen suggests that the rumen microbiome may play an important role in the conversion of primary to secondary bile acids in ruminants (Braun et al., 1989). Further studies are needed to explore the mechanisms by which bile acids circulate in ruminants. Uridine 5′-monophosphate consists of phosphoric acid, ribose, and uracil, the latter of which is a nucleobase for RNA biosynthesis. The enriched pyrimidine metabolism in the cattle rumen might explain the increased uridine 5′-monophosphate therein. The enhanced nucleotide metabolism pathway in the cattle rumen microbiome may provide more substrates for replication and repair, translation, and transcription. The upregulated starch and sucrose metabolism in the cattle rumen might be attributable to the higher concentration of glucose-6-phosphate therein. Taken together, metabolite alterations were consistent with the metagenome results, and they jointly corroborate the adaptation of yak to high-altitude plateau environments.
As demonstrated in the literature (Hugenholtz and de Vos, 2018; Odamaki et al., 2016; Woff et al., 2017), many factors, including feed, age, and management, can affect the gut microbiome composition and its functions in animals. In recent years, many studies found that gut microbiota was also affected by host genetics (Fan et al., 2020; Li et al., 2019b). Thus, it is possible that variations among different species may result from their mother/parent, which may partly explain why the microorganism composition of the dzomo lies between yak and cattle. Because yak are not domesticated, and the yak used in the present study were not confined, we could not weigh each of the study animals to obtain their exact bodyweight or track the specific volume of feed that each animal consumed. Differences in body weight and feed consumption might be confounding factors that could have affected the results. Apparently, the yak rumen microbiome features, as determined by metataxonomics, enzymatic assays, metagenomics, and metabolomics, can help the host to adapt to its harsh environment, especially given the poor diet available, but it remains to be determined how the body of the yak affects its rumen microbiome. Future studies are required to explore the adaptation relationship between high-altitude animals and the symbiotic microbiome. Such information will be useful to understand the adaptability of high-altitude animals to various environments and their domestication.
5. Conclusion
Our study revealed a clear difference in the composition, functions, and metabolome of the rumen microbiome among yak, dzomo, and Qaidam cattle. Although the relative abundance of the genes that code for amylases and their associated CBM families were lower in yak than in cattle, the yak rumen microbiome increased the abundance of cellulolytic bacteria and the genes encoding cellulase and hemicellulase, making yak better adapted to digest their roughage-based diets. Besides, the yak rumen microbiome had more succinate-producing and -utilizing bacterial species, supporting more pyruvate fermentation to propionate. These findings can enhance our understanding of how the yak rumen microbiome aids in yak adaptation to the poor diet in high-altitude plateau environments, and provide a foundation for further studies on microbial roles in the physiology and health of agricultural animals.
Author contributions
Junhu Yao, Zhongtang Yu, Xinhua Chen and Yangchun Cao designed the study. Congcong Zhao, Yu Qiao and Zhanhong Cui collected rumen samples. Fan Zhang, Yong Li and Lamei Wang analyzed the rumen samples for volatile fatty acids. Congcong Zhao, Jing Wang and Dangdang Wang performed the enzyme assays for rumen fibrolytic enzyme activities. Lamei Wang did the DNA extraction, Congcong Zhao, Shanlin Ke, Yangchun Cao and Wenjuan Sun performed the bioinformatics and statistical analysis. Lamei Wang, Yangchun Cao, Ákos Kenéz, Wei Xu and Jianhua Zhao performed rumen metabolomic analysis. Congcong Zhao and Zhongtang Yu wrote the manuscript, and all other authors revised the manuscript. All authors read and approved the final manuscript.
Declaration of competing interest
We declare that we have no financial and personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and/or company that could be construed as influencing the content of this paper.
Acknowledgements
This study was supported by the National Key Research and Development Projects of China (2021YFD1300301) and the Shaanxi Provincial Key Research and Development Program (2021NY-019). The authors are grateful to Tong Wang, Tao Zhang, and Gaofeng Liang for their assistance in the field sampling. We are grateful to Jing Shen, Shengru Wu, and Zongjun Li for their advice and technical assistance.
Footnotes
Peer review under responsibility of Chinese Association of Animal Science and Veterinary Medicine.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aninu.2022.07.014.
Contributor Information
Zhongtang Yu, Email: yu.226@osu.edu.
Yangchun Cao, Email: caoyangchun@126.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ahmad A.A., Yang C., Zhang J.B., Kalwar Q., Liang Z.Y., Li C., et al. Effect of dietary energy levels on rumen fermentation, microbial diversity and feed efficiency of yak (Bos grunniens) Front Microbiol. 2020;11:625. doi: 10.3389/fmicb.2020.00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison M.J., Bryant M.P., Katz I., Keeney M. Metabolic function of branched-chain volatile fatty acids, growth factors for ruminococci. II. Biosynthesis of higher branched-chain fatty acids and aldehydes. J Bacteriol. 1962;83:1084–1093. doi: 10.1128/jb.83.5.1084-1093.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun U., Hausammann K., Forrer R. Reflux of bile acids from the duodenum into the rumen of cows with a reduced intestinal passage. Vet Rec. 1989;124:373–376. doi: 10.1136/vr.124.14.373. [DOI] [PubMed] [Google Scholar]
- Cao Y.C., Wang D.D., Wang L.M., Wei X.S., Li X.Y., Cai C.J., et al. Physically effective neutral detergent fiber improves chewing activity, rumen fermentation, plasma metabolites, and milk production in lactating dairy cows fed a high-concentrate diet. J Dairy Sci. 2021;104:5631–5642. doi: 10.3168/jds.2020-19012. [DOI] [PubMed] [Google Scholar]
- Cui G.X., Yuan F., Degen A.A., Liu S.M., Zhou J.W., Shang Z.H., et al. Composition of the milk of yak raised at different altitudes on the Qinghai Tibetan Plateau. Int Dairy J. 2016;59:29–35. [Google Scholar]
- Dai X., Tian Y., Li J.T., Su X.Y., Wang X.W., Zhao S.G., et al. Metatranscriptomic analyses of plant cell wall polysaccharide degradation by microorganisms in the cow rumen. Appl Environ Microbiol. 2015;81:1375–1386. doi: 10.1128/AEM.03682-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difford G.F., Plichta D.R., Løvendahl P., Lassen J., Noel S.J., Højberg O., et al. Host genetics and the rumen microbiome jointly associate with methane emissions in dairy cows. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durmowicz A.G., Hofmeister S., Kadyraliev T.K., Aldashev A.A., Stenmark K.R. Functional and structural adaptation of the yak pulmonary circulation to residence at high altitude. J Appl Physiol. 1993;74:2276–2285. doi: 10.1152/jappl.1993.74.5.2276. [DOI] [PubMed] [Google Scholar]
- Fan P.X., Bian B.L., Teng L., Nelson C.D., Driver J., Elzo M.A., et al. Host genetic effects upon the early gut microbiota in a bovine model with graduated spectrum of genetic variation. ISME J. 2020;14:302–317. doi: 10.1038/s41396-019-0529-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L.M., Niu B.J., Zhu Z.W., Wu S.T., Li W.Z. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J.Q., Long K.R., Ma J.D., Zhang J.W., He D.F., Jin L., et al. Comparative analysis of the microRNA transcriptome between yak and cattle provides insight into high-altitude adaptation. PeerJ. 2017;5:e3959. doi: 10.7717/peerj.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess M., Sczyrba A., Egan R., Kim T., Chokhawala H., Schroth G., et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Nature. 2011;331:463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- Hugenholtz F., de Vos W.M. Mouse models for human intestinal microbiota research: a critical evaluation. Cell Mol Life Sci. 2018;75:149–160. doi: 10.1007/s00018-017-2693-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeček Š., Majzlová K., Svensson B., MacGregor E. The starch-binding domain family CBM41-an in-silico analysis of evolutionary relationships. Proteins. 2017;85:1480–1492. doi: 10.1002/prot.25309. [DOI] [PubMed] [Google Scholar]
- Jia W., Xie G.X., Jia W.P. Bile acid–microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol. 2018;15:111–128. doi: 10.1038/nrgastro.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamke J., Kittlelmann S., Soni P., Li Y., Tavendale M., Ganesh S., et al. Rumen metagenome and metatranscriptome analyses of low methane yield sheep reveals a Sharpea enriched microbiome characterised by lactic acid formation and utilisation. Microbiome. 2016;4:56. doi: 10.1186/s40168-016-0201-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E., Fredrik B., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. Obesity alters gut microbial ecology. P Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Li D.H., Liu C.M., Luo R.B., Sadakane K., Lam T. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2014;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- Li F., Li Z.J., Li S.X., Ferguson J.D., Cao Y.C., Yao J.H., et al. Effect of dietary physically effective fiber on ruminal fermentation and the fatty acid profile of milk in dairy goats. J Dairy Sci. 2014;97:2281–2290. doi: 10.3168/jds.2013-6895. [DOI] [PubMed] [Google Scholar]
- Li F.Y., Hitch T.C.A., Chen Y., Creevey C.J., Guan L.L. Comparative metagenomic and metatranscriptomic analyses reveal the breed effect on the rumen microbiome and its associations with feed efficiency in beef cattle. Microbiome. 2019;7:6. doi: 10.1186/s40168-019-0618-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.Y., Li C.X., Chen Y.H., Liu J.H., Zhang C.Y., Irving B., et al. Host genetics influence the rumen microbiota and heritable rumen microbial features associate with feed efficiency in cattle. Microbiome. 2019;7:92. doi: 10.1186/s40168-019-0699-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R.Q., Li Y.R., Kristiansen K., Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Lin L.M., Xei F., Sun D.M., Liu J.H., Zhu W.Y., Mao S.Y. Ruminal microbiome-host crosstalk stimulates the development of the ruminal epithelium in a lamb model. Microbiome. 2019;7:83–98. doi: 10.1186/s40168-019-0701-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Wu H., Liu S.J., Chai S.T., Meng Q.X., Zhou Z.M. Dynamic alterations in yak rumen bacteria community and metabolome characteristics in response to feed type. Front Microbiol. 2019;10:1116. doi: 10.3389/fmicb.2019.01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machovic M., Janecek S. Domain evolution in the GH13 pullulanase subfamily with focus on the carbohydrate-binding module family 48. Biologia. 2008;63:1057–1068. [Google Scholar]
- Miller G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Biochem. 1959;31:426–428. [Google Scholar]
- Moraïs S., Mizrahi I. Islands in the stream: from individual to communal fiber degradation in the rumen ecosystem. FEMS Microbiol Rev. 2019;43:362–379. doi: 10.1093/femsre/fuz007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulin L., Munive A., Dreyfus B., Boivin-Masson C. Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature. 2001;411:948–950. doi: 10.1038/35082070. [DOI] [PubMed] [Google Scholar]
- Myer P.R., Wells J.E., Smith T.P.L., Kuehn L.A., Freetly H.C. Cecum microbial communities from steers differing in feed efficiency. J Anim Sci. 2015;93:5327–5340. doi: 10.2527/jas.2015-9415. [DOI] [PubMed] [Google Scholar]
- Noguchi H., Park J., Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 2006;34:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odamaki T., Kato K., Sugahara H., Hashikura N., Takahashi S., Xiao J.Z., et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 2016;16:90. doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz H.A., Anderson C.L., Muller M.J., Kononoff P.J., Fernando S.C. Rumen bacterial community composition in Holstein and Jersey cows is different under same dietary condition and is not affected by sampling method. Front Microbiol. 2016;7:1206. doi: 10.3389/fmicb.2016.01206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perea K., Perz K., Olivo S.K., Williams A., Lachman M., Ishaq S.L., et al. Feed efficiency phenotypes in lambs involve changes in ruminal, colonic, and small-intestine-located microbiota. J Anim Sci. 2017;95:2585–2592. doi: 10.2527/jas.2016.1222. [DOI] [PubMed] [Google Scholar]
- Ryle M., Orskov E.R. Springer; Netherlands: 1990. Energy nutrition in ruminants. [Google Scholar]
- Shabat S.K.B., Sasson G., Doron-Faigenboim A., Durman T., Yaacoby S., Berg Miller M.E., et al. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 2016;10:2958–2972. doi: 10.1038/ismej.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao B., Long R., Ding Y., Wang J., Ding L., Wang H. Morphological adaptations of yak (Bos grunniens) tongue to the foraging environment of the Qinghai-Tibetan Plateau. J Anim Sci. 2010;88:2594–2603. doi: 10.2527/jas.2009-2398. [DOI] [PubMed] [Google Scholar]
- Shi W.B., Moon C.D., Leah S.C., Kang D., Froula J., Kittelmann S., et al. Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res. 2014;24:1517–1525. doi: 10.1101/gr.168245.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sichert A., Corzett C.H., Schechter M.S., Unfried F., Markert S., Becher D., et al. Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat Microbiol. 2020;5:1026–1039. doi: 10.1038/s41564-020-0720-2. [DOI] [PubMed] [Google Scholar]
- Spor A., Koren O., Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- Stewart R.D., Auffret M.D., Warr A., Wiser A.H., Press M.O., Langford K.W., et al. Assembly of 913 microbial genomes from metagenomic sequencing of the cow rumen. Nat Commun. 2018;9:870–880. doi: 10.1038/s41467-018-03317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase H., Tumennasan K., Hiratsuka K., Chandley A.C., Hotta Y. Fertility investigation in F1 hybrid and backcross progeny of cattle (Bos taurus) and yak (B. gruniens) in Mongolia. : II. Little variation in gene products studied in male sterile and fertile animals. Niigata J Health Welf. 2002;2:42–52. [Google Scholar]
- Tremaroli V., Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Weir E.K., Tucker A., Reeves J.T., Will D.H., Grover R.F. The genetic factor influencing pulmonary hypertension in cattle at high altitude. Cardiovasc Res. 1974;8:745–749. doi: 10.1093/cvr/8.6.745. [DOI] [PubMed] [Google Scholar]
- Woff S.M., Ellison M.J., Hao Y., Cockrum R.R., Austin K.J., Baraboo M., et al. Diet shifts provoke complex and variable changes in the metabolic networks of the ruminal microbiome. Microbiome. 2017;5:60. doi: 10.1186/s40168-017-0274-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J.G., Sinelnikov I.V., Han B., Wishart D.S. MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Mao X.Z., Huang J.J., Ding Y., Wu J., Dong S., et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–W322. doi: 10.1093/nar/gkr483. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin J.W., Chai Z.X., Zhang C.F., Zhang Q., Zhong J.C. Transcriptome profiles revealed the mechanisms underlying the adaptation of yak to high-altitude environments. Sci Rep. 2019;9:7558. doi: 10.1038/s41598-019-43773-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin J.W., Chai Z.X., Zhang C.F., Zhang Q., Zhu Y., Cao H.W., et al. Comparing the microbial community in four stomach of dairy cattle, yellow cattle and three yak herds in Qinghai-Tibetan plateau. Front Microbiol. 2019;10:1547. doi: 10.3389/fmicb.2019.01547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M.Y., Sun H.Z., Wu X.H., Liu J.X., Guan L.L. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome. 2020;8:64. doi: 10.1186/s40168-020-00819-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y.F., Lin L.M., Hu F., Zhu W.Y., Mao S.Y. Disruption of ruminal homeostasis by malnutrition involved in systemic ruminal microbiota-host interactions in a pregnant sheep model. Microbiome. 2020;8:138. doi: 10.1186/s40168-020-00916-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.J. Response of forage fiber degradation by ruminal microorganisms to branched-chain volatile fatty acids, amino acids, and dipeptides. J Dairy Sci. 2002;85:1183–1190. doi: 10.3168/jds.S0022-0302(02)74181-7. [DOI] [PubMed] [Google Scholar]
- Yang C.J., Mao S.Y., Long L.M., Zhu W.Y. Effect of disodium fumarate on microbial abundance, ruminal fermentation and methane emission in goats under different forage: concentrate ratios. Animal. 2012;6:1788–1794. doi: 10.1017/S1751731112000857. [DOI] [PubMed] [Google Scholar]
- Yang H.J., Yue Q. Effect of glucose addition and N sources in defined media on fibrolytic activity profiles of Neocallimastix sp. YQ1 grown on corn stover. J Anim Physiol Anim Nutr. 2012;96:554–562. doi: 10.1111/j.1439-0396.2011.01177.x. [DOI] [PubMed] [Google Scholar]
- Yang W.Z., Beauchemin K.A. Physically effective fiber: method of determination and effects on chewing, ruminal acidosis, and digestion by dairy cows. J Dairy Sci. 2006;89:2618–2633. doi: 10.3168/jds.S0022-0302(06)72339-6. [DOI] [PubMed] [Google Scholar]
- Yue Q., Yang H.J. Feruloyl and acetylesterase production of an anaerobic rumen fungus Neocallimastix sp. YQ2 effected by glucose and soluble nitrogen supplementations and its potential in the hydrolysis of fibrous feedstuffs. Anim Feed Sci Technol. 2009;153:263–277. [Google Scholar]
- Zebeli Q., Ghareeb K., Humer E., Metzler-Zebeli B., Besenfelder U. Nutrition, rumen health and inflammation in the transition period and their role on overall health and fertility in dairy cows. Res Vet Sci. 2015;103:126–136. doi: 10.1016/j.rvsc.2015.09.020. [DOI] [PubMed] [Google Scholar]
- Zebeli Q., Tafaj M., Steingass H., Metzler B., Drochner W. Effects of physically effective fiber on digestive processes and milk fat content in early lactating dairy cows fed total mixed rations. J Dairy Sci. 2006;89:651–668. doi: 10.3168/jds.S0022-0302(06)72129-4. [DOI] [PubMed] [Google Scholar]
- Zeng Y., Xu J.Y., Fu X.P., Tan M., Liu F., Zheng H.C., et al. Effects of different carbohydrate-binding modules on the enzymatic properties of pullulanase. Int J Biol Macromol. 2019;137:973–981. doi: 10.1016/j.ijbiomac.2019.07.054. [DOI] [PubMed] [Google Scholar]
- Zhang H.L., Chen Y., Xu X.L., Yang Y.X. Effects of branched-chain amino acids on in vitro ruminal fermentation of wheat straw. Asian-Australas J Anim Sci. 2013;26:523–528. doi: 10.5713/ajas.2012.12539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Yang M., Cai D.Y., Hao Y.J., Zhao X., Zhu Y.H., et al. Composition, coagulation characteristics, and cheese making capacity of yak milk. J Dairy Sci. 2020;103:1276–1288. doi: 10.3168/jds.2019-17231. [DOI] [PubMed] [Google Scholar]
- Zhang Z.G., Xu D.M., Wang L., Hao J.J., Wang J.F., Zhou X., et al. Convergent evolution of rumen microbiomes in high-altitude mammals. Curr Biol. 2016;26:1873–1879. doi: 10.1016/j.cub.2016.05.012. [DOI] [PubMed] [Google Scholar]
- Zhou J.W., Liu H., Zhong C.L., Degend A.A., Yanga G., Zhang Y., et al. Apparent digestibility, rumen fermentation, digestive enzymes and urinary purine derivatives in yak and Qaidam cattle offered forage-concentrate diets differing in nitrogen concentration. Livest Sci. 2018;208:14–21. [Google Scholar]
- Zhou Z.C., Tran P.Q., Kieft K., Anantharaman K. Genome diversification in globally distributed novel marine Proteobacteria is linked to environmental adaptation. ISME J. 2020;14:2060–2077. doi: 10.1038/s41396-020-0669-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.