Abstract
FtsZ, the ancestral homologue of eukaryotic tubulins, assembles into the Z ring, which is required for cytokinesis in prokaryotic cells. Both FtsZ and tubulin have a GTPase activity associated with polymerization. Interestingly, the ftsZ2 mutant is viable, although the FtsZ2 mutant protein has dramatically reduced GTPase activity due to a glycine-for-aspartic acid substitution within the synergy loop. In this study, we have examined the properties of FtsZ2 and found that the reduced GTPase activity is not enhanced by DEAE-dextran-induced assembly, indicating it has a defective catalytic site. In the absence of DEAE-dextran, FtsZ2 fails to assemble unless supplemented with wild-type FtsZ. FtsZ has to be at or above the critical concentration for copolymerization to occur, indicating that FtsZ is nucleating the copolymers. The copolymers formed are relatively stable and appear to be stabilized by a GTP-cap. These results indicate that FtsZ2 cannot nucleate assembly in vitro, although it must in vivo. Furthermore, the stability of FtsZ-FtsZ2 copolymers argues that FtsZ2 polymers would be stable, suggesting that stable FtsZ polymers are able to support cell division.
FtsZ is essential for cell division in bacteria (18). During the cell division cycle, it assembles into the Z ring, a cytoskeletal element that is at the leading edge of the invaginating septum (5). One role of the Z ring is to recruit additional division proteins to the division site to form the septal ring that carries out cell division in bacteria (18, 19, 30). An additional role for the Z ring may be to provide the force for constriction of the septum. Such a force could arise from depolymerization of FtsZ filaments or from motor proteins acting upon FtsZ filaments (6).
The remarkable functional and structural similarity between FtsZ and tubulin suggests that FtsZ is the ancestral homologue of tubulin, despite sharing only 10 to 18% amino acid identity (10, 11, 17, 23). FtsZ assembles into protofilaments that are structural analogues of the protofilaments present in the walls of microtubules (12). It can also assemble into a variety of additional structures, including sheets, tubes, and rings, depending upon the in vitro conditions. Like tubulin, FtsZ displays a GTPase activity that is concentration dependent, suggesting that the GTPase activity is stimulated during assembly (8, 16, 33, 39).
Assembly of the Z ring is a critical step during cell division. SulA, a division inhibitor produced following DNA damage, blocks division by preventing Z ring formation (4). In vitro SulA blocks both FtsZ's GTPase activity and FtsZ's assembly, providing a mechanism for its ability to prevent formation of the Z ring (21, 35).
Several ftsZ mutations have been isolated that confer resistance to SulA (2). These mutations are dominant to the wild type, so that merodiploids are resistant to SulA. Two of the mutant proteins have been studied in some detail. FtsZ114 supports viability and assembles into the Z ring in vivo even in the presence of SulA (2). In vitro FtsZ114 has one-half the wild-type GTPase activity and polymerizes efficiently (21). Both of these activities are resistant to SulA. Consistent with this, FtsZ114 does not bind SulA in yeast two-hybrid studies (14). The other mutant, FtsZ2, has dramatically reduced GTPase activity, but still supports growth, provided the ftsZ2 gene is present on a low-copy plasmid that supplies about twice the level of the chromosomal locus (3, 7). In vitro FtsZ2 assembles in the presence of DEAE-dextran (23), and this assembly is resistant to SulA (35).
The ftsZ2 mutation results in a glycine-for-aspartic acid substitution at residue 212 (2). This residue is part of the synergy loop likely to be directly involved in the GTPase activity during assembly of FtsZ (7, 11, 27, 38). Several additional mutations that alter other conserved residues within this loop are lethal and lead to loss of GTPase activity without loss of GTP binding (7, 38). Based upon analogy with tubulin, during assembly, the synergy loop on the incoming FtsZ is juxtaposed to the γ-phosphate of GTP on the terminal subunit of the polymer, forming a catalytic site at the interface of the two subunits.
In tubulin, the position corresponding to the altered residue in FtsZ2 is thought to play a critical role in differentiating the ability of α- and β-tubulin to induce GTPase activity (27). β-Tubulin contains a lysine at this position and is unable to stimulate hydrolysis of GTP bound to α-tubulin. As a result there is a nonexchangeable GTP bound at the dimer interface. In contrast, α-tubulin, which has a glutamate at this position, induces hydrolysis of GTP bound to the β-tubulin subunit during assembly into microtubules. It's possible that a negative charge at this position is necessary for the synergy loop to induce GTP hydrolysis on a neighboring subunit during assembly.
To more fully understand the mechanism of GTP hydrolysis and its role during assembly and cell division, we have further characterized FtsZ2. We find that the weak GTPase activity of FtsZ2 is not stimulated by increasing the concentration of FtsZ2 or by forcing assembly, indicating that the catalytic site is defective. Furthermore, FtsZ2 is unable to polymerize in vitro, but copolymerizes efficiently with FtsZ, provided FtsZ is present above the critical concentration for polymerization. The copolymers are relatively stable, suggesting that FtsZ2 polymers would be stable. Furthermore, these copolymers are stabilized by a GTP-cap. These results are discussed in terms of assembly of FtsZ and the role of the Z ring during cell division.
MATERIALS AND METHODS
Overexpression and purification of proteins.
The plasmids pKD126 and pXYF222 were used for overexpression of FtsZ and FtsZ2, respectively. Their construction has been described previously (7). These plasmids contain ftsZ and ftsZ2 with their ribosome binding sites cloned downstream of the tac promoter in the expression vector pJF118HE. For overexpression of FtsZ-GFP, the green fluorescent protein (GFP) gene, gfp (mut2), was amplified with primers with XbaI and HindIII sites added to the 5′ and 3′ primers, respectively. The GFP fragment was cloned at the poly-linker site of pBAD18 (13). ftsZ was amplified along with its ribosome binding site by using primers that had SacI and XbaI sites added to the 5′ and 3′ primers, respectively. The ftsZ fragment was then cloned downstream of the ara promoter and was in frame with the 5′ end of gfp. The resulting plasmid was designated pJC104. pKD126 and pJC104 were transformed into Escherichia coli W3110, and pXYF222 was transformed into E. coli JFL101 (7). Overnight cultures were diluted 100-fold into Luria-Bertani broth containing ampicillin at 100 μg/ml and grown at 37°C until an optical density at 590 nm of between 0.3 and 0.4 was reached. Cultures were then induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG; 0.1% arabinose in the case of pJC104) for 3 h, harvested, washed, and stored frozen as described previously (24). All three proteins were purified as described previously (25). Protein concentration was determined by using the Bio-Rad protein assay reagent with bovine serum albumin as a standard.
FtsZ polymerization and GTPase assays.
FtsZ polymerization assays by electron microscopy and centrifugation were performed as described previously (24). The amounts of FtsZ-GFP and FtsZ2 in copolymers were determined by analyzing pellets obtained by centrifugation and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Following staining with Coomassie blue digital images of the gels were captured with a charge-coupled device camera, and bands were quantitated with Alpha Innotech software. Copolymerization experiments were done with a fixed concentration of FtsZ-GFP and increasing concentrations of FtsZ2. For each concentration of FtsZ2, control experiments were done with FtsZ2 alone with GTP added. The amount in the pellet was subtracted from the copolymerization experiments to obtain the amount of FtsZ2 that had copolymerized with FtsZ-GFP. Static light scattering assays for polymerization were done as described elsewhere (25), except that a slit width of 3 nm was used. GTPase assays have been described before (22).
Analytical ultracentrifugation.
Sedimentation equilibrium experiments were performed in a Beckman Optima XL-A at 20°C and different rotor speeds. Each protein, in a total volume of 100 μl, was loaded at different concentrations into six-channel 12-mm-path-length centerpieces, and the radial protein distribution was determined by UV A280 by consecutive automated scans acquired at each 0.001 cm of radial spacing. Data sets were collected after reaching thermodynamic equilibrium, judged to be achieved when overlaid scans taken 24 h apart were superimposable. Sedimentation data were analyzed by nonlinear least-squares methods (15) with different models running under SigmaPlot software (SPSS, Inc.). The second moment mass Mw (weight-average molecular weight) was estimated by fitting the data to a single-solute model by using the following equation:
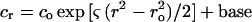 |
1 |
where cr is the concentration of the solute at arbitrary radial position r, and co at the reference radial position, ro, and base is an error term for the nonsedimenting material. The parameter ς is the reduced molecular weight, and is defined as follows:
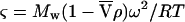 |
2 |
where R is the universal gas constant, T is the kelvin temperature, and ω is the rotor angular velocity, 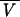 is the protein partial specific volume, and ρ is the solvent density. The partial specific volume of FtsZ,
is the protein partial specific volume, and ρ is the solvent density. The partial specific volume of FtsZ, 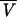 , and solvent density, ρ, were calculated to be 0.7396 ml/g at 25°C and 1.006 at 20°C, respectively, by using the program SEDNTERP, which is available from www.cauma.uthscsa.edu/software/. Adjustments for temperature differences were calculated by using the following equation (9):
, and solvent density, ρ, were calculated to be 0.7396 ml/g at 25°C and 1.006 at 20°C, respectively, by using the program SEDNTERP, which is available from www.cauma.uthscsa.edu/software/. Adjustments for temperature differences were calculated by using the following equation (9):
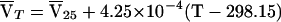 |
3 |
where 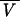 T is the partial specific volume at temperature T (in kelvins) and
T is the partial specific volume at temperature T (in kelvins) and 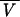 25 is the partial specific volume at 25°C. To determine dissociation constants, the data were analyzed with an indefinite self-association (isodesmic) model (1, 36), in which the free energy change is the same for the addition of a monomer to any oligomer. In this case, the total concentration, ct, of a reversibly self-associating protein can be related to the monomer concentration by
25 is the partial specific volume at 25°C. To determine dissociation constants, the data were analyzed with an indefinite self-association (isodesmic) model (1, 36), in which the free energy change is the same for the addition of a monomer to any oligomer. In this case, the total concentration, ct, of a reversibly self-associating protein can be related to the monomer concentration by
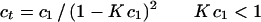 |
4 |
where the distribution of the monomer c1 at any radial distance is given by equation 1. In this relationship, 1/K is the isodesmic dissociation constant, Kd, expressed in milligrams per milliliter. Thus, the radial distribution of the total concentration can be obtained by substituting equation 1 into equation 4, and the experimental data were fit with the expression
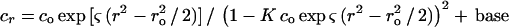 |
5 |
RESULTS
FtsZ2 does not polymerize in the absence of DEAE-dextran.
We have previously shown that FtsZ2 binds GTP as well as wild-type FtsZ and polymerizes in the presence of GTP and DEAE-dextran (23). Furthermore, we have shown that FtsZ2 hydrolyzes GTP poorly, with less than 1% of the activity of wild-type FtsZ (7). In those studies, however, FtsZ2 was purified by a procedure that rendered wild-type FtsZ polymerization dependent upon DEAE-dextran. A modification of the purification procedure yielded a purified FtsZ that contained 0.7 mol of GDP per mol of FtsZ. Polymerization of this FtsZ required GTP but occurred independently of DEAE-dextran (24). To further study FtsZ2 polymerization, FtsZ2 was purified by the latter purification procedure.
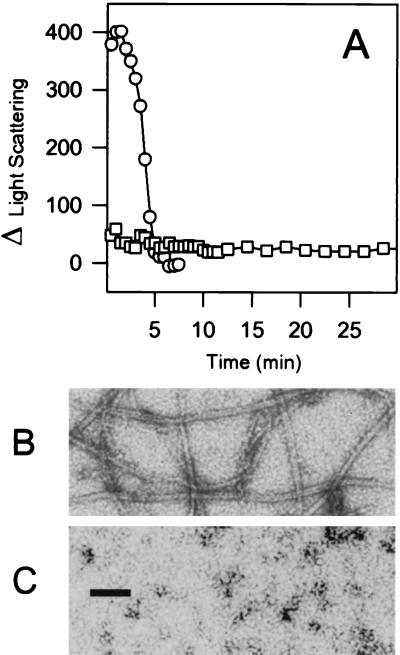
FtsZ2 purified by the latter method also contained 0.7 mol of GDP bound per mol of FtsZ2 (data not shown). To determine if this preparation of FtsZ2 could polymerize in the absence of DEAE-dextran, we used light scattering and electron microscopy. FtsZ or FtsZ2 was incubated at 500 μg/ml (12.5 μM) in polymerization buffer for 8 min, and polymerization was initiated by addition of 0.1 mM GTP (Fig. 1A). Immediately upon addition of GTP, the net increase in light scattering for FtsZ was about 400, while that for FtsZ2 was about 20. The light scattering signal for FtsZ rapidly decreased, reaching the baseline within 5 min. This decline is due to net disassembly following GTP exhaustion (24). With FtsZ2, the small increase in light scattering remained constant for at least 40 min. Samples for electron microscopy were examined for FtsZ2 polymer formation 5 and 30 min after GTP addition. No polymer formation was observed at either time point (Fig. 1C, 30-min time point). Even at 1 mg of FtsZ2 per ml, no polymers were detected. In contrast, electron microscopy of the FtsZ sample taken during the steady-state phase of polymerization revealed abundant polymers with variable numbers of protofilaments as reported previously (Fig. 1B) (24). We conclude that FtsZ2 is unable to polymerize under these conditions, which promote rapid assembly of FtsZ.
FIG. 1.
Assay of FtsZ and FtsZ2 polymerization by 90° angle light scattering. Reaction mixtures (300 μl) containing FtsZ (open circles) or FtsZ2 (open squares) at a concentration of 500 μg/ml were incubated in polymerization buffer at 30°C for 8 min, after which, 0.1 mM GTP was added to initiate polymerization. (A) The net change in light scattering following GTP addition was plotted against time. The reaction mixtures for FtsZ (B) and FtsZ2 (C) were also examined by electron microscopy at 1 min. The black bar in panel C represents 100 μm.
FtsZ2 cannot self-activate its GTPase activity.
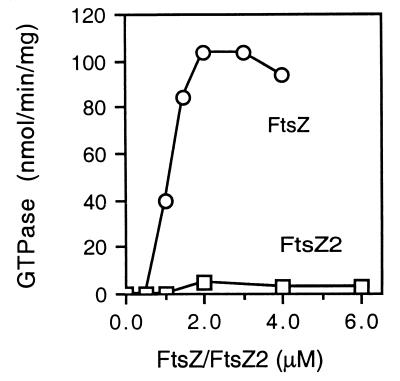
Studies with FtsZ from several bacteria have revealed that it's GTPase activity is stimulated by increasing concentrations of FtsZ (16, 33, 39). This concentration-dependent stimulation of enzymatic activity is a typical feature of self-associating proteins. Although FtsZ2 has low GTPase activity, we nonetheless tested whether this low specific activity changes with FtsZ2 concentration. The GTPase activities of FtsZ and FtsZ2 were measured over a concentration range of 1 to 6 μM, and the specific activity at each concentration was plotted against the concentration of the protein (Fig. 2). FtsZ's GTPase displays low specific activity at 0.5 μM; however, the specific activity increases dramatically between 0.5 and 3 μM and remains high thereafter. These results are similar to other reports on FtsZ from various bacteria (16, 33, 39). On the other hand, the specific activity of FtsZ2, which is comparable to the specific activity of FtsZ at 0.5 μM, did not manifest a significant change over a similar range of concentrations. Thus, FtsZ2 does not self-activate its basal GTPase activity, but this may be due to its failure to assemble under these conditions.
FIG. 2.
Assay of the GTPase activities of FtsZ and FtsZ2 at different protein concentrations. The GTPase activities of FtsZ and FtsZ2 were measured at different protein concentrations in polymerization buffer at 30°C with 0.2 mM [γ-32P]GTP. The specific activity was plotted against protein concentration.
To test whether the failure of FtsZ2 to self-activate its GTPase activity is due to its inability to assemble under these conditions, we assayed the GTPase activity in the presence of DEAE-dextran, which promotes polymerization of FtsZ2. The GTPase activities of FtsZ and FtsZ2 were measured at 200 μg/ml (5 μM), with and without 50 μg of DEAE-dextran per ml. As can be seen in Fig. 3B, this concentration of DEAE-dextran promotes polymerization of FtsZ2 into tubular structures as reported previously (23, 35). However, this concentration of DEAE-dextran does not stimulate the GTPase activity of FtsZ2 nor affect that of FtsZ (Fig. 3A). Thus, even though DEAE-dextran promotes polymerization of FtsZ2, it does not stimulate its GTPase activity. This result argues that FtsZ2 has a defective catalytic site.
FIG. 3.
DEAE-dextran does not affect the GTPase activities of FtsZ or FtsZ2. (A) The GTPase activities of FtsZ or FtsZ2 at a concentration of 200 μg/ml were measured in polymerization buffer at 30°C with and without 50 μg of DEAE-dextran per ml with 1 mM [γ-32P]GTP. Open circles, FtsZ; solid circles, FtsZ plus DEAE-dextran; open squares, FtsZ2; solid squares, FtsZ2 plus DEAE-dextran. (B) Assay of FtsZ2 polymerization in the presence of DEAE-dextran by electron microscopy. FtsZ2 at a concentration of 200 μg/ml was incubated in polymerization buffer at 30°C with 1 mM GTP and 50 μg of DEAE-dextran per ml. Samples were taken at 5 and 60 min for electron microscopy. Both samples were similar, so only the 60-min sample is shown. The black bar represents 100 μm.
Sedimentation equilibrium studies with FtsZ and FtsZ2.
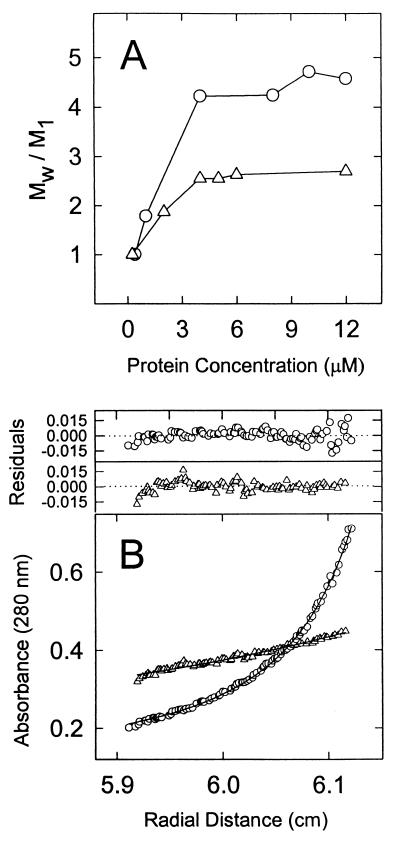
The observations presented above that FtsZ2 does not polymerize in vitro (in the absence of DEAE-dextran) nor self-activate its GTPase activity led us to analyze the self-association properties of FtsZ2 by sedimentation equilibrium. Single-component sedimentation analyses were carried out in 20 mM HEPES–NaOH–150 mM KCl (pH 7.2) over the concentration range 0.25 to 12 μM in the absence of GTP. Diagnostics for FtsZ and FtsZ2 complexity consisting of a plot of the state of association (Mw/M1) versus protein concentration are shown in Fig. 4A. Mw/M1 values (where M1 = monomer mass and Mw = best fit mass of the predominant species) increase continuously with protein concentration until 4 μM, at which the values plateau at 4 for FtsZ and 2 for FtsZ2. The results of the self-association analysis of FtsZ are similar to results reported elsewhere, except that we see self-association at lower FtsZ concentrations (33). Importantly, the average size of FtsZ2 is a dimer under conditions in which the average size of FtsZ is a tetramer, indicating that FtsZ2 has a reduced ability to self-associate. This result indicates that FtsZ2 has a weaker longitudinal bond than FtsZ.
FIG. 4.
Sedimentation equilibrium centrifugation of FtsZ and FtsZ2. (A) Concentration-dependent formation of FtsZ (open circles) and FtsZ2 (open triangles) oligomers is presented as the association state (Mw/M1) against cell loading concentration. (B) Isodesmic fit (solid lines) to the FtsZ distributions at an initial concentration of 10 μM (open circles) and in the presence of 4 M guanidinium hydrochloride (open triangles) are shown. The top panel shows the distribution of the residuals from each curve fit. All samples were centrifuged at 10,000 rpm for 48 h.
In order to define the relative affinity for each complex, different protein distributions with concentrations >10 μM were analyzed by using a multicomponent isodesmic model. The FtsZ tetramer was characterized by a dissociation constant (Kd) of 2.5 ± 0.3 μM. The complex was abolished in the presence of 4 M guanidinium hydrochloride, generating a 40-kDa species expected for FtsZ monomers. This result indicates that the oligomerized FtsZ is a noncovalent reversible mass-action equilibrium association (Fig. 4B). Interestingly, FtsZ2 was found to have a lower affinity (Kd of 5.7 ± 0.8 μM) for the formation of the dimeric complex.
Copolymerization of FtsZ2 and FtsZ.
Although FtsZ2 does not polymerize in vitro (unless DEAE-dextran is provided), in vivo it is able to support growth and division because cells containing only FtsZ2 are viable (3). Such cells have an increased average cell length, suggesting FtsZ2 has impaired function. Furthermore, a merodiploid strain is resistant to SulA due to FtsZ2, and the aberrant morphology of an ftsZ2 mutant is largely corrected by the wild-type FtsZ, suggesting FtsZ and FtsZ2 are codominant.
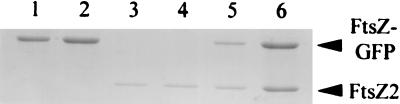
The inability of FtsZ2 to form larger oligomers (average size of a dimer versus a tetramer for wild-type FtsZ) in sedimentation studies indicated a deficiency in self-association that might be a barrier for FtsZ2 polymerization. If so, then addition of FtsZ to FtsZ2 might promote copolymer formation by nucleating assembly. We therefore tested if FtsZ2 would copolymerize with FtsZ by using a centrifugation assay. In order to distinguish between FtsZ and FtsZ2 in the pellet, we used FtsZ tagged at the C-terminal end with GFP (FtsZ-GFP, Mw of 75 kDa). In reactions containing FtsZ-GFP at 200 μg/ml (2.75 μM), there was more FtsZ-GFP in the pellet with GTP than in that with GDP (Fig. 5, lanes 1 and 2). FtsZ-GFP gives a higher background with GDP than FtsZ; however, electron microscopy results confirmed that only the reaction with GTP contained polymers (data not shown). FtsZ2 at 200 g/ml (5 μM) did not polymerize (Fig. 5, lanes 3 and 4), whereas incubation of 200 μg of FtsZ2 per ml with 200 μg of FtsZ per ml allowed polymerization, as demonstrated by the increase in FtsZ2 in the pellet (Fig. 5, lanes 5 and 6). This result demonstrates that FtsZ can induce copolymerization of FtsZ2 and suggests that the barrier to FtsZ2 polymerization in this in vitro system is the nucleation step.
FIG. 5.
Copolymerization of FtsZ-GFP and FtsZ2 assayed by centrifugation and SDS-PAGE. Reaction mixtures (100 μl) containing 200 μg of FtsZ-GFP (lanes 1 and 2), FtsZ2 (lanes 3 and 4), or FtsZ-GFP plus FtsZ2 (lanes 5 and 6) per ml were incubated in polymerization buffer containing 1 mM MgCl2 and either 1 mM GDP (lanes 1, 3, and 5) or 1 mM GTP (lanes 2, 4, and 6). After incubation for 2 min at 30°C, the samples were centrifuged at 80,000 rpm for 15 min. The pellets were suspended in 100 μl of SDS sample buffer, and 20-μl aliquots were run on SDS-PAGE gels. The relative spot density units were for FtsZ-GFP, 19250 (lane 1) and 26100 (lane 2). The units for FtsZ2 were 8,470 (lane 3), 9,240 (lane 4), 10,010 (lane 5), and 23,100 (lane 6). Only the latter represents a significant increase over that seen in lanes 3 to 5.
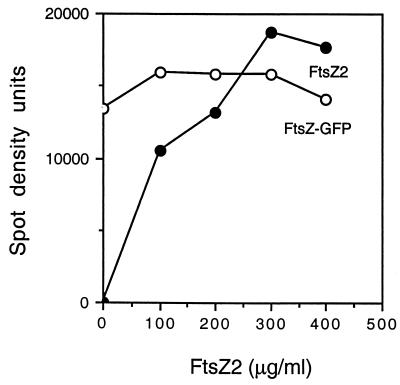
To determine the efficiency of incorporation of FtsZ2 into the copolymers, the concentration of FtsZ-GFP in the reaction mixtures was fixed, and FtsZ2 was varied. The amounts of FtsZ and FtsZ2 in the copolymers were assessed by SDS-PAGE and densitometry. The FtsZ-GFP in the reaction was 150 μg/ml (2 μM), and the amount of FtsZ2 varied from 0 to 400 μg/ml (10 μM). The data in Fig. 6 show that the amount of FtsZ-GFP in the polymers was about constant, whereas the amount of FtsZ2 increased with increasing FtsZ2. At a molar ratio of FtsZ2 to FtsZ-GFP of 3.75 (the highest concentration of FtsZ2 tested), the molar ratio in the copolymer was approximately 1.25. This indicates that FtsZ2 is less efficient at assembly than FtsZ-GFP.
FIG. 6.
Efficiency of assembly of FtsZ2 into the copolymers. FtsZ2 was incubated at 30°C in polymerization buffer containing 1 mM MgCl2 and1 mM GTP. After 3 min, 2 μM FtsZ-GFP (130 μg/ml) was added to each of the reaction mixtures, and incubation continued for 5 min. The reaction mixtures were centrifuged at 80,000 rpm for 15 min, the pellets were suspended in 100 μl of SDS sample buffer, and 20 μl was analyzed by SDS-PAGE. The amounts of FtsZ-GFP and FtsZ2 in the copolymers were determined by densitometry.
FtsZ2 and FtsZ copolymers have increased stability.
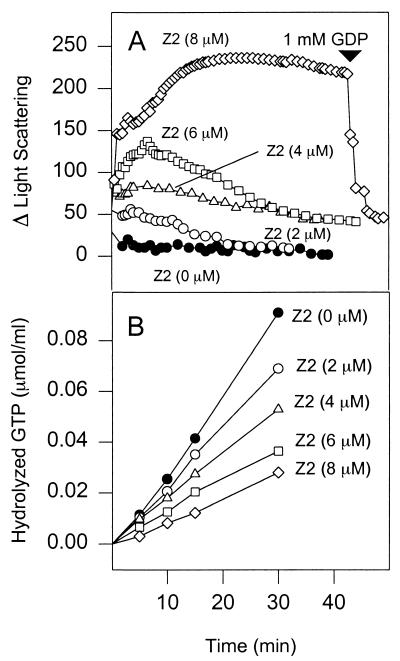
Since FtsZ2 is deficient in GTPase activity, we suspected that the copolymers might be more stable. The stability of the copolymers was assessed by light scattering. In these experiments, increasing concentrations of FtsZ2 were incubated with a constant concentration of FtsZ. The FtsZ concentration was 100 μg/ml (2.5 μM), which is near the critical concentration for polymerization (24, 26). At this concentration, the addition of 0.1 mM GTP causes only a slight increase in light scattering (Fig. 7A). The addition of FtsZ (100 μg/ml) to reaction mixtures containing FtsZ2 preincubated with 0.1 mM GTP resulted in significant increases in light scattering. The increase in light scattering correlated with the amount of FtsZ2, indicating that polymer mass was proportional to the amount of FtsZ2 added. Significantly, the copolymers formed with increasing concentrations of FtsZ2 become increasingly stable. With 2 and 4 μM FtsZ2, the copolymers depolymerize in about 20 and 40 min, respectively. At 6 and 8 μM FtsZ2, the change in light scattering and the stability of the copolymers further increased, and only a small decrease in light scattering is observed over the length of the experiment. These results are in contrast to polymers formed with FtsZ and 0.1 mM GTP, which depolymerize within 5 min due to GTP exhaustion (Fig. 1).
FIG. 7.
FtsZ and FtsZ2 copolymers have increased stability. (A) Various concentrations of FtsZ2 were incubated in polymerization buffer with 0.1 mM GTP at 30°C for 5 min (300 μl), and polymerization was initiated by adding FtsZ at 100 μg/ml (2.5 μM). The net change in light scattering was plotted against time. To one of the reaction mixtures was added 1 mM GDP, as indicated by the arrow. (B) Effect of varying the FtsZ2 concentration on the GTPase activity of FtsZ. The GTPase activity of FtsZ at a concentration of 100 μg/ml was measured without and with various concentrations of FtsZ2 in polymerization buffer at 30°C with 0.2 mM [γ-32P]GTP.
Copolymer stability correlates with decreased GTPase activity.
The stability of the copolymers indicates that they have decreased GTPase activity resulting in the persistence of the GTP in the polymer. This would be expected, since FtsZ2 molecules are unable to trigger GTP hydrolysis as they assemble into the polymer. Also, the stabilized polymers would trap FtsZ, not allowing it to rapidly cycle through rounds of polymerization. When the reactions were checked for GTPase activity, we found decreasing activity with increasing FtsZ2 concentration (Fig. 7B). At molar ratios of FtsZ2 to FtsZ of 2.4:1 and 3.6:1, the levels of inhibition were 50 and 75%, respectively.
We and others have observed that increased bundling of FtsZ protofilaments correlates with a decrease in GTPase activity (16, 26, 41). It may be that longer-lived polymers have more time to associate laterally resulting in bundling. Since the GTPase activity slowed down with increasing FtsZ2 concentrations, we suspected that the copolymers are likely to be bundled. Electron microscopic examination of copolymers present at 40 min after GTP addition from the reaction mixture containing 8 μM FtsZ2 showed more bundling than FtsZ alone (data not shown). The formation of these large bundles is presumably responsible for the slow increase in light scattering observed after the rapid initial rise (Fig. 7A).
Copolymers are destabilized by GDP.
Since FtsZ2 is deficient in GTP hydrolysis, there would be an increasing number of GTP-containing subunits in the copolymer as the ratio of FtsZ2 to FtsZ increases. This would lead to more stable polymers if GTP-containing subunits do not readily dissociate from the polymer ends, functioning as a stabilizing GTP-cap as in microtubules (20). We therefore tested the stability of copolymers formed at an FtsZ2/FtsZ ratio of 3.6:1 in the presence of GDP. As seen in Fig. 7A, addition of 1 mM GDP leads to rapid depolymerization. This suggests that the GDP exchanges with GTP bound to FtsZ at the growing end, leading to loss of that subunit. Repetition of this process as nonhydrolyzed GTP is exposed during subunit loss would lead to complete disassembly. We infer that the copolymers are indeed stabilized by a GTP-cap.
FtsZ needs to be above the critical concentration to stimulate copolymerization of FtsZ2.
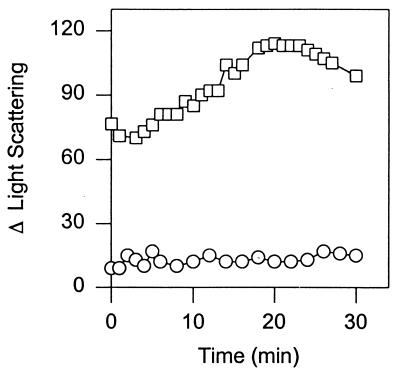
In the experiments we described above, we observed that FtsZ induced efficient copolymerization of FtsZ2 when FtsZ was added at 100 μg/ml. To analyze the requirement for FtsZ in more detail, we tested the concentration of FtsZ required to induce FtsZ2 assembly. Addition of FtsZ at or above the critical concentration to a reaction mixture of FtsZ2 incubated with GTP led to assembly; however, addition of FtsZ below this level did not stimulate assembly. As shown in Fig. 8, addition of FtsZ at 40 μg/ml to a reaction mixture containing 300 μg of FtsZ2 per ml did not lead to an increase in the light scattering signal, whereas addition of FtsZ at 100 μg/ml led to copolymerization. This result suggests that FtsZ has to be in a higher oligomeric form, which is present at 100 μg/ml and above, but not present at 40 μg/ml, to stimulate FtsZ2 copolymerization.
FIG. 8.
Copolymerization of FtsZ and FtsZ2 requires FtsZ to be above the critical concentration. FtsZ at 40 μg/ml (open circles) or 100 μg/ml (open squares) was added to a reaction mixture containing 0.1 mM GTP and FtsZ2 at 300 μg/ml.
DISCUSSION
FtsZ has a GTPase activity that is associated with assembly and required for the dynamics of FtsZ polymers (24). In this study, we have investigated the properties of FtsZ2 that can support cell division despite having a dramatically reduced GTPase activity. We found that FtsZ2 is unable to assemble in vitro; however, it copolymerized upon addition of FtsZ, provided FtsZ is above the critical concentration. This supports a model for cooperative assembly of FtsZ polymers. Significantly, the stability of the copolymers increased with increasing FtsZ2 incorporation, implying FtsZ2 polymers, if formed, would be stable. Since FtsZ2 can support viability, our results suggest that stable FtsZ filaments are able to function in cell division. This result has important implications for the role of the Z ring in cell division, because it argues that constriction of the Z ring can occur through forces acting on FtsZ filaments.
FtsZ2 was isolated as an allele of ftsZ that was resistant to the cell division inhibitor SulA (2). It was isolated in the presence of a second copy of ftsZ, but was subsequently found to support cell division, provided the level is slightly elevated (3). Cells containing only FtsZ2 produce minicells, because ftsZ2 is resistant to inhibition by the min system, and have an altered septal morphology. It was thus surprising that FtsZ2 had little GTPase activity compared to wild-type FtsZ (7, 35). Although FtsZ2 has a deficiency in self-association, which could account for the failure of its GTPase to undergo a concentration-dependent activation, we observed that the GTPase activity was not stimulated even when assembly was forced by the addition of DEAE-dextran. This result argues that FtsZ2 has a defective catalytic site and confirms that the synergy loop is important for GTPase activity as previously suggested (11). This deficiency in FtsZ2's GTPase activity correlates with the differential ability of α- and β-tubulin to promote GTP hydrolysis. The similarity in the GTPase mechanism between FtsZ and tubulins further strengthens an ancestral relationship among this family of proteins.
To try and examine directly the effect of the ftsZ2 mutation on polymer dynamics, we attempted to assemble FtsZ2 in the absence of DEAE-dextran. Purified FtsZ2, however, could not assemble under our conditions, in which wild-type FtsZ readily assembles. However, FtsZ2 readily copolymerized with FtsZ, suggesting that FtsZ2 is primarily deficient in nucleation of assembly in our in vitro conditions. This result suggests that some factor in vivo, mimicked by DEAE-dextran in vitro, promotes nucleation of FtsZ2 polymerization. Significantly, FtsZ only stimulated FtsZ2 copolymer formation when present above the critical concentration, suggesting that it was an oligomeric species of FtsZ present at or above the critical concentration, which nucleates FtsZ2 assembly.
Analysis of the copolymers revealed that the FtsZ2/FtsZ ratio approached 1.25:1 stoichiometry as the concentration of FtsZ2 in the reaction approached 3.6 times the FtsZ concentration. Although the nucleation deficiency of FtsZ2 is the major barrier to polymerization in vitro, this result indicates that addition of FtsZ2 to filament ends is less efficient than addition of FtsZ.
The inability of FtsZ2 to nucleate assembly and add efficiently to filament ends correlated with a deficiency in FtsZ2 self-association revealed in sedimentation and electron microscopy studies. The sedimentation studies revealed that FtsZ assembled into larger oligomers than FtsZ2, suggesting that FtsZ2 forms a weaker longitudinal bond. We speculate that the weaker self-association under these conditions may reflect a weaker association under polymerizing conditions, explaining the inability of FtsZ2 to polymerize. Also, electron microscopy of FtsZ2 failed to reveal any assembly of FtsZ2, whereas FtsZ readily assembled into long polymers above the critical concentration and short polymers near the critical concentration (24; data not shown).
Recently, there have been two reports analyzing FtsZ self-association by sedimentation under nonpolymerizing conditions (in the presence of GDP) (28, 33). Rivas et al. (28) failed to observe self-association in the absence of Mg2+. In contrast, our results obtained in the absence of Mg2+ are similar to the report of Sossong et al. (33), who observed a concentration-dependent formation of short oligomers of FtsZ in the absence of Mg2+. A difference between their results and ours is that we see self-association at lower concentrations of FtsZ. We observed self-association over a concentration range of 0.5 to 5 μM FtsZ and in the absence of Mg2+. Interestingly, this is the same concentration range over which we (Fig. 2) and others have observed the self-activation of FtsZ GTPase (16, 39). Sossong et al. (33) observed GTPase activity only above 2 μM.
Nucleation of FtsZ polymers has not been studied in detail, in part because the physiologically relevant polymer is unknown and the assembly is quite rapid without a significant lag phase. Nucleated assembly of rod structures can be thought of as a combination of two linear assembly pathways (34). The first is an assembly pathway to generate a nucleation species, whereas the second pathway adds subunits to the nucleation species. For microtubule assembly, the nucleation species is a two-stranded sheet composed of seven α/β-tubulin dimers (37). Recently cooperative assembly of FtsZ polymers was questioned by Romberg et al. (29), who observed only short protofilaments under their assembly conditions. Instead, they suggested that FtsZ assembly might be an isodesmic, nonnucleated assembly process that does not display a critical concentration. However, under our assembly conditions (in the absence of such bundling factors as DEAE-dextran), FtsZ always assembles into a mixture of single protofilaments, pairs of protofilaments, and polymers containing more than two protofilaments that are of considerable length, indicating cooperative assembly (Fig. 1B) (24). Furthermore, we found that the assembly displays a critical concentration (24, 26), which was also observed by Xu and Margolin (41) and by White et al. for FtsZ from Mycobacterium tuberculosis (40). Presumably, the conditions employed by Romberg et al. (29) prevent pairs of protofilaments from forming. Thus, they are only observing the first stage of assembly, the formation of protofilaments, which would be an isodesmic assembly reaction and hence explains their failure to observe a critical concentration. The copolymerization results with FtsZ2 further support cooperative assembly, since FtsZ-aided assembly of FtsZ2 must involve at least a pair of protofilaments. The lateral interactions among subunits in such a structure would aide FtsZ2 assembly by allowing the weaker longitudinal bond between between FtsZ2 subunits to be overcome. In contrast, an isodesmic assembly of single protofilaments would not lead to coassembly.
The copolymers formed between FtsZ and FtsZ2 became increasingly stable as the ratio of FtsZ2 to FtsZ increased. This would be expected if incorporation of FtsZ2 into the copolymer does not lead to GTP hydrolysis and GTP-containing subunits do not readily dissociate from the ends of the polymer. This result, suggesting that stability is regulated by a mechanism similar to a GTP-cap is supported by our observation that the copolymers are rapidly and completely destabilized by the addition of GDP. This result is expected if GTP bound at the exposed end of the polymer is rapidly exchanged with the excess GDP, resulting in loss of that subunit. Additional evidence for a GTP-cap comes from our previous observation that FtsZ polymers formed in the absence of Mg2+, which blocks GTPase activity, results in stable polymers (26). Also, GTPγS, which itself cannot promote polymer growth, can stabilize preformed polymers as long as Ca2+ is present (32). Stabilization is not seen in the absence of Ca2+, suggesting that bundling may be required.
Recently, Scheffers et al. (31) isolated three FtsZ mutants, containing D212 substituted with either glutatmate, cystine, or asparagine. The effects of these substitutions on viability were not reported. The two mutant proteins that lacked a negative charge at position 212 had bound GTP and underwent divalent cation-induced polymerization. FtsZ2 also lacks the negative charge at this position; however, it has bound GDP and clearly does not undergo cation-induced polymerization. Thus, different amino acid substitutions at this position lead to different effects on polymerization.
An extrapolation of the relative stability of FtsZ-FtsZ2 copolymers is that FtsZ2 polymers would be stable. Although we have been unable to get FtsZ2 to polymerize in our in vitro system without DEAE-dextran, it must polymerize in vivo as it localizes to the division site and functions in division (3). Consistent with our expectations, cells containing only FtsZ2 form Z rings; however, all cells contain FtsZ2 at the poles suggesting the Z ring does not disassemble in this mutant (data not shown). Such an in vivo result, along with our in vitro results indicating that FtsZ2 lacks GTPase activity and leads to stable polymers, suggests that a stable Z ring is able to participate in division. However, the septa are often twisted, indicating some effect on septation (3). It has been suggested that the energy for invagination of the septum comes either from the depolymerization of FtsZ filaments or from a force generating a protein acting on FtsZ filaments (6). Our results suggest that a force acting on stable FtsZ polymers is able to drive invagination of the septum.
ACKNOWLEDGMENT
This work was supported in part by grant GM29764 from the National Institutes of Health.
REFERENCES
- 1.Adams E T J, Lewis M S. Sedimentation equilibrium in reacting systems. VI. Some applications to indefinite self-associations. Studies with beta-lactoglobulin A. Biochemistry. 1968;7:1044–1053. doi: 10.1021/bi00843a025. [DOI] [PubMed] [Google Scholar]
- 2.Bi E, Lutkenhaus J. Analysis of ftsZ mutations that confer resistance to the cell division inhibitor SulA (SfiA) J Bacteriol. 1990;172:5602–5609. doi: 10.1128/jb.172.10.5602-5609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bi E, Lutkenhaus J. Isolation and characterization of ftsZ alleles that affect septal morphology. J Bacteriol. 1992;174:5414–5423. doi: 10.1128/jb.174.16.5414-5423.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi E, Lutkenhaus J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J Bacteriol. 1993;175:1118–1125. doi: 10.1128/jb.175.4.1118-1125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bi E F, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 6.Bramhill D. Bacterial cell division. Annu Rev Cell Dev Biol. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- 7.Dai K, Mukherjee A, Xu Y, Lutkenhaus J. Mutations in ftsZ that confer resistance to SulA affect the interaction of FtsZ with GTP. J Bacteriol. 1994;176:130–136. doi: 10.1128/jb.176.1.130-136.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 9.Durchschlag H. Specific volumes of biological macromolecules and some other molecules of biological interest. In: Hinz H J, editor. Thermodynamic data for biochemistry and bio/technology. New York, N.Y: Springer-Verlag; 1986. pp. 45–128. [Google Scholar]
- 10.Erickson H P. FtsZ, a prokaryotic homolog of tubulin? Cell. 1995;80:367–370. doi: 10.1016/0092-8674(95)90486-7. [DOI] [PubMed] [Google Scholar]
- 11.Erickson H P. Atomic structures of tubulin and FtsZ. Trends Cell Biol. 1998;8:133–137. doi: 10.1016/s0962-8924(98)01237-9. [DOI] [PubMed] [Google Scholar]
- 12.Erickson H P, Taylor D W, Taylor K A, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang J, Cao C, Lutkenhaus J. Interaction between FtsZ and inhibitors of cell division. J Bacteriol. 1996;178:5080–5085. doi: 10.1128/jb.178.17.5080-5085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson M L, Correia J J, Baty L T, Williams R C J. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys J. 1981;36:575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu C, Stricker J, Erickson H P. FtsZ from Escherichia coli, Azotobacter vinelandii, and Thermotoga maritima—quantitation, GTP hydrolysis, and assembly. Cell Motil Cytoskelet. 1998;40:71–86. doi: 10.1002/(SICI)1097-0169(1998)40:1<71::AID-CM7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 17.Lutkenhaus J. FtsZ ring in bacterial cytokinesis. Mol Microbiol. 1993;9:403–409. doi: 10.1111/j.1365-2958.1993.tb01701.x. [DOI] [PubMed] [Google Scholar]
- 18.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 19.Margolin W. Themes and variations in prokaryotic cell division. FEMS Microbiol Rev. 2000;24:531–548. doi: 10.1111/j.1574-6976.2000.tb00554.x. [DOI] [PubMed] [Google Scholar]
- 20.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 21.Mukherjee A, Cao C, Lutkenhaus J. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2885–2890. doi: 10.1073/pnas.95.6.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee A, Dai K, Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci USA. 1993;90:1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukherjee A, Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee A, Lutkenhaus J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998;17:462–469. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukherjee A, Lutkenhaus J. Purification, assembly, and localization of FtsZ. Methods Enzymol. 1998;298:296–305. doi: 10.1016/s0076-6879(98)98026-0. [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee A, Lutkenhaus J. Analysis of FtsZ assembly by light scattering and determination of the role of divalent metal cations. J Bacteriol. 1999;181:823–832. doi: 10.1128/jb.181.3.823-832.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nogales E, Downing K H, Amos L A, Lowe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol. 1998;5:451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- 28.Rivas G, Lopez A, Mingorance J, Ferrandiz M J, Zorrilla S, Minton A P, Vicente M, Andreu J M. Magnesium-induced linear self-association of the FtsZ bacterial cell division protein monomer. The primary steps for FtsZ assembly. J Biol Chem. 2000;275:11740–11749. doi: 10.1074/jbc.275.16.11740. [DOI] [PubMed] [Google Scholar]
- 29.Romberg L, Simon M, Erickson H P. Polymerization of Ftsz, a bacterial homolog of tubulin. Is assembly cooperative? J Biol Chem. 2001;276:11743–11753. doi: 10.1074/jbc.M009033200. [DOI] [PubMed] [Google Scholar]
- 30.Rothfield L, Justice S, Garcia-Lara J. Bacterial cell division. Annu Rev Genet. 1999;33:423–438. doi: 10.1146/annurev.genet.33.1.423. [DOI] [PubMed] [Google Scholar]
- 31.Scheffers D J, de Wit J G, Den Blaauwen T, Driessen A J. Substitution of a conserved aspartate allows cation-induced polymerization of FtsZ. FEBS Lett. 2001;494:34–37. doi: 10.1016/s0014-5793(01)02310-9. [DOI] [PubMed] [Google Scholar]
- 32.Scheffers D J, Den Blaauwen T, Driessen A J. Non-hydrolysable GTP-gamma-S stabilizes the FtsZ polymer in a GDP-bound state. Mol Microbiol. 2000;35:1211–1219. doi: 10.1046/j.1365-2958.2000.01791.x. [DOI] [PubMed] [Google Scholar]
- 33.Sossong T M, Jr, Brigham-Burke M R, Hensley P, Pearce K H., Jr Self-activation of guanosine triphosphatase activity by oligomerization of the bacterial cell division protein FtsZ. Biochemistry. 1999;38:14843–14850. doi: 10.1021/bi990917e. [DOI] [PubMed] [Google Scholar]
- 34.Timashef S N. The self-assembly of long rodlike structures. In: Frieden C, Nichol L W, editors. Protein-protein interactions. New York, N.Y: John Wiley & Sons; 1981. pp. 315–336. [Google Scholar]
- 35.Trusca D, Scott S, Thompson C, Bramhill D. Bacterial SOS checkpoint protein SulA inhibits polymerization of purified FtsZ cell division protein. J Bacteriol. 1998;180:3946–3953. doi: 10.1128/jb.180.15.3946-3953.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Holde K E, Rossetti G P. A sedimentation equilibrium study of the association of purine in aqueous solutions. Biochemistry. 1967;6:2189–2194. doi: 10.1021/bi00859a041. [DOI] [PubMed] [Google Scholar]
- 37.Voter, W. A., and H. P. Erickson. 1984. The kinetics of microtubule assembly. Evidence for a two-stage nucleation mechanism. J. Biol. Chem. 10430–10438. [PubMed]
- 38.Wang X, Huang J, Mukherjee A, Cao C, Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J Bacteriol. 1997;179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Lutkenhaus J. The FtsZ protein of Bacillus subtilis is localized at the division site and has GTPase activity that is dependent upon FtsZ concentration. Mol Microbiol. 1993;9:435–442. doi: 10.1111/j.1365-2958.1993.tb01705.x. [DOI] [PubMed] [Google Scholar]
- 40.White E L, Ross L J, Reynolds R C, Seitz L E, Moore G D, Borhani D W. Slow polymerization of Mycobacterium tuberculosis FtsZ. J Bacteriol. 2000;182:4028–4034. doi: 10.1128/jb.182.14.4028-4034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu X C, Margolin W. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J. 1997;16:5455–5463. doi: 10.1093/emboj/16.17.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]