Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)-positive pet owners are reported to be a risk factor for infection of their pets; however, the influence of the viral load and associated risks has not been fully established. This study aimed to assess potential association of viral load in owners with the presence of SARS-CoV-2 infection in their dogs. Of 20 SARS-CoV-2-positive pet owners from 13 families in Curitiba, Brazil, 5 of 22 (22.7%) dogs were positive for SARS-CoV-2. Viral presence was detected in oropharyngeal samples for 2 of 5 (40.0%) dogs at 8 and 9 days after the first positive sample. Detection of SARS-CoV-2 in these dogs was associated with higher viral loads in the owners and close owner contact. All 5 RT-qPCR-positive dogs had antibodies to at least one viral protein tested in the serological assay. Molecular detection of SARS-CoV-2 in dogs was statistically associated with clinical signs in owners such as cold, cough, or diarrhea (P = 0.039), number of positive persons in the household (P = 0.002), and higher viral load (P = 0.039). Such findings serve as a warning for risks of human to dog infection, mainly due to sharing beds and other close interactions without protection. In conclusion, people with coronavirus disease 2019 (COVID-19), particularly in households with multiple residents and high viral load, should take the same preventive measures when interacting with their dogs during self-isolation as they do with people.
Keywords: Coronavirus, Pets, Transmission
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is described as a positive-sense single-stranded RNA virus causing the coronavirus disease 2019 (COVID-19) in humans (Zhu et al., 2020). While primarily a human infection, SARS-CoV-2 has been reported in other animal species such as cats, dogs, non-human primates, and minks (Abdel-Moneim and Abdelwhab, 2020). In dogs, the infection has been associated with close owner interaction, with or without associated clinical signs (de Morais et al., 2020).
Dogs experimentally infected by SARS-CoV-2 intranasal inoculation resulted in viral particles found in 2 of 4 rectal swabs, with no detection in internal organs after 4 days, and antibodies in 2 of 4 dogs after 14 days (Shi et al., 2020). Experimental studies have demonstrated seroconversion in dogs exposed to infected humans and an absence of viral shedding that suggested low susceptibility to SARS-CoV-2 (Bosco-Lauth et al., 2020).
Despite SARS-CoV-2 infected owners reportedly being risk factors for pet infection, viral load as an associated risk factor has not been fully established. As part of an ongoing multicentric study of SARS-CoV-2 in infected owners and their pets, this study aimed to assess the potential association of owner viral load with detection of SARS-CoV-2 in dogs of southern Brazil.
2. Materials and method
This study was conducted from October 2020 through June 2021 and assessed dogs of owners who tested positive for SARS-CoV-2 infection by RT-qPCR assay at the Federal University of Paraná after invitation, signed consent, and voluntary participation of owners and household members (Table S1). An epidemiological questionnaire was given to owners. Dogs were sampled via oropharyngeal and rectal swabs by a certified veterinarian after gentle physical restrain. Blood samples were collected for serological analysis on day 0 and after 7 days. This study was approved by the Ethics Committee in Animal Use (protocol number 4879280420).
2.1. SARS-CoV-2 detection by RT-qPCR
The nucleic acid extraction was performed by an automated system using commercially available magnetic beads (Mvxap016Fast, Extracta 32, Loccus Co, Cotia, SP, Brazil) following the manufacturer's protocol. One-step multiplex RT-qPCR was performed to detect three specific target genes of SARS-CoV-2 (N, RdRP, and E genes) using a commercial kit (Allplex™ 2019-nCoV, Seegene, Seul, South Korea), following the manufacturer's recommendations. Cyclic threshold (Ct) values ≤40 correspond to a detectable result, while Ct values >40 or undetectable. Also, given that the kit used is a multiplex assay for 3 different target genes of SARS-CoV-2, the interpretation of the results is described in Table S2. The amplification of all three genes, E and N, E and RdRP, RdRp and N, only N or only RdRP, means 2019-nCoV (SARS-CoV-2) RNA is detected, and the sample is positive. If there is only amplification of gene E, as occurred for the first dog case (1.D1), confirmation using a different method is necessary. The 1.D1 case was confirmed with a different commercial kit to validate the results, eliminate the possibility of a false positive, and show effectiveness of the first kit at detecting the virus in canine samples. Diagnostic confirmation for the first case to validate the results using a second commercial RT-qPCR reaction kit (EuroRealTime SARS-CoV-2 kit, Euroimmun, Lübeck, Germany) for the detection of SARS-CoV-2 nucleic acids, following the manufacturer's recommendations. All swab samples were also tested at a private animal laboratory to ensure positivity (TECSA, Technology in Animal Health, Brazil). Furthermore, all dog samples were tested and found negative for Canine Adenovirus type 2, Bordetella bronchiseptica, Canine herpesvirus-1, and Canine Distemper virus.
2.2. Genotyping of Variants
Samples of positive dog households were further tested using a probe-based genotyping system to detect Variants of Concern, using a dropout scheme for Spike Δ69–70 and Orf1a Δ3675–3677 deletions as an outcome to distinguish B.1.1.7, B.1.351 or P.1, and wild type or other lineages, as previously established (Vogels et al., 2021). Due to the limitation of the technique, only samples that had Ct <30 in RT-qPCR were genotyped. Assays were performed using a commercial system (GoTaq® Probe 1-Step RT-qPCR System, Promega, Madison, WI, USA) with a CDC N1 target included and a Ct 28 threshold defined to evaluate the gene dropouts.
2.3. Antibody detection
The magnetic bead ELISA assay was performed to verify the presence of specific antibodies to SARS-CoV-2 proteins in RT-qPCR positive dogs. Day zero was the day of swab sample collection. The blood samples from the dogs were analyzed for the presence of IgG antibodies to viral proteins nucleocapsid (N) and spike (S) by magnetic bead-based ELISA immunoassay (Huergo et al., 2021). The magnetic bead immunoassay was performed using a 96-well with flat bottom commercial plate (Cralplast plate, Cotia, SP, Brazil), as previously established (Conzentino et al., 2021). Aliquots of antigen-loaded beads were resuspended in tris buffered saline with tween 20 (TBST) containing 1% milk, with 0.1 ml of the mix put into each well. Four microliters of dog serum were diluted in 0.2 ml of TBST and put into wells of a second 96-well plate. The magnetic beads were transferred to the sample plate and incubated with the dog sample (serum) for 2 min with gentle mixing. Beads were captured and loaded in two sequential washing steps for 30 s in 1× TBST. For fluorescent detection, beads were incubated for 2 min with 0.15 ml of FITC-conjugated anti-dog antibody diluted 1:100 in 1× TBST, followed by a second wash step for 30 s at 1× TBST. When the reactions were complete, the beads were removed, and reading was performed using a TECAN M Nano plate reader (TECAN) operating at top fluorescent readout. Excitation 545ηm (bandwidth 9ηm and 25 flashes) and emission 578ηm (bandwidth 20ηm, integration time 20 μs and Z position at 20,000 μm). Dog blood samples obtained before the pandemic were used as a negative control. The cut-off point was established through calculation (mean of pre-pandemic samples plus 3× standard deviation).
2.4. Statistical analysis
Statistical analysis was performed using Fisher's exact test to verify the association between the results of SARS-CoV-2 in dogs (positive and negative) and the qualitative variables of the questionnaire (Table S3). The Mann-Whitney test was applied to compare the results of SARS-CoV-2 in dogs (positive and negative) and the quantitative variables of the questionnaire. The tests were performed considering a bidirectional significance level (α) of 5%. IBM SPSS 26 (IBM corp., 2019) and Microsoft Excel 365® software were used as computational support.
3. Result
Out of 13 households with 22 dogs, 5 (22.7%) dogs tested positive and 17 (77.3%) dogs had negative results for SARS-CoV-2 by real-time RT-qPCR (Table S4). These were the first 5 reported dog cases in Brazil (OIE, 2021). Results and epidemiological data from all owners and their dogs are presented (Table 1 ).
Table 1.
Ct (Cycle threshold) comparisons between owners (O) and dogs (D) in households with positive dogs. Dogs that showed amplification of one or more genes of the SARS-CoV-2 virus underwent new sample collection after 7, 10 and/or 14 days to assess the virus persistence in positive dogs.
| AllPlex | EuroImmun | |||||||
|---|---|---|---|---|---|---|---|---|
| Animal | Day | Intern control | Gene E | RdRP | Gene N | Endogenous | SARS-CoV-2 | |
| Household 1 | 1.O1 | 1 | 22.65 | – | – | 35.67 | 35.32 | 35.25 |
| 1.O1 | 7 | 32.60 | – | – | – | – | – | |
| 1.D1 | 1 | 27.45 | – | – | 36.92 | 29.87 | 38.71 | |
| 1.D1 | 7 | 28.78 | – | – | – | 29.97 | – | |
| Household 2 | 2.O1 | 1 | – | 13.92 | 14.31 | 13.69 | – | – |
| 2.O2 | 1 | – | 19.61 | 20.43 | 18.85 | – | – | |
| 2.O3 | 1 | 30.26 | – | – | 32.78 | – | – | |
| 2.O4 | 1 | 28.36 | – | 32.74 | 30.41 | – | – | |
| 2.D1 | 1 | 26.60 | – | 34.17 | – | – | – | |
| 2.D2 | 1 | 28.27 | – | – | – | – | – | |
| 2.D3 | 1 | 27.79 | – | – | – | – | – | |
| 2.D4 | 1 | 27.42 | – | 32.72 | 32.97 | – | – | |
| 2.D5 | 1 | 27.67 | – | – | – | – | – | |
| 2.O1 | 10 | – | 21.64 | 22.30 | 21.82 | – | – | |
| 2.O2 | 10 | 34.21 | 24.88 | 25.81 | 24.44 | – | – | |
| 2.O3 | 10 | 29.94 | – | – | 36.06 | – | – | |
| 2.O4 | 10 | 29.98 | – | 38.48 | 31.35 | – | – | |
| 2.D1 | 10 | 25.65 | – | – | – | – | – | |
| 2.D4 | 10 | 26.35 | – | – | – | – | – | |
| Household 3 | 3.O1 | 1 | – | 14.09 | 14.39 | 13.98 | – | – |
| 3.O2 | 1 | – | 19.88 | 20.39 | 19.25 | – | – | |
| 3.D1 | 1 | 30.25 | – | – | 36.83 | – | – | |
| 3.D1 | 7 | 25.89 | – | 33.24 | 36.40 | – | – | |
| 3.D1 | 14 | 28.59 | – | – | – | – | – | |
| Household 4 | 4.O1 | 1 | 39.62 | – | – | 38.63 | – | – |
| 4.O2 | 1 | – | 17.27 | 18.62 | 16.39 | – | – | |
| 4.D1 | 1 | 28.57 | – | – | 35.58 | – | – | |
| 4.D1 | 7 | 25.84 | 34.71 | 37.16 | – | – | ||
| 4.D1 | 14 | 30.10 | – | – | – | – | – | |
O = Owner; D = Dog; −*≥40 (undetectable).
Case 01 was a four-year-old male French Bulldog with no pre-existing diseases that shared a bed with the owner. The owner, an adult male, had symptoms on November 10th and tested positive (saliva sample) on November 12, 2020. The owner reported that the dog had mild and transitory nasal discharge. On November 19th, the owner tested negative (saliva) and the dog tested positive (oropharyngeal swab).
Dog cases 02 and 03 were part of family with 5 dogs and 4 people. Two human adults tested positive on November 19th. The following day, all household members were tested; nasopharyngeal swab samples from the people and oropharyngeal and rectal swab samples from the dogs were used to test. All the people and 2 of the dogs tested positive. The dogs were mixed-breed adults that slept on the same bed as 2 of the owners. The dogs tested negative on samples collected on November 30th.
Case 04 was a dog owned by an elderly couple. One person tested positive on a sample from November 19th. The dog slept in the living room on a pet bed. Samples from the entire household were collected and results were positive on November 21st and 30th but negative on December 7th.
Case 05 was 1 dog that shared the bed with an owner couple. The couple had contact with the family of cases 02 and 03 when they were infected. Samples collected from this household on November 22nd and 30th, yielded positive results. On December 7th samples, all test results were negative.
The presence of SARS-CoV-2 RNA in 2 of 5 (40.0%) dogs was detected in oropharyngeal samples at 8 and 9 days after the first positive sampling.
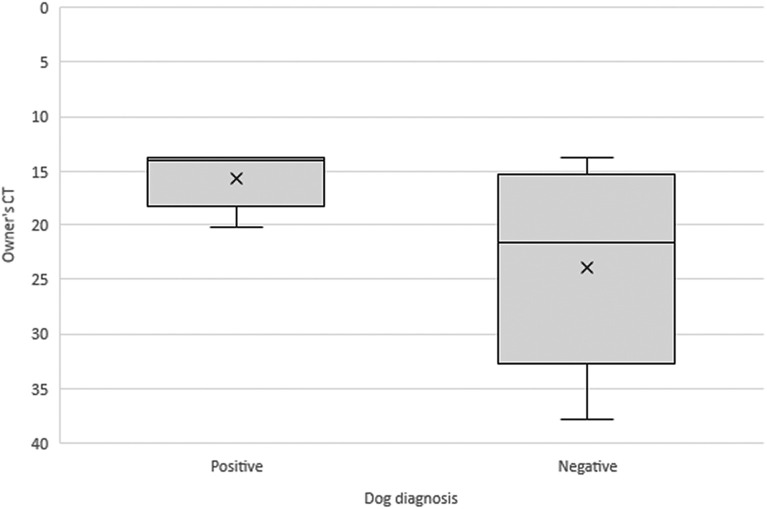
A significant association was observed between the presence of clinical signs such as nasal discharge, cough, or diarrhea and a positive test result for SARS-CoV-2 in dogs (P = 0.039). Among the positive dogs, 80.0% showed signs of nasal discharge, cough, or diarrhea, while 76.5% of the negative dogs did not show such clinical signs. In addition, a statistically significant difference was observed between the number of confirmed human cases in the household and the result of SARS-CoV-2 in dogs (P = 0.002) (Fig. S1). The median number of positive persons in the household was 2 for dogs with a positive test result compared to a median of 1 positive person for dogs with a negative test result. Furthermore, there was also a statistically significant difference between the owner viral load, which can be estimated using the Ct number, and the result of SARS-CoV-2 in dogs (P = 0.039) (Fig. 1 ). The median of the owner Ct value was lower in the group of dogs with a positive result (13.83) compared to the group of dogs with a negative result (21.57). In this analysis, household 1 was excluded because the owner's sample was not collected during the symptomatic period of the disease, making it an outlier.
Fig. 1.
The median of owners Ct was lower in the group of dogs with a positive result (13.83) compared to the group of dogs with a negative result (21.57). The Ct value is inversely proportional to the viral load presented in the sample. That is, the lower the Ct, the higher the owner's viral load.
At probe-based genotyping, the samples 2.H1, 2.H2, 3.H1, 3.H2, and 4.H2 were identified as wild strains, with no identification of the Spike Δ69–70 and Orf1a Δ3675–3677 deletions as results distinguishing B.1.1.7, B.1.351, or P.1. Samples 2.H3, 2.H4, 3.D1, 4.H1, and 4.D1, despite having CT > 30, were included in the genotyping protocol to demonstrate the limitation of the technique and were classified as invalid, as they only showed N1 gene amplification (Table S5). All dogs showed high Ct values on RT-qPCR. Therefore, it was not possible to genotype the canine samples due to the low viral load.
Based on the magnetic bead ELISA assay, all 5 positive dogs by RT-qPCR showed intense fluorescence for at least one target viral protein (N and S), demonstrating seroconversion and the presence of specific SARS-CoV-2 antibodies (Table S6). Dogs 1.D1 and 2.D1 presented increased fluorescence intensity for both viral proteins in the two tested samples (days 0 and 7), presenting higher titration of specific anti-SARS-CoV-2 antibodies. Dog 4.D1 showed an increase in antibody titer only in the sample collected after 7 days. Dog 3.D1 showed antibodies only to protein N and dog 2.D4 showed antibodies only to protein S (Fig. S2).
4. Discussion
The study presents the first 5 reports of naturally occurring SARS-CoV-2 infection in dogs of Brazil, which were immediately notified to the Brazilian Ministry of Agriculture and the World Organization for Animal Health (OIE, 2021). Higher likelihood of dog infection was associated with closer contact with owners (such as exchanging kisses and sleeping on the same bed), presenting clinical signs noticed by owners, and with owners with higher viral load.
Although results have demonstrated a positive correlation between SARS-CoV-2 infection in dogs and clinical onset, signs were mild and thus corroborate previous studies showing pet infections as mostly asymptomatic or self-resolving (Hamer et al., 2017). However, death of domestic animals has been observed during the time of SARS-CoV-2 diagnosis, including a dog with evidence of pre-existing severe chronic respiratory disease presented clinical worsening due to SARS-CoV-2 infection, leading to the animal's death (Carpenter et al., 2021). Moreover, rare cases where SARS-CoV-2 infection could cause serious consequences in healthy dogs, regardless of their clinical history have also been reported (Carpenter et al., 2021; Davis et al., 2017). Such a wide difference in the outcome of dog cases mirrors some human cases and remains to be fully established.
The presence of SARS-CoV-2 RNA in dogs of this study has been described in dogs of China, with 5 positive nasopharyngeal swabs following 13-days after the first detection (Sit et al., 2020). In addition, a recent longitudinal study with domestic animals and owners in Brazil has shown 3 of 8 (37.5%) dogs tested positive to SARS-CoV-2 RNA in nasopharyngeal and oropharyngeal samples for 14 to 31 days after the first positive sample (Calvet et al., 2021). In addition, the correlation between the number of infected household owners and dog infection corroborates previous studies of human-to-human transmission, particularly among children (Madewell et al., 2020), which may be extrapolated to the interspecies transmission when there is close owner-dog contact. Ct values in the study herein depended on the period when samples were collected from owners after the onset of symptoms. A previous review study on human SARS-CoV-2 infection has reported Ct values commonly below 30, and even below 20, in the first days of SARS-CoV-2 infection and symptoms, associated with high virus spreading and seriousness of human infection (Jaafar et al., 2020). Another study has suggested a low ability of SARS-CoV-2 to infect Vero cell line, when collected from patients with Ct higher than 24 and after 8 days of symptoms (Bullard et al., 2020). In general, positive individuals with high Ct values for weeks or months after symptom resolution may be result of non-infective viral remains (Bullard et al., 2020). However, as these studies assessed only human SARS-CoV-2 infection, owner role and transmission routes for companion animal infection over time remains to be fully established.
Occurrence of SARS-CoV-2 negative dogs from positive owners with low Ct herein may be explained by dog natural low susceptibility or lack of virus transmission, as even dogs experimentally infected have shown no nasal and oral virus shedding, with transient presence in feces (Bosco-Lauth et al., 2020; Shi et al., 2020).
Seronegative dogs of positive owners with low cT values herein may be consequence of early sera sampling of 7 days instead of 14 days, as dogs experimentally infected have shown seroconversion starting at the 14-day post infection, with titers between 1:40 and 1:80 against RBD and spike antigens (Bosco-Lauth et al., 2020).
The complex and multidimensional issues raised herein by the interface of human, animal, and environmental health have required a One Health Approach to better understand the epidemiology, prevention, and control of COVID-19 and SARS-CoV-2 transmission. Further multidisciplinary and comparative research should also be carried out to understand COVID-19 transmission and prevent future pandemics, particularly the role of humans and animals sharing the environment.
In summary, despite low sample numbers, molecular detection of SARS-CoV-2 in dogs was statistically associated with clinical signs such as nasal discharge, cough, or diarrhea, number of positive persons in the household, and higher viral load (low Ct). Such findings should raise awareness of human-to-dog infection risk, especially when sharing beds or other closer interactions without protection. In conclusion, COVID-19 infected persons, particularly with multiple residents, should take the same preventive person-to-person precautions with their dogs during self-isolation. In addition, dogs presenting mild clinical signs may likely indicate infection. Finally, as human-animal-environment interactions may play an important role in cross-species infection and transmission, One Health Approach should always be conducted in zoonotic infections.
Funding
Present research was funded through the Brazilian National Council for Scientific and Technological Development (CNPq) (2020-1/402341).
Declaration of Competing Interest
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Acknowledgments
The authors kindly thank faculties and technicians working at the Molecular and Serological Diagnostic Laboratories of Federal University of Parana, and Suzanne Pratt for editing and improving the article.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.rvsc.2022.10.006.
Appendix A. Supplementary data
Supplementary material
References
- Abdel-Moneim A.S., Abdelwhab E.M. 2020. Evidence for SARS-CoV-2 Infection of Animal Hosts. Pathog. (Basel, Switzerland) p. 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco-Lauth A.M., Hartwig A.E., Porter S.M., Gordy P.W., Nehring M., Byas A.D., VandeWoude S., Ragan I.K., Maison R.M., Bowen R.A. Experimental infection of domestic dogs and cats with SARS-CoV-2: Pathogenesis, transmission, and response to reexposure in cats. Proc. Natl. Acad. Sci. 2020;117 doi: 10.1073/pnas.2013102117. 26382 LP – 26388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L., Boodman C., Bello A., Hedley A., Schiffman Z., Doan K., Bastien N., Li Y., Van Caeseele P.G., Poliquin G. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin. Infect. Dis. 2020;71(10):2663–2666. doi: 10.1093/cid/ciaa638. 2020; 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvet G.A., Pereira S.A., Ogrzewalska M., Pauvolid-Corrêa A., Resende P.C., de Souza Tassinari W., de Pina Costa A., Keidel L.O., da Rocha A.S.B., da Silva M.F.B., Dos Santos S.A., Lima A.B.M., de Moraes I.C.V., Mendes Junior A.A.V., das Chagas Souza T., Martins E.B., Ornellas R.O., Corrêa M.L., da Silva Antonio I.M., Guaraldo L., do Couto Motta F., Brasil P., Siqueira M.M., Gremião I.D.F., Menezes R.C. Investigation of SARS-CoV-2 infection in dogs and cats of humans diagnosed with COVID-19 in Rio de Janeiro, Brazil. PLoS One. 2021;16 doi: 10.1371/journal.pone.0250853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A., Ghai R., Gary J., Ritter J., Carvallo F., Diel D., Martins M., Murphy J., Schroeder B., Brightbill K., Tewari D., Boger L., Gabel J., Cobb R., Hennebelle J., Stanton J., McCullough K., Mosley Y.-Y., Naikare H., Radcliff R., Parr B., Balsamo G., Robbins B., Smith D., Slavinski S., Williams C., Meckes D., Jones D., Frazier T., Steury K., Rooney J., Torchetti M., Wendling N., Currie D., Behravesh C.B., Wallace R. 2021. Determining the Role of Natural SARS-CoV-2 Infection in the Death of Ten Domestic Pets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzentino M.S., Santos T.P.C., Selim K.A., Wagner B., Alford J.T., Deobald N., Paula N.M., Rego F.G.M., Zanette D.L., Aoki M.N., Nardin J.M., Huergo M.C.C., Reis R.A., Huergo L.F. Ultra-fast, high throughput and inexpensive detection of SARS-CoV-2 seroconversion using Ni2+ magnetic beads. Anal. Biochem. 2021;631 doi: 10.1016/j.ab.2021.114360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.F., Rankin S.C., Schurer J.M., Cole S., Conti L., Rabinowitz P. vol. 4. 2017. Checklist for One Health Epidemiological Reporting of Evidence (COHERE). One Heal. (Amsterdam, Netherlands) pp. 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Morais H.A., dos Santos A.P., do Nascimento N.C., Kmetiuk L.B., Barbosa D.S., Brandão P.E., Guimarães A.M.S., Pettan-Brewer C., Biondo A.W. Natural infection by SARS-CoV-2 in companion animals: a review of case reports and current evidence of their role in the epidemiology of COVID-19. Front. Vet. Sci. 2020;27(7) doi: 10.3389/fvets.2020.591216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer Sarah A., Pauvolid-Corrêa Alex, Zecca Italo B., Edward Davila L., Auckland D., Roundy Christopher M., Tang Wendy, Mia Torchetti M.L.K., Jenkins-Moore Melinda, Mozingo Katie, Akpalu Yao, Ghai Ria R., Spengler J.R., Behravesh Casey Barton, Fischer Rebecca S.B., Hamer G.L. Natural SARS-CoV-2 infections, including virus isolation, among serially tested cats and dogs in households with confirmed human COVID-19 cases in Texas, USA. J. Chem. Inf. Model. 2017;53:21–25. [Google Scholar]
- Huergo L.F., Selim K.A., Conzentino M.S., Gerhardt E.C.M., Santos A.R.S., Wagner B., Alford J.T., Deobald N., Pedrosa F.O., de Souza E.M., Nogueira M.B., Raboni Nia M., Souto Dênio, Rego F.G.M., Zanette D.L., Aoki M.N., Nardin J.M., Fornazari B., Morales H.M.P., Borges Vânia A., Nelde A., Walz J.S., Becker M., Schneiderhan-Marra N., Rothbauer U., Reis R.A., Forchhammer K. Vol. 6. 2021. Magnetic Bead-Based Immunoassay Allows Rapid, Inexpensive, and Quantitative Detection of Human SARS-CoV-2 Antibodies; pp. 703–708. [DOI] [PubMed] [Google Scholar]
- Jaafar R., Aherfi S., Wurtz N., Grimaldier C., Van Hoang T., Colson P., Raoult D., La Scola B. Clin. Infect. Dis. 2020;72(11) doi: 10.1093/cid/ciaa1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madewell Z.J., Yang Y., Longini I.M., Halloran M.E., Dean N.E. Household transmission of SARS-CoV-2: a systematic review and meta-analysis. JAMA Netw. Open. 2020 doi: 10.1001/jamanetworkopen.2020.31756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OIE, W.O. for A.H . 2021. COVID-19 - Events in Animals [WWW Document] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J., Zhang X., Guan Y., Tan W., Wu G., Chen H., Bu Z., Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sit T.H.C., Brackman C.J., Ip S.M., Tam K.W.S., Law P.Y.T., To E.M.W., Yu V.Y.T., Sims L.D., Tsang D.N.C., Chu D.K.W., Perera R.A.P.M., Poon L.L.M., Peiris M. Infection of dogs with SARS-CoV-2. Nature. 2020;586:776–778. doi: 10.1038/s41586-020-2334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogels C.B.F., Breban M.I., Ott I.M., Alpert T., Petrone M.E., Watkins A.E., Kalinich C.C., Earnest R., Rothman J.E., de Jesus J.G., Claro I.M., Ferreira G.M., Crispim M.A.E., Singh L., Tegally H., Anyaneji U.J., Hodcroft E.B., Mason C.E., Khullar G., Metti J., Dudley J.T., MacKay M.J., Nash M., Wang J., Liu C., Hui P., Murphy S., Neal C., Laszlo E., Landry M.L., Muyombwe A., Downing R., Razeq J., de Oliveira T., Faria N.R., Sabino E.C., Neher R.A., Fauver J.R., Grubaugh N.D., da Silva Sales F.C., Ramundo M.S., Candido D.S., Silva C.A.M., de Pinho M.C., de Moura Coletti T., dos Santos Andrade P., de Souza L.M., Rocha E.C., Gomes Jardim A.C., Manuli E., Gaburo N., Granato C., Levi J.E., Costa S., de Souza W.M., Salum M.A., Pereira R., de Souza A., Matkin L.E., Nogueria M.L., Levin A.S., Mayaud P., Alexander N., Souza R., Acosta A.L., Prete C., Quick J., Brady O., Messina J., Kraemer M., da Cruz Gouveia N., Oliva I., de Souza M., Lazari C., Alencar C.S., Thézé J., Buss L., Araujo L., Cunha M.S., Loman N.J., Pybus O.G., Aguiar R.S., Wilkinson E., Msomi N., Iranzadeh A., Fonseca V., Doolabh D., San E.J., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao M., Korsman S., Davies M.A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C.K., Sewell B.T., Lourenço J., Kosakovsky Pond S.L., Weaver S., Giovanetti M., Alcantara L.C.J., Martin D., Bhiman J.N., Williamson C. Multiplex qPCR discriminates variants of concern to enhance global surveillance of SARS-CoV-2. PLoS Biol. 2021;19:1–12. doi: 10.1371/journal.pbio.3001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material