Abstract
Drug susceptibility testing of M. tuberculosis is rooted in a binary susceptible/resistant paradigm. While there are considerable advantages in measuring the minimum inhibitory concentrations (MICs) of a panel of drugs for an isolate, it is necessary to measure the epidemiological cut-off values (ECOFF/ECVs) to permit comparison with qualitative data. Here we present ECOFF/ECVs for 13 anti-tuberculosis compounds, including bedaquiline and delamanid, derived from 20 637 clinical isolates collected by 14 laboratories based in 11 countries on five continents. Each isolate was incubated for 14 days on a dry 96-well broth microdilution plate and then read. Resistance to most of the drugs due to prior exposure is expected and the MIC distributions for many of the compounds are complex, and therefore a phenotypically wild-type population could not be defined. Since a majority of samples also underwent genetic sequencing, we defined a genotypically wild-type population and measured the MIC of the 99th percentile by direct measurement and via fitting a Gaussian using interval regression. The proposed ECOFF/ECVs were then validated by comparing with the MIC distributions of high-confidence genetic variants that confer resistance and with qualitative drug susceptibility tests obtained via the Mycobacterial Growth Indicator Tube (MGIT) system or Microscopic-Observation Drug Susceptibility (MODS) assay. These ECOFF/ECVs will inform and encourage the more widespread adoption of broth microdilution: this is a cheap culture-based method that tests the susceptibility of 12–14 antibiotics on a single 96-well plate and so could help personalise the treatment of tuberculosis.
Short abstract
International tuberculosis (TB) consortium proposes epidemiological cut-off values for a 96-well broth microdilution plate containing 13 anti-TB drugs using a dataset of 15 211 M. tuberculosis samples https://bit.ly/3vpOzIk
Introduction
Mycobacterium tuberculosis kills more people worldwide than any other single pathogen, severe acute respiratory syndrome coronavirus 2 excepted [1]. Despite its impact on global health, antibiotic susceptibility testing (AST) for M. tuberculosis has lagged behind other bacterial diseases due to its slow growth rate, difficulty in culturing and its low prevalence in high-income countries. The consequence is that most patients in the world receive empiric, or semi-empiric, treatment, which reduces the chance of treatment success and risks the amplification of resistance where too few effective drugs are prescribed.
The dramatic reduction in genetic sequencing costs has enabled genetics-based AST where the genome of a pathogen is sequenced and then examined for known variants that confer resistance to specific antibiotics. M. tuberculosis is well suited to this approach [2–10] and several public health bodies have adopted whole-genome sequencing as their standard AST method [11]. Although PCR platforms can deliver universal AST in its narrowly defined sense [12], genome sequencing is the only approach that can realistically deliver comprehensive AST in settings where phenotyping remains too expensive and too infrastructure dependent, and comprehensive AST is the only way to optimise treatment regimens and outcomes.
The Comprehensive Research Prediction for Tuberculosis: an International Consortium (CRyPTIC) research project has collected 20 637 clinical M. tuberculosis samples from across the world. The primary aim of the project is to identify mutations in the M. tuberculosis genome that confer phenotypic resistance to a wide range of antibiotics. The CRyPTIC project measured minimum inhibitory concentrations (MICs) of each drug to permit quantitative analyses, associating mutations with MIC values with a view to using genome sequencing data to personalise drug regimens and doses. From the start the CRyPTIC project has taken a data-driven approach whereby all analyses are algorithmic, hence the allocation of a sample to a subgroup requires little or no expert, and hence subjective, intervention. This has the virtue of ensuring the results are reproducible.
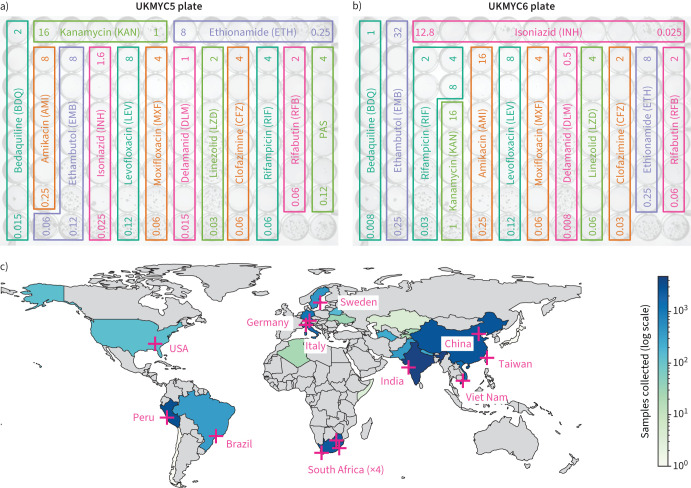
The most practical and affordable means of determining MICs at scale was to use a pre-prepared 96-well 7H9 broth microdilution plate based on the Thermo Fisher Sensititre MYCOTB MIC plate [13–18], but including the new or repurposed antibiotics that feature in current World Health Organization (WHO) guidance [19]. The CRyPTIC project designed a variant of the MYCOTB plate, called UKMYC5, that contains 14 antibiotics, including bedaquiline (BDQ), delamanid (DLM), clofazimine (CFZ) and linezolid (LZD) but not pyrazinamide (PZA) (figure 1a). Based on a multi-laboratory study that examined the inter- and intra-laboratory reproducibility of the UKMYC5 plate, and determined the optimum reading methods and incubation period [17], CRyPTIC subsequently modified the design by removing para-aminosalicylic acid (PAS) and extending/changing the concentration of certain drugs, leading to the 13-drug UKMYC6 plate (figure 1b).
FIGURE 1.
The CRyPTIC Consortium has collected 20 637 clinical tuberculosis (TB) samples worldwide. a, b) Layout and concentrations of the anti-TB drugs on the a) UKMYC5 and b) UKMYC6 microdilution 96-well plates. All concentrations are in mg·L−1; for clarity only the first and last concentrations in each doubling series are given. The two unlabelled wells in the bottom right-hand corner contain no antibiotic and are therefore positive controls. Note that all doubling dilution series are based around 1 mg·L−1 with the exception of INH which is based around 0.1 mg·L−1. c) Geographical distribution of CRyPTIC laboratories and samples collected. 14 laboratories from 11 countries collected data from 27 countries. Each country is coloured depending on the number of originating samples using a logarithmic scale. PAS: para-aminosalicylic acid.
In this article we propose epidemiological cut-off values (ECOFF/ECVs) for the UKMYC series of plates to enable subsequent research on this dataset. The ECOFF/ECV is the highest MIC observed within a phenotypically wild-type (pWT) population, usually defined as the MIC which encompasses 99% of that population [20], and allows interpretation of an MIC value as “susceptible” or “resistant”, which is crucial to the decision on whether to prescribe a drug. The standard approach requires uncensored MICs and assumes that the pWT population can be readily identified, either because the population has been minimally exposed to the drug or because the MIC distribution is strongly bimodal. These conditions are not universally met in our dataset and we shall therefore identify a genotypically wild-type (gWT) population from which we can either measure the ECOFF/ECV directly or via a Gaussian fitted using interval regression, a statistical technique that can fit to censored data. We have made Python code publicly available that enables anyone to reproduce most of the figures and tables in a web browser window [21]. Although ECOFF/ECVs have been proposed for the MYCOTB microdilution plate using 385 strains from South Africa [22], here we are able to draw upon a far larger and more geographically diverse M. tuberculosis dataset.
Results
AST was performed on 20 637 isolates to 13 anti-tuberculosis (TB) drugs using either the UKMYC6 (12 672 (61%)) or the UKMYC5 (7965 (39%)) plate design (table 1, and figure 1a and b). These data were generated in 14 CRyPTIC laboratories based in 11 countries on five continents (figure 1c and supplementary table S1). The isolates themselves were collected from 27 countries, with 19 countries contributing 10 or more isolates and 15 countries contributing 100 or more isolates (supplementary table S2). Due to differences between the laboratories, it was not possible to collect clinical outcome data for the samples. Quality control processes detected that one laboratory developed a problem inoculating the plates (these plates were removed) and that another laboratory never managed to inoculate successfully (all their plates were excluded). Excluded these left 17 054 plates.
TABLE 1.
Number of isolates collected, split by the two microtitre plate designs used
| Total | UKMYC6 | UKMYC5 | |
| Isolates collected | 20 637 | 12 672 | 7965 |
| Readable plates | 17 054 | 10 010 | 7044 |
| Readable plates with images | 15 138 | 9272 | 5866 |
| Readable plates with genetics | 12 362 | 6019 | 6343 |
| Readable plates with genetics and images | 10 938 | 5552 | 5386 |
| Readable plates with images and passing QA | 11 801# | 6896# | 4904# |
| Readable plates with genetics, images and passing QA | 8553# | 4027# | 4526# |
| Readable plates with genetic, images, passing QA and genotypically wild-type | 3328# | 1606# | 1722# |
QA: quality assurance. #: average number of plates across all drugs. This table can be reproduced online [21].
Of these, 12 362 also had their whole genome sequenced (Methods and table 1), allowing us to infer species and lineage information using SNP-IT [23]. All isolates belonged to the M. tuberculosis complex (MBTC), with the majority (12 348 (99.9%)) confirmed as M. tuberculosis (supplementary table S3), of which the majority belonged to either Lineage 2 (35%) or 4 (50%) (supplementary table S4) with the expected geographic distribution (supplementary table S5 and supplementary figure S1) [24].
A previous study demonstrated that MICs measured by a single laboratory scientist after 14 days incubation of the UKMYC5 plate using either a Thermo Fisher Vizion instrument or a mirrored-box were reproducible and accurate [17]. As a further reproducibility check we pooled the MIC measurements of the H37Rv reference strain that were taken as part of our quality control process (supplementary figure S2 and supplementary table S6); the histograms for both UKMYC plates showed that the majority of MICs measured by the laboratory scientists for many, but not all, of the drugs lay within one doubling dilution of the mode. Since the magnitude of MIC measurement error is anticipated to be much greater than the error in the genetic sequencing, we constructed an MIC quality assurance (QA) process to minimise the measurement error of the MICs (supplementary figure S3). This measured each MIC using up to three independent methods and only MICs where two of these methods concur are allowed into the final dataset. Overall, 77% of all MIC measurements passed the MIC QA process.
MIC histograms are different for different drugs
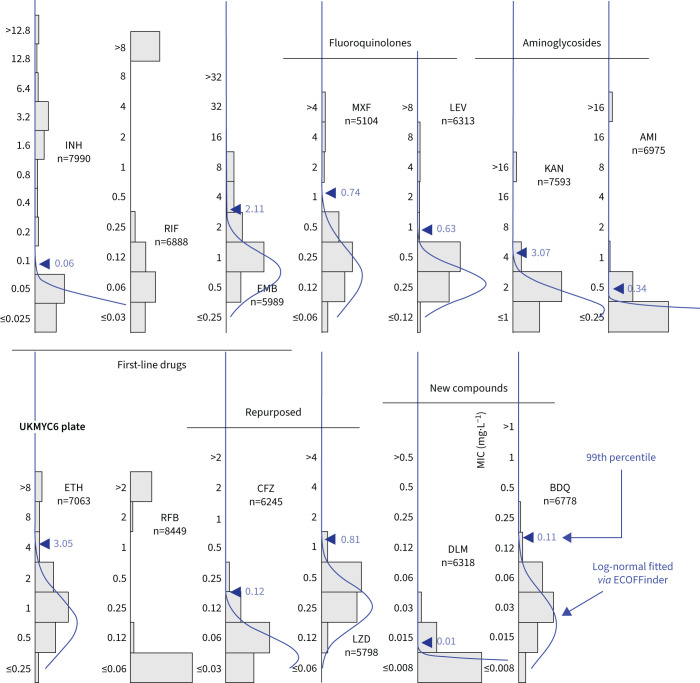
As expected, the MIC histograms differ between drugs (figure 2, and supplementary figures S5 and S6); the MICs for some compounds form bimodal distributions (isoniazid (INH), kanamycin (KAN), amikacin (AMI), rifampicin (RIF) and rifabutin (RFB)) and therefore conform to the classical binary paradigm whereby an isolate is either “resistant” or “susceptible”. CRyPTIC aimed for half the isolates collected to be multidrug resistant (MDR), and the MIC histograms for INH and RIF are consistent with this. Given this bias towards MDR in the dataset, one would expect appreciable resistance to ethambutol (EMB), ethionamide (ETH), both fluoroquinolones and both aminoglycosides. Both drugs belonging to the latter class indeed have a subset of isolates with very high MICs. The MIC histograms for the remaining compounds (EMB, ETH, moxifloxacin (MXF) and levofloxacin (LEV)) are not bimodal, hence it is unclear whether they can be adequately described by two log-normal distributions. Since the remaining drugs on the plates (BDQ, DLM, CFZ and LZD) have not yet been widely used, and for some countries were not even available to treat TB, one expects little resistance in the dataset and hence it is likely these MIC histograms are “phenotypically wild-type” (pWT). All the MIC histograms are truncated/censored at either one or both ends and some are severely truncated with the mode MIC occurring in the lowest dilution (AMI, RFB and DLM). Our large dataset allows us to use reproducible, algorithmic approaches for estimating the 99th percentile of the wild-type population.
FIGURE 2.
Minimum inhibitory concentration (MIC) histograms for the 13 antibiotics on the UKMYC6 plate. Only MICs which have passed the quality assurance process described in the Methods are shown. ECOFFinder was used to fit a log-normal distribution to each histogram; this is drawn in blue and the resulting 99th percentile is labelled. ECOFFinder was unable to fit a log-normal to both RIF and RFB. See figure 1 for drug abbreviations. See supplementary figure S6 for the UKMYC5 histograms and the TSV file in the supplementary material for the numerical data. The histograms can be reproduced online [21].
Iteratively fitting a log-normal distribution
ECOFFinder is a heuristic approach that attempts to iteratively fit a log-normal distribution to the MIC histogram, and is recommended by both the European Committee on Antimicrobial Susceptibility Testing (EUCAST) and the Clinical and Laboratory Standards Institute (CLSI) [20, 25]. EUCAST advise that ECOFFinder should not be applied to truncated data, but here we apply it to demonstrate how it performs for different levels of censored data. Distributions derived using ECOFFinder (figure 2, supplementary figure S6 and table 2) describe our data well where the MIC histogram is minimally truncated (ETH, LZD and BDQ); however, where the MIC histogram is heavily truncated (AMI, RFB and DLM) the resulting log-normal distribution does not fit the MIC histogram and where the mode MIC is resistant (RIF) it fails to perform a fit at all. In addition, since ECOFFinder requires a single consistent MIC distribution and our dataset is composed of two plate designs, two ECOFF/ECVs are returned for each drug. For many drugs these are very similar, but for EMB and DLM the estimates are almost a doubling dilution different.
TABLE 2.
99th percentiles of the wild-type population as determined by three different algorithmic approaches and the resulting proposed epidemiological cut-off values (ECOFF/ECVs) for the 13 drugs on the UKMYC6/5 plates
| ECOFFinder | Direct measurement on gWT (mg · L −1 ) | Interval regression on gWT (mg · L −1 ) (both) | Proposed ECOFF/ECV (mg · L −1 ) | |||
| UKMYC6 | UKMYC5 | UKMYC6 | UKMYC5 | |||
| Isoniazid | 0.06 | 1.6 | 0.1 | 0.25 | 0.1 | |
| Rifampicin | 0.08 | 0.5 | 0.25 | 0.52 | 0.5 | |
| Ethambutol | 2.1 | 4.0 | 4 | 4 | 3.7 | 4 |
| Moxifloxacin | 0.74 | 0.75 | 1 | 2 | 1.4 | 1 |
| Levofloxacin | 0.63 | 0.72 | 1 | 4 | 1.3 | 1 |
| Kanamycin | 3.1 | 2.9 | 8 | 4 | 7.0 | 4 |
| Amikacin | 0.34 | 0.31 | 1 | 1 | 1.6 | 1 |
| Ethionamide | 3.1 | 3.0 | 4 | 4 | 4.0 | 4 |
| Rifabutin | 0.12 | 0.12 | 0.09 | 0.12 | ||
| Clofazimine | 0.12 | 0.078 | 0.25 | 0.5 | 0.34 | 0.25 |
| Linezolid | 0.81 | 0.95 | 1 | 1 | 1.6 | 1 |
| Delamanid | 0.010 | 0.019 | 0.12 | 0.12 | 0.11 | 0.12 |
| Bedaquiline | 0.11 | 0.11 | 0.25 | 0.25 | 0.20 | 0.25 |
gWT: genotypically wild-type.
To overcome the problem that our MIC histograms are truncated we applied interval regression, an established statistical method for fitting normal distributions to truncated data [26, 27] which, unlike conventional maximum-likelihood algorithms, takes into account that observations are properly represented by intervals. The entire dataset containing measurements from both plate designs can then be considered simultaneously, resulting in a single pair of log-normal distributions that describe the MIC histograms on both plate designs (supplementary figure S7). The model fails to converge for KAN and EMB, and for several drugs the second distribution has a variance much larger than the MIC range, which is nonsensical (AMI, ETH, RFB, CFZ, LZD, DLM and BDQ), although for the new and repurposed compounds this is understandable since we do not expect many resistant isolates. Where the two distributions describe the data reasonably well (INH, RIF, MXF and LEV), they are well separated, as defined by the 99th percentile of the lower distribution (ECOFF/ECV) being smaller than the 1st percentile of the upper distribution (the non-wild-type cut-off value (NCOFF)), with the exception of INH where NCOFF<ECOFF.
Defining a gWT population
Using these approaches we were not able to produce acceptable results when the MIC histogram is truncated and/or is not clearly bimodal. In the latter case it is probable that the overall MIC histogram is a convolution of several smaller, narrower distributions. Genetics offers a way to disentangle these subpopulations: one can predict genetically the susceptibility of strains to most, but not all, of the 13 anti-TB compounds of interest [6–8]. Note that we were unable to use the newer and more comprehensive genetic catalogue released by the WHO since its derivation set included these samples [9, 10]. We predicted the antibiogram for the first-line (INH, RIF, EMB and also PZA; see Discussion) and second-line (AMI, KAN, LEV, MXF and ETH) compounds (Methods). No predictions were made for the other anti-TB compounds on the plate since the association between genetics and their resistance is poorly understood at present.
We defined an isolate as being gWT if it is predicted to be susceptible to the four first-line compounds and not resistant to the second-line compounds (see [6] for the distinction). The laxer criterion for the second-line compounds allowed for the fact that our understanding for these drugs is less complete. Epidemiologically, it is the case that if an isolate is susceptible to all four first-line antibiotics, it is also likely to be susceptible to second-line antibiotics (except perhaps for prior fluoroquinolone exposure or deeply rooted second-line resistance mutations). To contribute, a laboratory had to have collected susceptible samples which had undergone whole-genome sequencing and also had a high-quality photograph taken of the UKMYC plate after 14 days incubation so we could run the QA process. As a result, isolates from only nine CRyPTIC laboratories made up this dataset (supplementary table S8), and the number of confirmed MICs varied between 2594 and 4078 by drug, with a mean of 3263 (supplementary table S9). Visually, the resulting MIC histograms are simpler and more likely to be adequately described by a single log-normal distribution (supplementary figure S8).
Directly measuring the ECOFF/ECV from the gWT population
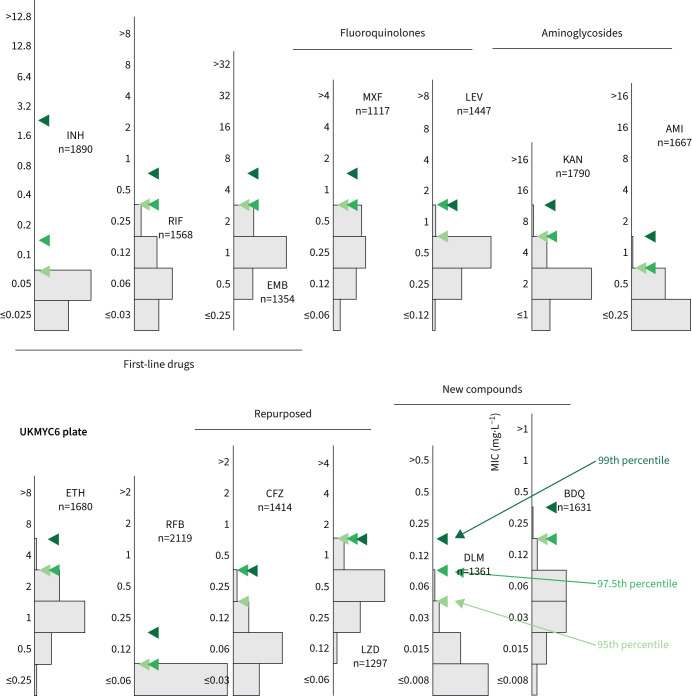
Directly determining the MIC of the 99th percentile from the gWT population is an attractive option since it requires no further assumptions. This is not usually possible since typically either one cannot discern the wild-type population and/or there are an insufficient number of isolates. The large size of our dataset and the inclusion of genetic information enables us to directly measure the ECOFF/ECV (figure 3 and supplementary figure S9). Our dataset is enriched for resistance, hence the proportion of resistant samples misclassified as susceptible due to sample mislabelling is likely to be of the order of a few percentage points, even after we have removed some putative mislabelled samples (Methods). This makes directly identifying the 99th percentile challenging, hence we shall also consider the 97.5th and 95th percentiles.
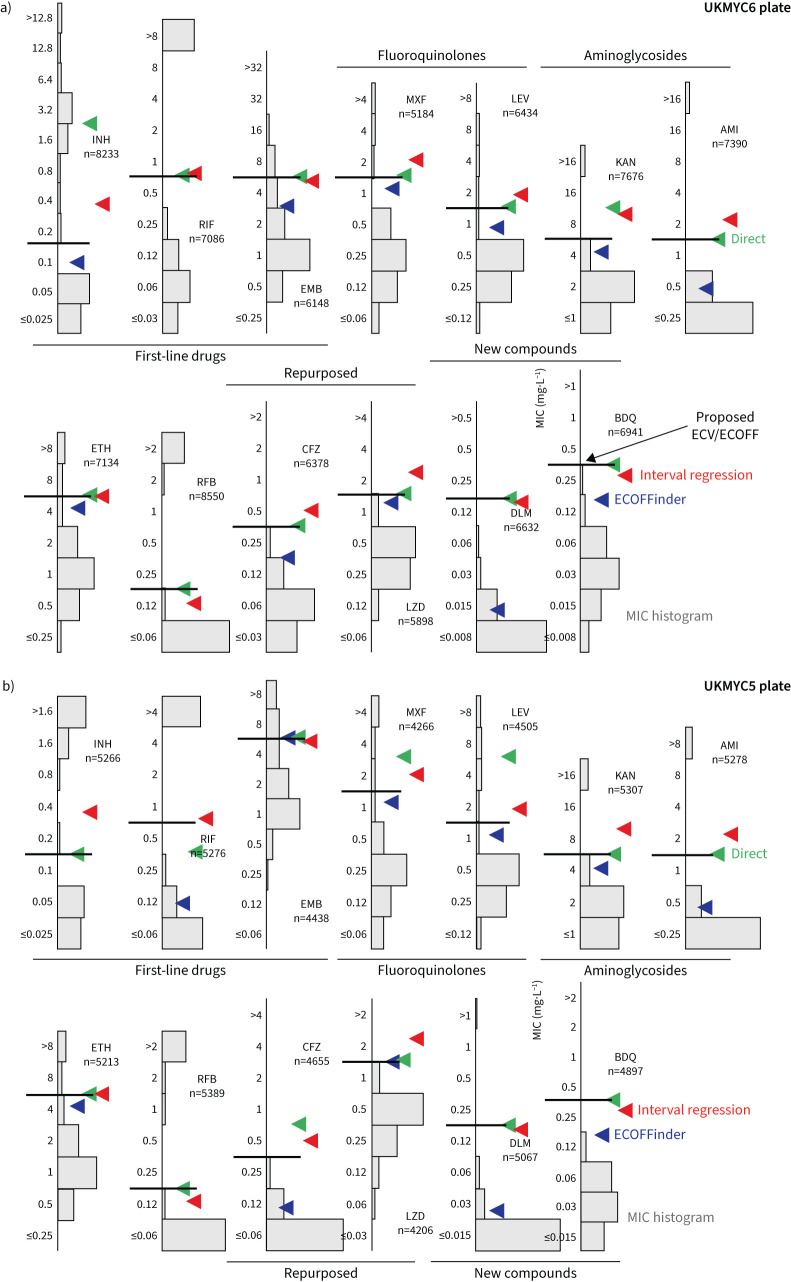
FIGURE 3.
Directly measuring the epidemiological cut-off values (ECOFF/ECVs) from the genotypically wild-type population on the UKMYC6 plate. To illustrate the sensitivity to the precise percentile used in the definition, the 95th, 97.5th and 99th percentiles are all shown. See figure 1 for drug abbreviations. The analysis and figure can be reproduced online [21].
All three percentiles for the MIC histograms of the gWT population are at most two doubling dilutions apart, except for LEV (UKMYC5) and INH (UKMYC6). The latter has an appreciable number of isolates that, despite being classified as gWT, have elevated MICs. These are likely due to some remaining samples that were mislabelled and illustrates the difficulty in using the 99th percentile to define an ECOFF/ECV due to its sensitivity to errors in the dataset, especially when the prevalence of resistance is high, as in the case for INH in our dataset. We shall take forward the values for the 99th percentiles (table 2) but will bear in mind that a high amount of variation may indicate strain mislabelling.
Interval regression takes account of the truncated distributions
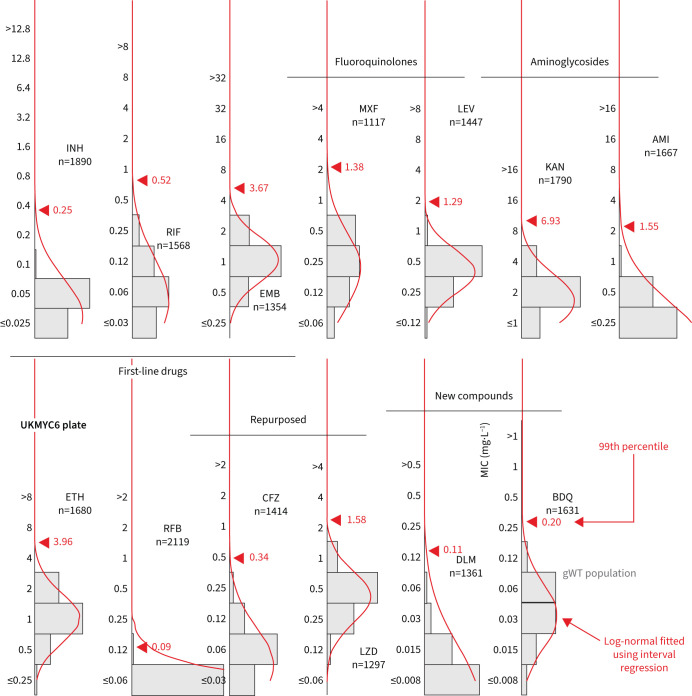
To avoid the identification of the 99th percentile being disproportionately affected by a small number of mislabelled resistant samples one usually fits a log-normal distribution to the pWT (here gWT) population and then calculates from the resulting function the MIC of the 99th percentile. We cannot apply ECOFFinder here since its heuristic requires the presence of non-susceptible isolates in the distribution, so we instead simultaneously fit a single log-normal distribution using interval regression to the MIC histograms from both plate designs (figure 4, supplementary figure S10 and table 2). With the exceptions of INH, RFB and DLM, the resulting log-normal distributions describe the MIC histograms well, even when there is moderate truncation due to the plate design. The RFB MIC distribution is, however, extremely truncated and hence there are insufficient data to perform a fit; the concentration range for this drug should be lowered in future designs.
FIGURE 4.
Interval regression is able to fit a log-normal distribution to the minimum inhibitory concentration (MIC) histograms of the genotypically wild-type (gWT) isolates for all 13 drugs on the UKMYC6 plate. Data from both plate designs were considered simultaneously, hence the resulting distributions are those the algorithm considers to best describe both the UKMYC5 (supplementary figure S10) and UKMYC6 datasets. See figure 1 for drug abbreviations. See the TSV file in the supplementary material for the numerical data. The data can be reproduced online [21].
Proposed ECOFF/ECVs
We infer that direct measurement is the most reliable method since it makes the fewest assumptions. However, for drugs where there is variation of more than a doubling dilution between the 95th, 97.5th and 99th percentiles, which may indicate the gWT population includes a small but unknown number of isolates with elevated MICs, we shall place greater weight on the result obtained by interval regression. When the MIC histogram is not heavily truncated, we shall also include the 99th percentile reported by ECOFFinder. Note that to convert an MIC that is reported as a real number into an ECOFF/ECV it should be rounded up to the next value in the doubling dilution series. All these data and the resulting ECOFF/ECVs are shown in figure 5 and table 2.
FIGURE 5.
99th percentiles of the wild-type populations for the 13 drugs on the a) UKMYC6 and b) UKMYC5 plate designs as calculated by ECOFFinder, direct measurement and interval regression. The epidemiological cut-off values (ECOFF/ECVs) are drawn on each graph as a horizontal line. See figure 1 for drug abbreviations.
The 99th percentile determined by direct measurement for INH for the UKMYC6 dataset was discounted due to the large range of MICs spanned by the percentiles; this is most likely due to resistant samples misidentified as susceptible due to laboratory mislabelling. INH has the highest prevalence of resistance in the dataset and hence would be affected most. Our starting point is therefore the corresponding value for the UKMYC5 dataset (0.1 mg·L−1). The ECOFFinder results were ignored for INH since the gWT MIC histogram is truncated on both plate designs and hence the fits were poor. Visually, the gWT MIC histograms (supplementary figure S8) do not appear to follow a log-normal distribution and consequently interval regression overestimates the 99th percentile (figure 4 and supplementary figure S10). The ECOFF/ECV of 0.1 mg·L−1 for INH is therefore less well supported than the values for the remaining drugs.
Direct measurement of the 99th percentiles for RIF were 0.5 and 0.25 mg·L−1 for the UKMYC6 and UKMYC5 datasets, respectively. Again, ECOFFinder was not used due to concerns about truncation. Visually, the gWT MIC histogram appears more normal in character and the 99th percentile derived from the interval regression fit is 0.52 mg·L−1. Our proposed consensus ECOFF/ECV for RIF is hence 0.5 mg·L−1. Direct measurement produced a consistent value of 4 mg·L−1 for EMB, which is supported by interval regression and ECOFFinder for the UKMYC5 dataset (the concentration range on the UKMYC6 plate was more truncated). Both fluoroquinolones behaved similarly: direct measurement gave a value of 1 mg·L−1 for the 99th percentile for both compounds for the UKMYC6 dataset, but 2 and 4 mg·L−1 for the UKMYC5 dataset (MXF and LEV, respectively). These values for the latter dataset were very sensitive to the exact percentile used in the definition (supplementary figure S9), again suggesting that these gWT populations may contain a small number of mislabelled resistant samples. Both drugs have the same concentration range on both plate designs, are only moderately truncated and hence ECOFFinder would be expected to give reasonable results. These, along with the result of the interval regression (figure 5), result in an ECOFF/ECV of 1 mg·L−1 for both fluoroquinolones.
Direct measurement indicates the 99th percentile for KAN is 8 and 4 mg·L−1 for the UKMYC6 and UKMYC5 datasets, respectively, while it produces the consistent value of 1 mg·L−1 for AMI. The MIC histogram of the latter is too truncated for ECOFFinder to function correctly and visually the log-normal fitted by interval regression appears to have overestimated the 99th percentile as 1.6 mg·L−1; hence, we propose an ECOFF/ECV for AMI of 1 mg·L−1. The KAN MIC histograms are less truncated and interval regression better describes the gWT population; these data support an ECOFF/ECV of 4 mg·L−1 for KAN. For ETH, direct measurement produces a consistent value of 4 mg·L−1, which is supported by interval regression and ECOFFinder. The MIC histogram of RFB is extremely truncated and hence only direct measurement is likely to be effective; it estimates 0.12 mg·L−1 to be the 99th percentile for both datasets, which is therefore our ECOFF/ECV.
Direct measurement yields consistent values of 0.25 and 1 mg·L−1 for BDQ and LZD, respectively, with each supported by both interval regression and ECOFFinder. Our ECOFF/ECV for DLM is 0.12 mg·L−1 since this is the direct measurement which is the same for both datasets and it is supported by interval regression. Lastly, direct measurement for CFZ suggests the 99th percentiles are 0.25 and 0.5 mg·L−1 for the UKMYC6 and UKMYC5 datasets, respectively. The latter has more variation and hence we propose its ECOFF/ECV is 0.25 mg·L−1.
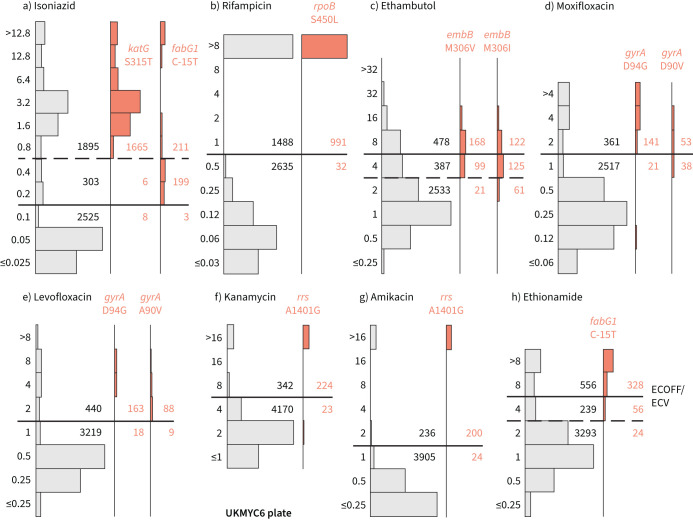
Comparison against genetic variants known to confer resistance
Using the subset of the isolates with genetic information, we can examine our proposed ECOFF/ECVs by plotting the MIC histograms of several genetic variants that are widely accepted to confer resistance to key anti-TB drugs (figure 6 and supplementary figure S11). The rpoB S450L and katG S315T single nucleotide polymorphisms (SNPs) substantially increase the MICs of RIF and INH, respectively, and the majority (96.9% and 99.5%) of isolates with these mutations had MICs greater than the ECOFF/ECV. The C-15T mutation in the promoter of the fabG1/inhA operon was associated with borderline (0.2 mg·L−1) INH MICs unless present in combination with a katG S315T mutation (MIC >1.6 mg·L−1), as observed elsewhere [22, 28]. It is likely that this promoter mutation, and others like it, are responsible for the small peak in the MIC histogram observed for INH at 0.2 mg·L−1.
FIGURE 6.
Minimum inhibitory concentrations (MICs) of isolates containing genetic variants known to confer resistance to different drugs tend to lie above the epidemiological cut-off value (ECOFF/ECV) on the UKMYC6 plate: a) isoniazid, b) rifampicin, c) ethambutol, d) moxifloxacin, e) levofloxacin, f) kanamycin, g) amikacin and h) ethionamide. The number of isolates lying above and below the ECOFF/ECV is annotated. The dashed line indicates the margin of a proposed “borderline” category for isoniazid, ethambutol and ethionamide. The same analysis has been repeated on the UKMYC5 dataset (supplementary figure S11) and can be reproduced online [21].
Substituting isoleucine or valine at position 306 in the embB gene was associated with elevated EMB MICs; however, the increase in MIC is much less than observed for either of the RIF or INH resistance-conferring mutations mentioned earlier, leading to only 58.3% and 39.6% of isolates containing these mutations, respectively, having an MIC above the ECOFF/ECV. This is expected since it is known that isolates containing these variants can have variable or discordant MGIT results [29]. For both fluoroquinolones, the gyrA D94G mutation increases the MIC more than the gyrA A90V mutation [30]; however, for LEV the wild-type and non-wild-type populations appear slightly better separated, with the result that for these mutations 90.0% and 90.7% of isolates lie above the ECOFF/ECV, while for MXF the equivalent values are 87.0% and 58.2%. The majority of isolates (90.7% and 89.3%) with the A1401G mutation in the rrs gene have an MIC above the ECOFF/ECV for KAN and AMI, respectively. Finally, while the C-15T mutation in the promoter of the fabG1/inhA operon increases the MIC of ETH more than it does INH, only 80.4% of isolates with this variant lie above the ECOFF/ECV.
A role for a borderline category?
The ECOFF/ECV merely defines an MIC below which the majority of the “wild-type” isolates should lie. It does not necessarily follow that the majority of non-wild-type isolates have an MIC above the ECOFF/ECV, and therefore care needs to be taken when using an ECOFF/ECV to define susceptibility and resistance. Although for most drugs an MIC below or equal to the ECOFF/ECV can be categorised as “susceptible” and those with MICs above the ECOFF/ECV are “resistant”, the MIC histograms of INH, EMB and ETH are more complex. There is genetic evidence (figure 6) that this is due to a multitude of genetic variants, each with a different effect on the MIC. We therefore propose a third category, “borderline”, for INH, EMB and ETH (figure 6 and table 3).
TABLE 3.
Proposed epidemiological cut-off values (ECOFF/ECVs) and suggested borderline minimum inhibitory concentrations (MICs) for three compounds
| ECOFF/ECV (mg · L −1 ) | Borderline MIC (mg · L −1 ) | |
| Isoniazid | 0.1 | 0.2, 0.4 |
| Rifampicin | 0.5 | |
| Ethambutol | 4 | 4 |
| Moxifloxacin | 1 | |
| Levofloxacin | 1 | |
| Kanamycin | 4 | |
| Amikacin | 1 | |
| Ethionamide | 4 | 4 |
| Rifabutin | 0.12 | |
| Clofazimine | 0.25 | |
| Linezolid | 1 | |
| Delamanid | 0.12 | |
| Bedaquiline | 0.25 |
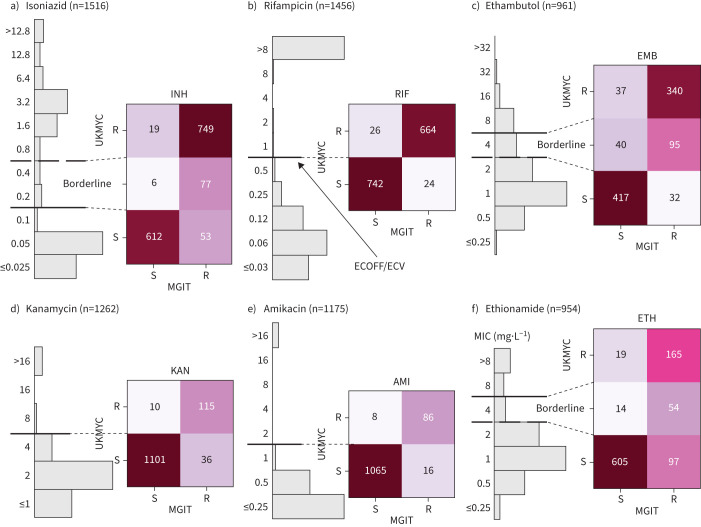
Validation by comparison with MGIT and MODS results
The resistance of a subset of isolates was independently tested to a range of compounds using either the Mycobacterial Growth Indicator Tube (MGIT) system or Microscopic-Observation Drug Susceptibility (MODS) assay [31]. We can therefore validate our MIC-based categorisation by directly comparing between the binary (or ternary) phenotype derived from an MIC and the result from one of these well-established clinical microbiology methods (figure 7, supplementary figure S12 and supplementary table S10).
FIGURE 7.
Binary (or ternary) classification derived from the minimum inhibitory concentration (MIC) using the epidemiological cut-off values (ECOFF/ECVs) and MIC-based categorisation in table 3 agrees well with Mycobacterial Growth Indicator Tube (MGIT) results for the samples for a) isoniazid (INH), b) rifampicin (RIF), c) ethambutol (EMB), d) kanamycin (KAN), e) amikacin (AMI) and f) ethionamide (ETH). S: susceptible; R: resistant. The data and figure panels can be reproduced online [21].
The agreement between MGIT and UKMYC is good, with a sensitivity of 93.4% and a specificity of 97.0% (supplementary table S10). Since the borderline category lies above the ECOFF/ECV, it is interpreted as providing a way of discriminating between isolates with a moderately elevated MIC and those with a high MIC. For RIF, the agreement between MGIT and UKMYC is excellent, with a sensitivity of 96.5% and a specificity of 96.6%. The borderline category for EMB provides a “buffer zone” since isolates with an MIC of 4 mg·L−1 are only 70.4% resistant according to MGIT. Ignoring these isolates, the sensitivities and specificities are 91.4% and 91.9%, respectively, for EMB. Since the “borderline” category in this case lies below the ECOFF/ECV, these isolates would otherwise be classified as “susceptible” but in reality have a mixed character. Hence, not assigning a borderline category would result in the sensitivities and specificities becoming 72.8% and 92.5%, respectively.
The aminoglycosides behave similarly to one another, with sensitivities and specificities of 76.2% and 99.1% for KAN and 84.3% and 99.3% for AMI, respectively. The “borderline” category for ETH (MIC 4 mg·L−1) is 79.4% resistant according to MGIT. Excluding these isolates, the sensitivity is 63.0% and 97.0%, respectively. Limited numbers of isolates were tested for MXF or LEV resistance using MGIT (supplementary figure S12). Although large number of isolates were tested for CFZ and LZD resistance by MGIT (supplementary figure S12), the low prevalence of resistance ensures no useful conclusions can be drawn.
A different set of samples were tested in parallel using the MODS assay (supplementary figure S13 and supplementary table S11). The sensitivities and specificities for INH (n=1888) and RIF (n=1857) were 95.3% and 98.9% and 95.1% and 99.2%, respectively.
Discussion
We have proposed ECOFF/ECVs for research use for 13 different anti-TB compounds for the UKMYC series of broth microdilution plates using an aggregated dataset of 20 637 TB samples collected worldwide by 14 CRyPTIC laboratories based in 11 countries on five continents. The UKMYC6 plate design (figure 1b) not only contains the first-line drugs RIF, INH and EMB, but also all of the Group A medicines, one of the two Group B medicines (CFZ) and five of the seven Group C medicines recommended by the WHO for treating cases of MDR-TB [19]. As such these plates offer near comprehensive phenotypic AST as well as a standardised, scalable phenotype that, once analysed with linked genomic data, will inform clinical decisions where routine diagnostics switch to genome sequencing.
We caution that while the ECOFF/ECVs proposed herein have been derived using the largest collection of M. tuberculosis samples to date, the methods do not conform with those laid out by EUCAST. That said, our analyses illustrate that the EUCAST definition of an ECOFF/ECV as the 99th percentile of the wild-type population [20] is difficult to apply in practice since firstly it is not always possible to define which isolates are “phenotypically wild-type” (pWT) without engaging in a circular argument. We were able to avoid this here by defining a “genotypically wild-type” (gWT) population. The second problem is that using the 99th percentile to define the ECOFF/ECV places a very stringent upper limit on the total error rate, which becomes harder to meet as the prevalence of resistance in any dataset increases. Despite our efforts, we see evidence that our gWT populations for some compounds contain >1% resistant isolates for some drugs which, for example, hampers the use of direct measurement. In contrast, the CLSI have a less-stringent definition for the ECOFF/ECV which avoids this issue [32], but in turn can create inconsistencies between studies. As suggested elsewhere, using a lower percentile (e.g. 97.5th) could help [22].
The CLSI recently proposed breakpoints for the MYCOTB plate [33]. There are a few minor differences: our proposed ECOFF/ECV for RIF is one doubling dilution lower at 0.5 mg·L−1. The impact of this is difficult to assess due to the paucity of isolates with MICs of 0.5 and 1.0 mg·L−1. For INH, although it is not possible to make an exact comparison between the ECOFF/ECVs since the doubling dilution series used on the UKMYC plates and by the CLSI are different, the value proposed by the CLSI (0.12 mg·L−1) is close the value proposed here (0.1 mg·L−1). The CLSI breakpoints for EMB exactly agree with the MIC-based classification adopted by CRyPTIC. Our ECOFF/ECVs are different to those of a recent MYCOTB study [22]; however, we note that the number of samples was modest (n=385) and originated from a single country. In addition, ECOFF/ECVs were determined using ECOFFinder, which given the truncated nature of the MIC histograms for many of the drugs, is not now advised and may have biased some of the results. Our ECOFF/ECVs for RIF and INH are, however, consistent with a recent recommendation made by the WHO, albeit for MGIT.
Critical concentrations for several of the drugs on the UKMYC plates (which are inoculated with 7H9 growth media) exist for M. tuberculosis grown in other growth media, such as Löwenstein–Jensen, 7H10 and 7H11, and also other AST methods, such as BACTEC MGIT 960 [34–36]. Caution must, of course, be applied when comparing cut-offs derived using fundamentally different growth media and AST methods.
The ECOFF/ECVs for nine drugs proposed by a series of 7H10 agar studies all either agree or are one doubling dilution different to our proposed ECOFF/ECVs for the UKMYC plates [37–39]. More recently, there has been a push to set breakpoints for the new compounds DLM and BDQ so that AST can be performed for these important drugs [40]. An early MGIT study using 194 isolates proposed an ECOFF/ECV for DLM of 0.125 mg·L−1 [41], which is identical to our value. An ECOFF/ECV of 0.125 mg·L−1 for BDQ on broth microdilution plates was proposed [42]; however, the 95th percentile of the wild-type population was used to define the ECOFF/ECV since CLSI guidelines were followed [32] and ECOFFinder was used despite the truncated nature of the MIC histograms. This value was supported by two subsequent studies, the first of which showed that that the sensitivity and specificity is maximised with an ECOFF/ECV of 0.12 mg·L−1 compared with 0.25 mg·L−1 [43]. The second study confirmed this value, but also stated that the 99th percentile of the wild-type population was 0.25 mg·L−1 [44]. This illustrates that the exact value can be difficult to pin down when different ECOFF/ECV definitions are used; hopefully, the number and diversity of samples in our study will help resolve this important question. Lastly, the ECOFF/ECVs proposed here lie within the range of breakpoints recommended by the WHO for different growth media, with the exception of CFZ for which the WHO recommends a cut-off of 1 mg·L−1 in MGIT [35, 36].
Deriving ECOFF/ECVs from MICs relies on several assumptions, foremost that applying a binary resistant/susceptible classification to a clinical infection is a reasonable and helpful way to proceed. That simplifying the description of the results of clinical microbiology investigations helps interpretation is not in doubt [45]; however, problems with reproducibility can arise depending on the character of the underlying MIC histogram. If the MIC histogram is “bimodal” (i.e. has two narrow peaks separated by an interval greater than their individual variance) then placing the ECOFF/ECV between the peaks leads to a helpful and reproducible classification system [46]. On the UKMYC series of plates, the only drugs that conform to this ideal are the rifamycins and the aminoglycosides; the other compounds either have more complex distributions (INH, EMB, MXF, LEV and ETH) or resistance is not sufficiently prevalent for us to fully characterise their MIC distributions (CFZ, LZD, DLM and BDQ).
There is a further implicit (weak) assumption that the “susceptible” and “resistant” subpopulations can each be described by a single MIC distribution, which is not necessarily true, as exemplified by the effect of the fabG1 promoter mutations on INH (figure 6a) [28]. In addition, this assumption implies that different lineages behave similarly when exposed to an antibiotic, which is unlikely to be true [47]. Finally, the wild-type distribution is usually assumed to be log-normal; however, our data do not support this for all drugs (e.g. LEV; figure 4) resulting in the log-normal distributions fitted by interval regression apparently overestimating the 99th percentile. Should this turn out to be generally true, this would invalidate methods based on fitting such distributions, making direct measurement more appealing [25].
One can deconstruct the error in determining an ECOFF/ECV using a microtitre plate into sample selection biases, data entry and labelling errors, inoculation and incubation errors, measurement errors, errors in defining the wild-type population, uncertainties arising from censored data, and errors in fitting a curve to the resulting MIC histogram. In addition to the obvious benefits in collecting such a large and diverse dataset (table 1, figure 1 and supplementary table S2), we have been careful to minimise measurement errors (supplementary figure S3) and have also used a principled method to attempt to remove some putative mislabelled samples (Methods). By defining a gWT population and either applying interval regression to fit normal distributions (figure 4) or directly measuring the 99th percentile (figure 5), we have also minimised the final two sources of error. Despite these steps, further sources of error no doubt remain. Another key weakness of this study is the lack of PZA, which due to its preference for acidic conditions, is currently unable to be successfully incorporated onto broth microdilution plates, although there is hope that this could be rectified in future [48].
The debate about how to define and calculate ECOFF/ECVs will continue and new approaches will be suggested [49–52]. However it evolves, larger and more geographically diverse TB datasets, such as presented here, will bring more confidence and rigour to the AST of clinical TB samples. We hope also, that as clinical microbiology transitions into a data-driven science, our proposed method of directly measuring the required percentile of the gWT population will gain traction due to its simplicity and reproducibility as the genetics of M. tuberculosis resistance becomes better understood and accepted.
Although the main objective of the CRyPTIC project is to map the genetic variations in the M. tuberculosis genome that confer resistance to many antibiotics, the sheer number of samples collected provides a body of evidence to support the use of 7H9 broth microdilution plates in clinical mycobacteriology. Applications potentially include AST for samples that are predicted to be MDR or extensively drug resistant (XDR) by the GeneXpert RIF/MDR assay system, or surveying the prevalence of different patterns of resistance by region or country, allowing regional regimens to be designed and their impact monitored. Finally, even in settings which adopt genetics-based clinical microbiology [11], it would be prudent to maintain culture-based testing not only to identify new genetic variants as they arise but also to continuously monitor the performance of the genetic resistance catalogue which is likely to change over time as such catalogues are only likely partly causal.
In future work the CRyPTIC project will apply the ECOFF/ECVs proposed here not only to further optimise a genetic catalogue for the first-line anti-TB compounds [6] but also to extend coverage to second-line, repurposed and new compounds, with the aim of covering as many of the drugs recommended by the WHO for treating MDR- and XDR-TB [19]. Clearly the numerical data being collected by the CRyPTIC Consortium also lends itself to the development of a genetic catalogue for anti-TB compounds that can make quantitative predictions; such a catalogue would naturally take account of additivity, epistasis and non-linear effects. Finally, the tools and data-driven approaches developed here could be applied to other pathogens, especially other mycobacteria.
Methods
Ethics review
Approval for the CRyPTIC study was obtained by the Taiwan Centers for Disease Control Institutional Review Board (106209), University of KwaZulu Natal Biomedical Research Ethics Committee (BE022/13), University of Liverpool Central University Research Ethics Committees (2286), Institutional Research Ethics Committee of The Foundation for Medical Research, Mumbai (FMR/IEC/TB/01a/2015 and FMR/IEC/TB/01b/2015), Institutional Review Board of P.D. Hinduja Hospital and Medical Research Centre, Mumbai (915-15-CR (MRC)), and Scientific Committee of the Institute Adolfo Lutz (CTC-IAL 47-J/2017), and in the Ethics Committee (CAAE: 81452517.1.0000.0059) and Ethics Committee review by Universidad Peruana Cayetano Heredia (Lima, Peru) and London School of Hygiene & Tropical Medicine (London, UK). No ethics approval was required for the remaining laboratories since at no time was any patient-identifiable information shared with the CRyPTIC Consortium.
Sample selection
The CRyPTIC project aimed for around half the samples collected to be susceptible to the first-line compounds with the remainder MDR/XDR. There was, however, large variation between the different participating laboratories.
Incubation and inoculation protocol
Each laboratory followed a standard operating protocol laid out by the CRyPTIC Consortium, which was similar to that described previously [17]. Clinical samples were subcultured either using Löwenstein–Jensen tubes, 7H10 agar plates or MGIT tubes. The protocol specified that first a suspension at 0.5 McFarland standard in saline Tween with glass beads (Thermo Fisher Scientific, Waltham, MA, USA) from 20–25-day-old colonies be prepared. These were then diluted 100-fold by adding 100 µL suspension to 10 mL enriched 7H9 broth [17]. A semi-automated Sensititre Autoinoculator (Thermo Fisher Scientific) was used to dispense 100 µL inoculum (1.5×105 CFU·mL−1, with approximate range from 5×104 to 5×105 CFU·mL−1) into a well of a UKMYC5/6 microdilution plate. The plate was then sealed using transparent plastic provided by the manufacturer. The UKMYC5 and UKMYC6 microdilution plates were designed by the CRyPTIC Consortium and manufactured by Thermo Fisher Scientific. The drugs included and their concentrations are described in figure 1. DLM and BDQ pure substances were provided by Otsuka Pharmaceutical (Tokyo, Japan) and Jannsen Pharmaceuticals (Beerse, Belgium), respectively. H37Rv ATCC 27294 was used to perform periodic quality control runs since it is susceptible to all the drugs on both plate designs.
Measurement of MICs after 14 days incubation
In each laboratory a scientist read each plate after 14 days incubation using a Thermo Fisher Sensititre Vizion digital MIC viewing system, with results entered via a bespoke web portal, CliRes2 (https://clires2.oucru.org). In those cases where this was not possible, spreadsheets were sent. A photograph was also taken using the Vizion system and also stored in CliRes2. Two laboratories used a mirrored-box to read the plates and one of these also took a photograph using a DSLR camera. A plate was marked as invalid if it did not have adequate bacterial growth in both positive control wells. A small subset of plates with poor growth at day 14 were incubated for a further week and then read again.
Other AST measurements
Where available, the results of standard AST tests conducted by the participating laboratory were also entered via the CliRes2 online portal or, in some cases, shared via spreadsheet. The methods used were mainly either the MGIT system or MODS assay [31]. All MGIT tests used standard critical concentrations; for the MXF results only those with a critical concentration of 0.5 mg·L−1 were included.
Genetic sequencing and interpretation
Sequencing arrangements differed slightly between each CRyPTIC participating laboratory. All sequencing was performed using Illumina machines and hence the input to our genetic sequencing pipelines was a matched pair of FASTQ files containing the short reads. Data integrity was ensured throughout by tracking the MD5SUM hashs of the FASTQ files.
Human and HIV reads were removed from the raw sequence data as follows. Reads were mapped to the reference genome H37Rv, the human genome version GRC38, the HIV reference NC_001802.1, various other viral genomes (so that, if any reads mapped to HIV, one would only know that they mapped to some virus) and nasopharyngeal flora genomes from the human microbiome project, using the BWA-MEM aligner. First, a read pair was kept if either read matched H37Rv, then removed if either read matched one of the other genomes and finally kept if both reads were unmapped.
Variants were initially called using SAMtools and Cortex, two variant callers with orthogonal strengths (SAMtools is a high sensitivity SNP caller; Cortex is a high specificity SNP and insertion/deletion caller). These calls were then passed to the adjudication software minos, which produces a graph representation of the reference genome plus conflicting calls from the two callsets and then remaps reads to the graph to adjudicate statistically. This adjudication process and the performance of the combined SAMtools/Cortex callset are documented [53]. All of this process, including versions of SAMtools and Cortex and the reference genomes for filtering, is encapsulated in Clockwork version 0.8.3 [54].
Samples were excluded from the dataset if they had either >100 000 unfiltered SAMtools variant calls (a weak filter applied to detect samples contaminated with the wrong species) or an average read coverage of ≤15 when mapped to reads covering the H37Rv reference. The samples that pass these criteria and have paired phenotype data are named the GPI (geno–pheno intersection). Variant calls were removed if they overlapped a set of masked positions as previously defined [55]. This mask consists of 324 971 positions from the H37Rv reference with self-blast matches (https://github.com/iqbal-lab-org/cryptic_tb_callable_mask/commit/43ec21319209b23f648f32e4868bdf07cf09f2a0).
Version 3 of the H37Rv strain (NC_000962.3) was used as the TB reference genome throughout. The resulting VCF files were then transferred to the CRyPTIC data warehouse where they were interpreted.
Genetic resistance catalogue
A hybrid TB genetic resistance catalogue was constructed by merging two published catalogues. The first more recent catalogue contained rows for the four first-line drugs (INH, RIF, EMB and PZA) [6]. The second also contained rows for MXF, LEV, streptomycin (STM), ofloxacin (OFX), AMI, KAN, capreomycin (CAP), ETH, LZD, CFZ, DLM, BDQ, RFB, prothionamide (PTO) and PAS [8]. Since each catalogue was constructed with respect to version 2 of the H37Rv M. tuberculosis reference genome, they were first translated to version 3 of the reference. These catalogues are freely available to download [56], and use a standard grammar, GARC, that is both machine and human readable. To avoid putting as few assumptions into downstream code as possible, default rules are included that, for example, specify that non-synonymous amino acid mutations that match no other row have an unknown effect. The hybrid catalogue was constructed by taking the rows for the first-line compounds from the first catalogue and rows for all other drugs from the second. This catalogue, called CRyPTICv1.31, is freely available for download [56] and is also provided in the attendant repository [21].
Genetic analysis
Each sample VCF was compared to a reference genome object using v0.1.0 of the Python gumpy module [57], thereby creating a table of genetic variants (both SNPs and insertions/deletions). Both the individual SNPs were stored and also their aggregated effect on any amino acids in the coding region of any gene. An intergenic region of up to 100 bases upstream of the start codon was assumed to be the promoter sequence and hence was associated with the gene. This list of variants was then parsed by a second bespoke Python module, piezo, that reads the hybrid catalogue and understands the GARC grammar, and so returns a resistant, susceptible or unknown prediction for each drug in the catalogue [58]. The species and, if M. tuberculosis, lineage of all samples was determined by SNP-IT [23].
Data warehousing
With the exception of the compressed FASTQ files, all data (VCFs, images, MIC metadata, genetic variants and catalogue predictions) were aggregated and stored in a hierarchical file system using the Python datreant 1.0.2 module [59], which allowed for data discovery, tagging and filtering. Updates were performed by inhouse Python scripts. Plate metadata was downloaded from CliRes2 using the zeep Python SOAP client.
QA of MIC readings
Central to our QA process is the photograph taken of the plate after 14 days of incubation using the Vizion instrument by the laboratory scientist. Images were deduplicated by checking the MD5SUM was unique. The remaining images were first read by bespoke software, AMyGDA [60, 61], which detects the locations of the wells and, by measuring the growth in each well, estimates an MIC for all drugs. For 54.7% of all measurements the MICs measured by the laboratory scientist and AMyGDA were identical (supplementary figures S4 and S5a), and therefore passed the QA process.
Images of the 45.3% of cases where these two methods disagreed were uploaded to a Citizen Science project, hosted by the Zooniverse platform, called BashTheBug [62]. Each image was classified by at least 11 different volunteers and the median reading was taken to be the consensus. In 38.1% of the images sent (17.3% of the total) the consensus MIC agreed with the MIC measured by the laboratory scientist using the Vizion instrument. Visual inspection of a random subset (supplementary figure S5b) suggested that these were mostly cases where AMyGDA incorrectly estimated the MIC, usually because the growth was too small to be programmatically detected.
For a smaller proportion (12.0% of the images completed by BashTheBug; 5.4% of the total) (supplementary figure S3), the BashTheBug consensus agreed with the MIC measured by AMyGDA. Visual inspection of a random subset (supplementary figure S5c) indicated that, for the most part, these were errors made by the laboratory scientist. An error rate of 5.4% for a subjective laboratory-based measurement is reasonable, and catching and correcting these errors is the main goal of this QA process. Overall, therefore, we have a high degree of confidence in 77.4% of the MIC measurements since two or more independent methods concur on the value. Finally, in 22.6% of cases all three methods gave a different answer (supplementary figure S5d); these are excluded from further analysis. All these proportions are averaged over all drugs; there is significant variation between drugs (supplementary tables S7 and S8).
Putative mislabelled samples
Some 44 samples were assumed to be mislabelled samples as defined by being gWT but having both an INH MIC ≥1.6 mg·L−1 and an RIF MIC ≥4 mg·L−1. This corresponds to 0.8% of the dataset, which is likely an underestimate. All 44 samples were removed.
ECOFFinder
A version of ECOFFinder (ECOFFinderXL2011forMac.xlxs) that worked on Microsoft Excel running on Apple Mac computers was provided by Claudio Köser (University of Cambridge, Cambridge, UK) [25].
Interval regression
The intreg function in Stata version 15.1 (StataCorp, College Station, TX, USA) was used.
Data analysis and graphs
All data analysis, with the exception of the interval regressions and ECOFFinder, was performed using Python 3.8 in conjunction with Pandas 1.2.1 [63] and NumPy 1.19.5 [64]. Graphs were plotted using Matplotlib 3.3.4 [65] and GeoPandas 0.8.2 (https://geopandas.org).
Reproducibility
The raw data (photographs of 96-well plates, genetic variant call files) along with a series of data tables related by a schema can be downloaded from the European Bioinformatics Institute at http://ftp.ebi.ac.uk/pub/databases/cryptic/. In addition, one can reproduce nearly all the tables and figures, along with the supplementary data, in a browser window (i.e. no installation required) using Python code we have made publicly available [21].
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material ERJ-00239-2022.Supplement (3.4MB, pdf)
Supplementary data file ERJ-00239-2022.Data (14.5KB, tsv)
Shareable PDF
Acknowledgements
We are grateful to Claudio Köser (University of Cambridge, Cambridge, UK) for providing the ECOFFinder programme and for helpful comments, the EUCAST ESGMYC subcommittee chaired by Emmanuelle Cambau, especially John Turnidge, for helpful discussions, and all the BashTheBug volunteers for the time and energy they have contributed. We thank Faisal Masood Khanzada and Alamdar Hussain Rizvi (NTRL, Islamabad, Pakistan), Angela Starks and James Posey (Centers for Disease Control and Prevention, Atlanta, GA, USA), and Juan Carlos Toro and Solomon Ghebremichael (Public Health Agency of Sweden, Solna, Sweden).
Footnotes
CRyPTIC Consortium members and affiliations: Philip W. Fowler (University of Oxford, Oxford, UK), Ivan Barilar (Research Center Borstel, Borstel, Germany), Simone Battaglia (IRCCS San Raffaele Scientific Institute, Milan, Italy), Emanuele Borroni (IRCCS San Raffaele Scientific Institute, Milan, Italy), Angela Pires Brandao (Oswaldo Cruz Foundation, Rio de Janeiro, Brazil and Institute Adolfo Lutz, São Paulo, Brazil), Alice Brankin (University of Oxford, Oxford, UK), Andrea Maurizio Cabibbe (IRCCS San Raffaele Scientific Institute, Milan, Italy), Joshua Carter (Stanford University School of Medicine, Stanford, CA, USA), Daniela Maria Cirillo (IRCCS San Raffaele Scientific Institute, Milan, Italy), Pauline Claxton (Scottish Mycobacteria Reference Laboratory, Edinburgh, UK), David A. Clifton (University of Oxford, Oxford, UK), Ted Cohen (Yale School of Public Health, Yale, CT, USA), Jorge Coronel (Universidad Peruana Cayetano Heredia, Lima, Peru), Derrick W. Crook (University of Oxford, Oxford, UK), Viola Dreyer (Research Center Borstel, Borstel, Germany), Sarah G. Earle (University of Oxford, Oxford, UK), Vincent Escuyer (Wadsworth Center, New York State Dept of Health, Albany, NY, USA), Lucilaine Ferrazoli (Institute Adolfo Lutz, São Paulo, Brazil), George Fu Gao (Chinese Center for Disease Control and Prevention, Beijing, China), Jennifer Gardy (Bill & Melinda Gates Foundation, Seattle, WA, USA), Saheer Gharbia (UK Health Security Agency, London, UK), Kelen Teixeira Ghisi (Institute Adolfo Lutz, São Paulo, Brazil), Arash Ghodousi (IRCCS San Raffaele Scientific Institute, Milan, Italy and Vita-Salute San Raffaele University, Milan, Italy), Ana Luíza Gibertoni Cruz (University of Oxford, Oxford, UK), Louis Grandjean (University College London, London, UK), Clara Grazian (University of New South Wales, Sydney, Australia), Ramona Groenheit (Public Health Agency of Sweden, Solna, Sweden), Jennifer L. Guthrie (The University of British Columbia, Vancouver, BC, Canada and Public Health Ontario, Toronto, ON, Canada), Wencong He (Chinese Center for Disease Control and Prevention, Beijing, China), Harald Hoffmann (SYNLAB Gauting, Munich, Germany and Institute of Microbiology and Laboratory Medicine, IMLred, WHO-SRL Gauting, Gauting, Germany), Sarah J. Hoosdally (University of Oxford, Oxford, UK), Martin Hunt (EMBL-EBI, Hinxton, UK and University of Oxford, Oxford, UK), Zamin Iqbal (EMBL-EBI, Hinxton, UK), Nazir Ahmed Ismail (National Institute for Communicable Diseases, Johannesburg, South Africa), Lisa Jarrett (Public Health England, Birmingham, UK), Lavania Joseph (National Institute for Communicable Diseases, Johannesburg, South Africa), Ruwen Jou (Taiwan Centers for Disease Control, Taipei, Taiwan), Priti Kambli (Hinduja Hospital, Mumbai, India), Rukhsar Khot (Hinduja Hospital, Mumbai, India), Jeff Knaggs (EMBL-EBI, Hinxton, UK and University of Oxford, Oxford, UK), Anastasia Koch (University of Cape Town, Cape Town, South Africa), Donna Kohlerschmidt (Wadsworth Center, New York State Dept of Health, Albany, NY, USA), Samaneh Kouchaki (University of Oxford, Oxford, UK and University of Surrey, Guildford, UK), Alexander S. Lachapelle (University of Oxford, Oxford, UK), Ajit Lalvani (Imperial College London, London, UK), Simon Grandjean Lapierre (Université de Montréal, Montreal, QC, Canada), Ian F. Laurenson (Scottish Mycobacteria Reference Laboratory, Edinburgh, UK), Brice Letcher (EMBL-EBI, Hinxton, UK), Wan-Hsuan Lin (Taiwan Centers for Disease Control, Taipei, Taiwan), Chunfa Liu (Chinese Center for Disease Control and Prevention, Beijing, China), Dongxin Liu (Chinese Center for Disease Control and Prevention, Beijing, China), Kerri M. Malone (EMBL-EBI, Hinxton, UK), Ayan Mandal (The Foundation for Medical Research, Mumbai, India), Mikael Mansjö (Public Health Agency of Sweden, Solna, Sweden), Daniela Matias (Public Health England, Birmingham, UK), Graeme Meintjes (University of Cape Town, Cape Town, South Africa), Flávia de Freitas Mendes (Institute Adolfo Lutz, São Paulo, Brazil), Matthias Merker (Research Center Borstel, Borstel, Germany), Marina Mihalic (Institute of Microbiology and Laboratory Medicine, IMLred, WHO-SRL Gauting, Gauting, Germany), James Millard (Africa Health Research Institute, Durban, South Africa), Paolo Miotto (IRCCS San Raffaele Scientific Institute, Milan, Italy), Nerges Mistry (The Foundation for Medical Research, Mumbai, India), David Moore (London School of Hygiene & Tropical Medicine, London, UK and Universidad Peruana Cayetano Heredia, Lima, Peru), Kimberlee A. Musser (Wadsworth Center, New York State Dept of Health, Albany, NY, USA), Dumisani Ngcamu (National Institute for Communicable Diseases, Johannesburg, South Africa), Hoang Ngoc Nhung (Oxford University Clinical Research Unit, Ho Chi Minh City, Viet Nam), Stefan Niemann (Research Center Borstel, Borstel, Germany and German Center for Infection Research (DZIF), Hamburg-Lübeck-Borstel-Riems, Germany), Kayzad Soli Nilgiriwala (The Foundation for Medical Research, Mumbai, India), Camus Nimmo (University College London, London, UK), Nana Okozi (National Institute for Communicable Diseases, Johannesburg, South Africa), Rosangela Siqueira Oliveira (Institute Adolfo Lutz, São Paulo, Brazil), Shaheed Vally Omar (National Institute for Communicable Diseases, Johannesburg, South Africa), Nicholas Paton (National University of Singapore, Singapore), Timothy E.A. Peto (University of Oxford, Oxford, UK), Juliana Maira Watanabe Pinhata (Institute Adolfo Lutz, São Paulo, Brazil), Sara Plesnik (Institute of Microbiology and Laboratory Medicine, IMLred, WHO-SRL Gauting, Germany), Zully M. Puyen (Instituto Nacional de Salud, Lima, Peru), Marie Sylvianne Rabodoarivelo (Institut Pasteur de Madagascar, Antananarivo, Madagascar), Niaina Rakotosamimanana (Institut Pasteur de Madagascar, Antananarivo, Madagascar), Paola M.V. Rancoita (Vita-Salute San Raffaele University, Milan, Italy), Priti Rathod (Public Health England, Birmingham, UK), Esther Robinson (Public Health England, Birmingham, UK), Gillian Rodger (University of Oxford, Oxford, UK), Camilla Rodrigues (Hinduja Hospital, Mumbai, India), Timothy C. Rodwell (FIND, Geneva, Switzerland and University of California San Diego, CA, USA), Aysha Roohi (University of Oxford, Oxford, UK), David Santos-Lazaro (Instituto Nacional de Salud, Lima, Peru), Sanchi Shah (The Foundation for Medical Research, Mumbai, India), Thomas Andreas Kohl (Research Center Borstel, Borstel, Germany), Grace Smith (Public Health England, Birmingham, UK and UK Health Security Agency, London, UK), Walter Solano (Universidad Peruana Cayetano Heredia, Lima, Peru), Andrea Spitaleri (IRCCS San Raffaele Scientific Institute, Milan, Italy and Vita-Salute San Raffaele University, Milan, Italy), Philip Supply (University of Lille, CNRS, Inserm, CHU Lille, Institut Pasteur de Lille, U1019, UMR 9017, CIIL – Center for Infection and Immunity of Lille, Lille, France), Utkarsha Surve (Hinduja Hospital, Mumbai, India), Sabira Tahseen (National TB Reference Laboratory, National TB Control Program, Islamabad, Pakistan), Nguyen Thuy Thuong (Oxford University Clinical Research Unit, Ho Chi Minh City, Viet Nam), Guy Thwaites (Oxford University Clinical Research Unit, Ho Chi Minh City, Viet Nam and University of Oxford, Oxford, UK), Katharina Todt (Institute of Microbiology and Laboratory Medicine, IMLred, WHO-SRL Gauting, Gauting, Germany), Alberto Trovato (IRCCS San Raffaele Scientific Institute, Milan, Italy), Christian Utpatel (Research Center Borstel, Borstel, Germany), Annelies Van Rie (University of Antwerp, Antwerp, Belgium), Srinivasan Vijay (University of Edinburgh, Edinburgh, UK), Timothy M. Walker (University of Oxford, Oxford, UK and Oxford University Clinical Research Unit, Ho Chi Minh City, Viet Nam), A. Sarah Walker (University of Oxford, Oxford, UK), Robin Warren (Stellenbosch University, Cape Town, South Africa), Jim Werngren (Public Health Agency of Sweden, Solna, Sweden), Maria Wijkander (Public Health Agency of Sweden, Solna, Sweden), Robert J. Wilkinson (Wellcome Centre for Infectious Diseases Research in Africa, Cape Town, South Africa, Francis Crick Institute, London, UK and Imperial College London, London, UK), Daniel J. Wilson (University of Oxford, Oxford, UK), Penelope Wintringer (EMBL-EBI, Hinxton, UK), Yu-Xin Xiao (Taiwan Centers for Disease Control, Taipei, Taiwan), Yang (University of Oxford, Oxford, UK), Zhao Yanlin (Chinese Center for Disease Control and Prevention, Beijing, China), Shen-Yuan Yao (National Institute for Communicable Diseases, Johannesburg, South Africa) and Baoli Zhu (Institute of Microbiology, Chinese Academy of Sciences, Beijing, China).
Author contributions: All contributed laboratories, collected samples and provided data. D.W. Crook, T.E.A. Peto, S.J. Hoosdally, A.L. Gibertoni Cruz, A.S. Walker, T.M. Walker, P.W. Fowler and D.M. Cirillo designed the study. P.W. Fowler, S.J. Hoosdally and A.L. Gibertoni Cruz retrieved and analysed the MIC data. Z. Iqbal, M. Hunt, J. Knaggs and P.W. Fowler analysed all the genetic information. P.W. Fowler, A.S. Walker and T.M. Walker performed all analysis. P.W. Fowler wrote the manuscript with all partners offering feedback.
Conflict of interest: E. Robinson is employed by Public Health England and holds an honorary contract with Imperial College London. I.F. Laurenson is Director of the Scottish Mycobacteria Reference Laboratory. S. Niemann receives funding from German Center for Infection Research, Excellenz Cluster Precision Medicine in Chronic Inflammation, Leibniz Science Campus Evolutionary Medicine of the LUNG (EvoLUNG)tion EXC 2167. P. Supply is a consultant at Genoscreen. T.C. Rodwell is funded by the National Institutes of Health and Dept of Defense, and receives salary support from the non-profit organisation FIND. T.C. Rodwell is a co-founder, board member and shareholder of Verus Diagnostics Inc., a company that was founded with the intent of developing diagnostic assays. Verus Diagnostics was not involved in any way with data collection, analysis or publication of the results. T.C. Rodwell has not received any financial support from Verus Diagnostics. The University of California San Diego (UCSD) Conflict of Interest office has reviewed and approved T.C. Rodwell's role in Verus Diagnostics Inc. T.C. Rodwell is a co-inventor of a provisional patent for a TB diagnostic assay (provisional patent: 63/048.989). T.C. Rodwell is a co-inventor on a patent associated with the processing of TB sequencing data (European Patent Application 14840432.0 and USSN 14/912,918). T.C. Rodwell has agreed to “donate all present and future interest in and rights to royalties from this patent” to UCSD to ensure that he does not receive any financial benefits from this patent. S. Shah is working and holding employee stock ownership plans at HaystackAnalytics Pvt Ltd (Product: Using whole genome sequencing for drug-susceptibility testing for Mycobacterium tuberculosis). G.F. Gao is listed as an inventor on patent applications for RBD-dimer-based coronavirus vaccines. The patents for RBD-dimers as protein subunit vaccines for SARS-CoV-2 have been licensed to Anhui Zhifei Longcom Biopharmaceutical Co. Ltd, China.
Support statement: This work was supported by a Wellcome Trust/Newton Fund-MRC Collaborative Award (200205/Z/15/Z) and the Bill & Melinda Gates Foundation Trust (OPP1133541). Oxford CRyPTIC Consortium members are funded/supported by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre (BRC); the views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Dept of Health, and the NIHR Health Protection Research Unit in Healthcare Associated Infections and Antimicrobial Resistance, a partnership between Public Health England and the University of Oxford; the views expressed are those of the authors and not necessarily those of the NIHR, Public Health England or the Dept of Health and Social Care. J. Millard is supported by the Wellcome Trust (203919/Z/16/Z). Z. Yanlin is supported by the National Science and Technology Major Project, China (2018ZX10103001). K.M. Malone is supported by EMBL's EIPOD3 programme funded by the European Union's Horizon 2020 research and innovation programme under Marie Skłodowska Curie Actions. T.C. Rodwell is funded in part by funding from Unitaid (2019-32-FIND MDR). R. Siqueira Oliveira is supported by FAPESP (17/16082-7). L. Ferrazoli received financial support from FAPESP (2012/51756-5). B. Zhu is supported by the National Natural Science Foundation of China (81991534) and the Beijing Municipal Science and Technology Commission (Z201100005520041). N.T. Thuong is supported by the Wellcome Trust International Intermediate Fellowship (206724/Z/17/Z). G. Thwaites is funded by the Wellcome Trust. R. Warren is supported by the South African Medical Research Council. J. Carter is supported by the Rhodes Trust and Stanford Medical Scientist Training Program (T32 GM007365). A. Lalvani is supported by the NIHR Health Protection Research Unit in Respiratory Infections at Imperial College London. S. Grandjean Lapierre is supported by the Fonds de Recherche en Santé du Québec. C. Nimmo is funded by the Wellcome Trust (203583/Z/16/Z). A. Van Rie is supported by Research Foundation Flanders (FWO) under grant number G0F8316N (FWO Odysseus). G. Meintjes was supported by the Wellcome Trust (098316, 214321/Z/18/Z and 203135/Z/16/Z), and the South African Research Chairs Initiative of the Dept of Science and Technology and National Research Foundation (NRF) of South Africa (64787). The funders had no role in the study design, data collection, data analysis, data interpretation or writing of this report. The opinions, findings and conclusions expressed in this manuscript reflect those of the authors alone. L. Grandjean was supported by the Wellcome Trust (201470/Z/16/Z), the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (1R01AI146338), the GOSH Charity (VC0921) and the GOSH/ICH BRC (www.nihr.ac.uk). A. Brankin is funded by the NDM Prize Studentship from the Oxford Medical Research Council Doctoral Training Partnership and the Nuffield Dept of Clinical Medicine. D.J. Wilson is supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (101237/Z/13/B), and by the Robertson Foundation. A.S. Walker is an NIHR Senior Investigator. T.M. Walker is a Wellcome Trust Clinical Career Development Fellow (214560/Z/18/Z). A.S. Lachapelle is supported by the Rhodes Trust. R.J. Wilkinson receives funding from the Francis Crick Institute, which is supported by the Wellcome Trust (FC0010218), UKRI (FC0010218) and CRUK (FC0010218). T. Cohen has received grant funding and salary support from the National Institutes of Health (NIH), Centers for Disease Control and Prevention (CDC), US Agency for International Development (USAID) and Bill & Melinda Gates Foundation. The computational aspects of this research were supported by the Wellcome Trust Core Award (203141/Z/16/Z) and the NIHR Oxford BRC. Parts of the work were funded by the German Center of Infection Research (DZIF). The Scottish Mycobacteria Reference Laboratory is funded through National Services Scotland. The Wadsworth Center contributions were supported in part by cooperative agreement number U60OE000103 funded by the CDC through the Association of Public Health Laboratories and NIH/NIAID grant AI-117312. Additional support for sequencing and analysis was contributed by the Wadsworth Center Applied Genomic Technologies Core Facility and the Wadsworth Center Bioinformatics Core. SYNLAB Holding Germany GmbH for its direct and indirect support of research activities in the Institute of Microbiology and Laboratory Medicine Gauting. N. Rakotosamimanana thanks the Programme National de Lutte contre la Tuberculose de Madagascar.
References
- 1.World Health Organization . Global tuberculosis report 2000. 2020. www.who.int/publications/i/item/9789240013131 Date last accessed: 8 March 2022.
- 2.Enkirch T, Werngren J, Groenheit R, et al. . Systematic review of whole-genome sequencing data to predict phenotypic drug resistance and susceptibility in Swedish Mycobacterium tuberculosis isolates, 2016 to 2018. Antimicrob Agents Chemother 2020; 64: e02550-19. doi: 10.1128/AAC.02550-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gygli SM, Keller PM, Ballif M, et al. . Whole-genome sequencing for drug resistance profile prediction in Mycobacterium tuberculosis. Antimicrob Agents Chemother 2019; 63: e02175-18. doi: 10.1128/AAC.02175-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ransom EM, Potter RF, Dantas G, et al. . Genomic prediction of antimicrobial resistance: ready or not, here it comes! Clin Chem 2020; 66: 1278–1289. doi: 10.1093/clinchem/hvaa172 [DOI] [PubMed] [Google Scholar]
- 5.Phelan J, O'Sullivan DM, Machado D, et al. . The variability and reproducibility of whole genome sequencing technology for detecting resistance to anti-tuberculous drugs. Genome Med 2016; 8: 132. doi: 10.1186/s13073-016-0385-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The CRyPTIC Consortium and the 100,000 Genomes Project . Prediction of susceptibility to first-line tuberculosis drugs by DNA sequencing. N Engl J Med 2018; 379: 1403–1415. doi: 10.1056/NEJMoa1800474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker TM, Kohl TA, Omar SV, et al. . Whole-genome sequencing for prediction of Mycobacterium tuberculosis drug susceptibility and resistance: a retrospective cohort study. Lancet Infect Dis 2015; 15: 1193–1202. doi: 10.1016/S1473-3099(15)00062-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miotto P, Tessema B, Tagliani E, et al. . A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Eur Respir J 2017; 50: 1701354. doi: 10.1183/13993003.01354-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.World Health Organization . Catalogue of mutations in Mycobacterium tuberculosis complex and their association with drug resistance. 2021. www.who.int/publications/i/item/9789240028173 Date last accessed: 8 March 2022.
- 10.Walker TM, Miotto P, Köser CU, et al. . The 2021 WHO catalogue of Mycobacterium tuberculosis complex mutations associated with drug resistance: a genotypic analysis. Lancet Microbe 2022; 3: e265–e273. doi: 10.1016/S2666-5247(21)00301-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walker TM, Cruz AL, Peto TE, et al. . Tuberculosis is changing. Lancet Infect Dis 2017; 17: 359–361. doi: 10.1016/S1473-3099(17)30123-8 [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization . Report of the 16th meeting of the strategic and technical advisory group for tuberculosis. 2016. www.who.int/publications/m/item/report-of-the-16th-meeting-of-the-strategic-and-technical-advisory-group-for-tb Date last accessed: 8 March 2022.
- 13.Abuali MM, Katariwala R, LaBombardi VJ. A comparison of the Sensititre MYCOTB panel and the agar proportion method for the susceptibility testing of Mycobacterium tuberculosis. Eur J Clin Microbiol Infect Dis 2012; 31: 835–839. doi: 10.1007/s10096-011-1382-z [DOI] [PubMed] [Google Scholar]
- 14.Lee J, Armstrong DT, Ssengooba W, et al. . Sensititre MYCOTB MIC plate for testing mycobacterium tuberculosis susceptibility to first- and second-line drugs. Antimicrob Agents Chemother 2014; 58: 11–18. doi: 10.1128/AAC.01209-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu X, Ma YF, Jiang GL, et al. . Sensititre MYCOTB MIC plate for drug susceptibility testing of Mycobacterium tuberculosis complex isolates. Int J Tuberc Lung Dis 2016; 20: 329–334. doi: 10.5588/ijtld.15.0573 [DOI] [PubMed] [Google Scholar]
- 16.Xia H, Zheng Y, Zhao B, et al. . Assessment of a 96-well plate assay of quantitative drug susceptibility testing for Mycobacterium tuberculosis complex in China. PLoS One 2017; 12: e0169413. doi: 10.1371/journal.pone.0169413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rancoita PMV, Cugnata F, Gibertoni Cruz AL, et al. . Validating a 14-drug microtiter plate containing bedaquiline and delamanid for large-scale research susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother 2018; 62: e00344-18. doi: 10.1128/AAC.00344-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruesen C, Riza AL, Florescu A, et al. . Linking minimum inhibitory concentrations to whole genome sequence-predicted drug resistance in Mycobacterium tuberculosis strains from Romania. Sci Rep 2018; 8: 9676. doi: 10.1038/s41598-018-27962-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health Organization . Rapid Communication: key changes to treatment of multidrug-and rifampicin-resistant tuberculosis (MDR/RR-TB). 2018. www.who.int/tb/publications/2018/WHO_RapidCommunicationMDRTB.pdf Date last accessed: 8 March 2022.
- 20.European Committee for Antimicrobial Susceptibility Testing . MIC distributions and epidemiological cut-off value (ECOFF) setting. EUCAST SOP 10.0. 2017. www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/EUCAST_SOPs/EUCAST_SOP_10.0_MIC_distributions_and_epidemiological_cut-off_value__ECOFF__setting_20171117.pdf Date last accessed: 8 March 2022.
- 21.The CRyPTIC Consortium . Data and code to reproduce the figures and tables in the CRyPTIC ECOFF/ECV paper. 2022. www.github.com/fowler-lab/cryptic-ecoffs Date last accessed: 8 March 2022.
- 22.Ismail NA, Ismail F, Joseph L, et al. . Epidemiological cut-offs for Sensititre susceptibility testing of Mycobacterium tuberculosis: interpretive criteria cross validated with whole genome sequencing. Sci Rep 2020; 10: 1013. doi: 10.1038/s41598-020-57992-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipworth S, Jajou R, de Neeling A, et al. . SNP-IT tool for identifying subspecies and associated lineages of Mycobacterium tuberculosis complex. Emerg Infect Dis 2019; 25: 482–488. doi: 10.3201/eid2503.180894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gagneux S. Ecology and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol 2018; 16: 202–213. doi: 10.1038/nrmicro.2018.8 [DOI] [PubMed] [Google Scholar]
- 25.Turnidge J, Kahlmeter G, Kronvall G. Statistical characterisation of bacterial wild-type MIC value distributions and the determination of epidemiological cut-off values. Clin Microbiol Infect 2006; 12: 418–425. doi: 10.1111/j.1469-0691.2006.01377.x [DOI] [PubMed] [Google Scholar]
- 26.Amemiya T. Regression analysis when the dependent variable is truncated normal. Econometrica 1973; 41: 997. doi: 10.2307/1914031 [DOI] [Google Scholar]
- 27.Hurd M. Estimation in truncated samples when there is heteroscedasticity. J Econom 1979; 11: 247–258. doi: 10.1016/0304-4076(79)90039-3 [DOI] [Google Scholar]
- 28.Ghodousi A, Tagliani E, Karunaratne E, et al. . Isoniazid resistance in Mycobacterium tuberculosis is a heterogeneous phenotype composed of overlapping MIC distributions with different underlying resistance mechanisms. Antimicrob Agents Chemother 2019; 63: e00092-19. doi: 10.1128/AAC.00092-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahmad S, Mokaddas E, Al-Mutairi N, et al. . Discordance across phenotypic and molecular methods for drug susceptibility testing of drug-resistant Mycobacterium tuberculosis isolates in a low TB incidence country. PLoS One 2016; 11: e0153563. doi: 10.1371/journal.pone.0153563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farhat MR, Jacobson KR, Franke MF, et al. . Gyrase mutations are associated with variable levels of fluoroquinolone resistance in Mycobacterium tuberculosis. J Clin Microbiol 2016; 54: 727–733. doi: 10.1128/JCM.02775-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore DAJ, Evans CA, Gilman RH, et al. . Microscopic-observation drug-susceptibility assay for the diagnosis of TB. N Engl J Med 2006; 355: 1539–1550. doi: 10.1056/NEJMoa055524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clinical and Laboratory Standards Institute . M23: Development of In Vitro Susceptibility Testing Criteria and Quality Control Parameters. 5th Edn. Wayne, CLSI, 2018. [Google Scholar]
- 33.Clinical and Laboratory Standards Institute . M62: Performance Standards for Susceptibility Testing of Mycobacteria, Nocardia spp., and Other Aerobic Actinomycetes. 1st Edn. Wayne, CLSI, 2018. [PubMed] [Google Scholar]
- 34.World Health Organization . Updated interim critical concentrations for first-line and second-line DST. 2012. www.stoptb.org/wg/gli/assets/documents/Updated critical concentration table_1st and 2nd line drugs.pdf Date last accessed: 8 March 2022.
- 35.World Health Organization . Technical report on critical concentrations for drug susceptibility testing of medicines used in the treatment of drug-resistant tuberculosis. 2018. www.apps.who.int/iris/bitstream/handle/10665/260470/WHO-CDS-TB-2018.5-eng.pdf Date last accessed: 8 March 2022.
- 36.World Health Organization . Technical report on critical concentrations for drug susceptibility testing of isoniazid and the rifamycins (rifampicin, rifabutin and rifapentine). 2021. www.who.int/publications/i/item/technical-report-on-critical-concentrations-for-drugsusceptibility-testing-of-isoniazid-and-therifamycins-(rifampicin-rifabutin-and-rifapentine) Date last accessed: 8 March 2022.
- 37.Schön T, Juréen P, Giske CG, et al. . Evaluation of wild-type MIC distributions as a tool for determination of clinical breakpoints for Mycobacterium tuberculosis. J Antimicrob Chemother 2009; 64: 786–793. doi: 10.1093/jac/dkp262 [DOI] [PubMed] [Google Scholar]
- 38.Juréen P, Angeby K, Sturegard E, et al. . Wild-type MIC distributions for aminoglycoside and cyclic polypeptide antibiotics used for treatment of Mycobacterium tuberculosis infections. J Clin Microbiol 2010; 48: 1853–1858. doi: 10.1128/JCM.00240-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schön T, Juréen P, Chryssanthou E, et al. . Wild-type distributions of seven oral second-line drugs against Mycobacterium tuberculosis. Int J Tuberc Lung Dis 2011; 15: 502–509. doi: 10.5588/ijtld.10.0238 [DOI] [PubMed] [Google Scholar]
- 40.Köser CU, Maurer FP, Kranzer K. ‘Those who cannot remember the past are condemned to repeat it’: drug-susceptibility testing for bedaquiline and delamanid. Int J Infect Dis 2019; 80S: S32–S35. doi: 10.1016/j.ijid.2019.02.027 [DOI] [PubMed] [Google Scholar]
- 41.Schena E, Nedialkova L, Borroni E, et al. . Delamanid susceptibility testing of Mycobacterium tuberculosis using the resazurin microtitre assay and the BACTEC MGIT 960 system. J Antimicrob Chem 2016; 71: 1532–1539. doi: 10.1093/jac/dkw044 [DOI] [PubMed] [Google Scholar]
- 42.Ismail NA, Omar SV, Joseph L, et al. . Defining bedaquiline susceptibility, resistance, cross-resistance and associated genetic determinants: a retrospective cohort study. EBioMedicine 2018; 28: 136–142. doi: 10.1016/j.ebiom.2018.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaniga K, Aono A, Borroni E, et al. . Validation of bedaquiline phenotypic drug susceptibility testing methods and breakpoints: a multilaboratory, multicountry study. J Clin Microbiol 2020; 58: e01677-19. doi: 10.1128/JCM.01677-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ismail NA, Aono A, Borroni E, et al. . A multimethod, multicountry evaluation of breakpoints for bedaquiline resistance determination. Antimicrob Agents Chemother 2020; 64: e00479-20. doi: 10.1128/AAC.00479-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kahlmeter G, Cantón R, Giske CG, et al. . Re: In the name of common sense: EUCAST breakpoints and potential pitfalls. National dissemination of EUCAST guidelines is a shared responsibility. Clin Microbiol Infect 2020; 26: 1692–1693. doi: 10.1016/j.cmi.2020.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schön T, Matuschek E, Mohamed S, et al. . Standards for MIC testing that apply to the majority of bacterial pathogens should also be enforced for Mycobacterium tuberculosis complex. Clin Microbiol Infect 2019; 25: 403–405. doi: 10.1016/j.cmi.2019.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farhat MR, Sixsmith J, Calderon R, et al. . Rifampicin and rifabutin resistance in 1003 Mycobacterium tuberculosis clinical isolates. J Antimicrob Chemother 2019; 74: 1477–1483. doi: 10.1093/jac/dkz048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi W. Activity of pyrazinamide against Mycobacterium tuberculosis at neutral pH in PZA-S1 minimal medium. Antibiotics 2021; 10: 909. doi: 10.3390/antibiotics10080909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Michael A, Kelman T, Pitesky M. Overview of quantitative methodologies to understand antimicrobial resistance via minimum inhibitory concentration. Animals 2020; 10: 1405. doi: 10.3390/ani10081405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kahlmeter G, Turnidge J, Brown D. EUCAST General Consultation on “Considerations in the numerical estimation of epidemiological cutoff (ECOFF) values”. 2018. www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Consultation/2018/ECOFF_procedure_2018_General_Consultation_20180531.pdf Date last accessed: 8 March 2022.
- 51.Zabeti H, Dexter N, Libbrecht M, et al. . An interpretable classification method for predicting drug resistance in M. tuberculosis. bioRxiv 2020; preprint [ 10.1101/2020.05.31.115741]. doi: 10.1101/2020.05.31.115741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grazian C. Estimating MIC distributions and cutoffs through mixture models: an application to establish M. tuberculosis resistance. bioRxiv 2019; preprint [ 10.1101/643429]. doi: 10.1101/643429 [DOI] [Google Scholar]
- 53.Hunt M, Letcher B, Hall MB, et al. . Minos: principled variant adjudication and joint genotyping using genome graphs. bioRxiv 2021; preprint [ 10.1101/2021.09.15.460475]. [DOI] [Google Scholar]
- 54.Hunt M. Clockwork: pipelines for processing bacterial sequence data (Illumina only) and variant calling. 2021. www.github.com/iqbal-lab-org/clockwork Date last accessed: 8 March 2022.
- 55.Walker TM, Lalor MK, Broda A, et al. . Assessment of Mycobacterium tuberculosis transmission in Oxfordshire, UK, 2007–12, with whole pathogen genome sequences: an observational study. Lancet Respir Med 2014; 2: 285–292. doi: 10.1016/S2213-2600(14)70027-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fowler PW. Tuberculosis AMR catalogues in a standard grammar. 2021. www.github.com/oxfordmmm/tuberculosis_amr_catalogues Date last accessed: 8 March 2022.
- 57.Fowler PW. gumpy: genetics with numpy. 2020. www.github.com/oxfordmmm/gumpy Date last accessed: 8 March 2022.
- 58.Fowler PW. piezo: predicting the effect of a genetic mutation on an antibiotic. 2021. www.github.com/oxfordmmm/piezo Date last accessed: 8 March 2022.
- 59.Dotson DL, Seyler SL, Linke M, et al. . datreant: persistent, Pythonic trees for heterogeneous data. 2016. http://conference.scipy.org/proceedings/scipy2016/david_dotson.html Date last accessed: 8 March 2022.
- 60.Fowler PW, Gibertoni Cruz AL, Hoosdally SJ, et al. . Automated detection of bacterial growth on 96-well plates for high-throughput drug susceptibility testing of Mycobacterium tuberculosis. Microbiology 2018; 164: 1522–1530. doi: 10.1099/mic.0.000733 [DOI] [PubMed] [Google Scholar]
- 61.Fowler PW. AMyGDA. 2020. www.github.com/philipwfowler/amygda Date last accessed: 8 March 2022.
- 62.Fowler PW, Wright C, Spiers-Bowers H, et al. . A crowd of BashTheBug volunteers reproducibly and accurately measure the minimum inhibitory concentrations of 13 antitubercular drugs from photographs of 96-well broth microdilution plates. eLife 2022; 11: e75046. doi: 10.7554/eLife.75046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McKinney W. Data structures for statistical computing in Python. 2010. http://conference.scipy.org/proceedings/scipy2010/mckinney.html Date last accessed: 8 March 2022.
- 64.Harris CR, Millman KJ, van der Walt SJ, et al. . Array programming with NumPy. Nature 2020; 585: 357–362. doi: 10.1038/s41586-020-2649-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng 2007; 9: 90–95. doi: 10.1109/MCSE.2007.55 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary material ERJ-00239-2022.Supplement (3.4MB, pdf)
Supplementary data file ERJ-00239-2022.Data (14.5KB, tsv)
This one-page PDF can be shared freely online.
Shareable PDF ERJ-00239-2022.Shareable (231KB, pdf)