Abstract
We compared the performance of ID NOW™ COVID-19 assay nasal swabs with RT-PCR of nasopharyngeal swabs for SARS-CoV-2 in an outbreak setting, determining whether addition of RT-PCR of residual nasal swabs (rNS) (post ID NOW™ elution) would increase overall analytic sensitivity. Devices were placed at 2 long term and 1 acute care sites and 51 participants were recruited. Prospective paired nasopharyngeal and nasal samples were collected for RT-PCR and ID NOW™. ID NOW™ had a positive and negative categorical agreement of 86% and 93% compared to RT-PCR of nasopharyngeal swabs. Sensitivity and specificity of the ID NOW™ was 86% and 100%, positive and negative predictive value was 100% and 95% (COVID-19 positivity rate: 8%). Addition of rNS RT-PCR increased the positive and negative categorical agreement to 93% and 97%. Based on these results, we propose an alternative workflow which includes complementary testing of rNS on a secondary assay.
Keywords: ID NOW, sequencing, outbreak, COVID-19
1. Introduction
With the emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the continuing pandemic, there has been a global push to increase the availability and capacity of testing. In Vancouver, Canada, the majority of testing takes place in centralized laboratories using a combination of laboratory developed (LDT) reverse transcription real-time PCR (RT-PCR), high throughput commercial assays and limited decentralized rapid testing (i.e. GeneXpert® System, BioFire® FilmArray® panels).
In response to the pandemic, new strategies are being employed to rapidly respond to outbreak clusters, specifically in long-term care (LTC) settings. This need is evident in Canada where the attack rate is estimated at 37% and current case fatality rate in LTC is 26% to 29% (among the highest globally) representing 82% of coronavirus disease “2019″ (COVID-19) deaths nationally (average of 38% in developed nations) [1]. As near-patient testing becomes available, the paucity of published practical experience raises concerns about the feasibility of implementation and test performance in LTC.
The ID NOW™ COVID-19 assay (Abbott, IL) is an isothermal nucleic acid amplification near-patient test that is marketed as providing a qualitative result (positive, negative or invalid) in 15 minutes. The trade-off for this rapid result based on the manufacturer's protocol includes an absence of cycle threshold (CT) value, limited throughput and the unavailability of residual clinical sample for additional testing such as whole genome sequencing (WGS) for phylogenetic studies. The manufacturer reports a sensitivity and specificity of 93.3% and 98.4% respectively [2]; meanwhile published reports range from 71.7% to 89.1% sensitivity and 97.9% to 100% specificity as compared to nasopharyngeal swab testing using the CDC COVID-19 RT-PCR assay [3], [4], [5] or the Abbott RealTime SARS-CoV-2 assay performed on the Abbott m2000 system [6].
Dry nasal swabs are the recommended specimen type on this instrument; this provides an advantage in collection simplicity and tolerability to patients with cognitive impairment, which is particularly important in LTC [7], additionally throat and nasopharyngeal specimens are Health Canada and FDA approved. The product insert instructs the users to mix the direct nasal swab into the elution buffer for 10 seconds, then to discard the swab into a biohazard waste container. As previously published, ID NOW™ COVID-19 assay performance is decreased in specimens transported in Viral Transport Medium (VTM). One study reported a positive predictive agreement (sensitivity) of 66.7% [8] using these conditions compared to RT-PCR of nasopharyngeal swabs.
We describe the deployment of Abbott ID NOW™ instruments to 3 ongoing outbreaks to evaluate their performance as near-patient tests in both LTC and acute care settings. Nasal swabs were collected for direct ID NOW™ testing and compared to nasopharyngeal swabs concurrently collected and tested by RT-PCR as a reference standard. A new workflow is proposed to address concerns with the negative predictive value of this assay, where the nasal swab is tested using the ID NOW™ COVID-19 assay and the same residual nasal swab (rNS), postelution, is not discarded and then tested on a more sensitive secondary platform precluding the need for a nasopharyngeal swab. This pilot study provides a proof of concept to enable insights in the settings where this rapid test could be used to further strengthen public health response in outbreak situations.
2. Methods
During the week of November 22 to 28, 2020 the ID NOW™ was deployed to 3 outbreak settings (2 LTC and 1 acute care (AC) ward) in Vancouver, Canada. The 3 sites were selected after evidence of transmission was established:
-
•
LTC 1 – Ongoing large-scale outbreak with >80 positive residents and health care workers (HCW) in a 200 bed center in the second week of ongoing transmission and case detection. At the time of ID NOW™ deployment, a wide range of infection control measures had been enacted.
-
•
LTC 2 – Ring screening of asymptomatic residents following exposure (within 24–48 hours) to an initial positive resident case.
-
•
AC 1 – Ongoing outbreak with 4 positive patients in the first week in a hospital ward (30 patients, General Internal Medicine). ID NOW™ instruments were deployed onto the unit for near-patient testing.
The study participants were all adults (>18 years old) with potential exposure to COVID-19 as part of an outbreak in a LTC facility or in an AC setting. Patients were selected for testing based on current public health screening recommendations (symptomatic patients or known contacts) and/or identified by their care team after developing new symptoms consistent with COVID-19 (BC CDC Adult and Pediatric Testing Recommendations) [9] upon daily symptoms check and asked to provide an oral consent. HCWs self-identified on the basis of symptoms and were swabbed on demand or as part of ring screening for asymptomatic contacts. All included participants were tested for COVID-19 in parallel with a nasopharyngeal swab (NPS) to be tested by RT-PCR. Patients who were unable to provide consent or too agitated were excluded from this study. This study was reviewed and ethics were waved as a quality initiative by University of British Columbia Ethics Board.
Once consent was obtained from the patient or surrogate decision maker, a NPS (Yocon swab and VTM (Modified Hank's Solution), Yocon Biology, Beijing, China) was collected followed by a bilateral nasal swab by the study team (Puritan Sterile Foam Tipped Applicator; ID NOW™ Kit). The onset of symptoms was confirmed with the treating team prior to specimen collection and documented. Direct nasal swabs were tested immediately on site (<30 minutes from collection) using the ID NOW™ assay. The patients, the treating team and HCWs remained blinded to the ID NOW™ results. The post-elution nasal swab (rNS) was transported dry in its retained original packaging to the laboratory the same day and transferred into 3ml of VTM to be tested within 24 hours by RT-PCR.
The laboratory developed RT-PCR test, designed by the British Columbia Centre for Disease Control, targets the envelope (E) gene and the RNA-dependent RNA polymerase (RdRp) gene of SARS-CoV-2 [10,11]; results are considered positive with CT values <38 for both targets. The results from the ID NOW™ were compared to the results of paired NPS testing on the RT-PCR assay to determine agreement. Additionally, the ID NOW™ and rNS results were compared to the paired NPS tested by RT-PCR.
3. Results and discussion
A total of 51 individuals, including 39 LTC residents, 22 hospital patients and 3 HCWs (Table 1 ) were screened for COVID-19 using the ID NOW™ as a near-patient test and a paired NPS for routine RT-PCR testing. The study participants included 33 females (65%) and 18 males (35%). The LTC population was predominately female while evenly split in the AC ward. At the time of swabbing, LTC1 participants were symptomatic (<7 days), LTC2 participants were asymptomatic contacts and AC1 participants were mixed. Overall, there were 25 symptomatic (49%) and 26 asymptomatic (51%) participants. A total of 14 participants tested positive by NPS, with a range of detectable CT values from 16.26 to 37.4 for the 2 gene targets (Supplementary Table 1).
Table 1.
Participants demographics from the 3 testing sites.
| Total | LTC 1 | LTC 2 | AC1 | |||||
|---|---|---|---|---|---|---|---|---|
| Sex | ||||||||
| Female | 33 | 64.7% | 9 | 90% | 13 | 68% | 11 | 50% |
| Male | 18 | 35.3% | 1 | 10% | 6 | 32% | 11 | 50% |
| Age | ||||||||
| Mean | 80 | 80 | 84 | 77 | ||||
| Median | 84 | 89 | 86 | 75 | ||||
| Range | 21–102 | 29–102 | 65–101 | 21–100 | ||||
| Resident / HCW | ||||||||
| Resident | 48 | 94.1% | 8 | 80% | 19 | 100% | 21 | 93% |
| HCW | 3 | 5.9% | 2 | 20% | 0 | 0% | 1 | 7% |
| Clinical presentation | ||||||||
| Symptomatic | 25 | 49.0% | 10 | 100% | 0 | 0% | 15 | 68% |
| Asymptomatic | 26 | 51.0% | 0 | 0% | 19 | 100% | 7 | 32% |
Comparing the ID NOW™ to the NPS, the positive percent agreement (PPA) was 86% (CI: 57 - 98%) (12/14 samples) and the negative percent agreement (NPA) was 95% (CI: 86%–99%) (37/39 samples) (Table 2 ). There were two discordant nasal swabs. The first was on a patient with severe dementia that resulted in difficult collection. The second discordant swab was on a patient that tested positive near the limit of detection (LOD) for the NPS; as per the manufacturer, ID NOW™ performs best on samples with a CT< 33.
Table 2.
Results and agreement between ID NOW™ COVID-19 nasal swab alone and combined with the residual nasal swab (rNS) RT-PCR compared to the nasopharygeal swab (NPS) RT-PCR; Brackets include confidence internals at 95%.
| NPS | ID NOW | Agreement | ID NOW ± rNS | Agreement | |
|---|---|---|---|---|---|
| LTC 1 | |||||
| Positive | 7 | 5 | 71% (29%–96%) | 6 | 83% (42%–99%) |
| Negative | 3 | 5 | 60% (15%–95%) | 4 | 80% (19%–99%) |
| LTC 2 | |||||
| Positive | 0 | 0 | 100% | 0 | 100% |
| Negative | 19 | 19 | 100% (97%–100%) | 19 | 100% (97%–100%) |
| AC 1 | |||||
| Positive | 7 | 7 | 100% (97%–100%) | 7 | 100% (97%–100%) |
| Negative | 15 | 15 | 100% (97%–100%) | 15 | 100% (97%–100%) |
| Total | |||||
| Positive | 14 | 12 | 86% (57%–98%) | 13 | 93% (66%–99%) |
| Negative | 37 | 39 | 95% (83%–99%) | 38 | 97% (86%–99%) |
ID NOW™ results combined with the rNS RT-PCR results showed a 93% (CI: 66%–99%) (13/14) positive and 97% (CI: 83%–99%) (38/39) negative percent agreement (Table 2) when compared to the NPS RT-PCR. For the single discordant result obtains for the rNS, the paired NPS was at the LOD for the assay (negative on the rNS, Patient 14, (Supplementary Table 1). The addition of the rNS allowed for the detection of Patient 4 that was negative on ID NOW™. This false negative result might be attributable to a poorly collected nasal swab; the rNS was positive close to the limit of detection (LOD) while the NPS was positive. This discrepancy could also be due to the disease evolution where the NS cannot be compared to NPS (Supplementary Table 1).
A calculation of the testing performance of the ID NOW࣪ compared to the RT-PCR as a reference standard showed a sensitivity and specificity of 85.7% and 100% respectively, and PPV and NPV of 100% and 94.9% (Table 3 ). While a small sized data set, this pilot study is an early indication that this platform has an acceptable performance as a near-patient assay [3,4,8].
Table 3.
(A) Test performance characteristics of ID NOW™ COVID 19 nasal swab compared to the nasopharyngeal swab RT-PCR (B) test performance of residual nasal swab RT-PCR compared to nasopharyngeal swab RT-PCR.
| Nasopharyngeal LDT RT-PCR |
Nasopharyngeal LDT RT-PCR |
||||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||||
| ID NOW: COVID 19 | Positive | 12 | 0 | Residual Nasal | Positive | 13 | 0 |
| Negative | 2 | 37 | LDT RT-PCR | Negative | 1 | 38 | |
| Sensitivity | 0.86 | Sensitivity | 0.93 | ||||
| Specificity | 1.00 | Specificity | 1.00 | ||||
| PPV | 1.00 | PPV | 1.00 | ||||
| NPV | 0.95 | NPV | 0.97 | ||||
The performance characteristics for the ID NOW࣪ when combined with the rNS RT-PCR, results in a sensitivity of 93% (83% for the ID NOW™ alone), and specificity was 100%, PPV was 100% and NPV was 97% (95% for ID NOW™ alone) when compared to the NPS as a reference (Table 3).
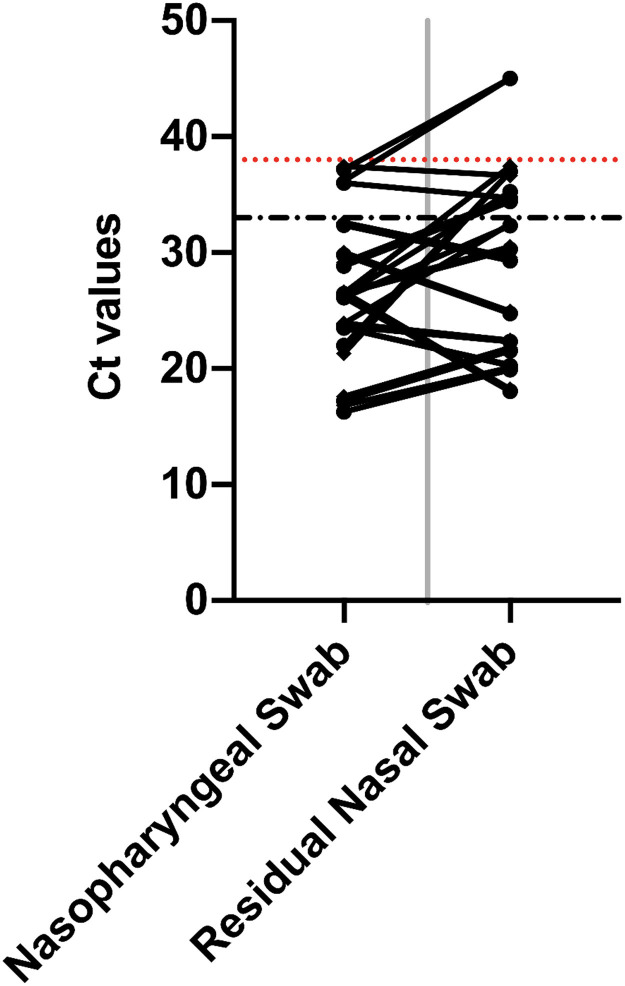
Unique to this study is the use of the rNS as a secondary detection method by RT-PCR. It demonstrated better categorical agreement with the NPS RT-PCR compared to the ID NOW™ alone. An analysis of the CT values for the 2 gene targets of the LDT RT-PCR showed that they were detected on average at similar values: RdRp average of 25.74 for NPS compared to 27.71 for the rNS (average ΔCT of 1.97, range -8.1 to 14.98) and E gene average of 25.83 for NPS compared to 28.17 for the rNS (average Δ 2.34 (Range -8.37 to 15.68) with no statistical difference when comparing the NPS and rRS for the 2 gene targets (RdRp p 0.48, E Gene p 0.45) (Fig. 1 , Supplementary Table 1). The shift in CT averages can be attributed to a dilution effect as the nasal swab is first eluted in the manufacture's assay buffer then a second time in VTM prior to the LDT RT-PCR.
Fig. 1.
The CT results of the RdRp and E Gene detected by LDT RT-PCR from the diagnostic nasopharyngeal swab (NPS) compared to the residual nasal swab (rNS); (Red Hatch line – LOD of LDT RT-PCR (CT 37), Black Hatch Line – LOD of ID NOW™ COVID 19 Assay) (CT 33 as per manufacturer); CT 45 represents non detection of target. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In the 3 situations used in this evaluation (ring screening at LTC, on-going outbreak at LTC, and on-going outbreak in an AC), we found that the ID NOW™ performed similarly to published studies that employed a prospective design [3,4,6]. This near-patient strategy provides a reduction in diagnostic delays, thereby allowing rapid decisions with respect to cohorting and prompt implementation of infection control measures. Furthermore, the combined results of direct NS and rNS demonstrated good agreement with traditional NPS in patients who recently developed symptoms. This new workflow alleviates the need for repeat NPS testing for result confirmation of positive patients. This assay requires minimal technical training, allowing for deployment and training of staff on site where its greatest benefit would be found.
The strength of this study is the prospective placement of the devices at the sites of outbreak as a near-patient test, thereby providing results, without transport delays, within 15 minutes. The study included symptomatic patients (< 7 days), as well as asymptomatic patients identified as high-risk exposure. Our proposed workflow (See supplementary Fig. 1) would provide an actionable positive result at the time of initial testing. Preserving and retesting the rNS by RT-PCR provides CT values and specimen for additional analysis such as WGS without the need for an additional NPS collection. WGS for phylogenetic outbreak investigation was successfully done on a few of the rNS [data not shown]. In situations where negative results require additional adjudication, the rNS RT-PCR would provide that result without the need for additional NPS collection. In terms of patient experience and ease of collection, the nasal swab is preferable to the NPS.
The limitation of the study is the small sample size of tested patients. The primary drawback of our proposed workflow is the requirement for sterile packaging as well as VTM for the residual nasal swab transportation, which is not provided by the manufacturer.
This study has demonstrated that despite the ID NOW™ COVID19 manufacturers’ protocol the nasal swab post-elution is still quite valuable and should be considered for complementary testing in the diagnostic process and could address some of the sensitivity concerns.
Declaration of competing interest
DFS reports honoraria outside of the submitted work from Alimentiv Inc, Pfizer, Amgen, Merck and Diaceutics, and stock ownership from Satisfai Health Inc. The authors report no conflicts of interest relevant to this article.
Author contributions
Clayton MacDonald: Formal analysis, Investigation. Writing - Original Draft. Claudine Desruisseaux: Investigation, Writing - Review and Editing. Eric Eckbo: Investigation, Writing - Review and Editing. Lisa Li: Investigation, Writing - Review and Editing. Kerstin Locher: Methodology, Writing - Review and Editing. Titus Wong: Writing - Review and Editing. Jennifer Grant: Writing - Review and Editing. Valery Lavergne: Writing - Review and Editing. David F. Schaeffer: Writing - Review and Editing. Linda M. N. Hoang: Writing - Review and Editing. Marthe Charles: Conceptualization, Methodology, Formal Analysis, Writing - Original Draft.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.diagmicrobio.2022.115832.
Appendix. Supplementary materials
References
- 1.Hsu A, Lane N. Report: understanding the impact of COVID-19 on residents of Canada's long-term care homes — ongoing challenges and policy responses2020. Article in LTCcovid.org, International Long-Term Care Policy Network, CPEC-LSE, 4 June 2020.
- 2.Abbot. Abbott releases ID NOW™ COVID-19 interim clinical study results from 1,003 people to provide the facts on clinical performance and to support public health. Abbot Online: Abbot Media; 2020.
- 3.Moore NM, Li H, Schejbal D, Lindsley J, Hayden MK. Comparison of two commercial molecular tests and a laboratory-developed modification of the CDC 2019-nCoV reverse transcriptase PCR assay for the detection of SARS-CoV-2. J Clin Microbiol. 2020;58(8) doi: 10.1128/JCM.00938-20. e00938-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smithgall MC, Scherberkova I, Whittier S, Green DA. Comparison of Cepheid Xpert Xpress and Abbott ID Now to Roche cobas for the rapid detection of SARS-CoV-2. J Clin Virol. 2020;128 doi: 10.1016/j.jcv.2020.104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stokes W, Berenger BM, Takshveer S, Adeghe I, Schneider A, King DT, et al. Acceptable performance of the Abbott ID NOW among symptomatic individuals with confirmed COVID-19. medRxiv. 2020 doi: 10.1101/2020.12.24.20248786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrington A, Cox B, Snowdon J, Bakst J, Ley E, Grajales P, et al. Comparison of Abbott ID Now and Abbott m2000 methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from symptomatic patients. J Clin Microbiol. 2020;58(8):e00798–20. doi: 10.1128/JCM.00798-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frazee BW, Rodríguez-Hoces de la Guardia A, Alter H, Chen CG, Fuentes EL, Holzer AK, et al. Accuracy and discomfort of different types of intranasal specimen collection methods for molecular influenza testing in emergency department patients. Ann Emerg Med. 2018;71(4):509–517. doi: 10.1016/j.annemergmed.2017.09.010. e1. [DOI] [PubMed] [Google Scholar]
- 8.Basu A, Zinger T, Inglima K, Woo K-M, Atie O, Yurasits L, et al. Performance of Abbott ID Now COVID-19 rapid nucleic acid amplification test using nasopharyngeal swabs transported in viral transport media and dry nasal swabs in a New York City Academic Institution. J Clin Microbiol. 2020;58(8) doi: 10.1128/JCM.01136-20. e01136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.BC CDC Online (Available from: http://www.bccdc.ca/Health-Professionals-Site/Documents/BCCDC_PHL_Updated_nCoV_Lab_Guidance.pdf) Accessed: November 15, 2020.
- 10.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gadkar VJ, Goldfarb DM, Young V, Watson N, Hoang L, Lee T, et al. Gargle-direct: extraction-free detection of SARS-CoV-2 using real-time PCR (RT-qPCR) of saline gargle rinse samples. medRxiv. 2020 2020.10.09.20203430. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.