Abstract
Genetic predisposition and disruption of host gut microbiota and immune system can result in inflammatory bowel disease (IBD). Here, we show that miRNA-149-5p (miR-149-5p) and miRNA-149-3p (miR-149-3p) play crucial roles in IBD. Mice lacking miR-149-3p were considerably more susceptible to dextran sulfate sodium (DSS)-induced colitis than wild-type (WT) mice, accompanied by more serious inflammatory symptoms and increased gene expression of certain inflammatory cytokines. Both miR-149-5p and miR-149-3p suppressed colon inflammatory response in vitro and in vivo. Furthermore, we found significant differences in the composition of the gut microbiota between WT and miR-149-3p−/− mice by 16S rRNA sequencing. Co-housing endowed susceptibility to WT mice against DSS-induced colitis compared with the WT control group. However, susceptibility of miR-149-3p−/− mice against DSS-induced colitis was still present after antibiotic treatment. These findings suggest that the deletion of miR-149-3p altered gut microbiota and influenced pathogenesis of intestinal inflammation, but sensitivity of miR-149-3p−/− mice to DSS-induced colitis is not conferred by microbiota. In addition, we identified the roles of miR-149-5p and miR-149-3p in colon inflammation, which may serve as an attractive therapeutic tool for colitis or IBD, and even colitis-associated carcinoma.
Keywords: MT: Non-coding RNAs, miR-149-3p, miR-149-5p, colitis, IBD, gut microbiota
Graphical abstract

Feng et al. demonstrated that the deletion of miR-149-3p altered the gut microbiota, which may be one of the factors that aggravate DSS-induced colitis in miR-149-3p−/− mice. miR-149-5p and miR-149-3p suppressed colon inflammation in vitro and in vivo, which may serve as an attractive therapeutic tool for IBD.
Introduction
Inflammatory bowel disease (IBD) is generally classified into ulcerative colitis (UC) and Crohn’s disease (CD), and is a chronic heterogeneous group of diseases in the gastrointestinal tract.1,2 From a clinical perspective, IBD is not a life-threatening disease in many patients, but it is a life-long burden that frequently relapses.3 Multiple inflammatory pathways, genetic variations, and the gut microbiota have been identified as the main etiopathogenesis of IBD.2,4,5 A growing number of investigations have revealed that patients with IBD have increased risk of colorectal cancer (CRC).6 Thus, it remains imperative to understand the pathogenetic mechanisms of IBD and develop effective strategies for the management of IBD.
Adenosine monophosphate-activated kinase (AMPK), as a heterotrimeric complex, is a pivotal molecule that maintains energy metabolism.7 There is growing evidence supporting that AMPK phosphorylation (Thr-172) in the α-subunit plays an extensive role in prevention of diseases, including diabetes and multiple cancers.8, 9, 10, 11, 12 Recent studies support that activation of AMPK may function as a protector in gut epithelial integrity, barrier function, and gut immunity.13, 14, 15, 16, 17 Consistent with this notion, the gut microbial metabolite butyrate may contribute to maintaining the intestinal barrier via activation of AMPK.18 In response to dextran sulfate sodium (DSS)-induced colitis, metformin may impair pathological damages in the intestine in an AMPK-dependent manner.7 These past studies suggest that AMPK activation can be a therapeutic strategy for intestinal disease and IBD.
Activated NF-κB is a ubiquitous nuclear transcription factor in multiple immune diseases, including IBD.19,20 The transcription factors of the NF-κB family, RelA, RelB, c-Rel, NF-κB1 (p50), and NF-κB2 (p52), form homo- and heterodimers, and then regulate the expression of the genes with κB sites in their promoter region. In response to diverse stimuli, including tumor necrosis factor (TNF) receptor superfamily members (lipopolysaccharide [LPS], TNF, double-stranded RNA and others), pattern-recognition receptors, various cytokine receptors, T cell receptor and B cell receptor,21 IKBα is phosphorylated by a multi-subunit IκB kinase complex, resulting in nuclear translocation of canonical NF-κB members and induction of the expression of proinflammatory genes.22,23 In IBD, activated NF-κB upregulating the expression of many proinflammatory cytokines and chemokines, is highly dynamic in the IBD pathogenesis and may serve as a promising therapeutic target or diagnostic marker.24, 25, 26, 27, 28, 29 In addition, some inhibitors for the NF-κB signaling pathway have been used for alleviating IBD in vivo.30,31 Thus, discovering novel therapeutic targets that block NF-κB activation would be valuable for developing novel appropriate therapeutic strategies for IBD.
The gut microbiota, which colonize the gastrointestinal tract of the host, is a major regulatory factor in intestinal diseases.5 Generally, the gut microbiota play a role in maintenance of physiology status through regulating genes, proteins, or metabolites, such as the major metabolite short-chain fatty acid formed by anaerobic fermentation, which is normally absorbed by the intestinal mucosa as an energy source, or alternatively modulates gene expression to affect host physiology.5,32, 33, 34 Typically, butyrate is involved in inhibiting histone deacetylases and NF-κB activation, activating HIF-1 and SP-1, increasing intestinal epithelial barrier function and reducing inflammation.33,34 MicroRNAs (miRNAs) are short (about 18–25 bp) and non-coding RNAs. Generally, the guide strand directly mediates the RNA-induced silencing complex (RISC, miRNA duplex binds with Argonaute) to the 3′ UTR complementary region of the target mRNAs, and binds to the complementary region by incomplete complement or complete complement, thereby causing translation inhibition or degradation of the target mRNAs.35,36 Functional studies indicate that miRNAs are post-transcriptional regulators of inflammation and cancer through targeting mRNAs.37 Both the gut microbiota and miRNAs may function as the regulators in intestinal inflammation. Furthermore, miRNAs may regulate intestinal disease, accompanied with changes in the intestinal microbial composition. For example, Johnston et al. reported that loss of miRNA-21 reduced the susceptibility of mice to DSS-induced colitis via influencing the gut microbiota.38 Runtsch et al. found that miRNA-146a deficiency in mice heightens resistance to DSS-induced colitis and shapes the gut microbiota.39 Therefore, miRNAs could represent a potential target for treatment of IBD and inflammation-associated colon carcinogenesis.
miRNA-149-5p (miR-149-5p) and miRNA-149-3p (miR-149-5p) are located in the first intron of the glypican-1 (GPC1) gene on the genome. After miRNA processing and synthesis, the sense strand is processed and matured into miR-149-5p (has: UCUGGCUCCGUGUCUUCACUCCC), while the antisense strand is processed to mature miR-149-3p (has: AGGGAGGGACGGGGGCUGUG) and is highly conserved across species.40 Some reports on the functions of miR-149-3p and miR-149-5p in different diseases, including cancer and inflammation, have been published. Previous studies have indicated that miR-149-5p plays a tumor-suppressing role in CRC.41,42 Also, the function of miR-149-3p has been clarified as a suppressor in breast cancer and bladder cancer, and recent research has reported that enterotoxigenic Bacteroides fragilis participates in cancer and inflammation by regulating exosome-packaged miR-149-3p.43, 44, 45 In addition, our recent publication supported the role of miR-149-3p in liver inflammation by negatively regulating STAT3-mediated cell signaling.46 Based on this, we reasoned that it would be important to identify the role of miR-149-3p in colitis and colon carcinogenesis. In this study, we showed that deletion of miR-149-3p in mice (miR-149-3p−/−) reduced resistance to DSS administration compared with WT mice. The survival rate of miR-149-3p−/− mice was low after DSS administration, and the colitis characteristics were more serious in miR-149-3p−/− mice. Also, miR-149-3p deficiency influenced the composition of gut microbiota. In addition, we revealed that miR-149-3p activated AMPK signaling and inhibited NF-κB signaling in vitro and in vivo. Overall, this study showed that the deletion of miR-149-3p in mice shaped the gut microbiota composition and increased sensitivity to DSS-induced colitis.
Results
miR-149-3p−/− mouse colon tissue displayed increased level of proinflammatory genes
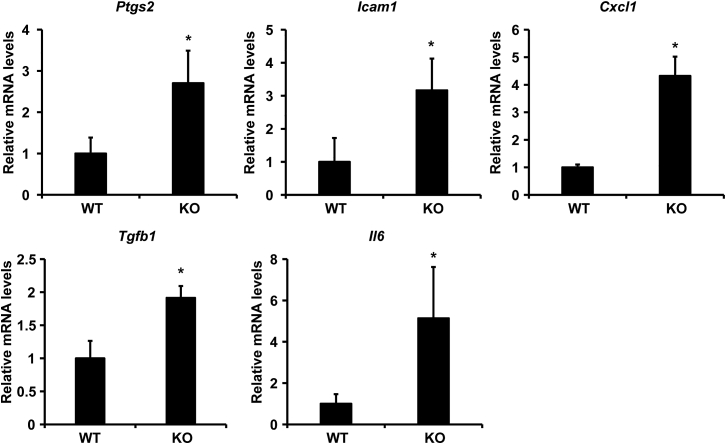
Our recent report supported the role of miR-149-3p in liver inflammation.46 To verify the effects of miR-149-3p on colonic inflammatory response, of which outcomes are affected by miR-149-3p deletion, we firstly examined the mRNA levels of proinflammatory genes derived from WT and miR-149-3p−/− (KO) mouse colons. Deficiency of miR-149-3p in the colon showed upregulated expression levels of inflammation-related genes compared with WT controls (Figure 1). These upregulated genes include cyclooxygenase-2 (Ptgs2), intercellular cell adhesion molecule-1 (Icam1), C-X-C motif chemokine (Cxcl1), transforming growth factor-β1 (Tgfb1), and interleukin-6 (Il6) (∗p < 0.05). These results indicate that miR-149-3p deletion may cause colonic inflammation.
Figure 1.
Deficiency of miR-149-3p increased expression of proinflammatory genes in mouse colon tissues
n = 5. WT, wild-type mice; KO, miR-149-3p−/− mice. ∗p < 0.05 versus the WT groups.
miR-149-3p−/− mice were more sensitive to DSS toxicity
Next, we explored the function of miR-149-3p in a long-term DSS-induced mouse model. WT mice and miR-149-3p−/− mice were supplied with 2% DSS for 7 consecutive days followed by 7 days of drinking water. Unexpectedly, all miR-149-3p−/− mice died in the first cycle, while all WT mice survived (Figure 2A).
Figure 2.
miR-149-3p−/− (KO) mice are more sensitive to DSS-induced colitis
(A) Percentage of death from WT (n = 8) and miR-149-3p−/− (KO) mice (n = 17), observed for 11 days after administration of 2% DSS (2% DSS for 7 consecutive days followed by drinking water). (B) WT and miR-149-3p−/− (KO) male mice at 8 weeks of age were weighed on day 0 and given drinking water (control group) or 2% DSS (w/v) ad libitum for 7 days (n = 5). Disease activity index (DAI) of each mouse was calculated every other day. (C) After 7 days of DSS administration, the weight of mice was recorded and weight reduction was calculated. (D–F) The colon of WT and KO was removed and photographed (D), and colon length (E), and colon weight (F) of each mouse were recorded. (G) Representative hematoxylin and eosin staining of colonic tissues from water- and 2% DSS-treated WT and KO mice are shown (magnification, ×200). (H) Relative mRNA levels of proinflammatory genes of colon from WT and KO mice after DSS administration. WT, wild-type mice; KO, miR-149-3p−/− mice. ∗p < 0.05.
Next, based on the above mortality differences, we determined whether the genetic deletion of miR-149-3p affected DSS-induced colitis. WT and miR-149-3p−/− mice were treated with drinking water (control group) or 2% DSS (w/v) ad libitum for 7 days. Mice were weighed on day 0 and disease activity index (DAI) of each mouse was tested every other day. In detail, DAI = 0, normal stool, no blood, and no body weight reduction; DAI = 1, 1%–3% body weight reduction; DAI = 2, loose stool, blood visible in stool, and 3%–6% body weight reduction; DAI = 3, 6%–9% body weight reduction; and DAI = 4, diarrhea-type stools and gross bleeding, and >9% body weight reduction. After 7 days of DSS administration, weight reduction, colon length, and colon weight of each mouse were recorded and the DAI scores were calculated. We found that miR-149-3p−/− mice had more severe colitis symptoms in response to DSS-induced acute colitis than WT mice. Compared with WT mice (0.2 ± 0.4), miR-149-3p−/− mice showed a significantly increased DAI score on day 5 (1.67 ± 0.47) (Figure 2B). After 7 days of DSS administration, deficiency of miR-149-3p resulted in an approximately 2.9-fold weight reduction relative to WT mice (Figure 2C). Moreover, the miR-149-3p−/− colons were shorter (4.40 ± 0.78 cm) and lighter (0.13 ± 0.02 g) in response to DSS treatment compared with WT mice (6.22 ± 0.34 cm; 0.18 ± 0.02 g) (Figures 2D–2F).
From histological analysis of colonic sections, the colons of control KO mice had reduced mucosal thickness, reduced and uneven distribution of functional layers, hyperplasia of the basal layer, and proliferation of fibroblasts in the mucosal stroma and submucosa with lymphocyte infiltration compared with the WT control group (Figure 2G). Mucosal space was enlarged and the crypt structure was deformed in DSS-treated WT colons. In contrast, DSS-treated miR-149-3p−/− colons had a disintegrative loss of lamina propria and the crypt structure was no longer visible, accompanied by inflammatory cell infiltration. Overall, deficiency of miR-149-3p resulted in more serious colon damage than WT mice in DSS-induced colitis.
To further address whether deficiency of miR-149-3p promotes colonic inflammation after DSS treatment, we examined expression of the proinflammatory cytokines in mouse colons. We found that DSS administration significantly upregulated interleukin-2 (Il2), interleukin-6 (Il6), monocyte chemotactic protein-1 (Ccl2), and transforming growth factor-β2 (Tgfb2) (∗p < 0.05) in miR-149-3p−/− colons (Figure 2H). Overall, these data indicate that deficiency of miR-149-3p promoted incidence of colonic inflammation and colitis in DSS-induced mouse model.
miR-149-3p−/− mouse colon tissues displayed elevated NF-κB activity and reduced AMPK activity
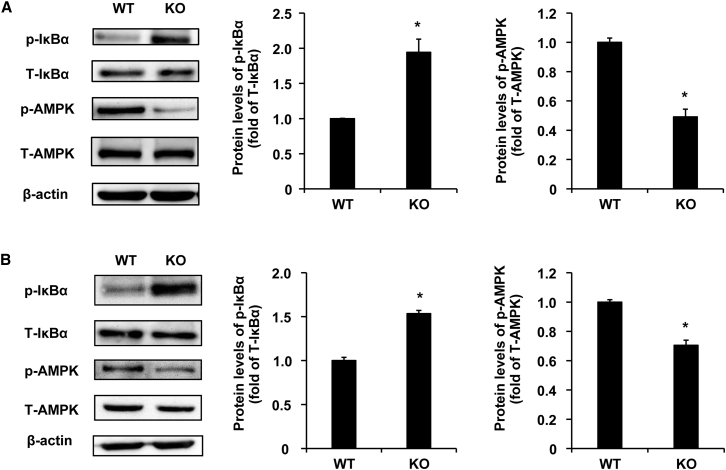
Our previous work explored the role of miR-149-3p in the mouse liver and found that miR-149-3p−/− mice had enhanced LPS-induced STAT3 phosphorylation levels compared with WT liver.46 In the current study, we found that some proinflammatory genes such as Ptgs2, Icam1, Cxcl1, Tgfb2, and Il6 were upregulated in the colon of miR-149-3p−/− mice (∗p < 0.05) (Figure 1), and the expression of these genes is regulated by NF-κB signaling. To understand the mechanism of miR-149-3p in the regulation of colonic inflammatory responses, we performed immunoblot analysis to examine the phosphorylation levels of IκBα in the colon of 8-week-old miR-149-3p−/− mice. We observed that the phosphorylation levels of IκBα were significantly elevated in miR-149-3p−/− mice colons compared with WT mice (Figure 3A).
Figure 3.
miR-149-3p−/− mouse colon tissues display elevated NF-κB activity and reduced AMPK activity
(A) The phosphorylated IκBα (p-IκBα) levels are upregulated and phosphorylated AMPK (p-AMPK) levels are downregulated in young KO colon compared with WT colon. Total IκBα (T-IκBα) and total AMPK (T-AMPK) were used for normalization of protein levels (n = 5). (B) Immunoblot analysis for the phosphorylated IκBα (p-IκBα) and phosphorylated AMPK (p-AMPK) in total protein pools from aged WT and miR-149-3p−/− mouse (KO) colon. Total IκBα (T-IκBα) and total AMPK (T-AMPK) were used for normalization of protein levels (n = 5). WT, wild-type mice; KO, miR-149-3p−/− mice. ∗p < 0.05.
Previous studies have addressed the protective role of activated AMPK in gut epithelial integrity and barrier function.13,14 Here, we examined the phosphorylation levels of AMPK in 8-week-old miR-149-3p−/− mouse colons. Deficiency of miR-149-3p resulted in 2.3-fold reduction in the phosphorylation levels of AMPK relative to the WT group (Figure 3A). Moreover, the phosphorylation levels of IκBα were significantly elevated and the phosphorylation levels of AMPK were significantly weakened in the colons of aged miR-149-3p−/− mice (15 months old) (Figure 3B). These results suggested that the deletion of miR-149-3p may impair barrier function and aggravate colitis through activating NF-κB signaling and reducing AMPK activity.
miR-149-5p and miR-149-3p regulated colon inflammatory response in vivo
To investigate the roles of miR-149-5p and miR-149-3p in colon inflammatory development, Firstly, we examined the responses of miR-149-3p and miR-149-5p to LPS-induced colonic inflammation. LPS can induce systemic inflammation47,48 and is also used for colonic inflammation model in vivo.49 Mice were given miRNA agomirs by a tail vein injection for 48 h. And then mice were fasted overnight and were injected intraperitoneally with LPS (30 mg/kg body weight). At 6 h after LPS exposure, mice were sacrificed (Figure S1A). In our present study, LPS increased the mRNA levels of proinflammatory genes Cxcl10, Il6, Ccl2, Mmp13, Ccl5, and Mmp12 (∗p < 0.05). miR-149-5p agomirs repressed the expression of LPS-induced Cxcl10, Il6, Ccl2, and Mmp13 in mouse colons (Figure S1B). Moreover, miR-149-3p agomirs downregulated LPS-induced Il6, Ccl5, Mmp13, Mmp12, and Ccl2 levels (∗p < 0.05) (Figure S1C). These results collectively demonstrate that both miR-149-5p and miR-149-3p suppressed LPS-induced colon inflammatory responses in vivo.
Next, WT mice were injected with miR-149-3p agomirs or miR-149-5p agomirs before DSS-induced colon inflammatory response. In the DSS-induced colitis model, WT mice were administered miRNA agomirs on days 0 and 4, and 2% DSS was administered from day 1 to induce colitis, and the inflammatory response was analyzed on day 7 (Figure 4A). Both WT mice treated with miR-149-3p agomirs or miR-149-5p agomirs had significantly lower DAI scores (1.83 ± 0.41 or 1.71 ± 0.49) compared with WT mice treated with negative control (NC) agomirs (3.00 ± 0.45) on day 7 (Figure 4B). And miR-149-3p agomir or miR-149-5p agomir treatment alleviated weight lost induced by DSS (Figure 4C). Moreover, the colon lengths and weights in mice treated with miR-149-3p agomirs (5.82 ± 0.59 cm; 0.16 ± 0.01 g) or miR-149-5p agomirs (6.27 ± 0.83 cm; 0.17 ± 0.01 g) were longer and heavier, respectively, in response to DSS treatment compared with those of the NC agomir with DSS-treated group (4.77 ± 0.01 cm; 0.11 ± 0.01 g) (Figures 4D–4F). Hematoxylin and eosin (H&E) staining to analyze the histopathological changes of mouse colons, compared with the NC agomirs with DSS treatment, both miR-149-3p and miR-149-5p agomirs reduced the area of inflammatory cell infiltration (Figure 4G). Moreover, both miR-149-3p and miR-149-5p agomirs significantly inhibited Ptgs2, Cxcl1, Icam1, Nos2, Cxcl10, Ccl2, Mmp13, Mmp2, Tgfb1, and Tgfb2 mRNA levels induced by DSS, and miR-149-5p agomirs also inhibited Tnfa induced by DSS (Figure S2). These results indicated that overexpression of both miR-149-3p and miR-149-5p could alleviate DSS-induced colitis in vivo.
Figure 4.
miR-149-5p and miR-149-3p agomirs regulate DSS-induced colitis in vivo
(A) Schematic diagram of the experimental process. WT mice were administered miRNA agomirs on days 0 and 4, and 2% DSS was administered from day 1 to induce colitis, and the inflammatory response was analyzed on day 7. (B) Disease activity index (DAI) was calculated every other day. (C) After 7 days of DSS administration, weight reduction (g) was calculated. (D) After 7 days of DSS administration, the mouse colons were removed and photographed, and colon length (E) and colon weight (F) of each mouse were recorded. (G) Representative hematoxylin and eosin staining of colonic tissues from WT-Control, NC agomir (DSS), miR-149-3p agomirs (DSS), and miR-149-5p agomirs (DSS) are shown (magnification, ×100).
miR-149-5p suppressed colon inflammatory responses in vitro
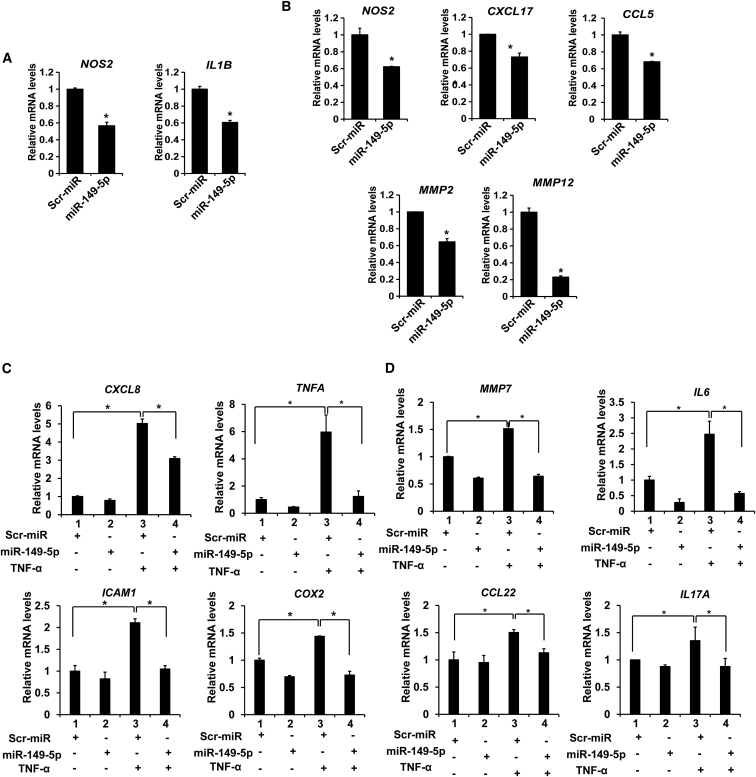
To further establish the role of miR-149-5p in colitis, we firstly measured mRNA levels of proinflammatory genes after the transfection of miR-149-5p mimics (50 nM, 24-h treatment) into Caco-2 and SW480 cells. miR-149-5p decreased NOS2 and IL1B (∗p < 0.05) expression levels in Caco-2 cells, and suppressed NOS2, CXCL17, CCL5, MMP2, and MMP12 mRNA expression (∗p < 0.05) in SW480 cells (Figures 5A and 5B). These results suggest that miR-149-5p suppressed the inflammatory response of colon adenocarcinoma cells.
Figure 5.
miR-149-5p suppressed inflammatory response in colon adenocarcinoma cells
(A) miR-149-5p mimics suppressed the proinflammatory gene expression in Caco2 cells (n = 3). (B) miR-149-5p mimics significantly decreased the expression of the proinflammatory genes and chemokines in SW480 cells. ∗p < 0.05 (n = 3). (C) Relative mRNA levels of inflammatory genes induced by TNF-α were reduced upon miR-149-5p mimics in Caco2 cells (n = 3). (D) miR-149-5p decreased the mRNA levels of the inflammatory genes induced by TNF-α in SW480 cells (n = 3). ∗p < 0.05.
Then we used TNF-α as a stimulator to induce inflammation response in Caco-2 and SW480 cells50 and further examined the effect of miR-149-5p mimics on TNF-α-induced inflammation in colon adenocarcinoma cells. Cells were transfected with miRNA mimics or control mimics. Next, cells were incubated with TNF-α (10 ng/mL) for 6 h and then harvested for RNA extraction. We found that miR-149-5p mimics suppressed TNF-α-induced expression of CXCL8, TNFA, ICAM1, and COX2 (∗p < 0.05) in Caco-2 cells (Figure 5C), and inhibited TNF-α-induced MMP7, IL6, CCL22, and IL17A (∗p < 0.05) in SW480 cells (Figure 5D).
miR-149-3p antagonized the NF-κB cell signaling pathway and activated AMPK in vitro
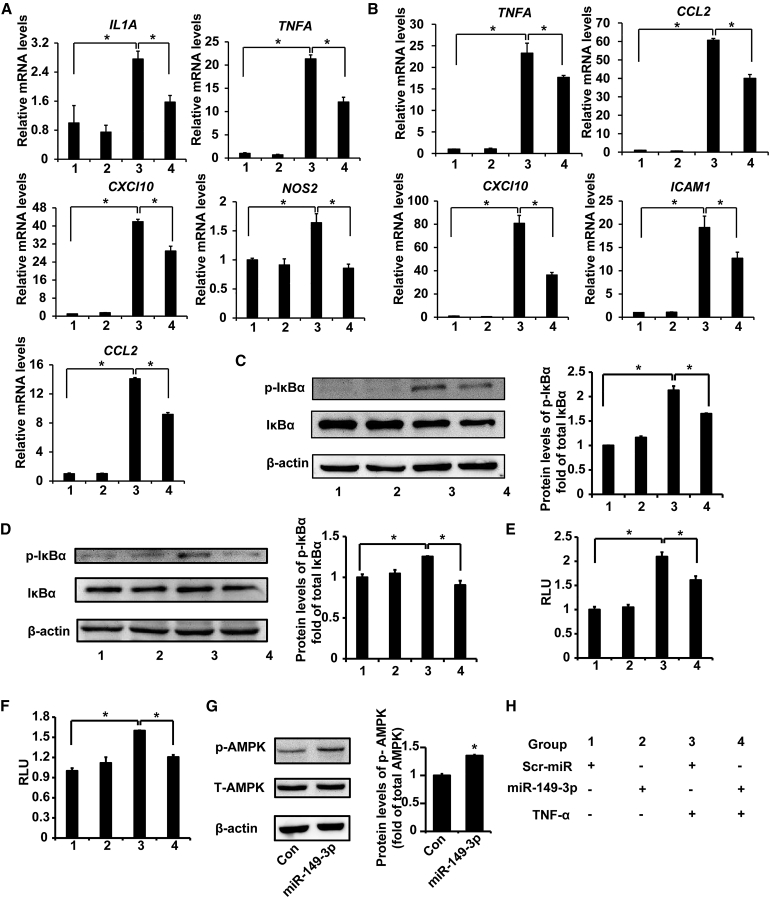
To analyze the effect of miR-149-3p on colon inflammation in colon adenocarcinoma cells, relative expression of proinflammatory genes in colon adenocarcinoma cells was determined after transfection of miRNA mimics. We found that miR-149-3p mimics caused a significant downregulation of expression of multiple genes induced by TNF-α compared with NC mimics (Scramble-miRNA mimics [Scr-miR]) in Caco-2 cells, including IL1A, TNFA, CXCl10, NOS2, and CCL2 mRNAs (∗p < 0.05) mediated by NF-κB (Figure 6A). It is known that NF-κB signaling is one of key regulators for IBD.51 Here, the overexpression of p65 was used to induce NF-κB signaling pathway. The expression of inflammatory genes such as IL1A, TNFA, ICAM1, CXCL8, and CCL2 (∗p < 0.05), induced by p65 overexpression, were also reduced in Caco-2 cells post-transfection with miR-149-3p mimics (Figure S3A). Similarly, miR-149-3p mimics reduced TNF-α-induced mRNA levels of TNFA, CCL2, CXCl10, and ICAM1 (∗p < 0.05) in SW480 cells (Figure 6B).
Figure 6.
miR-149-3p antagonized the NF-κB cell signaling pathway and activated AMPK in colon adenocarcinoma cells
(A) Relative mRNA levels of NF-κB-mediated inflammatory cytokines induced by TNF-α are decreased upon miR-149-3p mimics in Caco2 cells (n = 3). (B) Relative mRNA levels of NF-κB-mediated inflammatory cytokines induced by TNF-α are reduced upon miR-149-3p mimics in SW480 cells (n = 3). (C) miR-149-3p mimics suppressed TNF-α-induced p-IκBα in Caco2 cells. Caco2 cells were induced with TNF-α (10 ng/mL) for 1 h after being transfected with miR-149-3p mimics or control mimics (Scr-miR) (n = 3). (D) miR-149-3p mimics suppressed TNF-α-induced p-IκBα in SW480 cells. (E–F) miR-149-3p mimics lessened the luciferase signal of NF-κB activity induced by TNF-α in Caco2 (E) and SW480 cells (F) (n = 3). RLU, relative luciferase units. (G) miR-149-3p mimics increased the phospho-AMPK (p-AMPK) levels in SW480 cells (n = 3). (H) Group information. ∗p < 0.05.
To investigate whether miR-149-3p suppresses NF-κB signaling in colon cells, we transfected miR-149-3p mimics and measured p-IκBα levels in Caco-2 and SW480 cells. TNF-α (10 ng/mL) was utilized to induce IκBα phosphorylation, and incubation time was 30 min and 1 h in Caco-2 and SW480 cells, respectively. miR-149-3p mimics suppressed TNF-α-stimulated p-IκBα by about 23% in Caco-2 cells (Figure 6C). Similarly, miR-149-3p mimics in SW480 cells also blocked TNF-α-induced p-IκBα by about 27% (Figure 6D).
To further investigate the effect of miR-149-3p mimics on NF-κB transcriptional activity, we monitored the luciferase report in Caco-2 and SW480 cells. Cells were transiently transfected with 1 μg/well of NF-κB-LUC plasmid (3× NF-κB binding sites under CMV promoter) and control thymidine kinase-driven Renilla luciferase plasmid phRL-TK in combination with 50 nM control mimics (Scr-miR) or miR-149-3p mimics. Eighteen hours post-transfection, cells were treated with TNF-α (10 ng/mL) for 6 h. The luciferase activity was evaluated by the proportion of firefly luciferase to Renilla luciferase. In the luciferase assay, colon adenocarcinoma cells were treated with TNF-α for 6 h to activate the NF-κB pathway, while the activation was suppressed by miR-149-3p mimics in Caco-2 and SW480 cells (∗p < 0.05) (Figures 6E and 6F). Moreover, to confirm a direct effect of miR-149-3p on NF-κB transactivation induced by p65 overexpression, p65 overexpression plasmid was co-transfected with 50 nM miRNA mimics to activate the NF-κB reporter.21,52 p65 overexpression led to 12.5- and 7.2-fold greater NF-κB reporter activity (∗p < 0.05) in Caco-2 and SW480 cells, respectively. miR-149-3p mimics inhibited p65-induced NF-κB transactivity (∗p < 0.05) (Figures S3B and S3C). These findings strongly support the notion that miR-149-3p can antagonize NF-κB transactivity.
We next investigated whether miR-149-3p could activate AMPK in colon adenocarcinoma cells after transfection with miR-149-3p mimics in Caco-2 and SW480 cells. In Caco-2 cells, miR-149-3p mimics did not activate AMPK at the protein levels (data not shown). In contrast, miR-149-3p mimics promoted the phosphorylation of AMPK in SW480 cells (Figure 6G). This result illustrates that the role of miR-149-3p in activating AMPK in colon adenocarcinoma cells may be cell-type dependent.
miR-149-3p−/− mice shaped the microbiota composition
The role of the gut microbiota in host health and disease cannot be ignored.5 To characterize the role of miR-149-3p deletion on the gut microbiome composition, WT and miR-149-3p−/− males at 4 weeks were co-housed and single-housed at the same conditions for 4 weeks. Then the detected bacteria were assigned into one of 13 phyla. As reported previously, Bacteroidetes and Firmicutes represented the dominant bacteria at the phylum level (Figure S4).53 Moreover, Verrucomicrobia, represented by the Akkermansia genus, was more common in some WT control mice (DA) than in the miR-149-3p−/− mice (DB), but there were individual differences within the groups (Figure S4).
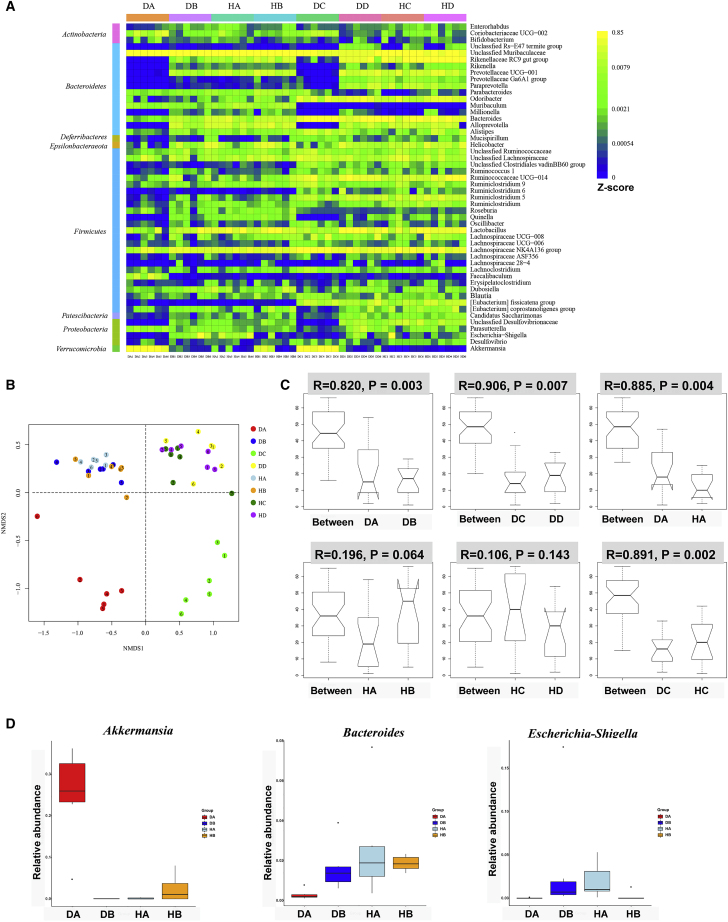
Next, we selected the top 50 dominant genera to perform heatmap analysis (Figure 7A). There were visible differences between the WT (DA) and miR-149-3p−/− control groups (DB), with miR-149-3p−/− mice having a higher proportion of Escherichia-Shigella, Bacteroides, Alloprevotella, Quinella, unclassified Desulfovibrionaceae and Alistipes, and a lower abundance of Akkermansia, Faecalibaculum, and Coriobacteriaceae_UCG-002 (Figure 7A). Combined with nonmetric multidimensional scaling (NMDS) analysis based on weighted UniFrac distances, the gut microbial community structure similarities and differences of all samples were visualized (Figure 7B). We found that the WT control group and miR-149-3p−/− control group displayed a clear and distinct separation on the heatmap; moreover, the DSS-treated WT group (DC) and DSS-treated miR-149-3p−/− group (DC) showed a distinct location on the heatmap. Regardless of whether DSS was administered or not, the gut microbiota structure and composition were different in the WT and miR-149-3p−/− groups. Following further observation, the microbial composition and abundance were altered in response to DSS administration in the WT (DA versus DC) and miR-149-3p−/− group (DB versus DD). Having observed microbiota differences among 24 samples in four groups, we further explored the community differences and differential species between the two groups based on ANOSIM test analysis. The comparison of communities between WT (DA) and miR-149-3p−/− control group (DB) showed significance (DA versus DB, R = 0.82, p = 0.003), and the significance between the two groups increased after DSS treatment (DC versus DD, R = 0.906, p = 0.007) according to the ANOSIM test (Figure 7C). Colitis-related genera, such as Akkermansia, Bacteroides, and Escherichia-Shigella, showed significant differences between the WT (DA) and KO group (DB) (Figure 7D). These results reveal that the deficiency of miR-149-3p shaped the gut microbiota community and structure.
Figure 7.
Deficiency of miR-149-3p in mice shaped the gut microbiota composition
To allow natural transfer of the microbiota, WT and miR-149-3p−/− males at 4 weeks were co-housed. As a control, single type control group was fed under the same conditions as when the mice reached 8 weeks, then 2% of DSS was given for 7 days, and fecal microbiota were detected by Illumina Miseq (n = 6). (A) Heatmap revealed homogenous patterns of the top 50 dominant genus among 48 sample different groups. Heatmap analysis was performed using the gplots package of R (http://www.r-project.org/) based on data of operational taxonomic units (OTUs). The "visible difference" here refers to the difference in relative abundance of top 50 dominant genus. The Z score value is defined as the number of sequences affiliated with that OTU divided by the total number of sequences per sample. The abundance is determined by the color key corresponding to the Z score value. (B) Nonmetric multidimensional scaling (NMDS) analysis revealed clustering of samples. (C) Analysis of similarities (Anosim) demonstrated similarities between two groups. (D) Relative abundance of Akkermansia, Bacteroides, and Escherichia-Shigella genus was shown among WT, KO, CH-WT, and CH-KO groups. DA, WT control group (WT); DB, miR-149-3p−/− control group (KO); HA, WT mice co-housed with miR-149-3p−/− mice (CH-WT); HB, miR-149-3p−/− mice co-housed with WT mice (CH-KO); DC, DSS-treated WT control group (WT (DSS)); DD, DSS-treated miR-149-3p−/− control group (WT (DSS)); HC, DSS-treated CH-WT group (CH-WT (DSS); HD, DSS-treated CH-KO group (CH-KO (DSS)).
Co-housing with KO mice increased susceptibility of WT mice against DSS-induced colitis
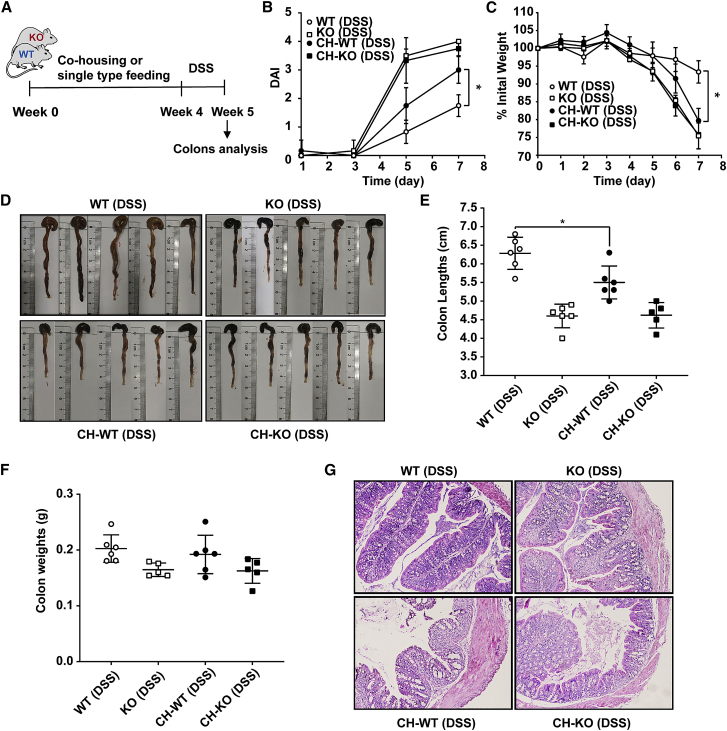
Deficiency of miR-149-3p in mice caused some alterations in the composition of the microbiota compared with WT controls, thus we tested whether natural transfer of the gut microbiota would affect the sensitivity of WT or KO mice to inflammatory infections. Four-week-old WT and miR-149-3p−/− males were co-housed to allow natural transfer of the microbiota. In addition, mice from single type control group were fed under the same conditions for 4 weeks. Then, 2% of DSS was administrated for 7 days, and weight, colon length, and colon weight were analyzed for each mouse (Figure 8A).
Figure 8.
WT mice co-housed with miR-149-3p−/− mice (CH-WT) endow susceptibility to DSS-induced colitis
(A) Schematic diagram of the experimental process. WT and miR-149-3p−/− males at 4 weeks were co-housed for 4 weeks. As a control, a single type control group was fed under the same conditions, and then 2% of DSS was given for 7 days (n = 6). (B) Disease activity index (DAI) was calculated every other day. (C) Weight was determined daily. (D) After 7 days of DSS administration, the colon of WT and KO mice was removed and photographed, and colon length (E) and colon weight (F) of each mouse were measured and recorded. (G) Representative hematoxylin and eosin staining of colonic tissues from the different groups (WT (DSS), KO (DSS), CH-WT (DSS), CH-KO (DSS)) are shown (magnification, ×100). WT (DSS), wild-type mice with DSS treatment; KO (DSS), miR-149-3p−/− mice with DSS treatment; CH-WT (DSS), WT mice co-housed with miR-149-3p−/− mice after DSS treatment; CH-KO, miR-149-3p−/− mice co-housed with WT mice after DSS treatment.
After 7 days of DSS administration, WT mice co-housed with miR-149-3p−/− mice (CH-WT) had a significantly increased DAI score (3.00 ± 0.47) compared with WT mice housed with other WT mice (1.75 ± 0.38) (Figure 8B), and CH-WT mice had a higher percentage of weight reduction relative to the WT control (Figure 8C). Moreover, the colon lengths (5.50 ± 0.44 cm) of CH-WT mice were shorter in response to DSS administration compared with those of the WT controls (6.48 ± 0.43 cm), while the changes in the weight of the colons were not significantly different (Figures 8D–8F). Next, we used H&E staining to analyze the histopathological changes of WT, KO, CH-WT, and CH-KO mouse colons. Compared with the DSS-treated WT control mice, CH-WT colons had a disintegrative loss of lamina propria and the crypt structure, accompanied by a larger area of inflammatory cell infiltration (Figure 8G). Then, examination of the proinflammatory cytokine expression revealed that CH-WT (DSS) mice exhibited significantly upregulated mRNA levels of Cxcl1, Icam1, Il1a, Il6, Ccl2, Tnfa, Tgfb1, Tgfb2, Mmp13, Mmp2, and Mmp12s in the colons compared with WT (DSS) mice (Figure S5). These subtle phenotypic results and inflammatory factor analysis support that CH-WT mice become more susceptible against DSS-induced colitis, and suggest that natural transfer of fecal microbiota from KO mice endows some susceptibility to WT mice against DSS-induced colitis.
Next, we analyzed the changes of the structure and composition of fecal microbiota in the natural transfer group compared with the group without natural transfer. As expected, when compared with the WT control group (DA), members from Verrucomicrobia were reduced in CH-WT mice (HA) at the phylum level (Figure S4). From a heatmap including the top 50 dominant genera, relative abundance of Akkermansia (phylum Verrucomicrobia) and Escherichia-Shigella (phylum Bacteroidetes) were significantly lower and higher, respectively, in CH-WT mice (Figure 7A). After NMDS analysis, all groups of mice clustered along NMDS1 and NMDS2 (Figure 7B). Although individual samples showed some overlap, the CH-WT mice (HA) and WT control (DA) groups were clearly separated, while CH-WT mice (HA), CH-KO mice (HB), and KO control (DB) groups showed similar and intersected locations. After DSS administration, there were similar clusters among the WT control (DSS) group (DC), KO control (DSS) group (DD), CH-WT (DSS) mice (HC), and CH-KO (DSS) group (HD) along NMDS1 (Figure 7B). ANOSIM test analysis between the WT control (DA) and CH-WT (HA) groups showed significance (DA versus HA, R = 0.885, p = 0.004). Notably, ANOSIM test of the CH-WT mice (HA) and CH-KO mice (HB) was very low (HA versus HB, R = 0.196, p = 0.064). After DSS administration, the microbiota composition between the WT control (DSS) group (DC) and CH-WT (DSS) mice (HC) changed significantly (DC versus HC, R = 0.891, p = 0.002). The CH-WT (DSS) mice group (HC) and CH-KO (DSS) group (HD) was more similar (HC versus HD, R = 0.106, p = 0.143) (Figure 7C). These findings suggested that the WT mouse microbiota were infected by the microbiota of KO mice after natural transfer of fecal microbiota. More noteworthy, the relative abundance in WT mice of the Akkermansia genus decreased significantly, while the Escherichia-Shigella and Bacteroides genera increased significantly after co-housing (Figure 7D). The diversity and composition of fecal microbiota supported this pathological phenotype, which indicates that the changes of the gut microbiota could participate in the process of DSS-induced colitis.
Susceptibility of miR-149-3p−/− mice against DSS-induced colitis was still present after antibiotic treatment
To determine whether the response of miR-149-3p−/− mice to DSS-induced colitis depends on the gut microbiota, WT and miR-149-3p−/− mice were pretreated with antibiotics before DSS administration. Mice were co-administrated for 2 weeks with four antibiotics in drinking water: vancomycin (0.5 g/L), ampicillin (1 g/L), kanamycin (1 g/L), and metronidazole (1 g/L). Then, these mice were treated with 2% DSS ad libitum and the mouse body weight was measured and fecal occult blood test was performed (Figure 9A). Mice were sacrificed on day 7 after DSS induction, and colons were collected and measured (Figure 9A). The DAI index did not show a significant difference between WT-DSS (2.20 ± 0.80) and KO-DSS (2.29 ± 0.43) mice with antibiotic pretreatment when DSS was administered for 5 days. Seven days after DSS treatment, the difference between the WT-DSS (2.40 ± 0.60) and KO-DSS (4.00 ± 0.00) groups was apparent (Figure 9B). In addition, after 7 days of DSS administration, deficiency of miR-149-3p still resulted in approximately 2.9-fold weight reduction (∗p < 0.05) relative to WT mice following antibiotic pretreatment (Figure 9C). The difference in colon length and weight between the WT and miR-149-3p−/− mice was already present after antibiotic pretreatment after DSS induction. Compared with WT-DSS (6.80 ± 0.25 cm, 0.18 ± 0.01 g), the miR-149-3p−/− colons (KO-DSS) remained shorter (5.11 ± 0.43 cm) and lighter (0.14 ± 0.01 g) in response to DSS treatment (Figures 9D–9F). Furthermore, deficiency of miR-149-3p still caused more serious damage with a disintegrative loss of the lamina propria upon pretreatment with antibiotics in histological analysis (Figure 9G). Overall, these phenotypes suggest that susceptibility of miR-149-3p−/− mice against DSS-induced colitis is not mainly dependent on the gut microbiota.
Figure 9.
Susceptibility of miR-149-3p−/− mice against DSS-colitis is still present after antibiotic treatment
(A) Schematic diagram of the experimental process. WT mice and miR-149-3p−/− mice at age 6 weeks were pretreated with antibiotics mix for 2 weeks, then 2% DSS was administered for 7 days (n = 5–6). (B) Disease activity index (DAI) was calculated every other day. (C) Weight reduction was calculated. (D) The colons of WT and KO mice were removed and photographed. Colon length (E) and colon weight (F) of each mouse were recorded. (G) Representative hematoxylin and eosin staining of colonic tissues from antibiotic-treated WT and KO mice with/without DSS administration (magnification, ×100). ∗p < 0.05, WT, wild-type; KO, miR-149-3p−/−.
Discussion
IBD, including colorectal inflammation, is an idiopathic disease, and a growing number of investigations suggest that IBD is involved in the development of colitis-associated tumorigenesis.54 DSS-mediated colitis is a mature chemically induced colitis as well as a colon cancer model in animals; accumulating evidence shows that clinical and histological features from DSS-induced colitis is similar with human UC, and that this model can be used for exploring molecular mechanisms and identifying novel therapeutic targets for repressing human colitis.55,56 This study demonstrates that the deletion of miR-149-3p enhanced DSS-induced colitis in a mouse model, whereby both miR-149-5p and miR-149-3p could suppress colonic inflammation in vivo and in vitro. Furthermore, we found that deficiency of miR-149-3p reshaped the gut microbiota. Our results indicate that both miR-149-5p and miR-149-3p may be the suppressors for development of colitis.
miRNAs have been described as molecules that regulate colonic inflammation and colitis-associated tumorigenesis.57, 58, 59 For example, miR-148a has a role as a suppressor in colitis and colitis-associated tumorigenesis through antagonizing the NF-κB and STAT3 signaling.57 Furthermore, deficiency of miRNA-21 increased resistance to DSS-induced colitis by shaping the gut microbiota.38 Previous studies have suggested that miR-149-5p has a role as a tumor suppressor in diverse human cancers, including CRC.41,42,60,61 Previous reports have shown that miR-149-3p has an important role in human melanoma and T cell acute lymphoblastic leukemia.62,63 Our recent report identified the role of miR-149-3p in liver inflammation and liver cancer.46,64 In addition, miR-149-3p was significantly increased in the blood of active CD patients and has potential as a marker in differentiate UC from CD.35,65 Cao et al. reported that plasma exosomes of IBD and CRC patients had reduced miR-149-3p, and enterotoxigenic Bacteroides fragilis promoted colon cancer cell proliferation depended on down-regulating miR-149-3p.45 In the present study, we further confirmed that miR-149-3p may inhibit colitis response and our findings show that miR-149-3p may function as a suppressor in colitis development by negatively regulating NF-κB activation and upregulating AMPK activation.
The constitutive activation of NF-κB is involved in colitis-associated tumorigenesis and CRC development,57 and blocking of NF-κB activation may be a potential treatment approach for suppressing colon inflammation and tumorigenesis.66 Moreover, the natural inhibitors of NF-κB are considered a safe strategy for IBD treatment.51 It seems that inhibition of NF-κB is a potential therapeutic strategy for treatment of colonic inflammation, and even colitis-associated tumorigenesis. In this study, we found that the deletion of miR-149-3p elevated the phosphorylation of IκBα and inflammatory responses in vivo, and miR-149-3p mimics decreased the phosphorylation of IκBα, NF-κB transactivity, and some NF-κB-mediated inflammatory cytokines in vitro. These results support a suppressing function of miR-149-3p against colitis, and suggest that miR-149-3p may be a potential therapeutic target for treating colitis and even colitis-associated cancer via inhibiting activation of NF-κB. Recent reports have suggested that activated AMPK exhibited antitumor activity in human cancers, including CRC.67,68 Sun et al. demonstrated that specific deletion of AMPKα1 villin is more susceptible to DSS-induced colitis, and revealed that activated AMPK enhances intestinal barrier function.13 Furthermore, several previous studies suggest a restoring intestinal barrier effect of metformin against experimental colitis and colitis-associated cancer via activation of AMPK.7,13,69 Herein, we discovered that AMPK activation in miR-149-3p−/− colon tissue is significantly suppressed in the DSS-untreated and DSS-treated mice. Meanwhile, miR-149-3p mimics enhanced the phosphorylation levels of AMPK in SW480 cells. Our results support one conceivable mechanism by which miR-149-3p attenuates colitis.
We noted that deficiency of miR-149-3p in mouse colons is sensitive to the specific sets of inflammatory genes compared with WT control, and that miR-149-3p or 5p repressed specific sets of inflammatory genes, but not all the inflammatory-related genes in response to LPS, TNF-α, or DSS. This phenomenon has also been observed for other miRNAs in inflammatory responses. Yin et al. demonstrated that a cohort of inflammatory genes was sensitive to anti-miR-217-mediated repression in cells induced by ethanol + LPS but the other set of the inflammatory genes was sensitive to miR-217 treatment in vivo.70 Similar results were obtained in that miR-182 mediated the functions on inflammatory gene expression.71 One possibility is that the transrepression programs that are mediated by miRNAs are regulated in a signal-specific manner. In addition, the transrepression pathways themselves may be subject to further regulation and can be overridden by specific signals in a gene-specific manner. It will be interesting to define the mechanism by which specific miRNA inhibits the inflammatory genes in a gene-specific manner.
As an “integral moderator” in the gastrointestinal tract, the gut microbiota dysbiosis is involved in a series of disorders, including DSS-induced colitis.5,72 miRNAs have previously been reported to affect the gut microbiota and thus may functionally regulate host heath.73 However, it is unknown whether the deletion of miR-149-3p is capable of shaping the gut microbiota. In the present study, we observed a significant difference in fecal microbiota composition between WT and miR-149-3p−/− mice. The miR-149-3p−/− mouse group was characterized by increased relative abundance of inflammatory-associated microbiota and decreased relative abundance of beneficial microbes. Escherichia-Shigella are generally considered to be inflammatory drivers, as they contain mucosa-associated Escherichia coli, which is the main pathogens associated with infectious diarrhea.74 In addition, the Bacteroides genus, which includes the enterotoxigenic Bacteroides fragilis, is also considered to be the main risk factor of IBD and CRC.75 Similarly, some anti-inflammatory bacteria were reduced in miR-149-3p−/− mice. Most notably, the Akkermansia genus has the most significant difference between WT and miR-149-3p−/− mice. Moreover, the relative abundance of the Akkermansia genus decreased significantly after co-housing. Akkermansia are next-generation beneficial microbes, and the extracellular vesicles of Akkermansia have been reported to be effective against DSS-induced colitis.76, 77, 78 In addition, Png et al. described that abundance of Akkermansia muciniphila decreased in patients with IBD.79 Recently, Zhai et al. identified the anti-inflammatory function of two A. muciniphila strains against DSS-induced colitis, and further suggested the potential effect of A. muciniphila on treatment of IBD.80 These altered gut microbiota compositions may be involved in the development of DSS-induced colitis. All the data have suggested that miR-149-3p shaped the microbiota composition, and that altered bacteria may have opportunistically aggravated DSS-induced colitis.
Next, to verify a hypothetical mechanism that miR-149-3p−/− mice confers enhanced DSS-induced colitis dependent on the gut microbiota, we pretreated mice with antibiotics prior to DSS administration. However, the susceptibility phenotype previously observed in the miR-149-3p−/− mice group was still present after antibiotic treatment according to DAI score, colon length and weight, and histological analysis. This finding confirms that sensitivity of miR-149-3p−/− mice to DSS-induced colitis is not conferred by the microbiota.
In summary, this study defines an essential role of miR-149-5p and miR-149-3p in regulating colitis development. We demonstrate that deficiency of miR-149-3p shaped the gut microbiota, which may be one of the mediators that aggravate DSS-induced colitis in miR-149-3p−/− mice. We reveal that genetic deletion of miR-149-3p promotes DSS-induced colitis response, thus establishing a foundation for evaluating miR-149-5p and miR-149-3p initiators as potential adjuvant therapies for colitis or IBD, and even colitis-associated carcinoma.
Materials and methods
Antibodies and reagents
Antibodies used for western blot analysis are shown in Table S1. TNF-α was from PeproTech (Cranbury, NJ). Vancomycin, ampicillin, kanamycin, and metronidazole were purchased from Solarbio (Beijing, China).
Animals
miR-149-3p−/− mice were generated with a C57B/6 background, as described previously,46 and wild-type C57B/6 mice were purchased from Beijing Experimental Animal Center (license no. SCXK (Jing) 2002-0003). All experiments were subject to ethical approval by the Ethics Committee of Henan University (permit no. 2020-001X) in China. All experiments followed the NIH guidelines for the care and use of laboratory animals.
For the resistance assay to DSS toxicity, WT and miR-149-3p−/− males at age 8 weeks were both randomly divided into two groups. DSS (36–50 kDa; MP Biomedicals, Canada) was dissolved and diluted to 2% w/v concentration with drinking water. Mice were provided with drinking water (control group) or DSS ad libitum, and water was replaced with fresh DSS water every other day. After DSS provision for 7 days, the DSS water was swapped with normal drinking water.
For initiating DSS-induced colitis, WT and miR-149-3p−/− males were treated in the same manner as above. Mouse body weight was measured, mortality rate was examined and fecal occult blood test was performed and recorded every other day. DAI of each mouse was calculated every other day according to the report of Johnston et al.38 After 7 days of DSS administration, mice were sacrificed, and the colon was removed to measure length and weight and collected for further analysis.
For the role of miRNAs in LPS-induced colon inflammatory response, 8-week-old male mice were given miRNA agomirs (Ribo-bio, Guangzhou, P.R. China) by a tail vein injection for 48 h. And then mice were fasted overnight and injected intraperitoneally with LPS (30 mg/kg body weight, St. Louis, MO). At 6 h after LPS exposure, mice were sacrificed.
To allow natural transfer of the microbiota, WT and miR-149-3p−/− males at age 4 weeks were co-housed. As a control, mice in a single type control group were fed under the same conditions. When the mice reached age 8 weeks, 2% DSS was given for 7 days, then body weight was measured and fecal occult blood test was performed every other day, and length and weight of colons were measured. On the fourth day of DSS administration, mouse feces were collected. The fecal pellets of each mouse were collected in a 1.5 mL sterile separation tube using the clean catch method and stored at −80°C.
Antibiotic treatment of mice was performed according to the description of Johnston et al.38 Mice were co-administrated for 2 weeks with four antibiotics in drinking water: vancomycin (0.5 g/L), ampicillin (1 g/L), kanamycin (1 g/L), and metronidazole (1 g/L). Then, these mice were treated with 2% DSS ad libitum and the mouse body weight was measured and fecal occult blood test was performed. The mice were sacrificed after 7 days of DSS induction, and length and weight of colon were measured.
Cell culture and transient transfection
miRNA mimics were purchased from RiboBio (Guangzhou). Human colon adenocarcinoma Caco-2 and SW480 cells were purchased from China Infrastructure of Cell Line Resource, and cultured in Dulbecco’s modified essential medium (with L-glutamine) (Corning, NY) with 10% fetal bovine serum (Gibco, NY) and 1% penicillin-streptomycin (Caisson Laboratories, North Logan). Cells were pre-plated into six-well plates and then transfected with miRNA mimics (50 nM) or siRNA mimics (50 nM) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Next, cells were incubated with TNF-α (10 ng/mL, PeproTech, Cranbury, NJ) for 6 h and then harvested for RNA extraction. For induction of p65, cells were co-transfected with miR-149-3p mimics (50 nM) or NC mimics (Scr-miR) and p65 expression plasmid (200 ng/mL). After 24 h, cells were harvested for RNA extraction and quantitative real-time PCR (qRT-PCR) analysis.
Dual-luciferase reporter assay
For the luciferase assay, cells were transiently transfected with 1 μg/well of NF-κB-LUC plasmid (provided by Dr. Peter Tontonoz and Dr. Bruce Blumberg, UCLA, Los Angeles, CA) and 100 ng/well of the control thymidine kinase-driven Renilla luciferase plasmid phRL-TK (provided by Akio Kruoda, City of Hope, Duarte, CA) in combination with 50 nM control mimics (Scr-miR) or miR-149-3p mimics. The luciferase activity was evaluated by the proportion of firefly luciferase to Renilla luciferase. Eighteen hours post-transfection, cells were treated with TNF-α (10 ng/mL) for 6 h. Then, luciferase activity was measured according to the instructions of the Dual-Luciferase Reporter Assay System (Promega, MD). If p65 overexpression was used for inducing NF-κB, then 100 ng/well p65 plasmid (provided by Xufeng Chen, City of Hope) was co-transfected with 50 nM miRNA mimics.
qRT-PCR
Total RNA was extracted from cells and colons using TRIzol Reagent (Invitrogen, Carlsbad, CA), following the detailed procedures that have been described previously.48,81 Amplification of human β-actin or mouse 36B4 was used as an internal reference for normalization of gene expression. Relative mRNA expression levels are described in our results. Primer sequences are shown in Table S2.
Immunoblot analysis
Proteins were isolated from cells or mouse colons, and then SDS-PAGE was performed to separate proteins as described previously.21,47 Bands on blots were visualized, and protein expression was analyzed with a computerized digital imaging system using Tanon software.
Histology
Fresh colon sections of each mouse underwent formalin fixation and paraffin embedding, and then 3-μm-thick longitudinal sections were visualized following H&E staining.
16S sequencing
Genomic DNA extraction from fecal microbiota was performed using a QIAamp DNA stool mini kit according to the manufacturer’s instructions. The hypervariable V3-V4 region of 16S rRNA was sequenced using paired-end sequencing in Illumina Miseq. The amplification primers used were 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806 (5′-GGACTACHVGGGTWTCTAAT-3′). Raw reads were processed using QIIME and sequences with ≥97% homology were assigned to the same operational taxonomic units (OTUs).82 The representative read of each OTU was annotated using Silva database (Release 132).83 NMDS and analysis of similarities (ANOSIM) were calculated using the vegan package implemented in R.
Statistics
All data represent at least three independent experiments and are expressed as the mean ± SEM. The Student’s t test and two-way analysis of variance (ANOVA), followed by Bonferroni’s post-hoc test, were performed. A p value <0.05 was considered significant.
Data availability statement
All data generated or analyzed during this study are included in this published article and its supplemental information files.
Acknowledgments
This work is supported by the National Natural Science Foundation of China (grant nos. 81970551 and 81672433), the National Key R&D Program of China (2021YFC2103900), the Fundamental Research Funds for the Central Universities (grant no. PT2001) (to Y.-D.W.), the National Natural Science Foundation of China (grant nos. 81970726 and 81472232), the Key Program for Science & Technology from Henan Education Department (grant no. 21A310001) (to W.-D.C.).
Author contributions
Y.-D.W. and W.-D.C. initiated the ideas for the project. Q.F., Y.L., Y.-D.W., and W.-D.C. conceived and designed the study. Q.F., Y.L., H.Z., Z.W., X.N., D.Y., and L.H. performed the experiments and analyzed data. Q.F. wrote the paper. Q.F., Y.L., Y.-D.W., and W.-D.C. reviewed and edited the manuscript. All authors read and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.09.018.
Contributor Information
Wei-Dong Chen, Email: wdchen666@163.com.
Yan-Dong Wang, Email: ydwang@mail.buct.edu.cn.
Supplemental information
References
- 1.Lin J., Cao Q., Zhang J., Li Y., Shen B., Zhao Z., Chinnaiyan A.M., Bronner M.P. MicroRNA expression patterns in indeterminate inflammatory bowel disease. Mod. Pathol. 2013;26:148–154. doi: 10.1038/modpathol.2012.131. [DOI] [PubMed] [Google Scholar]
- 2.Jeengar M.K., Thummuri D., Magnusson M., Naidu V.G.M., Uppugunduri S. Uridine ameliorates dextran sulfate sodium (DSS)-Induced colitis in mice. Sci. Rep. 2017;7:3924. doi: 10.1038/s41598-017-04041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wawrzyniak M., Scharl M. Genetics and epigenetics of inflammatory bowel disease. Swiss Med. Wkly. 2018;148:w14671. doi: 10.4414/smw.2018.14671. [DOI] [PubMed] [Google Scholar]
- 4.Zaid Y., Senhaji N., Bakhtaoui F.Z., Serrano A., Serbati N., Karkouri M., Badre W., Oudghiri M., Martin J., Nadifi S. The PTPN22 C1858T (R620W) functional polymorphism in inflammatory bowel disease. BMC Res. Notes. 2018;11:783. doi: 10.1186/s13104-018-3875-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng Q., Chen W.D., Wang Y.D. Gut microbiota: an integral moderator in health and disease. Front. Microbiol. 2018;9:151. doi: 10.3389/fmicb.2018.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho Y.A., Lee J., Oh J.H., Chang H.J., Sohn D.K., Shin A., Kim J. Vitamin D receptor FokI polymorphism and the risks of colorectal cancer, inflammatory bowel disease, and colorectal adenoma. Sci. Rep. 2018;8:12899. doi: 10.1038/s41598-018-31244-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L., Wang J., You Q., He S., Meng Q., Gao J., Wu X., Shen Y., Sun Y., Wu X., Xu Q. Activating AMPK to restore tight junction assembly in intestinal epithelium and to attenuate experimental colitis by metformin. Front. Pharmacol. 2018;9:761. doi: 10.3389/fphar.2018.00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardie D.G., Schaffer B.E., Brunet A. AMPK: an energy-sensing pathway with multiple inputs and outputs. Trends Cell Biol. 2016;26:190–201. doi: 10.1016/j.tcb.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viollet B., Horman S., Leclerc J., Lantier L., Foretz M., Billaud M., Giri S., Andreelli F. AMPK inhibition in health and disease. Crit. Rev. Biochem. Mol. Biol. 2010;45:276–295. doi: 10.3109/10409238.2010.488215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox M.M., Phoenix K.N., Kopsiaftis S.G., Claffey K.P. AMP-activated protein kinase α 2 isoform suppression in primary breast cancer alters AMPK growth control and apoptotic signaling. Genes Cancer. 2013;4:3–14. doi: 10.1177/1947601913486346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopsiaftis S., Hegde P., Taylor J.A., 3rd, Claffey K.P. AMPKα is suppressed in bladder cancer through macrophage-mediated mechanisms. Transl. Oncol. 2016;9:606–616. doi: 10.1016/j.tranon.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu F., Liu F., Dong L., Yang H., He X., Li L., Zhao L., Jin S., Li G. miR-1273g silences MAGEA3/6 to inhibit human colorectal cancer cell growth via activation of AMPK signaling. Cancer Lett. 2018;435:1–9. doi: 10.1016/j.canlet.2018.07.031. [DOI] [PubMed] [Google Scholar]
- 13.Sun X., Yang Q., Rogers C.J., Du M., Zhu M.J. AMPK improves gut epithelial differentiation and barrier function via regulating Cdx2 expression. Cell Death Differ. 2017;24:819–831. doi: 10.1038/cdd.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pongkorpsakol P., Buasakdi C., Chantivas T., Chatsudthipong V., Muanprasat C. An agonist of a zinc-sensing receptor GPR39 enhances tight junction assembly in intestinal epithelial cells via an AMPK-dependent mechanism. Eur. J. Pharmacol. 2019;842:306–313. doi: 10.1016/j.ejphar.2018.10.038. [DOI] [PubMed] [Google Scholar]
- 15.Olivier S., Diounou H., Pochard C., Frechin L., Durieu E., Foretz M., Neunlist M., Rolli-Derkinderen M., Viollet B. Intestinal epithelial AMPK deficiency causes delayed colonic epithelial repair in DSS-induced colitis. Cells. 2022;11:590. doi: 10.3390/cells11040590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahara M., Takaki A., Hiraoka S., Takei K., Yasutomi E., Igawa S., Yamamoto S., Oka S., Ohmori M., Yamasaki Y., et al. Metformin ameliorates chronic colitis in a mouse model by regulating interferon-γ-producing lamina propria CD4(+) T cells through AMPK activation. Faseb. J. 2022;36:e22139. doi: 10.1096/fj.202100831RR. [DOI] [PubMed] [Google Scholar]
- 17.Banskota S., Wang H., Kwon Y.H., Gautam J., Gurung P., Haq S., Hassan F.M.N., Bowdish D.M., Kim J.A., Carling D., et al. Salicylates ameliorate intestinal inflammation by activating macrophage AMPK. Inflamm. Bowel Dis. 2021;27:914–926. doi: 10.1093/ibd/izaa305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng L., Li Z.R., Green R.S., Holzman I.R., Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J. Nutr. 2009;139:1619–1625. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wullaert A. Role of NF-κB activation in intestinal immune homeostasis. Int. J. Med. Microbiol. 2010;300:49–56. doi: 10.1016/j.ijmm.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Read S.A., Douglas M.W. Virus induced inflammation and cancer development. Cancer Lett. 2014;345:174–181. doi: 10.1016/j.canlet.2013.07.030. [DOI] [PubMed] [Google Scholar]
- 21.Liu T., Zhang L., Joo D., Sun S.C. NF-κB signaling in inflammation. Signal Transduct. Targeted Ther. 2017;2:17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y.D., Chen W.D., Yu D., Forman B.M., Huang W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor κ light-chain enhancer of activated B cells (NF-κB) in mice. Hepatology. 2011;54:1421–1432. doi: 10.1002/hep.24525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q., Yang G., Feng M., Zheng S., Cao Z., Qiu J., You L., Zheng L., Hu Y., Zhang T., Zhao Y. NF-κB in pancreatic cancer: its key role in chemoresistance. Cancer Lett. 2018;421:127–134. doi: 10.1016/j.canlet.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 24.Gabriele M., Pucci L., Árvay J., Longo V. Anti-inflammatory and antioxidant effect of fermented whole wheat on TNFα-stimulated HT-29 and NF-κB signaling pathway activation. J. Funct.Foods. 2018;45:392–400. [Google Scholar]
- 25.Papoutsopoulou S., Burkitt M.D., Bergey F., England H., Hough R., Schmidt L., Spiller D.G., White M.H.R., Paszek P., Jackson D.A., et al. Macrophage-specific NF-κB activation dynamics can segregate inflammatory bowel disease patients. Front. Immunol. 2019;10:2168. doi: 10.3389/fimmu.2019.02168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shetty A., Forbes A. Pharmacogenomics of response to anti-tumor necrosis factor therapy in patients with Crohn's disease. Am. J. PharmacoGenomics. 2002;2:215–221. doi: 10.2165/00129785-200202040-00001. [DOI] [PubMed] [Google Scholar]
- 27.Szatkowski P., Krzysciak W., Mach T., Owczarek D., Brzozowski B., Szczeklik K. Nuclear factor-κB - importance, induction of inflammation, and effects of pharmacological modulators in Crohn's disease. J. Physiol. Pharmacol. 2020;71:1–13. doi: 10.26402/jpp.2020.4.01. [DOI] [PubMed] [Google Scholar]
- 28.McDaniel D.K., Eden K., Ringel V.M., Allen I.C. Emerging roles for noncanonical NF-κB signaling in the modulation of inflammatory bowel disease pathobiology. Inflamm. Bowel Dis. 2016;22:2265–2279. doi: 10.1097/MIB.0000000000000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merga Y.J., O'Hara A., Burkitt M.D., Duckworth C.A., Probert C.S., Campbell B.J., Pritchard D.M. Importance of the alternative NF-κB activation pathway in inflammation-associated gastrointestinal carcinogenesis. Am. J. Physiol. Gastrointest. Liver Physiol. 2016;310:G1081–G1090. doi: 10.1152/ajpgi.00026.2016. [DOI] [PubMed] [Google Scholar]
- 30.Herrington F.D., Carmody R.J., Goodyear C.S. Modulation of NF-κB signaling as a therapeutic target in autoimmunity. J. Biomol. Screen. 2016;21:223–242. doi: 10.1177/1087057115617456. [DOI] [PubMed] [Google Scholar]
- 31.Müller E.K., Białas N., Epple M., Hilger I. Nanoparticles carrying NF-κB p65-specific siRNA alleviate colitis in mice by attenuating NF-κB-Related protein expression and pro-inflammatory cellular mediator secretion. Pharmaceutics. 2022;14:419. doi: 10.3390/pharmaceutics14020419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodríguez-Nogales A., Algieri F., Garrido-Mesa J., Vezza T., Utrilla M.P., Chueca N., Garcia F., Olivares M., Rodríguez-Cabezas M.E., Gálvez J. Differential intestinal anti-inflammatory effects of Lactobacillus fermentum and Lactobacillus salivarius in DSS mouse colitis: impact on microRNAs expression and microbiota composition. Mol. Nutr. Food Res. 2017;61:1700144. doi: 10.1002/mnfr.201700144. [DOI] [PubMed] [Google Scholar]
- 33.Louis P., Flint H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 34.Parada Venegas D., De la Fuente M.K., Landskron G., González M.J., Quera R., Dijkstra G., Harmsen H.J.M., Faber K.N., Hermoso M.A. Short chain fatty acids (SCFAs)-Mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 2019;10:1486. doi: 10.3389/fimmu.2019.01486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung H., Kim J.S., Lee K.H., Tizaoui K., Terrazzino S., Cargnin S., Smith L., Koyanagi A., Jacob L., Li H., et al. Roles of microRNAs in inflammatory bowel disease. Int. J. Biol. Sci. 2021;17:2112–2123. doi: 10.7150/ijbs.59904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng Y., Croce C.M. The role of MicroRNAs in human cancer. Signal Transduct. Targeted Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Connell R.M., Rao D.S., Baltimore D. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 38.Johnston D.G.W., Williams M.A., Thaiss C.A., Cabrera-Rubio R., Raverdeau M., McEntee C., Cotter P.D., Elinav E., O'Neill L.A.J., Corr S.C. Loss of MicroRNA-21 influences the gut microbiota, causing reduced susceptibility in a murine model of colitis. J. Crohns Colitis. 2018;12:835–848. doi: 10.1093/ecco-jcc/jjy038. [DOI] [PubMed] [Google Scholar]
- 39.Runtsch M.C., Hu R., Alexander M., Wallace J., Kagele D., Petersen C., Valentine J.F., Welker N.C., Bronner M.P., Chen X., et al. MicroRNA-146a constrains multiple parameters of intestinal immunity and increases susceptibility to DSS colitis. Oncotarget. 2015;6:28556–28572. doi: 10.18632/oncotarget.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chamorro-Jorganes A., Araldi E., Rotllan N., Cirera-Salinas D., Suárez Y. Autoregulation of glypican-1 by intronic microRNA-149 fine tunes the angiogenic response to FGF2 in human endothelial cells. J. Cell Sci. 2014;127:1169–1178. doi: 10.1242/jcs.130518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang F., Ma Y.L., Zhang P., Shen T.Y., Shi C.Z., Yang Y.Z., Moyer M.P., Zhang H.Z., Chen H.Q., Liang Y., Qin H.L. SP1 mediates the link between methylation of the tumour suppressor miR-149 and outcome in colorectal cancer. J. Pathol. 2013;229:12–24. doi: 10.1002/path.4078. [DOI] [PubMed] [Google Scholar]
- 42.Wang A.L., Li Y., Zhao Q., Fan L.Q. Formononetin inhibits colon carcinoma cell growth and invasion by microRNA149mediated EphB3 downregulation and inhibition of PI3K/AKT and STAT3 signaling pathways. Mol. Med. Rep. 2018;17:7721–7729. doi: 10.3892/mmr.2018.8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dong Y., Chang C., Liu J., Qiang J. Targeting of GIT1 by miR-149∗ in breast cancer suppresses cell proliferation and metastasis in vitro and tumor growth in vivo. OncoTargets Ther. 2017;10:5873–5882. doi: 10.2147/OTT.S144280. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 44.Yang D., Du G., Xu A., Xi X., Li D. Expression of miR-149-3p inhibits proliferation, migration, and invasion of bladder cancer by targeting S100A4. Am. J. Cancer Res. 2017;7:2209–2219. [PMC free article] [PubMed] [Google Scholar]
- 45.Cao Y., Wang Z., Yan Y., Ji L., He J., Xuan B., Shen C., Ma Y., Jiang S., Ma D., et al. Enterotoxigenic bacteroidesfragilis promotes intestinal inflammation and malignancy by inhibiting exosome-packaged miR-149-3p. Gastroenterology. 2021;161:1552–1566.e12. doi: 10.1053/j.gastro.2021.08.003. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Q., Su J., Wang Z., Qi H., Ge Z., Li Z., Chen W.D., Wang Y.D. MicroRNA-149∗ suppresses hepatic inflammatory response through antagonizing STAT3 signaling pathway. Oncotarget. 2017;8:65397–65406. doi: 10.18632/oncotarget.18541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Y.D., Chen W.D., Wang M., Yu D., Forman B.M., Huang W. Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology. 2008;48:1632–1643. doi: 10.1002/hep.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y.D., Chen W.D., Yu D., Forman B.M., Huang W. The G-protein-coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor kappa light-chain enhancer of activated B cells (NF-kappaB) in mice. Hepatology. 2011;54:1421–1432. doi: 10.1002/hep.24525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gao R., Tian S., Wang J., Zhu W. Galacto-oligosaccharides improve barrier function and relieve colonic inflammation via modulating mucosa-associated microbiota composition in lipopolysaccharides-challenged piglets. J. Anim. Sci. Biotechnol. 2021;12:92. doi: 10.1186/s40104-021-00612-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Erdman S.E., Rao V.P., Poutahidis T., Rogers A.B., Taylor C.L., Jackson E.A., Ge Z., Lee C.W., Schauer D.B., Wogan G.N., et al. Nitric oxide and TNF-α trigger colonic inflammation and carcinogenesis in Helicobacter hepaticus-infected, Rag2-deficient mice. Proc. Natl. Acad. Sci. USA. 2009;106:1027–1032. doi: 10.1073/pnas.0812347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tambuwala M.M. Natural nuclear factor kappa beta inhibitors: safe therapeutic options for inflammatory bowel disease. Inflamm. Bowel Dis. 2016;22:719–723. doi: 10.1097/MIB.0000000000000655. [DOI] [PubMed] [Google Scholar]
- 52.Zhou C., Tabb M.M., Nelson E.L., Grün F., Verma S., Sadatrafiei A., Lin M., Mallick S., Forman B.M., Thummel K.E., Blumberg B. Mutual repression between steroid and xenobiotic receptor and NF-kappaB signaling pathways links xenobiotic metabolism and inflammation. J. Clin. Invest. 2006;116:2280–2289. doi: 10.1172/JCI26283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Milani C., Duranti S., Bottacini F., Casey E., Turroni F., Mahony J., Belzer C., Delgado Palacio S., Arboleya Montes S., Mancabelli L., et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 2017;81 doi: 10.1128/MMBR.00036-17. 000366-e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terzić J., Grivennikov S., Karin E., Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114.e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 55.Mo J.S., Han S.H., Yun K.J., Chae S.C. MicroRNA 429 regulates the expression of CHMP5 in the inflammatory colitis and colorectal cancer cells. Inflamm. Res. 2018;67:985–996. doi: 10.1007/s00011-018-1194-z. [DOI] [PubMed] [Google Scholar]
- 56.Elson C.O., Sartor R.B., Tennyson G.S., Riddell R.H. Experimental models of inflammatory bowel disease. Gastroenterology. 1995;109:1344–1367. doi: 10.1016/0016-5085(95)90599-5. [DOI] [PubMed] [Google Scholar]
- 57.Zhu Y., Gu L., Li Y., Lin X., Shen H., Cui K., Chen L., Zhou F., Zhao Q., Zhang J., et al. miR-148a inhibits colitis and colitis-associated tumorigenesis in mice. Cell Death Differ. 2017;24:2199–2209. doi: 10.1038/cdd.2017.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koukos G., Polytarchou C., Kaplan J.L., Morley-Fletcher A., Gras-Miralles B., Kokkotou E., Baril-Dore M., Pothoulakis C., Winter H.S., Iliopoulos D. MicroRNA-124 regulates STAT3 expression and is down-regulated in colon tissues of pediatric patients with ulcerative colitis. Gastroenterology. 2013;145:842–852.e2. doi: 10.1053/j.gastro.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He C., Yu T., Shi Y., Ma C., Yang W., Fang L., Sun M., Wu W., Xiao F., Guo F., et al. MicroRNA 301A promotes intestinal inflammation and colitis-associated cancer development by inhibiting BTG1. Gastroenterology. 2017;152:1434–1448.e15. doi: 10.1053/j.gastro.2017.01.049. [DOI] [PubMed] [Google Scholar]
- 60.Ren F.J., Yao Y., Cai X.Y., Cai Y.T., Su Q., Fang G.Y. MiR-149-5p: an important miRNA regulated by competing endogenous RNAs in diverse human cancers. Front. Oncol. 2021;11:743077. doi: 10.3389/fonc.2021.743077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He Y., Yu D., Zhu L., Zhong S., Zhao J., Tang J. miR-149 in human cancer: a systemic review. J. Cancer. 2018;9:375–388. doi: 10.7150/jca.21044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin L., Hu W.L., Jiang C.C., Wang J.X., Han C.C., Chu P., Zhang L.J., Thorne R.F., Wilmott J., Scolyer R.A., et al. MicroRNA-149∗, a p53-responsive microRNA, functions as an oncogenic regulator in human melanoma. Proc. Natl. Acad. Sci. USA. 2011;108:15840–15845. doi: 10.1073/pnas.1019312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fan S.J., Li H.B., Cui G., Kong X.L., Sun L.L., Zhao Y.Q., Li Y.H., Zhou J. miRNA-149∗ promotes cell proliferation and suppresses apoptosis by mediating JunB in T-cell acute lymphoblastic leukemia. Leuk. Res. 2016;41:62–70. doi: 10.1016/j.leukres.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 64.Feng Q., Zhang H., Nie X., Li Y., Chen W.D., Wang Y.D. miRNA-149∗ suppresses liver cancer progression by down-regulating TRADD protein expression. Am. J. Pathol. 2020;190:469–483. doi: 10.1016/j.ajpath.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 65.Wu F., Guo N.J., Tian H., Marohn M., Gearhart S., Bayless T.M., Brant S.R., Kwon J.H. Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn's disease. Inflamm. Bowel Dis. 2011;17:241–250. doi: 10.1002/ibd.21450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Allen I.C., Wilson J.E., Schneider M., Lich J.D., Roberts R.A., Arthur J.C., Woodford R.M.T., Davis B.K., Uronis J.M., Herfarth H.H., et al. NLRP12 suppresses colon inflammation and tumorigenesis through the negative regulation of noncanonical NF-κB signaling. Immunity. 2012;36:742–754. doi: 10.1016/j.immuni.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Din F.V.N., Valanciute A., Houde V.P., Zibrova D., Green K.A., Sakamoto K., Alessi D.R., Dunlop M.G. Aspirin inhibits mTOR signaling, activates AMP-activated protein kinase, and induces autophagy in colorectal cancer cells. Gastroenterology. 2012;142:1504–1515.e3. doi: 10.1053/j.gastro.2012.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dai C., Zhang X., Xie D., Tang P., Li C., Zuo Y., Jiang B., Xue C. Targeting PP2A activates AMPK signaling to inhibit colorectal cancer cells. Oncotarget. 2017;8:95810–95823. doi: 10.18632/oncotarget.21336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang S.Q., Cui S.X., Qu X.J. Metformin inhibited colitis and colitis-associated cancer (CAC) through protecting mitochondrial structures of colorectal epithelial cells in mice. Cancer Biol. Ther. 2018;20:338–348. doi: 10.1080/15384047.2018.1529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yin H., Liang X., Jogasuria A., Davidson N.O., You M. miR-217 regulates ethanol-induced hepatic inflammation by disrupting sirtuin 1–lipin-1 signaling. Am. J. Pathol. 2015;185:1286–1296. doi: 10.1016/j.ajpath.2015.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blaya D., Coll M., Rodrigo-Torres D., Vila-Casadesús M., Altamirano J., Llopis M., Graupera I., Perea L., Aguilar-Bravo B., Díaz A., et al. Integrative microRNA profiling in alcoholic hepatitis reveals a role for microRNA-182 in liver injury and inflammation. Gut. 2016;65:1535–1545. doi: 10.1136/gutjnl-2015-311314. [DOI] [PubMed] [Google Scholar]
- 72.Cao G., Wang K., Li Z., Tao F., Xu Y., Lan J., Chen G., Yang C. Bacillus amyloliquefaciens ameliorates dextran sulfate sodium-induced colitis by improving gut microbial dysbiosis in mice model. Front. Microbiol. 2018;9:3260. doi: 10.3389/fmicb.2018.03260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu S., da Cunha A.P., Rezende R.M., Cialic R., Wei Z., Bry L., Comstock L.E., Gandhi R., Weiner H.L. The host shapes the gut microbiota via fecal MicroRNA. Cell Host Microbe. 2016;19:32–43. doi: 10.1016/j.chom.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu W., Zhang Y., Qiu B., Fan S., Ding H., Liu Z. Quinoa whole grain diet compromises the changes of gut microbiota and colonic colitis induced by dextran Sulfate sodium in C57BL/6 mice. Sci. Rep. 2018;8:14916. doi: 10.1038/s41598-018-33092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tjalsma H., Boleij A., Marchesi J.R., Dutilh B.E. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat. Rev. Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 76.Cani P.D., de Vos W.M. Next-generation beneficial microbes: the case of Akkermansia muciniphila. Front. Microbiol. 2017;8:1765. doi: 10.3389/fmicb.2017.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Derrien M., Belzer C., de Vos W.M. Akkermansia muciniphila and its role in regulating host functions. Microb. Pathog. 2017;106:171–181. doi: 10.1016/j.micpath.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 78.Kang C.S., Ban M., Choi E.J., Moon H.G., Jeon J.S., Kim D.K., Park S.K., Jeon S.G., Roh T.Y., Myung S.J., et al. Extracellular vesicles derived from gut microbiota, especially Akkermansia muciniphila, protect the progression of dextran sulfate sodium-induced colitis. PLoS One. 2013;8:e76520. doi: 10.1371/journal.pone.0076520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Png C.W., Lindén S.K., Gilshenan K.S., Zoetendal E.G., McSweeney C.S., Sly L.I., McGuckin M.A., Florin T.H.J. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 80.Zhai R., Xue X., Zhang L., Yang X., Zhao L., Zhang C. Strain-specific anti-inflammatory properties of two Akkermansia muciniphila strains on chronic colitis in mice. Front. Cell. Infect. Microbiol. 2019;9:239. doi: 10.3389/fcimb.2019.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo C., Qi H., Yu Y., Zhang Q., Su J., Yu D., Huang W., Chen W.D., Wang Y.D. The G-protein-coupled bile acid receptor Gpbar1 (TGR5) inhibits gastric inflammation through antagonizing NF-kappaB signaling pathway. Front. Pharmacol. 2015;6:287. doi: 10.3389/fphar.2015.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplemental information files.