Abstract
The Arc (anoxic redox control) two-component signal transduction system of Escherichia coli, which comprises the tripartite ArcB sensor kinase and the ArcA response regulator, modulates the expression of numerous operons in response to redox conditions of growth. We demonstrate that the arcA and arcB genes of Haemophilus influenzae specify a two-component system. The Arc proteins of the two bacterial species sufficiently resemble each other that they can participate in heterologous transphosphorylation in vitro. Moreover, the Arc system of H. influenzae mediates transcriptional control according to the redox condition of growth both autologously in its own host and homologously in E. coli, indicating a high degree of functional conservation of the signal transduction system. The H. influenzae ArcB, however, lacks the PAS domain present in the region of E. coli ArcB linking the transmembrane to the cytosolic catalytic domains. Because the PAS domain participates in signal reception in a variety of sensory proteins, including sensors of molecular oxygen and redox state, a similar role was previously ascribed to it in ArcB. Our results demonstrate that the ArcB protein of H. influenzae mediates signal transduction in response to redox conditions of growth despite the absence of the PAS domain.
Two-component signal transduction systems, which consist of a sensor kinase and a response regulator, are highly conserved in nature and mediate adaptations to a variety of environmental changes. Although these systems have been found in some eukaryotes such as plant and yeast species, they are more prevalent in prokaryotes. The prokaryotic systems have been reported to regulate diverse processes that include energy metabolism, symbiotic nitrogen fixation, chemotaxis, cell division, sporulation, and pathogenic interactions with both plant and animal hosts (reviewed in references 3, 16, 30, and 37).
The Arc (anoxic redox control) two-component signal transduction system of Escherichia coli, which comprises the ArcB sensor kinase (see Fig. 1) and the ArcA response regulator, regulates the expression of more than 30 operons (the Arc modulon) in response to redox conditions of growth (18, 24). ArcB consists of a transmembrane domain linked to three cytosolic domains: an N-terminal transmitter domain (H1) with a conserved His292 residue, a central receiver domain (D1) with a conserved Asp576, and a C-terminal secondary transmitter domain (H2) with a conserved His717. In contrast, ArcA is a typical cytoplasmic response regulator possessing an N-terminal receiver domain with a conserved Asp54 and a C-terminal helix-turn-helix DNA binding domain. Under reducing conditions, ArcB undergoes autophosphorylation, a process enhanced by certain anaerobic metabolites such as d-lactate, acetate, and pyruvate (11), and transphosphorylates ArcA via a His292 → Asp576 → His717 → Asp54 phosphorelay (13, 19, 21). Under oxidizing conditions, ArcB autophosphorylation is inhibited by the quinone electron carriers (12). Dephosphorylation of phosphorylated ArcA (ArcA-P) occurs by a reverse phosphoryl group transfer to His717 of H2 and subsequently to Asp576 of D1, where the release of Pi takes place (10). Signal transduction by phosphorelay has also been reported for the Kin/Spo system of Bacillus subtilis (7), the BvgS/BvgA system of Bordetella pertussis (36), the TorS/TorR system of E. coli (20), and the Sln1p/Ypd1p/Ssk1p system of Saccharomyces cerevisiae (33). It is conjectured that this complex phosphotransfer mechanism in tripartite sensor kinases allows multiple inputs in the signal transduction process.
FIG. 1.
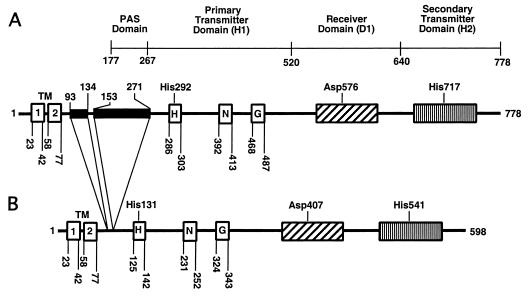
Schematic representation of the ArcB sensor kinase. (A) The E. coli protein; (B) the H. influenzae protein. The two N-terminal transmembrane segments (TM) were predicted on the basis of a hydrophobicity plot. The linker region of the E. coli protein (residues 78 to 267) contains a PAS domain (residues 177 to 267) (27). Black boxes represent sequences absent in H. influenzae ArcB. H1 (the primary transmitter domain) is shown with the catalytic determinants H, N, and G (29). The conserved site of autophosphorylation at His292 of the E. coli ArcB sequence corresponds to a conserved His131 of the H. influenzae ArcB sequence. G resembles the nucleotide-binding motif. In the receiver domain (D1), the conserved transphosphorylation site at Asp576 in E. coli ArcB corresponds to the conserved Asp407 of H. influenzae ArcB. In the secondary transmitter domain (H2), the conserved transphosphorylation site at His717 of E. coli ArcB corresponds to the conserved His541 of H. influenzae ArcB.
In contrast to the distinct periplasmic domains found in many other membrane-bound sensor kinase proteins, ArcB contains only a short periplasmic bridge of 16 amino acid residues separating its two transmembrane segments. The finding that the amino acid sequence of the transmembrane segments is unimportant for signal transduction led to the hypothesis that this portion of the protein serves only as a tethering device and not as a signal reception domain (22). Recently, Matsushika and Mizuno (27) suggested that the PAS domain, situated in the linker region between the transmembrane and cytosolic domains, is required for sensing the cellular redox condition. The PAS motif is present in a broad family of proteins from all kingdoms of life, including the eukaryotic transcriptional regulators that control the circadian clock (reviewed in reference 35). In prokaryotes, PAS-containing proteins have been demonstrated to bind various small molecules such as heme, flavin, and a 4-hydroxycinnamyl chromophore to sense molecular oxygen, redox potential, and light, respectively. It is particularly noteworthy that the PAS domain of the Aer energy taxis protein of E. coli binds a flavin adenine dinucleotide to sense changes in cellular redox (5, 34). Also notable is that the FixL sensor kinase protein of Rhizobium meliloti contains a heme-binding PAS domain that controls autophosphorylation in response to molecular oxygen (14, 23). Intriguingly, the ArcB homolog in Haemophilus influenzae lacks the PAS domain. In the present study, we characterize by a genetic and biochemical approach the signal transduction pathway of the Arc two-component system in H. influenzae.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
H. influenzae Rd strain KW20 was grown at 35°C in brain heart infusion agar or broth supplemented with 10 μg of nicotinamide adenine dinucleotide/ml and 10 μg of hemin/ml (sBHI). E. coli strain ECL5020 (arcA::Tetr) was obtained by infecting strain ECL5002 with λ1098, which carries a mini-Tn10-tet, and isolating Tetr and toluidine blue-sensitive colonies. The insertion of tet into arcA was confirmed by PCR and by DNA sequencing. Strain ECL5038 was constructed by P1 transduction of an arcA::Tetr allele from ECL5020 into strain ECL5013. Luria-Bertani (LB) broth and LB agar (17 g/liter) were used for routine growth. Ampicillin, tetracycline, kanamycin, and chloramphenicol were provided at final concentrations of 50, 12, 40, and 20 μg/ml, respectively. For the β-galactosidase activity assay, the Φ(lldP-lacZ)-bearing strains were cultured in buffered LB broth containing 0.1 M MOPS (morpholinepropanesulfonic acid) (pH 7.4), 20 mM d-xylose, and 20 mM l-lactate. When used, potassium nitrate, dimethyl sulfoxide, and trimethylamine N-oxide were added to a concentration of 20 mM. Toluidine blue sensitivity was tested by spreading ∼102 cells on a section of dye-containing agar plates (10 g of tryptone/liter, 8 g of NaCl/liter, 15 g of Bacto Agar/liter, 2 mg of toluidine blue/ml). Dye sensitivity was scored after overnight incubation at 37°C.
Recombinant DNA techniques and PCR.
Chromosomal and plasmid DNA were isolated using the Wizard genomic DNA purification kit (Promega) and the Qiaprep spin miniprep kit (Qiagen), respectively. DNA fragments were recovered from agarose gels using the Qiaquick gel extraction kit (Qiagen). The oligonucleotides used in this study were synthesized by Integrated DNA Technologies Inc. PCRs were carried out using the TaqPlus precision PCR system (Stratagene). The PCR products were purified using the QIAquick PCR purification kit (Qiagen). Sequence verification of PCR-amplified DNA was performed by the University of Michigan Sequencing Core Facility and by the Micro Core Facility of the Department of Microbiology and Molecular Genetics of Harvard Medical School.
Construction of vectors expressing ArcA and ArcB of H. influenzae.
To construct the plasmid pArcAHi, a 3.1-kb fragment containing the arcA gene of H. influenzae (arcAHi) was PCR amplified from chromosomal DNA of H. influenzae Rd strain KW20 with primers ArcA2466 (5′-GGAAGATCTGGTTCACGAGTTTGTGCCGCTGC-3′) and ArcA5582 (5′-GGAAGATCTCGATGCCACCAGCCCAGCAATC-3′). The PCR products were digested with BglII and cloned into BamHI-digested pACYC184. To construct the plasmid pArcBHi, a 2.2-kb fragment containing the arcBHi gene was PCR amplified by using chromosomal DNA of H. influenzae Rd strain KW20 with primers HIAB-N (5′-ACTGAATTCTGGATATGGTAAATCGGG-3′) and HIAB-3′ (5′-CCCGGATCCATGCACCCATTTTAAGCCTC-3′). The PCR products were digested with EcoRI and BamHI and cloned into EcoRI-BamHI-digested pBR322. To construct the plasmid pQE30ArcAHi, a 0.7-kb arcAHi fragment was PCR amplified by using pArcAHi as a template with primers HIA-5′ (5′-CCCGGATCCCATATGACTACTCCAAAAATTCTCGTTGTTGAAA-3′) and HIA-3′ (5′-CCCGGATCCCTGCAGTGCGAATTCTAACAAAACAGG-3′). The PCR products were digested with BamHI and PstI and cloned into BamHI-PstI-digested pQE30. To construct the plasmid pQE30ArcBHi80-599, a 1.6-kb arcBHi fragment was PCR amplified by using pArcBHi as a template with primers HIB-5′ (5′-CCCGGATCCCATATGCTTGAACATTCTCGTCTTG-3′) and HIB-3′ (5′-CCCGGATCCATGCATCCCATTTTAAGCCTC-3′). The PCR products were digested with BamHI and NsiI and cloned into BamHI-PstI-digested pQE30. Plasmids pQE30ArcBEc78-778 and pQE30ArcAEc, expressing the ‵ArcB and ArcA proteins of E. coli (‵ArcBEc and ArcAEc), respectively, have been described previously (13).
Purification of His6-tagged proteins.
E. coli M15 cells cotransformed with pREP4 and the appropriate pQE30 derivative were grown in 1 liter of LB broth supplemented with ampicillin (100 μg/ml) and kanamycin (25 μg/ml). The expression of the His-tagged proteins was induced at the midexponential phase (optical density at 600 nm of ∼0.7) by the addition of 2 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cultures were harvested 4 h after induction. Protein purification was performed at 4°C under nondenaturing conditions as described previously (13).
Phosphorylation and transphosphorylation assays.
Phosphorylation assays were carried out at room temperature in the presence of 40 μM [γ-32P]ATP (specific activity, 2 Ci/mmol; New England Nuclear)–33 mM HEPES (pH 7.5)–50 mM KCl–5 mM MgCl2–1 mM dithiothreitol–0.1 mM EDTA–10% glycerol. Where indicated, purified ArcB and ArcA peptides were used at 50 and 100 pmol, respectively. The reactions were initiated by the addition of [γ-32P]ATP and terminated by the addition of an equal volume of 2× sodium dodecyl sulfate (SDS) sample buffer, and the reaction products were immediately subjected to SDS-polyacrylamide gel electrophoresis on 12% polyacrylamide gels. The radioactivity of the proteins resolved in the gels was determined qualitatively by autoradiography of the dried gels with X-OMAT AR (Kodak) film or quantitatively with a PhosphorImager (Molecular Dynamics).
β-Galactosidase activity assay.
Aerobic cultures of 5 ml each were grown in 250-ml baffled flasks at 37°C with shaking (300 rpm), whereas anaerobic cultures were grown in closed 5-ml test tubes, which were filled to the brim and stirred with a small magnetic bar. β-Galactosidase activity was assayed with exponentially growing cultures as described previously (28).
Construction of recombinant H. influenzae strains.
A deletion in the H. influenzae arcA gene was created as follows: primers, ArcA2466 and ArcAoutC (5′-TGGAAAATGACGCGTAAAGTGATTGTTCTACATAAAAATTC-3′) were used to PCR amplify a product of 1.3 kb from strain KW20. Primers ArcA5582 and ArcAoutN (5′-ACGCGTCATTTTCCATCCTTATACTTATTTTG-3′) were used to PCR amplify a 1.15-kb product from H. influenzae KW20. These two PCR products were used as primers or templates along with ArcA5582 and ArcA2466 to PCR amplify a 2.45-kb stitched product. The 2.45-kb PCR product was digested with BglII and cloned into the BamHI site of pBluescript KS+ (Stratagene) to generate pDAA9. A 1.2-kb MluI fragment containing a kanamycin cassette from pENT3 (2) was cloned into the MluI site of pDAA9 to create pDAAK1. Plasmid pDAAK1, carrying a marked deletion of the arcA gene, was introduced in single copy onto the H. influenzae Rd (ATCC 9008) chromosome by homologous recombination to generate strain RAA6. To complement the arcA deletion in H. influenzae, a fragment containing the arcA gene and its promoter region was PCR amplified from KW20 using primers ArcA5 (5′-CGCAGATCTACGCGTGATGCTCGAAATTCTATCACAAAG-3′) and ArcA3 (5′-CGCAGATCTACGCGTGGAATTTTTATGTAGAACAATCAC-3′) to generate a 942-bp PCR product. This product was digested with MluI and cloned into the AscI site of pXT10 to create pXTAA. The pXT10 plasmid contains a tetracycline resistance gene and a unique AscI restriction site and is flanked by the xyl locus of H. influenzae for the introduction of complementing genes into the chromosome by homologous recombination (E. J. Rubin, J. J. Mekalanos, and B. J. Akerley, unpublished results). The plasmid pXTAA and the empty vector pXT10 were introduced in single copy onto the H. influenzae chromosome by homologous recombination into the d-xylose utilization locus to create strains RAA6C and RAA6V, respectively.
Northern hybridization analysis.
Total RNA from H. influenzae strain Eagan was obtained from cultures grown to an optical density at 600 nm of ∼0.3 under aerobic conditions (50 ml of sBHI containing 20 mM l-lactate in a 250-ml Erlenmeyer flask shaken at ∼200 rpm) or anaerobic conditions (∼50 ml of sBHI containing 20 mM l-lactate in sealed 50-ml conical tubes shaken at ∼200 rpm). RNA was isolated by using TRIzol Reagent (Life Technologies), treated with DNase I, and phenol extracted. For Northern blotting, 10 μg of total RNA was separated by electrophoresis on a 1.5% agarose gel containing 2 M formaldehyde and transferred to a nylon membrane (Nytran; Amersham). The probes for the lctD and rRNA transcripts were PCR products amplified by using primers 5′-ATGATTATTTCATCAGCTAG and 5′-AAGTTTACTTAGATCAACC for lctD and 5′-ACGCGTCATCAAATCTCCTAAAACATTATT and 5′-CGGTTTGGCGATTGCGGAAG for rRNA. The PCR products were labeled using the ECL direct nucleic acid labeling and detection system (Amersham). Washing and hybridization were performed according to the manufacturer's instructions.
RESULTS
The putative ArcB of H. influenzae lacks the PAS domain.
A search of the available complete and incomplete bacterial genome sequences revealed sequences in Vibrio cholerae, Salmonella enterica serovar Typhimurium, Yersinia pestis, and H. influenzae that have high levels of identity to E. coli ArcB (4, 6, 15). The putative ArcB sequence of H. influenzae (ArcBHi), lacks the regions corresponding to amino acids 93 to 134 and 153 to 271 of E. coli ArcB, which lie between the transmembrane regions and the primary transmitter domain (Fig. 1 and 2). The stretch of amino acids from positions 153 to 271 of E. coli ArcB contains the PAS domain (35, 38). If the arcBHi open reading frame is correctly identified, the gene product (ArcBHi) should catalyze the transphosphorylation of the putative cognate response regulator ArcAHi.
FIG. 2.
Predicted ArcB proteins in other bacterial species. Partial CLUSTALW alignment of the E. coli (EC) ArcB to homologues in S. enterica serovar Typhimurium (ST), Y. pestis (YP), V. cholerae (VC), and H. influenzae (HI) is shown. Regions corresponding to amino acids 93 to 134 and 153 to 271 in E. coli are absent in H. influenzae but are present in the other three bacteria.
Autophosphorylation and transphosphorylation of ArcAHi by ArcBHi.
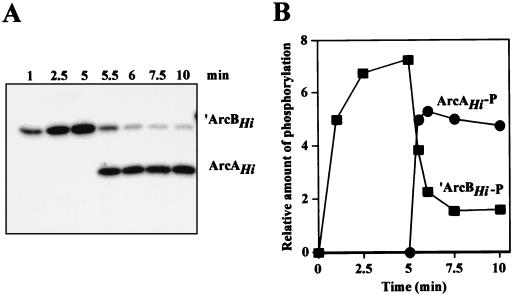
For convenience, we overexpressed and purified His6-tagged ArcAHi and His6-tagged ArcBHi with a deletion encompassing amino acid residues 1 to 79, which constitute the transmembrane segments (hereafter referred to as ‵ArcBHi) under nondenaturing conditions (see Materials and Methods). Previous studies of several sensor kinases have shown that in vitro autophosphorylation and subsequent transphosphorylation of the cognate response regulator protein occur efficiently despite the removal of the transmembrane domain (1, 9, 17, 19, 25). Purified ‵ArcBHi was first incubated with [γ-32P]ATP, and the time course of protein phosphorylation was monitored (Fig. 3). Autophosphorylation of ‵ArcBHi was indicated by its incorporation of increasing amounts of 32P over time. Purified ArcAHi was added at 5 min, and samples were taken during an additional 5-min period. After the addition of ArcA to the reaction mixture, a rapid loss of 32P from ArcB occurred concomitantly with the phosphorylation of ArcA. No phosphorylation of ArcA was observed in the absence of ArcB (data not shown). These results demonstrated that the H. influenzae arcB and arcA genes encode proteins of a two-component system.
FIG. 3.
Phosphorylation of H. influenzae ArcB and ArcA in vitro. (A) Kinetics of autophosphorylation of ArcBHi and transphosphorylation of ArcAHi in the presence of [γ-32P]ATP. ArcA was added immediately after the 5-min time point. At each time point, samples were withdrawn and analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography. (B) Quantitation of the relative amounts of radioactivity incorporated in the Arc proteins by using a PhosphorImager.
‵ArcBHi and ‵ArcBEc catalyze phosphorylation of heterologous ArcA proteins.
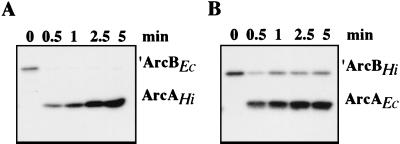
Since the Arc proteins of H. influenzae and E. coli are predominantly conserved, we next tested whether they could participate in heterologous phosphate transfer. ‵ArcBEc was incubated with ArcAHi and ‵ArcBHi was incubated with ArcAEc in the presence of labeled ATP (Fig. 4). Both ArcB kinases efficiently catalyzed the phosphorylation of ArcA from the heterologous species. Peak accumulation of phosphorylated ArcA occurred earlier in the reaction containing ‵ArcBHi than in the reaction containing ‵ArcBEc; however, we cannot ascribe this effect to increased activity of ‵ArcBHi, as it could be specific to heterologous ArcB-ArcA interactions. While we cannot rule out differences in the specific activities of the E. coli and H. influenzae ArcB sensor kinases, these results indicated that the two enzymes share the same substrate specificity in the transphosphorylation reaction. Because sensor kinases have been shown to exhibit a high degree of discrimination against noncognate response regulators (13, 31, 32), the ability of these ArcB proteins to efficiently phosphorylate heterologous ArcA proteins provides further evidence that the two Arc systems have equivalent biochemical functions.
FIG. 4.
Interspecies phosphorylation by Arc proteins of E. coli and H. influenzae. The kinetics of phosphorylation of purified ArcAHi by ‵ArcBEc (A) and ArcAEc by ‵ArcBHi (B) were monitored in reaction mixtures containing [γ-32P]ATP. ArcA was added immediately after 0 min.
Functional complementation of E. coli strains containing arcB and arcA null mutations by the corresponding wild-type H. influenzae alleles.
The ability of the arc genes of H. influenzae to substitute for the E. coli genes in a physiological assay was then addressed. In E. coli, arcA and arcB are both required for resistance to toluidine blue (18). We therefore used this phenotype to evaluate the complementation of E. coli mutants (data not shown). The wild-type strain formed colonies on toluidine blue, whereas the arcA, the arcB, and the arcA arcB double mutants of E. coli exhibited pronounced defects in colony formation. The complementation of E. coli arcA or arcB mutants with the corresponding allele of H. influenzae conferred toluidine blue resistance. Furthermore, the dye resistance of the E. coli arcA arcB double mutant was restored by the combined presence of the arcA+ and arcB+ genes of H. influenzae.
Signaling response and transcriptional control by the H. influenzae Arc system expressed in E. coli.
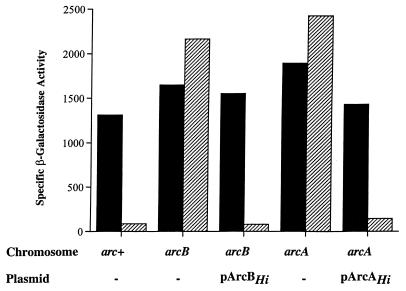
We next examined whether the Arc system of H. influenzae can sense changes in redox conditions of growth and modulate the expression of an ArcA-P target operon when expressed in E. coli. For a reporter, we used a transcriptional fusion of lacZ to the ArcA-P repressible promoter of the E. coli lldPRD operon in which the D gene encodes l-lactate dehydrogenase. The fusion construct Φ(lldP-lacZ) was integrated into the chromosome, and its expression was monitored during the growth of mutants under aerobic and anaerobic conditions. As illustrated in Fig. 5, the level of Φ(lldP-lacZ) expression in the wild-type strain was high in cells grown aerobically but was dramatically repressed during anaerobic growth. In contrast, anaerobic repression of Φ(lldP-lacZ) did not occur in mutant strains lacking ArcA, ArcB, or both (data not shown). When the E. coli mutant strains were provided with the homologous H. influenzae arc genes, anaerobic repression of Φ(lldP-lacZ) expression was restored.
FIG. 5.
Aerobic and anaerobic expression levels of Φ(lldP-lacZ) in different genetic backgrounds. Solid bars, aerobic levels of β-galactosidase activity; hatched bars, anaerobic levels of β-galactosidase activity.
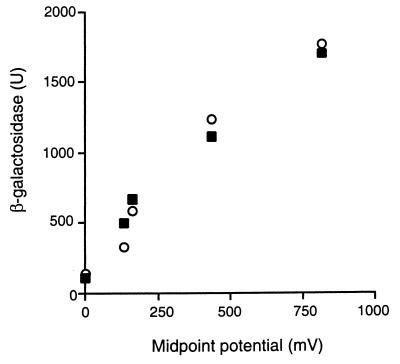
The responses of the two ArcB sensors to various external electron acceptors were then compared. E. coli reporter strains carrying arcB genes from either E. coli or H. influenzae were grown in buffered LB medium (see Materials and Methods) either fermentatively or with O2 (midpoint redox potential [E0′] = 880 mV), nitrate (E0′ = 420 mV), dimethyl sulfoxide (E0′ = 160 mV), or trimethylamine N-oxide (E0′ = 130 mV) as an exogenous electron acceptor, and the expression of Φ(lldP-lacZ) was analyzed. The two ArcB sensors responded in similar manners, as indicated by the increases in the expression of Φ(lldP-lacZ) proportional to the midpoint potential of the supplemented electron acceptor (Fig. 6). The redox regulation mediated by the H. influenzae arc genes demonstrated cross-species regulation and, more importantly, the ability of the PAS-less ArcBHi to sense and mediate responses to changes in redox conditions of growth.
FIG. 6.
Comparison of the responses of ArcBEc and ArcBHi sensors to various external electron acceptors. E. coli strains carrying Φ(lldP-lacZ) and arcB genes from either E. coli (closed squares) or H. influenzae (open circles) were grown aerobically (E0′ = 880 mV) or anaerobically in the presence or absence of nitrate (E0′ = 420 mV), dimethyl sulfoxide (E0′ = 160 mV), and trimethylamine N-oxide (E0′ = 130 mV). β-Galactosidase activity is plotted against the midpoint potential of the electron acceptors. Cultures and assays were performed with three independent samples per point, and variation was less than 10% between replicate samples.
Arc-dependent redox regulation in H. influenzae.
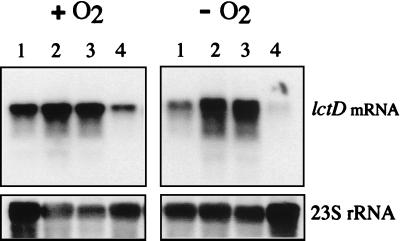
To examine Arc-dependent regulation in H. influenzae, we investigated changes in transcription levels of a candidate Arc-regulated gene, lctD (The Institute for Genomic Research gene name HI1739.1), in response to changes in redox conditions of growth. The promoter of the lctD gene possesses a putative ArcA recognition site, and the lctD gene product is 72% identical at the amino acid level to the l-lactate dehydrogenase of E. coli encoded by the lldD gene (8). Northern blots containing RNA from H. influenzae cultures grown anaerobically or aerobically were analyzed with an H. influenzae lctD-specific probe (Fig. 7). Levels of lctD mRNA were significantly higher in cells grown aerobically than in cells grown anaerobically (Fig. 7, lanes 1), which indicates that lctD expression is modulated in response to changes in culture aeration. Deletion of arcAHi prevented the anaerobic repression of lctD (lanes 2). However, when arcAHi was reintroduced in single copy into the H. influenzae chromosome (see Materials and Methods), the anaerobic repression of lctD was fully restored. Therefore, the derepression of lctD in the mutant strain was caused by the arcA deletion and was not due to a spontaneous unlinked mutation or to transcriptional polarity effects. These results indicate that the Arc systems of E. coli and of H. influenzae play similar transcriptional regulatory roles in response to redox conditions.
FIG. 7.
Expression of lctD mRNA in H. influenzae. A Northern blot containing total RNA extracted from aerobically (+O2) or anaerobically (−O2) grown H. influenzae cultures was hybridized with an lctD-specific probe (upper panels). The filter was reprobed with a 23S rRNA specific-probe (lower panels). Lanes 2 to 4 contain RNA samples from H. influenzae strains bearing the following arc alleles: ΔarcA::Kanr (strain RAA6) (lanes 2); ΔarcA::Kanr and ΔxylA::Tetr (strain RAA6V) (lanes 3); and ΔarcA::Kanr, ΔxylA::Tetr, and arcA+ (strain RAA6C) (lanes 4). Lanes 1 contain RNA samples from the wild-type strain H. influenzae Rd.
DISCUSSION
The ability of ArcBHi and ArcAHi to mediate anaerobic repression of the lld operon of E. coli as well as of the lctD gene of H. influenzae provides incontrovertible evidence that the Arc two-component systems of the two species play essentially similar roles in signal transduction. The signaling response is not exclusive to the lld operon, as ArcBHi expressed in E. coli has also been shown to mediate redox responsive control over succinate dehydrogenase expression (26). ArcBHi lacks the PAS domain, and yet, under a range of redox conditions, the sensor kinase is capable of mediating responses similar to those of ArcBEc. Therefore, it is reasonable to conclude that the H. influenzae protein senses these signals by a PAS-independent mechanism. The fact that ArcBHi alone can functionally replace ArcBEc in the E. coli host also excludes the possibility that ArcBHi works only in conjunction with an unidentified PAS-containing protein in H. influenzae for redox sensing. Theoretically, a cryptic domain fulfilling the function of the PAS domain could exist elsewhere in ArcBHi. Nevertheless, it is clear that the PAS itself is nonessential for signaling by the H. influenzae sensor which lacks this motif.
In a previous study, it was proposed that the PAS domain may play a role in signal reception based on the characterization of several mutant E. coli ArcB proteins. A deletion spanning the PAS domain of ArcB abrogated its ability to repress the sdh operon anaerobically in E. coli. Moreover, one out of four tested substitutions of conserved amino acid residues in the PAS domain, i.e., the replacement of Asn181 with Ala, affected in vivo signaling by ArcB (27). However, this mutant protein was also inactive in vitro, making it difficult to discern whether the observed in vivo defect is at the level of signal reception or is due to a negative effect on the catalytic activity of ArcB. The complex structure of ArcB makes it likely that a small local change can affect the function of a critical remote site.
The presence of the PAS domain in the ArcB sequences of E. coli, S. enterica serovar Typhimurium, Y. pestis, and V. cholerae suggests that the domain suffered a deletion during the evolution of H. influenzae. This pathogen is a fastidious gram-negative facultative anaerobe that colonizes the human respiratory tract and bloodstream and causes a diverse set of diseases, including otitis media, community-acquired pneumonia, and meningitis. It is possible that the PAS domain in the ArcB proteins of other bacterial species plays a sensory role that is critical only for cells that must adapt to a broader range of environmental conditions, such as growth outside of a host, but is superfluous for an obligate parasite.
ACKNOWLEDGMENTS
We acknowledge The Institute for Genomic Research (http://www.tigr.org) for providing access to the database for finished and unfinished microbial genomes.
This work was supported by a Faculty Research grant from the Horace H. Rackham School of Graduate Studies to B.J.A., U.S. Public Health Service grant GM40993 from the NIGMS, NIH, to E.C.C.L, and a fellowship from the American Cancer Society to S.M.W.
REFERENCES
- 1.Aiba H, Mizuno T, Mizushima S. Transfer of phosphoryl group between two regulatory proteins involved in osmoregulatory expression of the ompF and ompC genes in Escherichia coli. J Biol Chem. 1989;264:8563–8567. [PubMed] [Google Scholar]
- 2.Akerley B J, Rubin E J, Camilli A, Lampe D J, Robertson H M, Mekalanos J J. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc Natl Acad Sci USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett J F, Hoch J A. Two-component signal transduction as a target for microbial anti-infective therapy. Antimicrob Agents Chemother. 1998;42:1529–1536. doi: 10.1128/aac.42.7.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson D A, Karsch-Mizrachi I, Lipman D J, Ostell J, Rapp B A, Wheeler D L. GenBank. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibikov S I, Barnes L A, Gitin Y, Parkinson J S. Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli. Proc Natl Acad Sci USA. 2000;97:5830–5835. doi: 10.1073/pnas.100118697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 7.Burbulys D, Trach K A, Hoch J A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 8.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 9.Forst S, Delgado J, Inouye M. Phosphorylation of OmpR by the osmosensor EnvZ modulates expression of the ompF and ompC genes in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:6052–6056. doi: 10.1073/pnas.86.16.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Georgellis D, Kwon O, De Wulf P, Lin E C. Signal decay through a reverse phosphorelay in the Arc two-component signal transduction system. J Biol Chem. 1998;273:32864–32869. doi: 10.1074/jbc.273.49.32864. [DOI] [PubMed] [Google Scholar]
- 11.Georgellis D, Kwon O, Lin E C. Amplification of signaling activity of the arc two-component system of Escherichia coli by anaerobic metabolites. An in vitro study with different protein modules. J Biol Chem. 1999;274:35950–35954. doi: 10.1074/jbc.274.50.35950. [DOI] [PubMed] [Google Scholar]
- 12.Georgellis D, Kwon O, Lin E C. Quinones as the redox signal for the arc two-component system of bacteria. Science. 2001;292:2314–2316. doi: 10.1126/science.1059361. [DOI] [PubMed] [Google Scholar]
- 13.Georgellis D, Lynch A S, Lin E C C. In vitro phosphorylation study of the Arc two-component signal transduction system of Escherichia coli. J Bacteriol. 1997;179:5429–5435. doi: 10.1128/jb.179.17.5429-5435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilles-Gonzalez M A, Ditta G S, Helinski D R. A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature. 1991;350:170–172. doi: 10.1038/350170a0. [DOI] [PubMed] [Google Scholar]
- 15.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, Gill S R, Nelson K E, Read T D, Tettelin H, Richardson D, Ermolaeva M D, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann R D, Nierman W C, White O. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoch J A. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 17.Igo M M, Ninfa A J, Silhavy T J. A bacterial environmental sensor that functions as a protein kinase and stimulates transcriptional activation. Genes Dev. 1989;3:598–605. doi: 10.1101/gad.3.5.598. [DOI] [PubMed] [Google Scholar]
- 18.Iuchi S, Lin E C. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci USA. 1988;85:1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iuchi S, Lin E C C. Purification and phosphorylation of the Arc regulatory components of Escherichia coli. J Bacteriol. 1992;174:5617–5623. doi: 10.1128/jb.174.17.5617-5623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jourlin C, Ansaldi M, Mejean V. Transphosphorylation of the TorR response regulator requires the three phosphorylation sites of the TorS unorthodox sensor in Escherichia coli. J Mol Biol. 1997;267:770–777. doi: 10.1006/jmbi.1997.0919. [DOI] [PubMed] [Google Scholar]
- 21.Kwon O, Georgellis D, Lin E C C. Phosphorelay as the sole physiological route of signal transmission by the Arc two-component system of Escherichia coli. J Bacteriol. 2000;182:3858–3862. doi: 10.1128/jb.182.13.3858-3862.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwon O, Georgellis D, Lynch A S, Boyd D, Lin E C C. The ArcB sensor kinase of Escherichia coli: genetic exploration of the transmembrane region. J Bacteriol. 2000;182:2960–2966. doi: 10.1128/jb.182.10.2960-2966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lois A F, Weinstein M, Ditta G S, Helinski D R. Autophosphorylation and phosphatase activities of the oxygen-sensing protein FixL of Rhizobium meliloti are coordinately regulated by oxygen. J Biol Chem. 1993;268:4370–4375. [PubMed] [Google Scholar]
- 24.Lynch A S, Lin E C C. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J Bacteriol. 1996;178:6238–6249. doi: 10.1128/jb.178.21.6238-6249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makino K, Shinagawa H, Amemura M, Kawamoto T, Yamada M, Nakata A. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J Mol Biol. 1989;210:551–559. doi: 10.1016/0022-2836(89)90131-9. [DOI] [PubMed] [Google Scholar]
- 26.Manukhov I V, Bertsova Y V, Trofimov D Y, Bogachev A V, Skulachev V P. Analysis of HI0220 protein from Haemophilus influenzae, a novel structural and functional analog of ArcB protein from Escherichia coli. Biochemistry (Moscow) 2000;65:1321–1326. [PubMed] [Google Scholar]
- 27.Matsushika A, Mizuno T. Characterization of three putative sub-domains in the signal-input domain of the ArcB hybrid sensor in Escherichia coli. J Biochem (Tokyo) 2000;127:855–860. doi: 10.1093/oxfordjournals.jbchem.a022679. [DOI] [PubMed] [Google Scholar]
- 28.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 29.Parkinson J S. Genetic approaches for signaling pathways and proteins. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: ASM Press; 1995. pp. 9–24. [Google Scholar]
- 30.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 31.Pernestig A K, Melefors O, Georgellis D. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in Escherichia coli. J Biol Chem. 2001;276:225–231. doi: 10.1074/jbc.M001550200. [DOI] [PubMed] [Google Scholar]
- 32.Perraud A L, Kimmel B, Weiss V, Gross R. Specificity of the BvgAS and EvgAS phosphorelay is mediated by the C-terminal HPt domains of the sensor proteins. Mol Microbiol. 1998;27:875–887. doi: 10.1046/j.1365-2958.1998.00716.x. [DOI] [PubMed] [Google Scholar]
- 33.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 34.Repik A, Rebbapragada A, Johnson M S, Haznedar J O, Zhulin I B, Taylor B L. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol Microbiol. 2000;36:806–816. doi: 10.1046/j.1365-2958.2000.01910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor B L, Zhulin I B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uhl M A, Miller J F. Integration of multiple domains in a two-component sensor protein: the Bordetella pertussis BvgAS phosphorelay. EMBO J. 1996;15:1028–1036. [PMC free article] [PubMed] [Google Scholar]
- 37.Yuk M H, Cotter P A, Miller J F. Genetic regulation of airway colonization by Bordetella species. Am J Respir Crit Care Med. 1996;154:S150–S154. doi: 10.1164/ajrccm/154.4_Pt_2.S150. [DOI] [PubMed] [Google Scholar]
- 38.Zhulin I B, Taylor B L, Dixon R. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem Sci. 1997;22:331–333. doi: 10.1016/s0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]