Abstract
Dose limiting cardiotoxicity remains a major limiting factor in the clinical use of several cancer chemotherapeutics including anthracyclines and the antimetabolite 5-fluorouracil (5-FU). Prior work has demonstrated that chemotherapeutics increase expression of R7 family regulator of G protein signaling (RGS) protein-binding partner Gβ5, which drives myocyte cytotoxicity. However, though several R7 family members are expressed in heart, the exact role of each protein in chemotherapy driven heart damage remains unclear. Here, we demonstrate that RGS11, downregulated in the human heart following chemotherapy exposure, possesses potent anti-apoptotic actions, in direct opposition to the actions of fellow R7 family member RGS6. RGS11 forms a direct complex with the apoptotic kinase CaMKII and stress responsive transcription factor ATF3 and acts to counterbalance the ability of CaMKII and ATF3 to trigger oxidative stress, mitochondrial dysfunction, cell death, and release of the cardiokine neuregulin-1 (NRG1), which mediates pathological intercommunication between myocytes and endothelial cells. Doxorubicin triggers RGS11 depletion in the murine myocardium, and cardiac-specific OE of RGS11 decreases doxorubicin-induced fibrosis, myocyte hypertrophy, apoptosis, oxidative stress, and cell loss and aids in the maintenance of left ventricular function. Conversely, RGS11 knockdown in heart promotes cardiac fibrosis associated with CaMKII activation and ATF3/NRG1 induction. Indeed, inhibition of CaMKII largely prevents the fibrotic remodeling resulting from cardiac RGS11 depletion underscoring the functional importance of the RGS11-CaMKII interaction in the pathogenesis of cardiac fibrosis. These data describe an entirely new role for RGS11 in heart and identify RGS11 as a potential new target for amelioration of chemotherapy-induced cardiotoxicity.
Keywords: RGS proteins, Chemotherapy, Cardiotoxicity, Oxidative stress, Cell death
Graphical abstract
1. Introduction
According to the International Agency for Research on Cancer (IARC), there were 9.5 million deaths from cancer in 2018, numbers expected to rise in the coming decades due to the aging population and increasing prevalence of risk factors for malignant transformation. Irrespective of the specific malignancy, chemotherapy remains a critical component of cancer therapy and has been shown to significantly improve patient survival over a 20-year period [1]. However, the clinical utility of many of these drugs is limited by life-threatening side effects, which include chemotherapy-induced cardiotoxicity characterized by compromised left ventricular function, structural heart damage, conduction deficits, and/or vascular abnormalities. The most common causative agents are anthracyclines (e.g. doxorubicin) and the antimetabolite 5-fluorouracil (5-FU) with incidence rates of doxorubicin- and 5-FU-dependent heart damage estimated at 9% [2] and 1–19% [3], respectively. Though administration schedule, patient age, cancer type, and cumulative drug dose influence risk of cardiotoxicity, impacts of chemotherapy on the heart may not be appparent for or even years after the initial drug exposure [4]. Further, there are no adjuvants available to treat or prevent chemotherapy-driven cardiotoxicity. Thus, detection, risk assessment, and treatment of chemotherapeutic-dependent cardiac damage remain areas of active research.
Though several intracellular signaling cascades are altered in cardiomyocytes following chemotherapy exposure, several common cytotoxic mechanisms have emerged. Among the most frequently described are free radical generation, oxidative stress, mitochondrial dysfunction, and the result initiation of apoptotic cascades as well as disruptions in calcium (Ca2+) homeostasis and activation of cellular stress responses [5]. Myocytes are particularly sensitive to oxidative damage due to their high metabolic demand and mitochondrial volume [6] and chemotherapeutic drugs such as doxorubicin triggers oxidative stress via several mechanisms including secondary to mitochondrial dysfunction [7,8] and via activation of NADPH oxidase (Nox) complexes [9,10]. Ultimately, accumulating reactive oxygen species (ROS) overwhelm the cells antioxidant defense and lead to induction of intrinsic mitochondrial cell death pathways [11,12]. ROS generation also drives induction of Ca2+/Calmodulin-dependent protein kinase (CaMKII), which has been shown to contribute to Ca2+ leak from the sarcoplasmic reticulum and Ca2+ overload in cardiomyocytes following anthracycline exposure [13,14]. Though its function remains unclear, the profile of signal transducers recruited in the myocardium following doxorubicin treatment includes activating transcription factor 3 (ATF3) [15], a hub in the adaptive stress response that has been implicated in doxorubicin-induced cytotoxicity [16,17].
Though 5-FU metabolites also damage myocyte mitochondria, 5-FU also targets vascular endothelial cells (ECs) and has been specifically associated with myocardial ischemia secondary to coronary spasm [18]. Mechanisms of 5-FU-induced vascular toxicity also involve ROS generation and mitochondrial-dependent apoptosis [[19], [20], [21], [22]] suggesting that 5-FU may have a shared molecular target in ECs and cardiomyocytes that drives oxidative stress. Alternatively, release of autocrine or paracrine factors from one cell type might propagate cellular stress signals throughout the cardiovascular system. Indeed, doxorubicin drives release of several paracrine factors from the vascular endothelium that can influence the viability and function of cardiomyocytes [23]. Among these cytokines is the cardioactive growth factor neuregulin-1 (NRG1) [23], which may protect against acute doxorubicin-induced cardiotoxicity [24,25]. Indeed, NRG1 haploinsufficiency exacerbated doxorubicin-dependent cardiac damage following a single drug bolus [26], though the role of NRG1 in the chronic phase where sustained growth factor production often turns maladaptive driving ongoing fibrotic remodeling and compromising cardiac contractility remains unclear. Regardless, it is likely that mitigation of chemotherapy-dependent cardiotoxicity might require the targeting of several critical myocyte-intrinsic and -extrinsic cytotoxic signals.
The atypical G protein Gβ5 is up-regulated in the myocardium in response to several cancer chemotherapeutics including doxorubicin and 5-FU, a result that can be replicated in isolated ventricular cardiomyocytes (VCM) or fibroblasts (VCF) [27]. Indeed, knockdown (KD) of Gβ5 in heart ameliorates doxorubicin-induced hypertrophy and fibrosis [27]. Rather than mediating signal transduction via G protein coupled receptors, Gβ5 forms a requisite co-stabilizing complex with members of the R7 subfamily of Regulators of G protein Signaling (RGS) proteins [28], 3 of which are expressed in heart (RGS6, RGS7, and RGS11) [29,30]. Like Gβ5, RGS6 is up-regulated following doxorubicin exposure [27,31], and the RGS6-Gβ5 complex has been shown to ATM serine/threonine kinase (ATM)-dependent apoptotic signaling [27,31,32]. However, the function of RGS11 in heart remains unknown.
We now demonstrate that chemotherapy exposure in human cancer patients is associated with down-regulation of RGS11 in heart, an outcome opposite to that observed for RGS6 in the murine myocardium [31]. Further, while knockout (KO) of RGS6 protects against chemotherapy-induced cardiac dysfunction [31], cardiac-specific RGS11 depletion exacerbates doxorubicin-dependent cardiotoxicity and RGS11 overexpression (OE) in heart is protective. Notably, RGS11 appears to have a unique function in modulating both myocyte-intrinsic and paracrine mechanisms of chemotherapy-driven cardiac damage.
2. Materials and methods
2.1. Materials
The source/catalog information for all reagents (Table S1), antibodies (Table S2), assay kits (Table S3), and cell lines (Table S4) can be found in Supplementary Tables 1–4. Table S2 provides information on antibody dilutions (immunoblotting, immunohistochemistry, and immunoprecipitation).
2.2. Animals
Male Swiss albino mice (25–30 g) were reared on a balanced laboratory diet as per NIN, Hyderabad, India and given tap water and food ad libitum. They were kept at 20 ± 2 °C, 65–70% humidity, and day/night cycle (12 h/12 h). For all animal experiments, analyses were performed by an observer blinded to drug treatment.
2.3. Chemotherapy drug treatment regimens
Eight-to ten-week-old animals were subjected to either the acute or chronic chemotherapeutic drug regimens. Mice on the chronic treatment regimen received multiple doses of doxorubicin (cumulative dose of 45 mg/kg i. p.; 9 mg/kg every other week), 5-FU (cumulative dose of 200 mg/kg i. p.; 40 mg/kg every other week), oxaliplatin (cumulative dose of 45 mg/kg i. p.; 9 mg/kg every other week), irinotecan (cumulative dose of 175 mg/kg i. p.; 35 mg/kg every other week), or saline over the period of 10 weeks. Mice on the acute treatment regimen received a single dose of doxorubicin (20 mg/kg i. p.), 5-FU (150 mg/kg i. p.), oxaliplatin (30 mg/kg i. p.), irinotecan (50 mg/kg i. p.), or saline administrated via i. p. injection. 9 weeks after the final drug dose mice were euthanized by cervical dislocation and blood/multiple tissues were collected for downstream analysis. A second cohort of mice treated with doxorubicin were given a single dose of doxorubicin (20 mg/kg i. p.) and sacrificed 2, 4 or 8 weeks later after performing cardiovascular phenotyping. Doxorubicin dosing for the acute treatment paradigm was chosen based on prior reports [33]. The dose of doxorubicin used for chronic administration was calculated from a typical doxorubicin treatment schedule (40–75 mg/m2 intravenously every 21 days)by adjusting for the relative body surface area of a human versus mouse [34]. To correct for the bioavailability of doxorubicin when administered via intraperitoneal injection versus intravenously (∼44%) [35] we altered the schedule to biweekly administration. Doses of 5-FU, oxaliplatin, and irinotecan were estimated based on the proportionate drug dose administered clinically.
2.4. In vivo RGS11KD in heart
To achieve cardiac KD of RGS11 1-week-old wild type (WT) mice received an intra-cardiac injection of 4.5 × 108 lentiviral vectors containing scramble or RGS11-targeted small hairpin RNA (shRNA) according to a previously published protocol [27]. Mice were then returned to their mothers until weaning and allowed to age to adulthood. At 8–10 weeks of age animals were exposed to chemotherapeutic drug treatment regimens as outlined above. RGS11KD was verified via immunoblotting and immunohistochemistry. Following shRNA administration, body weight (1X/week) and food intake (1–2X/week) were monitored. No notable alterations in animal weight, food intake or general wellbeing were found.
2.4.1. Cloning and construct generation
The full-length RGS11 and CaMKIIδ coding sequences were amplified by PCR from human blood cDNA according to our published method [36]. RGS11 and CaMKIIδ deletion & point mutation sequences were generated by overlapping primer-based PCR amplification and cloned into the pEGFP-N1 vector or pCMV-HA-N vectors, respectively. Information regarding the primers utilized for all construct generation has been included in Table S5.
To generate viral constructs for RGS11OE in vivo, the full-length mouse RGS11 sequence was isolated from mouse brain and cloned into the pMD20T vector as above. The lentiviral vector for mRGS11 was generated via subcloning into the pLenti CMV Puro DEST cloning vector (Addgene, Watertown, MA, USA) and packaged using the pMD2. G VSV-G envelope expressing plasmid (Addgene) and psPAX2 (Addgene). Lentiviral particles were generated in AC-16 cells as per a standard protocol. 70 μL of lentivirus containing 2 × 108 particles of either mRGS11-Lenti or a control empty vector virus were packaged for delivery with Invivofectamine 3.0 (ThermoFisher) and administered via intracardiac injection as described above. Mice received injections of saline or KN-93 (8 mg/kg, 2 doses every 4 days), a dose previously shown to alter the pathogenesis of heart failure following pressure overload [37], beginning 15 days after the viral injection. Animals were sacrificed 2 days after the final KN-93 injection and tissues isolated for downstream analyses.
2.5. Histology and immunohistochemistry
Paraffin-embedded formalin-fixed mouse and human heart & liver tissue sections were stained with Hematoxylin and Eosin (H&E) or Masson trichome (Sigma, St. Louis, MO) to detect tissue architecture or collagen deposition, respectively. Reagents were utilized as per the manufacturers’ protocols. Immunohistochemical staining of both mouse and human tissue sections was performed as per a standard protocol [32]. For RGS11, 4-Hydroxynonenal (4HNE), and CaMKII staining, 7–10 sections were stained from each animal with 5 pictures randomly selected from each slide. The blue-stained collagen in tissue section image stained with Masson trichrome was processed using the "Threshold" tool of ImageJ software (NIH, USA) and the fraction of the total area that was stained blue was quantified. Average myocyte area was estimated by quantifying apparent myocyte area with ImageJ. 7–10 sections were quantified per animal with 5 myocytes randomly selected per slide. Quantification was done by an individual blinded to group.
2.6. Immunoblotting
Tissues were rapidly dissected from mice and flash frozen in liquid nitrogen. Tissue homogenates and cell lysates were prepared in RIPA buffer containing protease and phosphatase inhibitors (Sigma), quantified, and probed as previously described [38]. Twenty μg of protein per sample was subjected to SDS-PAGE and immunoblotting using standard techniques. Immunoblots were developed using chemiluminescence method with HRP-labeled secondary antibodies. Antibody dilution and catalog information can be found in Table S2. Densitometric quantification of western blots was performed utilizing Image J software (NIH). Protein expression was normalized to loading control (β-Actin) and expressed relative to control conditions.
2.7. Cardiovascular phenotyping
We used two-dimensional echocardiography to determine cardiac function in vivo. Using an ultrasound system (Vivid S5 system, GEHealthcare, USA) in M-mode, we measured the left ventricular parameters (left ventricular end diastolic and systolic pressure; LVEDP and LVESP) and left ventricular ejection fraction (LVEF) on lightly sedated mice treated with chemotherapeutic drugs and/or RGS11OE as described earlier.
2.8. Murine VCM, VCF, and EC isolation and culture
Primary VCM and VCF were isolated from 8 to 10-week-old adult mice according to a published protocol [39]. Cardiac ECs were isolated from 8-week-old mice hearts following previously published protocol [40]. Briefly, mice hearts were rinsed with chilled PBS thoroughly to remove blood and minced into small pieces for EC culture. The pieces were transferred to 8 ml of DMEM containing 10% FBS, 0.16 mg/ml gentamycin, 4 mM l-glutamine and 0.3 units/ml of collagenase A and incubated at 37 °C for at least 60 min with periodical shaking. Cell clumps were separated initially through a sterile 18G needle, the cell population was filtered through a 30-μm filter, and the resultant single-cell suspension was washed thrice in DMEM. The solution was spun once more at 400×g for 6 min and cell pellet was collected. The cells were re-suspended in 5 ml of DMEM. Next, 8 μl of Dynabeads (Invitrogen) coated with a CD31 antibody was added to the cell suspension as per the manufacturer's instructions. After 15 min at room temperature, the unbound cells were removed from the mixture using a magnet. Dynabead-bound cells were further washed 5 times with DMEM and immuno-selected cells were plated either on petri dishes or chamber slides with HiEndoXLTM EC expansion medium (Himedia) supplemented growth factors.
2.9. Culture of human cell lines
The human cardiomyocyte cell line AC-16 (Merck, Darmstadt, Germany) was cultured in DMEM and 10% FBS (Gibco, Waltham, MA, USA) in a 37 °C incubator at 5% CO2. The human umbilical vein ECs (HUVEC, Himedia) cell line was cultured in HiEndoXLTM EC expansion medium with 3% FBS in a 37 °C incubator at 5% CO2.
2.10. Generation of RG11 KO AC-16 cells using CRISPR/Cas9
Guide RNA (gRNA) targeting human RGS11 gene exon 15 were designed using tools available from Integrated DNA technologies (IDT, Newark, NJ, USA). High on target and low off target gRNAs were chosen without a PAM sequence, cloned into the PX459 CRISPR system plasmid (Addgene) using standard methods and confirmed via sequencing. The resulting construct was transfected into AC-16 cells using lipofectamine 3000 (Thermo Fisher). Cells were re-plated 48 h post-transfection and subjected to puromycin selection. After 14 days, puromycin selected colonies were plated at 1 cell/well. 21 colonies were picked, and each colony was pelleted down separately for subsequent genomic DNA isolation by phenol/chloroform/isoamyl alcohol extraction for sequencing and protein detection by western blotting. We successfully knocked out RGS11 in one colony (#4). The T7 endonuclease 1 (T7E1) mismatch detection assay was used for validation (Fig. S1).
2.11. Drug treatment in cultured cells and conditioned media experiments
Isolated cells were not disturbed for at least 16–24 h before experimentation. Cells were then transduced with lentiviral vectors according to the manufacturer's instructions with RGS11-targeted or scramble shRNA (Santa Cruz Biotechnology) or shRNA for ATF3, RGS7, or CaMKIIδ (Santacruz Biotechnology). Full length RGS11, RGS11 deletion constructs, WT CaMKII, or CaMKII mutants were transfected where indicated. Cells were treated with doxorubicin (3 μM, 16 h), oxaliplatin (0.06 mM, 16 h) or 5-FU (500 μM, 16 h) in the presence or absence of pre-treatment with Ru360 (50 μM, 1 h), cyclosporin A (0.2 mM, 45 min), KN93 (50 μM, 1 h), or cI-1033 (2 μM, 1 h) to block mitochondrial calcium uniporter, mitochondrial permeability transition pore, CaMKII, or NRG1 receptors erbB2/erbB4, respectively, where indicated. N-acetyl cysteine (NAC; 5 mM, 12 h) was used as a non-specific ROS scavenger due to its function as a glutathione donor [41]. Doses of chemotherapeutic drugs used in cultured cells were chosen based on prior reports investigating the toxic actions of these drugs in human cardiomyocytes [23,[42], [43], [44]]. Inhibitor doses were chosen to exceed reported IC50 values for each target [45,46], while also ensuring minimal off target effects on myocyte viability [47]. To determine whether ECs, typically located adjacent to cardiomyocytes in the capillary rich myocardium are capable of releasing cardiotoxic factors capable of damaging myocytes, we employed a conditioned media-based paradigm. This experimental design allowed us to manipulate RGS11 expression in each cell type in isolation while also isolating the chemokine signal facilitating this EC-VCM communication. For conditioned media experiments, cells were initially cultured in standard culture media until cells were 80–85% confluent. The media was then replaced with serum-free DMEM, and drugs were added for 36 h. After replacing the media again and rinsing, cells were incubated for an additional 8 h in drug/serum-free media. This “conditioned media” was collected from donor cells and then used to replace standard media on recipient cells. Recipient cells were collected for further processing after 12–16 h.
2.12. Immunoprecipitation
AC-16 cells (3 × 106) were lysed, and protein concentration measured via BCA protein assay. 600 μg of protein was equilibrated in IP lysis buffer (50 mM Tris, 5 mM EDTA, 250 mM NaCl and 0.1% Triton X-100) and bait antibodies (GFP, RGS11, CaMKII, ATF3 or control mouse IgG) for 12 h on a rotor at 4 °C. 30 μl of Protein G sepharose beads (Abcam) were pre-cleared, equilibrated and then added to lysate. After a 2-h incubation, bead slurries were centrifuged and washed 3X with IP buffer. Immunocomplexes were eluted in non-reducing laemmli buffer at 95 °C and subjected to SDS-PAGE and immunoblotting with prey antibody (CaMKII or RGS11).
2.13. Measurement of ROS generation
ROS generation was estimated in the tissues and primary cells using the cell-permeable oxidation-sensitive probe, CM-H2DCFDA as described previously [27]. Briefly, cells were harvested by centrifugation, washed three times with ice-cold PBS, re-suspended in PBS and incubated with 5 μM CM-H2DCFDA (Sigma) for 20 min at 37 °C. After incubation cells were again washed and lysed in PBS with 1% Tween 20. ROS level was determined at the ratio of dichlorofluorescein excitation at 480 nm to emission at 530 nm. Imaging of CM-H2DCFDA retention in cells was done after 30 min of incubation. The CM-H2DCFDA assay is utilized as a general oxidative stress indicator and not as a detector of a specific oxidant due to known limitations of the probe.
2.14. ELISAs and enzymatic assays
A summary of commercially available kits used to measure superoxide dismutase (SOD), caspase-3 activity, levels of mitochondrial Ca2+, cell death (apoptosis; cytoplasmic histone-associated DNA fragments), NRG beta 1 and Terminal deoxynucleotidyl transferased UTP nick end labeling (TUNEL) is available in Table S3. Cells were harvested and samples processed according to the manufacturer's instructions.
2.15. Cell viability
The MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) reduction assay was used to monitor cell viability. 5 × 104 cells/well were seeded in 48 well plates with DMEM +10% FCS. The constructs were transfected into the cells using lipofectamine and cells were harvested after 36 h. The MTT (Sigma) solution was prepared at 1 mg/mL concentration in medium without phenol red, and 200 μL of MTT solution was added into each well. The cells were incubated for 2 h at 37 °C 200 μL of DMSO was then added into each well for solubilization of the formed formazan crystals. The optical density of the wells was determined at a wavelength of 550 nm (Biotek Instruments).
2.16. YASARA homology modelling of RGS11 & in-silico molecular docking and molecular dynamics (MD) simulations
As there is no reported experimental X-ray or solution NMR structure of RGS11 in the international protein data bank (PDB) repository (https://www.rcsb.org), the 3D structural model of RGS11 was modelled using software program YASARA (http://www.yasara.org). The amino-acid sequence of RGS11 was used as an input (in FASTA format) and the default homology modelling macro of YASARA structure was executed to generate the structural model of RGS11 [48].
The binding modes of RGS11 in complex with CaMKII were generated using the inbuilt VINA docking method in YASARA software [49]. The highest energy RGS11-CamKII complex was further evaluated for solution stability under biological conditions by performing 116.50 ns molecular dynamics (MD) simulation in explicit water solvent using YASARA Dynamics software (20.7.4.W.64 employing the AMBER14 force field) [50]. The MD simulation was performed as per the details and parameters described previously [51,52]. The MD snapshots were saved every 250 ps and 467 MD trajectories were generated during the MD simulations andanalyzed using YASARA macro “md_analysis.mcr”. Energy terms were calculated using AMBER14 force field parameters [53,54]. For the analysis of amino acid residues present on the protein-protein interaction interface, the solvent accessible surface area (SASA) calculations were performed using the InterProSurf Webserver (http://curie.utmb.edu/usercomplex.html) [55]. The analysis involves the SASA calculation of residues in the complex and in the isolated subunits; the residues showing significant change in the SASA value were considered most likely to be on the interaction interface. The CaMKII-RGS11 complex structure with highest binding energy was energy minimized (first using YASARA software) and used as an input for the SASA calculation.
2.17. Study approval
Mouse experiments were performed at the Aryakul College of Pharmacy & Research, Lucknow, India. Animals were procured after obtaining clearance from the college Animal Ethics Committee (1896/PO/Re/S/16/CPCSEA/2021/5) and were handled following International Animal Ethics Committee Guidelines and in agreement with the Guide for the Use and Care of Laboratory Animals (NIH). Post-mortem human tissue samples and serum samples were acquired from the Department of Forensic Medicine, Sagore Dutta Medical College & Hospital, Kolkata, West Bengal after obtaining the ethical clearance from the Centre of Biomedical Research Ethics Committee (Ref: IEC/CBMR/Corr/2020/16/6). Information on the sex, age, cause of death, and chemotherapy history for all heart autopsy samples is available in Table S6. Cancer patients had a treatment history that included 5-FU, anthracyclines, and/or oxaliplatin. Samples taken from individuals with who died of cancer-related complications (e.g., metastasis) were classified as “Chemotherapy – Fibrosis”. Samples taken from individuals who died of cardiac complications (e.g., fibrosis, atrial fibrillation, myocardial infarction, or heart failure) were classified as “Chemotherapy + Fibrosis”. Cardiac fibrosis in all samples was quantified utilizing Masson Trichrome staining.
2.18. Statistical analyses
Immunoblots generally contain a minimum N of 3 and experiments were repeated at least twice with data pooled from each set. Mouse data was generated from two independent cohorts. Data were analyzed by student's t-test or one- or two-way ANOVA with the Bonferroni post hoc adjustment as appropriate. Statistical analyses were performed using Prism software (San Diego, CA, USA). Results were considered significantly different at P < 0.05. Values are expressed as means ± S.E.M.
3. Results
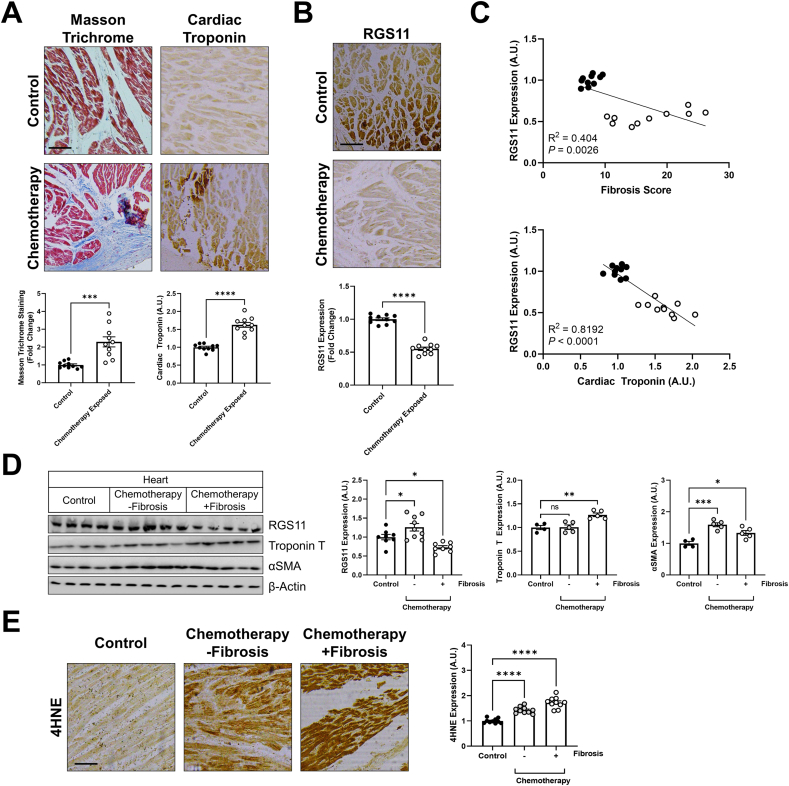
RGS11 down-regulation correlates with cardiac fibrosis in patients receiving cancer chemotherapy – We obtained cardiac tissue samples from patients exposed to cancer chemotherapy (Table S6) and unexposed controls. Chemotherapy exposure was associated with increased cardiac fibrosis and accumulation of cardiac troponin T, a marker of heart damage (Fig. 1A). Surprisingly, though doxorubicin triggers up-regulation of both Gβ5 and RGS6 in the murine myocardium [27,31], RGS11 levels in the heart were significantly lower in the chemotherapy group (Fig. 1B), and there was a significant negative correlation between RGS11 protein expression and both fibrosis and troponin levels (Fig. 1C). Similarly, RGS11 expression was lower in patients with a history of myocardial infarction (Fig. S2, Table S7). When we divided samples from chemotherapy patients based on history of cardiac complications, we noted a unique molecular signature in individuals with detectable cardiac remodeling that was characterized by low RGS11 and high Troponin T (Fig. 1D). In contrast, markers for myofibroblast differentiation (α smooth muscle actin, αSMA) (Fig. 1D) and oxidative stress (4HNE; Fig. 1E) were up-regulated irrespective of cardiac fitness. These observations stimulated our interest in a potential role for RGS11 as a cardioprotective molecule in the myocardium.
Fig. 1.
– RGS11 is downregulated in the human heart following chemotherapy exposure. We collected post-mortem heart tissue samples from individuals with a history of chemotherapeutic treatment with regimens containing 5-FU or an anthracycline.(A) Representative cardiac staining for fibrosis (Masson Trichrome) and immunohistochemistry of troponin T or (B) RGS11 in a control or chemotherapy exposed patients (n = 10) [scale bar = 100 μm]. (C) Correlation between RGS11 histoscores and fibrosis or cardiac troponin levels in control or chemotherapy exposed patients (n = 10). (D) Samples from chemotherapy-exposed patients were stratified based on cardiac health and classified into 2 groups: no heart phenotype and heart phenotype with fibrosis. Protein immunoreactivity was determined via western blotting in cardiac tissue (n = 4–10). β-Actin serves as a loading control for western blots. Immunoblots are accompanied by a densitometric quantification wherein expression is normalized to the corresponding control group. (E) Representative cardiac staining for 4HNE with quantification (n = 10) [scale bar = 100 μm]. Data were analyzed by student's t-test or one- or two-way ANOVA with Sidak's post-hoc test. *P < 0.05, **P < 0.01, ***P < 0.001 ****P < 0.0001. ns = not significant. Data are presented as mean ± SEM.
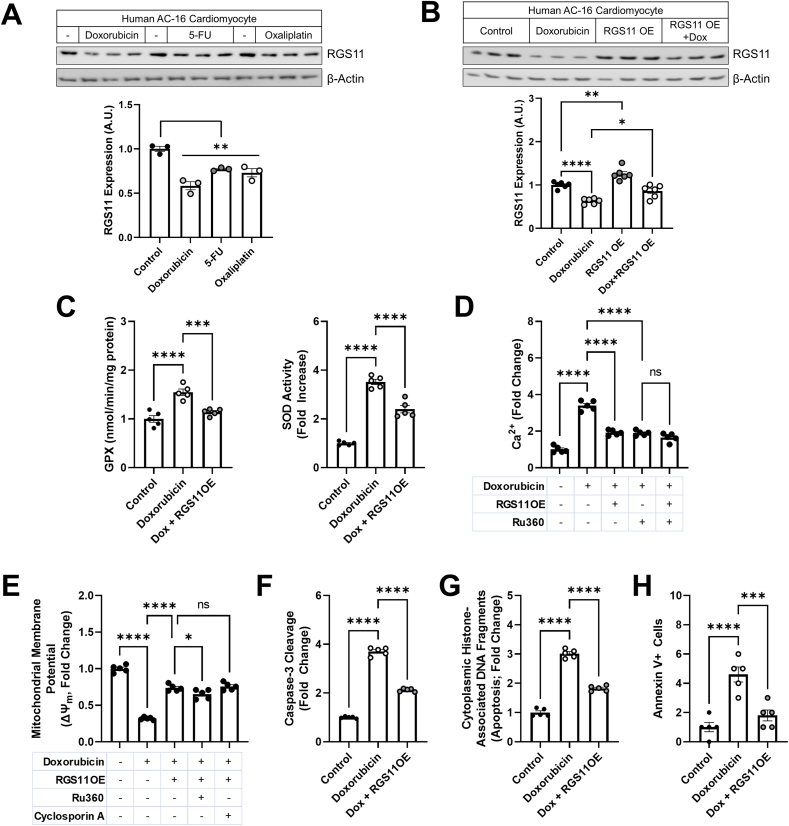
RGS11 OE protects human cardiomyocytes against chemotherapy-dependent mitochondrial dysfunction and cell death – Doxorubicin, 5-FU and oxaliplatin also decreased RGS11 protein expression in the human cardiomyocyte cell line AC-16 (Fig. 2A). Clues to the potential impact of RGS11 depletion on cardiomyocyte function were gleaned from studies of RGS11 binding partner Gβ5, which drives oxidative stress, mitochondrial dysfunction, and cell death in murine myocytes [27]. RGS11 appears to participate in these same signaling cascades, though with a diametrically opposite outcome. More specifically, RGS11 OE in AC-16 cells, which blocks doxorubicin-dependent RGS11 depletion (Fig. 2B), decreased doxorubicin-induced recruitment of antioxidant defense mechanisms such as glutathione peroxidase (GPX) and SOD (Fig. 2C) and protected the cells against mitochondrial calcium overload (Fig. 2D), loss of mitochondrial membrane potential (ΔψM) (Fig. 2E), caspase cleavage (Fig. 2F), and cell death (Fig. 2G and H). Similar results were obtained in cell treated with either 5-FU or oxaliplatin (Figs. S3A–S3C). In addition to promoting chemotherapeutic drug-dependent ROS generation, RGS11 depletion in cardiomyocytes also appears to be driven by accumulating oxidative stress as RGS11 down-regulation could be mitigated via glutathione supplementation with the donor NAC (Fig. S4).
Fig. 2.
RGS11 prevents doxorubicin-induced oxidative stress, mitochondrial dysfunction, and cell death in human cardiomyocytes. (A) Human AC-16 cardiomyocytes were treated with doxorubicin (3 μM, 16 h), 5-FU (500 μM, 16 h), or oxaliplatin (0.06 mM, 16 h) and immunoblotting performed probing for RSG11 (n = 3). (B–H) Control (scramble plasmid) or RGS11 OE AC-16 cells were treated with doxorubicin (3 μM, 16 h) ± pre-treatment with mitochondrial calcium uniporter inhibitor Ru360 (50 μM, 1 h) or mitochondrial permeability transition pore (mPTP) blocker cyclosporin A (0.2 mM, 45 min) where indicated. (B) Immunoblotting for RGS11 (n = 6).(C) GPX activity (n = 5) and SOD activity (n = 5). (D) Mitochondrial Ca2+ flux (n = 5). (E) Mitochondrial membrane potential (ΔψM; n = 5). (F) Caspase-3 cleavage (n = 5). (G) Apoptosis (cytoplasmic histone-associated DNA fragments; n = 5). (H) Annexin V+ cells (n = 5). β-Actin serves as a loading control for western blots. Immunoblots are accompanied by a densitometric quantification wherein expression is normalized to the corresponding control group. Data were analyzed by one-way ANOVA with Sidak's post-hoc test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns = not significant. Data are presented as mean ± SEM.
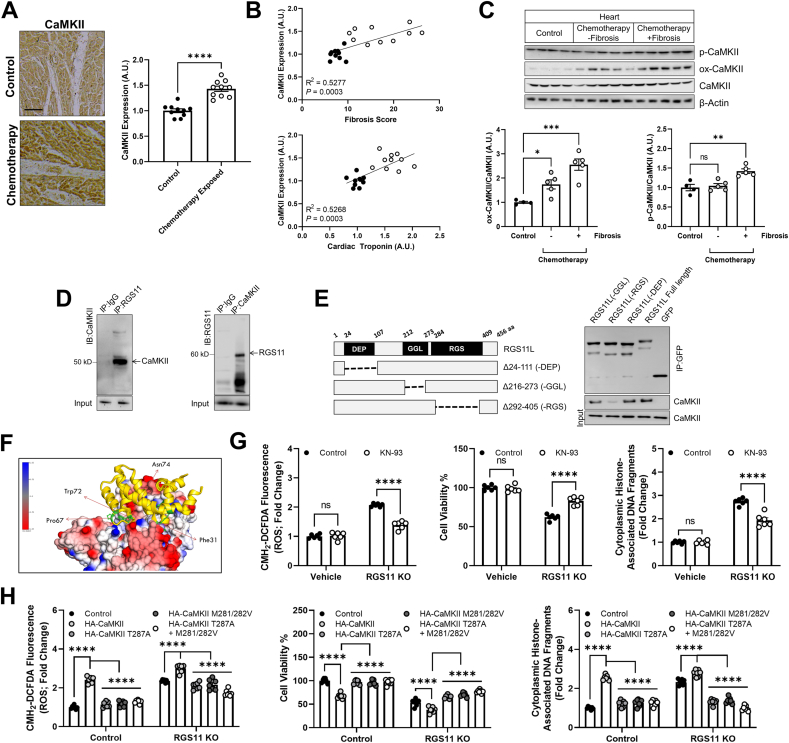
RGS11 interacts with CaMKII in heart and blocks CaMKII-dependent myocyte damage – In human chemotherapy patients, CaMKII levels were also elevated (Fig. 3A) and closely correlated with cardiac fibrosis (Fig. 3B). Notably, as we observed for RGS11, CaMKII phosphorylation but not oxidation levels were specifically altered in hearts from chemotherapy patients with detectable cardiac fibrosis (Fig. 3C). In prior work investigating the role of Gβ5 in chemotherapy-dependent cardiotoxicity, we noted that KD of Gβ5 was sufficient to prevent doxorubicin-dependent recruitment of both ATM and CaMKII [27]. While RGS6 has been linked to ATM regulation in myocytes [31], the R7 family member(s) responsible for CaMKII modulation remains unknown. In fact, RGS11 and CaMKII form a co-precipitable complex in cardiomyocytes (Fig. 3D) that can be abolished via deletion of the RGS domain in RGS11 (Fig. 3E). In silico modeling of the CaMKII-RGS11 complex revealed a tight association supported by hydrogen bonding and hydrophobic and ionic interactions (Fig. 3F, S5A-C). Doxorubicin, oxaliplatin, and 5-FU also increased CaMKII phosphorylation and oxidation in AC-16 cells (Fig. S6A), and CRISPR-mediated KO of RGS11 exacerbated doxorubicin-dependent ROS generation and cell death, effects reversed via treatment with the CaMKII inhibitor KN-93 (Fig. S6B). Either KN-93 pre-treatment, KD of the cardiac CaMKIIδ isoform or RGS11 OE also decreased cleavage of executioner caspase 3 and cell death in AC-16 cells exposed to doxorubicin (Fig. S6C), 5-FU (Fig. S6D), or oxaliplatin (Fig. S6E). Notably, CaMKII inhibition had no impact on cell death in cells containing excess RGS11 suggesting that RGS11 counteracts the ability of CaMKII to drive chemotherapy-dependent myocyte death. RGS11 KO alone also increased oxidative stress, compromised cell viability, and induced cell death in AC-16 cells via a mechanism sensitive to CaMKII inhibition (Fig. 3G). OE of WT CaMKII in AC-16 cells exacerbatedthe impact of RGS11 KO on metrics of cytotoxicity (Fig. 3H). However, expression of oxidation (M281/282 V)- or phosphorylation (T287A)-deficient CaMKII mutants failed to enhance cell death and, rather, mitigated the effect of RGS11 depletion on ROS generation and cell death (Fig. 3H). No differences in expression level of WT CaMKII vs phosphorylation- or oxidation-deficient mutants were observed in control or RGS11 KO cells (Fig. S7A), and, importantly, mutation of these sites failed to abolish CaMKII-RGS11 binding indicating that post-translational modification of CaMKII is not required for RGS11 recruitment to the CaMKII-containing macromolecular complex (Fig. S7B).
Fig. 3.
RGS11 forms a complex with CaMKII and blocks CaMKII-dependent cardiomyocyte damage. (A) Representative cardiac staining for CaMKII in a control or chemotherapy exposed patients [n = 10, scale bar = 100 μm]. (B) Correlation between CaMKII histoscores andfibrosis or cardiac troponin levels in control or chemotherapy exposed patients (n = 10). (C) Immunoblotting for phosphorylated and oxidized CaMKII protein expression in heart tissue from chemotherapy exposed patients or matched controls (n = 4–5). (D) Reciprocal co-immunoprecipitation of RGS11 and CaMKII from AC-16 cells. (E) Co-immunoprecipitation of CaMKII with RGS11 deletion constructs. (F) In silico modeling of the putative RGS11-CaMKII complex revealed key RGS11 residues predicted to support a direct interaction between RGS11 and CaMKII. (G) Control or RGS11 CRISPR KO AC-16 cells were treated with CaMKII inhibitor KN-93 (50 μM, 1 h). CM-H2-DCFDA fluorescence (total ROS), cell viability, apoptosis (cytoplasmic histone-associated DNA fragments; n = 5) were measured. (H) RGS11 KO AC-16 cells were transfected with control plasmid (vector) ± phosphorylation (T287A) or oxidation (M281/282 Vhourss)) deficient CaMKII (HA-tagged) where indicated. CM-H2-DCFDA fluorescence (total ROS), cell viability, apoptosis (cytoplasmic histone-associated DNA fragments; n = 7) were measured. β-Actin serves as a loading control for western blots. Immunoblots are accompanied by a densitometric quantification wherein expression is normalized to the corresponding control group. Data were analyzed by student's t-test or one- or two-way ANOVA with Sidak's post-hoc test. *P < 0.05, **P < 0.01,***P < 0.001, ****P < 0.0001. ns = not significant. Data are presented as mean ± SEM.
RGS7 is up-regulated following RGS11 depletion but fails to counteract the CaMKII phosphorylation, oxidative stress, or cell death resulting from RGS11 loss – As multiple R7 family RGS proteins are expressed in heart and, presumably, compete for a shared pool of Gβ5, we next investigated the impact of manipulating RGS11 levels on expression of other family members. We noted an increase in RGS7 in AC-16 cells following RGS11 KD or KO that likely accounts for intact Gβ5 expression (Fig. S8A). Consistent with the co-stabilizing relationship between the two proteins, Gβ5 levels did increase following RGS11 OE (Fig. S8A). The impact of RGS11 depletion on CaMKII activation (Fig. S8B), ROS generation (Fig. S8C), and cell death (Fig. S8D) could not, however, be counteracted by knocking down RGS7. Thus, the phenotypes observed in RGS11 deficient cells cannot be explained by compensatory actions of RGS7.
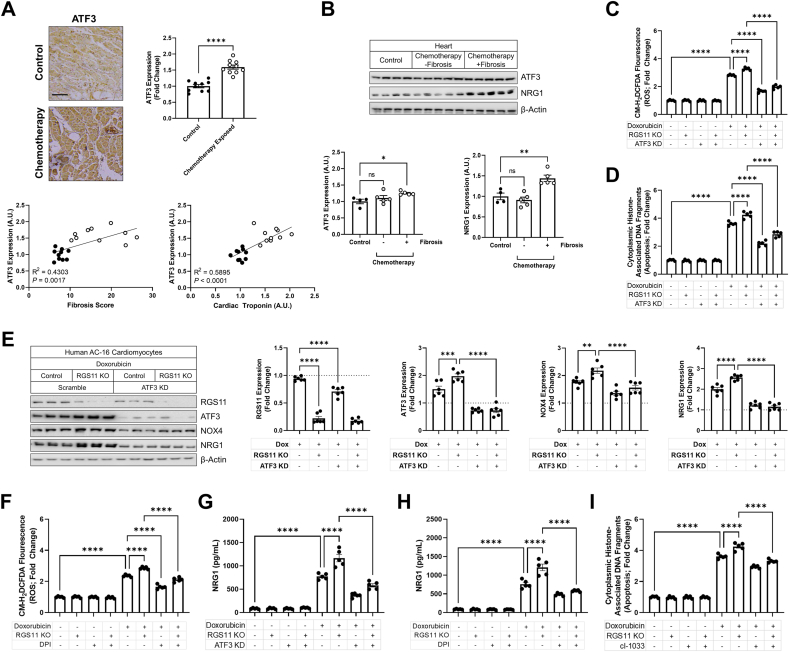
RGS11 prevents ATF3-dependent oxidative stress and cardiokine release – Robust up-regulation of the transcription factor ATF3 was also evident in the myocardium of chemotherapy patients particularly those will detectable cardiac fibrosis and elevated cardiac troponin (Fig. 4A). Like RGS11 and CaMKII, aberrations in ATF3 levels were unique to chemotherapy patients with detectable cardiac fibrosis (Fig. 4B). Given the correlation in protein levels, we hypothesized that RGS11, CaMKII, and ATF3 might be functionally linked. Doxorubicin, 5-FU, and oxaliplatin increased ATF3 expression in AC-16 cells (Fig. S9A), and ATF3 KD provided modest protection against chemotherapeutic-dependent caspase 3 cleavage and cell death but proved less efficacious than RGS11 OE (Figs. S9B–D). However, similar to results obtained following CaMKII inhibition, ATF3 depletion did prevent the increased sensitivity to doxorubicin-induced oxidative stress (Fig. 4C) and cell death (Fig. 4D) resulting from RGS11 KO. Consistent with RGS11 blocking ATF3 action, we noted that doxorubicin driven ATF3 induction increased in AC-16 cells lacking RGS11 (Fig. 4E). We identified two potential targets of ATF3 in chemotherapy exposed myocytes. Doxorubicin increased expression of cardiokine NRG1 as well as the constitutively active and transcriptionally regulated Nox isoform Nox4, effects that could be exacerbated by depleting RGS11 and prevented via ATF3 KD (Fig. 4E). RGS11 KO also triggered an ATF3-dependent increase in the Nox2 subunit gp91phox (Fig. S10A) whose activation is dependent on phosphorylation and membrane localization of accessory subunits. Either ATF3 KD or Nox inhibition with diphenyleneiodonium (DPI) decreased doxorubicin-dependent ROS generation (Fig. S10B) and NRG1 release (Fig. S10C) in AC-16 cells. Similarly, DPI counteracted the effect of RGS11 KO on doxorubicin driven oxidative stress (Fig. 4F). Either ATF3 KD (Fig. 4G) or DPI (Fig. 4H) also decreased NRG1 production from doxorubicin exposed RGS11 KO AC-16 cells. Together, these data indicate that ATF3/Nox drive chemotherapy-driven ROS generation and that RGS11 provides a break on this pathway. As a secondary consequence of increased oxidative stress, NRG1 is also released from chemotherapy-exposed AC-16 cells and functions in an autocrine manner to promote cell death. Indeed, the tyrosine kinase inhibitor cI-1033, which blocks NRG1 receptors erbB2 and erbB4, decreases doxorubicin-, 5-FU- and oxaliplatin-dependent apoptosis with RGS11 OE providing no additive benefit (Fig. S10D). Inhibition of NRG1 signal transduction also decreased cell death in doxorubicin treated RGS11 KO myocytes (Fig. 4I). Thus, RGS11 functions upstream of ATF3 to block Nox induction, oxidative stress, NRG1 release, and apoptosis following chemotherapy exposure.
Fig. 4.
RGS11 regulates ATF3 in cardiomyocytes. (A) Representative cardiac staining for ATF3 in a control or chemotherapy exposed patients [n = 10, scale bar = 100 μm] and correlation between ATF3 histoscores and fibrosis or cardiac troponin levels in control or chemotherapy exposed patients (n = 10). (B) Immunoblotting for ATF3 and NRG1 protein expression in heart tissue from chemotherapy exposed patients with or without fibrosis or matched controls (n = 4–5). (C–E) Control or RGS11 CRISPR KO AC-16 cells were treated with doxorubicin (3 μM, 16 h) ± ATF3 shRNA for 12 h. (C) CM-H2-DCFDA fluorescence (total ROS; n = 5). (D) Apoptosis (cytoplasmic histone-associated DNA fragments; n = 5). (E) Protein immunoreactivity was determined via western blotting (n = 6). (F–I) Control or RGS11 KO AC-16 cells were treated with doxorubicin (3 μM, 16 h) ± pre-treatment with ATF3 shRNA for 12 h or Nox blocker DPI (1 μM) or pan-ErbB blocker cI-1033 (2 μM). (F) CM-H2-DCFDA fluorescence (total ROS; n = 5). (G–H) NRG1 concentration in cell culture media (n = 5). (I) Apoptosis (cytoplasmic histone-associated DNA fragments; n = 5). β-Actin serves as a loading control for western blots. Immunoblots are accompanied by a densitometric quantification wherein expression is normalized to the corresponding control group. Data were analyzed by student's t-test or one- or two-way ANOVA with Sidak's post-hoc test. *P < 0.05, **P < 0.01,***P < 0.001, ****P < 0.0001. ns = not significant. Data are presented as mean ± SEM.
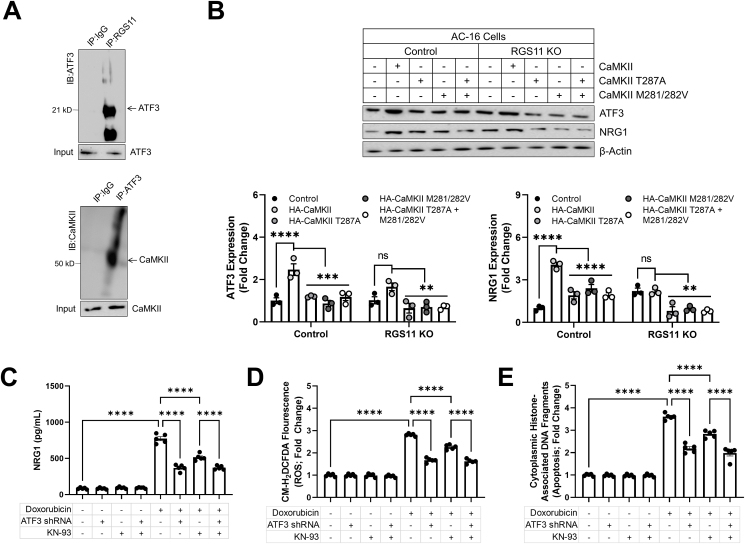
CaMKII regulates ATF3– We now had evidence that RGS11 negatively regulates both CaMKII and ATF3 in cardiomyocytes. However, it remained unclear if these mechanisms act in parallel or are, rather, a linear cascade wherein RGS11 dependent regulation of ATF3 requires CaMKII. In fact, ATF3 can be co-immunoprecipitated with both RGS11 and CaMKII suggesting that the three proteins exist in a complex in cardiomyocytes (Fig. 5A). OE of WT CaMKII increased ATF3 expression and NRG1 production in cardiomyocytes, effects that were absent for CaMKII mutants lacking the potential for oxidation or phosphorylation (Fig. 5B). Notably, CaMKII failed to significantly impact either ATF3 or NRG1 expression in cells lacking RGS11. Again, both CaMKII inhibition and ATF3 KD decreased doxorubicin dependent NRG1 production (Fig. 5C), ROS generation (Fig. 5D), and cell death (Fig. 5E), and, importantly, these interventions are non-additive suggesting that CaMKII and ATF3 lie in the same pathway.
Fig. 5.
RGS11-dependent regulation of ATF3 requires CaMKII. (A) Co-immunoprecipitation of ATF3 with RGS11or CaMKII in AC-16 cells. (B) Control or RGS11 (n = 3) CRISPR KO AC-16 cells were transfected with control plasmid (vector) ± CaMKII (HA-tagged) or phosphorylation (T287A) or oxidation (M281/282 V) deficient CaMKII (HA-tagged) where indicated. (C–E) Control (scramble shRNA) or ATF3 KD VCM were treated with doxorubicin (3 μM, 16 h) ± pre-treatment with CaMKII inhibitor KN-93 (50 μM, 1 h). (C) NRG1 concentration in cell culture media (n = 5). (D) CM-H2-DCFDA fluorescence (total ROS; n = 5). (E) Apoptosis (cytoplasmic histone-associated DNA fragments; n = 5). Immunoblots are accompanied by a densitometric quantification wherein expression is normalized to the corresponding control group. β-Actin serves as a loading control for western blots. Data were analyzed by student's t-test or one- or two-way ANOVA with Sidak's post-hoc test. *P < 0.05, **P < 0.01, ****P < 0.0001. ns = not significant. Data are presented as mean ± SEM.
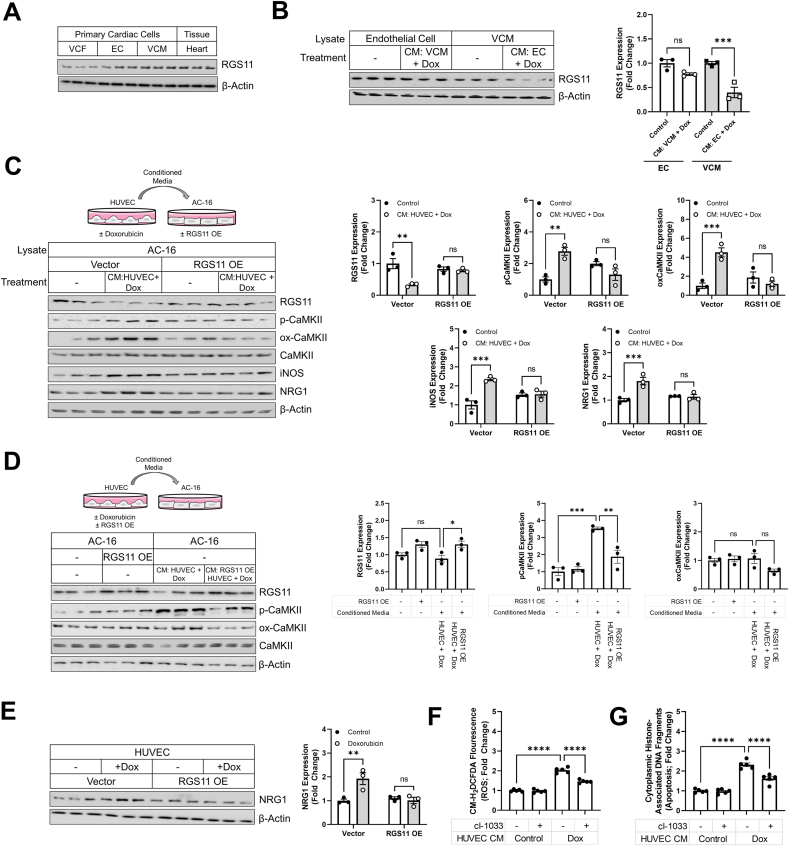
RGS11 modulates VCM to EC communication – While NRG1 is released from cardiomyocytes, the primary source of NRG1 in the cardiovascular system is the vascular endothelium. RGS11 levels are highest in VCM, but expression is also detectable in VCF and ECs (Fig. 6A). Thus, we next wished to establish if RGS11 played a functional role in non-cell autonomous regulation of myocyte viability. Intriguingly, we detected robust RGS11 down-regulation in VCM treated with conditioned media from doxorubicin treated EC (Fig. 6B). Consistent with a role for RGS11 in EC to VCM communication, RGS11 OE in AC-16 cardiomyocytes prevented CaMKII phosphorylation/oxidation, inducible nitric oxide synthase (iNOS) induction, and NRG1 production following transplantation of media from doxorubicin treated human ECs (HUVEC) (Fig. 6C). Thus, RGS11, acting in myocytes, counteracts the potentially deleterious action of paracrine factors released by EC. We next wished to establish whether RGS11 played a role in the production of these paracrine factors facilitating EC and VCM intercommunication. To do this we overexpressed RGS11 in HUVEC cells (Fig. 6D), treated the cells with doxorubicin, harvested the culture media, and transplanted the media onto cultures of AC-16 cells. RGS11 OE in HUVEC cells treated with doxorubicin decreased CaMKII activation in AC-16 cells following transplantation of conditioned media (Fig. 6D). Finally, having already established that RGS11 regulates NRG1 production from myocytes, we hypothesized that NRG1 might represent a viable candidate cardiokine mediating intercommunication between the myocardium and the endothelium. RGS11 OE decreased doxorubicin dependent NRG1 expression in HUVEC cells (Fig. 6E) and inhibition of NRG1 signaling was sufficient to decrease ROS generation (Fig. 6F) and cell death (Fig. 6G) in AC-16 cells exposed to culture media from doxorubicin treated HUVEC cells. Thus, RGS11-dependent NRG1 production plays a key role in propagating cellular stress signaling between ECs and myocytes.
Fig. 6.
RGS11/CaMKII regulate EC/VCM crosstalk. (A) RGS11 expression (immunoblotting) in heart tissue, VCM, VCF, and EC. (B) Isolated mice ECor VCM were treated with doxorubicin (3 μM, 12 h), culture media was removed and added to VCM and EC, respectively. Immunoblotting was performed to detect RGS11 (n = 3). (C) HUVEC cells were treated with doxorubicin (3 μM, 16 hVV). Culture media was removed and added to control or RGS11 OE AC-16 cells. Immunoblotting was performed to detect RGS11, p-CaMKII, ox-CaMKII, iNOS, and NRG1 (n = 3). (D) Control or RGS11 OE HUVEC cells were treated with doxorubicin (3 μM, 16 h). Culture media was removed and added to AC-16 cultures. Immunoblotting was performed to detect RGS11, p-CaMKII, and ox-CaMKII (n = 3). (E) Control, RGS11 OE HUVEC cells were treated with doxorubicin (3 μM, 16 h). Immunoblotting was performed to detect NRG1 (n = 3). (F–G) HUVEC cells were treated with doxorubicin (3 μM, 16 h) and the culture media was transplanted on AC-16 cell cultures ± pre-treatment with pan-ErbB blocker cI-1033 (2 μM, 1 h). (F) CM-H2-DCFDA fluorescence (total ROS; n = 5). (G) Apoptosis (cytoplasmic histone-associated DNA fragments; n = 5). β-Actin serves as a loading control for western blots. Immunoblots are accompanied by a densitometric quantification wherein expression is normalized to the corresponding control group. Data were analyzed by one- or two-way ANOVA with Sidak's post-hoc test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns = not significant. Data are presented as mean ± SEM.
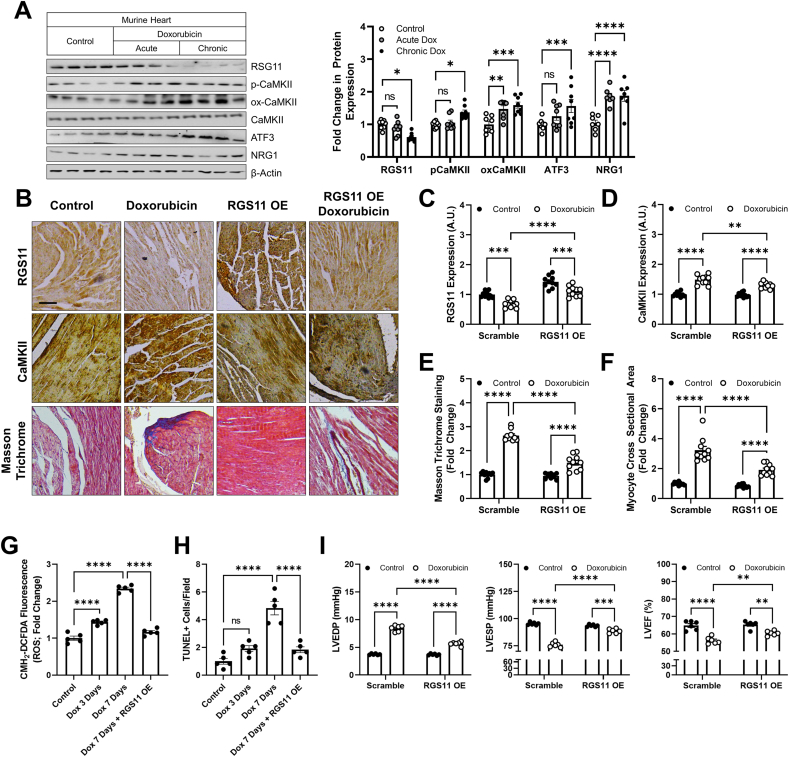
RGS11 OE mitigates doxorubicin-dependent hypertrophy, fibrosis, oxidative stress, cell death, and loss of ventricular function – Having detected robust down-regulation of RGS11 in the heart of chemotherapy patients and established a role for RGS11 in both myocyte-intrinsic and -extrinsic mechanisms of chemotherapeutic drug driven cardiotoxicity in vitro, we next sought to test whether manipulating RGS11 levels impacted cardiotoxicity in vivo. Doxorubicin, 5-FU, and oxaliplatin exposure leads to cardiac hypertrophy (Fig. S11A) accompanied by RGS11 depletion (Fig. S11B) and a molecular signature characterized by increased CaMKII phosphorylation and oxidation, ATF3 induction, and NRG1 production similar to that we observed in the human heart (Fig. S11C). RGS11 down-regulation was detectable in the heart 4 weeks after doxorubicin exposure but not at 2 weeks and was sustained through the 8-week time point (Fig. S11D). Importantly, the loss of cardiac function resulting from doxorubicin exposure followed an identical timeline (Fig. S11E). Further, changes in RGS11, ATF3, and pCaMKII were only detectable in animals exposed to multiple low doses of doxorubicin over an extended time (chronic) and were not seen in an acute treatment paradigm commonly employed to study mechanism(s) underlying chemotherapy-dependent cardiotoxicity (acute) (Fig. 7A). Thus, we utilized the chronic schedule for experiments aimed at establishing the role of RGS11 in the pathophysiology of doxorubicin-induced heart damage. Indeed, RGS11 OE in heart (Fig. 7B and C) decreased CaMKII expression (Fig. 7B and D), fibrotic remodeling (Fig. 7B and E), myocyte hypertrophy (Fig. 7B and F), ROS accumulation (Fig. 7G) and cell death (Fig. 7H) in mice exposed to doxorubicin. Importantly, RGS11 OE improved cardiac function measures including LVEDP, LVESP, and LVEF in mice administered doxorubicin (Fig. 7I).
Fig. 7.
Cardiac-specific RGS11 OE protects against doxorubicin cardiotoxicity in mice. (A) Mice were treated with doxorubicin (cumulative dose of 45 mg/kg i. p.) or saline control over 8 weeks (chronic) or mice were given a single dose of doxorubicin (20 mg/kg i. p.) or saline and sacrificed after 3 days (acute). Cardiac tissues were collected from both the sets for protein expression analyses of RGS11, p-CaMKII, ox-CaMKII, CaMKII and ATF3 (n = 8). (B–G) An RGS11 encoding viral construct or control (Ad-β-gal) was introduced into the myocardium. After 15 days, animals were given saline or doxorubicin (cumulative dose of 45 mg/kg i. p.) over 8 weeks. After 8 weeks cardiac phenotyping was performed, and tissues samples collected 1 week later for biochemical and histological analyses. (B) Representative images [scale bar = 100 μm] of RGS11 and CaMKII immunohistochemistry and cardiac fibrosis (Masson Trichrome staining). Quantification of (C) RGS11 and (D) CaMKII histoscores (n = 10). (E) Quantification of collagen deposition (fibrosis, n = 10). (F) Average myocyte cross sectional area (n = 10). (G) CM-H2-DCFDA fluorescence (total ROS; n = 5). (H) TUNEL positive cells (n = 5). (I) Cardiac parameters (n = 6). β-Actin serves as a loading control for western blots. Immunoblots are accompanied by a densitometric quantification wherein expression is normalized to the corresponding control group. Data were analyzed by two-way ANOVA with Sidak's post-hoc test. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns = not significant. Data are presented as mean ± SEM.
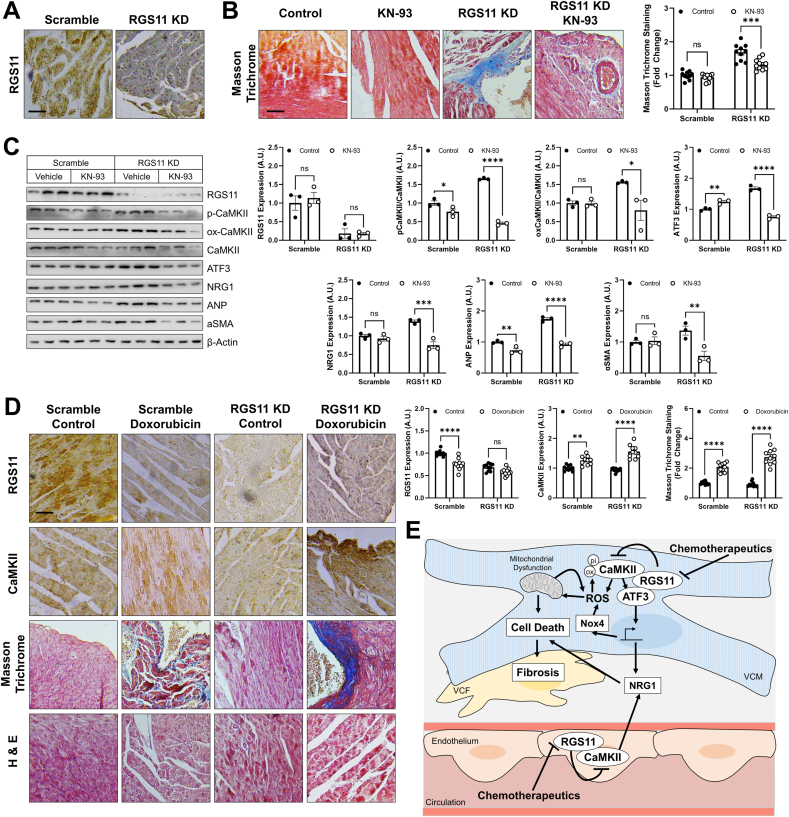
RGS11 KD in heart triggers cardiac fibrosis and exacerbates doxorubicin-driven cardiac remodeling – Given that RGS11 depletion correlated with measures of cardiac fibrosis in humans and mice, we hypothesized that KD of RGS11 might be sufficient to damage the heart. To test this hypothesis, we administered RGS11 shRNA to mice via intracardiac injection (Fig. 8A). RGS11 KD resulted in fibrotic collagen deposition (Fig. 8B) and led to increased CaMKII phosphorylation/oxidation as well as increased ATF3, NRG1, atrial natriuretic peptide (ANP), and αSMA expression in heart (Figs. 8C and S12A). Importantly, inhibition of CaMKII with KN-93 largely blocked the impact of RGS11 KD on cardiac fibrosis (Fig. 8B/C, S12A). Further, as we observed in cultured cardiac cells, RGS11 depletion in heart exacerbated doxorubicin-dependent CaMKII up-regulation, fibrosis, and NRG1 release into the circulation (Figs. 8D and S12B). These data indicate that RGS11 plays a key role in preventing CaMKII-dependent heart damage in vivo.
Fig. 8.
RGS11 KD in heart triggers CaMKII-dependent cardiac fibrosis. A viral construct encoding RGS11 shRNA or control (scramble) was introduced into the myocardium. After 15 days, animals were given saline or KN-93 (8 mg/kgi. p, 2 doses, 4 days apart) and tissues collected 2 days after the last injection for downstream analyses. (A) Immunostaining for RGS11 in cardiac tissue. (B) Representative images [scale bar = 100 μm] and quantification of cardiac fibrosis (Masson Trichrome staining; n = 10). (C) Immunoblotting for RGS11, p-CaMKII, ox-CaMKII, ATF3, NRG1, αSMA and ANP in cardiac tissue (n = 3). (D) A viral construct encoding RGS11 shRNA or control (scramble) was introduced into the myocardium. After 15 days, animals were given saline or doxorubicin (cumulative dose of 45 mg/kg i. p.) and tissues collected after 9 weeks for histological analyses. Representative images depicting RGS11 staining, CaMKII staining, Masson Trichrome staining (fibrosis), and H&E staining (myofibrillar architecture) in doxorubicin treated control and cardiac RGS11-KD mice [scale bar = 100 μm] and quantification. (E) Schematic depicting the role of RGS11 in regulation of CaMKII/ATF3 in cardiac myocytes and neighboring ECs. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns = not significant. Data are presented as mean ± SEM.
4. Discussion
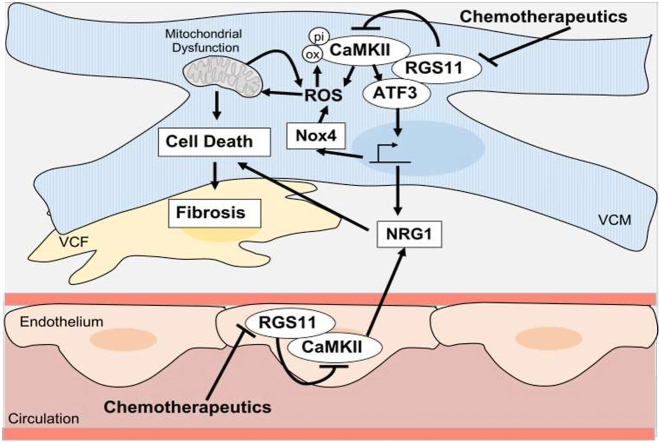
Despite decades of research, the pathophysiological mechanisms contributing to chemotherapy-induced heart damage remain elusive, and cardiotoxicity remains a key factor limiting cumulative chemotherapeutic drug dose. Given the number of cancer patients is only expected to rise in the coming decades, identifying novel means to circumvent off target actions of cancer chemotherapeutics is of paramount importance. Here, we demonstrate that the G protein regulator RGS11 is down-regulated in the myocardium of individuals with a history of cancer chemotherapy that included cardiotoxic agents such as 5-FU or anthracyclines. Notably, RGS11 depletion correlated with cardiac damage markers and was most pronounced in patients with detectable fibrotic remodeling. While other members of the R7 family of RGS proteins (e.g., RGS6 and RGS7) are also expressed in the heart, RGS11 has a unique function acting in both cardiomyocytes and neighboring ECs to prevent oxidative stress, mitochondrial dysfunction, and cell death (Fig. 8E). In heart, RGS11 forms a direct complex with the pro-apoptotic kinase CaMKII and transcription factor ATF3 both of which contribute to myocyte-intrinsic mechanisms of cytotoxicity and facilitate pathogenic crosstalk between the heart and the vascular endothelium. Cardiac-specific RGS11 OE provided marked protection against doxorubicin-dependent left ventricular dysfunction. On the other hand, RGS11 KD in heart was sufficient to drive fibrosis that could be mitigated via inhibition of CaMKII. Together these data demonstrate that RGS11 depletion in heart is a requisite step in the pathogenesis of chemotherapy-associated cardiotoxicity.
The atypical G protein Gβ5, which fails to dimerize with Gγ and, instead, forms a co-stabilizing complex with members of the R7 family of RGS proteins (RGS6, RGS7, RGS9, RGS11), has been previously implicated in regulation of multiple signal transduction cascades recruited upon exposure of cardiomyocytes to cancer chemotherapeutics [27]. More specifically, Gβ5KD in murine VCM blocks doxorubicin- and 5-FU-driven phosphorylation of both ATM and CaMKII. RGS6, which forms a direct complex with ATM [36] is required for activation of the ATM/p53 signaling cascade in VCM exposed to doxorubicin [31], but the R7 family member responsible for CaMKII regulation had yet to be identified. Indeed, the fact that Gβ5 expression is not completely absent from the hearts of RGS6−/− mice [56,57] indicates that another R7 family member is also present in the myocardium. Here we show that RGS11 forms a complex with CaMKII and blocks chemotherapeutic-dependent CaMKII activation. However, it is important to note that the impact of RGS11 depletion is opposite in direction to that observed for KD of Gβ5 or RGS6. The implications of this observation are two-fold. First, it is likely that another R7 family member collaborates with Gβ5 to increase CaMKII activity in response to cytotoxic drug exposure. One potential candidate is RGS7 whose expression increases in myocytes following RGS11 depletion. However, we demonstrated that RGS7 KD could not counteract the cytotoxic impact of RGS11 KO in myocytes. Second, as all R7 family members present in the heart require Gβ5 for stability, they must compete for a shared pool of available Gβ5. One potential hypothesis to explain the direct opposition of RGS6 and RGS11 action in heart might posit that RGS6/RGS11 exist in a dynamic equilibrium such that increased expression of one protein decreases the expression of the other. Such directly antagonistic actions of R7 family members might also explain the distinct phenotypes of Gβ5−/− vs Gβ5−/+ mice, which would have different levels of R7 family members depending on the relative affinity of each protein for Gβ5[58].
Doxorubicin-dependent oxidative stress, mitochondrial dysfunction, and cell death were all decreased in the presence of RGS11 OE. There are several potential sources of ROS in the failing myocardium including uncoupling of mitochondrial respiration, impaired antioxidant capacity, or stimulation of enzymatic sources such as xanthine oxidase, cyclooxygenase, nitric oxide synthase (NOS), and NAD(P)H oxidase (Noxs) [59,60]. Nox4 is unique amongst the members of the Nox superfamily as its activity is primarily controlled at the transcriptional level and it closely associates with the mitochondrial inner membrane [61]. Importantly, Nox4 is up-regulated in response to multiple hypertrophic factors [62] and represents a major source of superoxide production in the failing heart [63]. Though prior work has only tangentially linked activation of the adaptive stress responder ATF3 to Nox4 expression [64,65], our data demonstrate that ATF3 promotes Nox4 induction in response to chemotherapeutic drugs and that RGS11 antagonizes this process. Similarly, we noted that RGS11 depletion increased expression of the gp91phox subunit of the Nox2 complex, previously implicated in doxorubicin cardiotoxicity [10], in a ATF3-dependent manner. Indeed, inhibition of Nox activity completely prevented the enhanced ROS accumulation observed in RGS11 KO cardiomyocytes exposed to doxorubicin. Thus, while the ability of RGS11 to maintain mitochondrial Ca2+ homeostasis and prevent loss of ΔψM would be expected to decrease cellular oxidative stress, targeting of ATF3-dependent Nox induction likely also contributes.
The ability of the CaMKII inhibitor KN-93 to counteract the fibrotic remodeling evident following RGS11 KD in heart underscores the functional importance of the RGS11/CaMKII interaction as an essential break on myocardial fibrosis. Notably, RGS11 depletion in heart triggered both oxidation and phosphorylation of CaMKII, parallel signaling mechanisms that promote CaMKII activation in response to elevations in ROS [66] or intracellular Ca2+67, respectively. The synergistic impact of RGS11 KO and CaMKII OE on ROS accumulation and cell death can be blocked by mutation of either the key threonine residue on the CaMKII autoinhibitory domain (T286) responsible for auto-phosphorylation and catalytic activation in response to calmodulin binding [67] or the pair of methionine residues (M281/M282) in the regulatory domain allowing for enzyme activation in the absence of Ca2+/calmodulin [66]. Thus, regulation of CaMKII by both oxidation and phosphorylation are required for CaMKII-dependent cardiomyocyte apoptosis. Given that RGS11 directly impacts ROS generation in cardiomyocytes, the effect of RGS11 on CaMKII oxidation is not surprising. However, how RGS11 blocks CaMKII phosphorylation is less clear. Though R7/Gβ5 complexes have been shown to suppress GPCR-dependent intracellular Ca2+ elevations, RGS7/Gβ5 are also capable of facilitating Ca2+ influx via membrane-associated Ca2+ channels [68], providing a possible mechanism that may underlie RGS11-dependent inhibition of CaMKII phosphorylation. Given that RGS11 and CaMKII form direct complex, it is also possible that RGS11 binding masks autophosphorylation sites independent of both ROS and intracellular Ca2+.
CaMKII-driven, RGS11-modulated induction of ATF3 also promotes doxorubicin-dependent cardiomyocyte death in our model. However, the exact role of ATF3 in cardiac hypertrophy remains controversial. For example, doxorubicin triggers ATF3 induction in rat neonatal cardiomyocytes, and ATF3 OE in these cells provided protection against doxorubicin-induced apoptosis [69]. Similarly, ATF3 deficiency in mice promoted cardiac hypertrophy is response to pressure overload [70]. On the other hand, transgenic mice with cardiac-specific ATF3 OE display ventricular hypertrophy, fibrosis, and reduced cardiac contractility [71]. Thus, it is likely that ATF3 induction is necessary to protect the heart from pathological stimuli, but persistent ATF3 OE may be detrimental and accompanied by a switch in ATF3 action between transcription activation and repression underscoring the critical importance of both the magnitude and duration of ATF3 up-regulation. The role of RGS11 in this model might then be to direct ATF3 toward protective and away from pathological targets.
One critical ATF3 responsive gene is the growth factor NRG1 [72]. Like ATF3, NRG1 may be cardioprotective in certain contexts. Indeed, application of NRG1 to cultures of rat cardiomyocytes decreases doxorubicin-induced toxicity [25,73,74] and administration of recombinant NRG1 decreases cardiac damage following acute doxorubicin exposure in mice [24,75]. Similarly, KO of the NRG1 receptor erbB4 [76] or NRG1 haploinsufficiency exacerbates doxorubicin-dependent cardiotoxicity [26]. At first glance, our data appear to be in direct contradiction to this prior work. In human AC-16 cells, doxorubicin increases NRG1 release via an ATF3/CaMKII-dependent mechanism. Further, inhibition of NRG1 signaling with cI-1033 provided partial protection against doxorubicin-induced cell death. Reconciliation of these seemingly contradictory datasets may require extended analysis of the time course of chemotherapeutic-dependent heart damage, the specific cell types involved, and the status of several key molecules that act in concert to shift the balance between endurance and repair or resignation to a catastrophic cell fate in the face of stressful stimuli. For example, in mice, the molecular signatures associated with acute/high dose and chronic/low dose chemotherapeutic administration are quite distinct. While NRG1 levels are elevated in both cases, only the chronic, low dose doxorubicin treatment leads to CaMKII phosphorylation, ATF3 induction, and RGS11 depletion similar to that we observed in the human heart following chemotherapy exposure. Prior work investigating the role of NRG1/erbB4 in doxorubicin-induced heart damage have been confined to a single dose regimen that may not faithfully recapitulate the clinical condition for patients who receive several rounds of chemotherapy over the course of weeks or even months. Though increasing NRG1 action in the acute phase might prove beneficial in the short term, it is possible that prolonged application of exogenous NRG1 would be detrimental as has been observed for other pro-fibrotic factors such as transforming growth factor β1 (TGFβ1)[77].
RGS11 modulates NRG1 production and release from cardiomyocytes, which functions in an autocrine manner to increase myocyte death. However, this is another source of NRG1 in the cardiovascular system. Indeed, NRG1 is also generated in the vascular endothelium and facilitates intercommunication between the vasculature and heart [78]. Disruption of NRG1 signaling in ECs also exacerbates myocardial hypertrophy and fibrosis in mice [78]. Intriguingly, RGS11 OE impacts NRG1 release from ECs, propagation of cellular stress signals from doxorubicin-treated ECs to untreated myocytes, and the ability of myocytes to respond to these signals by increasing oxidative stress. Importantly, inhibition of erbBs prevented ROS generation and cell death in myocytes exposed to culture media of doxorubicin-treated ECs. Together, these data indicate that NRG1 released from ECs is detrimental to the viability of neighboring myocytes and that RGS11 functions to suppress this pathogenic intercellular communication.
There are no therapeutics currently approved for the treatment of chemotherapy-induced cardiotoxicity. Though typical heart failure therapies such as angiotensin converting enzyme (ACE) inhibitors, beta-blockers, and loop diuretics are commonly employed in these patients these interventions have been shown to normalize LVEF in only 42% of patients [79,80], underscoring the need for more efficacious interventions. RGS6 is required for the chemotherapeutic actions of Dox in cancer cells [81] and possess potent anti-tumor actions particularly in breast and bladder cancers whose therapeutic regimens often utilize doxorubicin and/or 5-FU [[82], [83], [84], [85]]. Thus, it is probable that, though inhibition of RGS6 may protect the heart against chemotherapy-induced damage, such an intervention might also compromise the therapeutic efficacy of chemotherapy. In contrast, there is only a single report implicating RGS11 in carcinogenesis where it appears to selectively influence cell migration in vitro [86]. Maintenance of RGS11 levels in heart might then prove a viable means to mitigate myocardial fibrosis and loss of ventricular integrity following extended exposure to cardiotoxic chemotherapeutic drugs without exacerbating carcinogenesis.
5. Conclusions
Given that the burden of cancer incidence and mortality is rapidly growing worldwide, the need for efficacious drugs to counteract malignant cell transformation and proliferation continues to expand. However, for several of the most efficacious anti-tumor agents, their use is limited by potentially life-threatening cardiotoxicity. Our data demonstrate that RGS11 depletion is both necessary and sufficient to drive chemotherapy-dependent heart damage. Thus, identifying a means to maintain RGS11 expression in heart might represent a viable means to counteract the detrimental impact of cancer chemotherapeutics on the myocardium.
Author contributions
Conception and design: K. Das, M. Basak, T. Mahata, M. Kumar, D. Kumar, N.C. Saha, K. Mondal, A. Stewart, B. Maity.
Acquisition of data: K. Das, M. Basak, T. Mahata, M. Kumar, D. Kumar, S. Biswas, P. Kumar, P. Das, M. Moniruzzaman, S. Chatterjee.
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): K. Das, M. Basak, M. Kumar, T. Mahata, S. Biswas, P. Kumar, N.C. Saha, K. Mondal, P. Das, A. Stewart, B. Maity.
Writing, review, and/or revision of the manuscript: A. Stewart, B. Maity.
Study supervision: A. Stewart and B. Maity.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgements
We acknowledge funds from CBMR, Department of Medical education, Uttar Pradesh Government (CBMR/IMR/0012/2021), Department of Biotechnology (DBT- BT/PR28635/MED/30/2145/2019), Indian Council of Medical Research (ICMR - 5/4/1–26/2020-NCD-I) India to BM. KD thanks UGC for the PhD research support (762/CSIR NET June 2019) and AcSIR for the Registration (No. 10BB22J71006). We also thank Dr. Sudipta Saha (Late), Assistant Professor, BBAU, Lucknow for helping in few of the mice experiments, Mr. Abhishek Singh Sengar from CBMR for the generation & validation of different constructs of CaMKII and Pharmaceutical Sciences Department of Aryakul College of Pharmacy & Research, Lucknow for assisting in majority of the mice experiments. Dr. Pranesh Kumar has recently moved to the Institute of Pharmaceutical Sciences, University of Lucknow, India.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2022.102487.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Shakir D.K., Rasul K.I. Chemotherapy induced cardiomyopathy: pathogenesis, monitoring and management. J. Clin. Med. Res. 2009:8–12. doi: 10.4021/jocmr2009.02.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henriksen P.A. Anthracycline cardiotoxicity: an update on mechanisms, monitoring and prevention. Heart. 2018;104:971–977. doi: 10.1136/heartjnl-2017-312103. [DOI] [PubMed] [Google Scholar]
- 3.Shiga T., Hiraide M. Cardiotoxicities of 5-fluorouracil and other fluoropyrimidines. Curr. Treat. Options Oncol. 2020;27 doi: 10.1007/s11864-020-0719-1. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grenier M.A., Lipshultz S.E. Epidemiology of anthracycline cardiotoxicity in children and adults. Semin. Oncol. 1998;25:72–85. [PubMed] [Google Scholar]
- 5.Rawat P.S., Jaiswal A., Khurana A., Bhatti J.S., Navik U. Doxorubicin-induced cardiotoxicity: an update on the molecular mechanism and novel therapeutic strategies for effective management. Biomed. Pharmacother. 2021;139 doi: 10.1016/j.biopha.2021.111708. [DOI] [PubMed] [Google Scholar]
- 6.Minotti G., Menna P., Salvatorelli E., Cairo G., Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol. Rev. 2004;56:185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- 7.Davies K.J., Doroshow J.H. Redox cycling of anthracyclines by cardiac mitochondria. I. Anthracycline radical formation by NADH dehydrogenase. J. Biol. Chem. 1986;261:3060–3067. [PubMed] [Google Scholar]
- 8.Doroshow J.H., Davies K.J. Redox cycling of anthracyclines by cardiac mitochondria. II. Formation of superoxide anion, hydrogen peroxide, and hydroxyl radical. J. Biol. Chem. 1986;261:3068–3074. [PubMed] [Google Scholar]
- 9.Deng S., Kruger A., Kleschyov A.L., Kalinowski L., Daiber A., Wojnowski L. Gp91phox-containing NAD(P)H oxidase increases superoxide formation by doxorubicin and NADPH. Free Radic. Biol. Med. 2007;42:466–473. doi: 10.1016/j.freeradbiomed.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y., et al. Nox2 NADPH oxidase promotes pathologic cardiac remodeling associated with Doxorubicin chemotherapy. Cancer Res. 2010;70:9287–9297. doi: 10.1158/0008-5472.CAN-10-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Childs A.C., Phaneuf S.L., Dirks A.J., Phillips T., Leeuwenburgh C. Doxorubicin treatment in vivo causes cytochrome C release and cardiomyocyte apoptosis, as well as increased mitochondrial efficiency, superoxide dismutase activity, and Bcl-2:Bax ratio. Cancer Res. 2002;62:4592–4598. [PubMed] [Google Scholar]
- 12.Wu S., et al. Adriamycin-induced cardiomyocyte and endothelial cell apoptosis: in vitro and in vivo studies. J. Mol. Cell. Cardiol. 2002;34:1595–1607. doi: 10.1006/jmcc.2002.2110. [DOI] [PubMed] [Google Scholar]
- 13.Sag C.M., Kohler A.C., Anderson M.E., Backs J., Maier L.S. CaMKII-dependent SR Ca leak contributes to doxorubicin-induced impaired Ca handling in isolated cardiac myocytes. J. Mol. Cell. Cardiol. 2011;51:749–759. doi: 10.1016/j.yjmcc.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tscheschner H., et al. CaMKII activation participates in doxorubicin cardiotoxicity and is attenuated by moderate GRP78 overexpression. PLoS One. 2019;14 doi: 10.1371/journal.pone.0215992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shuai Y., et al. Global gene expression profiles of MT knockout and wild-type mice in the condition of doxorubicin-induced cardiomyopathy. Toxicol. Lett. 2011;200:77–87. doi: 10.1016/j.toxlet.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 16.Park E.J., Kwon H.K., Choi Y.M., Shin H.J., Choi S. Doxorubicin induces cytotoxicity through upregulation of pERK-dependent ATF3. PLoS One. 2012;7 doi: 10.1371/journal.pone.0044990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasim M.S., Nessim C., Villeneuve P.J., Vanderhyden B.C., Dimitroulakos J. Activating transcription factor 3 as a novel regulator of chemotherapy response in breast cancer. Transl Oncol. 2018;11:988–998. doi: 10.1016/j.tranon.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayward R., Ruangthai R., Schneider C.M., Hyslop R.M., Strange R., Westerlind K.C. Training enhances vascular relaxation after chemotherapy-induced vasoconstriction. Med. Sci. Sports Exerc. 2004;36:428–434. doi: 10.1249/01.mss.0000117130.91142.38. [DOI] [PubMed] [Google Scholar]
- 19.Muneoka K., Shirai Y., Yokoyama N., Wakai T., Hatakeyama K. 5-Fluorouracil cardiotoxicity induced by alpha-fluoro-beta-alanine. Int. J. Clin. Oncol. 2005;10:441–443. doi: 10.1007/s10147-005-0516-7. [DOI] [PubMed] [Google Scholar]
- 20.Eskandari M.R., Moghaddam F., Shahraki J., Pourahmad J. A comparison of cardiomyocyte cytotoxic mechanisms for 5-fluorouracil and its pro-drug capecitabine. Xenobiotica. 2015;45:79–87. doi: 10.3109/00498254.2014.942809. [DOI] [PubMed] [Google Scholar]
- 21.Focaccetti C., et al. Effects of 5-fluorouracil on morphology, cell cycle, proliferation, apoptosis, autophagy and ROS production in endothelial cells and cardiomyocytes. PLoS One. 2015;10 doi: 10.1371/journal.pone.0115686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spasojevic I., Jelic S., Zakrzewska J., Bacic G. Decreased oxygen transfer capacity of erythrocytes as a cause of 5-fluorouracil related ischemia. Molecules. 2008;14:53–67. doi: 10.3390/molecules14010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luu A.Z., Chowdhury B., Al-Omran M., Teoh H., Hess D.A., Verma S. Role of endothelium in doxorubicin-induced cardiomyopathy. JACC Basic Transl Sci. 2018;3:861–870. doi: 10.1016/j.jacbts.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bian Y., et al. Neuregulin-1 attenuated doxorubicin-induced decrease in cardiac troponins. Am. J. Physiol. Heart Circ. Physiol. 2009;297:H1974–H1983. doi: 10.1152/ajpheart.01010.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An T., et al. Neuregulin-1 protects against doxorubicin-induced apoptosis in cardiomyocytes through an Akt-dependent pathway. Physiol. Res. 2013;62:379–385. doi: 10.33549/physiolres.932516. [DOI] [PubMed] [Google Scholar]
- 26.Liu F.F., et al. Heterozygous knockout of neuregulin-1 gene in mice exacerbates doxorubicin-induced heart failure. Am. J. Physiol. Heart Circ. Physiol. 2005;289:H660–H666. doi: 10.1152/ajpheart.00268.2005. [DOI] [PubMed] [Google Scholar]
- 27.Chakraborti S., et al. Atypical G protein beta 5 promotes cardiac oxidative stress. Apoptosis, and Fibrotic Remodeling in Response to Multiple Cancer Chemotherapeutics. Cancer Res. 2018;78:528–541. doi: 10.1158/0008-5472.CAN-17-1280. [DOI] [PubMed] [Google Scholar]
- 28.Chen C.K., et al. Instability of GGL domain-containing RGS proteins in mice lacking the G protein beta-subunit Gbeta5. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6604–6609. doi: 10.1073/pnas.0631825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kardestuncer T., Wu H., Lim A.L., Neer E.J. Cardiac myocytes express mRNA for ten RGS proteins: changes in RGS mRNA expression in ventricular myocytes and cultured atria. FEBS Lett. 1998;438:285–288. doi: 10.1016/s0014-5793(98)01319-2. [DOI] [PubMed] [Google Scholar]
- 30.Larminie C., et al. Selective expression of regulators of G-protein signaling (RGS) in the human central nervous system. Brain Res Mol Brain Res. 2004;122:24–34. doi: 10.1016/j.molbrainres.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 31.Yang J., et al. G-protein inactivator RGS6 mediates myocardial cell apoptosis and cardiomyopathy caused by doxorubicin. Cancer Res. 2013;73:1662–1667. doi: 10.1158/0008-5472.CAN-12-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pramanick A., et al. G protein beta 5-ATM complexes drive acetaminophen-induced hepatotoxicity. Redox Biol. 2021;43 doi: 10.1016/j.redox.2021.101965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Podyacheva E.Y., Kushnareva E.A., Karpov A.A., Toropova Y.G. Analysis of models of doxorubicin-induced cardiomyopathy in rats and mice. A modern view from the perspective of the pathophysiologist and the clinician. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.670479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nair A., Morsy M.A., Jacob S. Dose translation between laboratory animals and human in preclinical and clinical phases of drug development. Drug Dev. Res. 2018;79:373–382. doi: 10.1002/ddr.21461. [DOI] [PubMed] [Google Scholar]
- 35.Nagai K., Nogami S., Egusa H., Konishi H. Pharmacokinetic evaluation of intraperitoneal doxorubicin in rats. Pharmazie. 2014;69:125–127. [PubMed] [Google Scholar]
- 36.Mahata T., et al. Hepatic Regulator of G Protein Signaling 6 (RGS6) drives non-alcoholic fatty liver disease by promoting oxidative stress and ATM-dependent cell death. Redox Biol. 2021 doi: 10.1016/j.redox.2021.102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He Q., Cheng J., Wang Y. Chronic CaMKII inhibition reverses cardiac function and cardiac reserve in HF mice. Life Sci. 2019;219:122–128. doi: 10.1016/j.lfs.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 38.Stewart A., Maity B., Anderegg S.P., Allamargot C., Yang J., Fisher R.A. Regulator of G protein signaling 6 is a critical mediator of both reward-related behavioral and pathological responses to alcohol. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E786–E795. doi: 10.1073/pnas.1418795112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long C.S., Henrich C.J., Simpson P.C. A growth factor for cardiac myocytes is produced by cardiac nonmyocytes. Cell Regul. 1991;2:1081–1095. doi: 10.1091/mbc.2.12.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jelonek K., Walaszczyk A., Gabrys D., Pietrowska M., Kanthou C., Widlak P. Cardiac endothelial cells isolated from mouse heart - a novel model for radiobiology. Acta Biochim. Pol. 2011;58:397–404. [PubMed] [Google Scholar]
- 41.Chen F., et al. N-acetylcysteine reverses cardiac myocyte dysfunction in a rodent model of behavioral stress. J. Appl. Physiol. 1985;115:514–524. doi: 10.1152/japplphysiol.01471.2012. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maillet A., et al. Modeling doxorubicin-induced cardiotoxicity in human pluripotent stem cell derived-cardiomyocytes. Sci. Rep. 2016;6 doi: 10.1038/srep25333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weng K.C., Kurokawa Y.K., Hajek B.S., Paladin J.A., Shirure V.S., George S.C. Human induced pluripotent stem-cardiac-endothelial-tumor-on-a-chip to assess anticancer efficacy and cardiotoxicity. Tissue Eng. C Methods. 2020;26:44–55. doi: 10.1089/ten.tec.2019.0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szabo I., Zoratti M. The giant channel of the inner mitochondrial membrane is inhibited by cyclosporin A. J. Biol. Chem. 1991;266:3376–3379. [PubMed] [Google Scholar]
- 45.Wong M.H., et al. The KN-93 molecule inhibits calcium/calmodulin-dependent protein kinase II (CaMKII) activity by binding to Ca(2+)/CaM. J. Mol. Biol. 2019;431:1440–1459. doi: 10.1016/j.jmb.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Matlib M.A., et al. Oxygen-bridged dinuclear ruthenium amine complex specifically inhibits Ca2+ uptake into mitochondria in vitro and in situ in single cardiac myocytes. J. Biol. Chem. 1998;273:10223–10231. doi: 10.1074/jbc.273.17.10223. [DOI] [PubMed] [Google Scholar]
- 47.Hasinoff B.B., Patel D. The lack of target specificity of small molecule anticancer kinase inhibitors is correlated with their ability to damage myocytes in vitro. Toxicol. Appl. Pharmacol. 2010:132–139. doi: 10.1016/j.taap.2010.08.026. [DOI] [PubMed] [Google Scholar]
- 48.Venselaar H., et al. Homology modelling and spectroscopy, a never-ending love story. Eur. Biophys. J. 2010;39:551–563. doi: 10.1007/s00249-009-0531-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seeliger D., de Groot B.L. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010;24:417–422. doi: 10.1007/s10822-010-9352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krieger E., Vriend G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015;36:996–1007. doi: 10.1002/jcc.23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raj R., Agarwal N., Raghavan S., Chakraborti T., Poluri K.M., Kumar D. Exquisite binding interaction of 18 beta-Glycyrrhetinic acid with histone like DNA binding protein of Helicobacter pylori: a computational and experimental study. Int. J. Biol. Macromol. 2020;161:231–246. doi: 10.1016/j.ijbiomac.2020.06.039. [DOI] [PubMed] [Google Scholar]
- 52.Raj R., et al. Epigallocatechin gallate with potent anti-Helicobacter pylori activity binds efficiently to its histone-like DNA binding protein. ACS Omega. 2021;6:3548–3570. doi: 10.1021/acsomega.0c04763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dickson C.J., et al. Lipid 14: the amber lipid force field. J. Chem. Theor. Comput. 2014;10:865–879. doi: 10.1021/ct4010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hornak V., Abel R., Okur A., Strockbine B., Roitberg A., Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Negi S.S., Schein C.H., Oezguen N., Power T.D., Braun W. InterProSurf: a web server for predicting interacting sites on protein surfaces. Bioinformatics. 2007;23:3397–3399. doi: 10.1093/bioinformatics/btm474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wydeven N., Posokhova E., Xia Z., Martemyanov K.A., Wickman K. RGS6, but not RGS4, is the dominant regulator of G protein signaling (RGS) modulator of the parasympathetic regulation of mouse heart rate. J. Biol. Chem. 2014;289:2440–2449. doi: 10.1074/jbc.M113.520742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Posokhova E., Wydeven N., Allen K.L., Wickman K., Martemyanov K.A. RGS6/Gbeta5 complex accelerates IKACh gating kinetics in atrial myocytes and modulates parasympathetic regulation of heart rate. Circ. Res. 2010;107:1350–1354. doi: 10.1161/CIRCRESAHA.110.224212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Q., et al. Targeted deletion of one or two copies of the G protein beta subunit Gbeta5 gene has distinct effects on body weight and behavior in mice. FASEB. 2011;J25:3949–3957. doi: 10.1096/fj.11-190157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ide T., et al. Direct evidence for increased hydroxyl radicals originating from superoxide in the failing myocardium. Circ. Res. 2000;86:152–157. doi: 10.1161/01.res.86.2.152. [DOI] [PubMed] [Google Scholar]
- 60.Ide T., et al. Mitochondrial electron transport complex I is a potential source of oxygen free radicals in the failing myocardium. Circ. Res. 1999;85:357–363. doi: 10.1161/01.res.85.4.357. [DOI] [PubMed] [Google Scholar]
- 61.Shanmugasundaram K., Nayak B.K., Friedrichs W.E., Kaushik D., Rodriguez R., Block K. NOX4 functions as a mitochondrial energetic sensor coupling cancer metabolic reprogramming to drug resistance. Nat. Commun. 2017;997 doi: 10.1038/s41467-017-01106-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ago T., Kuroda J., Pain J., Fu C., Li H., Sadoshima J. Upregulation of Nox4 by hypertrophic stimuli promotes apoptosis and mitochondrial dysfunction in cardiac myocytes. Circ. Res. 2010;106:1253–1264. doi: 10.1161/CIRCRESAHA.109.213116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kuroda J., Ago T., Matsushima S., Zhai P., Schneider M.D., Sadoshima J. NADPH oxidase 4 (Nox4) is a major source of oxidative stress in the failing heart. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15565–15570. doi: 10.1073/pnas.1002178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Z., et al. IFI6 depletion inhibits esophageal squamous cell carcinoma progression through reactive oxygen species accumulation via mitochondrial dysfunction and endoplasmic reticulum stress. J. Exp. Clin. Cancer Res. 2020;39:144. doi: 10.1186/s13046-020-01646-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu S., et al. ATF3 contributes to brucine-triggered glioma cell ferroptosis via promotion of hydrogen peroxide and iron. Acta Pharmacol. Sin. 2021;42:1690–1702. doi: 10.1038/s41401-021-00700-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Erickson J.R., et al. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133:462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rich R.C., Schulman H. Substrate-directed function of calmodulin in autophosphorylation of Ca2+/calmodulin-dependent protein kinase II. J. Biol. Chem. 1998;273:28424–28429. doi: 10.1074/jbc.273.43.28424. [DOI] [PubMed] [Google Scholar]
- 68.Karpinsky-Semper D., Volmar C.H., Brothers S.P., Slepak V.Z. Differential effects of the Gbeta5-RGS7 complex on muscarinic M3 receptor-induced Ca2+ influx and release. Mol. Pharmacol. 2014;85:758–768. doi: 10.1124/mol.114.091843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nobori K., et al. ATF3 inhibits doxorubicin-induced apoptosis in cardiac myocytes: a novel cardioprotective role of ATF3. J. Mol. Cell. Cardiol. 2002;34:1387–1397. doi: 10.1006/jmcc.2002.2091. [DOI] [PubMed] [Google Scholar]
- 70.Zhou H., et al. Activating transcription factor 3 deficiency promotes cardiac hypertrophy, dysfunction, and fibrosis induced by pressure overload. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Okamoto Y., et al. Transgenic mice with cardiac-specific expression of activating transcription factor 3, a stress-inducible gene, have conduction abnormalities and contractile dysfunction. Am. J. Pathol. 2001;159:639–650. doi: 10.1016/S0002-9440(10)61735-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Epanchintsev A., et al. Defective transcription of ATF3 responsive genes, a marker for Cockayne Syndrome. Sci. Rep. 2020;10:1105. doi: 10.1038/s41598-020-57999-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.An T., et al. Neuregulin-1 attenuates doxorubicin-induced autophagy in neonatal rat cardiomyocytes. J. Cardiovasc. Pharmacol. 2013;62:130–137. doi: 10.1097/FJC.0b013e318291c094. [DOI] [PubMed] [Google Scholar]
- 74.Timolati F., et al. Neuregulin-1 beta attenuates doxorubicin-induced alterations of excitation-contraction coupling and reduces oxidative stress in adult rat cardiomyocytes. J. Mol. Cell. Cardiol. 2006;41:845–854. doi: 10.1016/j.yjmcc.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 75.Jay S.M., et al. An engineered bivalent neuregulin protects against doxorubicin-induced cardiotoxicity with reduced proneoplastic potential. Circulation. 2013;128:152–161. doi: 10.1161/CIRCULATIONAHA.113.002203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vasti C., et al. Doxorubicin and NRG-1/erbB4-deficiency affect gene expression profile: involving protein homeostasis in mouse. ISRN Cardiol. 2012;2012 doi: 10.5402/2012/745185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu R.M., Desai L.P. Reciprocal regulation of TGF-beta and reactive oxygen species: a perverse cycle for fibrosis. Redox Biol. 2015;6:565–577. doi: 10.1016/j.redox.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dugaucquier L., Feyen E., Mateiu L., Bruyns T.A.M., De Keulenaer G.W., Segers V.F.M. The role of endothelial autocrine NRG1/ERBB4 signaling in cardiac remodeling. Am. J. Physiol. Heart Circ. Physiol. 2020;319:H443–H455. doi: 10.1152/ajpheart.00176.2020. [DOI] [PubMed] [Google Scholar]
- 79.Cardinale D., et al. Anthracycline-induced cardiomyopathy: clinical relevance and response to pharmacologic therapy. J. Am. Coll. Cardiol. 2010;55:213–220. doi: 10.1016/j.jacc.2009.03.095. [DOI] [PubMed] [Google Scholar]
- 80.Cardinale D., Iacopo F., Cipolla C.M. Cardiotoxicity of anthracyclines. Front Cardiovasc Med. 2020;7:26. doi: 10.3389/fcvm.2020.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang J., Yang J., Maity B., Mayuzumi D., Fisher R.A. Regulator of G protein signaling 6 mediates doxorubicin-induced ATM and p53 activation by a reactive oxygen species-dependent mechanism. Cancer Res. 2011;71:6310–6319. doi: 10.1158/0008-5472.CAN-10-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maity B., Yang J., Huang J., Askeland R.W., Bera S., Fisher R.A. Regulator of G protein signaling 6 (RGS6) induces apoptosis via a mitochondrial-dependent pathway not involving its GTPase-activating protein activity. J. Biol. Chem. 2011;286:1409–1419. doi: 10.1074/jbc.M110.186700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang J., et al. RGS6 suppresses Ras-induced cellular transformation by facilitating Tip60-mediated Dnmt1 degradation and promoting apoptosis. Oncogene. 2014;33:3604–3611. doi: 10.1038/onc.2013.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maity B., Stewart A., O'Malley Y., Askeland R.W., Sugg S.L., Fisher R.A. Regulator of G protein signaling 6 is a novel suppressor of breast tumor initiation and progression. Carcinogenesis. 2013;34:1747–1755. doi: 10.1093/carcin/bgt128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang J., et al. RGS6 is an essential tumor suppressor that prevents bladder carcinogenesis by promoting p53 activation and DNMT1 downregulation. Oncotarget. 2016;7:69159–69172. doi: 10.18632/oncotarget.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yang S.H., et al. Overexpression of regulator of G protein signaling 11 promotes cell migration and associates with advanced stages and aggressiveness of lung adenocarcinoma. Oncotarget. 2016;7:31122–31136. doi: 10.18632/oncotarget.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.