Abstract
Background
Nosocomial outbreaks frequently occurred during the Coronavirus disease 2019 (COVID-19) pandemic; however, sharing experiences on outbreak containment is vital to reduce the related burden in different locations.
Objectives
This article aims at sharing a practical experience on COVID-19 outbreak containment, including contact tracing, screening of target population, testing including molecular analysis, and preventive modalities. It also provides an epidemiological and molecular analysis of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS‑CoV‑2) infection outbreak in a tertiary care hospital in Saudi Arabia.
Methods
The outbreak occurred in a non-COVID medical ward at a tertiary care hospital in Jeddah, Saudi Arabia, from 22nd March and 15th April 2021. The multidisciplinary outbreak response team performed clinical and epidemiological investigations. Whole-Genome Sequencing (WGS) was implemented on selected isolates for further molecular characterization.
Results
A total of eight nurses (20 % of the assigned ward nurses) and six patients (16.2 % of the ward admitted patients at the time of the outbreak) tested positive for the SARS-CoV-2 virus based on PCR testing. The outbreak investigation identified strong evidence of an epidemiologic link between the affected cases. WGS revealed a set of spike mutations and deletions specific to the Alpha variant (B.1.1.7 lineage). All the nurses had mild symptoms, and the fatality among the patients was 50 % (three out of the six patients).
Conclusions
The current nosocomial COVID-19 outbreak, caused by the Alpha variant, revealed multiple breaches in the adherence to the hospital infection control recommended measures. Containment strategies were successful in controlling the outbreak and limiting infection spread. Molecular analysis and genome sequencing are essential tools besides epidemiological investigation to inform appropriate actions, especially with emerging pathogens.
Keywords: SARS-CoV-2, Outbreak, Virus Evolution, Genome sequence, Molecular epidemiology
Introduction
As per WHO, COVID-19 is an infectious disease caused by the SARS-CoV-2 virus. As of May 4th, 2022, Saudi Arabia reported just above 750,000 confirmed COVID19 cases, with 9096 related deaths [1]. At the same time, there were more than 500 million confirmed COVID-19 cases globally, including over six million deaths, making this pandemic the most serious public health concern at the current time [2]. Since the emergence of the COVID19 pandemic, in December 2019, the nosocomial SARS-CoV-2 spread has been one of the major threats facing healthcare systems, leading to disruption of clinical services and negative consequences especially among high-risk patients (e.g., death, complications due to COVID-19 infection, long hospital stay). Dynamics of nosocomial SARS-COV-2 transmission had been variable; patients or health care workers (HCWs) could be the source of infection [3].
HCWs are at an increased risk for repeated exposure to SARS-CoV-2, and multiple outbreaks of COVID-19 among HCWs have been reported [4]. The risk of infection was associated with contact with patients who had an atypical presentation, exposure to infected co-workers, and non-compliance with precautionary measures[5]. In China, Li et al. reported a nosocomial COVID-19 outbreak, that affected 205 patients and 148 HCWs [6]. Another similar outbreak in Germany was reported by Kabesch et al. and included 562 health care workers. Both outbreaks were eventually controlled after applying escalated infection control measures, including symptoms monitoring of close contacts, universal masking, and physical distancing [7].
During SARS-CoV-2 pandemic, genome sequencing has become an essential infection control approach providing high-resolution discriminatory power to identify outbreaks and track dissemination of SARS-CoV-2 variants [8], [9]. Since early March 2020, more than 13 million SARS-CoV-2 genome sequences have been deposited into the GISAID database (https://www.gisaid.org/) contributed by clinicians and researchers globally to understanding the viral evolution [10]. The continuous SARS-CoV-2 evolution due to genetic mutations or viral recombination leads to the emergence of variants with different transmissibility, infection severity and vaccine effectiveness [11]. SARS-CoV-2 variants are mainly classified into three categories based on public health risk analysis which includes include Variants of interest (VOI), variants of public health importance or concern (VOC), and variants of high consequence (VOHC) [12]. For VOC, clear evidence has been reported indicating a significant impact on viral transmissibility infection severity and ability to overcome immunity [13]. These VOC includes Alpha variant (B.1.1.7) first detected in the UK, Beta (B.1.351) in South Africa, Gamma (P.1) in Brazil, Delta variant (B.1.617.2) in India and the Omicron (B.1.1.529) in South Africa [14].
Occurrence of healthcare-associated outbreaks depends on the compliance with infection control measures and is affected by the type of the spreading variant and vaccine coverage among patients and staff. The current study provides an epidemiological and molecular descriptive report of a SARS-CoV-2 infection outbreak in a tertiary care hospital in western Saudi Arabia. The study analysed the most potential contributing factors to the occurrence of the outbreak. It also involved documentation of the infection control measures applied to contain the outbreak and learning lessons for future interventions.
Methods and materials
This is a descriptive retrospective medical chart review study of a SARS-CoV-2 infection outbreak among health care providers and patients. The study was conducted at a referral tertiary care hospital in Jeddah, Saudi Arabia, with a capacity of over 600 beds. The outbreak took place between 22nd March and 15th April 2021, in a medical inpatient ward with a total capacity of 32 beds. Beds were distributed between seven 4-bedded rooms and four single-bed rooms; the ward included a lounge room for HCWs. This ward is assigned as a non-COVID ward, where all patients are screened before admission, and only negative cases are admitted to this ward.
The total nursing staff in the ward was 29, in addition to 11 rotating nurse interns. The nurses were distributed in 2 shifts per day, with nurses conducting handovers twice a day, each lasting 30–45 min. An assigned infection control practitioner visits the ward once daily to follow up the implementation of infection control measures.
Outbreak investigation
After detecting the first three COVID-19 confirmed cases, a multidisciplinary team was composed to investigate the possible outbreak and start implementing immediate control measures. The team was steered by the infection prevention and control department, composed of medical and operational leadership and representatives from nursing services, microbiology, bed management, and medicine departments.
A case definition was proposed to include HCWs and/or patients in the affected ward who developed fever or symptoms of respiratory illness (i.e., sudden onset of chest pain, body aches, or cough) with confirmed SARS-CoV-2 PCR result at any time following the emergence of the first positive case in the affected ward; or any HCWs or patient diagnosed with COVID-19 based on positive SARS-CoV-2 PCR result as a part of active screening during the same period.
The initial infection control measures included an active screening of all HCWs working at the ward for any manifestations suggestive of COVID19 infection, immediate implementation of a universal droplet and contact precautions inside the ward at all times, testing all admitted patients and HCWs working at the same ward for SARS-CoV-2 twice a week regardless of their symptoms, strict implementation of social distancing, especially during handover times, closure of HCWs’ lounge, immediate restriction of sitters and visitors, restricting the floating of nurses from and to the affected ward, and restricting patient transfer from and to the affected ward.
The outbreak investigation team collected data related to patient demographics, clinical manifestations, comorbidities and history of COVID-19 exposure and vaccination. A contact list was generated based on history of exposure. The unit was blocked for new admissions, and patients were actively discharged once they were stable, asymptomatic, and tested negative for COVID-19.
Screening of SARS-CoV-2 variants
A systematic retrospective screening of SARS-CoV-2 variants on samples recovered from the outbreak (n = 9/14), was performed using targeted sequencing approaches. Total RNA was extracted using the QIAamp Viral RNA (Qiagen) extraction kits, High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and used for cDNA synthesis. Targeted sequencing of the receptor-binding domain (RBD) of the spike protein was performed as part of the department's effort to monitor the dissemination of SARS-CoV-2 variants. The RBD region was amplified using the following primers (nCOV_F45 5′- TCTCAGCCTTTTCTTATGGACCT-3′), (nCOV_R46 5′- TGTGGGTATGGCAATAGAGTT −3′) and analysed by Sanger sequencing.
Whole-genome sequencing
Viral RNA for each sample was quantified by qPCR using the 2019-nCoV CDC EUA kit (IDT, USA) in a StepOn Real-Time PCR System (Thermo Fisher Scientific), and those exhibiting a CT value below 25 (n = 5/14) were selected for WGS. Prior to WGS, multiplex PCR reactions using a set of 58 pairs of overlapping primers were performed to enrich the entire SARS-CoV-2 genome. The amplified segments thus generated were equally pooled and purified with the Qiaquick PCR purification kit (QIAGEN) prior to library preparation with the Nextera XT library preparation DNA kit (Illumina) according to the manufacturer’s instructions.
Bioinformatics analysis
SARS-CoV-2 reads were filtered out by mapping to the viral reference genome sequence (GenBank accession: MN908947.3) using Bowtie2 (version 2.3.4.1) [15] and SAMtools (version 1.8) [16]. The aligned reads were then supplied to the trim component of the iVar software (version 1.3) [17] to remove the sequences of the primers used for amplification. Consensus sequences were called from the trimmed reads using the iVar consensus component with a minimum read coverage of 10 and a minimum base frequency of 85 %. The sequences were uploaded to Pangolin COVID-19 Lineage Assigner [18] and GISAID databases [10] for further genome sequencing analysis, including mutations detection, clade and lineages assignment [19].
Results
Timeline and total number of cases
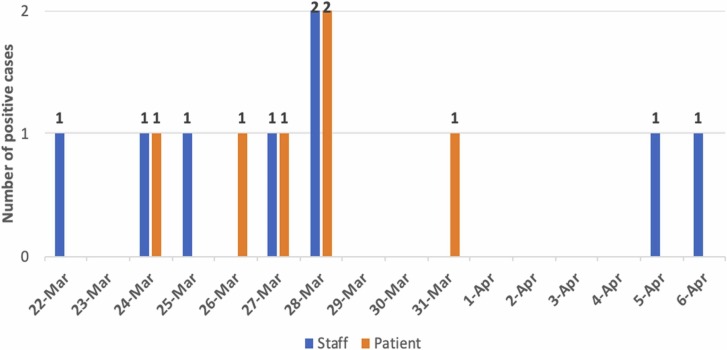
Between 22nd March and 6th April 2021, eight nurses (out of 40 nurses and nurse interns in the affected ward) and six patients (out of 37 admitted patients) were tested positive for SARS-CoV-2 virus based on PCR results, with an attack rate of 20 % and 16.2 %, respectively ( Fig. 1). Following that date, no more cases were reported as hospital-acquired transmitted, neither among patients nor among HCWs, for more than a week, considering the outbreak closed by 15th June 2021.
Fig. 1.
Diagnosis of COVID-19 cases over time.
Outbreak description
The index case was a staff nurse who developed a low-grade fever, diarrhea, and mild runny nose. She continued to work while symptomatic for four days before she was confirmed positive for SARS-CoV-2. She lives inside the medical city in the staff housing with no recent travel or exposure to suspected or confirmed COVID-19 cases. The second case was a nurse intern who works at the same unit and shifts with the first case. Her symptoms started one day after the first case and included low-grade fever, fatigue, and lower limb pain. SARS-CoV-2 PCR was done on March 22nd, 2021 and came back positive. She is single and lives with her family, and she disclosed no history of recent contact with a COVID-19 suspected or confirmed case in the community.
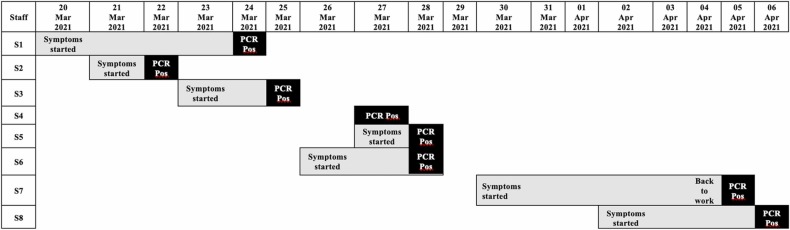
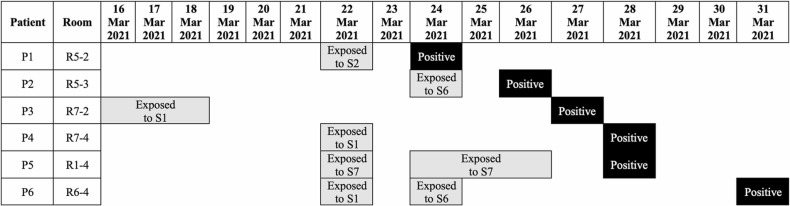
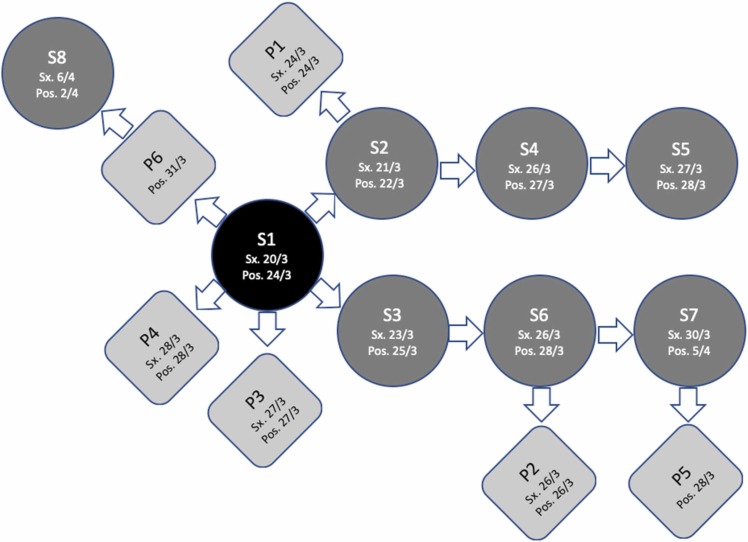
The third case from the same ward was an elderly male patient with comorbidities who developed a new low-grade fever and respiratory symptoms and was confirmed positive for SARS-CoV-2. He was admitted one week before detection of the first COVID-19 case in the ward, complaining of gastrointestinal bleeding, and his COVID-19 PCR screening was negative upon admission. During his hospital stay, he was looked after by the second infected nurse. At this point, a COVID-19 outbreak was declared at the unit, and case detection and surveillance testing for patients and healthcare staff were conducted. The screening measures identified additional six nurses ( Fig. 2) and five patients ( Fig. 3) infected with SARS-CoV-2. Fig. 4 depicts a summary of the epidemiological link between identified confirmed cases. Clinical characteristics and comorbidities of the infected patients are presented in ( Table 1). COVID19 PCR screening was expanded to include 29 physicians who visited the affected ward for consultations since the beginning of the outbreak, however, none of them was positive for SARS-CoV-2.
Fig. 2.
Symptom onset and Polymerase Chain Reaction (PCR) confirmed diagnosis among staff.
Fig. 3.
Exposure history and Polymerase Chain Reaction (PCR) confirmed diagnosis among patients.
Fig. 4.
Epidemiological link between confirmed staff and patient cases.
Table 1.
Clinical characteristics of patients infected with SARS-CoV-2.
| Patient | Age | Admission date | Clinical diagnosis | Positive swab | Swab at admission | Starting symptoms | Symptoms | COVID-19 Vaccination | Possible source | Outcome |
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | 83 | 14/03/2021 | Anaemia/ Gastrointestinal bleeding DM, COPD |
24/03/2021 | Yes: Negative | 24/03/2021 | Fever | No | Exposed to S2 |
Death 2/4/2021 Cause of Death: Respiratory failure, multiorgan failure |
| P2 | 66 | 16/03/2021 | Hepatocellular Carcinoma, Cirrhosis, Ascites | 26/03/2021 | Yes: Negative | 26/03/2021 | Fever | No | Exposed to S6 |
Death 4/4/2021 Cause of Death: Respiratory failure Hepatorenal syndrome |
| P3 | 89 | 08/03/2021 | Hypernatremia | 27/03/2021 | Yes: Negative | 25/03/2021 | Cough, SOB | No | Exposed to S1 | Recovered 12/4/2021 |
| P4 | 65 | 11/03/2021 | Stroke | 28/03/2021 | Yes: Negative | 28/03/2021 | Fever, SOB | No | Exposed to S1 and P3 | Recovered 25/42021 |
| P5 | 85 | 17/03/2021 | Gastric ulcers. Prostatic carcinoma | 28/03/2021 | Not done | No symptoms | No symptoms | No | Exposed to S7 |
Death 25/4/2021 Cause of Death: Candidemia. UTI. Respiratory failure |
| P6 | 56 | 29/10/2020 | Testicular Seminoma | 31/03/2021 | Yes: Negative | No symptoms | No symptoms | No | Exposed to S1 and S6 | Recovered 12/4/2021 |
SOB: Shortness of breath; S: Staff.
Infection control related practices
Revision of the adherence to the hospital's recommended measures against COVID19 revealed multiple breaches, including:
-
▪
Lack of screening for HCWs for respiratory symptoms before starting their duties.
Check points with Acute Respiratory Index (ARI score) screening is established in the hospital in all outpatient clinics, main hospital entrance, Emergency Department and Hemodialysis ward for all patients before being consulted, and those with ARI score ≥ 4 are isolated for further assessment.
Check points for HCWs´ respiratory symptoms are created during suspected intrahospital outbreaks in the ward or area affected. Otherwise, HCWs are counselled to self-report respiratory symptoms with immediate notification to their respective supervisor.
-
▪
Delay in HCWs’ self-reporting of clinical symptoms.
-
▪
Delay in testing symptomatic HCWs for SARS-CoV2.
-
▪
Gathering of HCWs during handover times without observing social distancing
-
▪
Failure to isolate symptomatic HCWs from their roommates at their accommodation.
Screening of the RBD of the spike protein
Sanger sequencing analysis of the RBD region of the spike protein identified the N501Y substitution in isolates (n = 9/14) recovered from the confirmed cases in this study. The detection of the N501Y showed that the strains in this study belong to one of the VOC, including the Alpha (B1.1.7), beta (1.351) and gamma (P.1) variants.
Alpha variant-defining mutations
Further characterization was performed using the genome sequencing data generated in this study. Genome sequence data were analysed using Pangolin to assign lineages, revealing that all the sequences in this study belong to Alpha (B.1.1.7.). The analysis showed that sequence har a set of spike mutations and deletions specific to the Alpha variant. This includes mainly mutations and deletions shown in Table 2 within the Spike protein, of which the N501Y is a key mutation enhancing the physical interactions between the virus and the human ACE2. Deletions of the Δ69/Δ70 and Δ144 amino-terminal domain (NTD) were also detected in the spike encoding region, which has been shown to decrease the sensitivity of some diagnostic assays and reduce susceptibility to neutralizing antibodies. Other mutations in the spike protein with potential clinical and biological importance (i.e., fA570D, D614G, P681H, T716I, S982A and D1118H have also been detected). Comparative analysis was performed on all SARS-CoV-2 genome sequences submitted to the GISAID database from Saudi Arabia. Of the 1396 SARS-COV-2 genomes, 31 genome sequences of Alpha B.1.1.7 were reported from Jeddah and Riyadh cities. The metadata of these reported cases was retrieved from GISAID databases suggesting that Alpha variants were reported in these cities mainly in April and May 2021, considering the paucity of SARS-CoV-2 genome sequencing data reported from Saudi Arabia ( Table 3).
Table 2.
List of non-synonymous mutations and deletions detected in all selected samples that are associated with the Alpha variant.
| Gene | Amino acid | Nucleotide |
|---|---|---|
| ORF1ab | T1001I | C3267T |
| A1708D | C5388A | |
| I2230T | T6954C | |
| S3675, G3676, F3677 deletion | 11288–11296 deletion | |
| P314L | C14408T | |
| Spike | H69, V70 deletion | 21765–21770 deletion |
| Y144 deletion | 21992–21994 deletion | |
| N501Y | A23063T | |
| A570D | C23271A | |
| D614G | A23403G | |
| P681H | C23604A | |
| T716I | C23709T | |
| S982A | T24506G | |
| D1118H | G24914C | |
| ORF8 | Q27stop | C27972T |
| R52I | G28048T | |
| K68 * | A28095T | |
| Y73C | A28111G | |
| N | D3L | G28280C |
| D3L | A28281T | |
| D3L | T28282A | |
| R203K | G28881A | |
| R203K | G28882A | |
| G204R | G28883C | |
| S235F | C28977T |
Table 3.
List of SARS-CoV-2 genome sequences deposited to GISAID database, reported from Saudi Arabia belong to Alpha variants.
| Isolates name | GISAID Accession ID | Collected date | Location | Host | Clade | Lineage |
|---|---|---|---|---|---|---|
| KAIMRC-S831* | EPI_ISL_14569355 | 04/04/2021 | Jeddah | Human | GRY | B.1.1.7 |
| KAIMRC-S838* | EPI_ISL_14569351 | 06/04/2021 | Jeddah | Human | GRY | B.1.1.7 |
| KAIMRC-S837* | EPI_ISL_14569352 | 06/04/2021 | Jeddah | Human | GRY | B.1.1.7 |
| KAIMRC-S836* | EPI_ISL_14569353 | 06/04/2021 | Jeddah | Human | GRY | B.1.1.7 |
| KAIMRC-S835* | EPI_ISL_14569354 | 06/04/2021 | Jeddah | Human | GRY | B.1.1.7 |
| DM-21–095140A | EPI_ISL_3236930 | 06/04/2021 | Jeddah | Human | GR | B.1.1.7 |
| KFSHRC_0105W | EPI_ISL_6699625 | 06/04/2021 | Jeddah | Human | GR | B.1.1.7 |
| KFSHRC8 | EPI_ISL_2151336 | 15/04/2021 | Riyadh | Human | GRY | B.1.1.7 |
| KFSHRC_0108W | EPI_ISL_6621331 | 22/04/2021 | Riyadh | Human | GRY | B.1.1.7 |
| KFSHRC_0112W | EPI_ISL_6626204 | 22/04/2021 | Riyadh | Human | GR | B.1.1.7 |
| DM-21–110634A | EPI_ISL_3237047 | 25/04/2021 | Jeddah | Human | GR | B.1.1.7 |
| DM-21–110332A | EPI_ISL_3237048 | 25/04/2021 | Jeddah | Human | GR | B.1.1.7 |
| KFSHRC_012 | EPI_ISL_6428137 | 02/05/2021 | Riyadh | Human | GR | B.1.1.7 |
| KFSHRC__014 | EPI_ISL_6437536 | 05/05/2021 | Riyadh | Human | GR | B.1.1.7 |
| KFSHRC_0120M | EPI_ISL_6700866 | 19/05/2021 | Jeddah | Human | GR | B.1.1.7 |
| KFSHRC_025A | EPI_ISL_6575670 | 20/05/2021 | Riyadh | Human | GR | B.1.1.7 |
| KFSHRC_016A | EPI_ISL_6632491 | 20/05/2021 | Riyadh | Human | GR | B.1.1.7 |
| KFSHRC_019A | EPI_ISL_6632876 | 20/05/2021 | Riyadh | Human | GR | B.1.1.7 |
| KFSHRC_048A | EPI_ISL_6575688 | 22/05/2021 | Riyadh | Human | GR | B.1.1.7 |
| KFSHRC_035A | EPI_ISL_6575678 | 24/05/2021 | Riyadh | Human | GR | B.1.1.7 |
| KFSHRC_0106W | EPI_ISL_6699777 | 2021–04 | Riyadh | Human | GR | B.1.1.7 |
| KFSHRC_030A | EPI_ISL_6700374 | 2021–05 | Riyadh | Human | GR | B.1.1.7 |
| KFSHRC_042A | EPI_ISL_6700686 | 2021–05 | Riyadh | Human | GR | B.1.1.7 |
| KFSHRC_033A | EPI_ISL_6914148 | 2021–05 | Riyadh | Human | GR | B.1.1.7 |
| KFSHRC_022A | EPI_ISL_7675055 | 2021–05 | Riyadh | Human | GRY | B.1.1.7 |
| KFSHRC 008 A | EPI_ISL_7987296 | 2021–05 | Riyadh | Human | GRY | B.1.1.7 |
| KFSHRC_0120J | EPI_ISL_7195005 | 2021–06 | Riyadh | Human | GR | B.1.1.7 |
| KFSHRC_0132F | EPI_ISL_6700204 | 2021–07 | Riyadh | Human | GR | B.1.1.7 |
| KFSHRC_0124L | EPI_ISL_7211807 | 2021–12 | Riyadh | Human | G | B.1.1.7 |
| KFSHRC_0121L | EPI_ISL_7285322 | 2021–12 | Riyadh | Human | GR | B.1.1.7 |
| KFSHRC_0117L | EPI_ISL_9291344 | 2021–12 | Riyadh | Human | GRY | B.1.1.7 |
Sequence data generated in this study.
Interventions for infection control
Following the detection of the first 3 COVID19 cases, a multidisciplinary team was composed to investigate the possible outbreak and to start implementing immediate control measures. The team was led by the infection prevention and control team. It included medical and operational leadership and a representative from nursing services, microbiology, bed management, and internal medicine departments. The initial infection control measures included an active screening of all HCWs working at the unit for any signs and symptoms suggestive of COVID19 before reporting to their duties, immediate implementation of universal droplet and contact precautions inside the unit at all times, emphasizing the adherence of universal mask policy, strictly follow correct hand hygiene practices, testing all admitted patients and staff working at the same unit for SARS-CoV-2 twice weekly regardless of their symptoms, strict implementation of social distancing especially during handover times, closure of staff Lounge inside the unit, immediate restriction of sitters and visitors from accessing the unit, blocking floating of nurses from and to ward six until further notice, blocking of patient transfer from and to ward six until outbreak controlled and aggressive cleaning and disinfection of all rooms and surfaces inside the unit. After the outbreak was controlled, routine COVID-19 testing and blocking of patient transfer were released, but all other control measures were kept in place in all hospital areas to prevent similar incidents in the future.
Discussion
Since the beginning of the COVID-19 pandemic, several reports of nosocomial transmission have been described [3], [20], [21]. The emergence of variants with higher transmissibility rates increases the risk of spreading within healthcare facilities, especially if infection prevention measures are not strictly applied. This report presents a COVID-19 nosocomial outbreak among HCWs and patients in one medical ward (non-COVID unit) between March and April 2021. Genome sequencing showed that the SARS-CoV-2 isolates involved in this outbreak belong to the Alpha (B.1.1.7.) variant.
The outbreak has affected a total of fourteen individuals; of those, eight were nursing staff and interns. When the sequence of events was analysed, we found out that the probable index case was a staff nurse (S1) who was diagnosed on March 24th, 2021, two days after the first confirmed report of a nurse intern (S2) on March 22nd, 2021. The probable index case was symptomatic while working, and her symptoms started earlier than the intern, who denied any contact with COVID-19 cases in the hospital, community, or among household members. The index case had been in contact in the workplace, while symptomatic, with a third staff case (another nurse intern) who shared the same shift and developed symptoms later (diagnosed on March 25th, 2021). Additionally, this index case had contact with three more patients, two of whom developed symptoms, and one was asymptomatic diagnosed during active screening.
After declaring an outbreak, secondary cases were reported among staff and patients due to previous exposure in the unit. A chronological chain of transmission was observed between each of the second and third staff cases and other staff. Patient cases were infected following contact with staff cases, except for one staff case who was exposed to an infected patient while asymptomatic.
There were neither hospital admissions nor reported deaths among the HCWs; however, three out of the six infected patients died (50 %). The HCWs were young staff with no comorbidities; they developed mild symptoms and clinically recovered. However, the patients had comorbidities that represented risk factors for ominous outcomes. None of the patients was vaccinated, although two nurses received one dose of AstraZeneca vaccine. Based on the updated COVID-19 Interim Practice Guidelines timely equivalent to the outbreak, HCWs diagnosed with COVID-19 were restricted from working and needed to be isolated for seven days (test date counted as day 1) and return to work on day eight if they were asymptomatic for at least the last two days. The discontinuation of isolation was not based on a negative PCR test.
In our analysis, we could notice variable transmission dynamics, HCWs to HCWs, HCWs to patients, and patients to HCWs. There was no clear patient-to-patient transmission; however, this transmission source cannot be ruled out because some of the patients were sharing the same room.
With a timeline observation of progressing COVID-19 case reports, a clear epidemiological link between the cases was considered. The samples were sent for SARS-CoV-2 screening of variants, initially identifying in the Receptor-Biding Domains (RBD) of the spike protein the N501Y mutation present in different variants of clinical concern. Then, the Whole Genome Sequencing (WGS) revealed that the isolates belonged to the Alpha variant (B.1.1.7 lineage), the same variant already identified in the UK. That confers the increasing binding affinity of the RBD region of SARS-CoV-2 to the Angiotensin-converting enzyme 2 receptors and, as a consequence, a high rate of transmissibility [22], [23]. This finding can explain the rapid and progressive spreading of the COVID-19 infection among HCWs and patients in one ward in a short period.
As an example of direct nosocomial transmission, there was an identical genome sequencing, 100 % matching between the isolates taken from the seventh staff case and the fifth patient, who were clinically linked.
On the other side, the genome sequencing of the last patient diagnosed on March 31st, 2021, who had contact with the index case, differed from the others. This patient did not have any other contact with COVID-19 cases outside the ward; therefore, this discrepancy might be related to the time-associated acquisition of new mutations from the same common source.
Several infection control-related breaches were identified as possible reasons for the dissemination of SRAS-CoV-2 infection in a single hospital ward, highlighting the importance of maintaining screening checkpoints for respiratory symptoms among HCWs. They need to be aware to immediately report if they suffer any COVID-19-related symptoms and to be assessed for further work restrictions to minimize the risk of transmission in the workplace [24].
Infection prevention and control measures are indispensable to control nosocomial transmission and outbreaks and essential to prevent their emergence. Laxity of compliance with preventive measures is one of the most important causes of nosocomial outbreaks, especially when facing novel variants with increased transmissibility capacity and virulence [25].
Despite the low vaccination coverage in the current report, it has been demonstrated that transmission of infection is possible even among fully vaccinated individuals [26]; therefore, universal masking, social distancing, and hand hygiene are basic protective measures that need to be reinforced and emphasized not only during the patient care but also during gathering and meetings in all facility areas.
This study may be limited by the inability to perform molecular testing of all infected samples. This was related to the time gap between declaring an outbreak and the decision to investigate for a specific viral mutation responsible for the outbreak.
Conclusions
The current report demonstrated a nosocomial COVID-19 outbreak caused by Alpha variant in Saudi Arabia. It revealed multiple breaches in the adherence to the hospital infection control and preventive measures. It also demonstrated successful containment strategies essential to limit the expansion of the infection and, finally, its control. Molecular characterization and genome sequencing are considered essential tools besides epidemiological investigation to inform the outbreak response teams of the appropriate actions to be implemented, especially with rapidly changing new pathogen.
Funding
MFA acknowledges funding from King Abdulaziz City for Science and Technology (KACST), Saudi Arabia (5–20–01–536–0003) as part of a research funding program (COVID-19 Research Grant Program) that aims to support R&D on COVID-19 to better understand and elucidate the Coronavirus pandemic through in-depth etiological and epidemiological studies, disease pathogenesis, data modelling, developing rapid diagnostics.
Data availability
The whole-genome sequence of the SARS-CoV-2 sequenced isolates in this study has been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the accession numbers OK081886-OK081890.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgements
We gratefully acknowledge all data contributors, i.e., the Authors and their Originating laboratories responsible for obtaining the specimens, and their Submitting laboratories for generating the genetic sequence and metadata and sharing via the GISAID Initiative, on which this research is based.
References
- 1.World Health Organization. Saudi Arabia - WHO Coronavirus (COVID-19) Dashboard [Internet]. World Heal. Organ. 2022 [cited 2022 May 4]. Available from: 〈https://covid19.who.int/region/emro/country/sa〉.
- 2.Organization W.H. COVID-19 Weekly Epidemiological Update Edition 58, published 21 September 2021. COVID-19 Wkly. Epidemiol. Updat. 2021. 2021.
- 3.Abbas M., Robalo Nunes T., Martischang R., Zingg W., Iten A., Pittet D., et al. Nosocomial transmission and outbreaks of coronavirus disease 2019: the need to protect both patients and healthcare workers. Antimicrob Resist Infect. Control BioMed Cent Ltd. 2021;10(1):1–13. doi: 10.1186/s13756-020-00875-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou R., Dana T., Buckley D.I., Selph S., Fu R., Totten A.M. Epidemiology of and risk factors for coronavirus infection in health care workers; a living rapid review. Ann Intern Med. 2020;173(2):120–136. doi: 10.7326/M20-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zabarsky T.F., Bhullar D., Silva S.Y., Mana T.S.C, Ertle M.T., Navas M.E., et al. What are the sources of exposure in healthcare personnel with coronavirus disease 2019 infection? Am J Infect Control. 2021;49(3):392–395. doi: 10.1016/j.ajic.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y., Peng S., Li L., Wang Q., Ping W., Zhang N., et al. Clinical and transmission characteristics of Covid-19 — a retrospective study of 25 cases from a single thoracic surgery department. Curr Med Sci. 2020;40(2):295–300. doi: 10.1007/s11596-020-2176-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabesch M., Roth S., Brandstetter S., Häusler S., Juraschko E., Weigl M., et al. Successful containment of Covid-19 outbreak in a large maternity and perinatal center while continuing clinical service. Pedia Allergy Immunol. 2020;31(5):560–564. doi: 10.1111/pai.13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meredith L.W., Hamilton W.L., Warne B., Houldcroft C.J., Hosmillo M., Jahun A.S., et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis. 2020;20(11):1263–1272. doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z., Azman A.S., Chen X., Zou J., Tian Y., Sun R., et al. Global landscape of SARS-CoV-2 genomic surveillance and data sharing. Nat Genet. 2022;54(4):499–507. doi: 10.1038/s41588-022-01033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khare S., Gurry C., Freitas L., Schultz M.B, Bach G., Diallo A., et al. GISAID’s role in pandemic response. China CDC Wkly. 2021;3:1049–1051. doi: 10.46234/ccdcw2021.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L., et al. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet Nat Publ Group. 2021;22(12):757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantón R., Ramos P.D.L., García-Botella A., García-Lledó A., Gómez-Pavón J., Del Castillo J.G., et al. New variants of SARS-CoV-2. Rev Esp Quimioter Soc Esp De Quimioter. 2021;34(5):419–428. doi: 10.37201/req/071.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thye A.Y.K., Law J.W.F., Pusparajah P., Letchumanan V., Chan K.G., Lee L.H., et al. Emerging SARS-CoV-2 variants of concern (VOCs): An impending global crisis. Biomedicines. 2021;9:1303. doi: 10.3390/biomedicines9101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi J.Y., Smith D.M. SARS-CoV-2 variants of concern. Yonsei Med J. 2021;62(11):961–968. doi: 10.3349/ymj.2021.62.11.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grubaugh N.D., Gangavarapu K., Quick J., Matteson N.L., De Jesus J.G., Main B.J., et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20(8):1–19. doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Toole Á., Scher E., Underwood A., Jackson B., Hill V., McCrone J.T., et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021;7(2):7. doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aksamentov I., Roemer C., Hodcroft E., Neher R.A. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw. 2021;6(67):3773. [Google Scholar]
- 20.Arons M.M., Hatfield K.M., Reddy S.C., Kimball A., James A., Jacobs J.R., et al. Presymptomatic SARS-CoV-2 infections and transmission in a skilled nursing facility. N Engl J Med. 2020;382(22):2081–2090. doi: 10.1056/NEJMoa2008457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sikkema R.S., Pas S.D., Nieuwenhuijse D.F., O’Toole Á., Verweij J.J., van der Linden A., et al. COVID-19 in health-care workers in three hospitals in the south of the Netherlands: a cross-sectional study. Lancet Infect Dis. 2020;20:1273–1280. doi: 10.1016/S1473-3099(20)30527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., et al. Nat. Rev. Microbiol. Nature Publishing Group; 2021. SARS-CoV-2 variants, spike mutations and immune escape; pp. 409–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CDC. Strategies to Mitigate Healthcare Personnel Staffing Shortages [Internet]. [cited 2022 May 6]. Available from: 〈https://www.cdc.gov/coronavirus/2019-ncov/hcp/mitigating-staff-shortages.html〉.
- 25.Asad H., Johnston C., Blyth I., Holborow A., Bone A., Porter L., et al. Health care workers and patients as Trojan horses: a COVID19 ward outbreak. Infect Prev Pr. 2020;2(3) doi: 10.1016/j.infpip.2020.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franco-Paredes C. Lancet Infect Dis. Elsevier Ltd,; 2022. Transmissibility of SARS-CoV-2 among fully vaccinated individuals. p. 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The whole-genome sequence of the SARS-CoV-2 sequenced isolates in this study has been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under the accession numbers OK081886-OK081890.