Abstract
The aim of the present study was to elucidate the significance of secreted protein acidic and cysteine rich (SPARC) expression in non-small cell lung cancer (NSCLC) in terms of clinicopathology, immune-cell infiltration and survival prognosis. A meta-analysis and bioinformatics analysis were performed using studies retrieved with PubMed, Web of Science, Wanfang Data and the Chinese National Knowledge Infrastructure databases. The meta-analysis suggested that, compared with normal tissues, SPARC expression was elevated in NSCLC tissues. The expression of SPARC was not significantly associated with TNM stage and lymph-node metastasis, and was positively associated with patient gender. Regarding the differential expression of SPARC and the relationship between expression levels and survival, the Oncomine database was consulted and Kaplan-Meier curves were drawn. It was indicated that SPARC mRNA expression levels were higher in NSCLC tissues than in normal tissues. Low expression of SPARC mRNA was negatively associated with overall survival, first progression survival and post-progression survival of patients. Further exploration of the relationship between SPARC expression and survival by univariate analysis indicated that TNM stage, lymph node metastasis, distant metastasis and depth of infiltration of lung cancer were negatively associated with patient prognosis. Cox multifactorial analysis suggested that SPARC expression levels and TNM stage were risk factors significantly affecting the prognosis of patients with NSCLC. Analysis with the GEPIA and UALCAN databases further indicated that the mRNA expression level of SPARC in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) was higher than that in normal lung tissue, and the SPARC expression levels were affected by factors such as the TNM stage of lung cancer. A lower the level of SPARC mRNA expression was associated with a better relative survival prognosis of patients. In the Human Protein Atlas database, the expression level of SPARC protein was higher in LUAD and LUSC than in normal lung tissue. In the Timer database, the expression level of SPARC was closely linked to immune cells related to the occurrence of lung cancer, and the degree of immune-cell infiltration and SPARC protein expression were closely related to the prognosis of patients with lung cancer. Immune cells were indicated to exhibit significant inhibition of DNA proliferation mutation mechanisms in lung cancer (P<0.05). In summary, SPARC expression may be used as a valuable indicator of prognosis in patients with NSCLC, which may provide new approaches for preventative treatment.
Keywords: SPARC, non-small cell lung cancer, meta-analysis, bioinformatics analysis, prognosis
Introduction
Lung cancer is one of the most common malignant tumor types. According to the latest statistics from the World Health Organization in 2021, 2.2 million patients are living with lung cancer worldwide, among which 1.8 million die; lung cancer ranks highest in terms of death rate from cancer and the patients are mostly male (1). Non-small cell lung cancer (NSCLC) is the most common type of lung cancer, accounting for 80–85% of cases. These mainly include lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). NSCLC mostly spreads along the trachea and alveolar wall, and as these areas are rich in blood vessels, local infiltration and hematogenous dissemination occur early, and it easily involves the pleura and causes pleural effusion, and numerous patients are diagnosed at this time; however, hematogenous and disseminated metastases have already occurred at that stage, and patients are no longer eligible for surgical treatment. Therefore, the 5-year survival rate of patients with advanced NSCLC is only ~10% (1). The treatment of advanced lung cancer mainly comprises chemotherapy and targeted therapy. Targeted therapy has been a hot research topic in recent years. Genetic testing of patients with NSCLC is used to select targeted therapeutic drugs. Current common targets include EGFR, echinoderm microtubule-associated protein-like 4-anaplastic lymphoma kinase, VEGF, programmed death receptor 1, HER2 and KRAS (2).
The secreted protein acidic and cysteine rich (SPARC) gene is located on the human chromosome Sq31.3-q32 and contains 10 exons. It is a small-molecule glycoprotein rich in cysteine and is closely related to cellular secretion. It is involved in various biological processes such as tumor angiogenesis and tissue repair and remodeling (3), and is able to regulate cell adhesion and proliferation through different signaling pathways. Members of the SPARC family all contain a special EC domain, and the EC domain contains the EF hand model. SPARC family members may be divided into different taxa: SPARC, Hevin, SMOC1 and follistatin-like protein, which are five taxa with sequence homology of different EC structural domains, of which the SPARC gene is the more important gene in the family (4). Studies have indicated that SPARC is abnormally highly expressed in liver cancer tissues and is related to the formation of microvessels in tumor tissues (5). In gastric cancer, SPARC regulates the epithelial-mesenchymal transition of gastric cancer through the Slug pathway, thereby promoting gastric cancer metastasis and leading to poor prognosis (6). Previous studies have indicated that the expression of SPARC in gastric cancer is correlated with the expression of E-cadherin, Slug and Vimentin, and the positive expression of SPARC is negatively correlated with the survival rate of patients with gastric cancer. Positive expression of E-cadherin is positively correlated with the survival rate of patients with gastric cancer (7). At the same time, it has been indicated that SPARC may affect the metastasis of lung cancer through the Wnt/β-catenin signaling pathway. The expression of β-catenin, c-myc and cyclin-D1 in the Wnt/β-catenin signaling pathway was increased in lung cancer tissues. After SPARC knockout, the expression of β-catenin, c-myc and cyclin-D1 decreased. It is inferred that SPARC is able to affect the epithelial-mesenchymal transition of NSCLC through the Wnt/β-catenin signaling pathway and promote invasion and metastasis of NSCLC. The high expression of SPARC in liver cancer and bladder cancer may inhibit apoptosis of tumor cells through the PI3K/AKT signaling pathway, while promoting the proliferation, invasion and metastasis of tumor cells. SPARC is an upregulated gene in CHD1L-induced hepatocellular carcinoma (8–11). In human NSCLC cells expressing SPARC induced by TGF-β, the expression of CDH1 did not change, while SPARC was strongly expressed in metastatic NSCLC. It is also expressed in triple-negative NSCLC and intraductal carcinoma in situ. The intensity of SPARC expression is related to the stage of breast ductal carcinoma in situ. The expression of SPARC in poorly differentiated ductal carcinoma in situ is stronger than that in well-differentiated cases, suggesting that SPARC may be related to the epithelial-mesenchymal transition of NSCLC (12).
Previous studies have indicated that SPARC is highly expressed in gastric cancer, pancreatic cancer, cervical cancer and liver cancer, but the role of SPARC in lung cancer remains elusive. The present study confirmed the relationship between SPARC mRNA and protein expression levels and survival prognosis, clinicopathological factors and immune infiltration mechanisms in NSCLC by meta-analysis and bioinformatics analysis. It provided a theoretical basis for the SPARC gene as a potential key factor in the occurrence and development of NSCLC.
Materials and methods
Literature search and data extraction
The PubMed (http://www.ncbi.nlm.nih.gov/pubmed), Web of Science (http://webofscience.com), Wanfang database (https://www.wanfangdata.com.cn/) and Chinese National Knowledge Infrastructure (CNKI) database (http://www.cnki.net/) were searched from inception until February 2022 by using the following key words: ‘SPARC’ and ‘lung cancer’ or ‘NSCLC’ or ‘lung adenocarcinoma’ or ‘lung squamous cell carcinoma’. The present study was conducted in strict accordance with the PRISMA guidelines (13). A total of 173 articles were screened after an initial search by reading abstracts and excluding duplicate studies. After further screening (SS and YRZ), we continued the careful screening of 26 articles for full-text reading. Ultimately, only 17 articles were included in the study of this paper and all case data were sourced from China. The study inclusion criteria were as follows: i) Patients with NSCLC; ii) immunohistochemical detection of SPARC; iii) articles containing SPARC expression and clinicopathological parameters; iv) none of the patients received chemotherapy or radiotherapy prior to surgery. The exclusion criteria were as follows: i) Abstracts, case reports, reviews and conference proceedings; ii) duplicate publications; iii) unclear diagnoses; iv) the expression of SPARC was investigated by western blot, reverse transcription-quantitative PCR, cDNA microarray or transcriptome sequencing.
Data extraction and quality assessment
As presented in Table I, the eligible paper information was extracted by two reviewers (GYM and ZGZ), including the first author's name, publication year, patient's country, antibody company, number of cases and controls, cancer risk and follow-up results. According to the Newcastle-Ottawa Oncomine scale (14), the quality of the studies was independently assessed by two reviewers. The criteria to judge the quality of the studies included sample selection, comparability and determination of results.
Table I.
Main characteristics of eligible studies.
| First author, year | Country | Antibody supplier | Total cases, n | Events, n | Risk of cancer | Follow-up outcome | Quality score | (Refs.) |
|---|---|---|---|---|---|---|---|---|
| Andriani et al, 2018 | Italy | NS | 57 | 28 | Raised | NS | 8 | (17) |
| Duan et al, 2017 | China | Invitrogen; Thermo Fisher Scientific, Inc. | 32 | 18 | Raised | Negative | 7 | (19) |
| Koumiya et al, 2016 | Japan | Abcam | 200 | 145 | Raised | NS | 7 | (20) |
| Koukourakis et al, 2003 | Greece | Abcam | 113 | 107 | Raised | NS | 9 | (28) |
| Zhang et al, 2012 | China | Fuzhou Maixin Biotech Co., Ltd. | 89 | 63 | Raised | NS | 8 | (25) |
| Zheng et al, 2014 | China | Bostik | 71 | 44 | Raised | NS | 8 | (29) |
| Zheng et al, 2015 | China | NS | 71 | 40 | Raised | NS | 8 | (30) |
| Huang et al, 2012 | China | R&D Systems, Inc. | 105 | 57 | Raised | NS | 8 | (26) |
| Xu et al, 2019 | China | Abcam | 90 | 54 | Raised | NS | 8 | (31) |
| Kurtul et al, 2014 | Turkey | BIOSS | 84 | NS | Raised | Negative | 8 | (22) |
| Fabrizio et al, 2020 | Italy | Cell Signaling Technology, Inc. | 21 | NS | Raised | NS | 7 | (15) |
| Yin et al, 2016 | China | NS | NS | NS | Raised | NS | 7 | (24) |
| Gao et al, 2020 | China | NS | NS | NS | Raised | NS | 8 | (16) |
| Li et al, 2018 | USA | NS | NS | NS | NS | NS | 7 | (18) |
| Ma et al, 2006 | China | NS | NS | NS | NS | NS | 7 | (27) |
| Zheng et al, 2016 | China | NS | NS | NS | Raised | Negative | 8 | (21) |
| Grant et al, 2014 | USA | Prolytix | 19 | 7 | Raised | NS | 8 | (23) |
The quality score is based on the Newcastle-Ottawa Oncomine scale. NS, not specified.
Meta-analysis
SPARC expression was estimated according to the clinicopathological parameters and the odds ratio (OR) and its 95% CI were used to express the effect size in patients with NSCLC. First, the χ2 test was used to assess the heterogeneity of the original study. When heterogeneity was not significant, i.e., P≤0.1, a fixed-effects model (Mantel-Haenszel method) was used. When P>0.1, a random-effects model (Der Simonian and Laird method) was used. I2 statistics were used to quantify the effect of heterogeneity at cutoffs of 25, 50 and 75%, respectively. Heterogeneity in the results was considered significant if P<0.1 and I2>50%. Meta-analysis was performed based on the random-effects model. If P>0.1 and I2<50%, it was considered that there was no significant heterogeneity among the study results and the fixed-effects model was used for meta-analysis. Funnel plots were used to evaluate publication bias, and Begg's test and Egger's test were used to evaluate whether the funnel plots were consistent. Furthermore, to perform a sensitivity analysis on the aggregated results, one study was deleted at a time and the impact of a single study on the results was thereby examined. In the sensitivity analysis, none of the studies affected the combined HR and OR, which indicated that the results were stable (results not shown). Meta-analysis was performed using RevMan 5.2 software and SPSS software (version 10.0; SPSS, Inc.) with the t-test. A two-sided P<0.05 was considered to indicate statistical significance.
Bioinformatics analysis
The prognostic value of SPARC mRNA expression in NSCLC was analyzed using the Kaplan-Meier plotter (KM-plotter; http://www.kmplot.com). The influence of the expression of SPARC on overall survival, recurrence-free survival, distant metastasis-free survival and post-progression survival in all patients was determined. The correlation of the expression of SPARC with clinicopathological features was also examined. To investigate SPARC gene expression, the Oncomine database (www.oncomine.org), an extensive database of tumor microarray data, including microarray and gene expression data, was used. The database may be used to analyze differences in gene expression and to classify clinical information about tumor patients. The expression differences of SPARC mRNA in cancer tissues and normal tissues were compared. The gene expression and clinicopathological data of SPARC were downloaded from The Cancer Genome Atlas (TCGA; www.cancer.gov) database using R software TCGA-assembler. The data were collated and SPARC mRNA expression in NSCLC was analyzed. Furthermore, the clinicopathological data and prognosis of tumor patients were analyzed. Cox hazard regression models were used to perform univariate and multivariate analyses. The model analyzed the effects of risk factors, hazard ratios and 95% CIs. The TCGA data and GTEx data from the GEPIA online analysis website (http://gepia.cancer-pku.cn/) were used to analyze the expression of SPARC mRNA in NSCLC tissues and normal lung tissues. SPARC mRNA expression was analyzed according to library online analysis and mining site exploration subpopulations by performing TCGA number analysis on the UALCAN online website (http://ualcan.path.uab.edu/). According to the mRNA expression value of each gene, cancer patients were automatically divided into a high expression group and low expression group for comparison. P<0.05 was considered to indicate a statistically significant difference. Based on the Human Protein mapping database [the Human Protein Atlas (HPA); https://www.proteinatlas.org/], the SPARC mRNA and protein levels in NSCLC and normal lung tissues were analyzed and compared. Representative immunohistochemical images of NSCLC tissues with varying levels of SPARC protein expression were downloaded. Based on the website of the Timer database (http://http://timer.cistrome.org/), the relationship between immune cells and survival prognosis in NSCLC, as well as the relationship between lung cancer and related immune cell infiltration and SPARC gene expression, were analyzed. The scores were mainly calculated for 35% and a survival time of 60 months. It was fitted automatically by software which uses the R language package and the main algorithms are based on genetic marker method and deconvolution. The R language package immunedeconv was used, which integrates the six algorithms TIMER, xCell, MCP-counter, CIBERSORT, EPIC and quanTIseq.
Statistical analysis
Revman Version 5.3 (Cochrane Collaboration) was adopted to perform the meta-analysis. Comparison between the case group and the control group was indicated by OR and 95% CI. I2 statistics were used to determine heterogeneity between study results. Publication bias was evaluated by funnel plots and the asymmetry of the funnel plot was tested by Begg's and Egger's tests. The Cox risk regression model was used for univariate and multivariate analysis. The model analyzed the impact of risk factors, risk ratios and 95% CI. P<0.05 was considered to indicate statistical significance. All data were analyzed using SPSS 19.0 software (IBM Corp).
Results
Literature search and publication bias
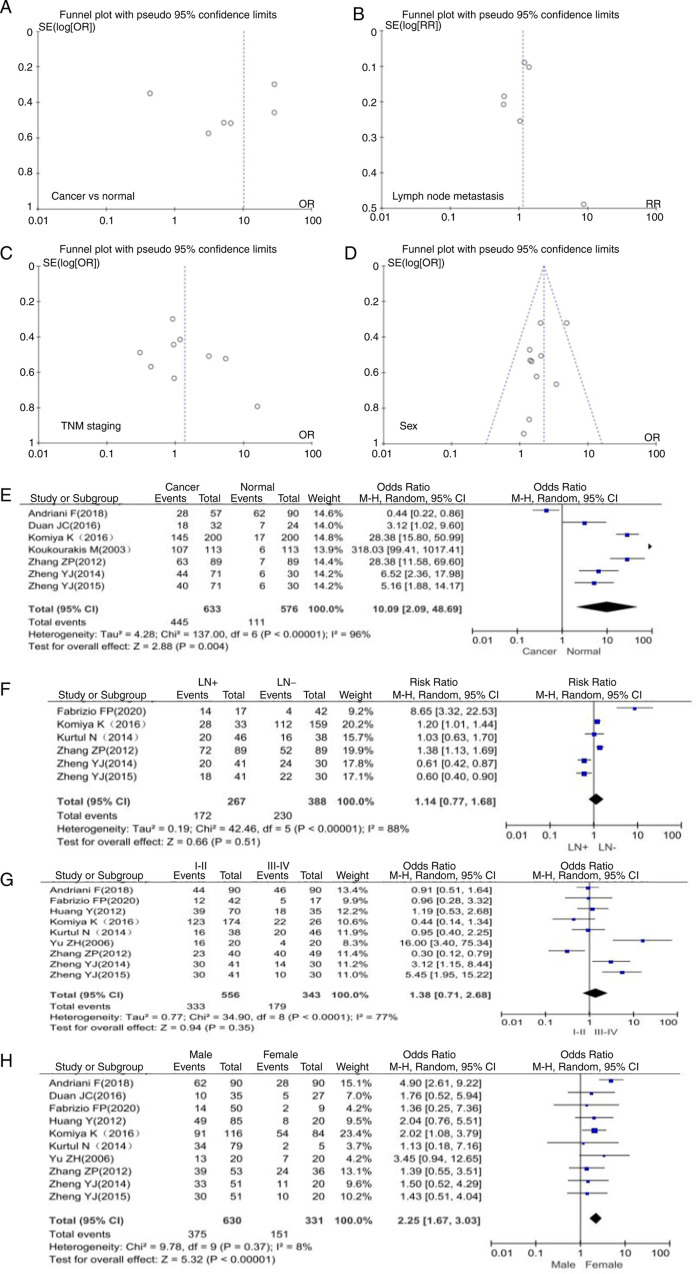
The final 17 articles included in the present study discussed the relationship between SPARC expression and clinicopathologically or prognostically relevant expression markers in NSCLC (Fig. 1) (15–31). Clinicopathological characteristics of NSCLC included histological grade, TNM stage, presence of lymph node metastasis and patient gender (Table I). As presented in Fig. 2, funnel plots were used to examine the heterogeneity among studies. The results of Egger's test indicated that there was no significant publication bias in the present meta-analysis. It was indicated that SPARC expression was able to distinguish between cancer and normal tissue, and was related to the parameters of lymph node metastasis, TNM stage and gender (Fig. 2A-D).
Figure 1.
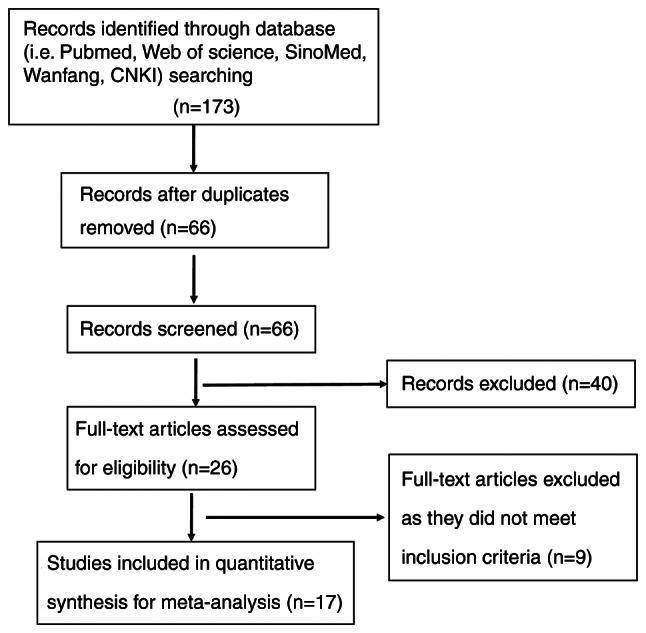
Flow diagram of article selection. CNKI, Chinese National Knowledge Infrastructure.
Figure 2.
Funnel plots of publication bias detected in NSCLC expressing SPARC. Publication bias was analyzed based on the relationship between SPARC expression and clinicopathological features of NSCLC. This included (A) the relationship between cancer and normal lung tissue, (B) lymph node metastasis, (C) TNM staging and (D) gender. Forest plot of SPARC expression and clinicopathological features of NSCLC. (E) Cancer and normal tissue, (F) lymph node metastasis, (G) TNM staging and (H) gender. SPARC, secreted protein acidic and cysteine rich; NSCLC, non-small cell lung cancer; SE, standard error; OR, odds ratio; M-H, Mantel-Haentzel; LN, lymph node involvement; df, degrees of freedom.
Association between SPARC expression and clinicopathological characteristics of patients with NSCLC according to the OR forest plot
A total of 7 articles included data on 633 patients with NSCLC and 576 normal controls. Compared with normal tissues, the expression of SPARC in lung cancer tissues was significantly upregulated (OR=10.09, 95% CI 2.09-48.69, P=0.004; Fig. 2E). The SPARC gene was expressed at higher levels in NSCLC than in normal lung tissue. There was high heterogeneity among study results (P=0.004, I2=96%), which may be due to potential selection bias among study subjects and no medical history in the control group. The present meta-analysis indicated that SPARC expression was not significantly associated with TNM stage and lymph node metastasis in patients with NSCLC (Fig. 2F and G). The expression of SPARC was closely related to the gender of the patients and males were more likely to develop NSCLC than females (OR=2.25, 95% CI 1.67-3.03, P<0.00001; Fig. 2H).
Relationship between SPARC expression and prognosis of NSCLC
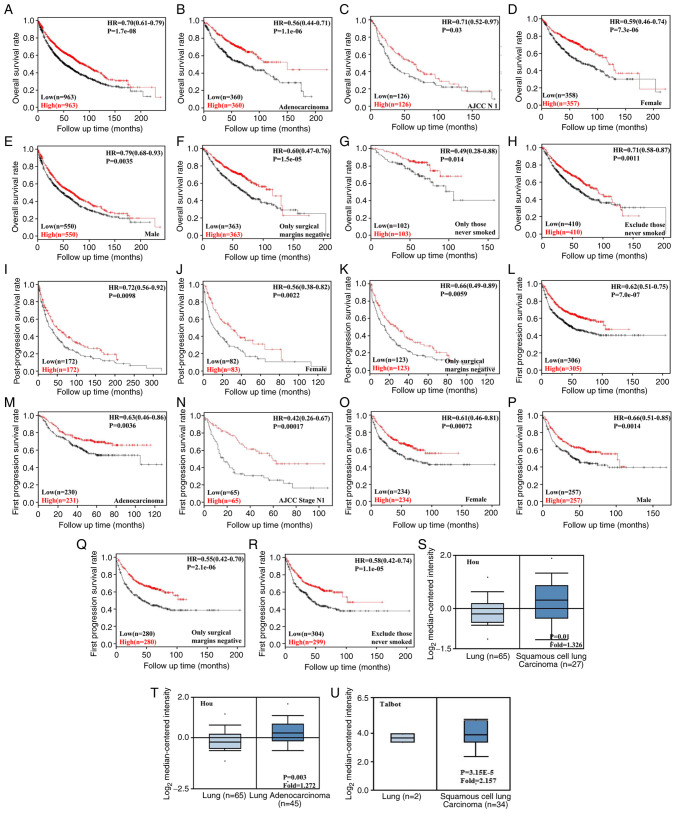
Using KM-Plotter, it was indicated that SPARC expression levels were positively correlated with overall survival of patients with different stages of NSCLC. In addition, there was a positive correlation with the survival rate of patients with different tumor types, gender and smoking history (P<0.05). Increased SPARC expression levels were positively associated with first progression survival and post-progression survival (P<0.05). Patients with NSCLC and high SPARC expression levels had a longer survival time (P<0.05). These data suggested that the SPARC gene is closely associated with patient survival prognosis (P<0.05, Fig. 3A).
Figure 3.
KM-Plotter (http://www.kmplot.com) was used to examine the effect of SPARC mRNA expression on prognosis of patients with NSCLC. (A) SPARC mRNA expression was positively associated with overall survival in patients with NSCLC. (B) Relationship between SPARC mRNA expression and overall survival in lung adenocarcinoma. (C) Relationship between SPARC mRNA expression and overall survival in AJCC N1. (D) Relationship between SPARC mRNA expression and overall survival in females. (E) Relationship between SPARC mRNA expression and overall survival in males. (F) Relationship between SPARC mRNA expression and overall survival when surgical margins are negative. (G) Relationship between SPARC mRNA expression and overall survival for patients who never smoked. (H) Relationship between SPARC mRNA expression and overall survival after excluding those who never smoked. (I) SPARC mRNA expression was positively associated with post-progression survival in patients with NSCLC. (J) Relationship between SPARC mRNA expression and post-progression survival in females. (K) Relationship between SPARC mRNA expression and post-progression survival when surgical margins were negative. (L) SPARC mRNA expression was positively associated with first progression survival in patients with NSCLC. (M) Relationship between SPARC mRNA expression and first progression survival in lung adenocarcinoma. (N) Relationship between SPARC mRNA expression and first progression survival in AJCC stage N1. (O) Relationship between SPARC mRNA expression and first progression survival in females. (P) Relationship between SPARC mRNA expression and first progression survival in males. (Q) Relationship between SPARC mRNA expression and first progression survival when surgical margins were negative. (R) Relationship between SPARC mRNA expression and first progression survival after excluding those who never smoked. Effects of SPARC mRNA expression on patients with NSCLC according to the Oncomine database (www.oncomine.org). The datasets within the Oncomine database were used for bioinformatics analysis to examine the expression of SPARC mRNA during the development of NSCLC. SPARC expression in NSCLC tissues was higher than that in normal lung tissues. Values in the box plots are expressed as the median (interquartile range). (S) SPARC mRNA expression in Lung tissue vs Squamous cell lung Carcinoma. (T) SPARC mRNA expression in lung tissue vs. lung adenocarcinoma. (U) SPARC mRNA expression in lung tissue vs. squamous cell lung carcinoma. SPARC, secreted protein acidic and cysteine rich; NSCLC, non-small cell lung cancer; HR, hazard ratio; AJCC, American Joint Committee on Cancer; KM, Kaplan-Meier.
The association between SPARC expression and the bioinformatics signature of NSCLC in the Oncomine database was then analyzed. The databases published by Hou and Talbot suggested that the mRNA expression of SPARC was higher in LUSC than in normal lung tissue; furthermore, according to the database by Hou, SPARC expression in LUAD was also higher than that in normal lung tissue (P<0.05, Fig. 3B).
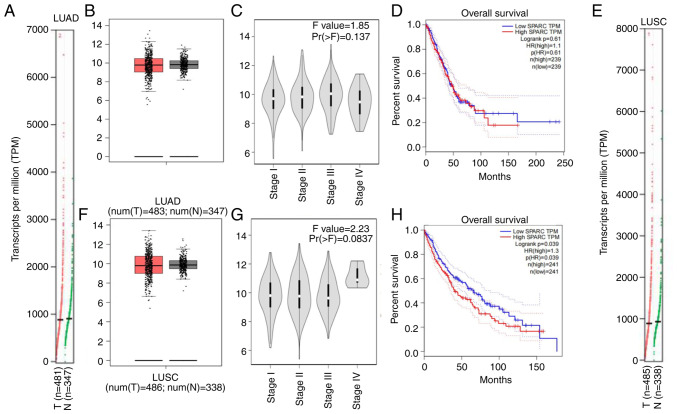
In the GEPIA database, the mRNA expression of SPARC in LUAD tissue was higher than that in normal lung tissue (Fig. 4A and B), SPARC expression was not significantly associated with the stage of LUAD, but when it was analyzed in Stage I to Stage III, SPARC expression levels were enhanced with the increase in stage; however, the level of SPARC expression was slightly decreased again in Stage IV (Fig. 4C). Kaplan-Meier analysis with the log-rank test suggested that among 239 patients with LUAD, the SPARC gene was not significantly associated with OS. When SPARC expression levels were reduced, the survival and prognostic practices of LUSC patients tended to be prolonged (Fig. 4D). Higher SPARC expression levels were associated with a shorter survival time of patients. The mRNA expression levels of SPARC were not significantly different between normal lung tissues and LUSC, with an increasing trend of expression in LUSC (Fig. 4E and F). This indicates that the mRNA expression levels of SPARC and protein expression levels in LUSC are in agreement with each other, suggesting that the analysis results are credible. SPARC expression is upregulated in LUSC. SPARC expression was associated with the staging of LUSC but this was not significant (P=0.0837). SPARC mRNA expression was higher in LUSC Stage IV than in other stages (Fig. 4G). Kaplan-Meier analysis with the log-rank test indicated that among 241 patients with LUSC, the expression levels of the SPARC gene had a significant influence on OS (P=0.039; Fig. 4H), with a higher expression level of SPARC mRNA indicating a shorter survival time of patients.
Figure 4.
Analysis in the GEPIA database. (A) Association between mRNA expression of SPARC and lung tissue and (B) comparison of SPARC gene expression in LUAD and normal lung tissue. (C) Association between stage grading of LUAD and SPARC expression. (D) Trend relationship between SPARC expression and OS in patients with LUAD; effect of SPARC expression level on LUAD patient survival. (E) Association between mRNA expression of SPARC gene and lung tissue and (F) comparison of SPARC gene expression in LUSC tissue and normal lung tissue. Values in the box plots are expressed as the median (interquartile range). (G) Association between stage grading of LUSC and SPARC expression. (H) Trend relationship between SPARC expression and OS in patients with LUSC; effect of SPARC expression level on LUSC patient survival. SPARC, secreted protein acidic and cysteine rich; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; TPM, transcripts per million; T, tumor tissue; N, normal tissue.
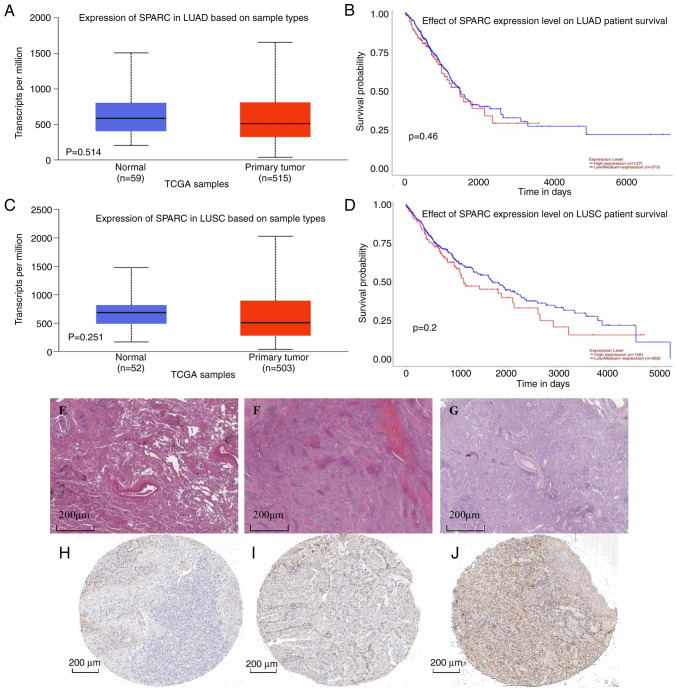
In the UALCAN database, the TCGA dataset was analyzed, comprising 515 LUAD tissues and 59 normal lung tissues. SPARC mRNA expression levels were upregulated in LUAD compared with normal lung tissue (P>0.05; Fig. 5A). The level of SPARC gene expression was not significantly associated with the prognosis of patients with LUAD (Fig. 5B). Furthermore, 503 LUSC tissues and 52 lung tissues included in the TCGA dataset were analyzed. In LUSC, SPARC mRNA expression levels were higher than those in normal lung tissue, but the differential expression was not significant (P>0.05; Fig. 5C). The SPARC gene expression level was not significantly associated with the prognosis of patients with LUSC (Fig. 5D), but the trends still indicated that patients with LUSC had a better prognosis when the SPARC mRNA expression level was lower. In the HPA database, the pathological morphology was compared among normal lung tissue, LUAD and LUSC (Fig. 5E-G). Screening analysis suggested that the expression level of SPARC protein in normal lung tissue was lower than that in LUAD. In cancer tissue, positivity of immunohistochemical staining was observed in the membrane and cytoplasm (Fig. 5H and I); SPARC protein was highly expressed in LUSC tissue and low in normal lung tissue, and positivity of immunohistochemical staining was present in the cytoplasm (Fig. 5J). This trend of expression was closely related to the mRNA expression level and the analytic results were consistent with this regard.
Figure 5.
(A-D) Analysis in the UALCAN database. (A) Expression of SPARC gene in different samples of LUAD. (B) Relationship between SPARC mRNA expression level and survival level of patients with LUAD. (C) Expression of SPARC gene in different LUSC specimens. Values in the box plots are expressed as the median (interquartile range). (D) Relationship between SPARC mRNA expression level and survival of patients with LUSC. (E-J) Analysis in the Human Protein Atlas database. H&E staining of (E) normal lung tissue, (F) LUAD and (G) LUSC. Immunohistochemical staining for SPARC in (H) normal lung tissue, (I) LUAD and (J) LUSC (scale bars, 200 µm). SPARC was positive in LUAD and LUSC. SPARC, secreted protein acidic and cysteine rich; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
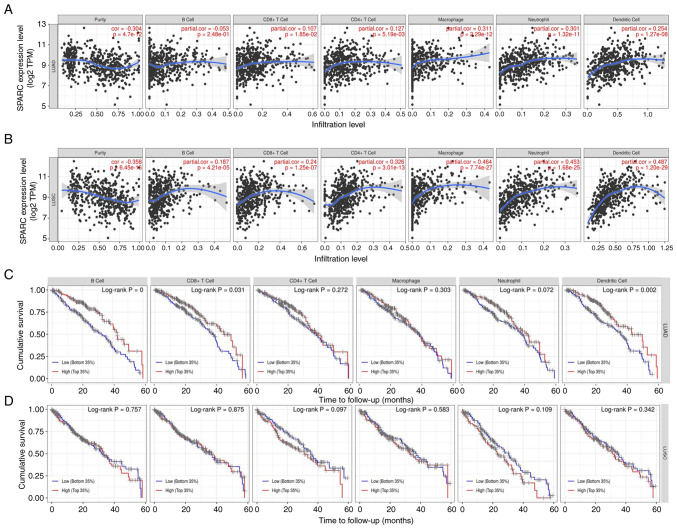
In the Timer database, the infiltration of immune cells closely related to lung cancer tissue was as follows: B cells, NK, CD4+T, CD8+T, monocytes and granulocytes all had different degrees of infiltration in NSCLC and normal lung tissue (Fig. 6A and B). The difference in the degree of infiltration of malignant tumor tissue was statistically significant (P<0.05). A correlation analysis of the proportion of liver immune cells in NSCLC patients according to SPARC levels was then performed. In LUAD, an inverse relationship between the proportion of B cells among immune cells and SPARC gene expression was observed. In LUSC, the eight most important immune cell types were positively correlated with the level of SPARC gene expression. Among immune cells, the infiltration of various cells was strongly correlated with the prognosis of patients with NSCLC. A higher degree of immune-cell infiltration was linked to a longer survival time of patients with LUAD and LUSC, with a positive association (Fig. 6C and D), among which B cells and dendritic cells exhibited the greatest association (P<0.05). It was observed that the expression of SPARC was closely related to the immune-cell infiltration mechanism in NSCLC.
Figure 6.
(A) Correlation of SPARC expression levels with immune cells (CD4+ T cells, macrophages, B cells, CD8+ T cells, neutrophils and dendritic cells) in LUAD. (B) Correlation of SPARC expression levels with immune cells (CD4+ T cells, macrophages, B cells, CD8+ T cells, neutrophils and dendritic cells) in LUSC. The infiltration level is displayed on the X-axis and the expression of SPARC on the Y-axis. (C) Relationship between the degree of immune-cell infiltration and survival prognosis of patients with LUAD. (D) Relationship between the degree of immune-cell infiltration and survival prognosis of patients with LUSC. The time to follow-up (months) is presented on the X-axis and the cumulative survival on the Y-axis. SPARC, secreted protein acidic and cysteine rich; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; TPM, transcripts per million; cor, correlation coefficient.
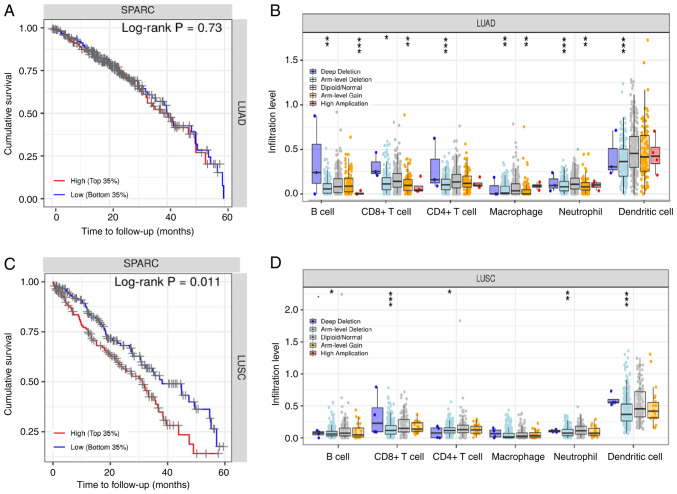
To further estimate the influence on SPARC on survival, patients with lung cancer were automatically divided into two groups based on SPARC protein expression levels: High expression and low expression. The SPARC gene had a non-significant effect on the survival rate of patients with LUAD (Fig. 7A). For LUAD copy number changes, deep deletions of immune cells (B cells, CD8+ T cells, neutrophils and dendritic cells) were greater than arm level deletions, diploid/normal, arm level gain and high amplification (Fig. 7B; Table SI). In LUSC, the SPARC expression level had a significant influence on survival and prognosis (P<0.05; Fig. 7C). In patients with LUSC, a higher the expression of SPARC protein was associated with a shorter survival time and unfavorable prognosis (P<0.05). In terms of LUSC copy number changes, deep deletions of immune cells (B cells, CD8+ T cells, neutrophils, and dendritic cells) were greater than arm level deletions, diploid/normal, arm level gain and high amplification, and the copy number trends of LUAD and LUSC were consistent (Fig. 7D; Table SI). Cox univariate analysis of TCGA data indicated that TNM stage, lymph node metastasis, distant metastasis and depth of invasion were negatively associated with patient prognosis (P<0.05; Table II). Cox multivariate analysis suggested that SPARC expression levels and TNM stage were risk factors affecting the prognosis of patients with NSCLC (P<0.05; Table III).
Figure 7.
(A) Association between SPARC gene expression and survival prognosis in patients with LUAD. (B) The relationship between copy number variation in LUAD and immune cell infiltration. (C) Association between SPARC gene expression and survival prognosis in patients with LUSC. (D) The relationship between copy number variation in LUSC and immune-cell infiltration. *P<0.05; **P<0.01; ***P<0.001 vs. diploid/normal immune cells. SPARC, secreted protein acidic and cysteine rich; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Table II.
Univariate analysis of prognostic risk factors in patients with lung cancer from TCGA database.
| Characteristics | Patients, n (%) | Hazard ratio (95% CI) | P-value |
|---|---|---|---|
| Sex | 1.090 (0.879-1.352) | 0.433 | |
| Male | 593 (59.8) | ||
| Female | 398 (40.2) | ||
| Age, years | 1.203 (0.940-1.541) | 0.142 | |
| <60 | 268 (27.0) | ||
| ≥60 | 724 (73.0) | ||
| TNM staging | 2.058 (1.639-2.584) | <0.001 | |
| I–II | 782 (79.9) | ||
| III–IV | 197 (20.1) | ||
| Depth of invasion | 1.486 (1.153-1.912) | 0.002 | |
| - | 285 (28.7) | ||
| + | 708 (71.3) | ||
| Lymph node metastasis | 1.695 (1.374-2.092) | <0.001 | |
| - | 636 (65.2) | ||
| + | 339 (34.8) | ||
| Distant metastasis | 2.001 (1.244-3.247) | 0.004 | |
| - | 375 (92.4) | ||
| + | 31 (7.6) |
CI, confidence interval; TNM, tumor-nodes-metastasis.
Table III.
Multivariate analysis of clinicopathological variables influencing the survival of patients with lung cancer in TCGA dataset.
| Clinicopathological parameter | Hazard ratio (95% CI) | P-value |
|---|---|---|
| SPARC expression | ||
| (+/-) | 0.697 (0.547-0.887) | 0.003 |
| Sex | ||
| (female/male) | 0.966 (0.954-1.242) | 0.786 |
| Age | ||
| (≥60 vs. <60 years) | 1.234 (0.926-1.645) | 0.152 |
| TNM stage | ||
| (III–IV/I–II) | 1.684 (1.230-2.306) | 0.001 |
| Depth of invasion | ||
| (+/-) | 1.316 (0.966-1.794) | 0.082 |
| Lymph node metastasis | ||
| (+/-) | 1.282 (0.978-1.681) | 0.072 |
| Distant metastasis | ||
| (+/-) | 1.273 (0.781-2.254) | 0.408 |
CI, confidence interval; TNM, tumor-nodes-metastasis; SPARC, secreted protein acidic and cysteine rich.
Discussion
The incidence of NSCLC is increasing year by year worldwide and most patients are at the advanced stage when they are diagnosed. At present, the clinical early diagnosis of lung cancer includes lung enhanced CT, position emission computed tomography and bronchoscopy. The improvement of diagnostic methods has increased the early diagnosis rate of lung cancer (32). The current treatment methods for lung cancer mainly include surgery, chemotherapy, targeted therapy, interventional therapy, immune cell infusion therapy, Traditional Chinese Medicine therapy and adjustment of living habits (33). At present, the five-year survival rate of lung cancer is ~10% and the invasion and metastasis of lung cancer are the main causes of death of patients. Finding new lung cancer targets may help improve the rate of early diagnosis, find new directions for treatment and improve the survival rate of patients. Among the multiple pathways involved in epithelial-mesenchymal transition are the Wnt/β-catenin signaling pathway and the PI3K/AKT signaling pathway (2,34).
SPARC is a highly conserved extracellular mesenchymal protein. Its main role is to prevent cell adhesion, regulate cell differentiation, prevent cell spreading, inhibit cell response to specific growth factors and regulate the production of extracellular matrix and MMPs; SPARC may also directly bind to VEGF, inhibit the VEGF pathway and prevent the binding of VEGF and its receptors to each other (35). At the same time, SPARC is able to bind to the platelet-derived growth factor and indirectly impair angiogenesis by downregulating MMPs and TGF-β1, thereby inhibiting tumors. invasion and metastasis (36). SPARC has an inhibitory role in various tumor types, including colorectal, pancreatic, prostate and ovarian cancer, while it has a promoting role in others such as liver cancer. In liver cancer and bladder cancer, highly expressed SPARC is able to inhibit tumor cell apoptosis through the PI3K/AKT signaling pathway, while promoting tumor cell proliferation, invasion and metastasis (37). SPARC overexpression is associated with poor prognosis in urothelial carcinoma. Alterations in the TGF-β signaling pathway may contribute to the dysregulation of SPARC, which in turn contributes to poor prognosis of adenocarcinoma. As a highly conserved and multi-domain proteoglycan, SPARC has an acidic N-terminal domain, including a chemosensitive N-terminal region, a follistatin homology region and a C-terminal Ca2+ binding region. SPARC is able to pass through the folic acid-containing structure. Domain and EC domain proteins have an effect on the activity of membrane-type MMP (38), and serum MMP-9 levels in patients with lung cancer with lymph node metastasis were significantly higher than in those without lymph node metastasis. SPARC may have a role in the treatment of NSCLC, which is mostly treated with chemotherapy regimens such as paclitaxel, and studies have indicated that high SPARC gene expression has an important impact on the drugs used to treat NSCLC (39–41). The experimental results of the present study also suggested that the protein expression level of SPARC was higher in NSCLC than in normal lung tissues and that patients in the high SPARC expression group had unfavorable prognosis, which is consistent with the trends indicated by previous studies (24,42). At present, the commonly used immunohistochemical indicators for the diagnosis and differential diagnosis of lung cancer mainly include NapsinA, TTF-1, P40 and CK5/6. However, since the above indicators cannot achieve a high level of specificity and sensitivity at the same time, they are generally used in combination for detection (43,44), and it is necessary to find more suitable detection indicators. In primary pancreatic cancer, aberrant methylation of the SPARC gene promoter region leads to silencing of gene expression. SPARC mRNA was indicated to be expressed in non-neoplastic pancreatic duct epithelial cells, but not found in pancreatic cancer cell lines, which indirectly suggests that silencing of the SPARC gene may be one of the processes leading to the occurrence of pancreatic cancer (15,45), which may be used for initial screening of early pancreatic cancer. SPARC increases the phosphorylation level of AKT in gliomas through the PI3K/AKT pathway and significantly inhibits the activity of EGF in ovarian cancer (46,47). In gastric cancer, SPARC expression was negatively correlated with the degree of differentiation, lymph node metastasis and Lauren classification. Cox analysis indicated that lymph node metastasis was an independent prognostic factor in patients with gastric cancer (48,49). Furthermore, the SPARC gene was observed to be overexpressed in oral squamous carcinoma and hepatocellular carcinoma and to have a significant effect on the prognosis of patients with hepatocellular carcinoma (50,51). Various studies reported that the mRNA expression level of SPARC in cancer tissue was significantly higher than that in normal adjacent tissue. SPARC was indicated to be highly expressed in NSCLC tissues (52), and its expression is related to the gender and TNM stage of patients. The Cox analysis performed in the present study suggested that TNM stage and lymph node metastasis were risk factors affecting the prognosis of patients. Furthermore, PCR results indicated that SPARC mRNA was highly expressed in NSCLC tissues. SPARC expression was increased in mouse models with an epithelial-mesenchymal transition phenotype and SPARC expression had an effect on lung function in both human bronchial epithelial cells and mouse models (53,54). The SPARC fragment binding with the present ligand protein inhibited tumor cell adhesion, while promoting spreading and stimulating tumor cell migration and invasion, suggesting an important effect of SPARC expression on lung cancer development and progression (55–57). Univariate analysis suggested that TNM stage, lymph node metastasis, distant metastasis and smoking history were associated with poor prognosis in patients with NSCLC. Cox proportional hazards regression model analysis indicated that SPARC expression and TNM stage were important factors affecting the survival time of patients with NSCLC. The expression of SPARC at the protein and mRNA levels indicated the same trend in NSCLC, and the expression levels of the two had a negative effect on survival and prognosis of patients. A linear relationship (positive or negative correlation) is expected for the following reasons: First, gene expression is regulated at different levels. Genes may have carcinogenic or tumor-inhibitory effects, while the regulation of the transcription level is only an intermediate link (58). Furthermore, post-transcriptional, translational and post-translational regulation all contribute to the expression of the final protein. Finally, mRNA degradation, protein degradation and modified folding may lead to inconsistent mRNA and protein expression levels for a given protein. In addition, the population selected in the dataset for clinical pathological data statistics and survival prognosis analysis may exhibit regional or genetic differences and certain differences in human physique and living environment are likely to be present. Increases in the serum levels of SPARC may occur as part of biological functions and processes such as tumors and obesity; of note, SPARC expression levels were upregulated in the lung during hypoxia induction in a mouse model, and the hypoxia-inducible factor 2A signaling pathway induces SPARC expression, which in turn has an effect on proliferation and apoptosis of tumor cells in humans (59–61). It has also been indicated that the SPARC signaling pathway induces albumin-bound paclitaxel, which enhances initial chemoresistance of patients with lung cancer and this may also be used to optimize individual gene therapy regimens, which are important for mechanistic studies, animal models and clinical treatment evaluation in NSCLC (62). At the same time, it was confirmed that the expression level of SPARC and the immune cells related to the occurrence and development of lung cancer interact with each other and the infiltration degree of immune cells is closely related to the prognosis of patients with NSCLC.
In summary, the available evidence suggested that SPARC protein expression is significantly associated with NSCLC and its different clinicopathological features, and is also positively correlated with SPARC mRNA expression. The expression levels of SPARC mRNA may be used to predict the corresponding protein levels, suggesting that SPARC may have an important role in the development of NSCLC and is closely associated with the molecular mechanisms of immune infiltration. Thus, the present systematic evaluation enhances the current knowledge on the pathogenesis of NSCLC and may provide a new target for screening and gene therapy of NSCLC. However, due to the limited number and quality of included studies, the above findings require to be validated by further high-quality, large-sample, rigorously designed studies. The present meta-analysis has several limitations. First, the potential publication bias comes from the fact that the published results were predominantly positive. Furthermore, the patients analyzed in the study were from Asia only. Due to the different level of medical development in different regions, the experimental methods to detect SPARC expression may also be different, which may affect the results. In addition, survival data were extracted from the survival curves, which may have affected the results. Finally, the small sample size may have affected the statistical weight of certain articles. SPARC protein expression was indicated to be upregulated during the development of NSCLC. Overall, SPARC had an inhibitory effect on lung cancer development and progression, while being associated with favorable prognosis. Therefore, SPARC protein expression may be used as a potential marker to predict the prognosis of patients with NSCLC and, combined with chemotherapeutic agents such as paclitaxel, it may be a new clinical treatment option, which is expected to provide a new therapeutic strategy.
In conclusion, SPARC was indicated to have a complex role in the development of tumors. Furthermore, SPARC expression levels were upregulated in patients with NSCLC. mRNA and protein expression of the SPARC gene were positively associated with immune-cell infiltration. In NSCLC, differences in SPARC expression levels and TNM stage may be used as independent prognostic factors. SPARC expression may be used as a marker to estimate the prognosis of tumor patients and is closely related to the molecular mechanisms of immune cells, which provides a new approach and idea for gene immunotherapy.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
All data used in this paper are from published articles and public data platforms, including KM plotter (kmplot.com), oncomine database (www.oncomine.org) and the TCGA database (www.cancer.gov).
Authors' contributions
GYM and ZGZ performed the meta-analysis and wrote the manuscript. SS and YRZ obtained all the data in this meta-analysis and made critical changes to the content of the article. GYM and ZGZ confirmed the authenticity of all the raw data. ZBG and WWB analyzed the data from KM-plotter and TCGA. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zhao J, Cheng M, Gai J, Zhang R, Du T, Li Q. SPOCK2 serves as a potential prognostic marker and correlates with immune infiltration in lung adenocarcinoma. Front Genet. 2020;11:588499. doi: 10.3389/fgene.2020.588499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun W, Feng J, Yi Q, Xu X, Chen Y, Tang L. SPARC acts as a mediator of TGF-β1 in promoting epithelial-to-mesenchymal transition in A549 and H1299 lung cancer cells. Biofactors. 2018;44:453–464. doi: 10.1002/biof.1442. [DOI] [PubMed] [Google Scholar]
- 3.Zhang S, Jin J, Tian X, Wu L. hsa-miR-29c-3p regulates biological function of colorectal cancer by targeting SPARC. Oncotarget. 2017;8:104508–104524. doi: 10.18632/oncotarget.22356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H, Gu XH, Zhou Y, Ge Z, Wang B, Siok WT, Wang G, Huen M, Jiang Y, Tan LH, Sun Y. A genome-wide association study identifies genetic variants associated with mathematics ability. Sci Rep. 2017;7:40365. doi: 10.1038/srep40365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang F, Zhang Y, Da J, Jia Z, Wu H, Gu K. Downregulation of SPARC expression decreases cell migration and invasion involving epithelial-mesenchymal transition through the p-FAK/p-ERK pathway in esophageal squamous cell carcinoma. J Cancer. 2020;11:414–420. doi: 10.7150/jca.31427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang S, Xiang X, Liu L, Yang H, Cen D, Tang G. Bioinformatics analysis of hub genes and potential therapeutic agents associated with gastric cancer. Cancer Manag Res. 2021;13:8929–8951. doi: 10.2147/CMAR.S341485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aghamaliyev U, Gaitantzi H, Thomas M, Simon-Keller K, Gaiser T, Marx A, Yagublu V, Araos J, Cai C, Valous NA, et al. Downregulation of SPARC is associated with epithelial-mesenchymal transition and low differentiation state of biliary tract cancer cells. Eur Surg Res. 2019;60:1–12. doi: 10.1159/000494734. [DOI] [PubMed] [Google Scholar]
- 8.Naczki C, John B, Patel C, Lafferty A, Ghoneum A, Afify H, White M, Davis A, Jin G, Kridel S, Said N. SPARC Inhibits metabolic plasticity in ovarian cancer. Cancers (Basel) 2018;10:385. doi: 10.3390/cancers10100385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Wang Y, Zheng G, Liu Y, Li J, Huang H, Xu C, Zeng Y, Zhang X, Qin J, et al. Follistatin-like 1 (FSTL1) interacts with Wnt ligands and Frizzled receptors to enhance Wnt/β-catenin signaling in obstructed kidneys in vivo. J Biol Chem. 2022;298:102010. doi: 10.1016/j.jbc.2022.102010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Zhou SJ, Xu Y, Liao QM, Zou YS, Pei H. CCAR2 promotes a malignant phenotype of osteosarcoma through Wnt/β-catenin-dependent transcriptional activation of SPARC. Biochem Biophys Res Commun. 2021;580:67–73. doi: 10.1016/j.bbrc.2021.09.070. [DOI] [PubMed] [Google Scholar]
- 11.Ye Z, Chen J, Hu X, Yang S, Xuan Z, Lu X, Zhao Q. SPOCK1: A multi-domain proteoglycan at the crossroads of extracellular matrix remodeling and cancer development. Am J Cancer Res. 2020;10:3127–3137. [PMC free article] [PubMed] [Google Scholar]
- 12.Bhat AA, Wani HA, Ishaq S, Waza AA, Malik RA, Shabir I, Jeelani S, Kadla S, Qureshie W, Masood A, Majid S. Promoter hypermethylation and its impact on expression of MGMT gene in the GIT malignant patients of Kashmiri origin. Cancer Invest. 2017;35:116–121. doi: 10.1080/07357907.2016.1271887. [DOI] [PubMed] [Google Scholar]
- 13.Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, Shamseer L, Tetzlaff JM, Akl EA, Brennan SE, et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. PLoS Med. 2021;18:e1003583. doi: 10.1371/journal.pmed.1003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 15.Fabrizio FP, Sparaneo A, Fontana A, Mazza T, Graziano P, Pantalone A, Parente P, Centra F, Orlando N, Trombetta D, et al. Potential prognostic role of SPARC methylation in non-small-cell lung cancer. Cells. 2020;9:1523. doi: 10.3390/cells9061523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao M, Hu Q, Li C. Effects of overexpression of cysteine-rich acidic secreted protein on the sensitivity of albumin binding paclitaxel in non-small cell lung cancer cells. Chin Med. 2020;15:5. [Google Scholar]
- 17.Andriani F, Landoni E, Mensah M, Facchinetti F, Miceli R, Tagliabue E, Giussani M, Callari M, De Cecco L, Colombo MP, et al. Diagnostic role of circulating extracellular matrix-related proteins in non-small cell lung cancer. BMC Cancer. 2018;18:899. doi: 10.1186/s12885-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Kabolizadeh P, Yan D, Qin A, Zhou J, Hong Y, Guerrero T, Grills I, Stevens C, Ding X. Improve dosimetric outcome in stage III non-small-cell lung cancer treatment using spot-scanning proton arc (SPArc) therapy. Radiat Oncol. 2018;13:35. doi: 10.1186/s13014-018-0981-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan J, Hao Y, Wan R, Yu S, Bai H, An T, Zhao J, Wang Z, Zhuo M, Wang J. Efficacy and safety of weekly intravenous nanoparticle albumin-bound paclitaxel for non-small cell lung cancer patients who have failed at least two prior systemic treatments. Thorac Cancer. 2017;8:138–146. doi: 10.1111/1759-7714.12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komiya K, Nakamura T, Nakashima C, Takahashi K, Umeguchi H, Watanabe N, Sato A, Takeda Y, Kimura S, Sueoka-Aragane N. SPARC is a possible predictive marker for albumin-bound paclitaxel in non-small-cell lung cancer. Onco Targets Ther. 2016;9:6663–6668. doi: 10.2147/OTT.S114492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng PF, Han FC, Guo QX, Wang J, Shan BB, Zhang F. Progress of SPARC and albumin-bound paclitaxel in non-small cell lung cancer. Chin J Cliicians (Electronic Edition) 2016;10:2136–2140. (In Chinese) [Google Scholar]
- 22.Kurtul N, Eroglu C, Unal D, Tasdemir EA, Orhan O, Zararsiz G, Baran M, Kaplan B, Kontas O. Prognostic value of SPARC expression in unresectable NSCLC treated with concurrent chemoradiotherapy. Asian Pac J Cancer Prev. 2014;15:8911–8916. doi: 10.7314/APJCP.2014.15.20.8911. [DOI] [PubMed] [Google Scholar]
- 23.Grant JL, Fishbein MC, Hong LS, Krysan K, Minna JD, Shay JW, Walser TC, Dubinett SM. A novel molecular pathway for Snail-dependent, SPARC-mediated invasion in non-small cell lung cancer pathogenesis. Cancer Prev Res (Phila) 2014;7:150–160. doi: 10.1158/1940-6207.CAPR-13-0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin H, Wang Y, Chen W, Zhong S, Liu Z, Zhao J. Drug-resistant CXCR4-positive cells have the molecular characteristics of EMT in NSCLC. Gene. 2016;594:23–29. doi: 10.1016/j.gene.2016.08.043. [DOI] [PubMed] [Google Scholar]
- 25.Zhang ZP, Wang Z, Liu XY, Shi M, Chen G, Zhang B, Li Z, Song L. Correlation of KLF4 and SPARC expression with the clinical characteristics of non-small cell lung cancer. Zhongguo Fei Ai Za Zhi. 2012;15:720–724. doi: 10.3779/j.issn.1009-3419.2012.12.05. (In Chinese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Zhang J, Zhao YY, Jiang W, Xue C, Xu F, Zhao HY, Zhang Y, Zhao LP, Hu ZH, et al. SPARC expression and prognostic value in non-small cell lung cancer. Chin J Cancer. 2012;31:541–548. doi: 10.5732/cjc.012.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma YG, Lou Y, Zhao HR. Study on expression and promoter methylation of Apaf-1 gene in non-small cell lung cancer. Chin J Cancer Prev Treat. 2006;13:116–119. (In Chinese) [Google Scholar]
- 28.Koukourakis MI, Giatromanolaki A, Brekken RA, Sivridis E, Gatter KC, Harris AL, Sage EH. Enhanced expression of SPARC/osteonectin in the tumor-associated stroma of non-small cell lung cancer is correlated with markers of hypoxia/acidity and with poor prognosis of patients. Cancer Res. 2003;63:5376–5380. [PubMed] [Google Scholar]
- 29.Zheng Y, Tang J, Zhang Z, Zhang Z, Zhang X, Feng P. Expression and significance of SPARC and TGFβ1 in lung cancer. Cancer Prev Treat Res. 2014;41:593–597. [Google Scholar]
- 30.Zheng YJ, Zhang ZH, Zhang XL, Tang JH, Zhang ZL, Feng P. Expression and significance of SPARC and CD105 in lung cancer tissues. Shandong Medicine. 2015;55:64–65. (In Chinese) [Google Scholar]
- 31.Xu J, Yang S, Gu X, Shen H, Wang L, Xu W, Fang L, Mao Y, Xu L, Chen Y, et al. SPARC correlates with unfavorable outcome and promotes tumor growth in lung squamous cell carcinoma. Exp Mol Pathol. 2019;110:104276. doi: 10.1016/j.yexmp.2019.104276. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 32.Yan J, Zhang J, Zhang X, Li X, Li L, Li Z, Chen R, Zhang L, Wu J, Wang X, et al. SPARC is down-regulated by DNA methylation and functions as a tumor suppressor in T-cell lymphoma. Exp Cell Res. 2018;364:125–132. doi: 10.1016/j.yexcr.2017.12.022. [DOI] [PubMed] [Google Scholar]
- 33.Wong SL, Sukkar MB. The SPARC protein: An overview of its role in lung cancer and pulmonary fibrosis and its potential role in chronic airways disease. Br J Pharmacol. 2017;174:3–14. doi: 10.1111/bph.13653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.John B, Naczki C, Patel C, Ghoneum A, Qasem S, Salih Z, Said N. Regulation of the bi-directional cross-talk between ovarian cancer cells and adipocytes by SPARC. Oncogene. 2019;38:4366–4383. doi: 10.1038/s41388-019-0728-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma J, Gao S, Xie X, Sun E, Zhang M, Zhou Q, Lu C. SPARC inhibits breast cancer bone metastasis and may be a clinical therapeutic target. Oncol Lett. 2017;14:5876–5882. doi: 10.3892/ol.2017.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu XZ, Guo ZY, Di Y, Yang F, Ouyang Q, Fu DL, Jin C. The relationship between SPARC expression in primary tumor and metastatic lymph node of resected pancreatic cancer patients and patients' survival. Hepatobiliary Pancreat Dis Int. 2017;16:104–109. doi: 10.1016/S1499-3872(16)60168-6. [DOI] [PubMed] [Google Scholar]
- 37.Murakawa M, Aoyama T, Miyagi Y, Kobayashi S, Ueno M, Morimoto M, Numata M, Yamamoto N, Tamagawa H, Yukawa N, et al. The impact of SPARC expression on the survival of pancreatic ductal adenocarcinoma patients after curative resection. J Cancer. 2019;10:627–633. doi: 10.7150/jca.28660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hung JY, Yen MC, Jian SF, Wu CY, Chang WA, Liu KT, Hsu YL, Chong IW, Kuo PL. Secreted protein acidic and rich in cysteine (SPARC) induces cell migration and epithelial mesenchymal transition through WNK1/snail in non-small cell lung cancer. Oncotarget. 2017;8:63691–63702. doi: 10.18632/oncotarget.19475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu G, Zhao L, Qin A, Grills I, Deraniyagala R, Stevens C, Zhang S, Yan D, Li X, Ding X. Lung stereotactic body radiotherapy (SBRT) using spot-scanning proton Arc (SPArc) therapy: A feasibility study. Front Oncol. 2021;11:664455. doi: 10.3389/fonc.2021.664455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li C, Hou X, Yuan S, Zhang Y, Yuan W, Liu X, Li J, Wang Y, Guan Q, Zhou Y. High expression of TREM2 promotes EMT via the PI3K/AKT pathway in gastric cancer: Bioinformatics analysis and experimental verification. J Cancer. 2021;12:3277–3290. doi: 10.7150/jca.55077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao L, Zhou Y, Li J, Wickens L, Conforti F, Rattu A, Ibrahim FM, Alzetani A, Marshall BG, Fletcher SV, et al. Bidirectional epithelial-mesenchymal crosstalk provides self-sustaining profibrotic signals in pulmonary fibrosis. J Biol Chem. 2021;297:101096. doi: 10.1016/j.jbc.2021.101096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertino EM, Williams TM, Nana-Sinkam SP, Shilo K, Chatterjee M, Mo X, Rahmani M, Phillips GS, Villalona-Calero MA, Otterson GA. Stromal caveolin-1 is associated with response and survival in a phase II trial of nab-paclitaxel with carboplatin for advanced NSCLC patients. Clin Lung Cancer. 2015;16:466–474. doi: 10.1016/j.cllc.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toyota K, Murakami Y, Kondo N, Uemura K, Nakagawa N, Takahashi S, Sueda T. Impact of secreted protein acidic and rich in cysteine (SPARC) expression on prognosis after surgical resection for biliary carcinoma. J Gastrointest Surg. 2017;21:990–999. doi: 10.1007/s11605-017-3407-0. [DOI] [PubMed] [Google Scholar]
- 44.Ma Y, Zhu J, Chen S, Ma J, Zhang X, Huang S, Hu J, Yue T, Zhang J, Wang P, et al. Low expression of SPARC in gastric cancer-associated fibroblasts leads to stemness transformation and 5-fluorouracil resistance in gastric cancer. Cancer Cell Int. 2019;19:137. doi: 10.1186/s12935-019-0844-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siddiqui MA, Gollavilli PN, Ramesh V, Parma B, Schwab A, Vazakidou ME, Natesan R, Saatci O, Rapa I, Bironzo P, et al. Thymidylate synthase drives the phenotypes of epithelial-to-mesenchymal transition in non-small cell lung cancer. Br J Cancer. 2021;124:281–289. doi: 10.1038/s41416-020-01095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang B, Zhang Z, Tang J, Tao H, Zhang Z. Correlation between SPARC, TGFβ1, Endoglin and angiogenesis mechanism in lung cancer. J Biol Regul Homeost Agents. 2018;32:1525–1531. [PubMed] [Google Scholar]
- 47.Wan S, Meyer AS, Weiler SME, Rupp C, Tóth M, Sticht C, Singer S, Thomann S, Roessler S, Schorpp-Kistner M, et al. Cytoplasmic localization of the cell polarity factor scribble supports liver tumor formation and tumor cell invasiveness. Hepatology. 2018;67:1842–1856. doi: 10.1002/hep.29669. [DOI] [PubMed] [Google Scholar]
- 48.Dimas DT, Perlepe CD, Sergentanis TN, Misitzis I, Kontzoglou K, Patsouris E, Kouraklis G, Psaltopoulou T, Nonni A. The prognostic significance of Hsp70/Hsp90 expression in breast cancer: A systematic review and meta-analysis. Anticancer Res. 2018;38:1551–1562. doi: 10.21873/anticanres.12384. [DOI] [PubMed] [Google Scholar]
- 49.Alcaraz LB, Mallavialle A, David T, Derocq D, Delolme F, Dieryckx C, Boissière-Michot F, Simony-Lafontaine J, du Manoir S, Huesgen PF, et al. Cathepsin D exacerbates SPARC-driven aggressiveness by limited proteolysis in triple-negative breast cancer. bioRxiv. 2020 [Google Scholar]
- 50.Poomsawat S, Kosanwat T, Meesakul O, Sanguansin S. Epithelial and fibroblast SPARC expression patterns in oral leukoplakia and oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol. 2022;134:e44–e50. doi: 10.1016/j.oooo.2021.10.019. [DOI] [PubMed] [Google Scholar]
- 51.Gao ZW, Liu C, Yang L, He T, Wu XN, Zhang HZ, Dong K. SPARC overexpression promotes liver cancer cell proliferation and tumor growth. Front Mol Biosci. 2021;8:775743. doi: 10.3389/fmolb.2021.775743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanità G, Armanetti P, Silvestri B, Carrese B, Calì G, Pota G, Pezzella A, d'Ischia M, Luciani G, Menichetti L, Lamberti A. Albumin-modified melanin-silica hybrid nanoparticles target breast cancer cells via a SPARC-dependent mechanism. Front Bioeng Biotechnol. 2020;8:765. doi: 10.3389/fbioe.2020.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kehlet SN, Manon-Jensen T, Sun S, Brix S, Leeming DJ, Karsdal MA, Willumsen N. A fragment of SPARC reflecting increased collagen affinity shows pathological relevance in lung cancer-implications of a new collagen chaperone function of SPARC. Cancer Biol Ther. 2018;19:904–912. doi: 10.1080/15384047.2018.1480887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xing P, Zhu Y, Shan L, Chen S, Hao X, Li J. The role of weekly nanoparticle albumin bound paclitaxel monotherapy as second line or later treatment for advanced NSCLC in China. Oncotarget. 2017;8:87442–87454. doi: 10.18632/oncotarget.21103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Croci GA, Au-Yeung RKH, Reinke S, Staiger AM, Koch K, Oschlies I, Richter J, Poeschel V, Held G, Loeffler M, et al. SPARC-positive macrophages are the superior prognostic factor in the microenvironment of diffuse large B-cell lymphoma and independent of MYC rearrangement and double-/triple-hit status. Ann Oncol. 2021;32:1400–1409. doi: 10.1016/j.annonc.2021.08.1991. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X, Xie J, Sun H, Wei Q, Nong G. miR-29a-3p regulates the epithelial-mesenchymal transition via the SPARC/ERK signaling pathway in human bronchial epithelial cells. Int J Mol Med. 2021;48:171. doi: 10.3892/ijmm.2021.5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alcaraz LB, Mallavialle A, David T, Derocq D, Delolme F, Dieryckx C, Mollevi C, Boissière-Michot F, Simony-Lafontaine J, Du Manoir S, et al. A 9-kDa matricellular SPARC fragment released by cathepsin D exhibits pro-tumor activity in the triple-negative breast cancer microenvironment. Theranostics. 2021;11:6173–6192. doi: 10.7150/thno.58254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paulsson J, Micke P. Prognostic relevance of cancer-associated fibroblasts in human cancer. Semin Cancer Biol. 2014;25:61–68. doi: 10.1016/j.semcancer.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 59.Ghanemi A, Yoshioka M, St-Amand J. Measuring exercise-induced secreted protein acidic and rich in cysteine expression as a molecular tool to optimize personalized medicine. Genes (Basel) 2021;12:1832. doi: 10.3390/genes12111832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghanemi A, Yoshioka M, St-Amand J. Secreted protein acidic and rich in cysteine as a molecular physiological and pathological biomarker. Biomolecules. 2021;11:1689. doi: 10.3390/biom11111689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Veith C, Vartürk-Özcan I, Wujak M, Hadzic S, Wu CY, Knoepp F, Kraut S, Petrovic A, Gredic M, Pak O, et al. SPARC, a novel regulator of vascular cell function in pulmonary hypertension. Circulation. 2022;145:916–933. doi: 10.1161/CIRCULATIONAHA.121.057001. [DOI] [PubMed] [Google Scholar]
- 62.Yuan S, Xu J, Zhou B, Zhou Y, Lang M, Cao J, Liu Z, Yang S, Gao S, Hao J. SOX8 affects tumoral SPARC expression by regulating EZH2 to attenuate effectiveness of albumin-bound paclitaxel in PDAC. Int J Biol Sci. 2022;18:911–922. doi: 10.7150/ijbs.64752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this paper are from published articles and public data platforms, including KM plotter (kmplot.com), oncomine database (www.oncomine.org) and the TCGA database (www.cancer.gov).