Abstract
Cold stress limits plant growth, geographical distribution, and crop yield. The MYC-type bHLH transcription factor ICE1 is recognized as the core positive regulator of the cold-stress response. However, how ICE1 protein levels are regulated remains to be further studied. In this study, we observed that a U-box-type E3 ubiquitin ligase, MdPUB23, positively regulated the cold-stress response in apple. The expression of MdPUB23 increased at both the transcriptional and post-translational levels in response to cold stress. Overexpression of MdPUB23 in transgenic apple enhanced sensitivity to cold stress. Further study showed that MdPUB23 directly interacted with MdICE1, promoting the ubiquitination-mediated degradation of the MdICE1 protein through the 26S-proteasome pathway and reducing the MdICE1-improved cold-stress tolerance in apple. Our results reveal that MdPUB23 regulates the cold-stress response by directly mediating the stability of the positive regulator MdICE1. The PUB23–ICE1 ubiquitination module may play a role in maintaining ICE1 protein homeostasis and preventing overreactions from causing damage to plants. The discovery of the ubiquitination regulatory pathway of ICE1 provides insights for the further exploration of plant cold-stress-response mechanisms.
Introduction
Plants are constantly subjected to various environmental stimuli during their growth and development, such as extreme temperature, drought, waterlogging, and high salinity [1–3]. To adapt to harsh environmental challenges, plants have evolved elaborate regulatory mechanisms [4–6]. Cold stress is one of the most important abiotic stresses affecting plant growth, geographical distribution, and crop yield. It affects plant metabolism by directly inhibiting the expression of genes related to metabolic enzymes [7–9]. Cold stress rapidly activates the expression of a series of transcription factors, among which C-repeat-binding factors (CBFs) are the most thoroughly studied. They directly induce the expression of downstream cold-responsive (COR) genes, regulating the response to cold stress [7–13]. Three CBF genes (AtCBF1–AtCBF3) were identified in Arabidopsis, and five (MdCBF1–MdCBF5), in apple [14–17]. In Arabidopsis, the CBF genes are regulated by many upstream transcription factors, including INDUCER OF CBF EXPRESSION1/2 (ICE1/2), CALMODULIN-BINDING TRANSCRIPTION ACTIVATOR1–5 (CAMTA1–5), MYB15, PHYTOCHROME-INTERACTING FACTOR3/4/7 (PIF3/4/7), PSEUDO RESPONSE REGULATORS (PRRs), CIRCADIAN CLOCK-ASSOCIATED1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), ETHYLENE INSENSITIVE3 (EIN3), CESTA, BRASSINAZOLERESISTANT1 (BZR1), and BRI1-EMS-SUPPRESSOR1 (BES1) [18–32]. ELONGATED HYPOCOTYL5 (MdHY5), MdMYB23, BASIC HELIX–LOOP-HELIX33 (MdbHLH33), and B-box37 (MdBBX37) have been identified as positive regulators of MdCBF genes in apple [33–37]. Among them, the ICE1–CBF–COR regulatory module plays a particularly essential role in the cold-stress response [11, 18, 38, 39].
ICE1 interacts with many cold-stress regulatory proteins to jointly regulate the cold-stress response. In Arabidopsis, ICE1 directly interacts with MYB15, a negative regulator of cold stress [19]. The jasmonic acid (JA)-signaling repressors JASMONATE ZIM-DOMAIN1 (JAZ1) and JAZ4 interact with ICE1, inhibiting the transcriptional activity of ICE1 [40]. HIGH OSMOTIC EXPRESSION1 (HOS1), MITOGEN-ACTIVATED PROTEIN KINASE3/6 (MPK3/6), and BRASSINOSTEROID-INSENSITIVE2 (BIN2) negatively regulate cold-stress tolerance through interaction with ICE1 and attenuate the protein stability of ICE1 [41–44]. By contrast, SAP and Miz1 (SIZ1) and OPEN STOMATA 1 (OST1) enhance the protein stability of ICE1 in the cold-stress response [28, 45]. Rice OsMAPK3 enhances the stability of OsICE1 by inhibiting OsICE1 degradation via OsHOS1 [46]. In banana fruit, JA-signaling regulators called MaMYC2s may mediate cold-stress tolerance by interaction with MaICE1 [47]. SEVEN IN ABSENTIA1 (MaSINA1) may negatively regulate the cold-stress response by reducing the protein stability of MaICE1 [48]. MdBBX37 and ABSCISIC ACID INSENSITIVE4 (MdABI4), as interaction partners of MdICE1, positively regulate the transcriptional activity of MdICE1 in apple [37, 49].
Post-translational modifications, such as ubiquitination, phosphorylation, methylation, and sumoylation, play key roles in the regulation of protein stability and biological activity [50–54]. Among them, ubiquitination has been well studied. The ubiquitination cascade requires the coordination of three components: E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes, and E3 ubiquitin ligases [55, 56]. In particular, E3 ubiquitin ligases play a decisive role in the specificity of target proteins [57, 58]. In Arabidopsis, more than 1400 genes encode E3 ubiquitin ligases [57, 59]. According to the characteristic domain of ubiquitin ligases and the mechanism of ubiquitin’s transfer to target proteins, E3 ubiquitin ligases are mainly divided into three categories: homologous to E6AP COOH terminus (HECT)-type E3 ubiquitin ligases, really interesting new gene (RING)-finger-type E3 ubiquitin ligases, and U-box-type E3 ubiquitin ligases [60–62]. Plant U-box-type E3 ubiquitin ligases (PUBs) play a broad role in the regulation of plant growth and development and stress responses [63–65]. In Arabidopsis, PUB2/4/12/13/22/23/24/25/26 are involved in plant immune regulation [66–70]. PUB11/22/23/46/48 mediate the drought-stress response [71–74]. PUB10/12/13/18/19/40 play key roles in plant responses to multiple hormonal signals [75–80]. A recent study showed that PUB25 and PUB26 positively regulated the cold-stress response by mediating MYB15 protein degradation [81]. In apples, MdPUB24 and MdPUB29 regulate fruit quality [82, 83]. In addition, MdPUB29 may also be involved in the regulation of the plant immune response to fungal pathogens [84]. However, the roles of PUB proteins in apple remain to be further studied.
In this study, we report that the U-box-type E3 ubiquitin ligase MdPUB23 is a negative regulator of the cold-stress response in apple. MdPUB23 interacts with MdICE1, a key regulator of cold stress, and negatively regulates the cold-stress response by promoting the protein stability of MdICE1. This study reveals a new post-translational regulatory mechanism that maintains the protein stability of ICE1 in the cold-stress response.
Results
MdPUB23 interacts with MdICE1
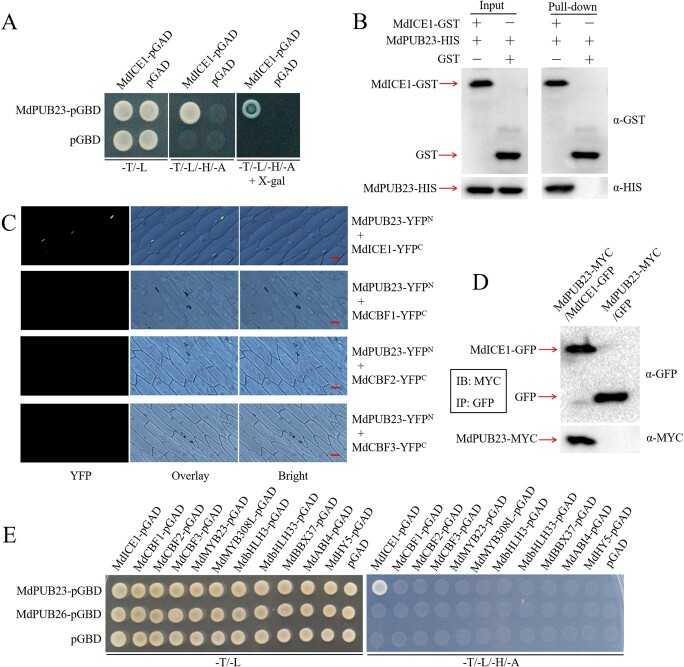
ICE1 is considered a key positive regulator of the cold-stress response [18, 38]. However, the post-translational regulation of the ICE1 protein has not been fully studied. To study the post-translational regulatory mechanism of MdICE1 in apple, the MdICE1-GFP protein was extracted from MdICE1-overexpressing apple calli (MdICE1-GFP) [37] and analysed by mass spectroscopy. The U-box E3 ubiquitin ligase MdPUB23 was isolated as a potential interaction partner of MdICE1. To test the interaction between MdPUB23 and MdICE1, we performed Y2H assays. The results showed that yeast cells transformed with MdPUB23 or MdICE1 alone could not grow on selective medium (−T/−L/−H/−A), and only the yeast cells transformed with MdPUB23 and MdICE1 could grow normally on selective medium (Fig. 1A), suggesting that MdPUB23 and MdICE1 interact with each other in yeast cells. Next, in vitro pull-down assays were performed to confirm the interaction. The purified MdPUB23-HIS fusion protein was pulled down using MdICE1-GST (Fig. 1B), indicating that MdPUB23 physically interacted with MdICE1 in vitro. We further verified the interaction between MdPUB23 and MdICE1 by BiFC assays. Fluorescence detection results showed that only onion epidermal cells co-expressed MdPUB23 and MdICE1 could produce a strong fluorescence signal (Fig. 1C). These data reveal that MdPUB23 interacts with MdICE1. Moreover, coimmunoprecipitation (Co-IP) assays showed that MdPUB23 interacted with MdICE1 in vivo (Fig. 1D). Further Y2H assays showed that MdPUB23 specifically interacted with MdICE1 in yeast cells (Fig. 1E).
Figure 1.
The interaction between MdPUB23 and MdICE1. (A) Y2H assays showing the interaction between MdPUB23 and MdICE1 proteins. The full-length MdICE1 and MdPUB23 were cloned into a pGAD424 or a pGBT9 vector, respectively. Yeasts grown in SD (-T/-L), SD (-T/-L/-H/-A), and SD (-T/-L/-H/-A + X-gal) media are indicated. (B) Pull down assays showing the interaction between MdPUB23 and MdICE1 in vitro. MdPUB23-HIS fusion protein was incubated with a cobalt chelate affinity resin containing the immobilized MdICE1-GST or GST protein. The protein mixtures were purified using a GST purification kit. (C) BiFC assays showing the interaction between MdPUB23 and MdICE1 proteins in nuclei of epidermal cells of onions. Bars = 10 μm. (D) Co-IP assays showing the interaction between MdPUB23 and MdICE1 proteins in vivo. (E) Y2H assays showing the interaction between MdPUB23 and a series of cold-stress-responsive proteins. Yeasts grown in SD (-T/-L), SD (-T/-L/-H/-A), and SD (-T/-L/-H/-A + X-gal) media are indicated. All experiments were repeated three times with similar results. A representative picture is shown here.
Identification of MdPUB23 encoding a U-box E3 ligase
Identification and amino acid sequence analysis revealed that the cDNA sequence of MdPUB23 comprised 1221 bp and encoded 406 amino acids with a predicted U-box motif in the N-terminal region (Fig. 2A). MdPUB23 showed high sequence similarity with PUB23 proteins in nine other species, especially the conserved U-box motif (Fig. 2B). Among them, MdPUB23 shared the lowest sequence identity with AtPUB23 (58.07%) and the highest sequence identity with PbPUB23 (97.04%). The phylogenetic tree analysis showed that MdPUB23 and PbPUB23 had the closest relationship (Fig. 2C), suggesting that there may be functional similarities between the two.
Figure 2.

Identification of MdPUB23. (A) The structure of the conserved regions of the MdPUB23 protein. The U-box domain (11–75 amino acids) is shown in the black box. (B) Multiple sequence alignment of MdPUB23 with PUB23 proteins from different species. PbPUB23, Pyrus x bretschneideri, XP_009348934.1; PpPUB23: Prunus persica, XP_007204425.1; FvPUB23: Fragaria vesca subsp. vesca, XP_004303406.1; RcPUB23: Rosa chinensis, XP_024190862.1; MnPUB23: Morus notabilis, XP_010106989.1; GaPUB23: Gossypium arboreum, XP_017610198.1; TcPUB23: Theobroma cacao, XP_007027990.1; VvPUB23: Vitis vinifera, XP_002267438.1; MdPUB23: Malus x domestica, MDP0000773851; AtPUB23: Arabidopsis thaliana, AT2G35930.1. The U-box domain is the red line. (C) Phylogenetic tree of above 10 PUB23 proteins (constructed with MEGA 5.0).
MdPUB23 reduces cold-stress tolerance
Because MdPUB23 acts as an interaction partner of MdICE1, we hypothesized that MdPUB23 might be involved in the cold-stress response. To confirm this hypothesis, we first detected the expression of MdPUB23 in response to cold stress. Previous transcriptome data showed that cold stress induced MdPUB23 expression (Fig. 3A) [34]. qRT-PCR results showed that the expression of MdPUB23 increased gradually after cold-stress treatment (4°C), and the expression was the highest after 6 h of cold-stress treatment (Fig. 3B). These results demonstrate that cold stress promotes the expression of MdPUB23.
Figure 3.

Cold-stress response of MdPUB23. (A) Transcriptome analysis showing the expression levels of 15 PUB genes after cold-stress treatment in apple. Apple seedlings were exposed to 4°C for 9 h, and seedlings grown at 24°C were used as controls. Total RNAs from plants treated with or without cold stress were extracted and used for transcriptome analyses (An et al., 2018a). (B) Gene expression analysis of MdPUB23 under cold-stress treatments. Thirty-day-old apple seedlings grown at 24°C were transferred to 4°C for 9 h and were collected to detect the expression of MdPUB23 using qRT-PCR. Control, apple seedlings without cold treatment; Cold treatment, apple seedlings with cold treatment. The expression level at 0 h was used as the reference and set to 1. Three biological replicates were carried out with three technical repeats. Error bars denote the standard deviations (SD).
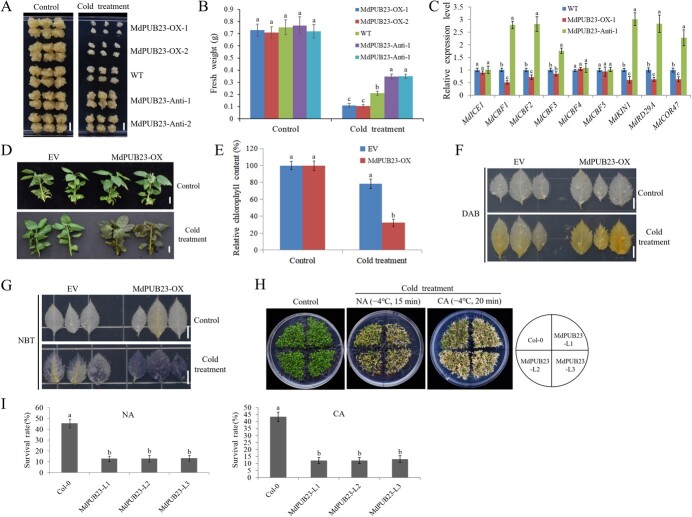
To identify the biological function of MdPUB23 in the cold-stress response, we generated stable MdPUB23 transgenic apple calli (MdPUB23-OX-1 and MdPUB23-OX-2, MdPUB23-overexpressing apple calli; MdPUB23-Anti-1 and MdPUB23-Anti-2, apple calli expressing the MdPUB23 antisense construct; Supplementary Fig. S1A). Although there was no significant difference in the growth status of wild-type (WT) and MdPUB23 transgenic apple calli at room temperature (24°C), after cold-stress treatment, the overexpression of MdPUB23 inhibited the growth of calli compared with the WT control, while the suppression of MdPUB23 expression contributed to the growth of calli (Fig. 4A–B). Gene expression analysis showed that the overexpression of MdPUB23 inhibited and the suppression of MdPUB23 increased the expression of MdCBF1 and MdCBF2 and its target genes (Fig. 4C). To evaluate its function in intact plant tissues, transient transgenic apple seedlings and leaves overexpressing MdPUB23 were generated and subjected to cold-stress treatment (Supplementary Fig. S1B, C).
Figure 4.
Cold-stress phenotypes of MdPUB23 transgenie apple calli, leaves, and Arabidopsis seedlings. (A) Cold-stress phenotypes of MdPUB23 transgenic apple calli. Apple calli were grown on medium at 24°C for 5 d and then treated at 4°C for another 10 d. The experiments were repeated three times with similar results, and each experiment contained 8–C10 calli subgroups per treatment. A representative picture is shown here. WT, wild-type control; MdPUB23-OX-1 and MdPUB23-OX-2, MdPUB23-overexpressing apple calli; MdPUB23-Anti-1 and MdPUB23-Anti-2, apple calli expressing the MdPUB23 antisense construct. Bars = 1 cm. Control, apple calli without cold treatment; Cold treatment, apple calli with cold treatment. (B) The fresh weight of apple calli with or without cold stress treatment shown in (A). Error bar denotes SD. Data are presented as mean ±C SD of three independent experiments each containing 8–10 calli subgroups per treatment. Different letters above the bars indicate significant differences (p <C 0.05) based on one-way ANOVA with Duncan's test. (C) Expression of the MdICE1 and MdCBF genes (MdCBF1, MdCBF2, MdCBF3, MdCBF4, and MdCBF5) and the CBF target genes (MdKIN1, MdRD29A, and MdCOR47) in MdPUB23 transgenic apple calli under cold-stress treatment shown in (A). qRT-PCR was performed in three biological replicates and three technical replicates. The value of WT was used as the reference and was set to 1. Error bar denotes SD. Different letters above the bars indicate significant differences (p < 0.05) based on one-way ANOVA with Duncan's test. (D) Cold-stress phenotypes of MdPUB23 transient transgenic apple seedlings. Apple seedlings were treated at 4°C for 5 d. The experiments were repeated three times with similar results, and each experiment contained 4–6 seedlings per treatment. A representative picture is shown here. EV, the empty-vector control; MdPUB23-OX, MdPUB23-overexpressing apple seedlings. Bars = 1 cm. Control, apple seedlings without cold treatment; Cold treatment, apple seedlings with cold treatment. (E) The relative chlorophyll content of apple seedlings with or without cold-stress treatment shown in (D). The value of EV without cold treatment was used as the reference and was set to 100%. Error bar denotes SD. Data are presented as mean ± SD of three independent experiments each containing 4–6 seedlings per treatment. Different letters above the bars indicate significant differences (p < 0.05) based on one-way ANOVA with Duncan's test. (F) DAB and (G) NBT staining of MdPUB23 transient transgenic apple leaves. Apple leaves were treated at 4°C for 10 h, and then, leaves were dyed with DAB and NBT. EV, the empty-vector control; MdPUB23-OX, MdPUB23-overexpressing apple leaves. Control, apple leaves without cold treatment; Cold treatment, apple leaves with cold treatment. The experiments were repeated three times with similar results, and each experiment contained 6–C9 apple leaves per treatment. Bars = 1 cm. A representative picture is shown here. (H) Cold-stress phenotypes of MdPUB23 transgenic Arabidopsis seedlings. Arabidopsis seedlings were grown on MS plates at 22°C for 8 d and then treated at −4°C for 15 min for non-acclimated plants (NA) and at −4°C for 20 min for acclimated plants (CA: 3 d at 4°C). Plants without cold-stress treatment were used as control. The experiments were repeated three times with similar results, and each experiment contained 3–5 plates of Arabidopsis seedlings per treatment. A representative picture is shown here. Col-0, wild-type control; MdPUB23-L1, L2, and L3, MdPUB23-overexpressing Arabidopsis seedlings. (I) Survival rates of MdPUB23 transgenic Arabidopsis seedlings under non-acclimated and acclimated conditions shown in (H). Error bar denotes SD. Data are presented as mean ± SD of three independent experiments each containing 3–5 plates of Arabidopsis seedlings per treatment. Different letters above the bars indicate significant differences (p <C 0.05) based on one-way ANOVA with Duncan's test.
After cold-stress treatment, the chlorophyll loss of transgenic apple seedlings was determined, and the transgenic apple leaves were stained with reactive oxygen species (ROS) dye to determine the degree of damage to the seedlings or leaves caused by the cold stress. The results showed that, compared with the empty-vector control (EV), the chlorophyll loss and ROS content in the MdPUB23 transgenic leaves were higher after cold-stress treatment (Fig. 4D–G), indicating that MdPUB23 negatively regulates cold-stress tolerance. Furthermore, we obtained MdPUB23 transgenic Arabidopsis seedlings and used them for cold-stress assays (Supplementary Fig. S1D). The survival rate of MdPUB23-overexpressing Arabidopsis was lower than that of the Col-0 after cold-stress treatment (Fig. 4H, I). These results indicate that MdPUB23 negatively regulates cold-stress tolerance.
MdPUB23 mediates the ubiquitination of MdICE1 in vitro and in vivo
Given that MdPUB23 encodes a E3 ubiquitin ligase and MdICE1 acts as an interaction partner of MdPUB23, we questioned whether MdPUB23 could mediate the ubiquitination of the MdICE1 protein. In vitro ubiquitination assays showed that only when ATP, ubiquitin, E1, E2, and MdPUB23-HIS were present at the same time would ubiquitin-modification bands appear for MdICE1-GST (Fig. 5A), suggesting that MdICE1 is a direct target of MdPUB23 ubiquitination. In addition, we detected the ubiquitination of MdICE1 by MdPUB23 in vivo. Apple calli of single transgenic MdICE1 and co-transgenic MdICE1 and MdPUB23 were prepared (Supplementary Fig. S1A and E). The MdICE1-GFP protein was immunocoprecipitated with GFP antibody and detected with GFP and ubiquitin antibodies. The results showed that the overexpression of MdPUB23 significantly increased the degree of polyubiquitination of MdICE1 (Fig. 5B), demonstrating that MdPUB23 mediates the ubiquitination of MdICE1 in vivo. Moreover, we found that cold stress enhanced MdICE1 ubiquitination and this process was dependent on MdPUB23 (Fig. 5C).
Figure 5.
Effect of MdPUB23 on level of ubiquitination of MdICE1. (A) Ubiquitination assays in vitro. MdPUB23-HIS was tested for E3 ubiquitin ligase activity in the presence and absence of ATP, ubiquitin, E1, E2, MdPUB23-HIS, and MdICE1-GST. The protein gel blot was analyzed using a GST antibody. (B) Ubiquitination analysis in vivo. MdICE1-GFP was immunoprecipitated using a GFP antibody from the three transgenic apple calli (GFP, MdICE1-GFP, and MdICE1-GFP/MdPUB23-OX). Immunoblotting using a ubiquitin (Ubi) antibody is shown on the left, and that using a GFP antibody is shown on the right. HIS fusion protein was used as the control. (C) Effect of cold stress on level of ubiquitination of MdICE1. MdICE1-GFP was immunoprecipitated using a GFP antibody from the two transgenic apple calli (MdICE1-GFP and MdICE1-GFP/MdPUB23-Anti) with or without cold-stress treatment for 1 h. Immunoblotting using a ubiquitin (Ubi) antibody is shown on the left, and that using a GFP antibody is shown on the right. Control, without cold-stress treatment; Cold treatment, with cold-stress treatment.
MdPUB23 promotes MdICE1 degradation
To assess whether MdPUB23 affected the protein stability of MdICE1, in vitro protein-degradation assays were performed. The MdICE1-GST fusion protein was incubated with total proteins extracted from WT and MdPUB23 transgenic apple calli. Western blotting analysis showed that the degradation rate for MdICE1-GST was higher in MdPUB23-overexpressing calli than in the WT control, while the suppression of MdPUB23 expression reduced the degradation rate for the MdICE1 (Fig. 6A). When the proteasome inhibitor MG132 was applied, degradation of MdICE1 was completely abolished (Fig. 6A), indicating that MdPUB23 promotes MdICE1 degradation via the 26S-proteasome pathway. Next, we detected the protein abundance of MdICE1 in the MdICE1 single and MdICE1–MdPUB23 co-transformed apple calli using a GFP antibody. The results showed that the overexpression of MdPUB23 decreased, while the suppression of MdPUB23 increased the abundance of the MdICE1 protein (Fig. 6B). These results suggest that MdPUB23 targets MdICE1 for ubiquitinated degradation. Moreover, we found that cold stress promoted MdICE1 degradation and this process was dependent on MdPUB23 (Fig. 6C and Supplementary Fig. S2).
Figure 6.

Effect of MdPUB23 on the stability of MdICE1 protein. (A) Protein-degradation assays in vitro. Total proteins extracted from wild-type (WT) and MdPUB23 transgenic apple calli (MdPUB23-OX: MdPUB23 overexpression; MdPUB23-Anti: MdPUB23 antisense suppression) with or without 100 μM MG132 treatments were incubated with the purified MdICE1-GST fusion protein. The samples were collected at the indicated times (0, 0.5, 1, 2, and 4 h). ACTIN was used as an internal reference. (B) MdICE1-GFP protein abundance in transgenic apple calli (MdICE1-GFP, MdICE1-GFP/MdPUB23-OX, and MdICE1-GFP/MdPUB23-Anti) was assessed by immunoblotting using a GFP antibody. (C) Effect of cold stress on the stability of MdICE1 protein. MdICE1-GFP and MdICE1-GFP/MdPUB23-Anti transgenic apple calli were treated with or without cold stress for 1 h. MdICE1-GFP protein abundance was assessed by immunoblotting using a GFP antibody. Control, without cold-stress treatment; Cold treatment, with cold-stress treatment.
MdPUB23 negatively regulates MdICE1-induced increase in cold-stress tolerance
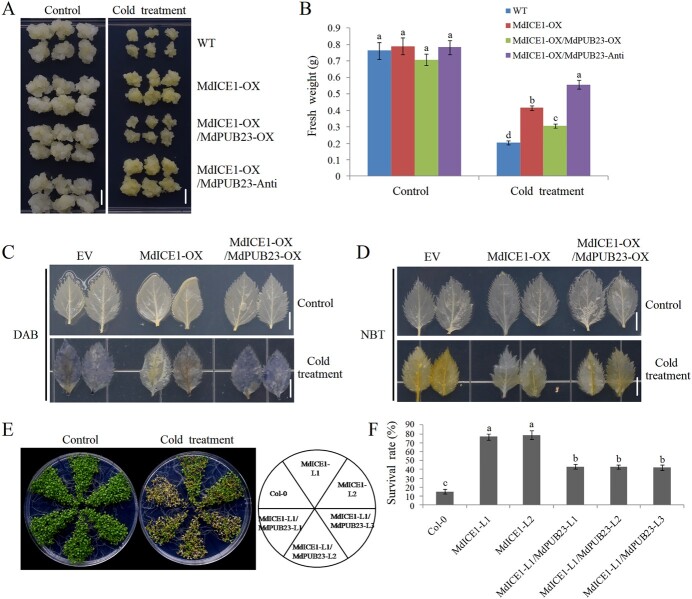
Because MdPUB23 mediates the ubiquitin degradation of MdICE1 and negatively regulates cold-stress tolerance, we further investigated whether MdPUB23 played a role in MdICE1-mediated cold-stress resistance. Apple calli co-transformed for MdICE1 and MdPUB23 were prepared and used for cold-stress assays (Supplementary Fig. 1E). After cold-stress treatment, the overexpression of MdPUB23 inhibited the growth of calli compared with the WT control, while the suppression of MdPUB23 expression contributed to the growth of calli (Fig. 4A–B). After cold-stress treatment, the overexpression of MdICE1 increased calli growth compared with the WT control, which was consistent with previous research results (Fig. 7A, B) [37]. The overexpression of MdPUB23 on the basis of MdICE1-OX inhibited MdICE1-mediated cold resistance, while the suppression of MdPUB23 expression further improved MdICE1-induced cold resistance (Fig. 7A–B), suggesting that MdPUB23 negatively regulates MdICE1-increased cold-stress tolerance. In parallel, we generated apple leaves and Arabidopsis seedlings co-transformed with MdICE1 and MdPUB23 (Supplementary Fig. 1F and G). As expected, cold-stress assays showed that the overexpression of MdPUB23 reduced MdICE1-induced cold-stress tolerance in apple leaves and Arabidopsis seedlings (Fig. 7C–F). Taking these data together, we conclude that MdPUB23 negatively regulates MdICE1-induced cold-stress tolerance by targeting MdICE1 for ubiquitinated degradation.
Figure 7.
Effects of MdPUB23 on MdICE1’s regulation of cold tolerance. (A) Cold-stress phenotypes of MdICE1-OX/MdPUB23-OX transgenic apple calli. WT, wild-type control; MdICE1-OX, MdICE1-overexpressing calli; MdICE1-OX/MdPUB23-OX, overexpression of MdPUB23 in the background of MdICE1-overexpressing calli; MdICE1-OX/MdPUB23-Anti, suppression of MdPUB23 in the background of MdICE1-overexpressing calli. Control, apple calli without cold treatment; Cold treatment, apple calli with cold treatment. Apple calli were grown on medium at 24°C for 5 d and then treated at 4°C for another 10 d. The experiments were repeated three times, and each experiment contained 8–10 calli subgroups per treatment. Bars = 1 cm. A representative picture is shown here. (B) The fresh weight of apple calli with or without cold-stress treatment shown in (A). Error bar denotes SD. Data are presented as mean ± SD of three independent experiments each containing 8–10 calli subgroups per treatment. Different letters above the bars indicate significant differences (p < 0.05) based on one-way ANOVA with Duncan's test. (C) DAB and (D) NBT staining of MdICE1 and MdPUB23 transient co-expressing apple leaves. Apple leaves were treated at 4°C for 1 d, and then, leaves were dyed with DAB and NBT. EV, the empty-vector control; MdICE1-OX, MdICE1-overexpressing apple leaves; MdICE1–OX/MdPUB23–OX, MdICE1 and MdPUB23 co-expressing apple leaves. Control, apple leaves without cold treatment; Cold treatment, apple leaves with cold treatment. The experiments were repeated three times with similar results, and each experiment contained 6–9 apple leaves per treatment. Bars = 1 cm. A representative picture is shown here. (E) Cold-stress phenotypes of MdICE1 or MdICE1–MdPUB23 transgenic Arabidopsis seedlings. Col-0, wild-type control; MdICE1-L1 and L2, MdICE1-overexpressing Arabidopsis seedlings; MdICE1-L1/MdPUB23-L1, MdICE1-L1/MdPUB23-L2, and MdICE1-L1/MdPUB23-L3, MdICE1 and MdPUB23 co-transformed Arabidopsis. Arabidopsis seedlings were grown on MS plates at 22°C for 8 d and then treated at −4°C for 1 h. The experiments were repeated three times, and each experiment contained 3–5 plates of Arabidopsis seedlings per treatment. A representative picture is shown here. (F) Survival rates of MdICE1 or MdICE1–MdPUB23 transgenic Arabidopsis seedlings shown in (E). Error bars denote standard deviations. Different letters above the bars indicate significant differences (p < 0.05) as obtained by one-way ANOVA tests.
Discussion
As one of the most productive and consumed fruits in the world, apples are popular for their rich nutritional value. In the process of apple cultivation and management, extreme-low-temperature disasters result in inestimable losses in fruit-tree production. Preventing and reducing the harm to fruit trees caused by extreme low temperatures is a prerequisite for stable and high yields for fruit trees. Therefore, it is of great practical significance to study the mechanism of the response to low-temperature stress in apple. In the present study, an apple U-box-type E3 ubiquitin ligase, MdPUB23, induced by cold stress at the transcriptional level (Fig. 3), acted as a negative regulator of the cold-stress response (Fig. 4).
ICE1 is recognized as a key regulator of the cold-stress response that enhances the cold-stress resistance of plants by directly mediating the expression of CBFs [18,38, 39]. The ICE1–CBF module plays an important role in the regulation of plant growth and development and the cold-stress response [11, 38, 39]. As a core regulator of the cold-stress response, the protein abundance of ICE1 remains dynamically stable in plants. In Arabidopsis, the protein kinases MPK3/6 and BIN2 negatively regulate the stability of the ICE1 protein, whereas OST1 increases the stability of ICE1 [28, 42–44]. In addition, the SUMO E3 ligase SIZ1 also enhances the protein stability of ICE in the cold-stress response [45]. In addition to phosphorylation and sumoylation, ubiquitination plays an essential role in the regulation of protein stability and activity of ICE1. The E3 ubiquitin ligase HOS1 targets ICE1 for degradation, thus negatively regulating the cold-stress response in Arabidopsis and rice [41, 46]. The banana fruit SINA E3 ubiquitin ligase MaSINA1 interacts with MaICE1 and attenuates its protein stability, reducing cold-stress tolerance [48]. Besides HOS1 and SINA1, no other E3 ubiquitin ligases have been found to regulate the protein stability of ICE1. Here, we used MdICE1 as the bait protein to obtain an E3 ubiquitin ligase, MdPUB23, which may be an interaction partner of MdICE1. The direct interaction between MdPUB23 and MdICE1 was verified by Y2H, pull-down, and BiFC assays (Fig. 1).
PUBs encode a class of E3 ubiquitin ligase proteins characterized by a specific U-box domain [65, 85]. As a typical U-box protein, MdPUB23 contains a U-box motif in the N-terminal region (Fig. 2A). Sequence alignment and phylogenetic tree analysis showed that MdPUB23 shared the highest sequence identity and the closest relationship with PbPUB23 (Fig. 2B–C). PUB E3 ubiquitin ligases are involved in the regulation of multiple stress responses, including the cold-stress response [63–65]. OsPUB2 and OsPUB3 positively regulate the low-temperature-stress response in rice [86]. In Vitis pseudoreticulata, VvPUB24 enhances cold-stress tolerance by alleviating the ubiquitin degradation of VvICE1 by VvHOS1 [87]. A recent study showed that Arabidopsis PUB25 and PUB26 enhanced the cold-stress response by promoting the degradation of MYB15, a negative regulator of cold stress [81]. However, the mechanism of the PUB-mediated cold-stress response remains unclear, especially in apple. Different from the results for V. pseudoreticulata showing that VvPUB24 interacts with VvICE1 but cannot directly regulate the stability of VvICE1 [87], we found that MdPUB23 directly interacted with MdICE1 to promote the ubiquitination degradation of MdICE1 in apple, thus negatively regulating MdICE1-induced cold-stress tolerance (Figs 5, 6 and 7), which suggests that the functions and regulatory mechanisms of proteins in the same family may be also different in different species. Previous studies on the functions of PUB23 have focused on plant immunity and drought stress [66, 71, 73]. The study of PUB23’s involvement in the cold-stress response in this work will further enrich the knowledge of its biological functions.
A hypothetical model is proposed to demonstrate the role of MdPUB23 in the cold-stress response (Fig. 8). Under cold-stress conditions, MdICE1 enhances cold-stress tolerance through the ICE1–CBF–COR transcription cascade pathway. In addition, cold stress can also promote the expression of MdPUB23, which targets the MdICE1 protein for degradation through the 26S-proteasome pathway, thus maintaining ICE1-protein homeostasis and preventing overreactions from causing damage to plants. This study reveals the ubiquitination regulation of the ICE1 protein, providing new insights for further enriching knowledge on and studying the regulatory pathways of plant cold-stress responses.
Figure 8.
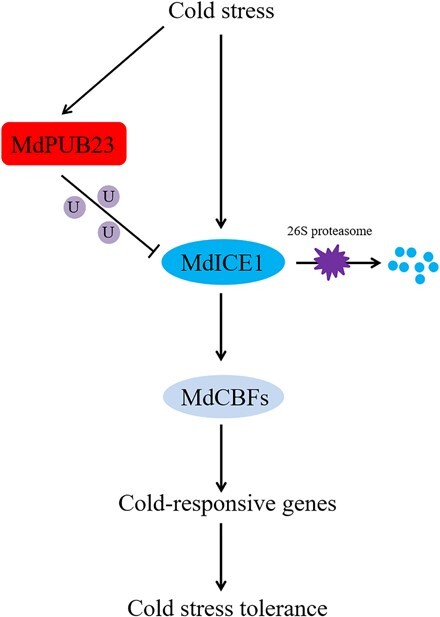
A hypothetical model of the role of MdPUB23 in cold-stress response. MdPUB23 degraded the MdICE1 protein through the ubiquitination pathway and reduced cold resistance. Under cold-stress conditions, MdPUB23 was inhibited and the MdICE1 protein was released from the MdPUB23–MdICE1 ubiquitin module. MdICE1 enhanced cold resistance through the ICE1–CBF–COR pathway.
Materials and methods
Plant materials
The plant materials used in this study included apple calli (Malus domestica, Orin), apple tissue culture seedlings (M. domestica, GL-3), and Arabidopsis seedlings (Arabidopsis thaliana, Col-0). The detached apple leaves were collected from apple tissue culture seedlings. The growth conditions of plant materials can be queried in previous studies in our laboratory [33].
Vector construction and genetic transformation
MdICE1 and MdPUB23 genes were cloned by using PCR technology from the apple tissue culture seedlings GL-3. To construct the MdICE1 and MdPUB23 overexpression recombinant plasmids, full-length MdICE1 and MdPUB23 were cloned into pCXSN-GFP and pRI101 vectors, respectively. The fragment of MdPUB23 was cloned into the pCXSN vector to construct the MdPUB23 suppression expression recombinant plasmid. Transgenic apple calli and leaves were obtained as previously described in our laboratory [37]. The primers used in this study are listed in Supplementary Table S1.
Screening the interacting proteins
The MdICE1-GFP protein was extracted from MdICE1-overexpressing apple calli and analysed by mass spectroscopy to screen the MdICE1-interacting proteins [88].
Gene expression analysis
TRIzol RNA extraction solution (Thermo Fisher Scientific, Waltham, MA, USA) and a PrimeScript™ RT kit (Takara, Dalian, China) were used for RNA extraction and reverse transcription, respectively, as previously described in our laboratory [88]. qRT-PCR analysis was performed to determine the expression levels of cold-stress-responsive genes.
Y2H, pull-down, BiFC, and co-IP assays
The interaction between MdPUB23 and MdICE1 was studied by Y2H, pull-down, BiFC, and Co-IP assays. Detailed experimental methods can be found in the Supplementary Experimental Procedures 1–4.
Cold-stress assays
Cold-stress assays of apple calli, detached apple leaves, and Arabidopsis seedlings were performed as previously described in our laboratory [37]. After cold-stress treatment, the fresh weight of the calli, reactive oxygen species (ROS) staining of the leaves, and survival rate of the Arabidopsis seedlings were determined.
ROS staining
The ROS of apple leaves after cold-stress treatment were dyed with diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) as previously described in our laboratory [37].
In vitro ubiquitination and protein-degradation assays
In vitro ubiquitination and protein-degradation assays were performed as previously described in our laboratory [37].
Accession numbers
MdPUB23, MD14G1040300 (MDP0000773851); MdPUB26, MDP0000448457; MdICE1, MDP0000662999; MdCBF1, HM992942; MdCBF2, MDP0000198054; MdCBF3, Genomic position: MDC023575.38:2048.0.2752; MdCBF4, MDP0000154764; MdCBF5, Genomic position: MDC001207.483:32385.0.33047; MdMYB23, MDP0000230141; MdMYB308L, MDP0000950559; MdbHLH3, MDP0000225680; MdbHLH33, MDP0000309179; MdBBX37, MDP0000157816; MdABI4, MD01G1155400; MdHY5, MDP0000586302; MdKIN1, MDP0000165526; MdRD29A, MDP0000598443; MdCOR47, MDP0000529003; MdACTIN, EB136338.
Acknowledgements
This work was financially supported by grants from the China postdoctoral Science Foundation (2022 M710086), the Natural Science Foundation of China (32172538), and the Open Project Programme of the State Key Laboratory of Crop Biology (2021KF06).
Author contributions
J.-P.A. conceived and designed the experiments. D.-R.W. and J.-P.A. performed the research. X.-W.Z., R.-R.X., G.-L.W., and C.-X.Y. analysed the data. J.-P.A. wrote the paper.
Data availability
All the data generated or analysed during this study are included in this published article.
Conflict of interest
The authors declare no competing interests.
Supplementary data
Supplementary data is available at Horticulture Research online.
Supplementary Material
Contributor Information
Da-Ru Wang, State Key Laboratory of Crop Biology, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An, 271018, Shandong, China.
Xiao-Wei Zhang, State Key Laboratory of Crop Biology, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An, 271018, Shandong, China.
Rui-Rui Xu, Key Laboratory of Biochemistry and Molecular Biology in Universities of Shandong, College of Biology and Oceanography, Weifang University, Weifang 261061, Shandong, China.
Gui-Luan Wang, State Key Laboratory of Crop Biology, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An, 271018, Shandong, China.
Chun-Xiang You, State Key Laboratory of Crop Biology, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An, 271018, Shandong, China.
Jian-Ping An, State Key Laboratory of Crop Biology, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An, 271018, Shandong, China.
References
- 1. Ahuja I, Vos RC, Bones AMet al. . Plant molecular stress responses face climate change. Trends Plant Sci. 2010;15:664–74. [DOI] [PubMed] [Google Scholar]
- 2. Gong Z, Xiong L, Shi Het al. . Plant abiotic stress response and nutrient use efficiency. Sci China Life Sci. 2020;63:635–74. [DOI] [PubMed] [Google Scholar]
- 3. Zhang H, Zhu J, Gong Zet al. . Abiotic stress responses in plants. Nat Rev Genet. 2022;23:104–19. [DOI] [PubMed] [Google Scholar]
- 4. Chen WJ, Zhu T. Networks of transcription factors with roles in environmental stress response. Trends Plant Sci. 2004;9:591–6. [DOI] [PubMed] [Google Scholar]
- 5. Zhu JK. Abiotic stress signaling and responses in plants. Cell. 2016;167:313–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kidokoro S, Shinozaki K, Yamaguchi-Shinozaki K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022;27:922–35. [DOI] [PubMed] [Google Scholar]
- 7. Chinnusamy V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007;12:444–51. [DOI] [PubMed] [Google Scholar]
- 8. Ding Y, Yang S. Surviving and thriving: how plants perceive and respond to temperature stress. Dev Cell. 2022;57:947–58. [DOI] [PubMed] [Google Scholar]
- 9. Zhang H, Zhao Y, Zhu JK. Thriving under stress: how plants balance growth and the stress response. Dev Cell. 2020;55:529–43. [DOI] [PubMed] [Google Scholar]
- 10. Chinnusamy V, Zhu JK, Sunkar R. Gene regulation during cold stress acclimation in plants. Methods Mol Biol. 2010;639:39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shi Y, Ding Y, Yang S. Molecular regulation of CBF Signaling in cold acclimation. Trends Plant Sci. 2018;23:623–37. [DOI] [PubMed] [Google Scholar]
- 12. Ding Y, Shi Y, Yang S. Molecular regulation of plant responses to environmental temperatures. Mol Plant. 2020;13:544–64. [DOI] [PubMed] [Google Scholar]
- 13. Song Y, Zhang X, Li Met al. . The direct targets of CBFs: in cold stress response and beyond. J Integr Plant Biol. 2021;63:1874–87. [DOI] [PubMed] [Google Scholar]
- 14. Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gilmour SJ, Zarka DG, Stockinger EJet al. . Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–42. [DOI] [PubMed] [Google Scholar]
- 16. Liu Q, Kasuga M, Sakuma Yet al. . Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively. Plant Cell. 1998;10:1391–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wisniewski M, Nassuth A, Teulières Cet al. . Genomics of cold hardiness in Woody plants. Crit Rev Plant Sci. 2014;33:92–124. [Google Scholar]
- 18. Chinnusamy V, Ohta M, Kanrar Set al. . ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003;17:1043–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Agarwal M, Hao Y, Kapoor Aet al. . A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem. 2006;281:37636–45. [DOI] [PubMed] [Google Scholar]
- 20. Franklin KA, Whitelam GC. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat Genet. 2007;39:1410–3. [DOI] [PubMed] [Google Scholar]
- 21. Doherty CJ, Van Buskirk HA, Myers SJet al. . Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell. 2009;21:972–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fursova OV, Pogorelko GV, Tarasov VA. Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene. 2009;429:98–103. [DOI] [PubMed] [Google Scholar]
- 23. Nakamichi N, Kusano M, Fukushima Aet al. . Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009;50:447–62. [DOI] [PubMed] [Google Scholar]
- 24. Dong MA, Farré EM, Thomashow MF. Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc Natl Acad Sci USA. 2011;108:7241–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee CM, Thomashow MF. Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2012;109:15054–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shi Y, Tian S, Hou Let al. . Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-a ARR genes in Arabidopsis. Plant Cell. 2012;24:2578–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim Y, Park S, Gilmour SJet al. . Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 2013;75:364–76. [DOI] [PubMed] [Google Scholar]
- 28. Ding Y, Li H, Zhang Xet al. . OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev Cell. 2015;32:278–89. [DOI] [PubMed] [Google Scholar]
- 29. Eremina M, Unterholzner SJ, Rathnayake AIet al. . Brassinosteroids participate in the control of basal and acquired freezing tolerance of plants. Proc Natl Acad Sci USA. 2016;113:E5982–e5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiang B, Shi Y, Zhang Xet al. . PIF3 is a negative regulator of the CBF pathway and freezing tolerance in Arabidopsis. Proc Natl Acad Sci USA. 2017;114:E6695–e6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kidokoro S, Yoneda K, Takasaki Het al. . Different cold-Signaling pathways function in the responses to rapid and gradual decreases in temperature. Plant Cell. 2017;29:760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li H, Ye K, Shi Yet al. . BZR1 positively regulates freezing tolerance via CBF-dependent and CBF-independent pathways in Arabidopsis. Mol Plant. 2017;10:545–59. [DOI] [PubMed] [Google Scholar]
- 33. An JP, Yao JF, Wang XNet al. . MdHY5 positively regulates cold tolerance via CBF-dependent and CBF-independent pathways in apple. J Plant Physiol. 2017;218:275–81. [DOI] [PubMed] [Google Scholar]
- 34. An JP, Li R, Qu FJet al. . R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 2018;96:562–77. [DOI] [PubMed] [Google Scholar]
- 35. Xu H, Wang N, Wang Yet al. . Overexpression of the transcription factor MdbHLH33 increases cold tolerance of transgenic apple callus. Plant Cell Tissue & Organ. Culture. 2018;134:131–40. [Google Scholar]
- 36. An JP, Wang XF, Zhang XWet al. . An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol J. 2020;18:337–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. An JP, Wang XF, Zhang XWet al. . Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytol. 2021;229:2707–29. [DOI] [PubMed] [Google Scholar]
- 38. Lee BH, Henderson DA, Zhu JK. The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell. 2005;17:3155–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tang K, Zhao L, Ren Yet al. . The transcription factor ICE1 functions in cold stress response by binding to the promoters of CBF and COR genes. J Integr Plant Biol. 2020;62:258–63. [DOI] [PubMed] [Google Scholar]
- 40. Hu Y, Jiang L, Wang Fet al. . Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor1 cascade and freezing tolerance in Arabidopsis. Plant Cell. 2013;25:2907–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dong CH, Agarwal M, Zhang Yet al. . The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc Natl Acad Sci USA. 2006;103:8281–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li H, Ding Y, Shi Yet al. . MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev Cell. 2017;43:630–642.e4. [DOI] [PubMed] [Google Scholar]
- 43. Zhao C, Wang P, Si Tet al. . MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev Cell. 2017;43:618–629.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ye K, Li H, Ding Yet al. . BRASSINOSTEROID-INSENSITIVE2 negatively regulates the stability of transcription factor ICE1 in response to cold stress in Arabidopsis. Plant Cell. 2019;31:2682–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Miura K, Jin JB, Lee Jet al. . SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. 2007;19:1403–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Z, Li J, Li Fet al. . OsMAPK3 phosphorylates OsbHLH002/OsICE1 and inhibits its Ubiquitination to activate OsTPP1 and enhances Rice chilling tolerance. Dev Cell. 2017;43:731–743.e5. [DOI] [PubMed] [Google Scholar]
- 47. Zhao ML, et al. Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ. 2013;36:30–51. [DOI] [PubMed] [Google Scholar]
- 48. Fan ZQ, Chen JY, Kuang JFet al. . The Banana fruit SINA ubiquitin ligase MaSINA1 regulates the stability of MaICE1 to be negatively involved in cold stress response. Front Plant Sci. 2017;8:995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. An JP, Xu RR, Liu Xet al. . Abscisic acid insensitive 4 interacts with ICE1 and JAZ proteins to regulate ABA signaling-mediated cold tolerance in apple. J Exp Bot. 2022;73:980–97. [DOI] [PubMed] [Google Scholar]
- 50. Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol. 2004;16:119–26. [DOI] [PubMed] [Google Scholar]
- 51. Miura K, Jin JB, Hasegawa PM. Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol. 2007;10:495–502. [DOI] [PubMed] [Google Scholar]
- 52. Ubersax JA, Ferrell JE Jr. Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol. 2007;8:530–41. [DOI] [PubMed] [Google Scholar]
- 53. Zhang H, Lang Z, Zhu JK. Dynamics and function of DNA methylation in plants. Nat Rev Mol Cell Biol. 2018;19:489–506. [DOI] [PubMed] [Google Scholar]
- 54. Chen X, Ding Y, Yang Yet al. . Protein kinases in plant responses to drought, salt, and cold stress. J Integr Plant Biol. 2021;63:53–78. [DOI] [PubMed] [Google Scholar]
- 55. Lyzenga WJ, Stone SL. Abiotic stress tolerance mediated by protein ubiquitination. J Exp Bot. 2012;63:599–616. [DOI] [PubMed] [Google Scholar]
- 56. Zhao Q, Tian M, Li Qet al. . A plant-specific in vitro ubiquitination analysis system. Plant J. 2013;74:524–33. [DOI] [PubMed] [Google Scholar]
- 57. Mazzucotelli E, Belloni S, Marone Det al. . The e3 ubiquitin ligase gene family in plants: regulation by degradation. Curr Genomics. 2006;7:509–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xu FQ, Xue HW. The ubiquitin-proteasome system in plant responses to environments. Plant Cell Environ. 2019;42:2931–44. [DOI] [PubMed] [Google Scholar]
- 59. Vierstra RD. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol. 2009;10:385–97. [DOI] [PubMed] [Google Scholar]
- 60. Hatakeyama S, Nakayama KI. U-box proteins as a new family of ubiquitin ligases. Biochem Biophys Res Commun. 2003;302:635–45. [DOI] [PubMed] [Google Scholar]
- 61. Morreale FE, Walden H. Types of ubiquitin ligases. Cell. 2016;165:248–248.e1. [DOI] [PubMed] [Google Scholar]
- 62. Toma-Fukai S, Shimizu T. Structural diversity of ubiquitin E3 ligase. Molecules. 2021;26:6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yee D, Goring DR. The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J Exp Bot. 2009;60:1109–21. [DOI] [PubMed] [Google Scholar]
- 64. Sharma M. Role of plant U-BOX (PUB) protein in stress and development. In Dr. Girdhar K. Pandey (ed.), Plant Stress. Global Science books 2013. [Google Scholar]
- 65. Trujillo M. News from the PUB: plant U-box type E3 ubiquitin ligases. J Exp Bot. 2018;69:371–84. [DOI] [PubMed] [Google Scholar]
- 66. Trujillo M, Ichimura K, Casais Cet al. . Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr Biol. 2008;18:1396–401. [DOI] [PubMed] [Google Scholar]
- 67. Lu D, Lin W, Gao Xet al. . Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science. 2011;332:1439–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhou J, Lu D, Xu Get al. . The dominant negative ARM domain uncovers multiple functions of PUB13 in Arabidopsis immunity, flowering, and senescence. J Exp Bot. 2015;66:3353–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ma A, Zhang D, Wang Get al. . Verticillium dahliae effector VDAL protects MYB6 from degradation by interacting with PUB25 and PUB26 E3 ligases to enhance Verticillium wilt resistance. Plant Cell. 2021;33:3675–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang Y, Wu Y, Zhong Het al. . Arabidopsis PUB2 and PUB4 connect signaling components of pattern-triggered immunity. New Phytol. 2022;233:2249–65. [DOI] [PubMed] [Google Scholar]
- 71. Cho SK, Ryu MY, Song Cet al. . Arabidopsis PUB22 and PUB23 are homologous U-box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell. 2008;20:1899–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Adler G, Konrad Z, Zamir Let al. . The Arabidopsis paralogs, PUB46 and PUB48, encoding U-box E3 ubiquitin ligases, are essential for plant response to drought stress. BMC Plant Biol. 2017;17:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhao J, Zhao L, Zhang Met al. . Arabidopsis E3 ubiquitin ligases PUB22 and PUB23 negatively regulate drought tolerance by targeting ABA receptor PYL9 for degradation. Int J Mol Sci. 2017;17:1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chen X, Wang T, Rehman AUet al. . Arabidopsis U-box E3 ubiquitin ligase PUB11 negatively regulates drought tolerance by degrading the receptor-like protein kinases LRR1 and KIN7. J Integr Plant Biol. 2021;63:494–509. [DOI] [PubMed] [Google Scholar]
- 75. Bergler J, Hoth S. Plant U-box armadillo repeat proteins AtPUB18 and AtPUB19 are involved in salt inhibition of germination in Arabidopsis. Plant Biol (Stuttg). 2011;13:725–30. [DOI] [PubMed] [Google Scholar]
- 76. Jung C, Zhao P, Seo JSet al. . PLANT U-BOX PROTEIN10 regulates MYC2 stability in Arabidopsis. Plant Cell. 2015;27:2016–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kong L, Cheng J, Zhu Yet al. . Degradation of the ABA co-receptor ABI1 by PUB12/13 U-box E3 ligases. Nat Commun. 2015;6:8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Seo DH, Ahn MY, Park KYet al. . The N-terminal UND motif of the Arabidopsis U-box E3 ligase PUB18 is critical for the negative regulation of ABA-mediated Stomatal movement and determines its Ubiquitination specificity for exocyst subunit Exo70B1. Plant Cell. 2016;28:2952–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhou J, Liu D, Wang Pet al. . Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proc Natl Acad Sci USA. 2018;115:E1906–e1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kim EJ, Lee SH, Park CHet al. . Plant U-Box40 mediates degradation of the Brassinosteroid-responsive transcription factor BZR1 in Arabidopsis roots. Plant Cell. 2019;31:791–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wang X, Ding Y, Li Zet al. . PUB25 and PUB26 promote plant freezing tolerance by degrading the cold Signaling negative regulator MYB15. Dev Cell. 2019;51:222–235.e5. [DOI] [PubMed] [Google Scholar]
- 82. Hu DG, Yu JQ, Han PLet al. . The regulatory module MdPUB29-MdbHLH3 connects ethylene biosynthesis with fruit quality in apple. New Phytol. 2019;221:1966–82. [DOI] [PubMed] [Google Scholar]
- 83. Wei Y, Jin J, Xu Yet al. . Ethylene-activated MdPUB24 mediates ubiquitination of MdBEL7 to promote chlorophyll degradation in apple fruit. Plant J. 2021;108:169–82. [DOI] [PubMed] [Google Scholar]
- 84. Han PL, Dong YH, Gu KDet al. . The apple U-box E3 ubiquitin ligase MdPUB29 contributes to activate plant immune response to the fungal pathogen Botryosphaeria dothidea. Planta. 2019;249:1177–88. [DOI] [PubMed] [Google Scholar]
- 85. Azevedo C, Santos-Rosa MJ, Shirasu K. The U-box protein family in plants. Trends Plant Sci. 2001;6:354–8. [DOI] [PubMed] [Google Scholar]
- 86. Byun MY, Cui LH, Oh TKet al. . Homologous U-box E3 ubiquitin ligases OsPUB2 and OsPUB3 are involved in the positive regulation of low temperature stress response in Rice (Oryza sativa L.). Front Plant Sci. 2017;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yao W, Wang L, Wang Jet al. . VpPUB24, a novel gene from Chinese grapevine, Vitis pseudoreticulata, targets VpICE1 to enhance cold tolerance. J Exp Bot. 2017;68:2933–49. [DOI] [PubMed] [Google Scholar]
- 88. An JP, Yao JF, Xu RRet al. . Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Environ. 2018;41:2678–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data generated or analysed during this study are included in this published article.