Abstract
An early step in the utilization of starch by Bacteroides thetaiotaomicron is the binding of starch to the bacterial surface. Four starch-associated outer membrane proteins of B. thetaiotaomicron that have no starch-degrading activity have been identified. Two of these, SusC and SusD, have been shown by genetic analysis to be required for starch binding. In this study, we provide the first biochemical evidence that these two proteins interact physically with each other. Both formaldehyde cross-linking and nondenaturing gel electrophoresis experiments showed that SusC and SusD interact to form a complex. Two other proteins encoded by genes in the same operon, SusE and SusF, proved not to be essential for starch utilization and actually decreased starch binding when they were present along with SusC and SusD. Consistent with this, nondenaturing gel analysis revealed that in a strain producing SusC, SusD, and SusE, the SusCD complex was partially destabilized. The strain producing SusC, SusD, and SusE also grew more slowly on starch than a strain producing SusC, SusD, SusE, and SusF (μmax, 0.29 and 0.37/h, respectively). Thus, SusE appears to interact with the SusCD complex. SusE also interacts with SusF, because SusE was less susceptible to proteinase K digestion when SusF was present, and nondenaturing gel analysis detected a complex formed by these two proteins. Our results indicate that SusC, SusD, SusE, and SusF form a protein complex in the outer membrane but that SusE and SusF are dispensable members of this complex.
Undigested polysaccharides are thought to be a major source of carbon and energy for members of the human colonic microflora. In an earlier survey, it was found that the most of the intestinal isolates that could degrade polysaccharides were members of the genus Bacteroides (12). The ability to utilize polysaccharides may explain why Bacteroides is one of the numerically predominant genera in the intestine. We have used the starch utilization system (Sus) of Bacteroides thetaiotaomicron as a model for investigating how Bacteroides species utilize polysaccharides.
Previously, we found that the starch-degrading enzymes of B. thetaiotaomicron are cell associated and that there are outer membrane proteins distinct from the enzymes that bind starch to the bacterial surface (11). Starch binding appears to be the first step in starch utilization. Bound starch is then digested by the degradative enzymes, and products of starch breakdown are internalized (14). This strategy for polymer breakdown helps the bacteria to degrade a large polymer into segments small enough to pass through outer membrane porins without losing the products of digestion to competitors located nearby. Eight genes that contribute to starch utilization have been identified; a regulatory gene, susR, and seven structural genes, susA through susG. Expression of the susA-through-susG genes is regulated by SusR, an activator that responds to the presence of maltose or larger oligosaccharides by increasing expression of the sus structural genes (5). The fact that maltose is an inducer of starch utilization gene expression makes it possible to study sus gene expression even in mutants that are unable to grow on starch.
The biochemical properties of some of the Sus structural proteins have been determined. SusA is a neopullulanase, a type of starch-degrading enzyme that can digest all three forms of starch: amylose (linear chains of α-1,4-linked glucose residues), amylopectin (amylose chains linked by α-1,6 linkages), and pullulan (maltotriose units linked in a linear chain by α-1,6 linkages). SusA is a soluble enzyme which appears to be located in the periplasmic space (4). When susA is disrupted, B. thetaiotaomicron can still grow on starch, but the growth rate is only 30% that of the wild type (4). Another starch-degrading enzyme is SusG, which is also a neopullulanase. SusG is an outer membrane protein and is essential for growth on starch (14). Disruption of susG abolishes growth on any form of starch. SusB breaks down the oligosaccharides released by SusA and SusG into glucose residues.
Much less is known about the biochemical characteristics and functions of SusC, SusD, SusE, and SusF. Previous work has shown that they are all outer membrane proteins with no detectable enzyme activity. Genetic analyses have suggested that they have a role in binding starch to the bacterial surface (11). Disruption of susC and susD abolished starch binding completely, and a disruption in susE was associated with reduced starch binding compared to that for the wild type. A mutant with a disruption in susF bound starch almost as well as the wild type. These results suggested that SusC and SusD are responsible for most of the starch-binding activity detected using intact wild-type cells but that SusE might make some additional contribution to binding. One hypothesis to explain these results is that SusC, SusD, SusE, and possibly SusF form a starch-binding complex in the outer membrane. Two findings indirectly supported the hypothesis that SusC and SusD interact. First, when SusC and SusD were provided individually, their susceptibilities to protease digestion increased. Second, neither SusC nor SusD alone bound to a starch column, but when present together they were retained on the column (13). This finding could also be explained, however by a different hypothesis—that one of the proteins modified the other to a form that bound starch. In this paper, we provide direct biochemical evidence for direct physical interactions between SusC and SusD and for an interaction between SusE and SusF.
The susA gene is in its own transcriptional unit, but susB through susG are arranged in an operon. This operon structure has made it difficult to assess whether the genes in the susB operon are essential for growth on starch, because disruptions in any of the genes upstream of susG have a polar effect on susG, a gene which is essential for starch utilization. In this study, we have determined which of the outer membrane proteins encoded in this region are essential for starch utilization.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All Escherichia coli strains used in this study were grown in Luria-Bertani (LB) broth or on LB agar at 37°C. B. thetaiotaomicron 5482, transposon-generated derivatives, and single disrupted mutants used in this study have been described previously (2, 4, 11). Cells were grown initially in a prereduced Trypticase-yeast extract-glucose (TYG) medium. For optimal induction of starch utilization genes, cells were transferred to a defined medium (7) containing maltose (0.3%) as the sole carbohydrate source. To test for growth on malto-oligosaccharides or starch, cells were inoculated into a defined medium with maltopentaose (G5), maltoheptaose (G7), amylopectin, or pullulan (0.3%) as a sole carbohydrate source. Antibiotic concentrations used in this study were as follows: for ampicillin, 50 μg/ml; for chloramphenicol, 20 μg/ml (E. coli) or 15 μg/ml (B. thetaiotaomicron); for erythromycin, 10 μg/ml; for gentamicin, 200 μg/ml; and for tetracycline, 1 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Description and/or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5αMCR | RecA− Gns | 6 |
| S17-1 | RecA− Gns | IncP RP4 inserted into the chromosome (17) |
| B. thetaiotaomicron | ||
| BT5482 | Wild type; Gnr (G2–G7)+ Starch+ | Anaerobe Laboratory, Virginia Polytechnic Institute, Blacksburg, Va. |
| BT4007 | Tcr Emr Gnr (G2–G7)+ Starch+ | B. thetaiotaomicron mutant with conjugative transposon, CTnDOT, carrying Tcr (tetQ) and Emr (ermF) genes; displays wild-type growth, since no sus genes are disrupted |
| Ms-2 | Emr Gnr SusA+ SusB+ SusC− SusD− SusE− SusF− SusG− G2+ G3+ G7− Starch− | TN4351-generated mutant displaying BTΩsusC phenotype, since susC is disrupted (2) |
| BTΩsusC | Tcr Gnr SusC− SusD− SusE− SusF− SusG− G2+ G3+ G7− Starch− | B. thetaiotaomicron mutant with a susC disruption created by suicide vector pBT-1 containing a PCR-generated 0.61-kbp internal fragment of susC (pBT-1-SC) (11) |
| BTΩsusD | Tcr Gnr SusC+ SusD− SusE− SusF− SusG− (G2–G7)+ Starch− | B. thetaiotaomicron mutant with a susD disruption created by suicide vector pBT-1 containing a PCR-generated 0.44-kbp internal fragment of susD (pBT-1-SD) (11) |
| BTΩsusE | Tcr Gnr SusC+ SusD+ SusE− SusF− SusG− (G2–G7)+ Starch− | B. thetaiotaomicron mutant with a susE disruption created by suicide vector pBT-1 containing a PCR-generated 0.49-kbp internal fragment of susE (pBT-1-SE) (11) |
| BTΩsusF | Tcr Gnr SusC+ SusD+ SusE+ SusF− SusG− (G2–G7)+ Starch− | B. thetaiotaomicron mutant with a susF disruption created by suicide vector pBT-1 containing a PCR-generated 0.53-kbp internal fragment of susF (pBT-1-SF) (11) |
| BTΩsusG | Tcr Gnr (G2–G7)+ Starch− | B. thetaiotaomicron mutant with a susG disruption created by suicide vector pBT-1 containing a PCR-generated 0.61-kbp internal fragment of susG (pBT-1-SG) (11) |
| Plasmids | ||
| pBT-1 | Knr Tcr | RSF1010-based suicide vector used to make insertional disruptions in Bacteroides spp. (18) |
| pNLY1::PsusA | Apr Cmr Cmr | pACYC-based shuttle vector containing the susA promoter used to express genes in |
| trans (14) | ||
| pSGC23A | Apr Cmr Cmr | pNLY1::PsusA containing a PCR-generated entire susG gene cloned downstream of the susA promoter (14) |
| pSDC27 | Apr Cmr Cmr | pNLY1::PsusA containing a PCR-generated entire susD gene cloned downstream of the susA promoter (13) |
Abbreviations: G2, maltose; G3, maltotriose; G7, maltoheptaose; Tc, tetracycline; Em, erythromycin; Gn, gentamicin; Ap, ampicillin; Kn, kanamycin; Cm, chloramphenicol; CTn, conjugative transposon. Boldfaced antibiotic resistance genes are expressed only in E. coli. Other resistance genes are expressed only in Bacteroides spp.
DNA methods.
Plasmids were isolated using a Wizard Plus DNA purification system (Promega Corp). Dephosphorylation reactions and restriction digestions were performed in accordance with the manufacturer's instructions (Bethesda Research Laboratories, Bethesda, Md., or New England Biolabs, Beverly, Mass.). E. coli DH5αMCR was transformed by the method of Lederberg and Cohen (9). Conjugations, where constructs generated in E. coli were transferred to Bacteroides recipients, were performed as described by Shoemaker et al. (15). Insertional and replicative shuttle vectors were mobilized from E. coli donors to Bacteroides recipients by the transfer function of RP4 integrated into the chromosome of S17-1 (17).
Chemicals.
14C-labeled starch (Nicotinia tabacum 1) was purchased from DuPont, NEN. Glucose, maltose, maltopentaose, maltoheptaose, amylopectin, pullulan, proteinase K, and phenylmethylsulfonyl fluoride (PMSF) were purchased from Sigma Corp. Formaldehyde was purchased from J. T. Baker Chemical Co.
Expression of susG in trans.
pSGC23A (14), a plasmid that carries susG cloned downstream of the susA promoter, had been used previously to characterize the activities of SusG independently of the other Sus proteins. pSGC23A was used to provide SusG in strains with disruptions in upstream sus genes. The E. coli strain that contained pSGC23A was mated to BTΩsusC, BTΩsusD, BTΩsusE, BTΩsusF, and BTΩsusG. The transconjugants were named BTΩsusC(pSGC23A), BTΩsusD(pSGC23A), BTΩsusE(pSGC23A), BTΩsusF(pSGC23A), and BTΩsusG(pSGC23A), respectively. These strains were tested for SusG production by Western blotting. The production of SusC, SusD, SusE, and SusF was also monitored by Western blot analysis. This was important because a previous study had shown that providing either the susA promoter or the susB promoter in trans on a multicopy plasmid (about 5 copies per cell) caused a decrease in expression of the chromosomal genes, presumably due to titration of SusR by the cloned promoter regions. The susA promoter is weaker than the susB promoter and exerts less of a titration effect when provided on the plasmid. This is the reason that the susA promoter was used instead of the susB promoter in the plasmid that provided susG in trans. These strains were tested for growth on starch and for starch-binding activity. Western blotting was done as described previously (10). Approximately 50 μg of protein in a membrane fraction was loaded in each lane of the gel. Antibodies bound to the protein were detected with biotinylated secondary antibodies, followed by treatment with streptavidin β-galactosidase reagent (Bethesda Research Laboratories). Membrane protein concentrations were determined using the DC protein assay kit in accordance with the manufacturer's instructions (Bio-Rad Laboratories, Hercules, Calif.).
Starch binding activity.
Binding of 14C-labeled starch to intact cells of B. thetaiotaomicron was carried out as described by Shipman et al. (13). Briefly, labeled starch was added to washed intact cells and incubated for 5 min at room temperature. Cells were then harvested by centrifugation and washed twice. Previous work has shown that the starch is bound irreversibly to the cells so that cells can be washed to reduce the nonspecific background. Under aerobic conditions, the cells do not internalize or accumulate starch except for that initially bound (1). Thus, in this assay, binding is uncoupled from uptake into the cytoplasm. B. thetaiotaomicron 4007 was used as the wild-type control because it contains the tetQ gene, which was used as a selectable marker in construction of the insertional disruption mutants. Thus, tetracycline could be added to the media used to grow all strains, including the wild-type control.
Binding values are reported in as micrograms of starch bound per milligram of cell protein. These values were obtained by multiplying the total counts per minute by a dilution factor, which was the ratio of labeled starch to total starch in each assay. That number was converted by an empirical constant based on observed counts per minute per a given amount of starch to disintegrations per minute (dpm), which allowed the total micrograms of starch bound to be calculated by using the reported values of 2.2 × 106 dpm per μg of starch. Experimental values were standardized by assaying the cell protein concentration after sonication.
Proteinase K treatment of cells to assess surface exposure of Sus proteins.
The surface exposure of Sus proteins was assessed as described by Shipman et al. (14). Briefly, proteinase K was added to washed and resuspended intact cells and incubated with the cells at 37°C for various times. PMSF (final concentration, 1 mM) was added to stop the reaction. Cells were sonicated, and proteins were detected on Western blots with antisera obtained as described previously (14). Antibodies bound to proteins were detected with secondary antibodies conjugated with horseradish peroxidase. To confirm that the cells survived the incubation with proteinase K intact, a protease-sensitive periplasmic marker, SusA, was also detected on Western blots at the initial and final time points. For all the experiments reported here, the concentration of SusA at the end of the incubation period was the same as that at the beginning, indicating that the outer membrane had not been breached.
Formaldehyde cross-linking experiments.
Cells were grown on 100 ml of defined medium plus maltose (0.3%) to an optical density at 650 nm of 0.5 and were pelleted by centrifugation at room temperature (22°C). The cell pellet was washed twice with 0.1 M potassium phosphate (KPi) buffer (pH 7.2) and suspended in 1/2 volume of KPi buffer. PMSF (14 μg/ml) was added to protect cells from protease. Formaldehyde was added to a final concentration of 1% (wt/vol). Samples were incubated at room temperature without shaking for 1 h. After the incubation, cells were harvested by centrifugation and washed twice with the KPi buffer, immediately. The pellets were solubilized in 2× concentrated electrophoresis sample buffer (8) and either heated at 65°C for 10 min to maintain the formaldehyde cross-linkings or boiled for 30 min to break the chemical cross-links; boiling for 10 min was not enough to break all the cross-links. Samples obtained from formaldehyde-treated cells or controls (100 μg of protein per lane) were subjected to Western blotting. Antibodies bound to proteins were detected with secondary antibodies conjugated with horseradish peroxidase.
Native gel electrophoresis.
Native gel electrophoresis was used to assess interactions among Sus outer membrane proteins. Polyacrylamide gels (6%) without the stacking gel were used to separate native proteins or protein complexes. The Sus outer membrane proteins were solubilized with 1.5% octyl-glucoside, and octyl-glucoside (0.75%) was added to the native gels and the electrophoresis buffer (25 mM Tris, 192 mM glycine) to ensure that the outer membrane proteins remained solubilized when they were electrophoresed. The samples were loaded on a native gel after being mixed with the sample buffer (62.5 mM Tris-HCL, 7.5% Ficoll type 400, 0.001% bromophenol blue [pH 6.8]) without boiling. The gels were electrophoresed in a cold room (4°C) for 8 to 10 h at 15 mA and then subjected to Western blotting. Antibodies bound to proteins were detected with secondary antibodies conjugated with horseradish peroxidase. A large amount of protein (600 μg), compared to that loaded on the sodium dodecyl sulfate (SDS) gels, had to be loaded on these gels to achieve reproducibly detectable bands on the Western blot. This much protein was probably needed because the poorer resolution of a nondenaturing gel makes the bands spread out more, and blotting was less efficient than with the SDS gels.
Denaturing gel electrophoresis.
The gel used in the cross-linking experiment was 12% acrylamide with a 5% stacking gel. The gel was 1.5 mm thick. For other experiments involving SDS-polyacrylamide gel electrophoresis (PAGE) 10% acrylamide gels were used. Electrophoresis was done either overnight at 5 mA or for 4 to 7 h at 15 mA. The amount of protein placed in each lane was 50 μg.
RESULTS
SusG was slightly overexpressed in the susG-complemented strains.
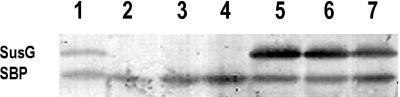
The susG gene, which is essential for growth on starch, is also the last gene in the susB operon. Accordingly, disruption of any gene upstream of susG makes the cells unable to grow on starch due to a polar effect of the insertion on susG. For this reason, it was necessary to express susG in trans in order to determine whether susD, susE, or susF is essential for starch utilization. To this end, pSGC23A, a plasmid containing the susG gene, was transferred into various sus gene disruption strains, including BTΩsusC, BTΩsusD, BTΩsusE, BTΩsusF, and BTΩsusG. pSGC23A has a copy number of about 5 per cell in Bacteroides. The level of SusG cells containing pSGC23A was about twofold higher than that in a wild-type strain with only a single copy of susG, but the levels of SusG in all of the disruption strains were the same (Fig. 1).
FIG. 1.
Western blot of SusG-complemented strains. Approximately 50 μg of membrane protein was loaded into each lane. All membrane fractions were obtained from cells grown on defined media that contained maltose (0.3 %, wt/vol). Lanes: 1, BT5482: 2, BTΩsusD: 3, BTΩsusE: 4, BTΩsusF: 5, BTΩsusD(pSGC23A): 6, BTΩsusE(pSGC23A); 7, BTΩsusF(pSGC23A). SBP is a streptavidin-binding protein of unknown function which is present in B. thetaiotaomicron extracts and is detected by the Western blot detection reagents even in the absence of antibody.
Only three of the five Sus outer membrane proteins are needed for starch utilization.
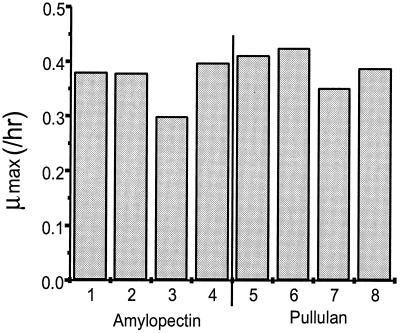
The abilities of the various disruption mutants to grow on starch (amylopectin and pullulan) were tested. The wild-type control strain used for comparison was BT4007; it contained pNLY1::PsusA, the shuttle vector into which susG was cloned to produce pSGC23A. The strain also contained a tetracycline resistance gene (tetQ). Thus, the control strain not only contained multiple copies of the susA promoter but also contained both antibiotic resistance genes that are present in the disruption mutant strains. The presence of the susA promoter in multiple copies and the need to select for two antibiotics combine to reduce the rate of growth on starch by about 10%. All the susG-complemented strains grew as well as the control on starch except for BTΩsusD(pSGC23A). This result indicated that SusD is essential for starch utilization, since BTΩsusE(pSGC23A), which has only one additional Sus protein (SusD) compared to BTΩsusD(pSGC23A), grew well on starch (Fig. 2). The fact that BTsusE(pSGC23A) grew on starch as well as the control did shows that only SusC, SusD, and SusG are needed for growth on starch and that SusE and SusF are dispensable. We designate SusC, SusD, and SusG the minimal starch utilization system. The growth rate of BTΩsusF(pSGC23A) on starch was lower than that of BTΩsusE(pSGC23A) or BTΩsusG(pSGC23A). BTΩsusF(pSGC23A) produces SusE but not SusF. This result suggested that SusE and SusF might be interacting.
FIG. 2.
Comparison of growth rates of SusG-complemented strains on starch. Two kinds of starch, amylopectin and pullulan, were used as substrates. Growth curves were done in triplicate, and the growth rates are averages of the triplicate experiments. Lanes 1 and 5, BT4007(pNLY1::PsusA); lanes 2 and 6, BTΩsusE(pSGC23A); lanes 3 and 7, BTΩsusF(pSGC23A); lanes 4 and 8; BTΩsusG(pSGC23A). Experiment-to-experiment variation was less than 10%.
We tested the various mutants for growth on malto-oligosaccharides such as maltopentaose (G5) and maltoheptaose (G7) to determine whether SusE and/or SusF affected malto-oligosaccharide utilization. BTΩsusE(pSGC23A) grew as well as the control or BTΩsusG(pSGC23A). Thus, SusE and/or SusF is not required for malto-oligosaccharide uptake. BTΩsusD(pSGC23A) grew on malto-oligosaccharides, but much more slowly than the other mutants or the wild type. BTΩsusD showed the same growth pattern as BTΩsusD(pSGC23A), indicating that SusG was not involved. Therefore, even though SusD is not essential for growth on malto-oligosaccharides, it affects the uptake of malto-oligosaccharides. This finding supports the hypothesis that SusC and SusD interact with each other in the outer membrane.
The starch-binding activities of cells containing the minimal starch utilization system were higher than that of the wild-type control.
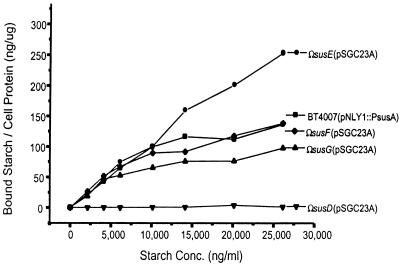
Starch-binding activities of the susG-complemented strains were compared (Fig. 3). BTΩsusG(pSGC23A) had binding activity similar to that of the wild-type control strain, BT4007(pNLY1::PsusA). BTΩsusD(pSGC23A), which produces only SusC and SusG, had no starch-binding activity. Thus, it appeared that SusC and SusG alone do not bind to starch, and the failure of this strain to grow on starch is probably due to its inability to bind starch. By contrast, BTΩsusE(pSGC23A), which carries the minimal starch utilization system, had starch-binding activity that was nearly twofold higher than that of the wild-type control. The growth rate of BTΩsusE(pSGC23A) was almost the same as that of the wild-type control. Thus, the higher starch-binding activity of BTΩsusE(pSGC23A) did not affect the growth rate of the strain.
FIG. 3.
14C-labeled starch binding by intact cells of B. thetaiotaomicron. 14C-labeled starch at 132.5 ng/ml was added to varying concentrations of cold amylopectin. The binding experiment was conducted in triplicate. Experiment-to-experiment variation was less than 20%.
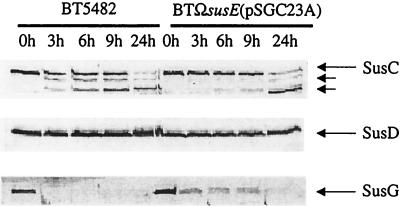
SusC in the strain carrying the minimal starch utilization system exhibited a pattern of proteolytic digestion different from that of SusC in the wild-type strain.
The proteolytic sensitivities of SusC, SusD, and SusG in the strain that carried the minimal starch utilization system was compared to that of these proteins in wild-type cells (Fig. 4). Since the starch-binding activity of the strain carrying the minimal system was higher than that of the wild type, it was possible that the topology of one or more of the Sus outer membrane proteins, in ΩsusE(pSGC23A) was different from that of the same protein in wild-type cells. SusC in the wild-type cells appeared to be degraded sooner than SusC in ΩsusE(pXGC23A), although it was also possible that the proteolytic fragments of SusC were stabilized compared to the wild type. There was no change in the proteolysis patterns of SusD. Previously, we had reported that SusD was not exposed on the cell surface in wild-type cells (13). The absence of SusE and SusF did not change this result. SusG in the susG-complemented strain was degraded rapidly, as was SusG in wild-type cells. The apparent difference between the proteolytic sensitivities of SusG in the two strain backgrounds is probably not significant. The higher initial level of SusG in the susG-complemented strain would be expected to cause a delay in the complete disappearance of SusG from this strain compared with the wild type. The fact that SusG in the susG-complemented strain was entirely degraded shows that all of the SusG produced from the gene carried on the plasmid was localized to the cell surface.
FIG. 4.
Immunoblots showing proteolytic sensitivities of SusC, SusD, and SusG in two strains, the wild type (BT5482) and the strain with the minimal starch utilization system [BTΩsusE(pSGC23A)]. Portions of a cell extract from each time point (100 μg) were loaded into each lane. The lanes are labeled according to the sampling time after addition of proteinase K. As expected, SusD was not degraded at all. This panel is provided to show that the outer membrane remained intact throughout the digestion period. Also, SusA, a periplasmic protein, was detected at the same concentration at all time points (data not shown). Electrophoresis conditions are described in Materials and Methods.
The results of formaldehyde cross-linking experiments show that SusC and SusD interact.
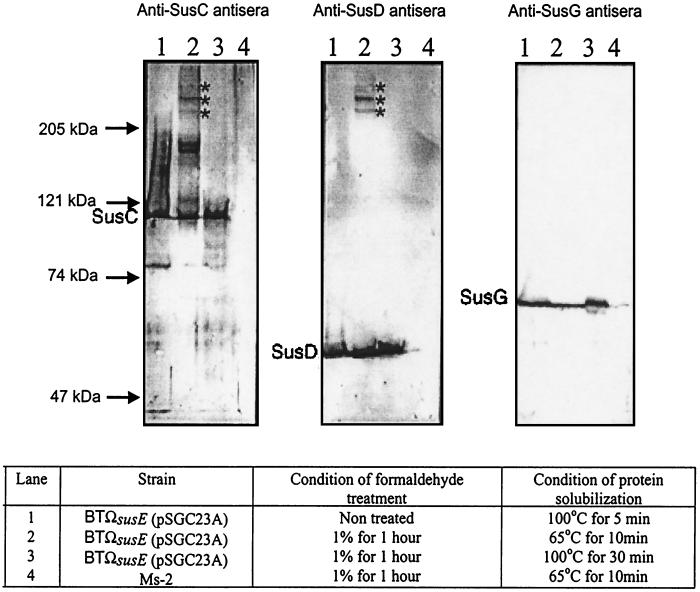
To examine the physical interactions of the outer membrane proteins of the minimal starch utilization system, a formaldehyde cross-linking experiment was performed (Fig. 5). The sizes of protein bands were calculated on the basis of the sizes of standard markers. Strain Ms-2, which produces no Sus outer membrane proteins, was used as a negative control. When the strain carrying the minimal system was treated with a 1% formaldehyde solution, three protein bands with molecular sizes higher than 250 kDa reacted with both anti-SusC and anti-SusD antisera. These appear to be complexes that contain both SusC and SusD; the band estimated to be migrating at approximately 270 kDa is the darkest. SusC is 115 kDa and SusD is 65 kDa, so a simple 1:1 complex of SusC and SusD would have been expected to be about 180 kDa. Thus, the complex that migrates at a molecular size above the 250-kDa band may contain 2 copies of SusC. There were two dark bands of about 150 to 160 kDa that reacted only with the anti-SusC antisera. These might have been dimers of SusC. If so, the complex runs as a smaller unit that the 230-kDa complex predicted from the molecular weight of SusC. We cannot rule out the possibility that SusC is binding to an as-yet-unidentified protein.
FIG. 5.
Immunoblots of the Sus outer membrane proteins of the minimal starch utilization system after formaldehyde cross-linking. Approximately 100 μg of protein from whole cells was loaded onto each lane. Sizes of molecular markers are given on the left. The Sus outer membrane proteins detected on the immunoblots are shown to the left of each immunoblot. Stars, cross-linked complexes. Electrophoresis conditions are described in Materials and Methods.
The protein bands at 250 to 290 kDa did not react with the anti-SusG antiserum. Thus, SusG appears not to form a complex with SusC and SusD. In some experiments, amylopectin (200 μg/ml) was incubated with the cells prior to the addition of formaldehyde to determine whether this large molecule might induce the formation of a complex between SusG and SusCD. Added amylopectin did not change the cross-linking pattern (data not shown). Thus, it seems that amylopectin bound to Sus proteins does not significantly change the interaction pattern.
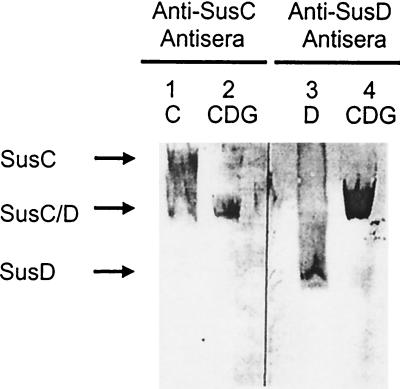
Native gel electrophoresis confirms the existence of the SusC/SusD complex.
Native gel electrophoresis was another approach used to test for interactions between SusC, SusD, and SusG. The gel contained 0.75% octyl-glucoside to keep the outer membrane proteins solubilized. Resolution of protein bands on a native gel is much poorer than on an SDS gel because of the lack of a stacking gel and the high concentration of protein applied to the gel. Nonetheless, the gel was able to resolve differences between SusC or SusD alone and the complex they formed. When both SusC and SusD were present, the anti-SusC and anti-SusD antisera detected the same band, which migrated differently than bands corresponding to SusC or SusD alone (Fig. 6). Anti-SusG antiserum did not detect the band corresponding to the SusC/SusD complex. These findings support the hypothesis that SusC and SusD interact with each other but provide no evidence for interaction of SusG with either SusC or SusD.
FIG. 6.
Immunoblots of the Sus outer membrane proteins separated by electrophoresis on a nondenaturing gel. The resolution of proteins on these gels is poorer than for SDS gels because of the lack of a stacking gel and the high concentration of protein applied to the gel. Membrane proteins and protein complexes were solubilized with 1.5% octyl-glucoside. The 6% polyacrylamide gel and the running buffer contained 0.75% octyl-glucoside to keep the proteins and protein complexes from aggregating nonspecifically. Approximately 600 μg of each membrane fraction was loaded onto each lane. Lane 1, BTΩsusD; lane 3, BTΩsusC(pSDC27); lanes 2 and 4, BTΩsusE(pSGC23A).
SusD alone migrated faster than the SusC/SusD complex, but this was not true for SusC. SusC alone migrated more slowly than the SusC/SusD complex. The putative isoelectric points of SusC and SusD are similar (5.7 and 5.2, respectively), so the proteins should migrate according to size in native gels. The slower migration of SusC alone could be due to formation of SusC complexes in the absence of SusD.
Effects of SusE and SusF.
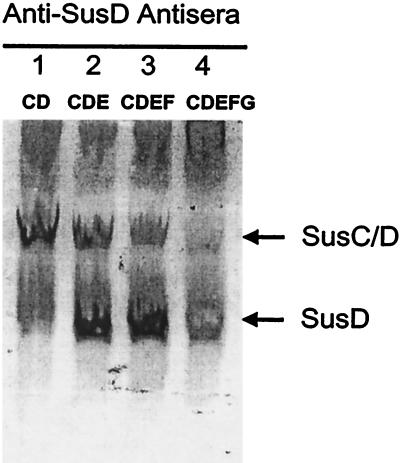
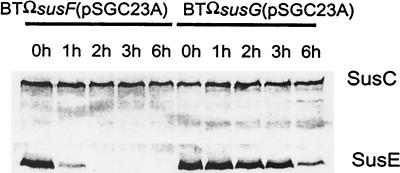
The finding that a strain that produced SusE but not SusF grew more slowly than the wild type raised the question of what effect SusE was exerting on SusC, SusD, and SusG. Also, restoring SusF to the strain restored growth to normal, an observation that raises the possibility of an interaction between SusE and SusF. Evidence that SusE without SusF might be destabilizing the SusC/SusD complex came from native gel analysis of the effect of adding back SusE and SusF to a strain that was producing SusC and SusD (Fig. 7). In the presence of SusE, less SusD was incorporated into the SusC/SusD complex. Restoration of SusF did not reverse this apparent destabilization of the SusC/SusD complex. Comparison of the proteolytic sensitivity of SusE in the absence of SusF with that of SusE in a strain that was producing SusF revealed that SusE in intact cells was degraded much more rapidly if SusF was absent than if SusF was present (Fig. 8). This result supports the hypothesis that SusE and SusF interact with each other in the outer membrane. Native gel experiments confirmed this finding (data not shown).
FIG. 7.
Immunoblots of the Sus outer membrane proteins separated by electrophoresis on a nondenaturing gel. Conditions were the same as those for Fig. 6. Approximately 600 μg of protein was loaded onto each lane. Lanes: 1, BTΩsusE; 2, BTΩsusF; 3, BTΩsusG; 4, BT5482.
FIG. 8.
Immunoblots showing changes in proteolytic sensitivity of SusC and SusE in two strains, BTΩsusF(pSGC23A) and BTΩsusG(pSGC23A). Strain BTΩsusF(pSGC23A) does not produce SusF; strain BTΩsusG(pSGC23A) produces SusF. Portions of cell extracts (100 μg of protein) were loaded onto each lane. Conditions were the same as those used for Fig. 4. The protein detected on each immunoblot is given to the right of the panel. Lanes are labeled according to the sampling time after addition of proteinase K. Note that these digestion times are shorter than those in Fig. 4, so that digestion of SusC is not detected.
DISCUSSION
Since the genes encoding the Sus outer membrane proteins are in the same operon, it seemed likely that the proteins they encode would be working together, possibly as part of a complex. The results of experiments reported here provide the first direct support for the hypothesis that SusC and SusD interact to form a complex and that this complex interacts with SusE. Since results of nondenaturing gel electrophoresis indicated that SusE and SusF interact with each other, the finding that SusE interacts with SusC/SusD suggests that all four proteins are present in the complex.
In the studies reported here, not only was SusE found not to be essential for binding, but bacteria lacking it and SusF actually exhibited increased ability to bind starch compared with the wild type. This is consistent with the results of nondenaturing gel experiments, which indicate that SusE destabilizes the SusC/SusD complex. Also, SusE in a strain producing SusC, SusD, SusE, and SusG was more susceptible to proteoloysis than it was in the wild type (Fig. 8), a finding that indicates there is some change in the interaction of SusE with the complex when SusG is present. In a previous study (14), SusE seemed to make a positive contribution to starch binding, because a strain that produced SusC, SusD, and SusE bound starch as well as the wild type, but a strain producing only SusC and SusD bound only about half as much starch. A difference between the previous binding experiments and those reported here is that the strains used in the present study contained multiple copies of susG. Thus, it is possible that the increased starch binding of the strain containing only SusC, SusD, and SusG was due as much to increased concentrations of SusG as to the absence of SusE and SusF. We still do not know the stoichiometry of different members of the complex. Results of cross-linking experiments and nondenaturing gel experiments suggest the possibility that there may be multiple copies of SusC in the complex. Some of the confusion about the role of SusE could be due to the fact that different strains had different relative levels of proteins in the complex.
One would expect SusG to interact with the SusC/SusD complex, but none of our assays detected any direct evidence for such a physical interaction. SusG did not cross-link with SusC or SusD, nor was it part of the complex detected on nondenaturing gels. Although these results are all negative, they raise the possibility that SusG is not interacting closely with SusCDEF. It may be that once a starch molecule is bound to the cell surface, it is so large that SusG only has to be nearby. Starch is very tightly bound to the surface of B. thetaitaomciron even in the absence of SusG, so SusG is not needed to tether the polysaccharide to the cell surface.
The B. thetaiotaomicron starch utilization complex appears to be quite different from the cellulosome complex of cellulolytic clostridia, which is also located on the surface of the bacterium. First, enzymes and cellulose binding proteins in the cellulosome complex are attached to a scaffolding protein (scaffoldin), and it seems that there are no interactions among the enzymes and cellulose binding proteins (3). However, the proteins in the starch utilization system seem to interact with each other. Second, the cellulosome complex is anchored on the cell surface and protrudes from the cell surface by 100 to 500 nm (16). In contrast, the proteins of the Bacteroides starch utilization system are embedded in the outer membrane.
ACKNOWLEDGMENTS
We thank Joseph Shipman for excellent technical advice on the proteolytic sensitivity experiments.
This work was supported by grant AI/GM 17876 from the National Institutes of Health.
REFERENCES
- 1.Anderson K L, Salyers A A. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch-binding sites and periplasmic starch-degrading enzymes. J Bacteriol. 1989;171:3192–3198. doi: 10.1128/jb.171.6.3192-3198.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson K L, Salyers A A. Genetic evidence that outer membrane binding of starch is required for starch utilization by Bacteroides thetaiotaomicron. J Bacteriol. 1989;171:3199–3204. doi: 10.1128/jb.171.6.3199-3204.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer E A, Chanzy H, Lamed R, Shoham Y. Cellulose, cellulases and cellulosome. Curr Opin Struct Biol. 1998;8:548–557. doi: 10.1016/s0959-440x(98)80143-7. [DOI] [PubMed] [Google Scholar]
- 4.D'Elia J N, Salyers A A. Contribution of a neopullulanase, a pullulanase, and an α-glucosidase to growth of Bacteroides thetaiotaomicron on starch. J Bacteriol. 1996;178:7173–7179. doi: 10.1128/jb.178.24.7173-7179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D'Elia J N, Salyers A A. Effect of regulatory protein levels on utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol. 1996;178:7180–7186. doi: 10.1128/jb.178.24.7180-7186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D. Studies on the transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 7.Kotarski S F, Salyers A A. Isolation and characterization of outer membranes of B. thetaiotaomicron grown on different carbohydrates. J Bacteriol. 1984;158:102–109. doi: 10.1128/jb.158.1.102-109.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laemmli U K. Cleavage of structure proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 9.Lederberg E M, Cohen S M. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974;119:1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reeves A R, D'Elia J N, Salyers A A. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J Bacteriol. 1996;178:823–830. doi: 10.1128/jb.178.3.823-830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeves A R, Wang G-R, Salyers A A. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol. 1997;179:643–649. doi: 10.1128/jb.179.3.643-649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salyers A A, Vercellotti J R, West S H E, Wilkins T D. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl Environ Microbiol. 1977;33:319–322. doi: 10.1128/aem.33.2.319-322.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shipman J A, Berleman J E, Salyers A A. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J Bacteriol. 2000;182:5365–5372. doi: 10.1128/jb.182.19.5365-5372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shipman J A, Cho K H, Siegel H A, Salyers A A. Physiological characterization of SusG: an outer membrane protein essential for starch utilization by Bacteroides thetaiotaomicron. J Bacteriol. 1999;181:7206–7211. doi: 10.1128/jb.181.23.7206-7211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shoemaker N B, Getty C, Guthrie E P, Salyers A A. Regions in Bacteroides plasmids pBFTM10 and pB8–51 that allow Escherichia coli-Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J Bacteriol. 1986;166:959–965. doi: 10.1128/jb.166.3.959-965.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoham Y, Lamed R, Bayer E A. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 1999;7:275–281. doi: 10.1016/s0966-842x(99)01533-4. [DOI] [PubMed] [Google Scholar]
- 17.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 18.Tancula E, Feldhaus M J, Bedzyk L A, Salyers A A. Location and characterization of genes involved in binding of starch to the surface of Bacteroides thetaiotaomicron. J Bacteriol. 1992;174:5609–5616. doi: 10.1128/jb.174.17.5609-5616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]