Abstract
In this study, we addressed the effects of N limitation in Bradyrhizobium japonicum for its association with soybean roots. The wild-type strain LP 3001 grew for six generations with a growth rate of 1.2 day−1 in a minimal medium with 28 mM mannitol as the carbon source and with the N source [(NH4)2SO4] limited to only 20 μM. Under these conditions, the glutamine synthetase (GS) activity was five to six times higher than in similar cultures grown with 1 or 0.1 mM (NH4)2SO4. The NtrBC-inducible GSII form of this enzyme accounted for 60% of the specific activity in N-starved rhizobia, being negligible in the other two cultures. The exopolysaccharide (EPS) and capsular polysaccharide (CPS) contents relative to cell protein were significantly higher in the N-starved cultures, but on the other hand, the poly-3-hydroxybutyrate level did not rise in comparison with N-sufficient cultures. In agreement with the accumulation of CPS in N-starved cultures, soybean lectin (SBL) binding as well as stimulation of rhizobial adsorption to soybean roots by SBL pretreatment were higher. The last effect was evident only in cultures that had not entered stationary phase. We also studied nodC gene induction in relation to N starvation. In the chromosomal nodC::lacZ fusion Bj110-573, nodC gene expression was induced by genistein 2.7-fold more in N-starved young cultures than in nonstarved ones. In stationary-phase cultures, nodC gene expression was similarly induced in N-limited cultures, but induction was negligible in cultures limited by another nutrient. Nodulation profiles obtained with strain LP 3001 grown under N starvation indicated that these cultures nodulated faster. In addition, as culture age increased, the nodulation efficiency decreased for two reasons: fewer nodules were formed, and nodulation was delayed. However, their relative importance was different according to the nutrient condition: in older cultures the overall decrease in the number of nodules was the main effect in N-starved cultures, whereas a delay in nodulation was more responsible for a loss in efficiency of N-sufficient cultures. Competition for nodulation was studied with young cultures of two wild-type strains differing only in their antibiotic resistance, the N-starved cultures being the most competitive.
The environments where most prokaryotic species are found in nature are often limited in nutrients, with a changing composition in both space and time. Many adaptations to these conditions are known, some of which result from high-affinity uptake systems, differentially expressed enzymes, and a variety of metabolic control mechanisms (27). Amidst bacterial processes that depend on the relative scarcity of one macronutrient, atmospheric N2 fixation in N-poor environments has caused considerable interest for more than a century (11, 39).
Many species of the family Rhizobiaceae are outstanding in that they fix N2 only in symbiosis with legume plants. This interaction starts in the soil with a specific plant-rhizobium molecular signal exchange involving plant flavonoids released into the root exudates, which induce the expression of the nod operons in the rhizobia. These genes encode the enzymes for the biosynthesis and release of a lipochitooligosaccharide known as Nod factor, which triggers nodule development in the inner root cortex with no requirement for the presence of active rhizobia (38). The nodule will provide the rhizobia with the nutrients and the microaerobic environment required for N2 fixation (38). In parallel with plant nodule organogenesis, rhizobia are chemoattracted to the root surface (21), attach to it in nonspecific (48) as well as in bacterial lectin-mediated specific (22, 29) ways, penetrate the root hairs, forming characteristic structures known as infection threads, and finally invade the developing nodule (49), where they subsequently differentiate into bacteroids, a distinct rhizobial form which is the only one able to reduce atmospheric N2 (38).
The process of infection and bacteroid differentiation is strongly dependent on the structure of the bacterial surface polysaccharide, which seems to play a role not only in recognition of rhizobia by the plant, but also as a signal to prevent the elicitation of plant defense activities arising against bacterial penetration (2, 24). In addition, some plant lectins, such as soybean lectin (SBL), are released into the rhizosphere (52, 55) and specifically stimulate rhizobial adsorption and infection (30, 55). Although the mechanism of plant lectin action remains obscure, its role in restricting rhizobium-host specificity range was demonstrated in studies with transgenic plants (15, 52).
Therefore, rhizobial adsorption, root hair infection, nodule formation, and nitrogen fixation are key steps of a complex process, each one contributing to a different level of symbiotic recognition and effectiveness. Throughout this process soil nitrogen sources like nitrate and ammonia (hereafter referred to as combined nitrogen) are required in limited amounts. It is well known that legumes possess a systemic regulatory control able to detect the presence of combined nitrogen in the rhizosphere and block nodulation in response (45), although the identity of the plant messenger that controls nodulation remains to be elucidated (47). Moreover, when combined nitrogen is administered to already nodulated plants, established nodules undergo senescence faster (33).
Other studies addressed the effect of changes in combined nitrogen availability from the rhizobial side. In Bradyrhizobium japonicum and Sinorhizobium meliloti the induction of nod genes is inhibited by high amounts of combined nitrogen (17, 56). On the other hand, induction of nod genes under N starvation conditions was weaker in mutants with mutations in the NtrBC two-component regulatory system of both species, although the extent to which this global regulatory system actually modulates nod gene expression was not fully established (56). Nodulation and nitrogen fixation were severely impaired in Rhizobium etli when bacterial N assimilation was artificially increased by introducing the Escherichia coli glutamate dehydrogenase gene (34). In addition, it was reported that N-limited R. etli formed more infection threads in common beans than either C-limited or exponentially growing cells (10). Indeed, the convenience of using a culture medium with a high C/N ratio instead of a low one to get better nodulation is well known in the industry of rhizobial inoculants for legume crops and was also related with an enhanced bacterial ability to initiate root hair infections (26). All these results indicate that, as for the plant counterpart, the presence of combined nitrogen in excess impairs key rhizobial symbiotic activities. On the other hand, limitation of combined nitrogen might stimulate the rhizobia for infection and nodulation (10). Although the latter could have important biotechnological implications, this has been less studied than the effects of N excess.
In B. japonicum, as in the other rhizobia, NH4+ assimilation proceeds through the high-affinity glutamine synthetase (GS)-glutamate synthase (GOGAT) cycle (12). In addition, these rhizobacteria possess two different GSs. GSI, encoded by the glnA gene, is similar to other prokaryotic GSs except that its synthesis is not regulated by the two-component system NtrBC, its activity being controlled by adenylyl transferase-dependent adenylylation. GSII, encoded by the glnII gene, is similar to the chloroplastic GS, and its synthesis is under the control of NtrBC (12, 32). When N supply is sufficient, GSI is partially active, depending on the number of adenylylated (inactive) subunits per GSI dodecamer. As cell demand for N compounds increases, GSI deadenylylates, and GSII synthesis is induced.
Both control systems respond to the relative amounts of 2-oxoglutarate and glutamine inside the cell (12, 34, 40). A high 2-oxoglutarate/glutamine ratio signals a limitation of N precursors, and thus deadenylylation of GSI and synthesis of GSII are promoted in order to increase NH4+ assimilation. These two control levels operating on different GS enzymes might help the rhizobia become highly efficient for scavenging, in their natural environments, the low levels of N sources that are needed to establish a productive nitrogen-fixing symbiosis, as already mentioned. On the other hand, when bacterial growth is N-limited, the relative C/N imbalance causing an abundance of C nutrients often results in changes of carbon fluxes, favoring the synthesis of polysaccharides and/or polyhydroxyalkanoates (31, 57). Since certain surface polysaccharides are involved in infection and nodulation efficiency, it is important to establish whether N-limiting conditions could favor their accumulation. In addition, this understanding could provide additional insight into the coordination of the C and N metabolic fluxes in rhizobia.
Here we present our study about the effects of N limitation on C accumulation in B. japonicum and its relation to the enhancement of its symbiotic association with soybean.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. japonicum LP 3001 and LP 3004 are Nod+ Fix+ spontaneous derivatives, spectinomycin (Sp) resistant and streptomycin (Sm) resistant, respectively, of strain E111 (29). Bj110-573 is a tetracycline (Tc)-resistant nodC::lacZ chromosomal fusion (16), kindly provided by Michael Göttfert (Dresden, Germany). Antibiotics were used at the following concentrations: Sp, 300 mg liter−1; Sm, 400 mg liter−1; Tc, 100 mg liter−1; cycloheximide (Ch), 50 mg liter−1. Bacteria were maintained on yeast extract-mannitol (YM) medium (54) with 30% (vol/vol) glycerol at −80°C.
For routine use, bacteria from the stocks were grown at 28°C in solid YM. For each experiment, a single rhizobial colony was cultured in 10 ml of liquid Götz minimal medium (19) with 28 mM mannitol as the carbon source and 1 mM (NH4)2SO4 as the nitrogen source and grown at 28°C on a rotary shaker at 180 rpm for a week. After this period, the culture was diluted 1:100 in fresh Götz medium and allowed to grow again for an additional 3 days under the same conditions. Next, this starter culture was diluted 1:50 in Erlenmeyer flasks containing a volume of the medium to be assayed equal to 20% of their capacity, and growth was continued in the same conditions for the indicated periods. These media were prepared by modifying the original Götz recipe, varying the N source, (NH4)2SO4, added as follows: GN1 medium contained 1 mM [the original (NH4)2SO4 concentration in the Götz medium]; GN10 contained 10 mM, GN0.1 contained 0.1 mM, and GN0 was prepared without N source addition. In other media, different nutrients were varied as follows: in GM56, the mannitol concentration (56 mM) was doubled, while in GCa1 (1 mM CaCl2) and GMg10 (10 mM MgSO4), 10 times higher nutrient concentrations were used. Growth was monitored by measuring the total biomass as optical density at 500 nm (OD500) and viable CFU, estimated by plate counts in solid YM from appropriate serial dilutions.
Determination of GS activity.
GSI and GSII activities were assayed with permeabilized cells (3). Cultures (300 ml) were halted by the addition of 3 ml of a 1% (wt/vol) solution of hexadecyltrimethylammonium bromide (CTAB). Cells were harvested by filtration through a 0.2-μm-pore membrane, washed once with 1% (wt/vol) KCl, and resuspended in 3 ml of 1% KCl. GS was measured by its γ-glutamyl transferase (γ-GT) activity (3). To discriminate between GSI and GSII, the activity was measured before and after a 1-h incubation at 50°C, which irreversibly inactivates GSII (14). The adenylylation state of GSI was determined by inhibiting the activity of adenylylated subunits in the presence of 60 mM MgCl2, and the mean number of adenylylated subunits was calculated by comparison with the activity measured in the absence of MgCl2 (46). Specific activity was expressed as nanomoles of γ-glutamyl hydroxamate produced per minute per milligram of protein (3).
Polysaccharide preparations.
Cultures (300 ml) of B. japonicum 3001 were centrifuged at 12,000 × g for 40 min. The supernatant was used for exopolysaccharide (EPS) preparation, and the cell pellet was used for the capsular polysaccharide (CPS). The EPS was precipitated with 3 volumes of ethanol 96% at −20°C, and after resuspension in 0.5 M NaCl, it was stored at −20°C for further analysis. To prepare the CPS (36), pelleted bacteria were washed with 0.5 M NaCl and shaken in a Beckman Omnimixer for 30 s at mean power. This suspension was centrifuged for 15 min at 14,000 rpm in a microcentrifuge, the supernatant was stored at −20°C, and the cell pellet was reserved for cell protein determination. EPS and CPS were viewed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by Alcian blue-silver staining (43).
Analytical determinations.
Protein concentration was determined as described (9) in supernatants from cells disrupted by ultrasonic treatment with a Sanyo Soniprep 150 using three 10-s pulses at mean power. Polysaccharides were determined with 0.2% (wt/vol) anthrone in 95% H2SO4, as described (51), using glucose as the standard. EPS was determined in the above-described culture supernatants, and CPS was quantified in the Omnimixer-shaken cell supernatants.
Fourier-transformed infrared spectroscopy (FTIR) analysis was carried out as follows: 200 μl of each bacterial sample was applied to an Si plate and dried at 50°C for 1 h. Transmission infrared absorption spectra were recorded between 4,000 and 500 cm−1 on a Spectrum One spectrometer (Perkin Elmer). The spectral resolution was 2 cm−1. To improve the signal-to-noise ratio, 500 scans were measured and averaged for each sample. Three replicates were made for each sample. The data were processed using the Perkin Elmer spectrum software.
nodC expression.
Strain Bj110-573 was cultivated as described above, and at the appropriate times, the culture was divided into 2-ml aliquots. For induction, genistein was added at a final concentration of 2 μM (1), and incubation was continued at the same temperature and shaking conditions for an additional 12 to 14 h. Then the cells were permeabilized with 0.1% (wt/vol) SDS and chloroform (35), and the β-galactosidase activity was determined using chlorophenol red galactopyranoside (CPRG) as the substrate (1). Activity is expressed in Miller units (35) except that CPRG hydrolysis was quantitated using absorbance at 574 nm.
SBL binding.
SBL was isolated from soybean Promax seeds (kindly provided by A. Perticari, INTA, Argentina) by affinity column chromatography on ε-amino-caproyl-N-acetyl-d-galactosamine agarose; its purity was evaluated as described (30), and an aliquot from this preparation was labeled with fluorescein isothiocyanate (FITC) (5). Rhizobia were incubated with 60 μg of FITC-SBL ml−1 for 30 min at 28°C without shaking. The percentage of fluorescent cells was recorded by fluorescence/phase-contrast microscopy in a Neubauer chamber as described (5).
Plant experiments.
Soybean Promax seeds were surface sterilized by immersion in 96% ethanol for 5 s and then in 20% (vol/vol) commercial bleach for 10 min, followed by six sterile distilled-water washes. Seeds were germinated on aqueous agar (1.5%, wt/vol) in darkness (29).
For adsorption experiments, the procedure already described was followed (30). Briefly, 10 4-day-old soybean plants were incubated for 1 h at 28°C and 50 rpm shaking in 50 ml of N-free Fåhraeus solution (18) with approximately 104 SBL-treated B. japonicum 3001 per ml. SBL treatment was performed by incubating the rhizobia for 12 h in Fåhraeus solution with or without SBL (10 μg ml−1). After incubation, the roots were washed to remove loosely adsorbed rhizobia. Next, the roots were embedded in solid YM supplemented with Sp and Ch to allow the firmly adsorbed rhizobia to develop microcolonies, which were counted with the aid of a dissecting microscope. Total counts of visible microcolonies on all primary roots, expressed as the percentage of the total number of CFU present during the incubation, represent the adsorption index (%A). Stimulation of adsorption is the difference in %A between protein-treated and control rhizobia as a percentage of the %A of control rhizobia (30, 55).
Nodulation profiles were obtained by inoculating 42 plants with approximately 5 × 104 rhizobia plant−1 in plastic growth pouches watered with a modified N-free Fåhraeus solution (6, 30). The nodule distribution along the primary roots was recorded after 10 days of growth in the greenhouse at 26°C/18°C day/night temperatures, with respect to the smallest emergent root hairs and the root tip positions at the time of inoculation. The distance between smallest emergent root hairs and root tip for each plant is the relative distance unit (RDU). Therefore, all the nodule positions in individual plants were expressed in RDU to compensate for the different elongation rates and magnitudes (in millimeters) of the RDU among individual plants (6).
Competitiveness was assayed using 1:1 mixtures of LP 3001 and LP 3004 grown in the above-described conditions and diluted to approximately 104 (low inoculum) or 106 (high inoculum) rhizobia of each strain ml−1 in bottles containing 800 ml of the modified N-free Fåhraeus plant nutrient solution (30). The rhizobium-containing solution was added to 1.5-liter vermiculite pots. Next, three soybean plantlets were aseptically transferred to each pot and left in the greenhouse, with watering as required. After 25 days, nodules from nine plants per treatment were excised and surface sterilized for 5 min with 20% (vol/vol) commercial bleach, followed by six washes with sterile distilled water. Surface-sterilized nodules were crushed individually, and their contents were plated onto YM replica plates with Ch and selective antibiotics to differentiate the strain occupying each nodule (30).
Nodule occupancy was then defined as the proportion of nodules occupied by each strain, inferred from the proportion of positive growths in the presence of each antibiotic. Nodules from which growth with both antibiotics was observed were considered to be occupied by both strains and therefore were counted twice. Since strains LP 3001 and LP 3004 are isogenic and did not differ in their competitiveness for nodule occupation when grown in the same conditions, statistical analysis was carried out by analysis of variance (8), and significant differences are referred to a 1:1 nodule occupancy as the null hypothesis.
RESULTS
Physiological and symbiotic characterization of B. japonicum LP 3001 grown in N-limited Götz media.
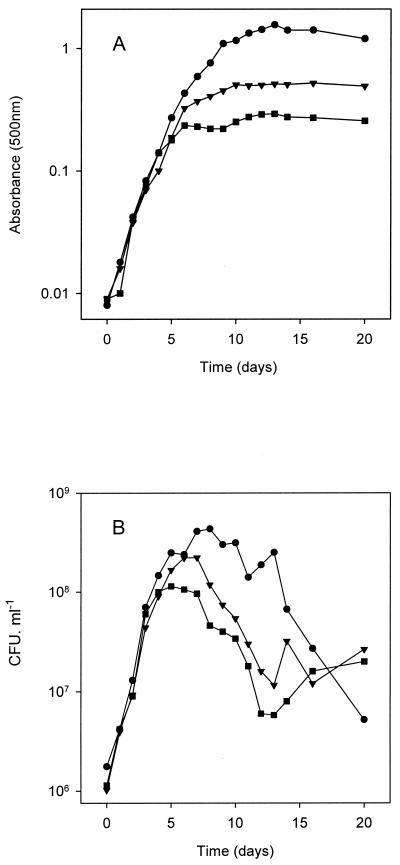
Growth of strain LP 3001 in GN0, GN0.1, and GN1 in batch cultures was studied by recording the OD500 and viable cell concentration every 24 h over a 20-day period. The resulting growth curves are shown in Fig. 1, being representative of three independent experiments with essentially the same results. During the first 5 days, all three cultures were in the exponential phase of growth despite the different N availability; afterwards the cultures entered the stationary phase: GN0 at day 6, GN 0.1 at day 10, and GN1 at day 13. After these time periods, the OD500 remained constant, although at higher readings for GN1 (around 1.42) than for GN0.1 (0.51) and GN0 (0.28), reflecting the N limitation for growth (Fig. 1A).
FIG. 1.
Growth of B. japonicum LP 3001 in GN0 (squares), GN0.1 (triangles), and GN1 (circles). (A) Biomass, as estimated by optical density at 500 nm. (B) Viable cell counts.
In a separate experiment, we evaluated whether N or some other factor was also limiting for growth in GN1. For this purpose, we grew LP 3001 cultures in GN10, GMg10, GCa1, and GM56, i.e., Götz media 10 times more concentrated in its N, Mg, or Ca source or two times more concentrated in its C source, keeping all the other constituents constant. In two different readings, the OD500 remained constant for GN10, GMg10, and GM56 with respect to the control grown in GN1; however, the OD500 readings for GCa1 were 1.8-fold higher than for the control in GN1 at late log phase (8 days of growth) and 1.5-fold greater at late stationary phase (20 days of growth), indicating that of the above four medium components, Ca was the limiting nutrient for growth in GN1.
Survival, as measured through the CFU counts, started to decrease when the cells began entering stationary phase (Fig. 1B). Total CFU drop could not be attributed to cell clumping, since clumps with more than three cells accounted for less than 2% of total units all during the growth period, as observed by microscopic inspection and in agreement with our previous results (29). The maximal CFU produced by each culture was 4.3 × 108 for GN1 at day 8, 2.2 × 108 for GN0.1 at day 7, and 1.1 × 108 for GN0 at day 5. Loss of survival was more pronounced in GN1, which showed a constant decay trend. On the other hand, culture survival in GN0 and GN0.1 was rather similar, diminishing during the first 6 days after entry into stationary phase with a similar slope as GN1, but then stabilizing during the next 10 days. At the end of the experiment, these later cultures had nearly 10-fold more viable bacteria than the GN1-grown cultures. We also observed that in the GN1 cultures, the pH diminished to 5.5 by 10 to 20 days of growth, while it remained relatively constant, 6.5, for GN0 and GN0.1. However, these differences in pH cannot explain the loss of viability in GN1, since this strain is able to grow within this pH range.
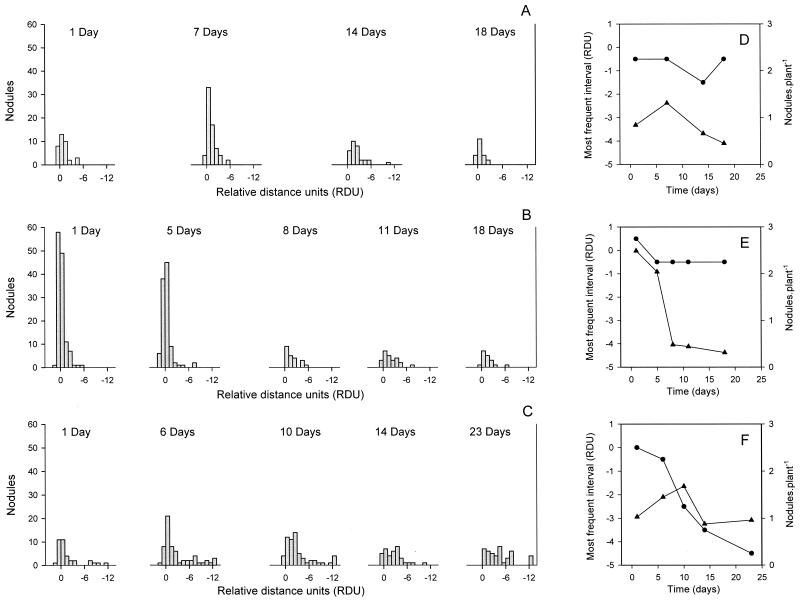
The nodulation profiles produced by B. japonicum depend on the culture age of the inoculum (7). Hence, we evaluated how the different media influenced this symbiotic parameter. We observed that in all three media, the younger cultures nodulated faster and produced more nodules than the older ones (Fig. 2A to C). While N-limited GN0 and GN0.1 cultures formed fewer nodules but with similar frequency distributions along the primary roots as the culture aged (Fig. 2D and E), in older GN1 cultures the frequency peak was displaced to younger regions of the primary roots, while the average number of nodules diminished less than in the N-limited cultures (Fig. 2F). In addition, cultures grown in GN0 or GN0.1 seemed to nodulate more efficiently than the GN1 culture, although this interpretation must be considered carefully, since each culture medium was tested in independent experiments, and thus the result is subjected to the influence of other variables such as the set of plants used or the daily light intensity received by the plants in the greenhouse (see below).
FIG. 2.
Nodulation profiles produced in soybean primary roots by B. japonicum LP 3001 grown for the indicated times (days) in GN0 (A and D), GN0.1 (B and E), or GN1 (C and F). In A, B, and C, the distribution of the nodules along the primary roots of 42 plants is shown as the number of nodules within intervals of 1 RDU from the root tip (RT) mark made at inoculation. The zero value corresponds to the root tip position; negative values represent those parts of the roots which grew after inoculation. In D, E, and F, the RDU interval where most nodules appeared (most frequent interval, circles) and the mean number of nodules per primary root, including those with zero nodules (triangles), are shown as a function of the culture age in days.
Taking the above results into account, we chose two different chronological ages for bacterial growth in all three media to continue our studies: 5 and 14 days old. At 5 days, cultures in GN0 were in late exponential phase and about to reduce their growth rate as a consequence of N limitation, while those in GN0.1 were probably in a much earlier state of N limitation, and those in GN1 were not limited at all (Fig. 1 and see below). On the other hand, this growth time should allow enough generations to be spanned so as to dissipate any physiological influence of the previous growth in GN1 carried over from the starter culture. In addition, all three cultures were around their optimal physiological state for root infection and nodulation (Fig. 2). At 14 days, all three cultures were in stationary phase under their respective nutrient limitation, and their viabilities were similar (Fig. 1B). Nevertheless, at this stage all three cultures already displayed a reduced nodulation efficiency in comparison with their respective exponential-phase cultures (Fig. 2).
Effects of N limitation on activity of GS and on carbon sink.
While 14-day-old GN0 and GN0.1 cultures were clearly N limited, up to 5 days of growth the cultures in all three media were in exponential phase (Fig. 1). This suggests that whatever the amount of N added to the medium, even a small amount of NH4+ carried over from the starter culture, estimated to be less than 40 μM final concentration in GN0, was enough to sustain growth for these first 5 days at a rate of 1.2 day−1 (Fig. 1). This growth rate agrees with previous reports of B. japonicum growth with mannitol as the carbon source (25). Rhizobia are able to satisfy their N needs in very scarce environments by means of their two GS enzymes (12), which together form a highly efficient N-assimilating system. Regulation is achieved by sensing the 2-oxoglutarate:glutamine ratio, which is indicative of the C/N relationship inside the cell. Therefore, similar growth rates of young cultures in GN0, GN0.1, and GN1 could be obtained through this metabolic control of N assimilation.
We studied GS activity in the 5- and 14-day-old LP 3001 cultures in GN0, GN0.1, and GN1 through the GS γ-GT specific activity (3). This activity can at the same time reveal the level of GSI adenylylation (in the presence of 60 mM Mg2+ adenylylated GSI is inhibited [46]) and the amount of activity due to GSII (which is irreversibly inactivated by incubation for 1 h at 50°C [14]). As shown in Table 1, GS γ-GT specific activity was rather similar for 5-day-old GN0.1 and GN1 cultures, while GN0 cultures had five to six times more activity, most of it due to GSII. In 14-day-old cultures, GS activity reflected N limitation: while in GN1 cultures GSI had a mean of 6.7 adenylylated subunits and GSII was at very low levels, in both N-limited cultures the GSI was less adenylylated (a mean of 3.3 adenylylated subunits in GN0.1 and 2.3 in GN0), while GSII was low in GN0.1 but raised in GN0. Thus, total GS activity in the 14-day-old GN0 and GN0.1 cultures was more than double that in GN1.
TABLE 1.
GSI and GSII activities in B. japonicum LP 3001 grown in GN0, GN0.1, and GN1 for 5 or 14 daysa
| Culture age (days) | Culture medium | GS sp act
|
Avg no. of adenylylated GSI subunits | |||
|---|---|---|---|---|---|---|
| Mean nmol of γ-glutamyl hydroxamate min−1 (mg of protein)−1 ± SD
|
% of total
|
|||||
| GSI | GSII | GSI | GSII | |||
| 5 | GN0 | 646.62 ± 33.76 | 1,161.66 ± 60.64 | 35.7 | 64.3 | 1.9 |
| GN0.1 | 275.69 ± 6.73 | 78.00 ± 1.90 | 77.9 | 22.1 | 1.0 | |
| GN1 | 254.60 ± 12.32 | 54.68 ± 2.65 | 82.5 | 17.5 | 1.3 | |
| 14 | GN0 | 1,172.93 ± 89.31 | 278.20 ± 21.2 | 80.8 | 19.2 | 2.3 |
| GN0.1 | 2,153.82 ± 124.97 | 140.98 ± 8.18 | 93.8 | 6.2 | 3.3 | |
| GN1 | 785.76 ± 64.57 | 77.04 ± 6.33 | 91.1 | 8.9 | 6.7 | |
The GS activities were measured as γ-GT specific activities and are also presented as percent of total activity. The average number of adenylylated subunits in GSI was deduced by incubations in the presence and absence of 60 mM Mg2+ (46). All determinations were done in quadruplicate.
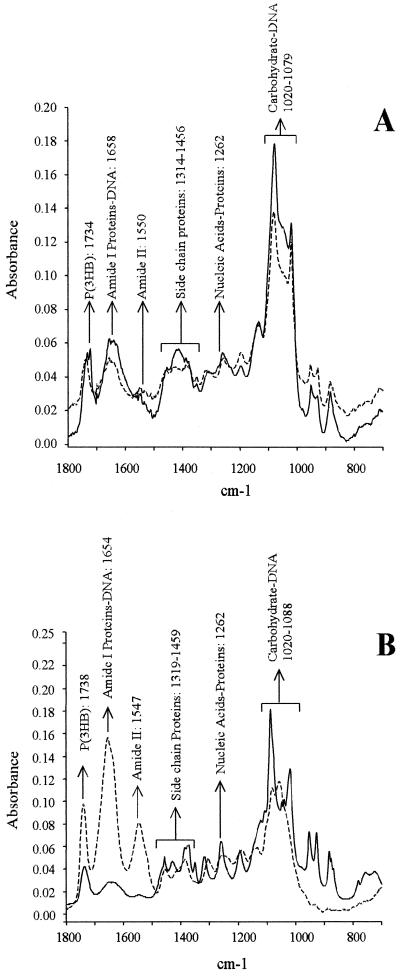
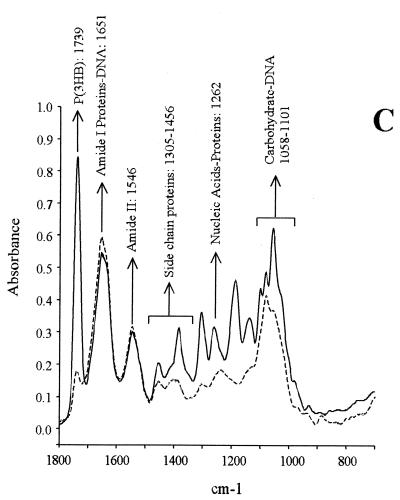
Unbalanced bacterial growth in the presence of an adequate carbon source often leads to an excess in reducing power and energy charge that favors an accumulation of polymers as polysaccharides and/or polyhydroxyalkanoates (31, 57). To study the accumulation of these polymers in B. japonicum, we analyzed whole cells by FTIR. The results (Fig. 3) showed a striking accumulation of polysaccharides relative to cellular proteins for the 5-day-old GN0 and the 14-day-old GN0 and GN0.1 cultures. Interestingly, three distinct peaks were observed after the carbohydrates for these cultures (at 850 to 950 cm−1), which indicated specific compositional changes due to N starvation. The nature of these peaks is currently under investigation. In addition, a sharp peak of poly(3-hydroxybutyrate) [P(3HB)] was observed in the 14-day-old GN1 culture, which accounted for five times more P(3HB) than after 5 days of growth, on the basis of cell protein. On the contrary, the polysaccharide content in the 14-day-old GN1 culture was not significantly different than in exponential phase. P(3HB) content did not increase in GN0 from exponential to stationary phase, whereas the GN0.1 culture was in an intermediate situation for both polysaccharide and P(3HB) increase.
FIG. 3.
FTIR spectra of 5-day-old (dashed lines) and 14-day-old (solid lines) B. japonicum LP 3001 grown in GN0 (A), GN0.1 (B), or GN1 (C).
Certain surface polysaccharides are known to affect symbiotic efficiency in B. japonicum (41). The extracellular polysaccharide can be obtained from the culture supernatant and from the cell surface by vigorous shaking. Although the polysaccharides extracted from these sources are indistinguishable in composition and repeating-unit structure, their location either free in the culture medium or bound to the cell surface led to them being named exopolysaccharide (EPS) and capsular polysaccharide (CPS), respectively (37).
In Table 2, we measured the EPS and CPS production of each culture on the basis of cell protein, which remained at a relatively constant level by OD500 unit, to relate polysaccharide and N accumulation. Both EPS and CPS were produced in significantly higher amounts by the N-starved cultures, namely the 5-day-old GN0 and 14-day-old GN0 and GN0.1. The sum of both polysaccharides was 10 to 20 times higher in these cultures than in the 14-day-old GN1 culture. In addition, the N-sufficient cultures produced more EPS and CPS at exponential growth phase than at stationary phase, in agreement with previous reports (28, 36) but contrary to the N-starved cultures, which tended to accumulate both polysaccharides at stationary phase. On the other hand, no differences were observed in the degree of polymerization for any of these EPS or CPS, as judged by SDS-PAGE (not shown).
TABLE 2.
Extracellular polysaccharide content in B. japonicum LP 3001 cultures in media with different N source concentrations at two ages
| Culture age (days) | Culture medium | Mean polysaccharide (mg/mg of cell protein) ± SD
|
|
|---|---|---|---|
| EPS | CPS | ||
| 5 | GN0 | 4.51 ± 0.55 | 1.78 ± 0.04 |
| GN0.1 | 3.13 ± 0.13 | 0.39 ± 0.05 | |
| GN1 | 1.06 ± 0.17 | 0.62 ± 0.02 | |
| 14 | GN0 | 10.88 ± 0.86 | 4.55 ± 0.26 |
| GN0.1 | 2.96 ± 0.06 | 3.12 ± 0.48 | |
| GN1 | 0.57 ± 0.02 | 0.07 ± 0.01 | |
Taken together, the above results indicated that (i) the 5-day-old GN0 and the 14-day-old GN0 and GN0.1 cultures were under N-starving conditions, (ii) at 14 days, the growth in GN0 and GN0.1 was limited by N, (iii) the GN1 cultures did not suffer from N starvation, whose growth at 14 days was not limited by N, and (iv) the 5-day-old GN0.1 culture could be in an early state of N starvation, as indicated by its higher extracellular polysaccharide content. Therefore, in the following sections we will refer to the 5-day-old GN0 and the 14-day-old GN0 and GN0.1 cultures as N-starved, and the term N-limited will be kept for the 14-day-old GN0 and GN0.1 cultures.
Symbiotic performance in relation to N limitation.
CPS contains the binding site for SBL (5), which stimulates adsorption of rhizobia to roots, as well as infection and competition for nodulation (30). Since N starvation strongly increased CPS production (Table 2), we tested whether, under those conditions, SBL binding to rhizobia and its further stimulation of adsorption were also enhanced by N starvation. To this end we incubated the different cultures of strain LP 3001 in the presence of FITC-labeled SBL for at least 30 min (5) and recorded the percentages of fluorescent cells. We also incubated cells for 12 h in the presence of unlabeled SBL and recorded the adsorption index to soybean roots. We observed that SBL binding was strongest to 5-day-old GN0 cells, intermediate to 14-day-old GN0 and GN0.1 cells, and lowest to the other cultures. The adsorption index increased by 276.0% in 5-day-old GN0 rhizobial cells and 58.3% in the 5-day-old GN0.1 culture, but was half-reduced in 14-day-old GN0 or GN0.1 cultures (Table 3). Cell clumping was not significant, as observed under the microscope.
TABLE 3.
SBL binding to and stimulation of adsorption of B. japonicum LP 3001 cultures from media with different N contents harvested at two agesa
| Culture age (days) | Culture medium | Fluorescent cells (%) | Mean adsorption index (%A) ± SD
|
|
|---|---|---|---|---|
| Without SBL | With SBL | |||
| 5 | GN0 | 27.0 (262) | 0.25 ± 0.04 | 0.94 ± 0.12 |
| GN0.1 | 1.5 (303) | 0.24 ± 0.04 | 0.38 ± 0.06 | |
| GN1 | 1.5 (305) | 0.33 ± 0.06 | 0.27 ± 0.03 | |
| 14 | GN0 | 6.5 (395) | 0.61 ± 0.19 | 0.30 ± 0.08 |
| GN0.1 | 8.5 (483) | 0.48 ± 0.08 | 0.26 ± 0.04 | |
| GN1 | 0.7 (706) | 0.11 ± 0.03 | 0.12 ± 0.03 | |
For SBL binding, rhizobia were incubated with 60 μg of FITC-SBL per ml for 30 min, and the percentage of fluorescent cells was recorded in a Neubauer chamber. Values are means from duplicate counts (means of total cells counted are in parentheses). For SBL stimulation of adsorption, rhizobia were incubated with or without (10 μg ml−1) for 12 h. Next, rhizobia were incubated with 10 soybean plants in Fåhraeus solution, and their adsorption indexes (%A) were calculated.
Large amounts of NH4+ in the culture medium prevent the induction of nodD and nodY in B. japonicum (56), and similar results were obtained in Sinorhizobium meliloti (17). To see whether the opposite is also valid, we studied nodC expression in B. japonicum under diverse degrees of N starvation. The strain Bj110-573 was cultured in GN0, GN0.1, or GN1 for 5 or 14 days in the same conditions as described for LP 3001 (reaching similar OD500 values), and nod gene expression was induced by 12 to 14 h in 2-ml culture aliquots with 2 μM genistein (1). Parallel aliquots were incubated in the same conditions without genistein as controls. Furthermore, we quantified the resulting β-galactosidase activity with CPRG as the substrate (1). As shown in Table 4, nodC gene expression was 2.7-fold higher in N-starved 5-day-old cultures, and the same trend was observed for N-limited 14-day-old cultures, where induction in GN1 was very poor, in contrast to GN0 or GN0.1. These results indicate that nodC gene expression was enhanced under N-starving conditions.
TABLE 4.
Induction of a nodC::lacZ fusion in B. japonicum Bj110-573 with 2 μM genisteina
| Culture age (days) | Culture medium | Mean β-galactosidase activity (Miller units) ± SD
|
|
|---|---|---|---|
| Without genistein | With genistein | ||
| 5 | GN0 | 110.3 ± 18.4 | 1,673.6 ± 18.4 |
| GN0.1 | 105.6 ± 22.2 | 605.5 ± 27.8 | |
| GN1 | 75.2 ± 6.8 | 629.0 ± 61.5 | |
| 14 | GN0 | 83.3 ± 5.5 | 594.4 ± 150.0 |
| GN0.1 | 66.7 ± 6.7 | 682.2 ± 103.3 | |
| GN1 | 77.8 ± 10.0 | 121.1 ± 13.3 | |
Cultures were grown in media with different N contents and harvested at two ages. CPRG was used as the substrate.
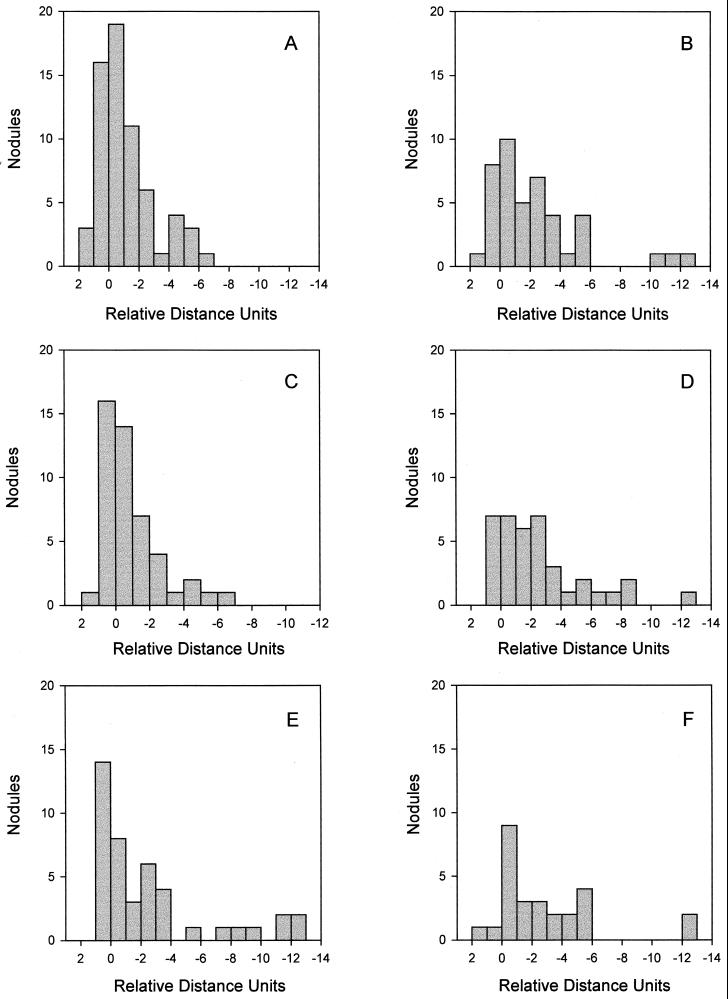
As mentioned before, nodulation efficiency seemed to be higher for young GN0 or GN0.1 cultures, but these results needed to be confirmed simultaneously in a single experiment. This is why we performed this analysis in an experiment with 5- and 14-day-old rhizobia grown in GN0, GN0.1, or GN1 and as shown in Fig. 4, we confirmed that young cultures nodulate faster and produce more nodules than older ones and that GN0- and GN0.1-grown cultures are more efficient in this sense than the GN1-grown for both young and old cultures. As observed before in Fig. 2, in this experiment 14-day-old N-limited cultures gave fewer nodules but with a similar distribution along the root as 5-day-old cultures for the same media. In the case of GN1 cultures, nodule distribution was again more displaced to younger regions of the roots inoculated with the older culture, although more nodules appeared near the root tip mark than in the previous experiment.
FIG. 4.
Nodulation profiles produced in soybean primary roots by B. japonicum LP 3001 grown in GN0 (A and B), GN0.1 (C and D), or GN1 (E and F) for 5 days (A, C, and E) or 14 days (B, D, and F). The distribution of the nodules along the primary roots of 42 plants is shown as the number of nodules within intervals of 1 RDU from the root tip (RT) mark made at inoculation. The zero value corresponds to the root tip position; negative values represent those parts of the roots which grew after inoculation.
Since 14-day-old cultures were the least efficient for early nodulation, we analyzed competitiveness with 5-day-old cultures only. The competition experiment was performed in vermiculite pots as already described (30), inoculating the plants with 1:1 mixtures of LP 3001 and LP 3004 isogenic rhizobia grown either in GN0, GN0.1, or GN1 in all possible combinations, at low (approximately 104 rhizobia ml−1) and high (approximately 106 rhizobia ml−1) inocula. As shown in Table 5, the N-starved GN0-grown rhizobia were significantly more competitive than the others at both inoculum doses. On the other hand, the GN0.1 culture was more competitive than the GN1 culture only at high inoculum doses, i.e., when competition was more stringent.
TABLE 5.
Relationship of nodule occupancies by strains LP 3001 and LP 3004 at low and high inoculum doses, grown in Götz medium with different concentrations of N source for 5 daysa
| Competition assay Growth media and inoculum dose (CFU ml−1) | Nodule occupancy |
|---|---|
| GN0 (3.1 × 104) vs GN0.1 (1.6 × 104) | 1.6:1 |
| GN0 (5.8 × 105) vs GN0.1 (8.0 × 105) | 1.5:1 |
| GN0 (3.1 × 104) vs GN1 (1.7 × 104) | 2.6:1 |
| GN0 (5.8 × 105) vs GN1 (7.8 × 105) | 1.8:1 |
| GN0.1 (1.1 × 104) vs GN1 (1.7 × 104) | 1.1:1 |
| GN0.1 (3.5 × 105) vs GN1 (7.8 × 105) | 2.3:1 |
The results were analyzed by analysis of variance. Relationships of 1.5:1 or more were statistically different than 1:1, with P < 0.01.
DISCUSSION
The ability of B. japonicum to survive and remain infective after months of incubation in deionized water is known (13), as well as its fitness under starvation conditions (23). Here we have shown that growth can be sustained for at least six generations with micromolar amounts of NH4+ as the N source in minimal medium (Fig. 1). On the other hand, the Götz medium, with a 2 mM NH4+ concentration and 28 mM mannitol as the carbon source (GN1), allowed the production of a considerable biomass and viable cells (Fig. 1) and did not produce any symptom of N starvation in B. japonicum. Thus, total biomass could not be increased in this medium by raising the NH4+ concentration 10-fold. In addition, GS, one of the main N-assimilating activities in free-living B. japonicum, was downregulated in stationary-phase GN1 cultures, GSI activity being half-repressed and GSII not being induced (Table 1), indicating a low 2-oxoglutarate/glutamine intracellular ratio (34, 40). Furthermore, extracellular polysaccharides did not rise from exponential to stationary phase in GN1 (Fig. 3 and Table 2), indicating that inside the cell C was not in excess over N.
The state of N starvation in the other cultures was indicated by the control of N assimilation by the GS enzymes, which appeared to be regulated in two consecutive levels. When available N was more than enough to satisfy the cell's needs (as in the 14-day-old GN1 culture), GSI was partially inactivated by adenylylation, and GSII activity was negligible; as the level of available N decreased, the first increase in GS activity was achieved through GSI deadenylylation, and only when N needs were extreme (as in the 5-day-old GN0 culture) was GSII activity significantly induced in addition to GSI complete deadenylylation (Table 1). Thus, total GS activity was significantly higher for N-starved cultures.
When rhizobial cultures enter stationary phase, electron transport could drop, leading to accumulation of carbon and reducing power, which might inhibit 2-oxoglutarate dehydrogenase (44). However, in B. japonicum a bypass through 2-oxoglutarate decarboxylase and succinic semialdehyde dehydrogenase was recently identified (20), whereby under certain circumstances the tricarboxylic acid cycle would be less impaired in both the free-living and bacteroid states. In addition, N starvation precludes protein synthesis, leading to a rise in energy charge if an appropriate C source is present. Here we observed whether those possible excesses in C, reducing power, and/or energy charge were directed to the synthesis of polysaccharides and P(3HB).
In N-limited cultures, the rise in EPS and CPS production with respect to total cell protein was much higher than accumulation of P(3HB). On the other hand, when growth was limited by another nutrient, C was preferentially channeled to P(3HB) synthesis (Fig. 3 and Table 2). In agreement with our results, S. meliloti was also found to produce more EPS than P(3HB) when growth was N limited (50). Since P(3HB) is intracellular, its accumulation should be limited by cell size and also by the kinetics of P(3HB) granule formation (31). Therefore, as the C/N ratio increases, C excess over P(3HB) storage capacity could be channeled to extracellular structures such as CPS and excreted into EPS. Although somewhat obvious, this cell compartmentation of C polymers does not seem to be the only explanation for the difference in C channeling in response to the C/N ratio.
P(3HB) is more reduced than mannitol, CPS, or EPS, and polysaccharide synthesis by overflow metabolism might involve a recycling of carbon through dehydrogenating pathways (42), thus contributing much less to the disposal of excess reducing power than P(3HB) synthesis does. This could indicate that N-limited cultures accumulated less reducing power, probably as a consequence of their lower biomass. In addition, the high NH4+ affinity of the GS enzymes, inferred from the ability of these rhizobia to grow at very low NH4+ concentrations, might allow the glutamate resulting from 2-oxoglutarate transamination to be taken up by the GS-GOGAT cycle at a rate sufficient to continue with NAD(P)H consumption (e.g., for glutamine synthesis) and to prevent, to some extent, the accumulation of acetyl-coenzyme A at stationary phase in N-limited cultures. By contrast, GN1 cultures could accumulate enough glutamate to cause product inhibition of GOGAT-catalyzed transamination because of its high NH4+ availability and low GS activity, which would lead to higher 2-oxoglutarate concentration and less NADPH consumption at this step.
Despite higher polysaccharide accumulation in N-starved cultures, we observed that the degree of EPS and CPS polymerization was the same in all six cultures, suggesting that the influence of N starvation on extracellular polysaccharide production was exerted at early biosynthetic steps.
Differences in EPS and CPS production might at least in part explain the different nodulation profiles produced by the stationary-phase cells (Fig. 2), if we assume that in either case only a subpopulation of the rhizobial cells in the culture are able to infect and nodulate the roots (53). When grown in N-starving conditions, the relative size of this subpopulation seemed to diminish as age increased, although individual cells retained their ability to nodulate fast: the reduction in the mean number of nodules per plant was 66% in GN0 and 87% in GN0.1, while most of the nodules appeared around the position occupied by the root tip at inoculation (Fig. 2D and E). On the contrary, the reduction in the mean number of nodules per plant with N-sufficient rhizobia was only 28% as age increased, but the most frequent position of these nodules was displaced 5 RDU towards the growing root apex (Fig. 2F), suggesting that individual bacterial cells were slower to nodulate. This indicates that these cells could require more time to initiate infections, perhaps because of changes in CPS (and EPS) content and composition that had to take place in the rhizosphere (4).
Cells with higher CPS content might bind more SBL, depending on the composition of these CPS (5). In contrast to previous findings employing N-sufficient cultures, we observed that under N-starving conditions the cultures possessed more EPS and CPS in stationary than in exponential phase (28, 36). Nevertheless, SBL binding was the highest in 5-day-old GN0 cultures (Table 3), suggesting that their CPS had a different composition than those of stationary-phase bacteria for the same media (5). Such a compositional change might have important symbiotic implications because (i) exoB mutants unable to synthesize UDP-galactose, the precursor of the SBL-binding monosaccharide unit, show delayed nodulation and impaired competitiveness (41) and (ii) preincubation of rhizobia in SBL preparations was shown to improve early preinfection and nodulation as well as competitiveness (30).
Stimulation of adsorption by the SBL was strongly enhanced by N starvation only in young cultures (Table 3), but there was some stimulation in the GN0.1 culture. The stimulation value obtained for 5-day-old GN0.1 cells approaches the values we obtained before with yeast extract-mannitol medium (30), which has an approximate total N concentration of 0.3 mM and 56 mM mannitol as the main carbon source. Therefore, we can suggest that SBL stimulation of rhizobial adsorption to soybean roots requires rhizobia grown in N-starving conditions (at least at an early starvation state), in partial correlation with the CPS rhizobial content. On the other hand, a contrary effect was observed in the 14-day-old cultures, where adsorption of the N-limited ones was partially inhibited by SBL binding (Tables 2 and 3). These results were not affected by differential cell clumping, as indicated by direct microscopic observation of the cells, in agreement with our previous results (29, 30). Although different incubation times of rhizobia with SBL were used to study SBL binding and stimulation of adsorption, this result might indicate that beyond or as a consequence of SBL binding, a certain mechanism(s) operative in exponential cells only (55) activated a higher adsorption. The existence of such a mechanism is suggested by the long incubation time required for an enhanced adsorption response (30).
In addition to plant lectin action on adsorption, another important preinfection event is the induction of the nod genes by root-exuded flavonoids. Induction of nodC gene expression by genistein was higher in the N-starved cultures (Table 4). This result adds to previous ones on ammonia inhibition of nodY and nodD gene expression in B. japonicum (56). On the other hand, the small nodC induction observed in the 14-day-old GN1 culture could be related to the inhibitory activity of a recently detected quorum-sensing signal molecule (G. Stacey, B. Zhang, Y. Chen, D. Xu, Y.-H. Lee, C. Bickley, D. Lohar, G. Liao, G. Copley, and J. Loh, Abstr. 13th Int. Congress Nitrogen Fixation, abstr. L36, 2001) which might not accumulate in the less dense GN0 or GN0.1 cultures.
Nodulation efficiency (Fig. 4) and competitiveness (Table 5) were also improved by N limitation. Nodulation profiles of young cultures correlated with SBL stimulation of adsorption, since the GN0 and GN0.1 cultures were the more effective and the only ones in which adhesiveness was stimulated by lectin pretreatment (Fig. 4 and Table 3). However, nodulation efficiency in 14-day-old N-limited cells was higher than in GN1-grown cells (Fig. 4), in contrast to stimulation of adsorption which was lacking in all three 14-day-old cultures (Table 3). The trend for nodulation profiles in stationary-phase cultures correlated better with EPS and CPS content (Table 2) and nodC induction (Table 4), both of which were higher in N-limited cultures. Competition for nodulation correlated well with the level of extracellular polysaccharide content and nodC induction (Tables 2, 4, and 5), the GN0 N-starved culture showing the highest response in these three traits, while the GN0.1 could compete against the GN1 culture only under a stringent competition represented by high inoculum doses. This range of competitive ability correlates with the effects of SBL in stimulating adsorption, in agreement with previous observations (30).
The results presented here indicate that rhizobial N starvation has a positive influence on the symbiosis of B. japonicum with soybean plants, through parallel effects on the EPS and CPS structure (41), nod gene induction (38), and SBL stimulation of adsorption (30), all of which resulted in increased nodulation efficiency and competitiveness.
ACKNOWLEDGMENTS
We are grateful to Alejandra Bosch for obtaining FTIR spectra and Augusto J. L. Pich-Otero for preparing the FTIR graphs. We are also indebted to an anonymous reviewer who suggested new experiments that helped to clarify the changes in EPS and CPS production and to Christina McCarthy for English revision.
This work was supported by the International Foundation for Science grant C/2736–1 to A.R.L. and by Universidad Nacional de La Plata. S.L.L.-G. and T.E.E.V. are supported by Consejo Nacional de Investigaciones Científicas y Técnicas, Argentina. G.F. is Emeritus at Universidad Nacional de La Plata, Argentina, and A.R.L. is a Professional at Comisión de Investigaciones Científicas de la Provincia de Buenos Aires, Argentina.
REFERENCES
- 1.Banfalvi Z, Nieuwkoop A, Schell M, Besl L, Stacey G. Regulation of nod gene expression in Bradyrhizobium japonicum. Mol Gen Genet. 1988;214:420–424. doi: 10.1007/BF00330475. [DOI] [PubMed] [Google Scholar]
- 2.Becker A, Pühler A. Production of expoplysaccharides. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae: molecular biology of model plant-associated bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 97–118. [Google Scholar]
- 3.Bender R A, Janssen K A, Resnick A D, Blumenberg M, Foor F, Magasanik B. Biochemical parameters of glutamine synthetase from Klebsiella aerogenes. J Bacteriol. 1977;129:1001–1009. doi: 10.1128/jb.129.2.1001-1009.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhuvaneswari T V, Bauer D W. Role of lectins in plant-microorganism interactions. III. Binding of soybean lectin to root-cultured rhizobia. Plant Physiol. 1978;62:71–74. doi: 10.1104/pp.62.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhuvaneswari T V, Pueppke S G, Bauer W D. Role of lectins in plant microorganism interactions. I. Binding of soybean lectin to rhizobia. Plant Physiol. 1977;60:486–491. doi: 10.1104/pp.60.4.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhuvaneswari T V, Turgeon B G, Bauer W D. Early events in the infection of soybean (Glycine max L. Merr) by Rhizobium japonicum. I. Localization of infectible root cells. Plant Physiol. 1980;66:1027–1031. doi: 10.1104/pp.66.6.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhuvaneswari B V, Mills K K, Crist D K, Evans W R, Bauer W D. Effects of culture age on symbiotic infectivity of Rhizobium japonicum. J Bacteriol. 1983;153:443–451. doi: 10.1128/jb.153.1.443-451.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Box G E P, Hunter W G, Hunter J S. Statistics for experimenters. New York, N.Y: John Wiley and Sons; 1978. [Google Scholar]
- 9.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 10.Brelles-Mariño G, Boiardi J L. Nitrogen limitation of chemostat-grown Rhizobium etli elicits higher infection-thread formation in Phaseolus vulgaris. Microbiology. 1996;142:1067–1070. doi: 10.1099/13500872-142-5-1067. [DOI] [PubMed] [Google Scholar]
- 11.Burris R H. 100 years of discoveries in biological N2 fixation. In: Bothe H, de Bruijn F J, Newton W E, editors. Nitrogen fixation: hundred years later. Stuttgart, Germany: Gustav Fisher; 1988. pp. 21–30. [Google Scholar]
- 12.Carlson T A, Martin G B, Chelm B K. Differential transcription of the two glutamine synthetase genes of Bradyrhizobium japonicum. J Bacteriol. 1987;169:5861–5866. doi: 10.1128/jb.169.12.5861-5866.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crist D K, Wyza R E, Mills K K, Bauer W D, Evans W R. Preservation of Rhizobium viability and symbiotic infectivity by suspension in water. Appl Environ Microbiol. 1984;47:895–900. doi: 10.1128/aem.47.5.895-900.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darrow R A, Knotts R. Two forms of glutamine synthetase in free-living root-nodule bacteria. Biochem Biophys Res Commun. 1977;78:554–559. doi: 10.1016/0006-291x(77)90214-5. [DOI] [PubMed] [Google Scholar]
- 15.Díaz C L, Spaink H P, Kijne J W. Heterologous rhizobial lipochitin oligosaccharides and chitin oligomers induce cortical cell divisions in red clover roots, transformed with the pea lectin gene. Mol Plant-Microbe Interact. 2000;13:268–276. doi: 10.1094/MPMI.2000.13.3.268. [DOI] [PubMed] [Google Scholar]
- 16.Dockendorff T C, Sanjuan J, Grob P, Stacey G. NolA represses nod gene expression in Bradyrhizobium japonicum. Mol Plant-Microbe Interact. 1994;7:596–602. doi: 10.1094/mpmi-7-0173. [DOI] [PubMed] [Google Scholar]
- 17.Dusha I, de Bruijn F J, Kondorosi A, Schell J. The Rhizobium meliloti early nodulation genes (nodABC) are nitrogen regulated: isolation of a mutant strain with efficient nodulation capacity on alfalfa in the presence of ammonium. Mol Gen Genet. 1989;219:89–96. [Google Scholar]
- 18.Fåhraeus G. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol. 1957;16:374–381. doi: 10.1099/00221287-16-2-374. [DOI] [PubMed] [Google Scholar]
- 19.Götz R, Limmer N, Ober K, Schmitt R. Motility and chemotaxis in two strains of Rhizobium with complex flagella. J Gen Microbiol. 1982;128:789–798. [Google Scholar]
- 20.Green L S, Li Y, Emerich D W, Bergersen F J, Day D A. Catabolism of α-ketoglutarate by a sucA mutant of Bradyrhizobium japonicum: evidence for an alternative tricarboxylic acid cycle. J Bacteriol. 2000;182:2838–2844. doi: 10.1128/jb.182.10.2838-2844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gulash M, Ames P, Larosiliere R C, Bergman K. Rhizobia are attracted to localized sites on legume roots. Appl Environ Microbiol. 1984;48:149–152. doi: 10.1128/aem.48.1.149-152.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho S C, Wang J L, Schindler M. Carbohydrate binding activities of Bradyrhizobium japonicum. I. Saccharide-specific inhibition of homotypic and heterotypic adhesion. J Cell Biol. 1990;111:1631–1638. doi: 10.1083/jcb.111.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Humbeck C, Thierfelder H, Gresshoff P M, Werner D. Competitive growth of slow growing Rhizobium japonicum against fast growing Enterobacter and Pseudomonas species at low concentrations of succinate and other substrates in dialysis culture. Arch Microbiol. 1985;142:223–228. [Google Scholar]
- 24.Kannenberg E L, Reuhs B L, Forsberg L S, Carlson R W. Lipopolysaccharides and K-antigens: their structures, biosynthesis and functions. In: Spaink H P, Kondorosi A, Hooykaas P J J, editors. The Rhizobiaceae: molecular biology of model plant-associated bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 119–154. [Google Scholar]
- 25.Karr D B, Liang R-T, Reuhs B L, Emerich D W. Altered exopolysaccharides of Bradyrhizobium japonicum mutants correlate with impaired soybean lectin binding, but not with effective nodule formation. Planta. 2000;211:218–226. doi: 10.1007/s004250000288. [DOI] [PubMed] [Google Scholar]
- 26.Kijne J W, Smit G, Díaz C L, Lugtenberg B J J. Lectin enhanced accumulation of manganese-limited Rhizobium leguminosarum cells on pea root hair tips. J Bacteriol. 1988;170:2994–3000. doi: 10.1128/jb.170.7.2994-3000.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolter R, Siegele D A, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 28.Law I J, Yamamoto Y, Mort A J, Bauer W D. Nodulation of soybean by Rhizobium japonicum mutants with altered capsule synthesis. Planta. 1982;154:100–109. doi: 10.1007/BF00387901. [DOI] [PubMed] [Google Scholar]
- 29.Lodeiro A R, Favelukes G. Early interactions of Bradyrhizobium japonicum and soybean roots: specificity in the process of adsorption. Soil Biol Biochem. 1999;31:1405–1411. [Google Scholar]
- 30.Lodeiro A R, López-García S L, Vázquez T E, Favelukes G. Stimulation of adhesiveness, infectivity, and competitiveness for nodulation of Bradyrhizobium japonicum by its treatment with soybean seed lectin. FEMS Microbiol Lett. 2000;188:177–184. doi: 10.1111/j.1574-6968.2000.tb09190.x. [DOI] [PubMed] [Google Scholar]
- 31.Madison L L, Huisman G W. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev. 1999;63:21–53. doi: 10.1128/mmbr.63.1.21-53.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin G B, Chapman K A, Chelm B K. Role of the Bradyrhizobium japonicum ntrC gene product in differential regulation of the glutamine synthetase II gene (glnII) J Bacteriol. 1988;170:12. doi: 10.1128/jb.170.12.5452-5459.1988. . 5452–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matamoros M, Baird L M, Escuredo P R, Dalton D A, Minchin F R, Iturbe-Ormaetxe I, Rubio M C, Moran J F, Gordon A J, Becana M. Stress-induced legume root nodule senescence: physiological, biochemical, and structural alterations. Plant Physiol. 1999;121:97–111. doi: 10.1104/pp.121.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendoza A, Leija A, Martínez-Romero E, Hernández G, Mora J. The enhancement of ammonium assimilation in Rhizobium etli prevents nodulation of Phaseolus vulgaris. Mol Plant-Microbe Interact. 1995;8:584–592. doi: 10.1094/mpmi-8-0584. [DOI] [PubMed] [Google Scholar]
- 35.Miller J. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 36.Mort A J, Bauer W D. Composition of the capsular and extracellular polysaccharides of Rhizobium japonicum: changes with culture age and correlations with binding of soybean seed lectin to the bacteria. Plant Physiol. 1980;66:158–163. doi: 10.1104/pp.66.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mort A J, Bauer W D. Application of two new methods for cleavage of polysaccharides into specific oligosaccharide fragments. Structure of the capsular and extracellular polysaccharides of Rhizobium japonicum that bind soybean lectin. J Biol Chem. 1982;257:1870–1875. [PubMed] [Google Scholar]
- 38.Mylona P, Pawlowski K, Bisseling T. Symbiotic nitrogen fixation. Plant Cell. 1995;7:869–885. doi: 10.1105/tpc.7.7.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newton W E. Nitrogen fixation in perspective. In: Pedrosa F O, Hungria M, Yates G, Newton W E, editors. Nitrogen fixation: from molecules to crop productivity. Dordrecht, The Netherlands: Kluwer Academic Publishers; 2000. pp. 3–8. [Google Scholar]
- 40.Ninfa A J, Atkinson M R, Kamberov E S, Feng J, Ninfa E G. Control of nitrogen assimilation by the NRI-NRII two-comoponent system of enteric bacteria. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 67–88. [Google Scholar]
- 41.Parniske M, Kosch K, Werner D, Müller P. ExoB mutants of Bradyrhizobium japonicum with reduced competitiveness for nodulation of Glycine max. Mol Plant-Microbe Interact. 1993;6:99–106. [Google Scholar]
- 42.Portais J-C, Tavernier P, Gosselin I, Barbotin J-N. Cyclic organization of the carbohydrate metabolism in Sinorhizobium meliloti. Eur J Biochem. 1999;265:473–480. doi: 10.1046/j.1432-1327.1999.00778.x. [DOI] [PubMed] [Google Scholar]
- 43.Reuhs B L, Carlson R W, Kim J S. Rhizobium fredii and Rhizobium meliloti produce 3-deoxy-d-manno-2-octolusonic acid-containing polysaccharides that are structurally analogous to group II K antigens (capsular polysaccharides) found in Escherichia coli. J Bacteriol. 1993;175:3570–3580. doi: 10.1128/jb.175.11.3570-3580.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salminen S O, Streeter J G. Factors contributing to the accumulation of glutamate in Bradyrhizobium japonicum bacteroids under microaerobic conditions. J Gen Microbiol. 1990;136:2119–2126. [Google Scholar]
- 45.Schubert S. Nitrogen assimilation by legumes — processes and ecological limitations. Fertil Res. 1995;42:99–107. [Google Scholar]
- 46.Shapiro B M, Stadtman E R. Glutamine synthetase (E. coli) Methods Enzymol. 1970;17A:910–922. [Google Scholar]
- 47.Sheng C, Harper J E. Shoot versus root signal involvement in nodulation and vegetative growth in wild-type and hypernodulating soybean genotypes. Plant Physiol. 1997;113:825–831. doi: 10.1104/pp.113.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smit G, Swart S, Lugtenberg B J J, Kijne J W. Molecular mechanisms of attachment of Rhizobium bacteria to plant roots. Mol Microbiol. 1992;6:2897–2903. doi: 10.1111/j.1365-2958.1992.tb01748.x. [DOI] [PubMed] [Google Scholar]
- 49.Stuurman N, Bras C P, Schlaman H R M, Wijfjes A H M, Bloemberg G, Spaink H P. Use of green fluorescent protein color variants expressed on stable broad-host-range vectors to visualize rhizobia interacting with plants. Mol Plant-Microbe Interact. 2000;13:1163–1169. doi: 10.1094/MPMI.2000.13.11.1163. [DOI] [PubMed] [Google Scholar]
- 50.Tavernier P, Portais J-C, Nava Saucedo J E, Courtois J, Courtois B, Barbotin J-N. Exopolysaccharide and poly-β-hydroxybutyrate coproduction in two Rhizobium meliloti strains. Appl Environ Microbiol. 1997;63:21–26. doi: 10.1128/aem.63.1.21-26.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trevelyan W E, Harrison J S. Studies on yeast metabolism. I. Fractionation and microdetermination of cell carbohydrates. Biochem J. 1952;50:298–310. doi: 10.1042/bj0500298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Rhijn P, Goldberg R B, Hirsch A M. Lotus corniculatus nodulation specificity is changed by the presence of a soybean lectin gene. Plant Cell. 1998;10:1233–1249. doi: 10.1105/tpc.10.8.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vesper S J, Bauer W D. Characterization of Rhizobium attachment to soybean roots. Symbiosis. 1985;1:139–162. [Google Scholar]
- 54.Vincent J M. A manual for the practical study of the root nodule bacteria. IBP handbook No. 15. Oxford, United Kingdom: Blackwell Scientific Publications; 1970. [Google Scholar]
- 55.Wall L G, Favelukes G. Early recognition in the Rhizobium meliloti-alfalfa symbiosis: root exudate factor stimulates root adsorption of homologous rhizobia. J Bacteriol. 1991;173:3492–3499. doi: 10.1128/jb.173.11.3492-3499.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang S-P, Stacey G. Ammonia regulation of nod genes in Bradyrhizobium japonicum. Mol Gen Genet. 1990;223:329–331. doi: 10.1007/BF00265071. [DOI] [PubMed] [Google Scholar]
- 57.Zevenhuizen L P T M. Cellular glycogen, β-1,2-glucan, poly-β-hydroxybuyric acid and extracellular polysaccharides in fast-growing species of Rhizobium. Antonie van Leeuwenhoek. 1981;47:481–497. doi: 10.1007/BF00443236. [DOI] [PubMed] [Google Scholar]