Abstract
Staphylococcus aureus is a gram-positive facultative aerobe that can grow in the absence of oxygen by fermentation or by using an alternative electron acceptor. To investigate the mechanism by which S. aureus is able to adapt to changes in oxygen concentration, we analyzed the transcriptional regulation of genes that encode the aerobic class Ib and anaerobic class III ribonucleotide reductase (RNR) systems that are responsible for the synthesis of deoxyribonucleotides needed for DNA synthesis. The S. aureus class Ib RNR nrdIEF and class III RNR nrdDG genes and their regulatory regions were cloned and sequenced. Inactivation of the nrdDG genes showed that the class III RNR is essential for anaerobic growth. Inhibition of aerobic growth by hydroxyurea showed that the class Ib RNR is an oxygen-dependent enzyme. Northern blot analysis and primer extension analysis demonstrated that transcription of class III nrdDG genes is regulated by oxygen concentration and was at least 10-fold higher under anaerobic than under aerobic conditions. In contrast, no significant effect of oxygen concentration was found on the transcription of class Ib nrdIEF genes. Disruption or deletion of S. aureus nrdDG genes caused up to a fivefold increase in nrdDG and nrdIEF transcription under anaerobic conditions but not under aerobic conditions. Similarly, hydroxyurea, an inhibitor of the class I RNRs, resulted in increased transcription of class Ib and class III RNR genes under aerobic conditions. These findings establish that transcription of class Ib and class III RNR genes is upregulated under conditions that cause the depletion of deoxyribonucleotide. Promoter analysis of class Ib and class III RNR operons identified several inverted-repeat elements that may account for the transcriptional response of the nrdIEF and nrdDG genes to oxygen.
Staphylococcus aureus is a gram-positive facultative aerobe and a major human pathogen (33, 39). In common with other facultative aerobes, S. aureus can grow in the absence of oxygen either by fermentation or by using an alternative terminal electron acceptor, such as nitrate. Several studies suggest that oxygen plays a role in the pathogenesis of S. aureus, in both its capacity to produce virulence factors and its ability to persist and grow in different and often hostile environmental niches (5, 6, 26, 37, 40, 53). The ability of S. aureus to adapt to extreme changes in external oxygen concentration implies the existence of one or more oxygen-sensing systems that regulate the expression of genes in the transition from aerobic to anaerobic growth. While considerable progress has been made in characterizing global regulators of anaerobic gene expression, for example, the FNR and Arc families of proteins (3, 14, 17, 42), relatively little is known about the regulatory systems that operate in S. aureus during anaerobiosis. Knowledge of these signal transduction systems is crucial for understanding how, in S. aureus, oxygen brings about changes in the expression of virulence genes. Several recent in vitro studies indicate that the oxygen concentration can affect the production of virulence factors (52, 53). Thus, the presence of oxygen is necessary for production by S. aureus of toxic shock syndrome toxin 1 through a two-component system, SrrA/SrrB, that is homologous to the ResD/ResE system of Bacillus subtilis that has been implicated in global regulation of aerobic and anaerobic respiratory metabolism. Other studies (7, 8) have shown that anaerobic conditions induce the expression in S. aureus and Staphylococcus epidermidis of ica-specific genes that encode the production of an extracellular polysaccharide, which mediates cell-cell adhesion and biofilm formation and may stimulate pathogenicity in vivo. More generally, anaerobiosis may act as an environmental cue in vivo for the production of virulence factors that enable the pathogen to adapt to low-oxygen tensions (46).
Because an essential feature of all facultative aerobic bacteria is the need to synthesize DNA under aerobic and anaerobic conditions, they must contain genes that determine enzymatic systems, ribonucleotide reductases (RNRs), that reduce all four ribonucleotides to deoxyribonucleotides in the presence or absence of oxygen. Moreover, the expression of these genes is likely to be controlled by one or more oxygen-sensing systems. To date, three classes of RNRs have been described (25). Class I RNRs are aerobic enzymes present in eukaryotes and in many bacteria. They consist of two homodimers present in an α2β2-subunit structure. In the bacterial class Ia reductases, the larger α chain (NrdA) is encoded by the nrdA gene and contains the catalytic site and binding sites for substrates and effectors; the smaller β chain (NrdB) is encoded by the nrdB gene and contains, in its active form, a stable ferric-tyrosyl free radical. In Escherichia coli, the nrdA and nrdB genes form an operon and are cotranscribed in a 3.2-kb mRNA (4, 18). Class Ib RNRs are confined to certain eubacteria. They possess the same α2β2-subunit structure as the class Ia RNRs but share only modest sequence identity and differ in some functional aspects (9, 23). The corresponding subunits in class Ib RNRs are encoded by the nrdE and nrdF genes. In E. coli and Salmonella enterica serovar Typhimurium, the nrdEF genes are transcribed together with two small open reading frames (ORFs), located immediately upstream of nrdE, in an mRNA of ∼4 kb (22). In E. coli, the proximal ORF, termed nrdI, codes for a protein that is reported to stimulate the activity of the class Ib RNR (21); the distal ORF, termed nrdH, functions as a hydrogen donor system with a higher specificity for the class Ib than the class Ia RNR (21). A similar organization of nrdHIEF genes occurs in the gram-positive bacterium Lactococcus lactis (23). Class I RNRs require molecular oxygen for radical formation, and therefore, these enzymes function only under aerobic conditions. Their source of reducing power comes from one or two small proteins, thioredoxin or glutaredoxin, each of which contains a pair of redox-active cysteines; thioredoxin is maintained in its reduced state by thioredoxin reductase, while glutaredoxin is kept reduced by glutathione and glutathione reductase, in both cases at the expense of NADPH. Class II RNRs are oxygen-independent enzymes that use adenosylcobalamin as the cofactor and are limited to some microorganisms.
Class III RNRs are expressed in strict anaerobes and in certain facultative anaerobes during growth under anaerobic conditions (25). The recently described structure of the phage T4 enzyme suggests a common origin for class I and class III RNRs with differences existing in the mechanism of radical initiation and the source of reducing power (32). The class III RNR, termed protein α, is in its active form a dimer and is encoded by the nrdD gene; it contains the active site for binding of substrates and allosteric effectors. The smaller β subunit, encoded by the nrdG gene, is an iron sulfur protein, also known as activase, that catalyzes the one-electron transfer from reduced flavodoxin to S-adenosylmethionine to generate a stable glycyl radical near the carboxy-terminal portion of the larger subunit (49). Exposure of the active complex to oxygen results in cleavage adjacent to the glycyl radical and removal of the carboxy-terminal 25 residues (27). In class III RNRs, formate can serve as the overall reductant and is oxidized to CO2 (35).
The work described in this paper commenced with the assumption that S. aureus contains genes coding for aerobic and anaerobic RNRs and that their expression is regulated in response to oxygen concentration. Inspection of the S. aureus genome databases revealed the presence of two gene clusters, one resembling nrdEF and another resembling nrdDG. Here we report the structural organization of the S. aureus class Ib and class III RNR gene clusters and analyze their transcription in response to changes in oxygen concentration.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The bacterial strains and plasmids used in this study are described in Table 1. S. aureus strains were grown at 37°C in tryptone soy broth (TSB; Difco) and brain heart infusion (Difco) supplemented with erythromycin (12 μg ml−1) and kanamycin (200 μg ml−1) where appropriate. Recombinants were selected on TSB agar plates containing antibiotics. Phage transductions were carried out with φ11 as described previously (36). E. coli was grown in Luria-Bertani medium with the addition of ampicillin (100 μg ml−1) or kanamycin (50 μg ml−1) as needed.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant characteristicsa | Source or reference(s) |
|---|---|---|
| Strains | ||
| E. coli XL-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqlacZΔM15 Tn10(Tcr)] | Laboratory stock |
| S. aureus | ||
| Oxford (NCTC6571) | Wild-type laboratory strain | Laboratory stock |
| RN4220 | Restriction mutant of 8325-4 (UV-cured of prophages) used as primary recipient for plasmids propagated in E. coli; spontaneous mutant with 11-bp deletion in rsbU gene | 16, 28 |
| SH1000 | Derivative of 8325-4 carrying the intact rsbU gene | Gift of S. Foster |
| MMA6 | RN4220 nrdD::(pMM-1) Emr | This study |
| MM1B | RN4220 nrdD::(pMM-3) Emr Kmr | This study |
| MM1C | RN4220 nrdDG::Ωkm-2 Kmr | This study |
| MM1C+ | MM1C (pMM-5) Emr Kmr | This study |
| MSA6 | φ11/MMA6 × SH1000b Emr | This study |
| MS1C | φ11/MM1C × SH1000, Kmr | This study |
| Plasmids | ||
| pUC18 | Cloning vector; lacZ Apr | MBI Fermentas |
| PMUTIN-4 | pUC18-based suicidal vector for gram-positive bacteria; Pspac−lacZ PpenP-lacI Apr Emr | 51 |
| pAUL-A | Temperature-sensitive shuttle vector; lacZ Emr | 43 |
| pBR322::Ωkm-2 | pBR322 carrying the Ωkm-2 cassette; Apr Kmr | 38 |
| pMM-1 | PacI-NarI-digested pMUTIN-4 containing ∼900-bp PacI-NarI PCR-amplified nrdD fragment; Apr Emr | This study |
| pMM-2 | Product of the triple ligation of the PacI-NarI-digested pMUTIN-4, ∼1-kb PacI-EcoRI PCR-amplified 5′ end of nrdD, and ∼1-kb EcoRI-NarI PCR-amplified 3′ end of nrdG fragments; Apr Emr | This study |
| pMM-3 | EcoRI-digested pMM-2 containing the Ωkm-2 cassette; Apr Emr Kmr | This study |
| pMM-4 | PstI-XbaI-digested pUC18 containing ∼2.8-kb PCR-amplified complete nrdDG operon of RN4220; Apr | This study |
| pMM-5 | HindIII-BamHI-digested pAUL-A containing the HindIII-BamHI fragment of pMM-4; Emr | This study |
Abbreviations: Apr, Emr, Kmr, and Tcr, resistance to ampicillin, erythromycin/lincomycin, kanamycin, and tetracycline, respectively.
Transduction with phage φ11 from donor × recipient.
S. aureus aerobic liquid cultures were grown at 37°C in an air orbital shaker at 250 rpm. For limiting oxygen conditions, the standard anaerobic growth conditions used for growth of cultures were agitation at 100 rpm in an orbital shaker at 37°C in TSB medium supplemented with cysteine (5.7 mM) to scavenge traces of oxygen and 0.001% resazurin as a redox indicator. Wheaton serum bottles (100-ml capacity) containing 60 ml of the above-described medium were purged with nitrogen gas for 4 min at a pressure of 0.75 atm prior to being autoclaved. Aerobic cultures were subcultured (0.5 ml) in 60 ml of the above-described medium and grown for 16 to 20 h to stationary phase (optical density at 600 nm [OD600], ∼2), and 2 ml was used to inoculate 60 ml of the same medium. Anaerobic growth of S. aureus colonies on plates was carried out in a sealed anaerobic jar (Oxoid) equipped with an AnaeroGen (Oxoid) sachet and employing an Anaerotest indicator strip (Merck) for verifying anaerobic conditions.
DNA manipulations.
For E. coli, preparation of plasmids, DNA manipulations, and transformation of competent cells were performed as previously described (41). For S. aureus, genomic DNA was prepared as described previously (36). Standard procedures were employed for restriction enzyme digestion, ligation, Southern blotting, and radiolabeling of oligonucleotides (41) unless otherwise stated. The nucleotide sequences of the DNA regions containing the S. aureus Oxford class Ib and class III RNR genes were determined from both strands by the dideoxy procedure with the ABI Prism 377 automatic sequencer (Perkin-Elmer Biosystems) and the Prism dye terminator cycle-sequencing kit (Applied Biosystems).
Construction of nrdDG insertion and deletion mutants.
To create a disruption of the S. aureus nrdD gene, an internal fragment (∼900 bp) of the RN4220 nrdD gene was amplified by PCR, using the forward primer (with an added PacI restriction site) 5′-GCTGTTAATTAAGAACAACATAGAAATATAG-3′ and the reverse primer (with an added NarI restriction site) 5′-TGAGGGCGCCCTGTAAATACTGAACCAAATG-3′, and ligated into the PacI-NarI-digested integration vector pMUTIN-4 to generate the plasmid pMM-1. After electroporation into S. aureus RN4220 (44), pMM-1 is expected to undergo a single reciprocal crossover event with the host genome, resulting in the insertion of the plasmid in the chromosomal nrdD gene. Transformants were selected for on TSB plates containing erythromycin (5). Integration of pMM-1 into the nrdD gene was confirmed in one transformant, termed MMA6, by Southern blot analysis, PCR, and DNA sequencing.
To obtain an internal deletion within the RN4220 nrdDG genes, a fragment containing 951 bp of the 5′ untranslated region of the nrdD gene and 50 bp of the upstream region (lacking the −35 site of the promoter region) was amplified by PCR using the forward primer (with an added PacI restriction site) 5′-GGGGTTAATTAAGTGGTATAAAGTAATGAGTAG-3′ and the reverse primer (with an added EcoRI restriction site) 5′-AAAAGAATTCAGTGTAACAACACCAAGATTAC-3′, and a fragment containing 309 bp of the 3′ portion of the nrdG gene and 650 bp of the downstream region was amplified by PCR using the forward primer (with an added EcoRI restriction site) 5′-TTTTGAATTCTGGGCTAAGTCTATTAGGTGG-3′ and the reverse primer (with an added NarI restriction site) 5′-CCCCGGCGCCATTAATACCAGTGATGATATC-3′. The two fragments were ligated with the ∼3.8-kb PacI-NarI fragment of pMUTIN-4 to give pMM-2 in E. coli XL1 Blue. The plasmid was cut with EcoRI and ligated with the 2.27-kb Ωkm-2 cassette (38) to give pMM-3. The resulting plasmid was electroporated into S. aureus RN4220, and transformants were selected for on TSB plates containing erythromycin and kanamycin. The expected single integration event between pMM-3 and the host chromosomal region was confirmed by PCR, and the strain was termed MM1B. To select for segregation of the wild-type nrdDG alleles, one transformant was propagated for 200 generations in TSB liquid medium containing kanamycin but lacking erythromycin and plated on TSB plates containing kanamycin, and the colonies were screened for loss of the erythromycin marker. Several kanamycin-resistant, erythromycin-sensitive clones were isolated, and one, termed MM1C, was shown by PCR and DNA sequencing to have the expected replacement of the∼1.1-kb internal portion of the nrdDG genes by the kanamycin cassette. The nrdD disrupted mutation (A6) and the nrdDG deletion mutation (1C) were introduced into S. aureus SH1000 (rsbU+) by φ11 phage transduction, and their presence was verified by PCR to give MSA6 and MS1C, respectively.
Complementation of nrdDG mutants.
For complementation of nrd mutants, an ∼2.8-kb DNA fragment containing the promoter and structural coding regions of the nrdDG genes was amplified by PCR, using a forward primer with an added PstI restriction site and a reverse primer with an added XbaI restriction site, and cloned into the vector pUC18 cut with PstI and XbaI. The resulting plasmid, pMM-4, was cut with HindIII and BamHI, and the fragment was ligated into the E. coli shuttle vector pAUL-A (43) to give pMM-5. S. aureus strain RN4220 containing the deletion-kanamycin cassette substitution mutation (1C) of the chromosomal nrdDG genes was electroporated with pMM-5, and transformants were selected for on erythromycin plates. One of the transformants, termed MM1C+, was tested for the presence of pMM-5 and its ability to grow under anaerobic conditions in liquid medium and on plates.
RNA extraction.
Total RNA was isolated as described previously (19) from S. aureus exponential-phase cultures grown in TSB medium at 37°C. Cells (50 mg [wet weight]) were lysed in 0.3 ml of TES buffer containing 100 μg of lysostaphin (Sigma) ml−1, and RNA was extracted using 1.5 ml of RNazol B (Tel-test). For reverse transcription (RT)-PCR and primer extension, residual DNA was removed by treatment with RQ1 RNase-free DNase (Promega). RNA concentrations were determined by A260 measurements, and RNA integrity was analyzed by agarose/formaldehyde gel electrophoresis (41).
Northern hybridization.
Quantitation of nrdIEF, orf1, and nrDG mRNA levels and sizes of transcripts in total RNA from S. aureus cultures grown under aerobic and anaerobic conditions was performed by Northern blot analysis. Internal fragments of the genes nrdD (nucleotides [nt] 1953 to 2149 of GenBank AJ292926), nrdE (nt 1699 to 1972 of GenBank 292927), and nrdF (nt 3711 to 4088 of GenBank 292927) were amplified by PCR and labeled with the DIG PCR synthesis kit (Roche Molecular Biochemicals). In some experiments, oligonucleotides labeled at the 3′ end were used as probes and labeled with the DIG oligonucleotide 3′-end DNA labeling kit (Roche Molecular Biochemicals). Oligonucleotide probes for orf1 and the nrdIEF genes (positions are from GenBank AJ292927) were as follows: orf1-rev, 5′-GATACCTTCCATTTGCTCAGTAC-3′, complementary to nt 617 to 639; nrdI-rev, 5′-TCCAAATCCAATAGTGCCAG-3′, complementary to nt 991 to 1011; nrdE-rev, 5′-CACAGCACCAGCACCAGGGCGTTGACC-3′, complementary to nt 3711 to 3735; and nrdF-rev, 5′-CGCGTGTATTTGCTCCATCATCGCC-3′, complementary to nt 1946 to 1972. Oligonucleotide probes for the nrdDG genes (positions are from GenBank 292926) were as follows: nrdD1-rev, 5′-CGTCAACGCGGTCAACCGTACAGCCACC-3′, complementary to nt 698 to 715; and nrdG-rev, 5′-CGCCACCTAATAGACTTAGCCC-3′, complementary to nt 2385 to 2406. RNA samples (10 μg) denatured in formaldehyde were loaded onto agarose gels, electrophoresed, and transferred to Sartolon membranes (Sartorius) essentially as described previously (41). Prehybridization, hybridization with the DIG-labeled nrd-specific probes in DIG-modified hybridization buffer plus 50% formamide solution, and detection with the CSPD chemiluminescence system were carried out according to the user's guide (Roche Molecular Biochemicals).
Primer extension.
Primer extension was carried out with avian myeloblastosis virus (AMV) reverse transcriptase (Promega). Synthetic oligonucleotide primers complementary to the N-terminal region were labeled at the 5′ end with [γ-32P]ATP by T4 polynucleotide kinase (41), and 0.5 to 1 pmol (100,000 to 200,000 cpm) was mixed with 40 μg of RNA in a final volume of 18.5 μl containing 4 μl of 5× AMV reverse transcriptase buffer and 2 μl of deoxynucleoside triphosphates (dNTPs; 1 mM final concentration). The mixture was heated to 80°C for 10 min and cooled, and polymerization was carried out at 42°C for 2 h with 20 U (1 μl) of AMV reverse transcriptase and 20 U (0.5 μl) of RNasin inhibitor (Promega). The reaction was stopped by the addition of 1 μl of 0.5 M EDTA. Free RNA was removed by incubation with 1 μl of heat-inactivated RNase A (Sigma) (10 mg ml−1) at 37°C for 30 min, and the DNA fragment was purified by phenol extraction and ethanol precipitation. The primer extension product was separated on a 6% denaturing polyacrylamide gel alongside sequencing reactions using the same oligonucleotide as a primer. The reverse primers used in reactions were as follows: orf1-rev, nrdI-rev, and nrdF-rev, described above, and nrdD2-rev, 5′-GCATCTGCAACATGCTTTGG-3′, complementary to nt 466 to 485 of GenBank AJ292926.
RT-PCR.
Reactions were performed using Moloney murine leukemia virus reverse transcriptase (Promega). Because S. aureus DNA has a low G+C content (∼40%), annealing of primers to RNA and the reverse transcriptase reaction were performed together at 37οC. Total RNA (10 μg) in an 18-μl final reaction volume containing 5 pmol of reverse primer and 1 mM dNTPs was denatured for 10 min at 80οC, 0.5 μl (100 U) of reverse transcriptase and 0.5 μl (20 U) of RNasin were added, and the mixture was incubated at 37οC for 2 h. Free RNA was removed by digestion with 1 μl of RNase A (10 mg ml−1), and the reaction was stopped by addition of 180 μl of TE (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). cDNA was phenol extracted, ethanol precipitated, and amplified in PCRs containing (in a final volume of 50 μl) 1 μl of the cDNA sample, 5 μl of 10× PCR buffer, 3 μl of 25 mM MgCl2, 1 μl of dNTPs (10 mM), 0.4 μl (2 U) of Taq DNA polymerase (MBI Fermentas), and 50 pmol of each primer. The mixture was heated for 3 min at 95°C; run in a thermal cycler for 29 rounds of 30 s at 94°C, 30 s at 45°C, and 40 s at 72°C; and completed by being heated for 10 min at 72°C. The forward and reverse primers were as follows: orf1-for, 5′-TGTACTGAGCAAATGGAAG-3′ (nt 616 to 634), and nrdI-rev, 5′-TCCAAATCCAATAGTGCCAG-3′ (nt 992 to 1011); nrdI-for, 5′-ACTGGCACTATTGGATTTGG-3′ (nt 991 to 1010), and nrdE-rev, 5′-CTTCTCTTCGTTTAGTGACC-3′ (nt 1273 to 1292); nrdE-for, 5′-TCTACACGTGAGTTAGCAAG-3′ (nt 3212 to 3231), and nrdF-rev, 5′-CCATCATCTGCTTGATGTG-3′ (nt 3632 to 3680) (the numbers refer to the positions of nucleotides in the DNA sequence of GenBank AJ292927). Control samples in which reverse transcriptase was omitted in RT-PCRs and in which genomic DNA was used as a template in PCRs were run in parallel with RT-PCRs.
Sequence analysis, database search, and deduced protein analysis.
Sequence entry, primary analysis, and ORF searches were performed using the National Center for Biotechnology Information server ORF Finder (http://www.ncbi.nim.nih.gov/gorf/.html) and the CloneManager 4.10 program. Primary sequences of S. aureus class Ib and class III RNRs were identified in databases of the University of Oklahoma Advanced Center for Genome Technology (http://www.genome.ou.edu/staph.html/) (strain NCTC8323), of The Institute for Genomic Research (TIGR [http://www.tigr.org/]) (strain COL), and of the Staphylococcus aureus Sequencing Group at the Sanger Centre (http://www.sanger.ac.uk /Projects/S_aureus/) (EMRSA-16 strain 252 and MSSA strain 476) using BLAST algorithms (BLASTn and tBLASTn) (1). Pairwise alignments were performed with the BESTFIT and GAP programs of the Wisconsin Genetics Computer Group package; multiple sequence alignments were made with the ClustalW program, version 1.84 (20) using the EMBL ClustalW server (http://www2.ebi.ac.uk/clustalw/?).
Other methods.
Signals from Northern blots and primer extension analysis were scanned and intensities were measured with the ImageMaster software system (Pharmacia).
Nucleotide sequence accession numbers.
The nucleotide sequences of the DNA regions containing the S. aureus Oxford class Ib and class III RNR genes have been deposited in the GenBank database with accession no. AJ292926 (class III RNR genes) and AJ292927 (class Ib RNR genes).
RESULTS
Chromosomal organization of the S. aureus class Ib and class III RNR gene clusters and comparison with other eubacteria.
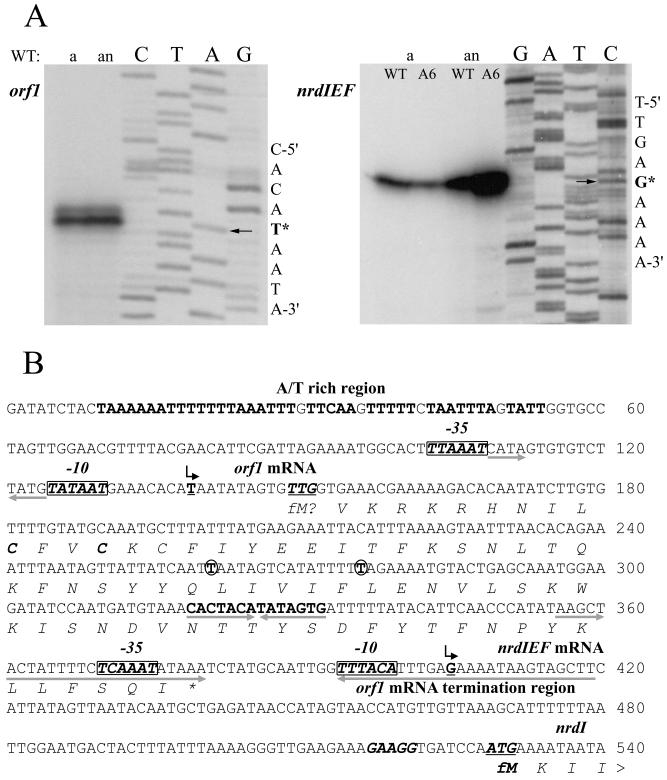
The nucleotide sequence of the S. aureus Oxford DNA region containing the class Ib RNR genes was determined; Fig. 1A shows the organization of genes in the nrdIEF operon. An identical organization occurs in the S. aureus strains NCTC8325, COL, EMRSA-16, MSSA (see Materials and Methods), N315, and Mu50 (29), whose genomes have been completely or nearly completely sequenced. The deduced amino acid sequences of the S. aureus NrdE and NrdF proteins share 62 and 54% sequence similarity with the corresponding E. coli homologs and 74 and 68% similarity with the corresponding L. lactis homologs. Immediately upstream of nrdE are located two short ORFs. The proximal ORF overlaps by 38 bp with nrdE and codes for a protein of 132 amino acids that is conserved in eubacteria; it shares 51 and 55% similarity with the E. coli and L. lactis NrdI homologs, respectively. The distal orf1 in Oxford codes for a putative 37-amino-acid protein that contains a pair of cysteines—CFVC—in the N-terminal portion resembling the redox-active domain present in NrdH and glutaredoxin-like proteins. S. aureus EMRSA-16 contains an identical ORF. The corresponding sequence of the S. aureus RN4220 orf1 (GenBank AJ312387) differs in two positions from that of Oxford; one nucleotide, a G, is replaced by a T, eliminating a TGA translational stop codon; another nucleotide downstream, a T, is deleted, resulting in an ORF coding for a putative 76-amino-acid protein. The same changes were found in the orf1 genes of strains NCTC8325, COL, and MSSA. Comparison of the deduced amino acid sequence of orf1 with those of NrdH and glutaredoxins failed to reveal any significant sequence similarity. A search of the S. aureus genome databases subsequently revealed an ORF, well separated from orf1, related to the L. lactis NrdH (GenBank X92690), with which it shares 60% similarity.
FIG. 1.
Organization of the class Ib and class III RNR genes in S. aureus and other gram-positive bacteria. (A) Class Ib RNR genes. Gene designations: nrdH, homolog of E. coli nrdH, which encodes a redoxin protein; nrdI, homolog of E. coli nrdI, which encodes a protein-stimulating class Ib RNR activity; nrdE and bnrdE, homologs of E. coli nrdE, which encodes the α subunit of class Ib RNR; nrdF and bnrdF, homologs of E. coli nrdF, which encodes the β subunit of class Ib RNR; ymzA, gene encoding a hypothetical protein containing a putative redox motif; grxS, gene encoding a glutaredoxin-like protein; orf1, putative ORF (see the text); other ORFs encode hypothetical proteins of unknown function. The positions of introns and regions encoding inteins are shown as grey boxes. (B) Class III RNR genes. Gene designations: nrdD, homolog of E. coli nrdD, which encodes the α subunit of class III RNR; nrdG, homolog of E. coli nrdG, which encodes the activating β protein of class III RNR. The directions of ORFs are indicated by arrows. See the text for other details.
Figure 1A shows a comparison of the S. aureus class Ib RNR gene cluster with that present in the genomes of several A/T-rich gram-positive bacteria. Southern blot analysis showed that S. aureus, like E. coli and L. lactis, possesses single copies of the nrdIEF genes (data not shown). In contrast, S. epidermidis, Streptococcus pyogenes, and B. subtilis all possess two class Ib RNR gene clusters. In S. epidermidis and B. subtilis, one of the copies appears to have originated from a phage, and there are other noticeable differences in their gene organizations.
The nucleotide sequence of the DNA region of S. aureus Oxford containing the anaerobic class III RNR genes was determined; Fig. 1B shows the organization of genes in the nrdDG operon. The same arrangement occurs in the genomes of each of the six S. aureus strains referred to above. Southern blot analysis showed the presence in S. aureus of a single copy of the nrdDG gene cluster (data not shown). The S. aureus nrdD and nrdG genes encode proteins of 616 and 178 amino acids, respectively. The deduced amino acid sequence of S. aureus NrdD shares 70 and 79% sequence similarity with the E. coli and L. lactis homologs, respectively; the deduced amino acid sequence of S. aureus NrdG shares 55 and 63% sequence similarity with the corresponding E. coli and L. lactis homologs. Alignment of the S. aureus NrdD sequence with the sequences of other bacterial NrdD proteins (data not shown) reveals that the S. aureus NrdD and S. epidermidis NrdD polypeptides lack an N-terminal segment (like the T4 phage homolog) of approximately 100 amino acids that determines a dATP binding allosteric site (2) that is present in the E. coli, L. lactis, and S. pyogenes NrdD proteins. This is shown schematically in Fig. 1B, which also shows that the nrdD and nrdG genes in S. aureus and S. epidermidis overlap (by 4 bp), while in L. lactis they are separated (by 2 bp). Analysis of the genomes of three streptococcal strains (only one is shown in Fig. 1B) revealed the presence of one or two ORFs separating the nrdD and nrdG genes.
The S. aureus nrdDG gene cluster is essential for anaerobic growth.
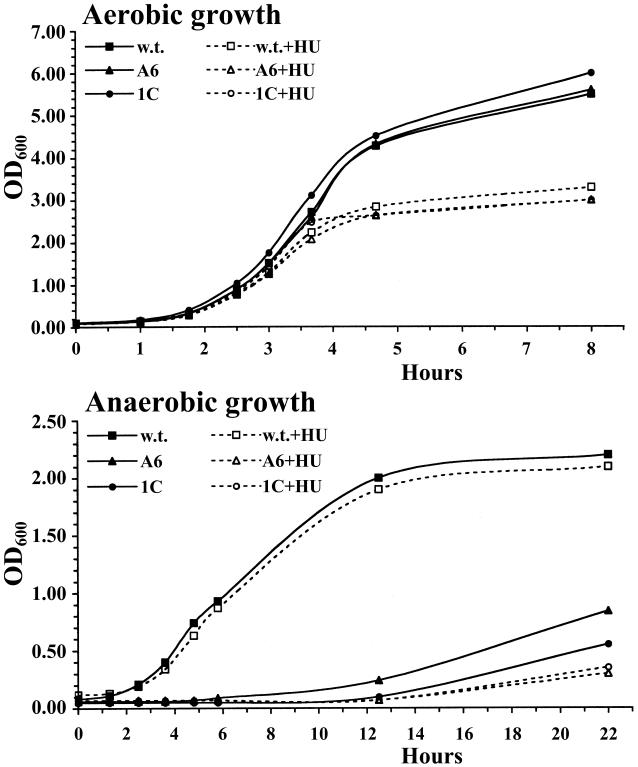
To determine whether the S. aureus nrdDG genes are essential for anaerobic growth, the nrdD gene was disrupted by the insertion of plasmid pMM-1, which carries an internal fragment of that gene. The correct integration of pMM-1 into the chromosome was verified by PCR analysis and Southern blotting (data not shown). Figure 2 shows the growth profiles of RN4220 and the MMA6 mutant in liquid medium. Under aerobic conditions, there was no discernible difference in the rates and extents of growth, as measured by the OD600, between the parent and mutant strains, which reached values of 6 to 7. Under standard anaerobic conditions, RN4220 grew more slowly (with a doubling time of 80 min compared with 40 min under aerobic conditions) and reached an OD600 of ∼2, whereas the MMA6 strain exhibited an extensive lag in growth, after which the OD very gradually increased under prolonged incubation (Fig. 2). The limited growth of the MMA6 mutant may be a consequence of the fact that the growth conditions in these experiments are not strictly anaerobic (microaerophilic) and reflect residual activity of the class Ib RNR. This view is supported by experiments in which the parent and the MMA6 mutant were spread on agar plates with or without 25 mM hydroxyurea (HU), a potent inhibitor of class I RNRs (24, 47), and were incubated in an anaerobic jar. In the absence of HU, the parent strain formed normal-size colonies whereas the MMA6 mutant grew slowly and formed small colonies. In the presence of HU, the parent strain formed normal-size colonies but the MMA6 mutant failed to form colonies. To corroborate these findings, we constructed a deletion mutant in which a portion of the nrdD and nrdG genes was replaced with a kanamycin cassette. The liquid growth profiles of the MM1C mutant are shown in Fig. 2. The growth profile of the MM1C mutant under aerobic conditions was essentially the same as that of the MM6 mutant; similarly, under standard anaerobic conditions, no significant difference was evident in the growth of the MM1C and MMA6 mutants. These observations establish that the S. aureus nrdDG genes are essential for anaerobic growth. They are similar to those described for E. coli and L. lactis, in which the growth deficiency of an nrdD mutant was only apparent when strict anaerobic growth conditions were employed (12, 50).
FIG. 2.
Growth of S. aureus RN4220 and class III RNR mutants under aerobic and anaerobic conditions in the presence or absence of HU. The OD600 was used to follow the growth of cultures in TSB medium at 37°C with or without 50 mM HU. w.t., RN4220; A6, nrdD disrupted mutant MMA6; 1C, nrdDG deletion mutant MM1C. The results are representative of three independent experiments. The gradual increase in the OD values of the MMA6 and MM1C mutants in the anaerobic cultures after extensive incubation may be due to the entrance of trace amounts of oxygen into the flasks over time.
Inhibition of the S. aureus class Ib RNR in vivo by HU limits aerobic growth.
Repeated attempts to isolate insertional mutants in the nrdIEF genes were unsuccessful, presumably due to the extreme sensitivity of the anaerobic RNR to oxygen. Alternatively, we took advantage of the inhibition of class I RNRs by HU to determine if the S. aureus class Ib RNR is required for aerobic growth. Preliminary experiments showed that 50 mM HU prevented growth (formation of colonies) of S. aureus in solid medium under aerobic conditions but did not affect anaerobic growth, while the MMA6 and MM1C mutants failed to grow in plates containing 50 mM HU under both aerobic and anaerobic growth conditions. In liquid culture, aerobically grown S. aureus cultures treated with 50 mM HU were partly inhibited in growth and reached final OD600 values of ∼2 to 3 (Fig. 2, top) compared to untreated cultures, which attained OD600 values of 6 to 7. The effect appears to be specific for the class Ib RNR, since the same concentration of HU did not affect the growth of RN4220 under anaerobic conditions (Fig. 2, bottom). When the MMA6 and MM1C mutants were grown aerobically in liquid medium containing 50 mM HU, the cultures reached the same OD as the parent strain with 50 mM HU (Fig. 2, top). However, under anaerobic growth conditions, this concentration of HU significantly extended the growth lag of the two mutants compared with that observed in the absence of HU (Fig. 2, bottom), presumably due to inhibition of residual activity of the class Ib RNR under microaerophilic conditions. These results indicate that the S. aureus class Ib RNR is necessary for normal growth under aerobic conditions.
Complementation of nrdDG
S. aureus MM1C was complemented in trans with the wild-type nrdDG alleles by introducing into that strain plasmid pMM-5, which carries an intact copy of the nrdDG gene cluster. MM1C contains the 1C deletion that replaces part of the nrdD and nrdG genes with a kanamycin cassette. Complementation was shown by the ability of pMM5 to permit growth on plates containing 50 mM HU (to inhibit residual activity from the class Ib RNR) in an anaerobic chamber, conditions under which the MM1C mutant is unable to form colonies (results not shown).
Northern blot analysis of nrdIEF and orf1 expression under aerobic and anaerobic conditions.
Northern blot analysis was used to monitor transcription of the nrdIEF genes in S. aureus RN4220. Figure 3A shows that an ∼3.9-kb mRNA transcript was detected in total RNA from cultures of RN4220 grown under aerobic and anaerobic conditions using probes designed for the nrdI, nrdE, and nrdF genes. Unexpectedly, the level of nrdIEF mRNA synthesized under anaerobic growth conditions was about the same (in some cases up to twofold more) as that synthesized under aerobic conditions. The same transcription pattern was observed for S. aureus SH1000 (data not shown). An orf1-specific probe detected an ∼0.27-kb mRNA, indicating that nrdIEF and orf1 are independently transcribed. To confirm these findings, RT-PCR was used to demonstrate that nrdIEF and orf1 are separately transcribed and that nrdI, nrdE, and nrdF are cotranscribed (Fig. 3B).
FIG. 3.
(A) Northern hybridization analysis of orf1 and nrdIEF transcripts in aerobic (a) and anaerobic (an) cultures of S. aureus RN4220. Total RNA was electrophoresed, blotted, and hybridized to orf1-rev, nrdI-rev, nrdE-rev, and nrdF-rev probes (see Materials and Methods). The sizes of transcripts in kilobases are shown on the left. The arrows indicate the positions of 16S and 23S rRNAs. (B) RT-PCR analysis to demonstrate that the nrdIEF genes are coordinately transcribed independently of orf1. At the top is a schematic showing the organization of the orf1-nrdIEF region and the positions of primers, indicated by solid arrows, used in the PCR analysis. The open arrows indicate the direction of transcription of genes. Lanes 4, 5, and 6, RT-PCR using total RNA from anaerobically (microaerophilically) grown culture as a template and the primer pairs nrdE-for and nrdF-rev, nrdI-for and nrdE-rev, and orf1-for and nrdI-rev, respectively, for amplification (see Materials and Methods); lanes 7, 8, and 9, RT-PCR using total RNA from an aerobic culture as a template with the same pairs of primers, respectively; lanes 11, 12, and 13, direct PCR using genomic DNA as a template with the same pairs of primers, respectively; lanes 1, 2, and 3, control PCR using total RNA as a template without reverse transcriptase; lane 10, DNA molecular size markers.
Primer extension analysis of orf1 and nrdIEF at low and high oxygen concentrations and structure of the promoter regions.
Primer extension was used to monitor transcription of the nrdIEF and orf1 genes in S. aureus RN4220 and in the MMA6 mutant in response to changes in oxygen concentration. This was found to be a more sensitive method for quantifying promoter activity than Northern blot analysis. The intensities of the signals determined in these and subsequent experiments reflect the levels of transcription initiating from the specific promoters. Figure 4A shows transcription of the nrdIEF and orf1 genes in cultures grown under aerobic and anaerobic conditions. In RN4220, the level of nrdIEF transcription was ∼1.5-fold higher under anaerobic conditions than under aerobic conditions (as measured by densitometry), in agreement with that found by Northern blot analysis. In the MMA6 mutant, under aerobic growth conditions, the level of transcription was about the same as that in the parent strain; under anaerobic conditions, the MMA6 mutant exhibited an ∼3- to 5-fold stimulation of transcription compared to that of the parent strain. Thus, the effect of the A6 mutation on transcription of nrdIEF genes results in up to a fivefold stimulation under anaerobic growth conditions compared to levels under aerobic conditions. A similar increase was seen in Northern blot analysis (data not shown). Transcription from the orf1 promoter in RN4220 was about the same under aerobic and anaerobic growth conditions (Fig. 3 and 4); in the MMA6 and MM1C mutants, it was stimulated severalfold under anaerobic conditions but not under aerobic conditions (data not shown).
FIG. 4.
(A) Primer extension analysis of orf1 and nrdIEF genes. Total RNA was isolated from aerobic (a) and anaerobic (an) cultures of RN4220 (WT) and the MMA6 mutant (A6). Primer extension was carried out with the primers orf1-rev and nrdI-rev (see Materials and Methods), and the products were separated by electrophoresis under denaturing conditions alongside sequencing reactions using the same primers. The arrows point to the nucleotides (labeled with an asterisk) of the orf1 and nrdIEF transcription start points. (B) Nucleotide sequence of the orf1 and nrdIEF promoter regions. Transcription start points are shown by bent arrows above the underlined boldface T nucleotide (for orf1) and G nucleotide (for nrdIEF) start sites. The nrdI ATG translational start codon (underlined) and its ribosomal binding site are shown in boldface italic letters; a putative orf1 translational start codon, TTG (underlined), and two in-frame cysteines are also shown in boldface italics. Putative −10 and −35 sequences are shown as boxed boldface italic letters. The pairs of opposing arrows placed beneath nucleotide sequences show three inverted-repeat sequences. The circled nucleotides are those that are changed in the orf1 of some S. aureus strains (see the text); another feature is a long A/T rich region (boldface roman letters) upstream of orf1.
Figure 4B shows the nucleotide sequence of the nrdIEF and orf1 promoter regions in S. aureus RN4220. Primer extension analysis identified a single strong transcription start site and several much weaker ones in the DNA region upstream of nrdIEF structural genes. The 5′ end of the major transcript maps 124 bp upstream of the predicted nrdI ATG start codon. A ribosomal binding site, GAAGG, is located 7 bp upstream of the start codon. The promoter region is A/T rich and contains two inverted repeats. A 64-bp interrupted partially symmetrical sequence is located from 110 to 173 bp upstream of the predicted nrdI translational start codon and overlaps the major transcription start site and the −10 and −35 promoter recognition sequences; a second, 14-bp imperfect symmetrical sequence, CACTACATATAGTG (12 matching residues out of 14), is located from 197 to 210 bp upstream of the start codon. The 5′ end of the orf1 transcript, ∼270 nt in size, maps 390 bp upstream of the predicted nrdI ATG start codon and terminates within the large 64-bp inverted repeat that spans the −10 and −35 sequences and the transcription start site of the major nrdIEF promoter. Inspection of the sequence downstream of the orf1 transcriptional start point failed to reveal a ribosomal binding site. A potential TTG translational start codon is indicated in Fig. 4B 10 nt downstream of the transcriptional start site. The orf1 promoter region contains an 8-bp inverted repeat interrupted by the sequence GTGTGTCT located between the −10 and −35 sequences. Further upstream, there is a 45-bp region containing 89% AT.
Northern blot analysis of nrdDG gene expression under aerobic and anaerobic conditions.
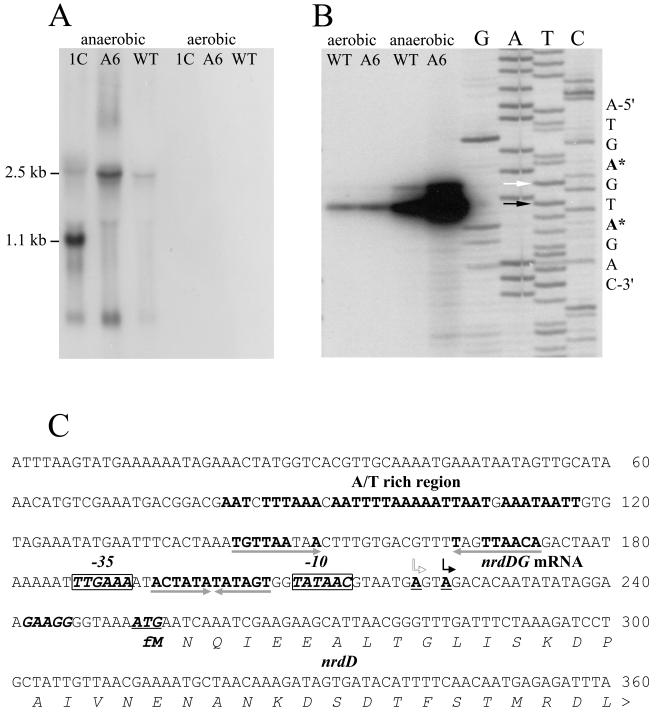
Northern blot analyses of nrdDG expression in total RNA from S. aureus RN4220 under anaerobic growth conditions using probes specific for nrdD and nrdG each revealed a signal corresponding to an mRNA of ∼2.4 kb, indicating coordinate transcription of the two genes (Fig. 5A). A transcript of the same size was detected in S. aureus SH1000 (data not shown). When the same nrdD probe was used to detect transcripts in the MMA6 and MM1C mutants, the levels were increased some three- to fourfold and four- to sevenfold above that of the parent, respectively (Fig. 5A). In the MMA6 disruptant mutant, the mRNA terminates within the pMUTIN-4 vector and results in a transcript of ∼2.5 kb; in the MM1C deletion mutant, the mRNA terminates within the stem-loop structure of the kanamycin resistance gene (oriented opposite to that of nrdDG) and results in a transcript of the expected size of ∼1.1 kb. No transcripts were detected in the MMA6 and MM1C mutants using an nrdG probe specific for the 3′ end of the gene, confirming that a single promoter transcribes both nrdDG genes. Under aerobic growth conditions, this transcript was not detected in either the parent or mutant strain.
FIG. 5.
(A) Northern hybridization analysis of nrdDG transcripts in aerobic and anaerobic cultures of S. aureus RN4220 and in MMA6 and MM1C mutants (WT, A6, and 1C, respectively). Total RNA was electrophoresed, blotted, and hybridized to the probe nrdD1-rev (see Materials and Methods). The sizes of transcripts in kilobases are shown on the left. (B) Primer extension analysis of nrdDG genes. Total RNA was isolated from aerobic and anaerobic cultures of RN4220 and the MMA6 mutant. Primer extension was carried out with the primer nrd2-rev (see Materials and Methods), and the products were separated by electrophoresis under denaturing conditions alongside sequencing reactions using the same primer. The arrows show the nucleotides (labeled with asterisks) of the major (solid arrow) and minor (open arrow) nrdDG transcription start points. (C) Nucleotide sequence of the nrdDG promoter region. Major and minor transcription start points are shown by solid and open bent arrows, respectively, above the underlined boldface A nucleotide start sites. The nrdD ATG translational start codon (underlined) and its ribosomal binding site are shown in boldface italic letters. Putative −10 and −35 sequences are shown as boxed boldface italic letters. The pairs of opposing arrows placed beneath nucleotide sequences show two inverted-repeat sequences. A long A/T-rich region (boldface roman letters) is located upstream of the −35 sequence.
Primer extension analysis of nrdDG at low and high oxygen concentrations and structure of the promoter region
Primer extension analysis of nrdDG expression in cultures of RN4220 and the MMA6 mutant grown under aerobic and anaerobic growth conditions is shown in Fig. 5B. Primer extension analysis revealed a major and a minor transcript under anaerobic conditions and the presence of a weak transcript under aerobic growth conditions. In RN4220, nrdDG transcription was ∼5- to 10-fold higher under anaerobic than under aerobic growth conditions. In the MMA6 mutant, a further three- to fourfold increase in nrdDG transcription occurred under anaerobic conditions compared with that of the parent strain. Compared to Northern blot analysis, the more sensitive primer extension method enabled detection of weak nrdDG transcripts under aerobic conditions; no significant difference in transcription was apparent between the MMA6 mutant and the parent strain.
Figure 5C shows the nucleotide sequence of the nrdDG promoter region. Primer extension analysis identified two transcription start sites in the DNA region upstream of nrdD. The 5′ end of the major transcript maps 29 bp upstream of the ATG start codon; a second site corresponding to the minor transcript maps 32 bp upstream of that codon. A ribosomal binding site, with the same sequence as that in front of the nrdI gene, is located 9 bp upstream of the start codon. The promoter region contains two inverted repeats, one an interrupted symmetrical sequence of 29 bp located upstream of the −35 sequence of the promoter and a second perfectly symmetrical sequence of 12 bp, ACTATATATAGT, located between the −35 and −10 sequences of that promoter. The latter sequence is very similar to the shorter of the two inverted repeats present in the nrdIEF promoter region (Fig. 4B). Upstream of the −35 sequence is a long A/T-rich (∼90%) region.
HU stimulates transcription of S. aureus class Ib and class III RNR genes.
HU inhibits the activity of class I RNR enzymes and was reported to stimulate in E. coli the transcription of class Ia RNR (nrdAB) (13) and class Ib RNR (nrdEF) genes (22, 34). To determine its effect on transcription of the S. aureus nrdIEF operon, total RNA was isolated from an aerobically grown culture that had been grown in the presence and absence of 50 mM HU and subjected to Northern blot analysis. HU treatment caused more than a 5- to 10-fold increase in transcription of the ∼3.9-kb nrdIEF mRNA. The same RNA preparation was used to measure the amount of nrdDG transcription, which, as shown above, is barely detectable in aerobically grown cultures. In the presence of HU, a massive increase in transcription of the nrdDG gene cluster occurred under aerobic growth conditions (data not shown). In other experiments, similar large effects were found for HU on transcription of the thioredoxin (trxA) and thioredoxin reductase (trxB) genes.
DISCUSSION
Class Ib oxygen-dependent RNRs are widespread among prokaryotic organisms. The prototype is the E. coli NrdEF enzyme that is encoded in an operon containing four genes, nrdHIEF. In S. aureus, the class Ib RNR operon comprises the nrdIEF genes only; an nrdH-like gene was located elsewhere in the genome (see below). Comparison of the S. aureus class Ib RNR gene cluster with that of its close relative, S. epidermidis, and those present in B. subtilis and S. pyogenes reveals other differences in gene organization (Fig. 1A). Thus, each of the last three gram-positive bacteria contains two class Ib RNR gene clusters. One S. epidermidis cluster (strain RP62A [TIGR database]) contains nrdIEF and orf1; a second contains an additional ORF of unknown function coding for 148 amino acids and located between nrdI and nrdE. Immediately downstream of nrdF is an ORF of unknown function coding for 200 amino acids followed by an ORF that codes for a putative 82-amino-acid glutaredoxin-like protein. The nrdE gene in this cluster is unusual in that it contains a sequence coding for a 385-amino-acid intein in the N-terminal portion and a 1,160-bp group I intron in the C-terminal portion. Group I introns were previously reported to be present in phage RNR genes; in the nrdB and nrdD/sunY genes of E. coli phage T4, which code for class Ia and class III RNRs (15, 48, 54); and in the bnrdE and bnrdF genes of the B. subtilis phage SPβ (31). Moreover, the nrdE gene of phage SPβ codes for an in-frame intein of 386 amino acids that is remarkably similar to the S. epidermidis intein and which shares the same splicing sites. These features suggest the atypical S. epidermidis class Ib RNR gene cluster may have originated by the insertion of a phage in its chromosome. Sequence analysis of the region containing this gene cluster supports this idea, as do the recent finding of multiple self-splicing introns in the genome of the S. aureus phage Twort (30). Both S. pyogenes class Ib RNR gene clusters are also unusual; one contains an ORF of unknown function located between nrdE and nrdF, and the other cluster has the order of the nrdI and nrdF genes inverted.
The E. coli and S. enterica serovar Typhimurium nrdHIEF genes, and presumably the corresponding L. lactis genes, are transcribed from a common promoter; S. aureus orf1, which immediately precedes nrdI, and nrdIEF are transcribed from separate promoters. Initially, we thought that orf1 might code for a small protein containing a redox-like domain—CFVC (Fig. 4B)—with a function similar to those of the E. coli and L. lactis NrdH proteins. However, several observations suggest that the ∼270-nt orf1 may determine a small nontranslated RNA molecule rather than a polypeptide. We could not identify a ribosomal binding site in front of the potential orf1 TTG or GTG translational start codons; also orf1 codon usage did not conform to that present in typical staphylococcal ORFs. Furthermore, comparison of orf1 nucleotide sequences from different S. aureus strains revealed two variants, one with a G or T nucleotide located 123 nt downstream of the transcription start site, potentially creating a TGA translational stop codon, and another with a T nucleotide present or absent at a position 138 nt downstream of the transcription start site, potentially creating a frame shift (Fig. 4B). Subsequently, we identified in the S. aureus genome databases an ORF, far removed from orf1, coding for a protein with significant similarity to the L. lactis NrdH redoxin protein. Structural analysis of the ∼270-bp orf1 transcript showed that it is capable of forming a molecule with considerable secondary structure. The 3′ end of the orf1 RNA molecule overlaps the DNA region containing the nrdIEF promoter and may fold to form a long stable double-stranded stem-loop structure (ΔG°, ∼30 kcal/mol). Possibly, an open form of the orf1 RNA molecule interferes with transcription from the nrdIEF promoter and thereby regulates its activity. Preliminary studies employing orf1-lacZ reporter gene fusions indicate that orf1 RNA is not translated (unpublished data).
Analysis of current prokaryotic genome databases shows that class III RNR genes are organized in eubacteria in fundamentally the same way. In S. aureus, the nrdD and nrdG genes overlap and form an operon. A similar situation is found in other gram-positive bacteria, S. epidermidis and Bacillus anthracis (TIGR), and in L. lactis and Enterococcus faecalis (TIGR) the two genes are separated by a few nucleotides and are presumably also cotranscribed. A notable exception is S. pyogenes, in which the nrdD and nrdG genes are separated by two ORFs that overlap one other and nrdG (10). One ORF codes for a 311-amino-acid protein that is similar to the S. enterica serovar Typhimurium MviM virulence factor (a putative oxidoreductase); the other ORF codes for a protein of unknown function. Neither ORF was identified in S. aureus genome databases. In some gram-negative bacteria, such as E. coli, S. enterica serovar Typhimurium, and Pseudomonas aeruginosa, the nrdD and nrdG genes are clustered but separated by more than 100 nt and may be transcribed from separate promoters.
Multiple inverted repeats are a feature of both the S. aureus nrdIEF and nrdDG promoter regions. To date, promoter identification and transcription analysis of class Ib RNR genes have been described for the E. coli, S. enterica serovar Typhimurium, and B. subtilis class Ib RNR gene clusters (22, 45) but not for the class III nrdDG gene cluster. In this paper, we show that the S. aureus nrdIEF and nrdDG genes are cotranscribed from ςA-like promoters in ∼3.9- and ∼2.4-kb mRNAs and that orf1 is independently transcribed in an ∼0.27-kb mRNA. Inspection of the nrdIEF promoter region showed it to contain a 64-bp imperfect inverted repeat that spans the −10 and −35 recognition sequences; an identical inverted repeat was found in each of the S. aureus strains sequenced in genome projects. Upstream of the major promoter (within orf1) there is a nearly perfect 14-bp inverted repeat, CACTACATATAGTG, positioned in what is possibly another promoter. Interestingly, the 5′ untranslated region of the L. lactis nrdHIEF gene cluster contains a similar 14-bp inverted repeat (CACAACATATAGTG) ∼170 nt upstream of the nrdH ATG start codon. Sequence analysis of the nrdDG promoter region also revealed two inverted repeats. Remarkably, one of the inverted repeats, located between the −10 and −35 recognition sequences, is the fully symmetrical form of the 14-bp inverted-repeat sequence present in the nrdIEF promoter region. We identified the same inverted repeat in the S. epidermidis genome and a very similar one in the B. anthracis genome in the predicted nrdDG promoter regions. A second, shorter inverted repeat occurs further upstream (Fig. 5C). While the significance of the different inverted repeats within the nrdIEF and nrdDG promoter regions is not clear, they may be implicated in the regulation of the gene clusters and possibly in their response to changes in oxygen concentration. Moreover, the fact that an almost identical inverted repeat occurs in the S. aureus nrdIEF and nrdDG promoters suggests that they may share some common regulatory features.
The results presented here establish the existence in S. aureus of control mechanisms that regulate the transcription of the anaerobic nrdDG genes in response to the level of oxygen. Evidence for this is twofold. First, the class III RNR genes are transcribed almost exclusively under anaerobic conditions. In contrast, the class Ib RNR genes are transcribed at about the same level under aerobic and anaerobic conditions. Thus, a shift from high to low oxygen concentration during anaerobiosis has radically different effects on the transcription of class Ib and class III RNR genes. Similar observations were obtained with S. aureus strain SH1000, a derivative of 8325-4, the parent of RN4220 (28), showing that the findings presented in this work are independent of the genetic background of RN4220. S. aureus SH1000 contains an intact copy of the rsbU gene, which is necessary for stress-induced activation of ςB and is partly deleted in RN4220 (16). Second, nrdD A6 and nrdDG 1C mutations that abolish the anaerobic class III RNR activity cause an increase in transcription of the nrdDG genes encoding that RNR. If we assume that under anaerobic (or microaerophilic) conditions RNR activity is predominantly due to the class III RNR (the class Ib RNR is unable to function efficiently under these conditions due to lack of formation of the tyrosyl radical) and that inactivation of it greatly reduces the pool of dNTPs and thereby retards DNA synthesis, then the increased activity of the nrdDG promoter in the A6 and 1C mutants implies the existence of a mechanism that upregulates transcription of the class III RNR nrdDG genes in response to depletion of dNTPs. This view is consistent with earlier observations that showed that reduction of the intracellular concentration of deoxyribonucleotides and inhibition of DNA synthesis in different bacteria resulted in an increase in RNR activity (11). Moreover, under anaerobic (but not aerobic) conditions, the same mutations also increase the expression of the class Ib RNR genes. This result implies the existence of a general feedback, or compensatory, mechanism that controls transcription from both promoters in response to changes in the anaerobic RNR activity. The finding that inhibition of the class Ib RNR by HU resulted in increased transcription of the class Ib and class III RNR genes under aerobic conditions supports this idea.
Although the present study does not address the molecular nature of the systems that control transcription of the nrdDG genes in response to changes in oxygen concentration, it implies the existence of different genetic systems, possibly similar to those that code for the Fnr and Arc proteins that regulate the expression of many genes during anaerobiosis. Current studies are aimed at identifying these control systems.
In conclusion, we note that in view of the recent emergence of antibiotic resistance in S. aureus, the class III RNR may provide an attractive target for the development of antistaphylococcal drugs because it is essential for anaerobic growth, conditions that may favor pathogen colonization, and because of its absence in the mammalian cell.
ACKNOWLEDGMENTS
We are grateful to the University of Oklahoma Advanced Center for Genome Technology, TIGR, and the Sanger Centre for the use of preliminary S. aureus genomic sequence data. We thank Simon Foster, Steven Projan, and Orit Uziel for providing strains and plasmids.
This work was supported in part by the Constantiner Institute for Molecular Genetics at Tel Aviv University and the Israel Science Foundation (grant 787/01-1).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson J, Westman M, Hofer A, Sjöberg B M. Allosteric regulation of the class III anaerobic ribonucleotide reductase from bacteriophage T4. J Biol Chem. 2000;275:19443–19448. doi: 10.1074/jbc.M001490200. [DOI] [PubMed] [Google Scholar]
- 3.Bauer C E, Elsen S, Bird T H. Mechanisms for redox control of gene expression. Annu Rev Microbiol. 1999;53:495–523. doi: 10.1146/annurev.micro.53.1.495. [DOI] [PubMed] [Google Scholar]
- 4.Carlson J, Fuchs J A, Messing J. Primary structure of the Escherichia coli ribonucleoside diphosphate reductase operon. Proc Natl Acad Sci USA. 1984;81:4294–4297. doi: 10.1073/pnas.81.14.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan P F, Foster S J. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J Bacteriol. 1998;180:6232–6241. doi: 10.1128/jb.180.23.6232-6241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clements M O, Foster S J. Stress resistance in Staphylococcus aureus. Trends Microbiol. 1999;7:458–462. doi: 10.1016/s0966-842x(99)01607-8. [DOI] [PubMed] [Google Scholar]
- 7.Cramton S E, Gerke C, Schnell N F, Nichols W W, Götz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67:5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cramton S E, Ulrich M, Götz F, Doring G. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 2001;69:4079–4085. doi: 10.1128/IAI.69.6.4079-4085.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eliasson R, Pontis E, Jordan A, Reichard P. Allosteric regulation of the third ribonucleotide reductase (NrdEF enzyme) from Enterobacteriaceae. J Biol Chem. 1996;271:26582–26587. doi: 10.1074/jbc.271.43.26582. [DOI] [PubMed] [Google Scholar]
- 10.Ferretti J J, McShan W M, Adjic D, Savic D, Savic G, Lyon K, Primeaux C, Sezate S S, Surorov A N, Kenton S, Lai H, Lin S, Qian Y, Jia H G, Najar F Z, Ren Q, Zhu H, Song L, White J, Yuan X, Clifton S W, Roe B A, McLaughlin R E. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filpula D, Fuchs J A. Increased synthesis of ribonucleotide reductase after deoxyribonucleic acid inhibition in various species of bacteria. J Bacteriol. 1979;139:694–696. doi: 10.1128/jb.139.2.694-696.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garriga X, Eliasson R, Torrents E, Jordan A, Barbe J, Gibert I, Reichard P. nrdD and nrdG genes are essential for strict anaerobic growth of Escherichia coli. Biochem Biophys Res Commun. 1996;229:189–192. doi: 10.1006/bbrc.1996.1778. [DOI] [PubMed] [Google Scholar]
- 13.Gibert I, Calero S, Barbe J. Measurement of in vivo expression of nrdA and nrdB genes of Escherichia coli by using lacZ gene fusions. Mol Gen Genet. 1990;220:400–408. doi: 10.1007/BF00391745. [DOI] [PubMed] [Google Scholar]
- 14.Gostick D O, Griffin H G, Shearman C A, Scott C, Green J, Gasson M J, Guest J R. Two operons that encode FNR-like proteins in Lactococcus lactis. Mol Microbiol. 1999;31:1523–1535. doi: 10.1046/j.1365-2958.1999.01298.x. [DOI] [PubMed] [Google Scholar]
- 15.Gott J M, Shub D A, Belfort M. Multiple self-splicing introns in bacteriophage T4: evidence from autocatalytic GTP labeling of RNA in vitro. Cell. 1986;47:81–87. doi: 10.1016/0092-8674(86)90368-5. [DOI] [PubMed] [Google Scholar]
- 16.Guachino P, Engelmann S, Bischoff M. Sigma (B) activity depends on RsbU in Staphylococcus aureus. J Bacteriol. 2001;183:1843–1852. doi: 10.1128/JB.183.6.1843-1852.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guest J R, Green J, Irvine A S, Spiro S. The FNR modulon and FNR-regulated gene expression. In: Lin E C C, Lynch S A, editors. Regulation of gene expression in Escherichia coli. R. G. Austin, Tex: Landes; 1996. pp. 317–342. [Google Scholar]
- 18.Hanke P D, Fuchs J A. Characterization of the mRNA coding for ribonucleoside diphosphate reductase in Escherichia coli. J Bacteriol. 1983;156:1192–1197. doi: 10.1128/jb.156.3.1192-1197.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hart M E, Smeltzer M S, Iandolo J J. The extracellular protein regulator (xpr) affects exoprotein and agr mRNA levels in Staphylococcus aureus. J Bacteriol. 1993;175:7875–7879. doi: 10.1128/jb.175.24.7875-7879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 21.Jordan A, Åslund F, Pontis E, Reichard P, Holmgren A. Characterization of Escherichia coli NrdH. A glutaredoxin-like protein with a thioredoxin-like activity profile. J Biol Chem. 1997;272:18044–18050. doi: 10.1074/jbc.272.29.18044. [DOI] [PubMed] [Google Scholar]
- 22.Jordan A, Aragall E, Gibert I, Barbe J. Promoter identification and expression analysis of Salmonella typhimurium and Escherichia coli nrdEF operons encoding one of two class I ribonucleotide reductases present in both bacteria. Mol Microbiol. 1996;19:777–790. doi: 10.1046/j.1365-2958.1996.424950.x. [DOI] [PubMed] [Google Scholar]
- 23.Jordan A, Pontis E, Åslund F, Hellman U, Gibert I, Reichard P. The ribonucleotide reductase system of Lactococcus lactis. Characterization of an NrdEF enzyme and a new electron transport protein. J Biol Chem. 1996;271:8779–8785. doi: 10.1074/jbc.271.15.8779. [DOI] [PubMed] [Google Scholar]
- 24.Jordan A, Pontis E, Atta M, Krook M, Gibert I, Barbe J, Reichard P. A second class I ribonucleotide reductase in Enterobacteriaceae: characterization of the Salmonella typhimurium enzyme. Proc Natl Acad Sci USA. 1994;91:12892–12896. doi: 10.1073/pnas.91.26.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jordan A, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 1998;67:71–98. doi: 10.1146/annurev.biochem.67.1.71. [DOI] [PubMed] [Google Scholar]
- 26.Kass E H, Kendrick M I, Tsai Y C, Parsonnet J. Interaction of magnesium ion, oxygen tension, and temperature in the production of toxic-shock-syndrome toxin-1 by Staphylococcus aureus. J Infect Dis. 1987;155:812–815. doi: 10.1093/infdis/155.4.812. [DOI] [PubMed] [Google Scholar]
- 27.King D S, Reichard P. Mass spectrometric determination of the radical scission site in the anaerobic ribonucleotide reductase of Escherichia coli. Biochem Biophys Res Commun. 1995;206:731–735. doi: 10.1006/bbrc.1995.1103. [DOI] [PubMed] [Google Scholar]
- 28.Kreiswirth B N, Löfdahl S, Betley M J, O'Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 29.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Kobayashi N, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. Whole genome sequencing of meticillin-resistant Stapylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 30.Landthaler M, Shub D A. Unexpected abundance of self-splicing introns in the genome of bacteriophage Twort: introns in multiple genes, a single gene with three introns, and exon skipping by group I ribozymes. Proc Natl Acad Sci USA. 1999;96:7005–7010. doi: 10.1073/pnas.96.12.7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lazarevic V, Soldo B, Dusterhoft A, Hilbert H, Mauel C, Karamata D. Introns and intein coding sequence in the ribonucleotide reductase genes of Bacillus subtilis temperate bacteriophage SPβ. Proc Natl Acad Sci USA. 1998;95:1692–1697. doi: 10.1073/pnas.95.4.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Logan D T, Andersson J, Sjöberg B M, Nordlund P. A glycyl radical site in the crystal structure of a class III ribonucleotide reductase. Science. 1999;283:1499–1504. doi: 10.1126/science.283.5407.1499. [DOI] [PubMed] [Google Scholar]
- 33.Lowy F D. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 34.Monje-Casas F, Jurado J, Prieto-Alamo M J, Holmgren A, Pueyo C. Expression analysis of the nrdHIEF operon from Escherichia coli. Conditions that trigger the transcript level in vivo. J Biol Chem. 2001;276:18031–18037. doi: 10.1074/jbc.M011728200. [DOI] [PubMed] [Google Scholar]
- 35.Mulliez E, Ollagnier de Choudens S, Meier C, Cremonini M, Luchinat C, Trautwein A X, Fontecave M. Iron-sulfur interconversions in the anaerobic ribonucleotide reductase from Escherichia coli. J Biol Inorg Chem. 1999;4:614–620. doi: 10.1007/s007750050385. [DOI] [PubMed] [Google Scholar]
- 36.Novick R P. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 37.Ohlsen K, Koller K P, Hacker J. Analysis of expression of the alpha-toxin gene (hla) of Staphylococcus aureus by using a chromosomally encoded hla::lacZ gene fusion. Infect Immun. 1997;65:3606–3614. doi: 10.1128/iai.65.9.3606-3614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez-Casal J, Caparon M G, Scott J R. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Projan S L, Novick R P. The molecular basis of virulence. In: Cross K B, Archer G L, editors. Staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 55–81. [Google Scholar]
- 40.Ross R A, Onderdonk A B. Production of toxic shock syndrome toxin 1 by Staphylococcus aureus requires both oxygen and carbon dioxide. Infect Immun. 2000;68:5205–5209. doi: 10.1128/iai.68.9.5205-5209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Sawers G. The aerobic/anaerobic interface. Curr Opin Microbiol. 1999;2:181–187. doi: 10.1016/S1369-5274(99)80032-0. [DOI] [PubMed] [Google Scholar]
- 43.Schaferkordt S, Chakraborty T. Vector plasmid for insertional mutagenesis and directional cloning in Listeria spp. BioTechniques. 1995;19:720–722. , 724–725. [PubMed] [Google Scholar]
- 44.Schenk S, Laddaga R A. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;73:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- 45.Scotti C, Valbuzzi A, Perego M, Galizzi A, Albertini A M. The Bacillus subtilis genes for ribonucleotide reductase are similar to the genes for the second class I NrdE/NrdF enzymes of Enterobacteriaceae. Microbiology. 1996;142:2995–3004. doi: 10.1099/13500872-142-11-2995. [DOI] [PubMed] [Google Scholar]
- 46.Singh R D, Khullar M, Ganguly N K. Role of anaerobiosis in virulence of Salmonella typhimurium. Mol Cell Biochem. 2000;215:39–46. doi: 10.1023/a:1026545630773. [DOI] [PubMed] [Google Scholar]
- 47.Sinha N K, Snustad D P. Mechanism of inhibition of deoxyribonucleic acid synthesis in Escherichia coli by hydroxyurea. J Bacteriol. 1972;112:1321–1324. doi: 10.1128/jb.112.3.1321-1334.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sjöberg B M, Hahne S, Mathews C Z, Mathews C K, Rand K N, Gait M J. The bacteriophage T4 gene for the small subunit of ribonucleotide reductase contains an intron. EMBO J. 1986;5:2031–2036. doi: 10.1002/j.1460-2075.1986.tb04460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamarit J, Mulliez E, Meier C, Trautwein A, Fontecave M. The anaerobic ribonucleotide reductase from Escherichia coli. The small protein is an activating enzyme containing a [4Fe-4S]2+ center. J Biol Chem. 1999;274:31291–31296. doi: 10.1074/jbc.274.44.31291. [DOI] [PubMed] [Google Scholar]
- 50.Torrents E, Jordan A, Karlsson M, Gibert I. Occurrence of multiple ribonucleotide reductase classes in gamma-proteobacteria species. Curr Microbiol. 2000;41:346–351. doi: 10.1007/s002840010147. [DOI] [PubMed] [Google Scholar]
- 51.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 52.Yarwood J M, McCormick J K, Schlievert P M. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J Bacteriol. 2001;183:1113–1123. doi: 10.1128/JB.183.4.1113-1123.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yarwood J M, Schlievert P M. Oxygen and carbon dioxide regulation of toxic shock syndrome toxin 1 production by Staphylococcus aureus MN8. J Clin Microbiol. 2000;38:1797–1803. doi: 10.1128/jcm.38.5.1797-1803.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young P, Ohman M, Xu M Q, Shub D A, Sjöberg B M. Intron-containing T4 bacteriophage gene sunY encodes an anaerobic ribonucleotide reductase. J Biol Chem. 1994;269:20229–20232. [PubMed] [Google Scholar]