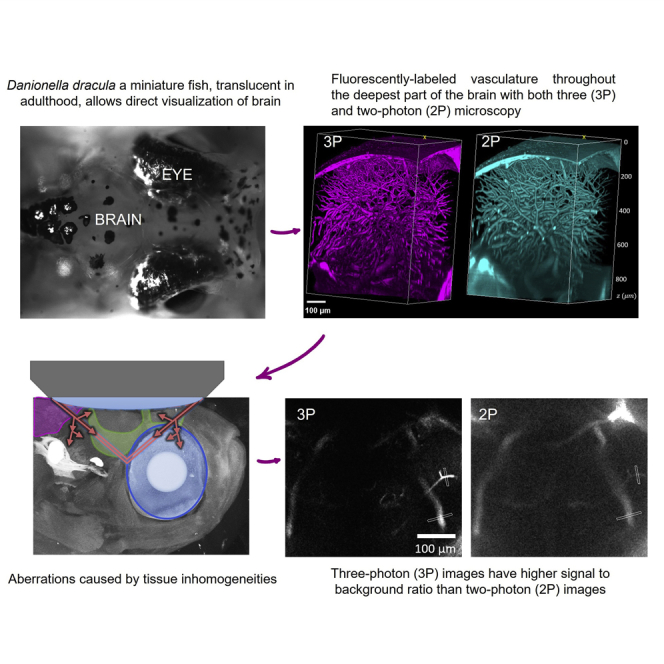
Summary
Although optical microscopy has allowed scientists to study the entire brain in early developmental stages, access to the brains of live, adult vertebrates has been limited. Danionella, a genus of miniature, transparent fish closely related to zebrafish has been introduced as a neuroscience model to study the adult vertebrate brain. However, the extent of optically accessible depth in these animals has not been quantitatively characterized. Here, we show that both two- and three-photon microscopy can access the entire depth and rostral-caudal extent of the adult wildtype Danionella dracula brain without any modifications to the animal other than mechanical stabilization. Three-photon microscopy provides higher signal-to-background ratio and optical sectioning of fluorescently labeled vasculature through the deepest part of the brain, the hypothalamus. Hence, we use multiphoton microscopy to penetrate the entire adult brain within the geometry of this genus’ head structures and without the need for pigment removal.
Subject areas: Biological sciences, Biotechnology, Biological sciences research methodologies
Graphical abstract
Highlights
-
•
The entire adult brain of Danionella dracula is accessible with multiphoton microscopy
-
•
Two-photon microscopy has low signal-to-background ratio in deep regions of the brain
-
•
Three-photon microscopy overcomes aberrations induced by tissue inhomogeneity
Biological sciences; Biotechnology; Biological sciences research methodologies
Introduction
Understanding the functional organization of the brain requires studying complex spatiotemporal relationships between neurons and how they interact to give rise to behavior. High scattering and absorption of tissue, including that of the pigmented skin and skull, and the head size of most commonly-used vertebrate species (Takasaki et al., 2020; Theer and Denk, 2006; Wang et al., 2020b) currently limits high-resolution optical imaging at the greatest depths of the adult brain (Horton et al., 2013; Takasaki et al., 2020; Wang et al., 2018a). Small animals such as zebrafish larva and frog tadpoles allow non-invasive, in vivo access to brains due to both the relative transparency of their skin and late ossification of the skull (Ahrens et al., 2012; Andalman et al., 2019; Cox and Fetcho, 1996; Fetcho and O’Malley, 1995; He et al., 2016; Higashijima et al., 2003; Koyama et al., 2016; McArthur et al., 2020; Vladimirov et al., 2014). However, these organisms often lack the behavioral complexity that develops in adulthood, especially in social contexts such as courtship, copulation, and resource defense (Rubenstein and Alcock, 2019). Multiphoton microscopy (MPM) enables deep optical access into scattering biological tissues(Cheng et al., 2019; Denk et al., 1990; Horton et al., 2013; Liu et al., 2019; Wang et al., 2020a). By using longer excitation wavelengths compared to one-photon excitation (e.g., confocal microscopy), MPM increases the penetration depth of photons, and the nonlinear excitation improves the contrast by suppressing the out-of-focus fluorescence. Two-photon microscopy (2PM) has become the gold standard of deep in vivo imaging and has greatly advanced our understanding of biological systems (Denk et al., 1990; Helmchen and Denk, 2005). More recently, the advent of three-photon microscopy (3PM) has enabled unprecedented imaging depths, including through the intact skull of adult mice, zebrafish, and Danionella minnows that are closely related to zebrafish (Chow et al., 2020; Horton et al., 2013; Huang et al., 2020; Wang et al., 2018b).
Three-photon microscopy allows penetration through the skull and scales of the adult zebrafish head, enabling a new range of questions in this well-established model animal. For example, this permitted structural and functional imaging through the entire telencephalon and deeper into the optic tectum and cerebellum than was possible before (Chow et al., 2020). However, the adult zebrafish brain approaches 1.5 mm in thickness (Friedrich et al., 2013) below the surface of the skull at its greatest depth in the hypothalamus (∼50% greater than the Danionella adults studied here), making imaging at its greatest depth not yet possible given the current limitations of optical microscopy (Chow et al., 2020).
D. dracula adults studied here reach approximately 11–18 mm in length and have a transparent body with a poorly ossified skull above the brain(Conway et al., 2021; Tatarsky et al., 2021), making them especially amenable to optical imaging. Although we recently demonstrated imaging of D. dracula brain vasculature with 3PM, the performance of this technique was not characterized (Chow et al., 2020) and optical imaging with 2PM, which is currently a more widely used technique, has remained relatively shallow (Penalva et al., 2018; Schulze et al., 2018). As reported here, we found that either 2PM or 3PM are capable of penetrating the entire brain of this wild-type, pigmented vertebrate without the need for any modifications to the animal other than mechanical stabilization. However, a rigorous quantitative assessment shows that 3PM is superior to 2PM for the deepest brain regions, which are within the hypothalamus. This is especially important as the hypothalamus has gained increasing attention for its critical role in the performance of a range of social behaviors (e.g., (Karigo et al., 2021; Tripp et al., 2018)). We compare 2PM at 920 nm to 3PM at 1280 nm excitation wavelength by imaging fluorescein-labeled vasculature through the deepest part of the adult brain. We additionally compare 2PM and 3PM images obtained with the same excitation beam at 1280 nm by simultaneous labeling of the blood vessels with fluorescein and Alexa Fluor 680. Our results indicate that the higher order of excitation in 3PM is greatly beneficial for maintaining high contrast and optical sectioning at greater depths compared to 2PM.
Results
2PM versus 3PM images of blood vessels through the entire depth of the adult brain
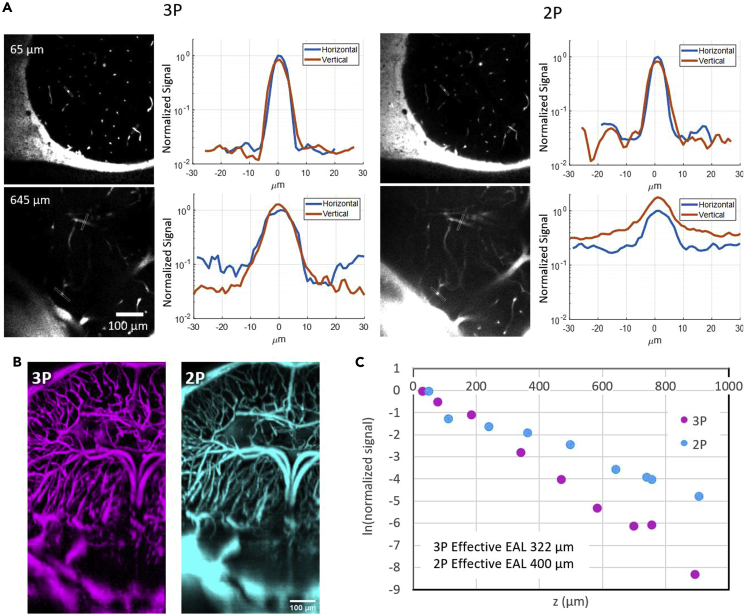
We labeled the vasculature of adult D. dracula with dextran-coupled fluorescein and collected 2PM and 3PM images using 920 and 1280 nm excitation wavelengths, respectively (see STAR Methods). Using the white-light path of the microscope (see Figure 6C), we chose an imaging region at the boundary of the cerebellum and midbrain optic tectum to ensure that the deepest part of the brain (hypothalamus) was within the field-of-view (FOV). To determine the bottom of the brain, we collected the second harmonic generation (SHG) signal generated by the 1280 nm excitation wavelength. The bone at the bottom of the brain was clearly identifiable in the SHG channel (Figure 1) (for gross morphology, see [23]]. To compare the performance of 2PM and 3PM, we collected images through the entire depth of the brain at the cerebellar-tectal boundary. The powers used to obtain the deepest images were 14 and 16 mW for 2PM and 3PM, respectively, for similar pixel values (within ∼25%). 3PM maintained high signal-to-background ratio (SBR) throughout the entire depth of the brain, up to 945μm at the cerebellar-tectal boundary (Figure 2A). In comparison, 2PM images had substantially lower SBR in the deep regions of the brain (Figures 2A and 2B). We also collected SHG and third harmonic generation (THG) of the skin to visualize the distribution of pigments above the brain (Figure 2C). There was no significant SHG from the pigments; however, the THG signal allows us to compare the location of our fluorescence images to the white light image of the head for verification of the location where imaging was done.
Figure 6.
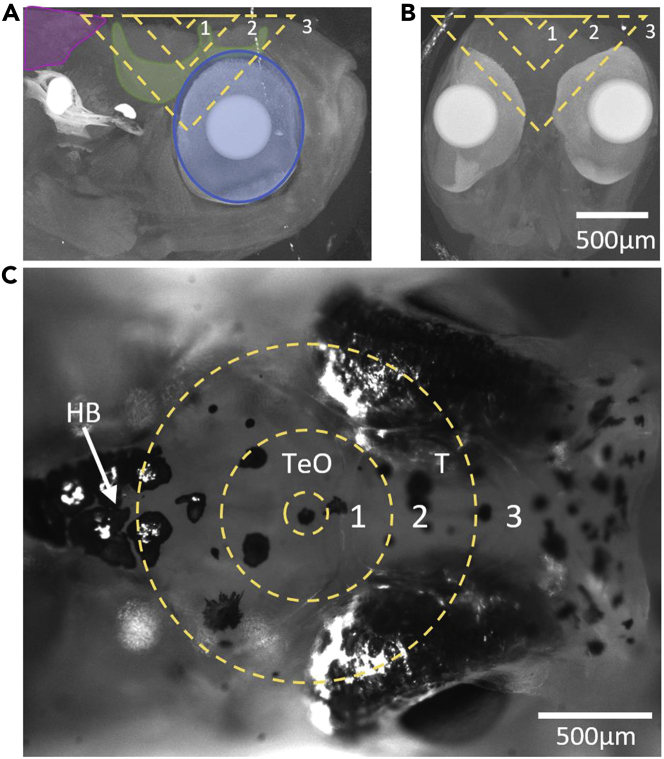
Illustration of head structures that fall within the cone of light for deep imaging in the adult D. dracula brain
The dashed lines represent the cone of light for an NA of 1 at depths of 100, 400, and 800 μm labeled as 1, 2, and 3, respectively.
(A) Maximum projection of computed tomography (CT) images of the head that contain the right half of the brain. Muscle (magenta), bone (green), and eye pigments (blue) are outlined in the vicinity of the cone of light.
(B) Maximum projection of CT images of the head that contain the brain.
(C) White light image of the adult head. Abbreviations: T, telencephalon; TeO, optic tectum; HB, hindbrain.
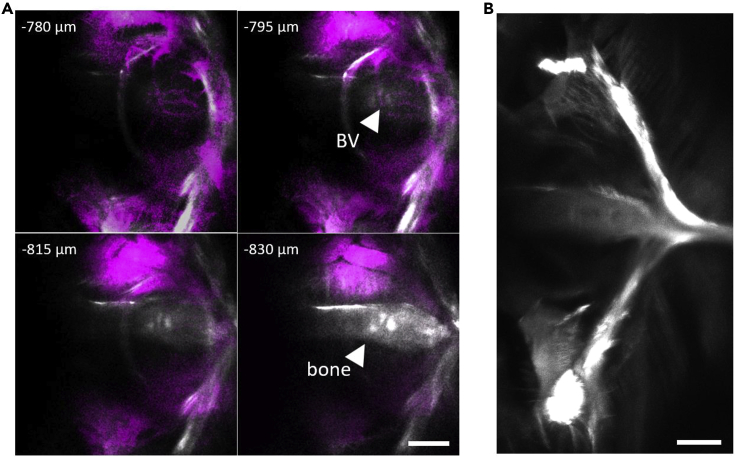
Figure 1.
Structural information obtained by SHG enables determining the bottom of the brain
(A and B) Determining the bottom of the brain using SHG from the bone under the brain.
(A) Frames at four depths near the bottom containing three-photon excited fluorescence in magenta and SHG in white. Bone structure in SHG channel is clearly visible at 830 μm (arrowhead). Bottom of the brain in this example is 795 μm, which is the top right image where clear blood vessel (BV) structure of the brain is visible adjacent to bone.
(B) SHG of entire bone under the brain at ∼850 μm depth (tiling of two FOVs; structure matches previous macroscopic characterization of D. dracula bone, see (Conway et al., 2021)). Scale bars indicate 100 μm.
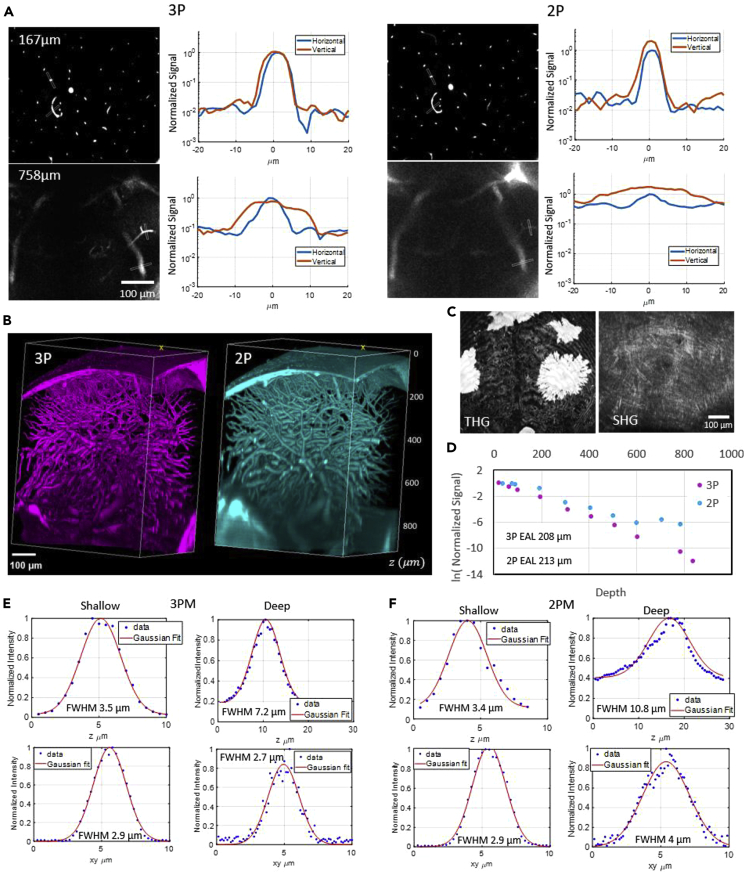
Figure 2.
Characterization of 2PM and 3PM images through the deepest part of the adult D. dracula brain–the cerebellum-midbrain boundary
Excitation wavelengths of 920 nm at 2 MHz repetition rate and 1280 nm at 333 kHz repetition rate were used for 2PM and 3PM, respectively.
(A) Signal-to-background ratio comparison of horizontal and vertical blood vessels for 2PM and 3PM images at two depths inside the brain. In each line profile plot, the values are normalized to the maximum brightness of the horizontal blood vessel.
(B) Representative 3D reconstruction of 3PM (left) and 2PM (right) images of fluorescein-labeled blood vessels in the midbrain.
(C) Maximum projection of third harmonic generation (THG, left) and second harmonic generation (SHG, right) of all frames containing the skin. Signals were generated by 1280 nm excitation. Pigments produce very bright signal in the THG channel.
(D) Characterization of effective attenuation length inside the brain for 2PM and 3PM excitation wavelengths as described in the STAR Methods section.
(E) Axial (z) and lateral (xy) width of small blood vessel characterized in shallow (∼100 μm) and deep (∼600 μm) regions with 3PM.
(F) Axial (z) and lateral (xy) width of small blood vessel characterized in shallow (∼100 μm) and deep (∼600 μm) regions with 2PM.
We measured the lateral and axial width of a small horizontal blood vessel in shallow (∼100 μm) and deep (∼600 μm) regions. In shallow regions, we found the lateral width to be 2.9 μm for both 3PM and 2PM and the axial width to be 3.4 and 3.5 μm for 3PM and 2PM, respectively. In the deep regions, we found the lateral width to be 2.7 and 4 μm and the axial width to be 7.2 and 10.8 μm for 3PM and 2PM, respectively (Figures 2E and 2F).
In addition, we compared the brightness of horizontal to vertical blood vessels for both 3PM and 2PM (Figure 2A). Although 3PM images maintained equal brightness for horizontal and vertical blood vessels, the brightness of vertical blood vessels was higher than that of horizontal ones in 2PM images. The relatively higher brightness in vertical blood vessels when compared to the horizontal ones in the 2P images can be attributed to distortions in the point spread function (PSF), causing elongation of the focal volume in the axial direction and therefore increasing the excitation volume (Wang et al., 2018b). With NA = 1, as in our system, the fluorescence generated within a 5 μm thick slab around the focal plane is ∼89% of the total fluorescence from an infinite volume for 2P excitation (920 nm) and ∼99% for 3P excitation (1280 nm). Therefore, the fluorescence generated by vertical blood vessels is expected to be approximately 1.1 times that generated by the horizontal ones in 2PM images. However, the brightness in vertical blood vessels was measured to be ∼2 times the brightness of horizontal ones for 2P excitation, indicating degraded axial confinement of 2PM. This observation is further supported by the effective attenuation length (EAL) values from these images which are measured to be 208 and 213 μm for 1280 and 920 nm excitation wavelengths, respectively (Wang et al., 2018b).
Although most of our experiments were done with low repetition rate lasers, we also used an 80 MHz laser for 2PM excitation to evaluate the power levels necessary for imaging through the brain. We collected 2PM and 3PM images of three adult fish through the deepest part of the brain using 80 MHz repetition rate for 2PM excitation. Differences in horizontal and vertical blood vessel brightness were observed, indicating PSF degradation (Figure S1). We measured the EAL (see STAR Methods) to be ∼267 μm at 920 nm and ∼300 μm at 1280 nm. To image (with similar pixel brightness values) the deepest part of the brain average power levels of 213 and 18 mW were used for 2PM and 3PM, respectively. Fish had healthy body color and movement immediately after imaging and for at least one week after imaging.
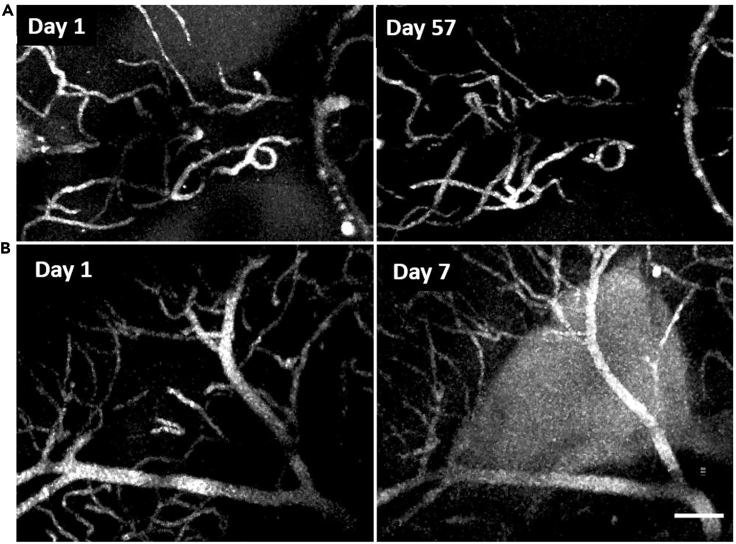
The optical accessibility in adulthood opens the feasibility of longitudinal imaging of the same animal over any period of its life. As an initial demonstration, we followed a first round of vasculature imaging by a second one two months later (Figure 3A) in one animal and one week later (Figure 3B) in a second animal. Note that we did not inject the animal with any dye on the day of the second time point. Of interest, there seems to be some dye leakage in the ventricles at the one-week timepoint, but in the animal where we imaged two months later, we do not observe a similar dye leakage.
Figure 3.
Longitudinal imaging of midbrain vasculature
(A and B) Maximum projection of a subset of vasculature in one animal at two timepoints separated by 57 days (A) and in a second animal separated by 7 days (B). Images in A are within the cerebellar-tectal boundary and in D within the optic tectum. Scale bar in lower right frame represents 100 μm for all frames.
3PM improves image quality by suppressing side lobes of the PSF
To delineate the effects of a longer wavelength and the higher order nonlinear excitation, we compared 2PM and 3PM both with 1280 nm excitation by injecting blood vessels with a combination of Alexa Fluor 680 and Fluorescein (both dextran coupled). Maximum powers used to obtain images through the deepest part of the brain were 1.2 and 34 mW for 2PM and 3PM, respectively, with pixel values about 50% higher in 2PM images.
In deep 2PM images, the brightness difference between horizontal and vertical blood vessels was observed (Figure 4A), indicating degradation of the PSF. This effect is also observable as blurriness of the features in the deep regions of the maximum projection of the 2PM, but not the 3PM stack (Figure 4B). The EAL value obtained from 2PM images was larger than the value obtained by analyzing the 3PM images, further verifying PSF degradation of 2PM (Wang et al., 2018b) (Figure 4C). We measured the lateral and axial width of a small horizontal blood vessel in shallow (∼100 μm) and deep (∼690 μm) regions by collecting the 2PM and 3PM signals from the same blood vessel (Figure S2). In shallow regions, we found the lateral width to be 2.6 and 3.3 μm and the axial width to be 4.2 and 6.5 μm for 3PM and 2PM, respectively. In deep regions, we found the lateral width to be 5.8 and 7.3 μm and the axial width to be 8 and 12.1 μm for 3PM and 2PM, respectively (Figure S2).
Figure 4.
Characterization of 2PM and 3PM (both with 1280 nm excitation) images through shallow and deep regions of the adult D. dracula brain
(A) Brightness comparison of horizontal and vertical blood vessels for 3PM and 2PM images at various depths inside the brain. In each line profile plot, the values are normalized to the maximum brightness of the horizontal blood vessel.
(B) Maximum projection of a column containing the deepest part of the brain.
(C) Characterization of effective attenuation length inside the brain for 2PM and 3PM excitation wavelengths as described in the STAR Methods section. EAL is measured to be 400 μm with 2PM images and 322 μm with 3PM.
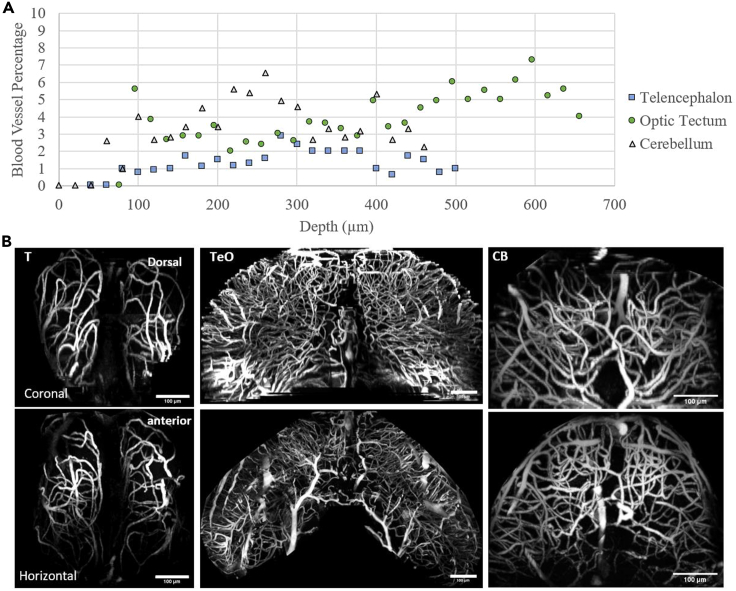
To determine the source of the background in deep images, we quantified the volume distribution of blood vessels in different regions of the brain, which are the main source of fluorescence in our images. The quantification of blood vessel distribution (Figure 5) shows that in all regions of the brain, blood vessels occupy less than ten percent of the volume. Based on the effective attenuation lengths obtained from tissue and the fluorophore distribution in blood vessels (≤10% fluorescent labeling density), the theoretically predicted SBR at all experimental depths (Figures 2 and 4) should be much higher than what we measured for both 2PM and 3PM (Akbari et al., 2022; Takasaki et al., 2020; Theer and Denk, 2006). Therefore, fluorophore distribution alone cannot explain the low SBR levels observed in the deep 2PM images.
Figure 5.
Characterization of blood vessel density in different regions of the brain
(A) Blood volume percentage in telencephalon, optic tectum, and cerebellum characterized at various depths (see STAR Methods).
(B) Maximum projection of vasculature in telencephalon (T), optic tectum (TeO), and cerebellum (CB). Coronal and horizontal views of vasculature are shown in top and bottom rows, respectively. Dorsal and anterior direction of each row is marked on the leftmost image.
Discussion
We demonstrate full optical accessibility in the adult brain of a teleost fish by imaging the deepest part of the brain (Figures 1A, 2B, and 4B) in the hypothalamus, and other entire regions such as the telencephalon and the cerebellum (Figure 5B). Although both 2PM and 3PM can penetrate deep inside the brain, only 3PM maintains high contrast through the deepest part. By comparing both 2PM and 3PM with the excitation wavelength of 1280 nm, we show that using longer wavelengths for 2PM is not sufficient for improving the image contrast in the deepest brain regions.
To determine the source of decline in the image contrast (e.g., SBR), we first assessed the distribution of fluorophores (e.g., blood volume fraction), which suggested that the background cannot be because of fluorophore distribution alone as the blood volume fraction is below 10% in all regions. In small aquatic vertebrates such as the D. dracula adults studied here (and zebrafish), fluorescence generated by pigmentation, particularly from the eyes, also contributes to the background (Figures S3 and S4). We found that the brightness of the fluorescence from the pigments is similar to that from the blood vessels (Figure S4) Although zebrafish genetic lines that remove pigments can help reduce such fluorescence, the complete removal of pigmentation can cause health issues (e.g., blindness) and therefore can limit applicability to biological questions (Holmgren and Sheets, 2021; Page-McCaw et al., 2004). Although decreasing fluorophore volume fraction (e.g. pigment removal) can decrease the uniformly distributed background (e.g., bulk background (Wang et al., 2020a)), the skin does not occupy a large volume in the cone of light. Therefore, pigment is not the main source of bulk background generation and removal of the pigment from the skin is not expected to drastically increase SBR levels. In addition, the differences in brightness of horizontal and vertical blood vessels cannot be explained by the fluorescent labeling density.
Aberrations in a sample can distort the PSF, leading to localized brightness differences and therefore, background generation that would depend on the specific location in the field of view (i.e., defocus background (Wang et al., 2020a)). For example, the baseline of the deep 2PM image is different between horizontal and vertical blood vessels in Figure 4A, indicating a potential localized background contribution. In small animals such as D. dracula, tissue inhomogeneities throughout the cone of light can lead to degradation of the PSF, producing defocus background in 2PM images that significantly degrade the SBR. Due to its higher order of excitation, 3PM can maintain a sharper focus through highly aberrating samples. Although scattering and aberration are mainly caused by a single layer of dense bone in through-skull imaging of the mouse brain (Wang et al., 2018b), aberrations in Danionella likely result from multiple tissue types that enter the cone of light with deep imaging, including skin with a variety of pigments, bone surrounding the perimeter of the brain and over the midbrain-telencephalon boundary (Conway et al., 2021), curvature and pigmentation of the eye (particularly at telencephalic levels), and muscle over the hindbrain region (Figure 6). The 3PM images have higher SBR because the higher order of excitation (compared to 2PM) causes the excitation volume to be more localized to the focus depth (e.g., suppression of the side lobes of the PSF caused by aberration), therefore, decreasing the excitation in out-of-focus regions.
Although we demonstrated imaging through the entire depth of the brain of an adult vertebrate, D. dracula, miniaturization is a widespread phenomenon with many examples in fish (Hanken and Wake, 1993; Nelson et al., 2016) and amphibians (Hanken and Wake, 1993; Perez-Martinez and Leal, 2021), making the method described here potentially applicable to hundreds of vertebrate species. Fishes alone account for more than half of all extant vertebrate species (Nelson et al., 2016) and most do not currently have readily available genetic lines. Nonetheless, diverse fish species are increasingly important for comparative studies addressing how divergent patterns of brain organization contribute to the variation and evolution of complex behavioral repertoires (Bukhari et al., 2019; Dunlap et al., 2021; Isa et al., 2021; Jaggard et al., 2020; Jeffery, 2020; McArthur et al., 2020). Deep, high-resolution imaging in small animal species like the one studied here thus provides the potential for investigating the functional organization of all brain regions of an adult vertebrate. The non-invasive nature of 2P and 3P imaging shown here also enables longitudinal imaging of the brain, in this case vasculature, in the same animal at different life history stages of such species.
In conclusion, we compare 2PM and 3PM performance for imaging through the entire depth of an adult vertebrate brain, in this case D. dracula. The results show that imaging throughout the entire adult brain is achievable, making regions that are not possible to visualize in adult zebrafish (Chow et al., 2020), now accessible in D. dracula and likely others in this genus. The small size of the adult D. dracula brain is particularly exciting for implementing whole-brain, in vivo volumetric imaging as well as detailed investigation of the relationship between brain activity and neurovascular circulation under both normal and pathological conditions with high temporal resolution. With MPM enabling optical access to adult brains and a repertoire of methods that allow observation of larval brains (Ahrens and Engert, 2015; McArthur et al., 2020), aquatic vertebrates such as Danionella and other genera with small species (Chow et al., 2020; Hanken and Wake, 1993; Huang et al., 2020) provide exciting opportunities for studying brain organization over the lifetime of individual vertebrates for the first time.
Limitations of the study
Although this study characterizes the optically accessible depth inside the brain in these animals, it does not demonstrate imaging of brain activity. Imaging calcium activity indicators might require higher power levels due to the requirement for high temporal resolution; however, based on the power levels used in experiments with 80MHz laser repetition rate, these animals can tolerate the higher power levels required for functional brain imaging.
In addition, although we demonstrate longitudinal imaging, the long term effects of anesthesia and imaging on the brain are not clear and are important to consider for answering biological questions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Experimental models: Organisms/strains | ||
| Danionella dracula | Breeding colony, Bass Lab, Cornell University |
minnows |
| Software and algorithms | ||
| MATLAB | Mathworks Inc. | R2020b |
| FIJI | ImageJ | https://imagej.net/Fiji |
| Excel | Microsoft | 2018 |
| Other | ||
| Multiphoton Microscope | Thorlabs Inc. | Bergamo II system B242 |
| Objective Lens | Olympus | XLPLN25XWMP2 |
| Ti:Sapphire Laser | Coherent | Chameleon |
| Non-collinear optical parametric amplifier | Spectra Physics | NOPA |
| Laser amplifier | Spectra Physics | Spirit 1030-70 |
| Fluorescent dye | ThermoFisher | Alexa Fluor 680, 70,000 MW |
| Fluorescent dye | ThermoFisher | Fluorescein, 70,000 MW |
| Mountable putty | Loctite | 1865809 |
| Water pump | Elemental Scientific | ESI MP2 |
| Anesthetic | Sigma-Aldrich | Benzocaine |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Najva Akbari (na395@cornell.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
In vivo imaging of fluorescently labeled vasculature
Following methods reported earlier (Chow et al., 2020), adult D. dracula were anesthetized in 0.0025-0.004% benzocaine solution and their vasculature labeled via injection of a 10% solution of dextran fluorescein (70,000 molecular weight, Thermo Fisher) and/or dextran Alexa Fluor 680 (70,000 molecular weight, Thermo Fisher) into blood vessels in a highly vascularized region caudal to the operculum and rostral to the heart. Fish were stabilized by positioning them on a mountable putty (Loctite 1865809). Fish were perfused through the mouth and over the gills at a rate of 1 mL min−1 with an ESI MP2 Peristaltic Pump (Elemental Scientific) with well-oxygenated temperature-controlled fish system water containing 0.00185% benzocaine (4-L reservoir heated with a Top Fin Betta Aquarium Heater set to 25°C). All procedures were in accord with the US National Institutes of Health guidelines for animal use in experiments and were approved by Cornell University’s Institutional Animal Care and Use Committee. We have imaged fluorescently-labeled vasculature in 14 D. dracula. All of these animals survived the experiments and we were able to visualize the entire depth of the brain in all of them.
CT (computed tomography)
Adult D. dracula were euthanized via deep anesthesia in 0.03% benzocaine solution. Whole fish were fixed in 4% paraformaldehyde at 4°C for 24 h and then stained with 1% iodine metal and 2% potassium iodide. The stained fish was scanned at 120 kV per 10 W on Zeiss Versa 520, using the 4X objective and a resolution of 2.7 μm. The machine took 2,401 exposures of 0.7 s each, and the CT data was reconstructed using the standard Zeiss reconstruction software.
Method details
Imaging setup
Images were obtained using a commercially available multiphoton microscope (Bergamo II system B242, Thorlabs Inc.) with a high numerical aperture objective lens (XLPLN25XWMP2, Olympus, NA 1.05). The back aperture was approximately backfilled for both 920 nm and 1280 nm excitation beams. The excitation source was a non-collinear optical parametric amplifier (NOPA, Spectra-Physics) pumped by an amplifier (Spirit 1030-70, Spectra-Physics). To control the excitation power, a half waveplate and a polarization beam splitter were used. A two-prism (SF11 glass) compressor was used to compensate for the normal dispersion of the optics of the light source and the microscope, including the objective lens. The full width at half maximum (FWHM) pulse width after the objective lens was measured to be 120 fs at 920 nm path and 50 fs at 1280 nm, assuming sech2 pulse intensity profile. For 3PM and 2PM, the excitation wavelength was 1280 nm and 920 nm, respectively. For deep imaging, low repetition rates of 333 kHz and 2 MHz were used for 3PM and 2PM, respectively. The FWHM pulse width was measured to be 120 fs at 920 nm path and 50 fs at 1280 nm, assuming sech2 pulse intensity profile. For some experiments a mode-locked Ti:Sapphire laser (Chameleon, Coherent) at 920 nm was used for 2PM excitation. The FWHM pulse width was measured to be 90 fs, assuming sech2 pulse intensity profile. Images were collected at approximately 1 frame per second over a field-of-view (FOV) of 539 μm by 539 μm with 512 by 512 pixels.
Image processing
Images were processed using ImageJ software. A 1-pixel radius median filter was applied to all images. For 3D reconstruction, the images were scaled by the ratio of z step size to the pixel size and were visualized using ‘Volume Viewer’ feature of the software. For line profiles, a 5-pixel wide line was drawn on the feature of interest. For resolution characterization line profiles were acquired using ImageJ and a Gaussian fit was obtained using MATLAB.
Characterization of effective attenuation lengths (EALs)
For EAL characterization a 10-pixel wide line was drawn on blood vessels that had the top 1% of brightest pixel values. The brightest value of the line profile was used as the signal value for that depth. In areas where the autofluorescence of other features dominated the brightest pixel values, the image was cropped to only contain blood vessels and then the top 1% of brightest pixel values were chosen as the value for that depth. The signal value was then normalized to the square or cube of the power used at the corresponding depth, depending on the order of excitation. The EALs and the reported maximum powers are representative of our other measurements.
Characterization of blood vessel distribution
Images were processed as described previously (see ‘image processing’). Areas corresponding to different regions (telencephalon, optic tectum, and cerebellum) were identified based on relative location in the brain and the distribution of vasculature. Areas of interest were outlined in ImageJ software and an appropriate threshold was applied to mask the blood vessels. Area fraction measurement function of ImageJ was used to obtain percentage of blood vessel for each frame, which likely over-estimates the blood volume concentration because the axial resolution (approximately 7 μm in our experiments) is comparable to or larger than the size of the capillary vessels. Therefore, the contribution to background fluorescence is less than the amount represented from images.
Quantification and statistical analysis
There is no statistical analysis in this article.
Additional resources
We have no relevant resources.
Acknowledgments
Thank you to Dr. Eric Schuppe for comments on the manuscript. We thank the Cornell Institute of Biotechnology for acquiring CT images of the adult D. dracula head. This work was funded by National Science Foundation (DBI-1707312, IOS1656664), National Institutes of Health (U01NS103516), and Cornell Neurotech Mong Fellowship.
Author contributions
Conceptualization, N.A., R.L.T, C.X., and A.H.B.; Software, N.A.; Methodology, N.A., R.T., K.E.K., A.H.B., and C.X.; Visualization, N.A.; Supervision, J.R.F., C.X., and A.H.B.; Writing – Original Draft, N.A.; Writing – Review & Editing, N.A., R.L.T, K.E.K., J.R.F., A.H.B., and C.X.
Declaration of interests
The authors declare no competing interests.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105191.
Supplemental information
Data and code availability
Original data reported in this study will be shared by the lead contact on request.
Any additional information required to reanalyze the data reported in this article is available from the lead contact on request.
This article does not report original code.
References
- Ahrens M.B., Engert F. Large-scale imaging in small brains. Curr. Opin. Neurobiol. 2015;32:78–86. doi: 10.1016/j.conb.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens M.B., Li J.M., Orger M.B., Robson D.N., Schier A.F., Engert F., Portugues R. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature. 2012;485:471–477. doi: 10.1038/nature11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari N., Rebec M.R., Xu C., Xia F., Xu C. Imaging deeper than the transport mean free path with multiphoton microscopy. Biomed. Opt Express. 2022;13:452–463. doi: 10.1364/BOE.444696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andalman A.S., Burns V.M., Lovett-Barron M., Broxton M., Poole B., Yang S.J., Grosenick L., Lerner T.N., Chen R., Benster T., et al. Neuronal dynamics regulating brain and behavioral state transitions. Cell. 2019;177:970–985.e20. doi: 10.1016/j.cell.2019.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari S.A., Saul M.C., James N., Bensky M.K., Stein L.R., Trapp R., Bell A.M. Neurogenomic insights into paternal care and its relation to territorial aggression. Nat. Commun. 2019;10:4437. doi: 10.1038/s41467-019-12212-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.-T., Lett K.M., Schaffer C.B. Surgical preparations, labeling strategies, and optical techniques for cell-resolved, in vivo imaging in the mouse spinal cord. Exp. Neurol. 2019;318:192–204. doi: 10.1016/j.expneurol.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow D.M., Sinefeld D., Kolkman K.E., Ouzounov D.G., Akbari N., Tatarsky R., Bass A., Xu C., Fetcho J.R. Deep three-photon imaging of the brain in intact adult zebrafish. Nat. Methods. 2020;17:605–608. doi: 10.1038/s41592-020-0819-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway K.W., M Kubicek K., Britz R. Extreme evolutionary shifts in developmental timing establish the miniature Danionella as a novel model in the neurosciences. Dev. Dyn. 2021;250:601–611. doi: 10.1002/dvdy.280. [DOI] [PubMed] [Google Scholar]
- Cox K.J., Fetcho J.R. Labeling blastomeres with a calcium indicator: a non-invasive method of visualizing neuronal activity in zebrafish. J. Neurosci. Methods. 1996;68:185–191. doi: 10.1016/0165-0270(96)00067-2. [DOI] [PubMed] [Google Scholar]
- Denk W., Strickler J.H., Webb W.W. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Dunlap K.D., Koukos H.M., Chagnaud B.P., Zakon H.H., Bass A.H. Vocal and electric fish: revisiting a comparison of two teleost models in the neuroethology of social behavior. Front. Neural Circuits. 2021;15:713105. doi: 10.3389/fncir.2021.713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetcho J.R., O’Malley D.M. Visualization of active neural circuitry in the spinal cord of intact zebrafish. J. Neurophysiol. 1995;73:399–406. doi: 10.1152/jn.1995.73.1.399. [DOI] [PubMed] [Google Scholar]
- Friedrich R.W., Genoud C., Wanner A.A. Analyzing the structure and function of neuronal circuits in zebrafish. Front. Neural Circuits. 2013;7:71. doi: 10.3389/fncir.2013.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanken J., Wake D.B. Miniaturization of body size: organismal consequences and evolutionary significance. Annu. Rev. Ecol. Syst. 1993;24:501–519. [Google Scholar]
- He H.-Y., Shen W., Hiramoto M., Cline H.T. Experience-dependent bimodal plasticity of inhibitory neurons in early development. Neuron. 2016;90:1203–1214. doi: 10.1016/j.neuron.2016.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmchen F., Denk W. Deep tissue two-photon microscopy. Nat. Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- Higashijima S.i., Masino M.A., Mandel G., Fetcho J.R. Imaging neuronal activity during zebrafish behavior with a genetically encoded calcium indicator. J. Neurophysiol. 2003;90:3986–3997. doi: 10.1152/jn.00576.2003. [DOI] [PubMed] [Google Scholar]
- Holmgren M., Sheets L. Influence of Mpv17 on hair-cell mitochondrial homeostasis, synapse integrity, and vulnerability to damage in the zebrafish lateral line. Front. Cell. Neurosci. 2021;15:693375. doi: 10.3389/fncel.2021.693375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton N.G., Wang K., Kobat D., Clark C.G., Wise F.W., Schaffer C.B., Xu C. In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat. Photonics. 2013;7:205–209. doi: 10.1038/nphoton.2012.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K.-H., Rupprecht P., Frank T., Kawakami K., Bouwmeester T., Friedrich R.W. A virtual reality system to analyze neural activity and behavior in adult zebrafish. Nat. Methods. 2020;17:343–351. doi: 10.1038/s41592-020-0759-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isa T., Marquez-Legorreta E., Grillner S., Scott E.K. The tectum/superior colliculus as the vertebrate solution for spatial sensory integration and action. Curr. Biol. 2021;31:R741–R762. doi: 10.1016/j.cub.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaggard J.B., Lloyd E., Yuiska A., Patch A., Fily Y., Kowalko J.E., Appelbaum L., Duboue E.R., Keene A.C. Cavefish brain atlases reveal functional and anatomical convergence across independently evolved populations. Sci. Adv. 2020;6:eaba3126. doi: 10.1126/sciadv.aba3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery W.R. Astyanax surface and cave fish morphs. EvoDevo. 2020;11:14. doi: 10.1186/s13227-020-00159-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karigo T., Kennedy A., Yang B., Liu M., Tai D., Wahle I.A., Anderson D.J. Distinct hypothalamic control of same- and opposite-sex mounting behaviour in mice. Nature. 2021;589:258–263. doi: 10.1038/s41586-020-2995-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama M., Minale F., Shum J., Nishimura N., Schaffer C.B., Fetcho J.R. A circuit motif in the zebrafish hindbrain for a two alternative behavioral choice to turn left or right. Elife. 2016;5:e16808. doi: 10.7554/eLife.16808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Deng X., Tong S., He C., Cheng H., Zhuang Z., Gan M., Li J., Xie W., Qiu P., Wang K. In vivo deep-brain structural and hemodynamic multiphoton microscopy enabled by quantum dots. Nano Lett. 2019;19:5260–5265. doi: 10.1021/acs.nanolett.9b01708. [DOI] [PubMed] [Google Scholar]
- McArthur K.L., Chow D.M., Fetcho J.R. In: The Zebrafish in Biomedical Research, American College of Laboratory Animal Medicine. Cartner S.C., Eisen J.S., Farmer S.C., Guillemin K.J., Kent M.L., Sanders G.E., editors. Academic Press; 2020. Chapter 46 - zebrafish as a model for revealing the neuronal basis of behavior ∗; pp. 593–617. [DOI] [Google Scholar]
- Nelson J.S., Grande T.C., Wilson M.V.H. John Wiley & Sons; 2016. Fishes of the World. [Google Scholar]
- Page-McCaw P.S., Chung S.C., Muto A., Roeser T., Staub W., Finger-Baier K.C., Korenbrot J.I., Baier H. Retinal network adaptation to bright light requires tyrosinase. Nat. Neurosci. 2004;7:1329–1336. doi: 10.1038/nn1344. [DOI] [PubMed] [Google Scholar]
- Penalva A., Bedke J., Cook E.S.B., Barrios J.P., Bertram E.P.L., Douglass A.D. Establishment of the miniature fish species Danionella translucida as a genetically and optically tractable neuroscience model. bioRxiv. 2018 doi: 10.1101/444026. Preprint at. [DOI] [Google Scholar]
- Perez-Martinez C.A., Leal M. Lizards as models to explore the ecological and neuroanatomical correlates of miniaturization. Beyond Behav. 2021;158:1121–1168. doi: 10.1163/1568539X-bja10104. [DOI] [Google Scholar]
- Rubenstein D.R., Alcock J. Eleventh Edition. Oxford University Press; 2019. Animal Behavior. ISBN: 9781605358949. [Google Scholar]
- Schulze L., Henninger J., Kadobianskyi M., Chaigne T., Faustino A.I., Hakiy N., Albadri S., Schuelke M., Maler L., Del Bene F., Judkewitz B. Transparent Danionella translucida as a genetically tractable vertebrate brain model. Nat. Methods. 2018;15:977–983. doi: 10.1038/s41592-018-0144-6. [DOI] [PubMed] [Google Scholar]
- Takasaki K., Abbasi-Asl R., Waters J. Superficial bound of the depth limit of two-photon imaging in mouse brain. eNeuro. 2020;7 doi: 10.1523/ENEURO.0255-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatarsky R.L., Guo Z., Campbell S.C., Kim H., Fang W., Perelmuter J.T., Schuppe E.R., Reeve H.K., Bass A.H. Acoustic and postural displays in a miniature and transparent teleost fish, Danionella dracula. J. Exp. Biol. 2021;225:jeb244585. doi: 10.1101/2021.11.10.468077. [DOI] [PubMed] [Google Scholar]
- Theer P., Denk W. On the fundamental imaging-depth limit in two-photon microscopy. J. Opt. Soc. Am. Opt Image Sci. Vis. 2006;23:3139–3149. doi: 10.1364/JOSAA.23.003139. [DOI] [PubMed] [Google Scholar]
- Tripp J.A., Feng N.Y., Bass A.H. Behavioural tactic predicts preoptic-hypothalamic gene expression more strongly than developmental morph in fish with alternative reproductive tactics. Proc. Biol. Sci. 2018;285:20172742. doi: 10.1098/rspb.2017.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladimirov N., Mu Y., Kawashima T., Bennett D.V., Yang C.-T., Looger L.L., Keller P.J., Freeman J., Ahrens M.B. Light-sheet functional imaging in fictively behaving zebrafish. Nat. Methods. 2014;11:883–884. doi: 10.1038/nmeth.3040. [DOI] [PubMed] [Google Scholar]
- Wang M., Wu C., Sinefeld D., Li B., Xia F., Xu C. Comparing the effective attenuation lengths for long wavelength in vivo imaging of the mouse brain. Biomed. Opt. Express. 2018;9:3534–3543. doi: 10.1364/BOE.9.003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Ouzounov D.G., Wu C., Horton N.G., Zhang B., Wu C.-H., Zhang Y., Schnitzer M.J., Xu C. Three-photon imaging of mouse brain structure and function through the intact skull. Nat. Methods. 2018;15:789–792. doi: 10.1038/s41592-018-0115-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Wang T., Xu C. Three-photon neuronal imaging in deep mouse brain. Optica. 2020;7:947–960. doi: 10.1364/OPTICA.395825. [DOI] [Google Scholar]
- Wang T., Wu C., Ouzounov D.G., Gu W., Xia F., Kim M., Yang X., Warden M.R., Xu C. Quantitative analysis of 1300-nm three-photon calcium imaging in the mouse brain. Elife. 2020;9:e53205. doi: 10.7554/eLife.53205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original data reported in this study will be shared by the lead contact on request.
Any additional information required to reanalyze the data reported in this article is available from the lead contact on request.
This article does not report original code.