Abstract
In vitro mariner transposon mutagenesis of Streptococcus pneumoniae chromosomal DNA was used to isolate regulatory mutants affecting expression of the comCDE operon, encoding the peptide quorum-sensing two-component signal transduction system controlling competence development. A transposon insertion leading to increased comC expression was found to lie directly upstream from the S. pneumoniae clpP gene, encoding the proteolytic subunit of the Clp ATP-dependent protease, whose expression in Bacillus subtilis is controlled by the CtsR repressor. In order to examine clp gene regulation in S. pneumoniae, a detailed analysis of the complete genome sequence was performed, indicating that there are five likely CtsR-binding sites located upstream from the clpE, clpP, and clpL genes and the ctsR-clpC and groESL operons. The S. pneumoniae ctsR gene was cloned under the control of an inducible promoter and used to demonstrate regulation of the S. pneumoniae clpP and clpE genes and the clpC and groESL operons by using B. subtilis as a heterologous host. The CtsR protein of S. pneumoniae was purified and shown to bind specifically to the clpP, clpC, clpE, and groESL regulatory regions. S. pneumoniae ΔctsR, ΔclpP, ΔclpC, and ΔclpE mutants were constructed by gene deletion/replacement. ClpP was shown to act as a negative regulator, preventing competence gene expression under inappropriate conditions. Phenotypic analyses also indicated that ClpP and ClpE are both required for thermotolerance. Contrary to a previous report, we found that ClpC does not play a major role in competence development, autolysis, pneumolysin production, or growth at high temperature of S. pneumoniae.
The regulatory pathways leading to the development of competence for DNA uptake in the gram-positive bacteria Bacillus subtilis and Streptococcus pneumoniae are strikingly similar. Proteins required for DNA binding and transport, encoded by the so-called late competence genes, are well conserved in the two bacteria (12), and the initial regulatory events involve extracellular peptide-signaling systems in both cases (57). Competence of S. pneumoniae for DNA transformation is controlled by a peptide quorum-sensing signal transduction pathway including the ComC-derived competence-stimulating peptide, the ComD membrane-bound histidine kinase, and the ComE response regulator, all of which are encoded by the comCDE operon (5, 19, 42). In B. subtilis, an unrelated extracellular peptide derived from the ComX polypeptide activates the ComP/ComA two-component system that is encoded by the comPA operon lying directly downstream from comX (25, 57).
Major differences exist, however, in the intermediate steps between the quorum-sensing device and the specific synthesis of competence proteins involved in DNA uptake and processing. In S. pneumoniae, this link is provided by a specific sigma factor, ComX, whose synthesis is dependent on the ComD/ComE two-component system (26). The S. pneumoniae ComX sigma factor is, in turn, required for the competence-specific expression of late com genes (26). In B. subtilis, no competence-specific sigma factor exists and the link between quorum sensing and late competence gene expression instead requires the release of the ComK transcription activator from targeted proteolysis by the ClpCP ATP-dependent protease (34, 59).
Clp ATP-dependent proteases are involved in regulation by proteolysis in several bacteria (45) and consist of a proteolytic subunit, ClpP, on which substrate specificity is conferred through association with ATPase subunits (ClpA, ClpC, and ClpX), which include members of the ubiquitous Hsp100 family (52). ComK synthesis in B. subtilis involves a complex network of two-component systems and global regulators, in which the general stress response genes clpC and clpP play essential roles (34, 35, 37). clpC and clpP of B. subtilis are both members of the class III group of heat shock genes, whose expression is controlled by the CtsR repressor (11).
There is growing evidence indicating that Clp proteins play an important role in the survival and virulence of pathogens during host infection. The clpP gene was isolated during a signature-tagged mutagenesis screen for virulence genes of Salmonella enterica serovar Typhimurium (21). In Yersinia enterocolitica, ClpP has been shown to modulate transcription of the adhesion invasion locus (ail) (41). Clp ATPases have also been shown to be involved in virulence, including ClpX of Staphylococcus aureus (31) and ClpB of Leishmania sp. (52). Furthermore, patients with leprosy or tuberculosis have antibodies specifically directed against mycobacterial ClpC (33). In Listeria monocytogenes, ClpP and the ClpC and ClpE Hsp100 ATPases are all required for stress survival, growth at high temperature, and virulence (13, 40, 47, 48).
In S. pneumoniae, a gene encoding an Hsp100-type Clp ATPase was isolated during a large-scale identification of virulence genes using the signature-tagged mutagenesis technique (44). Virulence of the S. pneumoniae mutant was significantly affected, as shown by using a mouse septicemia model (44). However, despite their ubiquity in bacteria and their important role in virulence, little is known about the regulation and function of clp genes in pathogens other than L. monocytogenes, where many of the clp genes have been shown to be controlled by the CtsR repressor (39). Elucidation of the regulatory pathways controlling clp gene expression is therefore likely to be important for our understanding of the virulence of gram-positive pathogens.
We show here that ClpP of S. pneumoniae plays a role in the maintenance of low levels of comCDE expression under conditions that do not support competence development. We also show that expression of the S. pneumoniae clpP and clpE genes and clpC and groESL operons is heat inducible and controlled directly by the CtsR repressor. Phenotypic analyses indicate that, unlike in B. subtilis and contrary to a previous report (4), ClpC is not involved in control of the expression of S. pneumoniae competence genes.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and transformation.
Escherichia coli K-12 strain TG1 (14) was used for cloning experiments, and E. coli strain BL21 λ DE3 (54) was used for protein overexpression and purification. E. coli strains were grown in LB medium (50) and transformed by electroporation with selection on LB plates supplemented with ampicillin (100 μg/ml).
The B. subtilis and S. pneumoniae strains used in this work are listed in Table 1. The B. subtilis strains used in this study were derivatives of B. subtilis 168 trpC2 and were grown in LB medium. They were transformed and selected by using plasmid or chromosomal DNA as previously described (35).
TABLE 1.
B. subtilis and S. pneumoniae strains used in this study
| Strain | Relevant genotype | Source or referencea |
|---|---|---|
| B. subtilis | ||
| QB4991 | trpC2 ΔctsR amyE::(′lacZ aphA3) | 11 |
| QB8068 | trpC2 ΔctsR amyE::(′lacZ cat) | pAC5→QB4991 |
| QB8069 | trpC2 ΔctsR amyE::(′lacZ aphA3) thrC::(PxylA-ctsR spc) | pxyl-ctsR-Spn→QB4991 |
| QB8070 | trpC2 ΔctsR amyE::(clpP′-bgaB cat) thrC::(PxylA-ctsR spc) | pDL-clpP-Spn→QB8069 |
| QB8071 | trpC2 ΔctsR amyE::(clpC′-bgaB cat) thrC::(PxylA-ctsR spc) | pDL-clpC-Spn→QB8069 |
| QB8132 | trpC2 ΔctsR amyE::(groES′-bgaB aphA3) | pDK-groE-Spn→QB8068 |
| QB8133 | trpC2 ΔctsR amyE::(groES′-bgaB aphA3) thrC::(PxylA-ctsR spc) | pxyl-ctsR-Spn→QB8132 |
| QB8134 | trpC2 ΔctsR amyE::(clpE′-bgaB aphA3) | pDK-clpE-Spn→QB8068 |
| QB8135 | trpC2 ΔctsR amyE::(clpE′-bgaB aphA3) thrC::(PxylA-ctsR spc) | pxyl-ctsR-Spn→QB8134 |
| S. pneumoniae | ||
| R6 | Subclone of R36A original isolate | Laboratory stock |
| R800 | R6 derivative | 27 |
| R348 | ebg::spc comC::pXF520 | 29 |
| R354 | ebg::kan comC::pXF520 | 29 |
| R461 | ebg::kan comC::pXF520 spc93::clpP | This study |
| R638 | spc93::clpP | R461→R800 |
| SP2000 | ebg::spc comC::pXF520 ΔclpP::aphA3 | ΔclpP DNA→R348 |
| SP2001 | ebg::spc comC::pXF520 ΔclpC::aphA3 | ΔclpC DNA→R348 |
| SP2002 | ebg::spc comC::pXF520 ΔclpE::aphA3 | ΔclpE DNA→R348 |
| SP2003 | ebg::spc comC::pXF520 ΔctsR::aphA3 | ΔctsR DNA→R348 |
| R895 | ssbB::pR424 | pR424→R800 |
| R1053 | ssbB::pR424 ΔclpE::aphA3 | SP2002→R895 |
| R1054 | ssbB::pR424 spc93::clpP | R461→R895 |
| R1055 | ssbB::pR424 ΔctsR::aphA3 | SP2003→R895 |
| R1056 | ssbB::pR424 ΔclpC::aphA3 | SP2001→R895 |
Arrows indicate construction by transformation with chromosomal or plasmid DNA or PCR-generated DNA fragments.
S. pneumoniae strains were grown in brain heart infusion (BHI; Difco) or CAT (46) medium. Expression of late competence genes was examined during growth in C+Y medium (28) as described previously (1). Transformation of S. pneumoniae was performed as described previously (1, 28), by using precompetent cells treated with synthetic competence-stimulating peptide 1 (25 ng/ml) to induce competence. Transformants were selected by plating on D medium agar plates supplemented with 4% horse blood (1) with kanamycin at 250 μg/ml or spectinomycin at 100 μg/ml.
In vitro mariner mutagenesis.
Mutagenesis of S. pneumoniae chromosomal DNA was performed as previously described (29). Briefly, plasmid pR412 was used as the source for the 1,146-bp spc mariner minitransposon, which carries the spc spectinomycin resistance-encoding gene (29). Plasmid pR412 was incubated with chromosomal DNA from S. pneumoniae strain R800 in the presence of purified Himar1 transposase, leading to random insertion of the minitransposon within the chromosomal DNA (29). Gaps in the transposition products were repaired, and the resulting in vitro-generated transposon insertion library was used to transform S. pneumoniae (29).
DNA manipulations and general experimental procedures.
Standard procedures were used to extract plasmids from E. coli (50). Chromosomal DNA of B. subtilis was isolated as previously described (35). Chromosomal DNA of S. pneumoniae was isolated by using the B. subtilis protocol, excepted for the lysis step, which was performed by resuspending cells in 0.1 ml of SEDS solution (NaCl at 0.15 M, EDTA at 0.15 M, deoxycholate [DOC] at 0.01%, sodium dodecyl sulfate at 0.02%) and incubating them for 5 min at 37°C. Amplification of DNA was performed by the PCR technique (38, 49) using Pwo polymerase (Roche) and S. pneumoniae R6 or R800 chromosomal DNA. Nucleotide sequences were determined by the dideoxy-chain termination method (51) using modified T7 DNA polymerase (55) (Amersham-Pharmacia).
Plasmids and plasmid constructions.
The plasmids used in this study are listed in Table 2, and the oligonucleotides used are listed in Table 3. Plasmid pXT (10), a derivative of pDG1731 (18), was used to express genes under control of the xylose-inducible promoter PxylA. A BamHI/EcoRI DNA fragment corresponding to the coding sequence of S. pneumoniae R6 ctsR was generated by PCR using oligonucleotides AC7(−25) and AC8(+469). Oligonucleotide positions are given relative to the translation initiation codon. This fragment was cloned into the respective sites of pXT to yield plasmid pxyl-ctsR-Spn, which allows ctsR expression under control of the xylose-inducible promoter, with integration as a single copy at the thrC locus. This plasmid was introduced into strain QB4991 or derivatives of QB8068, in which the endogenous ctsR gene is deleted, to give strains QB8069, QB8133, and QB8135 (see Table 1 for details). QB8068 is a derivative of strain QB4991 in which the Enterococcus faecalis aphA3 Kmr gene (58) at the amyE locus was replaced with the pC194 cat chloramphenicol resistance gene by transformation with plasmid pAC5 (30).
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pDG1731 | Plasmid allowing integration at the thrC locus | 18 |
| pXT | pDG1731 derivative allowing gene expression from the PxylA xylose-inducible promoter | 10 |
| pxyl-ctsR-Spn | pXT derivative carrying the ctsR coding sequence of S. pneumoniae | This study |
| pDL | Plasmid allowing integration at the amyE locus, and transcriptional fusion with bgaB | 61 |
| pAC5 | Plasmid allowing integration at the amyE locus, with the pC194 cat gene | 30 |
| pAC7 | Plasmid allowing integration at the amyE locus, with the E. faecalis aphA3 cassette | 60 |
| pDK | pAC7 derivative carrying the bgaB gene | This study |
| pDL-clpC-Spn | pDL derivative carrying a clpC′-bgaB fusion | This study |
| pDL-clpP-Spn | pDL derivative carrying a clpP′-bgaB fusion | This study |
| pDK-clpE-Spn | pDK derivative carrying a clpE′-bgaB fusion | This study |
| pDKgroES-Spn | pDK derivative carrying a groE′-bgaB fusion | This study |
| pET28a | Vector for overexpression of His-tagged proteins | Novagen |
| pETCtsR-Spn | pET28a derivative for overproduction of CtsR | This study |
| p5.00 | Plasmid carrying the luc reporter gene | Martin Stieger |
| pEVP3 | Plasmid containing the Cmr-encoding gene | 7 |
| pR412 | Plasmid carrying the mariner minitransposon | 29 |
| pR422 | p5.00 derivative with the luc gene under control of the ssbB promoter | This study |
| pR424 | pEVP3 derivative carrying the ssbB′-luc fusion from pR422, associated with the Cmr-encoding gene | This study |
TABLE 3.
Oligonucleotides used in this study
| Name | Sequence | Description |
|---|---|---|
| TM289 | GAAGAATTCGAAAAAGAAGAATGACTTGG | PclpP′-bgaB fusion |
| TM290 | GGAGGATCCTTTTGAGTTTTAATTTTGTTGG | PclpP′-bgaB fusion |
| TM291 | GAAGAATTCCACGCTTGGTATCTTGAGATTCAC | PclpC′-bgaB fusion |
| TM292 | GGAGGATCCTCTCTTTAAACCTTGACCTTG | PclpC′-bgaB fusion |
| AC6 | CTCCTCGAGCTTCCCTTTTCTATCTACCTC | CtsR overproduction |
| AC7 | GGAGGATCCGTTTAAAGAGAGAGGTGGGTTTGT | PxylA′-ctsR fusion |
| AC8 | GAAGAATTCAATAGTTCATCTTACTTCCCT | PxylA′-ctsR fusion |
| AC9 | GGTGGTCTCCCATGAGATTTAAAAATACATCGGATCATA | CtsR overproduction |
| AC22 | TCGAAAAGAAGAATGACTTGG | PclpP footprint |
| AC23 | TCCTTTTGAGTTTTAATTTTGTTGG | PclpP footprint |
| AC24 | GAATTAGGCTTAGATAAGTAG | PclpC footprint |
| AC25 | CCAGATTGATCTAAAATCGCC | PclpC footprint |
| AC72 | CGTAAGAACGTTCTCCACGGCTTGTTTG | Primer extension on clpP |
| AC80 | ATTGAATTCATCGCAATGGAAATTTACGAAC | PclpE′-bgaB fusion |
| AC81 | AAAGGATCCATCTACCTCATTTCTTTCTTTAGCC | PclpE′-bgaB fusion |
| AC82 | GTGAGAATTCTGCAGGCCAAGATTTGGCAG | PgroE′-bgaB fusion |
| AC83 | TCTGGATCCCTCCATAATGAGATAG | PgroE′-bgaB fusion |
| AC84 | GTTAGAATTCGCTTCTTGGGGTAT | aphA3 cassette |
| AC85 | TAGGGATCCAAATCTAGGTACTAA | aphA3 cassette |
| AC86 | TCCCCATGGACTTAGCGGTGGGATG | ctsR deletion |
| AC87 | ATCGAATTCCAAACCCACCTCTCTC | ctsR deletion |
| AC88 | AAGGGATCCAAGATGAACTATTCAAAAGC | ctsR deletion |
| AC89 | GTACCATGGCGTTGGCGTAAAGCC | ctsR deletion |
| AC90 | AAACCATGGAATGAATGTATCGAAAGTGCC | clpC deletion |
| AC91 | GTACGAATTCGTTGGCGTAAAGCC | clpC deletion |
| AC92 | TGGGGATCCGGATATTCGTTTTGACCAGG | clpC deletion |
| AC93 | ATGCCATGGCAAATTTTAACTGGCCTGC | clpC deletion |
| AC94 | TGACCATGGTTCCAGCTGCTAAAGTTGGC | clpP deletion |
| AC95 | TTTGAATTCTAATTTTGTTGGTCAAATG | clpP deletion |
| AC96 | ACTGGATCCGCGCCCAGGAAACACTTG | clpP deletion |
| AC97 | GCTACCATGGCAAGCGCCACAAACGATAG | clpP deletion |
| AC98 | CCTCCATGGTAAAATAGTAACGATAAG | clpE deletion |
| AC99 | CCTGAATTCTTTAAAGGTCAAAAATAG | clpE deletion |
| AC100 | GCAAGGGATCCATTCAGATTAAATCTGCC | clpE deletion |
| AC101 | ATACCATGGATAATGCAAGATTCC | clpE deletion |
| AC112 | TTTTCATTGTAACAACTTCTCAAAGC | PclpE footprint |
| AC113 | CCATTGAGATTGGTGTAAAGATG | PclpE footprint |
| AC114 | GATTTGGCAGATTTGGTCTTGG | PgroE footprint |
| AC115 | CGGTCCCCTAATGGTTTCAAC | PgroE footprint |
| AC124 | AGCCTGCAAGGACAAAGCCTCC | Primer extension on groE |
| MP122 | CGCGGATCCGGTGTAGACGTTAAACGTCC | ssbB′-luc fusion |
| MP158 | GCCGCGAAGCTTCTCAGGATATTGCAGATAC | ssbB′-luc fusion |
Plasmid pDL (61) was used to construct transcriptional fusions between the promoter region of clpC (clpC′-bgaB) or clpP (clpP′-bgaB) and the Bacillus stearothermophilus bgaB gene, encoding a thermostable β-galactosidase (22), with subsequent integration at the amyE locus. Plasmid pDK is a derivative of pAC7 (60) in which the E. coli lacZ gene is replaced with the B. stearothermophilus bgaB gene from plasmid pDL. Plasmid pDK was used to construct transcriptional fusions between the promoter region of clpE (clpE′-bgaB) or the groESL operon (groE′-bgaB) and the bgaB gene. Transcriptional fusions in pDL or pDK were constructed by using EcoRI/BamHI DNA fragments generated by PCR using oligonucleotides TM291(−345) and TM292(−15), TM289(−171) and TM290(−8), AC80(−260) and AC81(−3), and AC82(−210) and AC83(−9), corresponding to the clpC, clpP, clpE, and groESL promoter regions, respectively. Positions are given relative to the translation initiation codon. These fragments were cloned into the respective sites of plasmid pDL or pDK to produce plasmids pDLclpC-Spn, pDLclpP-Spn, pDKclpE-Spn, and pDKgroE-Spn, respectively. Linearization of these plasmids at the unique PstI site and transformation of the B. subtilis QB8068 or QB8069 strain with selection for chloramphenicol or Kmr yielded strains QB8071 (clpC′-bgaB), QB8070 (clpP′-bgaB), QB8134 (clpE′-bgaB), and QB8132 (groES′-bgaB).
CtsR was overexpressed by using pETCtsR-Spn, a derivative of pET28a (Novagen) in which a 478-bp BsaI/XhoI DNA fragment corresponding to the ctsR coding sequence, generated by PCR using oligonucleotides AC6 and AC9, was cloned between the NcoI and XhoI sites of plasmid pET28a. This allows the creation of a translational fusion adding six histidine residues to the carboxy terminus of the protein and placing expression of the gene under the control of a T7 promoter.
clpC, clpP, clpE, and ctsR deletion/replacement mutants were constructed by first performing a ligation between DNA fragments (∼500 bp) corresponding to the chromosomal DNA regions immediately upstream and downstream from each gene with an 877-bp EcoRI/BamHI DNA fragment generated by PCR using oligonucleotides AC84 and AC85 that carries the aphA3 Kmr gene deprived of its transcription initiation and termination signals. Fragments corresponding to the regions upstream and downstream from clpC, clpP, clpE, and ctsR were generated by PCR using oligonucleotides AC90 and AC91 and AC92 and AC93; AC94 and AC95 and AC96 and AC97; AC98 and AC99 and AC100 and AC101; and AC86 and AC87 and AC88 and AC89, respectively. Each resulting ligation was used as a template for PCR amplification using the external oligonucleotides (e.g., AC90 and AC93 for clpC). Products were purified following gel electrophoresis using the QIAquick Gel Extraction Kit (Qiagen). Purified DNA fragments were used directly for transformation of S. pneumoniae with selection for Kmr, and complete deletion of each gene was verified by PCR using additional oligonucleotides located further upstream and downstream from the original fragments.
Construction of an ssbB′-luc transcriptional fusion was carried out in two steps. The luc gene was placed under control of the ssbB promoter by construction of plasmid pR422 as follows. A DNA fragment overlapping the 5′ end of the ssbB gene was amplified from S. pneumoniae R800 chromosomal DNA by PCR using oligonucleotides MP122 and MP158 and digested with BamHI-HindIII to generate a 246-bp fragment. This fragment was cloned into a 9,004-bp-long BamHI-HindIII fragment from plasmid p5.00, which confers erythromycin resistance and carries the Photinus pyralis luc gene encoding firefly luciferase (53) to generate plasmid pR422 (Table 2). The ssbB′-luc transcriptional fusion was then associated with a chloramphenicol resistance-encoding gene to generate plasmid pR424 by cloning a 1,930-bp HindIII-SmaI fragment from plasmid pR422 containing the ssbB′-luc transcriptional fusion into a 2,022-bp HindIII-BsaAI fragment from plasmid pEVP3 (Table 2). Transformation of S. pneumoniae cells with plasmid pR424, with selection for chloramphenicol resistance, leads to integration of the plasmid at the ssbB locus by a single-crossover event.
β-Galactosidase and luciferase assays.
β-Galactosidase specific activities in S. pneumoniae were determined as described previously for B. subtilis (32, 35, 36), by using a Multiskan Ascent photometric microplate reader, and expressed as nanomoles of o-nitrophenyl-β-d-galactopyranoside per minute per milligram of protein. Cell lysis was performed by adding 0.005% DOC–0.01% sodium dodecyl sulfate (final concentrations).
For detection of luciferase activity, strains were first grown in CAT medium to an optical density at 550 nm (OD550) of 0.4. Cells were then resuspended in fresh 15% glycerol-containing CAT medium and frozen at −80°C. For inoculation, frozen cultures were thawed and diluted 1,500-fold in CAT medium and 280 μl was distributed among the wells of a 96-well Corning NBS plate. The cultures were incubated at 37°C in an Anthos LucyI luminometer. Approximately 3.5 h after inoculation and injection of 20 μl of a 10 mM luciferin solution in CAT medium, relative luminescence units (RLU) and OD492 were measured at 8-min intervals.
Overexpression and purification of CtsR.
pET-CtsR-Spn was introduced into the BL21 λ DE3 (plysE) strain (Novagen), in which the T7 RNA polymerase gene is under the control of the inducible lacUV5 promoter. The resulting strain was grown in LB medium at 30°C, and expression was induced during the exponential growth phase by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG; 0.1 mM). Purification of CtsR was performed by using immobilized metal affinity chromatography as previously described (11).
Gel mobility shift DNA-binding assays.
EcoRI/BamHI DNA fragments, corresponding to the promoter regions of the clpP and clpE genes and the clpC and groESL operons, were generated by PCR with oligonucleotides TM289 and TM290, AC80 and AC81, TM291 and TM292, and AC82 and AC83, respectively. Fragments were radioactively labeled with [α-32P]dATP using the Klenow fragment of DNA polymerase I (Gibco-BRL). Radiolabeling, DNA binding, and gel electrophoresis mobility shift assays were performed as previously described (11).
DNase I footprinting.
DNA fragments corresponding to the clpC, clpP, clpE, and groE promoter regions used for DNase I footprinting were prepared by PCR using 20 pmol of oligonucleotides AC24 and AC25, AC22 and AC23, AC112 and AC113, and AC114 and AC115, respectively. Labeling and DNase I treatment were performed as previously described (11).
RNA extraction and primer extension.
S. pneumoniae strains were grown in BHI medium at 37°C without shaking until the OD600 reached 0.6. Cells were pelleted and frozen immediately. Frozen cells were resuspended in 0.4 ml of water and disrupted with a FastPrep cell disintegrator (Bio 101, Inc.) for 30 s at 4°C by using 0.5 g of glass beads (106 μm; Sigma) in the presence of 0.4 ml of 4% Bentone MA (Rheox) and 0.5 ml of phenol-chloroform-isoamyl alcohol, pH 8.0 (Amresco). After centrifugation for 2 min at 20,817 × g supernatants were successively extracted with phenol-chloroform (1:1, vol/vol) and then chloroform-isoamyl alcohol (24:1, vol/vol). RNA was precipitated with isopropanol in the presence of 0.2 M NaCl and resuspended in 20 μl of water. RNA concentrations were determined by measuring the A260, and samples were stored at −20°C. Primer extensions were performed by incubating 20 μg of RNA, 1 pmol of oligonucleotide (previously labeled with [γ-32P]ATP [110 TBq/mmol] using T4 polynucleotide kinase), and 25 U of avian myeloblastosis virus reverse transcriptase (Roche). Oligonucleotides were chosen so as to hybridize approximately 30 bp downstream from the translation initiation codon (see Table 3). The corresponding DNA sequencing reactions were carried out by using the same oligonucleotides and PCR-amplified DNA fragments carrying the respective promoter regions.
Database comparisons and sequence analysis.
Computations were performed with the Genetics Computer Group sequence analysis software package (version 10.1; Genetics Computer Group, Inc., Madison, Wis.). Sequence comparisons with the GenBank database were accomplished with the National Center for Biotechnology Information BLAST2 (2) network service with the default parameter values provided. The complete S. pneumoniae type 4 genome sequence (56) was kindly made available by The Institute for Genomic Research (http://www.tigr.org).
RESULTS
Inactivation of clpP leads to overexpression of the comCDE operon in S. pneumoniae.
In vitro mariner transposon mutagenesis of S. pneumoniae chromosomal DNA was used to generate a library of mutants by transformation of strain R354, which carries a chromosomal comC′-lacZ transcriptional fusion (see Materials and Methods and reference 29). Mutants displaying a comCDEup or cup phenotype (29), i.e., increased β-galactosidase activity on 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)-T− plates (29), on which competence genes are normally not expressed, were chosen for further study. Chromosomal DNA was isolated from each mutant and used for backcross experiments by transforming the R354 parental strain with selection for spectinomycin resistance to ensure linkage of the cup phenotype with the transposon insertion. A total of 42 insertions were found to be distributed into nine different location groups, two of which have been previously characterized (29). A third class was represented by a single mutant, strain R461, in which the transposon was inserted directly upstream from a previously uncharacterized gene whose product shares 55% amino acid sequence identity with ClpP of B. subtilis, the proteolytic subunit of the Clp ATP-dependent protease (35). This suggested that ClpP may play a role in the early steps of competence development in S. pneumoniae. However, it remained to be determined whether the cup phenotype linked to the transposon insertion was due to overexpression or loss of expression of the clpP gene.
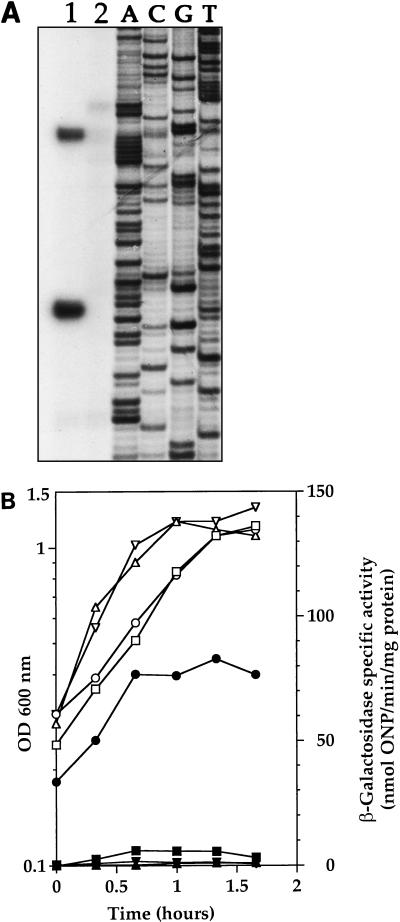
The transposon insertion upstream from clpP (spc93::clpP) was introduced into the wild type S. pneumoniae R800 strain by transformation with chromosomal DNA from strain R461 to give strain R638. Primer extension experiments were carried out to examine clpP expression by using total RNA isolated from strain R638 or the original wild-type parental strain, R800. As shown in Fig. 1A, clpP is expressed in strain R800 during growth in BHI medium and this expression is abolished in strain R638. Analysis of the nucleotide sequence of the region preceding the transcription initiation sites revealed likely −10 and −35 sequences for the lower signal (TTGACC N17 TATAAT; see Fig. 7B) sharing strong similarities with the consensus sequences of promoters recognized by the vegetative form of RNA polymerase holoenzyme, EςA. The mariner minitransposon was inserted with the duplication of a GA dinucleotide 65 bp upstream from the clpP translation initiation codon, between the −35 and −10 sequences (see Fig. 7B), consistent with the fact that clpP expression is correspondingly abolished. No consensus promoter-type sequences could be identified upstream from the uppermost signal, suggesting that this signal could be due to transcription from a promoter recognized by a minor sigma factor or to processing from a larger transcript.
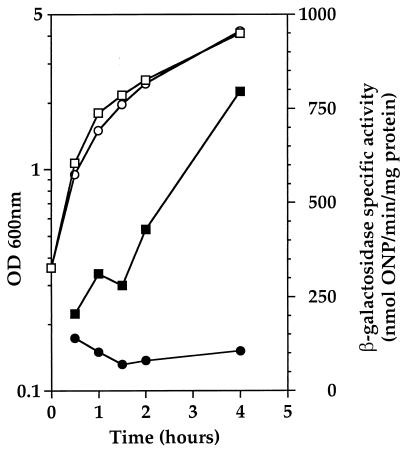
FIG. 1.
(A) clpP expression in strain R638 is drastically reduced. Primer extension analysis of clpP mRNA was performed with total RNAs isolated from parental strain R800 (lane 1) and spc93::clpP mutant strain R638 (lane 2). The corresponding DNA sequence is shown on the right. (B) Expression of a comC′-lacZ fusion is strongly increased in a ΔclpP mutant. Strains SP2000 (ΔclpP::apha3) (○, ●), SP2001 (ΔclpC::apha3) (▿, ▾), and SP2002 (ΔclpE::apha3) (▵, ▴) and parental strain R348 (□, ▪) were grown in BHI medium. Open symbols indicate the OD600, and solid symbols indicate β-galactosidase specific activity, expressed as nanomoles of ONP per minute per milligram of protein.
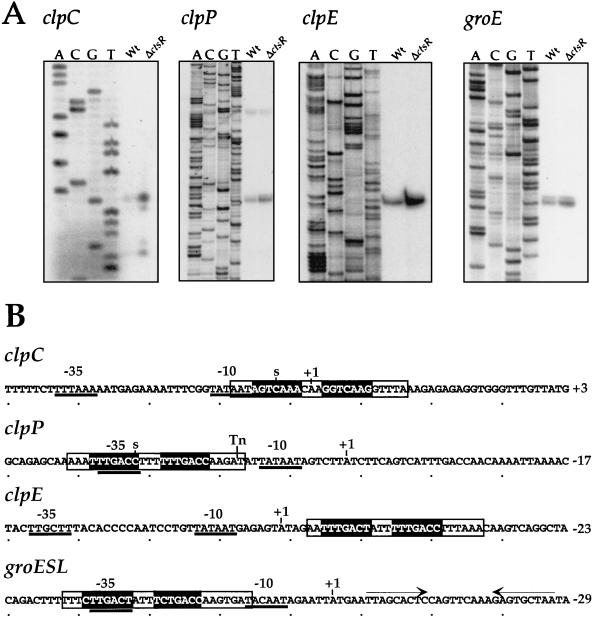
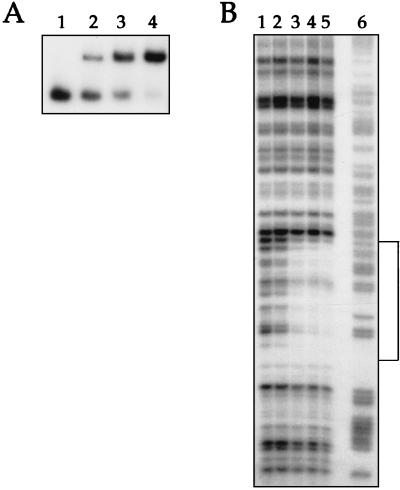
FIG. 7.
(A) Primer extension analysis of clpC, clpP, clpE, and groE mRNAs in the ΔctsR mutant. Total RNA isolated from wild-type (Wt) parental strain R348 or the ΔctsR mutant (SP2003) was used as the template for reverse transcriptase. The corresponding DNA sequences are shown on the left. (B) Nucleotide sequences of the clpC, clpP, clpE, and groE promoter regions. Potential −35 and −10 sequences are underlined, transcriptional start points are indicated by S and +1, CtsR heptad direct repeat operator sequences are shaded, the CIRCE operator sequence is indicated by inverted arrows, and regions protected by CtsR in DNase I footprint experiments are boxed. Tn indicates the transposon insertion site upstream from clpP in strain R461. Positions are numbered relative to the translation initiation codon.
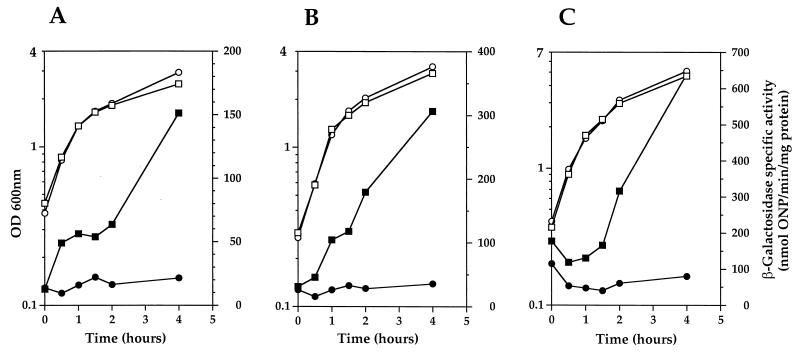
In order to confirm that ClpP negatively regulates comCDE expression in S. pneumoniae, a ΔclpP mutant (SP2000) was constructed by chromosomal replacement of the entire clpP coding sequence with the E. faecalis aphA3 Kmr gene through a double-crossover event. This was carried out by transforming S. pneumoniae strain R348, which carries a comC′-lacZ transcriptional fusion, with a PCR-generated DNA fragment containing the aphA3 Kmr-encoding gene and flanked by two 500-bp segments corresponding to the chromosomal regions immediately upstream and downstream of the clpP gene (see Materials and Methods).
As shown in Fig. 1B, expression of comC′-lacZ in strain R348 is very low during growth in BHI medium (approximately 4 nmol of ONP min−1 mg of protein−1) and is strongly increased (up to 24-fold) in strain SP2000 (ΔclpP::aphA3), confirming that ClpP negatively regulates comCDE expression.
Inactivation of clpC or clpE does not affect competence development or expression of the comCDE and ssbB genes in S. pneumoniae.
The fact that ClpP acts to negatively regulate expression of comCDE suggested that one of the Clp ATPase subunits may also act as a repressor. Analysis of the complete S. pneumoniae type 4 genome sequence (56) indicates that there are four genes encoding Clp ATPases, which we have designated clpC, clpE, clpL, and clpX, in accordance with established nomenclature (9, 52). ClpC, ClpE, and ClpL all belong to the Hsp100 family of Clp ATPases (9, 52).
In order to test whether the ClpC or ClpE ATPase plays a role in comCDE expression, ΔclpC and ΔclpE mutants of S. pneumoniae (strains SP2001 and SP2002, respectively) were constructed by transformation of strain R348 by the method described above for the ΔclpP mutant strain. As shown in Fig. 1B, comC′-lacZ expression in the ΔclpC and ΔclpE mutants was very low during growth in BHI medium and not significantly different from that in the R348 parental strain, indicating that, in contrast to ClpP, neither ClpC nor ClpE negatively regulates comCDE expression.
In order to examine the effects of ClpP, ClpC, and ClpE on late competence gene expression, chromosomal DNAs from strains R461 (spc93::clpP), SP2001 (ΔclpC::aphA3), and SP2002 (ΔclpE::aphA3) were used to introduce the corresponding mutations into S. pneumoniae strain R895 by transformation. The transposon insertion upstream from clpP, which practically abolishes expression of the gene (Fig. 1A), was used instead of the ΔclpP::aphA3 mutation, since strain SP2000 (ΔclpP::aphA3) was unable to grow in C+Y competence medium, a phenotype similar to that reported for the B. subtilis ΔclpP mutant (35), whereas strain R461 (spc93::clpP) was able to grow, albeit poorly (Fig. 2A), suggesting that residual expression of clpP occurred despite the transposon insertion within the promoter region.
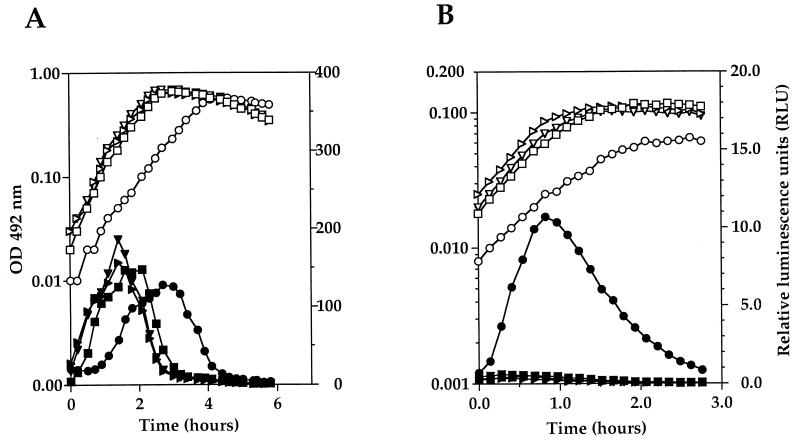
FIG. 2.
Expression of ssbB′-luc in strains R1053 (ΔclpE::apha3) (▹, ▸), R1054 (clpP::spc93) (○, ●), and R1056 (ΔclpC::apha3) (▿, ▾) and parental strain R895 (□, ▪). Strains were grown in C+Y competence-permissive medium (A) or CAT non-competence-permissive medium (B). Open symbols indicate the OD492, and solid symbols indicate RLU expressed as a function of time.
The resulting derivatives of strain R895 carry a chromosomal transcriptional fusion between the promoter of the ssbB gene, encoding single-stranded DNA-binding protein and known to be specifically induced during competence development (3, 43), and the P. pyralis luc gene, encoding firefly luciferase. The ssbB gene, also known as cilA (3), is specifically transcribed by RNA polymerase holoenzyme containing the ComX competence-specific sigma factor (26). Expression of ssbB′-luc has been shown to be directly correlated with the kinetics of transformation during competence development in S. pneumoniae (M. Prudhomme and J. P. Claverys, unpublished results).
As shown in Fig. 2A, no significant difference in the expression of ssbB′-luc was observed during growth in C+Y medium between the different clp mutants and the otherwise isogenic R895 reference strain, indicating that ClpP, ClpC, and ClpE are not required for competence development in S. pneumoniae under these conditions. Indeed, transformation assays during growth in C+Y medium for the S. pneumoniae ΔclpC::aphA3 mutant (strain SP2001) were not significantly different from that of the R348 parental strain (data not shown).
However, when cells were grown in CAT medium, in which derivatives of the R800 laboratory strain do not develop spontaneous competence, expression of ssbB′-luc was increased approximately 50-fold in strain R1054 (spc93::clpP), confirming the negative role of ClpP on competence gene expression in S. pneumoniae (Fig. 2B). Furthermore, unlike otherwise isogenic parental strain R354, the spc93 mutant (strain R461) developed spontaneous competence for transformation in CAT medium (data not shown), which is fully consistent with the isolation of the mutant on the basis of a cup phenotype on X-Gal-T− plates. The fact that the only transposon insertion at the clpP locus leading to a cup phenotype was found upstream from the gene and not within the coding sequence is not unexpected, since insertions disrupting the gene would be associated with a growth defect on X-Gal T− plates (29), a medium comparable to CAT, in which the ΔclpP::aphA3 mutant is also unable to grow.
CtsR of S. pneumoniae negatively regulates expression of clpP, clpE, and the clpC operon.
Analysis of the nucleotide sequence of the S. pneumoniae clpP promoter region revealed the existence of a likely operator site for the CtsR repressor of stress response genes, whose existence in S. pneumoniae was previously reported (11), suggesting that, as in B. subtilis, the clpP gene may belong to the CtsR regulon. A detailed DNA motif analysis of the complete S. pneumoniae type 4 genome sequence (56), carried out using the consensus CtsR heptad direct repeat operator sequence (A/GGTCAAA NAN A/GGTCAAA; 11), revealed only five candidate CtsR-binding sites.
These were located upstream from the clpE, clpL, and clpP genes, as well as the ctsR-clpC and groESL operons. In order to investigate regulation of the S. pneumoniae clp genes, the model gram-positive bacterium B. subtilis was used as a heterologous host. Regulation by CtsR of S. pneumoniae was studied in derivatives of B. subtilis strain QB4991, in which the entire B. subtilis ctsR gene is deleted (11).
The resulting strains contain the S. pneumoniae ctsR gene cloned under control of the PxylA xylose-inducible promoter and integrated as a single copy at the thrC locus, as well as transcriptional fusions between the promoter regions of the S. pneumoniae clpC (strain QB8071), clpP (strain QB8070), and clpE (strain QB8135) genes and the bgaB gene of B. stearothermophilus, which encodes a thermostable β-galactosidase, integrated as single copies at the amyE locus (see Materials and Methods).
Strains QB8071, QB8070, and QB8135 were grown at 37°C in LB medium in the presence or absence of xylose, and β-galactosidase activities were assayed (reported as nanomoles of ONP per minute per milligram of protein). As shown in Fig. 3A, clpC′-bgaB was weakly expressed (∼20 U of enzyme activity) in the presence of xylose when CtsR was produced and its expression was increased approximately eightfold in the absence of xylose. Expression of clpP′-bgaB and clpE′-bgaB fusions followed similar patterns (Fig. 3B and C), with basal levels of 40 U for clpP and 80 U for clpE in the presence of xylose, increasing approximately eightfold in the absence of xylose when ctsR was not expressed. These results clearly indicate that CtsR negatively regulates the S. pneumoniae clpC, clpP, and clpE genes.
FIG. 3.
Expression of clpC, clpP, and clpE is repressed by CtsR of S. pneumoniae. Expression of clpC′-bgaB (QB8071) (A), clpP′-bgaB (QB8069) (B), and clpE′-bgaB (QB8135) (C) in the presence (○, ●) or absence (□, ▪) of xylose. Cultures were grown in LB medium at 37°C to an OD600 of 0.3, and xylose was added to one-half of the culture at a final concentration of 20 mM. Open symbols indicate the OD600, and solid symbols indicate β-galactosidase specific activity, expressed as nanomoles of ONP per minute per milligram of protein.
CtsR binds specifically to the regions upstream from clpP, clpE, and the clpC operon.
An in vitro approach was used to demonstrate the direct interaction of S. pneumoniae CtsR with its target sites. For this purpose, the S. pneumoniae ctsR coding sequence was cloned into the pET28a vector, generating a carboxy-terminal translational fusion with six histidine residues. The resulting His-tagged CtsR protein (approximately 19 kDa) was then overproduced in E. coli and purified with an Ni-nitrilotriacetic acid agarose column (see Materials and Methods).
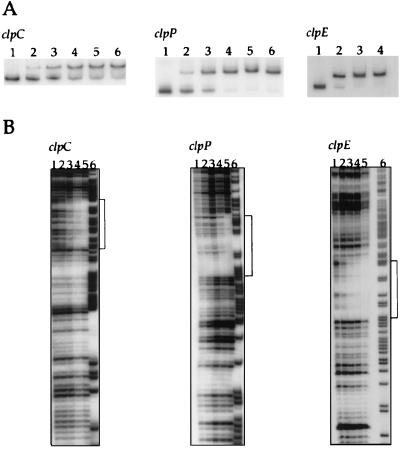
Purified S. pneumoniae CtsR was used in gel mobility shift DNA-binding assays with DNA fragments corresponding to the promoter regions of clpC, clpP, and clpE. Radiolabeled, PCR-generated DNA fragments corresponding to positions −139 to +65 (clpC), −171 to −9 (clpP), and −137 to +62 (clpE), relative to the respective translation initiation codons, were incubated with increasing amounts of CtsR. All DNA-binding assays were performed in the presence of an excess of nonspecific competitor DNA [1 μg of poly(dI-dC)]. As shown in Fig. 4A, CtsR bound specifically to all three radiolabeled promoter fragments, forming a single protein-DNA complex in each case, with complete displacement of the DNA fragments at the highest CtsR concentrations. These results indicate that CtsR of S. pneumoniae negatively regulates the expression of the clpC, clpP, and clpE genes by binding directly to their promoter regions.
FIG. 4.
CtsR binds specifically to the clpC, clpP, and clpE promoter regions. In gel mobility shift experiments (A), radiolabeled DNA fragments (10,000 cpm) corresponding to the clpC, clpP, and clpE promoter regions were incubated with increasing amounts of purified CtsR as follows: for clpC and clpP, lanes 1 to 6, 0, 10, 20, 40, 60, and 80 ng of CtsR, respectively; for clpE, lanes 1 to 4, 0, 20, 40, and 80 ng of CtsR, respectively. In DNase I footprinting analyses of CtsR binding (B), 50,000 cpm of each radiolabeled DNA fragment corresponding to the clpC, clpP, or clpE promoter region was incubated with increasing amounts of purified CtsR as follows: lanes 1 to 5, 0, 100, 200, 400, and 800 ng of CtsR, respectively; lane 6, G+A Maxam and Gilbert reaction of the corresponding DNA fragments. Regions protected by CtsR are shown by brackets.
In B. subtilis, CtsR binds to a highly conserved directly repeated sequence (A/GGTCAAA NAN A/GGTCAAA) that often overlaps the −35 and −10 sequences or the transcriptional start site of the controlled promoters (11). DNase I footprinting assays were performed on S. pneumoniae DNA fragments carrying the clpC, clpP, and clpE promoter regions to determine the extent of the protected region and the precise location of the CtsR-binding sites (Fig. 4B). When the nontemplate strand of the clpC DNA fragment was end labeled, CtsR protected a region extending from position −46 to position −21 (Fig. 4B). CtsR protected regions on the nontemplate strands extending from position −86 to position −62 for clpP and from position −58 to position −35 for clpE (Fig. 4B). All positions are given relative to the respective translational start sites.
The protected regions within the clpC, clpP, and clpE promoter sequences each contain the direct repeat CtsR operator site, in agreement with sequence analysis predictions (see Fig. 7B).
The groESL operon of S. pneumoniae is a novel member of the CtsR regulon.
As mentioned above, genome sequence analysis also revealed the existence of a potential CtsR-binding site upstream from the groESL operon, which encodes the classical chaperonins GroES and GroEL. This was somewhat surprising, since in B. subtilis and many other low-G+C gram-positive bacteria, the groESL operon belongs to the class I family of heat shock genes, known to be controlled by the HrcA repressor through its interaction with the highly conserved CIRCE operator sequence (TTAGCACTC-N9-GAGTGCTAA) (20). The CtsR regulons of B. subtilis and L. monocytogenes consist of genes encoding subunits of the Clp ATP-dependent protease (11, 39), and this would be the first example of a groESL operon regulated by CtsR. Inspection of the nucleotide sequence of the groESL upstream region revealed the presence of a highly conserved CIRCE operator sequence as well, located 16 bp downstream from the potential CtsR-binding site (see Fig. 7B). This tandem operator arrangement suggests that the S. pneumoniae groESL operon may be dually regulated by both HrcA and CtsR.
CtsR-dependent regulation of groESL expression was examined by using B. subtilis as a heterologous host as described above for the clp genes. Strain QB8133 carries a groES′-bgaB transcriptional fusion integrated at the amyE locus and the S. pneumoniae ctsR gene cloned under control of the PxylA xylose-inducible promoter at the thrC locus (see Materials and Methods). As shown in Fig. 5, groES′-bgaB was weakly expressed (∼100 U) in the presence of xylose when CtsR was produced and its expression was increased approximately eightfold in the absence of xylose, confirming the prediction that CtsR negatively regulates groESL expression.
FIG. 5.
groESL is under negative control of CtsR in vivo. Expression of groE′-bgaB (QB8133) in the presence (○, ●) or absence (□, ▪) of xylose. Cultures were grown in LB medium at 37°C to an OD600 of 0.3 and divided in two, and xylose was added to one of the cultures at a final concentration of 20 mM. Open symbols indicate the OD600, and solid symbols indicate β-galactosidase specific activity, expressed as nanomoles of ONP per minute per milligram of protein.
CtsR binds specifically to the regions upstream from the groESL operon.
Purified CtsR was used in gel mobility shift DNA-binding assays with a radiolabeled, PCR-generated DNA fragment corresponding to the promoter region of the groESL operon (positions −20 to +23 relative to the translation initiation codon). As shown in Fig. 6A, CtsR bound specifically to the radiolabeled promoter fragment, forming a single protein-DNA complex, with complete displacement of the DNA fragment at the highest CtsR concentration. DNase I footprinting assays were performed on the same DNA fragment to determine the extent of the protected region and the precise location of the CtsR-binding sites (Fig. 6B). When the nontemplate strand of the groESL DNA fragment was end labeled, CtsR protected a region extending from position −97 to position −73, relative to the translational start site, which contains the predicted direct repeat CtsR operator site (Fig. 6B and 7B).
FIG. 6.
CtsR binds specifically to the groESL promoter region. In gel mobility shift experiments (A), radiolabeled DNA fragments (10,000 cpm) were incubated with increasing amounts of purified CtsR. Lanes 1 to 4, 0, 20, 40, and 80 ng of CtsR, respectively. In DNase I footprinting experiments (B), 50,000 cpm of the radiolabeled DNA fragment corresponding to the promoter region was incubated with increasing amounts of purified CtsR. Lanes: 1 to 5, 0, 100, 200, 400, and 800 ng of CtsR, respectively; 6, G+A Maxam and Gilbert reaction of the corresponding DNA fragment. The region protected by CtsR is indicated by a bracket.
Expression of the clpP and clpE genes and the clpC and groESL operons is induced by heat shock.
Expression of CtsR-dependent genes is known to be induced under general stress conditions, including heat shock (9, 11, 23, 35, 39). In order to test whether the repression by S. pneumoniae CtsR also responds to heat shock, expression of the clpC′-bgaB, clpP′-bgaB, clpE′-bgaB, and groES′-bgaB transcriptional fusions was tested by using B. subtilis as a heterologous host in strains QB8071, QB8070, QB8135, and QB8133 during growth in LB medium in the presence of xylose at 37 or 48°C.
As shown in Table 4, all four genes were expressed at a low level at 37°C and strongly induced, from 20- to 30-fold, after a shift to 48°C, which is consistent with a CtsR-dependent stress response. This was confirmed in S. pneumoniae for the groESL operon at the mRNA level by primer extension experiments (data not shown), in agreement with a previous report showing that synthesis of GroEL is induced in response to heat shock (6).
TABLE 4.
Induction of clp′-bgaB and groES′-bgaB expression by heat shock
| Strain | Promoter | β-Galactosidase sp act (nmol of ONP/min/mg of protein)
|
Ratio | |
|---|---|---|---|---|
| 37°C | 48°C | |||
| QB8070 | PclpP | 35 | 1,200 | 34 |
| QB8071 | PclpC | 20 | 520 | 26 |
| QB8135 | PclpE | 80 | 1,600 | 20 |
| QB8133 | PgroES | 110 | 3,500 | 32 |
Genes of the CtsR regulon are derepressed in an S. pneumoniae ΔctsR mutant.
To confirm CtsR-dependent regulation of the clpP and clpE genes and the clpC and groESL operons in S. pneumoniae, a ΔctsR mutant of S. pneumoniae (strain SP2003) was constructed by transformation of strain R348 by chromosomal replacement of the entire ctsR coding sequence with the aphA3 Kmr-encoding gene through a double-crossover event. Primer extension experiments were performed to examine expression of CtsR-dependent genes by using total RNA isolated from ΔctsR::aphA3 mutant strain SP2003 or otherwise isogenic parental strain R348. As shown in Fig. 7A, expression of the clpP and clpE genes and the clpC and groESL operons is increased in the ΔctsR::aphA3 mutant during growth in BHI medium at 37°C, confirming their repression by CtsR in S. pneumoniae. The transcription start sites, the potential promoter sequences, the CtsR and HrcA operator sequences, and the regions protected by CtsR in DNase I footprinting experiments are indicated in Fig. 7B. The ΔctsR::aphA3 mutation had no effect on comCDE or ssbB expression in S. pneumoniae (data not shown), suggesting that competence regulation is not strongly affected by derepression of CtsR-dependent genes.
ClpP and ClpE are required for growth at high temperature.
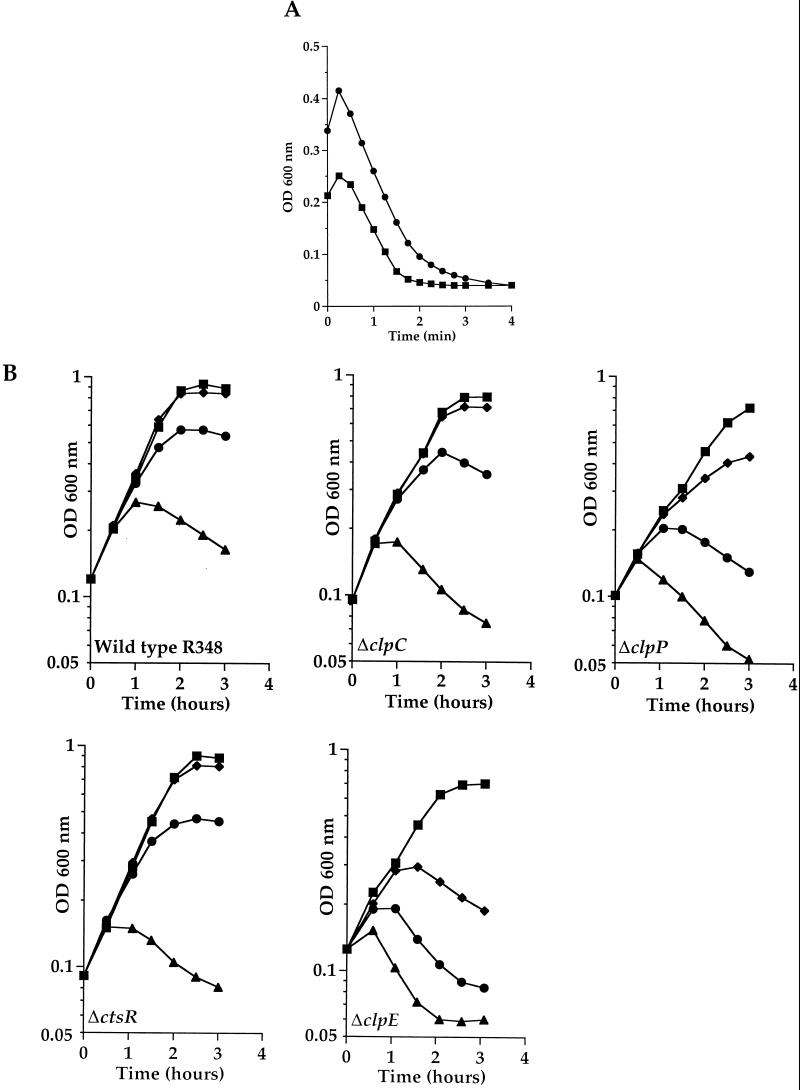
Mutations inactivating clp genes in B. subtilis are known to be highly pleiotropic, affecting cell motility, growth at high temperature, competence development, and sporulation (35, 37). A phenotypic analysis of the S. pneumoniae ΔctsR, ΔclpC, ΔclpP, and ΔclpE mutants and the R348 parental strain was carried out. All four mutants grew as did parental strain R348 as standard diplococcal cells in BHI medium at 37°C, with no cell filamentation or long-chain formation. No differences in penicillin- or DOC-induced autolysis were observed for the four mutants compared to the R348 parental strain, as shown in Fig. 8A for strain SP2001 (ΔclpC::aphA3), suggesting that production of the major autolysin, LytA, is not affected. All four mutants presented characteristic hemolytic halos on D-horse blood agar plates, comparable to that of the parental strain, suggesting that pneumolysin production is not affected.
FIG. 8.
(A) DOC-triggered autolysis is not affected in the ΔclpC mutant. Strain SP2001 (ΔclpC::aphA3) (●) and parental strain R348 (▪) were grown in BHI medium until mid-exponential phase before induction of autolysis by addition of DOC (0.05% final concentration). (B) Growth of the S. pneumoniae R348 wild-type strain and ΔclpC (SP2001), ΔclpP (SP2000), ΔctsR (SP2003), and ΔclpE (SP2002) mutant strains at 37°C (▪), 40°C (⧫),42°C (●), and 44°C (▴).
Growth at different temperatures was examined in BHI medium. Cultures growing exponentially at 37°C were divided and incubated at different growth temperatures. As shown in Fig. 8B, the R348 parental strain and the ΔctsR and ΔclpC mutants grew normally at 37 and 40°C, poorly at 42°C, and not at all at 44°C. In contrast, the ΔclpE and ΔclpP mutants presented a temperature-sensitive growth phenotype at 40, 42, and 44°C, suggesting that ClpE and ClpP are essential for adaptation to high temperatures. Our results also indicate that ClpC does not affect competence development in S. pneumoniae and that it is not involved in autolysis, cell filamentation, pneumolysin expression, or growth at high temperature, contrary to a recent report (4).
DISCUSSION
The Clp ATP-dependent protease plays an important role in regulation through proteolysis, in both E. coli (15, 17) and B. subtilis (34). We show here that ClpP of S. pneumoniae negatively regulates competence development by preventing expression of the comCDE operon under inappropriate conditions. Interestingly, the roles played by ClpP in competence development of B. subtilis and S. pneumoniae appear to be very different.
ClpP of B. subtilis provides a link between the initial quorum-sensing ComP/ComA signal transduction system and late com gene expression through targeted proteolysis of the ComK transcription activator (34, 59). ClpP is thus essential for competence gene expression, since in its absence, ComK is sequestered in an inactive form by the MecA/ClpC complex, preventing it from activating its own synthesis, as well as transcription of late com genes (34, 35, 59). In S. pneumoniae, however, the situation is quite different, since, as shown here, ClpP is not required for expression of late competence genes or competence development but, instead, acts negatively at the earliest stages of the competence regulatory pathway to prevent inappropriate expression of the genes encoding the peptide quorum-sensing system. This is consistent with the idea that competence in S. pneumoniae is induced in response to changes in environmental conditions (8).
Many of the Clp proteins (ClpA, ClpX, and ClpC) act as ATPase subunits of the ATP-dependent Clp protease by associating with the ClpP proteolytic subunit, on which they confer substrate specificity (16, 17, 59). As shown in this report, neither ClpC nor ClpE plays a role in controlling competence gene expression, suggesting that one of the remaining Clp ATPases present in S. pneumoniae, ClpX or ClpL, may associate with ClpP instead.
Apart from L. monocytogenes, little is known about clp gene regulation in pathogenic bacteria, despite the fact that many of these genes play important roles in virulence (39). A detailed analysis of the complete S. pneumoniae type 4 genome sequence (56; http://www.tigr.org) indicates that only three of the four different types of heat shock response regulatory mechanisms originally defined in B. subtilis (11, 20) coexist in S. pneumoniae. Among them are the class I heat shock genes, defined as the HrcA regulon (the dnaK and groESL operons). There are no class II heat shock genes, since the ςB stress sigma factor is not present in S. pneumoniae. We previously identified the ctsR gene of S. pneumoniae and several potential target genes from the genome sequence (11) and show here that class III regulation is present. Finally, class IV genes in B. subtilis are those whose induction by heat shock is not dependent on HrcA, ςB, or CtsR. Many of these genes are present in S. pneumoniae, such as clpX and ftsH, and one can speculate that they will also prove to be heat shock genes.
We have shown that expression of the S. pneumoniae clpP and clpE genes and clpC and groESL operons is heat inducible and controlled directly by the CtsR repressor. Although ClpP acts negatively on competence gene expression, this role appears to be restricted to growth conditions under which competence genes are not expressed. Indeed, in a ΔctsR mutant in which clpP expression is derepressed, competence gene expression is unaffected during growth in C+Y competence medium, suggesting that negative regulation by ClpP can no longer take place. Among the members of the CtsR regulon, the groESL operon of S. pneumoniae belongs to a new class of heat shock genes under dual regulation by both CtsR and HrcA and is preceded by operator sequences for both repressors. Accordingly, in a ΔctsR mutant, expression of groESL is not strongly increased, consistent with repression by HrcA.
Phenotypic analyses also indicate that, unlike in B. subtilis and contrary to a previous report (4), ClpC is not involved in controlling competence development, nor does it play a role in autolysis, pneumolysin production, or growth at high temperature of S. pneumoniae.
ClpP and ClpE of S. pneumoniae, on the other hand, have both been shown to be required for growth at high temperature, suggesting they may interact to form a Clp ATP-dependent protease. This is in contrast to B. subtilis, in which ClpC is required for growth at high temperature but ClpE is not (9, 37). As in L. monocytogenes, the ClpE and ClpC ATPases both appear to play a role in the virulence of S. pneumoniae. Indeed, the clpE gene encoding an Hsp100-type Clp ATPase was isolated during a large-scale identification of virulence genes using the signature-tagged transposon mutagenesis technique (44). An insertion inactivating the S. pneumoniae ctsR gene was isolated by using a similar approach (24), and the corresponding mutant was found to be highly attenuated in a murine respiratory tract infection model, which the authors attributed to a polar effect of the transposon insertion on the expression of the clpC gene that lies directly downstream. The precise role of the S. pneumoniae Clp proteins in competence development and virulence, however, remains to be determined and will be the subject of further investigation.
ACKNOWLEDGMENTS
We are grateful to Isabelle Derré and Bernard Martin for many helpful discussions and Georges Rapoport, in whose laboratory part of this work was carried out. We thank the Institute for Genomic Research for generously providing access to the complete S. pneumoniae type 4 genome sequence prior to publication and Martin Stieger for the kind gift of plasmid p5.00.
This work was supported by research funds from the European Commission (grants QLRK-2000-00543 to J.-P. Claverys and QLRT-1999-01455 to T. Msadek); the Centre National de la Recherche Scientifique, Institut Pasteur, Université Paris 7; the Ministère de la Défense (Direction Générale de l'Armement); and the Programme de Recherche Fondamentale en Microbiologie, Maladies Infectieuses et Parasitaires of the Ministère de la Recherche. Arnaud Chastanet was the recipient of a Ph.D. thesis fellowship from the Ministère de la Recherche.
REFERENCES
- 1.Alloing G, Granadel C, Morrison D A, Claverys J P. Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol Microbiol. 1996;21:471–478. doi: 10.1111/j.1365-2958.1996.tb02556.x. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell E A, Choi S Y, Masure H R. A competence regulon in Streptococcus pneumoniae revealed by genomic analysis. Mol Microbiol. 1998;27:929–939. doi: 10.1046/j.1365-2958.1998.00737.x. [DOI] [PubMed] [Google Scholar]
- 4.Charpentier E, Novak R, Tuomanen E. Regulation of growth inhibition at high temperature, autolysis, transformation and adherence in Streptococcus pneumoniae by clpC. Mol Microbiol. 2000;37:717–726. doi: 10.1046/j.1365-2958.2000.02011.x. [DOI] [PubMed] [Google Scholar]
- 5.Cheng Q, Campbell E A, Naughton A M, Johnson S, Masure H R. The com locus controls genetic transformation in Streptococcus pneumoniae. Mol Microbiol. 1997;23:683–692. doi: 10.1046/j.1365-2958.1997.2481617.x. [DOI] [PubMed] [Google Scholar]
- 6.Choi I H, Shim J H, Kim S W, Kim S N, Pyo S N, Rhee D K. Limited stress response in Streptococcus pneumoniae. Microbiol Immunol. 1999;43:807–812. doi: 10.1111/j.1348-0421.1999.tb02474.x. [DOI] [PubMed] [Google Scholar]
- 7.Claverys J P, Dintilhac A, Pestova E V, Martin B, Morrison D A. Construction and evaluation of new drug-resistance cassettes for gene disruption mutagenesis in Streptococcus pneumoniae, using an ami test platform. Gene. 1995;164:123–128. doi: 10.1016/0378-1119(95)00485-o. [DOI] [PubMed] [Google Scholar]
- 8.Claverys J P, Prudhomme M, Mortier-Barrière I, Martin B. Adaptation to the environment: Streptococcus pneumoniae, a paradigm for recombination-mediated genetic plasticity? Mol Microbiol. 2000;35:251–259. doi: 10.1046/j.1365-2958.2000.01718.x. [DOI] [PubMed] [Google Scholar]
- 9.Derré I, Rapoport G, Devine K, Rose M, Msadek T. ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis. Mol Microbiol. 1999;32:581–593. doi: 10.1046/j.1365-2958.1999.01374.x. [DOI] [PubMed] [Google Scholar]
- 10.Derré I, Rapoport G, Msadek T. The CtsR regulator of stress response is active as a dimer and specifically degraded in vivo at 37°C. Mol Microbiol. 2000;38:335–347. doi: 10.1046/j.1365-2958.2000.02124.x. [DOI] [PubMed] [Google Scholar]
- 11.Derré I, Rapoport G, Msadek T. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol Microbiol. 1999;31:117–132. doi: 10.1046/j.1365-2958.1999.01152.x. [DOI] [PubMed] [Google Scholar]
- 12.Dubnau D. DNA uptake in bacteria. Annu Rev Microbiol. 1999;53:217–244. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- 13.Gaillot O, Pellegrini E, Bregenholt S, Nair S, Berche P. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol Microbiol. 2000;35:1286–1294. doi: 10.1046/j.1365-2958.2000.01773.x. [DOI] [PubMed] [Google Scholar]
- 14.Gibson T J. Ph.D. thesis. Cambridge, England: University of Cambridge; 1984. [Google Scholar]
- 15.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 16.Gottesman S, Maurizi M R, Wickner S. Regulatory subunits of energy-dependent proteases. Cell. 1997;91:435–438. doi: 10.1016/s0092-8674(00)80428-6. [DOI] [PubMed] [Google Scholar]
- 17.Gottesman S, Wickner S, Maurizi M R. Protein quality control: triage by chaperones and proteases. Genes Dev. 1997;11:815–823. doi: 10.1101/gad.11.7.815. [DOI] [PubMed] [Google Scholar]
- 18.Guérout-Fleury A M, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- 19.Havarstein L S, Coomaraswamy G, Morrison D A. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci USA. 1995;92:11140–11144. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hecker M, Schumann W, Völker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 21.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 22.Hirata H, Fukazawa T, Negoro S, Okada H. Structure of a β-galactosidase gene of Bacillus stearothermophilus. J Bacteriol. 1986;166:722–727. doi: 10.1128/jb.166.3.722-727.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krüger E, Völker U, Hecker M. Stress induction of clpC in Bacillus subtilis and its involvement in stress tolerance. J Bacteriol. 1994;176:3360–3367. doi: 10.1128/jb.176.11.3360-3367.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau G W, Haataja S, Lonetto M, Kensit S E, Marra A, Bryant A P, McDevitt D, Morrison D A, Holden D W. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol Microbiol. 2001;40:555–571. doi: 10.1046/j.1365-2958.2001.02335.x. [DOI] [PubMed] [Google Scholar]
- 25.Lazazzera B A, Grossman A D. The ins and outs of peptide signaling. Trends Microbiol. 1998;6:288–294. doi: 10.1016/s0966-842x(98)01313-4. [DOI] [PubMed] [Google Scholar]
- 26.Lee M S, Morrison D A. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J Bacteriol. 1999;181:5004–5016. doi: 10.1128/jb.181.16.5004-5016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lefèvre J C, Claverys J P, Sicard A M. Donor deoxyribonucleic acid length and marker effect in pneumococcal transformation. J Bacteriol. 1979;138:80–86. doi: 10.1128/jb.138.1.80-86.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin B, García P, Castanié M P, Claverys J P. The recA gene of Streptococcus pneumoniae is part of a competence-induced operon and controls lysogenic induction. Mol Microbiol. 1995;15:367–379. doi: 10.1111/j.1365-2958.1995.tb02250.x. [DOI] [PubMed] [Google Scholar]
- 29.Martin B, Prudhomme M, Alloing G, Granadel C, Claverys J P. Cross-regulation of competence pheromone production and export in the early control of transformation in Streptococcus pneumoniae. Mol Microbiol. 2000;38:867–878. doi: 10.1046/j.1365-2958.2000.02187.x. [DOI] [PubMed] [Google Scholar]
- 30.Martin-Verstraete I, Débarbouillé M, Klier A, Rapoport G. Mutagenesis of the Bacillus subtilis “−12, −24” promoter of the levanase operon and evidence for the existence of an upstream activating sequence. J Mol Biol. 1992;226:85–99. doi: 10.1016/0022-2836(92)90126-5. [DOI] [PubMed] [Google Scholar]
- 31.Mei J M, Nourbakhsh F, Ford C W, Holden D W. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 32.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 33.Misra N, Habib S, Ranjan A, Hasnain S E, Nath I. Expression and functional characterisation of the clpC gene of Mycobacterium leprae: ClpC protein elicits human antibody response. Gene. 1996;172:99–104. doi: 10.1016/0378-1119(96)00053-4. [DOI] [PubMed] [Google Scholar]
- 34.Msadek T. When the going gets tough: survival strategies and environmental signaling networks in Bacillus subtilis. Trends Microbiol. 1999;7:201–207. doi: 10.1016/s0966-842x(99)01479-1. [DOI] [PubMed] [Google Scholar]
- 35.Msadek T, Dartois V, Kunst F, Herbaud M-L, Denizot F, Rapoport G. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol Microbiol. 1998;27:899–914. doi: 10.1046/j.1365-2958.1998.00735.x. [DOI] [PubMed] [Google Scholar]
- 36.Msadek T, Kunst F, Henner D, Klier A, Rapoport G, Dedonder R. Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU. J Bacteriol. 1990;172:824–834. doi: 10.1128/jb.172.2.824-834.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Msadek T, Kunst F, Rapoport G. MecB of Bacillus subtilis, a member of the ClpC ATPase family, is a pleiotropic regulator controlling competence gene expression and growth at high temperature. Proc Natl Acad Sci USA. 1994;91:5788–5792. doi: 10.1073/pnas.91.13.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mullis K B, Faloona F A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 39.Nair S, Derré I, Msadek T, Gaillot O, Berche P. CtsR controls class III heat shock gene expression in the human pathogen Listeria monocytogenes. Mol Microbiol. 2000;35:800–811. doi: 10.1046/j.1365-2958.2000.01752.x. [DOI] [PubMed] [Google Scholar]
- 40.Nair S, Fréhel C, Nguyen L, Escuyer V, Berche P. ClpE, a novel member of the HSP100 family, is involved in cell division and virulence of Listeria monocytogenes. Mol Microbiol. 1999;31:185–196. doi: 10.1046/j.1365-2958.1999.01159.x. [DOI] [PubMed] [Google Scholar]
- 41.Pederson K J, Carlson S, Pierson D E. The ClpP protein, a subunit of the Clp protease, modulates ail gene expression in Yersinia enterocolitica. Mol Microbiol. 1997;26:99–107. doi: 10.1046/j.1365-2958.1997.5551916.x. [DOI] [PubMed] [Google Scholar]
- 42.Pestova E V, Havarstein L S, Morrison D A. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol Microbiol. 1996;21:853–862. doi: 10.1046/j.1365-2958.1996.501417.x. [DOI] [PubMed] [Google Scholar]
- 43.Peterson S, Cline R T, Tettelin H, Sharov V, Morrison D A. Gene expression analysis of the Streptococcus pneumoniae competence regulons by use of DNA microarrays. J Bacteriol. 2000;182:6192–6202. doi: 10.1128/jb.182.21.6192-6202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polissi A, Pontiggia A, Feger G, Altieri M, Mottl H, Ferrari L, Simon D. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect Immun. 1998;66:5620–5629. doi: 10.1128/iai.66.12.5620-5629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porankiewicz J, Wang J, Clarke A K. New insights into the ATP-dependent Clp protease: Escherichia coli and beyond. Mol Microbiol. 1999;32:449–458. doi: 10.1046/j.1365-2958.1999.01357.x. [DOI] [PubMed] [Google Scholar]
- 46.Porter R D, Guild W R. Characterization of some pneumococcal bacteriophages. J Virol. 1976;19:659–667. doi: 10.1128/jvi.19.2.659-667.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rouquette C, de Chastellier C, Nair S, Berche P. The ClpC ATPase of Listeria monocytogenes is a general stress protein required for virulence and promoting early bacterial escape from the phagosome of macrophages. Mol Microbiol. 1998;27:1235–1246. doi: 10.1046/j.1365-2958.1998.00775.x. [DOI] [PubMed] [Google Scholar]
- 48.Rouquette C, Ripio M-T, Pellegrini E, Bolla J-M, Tascon R I, Vázquez-Boland J-A, Berche P. Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol Microbiol. 1996;21:977–987. doi: 10.1046/j.1365-2958.1996.641432.x. [DOI] [PubMed] [Google Scholar]
- 49.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual, second edition. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schirmer E C, Glover J R, Singer M A, Lindquist S. HSP100/Clp proteins: a common mechanism explains diverse functions. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 53.Stieger M, Wohlgensinger B, Kamber M, Rolf L, Keck W. Integrational plasmids for the tetracycline-regulated expression of genes in Streptococcus pneumoniae. Gene. 1999;226:243–251. doi: 10.1016/s0378-1119(98)00561-7. [DOI] [PubMed] [Google Scholar]
- 54.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 55.Tabor S, Richardson C C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci USA. 1987;84:4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tettelin H, Nelson K E, Paulsen I T, Eisen J A, Read T D, Peterson S, Heidelberg J, DeBoy R T, Haft D H, Dodson R J, Durkin A S, Gwinn M, Kolonay J F, Nelson W C, Peterson J D, Umayam L A, White O, Salzberg S L, Lewis M R, Radune D, Holtzapple E, Khouri H, Wolf A M, Utterback T R, Hansen C L, McDonald L A, Feldblyum T V, Angiuoli S, Dickinson T, Hickey E K, Holt I E, Loftus B J, Yang F, Smith H O, Venter J C, Dougherty B A, Morrison D A, Hollingshead S K, Fraser C M. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 57.Tortosa P, Dubnau D. Competence for transformation: a matter of taste. Curr Opin Microbiol. 1999;2:588–592. doi: 10.1016/s1369-5274(99)00026-0. [DOI] [PubMed] [Google Scholar]
- 58.Trieu-Cuot P, Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5"-aminoglycoside phosphotransferase type III. Gene. 1983;23:331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- 59.Turgay K, Hahn J, Burghoorn J, Dubnau D. Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 1998;17:6730–6738. doi: 10.1093/emboj/17.22.6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinrauch Y, Msadek T, Kunst F, Dubnau D. Sequence and properties of comQ, a new competence regulatory gene of Bacillus subtilis. J Bacteriol. 1991;173:5685–5693. doi: 10.1128/jb.173.18.5685-5693.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuan G, Wong S L. Isolation and characterization of Bacillus subtilis groE regulatory mutants: evidence for orf39 in the dnaK operon as a repressor gene in regulating the expression of both groE and dnaK. J Bacteriol. 1995;177:6462–6468. doi: 10.1128/jb.177.22.6462-6468.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]