Abstract
The energetic efficiency of microbial growth is significantly reduced in cultures growing under glucose excess compared to cultures growing under glucose limitation, but the magnitude to which different energy-dissipating processes contribute to the reduced efficiency is currently not well understood. We introduce here a new concept for balancing the total cellular energy flux that is based on the conversion of energy and carbon fluxes into energy equivalents, and we apply this concept to glucose-, ammonia-, and phosphate-limited chemostat cultures of riboflavin-producing Bacillus subtilis. Based on [U-13C6]glucose-labeling experiments and metabolic flux analysis, the total energy flux in slow-growing, glucose-limited B. subtilis is almost exclusively partitioned in maintenance metabolism and biomass formation. In excess-glucose cultures, in contrast, uncoupling of anabolism and catabolism is primarily achieved by overflow metabolism, while two quantified futile enzyme cycles and metabolic shifts to energetically less efficient pathways are negligible. In most cultures, about 20% of the total energy flux could not be assigned to a particular energy-consuming process and thus are probably dissipated by processes such as ion leakage that are not being considered at present. In contrast to glucose- or ammonia-limited cultures, metabolic flux analysis revealed low tricarboxylic acid (TCA) cycle fluxes in phosphate-limited B. subtilis, which is consistent with CcpA-dependent catabolite repression of the cycle and/or transcriptional activation of genes involved in overflow metabolism in the presence of excess glucose. ATP-dependent control of in vivo enzyme activity appears to be irrelevant for the observed differences in TCA cycle fluxes.
The very basis of microbial growth resides in balanced fluxes through anabolic and catabolic reactions. These metabolic fluxes are highly variable and change with the environmental conditions and the rate of growth, since faster-growing cells demand a higher rate of metabolism. To delineate these influences, metabolic flux responses are typically studied in chemostat cultures that are maintained under different nutrient limitations. When microorganisms are limited for their energy source (usually the carbon source), catabolism is tightly coupled to anabolism and high biomass yields on the carbon source are achieved (40). Compared to those with carbon (C) limitation, excess-C cultures exhibit generally high rates of carbon consumption and low yields of biomass and thus have a low energetic growth efficiency (11, 31, 32, 57). Most frequently excess-C cultures in chemostats are limited by other cellular macroelements such as nitrogen (N) and phosphorus (P) but also potassium, the predominant intracellular cation (11). Potassium or P limitation invokes usually a strong uncoupling of catabolic and anabolic processes that lead consequently to low biomass yields on the energy source, while N limitation has more moderate effects on cellular physiology (31, 68). Although mostly studied in the gram-negative Enterobacteria, these findings are consistent with data from gram-positive Bacillus spp. (4, 28, 32, 35).
The uncoupling of anabolism and catabolism in excess-C cultures reduces the energetic growth efficiency of these cultures, which is attributed to so-called overflow metabolism, metabolic shifts of carbon or electron flow to less efficient pathways, and/or a variety of energy-spilling reactions (40, 57). While the magnitude of ATP dissipation via energy-spilling reactions is not well understood, several such processes that catalyze net loss of energy though cyclic reactions are known at the molecular level. Such cycles are frequently referred to as futile, simply because no clear metabolic function of such apparent waste of energy could be envisaged (22, 30, 40). These futile cycles may result from ion or proton leakage across the cytoplasmic membrane that reduces the proton motive force and thus decreases the efficiency of respiratory ATP generation. Another type of futile cycling is based on metabolic reactions that act in an antagonistic fashion, so that one molecule of ATP is dissipated per cycled metabolite. While ion leakage may contribute significantly to energy spilling (30, 40), ATP-dissipating futile enzyme cycles are generally thought to be maintained at a relatively low level (5, 6, 8). In certain biological systems, however, futile enzyme cycling is extensive (22). The most prominent of these is probably thermogenesis in bumblebees during cold weather. More recently, metabolic flux analysis revealed that certain futile enzyme cycles may account for ATP dissipation in the order of one molecule of ATP per consumed molecule of glucose in microbial cultures (37; M. Emmerling, M. Dauner. A. Ponti, J. Fiaux, M. Hochuli, T. Szyperski, K. Wüthrich, J. E. Bailey, and U. Sauer, submitted for publication.).
Most enzymes that are involved in potential futile cycles are subject to complex allosteric control, so that their in vivo operation may be assessed only by isotope-labeling experiments (22). Such labeling experiments were traditionally conducted such that the distribution of label was indicative of one particular futile cycle (22, 59). A common problem of such analyses are corrections for alternative pathways and exchange reactions that may also affect label distribution. The currently most advanced form of metabolic flux analysis can quantitatively account for the use of alternative pathways and exchange reactions on the distribution of 13C label throughout the whole metabolic system of a cell (9, 50, 65). Hence, this methodology provides a holistic view on cellular metabolism and not only quantifies metabolic shifts and certain futile cycles but can also shed some light on the energetic aspects of growth. This is achieved by the use of comprehensive isotope isomer (isotopomer) models that encompass all relevant metabolic intermediates and appropriate computational tools for data analysis (9, 63, 64).
The primary focus of this work was to quantify the magnitude of energy dissipation via overflow metabolism, metabolic shifts, and futile enzyme cycling in C-limited and excess-C Bacillus subtilis chemostat cultures, by using isotopomer-balancing (9) and [U-13C6]glucose-labeling experiments (55). Thus, for the first time we report here intracellular carbon flux distributions in N- and P-limited B. subtilis.
MATERIALS AND METHODS
Strain.
Throughout this study, the recombinant, riboflavin-producing B. subtilis strain RB50::pRF69 was used. The host strain, RB50 (purA60 Azr-11 Dcr-15 MSr-46 RoFr-50 spo0A), contains several chemically introduced purine and riboflavin analog-resistant mutations (36). In RB50::pRF69, one copy of the constitutively expressed, recombinant B. subtilis rib operon pRF69 with the cat marker, was integrated in the native chromosomal rib operon. To increase gene dosage, the recombinant rib operon was amplified by chloramphenicol selection of RB50::pRF69 in serial batch cultures up to a concentration of 80 mg/liter. The resulting strain is designated RB50::(pRF69)n, where n refers to the number of amplified pRF69 rib operons. All cultures were inoculated from the same frozen stock. This amplification has no impact on the present study other than diverting 1 to 3% of substrate carbon into riboflavin.
Growth conditions and media.
All media were supplemented with 80 mg of chloramphenicol per liter. Frozen stocks were revived for 12 h in complex medium, containing (per liter) 5 g of glucose, 25 g of veal infusion broth, and 5 g of yeast extract. A 2.5-ml amount of this culture was transferred into 50 ml of minimal medium in a 500-ml baffled shake flask. After another 12 h, the whole culture was used for reactor inoculation. The minimal batch medium was similar to the chemostat medium but did not contain H2SO4 and was supplemented with 0.1 M sodium phosphate buffer (pH 6.8).
Chemostat cultures were grown at 38°C in a working volume of 1 liter in a 2-liter LH discovery 210 series reactor (Adaptive Biosystems) equipped with pH, dissolved-oxygen, temperature, optical-density, and foam probes. The N-limited medium contained (per liter) 8.5 g of glucose, 0.53 g of NH4Cl, 0.67 g of (NH4)2SO4, 2.54 g of Na2HPO4 · 12 H2O, 20.72 g of KH2PO4, 0.42 g of MgSO4 · 7 H2O, 46 mg of CaCl2 · H2O, 9 mg of FeSO4 · 7 H2O, and 40 ml of a trace element solution containing (per liter) 2.25 g of MnCl2 · 4 H2O, 1.32 g of ZnCl2, 0.34 g of CuCl2 · 2 H2O, 0.5 g of CoCl2 · 6 H2O, 0.5 g of Na2MoO4 · 2 H2O, and 1.25 g of AlCl3 · 6 H2O. To achieve P limitation at the low or high dilution rate (D), phosphate salts were added at the following concentrations (per liter): 0.00192 g of Na2HPO4 · 12 H2O and 0.01536 g of KH2PO4 or 0.032 g of Na2HPO4 · 2 H2O and 0.032 g of KH2PO4, respectively. Chloride salts were newly added at the following concentrations to ensure identical ionic strength in all experiments (per liter): 1.9 g of KCl and 0.45 g of NaCl or 1.68 g of KCl and 0.3 g of NaCl for high or low D, respectively.
For labeling experiments, the medium was replaced by an otherwise identical medium that contained the same glucose concentration as a mixture of 90% (wt/wt) natural glucose and 10% (wt/wt) [U-13C6]glucose (13C, >98%; Isotech, Miamisburg, Ohio). Biomass aliquots for nuclear magnetic resonance (NMR) analysis were withdrawn after 0.8 volume changes, so that 45% of the biomass was fractionally 13C labeled according to first-order washout kinetics.
Chemostat media were acidified to pH 2 to 3 by addition of H2SO4 (95 to 97%) and sterilized by passage through a 0.2-μm-pore-size filter. During the fermentation, the pH was maintained at 6.8 and the volume was kept constant by a weight-controlled pump. A constant airflow of 1 liter/min was achieved by a mass flowmeter, and the agitation speed was set to values between 600 and 1,000 rpm, ensuring dissolved oxygen levels well above 30% in all cases. All data reported are from cultures in physiological steady state, defined as at least five culture volume changes under the same conditions and stable optical density and off gas readings for at least three volume changes.
Analytical techniques.
Cell dry weight was determined from at least eight parallel 10-ml cell suspensions that were harvested by centrifugation, washed with distilled water, and dried at 80°C for 24 h to a constant weight. Cellular protein, RNA, and glycogen content were determined as described elsewhere (10). Glucose, succinate, pyruvate, and phosphoenolpyruvate concentrations were determined enzymatically with commercial kits (Beckman) or as described by Bergmeyer (3). Acetate, acetoin, and 2,3-butanediol concentrations were determined by gas chromatography (5890E; Hewlett-Packard) on a Carbowax MD-10 column (Macherey-Nagel) with butyrate as the internal standard. Additionally, organic acids, acetoin, and diacyl were determined by high-performance liquid chromatography on a Supelcogel C8 column (4.6 by 250 mm) (Sigma) with a diode array detector (Perkin-Elmer). Phosphoric acid (0.2 N) was used as mobile phase at a flow rate of 0.3 ml/min and 30°C. Concentrations of carbon dioxide and oxygen in the bioreactor feed and effluent gas were determined with a mass spectrometer (Prima 600; Fisons Instruments).
Hexosamine concentrations in the supernatant, originating from both peptidoglycan and teichoic acids, were determined colorimetrically (15), using glucosamine for calibration. Riboflavin concentrations were determined as the absorption at 444 nm (A444) in cell-free culture broth. The carbon, hydrogen, and nitrogen composition of dried biomass was determined with a CHN-elemental analyzer (EA1108; CE Instruments). Prior to analysis, cell pellets were washed with bidistilled H2O, dried in a freeze-dryer for 12 h, and stored over silica gel (Sigma) for another 12 h in an evacuated desiccator.
Intracellular concentrations of ATP and ADP were determined as described previously (60), using an ATP bioluminescence kit (HS II; Boehringer Mannheim). For this purpose, 10 ml of culture broth was rapidly (within 0.7 s) withdrawn with a syringe containing precooled glass beads (−20°C). Aliquots of 40 μl were transferred to a plastic cup and quenched by mixing with 160 μl of dimethyl sulfoxide. After addition of 800 μl of ice-cold 25 mM HEPES buffer (pH 7.75), samples were stored at −80°C and analyzed within 1 week.
Two-dimensional proton-detected heteronuclear single quantum 13C-1H correlation NMR spectroscopy ([13C,1H]-COSY) and data analysis were done exactly as described previously (48, 55) with amino acids obtained from hydrolyzed biomass that was harvested after the 13C-labeling experiments (9, 46, 48). For each sample, one spectrum was recorded for the aliphatic 13C-1H moieties and one spectrum was recorded for the aromatic 13C-1H groups. The relative abundances of 13C-13C scalar coupling multiplets in the [13C,1H]-COSY cross peaks were evaluated with the program FCAL (56; R. Glaser, 2.3.0. ed., 1999).
Biochemical reaction network.
To describe N and P limitation, the previously described comprehensive isotopomer model of B. subtilis central metabolism with an H+-to-ATP ratio of 4 was extended (10). Ammonium uptake in C- and P-limited (excess-N) cultures was assumed to proceed via energy-independent diffusion. Under N limitation, uptake was catalyzed by an NH4+/K+ antiporter (41) and K+ was replenished by a H+/K+ symporter (2). Intracellular ammonium assimilation was accomplished by glutamine synthetase and glutamate synthase (51). Phosphate uptake in C- and N-limited (excess-P) cultures was assumed to proceed via an energy-independent, low-affinity system. Uptake under P limitation was catalyzed by an ABC-type transporter that required one ATP per transported phosphate (41).
Building block requirements for biomass were calculated from experimental data and a previously developed structured model, in which the cell wall was assumed to consist of 45% peptidoglycan and 55% teichoic acids (10). Under P limitation, however, B. subtilis strongly reduces the P content by replacing teichoic acid with teichuronic acids (23), and thus an average chain length of 7.5 (34) with the repeating unit (GlcA)2-(GalNAc)2 (26) was assumed for P-limited cultures.
Estimation of intracellular carbon fluxes.
Metabolic flux analysis was based on three different data sets: (i) substrate uptake and product formation rates; (ii) macromolecular biomass composition; and (iii) relative abundances of the 13C-13C multiplets of 47 amino acid carbon positions in the [13C,1H]-COSY spectra. The requirement of metabolic precursor for biomass formation was deduced from the experimentally determined macromolecular composition and a previously published growth model (10).
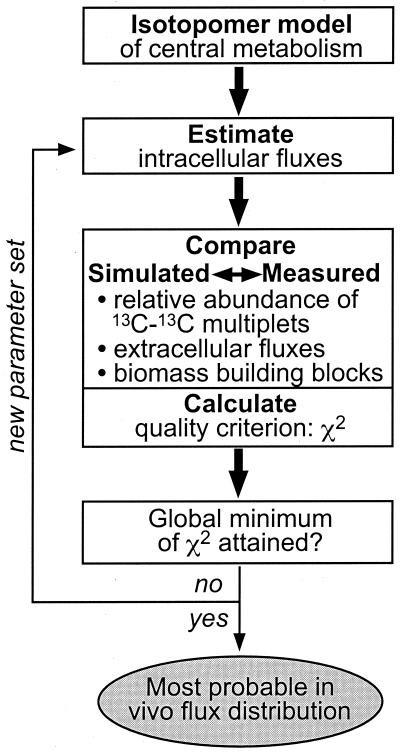
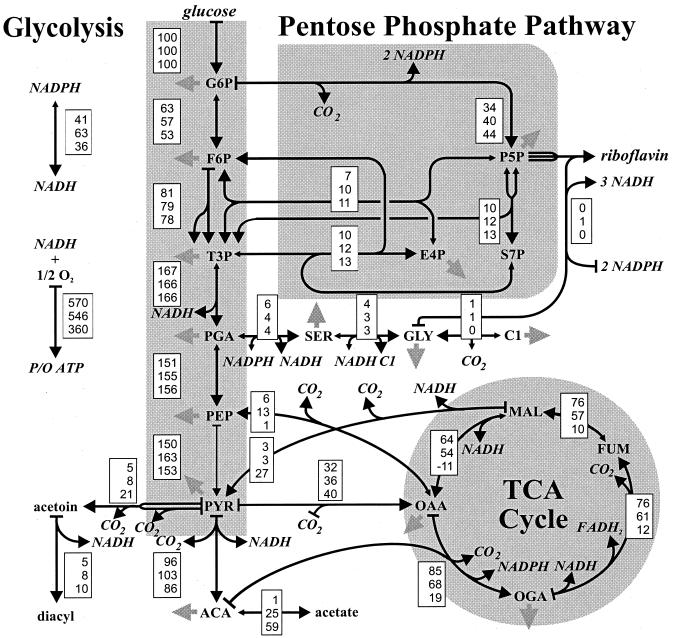
Intracellular carbon fluxes were then calculated as a best fit to the three data sets within an isotopomer model (9). This model enables rigorous accounting of the isotopomer pools through 25 metabolite equations, 502 isotopomer equations for metabolic intermediates, and 3,628 isotopomer equations for amino acids that balance all positional combinations of 12C and 13C in the considered compounds. Exchange fluxes via reversible reactions were quantitatively considered in the model. In the first step, the isotopomer balances of all metabolites (Fig. 1) were calculated from a randomly chosen flux distribution. Relative abundances of the 13C-13C multiplets in the amino acids are then simulated from this isotopomer distribution and compared to the determined multiplets. The quality of the fit to the multiplets and the physiological data is judged by the χ2-error criterion. Through an iterative process of flux estimation and signal fitting, a flux solution that corresponds to the minimal χ2 value is sought. This optimal solution represents the maximum-likelihood flux distribution in the investigated metabolic system that reflects both the physiological data and the [13C,1H]-COSY data.
FIG. 1.
Flow chart for the iterative flux estimation procedure within an isotopomer model.
A range-restricted evolutionary algorithm was used to identify the global error minimum by this iterative procedure (1). The final solution was obtained by restarting a modified direction-set search algorithm at the optimal flux solution that was identified by the evolutionary algorithm (38).
ATP balance.
Like the commonly used carbon balance, energy balances can be constructed for cellular processes that account also for the incorporation of energy source into cell material. The metabolic energy equivalent of glucose is the maximum amount of ATP that may be generated from complete oxidation of glucose to carbon dioxide. For aerobic organisms such as B. subtilis, this ATP generation is dependent on the coupling efficiency of respiration and ATP synthesis and is usually expressed as the so-called P-to-O (ATP per oxygen) ratio. B. subtilis contains a three-branched electron transport chain with different coupling efficiencies, but two of these branches are probably irrelevant in our well-aerated, glucose-grown cultures, i.e., the bd oxidase branch is primarily active at low oxygen tension (67), and the cytochrome c branch is inactive in glucose-grown cells (27, 66). Hence, oxidation of NADH in our cultures is probably catalyzed by the aa3 oxidase branch, which was also shown experimentally to be predominant during vegetative growth of B. subtilis (25, 42). Transport of one electron from NADH along this branch translocates two protons (25, 61), and four protons are needed to generate one ATP in the reaction catalyzed by ATP synthase (13, 43). Transport of one electron from FADH, however, translocates only one proton (49). Consequently, all cultures were assumed to exhibit a maximal P/O of unity (the exact value depends on the amount of FADH that is produced in the tricarboxylic acid [TCA] cycle), as was previously suggested for B. subtilis (10, 44). This P-to-O ratio corresponds to the generation of 1 ATP per NADH and 0.5 ATP per FADH, so that complete oxidation of glucose to carbon dioxide yields 15 ATP. The total cellular energy flux in ATP equivalents is therefore the specific glucose uptake rate multiplied by 15.
Excretion of metabolic by-products such as acetate causes an energetic loss to the cell because glucose cannot be oxidized completely to carbon dioxide, and hence less ATP is generated from the available glucose. To account quantitatively for the incorporation of glucose into products and biomass, we calculated the ATP equivalent of each product. First, the maximum ATP generation was calculated for complete conversion of glucose to each considered product and carbon dioxide at a P-to-O ratio of 1. The difference of this ATP amount to the 15 ATP that are potentially available from complete oxidation of glucose represents the ATP equivalents that are lost by formation of the considered product. For example, complete conversion of one molecule of glucose to two acetate and two carbon dioxide molecules yields 6.5 ATP molecules at the assumed P/O (16). Thus, formation of one molecule of acetate in glucose catabolism is equivalent to the loss of 4.25 ATP molecules. This calculation was done for each considered product and accounts also for all ATP and reducing equivalents that are generated in the conversion of glucose to each product.
RESULTS
Physiology of excess-C B. subtilis.
To elucidate the efficiency of B. subtilis growth when abundant carbon was available, we grew B. subtilis RB50::(pRF69)n under N or P limitation at dilution rates of 0.1 and 0.4 h−1 in chemostat culture. These rates were chosen such that the low value was below the critical D at which aerobic fermentation may occur (19, 52) and the high value was well below the maximum growth rate of RB50::(pRF69)n in this medium, 0.58 h−1. Residual glucose concentrations were 0.8 and 2.4 g/liter in N-limited culture and 7.2 and 1.8 g/liter in P-limited culture, at the low and high D, respectively, thus illustrating successful establishment of excess-C conditions. For C limitation we used previously reported glucose-limited chemostat experiments with RB50::(pRF69)n from the exact same stock culture (10).
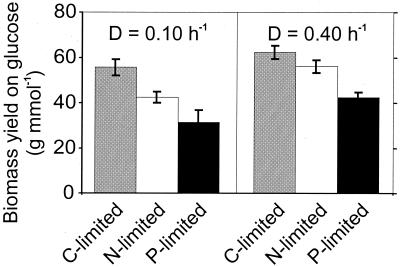
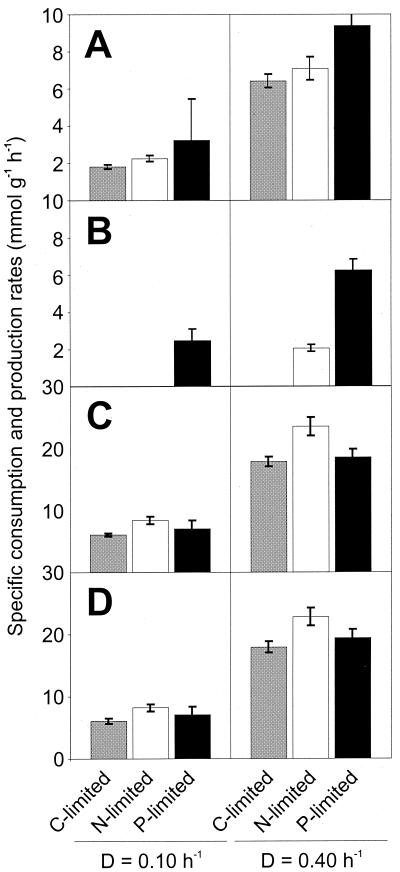
Most significantly, the biomass yield on glucose was highest in the C-limited culture and lowest in the P-limited culture, demonstrating clearly the low energetic efficiency in the latter (Fig. 2). Consequently, the specific glucose uptake rate was enhanced in excess-C cultures compared to C-limited cultures and most pronounced under P limitation (Fig. 3). As described also for other organisms (24), increased respiration is one response to C excess under N-limited conditions, illustrated here by higher specific rates of oxygen consumption and carbon dioxide formation (Fig. 3). Under P-limited conditions, in contrast, the rate of respiration is comparable to that under C-limited conditions. Thus, increased respiration does not appear to be a general response to C sufficiency in B. subtilis.
FIG. 2.
Biomass yields on glucose in C-, N-, or P-limited chemostat culture of B. subtilis. The data for C-limited cultures were taken from Dauner and Sauer (10).
FIG. 3.
Specific glucose (A) and oxygen (C) consumption rates as well as acetate (B) and carbon dioxide (D) production rates of B. subtilis during C-, N-, or P-limited chemostat cultivation. The data for C-limited cultures were taken from Dauner and Sauer (10).
Formation of low-molecular-weight by-products (excluding riboflavin) by so-called overflow metabolism was low in C-limited culture (10), moderate in N-limited culture, and extensive in P-limited culture (Table 1), as was described for Bacillus spp. (28, 35) and E. coli (31). The primary by-product was acetate, but diacyl and acetoin formation was also significant. The specific production rate of acetate was strongly influenced by the growth rate and was highest under P limitation (Fig. 3). Not generally considered classical products of overflow metabolism, extracellular protein and cell wall components contributed 1 to 4% of the carbon balance (data not shown). Recombinant riboflavin production contributed very little to the carbon balance and showed no pronounced response to the different environmental conditions (Table 1).
TABLE 1.
Relative contribution of metabolic (by)products to the carbon balance of C-, N-, or P-limited B. subtilis chemostat cultures at dilution rates of 0.1 and 0.4 h−1
| Compound | Fraction of carbon (%) recovered at indicated dilution rates in:
|
|||||
|---|---|---|---|---|---|---|
| N-limited chemostat
|
P-limited chemostat
|
C-limited chemostata
|
||||
| 0.1 h−1 | 0.4 h−1 | 0.1 h−1 | 0.4 h−1 | 0.1 h−1 | 0.4 h−1 | |
| Acetate | 0.2 | 5.9 | 20.7 | 20.2 | 0.2 | 0.4 |
| Acetoin | 0 | 0.2 | 6.5 | 6.9 | 0 | 0 |
| Diacyl | 6.5 | 4.8 | 8.6 | 6.4 | 2.9 | 1.7 |
| Riboflavin | 1.6 | 0.9 | 1.5 | 0.8 | 2.6 | 1.1 |
Data taken from Dauner and Sauer (10).
Biomass composition.
To obtain accurate information on biomass precursor requirements for subsequent metabolic flux analysis, we determined the relative contents of protein, RNA, and glycogen in our cultures. While glycogen was negligible under all conditions, the protein content was 55% ± 5% and 55% ± 3% in the N-limited culture and 65% ± 8% and 60% ± 3% in the P-limited culture at low and high D, respectively. The RNA contents of 6% ± 1% and 9% ± 1% in N-limited cultures and 4% ± 1% and 9% ± 1% in P-limited cultures at low and high D values, respectively, were low when compared to those in C-limited cultures (10). The elemental biomass compositions that were calculated from these macromolecular biomass data agreed well with data from experimental CHN analysis (Table 2), thus validating the results. The high protein content of the slow-growing, P-limited culture correlates with its high nitrogen content. Similarly, the low protein content of the fast-growing, N-limited culture correlates with its low nitrogen content.
TABLE 2.
Comparison of the experimentally determined elemental composition of B. subtilis biomass to the composition calculated from the macromolecular biomass data in N- and P-limited chemostat cultures
| Method of determination | Elemental composition of B. subtilis biomass at indicated dilution rates in:
|
|||
|---|---|---|---|---|
| N-limited chemostat
|
P-limited chemostat
|
|||
| 0.1 h−1 | 0.4 h−1 | 0.1 h−1 | 0.4 h−1 | |
| Experimentala | C1H1.49N0.22 | C1H1.63N0.22 | C1H1.61N0.23 | C1H1.59N0.24 |
| Calculatedb | C1H1.646N0.219O0.410P0.019S0.005 | C1H1.626N0.231O0.412P0.021S0.005 | C1H1.608 N0.235O0.364P0.008S0.006 | C1H1.594N0.239O0.387P0.012S0.005 |
Molecular weights are 24.83, 24.82, 23.70, and 24.23.
Molecular weights are 24.82, 24.92, 23.70, and 24.30.
Metabolic flux analysis.
To quantify the intracellular carbon flux distribution in excess-C cultures, labeling experiments with [U-13C]glucose were performed with cultures in physiological steady state. After about 0.8 volume changes, culture aliquots were harvested and hydrolyzed biomass was analyzed by [13C,1H]-COSY (55). The intracellular flux distribution was then calculated as the best fit to the relative abundances of 13C-13C coupling fine structure multiplets of 47 amino acid carbon positions (data not shown), the detailed building block requirements, and the extracellular fluxes (Fig. 1). Using the previously described, comprehensive isotopomer model of B. subtilis metabolism (9), we obtained very good fits to the data, which indicates that all data sets are consistent with each other.
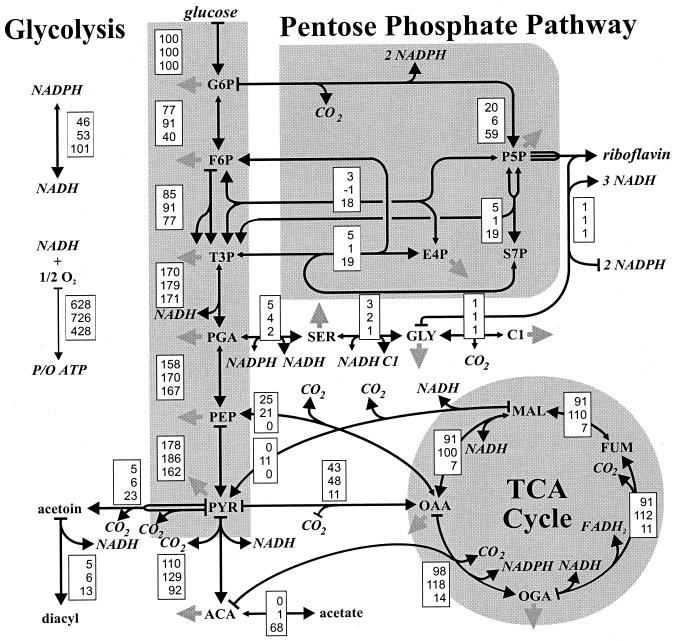
The calculated intracellular fluxes represent estimates for in vivo enzyme activities. To facilitate direct comparison of flux distributions under different conditions, we normalized the fluxes to the glucose uptake rate of each culture (Fig. 3). Generally, the intracellular flux distribution depends strongly on the choice of the limiting nutrient and the growth rate; and four major shifts in metabolic pathway usage were discernible (Fig. 4 and 5). First, the relative TCA cycle flux was extremely low under P limitation at either D, while the high TCA cycle flux in C- and N-limited cultures was markedly increased at the low D value. Second, a high relative malic enzyme flux of 27% was identified in the fast-growing P-limited culture (Fig. 5). Combined with the low TCA cycle flux under this condition, a flux distribution with reversed flux from oxaloacetate (OAA) to malate (MAL) via the MAL dehydrogenase was achieved, compared to the carbon flux usually encountered in aerobic cultures. Third, the relative PP pathway flux was low in the slow-growing N-limited culture, so that glucose catabolism proceeds almost exclusively through the glycolytic pathway. The split ratio between these two catabolic fluxes was otherwise relatively constant, with the possible exception of the slow-growing P-limited culture. Fourth, the gluconeogenic flux from OAA to phosphoenolpyruvate (PEP) catalyzed by PEP carboxykinase (16) was entirely absent in P-limited culture but was around 20 and 10% at low and high D, respectively, under C or N limitation.
FIG. 4.
Metabolic flux distribution in C-limited (top entry in the boxes), N-limited (middle), or P-limited (bottom) chemostat culture of B. subtilis at D = 0.1 h−1. Fluxes are relative to the specific glucose consumption rate of each culture shown in Fig. 3. Large arrowheads indicate the primary flux direction, and small arrowheads indicate that a reaction was considered reversible. Solid gray arrows indicate withdrawal of building blocks for biomass formation. For C-, N-, and P-limited cultures we recovered 107% ± 4%, 105% ± 5%, and 108% ± 6%, respectively, of the consumed carbon in the determined products. Abbreviations: G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; P5P, pentose phosphates; E4P; erythrose-4-phosphate; S7P, seduheptulose-7-phosphate; T3P, triose-3-phosphate; PGA, 3-phosphoglycerate; SER, serine; GLY, glycine; C1, methyl group bound to tetrahydrofolate; PEP, phosphoenolpyruvate; PYR, pyruvate; ACoA, acetyl-Coenzyme A; OGA, 2-oxoglutarate; FUM; fumarate; MAL, malate; OAA, oxaloacetate; and TCA cycle, tricarboxylic acid cycle. The data for C-limited cultures were taken from Dauner et al. (9).
FIG. 5.
Metabolic flux distribution in C-limited (top entry in the boxes), N-limited (middle), or P-limited (bottom) chemostat culture of B. subtilis at a D of 0.4 h−1. Fluxes are relative to the specific glucose consumption rate of each culture shown in Fig. 3. Large arrowheads indicate the primary direction of flux in a given reaction, and small arrowheads indicate that a reaction was considered reversible. Solid gray arrows indicate withdrawal of building blocks for biomass formation. For C-, N-, and P-limited cultures we recovered 97% ± 3%, 112% ± 7%, and 104% ± 7% %, respectively, of the consumed carbon in the determined products. The data for C-limited cultures were taken from Dauner et al. (9).
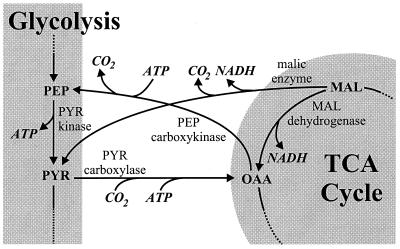
As a consequence of the above metabolic flux responses, two reaction sequences at the glycolysis-TCA cycle interface conclude ATP-dissipating futile cycles or bypass reactions (Fig. 6). The first is a cyclic flux from OAA via PEP and pyruvate (PYR) that promotes net loss of one ATP per turn and is catalyzed by PEP carboxykinase, PYR kinase, and PYR carboxylase. This futile enzyme cycle is active primarily in slow-growing N-limited cultures (Fig. 4 and 5), as was previously described for C-limited B. subtilis (9, 46). The second is a bypass of the MAL dehydrogenase reaction via PYR, catalyzed by malic enzyme and PYR carboxylase. Again, the result of this flux through the so-called PYR shunt is net loss of one ATP, compared to the direct conversion of MAL to OAA. The activity of the PYR shunt is mostly rather low, with the exception of the fast-growing P-limited culture. In this particular case, the reversed MAL dehydrogenase flux actually concludes a futile cycle (Fig. 5).
FIG. 6.
Futile cycle reactions at the interface of glycolysis and TCA cycle.
An important aspect of flux estimate interpretation is the extent to which the available data actually determine intracellular fluxes. To ensure that indeed a global error minimum was identified, the parameter search was started from at least five different starting points for each flux calculation. These solutions were always similar, and the flux solutions with the lowest χ2 values are presented in Fig. 4 and 5. These χ2 values were 119, 153, and 222 at low D and 151, 117, and 210 at high D for C, N, and P limitation experiments, respectively. Typically the 95% level of the χ2 significance test in such type of experiments is around 120 (9). Hence, the identified χ2 values are remarkably good for the analysis of a biological system (65) and indicate therefore a reliable description of the metabolic processes in B. subtilis.
The presented flux solutions were then subjected to further statistical error analysis based on a linear approximation of the model around the optimum flux solution (9, 65). For most fluxes we obtained 68% confidence intervals that were less than 10% of the estimated flux (data not shown). One exception were fluxes in the PEP-PYR-OAA triangle. The employed linearized statistical model is, however, not well suited to assess the statistical relevance of these particular fluxes, since the thus calculated confidence regions are much larger than those obtained by other approaches (9). Based on the almost-identical flux estimates that were obtained in five independent calculations, it appears, therefore, that the confidence intervals for the estimated fluxes in this triangle are similar to those of the other fluxes in the network.
A second exception with a higher confidence interval was the oxidative PP pathway flux for which 68% confidence intervals of about 25% were calculated. In the present analysis, the calculated transhydrogenase flux from NADPH to NADH (and vice versa) depends on the reported and well-established cofactor dependencies (10, 16, 18) in the biochemical reaction network (Fig. 4 and 5). As a consequence of higher confidence intervals for the oxidative PP pathway flux, the transhydrogenase flux is likewise not well determined. The net flux from NADPH to NADH via transhydrogenase, however, is with 68% confidence above 25% in all cases, except for the slow-growing, P-limited culture where this net flux is at least 56%. When the transhydrogenase reaction is omitted from the network, the χ2 values of the corresponding flux solution increase significantly from 119 to 167, 153 to 173, and 222 to 237 under C, N, and P limitations, respectively, at the low D. This indicates that indeed transhydrogenase fluxes from NADPH to NADH are consistent with all data.
Intracellular ATP and ADP concentrations.
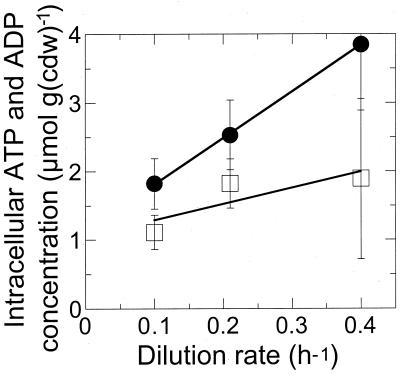
To investigate the direct impact of reduced energetic efficiency on the state of energy metabolism in our cultures, we determined the intracellular concentrations of two central intermediates of energy metabolism, ATP and ADP. The concentrations of both adenine nucleotides increased linearly with D in C-limited culture (Fig. 7). The ATP-to-ADP ratio that can be calculated from these values was about 2 but should be treated with care because the ADP concentrations have inherently large systematic errors, since they were determined as the difference in fluorescence between ATP and ATP plus ADP. The concentrations of ATP and ADP in N-limited culture were very similar to those of C-limited cultures (data not shown). As a consequence of the severe P limitation, the ATP and ADP concentrations in P-limited cultures were less than half of those found in C- and N-limited cultures (data not shown). These data show clearly that intracellular ATP concentrations depend on both the specific growth rate and the environmental conditions in B. subtilis chemostat cultures. Although ATP-to-ADP ratios are not well determined by the applied methodology, we have no indication that these ratios differ significantly in the investigated cultures.
FIG. 7.
Intracellular concentration of ATP (circles) and ADP (squares) in glucose-limited chemostat culture of B. subtilis.
DISCUSSION
Glucose catabolism in B. subtilis.
The TCA cycle in B. subtilis is subject to transcriptional control, since CcpA- and CcpC-dependent carbon catabolite repression (54) reduces the expression of several TCA cycle genes in the presence of glucose or other well-metabolized carbon sources (18, 21, 58). In C-limited B. subtilis chemostat culture, however, residual-glucose concentrations are apparently not sufficient to trigger or fully activate this regulatory response because TCA cycle fluxes are usually high (9, 46, 47). Despite residual-glucose concentrations of 0.8 and 2.4 g/liter, N-limited chemostat cultures exhibited high TCA cycle fluxes that were similar to those seen in C-limited culture. Thus, the apparent absence of catabolite repression in N-limited culture is probably due to the absence of glutamate and/or glutamine from the medium, which are synergistically required with glucose for full repression of TCA cycle enzymes (17, 39).
Despite the absence of glutamate and glutamine, TCA cycle fluxes were six- to eightfold lower in P-limited culture, with residual glucose concentrations of 2 to 7 g/liter, compared to C- or N-limited culture (Fig. 4 and 5). Although catabolite repression may play a role in this deactivation of the TCA cycle, CcpA-mediated transcriptional activation of genes involved in overflow metabolism (54) contributes probably significantly to these low TCA cycle fluxes by reducing the availability of PYR and ACA for fluxes into the TCA cycle. This view is supported by the extensive overflow metabolism in P-limited culture but not in N- or C-limited culture (Fig. 3 to 5).
In addition to transcriptional regulation, several TCA cycle enzymes are responsive to the energetic state of the cell (33), most prominently the major isoform of citrate synthase, which is a key enzyme of the cycle and is competitively inhibited by ATP (20). If ATP-dependent activation or reduced inhibition of enzymes contributes significantly to the control of TCA cycle activity, one would expect high TCA cycle fluxes in P-limited culture with twofold-lower intracellular ATP concentrations than those in C- or N-limited cultures. This is apparently not the case (Fig. 4 and 5), and thus it appears that indeed translational control and not ATP-dependent enzyme activity control is primarily responsible for down regulation of TCA cycle activity under P limitation. It is currently unclear why TCA cycle fluxes are high in N-limited culture and low in P-limited culture, but one explanation may be that intracellular levels of glutamate and glutamine are low under N limitation, so that CcpA-dependent control is not fully activated under this condition. Residual glucose concentration in N-limited cultures was probably sufficient, however, to activate expression of overflow genes (54), thus explaining intermediate acetate formation of these cultures (Fig. 3B).
Based on a different methodology for metabolic flux analysis that relied on significantly fewer NMR data, our lab estimated previously high oxidative PP pathway fluxes in glucose-limited chemostat cultures of B. subtilis (46). Although these experiments were done with a different strain, the results presented here (Fig. 4 and 5) and in a previous paper (9) suggest that oxidative PP pathway fluxes are usually less than 50% of the total glucose flux in chemostat cultures of B. subtilis. When environmental conditions cause a very strong uncoupling of anabolism and catabolism, such as shown here for the slow-growing, P-limited culture, the oxidative PP pathway flux may, however, exceed 50% (Fig. 4). Consequently, high transhydrogenase fluxes are needed to balance the reducing equivalents NADPH and NADH. Although in vitro transhydrogenase activity was observed in B. subtilis (10), conversion of NADPH into NADH may likewise be achieved by a carbon flux cycling through isoenzymes with different cofactor specificity, i.e., the glyceraldehyde 3-phosphate dehydrogenases (14) or malic enzyme (12). However, a significant contribution of the latter reaction is unlikely because it would also affect the 13C-labeling pattern.
Cellular ATP balances.
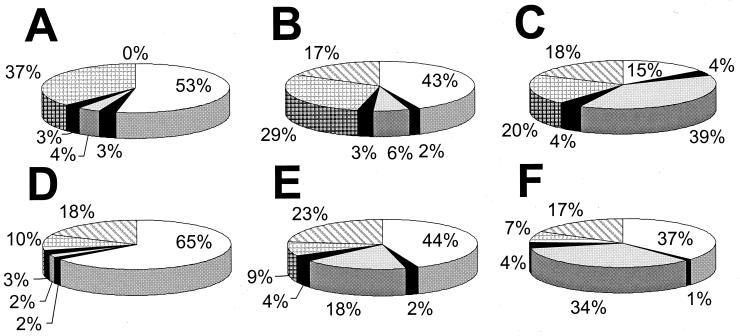
The presented flux results provide a holistic perspective on carbon metabolism and thus allow to construct an ATP balance that quantifies partitioning of the total energy flux in B. subtilis into different energy-consuming fluxes. To account also for the incorporation of glucose into biomass and metabolic (by-)products, all carbon fluxes were quantitatively converted into ATP equivalents as outlined before. The energetic equivalent of 1 mol of glucose in B. subtilis are 15 mol of ATP equivalents, which corresponds to a maximum P-to-O (ATP per oxygen) ratio of unity for respiratory ATP generation. Figure 8 depicts the partitioning of ATP equivalents into the cellular processes in B. subtilis considered here.
FIG. 8.
ATP balances of B. subtilis chemostat cultures under carbon-limited (A and D), nitrogen-limited (B and E), or phosphate-limited (C and F) conditions at D = 0.1 h−1 (A to C) and D = 0.4 h−1 (D to F). Balances are based on the total cellular energy flux in each culture, which are the specific glucose uptake rates of Fig. 3 multiplied by the maximum ATP that may be generated from glucose at a P-to-O ratio of unity, i.e., 15. The partioning of ATP equivalents into different metabolic processes includes the ATP equivalents of carbon-containing end products. The following processes were considered (in clockwise orientation): biomass formation (white); riboflavin biosynthesis (dark gray); overflow metabolism (light gray), i.e., the formation of acetate, diacyl, and acetoin; metabolic shifts from optimal pathway usage (black), including fluxes in the futile cycles of Fig. 6 and the PP pathway; maintenance metabolism (grid); and excess ATP (hatched), which indicates the remaining ATP equivalents of the total available in glucose after subtraction of the aforementioned ATP-consuming processes.
Generally, biomass formation was the most costly process and required between 40 and 70% of the available ATP equivalents, with the exception of the slow-growing P-limited culture. These energy requirements are strictly coupled to biosynthesis and polymerization and thus can be calculated in a straightforward fashion (10, 29, 45, 53). Recombinant riboflavin production accounted for only 2 to 4% of the total energy flux. The energetic relevance of overflow metabolism was low under C limitation and moderate under N limitation (Fig. 8). Under P limitation, however, overflow metabolism became the primary dissipating flux of ATP equivalents in B. subtilis. From the carbon fluxes presented in Fig.4 and 5 one can calculate dissipation of ATP equivalents via futile cycling in PEP carboxykinase and via metabolic shifts from energetically efficient to less efficient pathways, i.e., fluxes through the oxidative PP pathway and the PYR shunt. In contrast to what may be expected, dissipation of ATP equivalents via these metabolic shifts or futile cycles was not increased under excess-C conditions and the contribution to the ATP balance was below 4% in all cultures investigated (Fig. 8). Although the present flux analysis cannot quantify all potential futile cycles (e.g., glucokinase/glucose-6-phosphatase or phosphofructokinase/fructose-1,6-bisphosphatase), we found no evidence for significantly increased activity of futile cycles under excess-C conditions.
Lastly we considered dissipation of ATP equivalents in maintenance metabolism, which includes all energy expenditures that are not directly related to growth (40). Based on the maintenance coefficient of 0.66 mmol of glucose per g of biomass per h that was determined for glucose-limited, riboflavin-producing B. subtilis (47), one obtains a maintenance ATP requirement of 9.9 mmol of ATP per g of biomass per h at the assumed P/O of 1 for C-limited cultures. This value was also used for the excess-C cultures. The maintenance metabolism thus calculated becomes a major ATP-dissipating process at low D but contributes not more than 10% to the ATP balance at high D (Fig. 8). The assumed maintenance coefficient is significantly higher than the one obtained recently for the strain used here (10). These experiments were, however, not designed to accurately determine this coefficient, and newer data suggest that maintenance metabolism in the strain used here is indeed as high as was assumed above (N. Zamboni and U. Sauer, unpublished data).
The sum of all considered ATP equivalent-consuming processes in the slow-growing C-limited chemostat culture matched the available ATP equivalents from glucose exactly (Fig. 8). This demonstrates that all energetically relevant cellular processes were comprehensively considered in this particular case. Under all other conditions, however, a remarkably constant fraction of about 20% of the available ATP equivalents cannot be assigned to a specific ATP-consuming flux and is therefore referred to as excess ATP (Fig. 8). This value is an upper bound for the potential energetic contribution of all other ATP-dissipating processes that cannot be assessed from the present data, for example ion leakage (7, 40), nonquantified futile enzyme cycles, or growth rate-dependent maintenance requirements (40). The similarity of the estimated excess ATP fraction in C-limited and excess-C cultures at high D (Fig. 8D to F), suggests that there are no additional ATP-dissipating mechanisms that operate specifically under excess-C conditions in fast-growing B. subtilis. In excess-C cultures at low D, however, about 20% of the total energy flux may be dissipated by mechanisms that are specific to C sufficiency.
Any conclusion on cellular energetics depends to some extent on the assumed P/O. The above conclusions, however, are not critically dependent on a P/O of unity, since changes in P/O affect ATP production and consumption similarly. Specifically, the excess ATP equivalent values (Fig. 8) would be maximally 3% higher or lower when P/O varied within the biologically reasonable range for B. subtilis of 0.8 to 1.33, respectively. This P/O range is biologically reasonable because metabolic NADH is primarily oxidized via the aa3 oxidase branch (translocation of four protons per NADH) under the present conditions (25, 27, 42, 66, 67) and the H+-to-ATP ratio of ATP synthase is firmly established to be 4 (13, 43). Already a P/O of 1.33 would be unlikely, since it would, for example, require that the H+-to-ATP ratio of ATP synthase was only 3. In most cases, excess ATP would disappear at P/O between 0.3 and 0.4. Such P-to-O ratios, however, appear to be unrealistically low, so that probably the majority of the excess ATP fraction is indeed dissipated by processes that are not being considered at present.
The presented new concept for a cellular ATP balance that comprehensively considers the total energy flux from both carbon and ATP metabolism enables quantitative insights into cellular energy management. In particular, this concept attributes the previously observed discrepancy between theoretical and experimentally observed maximum biomass yields on ATP (see, for example, references 31, 53, and 62) to specific energy-dissipating processes.
ACKNOWLEDGMENTS
We thank Jocelyne Fiaux for analyzing the [13C,1H]-COSY spectra and for helpful discussions. Also we are grateful to one of the reviewers for helpful suggestions on catabolite repression.
Financial support was obtained through a scholarship from the Boehringer Ingelheim Fonds to M.D.
REFERENCES
- 1.Bäck T, Schwefel H-P. An overview of evolutionary algorithms for parameter optimization. Evol Comp. 1993;1:1–23. [Google Scholar]
- 2.Bakker E P. Alkali cation transport systems in prokaryotes. Boca Raton, Fla: CRC Press; 1993. [Google Scholar]
- 3.Bergmeyer H U. Methods of enzymatic analysis. IV. Deerfield Beach, Fla: VCH Publisher; 1985. [Google Scholar]
- 4.Brooke A C, Attwood M M, Tempest D W. Metabolic fluxes during the growth of thermotolerant methylotrophic Bacillus strains in methanol-sufficient chemostat cultures. Arch Microbiol. 1990;153:591–595. [Google Scholar]
- 5.Chambost J P, Fraenkel D G. The use of 6-labeled glucose to assess futile cycling in Escherichia coli. J Biol Chem. 1980;255:2867–2869. [PubMed] [Google Scholar]
- 6.Chao Y-P, Liao J C. Metabolic responses to substrate futile cycling in Escherichia coli. J Biol Chem. 1994;269:5122–5126. [PubMed] [Google Scholar]
- 7.Cook G M, Russell J B. Energy-spilling reactions of Streptococcus bovis and resistance of its membrane to proton conductance. Appl Environ Microbiol. 1994;60:1942–1948. doi: 10.1128/aem.60.6.1942-1948.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daldal F, Fraenkel D G. Assessment of a futile cycle involving reconversion of fructose 6-phosphate to fructose 1,6-bisphosphate during gluconeogenic growth of Escherichia coli. J Bacteriol. 1983;153:390–394. doi: 10.1128/jb.153.1.390-394.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dauner M, Bailey J E, Sauer U. Metabolic flux analysis with a comprehensive isotopomer model in Bacillus subtilis. Biotechnol Bioeng. 2001;76:144–156. doi: 10.1002/bit.1154. [DOI] [PubMed] [Google Scholar]
- 10.Dauner M, Sauer U. Stoichiometric growth model for riboflavin-producing Bacillus subtilis. Biotechnol Bioeng. 2001;76:132–143. doi: 10.1002/bit.1153. [DOI] [PubMed] [Google Scholar]
- 11.Dawson P S S. Continuous cultivation of microorganisms. Crit Rev Biotechnol. 1985;2:315–372. [Google Scholar]
- 12.Diesterhaft M D, Freese E. Role of pyruvate carboxylase, phosphoenolpyruvate carboxykinase, and malic enzyme during growth and sporulation of Bacillus subtilis. J Biol Chem. 1973;248:6062–6070. [PubMed] [Google Scholar]
- 13.Fillingame R H. Coupling H+ transport and ATP synthesis in F1 F0-ATP synthases: glimpses of interacting parts in a dynamic molecular machine. J Exp Biol. 1997;200:217–224. doi: 10.1242/jeb.200.2.217. [DOI] [PubMed] [Google Scholar]
- 14.Fillinger S, Boschi-Muller S, Azza S, Dervyn E, Branlant G, Aymerich S. Two glyceraldehyde 3-phosphate dehydrogenases with opposite physiological roles in a non-photosynthetic bacterium. J Biol Chem. 2000;275:14031–14037. doi: 10.1074/jbc.275.19.14031. [DOI] [PubMed] [Google Scholar]
- 15.Gerhardt P, Murray R G E, Wood W A, Krieg N R. Methods for general and molecular bacteriology. Washington, D.C.: ASM Press; 1994. [Google Scholar]
- 16.Gottschalk G. Bacterial metabolism. 2nd ed. New York, N.Y: Springer-Verlag; 1986. [Google Scholar]
- 17.Hanson R S, Cox D P. Effect of different nutritional conditions on the synthesis of tricarboxylic acid cycle enzymes. J Bacteriol. 1967;93:1777–1787. doi: 10.1128/jb.93.6.1777-1787.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hederstedt L. The Krebs citric acid cycle. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 181–197. [Google Scholar]
- 19.Hollywood N, Doelle H W. Effect of specific growth rate and glucose concentration on growth and glucose metabolism of Escherichia coli K-12. Microbios. 1976;17:23–33. [PubMed] [Google Scholar]
- 20.Jin S, Sonenshein A L. Characterization of the major citrate synthase of Bacillus subtilis. J Bacteriol. 1996;178:3658–3660. doi: 10.1128/jb.178.12.3658-3660.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jourlin-Castelli C, Mani N, Nakano M M, Sonenshein A L. CcpC, a novel regulator of the LysR family required for glucose repression of the citB gene in Bacillus subtilis. J Mol Biol. 2000;295:865–878. doi: 10.1006/jmbi.1999.3420. [DOI] [PubMed] [Google Scholar]
- 22.Katz J, Rognstad R. Futile cycling in glucose metabolism. Trends Biochem Sci. 1978;3:171–174. [Google Scholar]
- 23.Lang W K, Glassey K, Archibald A R. Influence of the phosphate supply on teichoic and teichuronic acid content of Bacillus subtilis cell walls. J Bacteriol. 1982;151:367–375. doi: 10.1128/jb.151.1.367-375.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsson C, von Stockar U, Marrison I, Gustafsson L. Growth and metabolism of Saccharomyces cerevisiae in chemostat cultures under carbon-, nitrogen-, or carbon- and nitrogen-limiting conditions. J Bacteriol. 1993;175:4809–4816. doi: 10.1128/jb.175.15.4809-4816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lauraeus M, Wikström M. The terminal quinol oxidases of Bacillus subtilis have different energy conservation properties. J Biol Chem. 1993;268:11470–11473. [PubMed] [Google Scholar]
- 26.Lifely M R, Tarelli E, Baddiley J. The teichuronic acid from the wall of Bacillus licheniformis ATCC 9945. Biochem J. 1980;191:305–318. doi: 10.1042/bj1910305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Taber H W. Catabolite regulation of the Bacillus subtilis ctaBCDEF gene cluster. J Bacteriol. 1998;180:6154–6173. doi: 10.1128/jb.180.23.6154-6163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martins M L L, Tempest D W. Metabolic response of Bacillus stearothermophilus chemostat cultures to a secondary oxygen limitation. J Gen Microbiol. 1991;137:1391–1396. [Google Scholar]
- 29.Neidhardt F C, Ingraham J L, Schaechter M. Physiology of the bacterial cell: a molecular approach. Sunderland, Mass: Sinauer Associates, Inc.; 1990. [Google Scholar]
- 30.Neijssel O M, Buurman E T, Teixeira de Mattos M J. The role of futile cycles in the energetics of bacterial growth. Biochim Biophys Acta. 1990;1018:252–255. doi: 10.1016/0005-2728(90)90260-b. [DOI] [PubMed] [Google Scholar]
- 31.Neijssel O M, Teixeira de Mattos M J, Tempest D W. Growth yield and energy distribution. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1683–1692. [Google Scholar]
- 32.Neijssel O M, Tempest D W. The physiology of metabolite over-production. Symp Soc Gen Microbiol. 1979;29:53–82. [Google Scholar]
- 33.Ohné M. Regulation of the dicarboxylic acid part of the citric acid cycle in Bacillus subtilis. J Bacteriol. 1975;122:224–234. doi: 10.1128/jb.122.1.224-234.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavlik J G, Rogers H J. Selective extraction of polymers from cell walls of Gram-positive bacteria. Biochem J. 1973;131:619–621. doi: 10.1042/bj1310619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pennock J, Tempest D W. Metabolic and energetic aspects of the growth of Bacillus stearothermophilus in glucose-limited and glucose-sufficient chemostat culture. Arch Microbiol. 1988;150:452–459. [Google Scholar]
- 36.Perkins J B, Sloma A, Hermann T, Theriault K, Zachgo E, Erdenberger T, Hannett N, Chatterjee N P, Williams II V, Rufo G A, Jr, Hatch R, Pero J. Genetic engineering of Bacillus subtilis for the commercial production of riboflavin. J Ind Microbiol Biotechnol. 1999;22:8–18. [Google Scholar]
- 37.Petersen S, de Graaf A A, Eggeling L, Möllney M, Wiechert W, Sahm H. In vivo quantification of parallel and bidirectional fluxes in the anaplerosis of Corynebacterium glutamicum. J Biol Chem. 2000;275:35932–35941. doi: 10.1074/jbc.M908728199. [DOI] [PubMed] [Google Scholar]
- 38.Press W H, Teukolsky S A, Vetterling W T, Flannery B P. Numerical recipes in C: the art of scientific computing. New York, N.Y: Cambridge University Press; 1995. [Google Scholar]
- 39.Rosenkrantz M S, Dingman D W, Sonenshein A L. Bacillus subtilis citB gene is regulated synergistically by glucose and glutamine. J Bacteriol. 1985;164:155–164. doi: 10.1128/jb.164.1.155-164.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell J B, Cook G M. Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol Rev. 1995;59:48–62. doi: 10.1128/mr.59.1.48-62.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saier M H J, Fagan M J, Hoischen C, Reizer J. Transport mechanisms. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 133–156. [Google Scholar]
- 42.Santana M, Kunst F, Hullo M F, Rapoport G, Danchin A, Glaser P. Molecular cloning, sequencing, and physiological characterization of the qox operon from Bacillus subtilis encoding the aa3-600 quinol oxidase. J Biol Chem. 1992;267:10225–10231. [PubMed] [Google Scholar]
- 43.Saraste M. Oxidative phosphorylation at the fin de siecle. Science. 1999;283:1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- 44.Sauer U, Bailey J E. Estimation of P-to-O ratio in Bacillus subtilis and its influence on maximum riboflavin yield. Biotechnol Bioeng. 1999;64:750–754. doi: 10.1002/(sici)1097-0290(19990920)64:6<750::aid-bit15>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 45.Sauer U, Cameron D C, Bailey J E. Metabolic capacity of Bacillus subtilis for the production of purine nucleotides, riboflavin, and folic acid. Biotechnol Bioeng. 1998;59:227–238. [PubMed] [Google Scholar]
- 46.Sauer U, Hatzimanikatis V, Bailey J E, Hochuli M, Szyperski T, Wüthrich K. Metabolic fluxes in riboflavin-producing Bacillus subtilis. Nat Biotechnol. 1997;15:448–452. doi: 10.1038/nbt0597-448. [DOI] [PubMed] [Google Scholar]
- 47.Sauer U, Hatzimanikatis V, Hohmann H-P, Manneberg M, van Loon A P G M, Bailey J E. Physiology and metabolic fluxes of wild-type and riboflavin-producing Bacillus subtilis. Appl Environ Microbiol. 1996;62:3687–3696. doi: 10.1128/aem.62.10.3687-3696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauer U, Lasko D R, Fiaux J, Hochuli M, Glaser R, Szyperski T, Wüthrich K, Bailey J E. Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J Bacteriol. 1999;181:6679–6688. doi: 10.1128/jb.181.21.6679-6688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schirawski J, Unden G. Menaquinone-dependent succinate dehydrogenase of bacteria catalyzes reversed electron transport driven by the proton potential. Eur J Biochem. 1998;257:210–215. doi: 10.1046/j.1432-1327.1998.2570210.x. [DOI] [PubMed] [Google Scholar]
- 50.Schmidt K, Nørregaard L C, Pedersen B, Meissner A, Duus J Ø, Nielsen J O, Villadsen J. Quantification of intracellular metabolic fluxes from fractional enrichment and 13C-13C coupling constraints on the isotopomer distribution in labeled biomass components. Metab Eng. 1999;1:166–179. doi: 10.1006/mben.1999.0114. [DOI] [PubMed] [Google Scholar]
- 51.Schreier H J. Biosynthesis of glutamine and glutamate and the assimilation of ammonia. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 281–298. [Google Scholar]
- 52.Snay J, Jeong J W, Ataai M M. Effects of growth conditions on carbon utilization and organic by-product formation in B. subtilis. Biotechnol Progr. 1989;5:63–69. [Google Scholar]
- 53.Stouthamer A H. The search for correlation between theoretical and experimental growth yields. In: Quayle J R, editor. Microbial biochemistry. Vol. 21. Baltimore, Md: University Park Press; 1979. pp. 1–48. [Google Scholar]
- 54.Stülke J, Hillen W. Regulation of carbon catabolism in Bacillus species. Annu Rev Microbiol. 2000;54:849–880. doi: 10.1146/annurev.micro.54.1.849. [DOI] [PubMed] [Google Scholar]
- 55.Szyperski T. Biosynthetically directed fractional 13C-labeling of proteinogenic amino acids: an efficient analytical tool to investigate intermediary metabolism. Eur J Biochem. 1995;232:433–448. doi: 10.1111/j.1432-1033.1995.tb20829.x. [DOI] [PubMed] [Google Scholar]
- 56.Szyperski T, Glaser R W, Hochuli M, Fiaux J, Sauer U, Bailey J E, Wüthrich K. Bioreaction network topology and metabolic flux ratio analysis by biosynthetic fractional 13C-labeling and two-dimensional NMR spectroscopy. Metab Eng. 1999;1:189–197. doi: 10.1006/mben.1999.0116. [DOI] [PubMed] [Google Scholar]
- 57.Teixeira de Mattos M J, Neijssel O M. Bioenergetic consequences of microbial adaptation to low-nutrient environments. J Biotechnol. 1997;59:117–126. doi: 10.1016/s0168-1656(97)00174-0. [DOI] [PubMed] [Google Scholar]
- 58.Tobisch S, Zühlke D, Bernhardt J, Stülke J, Hecker M. Role of CcpA in regulation of the central pathways of carbon catabolism in Bacillus subtilis. J Bacteriol. 1999;181:6996–7004. doi: 10.1128/jb.181.22.6996-7004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torres J C, Guixé V, Babul J. A mutant phosphofructokinase produces a futile cycle during gluconeogenesis in Escherichia coli. Biochem J. 1997;327:675–684. doi: 10.1042/bj3270675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tran Q H, Unden G. Changes in the proton potential and the cellular energetics of Escherichia coli during growth by aerobic and anaerobic respiration or by fermentation. Eur J Biochem. 1998;251:538–543. doi: 10.1046/j.1432-1327.1998.2510538.x. [DOI] [PubMed] [Google Scholar]
- 61.Trumpower B L, Gennis R B. Energy transduction by cytochrome complexes in mitochondrial and bacterial respiration: The enzymology of coupling electron transfer reactions to transmembrane proton translocation. Annu Rev Biochem. 1994;63:675–716. doi: 10.1146/annurev.bi.63.070194.003331. [DOI] [PubMed] [Google Scholar]
- 62.Walsh K, Koshland D E., Jr Determination of flux through the branch point of two metabolic cycles. J Biol Chem. 1984;259:9646–9654. [PubMed] [Google Scholar]
- 63.Wiechert W. 13C metabolic flux analysis. Metab Eng. 2001;3:195–206. doi: 10.1006/mben.2001.0187. [DOI] [PubMed] [Google Scholar]
- 64.Wiechert W, de Graaf A A. Bidirectional reaction steps in metabolic networks. I. Modeling and simulation of carbon isotopes labeling experiments. Biotechnol Bioeng. 1997;55:101–117. doi: 10.1002/(SICI)1097-0290(19970705)55:1<101::AID-BIT12>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 65.Wiechert W, Siefke C, de Graaf A A, Marx A. Bidirectional reaction steps in metabolic networks. II. Flux estimation and statistical analysis. Biotechnol Bioeng. 1997;55:118–135. doi: 10.1002/(SICI)1097-0290(19970705)55:1<118::AID-BIT13>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 66.Winstedt L, von Wachenfeldt C. Terminal oxidases of Bacillus subtilis strain 168: one quinol oxidase, cytochrome aa3 or cytochrome bd, is required for aerobic growth. J Bacteriol. 2000;182:6557–6564. doi: 10.1128/jb.182.23.6557-6564.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Winstedt L, Yoshida K-I, Fujita Y, von Wachenfeldt C. Cytochrome bd biosynthesis in Bacillus subtilis: characterization of the cydABCD operon. J Bacteriol. 1998;180:6571–6580. doi: 10.1128/jb.180.24.6571-6580.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yee L, Blanch H W. Defined media optimization for growth of recombinant Escherichia coli X90. Biotechnol Bioeng. 1993;41:221–230. doi: 10.1002/bit.260410208. [DOI] [PubMed] [Google Scholar]