Abstract
In response to heat stress, Bacillus subtilis activates the transcription of well over 100 different genes. Many of these genes are members of a general stress response regulon controlled by the secondary sigma factor, ςB, while others are under control of the HrcA or CtsR heat shock regulators. We have used DNA microarrays to monitor the global transcriptional response to heat shock. We find strong induction of known ςB-dependent genes with a characteristic rapid induction followed by a return to near prestimulus levels. The HrcA and CtsR regulons are also induced, but with somewhat slower kinetics. Analysis of DNA sequences proximal to newly identified heat-induced genes leads us to propose ∼70 additional members of the ςB regulon. We have also identified numerous heat-induced genes that are not members of known heat shock regulons. Notably, we observe very strong induction of arginine biosynthesis and transport operons. Induction of several genes was confirmed by quantitative reverse transcriptase PCR. In addition, the transcriptional responses measured by microarray hybridization compare favorably with the numerous previous studies of heat shock in this organism. Since many different conditions elicit both specific and general stress responses, knowledge of the heat-induced general stress response reported here will be helpful for interpreting future microarray studies of other stress responses.
DNA microarray technology provides a powerful tool for the analysis of global transcriptional responses elicited by various physical and chemical stresses. One challenge in this sort of analysis is to distinguish stress-specific responses from more general stress responses. For example, in Bacillus subtilis, many different stresses (including heat shock, osmotic stress, and energy stress) activate the large general stress response controlled by the ςB transcription factor (20, 44, 45). While others have used two-dimensional protein gels to classify cellular stress responses (55, 58), DNA-based methods have several advantages: they can be rapidly adapted to new organisms, they provide greater coverage of the genome, and data processing is comparatively easy to automate. Ultimately, it may be possible to integrate both technologies, at least for well-studied model organisms (19, 41, 56).
We have initiated a series of studies to characterize the global transcriptional responses of B. subtilis, a model gram-positive microorganism. Here, we document the heat-induced general stress response. Heat shock was chosen for this initial study since it is arguably the best-studied stress response in this organism and includes activation of the large general stress response under the control of ςB (20, 44). In addition, a subset of antibiotics that inhibit translation have been reported to induce heat shock genes in other organisms (57). It is anticipated that knowledge of transcriptional responses to antimicrobial compounds will be useful for both antibacterial discovery and characterization (47).
Analysis of the transcriptional profile of B. subtilis after heat shock clearly revealed the known heat shock regulons, including the large ςB-dependent general stress regulon (21, 44), together with several operons not previously anticipated to be heat inducible. Prominent among these are operons involved in arginine biosynthesis and transport and many candidate new members of the ςB regulon.
MATERIALS AND METHODS
Strains and growth conditions.
B. subtilis 168 strain MO945 was obtained from Niels Frandsen (GlaxoWellcome, Verona, Italy). It was grown in Bacto Mueller Hinton Broth (Difco, Detroit, Mich.). All experiments used baffled shake flasks. A 5-ml volume of medium in a 50-ml flask was inoculated and grown overnight at 37°C on a rotary platform (250 rpm). This culture was used to inoculate 50 ml of prewarmed medium (37°C) in a 500-ml flask to an optical density at 600 nm (OD600) of 0.05. The flask was shaken on a rotary platform (250 rpm) until an OD600 of 1.0 was attained. Samples (zero time) were taken from the 50-ml culture, and a 20-ml aliquot was transferred to a prewarmed 250-ml flask at 48°C which was incubated in a reciprocal-shaking water bath incubator at 48°C. A parallel identical experiment was performed with a prewarmed 250-ml flask at 37°C and incubation in a reciprocal-shaking water bath at 37°C. Samples were removed from these flasks for RNA extraction.
Sampling and RNA isolation.
Samples of the culture were rapidly removed into 2-ml tubes and centrifuged at 14,000 × g for 10 s, and the culture supernatant was rapidly removed. The tubes containing the cell pellet were placed in liquid nitrogen. The entire procedure from the start of the centrifugation to the obtaining of the frozen pellet took approximately 40 s. Total RNA was extracted from B. subtilis by disruption in phenol/guanidine isothiocyanate (TRIzol; Life Technologies, Rockville, Md.). Briefly, TRIzol and zirconium silica beads were added to each 2-ml tube containing frozen cell pellets. The tubes were shaken on a Mini-beadbeater-8 (BioSpec Products, Bartlesville, Okla.) for four 1-min cycles. Nucleic acid was precipitated, and residual DNA was removed with 4 U of RNase-free DNase I (Ambion, Austin, Tex.). After extraction with phenol-chloroform, precipitation and resuspension the RNA was quantitated with RiboGreen (Molecular Probes, Eugene, Oreg.).
Generation of ORF DNA and production of microarrays.
Oligonucleotide primers for all 4,100 open reading frames (ORFs) in the B. subtilis genome were purchased from Eurogentec (Seraing, Belgium). Full-length ORFs were made by PCR, with the following cycling conditions: 1 min of denaturing at 95°C, 45 s of annealing at 55°C, and 3.5 min of elongation at 72°C. All PCR products were purified with the QIAquick 96-well purification kit from Qiagen (Valencia, Calif.). The quality of the amplified sequences was checked by electrophoresis on a 1.5% agarose gel. The gels were digitally imaged, and the band sizes were entered into a database where the expected size was compared to the observed size. Additional data describing faint and multiple bands were also collected. In 481 cases, the PCRs failed to yield satisfactory products (no product, wrong size, additional bands, or faint bands) and oligonucleotide primers for selected genes were redesigned and obtained from Operon (Alameda, Calif.) or MWG Biotech (High Point, N.C.). Finally, over 90% (3,703 ORFs) of the B. subtilis genome was correctly amplified. Slide preparation and printing followed the procedures described by Wilson et al. (64). Briefly, amplicons were suspended in 6× SSC–15% DMSO and spotted onto poly-l-lysine-coated slides by using Telechem (Sunnyvale, Calif.) SMP5 spotting pins and an SPH16 printhead fitted to a Genemachines Omnigrid arrayer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Spot spacing was 230 μm. Slides were processed as previously described (64) and stored under N2.
cDNA labeling and slide hybridization.
Each fluorescently labeled cDNA probe was prepared from 6 μg of DNase I-treated total RNA by random hexamer [pd(N)6; Amersham Pharmacia Biotech, Piscataway, N.J.]-primed polymerization using reverse transcriptase (Superscript II RT; Life Technologies, Gaithersburg, Md.). Concentrations of nucleotides in the labeling reaction mixture were as follows: 0.5 mM dGTP, 0.5 mM dATP, 0.5 mM dTTP, and 0.05 mM dCTP. The final concentration of Cy3-dCTP or Cy5-dCTP (Fluorolink Cy dye- labeled dCTP; Amersham Pharmacia Biotech) was 0.04 mM. The final concentration of random hexamer was 0.06 mM. Unincorporated dye-labeled dCTP was removed by washing the probe in a microconcentrator (Microcon YM-30; Millipore, Bedford, Mass.).
Microarray slides were incubated for 30 min at 42°C with prehybridization solution (1% bovine serum albumin, 0.5% l-glutamate, 4× SSC), washed three times in double-distilled H2O, and dried by centrifugation at 50 × g for 90 s. For each hybridization, cDNA probe made from RNA from untreated cells (time zero sample) was mixed with probe made from RNA from heat-shocked cells. Each microarray received ∼22 μl of hybridization solution (4.2× SSC, 42% formamide, 0.17% SDS, 63 μg of salmon sperm DNA/μl) containing the two probes. The solution was applied by capillary action under a coverslip (LifterSlip; Erie Scientific Company, Portsmouth, N.H.) placed over the microarray. The whole assembly was sealed in a hybridization chamber (CMT Hybridization Chamber; Corning Incorporated, Corning, N.Y.) and submerged for 16 h in a 42°C water bath. Microarray slides were washed for 1 min in 1× SSC–0.05% SDS, 30 s in 0.06× SSC, and again for 1 min in 0.06× SSC. Slides were dried by centrifugation at 50 × g for 90 s and were immediately scanned and analyzed with a confocal laser scanner/software package (Axon GenePix 4000A/GenePix Pro 3.0; Axon Instruments, Inc., Foster City, Calif.).
Data analysis.
For analysis, any gene feature that had <80% of pixels >2 standard deviations above the local background in both channels was rejected. Ratios for levels of RNA (heat-shocked divided by time zero sample) were calculated using a ratio of medians method. Any gene feature wherein one channel was within one standard deviation of the local background was flagged as giving a potentially inaccurate ratio (indicated in the tables by values in italics; also indicated in supplemental material S2 [http://www.micro.cornell.edu/faculty.JHelmann.html]). Data normalization was based on the premise that the ratio of measured expression averaged over the entire set of sorted genes for which data was obtained is approximately equal to 1. We used a normalization method based on the geometric mean (average of the logarithmic measures of the ratios) rather than the arithmetic mean of ratios, as the geometric mean accounts for down- as well as up-regulation. Specifically, each ratio output from the scanner was multiplied by a factor of 2−[average of log2(ratios)]. A further explanation and proof of this normalization method are given in the supplemental material (S1 [http://www.micro.cornell.edu/faculty.JHelmann.html]).
To check for reproducibility in the cDNA preparation and hybridization steps, we tested the competitive hybridization of two cDNA samples both prepared from a culture grown at 37°C. Of the 2,033 gene signals detected, the overall range of ratios was quite small (1.47- to 0.62-fold range; 96% of the ratios were between 0.75 and 1.25). All experimental data were collected by the competitive hybridization of three independent cDNA preparations from each time point against the non-heat-shocked control sample (referred to as experiments 1 to 3). A comparison of 90 genes previously assigned to the heat shock stimulon showed that for genes where all three experiments yielded valid ratios, 46% of triplicate ratios yielded a coefficient of variation (CV) of <20% and 97% yielded a CV of <40%. The entire data set for all three experiments can be found in the supplemental material (Table S2 [http://www.micro.cornell.edu/faculty.JHelmann.html]).
Initial analysis focused on three overlapping sets of genes. For the first set, the induction profiles for all reported members of the heat shock stimulon were compiled from all three experiments. The resulting data from one hybridization experiment (no. 3) are presented in this study except where noted. This data set was chosen since this set of slides yielded a more complete data set than the other two replicates, with more than 3,000 genes detected with signals above background (compared to ∼2,600 genes for experiments 1 and 2). However, the overall transcriptional response in each set of hybridizations was very similar (e.g., see Fig. 4). For the second set, the 50 most highly induced genes from each of the nine data sets (three experiments with three time points each) were tabulated. The resulting list of 450 gene signals was found to result from 143 different genes (set 2), most of which appeared, as expected, in multiple experiments. For the third set, all 405 genes induced greater than twofold at the 3-min time point in experiment 3 were analyzed further. Since the ςB regulon was so large and was induced only transiently, many potential new members of this regulon were included in set 3 but were not found in the other two sets of genes.
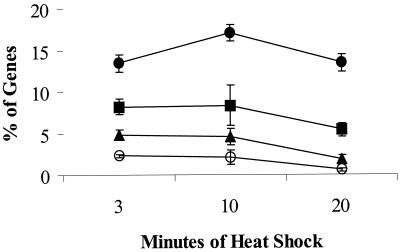
FIG. 4.
Overview of heat shock stimulon. The percentage of genes induced after heat shock was calculated for all three experiments for cells sampled at 3, 10, and 20 min after the shift from 37 to 48°C. The percentage of genes (±1 standard deviation) that are induced by at least 2-fold (closed circles), 3-fold (squares), 5-fold (triangles), or 10-fold (open circles) is shown. Percentages were determined by dividing the number of induced genes by the total number for which mRNA was detected at a level significantly above background (the average numbers of total genes [over all three time points] for which a signal was detected were 2,528 [experiment 1], 2,489 [experiment 2], and 3,204 [experiment 3]).
The lists of genes in sets 2 and 3 were analyzed to remove artifactual signals as judged by nonreproducibility in the induction. For example, in several cases, genes were initially included in set 2 (among the top 50 induced genes at one time point), but further analysis indicated that this was due to a single point which was flagged as possibly inaccurate as noted above. Moreover, there was often reliable data in the replicate experiments that clearly showed little or no induction of this same gene. By this criterion, 19 genes were removed from set 2. Of the remaining 124 genes which were reproducibly induced by heat shock, about half were members of known heat shock regulons (set 1). The remaining genes were visually analyzed to determine likely operon organization and inspected for the presence of candidate ςB-like promoter elements. A similar treatment was used for those genes in set 3. Those having a plausible match to the ςB consensus are listed in Table 4, and other heat-inducible genes of unknown regulation are included in the supplementary material (Table S3 [http://www.micro.cornell.edu/faculty.JHelmann.html]). The heat shock response of genes likely to be cotranscribed with strongly induced genes was also evaluated, and in many cases they demonstrated very similar folds induction and kinetics. This provides additional support for the observed regulation. Likely operon organization, DNA sequences, and current functional assignments were all obtained from the SubtiList database (37).
TABLE 4.
Heat shock-induced genes associated with candidate ςB-dependent promoters
| Genea | Sequence [GttTww---------------gGgwAw (sp12–15)] | 3 min | 10 min | 20 min | Function or commentsb |
|---|---|---|---|---|---|
| yaaI | ttcaaagttttttcattgc-ctaaaaaggctacatattaact | 11 | 2.4 | 1.9 | Isochorismatase homolog |
| yaaH | 12 | 2.5 | 2.0 | Similar to cortical fragment-lytic enzyme; N-acetylglucosaminidase (7) | |
| ybyB | caacaggtttagcaattt--ccaaaacgggaatgatacagga | 4.6 | 5.9 | 84 aa; unknown function (no signal in 2 expt) | |
| ycbP | aataaggtttaacttttt--acatttgaggaattatacataa | 5.3 | 4.3 | 2.0 | Membrane protein; divergent from cwlJ |
| ydaB | gaaatagttttacaagttatcttttttgggttaaaatgggtg | 5.5 | 1.6 | Predicted acid-CoA ligase (yfhL paralog) | |
| ydaC | aaaaatgtttcacatggaa-cgctgagaggaaattaactcaa | 19 | HC | ||
| ydaJ | ggtcctgttttcttaatg-ttcaaaaagggaaaaaaaagcta | 11 | 2.7 | 3.0 | No described homolog |
| ydaK | 4.8 | 0.81 | HC; similar in one domain to Myxococcus xanthus ActA response regulator (17) | ||
| ydaL | 5.3 | 1.1 | 1.2 | No described homolog | |
| ydaM | 4.4 | 0.71 | 0.94 | Similar to predicted glycosyl-transferases implicated in intercellular adhesion and biofilm formation in staphylococci (23) | |
| ydaN | 4.0 | 0.74 | No described homolog | ||
| ydbD | tttttcgtttatctttcta---tcgatcggaaatataaaaag | 17 | 23 | 7.1 | Identified as general stress protein (GS80) (2); 50% ID to Mn catalase from Lactobacillus plantarum |
| ydgC | atactcgtgagtaacatta---ctcgtgggtattatttttgg | 2.5 | 2.5 | 1.4 | Transcriptional regulator (TetR/AcrR family) |
| yerD | gctattgtttggaaagtgt-tctactgtggaaatggttacat | 8.4 | 3.3 | 1.8 | Similar to glutamate synthase (ferredoxin) |
| yfhF | aaacgcgttttcttttatt--acaatgaggtaaagtatattt | 11 | 1.9 | 2.4 | No described homolog, yfhFED operon |
| yfhD | 12 | 2.1 | No signal for yfhE; 36 aa | ||
| yfkE | tacaacgttttccaaaagcaggcaacctgaaaaaagcctata | 17 | 2.8 | 2.6 | Similar to H+/Ca2+ exchanger |
| yfkD | 18 | 3.2 | 2.8 | No described homolog | |
| yfkJ | atgaaggtttctttttaga-gaaataggggcaaagaataggg | 11 | 2.4 | 2.0 | Similar to protein-tyrosine phosphatase |
| yfkH | 9.4 | 2.6 | 2.1 | Similar to transporter (no signal for yfkI) | |
| yflSc | aacttagttaaggagtagaatggaaaaggggatcggaaaaca | 13 | 1.8 | 2.0 | 50% ID to 2-oxoglutarate/malate translocator (61) |
| ygxB | ccaaatgtataaataattt---cagccgggcagatttcatat | 16 | 4.4 | 1.5 | HC |
| yhaS | cataaagttttatatagtgaaaaagaagggatatcttgATGa | 4.0 | 5.9 | 2.3 | No described homolog |
| yhaT | 4.3 | 6.8 | 3.0 | YvrC paralog; HC | |
| yhaU | 3.0 | 4.9 | 2.4 | Similar to Na+/H+ antiporter | |
| yhcM | tataacggttaatttgtct-aacgagggggaaaatatgaata | 16 | 10 | 4.0 | No described homolog |
| yheK | ggaaaaggttaaTTGtgct-caaattcgggtagtagtgttgt | 30 | 16 | 8.4 | HC; renamed nhaX and proposed to be regulatory gene cotranscribed with nhaC encoding Na+/H+ antiporter (62) (see text) |
| yhxD | aaacatgtttttctgctta-tgctcaggggtacacatacgaa | 13 | 16 | 4.6 | YhdF (Table 2) paralog (41% ID) |
| yjcE | tgtgccgttttacaagaa-----acacgggtatcgcgtgctt | 17 | 1.7 | 2.1 | No described homolog; note suboptimal 10-bp spacer |
| yjgC | ttgtatgttttattgagtt-gttgtaagggaactgaaatagg | 18 | 11 | 3.5 | Formate dehydrogenase; divergent from yjgB |
| yjgD | 11 | 11 | 2.6 | HC | |
| yocB | agtcaggtttgatcgttt-ttaagagaggaaaaagaaaacta | 17 | 8.3 | 4.5 | No described homolog |
| ypuD | tttacggttttttattca-tgaaaaaaaggaataactcatat | 8.1 | 19 | 6.8 | No described homolog |
| yqgZ | taaatggtttaaatgaaa-aatgatccgggtagttattctac | 20 | 1.9 | 2.4 | HC |
| yqhB | acacatgttttatgagca-ttttcaggtggtatggaatgtag | 31 | 4.3 | 5.6 | HC, family of five paralogs similar to hemolysins |
| ytaB | tcgggggtttgatatttataagataaagggtaattaaataca | 14 | HC | ||
| yunG | gttctagtttttaaaatctcatcaacgtggtatcttttttta | 2.6 | 1.4 | 2.1 | No described homolog (possibly an operon with yunFEDC based on induction data) |
| yuzA | ataactgttttaataatt----catggaggaggttgcaaaac | 11 | 8.3 | 3.9 | HC |
| yvaA | agttaggttttaccattt-gatcaggagggtatatacttctg | 4.7 | 1.6 | 1.6 | Putative oxidoreductase; convergent with yvaB |
| yvaG | caatcagatttctgtcaa-taaataagaggaatcaaaaacgg | 10 | 2.9 | 1.5 | Similar to 3-oxoacyl-acyl-carrier protein reductase; possibly some transcription into downstream genes (yvaFEDCB) |
| yvaK | caaaacgtttttttctga-ttaaactgtggaaaactaaaatg | 3.6 | 0.72 | 1.0 | 70% identical to Bacillus stearothermophilus carboxylesterase (33) |
| yvaJ(rnr) | 5.1 | 1.3 | 1.2 | Exoribonuclease (40) | |
| yvbG | ataaaggtttaccgggaaatcgcctccgggtaaaagggtgga | 2.3 | 1.1 | 1.2 | HC |
| yvgN | ttaagcgtattattggtatcggctgagaggaatgtgagataa | 2.7 | 4.5 | 3.1 | Putative plant metabolite dehydrogenase, YtbE paralog |
| yvgO | tattgagattacaaatac-attgagcagggtatgcctgtagt | 9.2 | 7.1 | 3.0 | No described homolog (divergent from yvgN) |
| yvgW | gtttttgtttttcattgacactttcttggaaaacaacatata | 4.8 | 12 | 6.9 | Heavy-metal ATPase (downstream of yvgZYX) |
| yvgZ | acaaccgtttggacaatc-agtataatgggaattaatatcat | 2.0 | 1.6 | 1.5 | yvgY and yvgX weakly induced |
| ywdD | tcatctgtttcgctcttt---tcaggaaggaaagagtgagga | 2.7 | 4.3 | 2.4 | Possibly an operon with ywdEF ung, which are all also induced between two- and fourfold at 3′ end |
| ywiE | tacaaggtttatcgatta-gaaaaaagaggtaatacagaggt | 13 | 1.2 | 2.9 | Probable cardiolipin synthase; downstream genes ywjA and ywjB induced ∼3-fold, suggesting a possible operon structure |
| ywsC(pgsB) | agagaagtttggcttagt---cgattagggaagattatgtta | 3.4 | 1.4 | Capsular polyglutamate synthesis (5) | |
| ywtG | aaaaaggtttaatggccgg-aaaaagaggctaaaagatttct | 20 | 3.2 | 4.2 | 49% ID to CsbC |
| yxbG | tcgcatgtttatcactgca--catagcgggaagacaaataga | 22 | 12 | 8.0 | YcdF paralog (glucose-1-dehydrogenase) |
| yxlJd | acagccgttttttttgat--ctgcttcgggaatggtacaatg | 5.6 | 3.2 | Divergent from katX; similar to DNA-3-methyladenine glycosidase | |
| yxzFd | tagcatgtttaaggaagaggcaatcaggggaATGgttgagaa | 18 | 13 | 6.8 | Operon with yxlJ; start codon capitalized |
| yxnA | taaaaggggtaagaccct-tccggatggggtaatgtacaaaa | 11 | 6.2 | 3.5 | Similar to glucose-1-dehydrogenase |
| yycE | cttggggtttttttcatt--cgaaagatggaagaaatgacgt | 2.4 | 3.9 | No described homolog |
Downstream genes in (putative) operons are indented.
HC, hypothetically conserved; no described homolog, a unique protein found to date only in bacilli. Functional annotations are derived from the SubtiList database (37). ID, identity.
yflS is downstream of the strongly induced yflT gene, but our experiments did not detect induction of the intervening pel gene.
Data shown are from experiment 1.
Quantitative RT-PCR.
Taqman quantitative reverse transcription (RT)-PCR primers and probes were designed using Primer Express software (Applied Biosystems, Foster City, Calif.) and were synthesized by Applied Biosystems. 6FAM reporter dye and TAMRA quencher were affixed on the 5′ and 3′ ends of the probe, respectively. Primer and probe sequences were as follows: sigB sense primer, GATGAAGTCGATCGGCTCATAAG, antisense primer, CCCGCACAAGCGTTTCC, and probe, TTACCAAACAAAGCAAGATGAACAAGCGC; argB sense primer, TTGCTGAGCTTGCCAAACAC, antisense primer, CAAAAGACCGCCATCCTTACC, and probe, AATGCCCGCGGCTCGCAGT; dnaK sense primer, TGAGCTTGGCGACGGTGTA, antisense primer, GATGATCGATGATAACTTGGTCAAA, and probe, TTCGTTCAACTGCCGGCGACAA; ctsR sense primer, CAAGGTAATTTCAGAAAGAGAAGCAA, antisense primer, TTCTCGCTCTTAATTCATCACGTT, and probe, TAATGGACCGCTCAGTTTTACACATTGACTTACC. Reactions were performed using 50 ng of the DNase-treated total RNA, a 300 nM concentration of each Taqman primer, and 150 nM Taqman probe in a 50-μl volume. Controls lacking reverse transcriptase or template were used. Reactions were run on an ABI 7700 instrument (Applied Biosystems) using the following cycling parameters: reverse transcription at 48°C for 30 min, reverse transcriptase inactivation at 95°C for 10 min, 40 cycles of denaturation at 94°C for 15 s, and extension at 60°C for 1 min. Changes in expression were calculated from the displacement of the amplification curve of the heat-shocked sample from the time zero sample.
Determination of transcriptional orientation.
Transcriptional orientation for the elucidation of proximity effects in the transcription of yfkT was determined by Taqman RT-PCR using oppositely oriented primers. RT was carried out for 30 min at 48°C from 50 ng of DNase-treated total RNA with either 300 nM yfkT sense primer (TGACCAGAATGGCGCAGAT) or 300 nM antisense primer (CCAGCGTAAATGGAAGGAACA). The Taqman probe (6FAM-TTCCTATTTCCATTCGGCATCCTGGTC-TAMRA; 150 nM) and AmpliTaq Gold DNA polymerase (Applied Biosystems) were included in the reaction mixture. RNA was digested by the addition of an RNase A (5 μg; Roche)/RNase T1 (10 U; Ambion) mixture and incubated at 37°C for 1 h. Reverse transcriptase was inactivated, and Taq was activated at 95°C for 10 min. A 300 nM concentration of the opposing primer was then added, and the reaction was run on an ABI 7700 instrument with 40 cycles of denaturation at 94°C for 15 s and extension at 60°C for 1 min. Changes of expression in either orientation were calculated as described above.
RESULTS AND DISCUSSION
To develop a platform for monitoring global transcriptional responses in B. subtilis, we have amplified, using PCR, ∼90% of the ∼4,100 annotated ORFs and arrayed the resulting products on glass slides. In this report, we characterize the transcriptional response elicited by shifting a growing culture from 37 to 48°C, and we compare the resulting data with those obtained in the numerous previous studies of the heat shock stimulon in this organism (reviewed in references 19, 21, and 44).
Experimental design and array validation.
To measure gene expression under different conditions, total RNA was isolated and labeled by RT in the presence of either Cy3-dCTP or Cy5-dCTP in reactions primed with random hexamers. The resulting cDNAs were hybridized to glass slide microarrays as described in Materials and Methods. The relative hybridization of the two cDNA populations was ascertained by the relative fluorescence of the two fluorophores. The resulting data are expressed as the fold induction in the accompanying tables. While it is possible, by using appropriate normalizations, to convert fluorescence intensities to absolute transcript levels (63), we have not attempted such an analysis with these data.
Altogether, three sets of hybridization experiments were performed to measure heat shock-induced changes at 3, 10, and 20 min after shifting to 48°C (nine data sets). To control for possible variability in nucleoside incorporation, each experiment was performed at least once with the Cy3- and Cy5-labeled nucleosides reversed. As a practical matter, fold induction or repression could be confidently measured over a nearly 10,000-fold range (100-fold induction to 100-fold repression). However, for some genes, the fluorescence signal in one channel was near background and the fold induction or repression could not be confidently estimated.
In a typical experiment, hybridization signals were obtained, at levels significantly above background, for ∼70% of all genes under these growth conditions. This is comparable to results reported previously for Escherichia coli (4, 54, 63). When these signals are mapped onto the chromosome, several large clusters of apparently silent genes map to the integrated SPβ prophage, the skin element, and several other proposed prophages (data not shown). In a control experiment involving competitive hybridization of two cDNA samples both prepared from a culture grown at 37°C, no signal (of >2,000) varied by more than twofold (range, 1.47- to 0.62-fold). Thus, changes in cDNA populations well beyond this range are likely to reflect real differences in the corresponding RNA populations. The analysis described used data from one set of hybridizations (experiment 3), but similar results were obtained from the other two experiments (see Materials and Methods and supplemental material [http://www.micro.cornell.edu/faculty.JHelmann.html]), and reference is made to these results where needed. In order to independently confirm the veracity of the microarray results, the expression of four genes was also quantitated by real-time RT-PCR (22).
Overview of the heat shock stimulon.
To obtain an overview of the heat shock stimulon, we focused our analysis on three overlapping sets of genes. Set 1 included all previously described heat-inducible genes, set 2 included the 50 most strongly induced genes in all nine data sets (three replicate hybridization experiments with three time points each: 450 gene signals), and set 3 included all those genes induced at least twofold at the 3-min time point in experiment 3 (which yielded the most complete data set). Since the ςB regulon is very large and is induced transiently, many new candidate members of the ςB regulon appeared in set 3 but not in set 2. Our analysis identified many known members of the ςB and other heat shock regulons. However, we have also identified new heat shock genes, including many with candidate ςB-dependent promoters.
Consistent with existing nomenclature in B. subtilis (20, 21, 44), heat shock genes are assigned to several discrete classes: class I is the HrcA regulon, class II genes are ςB dependent, and class III genes are regulated by CtsR (and may also be regulated by ςB). We suggest that those genes for which the regulatory pathway is not yet characterized be designated class U heat shock genes (unknown regulation) rather than class IV, since the latter nomenclature will likely lead to confusion as additional regulons are defined.
The HrcA regulon (class I).
The HrcA protein is a transcriptional repressor of class I heat shock genes (50). This repressor binds to conserved cis-acting regulatory sequences known as CIRCE elements (67) and responds specifically to heat induction. In B. subtilis, HrcA is known to regulate the expression of two operons, the complex hrcA operon (24, 50) and the groEL-groES operon (34, 49). Strong and reproducible signals were not obtained for the groEL-groES operon in this study, so we focus our analysis on the hrcA operon.
Transcription initiating in the hrcA promoter region leads to the synthesis of an 8-kb primary transcript spanning seven genes that is rapidly processed into a complex family of smaller transcripts (24, 25). Previously, mRNA levels for all seven genes were measured at 5, 10, 15, and 30 min after the shift from 37 to 48°C (25). Our data are in reasonable agreement with those obtained previously (Table 1). We find the strongest induction for the first three genes in the operon, hrcA, grpE, and dnaK, with weaker effects on the downstream genes. In fact, in our studies, the three promoter distal genes were induced little if at all, with a maximal fold induction of ∼2-fold. This is consistent with the slot blot analysis, which demonstrated at most two- to fourfold induction for these genes (Table 1).
TABLE 1.
Induction of class I (HrcA-dependent) heat shock genes
| Geneb | Microarray results (fold induction at time indicated)
|
mRNA levels from slot blot analysisa
|
||||
|---|---|---|---|---|---|---|
| 3 min | 10 min | 20 min | 5 min | 10 min | 15 min | |
| hrcA | 6.0 | 8.6 | 2.6 | 10.5 | 11.0 | 4.0 |
| grpE | 3.1 | 5.8 | 2.2 | 7.0 | 7.0 | 2.0 |
| dnaKc | 2.6 (2.5) | 5.5 (3.1) | 2.3 (2.3) | 6.0 | 6.5 | 3.0 |
| dnaJ | 2.0 | 2.0 | 0.88 | 3.5 | 3.0 | 1.5 |
| yqeT | 1.7 | 1.5 | 0.67 | 3.5 | 3.0 | 1.5 |
| yqeU | 1.1 | 0.85 | 0.55 | 3.0 | 3.0 | 1.0 |
| yqeV | 1.4 | 1.1 | 0.79 | 2.5 | 2.5 | 1.0 |
| groEL | 3.1 | 6.9 | 4.1 | |||
| groES | 2.7 | |||||
Values were estimated to the nearest 0.5-fold. from a slot blot histogram (Fig. 2) (25).
Downstream genes in operons are indented.
Quantitative RT-PCR results are shown in parentheses.
The sigB regulon (class II heat shock genes).
Activation of ςB in response to heat stress is well documented, and it is estimated that the ςB regulon includes over 200 genes (19, 44, 45). Genes belonging to the ςB regulon are prominently represented among the genes of the heat shock stimulon, particularly at the 3-min time point. Because the mRNA levels for many ςB regulon genes return rapidly to pre-stimulus levels (see below), members of the ςB regulon were not well represented among the most strongly induced genes at later time points.
For purposes of discussion, we can divide known and putative members of the ςB regulon (class II heat shock genes) into three subcategories. Class IIA includes those genes for which a dependence on ςB has been documented by direct start site mapping (e.g., by primer extension), genetic experiments, or both (see reference 44). Class IIB includes genes previously postulated to be members of the ςB regulon, based on a promoter consensus search procedure (43). Class IIC includes additional heat shock genes identified in this study that are preceded by candidate ςB promoters. Many of these same genes were independently assigned to the ςB regulon based on transcriptional profiling experiments monitoring gene expression in response to ethanol stress and induction of sigB expression (45).
Class IIA.
Many of the well characterized genes belonging to the ςB regulon (44) are induced following heat shock (Table 2; Fig. 1 and 2). Comparison of their expression kinetics reveals a consistent pattern: in general, the ςB regulon is very rapidly activated in response to temperature shift with relative RNA levels (fold induction) rapidly increasing by the 3-min time point and often, though not always, declining by the 10-min time point. The transient nature of induction in response to heat stress is consistent with previous analyses of ςB-dependent transcripts, including sigB itself (6, 59), ctc (6), gspA (1), katB (13) and trxA (48).
TABLE 2.
Heat shock induction of selected ςB-dependent genesa
| Genec | Fold induction at time (min)b:
|
||
|---|---|---|---|
| 3 | 10 | 20 | |
| bmrU | 18 | 1.6 | 1.9 |
| bmr | 1.2 | 0.45 | 0.37 |
| bmrR | 2.0 | 1.6 | 1.2 |
| bofC | 5.7 | 0.86 | 1.3 |
| csbA | 4.2 | 1.1 | 1.4 |
| csbB | 7.3 | 0.56 | 1.2 |
| yfhO | 4.9 | 0.35 | 0.77 |
| csbC (yxcC) | 16 | 2.2 | 2.4 |
| csbD (ywmG) | 22 | 14 | 4.8 |
| csbX | 6.7 | 1.0 | 1.1 |
| ctc | 4.1 | 2.0 | 1.4 |
| dps | 8.1 | 2.5 | 2.5 |
| gsiB | 6.7 | 19 | 7.4 |
| gspA | 26 | 9.7 | 7.2 |
| katB | 25 | 9.0 | 4.4 |
| trxA | 1.6 | 2.6 | 2.3 |
| yacH | 17 | 27 | 3.6 |
| yacI | 18 | 30 | 4.5 |
| yacL | 5.0 | 4.3 | 1.4 |
| ycdF | 11 | 8.8 | 1.9 |
| ycdG | 22 | 7.9 | 3.8 |
| ydaP | 24 | 6.3 | 4.4 |
| yfkM | 10 | 7.5 | 3.6 |
| yflTd | 30 | 34 | 10 |
| yhdF | 23 | 15 | 6.2 |
| yhdG | 0.81 | 10 | 0.60 |
| yhdN | 14 | 5.2 | 3.7 |
| yjbC | 4.1 | 2.7 | 1.5 |
| yjbD | 2.9 | 4.2 | 3.0 |
| ykzA | 21 | 17 | 6.2 |
| yocK | 12 | 1.6 | 2.4 |
| ysdB | 4.3 | 1.9 | 1.4 |
| ytkL | 4.5 | 3.5 | 2.8 |
| ytxG | 8.0 | 2.0 | 1.4 |
| ytxH | 7.9 | 2.2 | 1.7 |
| ytxJ | 5.7 | 1.9 | 1.4 |
| yvyD | 2.3 | 1.3 | 1.4 |
| yxkO | 5.9 | 6.8 | —e |
Additional known ςB-dependent genes are shown graphically in Fig. 1 and 2. Three additional ςB-dependent genes (katX, gtaB, and opuE) were not included in the arrays used in this study.
All data are from one of three experiments (no. 3). Qualitatively similar results were obtained in each of the other two experiments except where noted. Numbers in italics are those flagged as inaccurate due to low signal in one channel.
Downstream genes in (putative) operons are indented.
yflTwas assigned to the ςB regulon based on proteome studies (60).
—, insufficient signal was obtained to estimate fold induction.
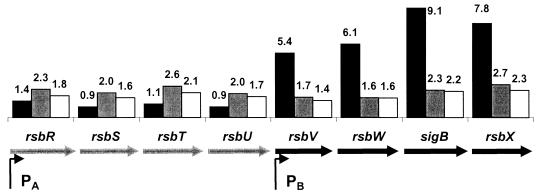
FIG. 1.
Heat induction of sigB operon genes (rsbR-S-T-U-V-W-sigB-rsbX). The sigB operon is illustrated schematically (genes are not to scale), and the fold induction by heat shock at 3 min (black), 10 min (grey), and 20 min (white) is superimposed on the operon structure. The operon is transcribed from an upstream ςA-dependent promoter (65) and from an internal, heat-inducible ςB-dependent promoter (6, 27). Measurements of sigB induction by RT-PCR yielded values of 11.8-fold (3 min), 3.0-fold (10 min), and 3.3-fold (20 min).
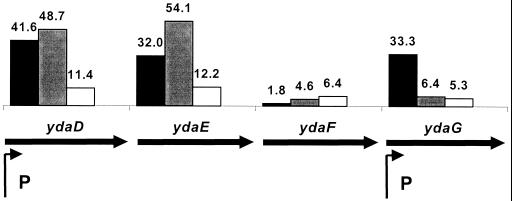
FIG. 2.
Heat induction of the ydaDEFG gene cluster. The ydaDEFG cluster of genes has been previously shown to be expressed from two ςB-dependent promoter elements as shown (42). Induction at 3, 10, and 20 min is shown as in Fig. 1.
The ςB regulon includes sigB itself, which is transcribed as part of a complex operon containing eight genes (Fig. 1). As expected, RNA levels corresponding to all four genes downstream of the internal ςB-dependent promoter are rapidly elevated after heat shock (∼5- to 9-fold), while RNA corresponding to the upstream genes is only slightly induced. Note that none of these genes are strongly induced, and none of them are represented among the top 50 induced genes, even at the 3-min time point. Like other members of the ςB regulon, mRNA levels return to near the pre-stress level by 10 and 20 min following heat shock. Previous measurements of sigB mRNA levels following heat shock revealed a maximal induction of >20-fold at 5 min, followed by reduced levels at 10, 15, and 20 min following temperature stress (59).
For comparison with the array data, we independently determined the degree of induction for the sigB mRNA using a quantitative RT-PCR approach (22). These experiments revealed consistent kinetics of induction with a maximal induction (at 3 min) of 12-fold. Note that this is somewhat higher than the induction measured using the arrays (ninefold). In a control experiment, using cells shifted from 37°C to another flask at 37°C, we also see a much weaker (∼2-fold) but still significant induction of sigB (data not shown). This may be a response to stresses associated with transfer of the cells. For example, removal from a well aerated flask using a glass pipette could allow a transient depletion of oxygen in the rapidly growing culture.
The ydaDEFG region of the chromosome has been previously analyzed (42) and found to have two ςB-dependent promoters giving rise to a complex family of transcripts as determined by Northern blot analysis. Our data support the suggestion, from Northern analysis, that ydaD and ydaE are cotranscribed as both are strongly and coordinately induced (Fig. 2). In addition, we find very strong induction of ydaG, but ydaF is induced much more modestly, and with slower kinetics. This is consistent with the presence of a prominent heat-induced transcript corresponding to the ydaG gene, and argues that there may be limited transcriptional readthrough from ydaDE into ydaF.
Two genes considered to be members of the ςB regulon that are not strongly induced by heat shock are bmrR and bmr, the two promoter distal genes in the bmrU operon (Table 2). However, bmrU is strongly and reproducibly induced. It had previously been suggested that bmrR and bmr may be cotranscribed with bmrU, but the published Northern blot analysis suggests that most transcription terminates after the bmrU gene (42). The lack of strong heat induction of the promoter distal genes, bmrR and bmr, is consistent with the idea that readthrough into these downstream genes does not greatly affect their expression.
Class IIB.
Using a consensus search approach, Petersohn et al. (43) identified 31 additional ςB-type promoters. In three cases (yhdF, yacL, ysdB), these promoters were confirmed by primer extension mapping, and these genes have therefore been added to class IIA (Table 2). An additional 25 sites were shown, using slot blot analysis, to be induced by ethanol. In all but four cases, this induction was apparent in the wild type but not in a sigB mutant strain (43). Three genes (yabJ, yhaR, and yqhZ) are not induced by ethanol (43), and we found that these genes did not respond to heat shock. Thus, these putative ςB promoters may represent false positives generated by an imperfect search algorithm.
We found strong heat shock induction for genes proximal to 16 of the proposed promoters (43), and the kinetics of induction are comparable to those of known members (class IIA) of the ςB regulon (Table 3). These results support the previous suggestion that these genes are part of the ςB regulon (43). Note that the yfhK gene is upstream of the ςW-dependent operon yfhLM, and this promoter may be responsible for the heat induction of those genes as well (26).
TABLE 3.
Fold induction of candidate ςB regulon genesa
| Geneb | 3 min | 10 min | 20 min |
|---|---|---|---|
| aldY | 17 | 11 | 18 |
| lctEc | 8.8 | 10 | 8.3 |
| ycnHc | 8.0 | 16 | 8.5 |
| ydaT | 26 | 17 | 4.9 |
| ydaS | 27 | 22 | 6.1 |
| ydhK | 14 | 1.9 | 1.5 |
| yfhK | 16 | 2.3 | 3.3 |
| yfhLd | 7.1 | 1.9 | 2.7 |
| yfhM | 7.7 | 1.9 | 2.4 |
| yflA | 21 | 3.1 | 3.5 |
| yjgB | 37 | 22 | 5.3 |
| yjgA | 3.6 | 2.8 | 1.7 |
| ykgA | 25 | 2.1 | 3.4 |
| ykgB | 2.6 | 1.7 | 1.5 |
| yoxC | 16 | 2.5 | 2.4 |
| yoxB | 12 | 1.2 | 1.5 |
| yoaA | 4.6 | 2.0 | 2.8 |
| yqhA | 12 | 0.82 | 1.8 |
| yqxLe | 17 | 2.3 | 3.8 |
| yvrE | 6.3 | 1.7 | 1.5 |
| ydbP | 1.4 | 1.4 | 1.4 |
| yoxA | 0.62 | 0.28 | 0.68 |
| ypuB | 1.9 | 1.2 | |
| ypuC | 2.8 | 1.4 | |
| yqhQ | 1.3 | 1.1 | 0.99 |
| yqhP | 1.6 | 1.3 | 1.2 |
| yqiS | 0.66 | 0.73 | 0.78 |
| yrvD | 1.7 | 0.65 | 1.1 |
Genes were previously identified by Petersohn et al. (43). No data were obtained for yotK and yycD.
Downstream genes in (putative) operons are indented.
Ethanol induction is not ςB dependent (43).
The yfhLM genes are also transcribed from a ςW-dependent promoter (26).
Data are from experiment 1 instead of experiment 3.
Five genes (ydbP, yoxA, ypuB, yqhQ, and yrvD) shown to be inducible by ethanol in a ςB-dependent manner (43) were not strongly induced by heat shock in our study. Nor was heat shock induction detected for yqiS, a gene induced by ethanol in both the wild-type and sigB mutant strains (Table 3). Finally, no data were obtained for yotK and yycD, as these genes were absent from the arrays used in these experiments. Additional experiments will be required to establish whether or not the putative ςB-dependent promoters associated with these eight genes are in fact functional.
Class IIC.
By sequence inspection, we propose 44 additional candidate ςB-dependent promoters (likely controlling ∼70 genes) proximal to newly identified heat shock genes (Table 4). In many cases, these candidate promoters are a good match to the ςB consensus (43, 44) in both the −35 and −10 recognition elements. Indeed, 19 of these operons were independently proposed to be candidate members of the ςB regulon, based on an analysis of genes induced by ethanol or by induction of ςB expression, and 11 of these same promoters were identified using a hidden Markov model (45). Thus, it is likely that many, although probably not all, of the genes we have identified represent new members of the ςB regulon. In some cases, for example, the candidate promoters we propose differ in potentially significant ways from the ςB consensus, and these may be nonfunctional, chance occurrences. Interestingly, several of these genes encode paralogs of known members of the ςB regulon (YwtG is 49% identical to CsbC; YxbG is 34% identical to YcdF; YdaB is 33% identical to YfhL; YxhD is 41% identical to YhdF). As a class, these candidate ςB regulon members include many predicted membrane proteins and transporters, functions consistent with the composition of the ςB regulon as a whole.
Mapping all the known ςB regulon members, together with additional likely members emerging from this study, onto the B. subtilis genome revealed three instances of clusters of transcriptional units. As many as nine ςB consensus elements are clustered around the ydaDEFG operon (Fig. 2). This cluster includes the ςB-dependent gsiB and ydaP genes (Table 2), the ydaTS and ydbD operons (Table 3), and the heat-induced ydaB, ydaC, and ydaJKLMN genes (Table 4). A second cluster occurs upstream of the comG operon (four promoters: yqxL, yqhB, yqhA, and yqgZ). The third cluster includes the yfkM, yfkJIH, yfkF, and yfkED operons. The vast majority of the remaining ςB-dependent operons are apparently isolated or are occasionally found in small clusters of two or three operons.
Finally, analysis of the yheK gene leads us to propose a revision to the existing genome annotation (37). This gene displays the characteristic ςB induction pattern, yet the best candidate ςB promoter is situated with the −35 region overlapping the assigned start codon (TTG). Sequence inspection identifies an alternative start site (ATG) at codon 19 of the yheK ORF. Furthermore, most YheK homologs lack the 18 additional amino acids that would result from initiation at the assigned TTG start codon. We therefore suggest that translation of YheK begins with the ATG codon at position 19 and that the indicated promoter element may therefore be physiologically relevant (this new translation start site was also chosen in the latest annotation of the SubtiList database; release R16.1). Note that this gene has been redesignated nhaX and is proposed to form an operon with the downstream gene nhaC (formerly yheL) (62). However, there are no published data to support the suggestion of an operon structure, and 130 bp separate the yheK (nhaX) and nhaC genes. Moreover, we did not observe heat induction for yheL.
The CtsR regulon (class III).
A subset of genes regulated by ςB is also controlled by another heat shock pathway under control of CtsR (10, 32). CtsR is encoded by the first gene in the ctsR operon, which is transcribed from both ςB- and ςA-dependent promoters. We noted strong induction of the ctsR operon in this study (Table 5), but unlike that of ςB-dependent heat shock genes, transcription of the ctsR operon peaked at the 10-min time point. The lower level of induction of the two promoter distal genes (sms and yacK) is consistent with recent data indicating that these genes are part of a separate, ςM-dependent operon (A. Moir, personal communication). CtsR also regulates clpP, clpE, and clpC (10), which are strongly induced at the 10-min time point. This is in agreement with previous mRNA measurements that document a peak induction of clpP of ∼28-fold between 6 and 9 min after a shift to 48°C (15). A similar pattern of induction was observed for clpE (9).
TABLE 5.
Fold induction of class III heat shock genes
| Genea | 3 min | 10 min | 20 min |
|---|---|---|---|
| ctsRb | 17 | 26 | 3.2 |
| yacH | 17 | 27 | 3.6 |
| yacI | 18 | 30 | 4.5 |
| clpC | 13 | 29 | 4.3 |
| sms | 5.1 | 9.6 | 2.2 |
| yacK | 4.2 | 7.2 | 1.8 |
| clpP | 6.4 | 21 | 6.8 |
| clpE | 63 | 88 | 25 |
| clpC | 13 | 29 | 4.3 |
Downstream genes in (putative) operons are indented.
Fold induction for ctsR obtained by RT-PCR was 22.2-fold (3 min), 19.8-fold (10 min), and 4.6-fold (20 min).
The AhrC regulon.
One of the most unexpected findings in this study was the exceptionally strong transcriptional induction of three operons involved in arginine biosynthesis and transport (Fig. 3). There was no induction at the 3-min time point, but by 10 min after heat shock, all three operons were induced at least 50-fold. Independent confirmation of argB induction by quantitative RT-PCR showed over 900-fold induction, suggesting that the microarray experiment may underestimate the change in expression. Since both the arginine biosynthesis operons are repressed by the AhrC arginine-sensing transcription factor (8, 53), it is possible that heat shock induced a transient arginine deprivation. Alternatively, the AhrC protein itself may be temperature labile (12). The yqiXYZ operon was recently shown to encode an arginine transport system (52), and it also displays the same magnitude and kinetics of induction as noted for the two biosynthetic operons (Fig. 3C).
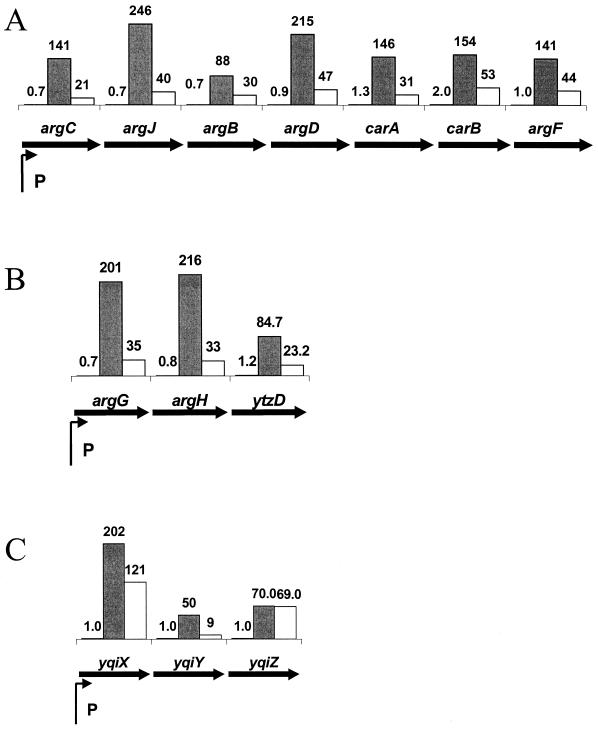
FIG. 3.
Heat induction of arginine biosynthesis and uptake genes. Transcription of the argC biosynthetic operon (A), argG biosynthetic operons (B), and the yqiX arginine transport system (C) (52) is illustrated. The biosynthetic operons are repressed in response to arginine by the AhrC regulatory protein (8, 53). The yqiXYZ operon has been renamed artPQM to be consistent with E. coli nomenclature (52). Values for argB obtained by RT-PCR were 0.9-fold (3 min), 920-fold (10 min), and 328-fold (20 min). In the microarray studies, signals were not detected reproducibly for the 3-min points, presumably due to low message levels. However, those signals that were detected (four signals) were near 1, so these values have been shown as 1.0 for illustration purposes.
Since AhrC is also required as a positive activator of arginine catabolic genes (14, 29, 35), we also looked at the effects of heat shock on transcription of the rocA, rocG, and rocD operons. These operons are rapidly repressed following temperature shift and are among the most dramatically repressed genes in our analysis (mRNA levels declined by 3- to 20-fold after 10 min). This is consistent with a rapid (within 10 min) functional inactivation of the AhrC transcriptional activator combined with a short mRNA half-life for these transcripts.
Other identified stress response genes (class U).
Many other genes have been identified as heat inducible in previous studies but are regulated by as-yet-unknown mechanisms. Several of these genes were also found to be heat induced in our study, as shown in Table 6. For example, we detect an ∼5-fold induction of both ykdA(htrA) and yvtA, two heat-inducible HtrA paralogs regulated by unknown mechanisms from similar promoter elements (38).
TABLE 6.
Class U (other): fold induction of known stress genes
| Genea | 3 min | 10 min | 20 min | Comment(s) and/or reference |
|---|---|---|---|---|
| ahpF | 1.5 | 1.6 | 1.8 | ahpC not on array (3); ahpC not on array (3) |
| clpX | 0.85 | 1.3 | 1.1 | 16 |
| ftsH | 1.1 | 1.5 | 1.4 | 11 |
| htpG | 3.2 | 5.8 | 5.5 | 51 |
| htrA | 4.9 | 3.1 | 2.9 | =ykdA (39) |
| lonA | 1.6 | 2.3 | 1.6 | 46 |
| yvtA | 4.8 | 1.9 | 2.1 | HtrA paralog (38) |
| yvtB | 5.0 | 1.7 | 1.7 | |
| ywcG | 4.7 | 6.9 | 3.3 | =nfrA (36) |
| ywcH | 3.3 | 4.5 | 2.0 |
Downstream genes in (putative) operons are indented.
An additional 66 members of the heat shock stimulon are not associated with obvious candidate promoter elements for ςB or obvious recognition sites for known heat shock regulators (see supplementary material; Table S3 at http://www.micro.cornell.edu/faculty.JHelmann.html). All of these genes showed reproducible heat induction of at least 3.5-fold (or are cotranscribed with induced genes). Since regulatory pathways for these genes are not known, we assigned them to class U. Interestingly, several of these genes encode transport functions, including the appDFABC operon, one of two oligopeptide uptake systems in B. subtilis (30, 31). Other transporters induced by heat shock include a choline ABC transporter (opuB operon), a putative Na+/nucleoside cotransporter (yutK) and a multidrug efflux homolog (yuxJ). We also note heat induction of a subset of the S-box regulon (18) including specifically those genes implicated in methionine biosynthesis. Additional work will be required to determine the mechanism and relevance of this heat induction.
Gene signals arising from proximity effects.
In addition to increased transcription due to heat shock, some of the signals detected in these experiments may arise from what we generically call proximity effects. For example, transcription termination at the end of operons is often less than 100% efficient, and these read-through transcripts may lead to signals corresponding to genes downstream of strongly induced heat shock genes. If the downstream gene is codirectional with the heat shock gene, these signals could be physiologically relevant. However, in some cases, the downstream gene is convergent with the heat shock gene and the transcript through this region is anti-sense. These are nevertheless detected using random hexamer priming and could give rise to spurious signals. Two likely examples that emerged in this study are the yfkQ operon and the yknA gene (Table 7). All four genes of the yfkQ operon showed some heat induction, but there was a clear gradient, with the largest apparent induction near the end of the operon. Since this operon is convergent with the strongly induced, ςB-dependent yflA gene (Table 3), this pattern is consistent with read-through transcription from yflA giving rise to (gradually diminishing) antisense RNA through this region.
TABLE 7.
Apparent heat induction of convergent operons due to readthrough transcription
| Genea | 3 min | 10 min | 20 min | Comment |
|---|---|---|---|---|
| yfkQ | 3.2 | 1.3 | 1.2 | yfkQ operon convergent with yflA |
| yfkR | 7.7 | 1.2 | ||
| yfkS | 11 | 1.9 | 2.4 | |
| yfkT | 20 | 3.2 | ||
| yknA | 8.9 | 4.2 | 2.3 | Convergent with ykzA |
Downstream genes in (putative) operons are indented.
To test this model, readthrough transcription from yflA into yfkT was measured by a modification of the standard Taqman quantitative RT-PCR protocol. RT was conducted with either a sense or an antisense primer, after which time the RNA was digested and reverse transcriptase was inactivated. The opposing primer was then added and quantitative PCR was carried out. RT with the sense primer (i.e., priming off the antisense strand) showed a 29-fold induction at the 3-min time point, whereas the antisense primer showed only a threefold induction (data not shown). This result is consistent with a proximity effect whereby the apparent induction of yfkT by heat shock is primarily due to read-through from yflA with the antisense strand of yfkT being transcribed. Similarly, the apparent induction of yknA may result from the fact that this gene is convergent with the strongly induced ykzA gene (Table 2). Although both the yknA gene and the yfkQ operon were considered good candidates for the ςB regulon on the basis of transcriptional profiling studies (45), our findings suggest that a reinterpretation of these data is in order. Similar proximity effects have been noted in microarray studies of E. coli (28, 66). It is possible to avoid this complication by using 3′-end, gene-specific primers for RT. However, as discussed in detail elsewhere (4), this approach does not uniformly label all mRNAs and therefore provides a more limited picture of the transcriptome.
Summary.
The transfer of B. subtilis from 37 to 48°C elicits a very large transcriptional response coordinated by several distinct transcription factors (19–21, 44). In the studies described here, we document the heat induction of hundreds of genes and independently confirm the microarray data for four genes by quantitative RT-PCR. Over 5% of the transcriptionally active genes are induced at least threefold, and well over 10% of the genome displays a measurable induction in response to heat shock (Fig. 4).
Activation of the ςB regulon is the single largest component of the heat shock response in B. subtilis (19, 44). We have measured the induction of 70 known or previously proposed members of the ςB regulon (Tables 2 and 3; Fig. 1 and 2) and identified another 72 candidate ςB regulon members (Table 4). Our heat shock data provides additional support for many, albeit not all, members of the ςB regulon proposed previously on the basis of consensus search procedures (43) and transcriptional profiling studies (45).
As expected, heat induction of the CtsR (Table 5) and HrcA (Table 1) regulons is apparent, and other known heat shock proteins (Table 6) are also induced. Finally, we can assign many new genes to the heat shock stimulon (Table S3 [http://www.micro.cornell.edu/faculty.JHeilmann.html]), though the factor(s) mediating their heat induction are not clear at present. Prominent among these genes are three operons involved in arginine biosynthesis and transport (Fig. 3). Induction of these genes may reflect an in vivo temperature lability of the AhrC regulatory protein, an idea supported by the decrease in expression of the AhrC-dependent arginine catabolic genes.
Our analysis provides further evidence of the power and utility of microarray approaches to defining bacterial stimulons and regulons. As we extend this work to include other stimulons, a thorough knowledge of the heat shock activated general stress response will be very useful in distinguishing specific from more general transcriptional responses.
ACKNOWLEDGMENTS
We thank Dave Huber for mathematical and software assistance, Young Kim for technical help, and Tarek Msadek for helpful comments on the manuscript.
This work was partially supported by grant MCB 9983656 from the National Science Foundation to J.D.H.
REFERENCES
- 1.Antelmann H, Bernhardt J, Schmid R, Hecker M. A gene at 333 degrees on the Bacillus subtilis chromosome encodes the newly identified sigma B-dependent general stress protein GspA. J Bacteriol. 1995;177:3540–3545. doi: 10.1128/jb.177.12.3540-3545.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelmann H, Bernhardt J, Schmid R, Mach H, Volker U, Hecker M. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis. 1997;18:1451–1463. doi: 10.1002/elps.1150180820. [DOI] [PubMed] [Google Scholar]
- 3.Antelmann H, Engelmann S, Schmid R, Hecker M. General and oxidative stress responses in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J Bacteriol. 1996;178:6571–6578. doi: 10.1128/jb.178.22.6571-6578.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arfin S M, Long A D, Ito E T, Tolleri L, Riehle M M, Paegle E S, Hatfield G W. Global gene expression profiling in Escherichia coli K12. The effects of integration host factor. J Biol Chem. 2000;275:29672–29684. doi: 10.1074/jbc.M002247200. [DOI] [PubMed] [Google Scholar]
- 5.Ashiuchi M, Soda K, Misono H. A poly-gamma-glutamate synthetic system of Bacillus subtilis IFO 3336: gene cloning and biochemical analysis of poly-gamma-glutamate produced by Escherichia coli clone cells. Biochem Biophys Res Commun. 1999;263:6–12. doi: 10.1006/bbrc.1999.1298. [DOI] [PubMed] [Google Scholar]
- 6.Benson A K, Haldenwang W G. The sigma B-dependent promoter of the Bacillus subtilis sigB operon is induced by heat shock. J Bacteriol. 1993;175:1929–1935. doi: 10.1128/jb.175.7.1929-1935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Fukuoka S, Makino S. A novel spore peptidoglycan hydrolase of Bacillus cereus: biochemical characterization and nucleotide sequence of the corresponding gene sleL. J Bacteriol. 2000;182:1499–1506. doi: 10.1128/jb.182.6.1499-1506.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czaplewski L G, North A K, Smith M C, Baumberg S, Stockley P G. Purification and initial characterization of AhrC: the regulator of arginine metabolism genes in Bacillus subtilis. Mol Microbiol. 1992;6:267–275. doi: 10.1111/j.1365-2958.1992.tb02008.x. [DOI] [PubMed] [Google Scholar]
- 9.Derre I, Rapoport G, Devine K, Rose M, Msadek T. ClpE, a novel type of HSP100 ATPase, is part of the CtsR heat shock regulon of Bacillus subtilis. Mol Microbiol. 1999;32:581–593. doi: 10.1046/j.1365-2958.1999.01374.x. [DOI] [PubMed] [Google Scholar]
- 10.Derre I, Rapoport G, Msadek T. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in gram-positive bacteria. Mol Microbiol. 1999;31:117–131. doi: 10.1046/j.1365-2958.1999.01152.x. [DOI] [PubMed] [Google Scholar]
- 11.Deuerling E, Mogk A, Richter C, Purucker M, Schumann W. The ftsH gene of Bacillus subtilis is involved in major cellular processes such as sporulation, stress adaptation and secretion. Mol Microbiol. 1997;23:921–933. doi: 10.1046/j.1365-2958.1997.2721636.x. [DOI] [PubMed] [Google Scholar]
- 12.Dion M, Charlier D, Wang H, Gigot D, Savchenko A, Hallet J N, Glansdorff N, Sakanyan V. The highly thermostable arginine repressor of Bacillus stearothermophilus: gene cloning and repressor-operator interactions. Mol Microbiol. 1997;25:385–398. doi: 10.1046/j.1365-2958.1997.4781845.x. [DOI] [PubMed] [Google Scholar]
- 13.Engelmann S, Lindner C, Hecker M. Cloning, nucleotide sequence, and regulation of katE encoding a sigma B-dependent catalase in Bacillus subtilis. J Bacteriol. 1995;177:5598–5605. doi: 10.1128/jb.177.19.5598-5605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardan R, Rapoport G, Debarbouille M. Role of the transcriptional activator RocR in the arginine-degradation pathway of Bacillus subtilis. Mol Microbiol. 1997;24:825–837. doi: 10.1046/j.1365-2958.1997.3881754.x. [DOI] [PubMed] [Google Scholar]
- 15.Gerth U, Kruger E, Derre I, Msadek T, Hecker M. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol Microbiol. 1998;28:787–802. doi: 10.1046/j.1365-2958.1998.00840.x. [DOI] [PubMed] [Google Scholar]
- 16.Gerth U, Wipat A, Harwood C R, Carter N, Emmerson P T, Hecker M. Sequence and transcriptional analysis of clpX, a class-III heat-shock gene of Bacillus subtilis. Gene. 1996;181:77–83. doi: 10.1016/s0378-1119(96)00467-2. [DOI] [PubMed] [Google Scholar]
- 17.Gronewold T M A, Kaiser D. The act operon controls the level and time of C-signal production for Myxococcus xanthus development. Mol Microbiol. 2001;40:744–756. doi: 10.1046/j.1365-2958.2001.02428.x. [DOI] [PubMed] [Google Scholar]
- 18.Grundy F J, Henkin T M. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 19.Hecker M, Engelmann S. Proteomics, DNA arrays and the analysis of still unknown regulons and unknown proteins of Bacillus subtilis and pathogenic gram-positive bacteria. Intl J Med Microbiol. 2000;290:123–134. doi: 10.1016/S1438-4221(00)80080-6. [DOI] [PubMed] [Google Scholar]
- 20.Hecker M, Schumann W, Volker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 21.Hecker M, Volker U. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the sigmaB regulon. Mol Microbiol. 1998;29:1129–1136. doi: 10.1046/j.1365-2958.1998.00977.x. [DOI] [PubMed] [Google Scholar]
- 22.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 23.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Goetz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 24.Homuth G, Masuda S, Mogk A, Kobayashi Y, Schumann W. The dnaK operon of Bacillus subtilis is heptacistronic. J Bacteriol. 1997;179:1153–1164. doi: 10.1128/jb.179.4.1153-1164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Homuth G, Mogk A, Schumann W. Post-transcriptional regulation of the Bacillus subtilis dnaK operon. Mol Microbiol. 1999;32:1183–1197. doi: 10.1046/j.1365-2958.1999.01428.x. [DOI] [PubMed] [Google Scholar]
- 26.Huang X, Gaballa A, Cao M, Helmann J D. Identification of target promoters for the Bacillus subtilis extracytoplasmic function sigma factor, sigma W. Mol Microbiol. 1999;31:361–371. doi: 10.1046/j.1365-2958.1999.01180.x. [DOI] [PubMed] [Google Scholar]
- 27.Kalman S, Duncan M L, Thomas S M, Price C W. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J Bacteriol. 1990;172:5575–5585. doi: 10.1128/jb.172.10.5575-5585.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khodursky A B, Peter B J, Cozzarelli N R, Botstein D, Brown P O, Yanofsky C. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:12170–12175. doi: 10.1073/pnas.220414297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klingel U, Miller C M, North A K, Stockley P G, Baumberg S. A binding site for activation by the Bacillus subtilis AhrC protein, a repressor/activator of arginine metabolism. Mol Gen Genet. 1995;248:329–340. doi: 10.1007/BF02191600. [DOI] [PubMed] [Google Scholar]
- 30.Koide A, Hoch J A. Identification of a second oligopeptide transport system in Bacillus subtilis and determination of its role in sporulation. Mol Microbiol. 1994;13:417–426. doi: 10.1111/j.1365-2958.1994.tb00436.x. [DOI] [PubMed] [Google Scholar]
- 31.Koide A, Perego M, Hoch J A. ScoC regulates peptide transport and sporulation initiation in Bacillus subtilis. J Bacteriol. 1999;181:4114–4117. doi: 10.1128/jb.181.13.4114-4117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kruger E, Hecker M. The first gene of the Bacillus subtilis clpC operon, ctsR, encodes a negative regulator of its own operon and other class III heat shock genes. J Bacteriol. 1998;180:6681–6688. doi: 10.1128/jb.180.24.6681-6688.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kugimiya W, Otani Y, Hashimoto Y. Molecular cloning and structure of the gene for esterase from a thermophilic bacterium, Bacillus stearothermophilus IFO 12550. Biosci Biotechnol Biochem. 1992;56:2074–2075. doi: 10.1271/bbb.56.2074. [DOI] [PubMed] [Google Scholar]
- 34.Li M, Wong S L. Cloning and characterization of the groESL operon from Bacillus subtilis. J Bacteriol. 1992;174:3981–3992. doi: 10.1128/jb.174.12.3981-3992.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller C M, Baumberg S, Stockley P G. Operator interactions by the Bacillus subtilis arginine repressor/activator, AhrC: novel positioning and DNA-mediated assembly of a transcriptional activator at catabolic sites. Mol Microbiol. 1997;26:37–48. doi: 10.1046/j.1365-2958.1997.5441907.x. [DOI] [PubMed] [Google Scholar]
- 36.Moch C, Schrogel O, Allmansberger R. Transcription of the nfrA-ywcH operon from Bacillus subtilis is specifically induced in response to heat. J Bacteriol. 2000;182:4384–4393. doi: 10.1128/jb.182.16.4384-4393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moszer I, Glaser P, Danchin A. SubtiList: a relational database for the Bacillus subtilis genome. Microbiology. 1995;141:261–268. doi: 10.1099/13500872-141-2-261. [DOI] [PubMed] [Google Scholar]
- 38.Noone D, Howell A, Collery R, Devine K M. YkdA and YvtA, HtrA-like serine proteases in Bacillus subtilis, engage in negative autoregulation and reciprocal cross-regulation of ykdA and yvtA gene expression. J Bacteriol. 2001;183:654–663. doi: 10.1128/JB.183.2.654-663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noone D, Howell A, Devine K M. Expression of ykdA, encoding a Bacillus subtilis homologue of HtrA, is heat shock inducible and negatively autoregulated. J Bacteriol. 2000;182:1592–1599. doi: 10.1128/jb.182.6.1592-1599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oussenko I A, Bechhofer D H. The yvaJ gene of Bacillus subtilis encodes a 3′-to-5′ exoribonuclease and is not essential in a strain lacking polynucleotide phosphorylase. J Bacteriol. 2000;182:2639–2642. doi: 10.1128/jb.182.9.2639-2642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Page M J, Amess B, Rohlff C, Stubberfield C, Parekh R. Proteomics: a major new technology for the drug discovery process. Drug Discov Today. 1999;4:55–62. doi: 10.1016/s1359-6446(98)01291-4. [DOI] [PubMed] [Google Scholar]
- 42.Petersohn A, Antelmann H, Gerth U, Hecker M. Identification and transcriptional analysis of new members of the sigmaB regulon in Bacillus subtilis. Microbiology. 1999;145:869–880. doi: 10.1099/13500872-145-4-869. [DOI] [PubMed] [Google Scholar]
- 43.Petersohn A, Bernhardt J, Gerth U, Hoper D, Koburger T, Volker U, Hecker M. Identification of sigma(B)-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J Bacteriol. 1999;181:5718–5724. doi: 10.1128/jb.181.18.5718-5724.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price C W. Protective function and regulation of the general stress response in Bacillus subtilis and related gram-positive bacteria. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 179–197. [Google Scholar]
- 45.Price C W, Fawcett P, Ceremonine H, Su Y, Murphy C K, Youngman P. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol Microbiol. 2001;41:757–774. doi: 10.1046/j.1365-2958.2001.02534.x. [DOI] [PubMed] [Google Scholar]
- 46.Riethdorf S, Volker U, Gerth U, Winkler A, Engelmann S, Hecker M. Cloning, nucleotide sequence, and expression of the Bacillus subtilis lon gene. J Bacteriol. 1994;176:6518–6527. doi: 10.1128/jb.176.21.6518-6527.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosamond J, Allsop A. Harnessing the power of the genome in the search for new antibiotics. Science. 2000;287:1973–1976. doi: 10.1126/science.287.5460.1973. [DOI] [PubMed] [Google Scholar]
- 48.Scharf C, Riethdorf S, Ernst H, Engelmann S, Volker U, Hecker M. Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J Bacteriol. 1998;180:1869–1877. doi: 10.1128/jb.180.7.1869-1877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt A, Schiesswohl M, Volker U, Hecker M, Schumann W. Cloning, sequencing, mapping, and transcriptional analysis of the groESL operon from Bacillus subtilis. J Bacteriol. 1992;174:3993–3999. doi: 10.1128/jb.174.12.3993-3999.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schulz A, Schumann W. hrcA, the first gene of the Bacillus subtilis dnaK operon, encodes a negative regulator of class I heat shock genes. J Bacteriol. 1996;178:1088–1093. doi: 10.1128/jb.178.4.1088-1093.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schulz A, Schwab S, Homuth G, Versteeg S, Schumann W. The htpG gene of Bacillus subtilis belongs to class III heat shock genes and is under negative control. J Bacteriol. 1997;179:3103–3109. doi: 10.1128/jb.179.10.3103-3109.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sekowska A, Robin S, Daudin J J, Henaut A, Danchin A. Extracting biological information from DNA arrays: an unexpected link between arginine and methionine metabolism in Bacillus subtilis. Genome Biol. 2001;2:0019.0011–0019.0012. doi: 10.1186/gb-2001-2-6-research0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith M C, Czaplewski L, North A K, Baumberg S, Stockley P G. Sequences required for regulation of arginine biosynthesis promoters are conserved between Bacillus subtilis and Escherichia coli. Mol Microbiol. 1989;3:23–28. doi: 10.1111/j.1365-2958.1989.tb00099.x. [DOI] [PubMed] [Google Scholar]
- 54.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.VanBogelen R A, Abshire K Z, Moldover B, Olson E R, Neidhardt F C. Escherichia coli proteome analysis using the gene-protein database. Electrophoresis. 1997;18:1243–1251. doi: 10.1002/elps.1150180805. [DOI] [PubMed] [Google Scholar]
- 56.VanBogelen R A, Greis K D, Blumenthal R M, Tani T H, Matthews R G. Mapping regulatory networks in microbial cells. Trends Microbiol. 1999;7:320–328. doi: 10.1016/s0966-842x(99)01540-1. [DOI] [PubMed] [Google Scholar]
- 57.VanBogelen R A, Neidhardt F C. Ribosomes as sensors of heat and cold shock in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:5589–5593. doi: 10.1073/pnas.87.15.5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.VanBogelen R A, Schiller E E, Thomas J D, Neidhardt F C. Diagnosis of cellular states of microbial organisms using proteomics. Electrophoresis. 1999;20:2149–2159. doi: 10.1002/(SICI)1522-2683(19990801)20:11<2149::AID-ELPS2149>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 59.Voelker U, Voelker A, Maul B, Hecker M, Dufour A, Haldenwang W G. Separate mechanisms activate sigma B of Bacillus subtilis in response to environmental and metabolic stresses. J Bacteriol. 1995;177:3771–3780. doi: 10.1128/jb.177.13.3771-3780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Volker U, Engelmann S, Maul B, Riethdorf S, Volker A, Schmid R, Mach H, Hecker M. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology. 1994;140:741–752. doi: 10.1099/00221287-140-4-741. [DOI] [PubMed] [Google Scholar]
- 61.Weber A, Menzlaff E, Arbinger B, Gutensohn M, Eckerskorn C, Fluegge U-I. The 2-oxoglutarate/malate translocator of chloroplast envelope membranes: molecular cloning of a transporter containing a 12-helix motif and expression of the functional protein in yeast cells. Biochemistry. 1995;34:2621–2627. doi: 10.1021/bi00008a028. [DOI] [PubMed] [Google Scholar]
- 62.Wei Y, Guffanti A A, Ito M, Krulwich T A. Bacillus subtilis YqkI is a novel malic/Na+-lactate antiporter that enhances growth on malate at low protonmotive force. J Biol Chem. 2000;275:30287–30292. doi: 10.1074/jbc.M001112200. [DOI] [PubMed] [Google Scholar]
- 63.Wei Y, Lee J M, Richmond C, Blattner F R, Rafalski J A, LaRossa R A. High-density microarray-mediated gene expression profiling of Escherichia coli. J Bacteriol. 2001;183:545–556. doi: 10.1128/JB.183.2.545-556.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson M, DeRisi J, Kristensen H H, Imboden P, Rane S, Brown P O, Schoolnik G K. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci USA. 1999;96:12833–12838. doi: 10.1073/pnas.96.22.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wise A A, Price C W. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor sigma B in response to environmental signals. J Bacteriol. 1995;177:123–133. doi: 10.1128/jb.177.1.123-133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zimmer D P, Soupene E, Lee H L, Wendisch V F, Khodursky A B, Peter B J, Bender R A, Kustu S. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc Natl Acad Sci USA. 2000;97:14674–14679. doi: 10.1073/pnas.97.26.14674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zuber U, Schumann W. CIRCE, a novel heat shock element involved in regulation of heat shock operon dnaK of Bacillus subtilis. J Bacteriol. 1994;176:1359–1363. doi: 10.1128/jb.176.5.1359-1363.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]