Abstract
The availability of the complete sequence of the Bacillus subtilis chromosome (F. Kunst et al., Nature 390:249–256, 1997) makes possible the construction of genome-wide DNA arrays and the study of this organism on a global scale. Because we have a long-standing interest in the effects of scoC on late-stage developmental phenomena as they relate to aprE expression, we studied the genome-wide effects of a scoC null mutant with the goal of furthering the understanding of the role of scoC in growth and developmental processes. In the present work we compared the expression patterns of isogenic B. subtilis strains, one of which carries a null mutation in the scoC locus (scoC4). The results obtained indicate that scoC regulates, either directly or indirectly, the expression of at least 560 genes in the B. subtilis genome. ScoC appeared to repress as well as activate gene expression. Changes in expression were observed in genes encoding transport and binding proteins, those involved in amino acid, carbohydrate, and nucleotide and/or nucleoside metabolism, and those associated with motility, sporulation, and adaptation to atypical conditions. Changes in gene expression were also observed for transcriptional regulators, along with sigma factors, regulatory phosphatases and kinases, and members of sensor regulator systems. In this report, we discuss some of the phenotypes associated with the scoC mutant in light of the transcriptome changes observed.
The transition state of Bacillus subtilis, which carries the cells from vegetative growth to stationary phase, is a stage in which the cell population is highly differentiated. In this stage, which is governed by the phosphorylation status of Spo0A, the cells can carry out a number of highly specialized functions (13). These functions include not only those controlling the initiation of the spore-forming process but also those responsible for competence and motility as well as the production of scavenging enzymes (e.g., AprE and NprE) and a host of secondary metabolites (33). By a not-yet-completely-elucidated mechanism, the fate of the individual cells within the population is determined by synthesis, secretion, processing, and uptake of signaling peptides used for cell-to-cell communication (29, 34). In addition, a number of other genes called transition state regulators play an important role in this differentiation process (28, 37). Hyperexpression and/or loss of function of any of these genes profoundly affects several aspects of the cell physiology.
One gene that belongs to this group of transition state regulators is scoC. Mutations in scoC have been isolated independently, by using different screening criteria, in at least three laboratories and are known as hpr (12), catA (14), and scoC (22). We suggest the adoption of the designation scoC for this gene and all its alleles to avoid confusion with the HPr system involved in sugar transport.
scoC (as the hpr allele) was first noted and defined by the study of B. subtilis mutants overproducing alkaline and neutral proteases as an approach to isolate sporulation-associated mutations. Other genetic lesions, catA (14) and scoC (22), which lead to a glucose-insensitive sporulation phenotype and exoprotease overproduction, were also mapped in the same chromosomal region. Later DNA sequencing showed that all of these mutants resulted from mutations in the same gene (27).
Northern blots to quantify subtilisin-specific mRNA suggested that the activity of the scoC gene was exerted at the level of transcription of the aprE promoter (8). Sequencing of the scoC locus and of its alleles showed that the observed phenotypes were due to a loss-of-function mutation of ScoC (27), indicating that ScoC acts as a negative regulator of transcription of aprE. This study also showed that overproduction of the gene product reduced sporulation by 3 to 4 orders of magnitude. Transcription of the scoC locus became constitutive by null mutations in the spo0A gene, suggesting that Spo0A is a negative regulator of scoC transcription. Purification of the ScoC protein demonstrated that it is a DNA binding protein. A consensus binding sequence, RATANTATY, was shown by footprint analysis to lie upstream of the nprE, aprE, and sinI genes (16).
It has been shown recently that scoC plays a direct role in the initiation of sporulation by acting as a repressor of the two major signaling peptide transport systems, opp and app (17). However, inactivation of the rapA gene, which rescues the sporulation defect of an opp mutant, does not rescue the sporulation defect caused by the presence of scoC in a multicopy plasmid (17). As pointed out by Koide et al. (17), this suggests the existence of other regulatory mechanisms by which the scoC gene controls sporulation. Furthermore, while scoC null mutations allow sporulation in the presence of glucose, they do not allow sporulation if both glucose and glutamine are present in the medium (33). In addition, the observation that scoC mutants affect alkaline phosphatase expression (4), motility (16), oxidative stress response (5), and competence (S. Causey and E. Ferrari, unpublished data) suggests that the ScoC gene product plays a major role in the physiology of B. subtilis. We have attempted to understand the effects of the scoC mutation in B. subtilis by comparing the gene expression patterns of wild-type cells and those of the scoC mutant using DNA microarrays.
MATERIALS AND METHODS
Strains and growth conditions.
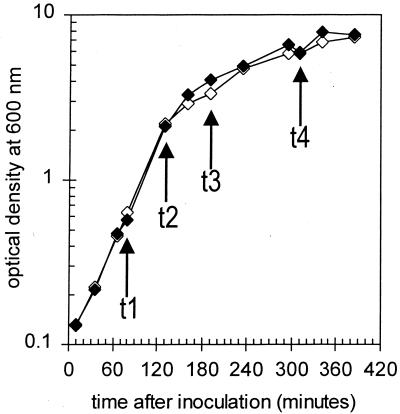
The strains used in this work are BG2815 scoC4 ΔnprE522 and the isogenic strain BG2822 ΔnprE522. The presence of the scoC4 mutation, a stop codon at position 32 (accession no. M20237), in strain BG2815 was verified by PCR and sequencing. The ΔnprE522 mutation has been described previously (41). Fresh cells were streaked onto Luria-Bertani medium–1.6% skim milk plates for overnight growth at 37°C. For each strain, single colonies were then inoculated into 10 ml of freshly prepared 2× SNB medium in a 125-ml flask. This medium contained the following (per liter): 16 g of Difco nutrient broth, 50 ml of 10% maltrin M150, and 40 ml of 25× SNB salts (25× salts contain [per liter] 3.7 g of CaCl2 · 2H2O, 9.6 mg of FeSO4 · 7H2O, 6 mg of MnCl2 · 4H2O, 25.0 g of KCl, and 3.26 g of MgSO4 · 7H2O). From this flask, two serial 1:3,000 dilutions were made, each into 10 ml of the same medium, for slow overnight growth (30°C at 150 rpm). The following day, the starter culture closest to logarithmic growth was used to inoculate the prewarmed (37°C) 75-ml primary culture in a 500-ml flask to an optical density (600 nm) of ∼0.015. From the primary culture, 25 ml was transferred to a separate 250-ml flask for growth measurements. Both primary and measurement flasks were shaken together at 37°C at 250 rpm. Figure 1 shows the growth curves for the wild-type and scoC4 cultures.
FIG. 1.
Growth curves for Bacillus strains BG2822 (wild type) (open diamonds) and BG2815 (scoC4) (filled diamonds). t1, t2, t3, and t4, time points used for RNA isolation and array analysis (log phase, early transition, late transition, and stationary phase, respectively).
Harvesting.
Cells were collected by pipetting onto a Millipore 0.8-μm mixed cellulose ester filter (catalog no. AAWPO4700) using a Millipore vacuum filtration apparatus (catalog no. 1004700). After filtration, the membrane circle with collected cells was quickly transferred to liquid nitrogen in a heavy-walled 200-ml beaker, allowing no more than 10 to 15 s to pass between pipetting and freezing. In the presence of liquid nitrogen, the filters were transferred to −80°C storage until used.
Each filter with a cellular sample was ground up for 3 min in a standard hand-held coffee grinder with 45 ml of finely crushed dry ice and 10 g of glass beads (∼110-μm Glasperlen; B. Braun). The powdered contents were then transferred via funnel to a 100-ml heavy-walled glass bottle (VWR Scientific Product Corporation). All equipment involved was prechilled with dry ice before use and maintained at dry-ice temperature throughout. The loosely capped bottle was then stored at −80°C overnight to allow the dry ice to sublime, leaving frozen cell powder and glass beads.
Preparation of RNA.
The following protocol was adapted from the work of Farrell (6). The frozen sample was rapidly transferred to a 50-ml Oakridge centrifuge tube with 5 ml of extraction buffer at room temperature (4 M guanidinium thiocyanate, 25 mM sodium citrate [pH 7], 0.5% sarcosyl, 100 mM β-mercaptoethanol). The sample was vortexed immediately for 45 s to ensure even mixing. One-tenth volume of 3 M sodium acetate (pH 5.5) was added and vortexed briefly. After centrifugation at 12,000 × g for 10 min at 4°C, the supernatant was transferred to a fresh ice-chilled tube. All remaining extraction steps were carried out on ice. An equal volume of 25:24:1 water-saturated phenol-chloroform-isoamyl alcohol was added, mixed for 1 min, and incubated for 5 min. The mixture was recentrifuged under the same conditions, and the aqueous phase was transferred to a new tube. The nucleic acid was precipitated in 0.75 volume of chilled isopropanol at −20°C for at least 1 h and then centrifuged at 20,000 × g for 20 min at 4°C. The isopropanol was removed completely, and the pellet was dissolved in 700 μl of extraction buffer. The sodium acetate addition, organic solvent extraction, and isopropanol precipitation were repeated. The second pellet was washed three times with 500 μl of ice-cold 70% ethanol (each for 10 min), followed by a final brief rinse with ice-cold 95% ethanol. The pellet was air dried for not more than 20 min and resuspended in 50 μl of diethyl pyrocarbonate (DEPC)-treated water with 1 U of RNase inhibitor (BM/Roche) per μl. The yield was determined by UV spectroscopy, and the crude RNA sample was stored at −80°C.
An adequate amount of crude RNA (∼250 μg) contaminated with genomic DNA was added to a 500-μl DNase I reaction mixture (40 mM Tris-HCl [pH 7.6], 6 mM MgCl2, 2 mM CaCl2, 1 U of RNase inhibitor per μl, 150 U of DNase I per 500 μl [BM/Roche]) and incubated for 30 min at 37°C. One-tenth volume of 3 M sodium acetate (pH 5.5) was added and mixed well, followed by organic solvent extraction, isopropanol precipitation, and ethanol washes as described above for the crude RNA preparation. After the pellet had been resuspended in DEPC-treated water with RNase inhibitor, the entire DNase I treatment and its subsequent purifications were repeated to remove all traces of genomic DNA. After final resuspension in 50 μl of DEPC-treated water with 1 U of RNase inhibitor per μl, the yield was determined by UV spectroscopy (with a 260/280 ratio of at least 1.8). RNA quality was also verified by 3% agarose gel electrophoresis (i.e., 23S and 16S rRNA band intensities at a ratio of 1.3:1 to 1.6:1). The purified total Bacillus RNA sample was stored at −80°C.
IVT controls and RT-PCR preparation.
Genes encoding Eryr (Staphylococcus aureus plasmid pE194, accession no. J01755-58), Bleor (S. aureus plasmid pUB110, accession no. M19465), Specr (Enterococcus faecalis, accession no. M69221), and green fluorescent protein (GFP) (plasmid pGFP; Clontech catalog no. 6090-1), start to stop codons inclusive, were PCR amplified from plasmid DNA using primers containing unique restriction sites for subsequent cloning into pBlueScript II KS(+) (Stratagene). The resulting plasmids were linearized at a unique site downstream of the coding sequence, and runoff in vitro transcription (IVT) reaction products were prepared from the upstream T3 promoter of the plasmid using the MEGAscript T3 IVT kit (Ambion catalog no. 1338) following the supplier's recommended protocol. Final transcript lengths were 735 nt (Eryr gene), 396 nt (Bleor gene), 768 nt (Specr gene), and 717 nt (GFP gene). IVT RNA was quantified by optical density at 260 nm (40 μg/ml = 1 optical density unit). A staggered IVT RNA mixture in RNase-free water was prepared: 4 nM Bleor gene, 1 nM GFP gene, 200 pM Specr gene, and 50 pM Eryr gene. In addition, three transcripts (amyE, citC, and sdhA) were assayed both by the microarray assay and by a reverse transcription (RT)-PCR assay to quantitate mRNA independently across two conditions (time or genotype). RT-PCR was performed with a Roche Molecular Biochemicals light cycler instrument (software version 3 and RNA amplification kit SYBR green I) according to the manufacturer's instructions. Primer pairs were chosen which gave a good dose response on dilutions of genomic DNA, of IVT standards, and of total RNA preparations used in chip assays. For RT-PCR, primer sequences were as follows (F denotes forward or sense strand; R denotes reverse or antisense strand): 280-bp amyE product, 5′-TGAAACGGTTCTTAGACAGG-3′ (F) and 5′-TGGGAGATATTCGACACG-3′ (R); 227-bp citC product, 5′-ATAAAACAGGTGAGYGGCTC-3′ (F) and 5′-ACGGAAGATGACCATATCAG-3′ (R); 243-bp sdhA product, 5′-CGATCATCCACTTATTAGACC-3′ (F) and 5′-GGCAGGTTCTGTCATCATC-3′ (R). Products were verified by melting curve analysis on the light cycler, by size determination on agarose gels, and by DNA sequencing.
Target preparation
RNA harvested from a given Bacillus strain and at a given time point was reverse transcribed into biotin-labeled cDNA by the method of de Saizieu et al. (3). Total RNA (25 μg) and 5.5 μl of the staggered control IVT mixture were incubated at 37°C overnight in a 100-μl reaction mixture: 1× GIBCO first-strand buffer (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2); 10 mM dithiothreitol; 40 μM random hexamer; 0.3 mM concentrations (each) of dCTP, dGTP and dTTP; 0.12 mM dATP; 0.3 mM biotin-dATP (NEN catalog no. NEL999); 2,500 U of SuperScript II reverse transcriptase. To remove RNA, the reaction was brought to 0.25 M NaOH and incubated at 65°C for 30 min. The reaction mixture was neutralized with HCl, and the nucleic acid was precipitated at −20°C in ethanol with 2.5 M ammonium acetate. The pellet was washed, air dried, resuspended in water, and quantitated by UV spectroscopy. The reaction yield was approximately 20 to 25 μg of biotin-labeled cDNA.
This cDNA (12 μg) was fragmented in 33 μl of 1× One-Phor-All buffer (Amersham-Pharmacia no. 27-0901-02) with 3.75 mU of DNase I at 37°C for 10 min. After the DNase had been heat killed, fragmentation was validated by running 2 μg of the fragmented cDNA on a 3% agarose gel. Biotin-containing cDNA routinely ranged in size from 25 to 125 nucleotides. The remaining 10 μg of cDNA was hybridized to an Affymetrix (Santa Clara, Calif.) Bacillus GeneChip array.
Array description
Probe sets on the custom B. subtilis expression array were designed from the wild-type (I168) B. subtilis sequence data of Kunst et al. (19) by Affymetrix and Genencor. The total of 4,107 open reading frames (ORF) were represented by the tiling of at least 20 probes pairs per ORF, each pair consisting of one perfectly matching complementary 25-mer and one control 25-mer with a centrally mismatched base. Genes longer than 1,500 bp were represented by additional probe sets, bringing the total number of ORF probe sets to 4,351. Probes sets for 40 tRNA genes were also tiled, as well as probe sets for over 40 control sequences. Affymetrix performed probe selection and array fabrication by published and proprietary methods (20, 40). The entire array of 454-by-454 25-μm features is bordered and interspersed by a standard oligonucleotide feature for the purposes of grid alignment and data analysis.
Hybridization, scanning, and data collection.
Hybridizations were performed as described in the Affymetrix expression analysis technical manual (Affymetrix) using reagent suppliers as suggested. Fragmented biotin-labeled cDNA (10 μg) was added to a 220-μl hybridization cocktail containing 100 mM MES [(N-morpholino)ethanesulfonic acid], 1 M Na+, 20 mM EDTA, 0.01% Tween 20, 5 mg of total yeast RNA per ml, 0.5 mg of bovine serum albumin per ml, 0.1 mg of herring sperm DNA per ml, and 50 pM control oligonucleotide (AFFX-B1). The cocktails were heated to 95°C for 5 min, cooled to 40°C for 5 min, and briefly microcentrifuged to remove particulates, and 200 μl was injected into each prewarmed prerinsed (1× MES buffer plus 5 mg of yeast RNA per ml) cartridge. The arrays were rotated at 40°C overnight.
The samples were removed, and the arrays were filled with nonstringent wash buffer consisting of 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) and 0.01% Tween 20 and washed on the Affymetrix fluidics station with protocol Euk-GE-WS2, using nonstringent and stringent (0.1 M MES, 0.1 M Na+, 0.01% Tween 20) wash buffers. Arrays were stained in three steps: streptavidin, antistreptavidin antibody tagged with biotin, and streptavidin-phycoerythrin conjugate.
Signal on the arrays were detected with a Hewlett Packard gene array scanner using 570-nm laser light with 3-μm pixel resolution. The signal intensities (also known as average differences) of the 4,351 ORF probe sets were scaled and normalized to a target value of 1,000 as described in the Microarray Suite 4.0 user guide (Affymetrix). The absolute analysis values for average difference and absolute call were collected for all scans; the values for difference call and fold change were also collected for all comparative analysis scans (scoC versus wild-type cells at matched time points).
Data analysis.
A replicate RNA sample was prepared for a given strain and time point. These two samples (R and R′) were prepared as identically as possible, followed by signal normalization in GeneChip 4.0. For each of the 4,351 ORF probe sets, we prepared a table of the average of the logs of the two replicate signals (ALS = 0.5 log R + 0.5 log R′) and the log ratio (LR = log R/R′). For this calculation, only the probe sets with two positive values were considered. The resulting table was sorted by the ALS. Then, in a series of sliding windows with a size of 201 ALS values, from one end of the table to the other, the mean ALS and the standard deviation for the corresponding LR (SDLR) values were determined. These values were saved as a look-up table listing SDLR in identical samples (i.e., scatter due to chance) as a function of ALS: in general, SDLR remained fairly constant when the ALS was >300 but increased as ALS decreased below 300. The plot of these look-up values is very consistent with a fitted equation based on a two-component error model for microarray-based data developed by Silicon Genetics, Redwood City, Calif. (B. Eynon and A. Conway, personal communication).
There were four experimental time points for both the scoC and the wild-type shake flask cultures (80, 130, 190, and 310 min after inoculation [t1 through t4, respectively]). For each time point, the scoC array was treated as the experimental array and the wild-type array was treated as the baseline array: GeneChip 4.0 software (Affymetrix) calculated fold change and difference change values in these comparative analyses (GeneChip software user guide). A gene was considered significantly changed between scoC and wild-type cells if for at least one of the four time points three conditions were met: (i) the difference change value was not “no change”; (ii) the absolute call for the gene in the strain with the higher expression level was listed as “present”; (iii) the fold change calculated by GeneChip 4.0 software was at least 4.37 times higher than SDLR for the ALS of the gene on scoC and wild type. A Z score of 4.37 corresponds to a Bonferroni-corrected confidence level of one in 80,000, or 0.05 divided by 4,000 genes. For large genes with multiple probe sets, at least one of the probe sets needed to be significant by the above criteria.
RESULTS
Quantitating transcriptional differences.
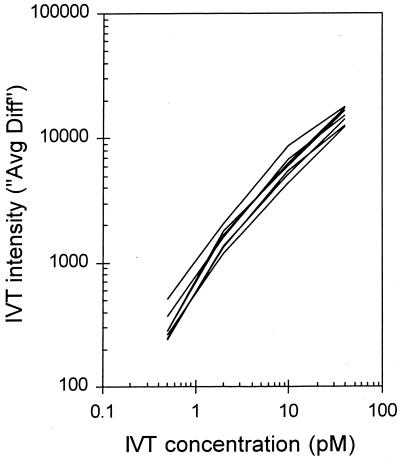
To determine the sensitivity and reproducibility of the microarray assay, several controls were performed and other indicators were monitored throughout the study. All RNA samples were spiked with a mixture of four in vitro transcripts at various concentrations to monitor the sensitivity and consistency of signal varying as a function of target concentration. Figure 2 shows the linearity of the IVT detection over several orders of magnitude in a complex mixture of total Bacillus RNA. To determine if the changes in expression assayed by the array were similar to those in other established methods, total RNA from two samples varying in growth phase or in genotype was quantitated by both RT-PCR and array hybridization (Table 1). For transcripts increasing, decreasing, and remaining unchanged under the conditions compared, the two assay methods were in close agreement.
FIG. 2.
Spiked control IVT signal as a function of concentration. Final concentrations of the four control in vitro transcripts in the 200-μl hybridization cocktail (assuming an equimolar reverse transcription from RNA template to biotin-labeled cDNA): 40 pM Bleor gene, 10 pM GFP gene, 2 pM Specr gene, and 0.5 pM Eryr gene. Plotted lines are for each of the eight microarray samples (two strains; four time points).
TABLE 1.
Changes in gene expression as determined independently by microarray and RT-PCR assaysa
| Target | Comparison | Change in expression (fold) inb:
|
|
|---|---|---|---|
| Chip assay | RT-PCR | ||
| amyE | scoC cells, t2 vs t3 | 4.6 ↑ | 4.8–5.0 ↑ |
| citC | scoC cells, t3 vs t4 | 7.2 ↓ | 5.3–7.1 ↓ |
| sdhA | t2, scoC vs wild-type cells | 1.0 | 0.99–1.0 |
The expressions of amyE, citC, and sdhA were quantitated independently by microarray and RT-PCR assays as described in the text.
↑ and ↓, gene expression increase and decrease between the compared conditions.
To determine array reproducibility, the intensities of all gene transcripts assayed across two samples, prepared as identically as possible, were compared. A Pearson correlation coefficient of 0.953 between replicates indicates good agreement. This value is slightly lower than that calculated for nylon membrane arrays reported recently (7). This is likely due to the higher signal values acquired from detection of radiolabeled targets on membranes relative to fluorescence-detected signals on glass microarrays used in this study (R. Caldwell, unpublished observation).
The percentage of ORF probe sets receiving an absolute call of “present” ranged from 67 to 74% on each array, with slightly more genes showing presence in the stationary than log phase. The percentage of ORF probe sets showing presence for at least one of the four time points in either scoC or the wild type is larger (>85%), indicating that a vast majority of transcripts are detectable at some point during growth by the GeneChip assay, though generally not at all time points.
General overview.
Across the four time points at which scoC cells were compared to wild-type cells, mRNAs transcribed from 560 genes (representing 14% of the Bacillus genome) met the significant-change criteria defined in Materials and Methods. These 560 most stringently selected genes fall into nearly all functional-class categories (Table 2), from genes coding for enzyme catalysis to those coding for proteins with a regulatory role.
TABLE 2.
Functional categories of 560 genes whose expression is significantly affected by the scoC4 mutation
| Classification | No. of genes
|
|
|---|---|---|
| Total | Affected | |
| 1. Cell envelope and cellular processes | ||
| 1.1. Cell wall | 108 | 12 |
| 1.2. Transport/binding proteins and lipoproteins | 388 | 76 |
| 1.3. Sensors (signal transduction) | 38 | 1 |
| 1.4. Membrane bioenergetics (electron transport and ATP synthesis) | 74 | 6 |
| 1.5. Mobility and chemotaxis | 55 | 29 |
| 1.6. Protein secretion | 20 | 2 |
| 1.7. Cell division | 21 | |
| 1.8. Sporulation | 152 | 37 |
| 1.9. Germination | 20 | 1 |
| 1.10. Transformation and competence | 21 | |
| 2. Intermediary metabolism | ||
| 2.1. Metabolism of carbohydrates and related molecules | ||
| 2.1.1. Specific pathways | 215 | 38 |
| 2.1.2. Main glycolytic pathways | 27 | 3 |
| 2.1.3. Tricarboxylic acid cycle | 18 | 3 |
| 2.2. Metabolism of amino acids and related molecules | 199 | 47 |
| 2.3. Metabolism of nucleotides and nucleic acids | 83 | 27 |
| 2.4. Metabolism of lipids | 82 | 9 |
| 2.5. Metabolism of coenzymes and prosthetic groups | 104 | 1 |
| 2.6. Metabolism of phosphate | 10 | 2 |
| 2.7. Metabolism of sulfur | 9 | |
| 3. Information pathways | ||
| 3.1. DNA replication | 24 | 3 |
| 3.2. DNA restriction/modification and repair | 39 | 1 |
| 3.3. DNA recombination | 19 | |
| 3.4. DNA packaging and segregation | 9 | |
| 3.5. RNA synthesis | ||
| 3.5.1. Initiation | 20 | 2 |
| 3.5.2. Regulation | 212 | 11 |
| 3.5.3. Elongation | 7 | 1 |
| 3.5.4. Termination | 4 | |
| 3.6. RNA modification | 21 | |
| 3.7. Protein synthesis | ||
| 3.7.1. Ribosomal proteins | 55 | 35 |
| 3.7.2. Aminoacyl-tRNA synthetases | 28 | 1 |
| 3.7.3. Initiation | 6 | 1 |
| 3.7.4. Elongation | 6 | 1 |
| 3.7.5. Termination | 3 | 1 |
| 3.8. Protein modification | 27 | 5 |
| 3.9. Protein folding | 8 | |
| 4. Other functions. | ||
| 4.1. Adaptation to atypical conditions | 71 | 15 |
| 4.2. Detoxification | 70 | 4 |
| 4.3. Antibiotic production | 28 | 3 |
| 4.4. Phage-related functions | 87 | 3 |
| 4.5. Transposon and IS | 10 | 2 |
| 4.6. Miscellaneous | 25 | |
| 5. Similar to unknown proteins | ||
| 5.1. From B. subtilis | 178 | 24 |
| 5.2. From other organisms | 477 | 36 |
| 6. No similarity | 1,049 | 106 |
For genes that have been assigned known or presumed functions, the largest group with altered transcriptional levels in the scoC mutant is a group of 76 genes coding for transport proteins, binding proteins, and lipoproteins, all of which are associated with the cell membrane. Forty-seven affected genes belong to the group involved in amino acid (and related molecule) metabolism, notably arginine, histidine, leucine, isoleucine, valine, and threonine biosynthetic genes. As one might expect, we identified changes in expression levels for several genes associated with sporulation, belonging to the SubtiList functional class 1.8 (25, 26) (http://genolist.pasteur.fr/SubtiList/). These changes in sporulation genes are summarized in Table 3. Other large functional groups affected by scoC include 29 genes associated with motility, 38 associated with carbohydrate metabolism (notably myoinositol and acetoin metabolism), 27 associated with metabolism of nucleotides and nucleosides (purine and pyrimidine biosynthetic genes), and 19 associated with adaptation to atypical conditions and detoxification. Thirty-five ribosomal proteins also showed a change in gene expression. The relevance of 35 out of 55 ribosomal proteins being transcribed at a higher level in the scoC mutant at time point t3 is not immediately obvious. There are 166 genes with unknown function or having no similarity to any protein in existing databases.
TABLE 3.
Sporulation genes affected by the scoC4 mutation
| Gene | Change (fold)a | Time pointb | Description |
|---|---|---|---|
| bofA | 4.4 | t4 | Inhibition of the pro-sigma-K processing machinery |
| cotE | 3.7 | t4 | Spore coat protein (outer) |
| cotJA | 4.6 | t4 | Polypeptide composition of the spore coat |
| cotJB | 4.1 | t4 | Polypeptide composition of the spore coat |
| cotJC | 4.4 | t4 | Polypeptide composition of the spore coat |
| cotY | 7.8 | t4 | Spore coat protein (insoluble fraction) |
| dacF | 6.1 | t4 | Penicillin-binding protein (putative Ala-Ala carboxypeptidase) |
| phrA | −3.0 | t2 | Phosphatase (RapA) inhibitor |
| rapA | −3.5 | t1 | Response regulator aspartate phosphatase |
| sinI | 2.3 | t2 | Antagonist of SinR |
| spmA | 4.8 | t4 | Spore maturation protein (spore core dehydration) |
| spmB | 4.7 | t4 | Spore maturation protein (spore core dehydration) |
| spoIID | 3.9 | t4 | Required for complete dissolution of the asymmetric septum |
| spoIIIAA | 5.7 | t4 | Mutants block sporulation after engulfment |
| spoIIIAB | 3.6 | t4 | Mutants block sporulation after engulfment |
| spoIIIAC | 3.3 | t4 | Mutants block sporulation after engulfment |
| spoIIIAD | 3.9 | t4 | Mutants block sporulation after engulfment |
| spoIIIAF | 3.5 | t4 | Mutants block sporulation after engulfment |
| spoIIIAG | 3.1 | t4 | Mutants block sporulation after engulfment |
| spoIIIAH | 3.3 | t4 | Mutants block sporulation after engulfment |
| spoIIP | 4.7 | t4 | Required for dissolution of the septal cell wall |
| spoIIQ | 2.9 | t4 | Required for completion of engulfment |
| spoIVA | 4.1 | t4 | Required for proper spore cortex formation and coat assembly |
| spoIVFA | 3.8 | t4 | Inhibition of spoIVFB |
| spoIVFB | 4.3 | t4 | Required for the processing of pro-sigma-K to active sigma-K |
| spoVB | 4.9 | t4 | Involved in spore cortex synthesis |
| spoVID | 6.1 | t4 | Required for assembly of the spore coat |
| spoVK | 4.8 | t4 | Disruption leads to the production of immature spores |
| spoVM | 4.5 | t4 | Required for normal spore cortex and coat synthesis |
| spoVR | 8.4 | t4 | Involved in spore cortex synthesis |
| sspA | 3.5 | t4 | Small acid-soluble spore protein (major alpha-type SASP) |
| sspC | 6.7 | t4 | Small acid-soluble spore protein (minor alpha/beta-type SASP) |
| sspE | 2.9 | t4 | Small acid-soluble spore protein (major gamma-type SASP) |
| tlp | 7.3 | t4 | Small acid-soluble spore protein (thioredoxin-like protein) |
| usd | 8.3 | t4 | Required for translation of spoIIID |
| yknT | 7.1 | t4 | Unknown; sporulation protein sigma-E- controlled |
| ykvU | 5.4 | t4 | Unknown; similar to spore cortex membrane protein |
| yrbA | 3.8 | t4 | Unknown; similar to spore coat protein |
A value greater than 0 indicates higher scoC mutant expression relative to wild type.
See Fig. 1.
Biochemical and genetic data are available for a number of genes for which scoC-specific expression changes are known, as determined by earlier studies. Table 4 shows that there is a substantial agreement between our observations and previously reported findings.
TABLE 4.
Correlation of findings in this study with findings in relevant literature
| Gene monitored | Change in scoC-specific expressiona
|
|
|---|---|---|
| This study | Literature (reference) | |
| hag | ↓ | Slight ↓b (23) |
| oppA | ∼1.5× ↑ | 2× ↑b (17) |
| oppBCDF | 2× ↑ | |
| appA | 2× ↑ | 2× ↑b (17) |
| appBCDF | 2× ↑ | |
| nprE and aprE | 2–3× ↑c (4) | |
| nprE | 2× ↑ | |
| aprE | 3× ↑ | |
| Alkaline phosphatased | ↑e (15) | |
| phoB | 5–10× ↑ | |
| sinI | 1.5× ↑ | 5× ↑ (33) |
Relative to the wild type. ↑ and ↓, increase and decrease.
Measured as a LacZ fusion.
Enzyme assay.
Alkaline phosphatase gene not known.
Qualitative assay, probably not a secreted enzyme (see Discussion).
Regulatory genes affected by ScoC.
The functional category of regulatory proteins shows a relatively large number of scoC-affected genes. Tables 5 and 6 list these genes and their changes in the scoC4 mutant relative to the wild type, as well as the genes or operons known or conjectured to be affected directly. This group includes not only gene products classified as transcriptional regulators (SubtiList functional class 3.5.2) but also sigma factors (as well as anti- and anti-anti-sigma factors), regulatory phosphatases and kinases, members of two-component sensor/regulator systems, and members of other categories (see “Sporulation and catabolite repression” below). The DNA sequence from −10 to −500 upstream of these regulatory genes was examined with the GeneSpring software package (Silicon Genetics) for the presence of potential ScoC binding sites (16). Tables 5 and 6 show the results of this search, in which many of the listed regulators exhibit potential ScoC sites. It is necessary to note that the sites identified in these tables were found by using a consensus ScoC binding site sequence which, because of its AT-rich nature and three ambiguous bases, is likely to appear in intergenic regions upstream of many genes and operons. Indeed, in more than 60% of these intergenic regions, we can identify potential ScoC binding site sequences. At this point, we do not have experimental evidence for ScoC binding in any of these regions.
TABLE 5.
Genes encoding transcriptional regulatory proteins (SubtiList functional class 3.5.2) whose expression is significantly changed in the scoC4 strain
| Regulator | Change (fold) | Time point | Description | Gene(s) controlled | Potential ScoC site(s)a | Position |
|---|---|---|---|---|---|---|
| comKb | −2.5 | t3 | Competence transcription factor | Competence and sigD- controlled genes | GATTTTATC | −96 |
| AATATCATT | −112 | |||||
| ykoM | −3.7 | t4 | Unknown; MarR family | ? | AAAATTATC | −155 |
| ykvE | −2.8 | t4 | Unknown; MarR family | ? | AAGAGTATC | −360 |
| pyrR | −3.6 | t1 | Transcriptional attenuation of the pyrimidine operon | pyrPBCADFE | ||
| cheY | −3.8 | t4 | Two-component response regulator | Flagellar, chemotaxis genes | AGTATTATC | −184 |
| cheB | −4.6 | t4 | MCP-glutamate methyltransferase | Chemotaxis genes | GAAATTATC | −35 |
| ytzE | −2.7 | t3 | Unknown; DeoR family | ? | AATAATATT | −232 |
| splA | 4.7 | t2 | Transcriptional regulator of the spore photoproduct lyase operon | splAB | AATCTTATC | −151 |
| rbsR | −3.3 | t4 | Transcriptional repressor of the ribose operon, LacI family | rbsKDACB | GATATTTTT | −78 |
| GATAGTCTT | −257 | |||||
| AATTTTATC | −415 | |||||
| AATAATTTT | −418 | |||||
| ywrC | −3.1 | t1 | Unknown; Lrp/AsnC family | ? | ATTATTATT | −69 |
| AAAATTATT | −72 | |||||
| GATATTTTC | −104 | |||||
| spoIIID | 5.6 | t4 | Transcriptional regulator of sigma-E- and sigma-K-dependent genes | Sporulation genes | ||
| hutP | −4.0 | t2 | Transcriptional activator of the histidine utilization operon | hutPHUIGM | ||
| yesNb | −2.6 | t4 | Unknown; similar to two-component response regulator | ? | ||
| cggR | −3.4 | t3 | Transcriptional repressor of gapA | gapA | AATATTGTC | −159 |
Sequences upstream of affected genes (−10 to −500 bp) were searched for the presence of the canonical ScoC binding site RATANTATY (R = A or G, Y = C or T, N = any base), allowing at most one mismatch, using the Find Regulatory Sequence function of the GeneSpring software (Silicon Genetics).
Both comK and yesN miss the strict 4.37ς significance cutoff (comK Z score = 4.31, yesN Z score = 4.15); however, the gene encoding the cognate histidine kinase of YesN (yesM) does pass the 4.37-ς threshold.
TABLE 6.
Genes encoding regulatory proteins not acting through transcription (nonmembers of SubtiList functional class 3.5.2) whose expression is significantly changed in the scoC4 strain
| Regulator | Change (fold) | Time point | Description | Protein affected | Potential ScoC site(s)a | Position |
|---|---|---|---|---|---|---|
| bofA | 4.4 | t4 | With SpoIVFA, inhibits processing of pro-ςK by spoIVFB | ςK | ||
| spoIVFA | 3.8 | t4 | With BofA, inhibits processing of pro-ςK by spoIVFB | ςK | GATAGAATC | −147 |
| GAAAATATT | −468 | |||||
| spoIVFB | 4.3 | t4 | Processing of pro-ςK | ςK | ||
| rapA | −3.5 | t1 | Response regulator aspartate phosphatase; dephosphorylates Spo0F-P | Spo0F | ||
| phrA | −3.6 | t1 | Inhibitor of the activity of phosphatase RapA | RapA | AAAAGTATC | −282 |
| GATAATCTT | −355 | |||||
| spo0M | 2.3 | t3 | Sporulation control gene; expression controlled by sigma-H | ? | AATATTATT | −125 |
| AATATTGTT | −240 | |||||
| sinI | 2.3 | t2 | Antagonist of SinR, transcriptional regulator of post-exponential-phase response genes | SinR | AATACGATT | −142 |
| AATAATATT | −200 | |||||
| AATACCATC | −289 | |||||
| usd | 8.3 | t4 | Required for translation of spoIIID | ? | ||
| yesM | −3.9 | t4 | Two-component sensor histidine kinase | YesN | ||
| yvyD | −3.5 | t1 | Similar to ς54 modulating factor from gram-negative organisms | ? | ||
| rsbV | −3.8 | t2 | Positive regulator of ςB activity (anti-anti-sigma factor) | ςB | GATATGATT | −271 |
| GATTCTATT | −387 | |||||
| rsbW | −3.5 | t2 | Negative regulator of ςB activity (anti-sigma factor) | ςB |
Sequences upstream of affected genes (−10 to −500 bp) were searched for the presence of the canonical ScoC binding site RATANTATY (R = A or G, Y = C or T, N = any base), allowing at most one mismatch, using the Find Regulatory Sequence function of the GeneSpring software (Silicon Genetics).
For genes in Tables 5 and 6 where the ScoC-controlled genes are known, the changes in the levels of expression for these affected genes are also observed to meet our statistical cutoff. The decrease in the transcription level of the antiterminator-encoding pyrR gene (3.6-fold) is mirrored by the larger (13- to 48-fold) decreases in the pyrimidine biosynthetic operon (pyrPBCADFE) in the scoC mutant. Likewise, the 4-fold down-regulation of the positive regulatory gene hutP at t2 leads to a concomitant 3- to 38-fold decrease in the histidine utilization genes (hutHUIGM). Interestingly, a 3.3-fold decrease at t4 in the rbsR transcript, originally annotated as encoding a transcriptional repressor of the ribose operon, is accompanied by a counterintuitive 3.8- to 5.1-fold decrease in ribose utilization (rbsKDACB) at the same time point. ScoC also appears to affect rbsR at the earliest time point (t1) in the opposite direction: rbsR transcription is increased 2.3-fold, while rbsKDACB also shows a 2.0- to 2.9-fold increase. This is consistent with the suggestion by Strauch (39) that rbsR may not act as a repressor in B. subtilis, as had been shown to be the case for its Escherichia coli ortholog.
Our data confirm the effect of scoC4 on increasing the expression of SinI, the antagonist of the transcriptional regulator SinR. Footprinting experiments have shown that ScoC binds to sinI upstream regulatory sequences (located at −131 and −200 from the sinI start codon). These binding sites correspond to loci lying 9 and 78 bp upstream from the sinI transcriptional start, as identified by Kallio et al. (16). The ScoC binding consensus at −131 was not found in our search of upstream sequences, as only one mismatch was allowed in the search for the RATANTATY motif. However, by our computer analyses, another additional ScoC-binding consensus was identified at −289.
Sporulation and catabolite repression.
In agreement with the sporulation control phenotype of scoC, several additional sporulation control genes are affected in the scoC4 null mutant (Table 7). These regulatory genes, which exert their effects through nontranscriptional means (e.g., bofA, spoIVFAB, spo0M, and usd), were not previously known to lie under ScoC control. It is important to note that the absence of ScoC lowers the level of rapA phosphatase transcription and that of its inhibitor, phrA, more than threefold compared to levels in the wild-type strain (Table 3).
TABLE 7.
Sporulation genes affected by scoC4 mutation and depending on both Spo0A and ςFa
| Gene | Change (fold)b | Time pointc | Description |
|---|---|---|---|
| phoB | 9.3 | t4 | Alkaline phosphatase III |
| ybaN | 5.8 | t4 | Unknown; similar to polysaccharide deacetylase |
| ymxH | 6.3 | t4 | Unknown; similar to unknown proteins |
| ywdL | 5.9 | t4 | Unknown |
| ymfJ | 4.2 | t4 | Unknown; similar to unknown proteins from B. subtilis |
| cotE | 3.7 | t4 | Spore coat protein (outer) |
| usd | 8.3 | t4 | Required for translation of spoIIID |
| prkA | 6.2 | t4 | Serine protein kinase |
| yodO | 4.4 | t4 | Unknown; similar to unknown proteins |
| ylaK | 4.3 | t4 | Unknown; similar to phosphate starvation-inducible protein |
| ywlB | 3.7 | t4 | Unknown |
| ykuS | 3.4 | t4 | Unknown |
| ytiA | 2.8 | t4 | Unknown; similar to unknown proteins |
| spoIIQ | 2.9 | t4 | Required for completion of engulfment |
| ytfJ | 3.5 | t4 | Unknown |
| yjbX | 4.5 | t4 | Unknown |
| ydjP | 5.7 | t4 | Unknown; similar to arylesterase |
| yeaA | 4.1 | t4 | Unknown |
| yqzG | 4.7 | t4 | Unknown |
| yfhE | 3.9 | t4 | Unknown |
| yhbH | 5.3 | t4 | Unknown; similar to unknown proteins |
| yjbA | 3.5 | t4 | Unknown |
| ydcC | 3.7 | t4 | Unknown |
| spoVR | 8.4 | t4 | Involved in spore cortex synthesis |
| yuzC | 6.8 | t4 | Unknown |
| ysxE | 4.6 | t4 | Unknown |
| spoIIID | 5.6 | t4 | Transcriptional regulator of ςE- and ςK-dependent genes |
| spoIID | 3.9 | t4 | Required for complete dissolution of the asymmetric septum |
| spoVID | 6.1 | t4 | Required for assembly of the spore coat |
| sspE | 2.9 | t4 | Small acid-soluble spore protein (major gamma-type) |
| glnQ | 4.4 | t4 | Glutamine ABC transporter (ATP-binding protein) |
Recent studies on whole-genome analysis of catabolite repression in B. subtilis describe the down-regulatory effect of glucose on the expression of a large number of genes (24, 42). The expression of some of these genes, such as gapB (−5-fold), pckA (−2-fold), cstA (−2-fold), msmX (−2-to −3-fold), acoA (−30-fold) and yesLM (−2-to −4-fold) are also down-regulated in the scoC mutant in this study. However, the transcription of some other genes repressed in presence of glucose, such as those of the iol and the opp operons, is increased in scoC-deficient strains.
A comparison of the transcriptional behavior of sporulation-associated genes in the scoC mutant with the membrane array results obtained by Fawcett et al. (7) suggests both a synergistic and an antagonistic effect between spo0A and scoC. In the present study, in at least one time point, the absence of ScoC lowered the transcriptional levels of the dpp, hut, and ybcPQST-ybdAB operons. These genes have been shown to depend on intact Spo0A for efficient transcription in the published membrane study (7). Similarly, the transcription of the yxbA, yxbB, and yxnB genes appears to require both Spo0A and ScoC (as determined from both studies).
Most of the scoC effects, however, suggest controlling effects in opposition between ScoC and Spo0A. Several of the genes reviewed by Stragier and Losick (36) that have a known or putative role in sporulation show a higher level of expression in the scoC4 strain than in the wild type (Table 3). Furthermore, more than 50% of the genes with unknown functions listed by Fawcett et al. (7), genes whose transcriptions are both Spo0A and ςF dependent, are transcribed at a higher level in the scoC4 mutant (Table 7). This list includes the yabP and yabQ genes, which have been identified as essential for sporulation (1, 7).
It is interesting that not all the degradative enzymes whose expression is associated with the stationary phase and which also appear to be under spo0A control (7) are repressed by ScoC. As an example, while the transcriptions of aprE, nprE, and nprB are elevated in scoC, the transcriptions of vpr, csn, and pel are not affected.
Nitrogen metabolism.
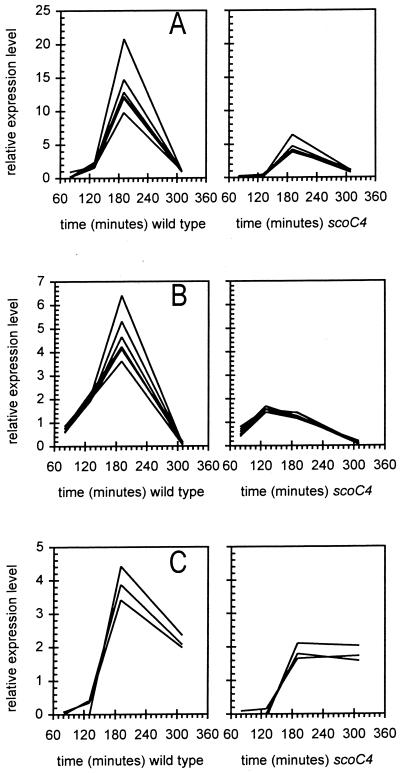
Microarray analysis reveals a number of changes in the transcriptional profile of several genes coding for enzymes involved in nitrogen utilization. The transcription of all the genes in the hut operon is decreased in the scoC4 mutant between 3- and 20-fold compared to that in the wild-type strain (Fig. 3A). Similarly, the sigL-specific transcription of the genes involved in the degradation of isoleucine and valine (2) is down-regulated in scoC4, as is the transcription of the ureABC genes (Figs. 3B and C). A striking observation is that these three scoC-affected operons are also regulated by codY (9, 32). Moreover, the transcriptional profile of these three operons demonstrates the typically tight clustering of transcriptional profiles expected for genes lying within operons, giving further confidence in the robustness of the expression assay.
FIG. 3.
Expression patterns for several (putative or known) operons as a function of time after inoculation. Intensities were normalized twice, to whole-array median intensities for all genes and to whole-gene median intensities for all arrays. (A) Histidine-degradative genes (hutPHUIGM); (B) Ile/Val degradative pathway genes (bkdR, ptb, bcd, buk, lpd, bkdA1, bkdA2, and bkdB; formerly known as yqiRSTUV bkdAA, bkdAB, and bkdB); (C) urea utilization genes (ureABC).
In addition, with further implications for the influence of scoC on genes involved in nitrogen metabolism, transcriptions levels of the operon glnQHMP (coding for the ABC glutamine transporter) are four- to sixfold higher in the mutant than in the wild type.
Motility and chemotaxis.
To ascertain the motility of the cells under our growth conditions, we looked at samples from cultures of both strains at the four time points. Wild-type strain motility ranged from approximately 60% at t1 to more than 90% at t4. In the scoC4 mutant culture, however, most of the cells were filamentous and only 5% (t1) to 20% (t4) of the cells were motile.
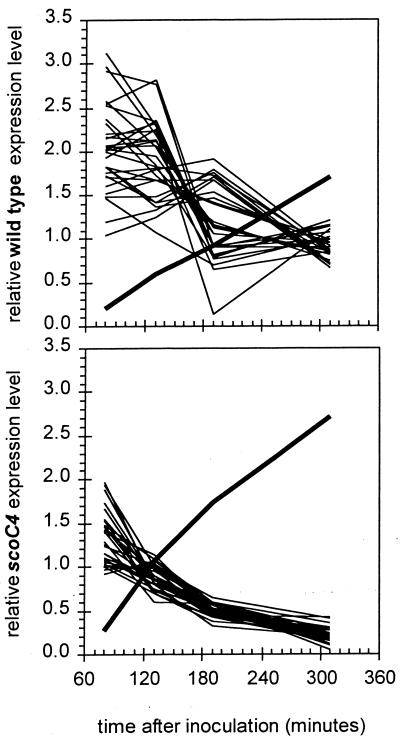
Figure 4 illustrates the transcriptional profiles for all the genes from flgB to cheD, in which a large number of motility and chemotaxis genes appear to be expressed at a lower level at the three later time points. Expression values were highest in mid-log phase (t1) and decreased progressively to the last time point analyzed in stationary phase (t4), while sinI transcription progressively increased with a steeper slope in the mutant than in the wild type. The transcription of the hag gene, which codes for flagellin, is also decreased in the scoC mutant. It is interesting that, as in the case of the hut, bdk, and ure operons, both codY and scoC are known to play a role in the expression of hag (23).
FIG. 4.
Expression patterns for the 30-gene flgB-to-sigD transcriptional cluster as a function of time after inoculation. The y axis shows the relative expression levels for 30 chemotaxis and flagellar-motility genes at position 145° to 146° in the B. subtilis genome (thin lines) and for sinI (thick line). Intensity was normalized as described for Fig. 3.
It is also interesting that the gene coding for McpC, a methyl-accepting protein mediating the carbohydrate chemotaxis in synergy with the phosphoenolpyruvate-dependent phosphotransferase system, is also down-regulated in scoC (10). A putative methyl-accepting chemotaxis protein, YfmS, is significantly down-regulated as well. mcpA and mcpB, although obviously down-regulated in scoC in our assay, failed to meet the most stringent requirements set by our statistical analysis (Z scores of only 4.1).
scoC4-dependent regulation of the yclF gene.
The list of 560 significant genes reflects Bacillus ORFs showing a significant expression variation in at least one of the four assayed time points (log phase, early transition, late transition, and stationary phase) between the wild type and the scoC4 transcriptomes. Most of the genes on this list vary significantly at only one or two time points. However, only one gene, yclF, shows significant change at all four time points. The yclF gene, encoding a hypothetical peptide/proton symporter orthologous to L. lactis DtpT (11, 18), is expressed at 6- to 20-fold-higher levels in scoC4 cells than in the isogenic wild-type cells. The Z scores associated with these measurements are greater than 9.0, indicating a high degree of significance in the difference between the two strains.
DISCUSSION
It has been recognized that, “[a]lthough transient in laboratory cultures, the transition state is probably the predominant metabolically active state of Bacillus in the natural environment of the soil where nutrients are usually limited” (38). In order to maximize the utilization of available resources, bacilli have developed complex regulatory circuits to allow functional diversification of the population and to fine tune and quickly adapt their metabolism to a constant state of nutrient flux. This communal organization ensures that a fraction of the population survives, either by the resumption of vegetative growth by exploiting new nutrient sources made available by the secretion of degradative enzymes, or by the entrance into sporulation, the ultimate survival strategy in a nutrient-depleted environment. To this end, while Spo0A acts as the master switch determining whether the cell continues vegetative proliferation or carries out stationary-phase functions, Bacillus has evolved a number of other regulators, called transition state regulators (38), with partially overlapping control functions. ScoC belongs to this group of regulators. However, the complete role of ScoC in the life of bacilli has not been fully elucidated.
In the present work, we have expanded the scope of our knowledge of scoC's overall regulatory role. To do so, we analyzed the B. subtilis transcriptome under defined laboratory conditions with the aid of species-specific oligonucleotide microarrays. The scoC4 mutant used in this experiment reveals transcriptional changes from isogenic wild-type cells in approximately 560 genes for at least one of the four time points tested.
The identification of these genes was based on a rigorous statistical treatment of the data. We required a 4.37-SDLR threshold for significance of an expression change between scoC and wild-type cells, relative to the expression changes assayed on replicate samples. This is a very stringent level based on a Bonferroni correction for a large (4,000+) set of comparisons. However, there may be biologically relevant changes in genes that do not quite meet this strict cutoff, which minimizes false positives at the cost of excluding true positives. Therefore, we also considered the expression changes for a subsidiary set of 121 ORFs that meet a 3.89-SDLR cutoff relative to the replicate data (corresponding to a 99.99% confidence threshold).
It is very important to consider that the significance of a given change in a gene's expression is a function of the intensity level at which that gene is detected on the microarray. At high signal levels, the scatter of the error of measurement is a small fraction of the measurement. In contrast, at low signal levels, the error of measurement tends to be dominated by an absolute value (32). Therefore, a smaller change occurring between scoC and wild-type cells might be more significant if the scanned gene signal is >10,000 than a larger change of expression if the gene signal is <100. Therefore, the Z score of a given expression change relative to the replicate scatter distribution is a better measure of significance than the gene expression change alone.
The reliability of the results obtained using this approach is supported by their agreement with data previously reported in the literature and summarized in Table 4. For example, the expression patterns for the opp operon genes in our experiments are similar to the results recently reported for an oppA-lacZ fusion analysis by Koide et al. (17). Furthermore, the scoC4-transcriptional level of aprE was substantially increased in the present study, as has been reported elsewhere (8). Also, the increase in the level of sinI-specific mRNA in scoC cells is in agreement with previously unpublished data (33). ScoC has been shown to also affect a previously unidentified alkaline phosphatase. From our expression data, it appears that this measured activity is due to PhoB (4).
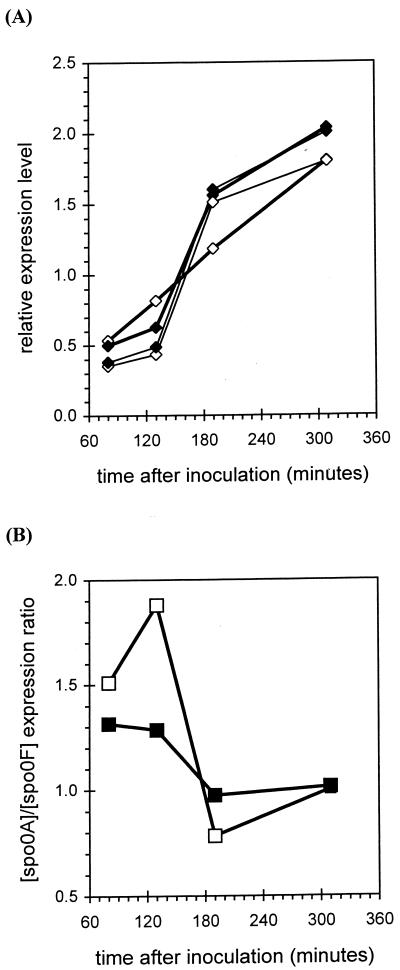
A major role attributed to scoC has been its effect on the sporulation process, coupled to catabolite repression (4, 14) by a not-yet-clarified mechanism. It has been reported that scoC controls the activity of SinR by transcriptionally modulating the gene expression of the SinR antagonist, sinI (15, 32). In addition, ScoC has been described as controlling the expression of the app and opp operons, whose gene products are responsible for the import of quorum-sensing signaling peptides (17). At first glance, while these are very important roles, it may not be sufficient to explain all the phenotypes associated with scoC. Data presented here show that the level of transcription of rapA and phrA in a scoC null mutant is diminished more than threefold. This is likely to affect the level of Spo0A phosphorylation, which, in turn, will affect the level of spo0A mRNA due to the fact that phosphorylated Spo0A activates its own transcription (13). Although our statistical analysis does not indicate a significant change in the absolute level of spo0A transcript between the wild type and mutant, there is a slight but noticeable difference in the level of spo0A transcript in the scoC strain at t2 relative to the level of spo0F transcription. While in all the strains we have tested so far spo0A mRNA rises sharply around T0, it has already increased considerably just before transition relative to the spo0F mRNA in the scoC mutant (Fig. 5). We also find that the transcription of bofA, spoIVFA and spoIVFb, involved in the processing of ςK, increases in the scoC strain after t3 (fourfold higher at t4).
FIG. 5.
Expression patterns for spo0A and spo0F transcripts in scoC and wild-type strains. (A) Relative expression levels for spo0A (thick lines) and spo0F (thin lines) from wild-type (closed diamonds) and scoC (open diamonds) cells. Expression levels were normalized as in Fig. 3. (B) Ratio of spo0A to spo0F expression levels for wild-type (closed squares) and scoC (open squares) cells.
However, while we find the level of rapA and phrA significantly reduced in the scoC strain, the expression levels of the other members of the Rap/Phr family, several of which are under Spo0A control (7), do not appear to be significantly affected. The expression of several of the other genes known to play an important role in the initiation of sporulation, such as spo0B, spo0F, abrB, kinA, kinB, kinC, or those encoding the sporulation-specific sigma factors, is likewise not significantly affected by scoC4.
In all, the results show that ScoC affects the level of expression of 38 sporulation genes as tabulated by Stragier and Losick (36). In addition, about 50% of the genes with unknown functions, identified by Fawcett et al. (7) as requiring both Spo0A and ςF for transcription, show positive shifts in expression in the scoC null mutation (Table 7). We show that ScoC has both negative (pckA and gapB) and positive (iol operon) effects on the expression of a number of genes involved in carbon metabolism (24, 42). This balancing act of both down- and up-regulation of genes may partially explain how scoC null mutations relieve the sporulation repression exerted by glucose. While these findings do not lead to a complete understanding of the mechanism by which scoC plays a role in sporulation, it certainly indicates a wider role for scoC in this process than previously appreciated.
B. subtilis has a varied repertoire of genes regulating nitrogen metabolism. The main regulatory genes identified to date are codY, tnrA, glnR, and glnA (9). In the absence of ScoC, it was a surprise to observe a very strong downward effect on the transcription of genes involved in the utilization of amino acids as a nitrogen source, such the hut and bkd operons. The fact that scoC also causes a decrease in the transcriptional level of the ureABC operon seems to suggest that there is at least a partial overlap in the regulation of this nitrogen-related group of genes by both CodY and ScoC. The possibility of a coregulatory function is strengthened by the down-regulation effect we have observed in scoC4 for other CodY-regulated genes, such as gabP, comK, rapA (31), the hag regulon (23), and, to a lesser extent, the dpp operon. However, there is no obvious scoC effect on srfA, rapC, and citB, which were reported as being regulated by codY (31). In addition, the expression of codY itself has an unchanging profile between the wild-type and scoC strains.
A strong (fivefold) expression increase was exerted in the scoC4 mutant on the glutamine transporter operon (glnQHMP). This may indicate a higher level and/or demand of glutamine, a key intermediary in nitrogen utilization in B. subtilis. While the scoC mutation represses nitrogen utilization by lowering the expression of the hut, bkd, and ure operons, it elevates the expression of the genes involved in the transport of the central player for nitrogen metabolism, glutamine. This may help explain the observation (33) that a strain carrying a mutation in scoC can affect catabolite repression exerted by glucose but not that exerted by glucose and glutamine together.
The slight but observable increase in the expression level of sinI is bound to antagonize SinR activity. It has been reported that sinR is a positive regulator of sigD (30), involved in motility functions. Therefore, a higher level of sinI, by inactivating sinR, would depress the motility of a scoC mutant by repressing sigD transcription. This may explain the observation that scoC mutants are somewhat less motile than wild-type cells (16). We have shown that a great many of the genes involved in motility and chemotaxis are indeed transcribed at lower levels in the scoC4 strain (Fig. 4).
Given the large number of genes affected by ScoC, as determined by comparing the scoC4 mutant and wild-type transcriptomes, it is perhaps not surprising that a large number of affected genes themselves encode regulatory proteins, thus transmitting the direct effects of ScoC to other genes. ScoC is known to be a pleiotropic regulatory protein, and we suggest that some of these effects are likely due to an indirect action mediated through other regulators (Tables 5 and 6). Potential ScoC DNA binding sites were found upstream of several regulatory genes, including ykoM, ykvE, cheY, cheB, rbsR, ywrC, and cggR, in which transcription appears to be decreased in the scoC loss-of-function mutant in at least one of the four time points assayed (Tables 5 and 6). The bidirectionality of these regulatory effects and of the effects seen on genes involved in nitrogen metabolism points to the interesting possibility that ScoC acts both in a negative and, perhaps directly or indirectly, in a positive manner.
The effect of the scoC mutation on comK expression (discussed above), together with the effect on the transcription of the opp operon, may explain the lower level of competence reached by a scoC mutant (Causey and Ferrari, unpublished). It is interesting that comK appears to have a putative scoC binding site at upstream position −147 (Table 5).
The yclF gene is unique in our set of significantly changed expressions in scoC. It is the only gene that showed an increased expression level at all four time points tested. In the promoter region of yclF, upstream of its ribosomal binding site, there are clear and canonical −10 and −35 ςA promoter sequences. Overlying the sequences of this region are two close matches to the ScoC-binding consensus sequence, RATANTATY (Fig. 6). These sequences are potential binding sites repressing yclF in wild-type cells relative to the scoC4 mutant, in which this repression would be relaxed. The function of the 492-amino-acid yclF protein in B. subtilis is unknown, but its amino acid sequence shows high similarity (E value = 10−106) to the 463-amino-acid Lactococcus lactis dtpT gene product, a novel di- and tripeptide transporter (11, 18). Both DtpT and YclF appear to be members of the PTR superfamily, peptide/proton symporters, which are structurally and functionally distinct from the multisubunit ABC peptide transporters, such as Opp and Dpp (35). A point mutation in dtpT leads to increases in protease gene transcription in L. lactis (21). Research aimed at investigating the role of yclF in B. subtilis is in progress.
FIG. 6.
The 280-bp promoter region of the yclF gene (and the yclG gene). Start codons are underlined and in bold capitals. R.B.S., ribosomal binding site. The −10 and −35 boxes of the yclF promoter are indicated. The ScoC-binding consensus sequences overlying these promoter elements are double underlined.
In this paper, we present a global analysis of the effects of the scoC4 null mutation on the B. subtilis transcriptome. This study has correlated known phenotypes of the scoC null mutation (lower motility, sporulation phenotype, development of competence, and degradative enzyme production) with changes detected in the expression pattern of a large number of genes with both known and unknown function. The data suggest (Table 7) that there are many additional genes involved in the complex network of sporulation genes, an observation echoed by the recent report by Fawcett et al. (7). In addition, our work suggests a close and unforeseen link of ScoC to the regulation of nitrogen metabolism. We propose that there may exist a strong interaction of scoC with the global nitrogen metabolism regulators, such as CodY. Finally, we have demonstrated that ScoC clearly plays a major role in various cellular functions of the life cycle of this bacterium. These observations allow us to paint a picture in which Spo0A is still the master switch of the transition phase, but the real differentiation occurring within the transition state population is mediated by a number of transition state regulators, one of which is scoC. By way of the global regulatory network implemented by these transition state regulators, the bacterial culture can swiftly achieve a redistribution of functions within its populations to carry out the most needed tasks. Elucidating the mechanistic details of how ScoC exercises control among the various processes it participates in will usher in an exciting era for the investigation of Bacillus growth and development.
ACKNOWLEDGMENTS
We thank Anita van Kimmenade, Mick Ward, and Mike Arbige for help and support throughout the study; Maria Diaz-Torres, Don Naki, and Jian Yao for helpful discussions; and Molly Schmid, Roopa Ghirnikar, Doug Crabb, Brian Schmidt, and Maggie Cervin for reviewing the manuscript.
REFERENCES
- 1.Asai K, Takamatsu H, Iwano M, Kodama T, Watabe K, Ogasawara N. The Bacillus subtilis yabQ gene is essential for formation of the spore cortex. Microbiology. 2001;147:919–927. doi: 10.1099/00221287-147-4-919. [DOI] [PubMed] [Google Scholar]
- 2.Debarbouille M, Gardan R, Arnaud M, Rapoport G. Role of BkdR, a transcriptional activator of the sigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J Bacteriol. 1999;181:2059–2066. doi: 10.1128/jb.181.7.2059-2066.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Saizieu A, Gardes C, Flint N, Wagner C, Kamber M, Mitchell T J, Keck W, Amrein K E, Lange R. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J Bacteriol. 2000;182:4696–4703. doi: 10.1128/jb.182.17.4696-4703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dod B, Balassa G, Raulet E, Jeannoda V. Spore control (Sco) mutations in Bacillus subtilis. II. Sporulation and the production of extracellular proteases and amylases by Sco mutants. Mol Gen Genet. 1978;163:45–56. [Google Scholar]
- 5.Dowds B C, Hoch J A. Regulation of the oxidative stress response by the hpr gene in Bacillus subtilis. J Gen Microbiol. 1991;137:1121–1125. doi: 10.1099/00221287-137-5-1121. [DOI] [PubMed] [Google Scholar]
- 6.Farrell R E., Jr . Protocol: guanidinium-acid-phenol extraction in RNA methodologies. 2nd ed. San Diego, Calif: Academic Press; 1996. p. 81. [Google Scholar]
- 7.Fawcett P, Eichenberger P, Losick R, Youngman P. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 2000;97:8063–8068. doi: 10.1073/pnas.140209597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrari E, Henner D J, Perego M, Hoch J A. Transcription of Bacillus subtilis subtilisin and expression of subtilisin in sporulation mutants. J Bacteriol. 1988;170:289–295. doi: 10.1128/jb.170.1.289-295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher S. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol Microbiol. 1999;32:223–232. doi: 10.1046/j.1365-2958.1999.01333.x. [DOI] [PubMed] [Google Scholar]
- 10.Garrity L F, Schiel S L, Merril R, Reitzer J, Saier M H, Ordal G W. Unique regulation of carbohydrate chemotaxis in Bacillus subtilis by the phosphoenolpyruvate-dependent phosphotransferase system and the methyl-accepting chemotaxis protein. J Bacteriol. 1998;180:4475–4480. doi: 10.1128/jb.180.17.4475-4480.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagting A, Kunji E R, Leenhouts K J, Poolman B, Konings W N. The di- and tripeptide transport protein of Lactococcus lactis. A new type of bacterial peptide transporter. J Biol Chem. 1994;269:11391–11399. [PubMed] [Google Scholar]
- 12.Higerd T B, Hoch J A, Spizizen J. Hyperprotease-producing mutants of Bacillus subtilis. J Bacteriol. 1972;112:1026–1028. doi: 10.1128/jb.112.2.1026-1028.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoch J A. Initiation of bacterial development. Curr Opin Microbiol. 1998;1:170–174. doi: 10.1016/s1369-5274(98)80007-6. [DOI] [PubMed] [Google Scholar]
- 14.Ito J, Spizizen J. Genetic studies of catabolite repression insensitive sporulation mutants of Bacillus subtilis. Colloq Int Cent Natl Rech Sci. 1973;227:81–82. [Google Scholar]
- 15.Jeannoda V, Balassa G. Spore control (Sco) mutations in Bacillus subtilis. IV. Synthesis of alkaline phosphatase during sporulation of Sco mutants. Mol Gen Genet. 1978;163:65–73. [Google Scholar]
- 16.Kallio P T, Fagelson J E, Hoch J A, Strauch M A. The transition state regulator Hpr of Bacillus subtilis is a DNA-binding protein. J Biol Chem. 1991;266:13411–13417. [PubMed] [Google Scholar]
- 17.Koide A, Perego M, Hoch J A. ScoC regulates peptide transport and sporulation initiation in Bacillus subtilis. J Bacteriol. 1999;181:4114–4117. doi: 10.1128/jb.181.13.4114-4117.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunji E R, Smid E J, Plapp R, Poolman B, Konings W N. Di-tripeptides and oligopeptides are taken up via distinct transport mechanisms in Lactococcus lactis. J Bacteriol. 1993;175:2052–2059. doi: 10.1128/jb.175.7.2052-2059.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunst F, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 20.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 21.Marugg J D, Meijer W, van Kranenburg R, Laverman P, Bruinenberg P G, de Vos W M. Medium-dependent regulation of proteinase gene expression in Lactococcus lactis: control of transcription initiation by specific dipeptides. J Bacteriol. 1995;177:2982–2989. doi: 10.1128/jb.177.11.2982-2989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milhaud P, Balassa G, Zucca J. Spore control (Sco) mutations in Bacillus subtilis. I. Selection and genetic mapping of Sco mutants. Mol Gen Genet. 1978;163:35–44. [Google Scholar]
- 23.Mirel D B, Estacio W F, Mathieu M, Olmsted E, Ramirez J, Marquez-Magana L M. Environmental regulation of Bacillus subtilis ςD-dependent gene expression. J Bacteriol. 2000;182:3055–3062. doi: 10.1128/jb.182.11.3055-3062.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno M S, Schneider B L, Maile R R, Weyler W, Saier M H. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol Microbiol. 2001;39:1366–1381. doi: 10.1111/j.1365-2958.2001.02328.x. [DOI] [PubMed] [Google Scholar]
- 25.Moszer I, Glaser P, Danchin A. SubtiList: a relational database for the Bacillus subtilis genome. Microbiology. 1995;141:261–268. doi: 10.1099/13500872-141-2-261. [DOI] [PubMed] [Google Scholar]
- 26.Moszer I. The complete genome of Bacillus subtilis: from sequence annotation to data management and analysis. FEBS Lett. 1998;430:28–36. doi: 10.1016/s0014-5793(98)00620-6. [DOI] [PubMed] [Google Scholar]
- 27.Perego M, Hoch J A. Sequence analysis and regulation of the hpr locus, a regulatory gene for protease production and sporulation in Bacillus subtilis. J Bacteriol. 1988;170:2560–2567. doi: 10.1128/jb.170.6.2560-2567.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perego M, Spiegelman G B, Hoch J A. Structure of the gene for the transition state regulator, AbrB: regulator synthesis is controlled by the spo0A sporulation gene in Bacillus subtilis. Mol Microbiol. 1988;2:689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- 29.Perego M. Self-signaling by Phr peptides modulates Bacillus subtilis development. In: Dunny G M, Winans S C, editors. Cell-cell signaling in bacteria. Washington, D.C.: American Society for Microbiology; 1999. pp. 243–258. [Google Scholar]
- 30.Rashid M H, Sekiguchi J. flaD (sinR) mutations affect SigD-dependent functions at multiple points in Bacillus subtilis. J Bacteriol. 1996;178:6640–6643. doi: 10.1128/jb.178.22.6640-6643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ratnayake-Lecamwasan M, Serror P, Wong K-W, Sonenshein A L. Bacillus subtilis codY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 2001;15:1093–1103. doi: 10.1101/gad.874201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rocke D M, Lorenzato S. A two-component model for measurement error in analytical chemistry. Technometrics. 1995;37:176–184. [Google Scholar]
- 33.Smith I. Regulatory proteins that control late-growth development. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C.: American Society for Microbiology; 1993. pp. 785–800. [Google Scholar]
- 34.Solomon J M, Lazazzera B A, Grossman A D. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 1996;10:2014–2024. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- 35.Steiner H Y, Naider F, Becker J M. The PTR family: a new group of peptide transporters. Mol Microbiol. 1995;16:825–834. doi: 10.1111/j.1365-2958.1995.tb02310.x. [DOI] [PubMed] [Google Scholar]
- 36.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 37.Strauch M A, Spiegelman G B, Perego M, Johnson W C, Burbulys D, Hoch J A. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 1989;8:1615–1621. doi: 10.1002/j.1460-2075.1989.tb03546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strauch M A, Hoch J A. Transition-state regulators: sentinels of Bacillus subtilis post-exponential gene expression. Mol Microbiol. 1993;7:337–342. doi: 10.1111/j.1365-2958.1993.tb01125.x. [DOI] [PubMed] [Google Scholar]
- 39.Strauch M A. AbrB modulates expression and catabolite repression of a Bacillus subtilis ribose transport operon. J Bacteriol. 1995;177:6727–6731. doi: 10.1128/jb.177.23.6727-6731.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wodicka L, Dong H, Mittmann M, Ho M H, Lockhart D J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 41.Yang M Y, Ferrari E, Henner D J. Cloning of the neutral protease gene of Bacillus subtilis and the use of the cloned gene to create an in vitro-derived deletion mutation. J Bacteriol. 1984;160:15–21. doi: 10.1128/jb.160.1.15-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida K, Kobayashi K, Miwa Y, Kang D-M, Matsunaga M, Yamaguchi H, Tojo S, Yamamoto M, Nishi R, Ogasawara N, Nakayama T, Fujita Y. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 2001;29:683–692. doi: 10.1093/nar/29.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]