Abstract
The mechanisms by which macrophages regulate energy storage remain poorly understood. We identify in a genetic screen a PDGF-family growth factor, Pvf3, produced by macrophages that is required for lipid storage in fat body cells of Drosophila larvae. Genetic and pharmacological experiments indicate that the mouse Pvf3 ortholog, PDGFcc, produced by adipose-tissue resident macrophages controls lipid storage in adipocytes in a Leptin- and Ccr2- independent manner. PDGFcc production is regulated by diet, and acts in a paracrine manner to control lipid storage in adipose tissues of newborn and adult mice. At the organismal level upon PDGFcc blockade, excess lipids are redirected toward thermogenesis in brown fat. These data identify a macrophage-dependent mechanism, conducive to the design of pharmacological interventions, that controls energy storage in metazoans.
One Sentence Summary:
Metazoan macrophages control lipid storage in fat cells through diet-regulated production of PDGF-family growth factors.
Introductory paragraph.
Daily and seasonal variation in caloric intake results in cycles of energy storage and expenditure (1). To accommodate these cycles, metazoans have evolved specialized fat-storing tissues dedicated to dynamic storage of energy (1–4) without the common lipotoxicity associated with fat accumulation in other cells (1, 2, 4). These adipose tissues consist of lipid-storing adipocytes, stromal cells, and immune cells such as macrophages. Landmark studies have firmly established that recruitment of monocytes into adipose and peripheral tissues of obese animals promotes inflammation, ectopic fat deposition in the liver, and insulin resistance, via production of inflammatory cytokines such as Tumor necrosis factor (TNF) by monocyte-derived macrophages (5–20). Genetic or pharmacological inhibition of the C-C chemokine receptor type 2 (Ccr2)-dependent recruitment of bone marrow-derived monocytes into tissues prevents or alleviates metabolic syndrome in obese animals (11, 15–18, 21). Thus, Ccr2-dependent macrophages are a therapeutic target in metabolic diseases associated with obesity. CCR2 blockade or deficiency, however, does not prevent weight gain, but only its inflammatory and metabolic complications. Recent studies in models of macrophage-deficient animals suggest that macrophages may also regulate mouse weight and adiposity (22–27). For example, Slc6a2 deficiency (26), which reduces clearance of sympathetic norepinephrine by neuron-associated macrophages in the adipose tissue leads to significant and sustained weight loss in obese mice. Tribbles homolog 1-deficient (Trib1–/–) mice which lack pro-adipogenic Trib1-dependent macrophages (22) are also lean and develop metabolic syndrome akin to lipodystrophy (22). Colony stimulating factor 1 receptor (CSF1R) blockade which depletes all macrophages keeps mice lean under a high-fat diet and protects them against metabolic syndrome in part due to the proposed role of microglia in regulating leptin sensitivity in the mediobasal hypothalamus (24, 25). Finally, Triggering receptor expressed on myeloid cells 2-deficient (Trem2–/–) mice (27) develop worse obesity and metabolic syndrome due to a global control of a lipid-associated macrophage program by Trem2 (27). Although the systemic effects of CSF1R blockade and germline deletion of Csf1r, Ccr2, Trib1, Slc6a2, or Trem2 combined with the developmental and functional heterogeneity of macrophages (3, 6, 22, 27–48) may confound the analysis of macrophage functions in these models, these data nevertheless suggest that macrophages are involved, directly or indirectly, in the regulation of energy storage. In this study, we therefore attempted to identify a molecular mechanism that mediate the direct control of adiposity and energy storage by macrophages.
Results
Csf1r-dependent lipid storage in newborn, hyperphagic, or high-fat diet fed adult mice.
Six-week-old adult wildtype C57Bl/6J and Ccr2-deficient mice fed a high-fat diet (45% kcal from fat, 4.73 kcal/g) and treated with the CSF1R tyrosine kinase inhibitor PLX5622 (PLX, 1200 mg PLX5622/kg of food) for 8 weeks stayed lean, with fat pads half the weight of controls (Fig 1A and fig. S1), contrary to untreated wildtype and Ccr2-deficient mice which became obese (11) (Fig 1A and fig. S2). White adipocytes from PLX-treated mice did not increase their BODIPY+ lipid content thus lacking the hypertrophy normally induced by high-fat diet (Fig 1 B and C). Mice treated with a blocking anti-CSF1R antibody (AFS98, 50 mg/kg, i.p., three times a week (49)) presented with a similar phenotype (Fig 1A and fig. S1 C to L), indicating that an off-target effect of the kinase inhibitor was unlikely. PLX treatment also reduced obesity and adipocyte hypertrophy in hyperphagic Leptin-receptor deficient mice (Lepr–/–, db/db) fed a control diet (10% kcal from fat, 3.85 kcal/g) (Fig 1 A and B and fig. S1 M and N). Thus, CSF1R blockade prevents fat storage and adipocyte hypertrophy in high-fat-diet-fed and hyperphagic mice, in a Ccr2- and, at least in part, Leptin receptor-independent manner.
Figure 1. Ccr2–independent macrophages control lipid storage in mouse adipocytes.
(A) Weight of epididymal (eWAT) and inguinal (iWAT) fat pads from 14-week-old C57Bl/6J mice, Ccr2–/– mice, and 12-week-old Lepr–/– mice all subjected to indicated diets and treatments for eight weeks. N= 12 to 20 mice per experimental group, from four independent experiments for C57Bl/6J mice, and n= 8 to 16 mice per experimental group, from three independent experiments for Ccr2–/– mice, and Lepr–/– mice. Dots represent individual mice. Statistics: P-values were obtained by comparing mean weight using one-way ANOVA with Sidak’s correction for multiple group comparison. Red lines indicate mean values. (B) Representative fluorescent whole-mount images stained for F4/80 and BODIPY of iWAT from 14-week-old C57Bl/6J mice and 12-week-old Lepr–/– (db/db) mice, all subjected to indicated diets and treatments for eight weeks. Scale bars are 40 μm. (C) Quantitative analysis of adipocyte size in iWAT of C57Bl/6J and Ccr2–/– mice from (A,B). N=10 C57Bl/6J and N=6 Ccr2–/– mice per experimental group, from two to three independent experiments. Hundred to two hundred adipocytes per fat pad were measured using bitplane Imaris image analysis software in three different areas. Dots represent percent of adipocytes per size intervals. Statistics: P-values were obtained by comparing mean adipocyte sizes using one-way ANOVA with Sidak’s correction for multiple group comparison. See also fig. S1 and S2.
The initial hypertrophy of adipocytes, which allows for fat storage during the first month of life, was abolished by global genetic deletion of Csf1r in Csf1rCre; Csf1rf/f mice (Fig 2A and fig. S3). Similarly, genetic deletion of Csf1r or Spi1 in resident macrophages (50, 51) (in Tnfrsf11aCre; Csf1rf/f mice and Tnfrsf11aCre; Spi1f/f mice) resulted in mice with small fat pads, corresponding to small Perilipin (PLIN1)+ adipocytes with little BODIPY+ lipid content (Fig 2A and fig. S4). By contrast, adipose tissue weight and morphology of adipocytes were normal in young Ccr2-deficient mice (52) (fig. S2 A, B, and D), as well as in mice carrying a Csf1r deletion in the fms-like tyrosine kinase 3 (Flt3)-expressing hematopoietic stem cell lineage (51, 53) (fig. S2 A, B and E). Thus, in addition to limiting adipocyte hypertrophy in adult hyperphagic and high-fat-diet-fed mice, Csf1r deficiency in the Ccr2-independent resident macrophage lineage may also prevent the initial hypertrophy of adipocytes in newborn mice. The adipocyte phenotype of genetic mutant animals, however, may be compounded by the effects of global macrophage deficiency during development.
Figure 2. Macrophages control lipid storage in adipocytes of newborn mice and fat body cells of Drosophila larvae.
(A) Representative fluorescent whole-mount images stained for F4/80 and BODIPY (Left) and quantitative analysis of adipocytes size (Right) for inguinal white adipose tissues (iWAT) from 4-week-old Csf1rCre; Csf1rf/f mice, Tnfrsf11aCre; Csf1rf/f mice and their littermate controls. Fat pads from six mice, obtained from three to six litters, were analysed for each group and hundred to two hundred adipocytes were measured using bitplane Imaris image analysis software in three different areas per fat pad. Dots represent percent of adipocytes per size intervals. Scale bars are 40 μm. Statistics: P-values were obtained by comparing mean adipocyte sizes using t-test. See also fig. S3 and S4. (B) Representative fluorescent images of L3 larvae depicting the expression pattern of hemocyte-specific drivers Hemolectin (Hml, Hml-gal4>uas-GFP) and Serpent (Srp, SrpHemo-mCherry) and the contact between hemocytes and fat body cells (c564-gal4>uas-GFP). Scale bars are 500 μm (whole larvae), 80 μm (larvae insets), and 10 μm (far right). (C) Quantitative analysis of buoyancy in wandering L3 larvae from SrpHemo-gal4>uas-reaper and control lines. N=6 independent experiments from three crosses, each with experimental groups of twenty larvae were analyzed. Statistics: Buoyancy (mean ± SD) was compared between groups using t-test. (D) Quantification of triglyceride level (TG) normalized to total protein measured in SrpHemo-gal4>uas-reaper L3 larvae and controls in N=11 independent experiments from four crosses, each with experimental groups of ten larvae, and in Hml-gal4>uas-reaper and controls in N=10 independent experiments, from four crosses, each with experimental groups of ten larvae. Dots represent experimental groups. Statistics: Mean ± SD TG level were compared between genotypes using one-way ANOVA with Sidak’s correction for multiple group comparison. (E) Quantitative analysis of BODIPY+ fat body cell size in wandering L3 SrpHemo-gal4>uas-reaper larvae and controls from N=6 experiments from three crosses, each with experimental groups of twenty larvae, and Hml-gal4>uas-reaper larvae and controls from N=9 experiments from four crosses, each with experimental groups of twenty larvae. For each replicate size of hundred fat body cells were measured using bitplane Imaris image analysis software. Dots represent percent of adipocytes per size intervals. Statistics: P-values were obtained by comparing mean fat body cell sizes between genotypes using t-test.
Macrophage-dependent lipid storage in Drosophila larvae.
We hypothesized that these adipocyte phenotypes may be attributable to a macrophage defect. Because fat-storing cells and macrophages are conserved across the animal kingdom, we generated macrophage-less Drosophila larvae by inducing apoptosis in hemocytes (Drosophila macrophages) using either SrpHemo-gal4>uas-reaper or Hml-gal4 >uas-reaper lines (54–56) (Fig 2 B to E and fig. S5). In both cases, we observed reduced buoyancy (57) in hemocyte-deficient wandering third-stage larvae (L3), corresponding to a ∼60% reduction in triglyceride content and fat body cell size as compared to control larvae (Fig 2 C to E). Genetic labeling of Drosophila hemocytes with Hml-gal4>uas-GFP (58) or SrpHemo-mCherry (56, 59) reporters also confirmed a close association between hemocytes and fat body cells, labeled with c564-gal4>uas-GFP+ (55, 60) in L3 larvae (Fig 2B). Thus, a genetic screen for macrophage-derived factors may reveal a conserved mechanism by which Drosophila and mouse macrophages control lipid storage and adipocyte hypertrophy.
Macrophage-derived PDGF-family growth factor Pvf3 controls lipid storage in Drosophila fat body cells.
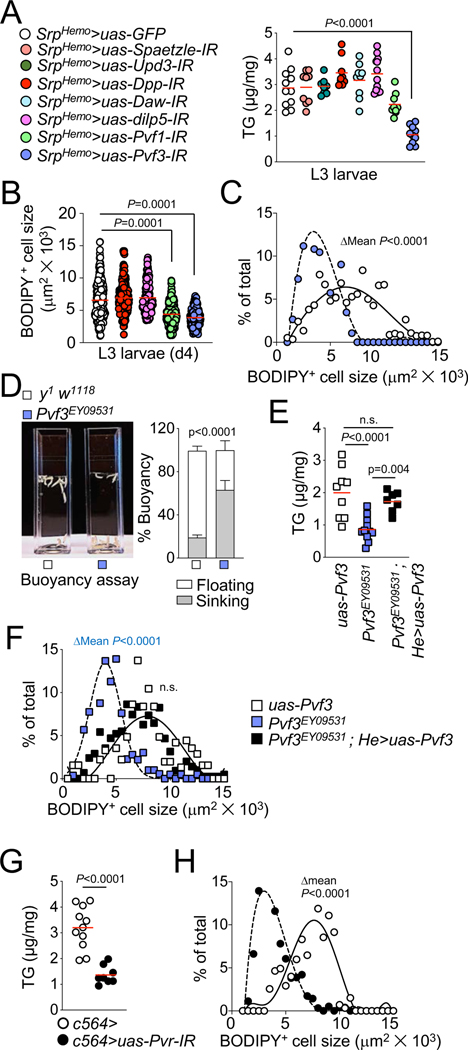
Drosophila larvae represent a highly tractable model for screening the role of conserved families of growth factors and cytokines produced by macrophages (58, 61, 62). Among these, hemocyte-specific RNAi of the Platelet-derived growth factor (PDGF)/ Vascular endothelial growth factor (VEGF)-family ortholog (61, 63) PDGF- and VEGF-related factor 3 (Pvf3) (SrpHemo-gal4>uas-pvf3-IR) decreased triglyceride content (Fig 3A) and fat body cell size by >50% (Fig 3 B and C and fig. S5 C). Hemocyte-specific RNAi of the closely related PDGF- and VEGF-related factor 1 (Pvf1), presented with a milder phenotype (Fig 3 A and B and fig. S5 C) .To confirm the role of Pvf3 and control for off-target effects of the RNAi constructs, we analyzed Pvf3 mutant flies (Pvf3EY09531, Fig 3 D to F). L3 Pvf3EY09531 larvae had reduced buoyancy (Fig 3D), a >50% reduction in triglyceride content (Fig 3E), and small fat body cells in comparison to controls (Fig 3F), whereas the genetic rescue of Pvf3 expression in Pvf3EY09531 larvae (Pvf3EY09531; He-gal4>uas-Pvf3 (64)) restored triglyceride content and fat body cell size (Fig 3 E and F). In addition, since all Drosophila PDGF-family orthologs share the same receptor, PDGF/VEGF-receptor (PVR) (61, 63), which is expressed by fat body cells (63, 65), we generated fat body-specific Pvr RNAi (c564-gal4>uas- Pvr-IR), which yielded L3 larvae with reduced triglyceride content and small fat body cells (Fig 3 G and H). These data strongly suggest that Drosophila hemocytes control fat body cell size and triglyceride storage via production of the PDGF-family growth factor Pvf3 by hemocytes, likely acting on Pvr-expressing fat body cells.
Figure 3. Hemocyte-derived PDGF-family growth factor Pvf3 controls lipid storage in Drosophila fat body cells.
(A) Quantification of triglyceride level (TG) normalized to total protein measured in wandering L3 larvae from flies with hemocyte-specific RNAi for the indicated genes and SrpHemo>uas-GFP control. For each RNAi, N= 8 to 12 experiments from four crosses were analyzed, each with experimental groups of ten larvae. Dots represent means of individual experiments. Statistics: P-values were obtained by comparing mean TG levels in mutant and SrpHemo>uas-GFP control lines using one-way ANOVA with Dunnett’s correction for multiple group comparison with a single control. Red lines indicate mean values. (B) Quantification of BODIPY+ fat body cell size in L3 larvae from flies with hemocyte-specific RNAi for the indicated genes and SrpHemo>uas-GFP control. For each RNAi, N= 4 to 7 experiments from three to six crosses were analyzed, each with experimental groups of ten larvae, and hundred cells per replicate were measured. Dot represents individual fat body cells. Statistics: P-values were obtained by comparing mean BODIPY+ fat body cell sizes using oneway ANOVA with Dunnett’s correction for multiple group comparison with a single control. Red lines depict mean values. (C) Analysis of BODIPY+ fat body cells size distribution in L3 SrpHemo-gal4>uas-pvf3-IR and control larvae in N=3 experiments from three crosses, each with experimental groups of ten larvae. Hundred fat body cells were measured using bitplane Imaris image analysis software for each replicate. Dots represent percent of fat body cells per size intervals. Statistics: P-values were obtained by comparing mean adipocyte sizes using t-test. (D) Quantitative analysis of buoyancy in wandering L3 larvae from Pvf3EY09531 and control uas-Pvf3 L3 larvae in N=6 independent experiments, from three crosses, each with experimental groups of twenty larvae. Statistics: Buoyancy (mean ± SD) was compared between groups using t-test. (E) Quantification of TG level normalized to total protein measured in Pvf3EY09531 , uas-Pvf3 (control), and Pvf3EY09531 ; He>uas-Pvf3 (rescue) L3 larvae. For each genotype N= 7 to 12 experiments from four crosses were analyzed, each with ten larvae per experimental group. Dots represents individual experimental groups. Statistics: Mean TG levels were compared using one-way ANOVA with Sidak’s correction for multiple group comparison. Red lines indicate mean values. (F) Analysis of BODIPY+ fat body cells size distribution in larvae from (E), in N=5 experiments per genotypes from three crosses, each with ten larvae per experimental group. Hundred fat body cells were examined using bitplane Imaris image analysis software for each replicate. Statistics: P-values were obtained by comparing mean adipocyte sizes using one-way ANOVA with Sidak’s correction for multiple group comparison as in (E). (G) Quantification of TG level normalized to total protein measured in L3 larvae from flies with fat body-specific RNAi of Pvr (the PVF receptor) and controls. For Pvf RNAi flies and controls N=8 and N=11 independent experiments, respectively, were performed, from four crosses, and with ten larvae per experiment. Dots represents individual experimental groups. Statistics: Mean TG levels were compared between the two genotypes using t-test. (H) Analysis of BODIPY+ fat body cell size distribution in L3 larvae from (G). N=4 experiments from four crosses, ten larvae per experiment, and hundred to hundred and fifty cells per replicate were analyzed using bitplane Imaris image analysis software. Dots represent percent of fat body cells per intervals of size. Statistics: Mean BODIPY+ fat body cell sizes were compared between the two genotypes using t-test. Red lines indicate mean values.
Macrophage-derived PDGFcc controls lipid storage in newborn and adult mice.
Among mouse Pvf orthologs, data from the ImmGen Consortium (66, 67) indicated that Platelet-Derived Growth Factor c (Pdgfc) is the most abundantly expressed PDGF/VEGF-family member in mouse fat-associated macrophages (Fig 4A). Pdgfc expression was upregulated in fat tissue of high-fat-diet-fed wild-ype and Ccr2-deficient mice as well as control-diet-fed hyperphagic Lepr–/– (db/db) mice in a CSF1R-dependent manner (Fig 4B). Additionally, Pdgfc expression was severely decreased in fat pads from 4-week-old Csf1rCre; Csf1rf/f mice (Fig 4C). We hypothesized that PDGFcc produced by macrophages may be the growth factor involved in adipocyte hypertrophy in mice. We therefore generated Csf1rCre; Pdgfcf/f mice (Fig 4 D to F and fig. S6 A to E). Adipocytes developed normally in Csf1rCre; Pdgfcf/f mice, based on Plin1 and Ucp1 expression (fig. S6E ), but white adipose tissue weight was decreased by 35% in 4-week-old Csf1rCre; Pdgfcf/f mice (Fig 4D) and adipocyte size and lipid content were reduced (Fig 4E). In contrast to Csf1r-deficient mice (fig. S3A), Csf1rCre; Pdgfcf/f mice have teeth and were not runted (Fig 4F and fig. S6A), which implied that macrophage functions were otherwise largely intact.
Figure 4. Macrophage-derived PDGFcc is required for adipocyte hypertrophy in mice.
(A) Normalized expression of Pdgf/Vegf family genes in murine macrophages from indicated tissues. Data were obtained from the Immgen database and the expression values were normalized by DESeq2. (B) qPCR analysis of Pdgfc transcripts normalized to Gapdh (2-ΔCt) in inguinal white adipose tissues (iWAT) from 14-week-old C57Bl/6J mice, Ccr2–/– mice, and 12-week-old Lepr–/– (db/db) mice all subjected to indicated diets and treatments for eight weeks. Fat pads from N= 8 to 15 mice from two to three independent experiments were analyzed for each group and experimental condition. Dots represent values for individual mice. Statistics: P-values were obtained by comparing mean expression values using one-way ANOVA with Sidak’s correction for multiple group comparison. Red lines indicate mean values. (C) qPCR analysis of Pdgfc transcripts, normalized to Gapdh (2-ΔCt), in iWAT of 4-week-old Csf1rCre; Csf1rf/f mice and littermate controls. Fat pads from five mice per genotype from three independent litters were analyzed. Dot represents values for individual mice. Statistics: P-values were obtained by comparing mean expression values between different genotypes using t-test. Red lines indicate mean values. (D) Weight of iWAT and interscapular brown adipose tissue (iBAT) from 4-week-old Csf1rCre; Pdgfcf/f mice and control littermates, N= 4 to 7 mice per genotype, from four litters. Dots represent individual mice. Statistics: P-values were obtained using one-way ANOVA with Sidak’s correction for multiple group comparison. Red lines indicate mean values. (E) Representative whole-mount images and quantitative analysis of adipocyte size of iWAT from Csf1rCre; Pdgfcf/f mice and control littermates at four weeks of life. Fat pads from N=5 mice from four independent litters were analyzed for each genotype. Hundred adipocytes were measured per fat pad using bitplane Imaris image analysis software, from three different areas. Dots represent percent of adipocytes per size interval. Scale bars are 40 μm. Statistics: P-values were obtained by comparing mean adipocyte sizes between different genotypes using t-test. (F) Representative photographs of teeth from N= 4 to 7 Csf1rCre; Pdgfcf/f mice and littermate controls at four weeks of life from four litters. (G) Weight gain of 12-week-old C57Bl/6J mice fed a 10% or 45% lipid diet and treated with anti-PDGFcc antibodies or Goat IgG for six weeks. N=9 mice per experimental group, from two independent experiments were analyzed. Mice were weighted weekly for six weeks. Dots and bars represent mean body weight ± SD. Statistics: P-values were obtained by comparing the mean body weights between experimental groups using two-way ANOVA with Sidak’s correction for multiple group comparison. (H) Weight of epididymal (eWAT) and inguinal (iWAT) fat pads from 12-week-old C57Bl/6J mice, N= 8 mice per experimental group, from two independent experiments. Dots represent individual mice. Statistics: P-values were obtained by comparing mean adipose tissue weights using one-way ANOVA with Sidak’s correction for multiple group comparison. Red lines indicate mean values. (I) Whole-mount images and quantitative analysis of adipocyte size for iWAT from 12-week-old C57Bl/6J mice. Fat pads from N= 9 mice from two independent experiments were analyzed for each group and experimental condition and hundred adipocytes were measured using bitplane Imaris image analysis software in three different areas per fat pad. Dots represent percent of adipocytes per size interval. Scale bars are 40 μm Statistics: P-values were obtained by comparing mean adipocyte sizes using one-way ANOVA with Sidak’s correction for multiple group comparison.
Furthermore, this suggested that the adipocyte phenotype was not compounded by a failure to thrive as is the case in mice with global Csf1r deficiency (fig. S3 A to D). To test the hypothesis that PDGFcc also controls lipid storage in adult mice, 6-week-old C57Bl/6J mice were treated with anti-PDGFcc-neutralizing antibodies (AF1447, 50 μg/mouse, i.p., three times a week) for 6 weeks (Fig 4 G to I and fig. S6 F to L). Mice treated with anti-PDGFcc antibodies and fed a high-fat diet remained lean (Fig 4G), with lower fat mass and smaller adipocytes than controls (Fig 4 H and I). Thus, the production of PDGFcc by macrophages appears to control lipid storage in the adipocytes of newborn mice. Additionally, PDGFcc blockade in adult mice prevents adipocyte hypertrophy in response to high-fat diet.
PDGFcc production by resident macrophages in response to high-fat diet in adults.
Flow cytometry, RNA-seq, and qPCR analyses revealed the presence of three main Csf1r-dependent F4/80+ myeloid subsets, large TIM4+ F4/80+ macrophages and two populations of small TIM4– F4/80+ cells, within visceral and sub-cutaneous white adipose tissue (Fig 5A and fig. S7 and S8). Pdgfc was preferentially expressed, and induced by high-fat diet, in white adipose tissue TIM4+ F4/80+ macrophages but not liver Kupffer cells or brown adipose tissue (iBAT) TIM4+ macrophages (Fig 5 B and C). By contrast, the production of Tnf and Il1b was restricted to small TIM4– F4/80+ cells, monocytes, and Kupffer cells (Fig 5C). White adipose tissue TIM4+ macrophages were present in Ccr2-deficient (52) and Flt3Cre; Csfr1rf/f (51, 53) mice, but absent in Tnfrsf11aCre; Csfr1rf/f mice (Fig 5D and fig. S2C and S4K). Whereas TIM4– F4/80+ monocyte/macrophages were absent or reduced in Ccr2-deficient (52) and Flt3Cre; Csfr1rf/f (51, 53) mice (Fig 5D and fig. S2C). Lineage-mapping analysis of Csf1r+ erythro-myeloid progenitors (EMP) labelled at embryonic day 8.5 showed post-natal labeling of large adipose tissue TIM4+ F4/80+ macrophages, but not of small TIM4– F4/80+ cells (Fig 5E and fig. S7D). Parabiosis experiments showed that TIM4+ F4/80+ cells do not exchange between parabionts in contrast to TIM4– F4/80+ cells and monocytes (Fig 5F). Thus, Pdgfc is preferentially expressed in embryo-derived white adipose tissue resident macrophages. Finally, immunofluorescence analysis confirmed that PDGFcc was undetectable in the fat pads of Tnfrsf11aCre; Csfr1rf/f mice (Fig 5G). Global transcriptional analysis (RNA-seq) further showed that TIM4+ and TIM4– cells cluster by population rather than diet in multidimensional scaling and hierarchical clustering of differentially expressed genes (fig. S8 and Data S1). Stable transcriptional signatures associated with TIM4+ resident macrophages primarily included homeostatic processes (Clusters 1 and 7) fig. S8 A to E), whereas TIM4– monocyte/macrophages highly expressed genes associated with innate immune response and cell proliferation (Clusters 2 and 5) (fig. S8 A to E). Thus, TIM4+ F4/80+ white adipose tissue resident macrophages (28–34) are the main PDGFcc-producing cells in mouse fat pads and respond to high-fat diet by increasing Pdgfc expression, in contrast to TIM4– F4/80+ Ccr2-dependent cells which respond to high-fat diet by expressing Tnf and Il1b.
Figure 5. Diet regulates PDGFcc production by adipose tissue resident macrophages.
(A) Representative tSNE color coded flow plot (Top) and May–Grunwald Giemsa staining (Bottom) of Csf1r-dependent F4/80+ macrophage subsets from inguinal white adipose tissue (iWAT) of C57Bl/6J mice. Fat pads from thirty wild-type mice were analyzed with similar results, see fig. S7. Scale bars are 10 μm. (B) qPCR analysis of Pdgfa, Pdgfb, Pdgfc, and Pdgfd transcripts, normalized to Gapdh and ActinB, in FACS-sorted iWAT macrophages from 4-week-old C57Bl/6J mice. N= 10 to 30 sorted macrophage samples from thirty mice were analyzed per group. Each dot represents a sorted sample from an individual mouse. Statistics: P-values were obtained by comparing mean expression values using one-way ANOVA with Sidak’s correction for multiple group comparison. Red lines indicate mean values. (C) qPCR analysis of Pdgfc, Tnf, and Il1b transcripts, normalized to Gapdh and ActinB, in iWAT macrophages, liver Kupffer cells, interscapular brown adipose tissue (iBAT) macrophages, and blood monocytes isolated by FACS from 14-week-old C57Bl/6J subjected to the indicated diets and treatments for eight weeks. N= 5 to 7 sorted macrophage samples and N=9 blood monocyte samples were analyzed per indicated treatment and diet. Each dot represents a sorted macrophage sample from an individual mouse. Statistics: P-values were obtained by comparing mean expression values using one-way ANOVA with Sidak’s correction for multiple group comparison. Red lines indicate mean values. (D) Representative tSNE color coded flow plot of Csf1r-dependent F4/80+ macrophage subsets from iWAT of Csf1rCre; Csf1rf/f mice, Flt3Cre; Csf1rf/f mice, Ccr2–/– mice, and Tnfrsf11aCre; Csf1rf/f mice. Fat pads from N= 4 to 9 mice per genotype were analyzed from at least three independent litters. (E) Analysis of YFP+ embryo-derived macrophages by flow cytometry of iWAT from 4-week-old Csf1rMeriCreMer; RosaLSL-YFP mice, pulse labeled at embryonic day (E) 8.5 with 4-hydroxy tamoxifen (OH-TAM). Fat pads from ten mice from eight independent litters were analyzed with similar results. (F) Analysis of macrophage dynamics in parabiotic pairs by flow cytometry of blood monocytes and adipose tissue macrophages from iWAT of CD45.1 / CD45.2 female parabiotic pairs maintained for eight weeks on 10% or 45% fat diet before analysis at 14-week-old. N=8 mice were analyzed, and each dot represents one mouse. (G) Fluorescent whole-mount images and quantitative analysis of PDGFcc+ F4/80+ cells from iWAT of 4-week-old Tnfrsf11aCre; Csf1rf/f mice and littermate control mice. Images are representative of N=5 fat pads from four independent litters. Scale bars are 40 μm. Fat pads from N=6 mice from four independent litters were analyzed for each genotype and cells were quantified from three different areas per fat pad. Each dot represents data from an individual mouse. Statistics: P-values were obtained by comparing mean values between different genotypes using t-test. Red lines indicate mean values.
PDGFcc blockade results in increased energy expenditure and hepatic storage.
To further investigate the consequence of PDGFcc blockade and the mechanisms that underlie leanness at the organismal level, we performed metabolic studies in high-fat-diet-fed mice. Food intake and fecal caloric density in anti-PDGFcc treated mice were comparable to controls (Fig 6 A and B and fig. S6H), indicating that PDGFcc blockade did not affect food intake or intestinal absorption, as observed similarly for mice treated with CSFR1 inhibitor (PLX5622) (fig. S1 H to J). In addition, PDGFcc blockade did not protect mice against high-fat-diet-induced hepatosteatosis (Fig 6C), inflammation, or insulin resistance (fig. S6 I to K). This was in sharp contrast to Ccr2-deficient mice and PLX-treated mice, which lack Ccr2-dependent inflammatory monocytes/macrophages and are protected against hepatosteatosis, inflammation, and insulin resistance (11, 15–18, 21) (fig. S 1 D to G and S 2 I to L). Thus, PDGFcc blockade decreases adiposity, without limiting food intake, intestinal absorption, or the metabolic syndrome, the latter being promoted by a different cell type (i.e. Ccr2-dependent monocyte/macrophages).
Figure 6. PDGFcc controls adipocyte hypertrophy and thermogenesis in mice.
(A, B) Cumulative food intake measured gravimetrically over 5 days (A) and fecal caloric density measured by bomb calorimetry on the last day of the experiment (B) in groups of 8-week-old C57Bl/6J mice fed a 45% lipid diet and treated with anti-PDGFcc antibodies or Goat IgG, N=8. Dots represent mean cumulative food intake ± SD in (A) and individual mice in (B). Statistics: Mean cumulative food intake and fecal caloric density were compared by t-test. (C) Representative H&E staining (Left) and quantification of triglycerides (TG) content per mg of tissue (right) of livers from 12-week-old C57Bl/6J mice (from Fig 4 G to I) fed a 10% or 45% lipid diet for six weeks and injected with anti-PDGFcc antibodies or with Goat IgG. N=8 mice per group, from two independent experiments were analyzed. Dots represent individual mice. Scale bars are 100 μm. Statistics: Mean values were compared by one-way ANOVA with Sidak’s correction for multiple group comparison. Red lines indicate mean values. (D) Quantification of TG content per mg of tissue from 4-week-old Csf1rcre; Pdgfcf/fmice and control littermates. N= 5 to 6 mice from three litters were analyzed. Dots represent individual mice. Statistics: Mean TG values were compared by t-test. Red lines indicate mean values. (E) Heatmap representation of Pdgfc, Plin1, Ucp1, Dio2, Prdm16, and Cidea transcripts expression (2-ΔCt, relative to Gapdh) in inguinal white adipose tissue (iWAT), epididymal white adipose tissue (eWAT), and interscapular brown adipose tissue (iBAT) from 12-week-old C57Bl/6J mice treated for six weeks with anti-PDGFcc antibodies or Goat IgG. Color scale is normalized to mean expression value of each transcript in fat tissues of mice fed a control diet (10% lipid) and treated with control Goat IgG. N= 5 to 10 mice per group were analyzed. Statistics: Mean expressions values were compared by one-way ANOVA with Sidak’s correction for multiple group comparison. * indicates P<0.01 as compared to control-diet-fed mice injected with Goat IgG controls, † indicates P<0.01 as compared to control-diet-fed mice injected with anti-PDGFcc neutralizing antibodies (⍺ PDGFcc), and # indicates P<0.001 as compared to high-fat-diet-fed (45% lipid) mice injected with Goat IgG controls. (F) Heatmap representation as in (E), from iWAT, eWAT, and iBAT from 14-week-old C57Bl/6J mice treated for eight weeks with PLX5622 or vehicle control. N= 5 to 10 mice per group were analyzed. Color scale is normalized as in (E) to transcripts in fat pads from controldiet-fed mice treated with vehicle control. Statistics: As in (E), * indicates P<0.01 as compared to control-diet-fed mice and treated with vehicle control, † indicates P<0.01 as compared to control-diet-fed mice and treated with PLX, @ indicates P <0.01 as compared to high-fat-diet-fed mice and treated with vehicle control. (G) Heatmap representation as in (E) from iWAT and iBAT from 4-week-old Csf1rcre; Csf1rf/f mice, Tnfrsf11acre; Csf1rf/f mice, Csf1rcre; Pdgfcf/f mice, and their littermate controls, N= 5 to 12 mice per group. Color scale is normalized to exoression in fat pads of Crenegative controls. Statistics: Mean expression values are compared by t-test. * indicates P<0.01 as compared to Cre-negative littermate controls. (H, I) Representative infrared images (H) and quantitative measurement of body temperature using the top 10% warmest pixels (I) of the interscapular area on day six of treatment in C57Bl/6J mice fed a 45% lipid diet and injected with anti-PDGFcc antibodies or Goat IgG, N=8 per condition. Dots represent individual mice. Statistics: Mean of experimental groups were compared by one-way ANOVA with Sidak’s correction for multiple group comparison. Red lines indicate mean values. (J, K) Analysis of Oxygen uptake (VO2) and Carbon Dioxide (VCO2) kinetics (J) and covariance corrected cumulative energy expenditure (EE, ANVOVA Corr.) (K) of C57Bl/6J mice fed a 45% lipid diet and injected with anti-PDGFcc antibodies or Goat IgG over a five-day period in a Promethion Metabolic Screening System. N=8 mice per experimental group were analyzed. Dots in J represent mean values over time, and dots in K represent individual mice. Statistics: Mean of experimental groups in K are compared using t-test. Red lines indicate mean values.
Six weeks of treatment with anti-PDGFcc under high-fat diet resulted in a 20% increase in ectopic storage of triglycerides in the liver as compared to controls (Fig 6C), although this was not observed in developing Csf1rCre; Pdgfcf/f mice (Fig 6D), suggesting that ectopic storage may not be the cause for reduced lipid storage in adipocytes. Indeed, although thermogenesis associated transcripts (68, 69) were not altered in the white adipose tissue (Fig 6 E to G), they were induced in the brown adipose tissue by high-fat diet and further increased by PDGFcc blockade and CSFR1 inhibition (Fig 6 E and F). Thermogenic transcripts were also induced in the brown adipose tissue from Csf1rCre; Pdgfcf/f mice as well as Csf1rCre; Csf1rf/f and Tnfrsf11aCre; Csf1rf/f mice (Fig 6G). Accordingly, PDGFcc blockade raised surface body temperature in the interscapular area (Fig 6 H and I) and increased total energy expenditure (Fig 6 J and K). Thus, reduced lipid storage in white adipocytes is associated with increased energy expenditure at the organismal level, at least in part in the form of thermogenesis by the brown adipose tissue, and with redistribution of excess lipids to the liver in conditions of high-fat diet. We therefore reasoned that PDGFcc may act either locally by promoting storage by adipocytes in a paracrine manner, and/or systemically by limiting expenditure/thermogenesis and lipid redistribution.
PDGFcc is required and sufficient for adipocyte hypertrophy in fat pad explants.
We next performed loss-of-function and rescue experiments in murine isolated fat pad explants. Wildtype neonatal (P4) murine epidydimal fat pad (eWAT) anlagen contain PDGFRα+ cells, capillaries and macrophages, but lack PLIN1+, BODIPY+ or LipidTox+ adipocytes (fig. S9A). PLIN1+ BODIPY+ adipocytes develop in eWAT explants cultured ex vivo for 10 days in conventional non-adipogenic medium (37) (Fig 7A and fig. S9B). We found that treatment of wildtype anlagen explants with anti-PDGFcc neutralizing antibodies (AF1447) resulted in the development of small PLIN1+ cells with little BODIPY + content, in comparison to anlagen treated with control Goat IgG (Fig 7 A to C and fig. S9C). Similarly, anlagen from Tnfrsf11aCre; Csf1rf/f mice also gave rise to small PLIN1+ cells that contain little to no lipids as evidenced by scant BODIPY staining as compared control littermates (Fig 7D to F and fig. S9D ). Addition of recombinant PDGFcc to Tnfrsf11aCre; Csf1rf/f fat-pads anlagen, however, rescued BODIPY staining and size of adipocytes to near wildtype levels (Fig 7 D to F ). These data demonstrate that PDGFcc produced by macrophages is required and sufficient for lipid storage ex vivo in isolated fat pads explants. PDGFcc appears to promote lipid storage at least in part via a paracrine effect on white adipocytes within fat pads. Thus, increased energy expenditure in vivo is likely to be secondary to the increased systemic availability of lipids that are not stored, although our data do not exclude the possibility that PDGFcc may also act systemically, for example on brown adipocytes to alter thermogenesis.
Figure 7. PDGFcc is both required and sufficient for adipocyte hypertrophy in isolated fat tissue.
(A-C) Representative whole-mount staining (A), quantitative analysis of BODIPY+ area (B), and quantitative analysis of adipocyte size (C), in wild-type epididymal white adipose tissue (eWAT) explants cultivated for ten days with Goat IgG or anti-PDGFcc neutralizing antibodies (⍺ PDGFcc) and stained for PERILIPIN (PLIN1), F4/80, and BODIPY, N=5. Dots represent individual explants in (B), and the % of adipocytes per size intervals in (C). For adipocyte size distribution, >10,000 cells were analyzed per explant using bitplane Imaris image analysis software. Scale bars are 20 μm. Statistics: Mean BODIPY+ area in (B) and adipocyte size in (C) were compared using t-test. Red lines indicate mean values. (D) Representative whole-mount staining of eWAT explants from Tnfrsf11aCre; Csf1rf/f mice and littermate Csf1rf/f controls. N= 10 to 15 explants from five litters were cultured for ten days ex vivo and stained for PLIN1, F4/80, and BODIPY. Dots represent individual explants. Scale bars are 20 μm. (E, F) Quantitative analysis of BODIPY+ area (E) and quantitative analysis of adipocyte size (F) using bitplane Imaris image analysis software in explants from Tnfrsf11aCre; Csf1rf/f mice and Csf1rf/f littermate controls. Explants were cultured with PBS or recombinant PDGFcc. N= 5 to 11 explants obtained from five litters were analyzed. Dots represent individual explants in (E) and the percent of adipocytes per intervals of size in (I). Statistics: Results from genotypes and treatment groups were compared by one-way ANOVA with Sidak’s correction for multiple group comparison. Red lines indicate mean values.
PDGFcc controls expression of negative regulators of lipid synthesis and storage.
PDGF-receptor signaling in mammals is complex and supports many cellular functions (70–74). To gain an initial insight into the molecular mechanisms that link PDGFcc to adipocyte hypertrophy, we performed an transcriptional analysis of inguinal white adipose tissue from eight-week-old wildtype mice fed a high-fat diet or a control diet and treated with anti-PDGFcc-neutralizing antibodies or Goat IgG for 2 weeks, a time point before dramatic changes in fat mass are observed. An unbiased analysis of hallmark pathways in iWAT of anti-PDGFcc antibody-treated mice and control animals using pre-ranked Gene Set Enrichment Analysis (GSEA) identified adipogenesis as the pathways most differentially regulated by PDGFcc blockade when controlling for diet effects (Fig 8A). By contrast, the hallmark fatty acid metabolism pathway which includes genes involved in lipid uptake, lipid oxidation, and lipid mobilization was not differentially regulated by anti-PDGFcc treatment (Data S2). Similarly, hallmark pathways glycolysis, MTORC1 signaling, or TGFb signaling were not differentially regulated by anti-PDGFcc treatment (Fig 8A and Data S2). An additional GSEA analysis of genes involved in lipid synthesis and storage (See Methods) identified a subset of genes up-regulated by PDGFcc blockade regardless of diet, which included several negative regulators of lipid synthesis and storage such as Insig1 (75), Ppara (76), and Idh2 (77) (Fig 8B) all of which are associated with obesity. These data altogether suggested that PDGFcc regulates lipid synthesis and storage, rather that lipid uptake, lipid mobilization, lipid oxidation, or extracellular matrix remodeling.
Figure 8. anti-PDGF regulates genes involved in lipid storage.
(A) Hallmark pathway analysis of RNAseq dataset obtained from whole inguinal white adipose tissues (iWAT) of 8-week-old mice treated with anti-PDGFcc antibodies or Goat IgG for two weeks, N=4 per condition. A selection of hallmark pathways significantly affected (FDR < 0.05) in untreated high-fat-diet-fed mice (HFU) as compared to untreated control-fat-fed mice (CFU), and in anti-PDGFcc antibody treated high-fat-diet-fed mice (HFT) as compared to untreated high-fat-diet-fed mice (HFU) are depicted. Statistics: P-values were estimated by the number of random gene sets with the same or more extreme enrichment scores divided by the total number of generated gene sets using the fgsea package. q-values were obtained via Benjamini-Hochberg multiple hypothesis correction. (B) Heatmap depicting row-based log2(fold change) z-scores is shown for leading-edge genes from the custom lipogenesis/lipid storage (See Methods) gene set in anti-PDGFcc antibody treated control-fat-fed mice (CFT) as compared to untreated control-fat-fed mice (CFU), anti-PDGFcc antibody treated high-fat-diet-fed mice (HFT) as compared to untreated high-fat-diet-fed mice (HFU), untreated high-fat-diet-fed mice group (HFU) as compared to untreated control-diet-fed mice group (CFU), and anti-PDGFcc antibody treated high-fat-diet-fed mice group (HFT) as compared to anti-PDGFcc antibody treated control-diet-fed mice group (CFT) are depicted. Crosses denote leading-edge genes. (C) qPCR analysis of Insig1 transcripts normalized to Perilipin (2-ΔCt) in iWAT from 14-week-old C57Bl/6J mice, Ccr2–/– mice, and 12-week-old Lepr–/– (db/db) mice all subjected to indicated diets and treatments for eight weeks. Fat pads from N=4 mice were analyzed for each group and experimental condition. Dots represent values for individual mice. Statistics: P-values were obtained by comparing mean expression values using one-way ANOVA with Sidak’s correction for multiple group comparison. Red lines indicate mean values. (D, E) qPCR analysis of Insig1 expression (2-ΔCt calculated relative to Perilipin) in explants from Csf1rf/f and Tnfrsf11aCre; Csf1rf/f littermates (D), and from wildtype explants cultured in the presence of Goat IgG or anti-PDGFcc neutralizing antibodies (⍺ PDGFcc) (E), as depicted in Fig 7. N=4 explants per experimental condition from two independent experiments were analyzed. Dots represent individual explants. Statistics: Results from (D) and (E) were compared using t-test. Red lines indicate mean values.
Insig1 restricts SREBP-mediated lipogenesis in mature adipocytes and blocks lipid storage (75), and Insig1-deficiency causes obesity in mice fed a control diet (78). We thus investigated the regulation of Insig1 by qPCR in white adipose tissue of wildtype, Ccr2-deficient, and Leptin receptor-deficient mice treated with PLX5622 or anti-PDGFcc blocking antibodies (Fig 8C). These experiments indicated that Insig1 is strongly upregulated in vivo by PLX5622 treatment and by PDGFcc blockade in all experimental models (Fig 8C). Additionally, a qPCR of isolated fat pad explants from Tnfrsf11aCre; Csf1rf/f mice and control littermates (Fig 8D), as well as wildtype mice treated with anti-PDGFcc neutralizing antibodies or Goat Ig (Fig 8E) indicated that Insig1 is upregulated by macrophage-deficiency and PDGFcc blockade in an adipose tissue autonomous manner. This analysis is consistent with the hypothesis that PDGFcc controls lipid synthesis and storage in white adipocytes.
Discussion
We report here observations which indicate that PDGF-family growth factors (PDGFcc and PVF3) produced by resident macrophages in response to dietary changes mediate the storage of lipids in adipocytes of hyperphagic and lipid-rich-diet-fed adult mice, developing mice, and Drosophila larvae. Based on our data, the lean phenotypes observed in the absence of macrophages or PDGFcc is unlikely to be due to decreased food intake, increased leptin signaling, intestinal malabsorption, or white adipose tissue browning. Rather, our findings indicate that the diet-regulated production of PDGFcc by macrophages is dispensable for adipocyte differentiation but controls their lipid content in a paracrine manner. Excess lipids which are not stored in the absence of PDGFcc appear to be redirected for thermogenesis or ectopic storage. This function of adipose-tissue resident macrophages (i.e. promoting lipid storage via the hypertrophy of specialized fat cells in response to nutrient intake) appears to be evolutionarily conserved in Drosophila and mice. Drosophila do not have fibroblasts or endothelial cells and Pvr silencing in fat-body cells phenocopies Pvf3 silencing in hemocytes, suggesting that the interaction between macrophages and fat cells may be a direct one at least in flies. Nevertheless, we acknowledge that PDGF-receptor signaling in mammals is complex and supports many cellular functions (70–74). Whether macrophages directly signal to adipocytes in mice, and which member(s) of the PDGF/VEGF-receptor family might mediate the effect of PDGFcc on mouse adipocytes are both unknown. An initial transcriptional analysis of white adipose tissue suggests that PDGFcc controls expression of negative regulators of lipid synthesis and storage, including Insig1, but more detailed studies are warranted. Overall, our data suggest that resident macrophages and specialized fat-storing cells constitute a functional unit in metazoans, whereby macrophages sense the organism’s nutritional state (79) and, when intake is high, signal to adipocytes for increased lipid storage through regulated paracrine production of PDGF-family growth factors (Fig 0).
Macrophage developmental heterogeneity frequently underlie specialized functions associated with distinct macrophage cell types. In the present study, our data clearly shows that Ccr2-independent TIM4+ resident macrophages are required and sufficient for lipid storage but not sufficient to promote diet-associated inflammation or insulin resistance. Whereas, TIM4– Ccr2-dependent monocyte/macrophages promote inflammation and the metabolic syndrome (11, 15–21), but are neither required nor sufficient for fat storage in white adipose tissue. Therefore our data do not support a model where obesity, inflammation, and insulin resistance in mice would all be controlled by the reprogramming of resident macrophages from a homeostatic into a pro-inflammatory role in the setting of metabolic stress. Instead our data strongly suggest that distinct developmental subsets (i.e. resident macrophages and recruited HSC derived monocyte/macrophages) perform distinct functions in the adipose tissue “niche” and are targeted independently by CCR2 and PDGFcc blockade. Altogether, the present study identifies a mechanism that controls adiposity (i.e. the expansion of fat stores) in metazoans in the context of a cellular and molecular “dissection” of macrophage functions, which may help explain and predict the effect of genetic variants and pharmacological interventions on lipid storage.
Supplementary Material
Acknowledgements.
This study is dedicated to the memory of Lucile Crozet. The authors thank J. Frampton, J. Pollard, and T. Boehm, for providing mouse strains, H. Ketchum for her technical assistance with the Csf1r–/– mice, and the molecular cytology core at MSKCC for their help with preparation of histological samples. The authors acknowledge all members of the Geissmann lab for helpful suggestions and editing the manuscript.
Funding:
This work was supported by NIH/NCI P30CA008748 to MSKCC, NIH/NIAID 1R01AI130345, NIH/NHLBI R01HL138090, Ludwig institute for Cancer research basic immunology grant and Cycle for Survival grants to F.G. This work was also supported by Leducq transatlantic network of excellence to C.K.G. and F.G., NIH/NCI F32CA225036 to N.C. and Alan and Sandra Gerry Metastasis and Tumor Ecosystems Center fellowship to PL.L.
Footnotes
Competing interests: F.G. and N.C. are co-inventors on a patent relating to use of PDGFcc, CSF1R, and CCR2 inhibitors in obesity. J.B.W. is a compensated consultant for SpringWorks Therapeutics.
Data and materials availability:
The mice and reagents are made available upon request. The raw data are available with the manuscript.
References
- 1.Rosen ED, Spiegelman BM, Adipocytes as regulators of energy balance and glucose homeostasis. Nature 444, 847–853 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rodeheffer MS, Birsoy K, Friedman JM, Identification of white adipocyte progenitor cells in vivo. Cell 135, 240–249 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Hotamisligil GS, Foundations of Immunometabolism and Implications for Metabolic Health and Disease. Immunity 47, 406–420 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wise A, Adipocyte number and size in hypothalamic obesity induced in weanling mice by gold thioglucose and bipiperidyl mustard. Nutr Metab 19, 291–298 (1975). [DOI] [PubMed] [Google Scholar]
- 5.Hirsch J, Batchelor B, Adipose tissue cellularity in human obesity. Clin Endocrinol Metab 5, 299–311 (1976). [DOI] [PubMed] [Google Scholar]
- 6.McLaughlin T, Ackerman SE, Shen L, Engleman E, Role of innate and adaptive immunity in obesity-associated metabolic disease. J Clin Invest 127, 5–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hotamisligil GS, Arner P, Caro JF, Atkinson RL, Spiegelman BM, Increased adipose tissue expression of tumor necrosis factor-alpha in human obesity and insulin resistance. J Clin Invest 95, 2409–2415 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hotamisligil GS, Shargill NS, Spiegelman BM, Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science 259, 87–91 (1993). [DOI] [PubMed] [Google Scholar]
- 9.Weisberg SP et al. , Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest 112, 1796–1808 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu H et al. , Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest 112, 1821–1830 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weisberg SP et al. , CCR2 modulates inflammatory and metabolic effects of high-fat feeding. J Clin Invest 116, 115–124 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suganami T, Ogawa Y, Adipose tissue macrophages: their role in adipose tissue remodeling. Journal of leukocyte biology 88, 33–39 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Surmi BK, Hasty AH, Macrophage infiltration into adipose tissue: initiation, propagation and remodeling. Future Lipidol 3, 545–556 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pekala P, Kawakami M, Vine W, Lane MD, Cerami A, Studies of insulin resistance in adipocytes induced by macrophage mediator. J Exp Med 157, 1360–1365 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J et al. , Silencing CCR2 in Macrophages Alleviates Adipose Tissue Inflammation and the Associated Metabolic Syndrome in Dietary Obese Mice. Mol Ther Nucleic Acids 5, e280 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan TJ et al. , Experimental evidence for the use of CCR2 antagonists in the treatment of type 2 diabetes. Metabolism 62, 1623–1632 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Obstfeld AE et al. , C-C chemokine receptor 2 (CCR2) regulates the hepatic recruitment of myeloid cells that promote obesity-induced hepatic steatosis. Diabetes 59, 916–925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito A et al. , Role of CC chemokine receptor 2 in bone marrow cells in the recruitment of macrophages into obese adipose tissue. J Biol Chem 283, 35715–35723 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Kanda H et al. , MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J Clin Invest 116, 1494–1505 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamei N et al. , Overexpression of monocyte chemoattractant protein-1 in adipose tissues causes macrophage recruitment and insulin resistance. J Biol Chem 281, 26602–26614 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Parker R et al. , CC chemokine receptor 2 promotes recruitment of myeloid cells associated with insulin resistance in nonalcoholic fatty liver disease. Am J Physiol Gastrointest Liver Physiol 314, G483–G493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satoh T et al. , Critical role of Trib1 in differentiation of tissue-resident M2-like macrophages. Nature 495, 524–528 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Pridans C et al. , Pleiotropic Impacts of Macrophage and Microglial Deficiency on Development in Rats with Targeted Mutation of the Csf1r Locus. J Immunol 201, 2683–2699 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valdearcos M et al. , Microglia dictate the impact of saturated fat consumption on hypothalamic inflammation and neuronal function. Cell Rep 9, 2124–2138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valdearcos M et al. , Microglial Inflammatory Signaling Orchestrates the Hypothalamic Immune Response to Dietary Excess and Mediates Obesity Susceptibility. Cell Metab 26, 185–197 e183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pirzgalska RM et al. , Sympathetic neuron-associated macrophages contribute to obesity by importing and metabolizing norepinephrine. Nat Med 23, 1309–1318 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaitin DA et al. , Lipid-Associated Macrophages Control Metabolic Homeostasis in a Trem2-Dependent Manner. Cell 178, 686–698 e614 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulz C et al. , A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Gomez Perdiguero E et al. , Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mass E et al. , Specification of tissue-resident macrophages during organogenesis. Science 353, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hassnain Waqas SF et al. , Adipose tissue macrophages develop from bone marrow-independent progenitors in Xenopus laevis and mouse. Journal of leukocyte biology 102, 845–855 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeyda M et al. , Newly identified adipose tissue macrophage populations in obesity with distinct chemokine and chemokine receptor expression. Int J Obes (Lond) 34, 1684–1694 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Silva HM et al. , Vasculature-associated fat macrophages readily adapt to inflammatory and metabolic challenges. J Exp Med 216, 786–806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coats BR et al. , Metabolically Activated Adipose Tissue Macrophages Perform Detrimental and Beneficial Functions during Diet-Induced Obesity. Cell Rep 20, 3149–3161 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kratz M et al. , Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab 20, 614–625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pollard JW, Trophic macrophages in development and disease. Nat Rev Immunol 9, 259–270 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han J et al. , The spatiotemporal development of adipose tissue. Development 138, 5027–5037 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Debels H et al. , Macrophages play a key role in angiogenesis and adipogenesis in a mouse tissue engineering model. Tissue Eng Part A 19, 2615–2625 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei S et al. , Modulation of CSF-1-regulated post-natal development with anti-CSF-1 antibody. Immunobiology 210, 109–119 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Lee YH, Petkova AP, Granneman JG, Identification of an adipogenic niche for adipose tissue remodeling and restoration. Cell Metab 18, 355–367 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wernstedt Asterholm I et al. , Adipocyte inflammation is essential for healthy adipose tissue expansion and remodeling. Cell Metab 20, 103–118 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biswas SK, Mantovani A, Orchestration of metabolism by macrophages. Cell Metab 15, 432–437 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Pang C et al. , Macrophage infiltration into adipose tissue may promote angiogenesis for adipose tissue remodeling in obesity. Am J Physiol Endocrinol Metab 295, E313–322 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hume DA, The Many Alternative Faces of Macrophage Activation. Front Immunol 6, 370 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lumeng CN, Deyoung SM, Bodzin JL, Saltiel AR, Increased inflammatory properties of adipose tissue macrophages recruited during diet-induced obesity. Diabetes 56, 16–23 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Nguyen MT et al. , A subpopulation of macrophages infiltrates hypertrophic adipose tissue and is activated by free fatty acids via Toll-like receptors 2 and 4 and JNK-dependent pathways. J Biol Chem 282, 35279–35292 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Odegaard JI et al. , Alternative M2 Activation of Kupffer Cells by PPARδ Ameliorates Obesity-Induced Insulin Resistance. Cell Metabolism 7, 496–507 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolf Y et al. , Brown-adipose-tissue macrophages control tissue innervation and homeostatic energy expenditure. Nat Immunol 18, 665–674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sudo T et al. , Functional hierarchy of c-kit and c-fms in intramarrow production of CFU-M. Oncogene 11, 2469–2476 (1995). [PubMed] [Google Scholar]
- 50.Percin GI et al. , CSF1R regulates the dendritic cell pool size in adult mice via embryo-derived tissue-resident macrophages. Nature Communications 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacome-Galarza CE et al. , Developmental origin, functional maintenance and genetic rescue of osteoclasts. Nature, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boring L et al. , Impaired monocyte migration and reduced type 1 (Th1) cytokine responses in C-C chemokine receptor 2 knockout mice. The Journal of Clinical Investigation 100, 2552–2561 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Christensen JL, Weissman IL, Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proceedings of the National Academy of Sciences of the United States of America 98, 14541–14546 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woodcock KJ et al. , Macrophage-derived upd3 cytokine causes impaired glucose homeostasis and reduced lifespan in Drosophila fed a lipid-rich diet. Immunity 42, 133–144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Franz A, Wood W, Martin P, Fat Body Cells Are Motile and Actively Migrate to Wounds to Drive Repair and Prevent Infection. Dev Cell 44, 460–470 e463 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bruckner K et al. , The PDGF/VEGF receptor controls blood cell survival in Drosophila. Dev Cell 7, 73–84 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Reis T, Van Gilst MR, Hariharan IK, A buoyancy-based screen of Drosophila larvae for fat-storage mutants reveals a role for Sir2 in coupling fat storage to nutrient availability. PLoS Genet 6, e1001206 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clark RI, Woodcock KJ, Geissmann F, Trouillet C, Dionne MS, Multiple TGFbeta superfamily signals modulate the adult Drosophila immune response. Curr Biol 21, 1672–1677 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gyoergy A et al. , Tools Allowing Independent Visualization and Genetic Manipulation of Drosophila melanogaster Macrophages and Surrounding Tissues. G3 (Bethesda) 8, 845–857 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kambris Z et al. , Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr Biol 16, 808–813 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Hoch RV, Soriano P, Roles of PDGF in animal development. Development 130, 4769–4784 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Brogiolo W et al. , An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol 11, 213–221 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Zheng H et al. , Premature remodeling of fat body and fat mobilization triggered by platelet-derived growth factor/VEGF receptor in Drosophila. FASEB J 31, 1964–1975 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Zettervall CJ et al. , A directed screen for genes involved in Drosophila blood cell activation. Proceedings of the National Academy of Sciences of the United States of America 101, 14192–14197 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leader DP, Krause SA, Pandit A, Davies SA, Dow JAT, FlyAtlas 2: a new version of the Drosophila melanogaster expression atlas with RNA-Seq, miRNA-Seq and sex-specific data. Nucleic Acids Res 46, D809–D815 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shay T et al. , Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proceedings of the National Academy of Sciences of the United States of America 110, 2946–2951 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heng TS, Painter MW, Immunological Genome Project C, The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 9, 1091–1094 (2008). [DOI] [PubMed] [Google Scholar]
- 68.Enerback S et al. , Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature 387, 90–94 (1997). [DOI] [PubMed] [Google Scholar]
- 69.de Jesus LA et al. , The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue. J Clin Invest 108, 1379–1385 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Onogi Y et al. , PDGFRbeta Regulates Adipose Tissue Expansion and Glucose Metabolism via Vascular Remodeling in Diet-Induced Obesity. Diabetes 66, 1008–1021 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Gao Z, Daquinag AC, Su F, Snyder B, Kolonin MG, PDGFRalpha/PDGFRbeta signaling balance modulates progenitor cell differentiation into white and beige adipocytes. Development 145, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun C, Berry WL, Olson LE, PDGFRalpha controls the balance of stromal and adipogenic cells during adipose tissue organogenesis. Development 144, 83–94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Haider N, Dusseault J, Larose L, Nck1 Deficiency Impairs Adipogenesis by Activation of PDGFRalpha in Preadipocytes. iScience 6, 22–37 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vaziri C, Faller DV, Down-regulation of platelet-derived growth factor receptor expression during terminal differentiation of 3T3-L1 pre-adipocyte fibroblasts. J Biol Chem 271, 13642–13648 (1996). [DOI] [PubMed] [Google Scholar]
- 75.Li J, Takaishi K, Cook W, McCorkle SK, Unger RH, Insig-1 “brakes” lipogenesis in adipocytes and inhibits differentiation of preadipocytes. Proceedings of the National Academy of Sciences of the United States of America 100, 9476–9481 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Costet P et al. , Peroxisome proliferator-activated receptor alpha-isoform deficiency leads to progressive dyslipidemia with sexually dimorphic obesity and steatosis. J Biol Chem 273, 29577–29585 (1998). [DOI] [PubMed] [Google Scholar]
- 77.Lee JH et al. , Isocitrate dehydrogenase 2 protects mice from high-fat diet-induced metabolic stress by limiting oxidative damage to the mitochondria from brown adipose tissue. Exp Mol Med 52, 238–252 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carobbio S et al. , Adaptive changes of the Insig1/SREBP1/SCD1 set point help adipose tissue to cope with increased storage demands of obesity. Diabetes 62, 3697–3708 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Remmerie A, Scott CL, Macrophages and lipid metabolism. Cell Immunol 330, 27–42 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maeda K et al. , Wnt5a-Ror2 signaling between osteoblast-lineage cells and osteoclast precursors enhances osteoclastogenesis. Nat Med 18, 405–412 (2012). [DOI] [PubMed] [Google Scholar]
- 81.Dai XM et al. , Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood 99, 111–120 (2002). [DOI] [PubMed] [Google Scholar]
- 82.Georg Gasteiger XF, Stanislav Dikiy, Sue Y. Lee, Alexander Y. Rudensky, Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 350 981 – 985 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ayala JE et al. , Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Dis Model Mech 3, 525–534 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Loening AM, Gambhir SS, AMIDE: a free software tool for multimodality medical image analysis. Mol Imaging 2, 131–137 (2003). [DOI] [PubMed] [Google Scholar]
- 85.Orr JS, Kennedy AJ, Hasty AH, Isolation of adipose tissue immune cells. J Vis Exp, e50707 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dobin A et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heinz S et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Law CW, Chen Y, Shi W, Smyth GK, voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ritchie ME et al. , A comparison of background correction methods for two-colour microarrays. Bioinformatics 23, 2700–2707 (2007). [DOI] [PubMed] [Google Scholar]
- 91.Ritchie ME et al. , Empirical array quality weights in the analysis of microarray data. BMC Bioinformatics 7, 261 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tripathi S et al. , Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe 18, 723–735 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hoffman GE, Schadt EE, variancePartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics 17, 483 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yates AD et al. , Ensembl 2020. Nucleic Acids Res 48, D682–D688 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chen S, Zhou Y, Chen Y, Gu J, fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liao Y, Smyth GK, Shi W, featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 97.Ritchie ME et al. , limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Korotkevich VSG, Sergushichev A, Fast gene set enrichment analysis. bioRxiv 060012 [ Preprint ]. 22 October 2019. [Google Scholar]
- 99.Liberzon A et al. , The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mitchell JA et al. , Gene indexing: characterization and analysis of NLM’s GeneRIFs. AMIA Annu Symp Proc, 460–464 (2003). [PMC free article] [PubMed] [Google Scholar]
- 101.Tennessen JM, Barry WE, Cox J, Thummel CS, Methods for studying metabolism in Drosophila. Methods 68, 105–115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mice and reagents are made available upon request. The raw data are available with the manuscript.