Abstract
N -glycolylneuraminic acid (Neu5Gc), and its precursor N-acetylneuraminic acid (Neu5Ac), commonly referred to as sialic acids, are two of the most common glycans found in mammals. Humans carry a mutation in the enzyme that converts Neu5Ac into Neu5Gc, and as such, expression of Neu5Ac can be thought of as a ‘human specific’ trait. Bacteria can utilize sialic acids as a carbon and energy source and have evolved multiple ways to take up sialic acids. In order to generate free sialic acid, many bacteria produce sialidases that cleave sialic acid residues from complex glycan structures. In addition, sialidases allow escape from innate immune mechanisms, and can synergize with other virulence factors such as toxins. Human-adapted pathogens have evolved a preference for Neu5Ac, with many bacterial adhesins, and major classes of toxin, specifically recognizing Neu5Ac containing glycans as receptors. The preference of human-adapted pathogens for Neu5Ac also occurs during biosynthesis of surface structures such as lipo-oligosaccharide (LOS), lipo-polysaccharide (LPS) and polysaccharide capsules, subverting the human host immune system by mimicking the host. This review aims to provide an update on the advances made in understanding the role of sialic acid in bacteria-host interactions made in the last 5–10 years, and put these findings into context by highlighting key historical discoveries. We provide a particular focus on ‘molecular mimicry’ and incorporation of sialic acid onto the bacterial outer-surface, and the role of sialic acid as a receptor for bacterial adhesins and toxins.
Keywords: sialic acid, N-acetylneuraminic acid, Neu5Ac, N-glycolylneuraminic acid, Neu5GC, adherence, adhesin, molecular mimicry, bacterial pathogen
Introduction
Sialic acids are members of the nonusolonic acids, nine-carbon sugars found in both eukaryotes and prokaryotes [1, 2]. N-glycolylneuraminic acid (Neu5Gc), and its precursor N-acetylneuraminic acid (Neu5Ac), are the two most abundant sialic acids found in mammals (Fig. 1). Although Neu5Gc is present in most mammals, humans have a mutation in the enzyme CMP-Neu5Ac hydroxylase (CMAH) [3–6], which converts Neu5Ac to Neu5Gc. Consequently, humans are only able to produce Neu5Ac de novo, although several studies have demonstrated that trace levels of Neu5Gc is present in human tissues [7–10]. It is thought that the Neu5Gc present in normal human tissue may come from dietary sources such as red meat and dairy products [10, 11], although novel pathways have been proposed that may reactivate the defective CMAH protein under certain conditions [12]. Neu5Ac is present in every human cell type, and it is involved in cell–cell adhesion, signalling and immunity, discriminating ‘self from ‘non-self’ [13]. Additionally, Neu5Ac plays a key role in mucus, where the high abundance of negative charges provides protection against bacterial adhesion and colonization [14].
Fig. 1.
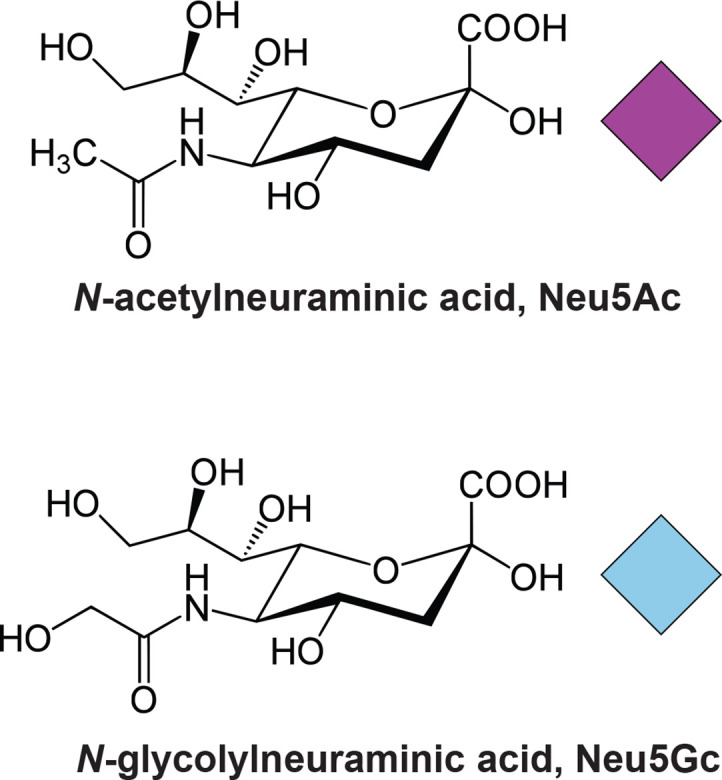
Structures of N-acetylneuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc), and the graphical representation of these glycans as per SNFG guidelines.
A number of bacterial species are able to directly synthesize Neu5Ac, including Escherichia coli [15–17], Campylobacter jejuni [18], Neisseria meningitidis [19], Streptococcus agalactiae [20] and Fusobacterium nucleatum [21]. In E. coli , two genes, neuB and neuC, are required for Neu5Ac biosynthesis [22–24]. NeuB catalyses the formation of Neu5Ac from N-acetyl-d-mannosamine and phosphoenolpyruvate [25], with NeuC responsible for the biosynthesis of the precursor of Neu5Ac, N-acetyl-d-mannosamine.
Bacteria are also able to synthesize additional nonusolonic acids (NulOs) that are structurally similar to sialic acid. Bacterial NulOs include Legionaminic acid, Pseudaminic acid, Fusaminic acid and Acinetaminic acid, with naming conventions typically following from the species they were first observed in; for example, Legionaminic acid was first observed in Legionella pneumophila [26], responsible for the respiratory infection Legionnaires disease, and Acinetaminic acid in the multi-drug-resistant pathogen Acinetobacter baumannii [27]. The structure, biosynthesis, and role of these NulOs were thoroughly reviewed recently [2], and as such, this review will focus on the role of Neu5Ac (‘sialic acid’) in bacterial–host interactions.
A quick trim isn’t good for the host: sialidases enhance bacterial virulence
Sialidases, also called neuraminidases, are enzymes that cleave sialic acid residues from complex glycan structures. This released sialic acid can then be acquired by bacteria to serve as a nutrient source, allow evasion of the immune response and the exposed underlying glycan structures may serve as points of adherence for bacterial cells. Action of viral sialidases, for example from Influenza A virus, can also serve to increase bacterial virulence; this may have been the case for the 1918 ‘Spanish flu’ pandemic, with many fatalities a result of secondary bacterial infections [28], mainly Streptococcus pneumoniae . The viral neuraminidase cleaves sialic acid from host glycan structures, increasing sialic acid and sialylated mucin availability. This promotes pneumococcal growth, proliferation and migration to the lungs [29]. S. pneumoniae itself expresses multiple neuraminidases, NanA, NanB and NanC, with diverse roles in pathobiology. NanA removal of terminal sialic acid from human glycan structures results in the exposure of galactose, which promotes pneumococcal biofilm formation [30]. NanA cleavage of sialic acid from the T-antigen [31], can lead to pneumococcal haemolytic uremic syndrome (HUS). NanA activity also modifies the immune response. For example, NanA desialylation of platelets can lead to platelet hyper-reactivity [32], leading to increased interaction with leukocytes, and increased inflammation; NanA cleavage of sialic acid activates TGF-β signalling leading to increased permeability of blood-brain-barrier and promoting meningitis [33]. A similar immune system modification effect is seen in the pig pathogen Glaesserella parasuis where desialylation leads to negative regulation of the lectin (glycan-binding protein) siglec-5 [34].
Synergy between multiple sialidases, and sialidases and other virulence factors, also potentiates virulence. For example, concerted action of the pneumococcal sialidases NanA and NanB leads to mucus escape – secreted NanB results in increased expression of NanA due to Neu5Ac stimulation of nanA transcription [35]. Expression of NanA results in increased cleavage of mucus, allowing S. pneumoniae to escape mucus binding and mucociliary clearance [35]. In Vibrio cholerae, the neuraminidase VcN hydrolyses sialic acids from complex gangliosides in the human gut, resulting in high amounts of the underlying GM1 structure. GM1 is the receptor for cholera toxin, Ctx [36, 37]. Thus, VcN-mediated cleavage of sialic acids from gangliosides potentiates the virulence of Ctx in the induction of diarrhoea.
Gardnerella vaginalis , a causative agent of bacterial vaginosis, employs multiple sialidases during pathogenesis [38, 39], with the activity of sialidases contributing to cellular invasion [40]. Similarly, the sialidase of the periodontal pathogen Porphyromonas gingivalis is required for attachment and invasion of oral epithelial cells by both P. gingivalis and other periodontal pathogens [41].
The sialidase NanI is important in Clostridium perfringens intestinal colonization [42] and supports survival in the presence of mucin [43], with NanI showing preference for glycan structures terminating in Neu5Ac [44]. Sialidases also allow the use of Neu5Ac as carbon sources in multiple Mycoplasma species [45, 46], potentiating colonization and persistence.
To snatch and dispatch: acquisition of sialic acid from the host, and consumption as a carbon source
Excellent reviews have been written recently describing bacterial sialic acid uptake [47], and how sialic acid is used as a carbon source [48]. Often, sialic acid released by sialidases/neuraminidases is used as a carbon source.
Multiple mechanisms exist in the bacterial domain to take up sialic acid from the environment. The first sialic acid transporter to be described was NanT in E. coli , a proton-coupled secondary transporter [49]. NanT protein homologues have been observed in multiple bacterial organisms, including Yersinia pestis , Tannerella forsythia and Bacteroides fragilis [47]. A further secondary transporter, named SiaT [47], a sodium solute symporter, exists in bacterial pathogens such as Clostridium perfringens [50] and Staphylococcus aureus [51]. Bacterial TRAP (tripartite ATP-independent periplasmic) transporters, made up of periplasmic substrate binding proteins and a cognate inner-membrane transporter complex [52] are also able to transport sialic acid. TRAP transporters are classed as secondary transporters as they use ion-electrochemical gradients to transport substrates [52]. The SiaPT TRAP transporter from Haemophilus influenzae was the first sialic acid TRAP transporter to be discovered [53, 54], is regulated by SiaR [55] and is essential for H. influenzae virulence [56]. Homologues of this system have been described in a diverse range of bacterial pathogens, including Vibrio cholerae [57] and Pasteurella multocida [58]. Like TRAP transporters, ABC (ATP-binding cassette) transporters also consist of periplasmic solute binding proteins and a cognate inner-membrane transporter complex. However, unlike TRAP transporters, ABC systems use ATP to directly transport solutes, and are classed as primary transporters [52]. Both Gram-negative and Gram-positive bacterial pathogens encode sialic acid ABC transporters, including Haemophilus ducreyi , and multiple Clostridial and Streptococcal species [59].
Following uptake, bacteria can use sialic acid in multiple ways. In addition to using sialic acid as a building-block for surface structures (reviewed in detail later in this review) a number of bacteria, both pathogens and commensals, use sialic acid directly as a carbon source. Catabolism of sialic acids, both Neu5Ac and Neu5Gc, results in formation of fructose-6-phosphate, which can then feed into glycolysis. The diversity of the systems for sialic acid catabolism and their roles are great and varied, and have been reviewed previously [48, 60]. However, a number of new examples have been described recently. V. cholerae motility is triggered by the presence of free Neu5Ac, and the sugar N-acetylglucosamine, which has a NHCOCH3 modification of the second carbon of the glucose ring. Catabolism of both these sugars into glucosamine-6-phosphate is the essential step in inducing motility [61]. The gut commensal Ruminococcus gnavus utilizes mucin as a nutrient source, with expression of multiple sialidases key to this utilization [62]. This illustrates that in addition to the pathogens described in the sialidase section above, members of the normal gut flora also utilise sialidases to release sialic acid as a nutrient source. Transcription of sialic acid catabolic genes is repressed by the NanR transcription factor in Corynebacterium glutamicum [63]. NanR is almost ubiquitously found in bacterial species that are able to catabolize sialic acid, indicating a possible common origin of the ability to break down sialic acids.
A wolf in sheep’s clothing: immune evasion and molecular mimicry
In addition to being able to cleave sialic acid to enhance virulence and to use free sialic acids as carbon and energy sources, many bacterial pathogens use sialic acids as a cell-surface decoration, and as a structural building block.
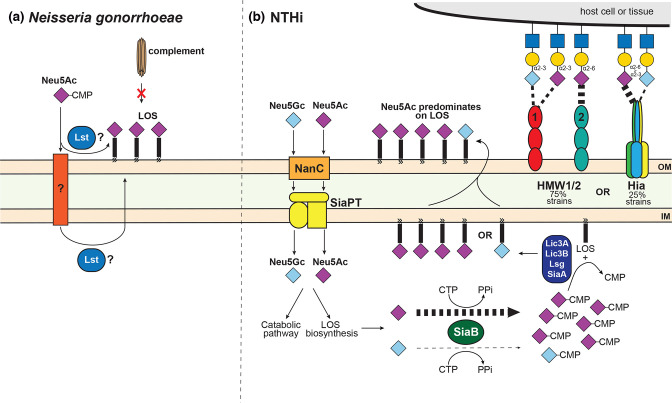
The key role for sialic acid in bacterial pathogenesis was first described in the sexually transmitted pathogen Neisseria gonorrhoeae, when it was discovered that CMP-Neu5Ac was the mystery substance that was acquired from the host, by a still as yet unknown mechanism, to render N. gonorrhoeae serum resistant [64, 65]. Addition of Neu5Ac in N. gonorrhoeae occurs at the terminus of lipo-oligosaccharide, LOS, a truncated version of LPS that lacks the extended O-antigen, with addition catalysed by the sialyltransferase Lst. Whether Lst is located in the cytoplasm or extracellularly is still not clear (Fig. 2). Neu5Ac addition to the LOS of N. gonorrhoeae prevents killing by complement [66]. Interestingly, addition of non-sialic acid NulOs to N. gonorrhoeae LOS by Lst resulted in an increased sensitivity to serum [67], illustrating the exquisite requirement of sialic acid for mediating this resistance and providing some insights into the mechanism of serum resistance. The increased Neu5Ac addition to the bacterial surface, which results in recruitment of more Factor H, inhibits the alternative complement pathway and decreases activation of the classical complement pathway [67]. N. gonorrhoeae LOS can also be sialylated on inner heptose residues, with loss of Neu5Ac on these residues attenuating virulence in a mouse model [68, 69]. A second aspect of sialic on the bacterial surface is the ‘cloaking’ effect of expressing molecules identical to host glycans. Such ‘molecular mimicry’ [70] of host structures allows bacterial pathogens to subvert the host immune response and survive.
Fig. 2.
The role of sialic acid in the pathobiology of (a) Neisseria gonorrhoeae and (b) nontypeable Haemophilus influenzae (NTHi). (a) The role of sialic acid in serum resistance was first described in Neisseria gonorrhoeae , with Neu5Ac-CMP the host-acquired molecule, which resulted in this resistance. Addition of Neu5Ac to the LOS of N. gonorrhoeae is catalysed by the enzyme Lst, with Neu5Ac-CMP transported by an as yet unidentified transporter. Whether Lst acts extracellularly, or in the cytoplasm, is also unclear. Incorporation of Neu5Ac into LOS also allows ‘molecular mimicry’ of the host by adding the human specific sugar to a key bacterial surface feature, masking the bacteria from the immune response. (b) NTHi is one of the best studied bacterial pathogens as regards use of sialic acid during pathobiology, and displays several examples of evolution of host-adaptation. Whist NTHi can take up both Neu5Ac and Neu5Gc with equal affinity via NanC and the TRAP transporter SiaPT, NTHi shows a marked preference for decorating LOS with Neu5Ac, with the bias being at activation of sialic acids for addition to LOS; the enzyme SiaB, which adds a CMP to sialic acid, shows 4000-fold higher affinity for Neu5Ac over Neu5Gc. In addition, the major NTHi adhesins HMW1/2 and Hia have both evolved to interact specifically with sialic acid containing receptors in the human airway, despite never being found in the same strains of NTHi. HMW 1 shows equal affinity for both α2,3-sialyllactosamine (2–3 SLN) containing either Neu5Ac or Neu5Gc, whereas HMW2 shows high affinity for α2,6-sialyllactosamine (2–6 SLN) containing Neu5Ac. Hia shows a marked preference for 2–6 SLN containing Neu5Ac, but also binds 2–6 SLN containing Neu5Gc, and 2–3 SLN. This convergent evolution of different adhesins for the same receptor means that all NTHi strains can colonize the human airway. The weight of the dashed line indicates specificity for particular sialic acid-containing structures.
The human-adapted pathogen non-typeable Haemophilus influenzae (NTHi), like N. gonorrhoeae , is a human-specific pathogen where the role of sialic acid in LOS has been extensively studied. Addition of sialic acid to the terminal position of LOS in NTHi promotes survival in vivo [71], and can occur via multiple enzymatic activities, including SiaA, LsgB, Lic3A and Lic3B. Growth in the presence of sialic acids also results in a decrease of complement-mediated killing; in this instance, increased sialic acid addition to LOS prevents binding of IgM antibodies, which results in decreased activation of the classical complement pathway [72]. In addition to using Neu5Ac as a substrate, LsgB can add the sugar ketodeoxyoctanoate (KDO) as a terminal structure of LOS [73]. The addition of KDO occurs when the concentration of free Neu5Ac is low, for example in certain in vivo nutrient-restricted niches and growth in a biofilm [73], and highlights the complex nature of NTHi LOS in immune-evasion. The underlying glycan structures of sialylated NTHi LOS also serve to protect the bacterium from serum IgM and complement [74]. By selectively inhibiting LOS sialyation, it is possible to increase serum sensitivity of NTHi, indicating a possible future treatment strategy for a major human pathogen [75]. Further, in an example of exquisite host-adaptation, NTHi has evolved preferential use of Neu5Ac over Neu5Gc [76] in decorating its LOS. Both Neu5Gc and Neu5Ac are taken up and used as sole carbon sources equally well, but Neu5Gc is incorporated much less efficiently into NTHi LOS. This bias occurs in the enzyme SiaB, a CMP-Neu5Ac synthetase, which activates Neu5Ac by addition of a CMP group. SiaB in NTHi has ∼4000-fold-higher catalytic efficiency for Neu5Ac than for Neu5Gc [76] (Fig. 2).
In addition to N. gonorrhoeae and NTHi, a number of other major human pathogens also decorate their LOS with Neu5Ac. For example, sialic acid found in the LOS of the gastric pathogens V. cholerae and Campylobacter jejuni [18, 57]. In C. jejuni , this molecular mimicry of host epitopes leads to the autoimmune disease Guillain–Barre syndrome, where anti-Neu5Ac antibodies, elicited by C. jejuni sialylated-LOS, attack the hosts own gangliosides containing these structures [77]. In the related organism Campylobacter coli , loss of LOS biosynthetic enzyme function means only a small fraction of strains (1%) are able to sialylate their LOS, meaning infection with C. coli is much less likely to lead to autoimmune disorders [78]. This is another nice example of niche adaptation: the ‘molecular mimicry’ benefits of LOS sialyation of a human pathogen ( C. jejuni ) results in severe disease sequelae for the host, whereas an organism that rarely infects humans ( C. coli ) does not need to sialylate its LOS in order to survive.
Several bacterial organisms express LPS, lipo-polysaccharide, instead of LOS. LPS contains the core LOS structure plus an extended polysaccharide, the O-antigen [79]. O-antigens typically contain Neu5Ac, and are critical virulence factors in many organisms such as Pseudomonas aeruginosa [80] and Salmonella enterica serovar Typhimurium [81]. The LPS of Vibrio vulnificus is decorated with sialic acid [82], with the inability to synthesize this molecule leading to a 300-fold lower chance of survival compared with a wild-type strain. In addition, sialic acid plays a critical role in V. vulnificus biofilm formation, motility, and protects the bacterium from the host immune response [83]. Fusobacterium nucleatum , a potential cancer-causing bacterium that inhabits the human oral cavity, incorporates sialic acid into its LPS [84], and sialylation of the LPS of P. gingivalis LPS reduces inflammation [85], allowing long-term periodontal colonization.
Many bacterial pathogens also express a capsule. Capsules are extracellular structures that are almost entirely glycan based, and serve to mask bacteria from the immune system. Serogroup B Neisseria meningitidis , in addition to decorating its LOS with sialic acid like the closely related organism N. gonorrhoeae [86], produces a capsule containing multiple Neu5Ac residues, as does E. coli K1. In both instances, these polysialic acid capsules confer resistance to complement mediated killing and phagocytosis [86–89]. Nearly all Streptococci express a polysaccharide capsule, but only Streptococcus agalactiae (Group B Strep, GBS), responsible for neonatal meningitis and urinary tract infections, and Streptococcus suis , a major pig and zoonotic pathogen, are known to contain sialic acid [90]. Sialylation of the GBS capsule is key to immune evasion [91, 92], likely through sialic acid on the GBS capsule binding to host Sia-recognizing Ig superfamily lectins (Siglecs). This blocks neutrophil and macrophage activation, and suppress platelet activation [93]. In S. suis , the role of capsular sialic acid is less clear. Not all S. suis capsular serotypes contain sialic acid (e.g. serotypes 2 and 14 contain sialic acid, serotype 9 does not) [94] but the interactions between strains expressing all three capsular types and host cells are similar and serotype 9 capsule is critical for bacterial survival in blood and development of clinical disease [94]. Confoundingly, sialic acid has been shown to be crucial for S. suis capsule synthesis, whereas for GBS, capsule synthesis may occur in the absence of a sialic acid moiety [95]. The linkage of sialic acid in the S. suis capsule occurs via an α2,6-linkage. These types of linkage are recognized by the haemagglutinins of both human and swine influenza viruses (demonstrating the zoonotic nature of S. suis infections). Because of this interaction, adherence to and invasion of airway epithelial cells is enhanced when S. suis encounters influenza virus-infected cells [96]. Adherence and invasion efficiency of GBS is also enhanced when cells were pre-infected by influenza A virus with α2–3 linked neuraminidase specificity [97]. These recent findings indicate understanding synergy between viral and bacterial pathogens is key to combatting disease.
Attaching to the host: sialic acid as a cellular receptor for bacterial adhesins
As one of the most abundant and wide-spread host surface molecules, sialic acid or glycan structures containing a sialic acid moiety, are ideal receptors for bacteria to bind to during colonization and disease.
NTHi, in addition to evading the host via addition of Neu5Ac onto LOS, expresses multiple adhesins that have high specificity for sialic acid (Fig. 2). NTHi express the adhesins HMW (HMW1 and HMW2) and Hia, but individual strains only encode genes for HMW1/2 (75 % of strains) [98], or Hia (25 % of strains) [99], with this discrete strain distribution likely the result of an ancient strain divergence event. HMW1 binds to α2,3-sialyllactosamine (2–3 SLN) [100] that contains terminal Neu5Ac or Neu5Gc via an α2,3 linkage [101]. HMW2, which is ~65 % identical to HMW1, binds the related glycan α2,6-sialyllactosamine (2–6 SLN), with high specificity for 2–6 SLN containing Neu5Ac [101]. Hia binds 2–6 SLN with a ~10-fold preference for Neu5Ac over Neu5Gc [102], with preference for these structures located in Hia binding domain 1 (BD1), the high-affinity substrate binding region of Hia [102]. Hia will also bind 2–3 SLN containing either a Neu5Ac or Neu5Gc with equal affinity, but this binding is not mediated by BD1. Intriguingly, 2–3 SLN is found mainly in the lower human respiratory tract, whereas 2–6 SLN is found throughout the entire respiratory tract, but predominates in the upper airway [103]. Thus, NTHi strains are able to colonize the entire human airway as strains have independently evolved the ability to bind human-airway-specific glycans via either the HMW1/2 proteins or the Hia protein. Moraxella catarrhalis is a respiratory tract pathogen often found in co-infections with NTHi. Binding of M. catarrhalis to mucus occurs via bacterial binding to Neu5Ac and Neu5Ac-linked glycan structures (e.g. α2–6 sialyllactose; Neu5Ac linked to lactose) within mucus, with this binding correlating with increased inflammation [104]. The human gastric pathogen Helicobacter pylori , responsible for stomach ulcers and gastric cancer, expresses multiple adhesins recognizing glycan structures containing terminal Neu5Ac residues. The SabA adhesin has specificity for terminal α2,3-linked Neu5Ac, including the sialyl Lewisxantigen [105] and gangliosides in the human stomach [106]. The H. pylori adhesin HP0721 specifically recognizes sialyllactose residues [107, 108] with a preference for Neu5Ac over Neu5Gc. Flagella and pili are also involved in binding to host sialic acid. For example, P. aeruginosa pili and flagellum interact with GM1 and GM2 gangliosides containing α2,3-linked Neu5Ac residues [109]. In an interesting example of sialylation not increasing adherence to the underlying glycans, the Ata adhesin of the multi-drug-resistant pathogen A. baumannii interacts with high-affinity to N-Acetylactosamine, which consists of a galactose and GlcNAc joined by a β1–4 linkage [110]. Sialylation of the GlcNAc with Neu5Ac to form sialyllactosamine did not change the affinity of the interaction [110]. This suggests that Neu5Ac may not be ‘seen’ by Ata, with the underlying structure of N-acetylactosamine, or the individual components of this structure, more important to binding, and that not all human pathogens have an affinity for sialic acids.
Many pathogenic Streptococci specifically recognize sialic acid-linked structures on multiple human cells and tissues. Streptococcus pyogenes (Group A Strep, GAS), a major human pathogen responsible for a throat and skin infections, and a number of severe invasive infections, interacts with sialic acid during colonization. This interaction occurs via the outer-surface M-protein [111], a major virulence factor in this bacterium, and which displays extreme variability between GAS strains. The M1 protein variant has varying affinity for ABO(H) blood group antigen structures [112]. Colonization of human oral epithelial cells by GAS expressing the M3 and M12 variants is also mediated by blood group antigens, as well as the sialic acid containing Lewis (Le) antigen. Removal of linkage-specific fucose, galactose, N-acetylgalactosamine and sialic acid, modulated GAS colonization [113] indicating a dependence on sialic acid, but that the sialic acid can be contained in multiple arrangements and structures. S. suis also adheres to sialic acid containing glycans via the adhesin SssP1, a fimbria-like structure, which promotes adherence to host cells [114]. Multiple Streptococcal species are able to cause endocarditis, an infection of the heart tissue that can be life threatening. Adherence to sialic acid containing structures via their Sialic acid-binding serine-rich repeat adhesins, which contain SLBRs (Siglec-like binding regions), is key to this interaction in Streptococcus gordonii and Streptococcus sanguinis [115]. Both species show a preference for α2–3 sialoglycan structures, with these structures present on multiple plasma proteins that have been cited as biomarkers for vascular disease [115]. The structure of these SLBRs is very similar to mammalian siglecs, but the glycan ligand they interact with quite different [116], suggesting it will be possible to target inhibitors to the bacterial proteins to treat infections. S. gordonii expresses a second adhesin that interacts with sialic acid, Hsa, with this protein having a role in biofilm formation in this species [117]. Streptococcus oralis , a human oral bacterium that can also cause meningitis, binds platelets via recognition of a terminal sialic acid by the adhesin Fap1 [118]. S. oralis also produces a neuraminidase, NanA, which cleaves this terminal sialic acid with the exposed underlying β1,4-linked galactose also bound by the Fap1 adhesin [118]. AsaA, a second sialic-acid binding adhesin characterized in S. oralis , helps bacteria bind to platelets and contributes to the development of endocarditis [119]. S. oralis subsp. dentisani produces a serine-rich repeat protein fibril, FapC, which binds sialic acid in saliva, and which aids oral colonization [120].
Mycoplasmas are host-adapted, reduced genome pathogens. Each vertebrate has its own specific Mycoplasma pathogens, where they cause a range of diseases. Mycoplasma genitalium , a human sexually transmitted pathogen, binds sialic acid containing receptors via the adhesin P110 [121], which together with protein P140 forms the Nap adhesion complex [122]. P110 binds α2–3- or α2–6-sialyllactose, with this interaction critical to initiating attachment to human cells [121]. Mycoplasma pneumoniae is a common cause of human respiratory tract infections, including bronchitis and atypical pneumonia. M. pneumoniae binds glycoprotein receptors having terminal sialic acid residues via the P1 adhesin protein [123], with no preference for either α2,3- or α2,6-sialyllactose. Mycoplasmas are also able to traverse surfaces via a characteristic ‘gliding’ motility, with M. pneumoniae specifically gliding via adherence to α2,6-sialyllactose [123]. Mycoplasma mobile , a fish pathogen, also uses sialylated glycans as a binding partner for gliding motility [124] via the adhesin Gli349 [125].
A sugar coating can kill you: sialic acid as receptors for bacterial toxins
Multiple bacterial species produce toxins, secreted protein effectors that damage host cells and tissues, releasing nutrients and potentiating virulence. Just like adhesins, bacterial toxins have evolved to recognise sialic acid residues as receptors to mediate host cell binding.
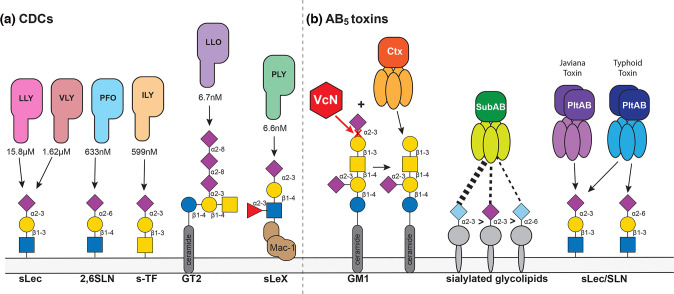
The CDCs, cholesterol-dependent cytolysins, are a major class of pore-forming toxin produced by Gram-positive bacterial pathogens [126]. In a recent study, all major bacterial CDCs were shown to bind host cell glycan structures as high-affinity receptors, and that some CDCs have no measurable binding affinity for cholesterol [127]. CDCs from C. perfringens (PFO), G. vaginalis (VGO), Listeria monocytogenes (LLO), Streptococcus pneumoniae (PLY), Streptococcus mitis (LLY) and Streptococcus intermedius (ILY) all have high affinity (in the nM-µM range) for glycan structures containing terminal Neu5Ac sialic acid residues [127] (Fig. 3), with the activity of these toxins blocked via addition of their corresponding glycan receptor.
Fig. 3.
Two major groups of bacterial toxins have evolved to specifically recognize sialic acid containing glycans. The CDCs (cholesterol dependent cytolysis), produced by Gram-positive bacteria, and the AB5 toxins, made by Gram-negative bacteria, both utilize host glycans as cellular receptors, with many specifically recognizing sialic acid containing glycan structures. Where available, the K D (dissociation constant) is shown for the glycan structure with the highest affinity binding to each toxin. The action of Ctx, produced by Vibrio cholerae , requires the action of the neuraminidase VcN to cleave Neu5Ac in order to expose the underlying glycan structure, which is the preferred receptor. Host-evolution is evident for the AB5 toxins, produced by Shiga-toxin producing E. coli (STEC; SubAB), and Salmonella spp. SubAB binds both Neu5Ac and Neu5Gc containing glycan structures, with a preference for Neu5Gc containing structures, demonstrating the broad mammalian host range of STEC; Typhoid toxin (PltAB), produced by the human-restricted pathogen Salmonella Typhi, binds Neu5Ac linked via an α2,6- or and α2,3-linkage, whereas Javiana toxin, produced by Salmonella Javiana, shows a marked preference for α2,3-linked Neu5Ac. These differences in receptor specificity strongly influence disease severity resulting from infections by these organisms.
Gram-negative bacteria produce a class of toxins known as the AB5 toxins, consisting of a multiprotein complex of A and B subunits [128]. The A subunits are the toxin effector, whereas the B subunits contain the cognate ligand binding function [128]. The subtilase AB5 toxin, SubAB, of Shiga toxin-producing E. coli (STEC) [129] and damage the internal organs, with STEC infection frequently resulting in acute kidney failure and haemolytic anaemia [130]. The highest affinity receptors for SubAB are sialylated glycolipids with terminal α2,3-Neu5Gc, to which the affinity is 20 times higher than to α2,3-Neu5Ac, and 30 times higher than to α2,6-Neu5Gc [131]. Although this toxin receptor preference indicates adaptation to non-human Neu5Gc expressing hosts, the lack of Neu5Gc-containing body fluid receptor competitors in humans may confer susceptibility to the gastrointestinal and systemic toxicities of SubAB [131]. In recent years, cellular receptors for other AB5 toxins have been demonstrated to be host glycan structures [132] (Fig. 3). Salmonella Typhi, the causative agent of typhoid fever in humans, produces the AB5 Typhoid toxin PltAB, in an A2B5 arrangement. The B subunit, binds Neu5Ac [133–135], is reported to bind to a galactose (Gal) β1–3/β1–4 linked N-acetylglucosamine (GlcNAc) or glucose, with either an α2,3- or α2,6-linkage to the terminal Neu5Ac [136]. In contrast, the PltB from nontyphoidal Salmonella (NTS) Salmonella Javiana displays a strong preference for α2,3-linked Neu5Ac [136]. This difference in receptor preference is consistent with the resulting disease presentation: S. Typhi causes life-threatening typhoid fever, with Typhoid toxin targeting multiple cell types including epithelial, endothelial and immune cells and involves both local and systemic infection; Javiana toxin binds epithelial cells found in the human gastro-intestinal (GI) tract [136], which display a high relative amount of α2,3-linked Neu5Ac over α2,6-linked Neu5Ac, and consequently results in self-limiting GI infections (non-typhoidal Salmonella infection).
Cholera toxin, Ctx, the AB5 toxin produced by Vibrio cholerae , also recognizes sialylated glycan structures, with specificity for GM1 polysialyated gangliosides [137]. However, as described in the sialidase section above, the action of the VcN sialidase is required for Ctx to bind to the underlying glycan structures, rather than sialic acid itself (Fig. 3). In a humanized mouse model of cholera using a CMAH deficient mouse that can only produce Neu5Ac, the action of Ctx is enhanced, indicating that VcN prefers Neu5Ac over Neu5Gc, and that Neu5Gc impedes VcN-mediated generation of the underlying glycans in gangliosides [138]. Human epithelial cells artificially expressing Neu5Gc were also less susceptible to Ctx binding and CtxA intoxication following VcN treatment. Therefore, it appears that V. cholerae evolved into a human-specific pathogen by adapting to the loss of Neu5Gc in humans [138].
Multiple additional types of bacterial toxin have evolved to specifically recognise and bind human sialic acid (Neu5Ac) containing glycan structures. For example, the Clostridium botulinum neurotoxin BoNT/DC specifically recognizes sialic acid moieties on the Sialyl-T antigen, but not to other moieties in gangliosides [139]. The E. coli heat-labile enterotoxin LT-IIb shows high-affinity for gangliosides with a neo-lacto core, that can contain either Neu5Gc or Neu5Ac residues [140].
Concluding remarks
Bacteria are able to utilize sialic acids in multiple ways. The advantage of being able to use an abundant carbon and energy source present in all mammalian hosts is clear. The concomitant ability to acquire sialic acids, and develop an ability to release this molecule from complex glycan structures – sialidases – goes hand in hand with the ability to use sialic acids in metabolism and biosynthesis. Using a highly prevalent molecule to bind to host cells and tissues makes a lot of evolutionary sense, and many bacterial pathogens have evolved multiple adhesins that specifically recognize host sialic acids. Co-evolution of specificity for Neu5Ac is evident in several human-adapted pathogens, and has likely occurred during loss of the ability of humans to make Neu5Gc, marking Neu5Ac as the ‘human specific’ sialic acid. The close interplay of multiple factors occurs during bacterial pathogenesis, with the synergy of Vibrio cholerae VcN sialidase and Ctx toxin a clear demonstration of human–bacterial co-evolution. The most pernicious way bacterial pathogens utilize sialic acid is as a means to evade and subvert the immune system, starkly illustrated by the incorporation of Neu5Ac in multiple outer-surface structures such as LOS, LPS and the polysaccharide capsule. This ‘molecular mimicry’ is perhaps best studied in NTHi and the pathogenic Neisseria . The bias for Neu5Ac over Neu5Gc in NTHi LOS incorporation occurs at a single point, the activation of sialic acid by the enzyme SiaB into the nucleotide sugar Neu5Ac-CMP for incorporation into LOS, in another stark illustration of co-evolution of bacteria and host.
An abundance of research is uncovering further roles for sialic acid in bacterial pathogenesis and host-bacterial interactions, building on initial characterisation of these systems in multiple bacterial pathogens. The advantage gained by the use of sialic acid in immune evasion and host binding means that it is a clear target for development of new antimicrobials; block the interactions required to colonize, or those needed for toxins to bind, and disease is prevented, or at the very least decreased. Many toxins and adhesins that recognize host sialic acid are key virulence factors, and highly expressed during pathogenesis. Therefore, in addition to being targets for novel antimicrobials, these proteins are also ideal vaccine candidates, and understanding their role is key to the development of rationally designed subunit vaccines against several major human pathogens.
Funding information
This work was funded by a National Health and Medical Research Council (NHMRC) Principal Research Fellowship (1138466) to M.P.J., and Ideas Grant (2003435) to M.P.J and C.J.D. We acknowledge support from National Institutes of Health (NIH; USA) R01 Grant DC015688.
Acknowledgements
We thank the editors for the invitation to submit a review to celebrate the 75th Anniversary of the Microbiology Society. We have tried to focus this review on developments in the last 5–10 years, and only cite literature older than this where necessary to put more recent findings into context, or describe key discoveries. We apologise to any authors whose work we have not included.
Author contributions
M.P.J. writing – review and editing. C.J.D. writing – review and editing. J.M.A. conceptualization; project administration; supervision; visualization; writing – original draft; writing – review and editing.
Conflicts of interest
The authors declare that there are no conflicts of interest
Footnotes
Abbreviations: CDC, cholesterol dependent cytolysin; CMAH, CMP-Neu5Ac hydroxylase; LOS, lipo-oligosaccharide; LPS, lipopolysaccharide; Neu5Ac, N-acetylneuraminic acid; Neu5Gc, N-glycolylneuraminic acid; NulO, nonusolonic acid.
References
- 1.Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem Rev. 2002;102:439–469. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- 2.McDonald ND, Boyd EF. Structural and biosynthetic diversity of nonulosonic acids (NulOs) that decorate surface structures in bacteria. Trends Microbiol. 2021;29:142–157. doi: 10.1016/j.tim.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irie A, Suzuki A. CMP-N-Acetylneuraminic acid hydroxylase is exclusively inactive in humans. Biochem Biophys Res Commun. 1998;248:330–333. doi: 10.1006/bbrc.1998.8946. [DOI] [PubMed] [Google Scholar]
- 4.Irie A, Koyama S, Kozutsumi Y, Kawasaki T, Suzuki A, et al. The molecular basis for the absence of N-glycolylneuraminic acid in humans. J Biol Chem. 1998;273:15866–15871. doi: 10.1074/jbc.273.25.15866. [DOI] [PubMed] [Google Scholar]
- 5.Muchmore EA, Diaz S, Varki A, et al. A structural difference between the cell surfaces of humans and the great apes. Am J Phys Anthropol. 1998;107:187–198. doi: 10.1002/(SICI)1096-8644(199810)107:2<187::AID-AJPA5>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 6.Chou HH, Takematsu H, Diaz S, Iber J, Nickerson E, et al. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc Natl Acad Sci U S A. 1998;95:11751–11756. doi: 10.1073/pnas.95.20.11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pham T, Gregg CJ, Karp F, Chow R, Padler-Karavani V, et al. Evidence for a novel human-specific xeno-auto-antibody response against vascular endothelium. Blood. 2009;114:5225–5235. doi: 10.1182/blood-2009-05-220400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hedlund M, Padler-Karavani V, Varki NM, Varki A, et al. Evidence for a human-specific mechanism for diet and antibody-mediated inflammation in carcinoma progression. Proc Natl Acad Sci U S A. 2008;105:18936–18941. doi: 10.1073/pnas.0803943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz SL, Padler-Karavani V, Ghaderi D, Hurtado-Ziola N, Yu H, et al. Sensitive and specific detection of the non-human sialic Acid N-glycolylneuraminic acid in human tissues and biotherapeutic products. PLoS One. 2009;4:e4241. doi: 10.1371/journal.pone.0004241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tangvoranuntakul P, Gagneux P, Diaz S, Bardor M, Varki N, et al. Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proc Natl Acad Sci U S A. 2003;100:12045–12050. doi: 10.1073/pnas.2131556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergfeld AK, Pearce OMT, Diaz SL, Pham T, Varki A, et al. Metabolism of vertebrate amino sugars with N-glycolyl groups: elucidating the intracellular fate of the non-human sialic acid N-glycolylneuraminic acid. J Biol Chem. 2012;287:28865–28881. doi: 10.1074/jbc.M112.363549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bousquet PA, Sandvik JA, Jeppesen Edin NF, Krengel U, et al. Hypothesis: Hypoxia induces de novo synthesis of NeuGc gangliosides in humans through CMAH domain substitute. Biochem Biophys Res Commun. 2018;495:1562–1566. doi: 10.1016/j.bbrc.2017.11.183. [DOI] [PubMed] [Google Scholar]
- 13.Blaum BS, Hannan JP, Herbert AP, Kavanagh D, Uhrín D, et al. Structural basis for sialic acid-mediated self-recognition by complement factor H. Nat Chem Biol. 2015;11:77–82. doi: 10.1038/nchembio.1696. [DOI] [PubMed] [Google Scholar]
- 14.Lewis AL, Lewis WG. Host sialoglycans and bacterial sialidases: a mucosal perspective. Cell Microbiol. 2012;14:1174–1182. doi: 10.1111/j.1462-5822.2012.01807.x. [DOI] [PubMed] [Google Scholar]
- 15.Barry GT, Goebel WF. Colominic acid, a substance of bacterial origin related to sialic acid. Nature. 1957;179:206. doi: 10.1038/179206a0. [DOI] [PubMed] [Google Scholar]
- 16.Rohr TE, Troy FA. Structure and biosynthesis of surface polymers containing polysialic acid in Escherichia coli . J Biol Chem. 1980;255:2332–2342. [PubMed] [Google Scholar]
- 17.Troy FA, McCloskey MA. Role of a membranous sialyltransferase complex in the synthesis of surface polymers containing polysialic acid in Escherichia coli. Temperature-induced alteration in the assembly process. J Biol Chem. 1979;254:7377–7387. [PubMed] [Google Scholar]
- 18.Linton D, Karlyshev AV, Hitchen PG, Morris HR, Dell A, et al. Multiple N-acetyl neuraminic acid synthetase (neuB) genes in Campylobacter jejuni: identification and characterization of the gene involved in sialylation of lipo-oligosaccharide. Mol Microbiol. 2000;35:1120–1134. doi: 10.1046/j.1365-2958.2000.01780.x. [DOI] [PubMed] [Google Scholar]
- 19.Gunawan J, Simard D, Gilbert M, Lovering AL, Wakarchuk WW, et al. Structural and mechanistic analysis of sialic acid synthase NeuB from Neisseria meningitidis in complex with Mn2+, phosphoenolpyruvate, and N-acetylmannosaminitol. J Biol Chem. 2005;280:3555–3563. doi: 10.1074/jbc.M411942200. [DOI] [PubMed] [Google Scholar]
- 20.Wessels MR, Pozsgay V, Kasper DL, Jennings HJ, et al. Structure and immunochemistry of an oligosaccharide repeating unit of the capsular polysaccharide of type III group B Streptococcus. A revised structure for the type III group B streptococcal polysaccharide antigen. J Biol Chem. 1987;262:8262–8267. doi: 10.1016/S0021-9258(18)47558-8. [DOI] [PubMed] [Google Scholar]
- 21.Lewis AL, Robinson LS, Agarwal K, Lewis WG, et al. Discovery and characterization of de novo sialic acid biosynthesis in the phylum Fusobacterium. Glycobiology. 2016;26:1107–1119. doi: 10.1093/glycob/cww068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zapata G, Crowley JM, Vann WF, et al. Sequence and expression of the Escherichia coli K1 neuC gene product. J Bacteriol. 1992;174:315–319. doi: 10.1128/jb.174.1.315-319.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vimr ER, Aaronson W, Silver RP, et al. Genetic analysis of chromosomal mutations in the polysialic acid gene cluster of Escherichia coli K1. J Bacteriol. 1989;171:1106–1117. doi: 10.1128/jb.171.2.1106-1117.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silver RP, Vann WF, Aaronson W, et al. Genetic and molecular analyses of Escherichia coli K1 antigen genes. J Bacteriol. 1984;157:568–575. doi: 10.1128/jb.157.2.568-575.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vann WF, Tavarez JJ, Crowley J, Vimr E, Silver RP, et al. Purification and characterization of the Escherichia coli K1 neuB gene product N-acetylneuraminic acid synthetase. Glycobiology. 1997;7:697–701. doi: 10.1093/glycob/7.5.697. [DOI] [PubMed] [Google Scholar]
- 26.Knirel YA, Rietschel ET, Marre R, Zähringer U, et al. The structure of the O-specific chain of Legionella pneumophila serogroup 1 lipopolysaccharide. Eur J Biochem. 1994;221:239–245. doi: 10.1111/j.1432-1033.1994.tb18734.x. [DOI] [PubMed] [Google Scholar]
- 27.Kenyon JJ, Marzaioli AM, De Castro C, Hall RM, et al. 5,7-di-N-acetyl-acinetaminic acid: A novel non-2-ulosonic acid found in the capsule of an Acinetobacter baumannii isolate. Glycobiology. 2015;25:644–654. doi: 10.1093/glycob/cwv007. [DOI] [PubMed] [Google Scholar]
- 28.Morens DM, Taubenberger JK, Fauci AS, et al. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198:962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siegel SJ, Roche AM, Weiser JN, et al. Influenza promotes pneumococcal growth during coinfection by providing host sialylated substrates as a nutrient source. Cell Host Microbe. 2014;16:55–67. doi: 10.1016/j.chom.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanchette KA, Shenoy AT, Milner J, 2nd, Gilley RP, McClure E, et al. Neuraminidase a-exposed galactose promotes Streptococcus pneumoniae biofilm formation during colonization. Infect Immun. 2016;84:2922–2932. doi: 10.1128/IAI.00277-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coats MT, Murphy T, Paton JC, Gray B, Briles DE, et al. Exposure of Thomsen-Friedenreich antigen in Streptococcus pneumoniae infection is dependent on pneumococcal neuraminidase A. Microbial Pathogenesis. 2011;50:343–349. doi: 10.1016/j.micpath.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kullaya V, de Jonge MI, Langereis JD, van der Gaast-de Jongh CE, Büll C, et al. Desialylation of platelets by pneumococcal neuraminidase A induces ADP-dependent platelet hyperreactivity. Infect Immun. 2018;86:10. doi: 10.1128/IAI.00213-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gratz N, Loh LN, Mann B, Gao G, Carter R, et al. Pneumococcal neuraminidase activates TGF-β signalling. Microbiology (Reading) 2017;163:1198–1207. doi: 10.1099/mic.0.000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song Y, Pan Q, Xiao J, Li W, Ma H, et al. Sialidase of Glaesserella parasuis augments inflammatory response via desialylation and abrogation of negative regulation of siglec-5. Infect Immun. 2021;89 doi: 10.1128/IAI.00696-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammond AJ, Binsker U, Aggarwal SD, Ortigoza MB, Loomis C, et al. Neuraminidase B controls neuraminidase A-dependent mucus production and evasion. PLoS Pathog. 2021;17:e1009158. doi: 10.1371/journal.ppat.1009158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King CA, Van Heyningen WE. Deactivation of cholera toxin by a sialidase-resistant monosialosylganglioside. J Infect Dis. 1973;127:639–647. doi: 10.1093/infdis/127.6.639. [DOI] [PubMed] [Google Scholar]
- 37.Holmgren J, Lönnroth I, Månsson J, Svennerholm L, et al. Interaction of cholera toxin and membrane GM1 ganglioside of small intestine. Proc Natl Acad Sci U S A. 1975;72:2520–2524. doi: 10.1073/pnas.72.7.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson LS, Schwebke J, Lewis WG, Lewis AL, et al. Identification and characterization of NanH2 and NanH3, enzymes responsible for sialidase activity in the vaginal bacterium Gardnerella vaginalis . J Biol Chem. 2019;294:5230–5245. doi: 10.1074/jbc.RA118.006221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hardy L, Jespers V, Van den Bulck M, Buyze J, Mwambarangwe L, et al. The presence of the putative Gardnerella vaginalis sialidase A gene in vaginal specimens is associated with bacterial vaginosis biofilm. PLoS One. 2017;12:e0172522. doi: 10.1371/journal.pone.0172522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Govinden G, Parker JL, Naylor KL, Frey AM, Anumba DOC, et al. Inhibition of sialidase activity and cellular invasion by the bacterial vaginosis pathogen Gardnerella vaginalis . Arch Microbiol. 2018;200:1129–1133. doi: 10.1007/s00203-018-1520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frey AM, Satur MJ, Phansopa C, Honma K, Urbanowicz PA, et al. Characterization of Porphyromonas gingivalis sialidase and disruption of its role in host–pathogen interactions. Microbiology. 2019;165:1181–1197. doi: 10.1099/mic.0.000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navarro MA, Li J, McClane BA, Morrell E, Beingesser J, et al. NanI sialidase is an important contributor to Clostridium perfringens type F strain F4969 intestinal colonization in mice. Infect Immun. 2018;86:12. doi: 10.1128/IAI.00462-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, McClane BA, Young VB. NanI sialidase can support the growth and survival of Clostridium perfringens strain F4969 in the presence of sialyated host macromolecules (Mucin) or Caco-2 cells. Infect Immun. 2018;86 doi: 10.1128/IAI.00547-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacMillan JL, Vicaretti SD, Noyovitz B, Xing X, Low KE, et al. Structural analysis of broiler chicken small intestinal mucin O-glycan modification by Clostridium perfringens . Poult Sci. 2019;98:5074–5088. doi: 10.3382/ps/pez297. [DOI] [PubMed] [Google Scholar]
- 45.Michaels DL, Moneypenny CG, Shama SM, Leibowitz JA, May MA, et al. Sialidase and N-acetylneuraminate catabolism in nutrition of Mycoplasma alligatoris . Microbiology. 2019;165:662–667. doi: 10.1099/mic.0.000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.May M, Brown DR. Genetic variation in sialidase and linkage to N-acetylneuraminate catabolism in Mycoplasma synoviae . Microb Pathog. 2008;45:38–44. doi: 10.1016/j.micpath.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas GH. Sialic acid acquisition in bacteria-one substrate, many transporters. Biochem Soc Trans. 2016;44:760–765. doi: 10.1042/BST20160056. [DOI] [PubMed] [Google Scholar]
- 48.Haines-Menges BL, Whitaker WB, Lubin JB, Boyd EF, et al. Host sialic acids: a delicacy for the pathogen with discerning taste. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MBP-0005-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez J, Steenbergen S, Vimr E, et al. Derived structure of the putative sialic acid transporter from Escherichia coli predicts a novel sugar permease domain. J Bacteriol. 1995;177:6005–6010. doi: 10.1128/jb.177.20.6005-6010.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walters DM, Stirewalt VL, Melville SB, et al. Cloning, sequence, and transcriptional regulation of the operon encoding a putative N-acetylmannosamine-6-phosphate epimerase (nanE) and sialic acid lyase (nanA) in Clostridium perfringens . J Bacteriol. 1999;181:4526–4532. doi: 10.1128/JB.181.15.4526-4532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olson ME, King JM, Yahr TL, Horswill AR. Sialic acid catabolism in Staphylococcus aureus . J Bacteriol. 2013;195:1779–1788. doi: 10.1128/JB.02294-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rosa LT, Bianconi ME, Thomas GH, Kelly DJ, et al. Tripartite ATP-Independent Periplasmic (TRAP) Transporters and Tripartite Tricarboxylate Transporters (TTT): from uptake to pathogenicity. Front Cell Infect Microbiol. 2018;8:33. doi: 10.3389/fcimb.2018.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Severi E, Randle G, Kivlin P, Whitfield K, Young R, et al. Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol Microbiol. 2005;58:1173–1185. doi: 10.1111/j.1365-2958.2005.04901.x. [DOI] [PubMed] [Google Scholar]
- 54.Allen S, Zaleski A, Johnston JW, Gibson BW, Apicella MA, et al. Novel sialic acid transporter of Haemophilus influenzae . Infect Immun. 2005;73:5291–5300. doi: 10.1128/IAI.73.9.5291-5300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnston JW, Zaleski A, Allen S, Mootz JM, Armbruster D, et al. Regulation of sialic acid transport and catabolism in Haemophilus influenzae . Mol Microbiol. 2007;66:26–39. doi: 10.1111/j.1365-2958.2007.05890.x. [DOI] [PubMed] [Google Scholar]
- 56.Jenkins GA, Figueira M, Kumar GA, Sweetman WA, Makepeace K, et al. Sialic acid mediated transcriptional modulation of a highly conserved sialometabolism gene cluster in Haemophilus influenzae and its effect on virulence. BMC Microbiol. 2010;10:48. doi: 10.1186/1471-2180-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gangi Setty T, Cho C, Govindappa S, Apicella MA, Ramaswamy S, et al. Bacterial periplasmic sialic acid-binding proteins exhibit a conserved binding site. Acta Crystallogr D Biol Crystallogr. 2014;70:1801–1811. doi: 10.1107/S139900471400830X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tatum FM, Tabatabai LB, Briggs RE. Sialic acid uptake is necessary for virulence of Pasteurella multocida in turkeys. Microb Pathog. 2009;46:337–344. doi: 10.1016/j.micpath.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 59.North RA, Horne CR, Davies JS, Remus DM, Muscroft-Taylor AC, et al. “Just a spoonful of sugar”: import of sialic acid across bacterial cell membranes. Biophys Rev. 2018;10:219–227. doi: 10.1007/s12551-017-0343-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM, et al. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev. 2004;68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reddi G, Pruss K, Cottingham KL, Taylor RK, Almagro-Moreno S, et al. Catabolism of mucus components influences motility of Vibrio cholerae in the presence of environmental reservoirs. PLoS ONE. 2018;13:e0201383. doi: 10.1371/journal.pone.0201383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crost EH, Tailford LE, Monestier M, Swarbreck D, Henrissat B, et al. The mucin-degradation strategy of Ruminococcus gnavus: The importance of intramolecular trans-sialidases. Gut Microbes. 2016;7:302–312. doi: 10.1080/19490976.2016.1186334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uhde A, Brühl N, Goldbeck O, Matano C, Gurow O, et al. Transcription of sialic acid catabolism genes in Corynebacterium glutamicum is subject to catabolite repression and control by the transcriptional repressor NanR. J Bacteriol. 2016;198:2204–2218. doi: 10.1128/JB.00820-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ward ME, Watt PJ, Glynn AA. Gonococci in urethral exudates possess a virulence factor lost on subculture. Nature. 1970;227:382–384. doi: 10.1038/227382a0. [DOI] [PubMed] [Google Scholar]
- 65.Nairn CA, Cole JA, Patel PV, Parsons NJ, Fox JE, et al. Cytidine 5’-monophospho-N-acetylneuraminic acid or a related compound is the low mr factor from human red blood cells which induces gonococcal resistance to killing by human serum. Microbiology. 1988;134:3295–3306. doi: 10.1099/00221287-134-12-3295. [DOI] [PubMed] [Google Scholar]
- 66.Wetzler LM, Barry K, Blake MS, Gotschlich EC. Gonococcal lipooligosaccharide sialylation prevents complement-dependent killing by immune sera. Infect Immun. 1992;60:39–43. doi: 10.1128/iai.60.1.39-43.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gulati S, Schoenhofen IC, Whitfield DM, Cox AD, Li J, et al. Utilizing CMP-sialic acid analogs to unravel Neisseria gonorrhoeae lipooligosaccharide-mediated complement resistance and design novel therapeutics. PLoS Pathog. 2015;11:e1005290. doi: 10.1371/journal.ppat.1005290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ram S, Gulati S, Lewis LA, Chakraborti S, Zheng B, et al. A novel sialylation site on Neisseria gonorrhoeae lipooligosaccharide links heptose II lactose expression with pathogenicity. Infect Immun. 2018;86 doi: 10.1128/IAI.00285-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lewis LA, Gulati S, Burrowes E, Zheng B, Ram S, et al. α-2,3-sialyltransferase expression level impacts the kinetics of lipooligosaccharide sialylation, complement resistance, and the ability of Neisseria gonorrhoeae to colonize the murine genital tract. mBio. 2015;6:e02465-14. doi: 10.1128/mBio.02465-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Apicella MA, Mandrell RE. Molecular mimicry as a factor in the pathogenesis of human neisserial infections: in vitro and in vivo modification of the lipooligosaccharide of Neisseria gonorrhoeae by N-acetylneuraminic acid. Pediatr Infect Dis J. 1989;8:901–902. doi: 10.1097/00006454-198912000-00033. [DOI] [PubMed] [Google Scholar]
- 71.Jackson MD, Wong SM, Akerley BJ, et al. Sialic Acid Protects Nontypeable Haemophilus influenzae from Natural IgM and Promotes Survival in Murine Respiratory Tract. Infect Immun. 2021;89:e00676-20. doi: 10.1128/IAI.00676-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oerlemans MMP, Moons SJ, Heming JJA, Boltje TJ, de Jonge MI, et al. Uptake of sialic acid by nontypeable Haemophilus influenzae increases complement resistance through decreasing IgM-dependent complement activation. Infect Immun. 2019;87:e00077-19. doi: 10.1128/IAI.00077-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Apicella MA, Coffin J, Ketterer M, Post DMB, Day CJ, et al. Nontypeable Haemophilus influenzae Lipooligosaccharide Expresses a Terminal Ketodeoxyoctanoate In Vivo, Which Can Be Used as a Target for Bactericidal Antibody. mBio. 2018;9:e01401-18. doi: 10.1128/mBio.01401-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jackson MD, Wong SM, Akerley BJ, Roy CR, et al. Underlying glycans determine the ability of sialylated lipooligosaccharide to protect nontypeable Haemophilus influenzae from serum IgM and complement. Infect Immun. 2019;87:11. doi: 10.1128/IAI.00456-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heise T, Langereis JD, Rossing E, de Jonge MI, Adema GJ, et al. Selective inhibition of sialic acid-based molecular mimicry in Haemophilus influenzae abrogates serum resistance. Cell Chemical Biology. 2018;25:1279–1285. doi: 10.1016/j.chembiol.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 76.Ng PSK, Day CJ, Atack JM, Hartley-Tassell LE, Winter LE, et al. Nontypeable Haemophilus influenzae has evolved preferential use of N-acetylneuraminic acid as a host adaptation. mBio. 2019;10:e00422-19. doi: 10.1128/mBio.00422-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lopes GV, Ramires T, Kleinubing NR, Scheik LK, Fiorentini ÂM, et al. Virulence factors of foodborne pathogen Campylobacter jejuni . Microb Pathog. 2021;161:105265. doi: 10.1016/j.micpath.2021.105265. [DOI] [PubMed] [Google Scholar]
- 78.Culebro A, Machado MP, Carriço JA, Rossi M, et al. Origin, evolution, and distribution of the molecular machinery for biosynthesis of sialylated lipooligosaccharide structures in Campylobacter coli . Sci Rep. 2018;8:3028. doi: 10.1038/s41598-018-21438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2:a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pier GB. Pseudomonas aeruginosa lipopolysaccharide: a major virulence factor, initiator of inflammation and target for effective immunity. Int J Med Microbiol. 2007;297:277–295. doi: 10.1016/j.ijmm.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Slauch JM, Lee AA, Mahan MJ, Mekalanos JJ, et al. Molecular characterization of the oafA locus responsible for acetylation of Salmonella typhimurium O-antigen: oafA is a member of a family of integral membrane trans-acylases. J Bacteriol. 1996;178:5904–5909. doi: 10.1128/jb.178.20.5904-5909.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lewis AL, Lubin J-B, Argade S, Naidu N, Choudhury B, et al. Genomic and metabolic profiling of nonulosonic acids in Vibrionaceae reveal biochemical phenotypes of allelic divergence in Vibrio vulnificus. Appl Environ Microbiol. 2011;77:5782–5793. doi: 10.1128/AEM.00712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lubin J-B, Lewis WG, Gilbert NM, Weimer CM, Almagro-Moreno S, et al. Host-like carbohydrates promote bloodstream survival of Vibrio vulnificus in vivo . Infect Immun. 2015;83:3126–3136. doi: 10.1128/IAI.00345-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Vinogradov E, St Michael F, Homma K, Sharma A, Cox AD, et al. Structure of the LPS O-chain from Fusobacterium nucleatum strain 10953, containing sialic acid. Carbohydr Res. 2017;440–441:38–42. doi: 10.1016/j.carres.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zaric SS, Lappin MJ, Fulton CR, Lundy FT, Coulter WA, et al. Sialylation of Porphyromonas gingivalis LPS and its effect on bacterial-host interactions. Innate Immun. 2017;23:319–326. doi: 10.1177/1753425917694245. [DOI] [PubMed] [Google Scholar]
- 86.Vogel U, Weinberger A, Frank R, Müller A, Köhl J, et al. Complement factor C3 deposition and serum resistance in isogenic capsule and lipooligosaccharide sialic acid mutants of serogroup B Neisseria meningitidis . Infect Immun. 1997;65:4022–4029. doi: 10.1128/iai.65.10.4022-4029.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moxon ER, Kroll JS. The role of bacterial polysaccharide capsules as virulence factors. Curr Top Microbiol Immunol. 1990;150:65–85. doi: 10.1007/978-3-642-74694-9_4. [DOI] [PubMed] [Google Scholar]
- 88.Silver R, Vimr E. The Bacteria, vol. 11. Molecular Basis of Bacterial Pathogenesis. New York, NY: Academic Press, Inc; 1990. [Google Scholar]
- 89.Hammerschmidt S, Birkholz C, Zähringer U, Robertson BD, van Putten J, et al. Contribution of genes from the capsule gene complex (cps) to lipooligosaccharide biosynthesis and serum resistance in Neisseria meningitidis . Mol Microbiol. 1994;11:885–896. doi: 10.1111/j.1365-2958.1994.tb00367.x. [DOI] [PubMed] [Google Scholar]
- 90.Segura M. Fisher scientific award lecture - the capsular polysaccharides of Group B Streptococcus and Streptococcus suis differently modulate bacterial interactions with dendritic cells. Can J Microbiol. 2012;58:249–260. doi: 10.1139/w2012-003. [DOI] [PubMed] [Google Scholar]
- 91.Weiman S, Dahesh S, Carlin AF, Varki A, Nizet V, et al. Genetic and biochemical modulation of sialic acid O-acetylation on group B Streptococcus: phenotypic and functional impact. Glycobiology. 2009;19:1204–1213. doi: 10.1093/glycob/cwp111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weiman S, Uchiyama S, Lin F-YC, Chaffin D, Varki A, et al. O-Acetylation of sialic acid on Group B Streptococcus inhibits neutrophil suppression and virulence. Biochem J. 2010;428:163–168. doi: 10.1042/BJ20100232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Uchiyama S, Sun J, Fukahori K, Ando N, Wu M, et al. Dual actions of group B Streptococcus capsular sialic acid provide resistance to platelet-mediated antimicrobial killing. Proc Natl Acad Sci U S A. 2019;116:7465–7470. doi: 10.1073/pnas.1815572116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Auger J-P, Payen S, Roy D, Dumesnil A, Segura M, et al. Interactions of Streptococcus suis serotype 9 with host cells and role of the capsular polysaccharide: Comparison with serotypes 2 and 14. PLoS One. 2019;14:e0223864. doi: 10.1371/journal.pone.0223864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Roy D, Takamatsu D, Okura M, Goyette-Desjardins G, Van Calsteren M-R, et al. Capsular sialyltransferase specificity mediates different phenotypes in Streptococcus suis and group B Streptococcus . Front Microbiol. 2018;9:545. doi: 10.3389/fmicb.2018.00545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Meng F, Wu NH, Nerlich A, Herrler G, Valentin-Weigand P, et al. Dynamic virus-bacterium interactions in a porcine precision-cut lung slice coinfection model: swine influenza virus paves the way for Streptococcus suis infection in a two-step process. Infect Immun. 2015;83:2806–2815. doi: 10.1128/IAI.00171-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tong J, Fu Y, Wu N-H, Rohde M, Meng F, et al. Sialic acid-dependent interaction of group B streptococci with influenza virus-infected cells reveals a novel adherence and invasion mechanism. Cell Microbiol. 2018;20 doi: 10.1111/cmi.12818. [DOI] [PubMed] [Google Scholar]
- 98.St Geme JW, Kumar VV, Cutter D, Barenkamp SJ. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae . Infect Immun. 1998;66:364–368. doi: 10.1128/IAI.66.1.364-368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barenkamp SJ, St Geme JW. Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable Haemophilus influenzae . Mol Microbiol. 1996;19:1215–1223. doi: 10.1111/j.1365-2958.1996.tb02467.x. [DOI] [PubMed] [Google Scholar]
- 100.St Geme JW. The HMW1 adhesin of nontypeable Haemophilus influenzae recognizes sialylated glycoprotein receptors on cultured human epithelial cells. Infect Immun. 1994;62:3881–3889. doi: 10.1128/iai.62.9.3881-3889.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Atack JM, Day CJ, Poole J, Brockman KL, Bakaletz LO, et al. The HMW2 adhesin of non-typeable Haemophilus influenzae is a human-adapted lectin that mediates high-affinity binding to 2-6 linked N-acetylneuraminic acid glycans. Biochem Biophys Res Commun. 2018;503:1103–1107. doi: 10.1016/j.bbrc.2018.06.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Atack JM, Day CJ, Poole J, Brockman KL, Timms JRL, et al. The Nontypeable Haemophilus influenzae major adhesin hia is a dual-function lectin that binds to human-specific respiratory tract sialic acid glycan receptors. mBio. 2020;11:e02714-20. doi: 10.1128/mBio.02714-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shinya K, Ebina M, Yamada S, Ono M, Kasai N, et al. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 104.Padra M, Benktander J, Padra JT, Andersson A, Brundin B, et al. Mucin binding to Moraxella catarrhalis during airway inflammation is dependent on sialic acid. Am J Respir Cell Mol Biol. 2021;65:593–602. doi: 10.1165/rcmb.2021-0064OC. [DOI] [PubMed] [Google Scholar]
- 105.Mahdavi J, Sondén B, Hurtig M, Olfat FO, Forsberg L, et al. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573–578. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Benktander J, Barone A, Johansson MM, Teneberg S, et al. Helicobacter pylori SabA binding gangliosides of human stomach. Virulence. 2018;9:738–751. doi: 10.1080/21505594.2018.1440171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Voland P, Weeks DL, Vaira D, Prinz C, Sachs G, et al. Specific identification of three low molecular weight membrane-associated antigens of Helicobacter pylori. Aliment Pharmacol Ther. 2002;16:533–544. doi: 10.1046/j.1365-2036.2002.01221.x. [DOI] [PubMed] [Google Scholar]
- 108.Bennett HJ, Roberts IS. Identification of a new sialic acid-binding protein in Helicobacter pylori. FEMS Immunol Med Microbiol. 2005;44:163–169. doi: 10.1016/j.femsim.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 109.Xia B, Sachdev GP, Cummings RD. Pseudomonas aeruginosa mucoid strain 8830 binds glycans containing the sialyl-Lewis x epitope. Glycoconj J. 2007;24:87–95. doi: 10.1007/s10719-006-9015-y. [DOI] [PubMed] [Google Scholar]
- 110.Tram G, Poole J, Adams FG, Jennings MP, Eijkelkamp BA, et al. The Acinetobacter baumannii autotransporter adhesin Ata recognizes host glycans as high-affinity receptors. ACS Infect Dis. 2021;7:2352–2361. doi: 10.1021/acsinfecdis.1c00021. [DOI] [PubMed] [Google Scholar]
- 111.Ryan PA, Pancholi V, Fischetti VA, et al. Group A streptococci bind to mucin and human pharyngeal cells through sialic acid-containing receptors. Infect Immun. 2001;69:7402–7412. doi: 10.1128/IAI.69.12.7402-7412.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.De Oliveira DMP, Hartley-Tassell L, Everest-Dass A, Day CJ, Dabbs RA, et al. Blood group antigen recognition via the group A streptococcal M protein mediates host colonization. mBio. 2017;8:e02237-16. doi: 10.1128/mBio.02237-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.De Oliveira DMP, Everest-Dass A, Hartley-Tassell L, Day CJ, Indraratna A, et al. Human glycan expression patterns influence Group A streptococcal colonization of epithelial cells. FASEB J. 2019;33:10808–10818. doi: 10.1096/fj.201900559R. [DOI] [PubMed] [Google Scholar]
- 114.Zhang Y, Lu P, Pan Z, Zhu Y, Ma J, et al. SssP1, a Streptococcus suis fimbria-like protein transported by the SecY2/A2 system, contributes to bacterial virulence. Appl Environ Microbiol. 2018;84:e01385-18. doi: 10.1128/AEM.01385-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bensing BA, Li Q, Park D, Lebrilla CB, Sullam PM, et al. Streptococcal Siglec-like adhesins recognize different subsets of human plasma glycoproteins: implications for infective endocarditis. Glycobiology. 2018;28:601–611. doi: 10.1093/glycob/cwy052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Di Carluccio C, Forgione RE, Bosso A, Yokoyama S, Manabe Y, et al. Molecular recognition of sialoglycans by streptococcal Siglec-like adhesins: toward the shape of specific inhibitors. RSC Chem Biol. 2021;2:1618–1630. doi: 10.1039/D1CB00173F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Oguchi R, Takahashi Y, Shimazu K, Urano-Tashiro Y, Kawarai T, et al. Contribution of Streptococcus gordonii Hsa adhesin to biofilm formation. Jpn J Infect Dis. 2017;70:399–404. doi: 10.7883/yoken.JJID.2016.492. [DOI] [PubMed] [Google Scholar]
- 118.Singh AK, Woodiga SA, Grau MA, King SJ, Pirofski L, et al. Streptococcus oralis neuraminidase modulates adherence to multiple carbohydrates on platelets. Infect Immun. 2017;85 doi: 10.1128/IAI.00774-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gaytán MO, Singh AK, Woodiga SA, Patel SA, An S-S, et al. A novel sialic acid-binding adhesin present in multiple species contributes to the pathogenesis of Infective endocarditis. PLoS Pathog. 2021;17:e1009222. doi: 10.1371/journal.ppat.1009222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ronis A, Brockman K, Singh AK, Gaytán MO, Wong A, et al. Streptococcus oralis subsp. dentisani produces monolateral serine-rich repeat protein fibrils, one of which contributes to saliva binding via sialic acid. Infect Immun. 2019;87:10. doi: 10.1128/IAI.00406-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Aparicio D, Torres-Puig S, Ratera M, Querol E, Piñol J, et al. Mycoplasma genitalium adhesin P110 binds sialic-acid human receptors. Nat Commun. 2018;9:4471. doi: 10.1038/s41467-018-06963-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Aparicio D, Scheffer MP, Marcos-Silva M, Vizarraga D, Sprankel L, et al. Structure and mechanism of the Nap adhesion complex from the human pathogen Mycoplasma genitalium . Nat Commun. 2020;11:2877. doi: 10.1038/s41467-020-16511-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Williams CR, Chen L, Driver AD, Arnold EA, Sheppard ES, et al. Sialylated receptor setting influences Mycoplasma pneumoniae attachment and gliding motility. Mol Microbiol. 2018;109:735–744. doi: 10.1111/mmi.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Morio H, Kasai T, Miyata M, et al. Gliding direction of Mycoplasma mobile . J Bacteriol. 2016;198:283–290. doi: 10.1128/JB.00499-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hamaguchi T, Kawakami M, Furukawa H, Miyata M, et al. Identification of novel protein domain for sialyloligosaccharide binding essential to Mycoplasma mobile gliding. FEMS Microbiol Lett. 2019;366:fnz016. doi: 10.1093/femsle/fnz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dal Peraro M, van der Goot FG. Pore-forming toxins: ancient, but never really out of fashion. Nat Rev Microbiol. 2016;14:77–92. doi: 10.1038/nrmicro.2015.3. [DOI] [PubMed] [Google Scholar]
- 127.Shewell LK, Day CJ, Jen FE-C, Haselhorst T, Atack JM, et al. All major cholesterol-dependent cytolysins use glycans as cellular receptors. Sci Adv. 2020;6:eaaz4926. doi: 10.1126/sciadv.aaz4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Beddoe T, Paton AW, Le Nours J, Rossjohn J, Paton JC, et al. Structure, biological functions and applications of the AB5 toxins. Trends Biochem Sci. 2010;35:411–418. doi: 10.1016/j.tibs.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang H, Paton JC, Paton AW. Pathologic changes in mice induced by subtilase cytotoxin, a potent new Escherichia coli AB5 toxin that targets the endoplasmic reticulum. J Infect Dis. 2007;196:1093–1101. doi: 10.1086/521364. [DOI] [PubMed] [Google Scholar]
- 130.Seyahian EA, Oltra G, Ochoa F, Melendi S, Hermes R, et al. Systemic effects of Subtilase cytotoxin produced by Escherichia coli O113:H21. Toxicon. 2017;127:49–55. doi: 10.1016/j.toxicon.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 131.Byres E, Paton AW, Paton JC, Löfling JC, Smith DF, et al. Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature. 2008;456:648–652. doi: 10.1038/nature07428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee S, Inzerillo S, Lee GY, Bosire EM, Mahato SK, et al. Glycan-mediated molecular interactions in bacterial pathogenesis. Trends Microbiol. 2021;30:254–267. doi: 10.1016/j.tim.2021.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Song J, Gao X, Galán JE, et al. Structure and function of the Salmonella Typhi chimaeric A(2)B(5) typhoid toxin. Nature. 2013;499:350–354. doi: 10.1038/nature12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Deng L, Song J, Gao X, Wang J, Yu H, et al. Host adaptation of a bacterial toxin from the human pathogen Salmonella Typhi. Cell. 2014;159:1290–1299. doi: 10.1016/j.cell.2014.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nguyen T, Lee S, Yang Y-A, Ahn C, Sim JH, et al. The role of 9-O-acetylated glycan receptor moieties in the typhoid toxin binding and intoxication. PLoS Pathog. 2020;16:e1008336. doi: 10.1371/journal.ppat.1008336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lee S, Yang Y-A, Milano SK, Nguyen T, Ahn C, et al. Salmonella typhoid toxin PltB subunit and its non-typhoidal salmonella ortholog confer differential host adaptation and virulence. Cell Host & Microbe. 2020;27:937–949. doi: 10.1016/j.chom.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Almagro-Moreno S, Boyd EF. Sialic acid catabolism confers a competitive advantage to pathogenic Vibrio cholerae in the mouse intestine. Infect Immun. 2009;77:3807–3816. doi: 10.1128/IAI.00279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Alisson-Silva F, Liu JZ, Diaz SL, Deng L, Gareau MG, et al. Human evolutionary loss of epithelial Neu5Gc expression and species-specific susceptibility to cholera. PLoS Pathog. 2018;14:e1007133. doi: 10.1371/journal.ppat.1007133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang S, Berntsson RP-A, Tepp WH, Tao L, Johnson EA, et al. Structural basis for the unique ganglioside and cell membrane recognition mechanism of botulinum neurotoxin DC. Nat Commun. 2017;8 doi: 10.1038/s41467-017-01534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zalem D, Ribeiro JP, Varrot A, Lebens M, Imberty A, et al. Biochemical and structural characterization of the novel sialic acid-binding site of Escherichia coli heat-labile enterotoxin LT-IIb. Biochem J. 2016;473:3923–3936. doi: 10.1042/BCJ20160575. [DOI] [PubMed] [Google Scholar]