Abstract
Alternaria alternata is a common species of fungus frequently isolated from plants as both an endophyte and a pathogen. Although the current definition of A. alternata rests on a foundation of morphological, genetic and genomic analyses, doubts persist regarding the scope of A. alternata within the genus due to the varied symbiotic interactions and wide host range observed in these fungi. These doubts may be due in large part to the history of unstable taxonomy in Alternaria, based on limited morphological characters for species delimitation and host specificity associated with toxins encoded by genes carried on conditionally dispensable chromosomes. This review explores the history of Alternaria taxonomy, focusing in particular on the use of nutritional mode and host associations in species delimitation, with the goal of evaluating A. alternata as it currently stands based on taxonomic best practice. Given the recombination detected among isolates of A. alternata, different symbiotic associations in this species should not be considered phylogenetically informative.
Keywords: taxonomy, systematics, symbiosis, fungi, species delimitation
Introduction
Fungi are widespread and exhibit broad variation in nutritional mode, living both as symbionts and as saprophytes on organic matter [1–3]. However, fungal ecology and evolution interface in varied ways; nutritional mode does not appear to be correlated with phylogeny for most fungi [4–6], while other lineages seem constrained to a single lifestyle [6, 7]. In this review, I examine the relationship between nutritional mode and systematics in Alternaria alternata, a filamentous Ascomycete occurring on plants as a pathogen and endophyte, and in soil as a saprophyte [8].
A. alternata is currently considered to be a cosmopolitan species with a wide host range [9–13]. Because A. alternata is pathogenic on many important crop plants [14] but also lives in asymptomatic symbiosis as an endophyte of many plants [13, 15–20], it is necessary for plant pathologists to know whether a given strain of A. alternata indicates a threat to food safety or simply the presence of an endophyte [21–23]. Given the prevalence of endophytic A. alternata [13, 24], it is also vital for conservationists to understand if changes to the abiotic environment might cause asymptomatic infections of A. alternata to shift into parasitism, leading to further stress to vulnerable plant populations [25–29]. These questions, however, are difficult to address due to a history of unstable taxonomy in the genus Alternaria [10, 12, 23, 30–38].
Many taxa described as ‘cosmopolitan’ are associated with long histories of unsatisfactory taxonomic revision because they lack readily recognizable characters, so their diversity is not captured in their taxonomy [39–46]. Microscopic organisms in particular are said to suffer from the ‘low morphology problem’ [40], where few morphological characters might be used to broadly delimit species that are then widely applied [43, 44, 46]. Typically, subsequent phylogenetic analyses based on DNA sequencing of these broadly delimited morphospecies have resolved more narrowly defined species with smaller geographical and host ranges, as well as physiological characters that had been previously overlooked that serve to diagnose the newly delimited species (see meta-analysis [41–47]). Unfortunately, for Alternaria we see the opposite case; phylogenetic analyses of sequence data have found fewer monophyletic groups than morphospecies, and agronomically relevant host-specific toxins (HSTs) do not segregate with individual lineages [10, 12, 34, 36, 37, 48, 49]. Without a stable basis for species delimitation in Alternaria, it is unclear if traits such as host interactions and nutritional modes are phylogenetically conserved, or if Alternaria spp. are truly cosmopolitan.
The purpose of this review is to explore how A. alternata has been historically delimited, and how its varied host interactions have influenced the concept of species in this lineage. I begin with a brief history of endophytes, how early research into fungal endophytes and Alternaria intersect, and the close phylogenetic association of endophytism and parasitism. Then, I trace the taxonomy and systematics of A. alternata from its original morphological description to its most recent re-description and phylogenetic characterization. In particular, I highlight evidence for genetic incongruence and recombination in this lineage, and how this may facilitate transfer of genetic elements encoding HSTs. Finally, I discuss current best practices in fungal species recognition, and attempt to apply them to A. alternata as it is currently defined.
A brief history of endophytes
History and use of the term endophyte
Today, the term endophyte is generally applied to organisms that live inside of plants without causing symptoms of infection [25, 50–52], but its original usage was very different. Heinrich Friedrich Link first coined the term ‘entophytae’ in 1809, meaning merely any organism living inside a plant. His definition was intended for pathogens, since it was assumed that healthy plants did not harbour internal symbionts [53]. When Galippe reported finding bacteria and fungi inside of healthy plants in 1887 and posited that they might have a beneficial role, the application of ‘endophyte’ expanded to include the potential for mutualists and commensals [54]. When rhizobia and mycorrhizae were discovered in the 1880s [55, 56], they were allowed under this definition of endophyte as well, although having their own unique labels meant that ‘endophyte’ was only sporadically applied to them. It was not until a century later in 1986 that George Carroll restricted the use of the term endophyte to micro-organisms that cause asymptomatic infections within plant tissues, excluding pathogens and mutualists such as mycorrhizae and rhizobia [50]. Aside from minor considerations over the possibility of latent pathogens residing in asymptomatic plants [25], or non-mycorrhizal endophytes providing mutualistic benefits to their hosts [57–60], Carroll’s definition of endophytes is widely accepted today.
Research on fungal endophytes as they are currently defined grew out of the independent investigations of livestock toxicosis, plant pathogens and leaf litter decomposition [61–65]. Other papers occasionally reported the presence of endophytes indirectly, especially surveys of internal moulds in visibly healthy seeds, but surface-sterilization was not widely applied in seed fungus research so the status of these moulds as endophytes is sometimes uncertain [66–68]. The first major effort to isolate endophytes began in response to ryegrass staggers affecting livestock in New Zealand [61]. Fungal endophytes had previously been observed on the grass Lolium perenne in the late 1800s [69–71], and a fungal endophyte was implicated in the case of ryegrass staggers [61], but toxicity was not definitively assigned to a species of Epichloë (classified as Neotyphodium at the time) on L. perenne until 1981 [72]. Other Epichloë spp. have been isolated from other grasses [73–80], and their particular lifestyle of systemic infection and vertical transmission eventually led to the division of endophyte research between vertically and horizontally transmitted endophytes. We are interested in horizontally transmitted endophytes for the purposes of this review.
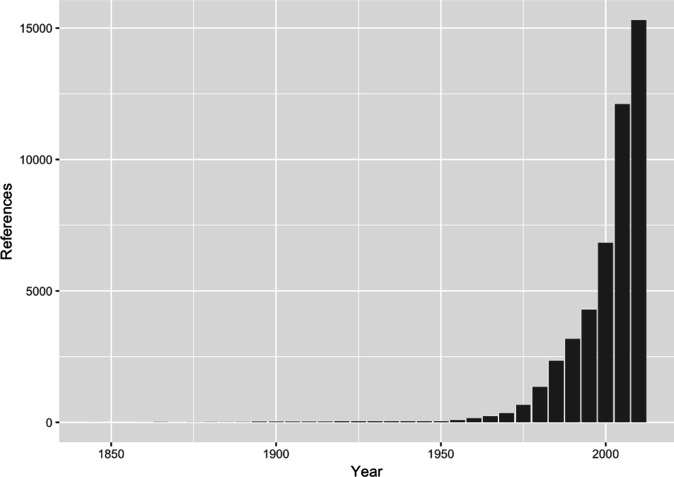
Horizontally transmitted endophytes were originally sampled from healthy leaf tissue as a control to compare either diseased or decomposing leaves [62–65]. One research group focused on the endophytes of Nicotiana spp., particularly the difference between the endophytic Alternaria spp. abundant on these plants and the pathogenic A. alternata that caused leaf spot of tobacco on the same plants [16, 81–84]. Meanwhile, research on leaf litter decomposition [63, 65, 85] grew into an extensive exploration of the diversity and host affinities of endophytes of gymnosperms and other trees, due in large part to the work of George Carroll and his collaborators throughout the 1970s and 1980s [86–94]. These studies were the beginning of a rapid increase in the pace of endophyte research (Fig. 1), which was facilitated by use of DNA sequencing for fungal identification in the 1990s and environmental sequencing in the early 2000’s.
Fig. 1.
The number of papers referring to endophytes has rapidly increased, particularly due to the proliferation of environmental DNA sequencing beginning in the early 2000s. Yearly reference counts were retrieved by searching Google Scholar with the term ‘endophyt’ and binned in 5 year intervals.
Studies reporting Alternaria strains as endophytes
The earliest report of Alternaria isolated from asymptomatic plant tissue seems to be from an investigation of mycorrhizae published in 1905, where the endophytic Alternaria strain was isolated instead of the mycorrhizal target ([95] as summarized in [96]). Alternaria was also commonly found in studies targeting other fungi in the 1910s, including systemic endophytes of liverwort thallus [97], contaminants of wheat seeds [98], and mycorrhizae associated with orchid roots [96]. Surveys of seed fungi have frequently reported infestation by Alternaria spp., generally either A. alternata or its synonym ‘A. tenuis’ [17, 66, 68, 99–101]. Paul Neergaard, a major name in early Alternaria taxonomy, was better known as a seed pathologist [102].
Among studies purposefully sampling endophytes, a strain of Alternaria was first isolated as one of the fast-growing fungi that overgrew the elusive Epichloë and complicated the search for the causal agent of ryegrass staggers [61]. Alternaria spp., likely A. alternata as currently defined, were the most abundant endophytes isolated from Nicotiana spp. by Spurr and Welty [16]. After pathogenicity assays confirmed that endophytic isolates were unable to cause tobacco leaf spot, Spurr and Welty speculated that the abundance of endophytic Alternaria isolates on tobacco was contributing to inconsistencies in species delimitation for the pathogenic A. alternata isolated from tobacco [16], which have since been renamed A. longipes [10].
Aside from tobacco, endophytic A. alternata has been reported from a variety of crop plants, including strawberry, apple, rapeseed, soybean, rice and citrus [48, 103–109]. Notably, Alternaria spp. including A. alternata are also considered pathogenic on these plants [14]. Incidence of isolation varies among host species and among different studies on the same host [110, 111]; occasionally Alternaria strains are the most abundant endophytes isolated [16, 24, 112], but usually they are detected in 1–10 % of samples [15, 17–19].
Phylogenetic analysis of endophytic lifestyle
Endophytism is common among Kingdom Fungi and has apparently evolved multiple times [4, 6]. In a phylogeny of the Ascomycota, frequent transitions between endophytism and parasitism were predicted by ancestral state reconstruction [4]. Transitions from parasitism to endophytism have been detected within individual genera, even multiple times [5, 7]. Transitions between endophytism and saprotrophy appear to be less common [4], and biotrophic pathogens do not appear to revert to other nutritional modes once they have evolved [6]. Ancestral state reconstructions of nutritional modes are complicated by uncertainty in assigning a species or isolate to a nutritional mode based on observations in nature, as well as species that encompass several different lifestyles [4].
It has also been proposed that endophytic fungi are pathogens or saprophytes waiting for ideal conditions to emerge [5, 113–115]. Indeed, some endophytes may also function as saprophytes since they have been isolated from both living and dead host tissue [116–120], and others are capable of causing disease on their endophytic host [121]. More generally, many fungi may cause endophytic or pathogenic infections depending on host identity, host condition and abiotic factors [26, 29, 115, 122–126]. Speciation based on endophytic specialization in asexual lineages arising from pathogens has occurred in vertically transmitted Epichloë on ryegrass [127], and may be occurring in an exclusively endophytic, asexual lineage of Verticillium dahliae on mustards [7], but it should not be considered the norm for horizontally transmitted endophytes.
Taxonomy and systematics of the genus Alternaria
Description of Alternaria amongst the ‘waste basket assemblage’
The first species in the genus Alternaria, A. tenuis was described from dead plant material by Nees von Esenbeck in 1816 [128], without sufficient characters for delimitation of phenotypically similar species or a surviving type specimen [13, 30, 31, 129]. The situation was complicated by Fries initially rejecting Nees' description while describing the highly similar genus Macrosporium, and then electing to synonymize A. tenuis under the name Torula alternata in 1819 [30, 130]. After Chevallier reported conflicting spore morphology between Alternaria and Torula [131], Fries finally accepted its generic status, but maintained that Macrosporium was a separate genus [31, 132, 133]. The distinction of these two genera was widely criticized due to overlap in morphological characters and host associations [30, 31, 129, 134, 135], and most species previously ascribed to Macrosporium are now categorized under Alternaria [9, 136].
Eventually the type species for Alternaria was formally established as Alternaria alternata (Fries) Keissler (1912), despite the apparent precedence of the name ‘A. tenuis’ for the same taxon as described by Nees [128]. This inconsistency is due to the status of ‘A. tenuis’ as an ‘invalid pre-starting date epithet’ based on the International Code of Botanical Nomenclature, and therefore illegal [32, 129]. As a result of this taxonomic technicality, ‘A. tenuis’ remained the more popular epithet applied to A. alternaria specimens for decades [31, 137], including one particularly egregious use of ‘A. tenuis auct. sensu str. cf. Neergaard (1945)’ by Yu, Mathur and Neergaard in 1982 [138]. Use of A. alternata was popularized by Emory Simmons, who spent decades cataloguing Alternaria spp. based on extremely fine morphological examinations beginning in the 1960s [32, 139]. However, ‘A. tenuis’ still persists in modern publications, likely due to GenBank records that still carry the old name [140].
Simmons, in addition to being the foremost expert on Alternaria morphology, was also extremely disdainful of Fries and his contributions to Alternaria taxonomy [141]. Simmons claimed that Fries had a low opinion of microfungi and relied on ‘published observations which often were as poor as or worse than his own,’ and that his classifications were no better than ‘historical fiction.’ [141]. In defence of Fries, fungi that lack a known sexual stage (‘anamorphs’ vs. sexual ‘teliomorphs’) are character-poor and have historically proven difficult to categorize [142]. Later authors declared that anamorphs such as Alternaria spp. were at best a confusing problem and at worst not worth characterizing, and lumped them all into the unnatural category of ‘Fungi Imperfecti’ [141–143]. Saccardo’s classification system for the Fungi Imperfecti, based on spore morphology and specific host interactions, was a practical solution for classifying plant pathogens that broke down almost immediately when every soil fungus without a known sexual stage was thrown into the ‘waste basket assemblage’ of Fungi Imperfecti [142]. By the time over 30 thousand ‘imperfect’ fungal species had been unceremoniously dumped into the bin, a great deal of inertia stood in the way of their ever being systematically reclassified [144].
Generic delimitation and species concepts in Alternaria: a problem with unsatisfying solutions
Unfortunately for the taxonomists of the 1900s, any alternative to Saccardo’s strategy for classifying microfungi without a known sexual phase was also inevitably morphological [142], and morphological classifications can be subjective [30]. Several camps of taxonomists (Elliott, Young, Wiltshire, Neergaard, Joly, Simmons, later Simmons, Roberts, Nishimura and colleagues) spent the bulk of the 1900s trying to circumscribe the genus Alternaria and catalogue its species using whatever physical and biochemical characters they could reliably detect. These camps rarely agreed.
The early period of generic circumscription featured arguments concerning synonymizing highly similar genera and the reliability of spore measurements and host ranges as informative characters [30, 145]. Elliott explicitly studied spore measurements by fixing several specimens and sending them to colleagues, and determined that variability between microscopists was greater than published intra-species variability [30]. Without an acceptable alternative forthcoming, species description based on spore measurements persisted [135, 146]. Species were frequently described, synonymized, and shuffled among genera including Alternaria, Macrosporium, Stemphylium and Pleospora (there was also, briefly, an expectation that all Alternaria spp. would eventually be revealed as anamorphs of Pleospora) [30, 32, 129, 134, 135, 137, 145, 147]. Disagreements over taxonomic classifications appear to have become somewhat heated (see in particular Joly’s take on Neergaard [147] and Simmons’s review of Joly [148]), although they may have been more civil in person than they were in print. The status of these genera could not satisfactorily be settled until the application of molecular methods [9, 21, 136]; in the meantime, the focus of taxonomists shifted to species delimitation.
Alternaria species can be broadly split into two categories: large spored species, which are relatively simple to diagnose due to their distinctive morphologies and stable host ranges, and small spored species, which are not [36, 149]. Among the small spored species, the group currently called A. infectoria was often treated separately in Alternaria taxonomic studies due to its sexual stage (otherwise not observed in Alternaria) identified as a species of Pleospora [21, 30]. The remainder of small spored Alternaria are currently classified as Section Alternaria [9–12, 136] and include A. alternata, A. arborescens and a wide variety of presumed host-specific plant pathogens that have been described as both species and as pathotypes or formae speciales of A. alternata [10, 34, 150–152]. Section Alternaria, being the figurative squeaky wheel, has received the bulk of the taxonomic revision.
Initially, it was difficult to positively identify or describe new species similar to A. alternata, the type species of Alternaria, because no type material was available for comparison [129]. This changed with the discovery of collections originally identified as A. tenuis by Nees that had been stored in Persoon’s herbarium, allowing Simmons to designate a formal epitype for A. alternata in 1967 [32]. Emboldened by the certainty of a good type specimen, Simmons proceeded to describe Alternaria spp. for the next 40 years [153–160], recognizing a total of 275 species with the publication of a monograph in 2007 [138]. These species descriptions were based on morphological observations (patterns of conidiophore development under tightly regulated culture conditions) and pathogenicity studies, going well beyond simple spore measurements [35], although some of the characters (particularly conidiophore development) were considered difficult to apply by non-experts [149]. Simmons' concepts of species were generally considered to be very narrowly defined [9, 36].
Opponents to Simmons' narrow species descriptions questioned the biological relevance and diagnostic value of the morphological characters he favoured [34]. Slifkin used electron microscopy to measure the variance of spore and conidiophore morphology within isolates and concluded that either these characters were not diagnostic, or that most of the species she examined should be synonymized [161]. Meanwhile, Nishimura and colleagues had begun describing pathogenic isolates as pathotypes of A. alternata, as these isolates were morphologically indistinguishable from non-pathogenic A. alternata aside from the production of key host-specific toxins (HSTs) that could be lost in long-term culture [33, 161–163]. Simmons, conversely, considered production of HSTs to be a basis for species differentiation [160]. More recent studies making use of DNA sequencing have supported a more broadly delimited A. alternata with pathotypes distinguished by HSTs, although the systematics of Section Alternaria is still the subject of study [10, 12].
Alternaria in the sequencing era (1995 to present)
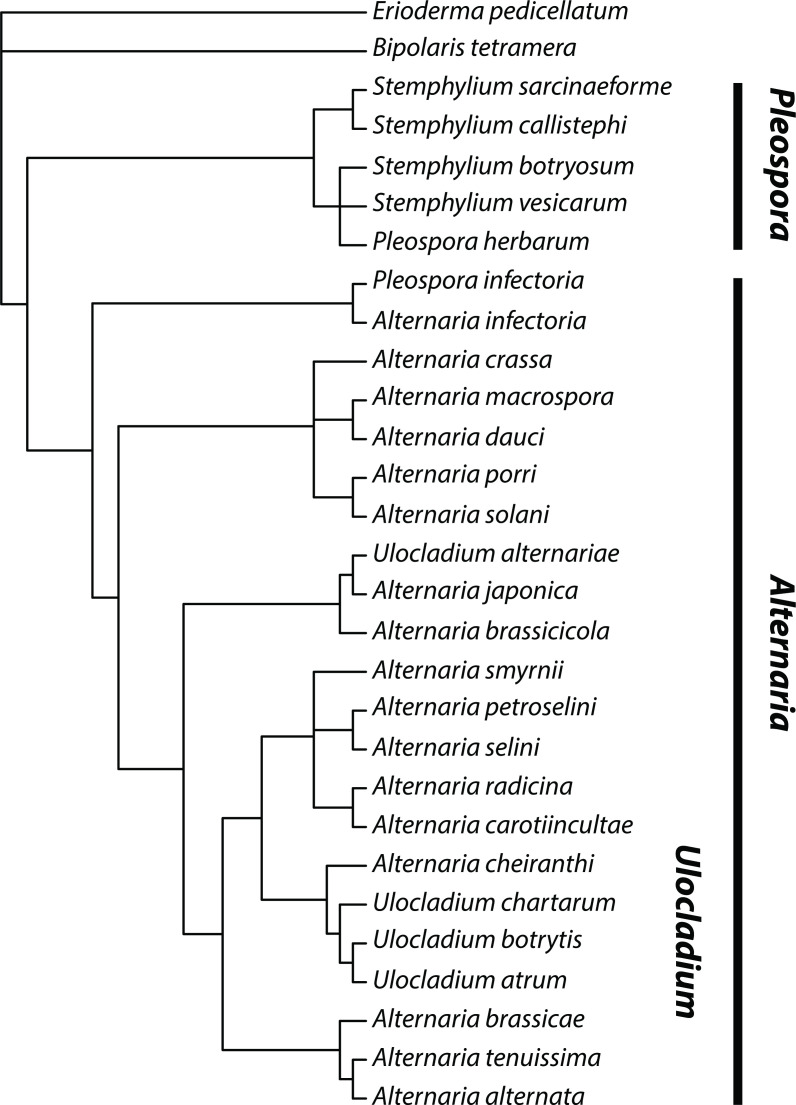
DNA sequencing to characterize isolates of Alternaria appears to have begun in earnest in 1995. Jasalavich et al. [164] reported the first molecular evidence that Alternaria belongs in the Pleosporaceae, and found that Alternaria was closely related to Stemphylium and Pleospora, in support of morphological evidence. In the same year, Kusaba and Tsuge [34] constructed a phylogeny of HST-producing Alternaria from ITS sequences and could not distinguish HST-producers from non-pathogenic A. alternata, in agreement with the recommendations of Nishimura et al. [33] and previous findings based on restriction fragment length polymorphisms (RFLPs) [165, 166]. Additional sequence-based phylogenetic studies found similar results, clustering several genera (Macrosporium, Stemphylium, Ulocladium, Lewia, Nimbya) closely with Alternaria and failing to support species status for many morphospecies (Fig. 2), particularly pathogens of citrus (Fig. 3) [21, 27, 36, 37, 149, 167].
Fig. 2.
Relationships between species of Alternaria, Ulocladium, Stemphylium and Pleospora as determined by Pryor and Gilbertson [21]. Ulocladium spp. are nested within the Alternaria lineage. Pleospora (here containing several Stemphylium spp.) appears as sister to Alternaria, except for a single individual assigned to ‘P. infectoria’ in the A. infectoria clade. Redrawn based on phylogeny constructed using neighbour-joining and parsimony analysis of nuclear ITS/5.8S rDNA sequences [21].
Fig. 3.
Spore size trait (large or small) mapped onto Alternaria spp. based on phylogeny constructed by Peever et al. [36] in an analysis of citrus pathogens. All small-spored isolates from citrus were indistinguishable from A. alternata using mitochondrial RNA large subunit (mtLSU) sequence data. Redrawn based on phylogeny constructed using maximum likelihood analysis of mtLSU sequences [36].
For several years, authors seemed hesitant to address the increasing number of genera closely aligned to Alternaria, unless it was to propose creating more. Lewia was established to house Pleospora spp. with Alternaria anamorphs [21], and Undifilum was established for swainsonine-producing mutualists of locoweeds [168]. Maintaining the status of these genera alongside a monophyletic Alternaria meant excluding all Alternaria spp. with a sexual stage; this would have required reclassifying the medically important and widely applied epithet A. infectoria (Fig. 4, Sect. Infectoriae) [149]. Lawrence et al. [169] proposed reclassifying Nimbya and Embellisia as Alternaria, since their status as separate genera was only supported by morphological characters, but later chose to discuss Alternaria as polyphyletic rather than come down on either side of including or excluding A. infectoria [136]. Finally, Woudenberg et al. redefined Alternaria to definitively include A. infectoria, as well as all allied genera that would otherwise be rendered paraphyletic (Fig. 4) [9].
Fig. 4.
Bayesian 50 % majority rule consensus tree based on three loci showing the phylogeny of the genus Alternaria as defined by Woudenberg et al. [9]. Sections proposed by Woudenberg et al. are shown at the right. Adapted from Fig. 1 of [9] with permission from the authors.
Recombination among lineages of Alternaria alternata
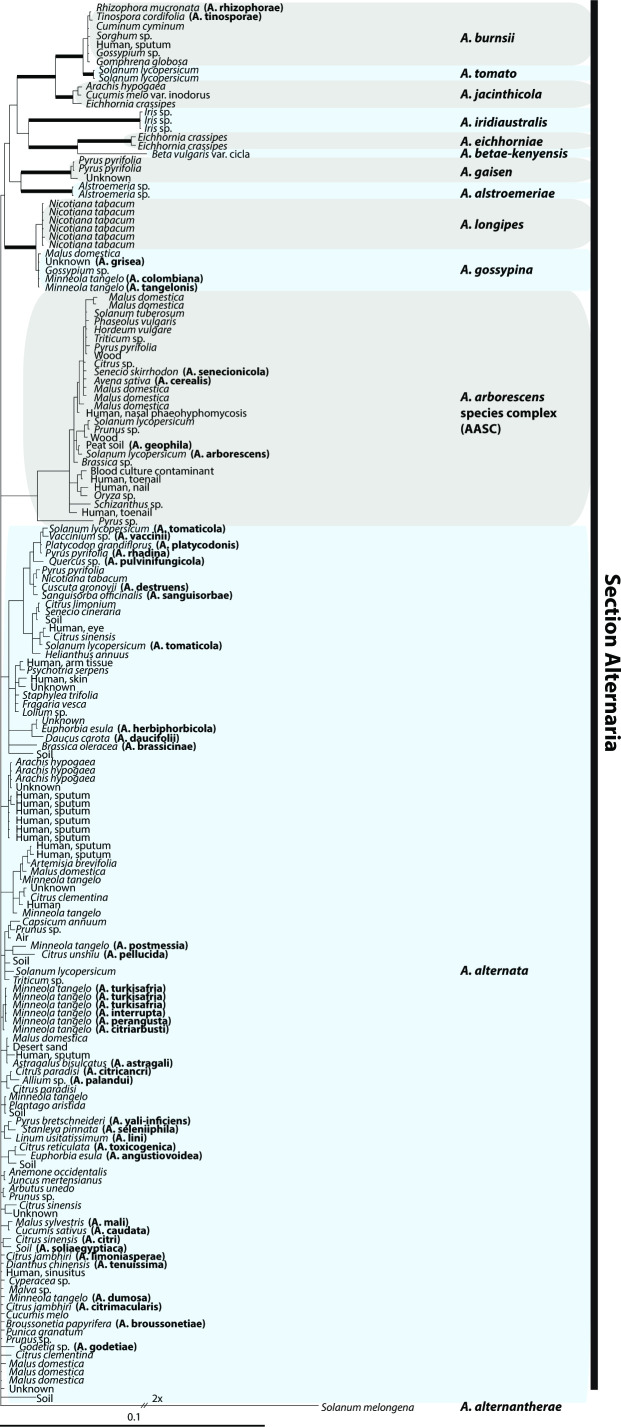
The redefinition of Alternaria by Woudenberg et al. resolved deeper systematic issues, but could not address A. alternata and its purported pathotypes beyond placing them into a single section, Sect. Alternaria [9]. Further efforts to characterize Sect. Alternaria have generally supported the use of pathotypes to describe isolates that produce HSTs rather than elevating them to species, and have synonymized more species under the name A. alternata (Fig. 5) [10, 11, 48]. Different studies have reported some support for an A. arborescens species complex separate from A. alternata based on morphological and molecular characters [10, 11, 14], but the entire section seems to be characterized by a common metabolic profile [14, 22, 23, 170]. Based on these shared metabolites, Patriarca et al. [23] suggested that Section Alternaria should be considered a threat to food safety regardless of species delimitation within the section.
Fig. 5.
Bayesian 50 % majority rule consensus tree based on seven loci, showing isolation sources of Alternaria spp. in Section Alternaria as re-defined by Woudenberg et al. [10]. Accessions with names synonymized by Woudenberg et al. are noted in parentheses. Retained species are delimited by highlight (grey or blue) with the epithet labelled at the right. Adapted from Fig. 2 of [10] with permission from the authors.
Several studies of taxa synonymized under A. alternata by Woudenberg et al. [10] have found evidence for gene tree discordance and recombination, suggesting that sexual or parasexual (nuclear fusion followed by haploidization) reproduction occurs in this lineage [38, 48, 49]. The presence of both mating type idiomorphs (highly diverged alleles) in roughly equal frequencies is considered good evidence for the maintenance of sexual reproduction in Ascomycetes [171], and both mating types have been sampled from A. alternata populations [12, 48]. Parasexual reproduction has been implicated as the mechanism of recombination in A. solani [172], and either method of genetic exchange could explain the signals of recombination observed in A. alternata [48].
Given the history of exchange across lineages, HSTs are not phylogenetically informative, but they do inform potential pathology. Horizontal exchange is facilitated by the location of HSTs on conditionally dispensable chromosomes (CDCs), which may segregate separately from the rest of the genome and can be lost or gained over time [173–180]. Recombination through sexual or parasexual reproduction may facilitate transfer of CDCs among lineages [12, 48, 181]. While screening for CDCs that carry HSTs seems to be the best course of action for the purposes of detecting economically important pathogenic isolates [12, 182], CDCs might be found in unexpected places. Strains capable of producing HSTs have been isolated as endophytes from non-target hosts [151, 183, 184], suggesting that specificity resides in the host-toxin interactions rather than as a characteristic of the toxin itself [150].
The concept of species in Fungi and current best practices
An essential goal of taxonomy and systematics is to reliably place individual organisms into categories such that we can make predictions about their structure and physiology. Taken to either extreme, too strict or too broad, and these categories become impractical: extremely strict categories will be highly predictive but rarely applicable, while broad categories can be widely applied but allow few meaningful predictions of biological form or function. Species, then, are constructs representing the smallest category of organisms allowing us to make biologically relevant predictions [185, 186]. The practical application of taxonomy is deciding how dissimilar two species need to be.
Underlying all modern concepts of species is the Evolutionary Species Concept [185, 187], the idea that species should be monophyletic groups and share both evolutionary history and unique, diagnosable characters [185–188]. Less theoretical species concepts help us to recognize species under this guiding principle, and establish boundaries between species. The most widely used methods of species delimitation in fungi have historically been the Morphological Species concept [189] and the Biological Species Concept [190, 191], particularly due to the requirement for morphological descriptions when describing new species of fungi [46]. Species defined solely by morphology tend to be too broad and can be confounded by convergent evolution [42, 44–46], whereas biological species cannot be defined in fungi without a sexual stage [186], but both of these concepts are valuable for species recognition if species are first defined by another method [46, 47]. Early definitions in the genus Alternaria were based entirely on morphology [30–32, 128–134].
Current best practice in fungal species delimitation is use of phylogenetic species based on Genealogical Concordance Phylogenetic Species Recognition (GCPSR) [171, 192]. By this method, species boundaries are defined at the point of transition between gene tree concordance and conflict [187, 193–196]. This method is broadly applicable despite the prevalence of asexual reproduction strategies in fungi because at least low levels of recombination occur even in largely clonal lineages, and purely asexual lineages are extremely rare (reviewed in [171]). Recent systematic treatments of the genus Alternaria and Section Alternaria have relied on GCPSR [9–11].
Conclusions
Woudenberg et al. [10] last defined A. alternata in 2015 based on whole-genome sequences as well as a multi-locus phylogeny, synonymizing most of its previous pathotypes and morphologically similar taxa (Fig. 5). CDCs carrying HSTs segregate independently among lineages within this species, likely through horizontal transfer or recombination via sexual or parasexual reproduction [173–180], and should not be considered diagnostic of monophyletic groups. More recently, authors have referred to this group as a species complex (or the ‘tenuissima clade’ [12]), but based on best practice in species delimitation, the re-definition by Woudenberg et al. [10] presents an adequate representation of the taxonomy of this lineage. A. alternata is, in fact, a cosmopolitan species with a wide host range encompassing multiple nutritional modes.
Given the phylogenetic basis of endophytism and varied lifestyles of many fungal species [26, 29, 115, 122–126], as well as several instances of pathogenicity in multiple lineages of A. alternata not associated with HSTs [27, 36, 37, 48, 49, 197], I suspect that all lineages of A. alternata are capable of both endophytism and at least mild pathogenicity. The broad suite of metabolites characteristic of Section Alternaria would likely support a capacity for varied host interactions and nutritional modes [14, 22, 23, 170]. In short, there is no evidence that particular lineages of A. alternata are genetically constrained to endophytism or parasitism on particular plants, aside from specificity imparted by production of HSTs.
Funding information
M.D. was supported by the Plant and Microbial Biology Graduate Program, University of Minnesota.
Acknowledgements
I would like to thank Georgiana May for helpful discussions that developed the framework of this review, and Corby Kistler and two anonymous reviewers for their valuable comments on the manuscript.
Author contributions
Mara DeMers - https://orcid.org/0000-0002-3782-5634
Conflicts of interest
The author declares that there are no conflicts of interest.
Footnotes
Abbreviations: CDC, conditionally dispensable chromosome; GCPSR, genealogical concordance phylogenetic species recognition; HST, host-specific toxin; ITS, internal transcribed spacer; mtLSU, mitochondrial large ribosomal subunit; rDNA, ribosomal DNA; RFLP, restriction fragment length polymorphism; sp., species (singular); spp., species (plural).
References
- 1.Arnold AE, Lutzoni F. Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology. 2007;88:541–549. doi: 10.1890/05-1459. [DOI] [PubMed] [Google Scholar]
- 2.Taylor DL, Hollingsworth TN, McFarland JW, Lennon NJ, Nusbaum C, et al. A first comprehensive census of fungi in soil reveals both hyperdiversity and fine-scale niche partitioning. Ecological Monographs. 2014;84:3–20. doi: 10.1890/12-1693.1. [DOI] [Google Scholar]
- 3.Sánchez-García M, Ryberg M, Khan FK, Varga T, Nagy LG, et al. Fruiting body form, not nutritional mode, is the major driver of diversification in mushroom-forming fungi. Proc Natl Acad Sci U S A. 2020;117:32528–32534. doi: 10.1073/pnas.1922539117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold AE, Miadlikowska J, Higgins KL, Sarvate SD, Gugger P, et al. A phylogenetic estimation of trophic transition networks for ascomycetous fungi: are lichens cradles of symbiotrophic fungal diversification? Syst Biol. 2009;58:283–297. doi: 10.1093/sysbio/syp001. [DOI] [PubMed] [Google Scholar]
- 5.Chaverri P, Samuels GJ. Evolution of habitat preference and nutrition mode in a cosmopolitan fungal genus with evidence of interkingdom host jumps and major shifts in ecology. Evolution. 2013;67:2823–2837. doi: 10.1111/evo.12169. [DOI] [PubMed] [Google Scholar]
- 6.Delaye L, García-Guzmán G, Heil M. Endophytes versus biotrophic and necrotrophic pathogens—are fungal lifestyles evolutionarily stable traits? Fungal Diversity. 2013;60:125–135. doi: 10.1007/s13225-013-0240-y. [DOI] [Google Scholar]
- 7.Wheeler DL, Dung JKS, Johnson DA. From pathogen to endophyte: an endophytic population of Verticillium dahliae evolved from a sympatric pathogenic population. New Phytol. 2019;222:497–510. doi: 10.1111/nph.15567. [DOI] [PubMed] [Google Scholar]
- 8.Thomma BPHJ. Alternaria spp.: from general saprophyte to specific parasite. Mol Plant Pathol. 2003;4:225–236. doi: 10.1046/j.1364-3703.2003.00173.x. [DOI] [PubMed] [Google Scholar]
- 9.Woudenberg JHC, Groenewald JZ, Binder M, Crous PW. Alternaria redefined. Stud Mycol. 2013;75:171–212. doi: 10.3114/sim0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woudenberg JHC, Seidl MF, Groenewald JZ, de Vries M, Stielow JB, et al. Alternaria section Alternaria: Species, formae speciales or pathotypes? Stud Mycol. 2015;82:1–21. doi: 10.1016/j.simyco.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Armitage AD, Barbara DJ, Harrison RJ, Lane CR, Sreenivasaprasad S, et al. Discrete lineages within Alternaria alternata species group: Identification using new highly variable loci and support from morphological characters. Fungal Biol. 2015;119:994–1006. doi: 10.1016/j.funbio.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 12.Armitage AD, Cockerton HM, Sreenivasaprasad S, Woodhall J, Lane CR, et al. Genomics evolutionary history and diagnostics of the Alternaria alternata species group including apple and asian pear pathotypes. Front Microbiol. 2019;10:3124. doi: 10.3389/fmicb.2019.03124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence DP, Rotondo F, Gannibal PB. Biodiversity and taxonomy of the pleomorphic genus Alternaria . Mycol Progress. 2015;15:3. doi: 10.1007/s11557-015-1144-x. [DOI] [Google Scholar]
- 14.El Gobashy SF, Mikhail WZA, Ismail AM, Zekry A, Moretti A, et al. Phylogenetic, toxigenic and virulence profiles of Alternaria species causing leaf blight of tomato in Egypt. Mycol Progress. 2018;17:1269–1282. doi: 10.1007/s11557-018-1442-1. [DOI] [Google Scholar]
- 15.Jacks H. A note on fungi isolated from plants. New Zealand J Agric Res. 2011;3:250–252. doi: 10.1080/00288233.1960.10418080. [DOI] [Google Scholar]
- 16.Spurr HW. Characterization of endophytic fungi in healthy leaves of Nicotiana spp. Phytopathology. 1975;65:417. doi: 10.1094/Phyto-65-417. [DOI] [Google Scholar]
- 17.Wallace HAH, Sinha RN. Microflora of stored grain in international trade. Mycopathologia. 1975;57:171–176. doi: 10.1007/BF00551424. [DOI] [PubMed] [Google Scholar]
- 18.Kowalski T, Kehr RD. Endophytic fungal colonization of branch bases in several forest tree species. Sydowia. 1992;44:137–168. [Google Scholar]
- 19.Cabral D, Stone JK, Carroll GC. The internal mycobiota of Juncus spp.: microscopic and cultural observations of infection patterns. Mycological Research. 1993;97:367–376. doi: 10.1016/S0953-7562(09)81140-4. [DOI] [Google Scholar]
- 20.Zamora P, Martinez-Ruiz C, Diez JJ. Fungi in needles and twigs of pine plantations from northern spain. Fungal Divers. 2008;30:171–184. [Google Scholar]
- 21.Pryor BM, Gilbertson RL. Molecular phylogenetic relationships amongst Alternaria species and related fungi based upon analysis of nuclear ITS and mt SSU rDNA sequences. Mycological Research. 2000;104:1312–1321. doi: 10.1017/S0953756200003002. [DOI] [Google Scholar]
- 22.da Cruz Cabral L, Rodriguero M, Stenglein S, Fog Nielsen K, Patriarca A. Characterization of small-spored Alternaria from Argentinean crops through a polyphasic approach. Int J Food Microbiol. 2017;257:206–215. doi: 10.1016/j.ijfoodmicro.2017.06.026. [DOI] [PubMed] [Google Scholar]
- 23.Patriarca A, da Cruz Cabral L, Pavicich MA, Nielsen KF, Andersen B. Secondary metabolite profiles of small-spored Alternaria support the new phylogenetic organization of the genus. Int J Food Microbiol. 2019;291:135–143. doi: 10.1016/j.ijfoodmicro.2018.11.022. [DOI] [PubMed] [Google Scholar]
- 24.DeMers MB, May G. Habitat-scale heterogeneity maintains fungal endophyte diversity in two native prairie legumes. Mycologia. 2021;113:20–32. doi: 10.1080/00275514.2020.1813487. [DOI] [PubMed] [Google Scholar]
- 25.Petrini O. Microbial Ecology of Leaves. New York: Springer; 1991. Fungal endophytes of tree leaves; pp. 179–197. [Google Scholar]
- 26.Redman RS, Dunigan DD, Rodriguez RJ. Fungal symbiosis from mutualism to parasitism: who controls the outcome, host or invader? New Phytol. 2001;151:705–716. doi: 10.1046/j.0028-646x.2001.00210.x. [DOI] [PubMed] [Google Scholar]
- 27.Rang J-C, Crous PW, Mchau GRA, Serdani M, Song S-M. Phylogenetic analysis of Alternaria spp. associated with apple core rot and citrus black rot in South Africa. Mycological Research. 2002;106:1151–1162. doi: 10.1017/S0953756202006524. [DOI] [Google Scholar]
- 28.Photita W, Lumyong S, Lumyong P, McKenzie EHC, Hyde KD. Are some endophytes of Musa acuminata latent pathogens? Fungal Divers. 2004;16:131–140. [Google Scholar]
- 29.van Kan JAL, Shaw MW, Grant-Downton RT. Botrytis species: relentless necrotrophic thugs or endophytes gone rogue? Mol Plant Pathol. 2014;15:957–961. doi: 10.1111/mpp.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elliott JA. Taxonomic characters of the genera Alternaria and Macrosporium . Am J Bot. 1917;4:439–476. doi: 10.1002/j.1537-2197.1917.tb05467.x. [DOI] [Google Scholar]
- 31.Tweedy BG, Powell D. The taxonomy of Alternaria and species of this genus reported on apples. Bot Rev. 1963;29:405–412. doi: 10.1007/BF02860826. [DOI] [Google Scholar]
- 32.Simmons EG. Typification of Alternaria, Stemphylium, and Ulocladium . Mycologia. 1967;59:67–92. doi: 10.1080/00275514.1967.12018396. [DOI] [PubMed] [Google Scholar]
- 33.Nishimura S, Sugihara M, Kohmoto K, Otani H. Two different phases in the pathogenicity of Alternaria pathogen causing black spot disease of Japanese pear. J Fac Agric Tottori University. 1978;13:1–10. [Google Scholar]
- 34.Kusaba M, Tsuge T. Phylogeny of Alternaria fungi known to produce host-specific toxins on the basis of variation in internal transcribed spacers of ribosomal DNA. Curr Genet. 1995;28:491–498. doi: 10.1007/BF00310821. [DOI] [PubMed] [Google Scholar]
- 35.Roberts RG, Reymond ST, Andersen B. RAPD fragment pattern analysis and morphological segregation of small-spored Alternaria species and species groups. Mycological Research. 2000;104:151–160. doi: 10.1017/S0953756299001690. [DOI] [Google Scholar]
- 36.Peever TL, Su G, Carpenter-Boggs L, Timmer LW. Molecular systematics of citrus-associated Alternaria species. Mycologia. 2004;96:119–134. doi: 10.1080/15572536.2005.11833002. [DOI] [PubMed] [Google Scholar]
- 37.Peever TL, Carpenter-Boggs L, Timmer LW, Carris LM, Bhatia A. Citrus black rot is caused by phylogenetically distinct lineages of Alternaria alternata . Phytopathology. 2005;95:512–518. doi: 10.1094/PHYTO-95-0512. [DOI] [PubMed] [Google Scholar]
- 38.Andrew M, Peever TL, Pryor BM. An expanded multilocus phylogeny does not resolve morphological species within the small-spored Altemrnaria species complex. Mycologia. 2009;101:95–109. doi: 10.3852/08-135. [DOI] [PubMed] [Google Scholar]
- 39.Thorpe JP, Sole-cava AM. The use of allozyme electrophoresis in invertebrate systematics. Zool Scripta. 1994;23:3–18. doi: 10.1111/j.1463-6409.1994.tb00368.x. [DOI] [Google Scholar]
- 40.Van Oppen MJH, Klerk H, Olsen JL, Stam WT. Hidden diversity in marine algae: some examples of genetic variation below the species level. J Mar Biol Ass. 2009;76:239–242. doi: 10.1017/S0025315400029192. [DOI] [Google Scholar]
- 41.Geiser DM, Pitt JI, Taylor JW. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus . Proc Natl Acad Sci U S A. 1998;95:388–393. doi: 10.1073/pnas.95.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Donnell K, Cigelnik E, Nirenberg HI. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia. 1998;90:465. doi: 10.2307/3761407. [DOI] [Google Scholar]
- 43.Klautau M, Russo CAM, Lazoski C, Boury-Esnault N, Thorpe JP, et al. Does cosmopolitanism result from overconservative systematics? a case study using the marine sponge Chondrilla nucula . Evolution. 1999;53:1414–1422. doi: 10.1111/j.1558-5646.1999.tb05406.x. [DOI] [PubMed] [Google Scholar]
- 44.Taylor JW, Jacobson DJ, Fisher MC. The evolution of asexual fungi: reproduction, speciation and classification. Annu Rev Phytopathol. 1999;37:197–246. doi: 10.1146/annurev.phyto.37.1.197. [DOI] [PubMed] [Google Scholar]
- 45.Agapow PM, Bininda-Emonds ORP, Crandall KA, Gittleman JL, Mace GM, et al. The impact of species concept on biodiversity studies. Q Rev Biol. 2004;79:161–179. doi: 10.1086/383542. [DOI] [PubMed] [Google Scholar]
- 46.Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D. Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi. Philos Trans R Soc Lond B Biol Sci. 2006;361:1947–1963. doi: 10.1098/rstb.2006.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geiser DM, Dorner JW, Horn BW, Taylor JW. The phylogenetics of mycotoxin and sclerotium production in Aspergillus flavus and Aspergillus oryzae . Fungal Genet Biol. 2000;31:169–179. doi: 10.1006/fgbi.2000.1215. [DOI] [PubMed] [Google Scholar]
- 48.Stewart JE, Thomas KA, Lawrence CB, Dang H, Pryor BM, et al. Signatures of recombination in clonal lineages of the citrus brown spot pathogen, Alternaria alternata sensu lato . Phytopathology. 2013;103:741–749. doi: 10.1094/PHYTO-08-12-0211-R. [DOI] [PubMed] [Google Scholar]
- 49.Stewart JE, Timmer LW, Lawrence CB, Pryor BM, Peever TL. Discord between morphological and phylogenetic species boundaries: incomplete lineage sorting and recombination results in fuzzy species boundaries in an asexual fungal pathogen. BMC Evol Biol. 2014;14:38. doi: 10.1186/1471-2148-14-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carroll GC. In: Microbiology of the Phyllosphere. Fokkema NJ, van den Heuvel J, editors. Cambridge: Cambridge University Press; 1986. The biology of endophytism in plants with particular reference to woody perennials; pp. 205–222. [Google Scholar]
- 51.Wilson D. Endophyte: the evolution of a term, and clarification of its use and definition. Oikos. 1995;73:274. doi: 10.2307/3545919. [DOI] [Google Scholar]
- 52.Hallmann J, Quadt-Hallmann A, Mahaffee WF, Kloepper JW. Bacterial endophytes in agricultural crops. Can J Microbiol. 1997;43:895–914. doi: 10.1139/m97-131. [DOI] [Google Scholar]
- 53.Hardoim PR, van Overbeek LS, Berg G, Pirttilä AM, Compant S, et al. The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol Mol Biol Rev. 2015;79:293–320. doi: 10.1128/MMBR.00050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Galippe V. Note sur la presence de micro-organismes dans les tissus vegetaux. Comptes Rendus Des Seances de La Societe de Biologie et de Ses Filiales. 1887;39:410–416. vol. [Google Scholar]
- 55.Frank BA. Ueber die auf wurzelsymbiose beruhende ernahrung gewisser baume durch unterirdische pilze. Ber Dtsch Bot Ges. 1885;3:128–145. [Google Scholar]
- 56.Beijerinck MW. Die bakterien der papilionaceenknollchen. Botanische Zeitung. 1888;46:740–750. [Google Scholar]
- 57.Omacini M, Chaneton EJ, Ghersa CM, Müller CB. Symbiotic fungal endophytes control insect host-parasite interaction webs. Nature. 2001;409:78–81. doi: 10.1038/35051070. [DOI] [PubMed] [Google Scholar]
- 58.Rubini MR, Silva-Ribeiro RT, Pomella AWV, Maki CS, Araújo WL, et al. Diversity of endophytic fungal community of cacao (Theobroma cacao L.) and biological control of Crinipellis perniciosa, causal agent of Witches’ Broom Disease. Int J Biol Sci. 2005;1:24–33. doi: 10.7150/ijbs.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giauque H, Hawkes CV. Climate affects symbiotic fungal endophyte diversity and performance. Am J Bot. 2013;100:1435–1444. doi: 10.3732/ajb.1200568. [DOI] [PubMed] [Google Scholar]
- 60.Hubbard M, Germida JJ, Vujanovic V. Fungal endophytes enhance wheat heat and drought tolerance in terms of grain yield and second-generation seed viability. J Appl Microbiol. 2014;116:109–122. doi: 10.1111/jam.12311. [DOI] [PubMed] [Google Scholar]
- 61.Neill JC. The endophyte of rye-grass (Lolium perenne) New Zealand Journal of Science and Technology, Section A. 1940;21:280–291. [Google Scholar]
- 62.Rayner RW. Latent infection in Coffea arabica L. Nature. 1948;161:245–246. doi: 10.1038/161245a0. [DOI] [Google Scholar]
- 63.Kendrick WB, Burges A. Biological aspects of the decay of pinus sylvestris litter. Nova Hedwigia. 1962;4:313–344. [Google Scholar]
- 64.Dickinson CH. Fungal colonization of pisum leaves. Can J Bot. 1967;45:915–927. doi: 10.1139/b67-093. [DOI] [Google Scholar]
- 65.Ruscoe QW. Mycoflora of living and dead leaves of Nothofagus truncata . Transactions of the British Mycological Society. 1971;56:463–474. doi: 10.1016/S0007-1536(71)80138-9. [DOI] [Google Scholar]
- 66.Bain DC. Fungi recovered from seed of sorghum vulgare pers. Phytopathology. 1950;40:521–522. [Google Scholar]
- 67.Ampratwum DB, McQuitty JB. Some physical factors affecting fungal population in stored wheat. Can J Plant Sci. 1970;50:47–51. doi: 10.4141/cjps70-007. [DOI] [Google Scholar]
- 68.Abdel-Hafez SII. Composition of the fungal flora of four cereal grains in Saudi Arabia. Mycopathologia. 1984;85:53–57. doi: 10.1007/BF00436702. [DOI] [PubMed] [Google Scholar]
- 69.Guerin D. Sur la presence d’un champignon dans l’ivraie. Journal Botany. 1898;12:230–238. [Google Scholar]
- 70.Hanausek TF. Vorlaufige mittheilung uber den von a vogl in der frucht von lolium temulentum entdeckten pilz. Berichte der Deutsche Botanische Gesellshaf. 1898;16:203. [Google Scholar]
- 71.Vogl A. Mehl und die anderen mehlprodukte der cerealien und leguminosen. Zeitschrift Nahrungsmittle Untersuchung Hyg Warenkunde. 1898;12:25–29. [Google Scholar]
- 72.Fletcher LR, Harvey IC. An association of a Lolium endophyte with ryegrass staggers. N Z Vet J. 1981;29:185–186. doi: 10.1080/00480169.1981.34839. [DOI] [PubMed] [Google Scholar]
- 73.Bacon CW, Porter JK, Robbins JD, Luttrell ES. Epichloë typhina from toxic tall fescue grasses. Appl Environ Microbiol. 1977;34:576–581. doi: 10.1128/aem.34.5.576-581.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.White JF. Widespread distribution of endophytes in the poaceae. Plant Dis. 1987;71:340. doi: 10.1094/PD-71-0340. [DOI] [Google Scholar]
- 75.Bush LP, Wilkinson HH, Schardl CL. Bioprotective alkaloids of grass-fungal endophyte symbioses. Plant Physiol. 1997;114:1–7. doi: 10.1104/pp.114.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cabral D, Cafaro MJ, Saidman BO, Lugo MA, Reddy PV, et al. Evidence supporting the occurrence of a new species of endophyte in some South American grasses. Mycologia. 1999;91:315. doi: 10.2307/3761376. [DOI] [Google Scholar]
- 77.Lane GA, Christensen MJ, Miles CO. In: Microbial Endophytes. Bacon CW, White JF, editors. New York: Marcel Dekker; 2000. Coevolution of fungal endophytes with grasses: The significance of secondary metabolites; pp. 341–388. [DOI] [Google Scholar]
- 78.Li C, Nan Z, Paul VH, Dapprich P, Liu Y. A new neotyphodium species symbiotic with drunken horse grass (Achnatherum inebrians) in China. Mycotaxon. 2004;90:141–147. [Google Scholar]
- 79.Gentile A, Rossi MS, Cabral D, Craven KD, Schardl CL. Origin, divergence, and phylogeny of epichloe endophytes of native Argentine grasses. Mol Phylogenet Evol. 2005;35:196–208. doi: 10.1016/j.ympev.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 80.Moon CD, Guillaumin JJ, Ravel C, Li C, Craven KD, et al. New neotyphodium endophyte species from the grass tribes stipeae and meliceae. Mycologia. 2007;99:895–905. doi: 10.3852/mycologia.99.6.895. [DOI] [PubMed] [Google Scholar]
- 81.Welty RE, Lucas GB, Fletcher JT, Yang H. Fungi isolated from tobacco leaves and brown-spot lesions before and after flue-curing. Appl Microbiol. 1968;16:1309–1313. doi: 10.1128/am.16.9.1309-1313.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Welty RE, Spurr HW. Isolation of alternaria alternata from Nicotiana species. Phytopathology. 1971;61:916 [Google Scholar]
- 83.Norse D. Fungi isoalted from surface-sterilized toabcco leaves. Transactions of the British Mycological Society. 1972;58:515–518. doi: 10.1016/S0007-1536(72)80104-9. [DOI] [Google Scholar]
- 84.Spurr Jr HW, RE Welty Incidence of tobacco leaf microflora in relation to brown spot disease and fungicidal treatment. Phytopathology. 1972;62:916. doi: 10.1094/Phyto-62-916. [DOI] [Google Scholar]
- 85.Millar CS. In: Biology of Plant Litter Decomposition. Dickinson CH, Pugh DJF, editors. Vol. 1. New York: Academic Press; 1974. Decomposition of coniferous leaf litter; pp. 105–108. vol. [Google Scholar]
- 86.Pugh GJF, Buckley NG. Aureobasidium pullulans: An endophyte in sycamore and other trees. Transactions of the British Mycological Society. 1971;57:227–231. doi: 10.1016/S0007-1536(71)80004-9. [DOI] [Google Scholar]
- 87.Bernstein ME, Carroll GC. Internal fungi in old-growth Douglas fir foliage. Can J Bot. 1977;55:644–653. doi: 10.1139/b77-079. [DOI] [Google Scholar]
- 88.Carroll FE, Muller E, Sutton BC. Preliminary studies on the incidence of needle endophytes in some European conifers. Sydowia. 1977;29:87–103. [Google Scholar]
- 89.Carroll GC, Carroll FE. Studies on the incidence of coniferous needle endophytes in the Pacific Northwest. Can J Bot. 1978;56:3034–3043. doi: 10.1139/b78-367. [DOI] [Google Scholar]
- 90.Petrini O, Muller E, Luginbuhl M. Pilze als Endophyten von grunen Pflanzen. Naturwissenschaften. 1979;66:262–263. doi: 10.1007/BF00571609. [DOI] [Google Scholar]
- 91.Petrini O, Muller E. Pilzliche endophyten von samenpflanzen am beispiel von Juniperus communis l. Sydowia. 1980;32:224–251. [Google Scholar]
- 92.Petrini O, Carroll GC. Endophytic fungi in foliage of some Cupressaceae in Oregon. Can J Bot. 1981;59:629–636. doi: 10.1139/b81-089. [DOI] [Google Scholar]
- 93.Petrini O, Stone J, Carroll FE. Endophytic fungi in evergreen shrubs in western Oregon: A preliminary study. Can J Bot. 1982;60:789–796. doi: 10.1139/b82-102. [DOI] [Google Scholar]
- 94.Fisher PJ, Anson AE, Petrini O. Fungal endophytes in Ulex europaeus and Ulex gallii . Transactions of the British Mycological Society. 1986;86:153–156. doi: 10.1016/S0007-1536(86)80128-0. [DOI] [Google Scholar]
- 95.Gallaud I. Etudes sur les mycorrhizas endotrophes. Revue generale de Botanique. 1905;17:5–48. [Google Scholar]
- 96.Rayner MC. Obligate symbiosis in Calluna vulgaris . Ann Bot. 1915;os-29:97–98. doi: 10.1093/oxfordjournals.aob.a089540. [DOI] [Google Scholar]
- 97.Clapp GL. The life history of Aneura pinguis . Botanical Gazette. 1912;54:177–193. doi: 10.1086/330898. [DOI] [Google Scholar]
- 98.Wollenweber HW. Identification of species of Fusarium occurring on the sweet potato, Ipomoea batatas . J Agric Res. 1914;2:1–286. [Google Scholar]
- 99.Flannigan B. Distribution of seed-borne micro-organisms in naked barley and wheat before harvest. Transactions of the British Mycological Society. 1974;62:51–58. doi: 10.1016/S0007-1536(74)80005-7. [DOI] [Google Scholar]
- 100.Koh CM, Lew J. Studies on the population of toxigenic fungi in foodstuffs. Yonsei Med J. 1974;15:74–91. doi: 10.3349/ymj.1974.15.2.74. [DOI] [PubMed] [Google Scholar]
- 101.Basak AB, Karim MR, Hoque MN, Biswas AP. Studies on the fungi associated with different varieties of wheat seeds grown in Bangladesh. Seed Research. 1987;15:71–75. [Google Scholar]
- 102.Neergaard P. Seed Pathology. London: Palgrave; 1977. [DOI] [Google Scholar]
- 103.Gourley CO. Microfungi of crowns and roots of apparently healthy dormant strawberry plants. Can J Bot. 1969;47:945–949. doi: 10.1139/b69-135. [DOI] [Google Scholar]
- 104.Penrose LJ. Phlyctaena vagabunda Desm., Pezicula malicorticis (Jackson) Nannf., and other fungi associated with apple twigs. Aust J Exp Agric. 1971;11:254. doi: 10.1071/EA9710254. [DOI] [Google Scholar]
- 105.Swinburne TR. Microflora of apple leaf scars in relation to infection by Nectria galligena . Transactions of the British Mycological Society. 1973;60:389–403. doi: 10.1016/S0007-1536(73)80024-5. [DOI] [Google Scholar]
- 106.Bhuiyan KA. Annual Research Review. Gazipur, Bangladesh: 1989. Prevalence of fungi association with Chilli seeds. [Google Scholar]
- 107.Fisher PJ, Petrini O. Fungal saprobes and pathogens as endophytes of rice (Oryza sativa L.) New Phytol. 1992;120:137–143. doi: 10.1111/j.1469-8137.1992.tb01066.x. [DOI] [Google Scholar]
- 108.Agarwal PC, Dev U, Singh B, Indra R, Khetarpal RK. Seed-borne fungi detected in consignments of soybean seeds (Glycine max) imported into India. EPPO Bulletin. 2006;36:53–58. doi: 10.1111/j.1365-2338.2006.00943.x. [DOI] [Google Scholar]
- 109.Zhang Q, Zhang J, Yang L, Zhang L, Jiang D, et al. Diversity and biocontrol potential of endophytic fungi in Brassica napus . Biological Control. 2014;72:98–108. doi: 10.1016/j.biocontrol.2014.02.018. [DOI] [Google Scholar]
- 110.Bettucci L, Saravay M. Endophytic fungi of Eucalyptus globulus: a preliminary study. Mycological Research. 1993;97:679–682. doi: 10.1016/S0953-7562(09)80147-0. [DOI] [Google Scholar]
- 111.Bettucci L, Alonso R, Tiscornia S. Endophytic mycobiota of healthy twigs and the assemblage of species associated with twig lesions of Eucalyptus globulus and E. grandis in Uruguay. Mycological Research. 1999;103:468–472. doi: 10.1017/S0953756298007205. [DOI] [Google Scholar]
- 112.Zhao Y, Xiong Z, Wu G, Bai W, Zhu Z, et al. Fungal endophytic communities of two wild Rosa varieties with different powdery mildew susceptibilities. Front Microbiol. 2018;9:2462. doi: 10.3389/fmicb.2018.02462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stanosz GR, Smith DR, Guthmiller MA, Stanosz JC. Persistence of Sphaeropsis sapinea on or in Asymptomatic shoots of red and jack pines. Mycologia. 1997;89:525. doi: 10.2307/3760986. [DOI] [Google Scholar]
- 114.Carroll GC. 16th International Botanical Congress. St. Louis, MO; 1999. The foraging ascomycete [abstract] p. 309. p. [Google Scholar]
- 115.Schulz B, Boyle C. The endophytic continuum. Mycol Res. 2005;109:661–686. doi: 10.1017/s095375620500273x. [DOI] [PubMed] [Google Scholar]
- 116.Promputtha I, Lumyong S, Dhanasekaran V, McKenzie EHC, Hyde KD, et al. A phylogenetic evaluation of whether endophytes become saprotrophs at host senescence. Microb Ecol. 2007;53:579–590. doi: 10.1007/s00248-006-9117-x. [DOI] [PubMed] [Google Scholar]
- 117.Promputtha I, Hyde KD, McKenzie EHC, Peberdy JF, Lumyong S. Can leaf degrading enzymes provide evidence that endophytic fungi becoming saprobes? Fungal Diversity. 2010;41:89–99. doi: 10.1007/s13225-010-0024-6. [DOI] [Google Scholar]
- 118.Porras-Alfaro A, Bayman P. Hidden fungi, emergent properties: endophytes and microbiomes. Annual Reviews. 2011;49:291–315. doi: 10.1146/annurev-phyto-080508-081831. [DOI] [PubMed] [Google Scholar]
- 119.U’Ren JM, Arnold AE. Diversity, taxonomic composition, and functional aspects of fungal communities in living, senesced, and fallen leaves at five sites across North America. PeerJ. 2016;4:e2768. doi: 10.7717/peerj.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhou J, Li X, Huang PW, Dai CC. Endophytism or saprophytism: Decoding the lifestyle transition of the generalist fungus Phomopsis liquidambari . Microbiol Res. 2018;206:99–112. doi: 10.1016/j.micres.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 121.Kuo HC, Hui S, Choi J, Asiegbu FO, Valkonen JPT, et al. Secret lifestyles of Neurospora crassa . Sci Rep. 2014;4:5135. doi: 10.1038/srep05135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fordyce C, Green RJ. Studies of the host specificity of verticillium albo-atrum var menthae. Phytopathology. 1960;50:635 [Google Scholar]
- 123.Schneider RW, Pendery WE. Stalk rot of corn: mechanism of predisposition by an early season water stress. Phytopathology. 1983;73:863. doi: 10.1094/Phyto-73-863. [DOI] [Google Scholar]
- 124.Álvarez-Loayza P, White JF, Jr, Torres MS, Balslev H, Kristiansen T, et al. Light converts endosymbiotic fungus to pathogen, influencing seedling survival and niche-space filling of a common tropical tree, Iriartea deltoidea. PLoS One. 2011;6:e16386. doi: 10.1371/journal.pone.0016386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wheeler DL, Johnson DA. Verticillium dahliae infects, alters plant biomass, and produces inoculum on rotation crops. Phytopathology. 2016;106:602–613. doi: 10.1094/PHYTO-07-15-0174-R. [DOI] [PubMed] [Google Scholar]
- 126.Lofgren LA, LeBlanc NR, Certano AK, Nachtigall J, LaBine KM, et al. Fusarium graminearum: pathogen or endophyte of North American grasses? New Phytol. 2018;217:1203–1212. doi: 10.1111/nph.14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Moon CD, Scott B, Schardl CL, Christensen MJ. The evolutionary origins of Epichloë endophytes from annual ryegrasses. Mycologia. 2019;92:1103–1118. doi: 10.1080/00275514.2000.12061258. [DOI] [Google Scholar]
- 128.Nees von Esenbeck CG. 1816 (“1817” in some copies) Das System der Pilze und Schwamme. :72. doi: 10.5962/bhl.title.110007. n.d. [DOI] [Google Scholar]
- 129.Wiltshire SP. The foundation species of Alternaria and Macrosporium . Transactions of the British Mycological Society. 1933;18:135–IN3. doi: 10.1016/S0007-1536(33)80003-9. [DOI] [Google Scholar]
- 130.Fries E. Macrosporium. Systema Mycologicum. 1819;3:373–375. [Google Scholar]
- 131.Chevallier FF. Flore generale des environs de Paris 1:32. 1826.
- 132.Fries E. Flora scania. Upsala. 1835:368. [Google Scholar]
- 133.Fries E. Summa vegetabilium scandinaviae. Sectio posterior. 1849;505 [Google Scholar]
- 134.Angell HR. Purple blotch of onion (Macrosporium porri Ell) J Agric Res. 1929;38:467–487. [Google Scholar]
- 135.Jackson CR, Weber GF. Morphology and Taxonomy of Alternaria Cucumerina . Mycologia. 1959;51:401. doi: 10.2307/3756059. [DOI] [Google Scholar]
- 136.Lawrence DP, Gannibal PB, Peever TL, Pryor BM. The sections of Alternaria: formalizing species-group concepts. Mycologia. 2013;105:530–546. doi: 10.3852/12-249. [DOI] [PubMed] [Google Scholar]
- 137.Neergaard P. Danish species of Alternaria and Stemphylium. London: Oxford University Press; 1945. p. 560. p. [Google Scholar]
- 138.Seung-Hun Y, Mathur SB, Neergaard P. Taxonomy and pathogenicity of four seed-borne species of Alternaria from sesame. Transactions of the British Mycological Society. 1982;78:447–458. doi: 10.1016/S0007-1536(82)80151-4. [DOI] [Google Scholar]
- 139.Simmons EG. CBS Biodiversity Series 6. CBS Fungal Biodiversity Centre. Utrecht, The Netherlands: 2007. Alternaria. An identification manual. [Google Scholar]
- 140.Kõljalg U, Nilsson HR, Schigel D, Tedersoo L, Larsson KH, et al. The Taxon Hypothesis Paradigm—on the unambiguous detection and communication of Taxa. Microorganisms. 2020;8:1910. doi: 10.3390/microorganisms8121910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Simmons EG. The theoretical bases for classification of the Fungi Imperfecti. Q Rev Biol. 1966;41:113–123. doi: 10.1086/404938. [DOI] [PubMed] [Google Scholar]
- 142.Goos RD. Classification of the fungi imperfecti. Proceedings of the Iowa Academy of Science. 1956;63:311–320. [Google Scholar]
- 143.Saccardo PA, Traverso GB, Trotter A. Patavii, Sumptibus P. A. Saccardo. 1882-1931. Sylloge fungorum omnium hucusque cognitorum. [DOI] [Google Scholar]
- 144.Bisby GR. An introduction to the taxonomy and nomenclature of Fungi. Kew, Surrey: The Commonwealth Mycological Institute; 1953. [Google Scholar]
- 145.Young PA. Tabulation of Alternaria and Macrosporium . Mycologia. 2018;21:155–166. doi: 10.1080/00275514.1929.12016948. [DOI] [Google Scholar]
- 146.Deshpande KB, Rajderkar NR. New species of Alternaria from Marathwada (India) Mycopathologia et Mycologia Applicata. 1964;23:277–280. doi: 10.1007/BF02048995. [DOI] [Google Scholar]
- 147.Joly P. Le Genre Alternaria [Encyclopedie Mycologique] Paris: P. Lechevalier; 1964. [Google Scholar]
- 148.Simmons EG, Joly P. Le Genre Alternaria, Encyclopedie Mycologique XXXIII. Mycologia. 1966;58:340. doi: 10.2307/3756979. [DOI] [Google Scholar]
- 149.de Hoog GS, Horré R. Molecular taxonomy of the Alternaria and Ulocladium species from humans and their identification in the routine laboratory. Mycoses. 2002;45:259–276. doi: 10.1046/j.1439-0507.2002.00747.x. [DOI] [PubMed] [Google Scholar]
- 150.Kohmoto K, Khan ID, Renbutsu Y, Taniguchi T, Nishimura S. Multiple host-specific toxins of Alternaria mali and their effect on the permeability of host cells. Physiological Plant Pathology. 1976;8:141–153. doi: 10.1016/0048-4059(76)90047-3. [DOI] [Google Scholar]
- 151.Nishimura S. Host-specific toxins from Alternaria alternata problems and prospects. Proc Jpn Acad, Ser B. 1980;56:362–366. doi: 10.2183/pjab.56.362. [DOI] [Google Scholar]
- 152.Tsuge T, Harimoto Y, Akimitsu K, Ohtani K, Kodama M, et al. Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata . FEMS Microbiol Rev. 2013;37:44–66. doi: 10.1111/j.1574-6976.2012.00350.x. [DOI] [PubMed] [Google Scholar]
- 153.Simmons EG. Alternaria themes and variations (22-26) Mycotaxon. 1986;25:287–308. [Google Scholar]
- 154.Simmons EG. Alternaria themes and variations (27-53) Mycotaxon. 1990;37:79–119. [Google Scholar]
- 155.Simmons EG. In: Alternaria: Biology, Plant Diseases and Metabolites. Chelkowski J, Visconti A, editors. Amsterdam: Elsevier Science Publishers; 1992. Alternaria taxonomy: Current status, viewpoint, challenge; pp. 1–35. [Google Scholar]
- 156.Simmons EG, Roberts RG. Alternaria themes and variations (73) Mycotaxon. 1993;48:109–140. [Google Scholar]
- 157.Simmons EG. Alternaria themes and variations (106-111) Mycotaxon. 1994;50:409–427. [Google Scholar]
- 158.Simmons EG. Alternaria themes and variations (112-144) Mycotaxon. 1995;55:55–163. [Google Scholar]
- 159.Simmons EG. Alternaria themes and variations (226-235). Classification of citrus pathogens. Mycotaxon. 1999;70:263–323. [Google Scholar]
- 160.Simmons EG. Alternaria themes and variations (236-243). Host-specific toxin producers. Mycotaxon. 1999;70:325–369. [Google Scholar]
- 161.Slifkin MK. Conidial wall structure and morphology of Alternaria spp. The Journal of the Elisha Mitchell Scientific Society December:231-236. 1971 [Google Scholar]
- 162.Ohtani H, Kohmoto K. In: Alternaria, Biology, Plant Diseases and Metabolites. Chelkowski J, Visconti A, editors. Amsterdam: Elsevier; 1992. Host-specific toxins of Alternaria species; pp. 123–156. [Google Scholar]
- 163.Nishimura S, Kohmoto K, Otani H, Ramachandran P, Tamura F. In: Plant Infection: The Physiological and Biochemical Basis. Asada Y, Bushnell WR, Ouchi S, Vance CP, editors. Berlin Heidelberg New York: Japan Scientific Societies Press, Tokyo/Springer-Verlag; 1982. Pathological and epidemiological aspects of Alternaria alternata infection depending on a host-specific toxin; pp. 199–214. [Google Scholar]
- 164.Jasalavich CA, Morales VM, Pelcher LE, Séguin-Swartz G. Comparison of nuclear ribosomal DNA sequences from Alternaria species pathogenic to crucifers. Mycological Research. 1995;99:604–614. doi: 10.1016/S0953-7562(09)80720-X. [DOI] [Google Scholar]
- 165.Adachi Y, Watanabe H, Tanabe K, Doke N, Nishimura S, et al. Nuclear Ribosomal DNA as a Probe for Genetic Variability in the Japanese Pear Pathotype of Alternaria alternata. Appl Environ Microbiol. 1993;59:3197–3205. doi: 10.1128/aem.59.10.3197-3205.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kusaba M, Tsuge T. Nuclear ribosomal DNA variation and pathogenic specialization in alternaria fungi known to produce host-specific toxins. Appl Environ Microbiol. 1994;60:3055–3062. doi: 10.1128/aem.60.9.3055-3062.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Serdani M, Kang JC, Andersen B, Crous PW. Characterisation of Alternaria species-groups associated with core rot of apples in South Africa. Mycological Research. 2002;106:561–569. doi: 10.1017/S0953756202005993. [DOI] [Google Scholar]
- 168.Pryor BM, Creamer R, Shoemaker RA, McLain-Romero J, Hambleton S. Undifilum, a new genus for endophytic embellisia oxytropis and parasitic helminthosporium bornmuelleri on legumes. Botany. 2009;87:178–194. doi: 10.1139/B08-130. [DOI] [Google Scholar]
- 169.Lawrence DP, Park MS, Pryor BM. Nimbya and Embellisia revisited, with nov. comb for Alternaria celosiae and A. perpunctulata. Mycol Progress. 2011;11:799–815. doi: 10.1007/s11557-011-0793-7. [DOI] [Google Scholar]
- 170.Ramires FA, Masiello M, Somma S, Villani A, Susca A, et al. Phylogeny and Mycotoxin Characterization of Alternaria Species Isolated from Wheat Grown in Tuscany, Italy. Toxins (Basel) 2018;10:E472. doi: 10.3390/toxins10110472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Taylor JW, Hann-Soden C, Branco S, Sylvain I, Ellison CE. Clonal reproduction in fungi. Proc Natl Acad Sci U S A. 2015;112:8901–8908. doi: 10.1073/pnas.1503159112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhao D, Fan S, Zhang D, Pan Y, Gu Q, et al. Parasexual reproduction in Alternaria solani: Simple sequence repeat molecular evidence for haploidization. Mycologia. 2021;113:949–955. doi: 10.1080/00275514.2021.1922243. [DOI] [PubMed] [Google Scholar]
- 173.Tanaka A, Shiotani H, Yamamoto M, Tsuge T. Insertional mutagenesis and cloning of the genes required for biosynthesis of the host-specific AK-toxin in the Japanese pear pathotype of Alternaria alternata . Mol Plant Microbe Interact. 1999;12:691–702. doi: 10.1094/MPMI.1999.12.8.691. [DOI] [PubMed] [Google Scholar]
- 174.Tanaka A, Tsuge T. Structural and functional complexity of the genomic region controlling AK-toxin biosynthesis and pathogenicity in the Japanese pear pathotype of Alternaria alternata . Mol Plant Microbe Interact. 2000;13:975–986. doi: 10.1094/MPMI.2000.13.9.975. [DOI] [PubMed] [Google Scholar]
- 175.Johnson LJ, Johnson RD, Akamatsu H, Salamiah A, Otani H, et al. Spontaneous loss of a conditionally dispensable chromosome from the Alternaria alternata apple pathotype leads to loss of toxin production and pathogenicity. Curr Genet. 2001;40:65–72. doi: 10.1007/s002940100233. [DOI] [PubMed] [Google Scholar]
- 176.Hatta R, Ito K, Hosaki Y, Tanaka T, Tanaka A, et al. A conditionally dispensable chromosome controls host-specific pathogenicity in the fungal plant pathogen Alternaria alternata . Genetics. 2002;161:59–70. doi: 10.1093/genetics/161.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Masunaka A, Tanaka A, Tsuge T, Peever TL, Timmer LW, et al. Distribution and Characterization of AKT Homologs in the Tangerine Pathotype of Alternaria alternata . Phytopathology. 2000;90:762–768. doi: 10.1094/PHYTO.2000.90.7.762. [DOI] [PubMed] [Google Scholar]
- 178.Masunaka A, Ohtani K, Peever TL, Timmer LW, Tsuge T, et al. An isolate of Alternaria alternata that is pathogenic to both tangerines and rough lemon and produces two host-selective toxins, ACT- and ACR-toxins. Phytopathology. 2005;95:241–247. doi: 10.1094/PHYTO-95-0241. [DOI] [PubMed] [Google Scholar]
- 179.Ma L-J, van der Does HC, Borkovich KA, Coleman JJ, Daboussi M-J, et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature. 2010;464:367–373. doi: 10.1038/nature08850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hu J, Chen C, Peever T, Dang H, Lawrence C, et al. Genomic characterization of the conditionally dispensable chromosome in Alternaria arborescens provides evidence for horizontal gene transfer. BMC Genomics. 2012;13:171. doi: 10.1186/1471-2164-13-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Berbee ML, Payne BP, Zhang G, Roberts RG, Turgeon BG. Shared ITS DNA substitutions in isolates of opposite mating type reveal a recombining history for three presumed asexual species in the filamentous ascomycete genus Alternaria . Mycol Res. 2003;107:169–182. doi: 10.1017/s0953756203007263. [DOI] [PubMed] [Google Scholar]
- 182.Johnson RD, Johnson L, Kohmoto K, Otani H, Lane CR, et al. A Polymerase Chain Reaction-Based Method to Specifically Detect Alternaria alternata Apple Pathotype (A. mali), the Causal Agent of Alternaria Blotch of Apple. Phytopathology. 2000;90:973–976. doi: 10.1094/PHYTO.2000.90.9.973. [DOI] [PubMed] [Google Scholar]
- 183.Pavon Moreno MA, Alonso IG, RM de S, Lacarra TG. Importancia del genero Alternaria como productor de micotoxinas y agente causal de enfermedades humanas. Nutr Hosp. 2012;27:1772–1781. doi: 10.3305/nh.2012.27.6.6017. [DOI] [PubMed] [Google Scholar]
- 184.Meena M, Samal S. Alternaria host-specific (HSTs) toxins: An overview of chemical characterization, target sites, regulation and their toxic effects. Toxicol Rep. 2019;6:745–758. doi: 10.1016/j.toxrep.2019.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Simpson GG. The species concept. Evolution. 1951;5:285–298. doi: 10.1111/j.1558-5646.1951.tb02788.x. [DOI] [Google Scholar]
- 186.Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, et al. Phylogenetic species recognition and species concepts in fungi. Fungal Genet Biol. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- 187.Mayden RL. A hierarchy of species concepts: the denouement in the saga of the species problem. In: Species: The units of diversity. Chapman & Hall; 1997. pp. 381–423. [Google Scholar]
- 188.Harrington TC, Rizzo DM. Structure and Dynamics of Fungal Populations. Dordrecht: Springer; 1999. Defining species in the fungi; pp. 43–71. [Google Scholar]
- 189.Hawksworth DL, Kirk PM, Sutton BC, Pegler DN. Ainsworth & Bisby’s dictionary of the fungi. Rev Inst Med trop S Paulo. 1996;38:272. doi: 10.1590/S0036-46651996000400018. [DOI] [Google Scholar]
- 190.Mayr E. Speciation phenomena in birds. The American Naturalist. 1940;74:249–278. doi: 10.1086/280892. [DOI] [Google Scholar]
- 191.Shear CL, Dodge BO. Life histories and heterothallism of the red bread-mold fungi of the Monilia sitophila group. J Agric Res. 1927;34:1019–1042. [Google Scholar]
- 192.Maharachchikumbura SSN, Chen Y, Ariyawansa HA, Hyde KD, Haelewaters D, et al. Integrative approaches for species delimitation in Ascomycota . Fungal Diversity. 2021;109:155–179. doi: 10.1007/s13225-021-00486-6. [DOI] [Google Scholar]
- 193.Cracraft J. In: Current Ornithology. Johnston RF, editor. New York, NY: Springer; 1983. Species concepts and speciation analysis; pp. 159–187. [Google Scholar]
- 194.Avise JC, Ball RM. In: Oxford Surveys in Evolutionary Biology. Futuyma D, Antonovics J, editors. Vol. 7. Oxford: Oxford University Press; 1990. Principles of genealogical concordance in species concepts and biological taxonomy; pp. 45–67. vol. [Google Scholar]
- 195.Baum DA, Shaw KL. Genealogical perspectives on the species problem. Experimental and Molecular Approaches to Plant Biosystematics. 1995;53:289–303. [Google Scholar]
- 196.Avise JC, Wollenberg K. Phylogenetics and the origin of species. Proc Natl Acad Sci U S A. 1997;94:7748–7755. doi: 10.1073/pnas.94.15.7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Garganese F, Schena L, Siciliano I, Prigigallo MI, Spadaro D, et al. Characterization of citrus-associated Alternaria species in Mediterranean areas. PLoS One. 2016;11:e0163255. doi: 10.1371/journal.pone.0163255. [DOI] [PMC free article] [PubMed] [Google Scholar]