Abstract
Streptococcus pyogenes (group A streptococcus [GAS]), a multiple-amino-acid-auxotrophic human pathogen, may face starvation for essential amino acids during various stages of the infection process. Since the response of GAS to such conditions is likely to influence pathogenetic processes, we set out to identify by transcriptional analyses genes and operons that are responsive to amino acid starvation and examined whether functionally meaningful response patterns can be ascertained. We discovered that GAS are capable of mounting a relA-independent amino acid starvation response that involves transcriptional modulation of a wide array of housekeeping genes as well as accessory and dedicated virulence genes. Housekeeping genes that were upregulated during starvation of both wild-type and relA mutant strains included the newly identified T-box members of the aminoacyl-tRNA synthetase genes, the genes for components of the tmRNA-mediated peptide tagging and proteolysis system for abnormal proteins (ssrA, smpB, clpP, and clpC), and the operons for the dnaK and groE groups of molecular chaperones. In addition to upregulation of the genes for oligopeptide permease (opp), intracellular peptidase (pepB), and the two-component regulator covRS reported previously (K. Steiner and H. Malke, Mol. Microbiol. 38:1004–1016, 2000), amino acid starvation stimulated the transcription of the growth phase-associated, virulence-regulatory fas operon, the streptolysin S operon (sag), and the gene for autoinducer-2 production protein (luxS). A prominent feature of operons exhibiting internal transcriptional termination (opp, fas, and sag) was starvation-promoted full-length transcription, a mechanism that improves the efficacy of these systems by increasing the level of coordinate transcription of functionally related genes. Based on these results, a regulatory network with feedback mechanisms is proposed that counteracts the stringent response, links the levels of key rate-limiting enzymes to virulence gene expression, and enables the organism in a dynamic way to take advantage of protein-rich environments provided by its human host. As several of the affected target genes are controlled by more than one regulator, fine modulation may result in accordance with the demands imposed by ecologically different colonization sites upon the adaptive capacity of the pathogen.
Streptococcus pyogenes (a group A streptococcus [GAS]) is a multiple-amino-acid-auxotrophic human pathogen that may encounter starvation for free amino acids when striving to propagate in its natural habitats during specific stages of the infection process. The organism is likely to face nutrient limitation when initially contacting the skin or throat as the primary infection sites. Tracing the migration path of organisms associated with impetigo, Ferrieri et al. (10) isolated GAS for an average period of 8 days from normal skin before skin lesions developed, and it took another 2 to 3 weeks before clonally identical organisms could be cultured from the upper respiratory tract. Although for streptococci found on the skin, the existence of a persistent carrier state seems unlikely, asymptomatic carriage is well established for organisms harbored by the oropharynx, and these appear to constitute the principal reservoir for GAS in the environment (40). The stasis-like carrier state may also characterize the physiological state of cells that not only adhere to epithelial cells but also succeed in invading them. Under such conditions, limited availability of essential nutrients may force the pathogen to transform its metabolism from a growth mode to a survival mode. Host nutritional deficiencies may also be encountered by GAS that persist at high cell densities in nidi of infection or initiate life-threatening invasive disease, such as streptococcal toxic shock syndrome, necrotizing fasciitis, and septicemia. Although the body sites affected in these cases are extremely rich in protein, free amino acids may be a limiting factor initially, and this condition would appear to trigger adaptive responses that complement the deficiencies. Given polyauxotrophy of the pathogen (4, 9) and the realization that as soon as the supply of an amino acid is restricted the corresponding aminoacyl-tRNA is immediately limited (7), amino acid availability becomes a central problem in order for the pathogen to survive and propagate. Since the response of GAS to such conditions is likely to influence pathogenetic processes, we set out to identify responsive genes and operons and examined whether functionally meaningful response patterns can be ascertained.
Our previous investigations along these lines have established that streptococci express the relA gene-determined hallmark features of the stringent response, i.e., (p)ppGpp-mediated inhibition of stable RNA synthesis in response to starvation for amino acids (30, 31, 45). Concomitant with prevention of wasteful macromolecular synthesis by a rapid shutdown of futile RNA synthesis, the functional significance of the streptococcal stringent factor consists in strongly supporting cell survival under nutritional stress conditions (45). Most significantly, conducting transcriptional analyses, we discovered that in addition to the stringent response, GAS are capable of mounting a relA-independent response to amino acid deprivation that involves transcriptional modulation of a wide array of dedicated as well as accessory virulence genes, among them genes encoding the oligopeptide (opp) and dipeptide (dpp) permeases, an intracellular oligopeptidase (pepB), and genes (covRS) functioning in global regulation of virulence factors (45). These observations suggest that GAS have evolved a stimulus response network that counteracts the stringent response and enables the pathogen to mount a dynamic response to the protein-rich environment provided by its human host. Guided by the prediction that gene regulation occurs by a positive mode when the environment of the organism demands a high expression level of the regulated gene, and vice versa (36), we now expand the breadth of the amino acid starvation response and arrive at a more comprehensive network of adaptive responses to a key environmental condition that GAS may encounter in association with their host.
MATERIALS AND METHODS
Bacterial strains and media.
The principle GAS strains used in these experiments were the wild-type M49 strain NZ131, obtained from J. J. Ferretti, and its erythromycin-resistant relA insertion mutant, NZ131relA::pFR1, constructed as described previously (45). Disruption of relA rendered the mutant incapable of accumulating (p)ppGpp in response to amino acid deprivation. The complete genomic sequence of S. pyogenes strain SF370 (M1) (9) allowed derivation of oligonucleotide primers for the synthesis of gene-specific PCR-generated DNA probes from NZ131 chromosomal DNA as a template (Table 1). Cultures of GAS were grown in ambient air at 37°C without agitation in brain heart infusion (BHI) broth (Difco) or in chemically defined medium (CDM) buffered with 26 mM morpholinepropanesulfonic acid (31). If appropriate, erythromycin, mupirocin (GlaxoSmithKline), and puromycin (Sigma) were added to the medium at 2.5, 0.4, and 200 μg ml−1, respectively.
TABLE 1.
Primers used to generate gene-specific PCR-derived probes from S. pyogenes NZ131 for RNA hybridizations
| Forward (F) or reverse (R) primer | SF370 genome coordinatesa |
|---|---|
| valSF | 1294547–1294566(C)b |
| valSR | 1293632–1293653 |
| ileSF | 1245196–1245218(C) |
| ileSR | 1243785–1243806 |
| ssrAF | 1065040–1065061 |
| ssrAR | 1065340–1065360(C) |
| smpBF | 406216–406235 |
| smpBR | 406617–406636(C) |
| clpPF | 334821–334843 |
| clpPR | 335341–335364(C) |
| clpCF | 1726643–1726663(C) |
| clpCR | 1725608–1725629 |
| hrcAF | 1460765–1460785(C) |
| hrcAR | 1460034–1460056 |
| dnaKF | 1459046–1459067(C) |
| dnaKR | 1457906–1457927 |
| dnaJF | 1456996–1457016(C) |
| dnaJR | 1455985–1456006 |
| groESF | 1724561–1724580(C) |
| groESR | 1724321–1724341 |
| groELF | 1723849–1723870(C) |
| groELR | 1722886–1722906 |
| sagAF | 598709–598731 |
| sagAR | 598972–598994(C) |
| fasBF | 210435–210457 |
| fasBR | 211345–211366(C) |
| fasCF | 211634–211658 |
| fasCR | 212514–212537(C) |
| fasXF | 213654–213675 |
| fasXR | 213955–213975(C) |
| luxSF | 1361940–1361962 |
| luxSR | 1362312–1362334(C) |
GenBank accession number AE004092.
C indicates the complementary strand.
Amino acid starvation protocol.
GAS cells were grown to an optical density at 600 nm of 0.35 to 0.40 in standard CDM or BHI broth and washed by centrifugation, and aliquots of the cultures were dispensed at the original cell density into experimental and control media. CDM lacking isoleucine and valine was used for amino acid starvation experiments, BHI broth containing mupirocin served to inhibit aminoacylation of tRNAIle, and BHI with puromycin was employed to stop all continuous polypeptide syntheses. Unless stated otherwise, cells were incubated in these media for 90 min before total-cell RNA was extracted and purified by centrifugation through a discontinuous CsCl gradient as described elsewhere (29).
RNA hybridization analysis.
Although, in general, all RNA analyses were performed using both slot blot and Northern hybridizations, results of the latter were given preference for analysis of the transcription pattern of operons. Gels to be used for Northern blotting were checked for possible DNA contamination, equal loading of the gel slots (5 or 10 μg of RNA per slot), and integrity of the RNA by ethidium bromide staining. Based on comparable cell densities, the staining procedure did not reveal any extensive rRNA degradation in either strain during the starvation period. The standard way of normalizing transcript abundances to total RNA was validated by alternative normalization of mRNA levels to fixed amounts of mga transcripts (∼2 kb). The mRNA of this multiple gene activator is fairly abundant and has a relatively long half-life (>10 min; our unpublished observations), and mga transcription is not responsive to amino acid limitation in either wild-type or relA mutant cells (45). Thus, data expressed relative to the amount of mga message are equivalent to those normalized to total RNA. The PCR-derived gene-specific probes (Table 1) were labeled with [α-32P]dATP using random oligonucleotide primers. RNA electrophoresis and hybridization were performed as described in detail elsewhere (29). Hybridizations were carried out at least twice with RNA isolated from independent experiments. In order to ensure that the adaptive response to nutritional stress had become manifest, stimulation of transcription of the covRS operon (45) was checked as an internal control. Hybridization signals were quantified by PhosphorImager analysis; where indicated, standard errors reflect the results of a minimum of three independent hybridizations.
RESULTS
Strain NZ131 responded to the omission of isoleucine and valine from CDM or the presence of mupirocin in BHI broth with an immediate halt in growth and by accumulating (p)ppGpp, with the latter response being manifest for at least 90 min when RNA was extracted from the cells for transcription analysis (45). In contrast, NZ131relA::pFR1 failed to accumulate this nucleotide under either condition. This pair of strains was therefore used to expand the range of genes responsive to amino acid starvation in order to ascertain cooperative effects of general and specific homeostatic processes that follow from nutritional stress and are initiated at the transcriptional level.
Aminoacyl-tRNA synthetases (AARS).
Inspection of the genomic sequence of strain SF370 led to the prediction that of the 19 AARS (there is no glutaminyl tRNA synthetase), at least 7 enzymes are encoded by genes (alaS, glyQS, ileS, pheST, thrS, trpS, and valS) that belong to the T-box antitermination family. The streptococcal members of this family have leader sequences containing an 18-nt stretch with excellent adherence to the consensus of the T box (AANNNNGG/AUGGU/AACCG/ACG, with the T box proper marked with italics). This sequence is located upstream of rho-independent transcription terminators in many AARS gene leaders from several gram-positive bacteria (16, 20, 28, 42). Structural models of the mRNA leaders of the AARS genes from SF370 exhibit all the conserved features of the secondary structure which are important for regulation (not shown). This analysis suggested that about one-third of the streptococcal AARS genes are regulated at the level of transcription termination, which, under amino acid starvation conditions, involves the interaction of cognate uncharged tRNAs with the leader RNA. This interaction is thought to stabilize the alternate antiterminator form of the leader, thus derepressing the expression of the corresponding AARS gene (20, 42).
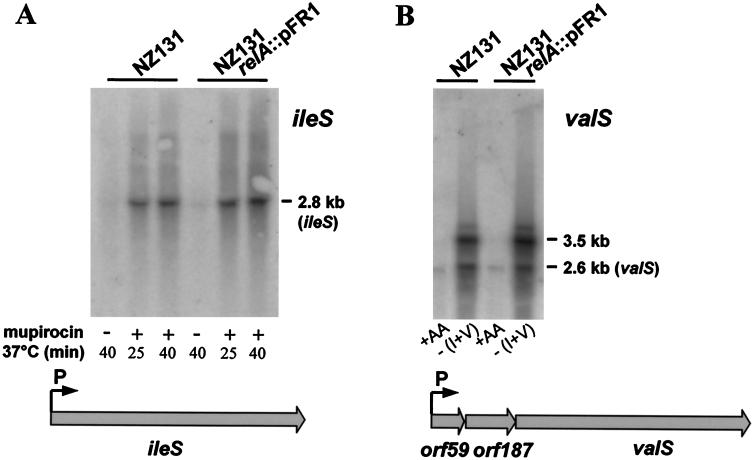
Experimental evidence for stimulation of ileS transcription by inhibition of aminoacylation of tRNAIle was provided by the finding that mupirocin increased readthrough transcription of the NZ131 ileS leader region terminator more than 10-fold, causing the pronounced synthesis of a 2.8-kb full-length transcript. This transcript was hardly detectable in unstarved control cells (Fig. 1A). Similar results were obtained by Northern hybridization analysis of valS transcription using RNA from cells grown in CDM from which valine was omitted (Fig. 1B). An interesting peculiarity of the latter system relates to the observation that the valS coding region was preceded by two open reading frames with homology to histone acetyl transferases (orf59) and tetratricopeptide repeat domain proteins (orf187), respectively. Since the leader sequence was localized between the promoter and the proximal orf59, the two peptides, seemingly unrelated to valS function, would thus appear to be encoded in the full-length transcript (3.5 kb) and coregulated with valS (Fig. 1B). Comparison of the transcript patterns of the wild type and the relA mutant shows that stimulation of readthrough transcription of the ileS and valS leader regions was independent of a functional relA gene (Fig. 1).
FIG. 1.
Stimulation of ileS (A) and valS (B) transcription in response to, respectively, mupirocin treatment (plus) and amino acid (AA) deprivation [minus (isoleucine plus valine)] of NZ131 wild-type and relA mutant cells. Control cells were incubated in BHI broth in the absence of mupirocin (minus) for the indicated period of time or grown in complete CDM (+AA). The sizes of the hybridizing bands in the Northern blots are consistent with the genetic organization of the two transcription units shown below the blots.
tmRNA-mediated peptide tagging and proteolysis system.
Like all members of the eubacterial kingdom studied so far, the GAS genome (9) contains all genes required for targeting truncated proteins for proteolysis. This system includes the following: (i) ssrA encoding a stable RNA, tmRNA, involved in a trans-translational process in which it functions both as an alanyl-tRNA and an mRNA for a C-terminal peptide tag [(A)AKNTNSYALAA] that is added to the partially synthesized polypeptide product of a damaged mRNA (22, 35, 47); the structural model of the streptococcal tmRNA (not shown) confirms that its tRNA-like portion contains the features known to specify a tRNA for alanine, namely, the GU base pair as the third base pair in the amino acid acceptor stem and the discriminator base A preceding the 3′ terminus CCA; (ii) smpB coding for a tmRNA-binding protein required for the formation of a stable tmRNA-ribosome complex (21); (iii) clpP, specifying an ATP-dependent, C-terminus-specific serine protease active on tmRNA-tagged polypeptides (13, 14); and (iv) clpC, clpL, and clpX, encoding regulatory ATPase subunits for ClpP (9, 11).
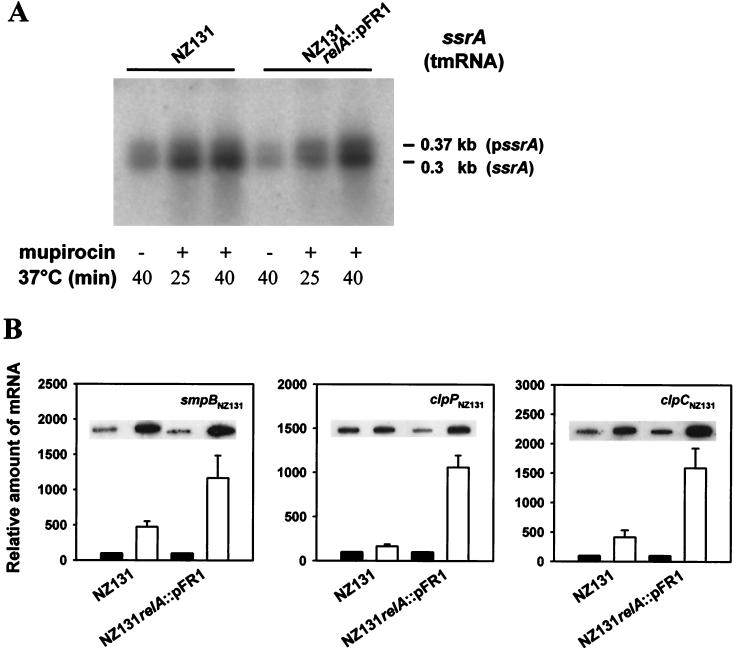
Northern hybridization with an ssrA-specific probe of RNA extracted from cells incubated for 25 or 40 min in mupirocin-containing BHI broth revealed that inhibition of isoleucyl-tRNA aminoacylation increased the amount of ssrA transcripts in wild-type and relA mutant cells in a similar manner (Fig. 2A). Thus, like tmRNA from Escherichia coli (47), the streptococcal ssrA gene does not appear to be subject to stringent control. Transcript abundance increased with the time of mupirocin treatment and this was true of both the amount of mature SsrA (0.3 kb) and its unprocessed precursor (0.37 kb). Starvation for isoleucine and valine of cells grown in CDM also increased the transcript abundance of the protein components of the system. Results of corresponding slot blot hybridizations of smpB-, clpP-, and clpC-specific mRNA revealed, respectively, fivefold, twofold, and fourfold increases for wild-type cells and 11-fold, 10-fold, and 15-fold increases for relA mutant cells relative to results for unstarved control cells (Fig. 2B). The greater effects observed with the relA mutant remain unexplained. Northern hybridizations (not shown) confirmed these results, showing increased amounts of a 0.68-kb monocistronic clpP-derived transcript and increased levels of a 2.9-kb dicistronic transcript corresponding to the ctsR-clpC operon detected in cells starved for isoleucine and valine. Using, in addition to a probe for clpC, a ctsR-specific probe, we also detected pronounced amounts of a 0.65-kb monocistronic transcript, apparently reflecting intraoperonic termination of ctsR transcription. In Bacillus subtilis, CtsR has recently been shown to control the transcription of clpP and clpC by targeting a directly repeated heptad sequence which is also present upstream of the S. pyogenes clp genes (5, 6, 9).
FIG. 2.
relA-independent transcriptional upregulation of the indicated genes of the tmRNA-directed peptide tagging and proteolysis system by inhibition of isoleucyl-tRNA synthetase or amino acid deprivation. (A) Northern hybridization of the precursor (pssrA) and processed form (ssrA) of tmRNA from mupirocin-treated (plus) and untreated (minus) control cells. (B) Slot blot hybridizations of RNA extracted from cells incubated in CDM deprived of isoleucine and valine (white columns) and control cells incubated in complete CDM (black columns).
Molecular chaperones.
The clp genes regarded above as components of the tmRNA-directed proteolysis system are well known to belong to the heat shock-inducible genes in E. coli (48) and B. subtilis (24, 25). We were prompted, therefore, to extend the above-described studies by including the classical heat-inducible genes of the dnaK and groE operons, the products of which function as molecular chaperones. In S. pyogenes SF370, both operons (hrcA-grpE-dnaK-dnaJ and groES-groEL) carry the CIRCE sequence as an operator site (9, 26) which, in B. subtilis, is known to bind the HrcA repressor (33, 34).
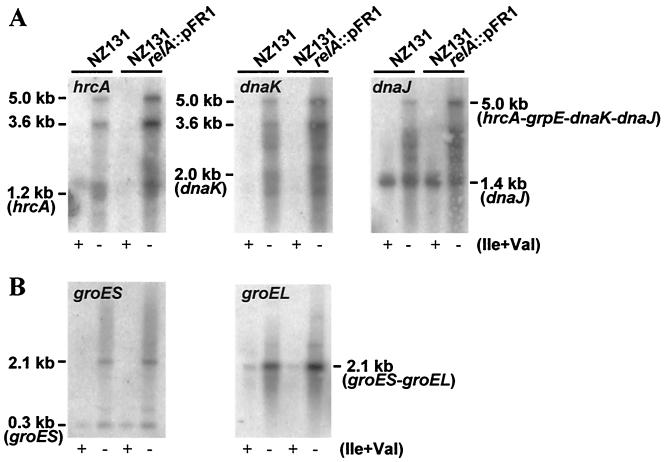
We found that the dnaK and groE operons responded strongly in a relA-independent manner to isoleucine and valine deprivation with cells incubated in CDM. Table 2 lists the relative increase of transcript abundances as determined in slot blot hybridizations with probes specific for the individual genes of the two operons. Corresponding Northern hybridizations provided additional resolution, showing that in addition to some degraded mRNA, the amounts of full-length transcripts (5.0 kb originating with the dnaK operon and 2.1 kb from the groE operon) could principally account for the increases of the mRNA amounts in response to nutritional stress. In unstarved control cells, only the groEL probe detected relatively small amounts of the full-length groES-groEL transcript (Fig. 3A and B).
TABLE 2.
Fold increase of transcript abundance of the dnaK and groE operons determined by slot blot hybridizations of RNA from amino acid-starved wild-type and relA mutant cells relative to results with unstarved control cells
| Strain | Fold increase for gene probe (mean ± SEM)
|
||||
|---|---|---|---|---|---|
| hrcA | dnaK | dnaJ | groES | groEL | |
| NZ131 | 10 ± 0.6 | 26 ± 5 | 25 ± 1.5 | 12 ± 0.6 | 10 ± 0.6 |
| NZ131relA::pFR1 | 14 ± 3 | 60 ± 6 | 27 ± 1.5 | 8 ± 0.3 | 38 ± 4 |
FIG. 3.
relA-independent transcriptional upregulation of the dnaK (A) and groE (B) operons following isoleucine and valine deprivation (minus) of cells. Control cells (plus) were incubated in complete CDM. The gene-specific probes used are indicated on the respective Northern blots.
Streptolysin S (SLS) operon.
Recently Nizet et al. (38) have identified the genetic locus responsible for the beta-hemolytic property of GAS. This locus represents a contiguous nine-gene operon consisting of the SLS prepropeptide structural gene (sagA), followed by eight genes putatively responsible for propeptide processing (sagBCD), cellular immunity (sagE), and transport of the hemolysin across the membrane (sagFGHI). Interestingly, operon transcription is internally disrupted by a factor-independent terminator downstream of sagA, resulting in the excess production of the SagA prepropeptide transcript relative to the full-length transcript. Actually, whereas the presence of a sagA transcript could be readily demonstrated by Northern analysis in strain NZ131 (2), the polycistronic message escaped detection by the Northern technique, albeit its existence at very low levels was suggested by the exploitation of a reverse transcription approach (38).
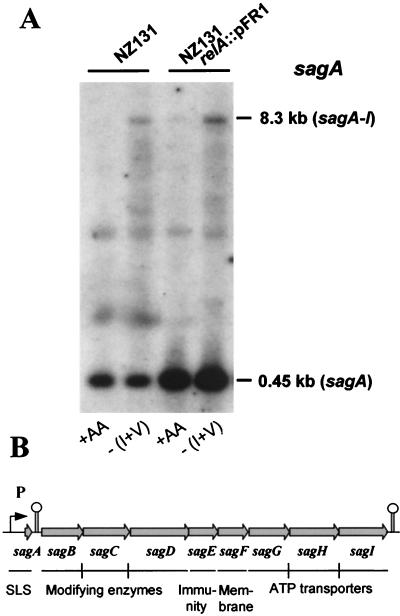
Knowing the importance of SLS as a virulence factor (2) and with the realization in mind that SagA requires the downstream gene products for maturation (38), we explored the possibility of whether environmental circumstances exist that could shift the differential abundance of mRNAs to higher levels of polycistronic messages. Using growth in CDM and imposition of isoleucine and valine starvation, we were able to demonstrate, by Northern hybridization with a sagA probe, the existence of prominent 8.3-kb full-length transcripts of the sag operon in total RNA extracted from amino acid-starved NZ131 cells (Fig. 4). These transcripts were likewise seen in starved wild-type and relA mutant cells but were present only in exceedingly small amounts in unstarved cells of the relA mutant (Fig. 4). Furthermore, amino acid starvation of either strain did not substantially alter the levels of the monocistronic sagA transcript (0.45 kb), which was present in high abundance in both starved and unstarved cells (Fig. 4). Thus, stimulation of readthrough transcription by amino acid starvation appears to be a striking feature of the regulatory sagA terminator. The alternate interpretations that amino acid starvation increases sag promoter activity or sag transcript stability are less likely, because the sagA transcript levels were unaffected under stress conditions (Fig. 4). Thus, in operons featuring internal terminators, environmental conditions are suggestive of stimulating readthrough transcription when they cause the ratio of the mRNA amounts of the terminator-proximal genes to the amounts of full-length transcripts to shift to values lower than those observed under control conditions.
FIG. 4.
(A) Stimulation of full-length transcription of the SLS (sag) operon probed with sagA in Northern hybridizations of RNA extracted from cells starved for isoleucine and valine [-(I+V)]. Control cells were grown in complete CDM (+AA). The indicated sizes of the hybridizing mRNA species are consistent with the map of the SLS operon shown in panel B.
The fas operon.
Very recently, Kreikemeyer et al. (23) described a locus, fasBCA, a homologue of staphylococcal agr loci, which regulates fibronectin and fibrinogen binding (encoded by fbp and mrp, respectively) and the expression of SLS and streptokinase (ska) in a growth-phase-dependent manner. The locus encodes two histidine protein kinases (fasBC) and a response regulator (fasA). Downstream of the fasBCA operon, these authors noticed, was a small separate gene (fasX) that apparently did not code for protein but gave rise to a distinct transcript required for fasBCA activity. Mutational analysis led them to conclude that the expression of fasX absolutely depends on FasA and that the two-component regulator controls transcription of the dependent genes indirectly via FasX (23). Despite the homology of fas to agr and the similarity between FasX and RNAIII of the staphylococcal agr regulon (39), Kreikemeyer et al. (23) found no evidence for fas being involved in quorum sensing.
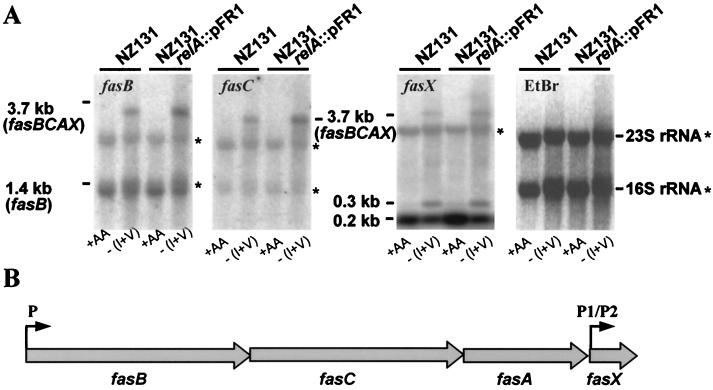
The obvious importance of the fas regulon for growth-phase-associated virulence gene regulation led us to study the transcription profile of this system in cells grown in complete CDM or cells incubated in CDM deprived of isoleucine and valine (Fig. 5). Northern blots probed with the proximal fasB gene probe detected a prominent 1.4-kb signal which apparently corresponded to monocistronically transcribed fasB gene and was present at about equal strength in starved and unstarved cells. In starved wild-type and relA mutant cells, the fasB gene probe detected an additional prominent band at 3.7 kb, a size corresponding closely to a polycistronic transcript originating with all four genes of the fas region. Our failure to detect a polycistronic transcript with the fasB probe (or any other probe specific for the remaining genes of the region) in unstarved early- to mid-exponential-phase cells may reflect insufficient buildup of transcript levels, given that transcription of the fasBCA operon was observed to peak in the postexponential phase (23). Consistent with this observation, the fasC probe detected only the 3.7-kb transcript, and that only in RNA from starved cells (Fig. 5). Results obtained with the fasX probe specific for the distal gene of the operon were most revealing and indistinguishable when wild-type and relA mutant cells were compared. First, fasX detected the 3.7-kb transcript in starved cells, establishing that all four genes can be cotranscribed (Fig. 5). Second, with unstarved control cells, the fasX probe gave only one strong hybridization signal at ∼0.2 kb, corresponding in size to a transcript initiated from the downstream promoter (P2) of the two potential fasX promoters noticed by Kreikemeyer et al. (23). The amount of the 0.2-kb transcript was diminished in starved cells in favor of a ∼0.3-kb transcript corresponding in size to fasX transcription initiated from the upstream promoter, P1. Taken together, these results provide evidence for differential transcription modes of the four fas genes, which depend on the availability of amino acids. Under unstarved conditions, fasB and fasX are preferentially transcribed in a monocistronic manner, fasB presumably from the promoter of the operon and fasX from the operon-internal P2 promoter. Under amino acid starvation conditions, cotranscription of the genes of the fasBCAX operon is strongly stimulated and, simultaneously, transcription of fasX from P1 is induced at the expense of that from P2. Thus, depending on the environmental circumstances, the regulatory fasX gene product may come in three forms which could have distinct activities. Although these results do not rule out the possibility that fasX is under FasBCA control (23), very low levels of the response regulator would appear to be sufficient for extensive fasX transcription (Fig. 5). In addition, considering that fasX can be cotranscribed with fasBCA, its expression does not appear to depend absolutely on FasA.
FIG. 5.
(A) relA-independent stimulation of full-length transcription of the fas operon following isoleucine and valine deprivation [-(I+V)] of cells. Control cells were incubated in complete CDM (+AA). The gene-specific probes used are indicated on the respective Northern blots. The ethidium bromide (EtBr)-stained gel demonstrates equal loading of the slots, integrity of the RNA, and association of the probe DNAs (fasB, fasC, and fasX) to various extents with 23S and 16S rRNA (asterisks). The indicated sizes of the hybridizing RNA species are consistent with the fas operon map shown in panel B.
Autoinducer-2 (AI-2) production protein.
The S. pyogenes SF370 genome (9) contains a gene, luxS, which is present in many gram-positive and gram-negative bacterial species. Studies of gram-negative organisms have shown that this gene is involved in synthesis of AI-2, a compound(s) different from the acyl-homoserine lactones but, like these, accumulating in the external environment as a function of cell density (46). Since the cues influencing AI-2 production appear to be important for virulence gene regulation (46), we examined whether amino acid starvation might be one such cue.
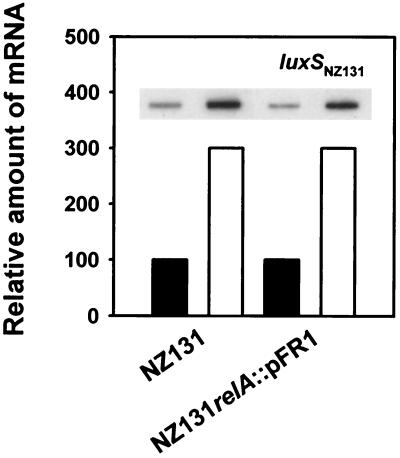
In Northern blots of NZ131 RNA, luxS showed up as specifying a 0.6-kb monocistronic transcript (not shown). Quantification of transcript levels by slot blot hybridization with cells grown in complete CDM and those subjected to deprivation of isoleucine and valine in the corresponding medium yielded approximately threefold-higher luxS transcript levels in the starved cells (Fig. 6). This finding indicates that luxS expression is responsive to nutritional stress.
FIG. 6.
relA-independent upregulation of luxS transcription during isoleucine plus valine starvation (white columns) of NZ131 cells analyzed by slot blot hybridization. Control RNA was extracted from cells grown in complete CDM (black columns).
The effect of puromycin on gene transcription.
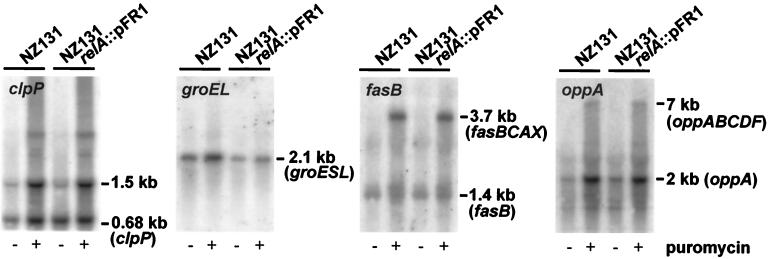
This tRNA analogue interferes with protein syntheses but leaves the ribosomal A site empty by causing the release and accumulation of truncated and misfolded puromycyl polypeptides (12, 13, 32, 37). Puromycin is well known to induce the heat shock response in E. coli (32) and, more recently, has also been shown to induce the synthesis of heat shock proteins, including ClpP, in the gram-positive organism Lactococcus lactis (11). In search of conditions that may overlap amino acid starvation circuits, we grew S. pyogenes NZ131 cells in BHI broth in the presence and absence of puromycin and determined the transcript levels of four selected operons under experimental and control conditions by Northern analysis.
Not surprisingly, we found elevated transcript levels of the clpP and groES-groEL genes in cells grown in the presence of puromycin (Fig. 7). Unexpectedly, however, the expression of the oligopeptide permease operon, opp, and that of the fas operon, both not classed with the heat-inducible operons, were strongly activated in puromycin-exposed cells as well (Fig. 7). We have previously shown that opp operon expression is responsive to amino acid starvation (45). Now we find a response pattern with puromycin treatment that is very similar to that with nutritional stress, featuring increased monocistronic expression of the proximal oppA gene encoding the oligopeptide-binding protein and stimulated full-length transcription of the entire operon, resulting additionally in elevated levels of the oppBCDF message that encodes the translocator proteins. Regarding stimulation of full-length operon transcription by the two stressors, basically the same observation was made for the fasBCAX operon (cf. Fig. 5 and 7). After pondering what amino acid starvation and puromycin exposure might have in common, we suggest that both stressors give rise to the accumulation of truncated, misfolded proteins.
FIG. 7.
Puromycin-induced upregulation (lanes marked by a plus sign) of the indicated genes or operons analyzed by Northern hybridization. Cells were exposed to puromycin for 40 min before RNA extraction; control RNA was extracted from cells grown in normal BHI broth (lanes marked by minus).
DISCUSSION
Together with previous results (45), the present findings show that amino acid starvation of GAS activates a global transcriptional response that is independent of (p)ppGpp accumulation and involves a broad array of genes responsible for housekeeping as well as accessory and dedicated virulence functions. The question is, does any of this matter in the actual life of the pathogen? We tend to answer this question in the affirmative, taking into consideration that blood plasma and the interstitial tissue fluids, in which the organism may find itself, contain roughly 10-fold-lower concentrations of free amino acids than laboratory CDM, which supports optimum growth (45). Considering further that at high growth rates a bacterial cell contains about 7 × 105 tRNAs and synthesizes protein at a rate of about 5 × 105 amino acids per s (7), a tRNA is discharged and recharged approximately every second. This means that restricting the availability of an amino acid immediately limits the corresponding aminoacyl-tRNA and, consequently, rapidly reduces protein synthesis. Under such circumstances, increased expression of the appropriate AARS would improve the tRNA charge state. Since no experiments have been carried out to study specifically the influence of amino acid deprivation and the stringent response on AARS expression in GAS, we present here experimental evidence for increased valS and ileS transcription in response to valine starvation and inhibition of isoleucyl-tRNA aminoacylation, respectively (Fig. 1). In addition, identification of seven AARS genes, including valS and ileS, as members of the T box family strongly suggests that at least five more AARS genes from GAS will show the same regulatory behavior.
The finding that the tmRNA-mediated tagging system for prematurely terminated polypeptides responds to amino acid starvation (Fig. 2) is novel and adds another molecular mechanism that uses a regulatory RNA molecule to systems responsive to nutritional stress. The genes which appear to be sufficient for the function of the system are scattered on the S. pyogenes chromosome and yet respond to amino acid deprivation in a coordinate fashion. Induction of this system would appear to be particularly advantageous to a polyauxotrophic organism in conditions where essentially all nascent polypeptide syntheses are interrupted, mRNAs may be seriously damaged, and nonfunctional protein fragments with conceivably harmful activities would otherwise accumulate. Thus, stimulated targeting of aberrant proteins for degradation and amino acid recycling may confer on S. pyogenes some extra robustness in order to cope with host nutritional deficiencies during specific stages of the infectious process.
The heat shock response can be induced by a variety of agents, many of which will denature proteins in vivo (49). Of interest in the present connection is the finding that E. coli heat shock proteins are also inducible by the stringent response (15). This does not appear to be the case for S. pyogenes (Table 2), an organism which lacks the ς32 subunit of RNA polymerase that mediates the induction of heat shock proteins in E. coli (48). Rather, the S. pyogenes dnaK and groE operons are highly likely to follow basically the regulatory mechanisms established for class I heat shock genes from B. subtilis (19). Based on the model proposed for this organism (33, 34), we suggest that GroE chaperones, which positively modulate the activity of the HrcA repressor (34), are titrated by amino acid starvation-induced misfolded proteins. This may transiently lead to lower levels of HrcA activity and thus higher transcription levels for the dnaK and groE operons. It is interesting that S. pyogenes SF370 appears to have no ςβ factor (9), which in B. subtilis controls the extensive class II heat shock genes of the general stress response through a complex regulatory network involving protein phosphorylation and an anti-sigma factor (reviewed in references 19 and 41). On the other hand, the negatively autoregulated ctsR gene, which in B. subtilis controls the ςB-independent class III genes (clpP, clpC, and clpE) (5, 6), is present in S. pyogenes (9). Induction of the B. subtilis clp genes has recently been reported to involve a putative heat-sensing domain which renders CtsR intrinsically temperature sensitive (6). Furthermore, CtsR turned out to be specifically degraded under stress conditions by the ClpCP protease (25). Targeting of CtsR for proteolysis by ClpCP is modulated by McsA and McsB, which are encoded in the clpC operon (25). Since homologues of these modulators cannot be convincingly identified in S. pyogenes and are certainly not encoded in its clpC operon, the mechanism which leads to derepression of this operon under stress conditions requires elucidation.
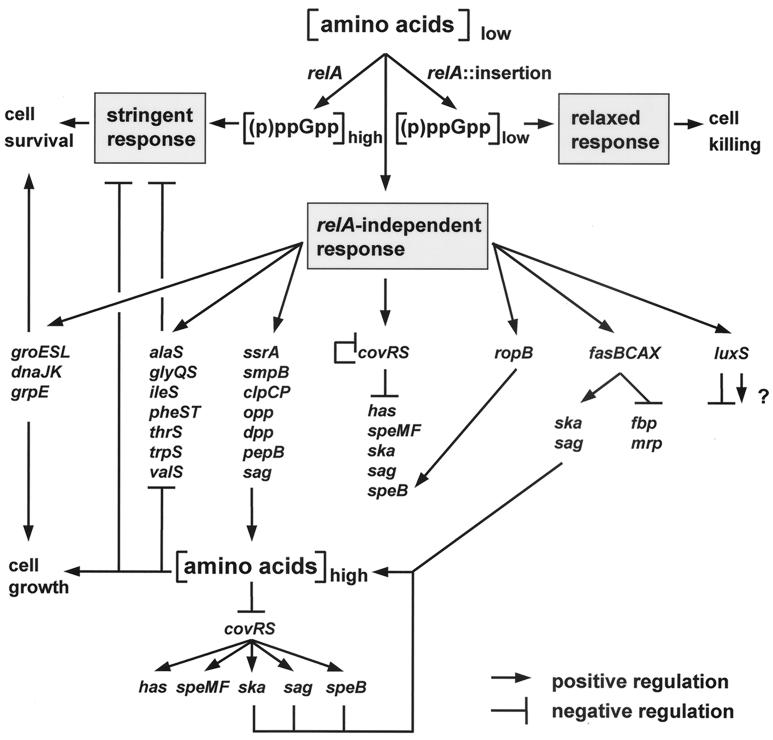
The present results, together with those reported previously (45), implicate several dedicated virulence genes of S. pyogenes in amino acid starvation-induced modulation of expression. Among these are two two-component signal transducers (covRS and fasBCAX), which couple as-yet-unidentified environmental cues to multiple effector outputs, all of which involve virulence factors (1, 8, 18, 23, 45). Furthermore, the transcription of at least one virulence regulator (RopB), which has only one known target gene (speB, which encodes the toxic cysteine protease), is stimulated by nutritional stress (45). Moreover, amino acid starvation strongly favors the coordinate transcription of the hallmark sag operon of GAS (Fig. 4). Whereas a rational basis can be provided for the stimulatory effect of amino acid deprivation on transcription of several housekeeping or accessory virulence genes, and, therefore, it becomes predictable at least to some extent, it appears that the corresponding regulatory behavior of the dedicated GAS virulence factors is largely unpredictable and therefore requires specific analysis. A case in point is mga, the transcription of which is not responsive to starvation (45). Given this situation, the study of system behavior in GAS may still have a long way to go. For the time being, we expand the regulatory network discussed previously (45), including additional circuits that support the notion of a dynamic system which enables GAS to take advantage of protein-rich host environments and to adjust their macromolecular syntheses in order to be compatible with the supply of amino acids. Although the model presented in Fig. 8 is self-explanatory, several novel aspects are worth discussing.
FIG. 8.
Amino acid starvation response network of S. pyogenes. Regulatory circuits included in this scheme but not based on present results were reported previously (1, 3, 8, 18, 23, 27, 45). The question mark indicates that the controlled genes are not yet known.
Identification of the amino acid starvation-responsive T-box family of AARS and tmRNA-directed peptide tagging and proteolysis system (Fig. 2) expands substantially the range of systems that counteract the stringent response. Conceivably, stimulated sag operon expression may also add to this effect, because SLS-effected host cell lysis could enrich certain microenvironments with substrates for the amino acid and peptide scavenging systems. Transient upregulation of the key rate-limiting enzymes that determine the intracellular amino acid pool (the oppBCDF and dppBCDE translocator complexes; see discussion in ref. 45) and the tRNA charge state (the AARS) will lead to diminished levels of CovR, the repressor of an array of at least five virulence genes (Fig. 8). Derepression of three of them, encoding streptokinase (ska), SLS (sag), and the SpeB protease (speB), may actuate an amplification loop as their activities increase the amounts of appropriate oligopeptides available for uptake into the cell. Thus, this circuitry links basic metabolic processes and the level of key rate-limiting enzymes to virulence gene expression. A recurrent theme in this connection is the starvation-induced upregulation of full-length operon transcription. Operons most notably affected in such a way include opp, dpp (45), sag (Fig. 4), and fas (Fig. 5). Regardless of the potentially involved mechanisms (increase of promoter activity, readthrough transcription, or transcript stability) that remain to be further explored, this regulatory phenomenon improves the efficacy of these systems by increasing the level of coordinate transcription of functionally related genes. This becomes most important in cases where the promoter-distal genes encode products, the amounts of which constitute bottlenecks for operon function. It is also interesting that several prominent virulence genes of GAS are subject to multiple control mechanisms. Notable examples are speB, ska, and sag, which are both positively and negatively regulated (8, 18, 23). Beyond that, both types of regulators are responsive to starvation (Fig. 8). The balance between these opposing actions under amino acid starvation conditions may favor repression, as is the case for ska and speB transcription (45), or derepression, exemplified by stimulated readthrough transcription of the sag operon (Fig. 4). Exploitation of such mechanisms may result in fine modulation of virulence factor expression in accordance with the demands imposed by ecologically different colonization sites upon the adaptive capacity of the pathogen. Given the disparity of genes responsive to amino acid starvation, the underlying mechanisms would seem to be similarly diverse. However, the recent discoveries that the pleiotropic transcriptional repressor CodY senses the intracellular levels of branched-chain amino acids and GTP in L. lactis (17) and B. subtilis (43), respectively, makes CodY a promising candidate in search of common regulatory components involved in the relA-independent responses to amino acid starvation. In the same vein, another such candidate protein might be the AI-2 synthase LuxS, the substrate of which has recently been shown to be S-ribosylhomocysteine, an amino acid derivative (44).
ACKNOWLEDGMENTS
We thank M. Völkel and U. Wrazidlo for excellent technical assistance and T. Henkin for helpful discussion of the T-box family of AARS. Mupirocin was a gift of GlaxoSmithKline.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Ma 1330/2-1) and the Fonds der Chemischen Industrie (10046).
REFERENCES
- 1.Bernish B, van de Rijn I. Characterization of a two-component system in Streptococcus pyogenes which is involved in regulation of hyaluronic acid production. J Biol Chem. 1999;274:4786–4793. doi: 10.1074/jbc.274.8.4786. [DOI] [PubMed] [Google Scholar]
- 2.Betschel S D, Borgia S M, Barg N L, Low D E, De Azavedo J C. Reduced virulence of group A streptococcal Tn916 mutants that do not produce streptolysin S. Infect Immun. 1998;66:1671–1679. doi: 10.1128/iai.66.4.1671-1679.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaussee M S, Ajdic D, Ferretti J J. The rgg gene of Streptococcus pyogenes NZ131 positively influences extracellular SPE B production. Infect Immun. 1999;67:1715–1722. doi: 10.1128/iai.67.4.1715-1722.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies H C, Karush F, Rudd J H. Effect of amino acids on steady state growth of a group A hemolytic streptococcus. J Bacteriol. 1965;89:421–427. doi: 10.1002/path.1700890156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derré I, Rapoport G, Msadek T. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in Gram-positive bacteria. Mol Microbiol. 1999;31:117–131. doi: 10.1046/j.1365-2958.1999.01152.x. [DOI] [PubMed] [Google Scholar]
- 6.Derré I, Rapoport G, Msadek T. The CtsR regulator of stress response is active as a dimer and specifically degraded in vivo at 37 degrees C. Mol Microbiol. 2000;38:335–347. doi: 10.1046/j.1365-2958.2000.02124.x. [DOI] [PubMed] [Google Scholar]
- 7.Emilsson V, Kurland C G. Growth rate dependence of global amino acid composition. Biochim Biophys Acta. 1999;1050:248–251. doi: 10.1016/0167-4781(90)90175-2. [DOI] [PubMed] [Google Scholar]
- 8.Federle M J, McIver K S, Scott J R. A response regulator that represses transcription of several virulence operons in the group A Streptococcus. J Bacteriol. 1999;181:3649–3657. doi: 10.1128/jb.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferretti J J, McShan W M, Ajdic D, Savic D, Lyon K, Sezate S, Suvorov A N, Clifton S, Kenton S, Lai H S, Lin S P, Najar F Z, Song L, White J, Yuan X, Roe B A, McLaughlin R. Complete genome sequence of an M1 strain of Streptoccus pyogenes. Proc Natl Acad Sci USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrieri P, Dajani A S, Wannamaker L W, Chapman S S. Natural history of impetigo. I. Site sequence of acquisition and familial patterns of spread of cutaneous streptococci. J Clin Investig. 1972;51:2851–2862. doi: 10.1172/JCI107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frees D, Ingmer H. ClpP participates in the degradation of misfolded protein in Lactococcus lactis. Mol Microbiol. 1999;31:79–87. doi: 10.1046/j.1365-2958.1999.01149.x. [DOI] [PubMed] [Google Scholar]
- 12.Goff S A, Goldberg A L. Production of abnormal proteins in E. coli stimulates transcription of lon and another heat shock gene. Cell. 1985;41:587–595. doi: 10.1016/s0092-8674(85)80031-3. [DOI] [PubMed] [Google Scholar]
- 13.Gottesmann S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 14.Gottesman S, Roche E, Zhou Y, Sauer R T. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grossman A D, Taylor W E, Burton Z F, Burgess R R, Gross C A. Stringent response in Escherichia coli induces expression of heat shock proteins. J Mol Biol. 1985;186:357–365. doi: 10.1016/0022-2836(85)90110-x. [DOI] [PubMed] [Google Scholar]
- 16.Grundy F J, Haldeman M T, Hornblow G M, Ward J M, Chalker A F, Henkin T M. The Staphylococcus aureus ileS gene, encoding isoleucyl-tRNA synthetase, is a member of the T-box family. J Bacteriol. 1997;179:3767–3772. doi: 10.1128/jb.179.11.3767-3772.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guédon E, Serror P, Ehrlich S D, Renault P, Delorme C. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol Microbiol. 2001;40:1227–1239. doi: 10.1046/j.1365-2958.2001.02470.x. [DOI] [PubMed] [Google Scholar]
- 18.Heath A, DiRita V J, Barg N L, Engleberg N C. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect Immun. 1999;67:5298–5305. doi: 10.1128/iai.67.10.5298-5305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hecker M, Schumann W, Völker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 20.Henkin T M. tRNA-directed transcription antitermination. Mol Microbiol. 1994;13:381–387. doi: 10.1111/j.1365-2958.1994.tb00432.x. [DOI] [PubMed] [Google Scholar]
- 21.Karzai A W, Susskind M M, Sauer R T. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keiler K C, Waller P R H, Sauer R T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 23.Kreikemeyer B, Boyle M D P, Leonard Buttaro B A, Heinemann M, Podbielski A. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol Microbiol. 2001;39:392–406. doi: 10.1046/j.1365-2958.2001.02226.x. [DOI] [PubMed] [Google Scholar]
- 24.Krüger E, Msadek T, Hecker M. Alternate promoters direct stress-induced transcription of the Bacillus subtilis clpC operon. Mol Microbiol. 1996;20:713–723. doi: 10.1111/j.1365-2958.1996.tb02511.x. [DOI] [PubMed] [Google Scholar]
- 25.Krüger E, Zühlke D, Witt E, Ludwig H, Hecker M. Clp-mediated proteolysis in gram-positive bacteria is autoregulated by the stability of a repressor. EMBO J. 2001;20:852–863. doi: 10.1093/emboj/20.4.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laport M S, de Castro A C D, Villardo A, Lemos J A C, Bastes M D F, Giambiagi-de Marval M. Expression of the major heat shock proteins DnaK and GroEL in Streptococcus pyogenes: a comparison to Enterococcus faecalis and Staphylococcus aureus. Curr Microbiol. 2001;42:264–268. doi: 10.1007/s002840110215. [DOI] [PubMed] [Google Scholar]
- 27.Levin J C, Wessels M R. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- 28.Luo D, Condon C, Grunberg-Manago M, Putzer H. In vitro and in vivo secondary structure probing of the thrS leader in Bacillus subtilis. Nucleic Acids Res. 1998;26:5379–5387. doi: 10.1093/nar/26.23.5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malke H, Steiner K, Gase K, Frank C. Expression and regulation of the streptokinase gene. Methods. 2000;21:111–124. doi: 10.1006/meth.2000.0982. [DOI] [PubMed] [Google Scholar]
- 30.Mechold U, Cashel M, Steiner K, Gentry D, Malke H. Functional analysis of a relA/spoT gene homolog from Streptococcus equisimilis. J Bacteriol. 1996;178:1401–1411. doi: 10.1128/jb.178.5.1401-1411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mechold U, Malke H. Characterization of the stringent and relaxed responses of Streptococcus equisimilis. J Bacteriol. 1997;179:2658–2667. doi: 10.1128/jb.179.8.2658-2667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller C G. Protein degradation and proteolytic modification. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 938–954. [Google Scholar]
- 33.Mogk A, Homuth G, Scholz C, Kim L, Schmid F X, Schumann W. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 1997;16:4579–4590. doi: 10.1093/emboj/16.15.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mogk A, Volker A, Engelmann S, Hecker M, Schumann W, Volker U. Nonnative proteins induce expression of the Bacillus subtilis CIRCE regulon. J Bacteriol. 1998;180:2895–2900. doi: 10.1128/jb.180.11.2895-2900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muto A, Ushida C, Himeno H. A bacterial RNA that functions as both a tRNA and an mRNA. Trends Biochem Sci. 1998;23:25–29. doi: 10.1016/s0968-0004(97)01159-6. [DOI] [PubMed] [Google Scholar]
- 36.Neidhardt F C, Savageau M A. Regulation beyond the operon. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1310–1324. [Google Scholar]
- 37.Nelson R J, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig E A. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1993;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 38.Nizet V, Beall B, Bast D J, Datta V, Kilburn L, Low D E, de Azavedo J C S. Genetic locus for streptolysin S production by group A Streptococcus. Infect Immun. 2000;68:4245–4254. doi: 10.1128/iai.68.7.4245-4254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novick R P. Pathogenicity factors and their regulation. In: Fischetti V A, Novick R P, Ferretti J J, Portnoy D A, Rood J I, editors. Gram-positive pathogens. Washington, D.C.: ASM Press; 2000. pp. 392–407. [Google Scholar]
- 40.Pichichero M E. Group A beta-hemolytic streptococcal infections. Pediatr Rev. 1998;19:291–302. doi: 10.1542/pir.19-9-291. [DOI] [PubMed] [Google Scholar]
- 41.Price C W. Protective function and regulation of the general stress response in Bacillus subtilis and related Gram-positive bacteria. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 179–197. [Google Scholar]
- 42.Putzer H, Grunberg-Manago M, Springer M. Bacterial aminoacyl-tRNA synthetases: genes and regulation of expression. In: Söll D, RajBhandary U, editors. tRNA: structure, biosynthesis, and function. Washington, D.C.: ASM Press; 1995. pp. 293–333. [Google Scholar]
- 43.Ratnayake-Lecamwasam M, Serror P, Wong K-W, Sonenshein A L. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 2001;15:1093–1103. doi: 10.1101/gad.874201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schauder S, Shokat K, Surette M G, Bassler B L. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41:463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- 45.Steiner K, Malke H. Life in protein-rich environments: the relA-independent response of Streptococcus pyogenes to amino acid starvation. Mol Microbiol. 2000;38:1004–1016. doi: 10.1046/j.1365-2958.2000.02203.x. [DOI] [PubMed] [Google Scholar]
- 46.Surette M G, Miller M B, Bassler B L. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams K P. The tmRNA website. Nucleic Acids Res. 1999;27:165–166. doi: 10.1093/nar/27.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yura T, Kanemori M, Morita M T. The heat shock response: regulation and function. In: Storz G, Hengge-Aronis R, editors. Bacterial stress response. Washington, D.C.: ASM Press; 2000. pp. 3–18. [Google Scholar]
- 49.Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]