Abstract
Considerable evidence suggests that self-renewal and differentiation of cancer stem-like cells (CSCs), a key cell population in tumorgenesis, can determine the outcome of disease. Though the development of microfluidics has enhanced the study of cellular lineage, it remains challenging to retrieve sister cells separately inside enclosed microfluidics for further analyses. In this work, we developed a photo-mechanical method to selectively detach and reliably retrieve target cells from enclosed microfluidic chambers. Cells cultured on carbon nanotube-polydimethylsiloxane composite surfaces can be detached using shear force induced through irradiation of a nanosecond-pulsed laser. This retrieval process has been verified to preserve cell viability, membrane proteins, and mRNA expression levels. Using presented method, we have successfully performed 96-plex single-cell transcriptome analysis on sister cells in order to identify the genes altered during self-renewal and differentiation, demonstrating phenomenal resolution in the study of cellular lineage.
Keywords: cell retrieval, cell detachment, carbon nanotube, photo-acoustics, microfluidics, single-cell transcriptome analysis
Intratumor heterogeneity imposes inevitable challenges to successful cancer treatment. Considerable evidence supports the presence of a sub-population of cancer stem-like cells (CSCs) marked by the capability to drive tumor growth and metastasis and contribute to treatment resistance in many cancers.1,2 During tumorgenesis, some CSCs differentiate to grow the bulk tumor, while others preserve stemness through self-renewal.3 Self-renewal is one key characteristic of CSCs to maintain unrestrained growth, metastatic capability, and drug resistance. Modulation of deregulated self-renewal pathways (SRPs) in CSCs is promising as therapeutics.4 Thus, it is imperative to understand the regulation of cellular determination to self-renew or differentiate. Although fluorescence-activated cell sorting (FACS) and Magnetic-activated cell sorting (MACS) instruments can sort CSCs based on cell surface markers or enzymatic activity,5,6 these methods merely identify current cell states, completely losing the information regarding cellular lineage; when trypsinizing cells for sorting, cells from different clones all mix together, precluding the identification of self-renewing and differentiating CSCs.
In contrast to conventional dish-based methods with such limited resolution, recent developments in microfluidics have enabled the isolation and monitoring of thousands of single cells on chip.7–11 Using these platforms, self-renewal and differentiation process of single CSCs can be monitored under microscope.12,13 Though the single-cell monitoring provides useful information, microfluidic systems still lack the capability to retrieve healthy individual cells for genotypic analysis, which is essential to elucidate the regulating pathways for cell renewal and differentiation in cancer development.1,14 Given some on-chip analyses can be performed,15–18 for full characterization of cell properties, it is critical to retrieve target cells for off-chip analyses since the off-chip analysis tools offer higher multiplexing capabilities, such as 96-plex quantitative real-time polymerase chain reaction (qRT-PCR) using Fluidigm C1 and Biomark, whole transcriptome analysis using sequencing, and 32-plex protein quantification using Fluidigm CyTOF.
Conventional cell detachment schemes, such as trypsinization or poly(N-isopropylacrylamide) (PNIPAAm)-based detachment,19 do not allow spatially localized removal of cultured cells from their substrate; instead, cells are detached from the entire substrate without specificity. The PALM CombiSystem developed by Zeiss can optically isolate and catapult the selected cells into a tube. However, this system works only on an open substrate with no cover frame. Therefore, it is not compatible for integration with microfluidics, which is an ideal platform for single-cell manipulation and monitoring. Alternatively, methods using capillary vacuum or localized trypsin exposure have been used for selective cell retrieval, but these are still limited to open substrates.20,21 Cell release through photo-degradation of a film substrate could provide better spatial resolution and allow integration with microfluidic devices, but this process generates acidic byproducts leading to toxicity and potentially affecting cell behavior and expression.22 Although detachable microrafts were introduced for the analysis of single cells, with this approach it would be very difficult to separate two sister cells carried on the same microrafts.23 Optical approach was reported using infrared laser irradiation directly to a carbon nanotube (CNT)-based substrate, but cell viability was very poor due to heat-induced cell necrosis.24 Recently, single-cell detachment was also demonstrated using laser-generated focused ultrasound (LGFU).25,26 The focused ultrasound could produce single-bubble cavitational disturbance at the microscale focal zone (<100 μm) to detach target cells cultured on a substrate. However, these works were also limited to open substrate applications. Pressure fields within a microfluidic channel, generated by surface acoustic waves, were capable of moving cells in suspension to form spheroids, but not detaching or controlling a single targeted cell.27
Here, we present a microfluidic system, which consists of a top frame for microfluidic guidance and a multi-functional bottom substrate made from a CNT-PDMS composite film,25 enabling complete single-cell retrieval with both spatial precision and high cell viability. The CNT-PDMS film allows (1) highly biocompatible cell culture, (2) easy incorporation with microfluidic fabrication, and most importantly (3) generation of an optically driven shear force (or micro-bubble) which breaks cell contact with the surface. Our approach enables physical detachment of a target single cell with better preservation of viability and membrane proteins as compared to a trypsin-treated reference. This process is highly reproducible within an enclosed microfluidic platform (>90% of detached single cells are successfully retrieved and available for secondary assays). Furthermore, we demonstrate that the cell detachment process does not affect gene expression, validating its use in single cell transcriptome analysis. Using the presented method, we compared the differences in 96 oncogene expression of self-renewing and differentiating sister cells, which were derived from one mother cell. The results support that differentiation of CSC induces more dramatic changes in gene expression. More interestingly, we found that two self-renewing sister cells still have differences in gene expression signature. The key genes altered during self-renewal and differentiation of CSCs were identified using the resolution of the presented approach.
Results and Discussion
Single Cell Capture Scheme
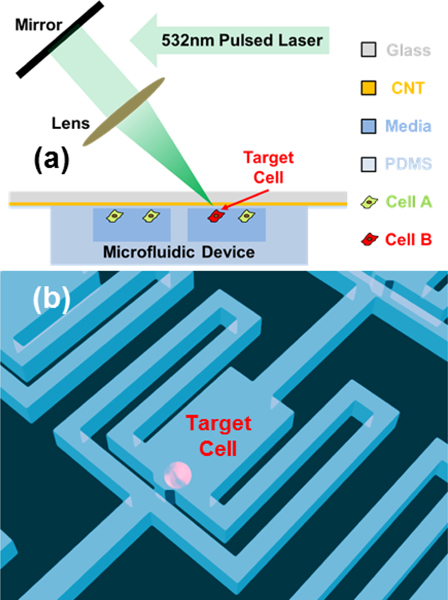
The cell detachment setup is illustrated in Fig. 1. To isolate single cells for lineage tracking and division monitoring prior to retrieval, we adapted our previously developed single-cell hydrodynamic capture scheme.10 Fig. 1(b) shows the schematic of a single cell capture chamber. MDA-MB-231 (breast cancer) cells are loaded from the inlet by gravity flow that is generated spontaneously due to a liquid height difference between the inlet and the outlet. No external pumps are required for cell loading or culture. In each micro-chamber, there are two fluidic paths: a central path and a serpentine path (Supplementary Fig. 1). Initially, the central path has a lower flow resistance so that the first cell entering the chamber tends to flow through the central path (Supplementary Fig. 1, Supplementary Movie 1). As the opening of the central path is smaller than the cell size (10 μm by 15 μm in our design), it sterically captures the cell. The captured cell will thus block the central path, increasing the central path resistance. Following cells will then preferentially flow through the serpentine path and be captured in the downstream micro-chambers (Supplementary Fig. 1, Supplementary Movie 2). Cell capture experiments have been performed with various cell types, and high cell capture rates (> 70%) have been achieved for all of these cell lines.10 This design has been used to create single cell capture arrays with greater than a thousand chambers.9–11
Fig. 1.
Schematic diagram of single-cell detachment setup. (a) The cells were cultured in the microfluidic chamber coated with CNT-PDMS composite. A short pulse laser is used to detach the target cell. (b) the single-cell hydrodynamical capture scheme in the microfluidic chamber.
Cell Detachment on a Plane Substrate without Microfluidic Chamber
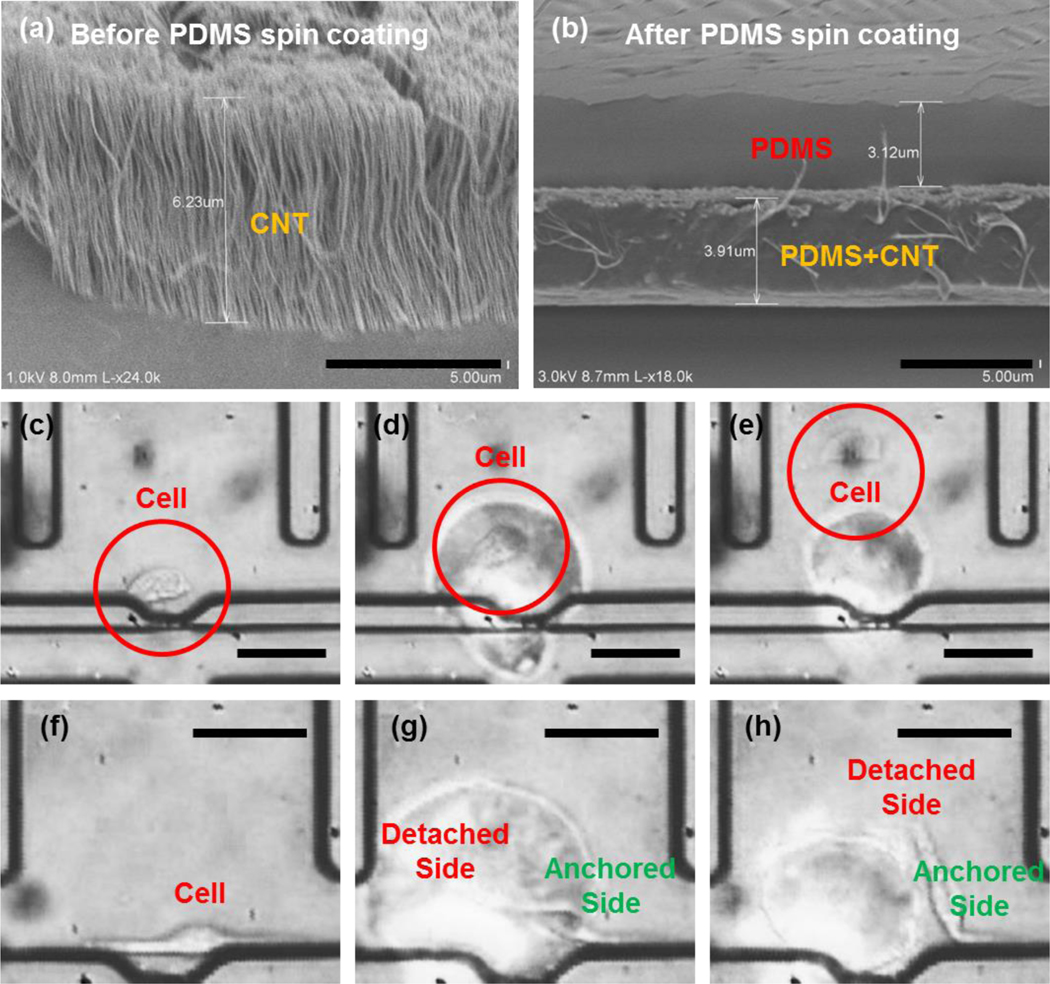
We first tested single-cell detachment by using a CNT-PDMS composite film coated on a glass substrate without microfluidic channels for single cell deployment.28 For fabrication of CNT-PDMS composite films, we used CNTs grown by a chemical vapor deposition (CVD) process, followed by spin-coating with PDMS (see Methods section). Although we used the high-temperature process to prepare a CNT-grown substrate separately from microfluidic frames, a low-temperature preparation technique using CNT gels was also reported,29 which is useful for direct fabrication of CNT-PDMS composite films on a microfluidic substrate. MDA-MB-231 cells were cultured on the CNT-PDMS composite film. Then, we applied pulsed laser irradiation (7-ns width) onto the CNT-PDMS film to produce a 1–2 micro-bubbles with a single pulse (Supplementary Fig. 2). In the CNT-PDMS composite, CNT strands mixed with the PDMS convert the incident optical energy into thermal energy. 25 CNTs allow rapid heat transfer to the surrounding PDMS, (e.g. ~0.5 ns for a 20-nm thick CNT strand which is much shorter than the laser pulse width).25,30 Note that the composite structure initially contains nanoscale porosity over the whole film. Therefore, the optical actuation can expand and merge nano-bubbles into a single bubble with greater dimension (~a few to tens μm in diameter).31,32 Thus, heat-induced bubbles are instantaneously generated from the inside of the PDMS and then ejected through the film surface. The targeted cell (Supplementary Fig. 2, Supplementary Fig. 3, and the Supplementary Video 3) is exposed to shear forces generated by the rupture of the surface as well as the lateral displacement created as the bubble expands. The shear stress we generated is expected to be higher than 600 Pa, which is enough to detach cells in literature.33 Since the small laser spot size (<10μm) creates a highly localized region where the shear force is applied, we can even detach a specific part of the single cell making focal contacts with the substrate. For example, an elongated single cell had only its left side detached, leaving the right side anchored (Fig 2g–f), demonstrating the specificity and resolution possible.
Fig. 2.
Selective single cell detachemnt by optical generation of shear forces on the CNT-PDMS film: (a) Scanning electron microscope (SEM) image of the CNT grown on quartz substrate (scale bar: 5 μm), (b) SEM image of CNTs (separate sample) after spin coating with PDMS (scale bar: 5 μm). (c-e) An example of laser detachment of a MDA-MB-231 cell: (c) before detachment, (d) immediately after pulsed laser irradiation, and (e) cell detached and flowed away from the original culture location (scale bar: 50 μm). Note that the cell is highly viable preserving its membrane without biochemical modification observed in the trypsinization (f-h) An example of partial detachment of a MDA-MB-231 cell: (f) before detachment, (g) after laser irradiation onto the left extension of the cell, and (h) the partially detached cell only having the physical contact on the right end (scale bar: 50 μm).
Selective Detachment of Cells in Microfluidic Chamber
Next, we integrated our detachment mechanism with our single cell capture design. Again, we used a focused nanosecond laser pulse to detach cells by generating a high shear force. Fig. 2 illustrates the detachment process of a single MDA-MB-231 cell (Supplementary Video 4). Initially (Fig. 2(c)), the cell was captured in the chamber and allowed to adhere onto the substrate for 24 hours. Figure 2(d) shows the cell detachment by the optically generated shear force. Driven by the reversed gravity flow, the detached cell travelled upward for retrieval in the outlet (Fig. 2(e)). The precise spatial resolution of the presented detachment scheme is clearly demonstrated in Fig. 2(f–h) and Supplementary Video 5. After focusing the irradiation to one side of a cell, we could partially detach the cell, leaving one side anchored and the other free. In addition to CNT, 20nm Au/Pd alloy, which has better uniformity and higher potential for scalable fabrication, can be used as an alternative light-absorbing layer.
Cell Retrieval Using Parallel Microfluidic Channel
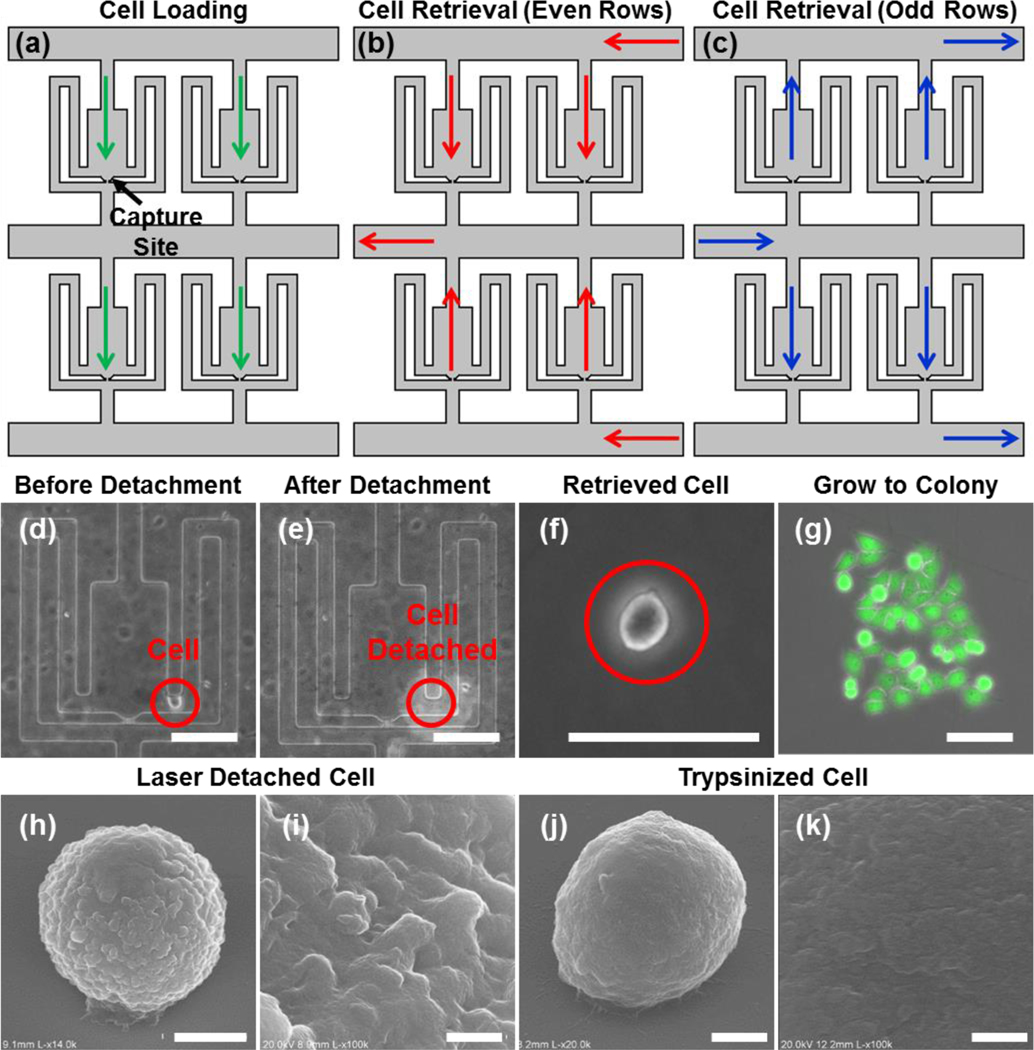
Once detached, the cells must be removed from the device for further analyses (e.g. mRNA qRT-PCR). For this purpose, we designed a microfluidic configuration that can achieve high retrieval yield while simultaneously avoiding undesired contamination from residual cells left in the inlet. In the cell loading phase, media flows from inlet to outlet (i.e. in Fig. 3, flowing from top to bottom), so that the cells can be captured at the capture sites (Fig. 3(a)). In the detachment phase, we first detach the cells from the chambers of even rows in the array before applying pressure from the right side along the horizontal path to collect the selected cells (Fig. 3(b), Supplementary Video 6). The detached cells in those chambers are guided upward and retrieved in the left outlet. Then, we can detach the cells in the chambers of the odd rows before applying the pressure from the left for collection (Fig. 3(c)). The detached cells in these chambers are guided upward and retrieved in the right outlet. Using the alternating parallel channels in an array, we can retrieve all the cells from either the left or right outlets. Consequently, the residual cells that may be retained in the inlet during cell loading do not contaminate the sample. Using this scheme, more than 90% of the detached cells can be successfully retrieved in the outlet. The detachment and retrieval process for four cells in the same chamber is demonstrated in Supplementary Fig. 4.
Fig. 3.
Cell retrieval and viability validation. (a-c) Three different flow schemes of the microfluidic device: (a) cell loading into the chambers for culturing and monitoring, (b) retrieving cells in the even rows, and (c) retrieving cells in the odd rows. (d-g) The recovery of a MDA-MB-231 cell: (d) before detachment, (e) right after detachment, (f) the retrieved cell, (g) proliferation after 4 days (scale bar: 100 μm). (h-k) Scanning electron microscope (SEM) images of laser detached and trypsinized cells: (h) the laser detached MDA-MB-231 cell (scale bar: 5 μm) and (i) its enlarged view (scale bar: 500 nm), (j) the trypsinized MDA-MB-231 cell (scale bar: 3 μm) and (k) its enlarged view (scale bar: 500 nm).
Validation of Retrieved Cell Viability
Due to the low thermal diffusivity of the PDMS layer (0.106 × 10−6 m2/sec) that insulates the cell culture area, the depth of the thermal penetration from the CNT after laser exposure is only ~27 nm for a 7-ns pulse width, well below the surface where the cells are cultured.25,30 Moreover, as we use a single laser pulse as needed with sufficiently long (more than a few seconds) intervals between each, there is negligible heating. This makes it unlikely that the optically generated heat affects cell viability. Fig. 3(d–g) demonstrate an example of a MDA-MB-231 cell was detached and then placed in 96-well plate to observe the effects of laser detachment. The retrieved single cell proliferated to ~20 cells 4 days after detachment. All of the cells were viable as confirmed by using LIFE/DEAD-staining (Fig. 3(g)). Thus, we confirmed that cell viability was maintained after recovery from laser induced cell detachment. We also quantitatively compared the viability of laser-detached cells and trypsinized cells. Remarkably, the laser detachment (viability: 85.3% ± 9.6%) scheme showed a comparable cell viability as the conventional trypsinization process (viability: 83.2% ± 8.7%). This indicates that our approach is highly reliable and suitable for applications requiring further culture of the cells after retrieval.
Moreover, we compared the surface topography of the detached cells using scanning electron microscope (SEM) (Fig. 3(h–k)). This revealed that the laser detachment approach preserves surface proteins, leaving the cell surface intact and rough (Fig. 3(h, i), while the trypsinization digested the membrane proteins significantly, smoothening the cell surface (Fig. 3(j, k).34, 35 Based on these results, we note that the laser detachment method provides an additional advantage as surface markers can be better preserved for immune-staining during downstream analysis.
Validation of Retrieved Cell Gene Expression
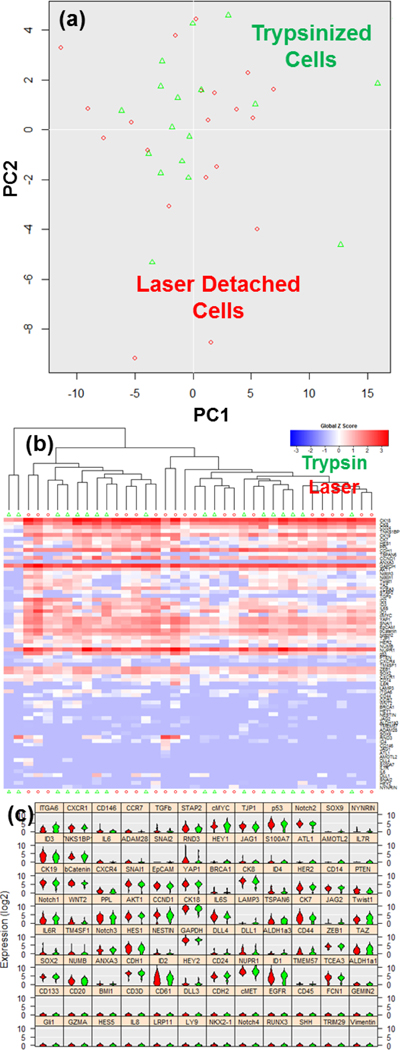
Although we have confirmed cell viability and preservation of surface proteins, the optically driven shear forces may cause cellular stress responses that alters gene expression.36 In order to determine if this was occurring, we characterized the mRNA expression of 96 genes comparing 20 laser-detached and 20 trypsinized T47D (breast cancer) cells using the Fluidigm C1 and Biomark HD. Fig. 4(a) illustrates the principal component analysis (PCA) plot of the expression data showing the characteristics of each single cell as a dot. The trypsizined cells are marked with green, while the laser detached ones are shown in red. In the plot, the two populations mix, meaning that no distinguishing feature (component) can be found to separate the two groups. In addition to PCA, we performed Hierarchical Clustering (HC) analysis (Fig. 4(b)) to group cells. In this manner, cells with similar expression will be clustered closely together. Again, the two populations mix, showing that cellular heterogeneity within group is larger than the difference between the two detachment methods. Fig. 4(c) is the violin plot of the trypsinized and the laser-detached cells. The cells detached by either method maintain typical T47D cell expression, such as high EPCAM and low Vimentin. No significant difference was found between the two populations. This result validates the presented method does not affect gene expression of single cells.
Fig. 4.
Single cell gene-expression data of trypsinized and laser detached T47D cells utilizing Fluidigm C1/Biomark HD for multiplexed gene expression analysis. (a) Principal component analysis (PCA) plot of single cell expression analysis for 20 trypsinized (green) and 20 laser detached (red) cells. Each dot represents a cell. (b) Heatmap hierarchical clustering of single cell expression analysis for 20 trypsinized (green triangle) and 20 laser detached (red circle) cells. In the heatmap, the red color indicates high gene expression, and the blue color indicates low gene expression. Two types of cells are mixed together, indicating no significant alteration in the gene expression when laser detachment was used, as compared to trypsinization. (c) The violin plots of gene expression for 20 trypsinized cells (green) and 20 laser detached cells (red). 96-gene expression of single cells was analyzed. The vertical axis indicates relative expression levels in log2 scale, and the horizontal axis indicates the distribution of cell population. The cells detached by both methods maintain typical T47D cell expression with no significant variation.
Comparison of Gene Expression Signatures Between Two Sister Cells
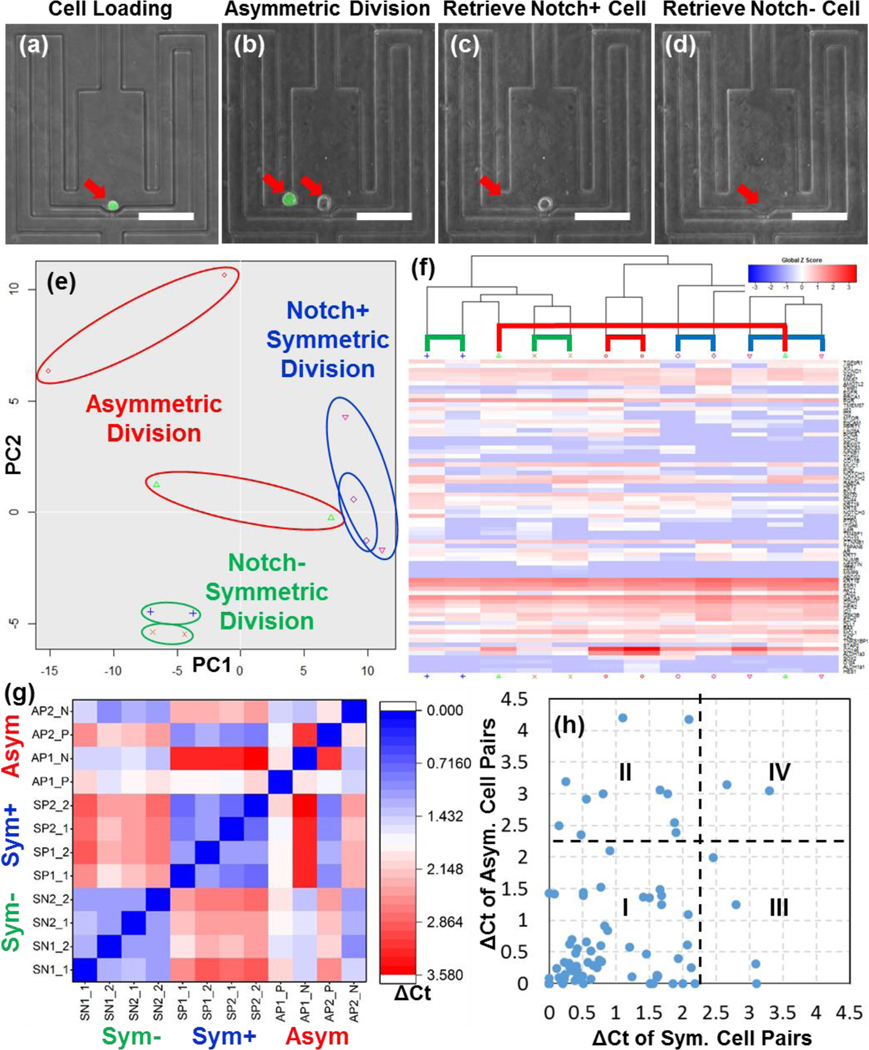
To study the gene expression signatures of two sister cells, we selectively retrieved symmetrically and asymmetrically divided Notch+ T47D cells. The Notch pathway is a signaling pathway that regulates cell self-renewal and differentiation. High Notch expression is related to stem-like properties and higher tumor initiating potential.37,38 To monitor Notch pathway activation, we transduced T47D cells with a lentiviral (pGreenFire1) Notch reporter containing multiple Notch response elements upstream from a minimal CMV promoter regulating destabilized GFP. We loaded Notch+ cells into the device using gravity flow for single cell capture (Fig. 5(a)). Then, we selected a cell that had asymmetrically divided into one Notch+ (green) and one Notch- (non-green) cell after 3 days (Fig. 5(b)). First, we retrieved the Notch+ cell (Fig. 5(c)). Due to precise cell detachment, the Notch- cell stayed at its original place. Then, the Notch- cell was retrieved (Fig. 5(d)), facilitating downstream analysis to compare the two sister cells.
Fig. 5.
Comparison of asymmetrically and symmetrically divided sister cells using single cell transcriptome analysis. (a) A single Notch+ T47D cell was captured in a chamber. (b) After 3 days, the Notch+ cell asymmetrically divided to two cells: i.e. Notch+ (green) and Notch- (non-green). (c) The Notch+ cell was selectively detached and retrieved first, leaving the Notch- cell alone in the chamber. (d) The Notch- cell was retrieved, leaving no cells. (e) Principal component analysis (PCA) plot of 2 pairs of asymmetrically divided sister cells (red), 2 pairs of symmetrically divided Notch+ sister cells (blue), and 2 pairs of symmetrically divided Notch- sister cells (blue). Each dot represents a cell. Comparison of the distances between the cell pairs indicates that the asymmetric division causes significant alteration in the gene expression profiles between two sister cells. (f) Heatmap hierarchical clustering of the asymmetrically and symmetrically divided cells (red - high gene expression, blue - low gene expression). The symmetrically divided cells are more likely to cluster together. (g) The pair-wise difference in the gene expression of 12 cells. The gene expression difference is defined as the average difference in cycle threshold (Ct) of all 96 genes. The blue color indicates low difference (closer expression profile), and the red color indicates high difference (more different expression profile). The results suggest that symmetric division generates two similar sister cells, while asymmetric division generates distinctive sister cells. (h) Identification of genes altered in symmetric and asymmetric division. Each dot represents a gene(horizontal axis - the average difference in Ct of symmetrically divided cell pairs, vertical axis - the average difference in Ct of asymmetrically divided cell pairs). The genes were categorized into 4 types.
The retrieved single cells were lysed, Reverse Transcribed (RT), and pre-amplified in tube, and then analyzed by Biomark HD multiplexed transcriptome analysis. The 96-gene panel (Supplementary Table 1) was chosen to identify the oncogenic signature of breast cancer cells.39 In addition to the asymmetrically divided Notch+ T47D cells, we retrieved symmetrically divided Notch+ (one Notch+ cell producing two Notch+ cells) and Notch- (one Notch- cell producing two Notch- cells) T47D cells for comparison. First, we examined and verified the gene expression profiles of Notch+ and Notch- T47D cells. As expected, significant differences were identified between these two populations (Supplementary Fig. 5–7). Then, we examined the similarities and differences between sister cells. Single-cell gene expression profiles of cell pairs were visualized using PCA (Fig. 5(d)) and hierarchical clustering (Fig. 5(e)). The violin plots of all cell pairs are shown in Supplementary Fig. 8. Fig. 5(f) illustrates the differences in gene expression between all pairs of cells. Here, the difference was quantified by calculating the averages of the difference in threshold cycle (delta cycle threshold (Ct) value) in the 96 genes. As expected, symmetrically divided sister cells showed similar expression profiles, indicated by the closer distance between the two sister cells on the PCA plot, while asymmetrically divided sister cells diverge in expression. A similar trend was observed on the HC plot by the proximity of symmetrically divided sister cells and when measuring the pair-wise distance of expression between all pairs of cells (Fig. 5(g)).
Identification of Genes Altered During Cell Divisions
Based on the gene expression differences between symmetrically and asymmetrically divided sister cells (Fig. 5(h)), the 96 genes were categorized into 4 types: (I) those unlikely to be changed both in symmetrical and asymmetrical division (the lower left corner), (II) those only likely to be changed in asymmetrical division (the upper left corner), (III) those only likely to be changed in symmetrical division (the lower right corner), and (IV) those likely to be changed both in symmetrical and asymmetrical division (the upper left corner). The detailed gene list is shown in Supplementary Table 2. The first type of genes are either not expressed due to the nature of T47D cells, such as Vimentin, or housekeeping genes, such as HPRT1.40 The second type of genes are closely associated with Notch regulation, such as HES1, HEY2, and HER2.41 The gene expression change correlates well with the phenotypic observation of Notch reporter intensity change, demonstrating the presented tool as a bridge between phenotypic monitoring and genotypic analyses. Interestingly, we also found genes with quite different expression levels between two symmetrically divided sister cells, such as Twist1 and BCL2.42 Thus, symmetric division, as defined by Notch pathway expression, does not guarantee conservation of genetic expression. Important oncogenes, such as ALDH1a1 and CXCR1,5,43 can be altered during both symmetric and asymmetric division in breast cancer stem-like cells. In this work, we directly measured the gene expression differences between sister cells that occurred during both symmetric and asymmetric division, which has not been previously demonstrated.
Conclusions
In this work, we developed a photo-mechanical actuation method to precisely detach and retrieve target cells. Thanks to the low thermal diffusivity of the PDMS and short laser pulse, the selective cell retrieval method does not alter cell viability, membrane proteins, and gene expression (mRNA). These features support the presented method as an ideal approach for retrieving target cells from the enclosed microfluidic chambers. Combining single-cell isolation, monitoring, detachment, and retrieval capabilities, we have successfully identified symmetric and asymmetric division events and retrieved the sister cells separately for 96-plex single-cell transcriptome analysis. Compared to conventional FACS sorting, which cannot distinguish self-renewing and differentiating CSCs, we can identify the cells with different lineage to identify critical self-renewal pathways in CSCs. Moreover, using single cell transcriptome analysis, we could identify key regulating genes altered during cell self-renewal and differentiation. Collectively, the proposed study represents a ground-breaking method to advance understanding of cell lineage can be widely applied to other studies of cellular heterogeneity.
Methods:
Substrate for Cell Detachment
In order to detach cells adhered on PDMS, here used as a culture substrate, we prepared a two-layer structure consisting of a bottom layer for light absorption and a top PDMS layer.25 For light absorption, we used a CNT layer that is grown by a high-temperature CVD process (Fig. 2(a)). CNT samples were fabricated in the single-zone CVD chamber on the thin-film catalyst comprising Fe (1 nm) and Al2O3 (10 nm) deposited on SiO2/Si or fused silica substrates at 775 °C under the mixed flowing of C2H4/H2/He.44 The density, length, and morphology were controlled by tuning the CVD parameters and growth time. The CNT layer was then spin-coated with PDMS for composite formation as shown in Fig. 2(b). In addition, we also prepared a 20-nm thick Au/Pd alloy (sputtered) as an alternative light-absorbing layer, instead of CNT. The Au/Pd alloy provided better uniformity and higher potential for scalable fabrication. PDMS, a commonly used microfluidic material, was selected as the polymer layer due to the advantages of low thermal conductance to isolate the cells above from the heat of the CNT surface, preserving cell viability and good bio-compatibility for cell culture. To obtain uniform coating of a thin PDMS layer on the CNT substrate, we diluted PDMS in hexane with a 1:1 ratio. A 3-μm PDMS layer could be achieved using a 6,000 r.p.m. spin coating speed (Fig. 2(b)). The detailed condition for spin-coating with PDMS is shown in Supplementary Fig. 9.
Device Fabrication
We used standard soft lithography to make PDMS layers and bond them to the substrate described in the previous section. Two masks were used to fabricate the multiple heights for the channel region (40 μm height) and the capture gap (15 μm height) as shown in the Supplementary Fig. 10. The PDMS layer and the substrate described in the previous section were bonded together after activation by oxygen plasma treatment (100 Watt, 60 second).
Cell Culture
MDA-MB-231 (human breast cancer cell line) and T47D (human breast cancer cell line) were cultured for the cell experiments. MDA-MB-231 and T47D cells were obtained from Dr. Wicha’s Lab (University of Michigan, MI, USA). MDA-MB-231 cells were cultured in DMEM (Gibco 11965) with 10% FBS (Gibco 10082) and 1% penicillin/streptomycin (Gibco 15070). T47D cells were cultured in RPMI (Gibco 11875) with 10% FBS (Gibco 10082) and 1% penicillin/streptomycin (Gibco 15140). All the cells were cultured in polystyrene culture dishes and passaged at or before cells reached 80% confluency.
Lentiviral transduction
T47D cells were transduced with pGreenFire1-Notch lentiviral (System Biosciences, Mountain View, CA) particles using standard protocols. Lentiviruses were prepared using 3rd generation helper plasmids to generate VSVG pseudotyped particles (roughly 1×107 units/mL) by the University of Michigan Vector Core. 500,000 T47D breast cancer cells/well (50,000/cm2) were plated in a 6-well plate, transduced the following day at a MOI of 10 for 24 hr. Transduction efficiency was ~90% at 1 week post transduction based on FACS analysis of eGFP from cells transduced with pGreenFire-CMV. GFP+ cells were collected by flow cytometry sorting using a MoFlo Astrios cytometer to insure that all cells contain the lentiviral vector. Following cell culture, GFP- cells were generated from GFP+ cells after reaching equilibrium.
Cell Loading
The fabricated microfluidic devices were put in a low pressure desiccator (0.4 atm) for 30 minutes, and then culture media was pipetted to the inlet and outlet to prime the device. Before the experiment, the devices were examined under microscope to make sure that there were no bubbles trapped in the device after priming. Cells were first harvested from a petri-dish with 0.05% Trypsin/EDTA and centrifuged at 100 x g for 5 minutes. Then, cells were re-suspended at 1×106 cells/mL in culture media, and 100 μL of the cell solution was pipetted into the inlet. The flow (0.04 μL/min) was generated spontaneously by gravity flow from the liquid height difference (5 mm, 50 Pa pressure) between the inlet and the outlet, so no external pump was required.10 Within 5 minutes, the cells were hydrodynamically captured in each chamber at single-cell resolution, and the cell solution was replaced with cell-free culture media.
Cell Culture On-chip
After loading of the cancer cells, the media in the inlet is replaced with regular cell culture media. Perfusion flow from inlet to outlets driven by gravity can provide nutrients and remove the wastes generated by cells, so cells can be healthily cultured on-chip.10 We exchange the cell culture media daily. We first take out all the residual media in the outlets and then inlet, and then we add 100 μL of fresh media into the inlet.
Cell detachment
Laser detachment was performed by using a nanosecond (3–7 ns) Q-switched Nd:YAG laser (Continuum,; 532 nm wavelength; 0.1 mJ/pulse). The laser beam was focused on a spot of 10 μm in diameter. After detachment, the cells were treated by LIVE/DEAD staining method (Life Technologies, calcein AM/ethidium homodimer-1, L3224) for 10 minutes in the incubator and examined under a microscope to quantify cell viability. For studying the morphology difference of laser-detached and trypsinized cells using SEM, the retrieved cells were first washed with PBS (Gibco, 10010) and then fixed by 2.5% Glutaraldehyde in 4°C overnight. The sample was then washed with PBS for 5 minutes twice, 5 minutes by 35% Ethanol, 5 minutes by 50% Ethanol, 5 minutes by 70% Ethanol, 5 minutes by 95% Ethanol, and 5 minutes by 100% Ethanol. The sample was then dried on a glass slide and coated with gold using sputter.
Image Acquisition
The microfluidic chips were imaged using an inverted microscope (Nikon TE2000). The bright-field and fluorescent images were taken with a 10x objective lens and a charge-coupled device (CCD) camera (Coolsnap HQ2, Photometrics). A FITC/TRITC filter set was used for the fluorescent imaging. Bright field imaging was performed using an exposure time shorter than 10 ms, and the fluorescent imaging was performed using an exposure time shorter than 100 ms, minimizing the phototoxic effect on cells. The microfluidic cell chamber array was scanned with a motorized stage (ProScan II, Prior Scientific). Before each scanning, the stage was leveled to ensure the image remained in the focus throughout the whole imaging area.
Single Cell Gene Expression Analysis
For the comparison between laser detachment and trypsinization, we loaded the cells into a device with 1,000 single-cell capture chambers and retrieved them by laser and 5-minute trypsinization. The retrieved cells were loaded onto the C1 (Fluidigm) chip and processed by the C1 instrument to isolate the single cells. All the chambers of C1 chip were examined under the IX83 fluorescent microscope to record the status of captured cells in each chamber. Single cells underwent lysis, RNA release, reverse transcription, and finally cDNA pre-amplification for 96 target gene transcripts in the C1 chip. Due to the number (96) of capture wells, the Fluidigm C1 chip does not capture every single cell that is loaded. However, as shown in Supplementary Fig. 11, the C1 platform does not have significant bias or preference toward certain population of single cells based on size or morphology, likely allowing capture of a reasonably representative sample. This is further supported by Supplementary Fig. 12, showing the good correlation between the average of single T47D cell (n = 28) expression data and bulk T47D (5,000 cells each sample, n = 3) expression. For the single-cell expression experiments of two sister cells, the retrieved single cell was transferred in 1 μL into a 200 μL tube. Cell lysis, reverse transcriptase (RT), and pre-amplification were performed in tube using Ambion® Single Cell-to-CT™ Kit (Thermal Fisher Scientific, 4458236). The pre-amplified cDNAs from each single cell were analyzed using the BioMark HD instrument that generates nearly 10,000 qPCR data-points in a single run using a 96 × 96 chip and TaqMan assays. Technical and biological replicates shown in Supplementary Fig. 13 have been performed to ensure reproducible single cell transcriptome analysis. Serial dilution experiments of total RNA, extracted from a T47D breast cancer cell line by using BioMark HD system and TaqMan assays, have been performed to confirm the experiment quantitatively and sensitively (Supplementary Fig. 14–17).
Data Analysis and Processing
Statistical analyses were performed using R (version 3.0). One-way ANOVA tests were used for all comparison studies. A significance level of p < 0.05 was used to consider statistical significance. * refers to P < 0.05, ** refers to P < 0.01, and *** refers to P < 0.001. Results are presented as mean ± SD. Measurements with high variability (such as gene expression levels) were compared on the log-scale. The gene expression data were normalized to GAPDH, a common housekeeper gene in the cell, and the un-normalized data were provided as supplementary information. For single-cell qRT-PCR data generated from the Fluidigm Biomark HD system, we used SINGuLAR v3.0 for data analysis, such as outlier detection, hierarchical clustering and principal component analysis. R package SingleCellAssay was used for improved statistical power in detecting differentially expressed genes.
Supplementary Material
Acknowledgements
This work was supported in part by the Department of Defense (W81XWH-12-1-0325) and in part by the National Institute of Health (1R21CA17585701, 1R21CA19501601A1). Y.-C. Chen acknowledges the support from Forbes Institute for Cancer Discovery. H. W. Baac acknowledges the support from the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (NRF-2014R1A1A2059612) and also from the Technology Innovation Program (No. 10052749) funded by the Ministry of Trade, Industry, and Energy of Korea. The Fluidigm C1/Biomark HD system was supported by SIG-NIH (S10OD16187). A. J. Hart acknowledges the support from Scalable Nanomanufacturing Program National Science Foundation (DMR-1120187). The Lurie Nanofabrication Facility of the University of Michigan (Ann Arbor, MI) is greatly appreciated for device fabrication. The authors also thank Dr. Buckanovich’s lab for providing cells.
Footnotes
Notes The authors declare no competing financial interest.
Associated Content
Supporting Information The Supporting Information is available free of charge on the ACS Publications website at DOI: XX.XXXX/acsnano.XXXXXXX. Information on experiments, fabrication, and single cell transcriptome analysis (PDF).
References
- 1.Reya T, Morrison SJ, Clarke MF & Weissman IL Stem Cells, Cancer, and Cancer Stem Cells. Nature. 2001, 414, 105–111. [DOI] [PubMed] [Google Scholar]
- 2.Maitland NJ & Collins AT Prostate Cancer Stem Cells: a New Target for Therapy. J Clin Oncol. 2008, 26, 2862–2870. [DOI] [PubMed] [Google Scholar]
- 3.O’Brien CA, Kreso A. & Jamieson CH Cancer Stem Cells and Self-Renewal. Clin Cancer Res. 2010, 16, 3113–3120. [DOI] [PubMed] [Google Scholar]
- 4.Borah A, Raveendran S, Rochani A, Maekawa T. & Kumar DS Targeting Self-Renewal Pathways in Cancer Stem Cells: Clinical Implications for Cancer Therapy. Oncogenesis. 2015, 4, e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA & Allan AL High Aldehyde Dehydrogenase and Expression of Cancer Stem Cell Markers Selects for Breast Cancer Cells with Enhanced Malignant and Metastatic Ability. JCell Mol Med. 2009, 13, 2236–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prestegarden L, Svendsen A, Wang J, Sleire L, Skaftnesmo KO, Bjerkvig R, Yan T, Askland L, Persson A, Sakariassen PØ & Enger PØ Glioma Cell Populations Grouped by Different Cell Type Markers Drive Brain Tumor Growth. Cancer Res. 2010, 70, 4274–4279. [DOI] [PubMed] [Google Scholar]
- 7.Hung PJ, Lee PJ, Sabounchi P, Lin R. & Lee LP Continuous Perfusion Microfluidic Cell Culture Array for High-Throughput Cell-Based Assays. Biotechnol Bioeng. 2005, 89, 1–8. [DOI] [PubMed] [Google Scholar]
- 8.Whitesides GM The Origins and the Future of Microfluidics. Nature. 2006, 442, 368–373. [DOI] [PubMed] [Google Scholar]
- 9.Véronique L, Vaninsberghe M, Sekulovic S, Knapp DJ, Wohrer S, Bowden W, Viel F, McLaughlin T, Jarandehei A, Miller M, Falconnet D, White AK, Kent DG, Copley MR, Taghipour F, Eaves CJ, Humphries RK, Piret JM & Hansen CL High-Throughput Analysis of Single Hematopoietic Stem Cell Proliferation in Microfluidic Cell Culture Arrays. Nat Methods. 2011, 8, 581–586. [DOI] [PubMed] [Google Scholar]
- 10.Chen YC Ingram, P.N., Fouladdel, S., McDermott, S.P., Azizi, E., Wicha, M.S. & Yoon, E. High-Throughput Single-Cell Derived Sphere Formation for Cancer Stem-Like Cell Identification and Analysis. Sci Rep. 2016, 6, 27301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng YH, Chen YC Brien R. & Yoon E. Scaling and Automation Of a High-Throughput Single-Cell-Derived Tumor Sphere Assay Chip. Lab Chip. 2016, 16, 3708–3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi YJ, Ingram PN, Yang K, Coffman L, Iyengar M, Bai S, Thomas DG, Yoon E. & Buckanovich RJ Identifying an Ovarian Cancer Cell Hierarchy Regulated by Bone Morphogenetic Protein 2. Proc Natl Acad Sci U S A. 2015, 112, E6882–6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Z, Chen YC, Cheng YH, Luan Y. & Yoon E. Microfluidics 3D Gel-Island Chip for Single Cell Isolation and Lineage-Dependent Drug Responses Study. Lab Chip. 2016, 16, 2504–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shackleton M, Quintana E, Fearon ER, Morrison SJ Heterogeneity in Cancer: Cancer Stem Cells Versus Clonal Evolution. Cell. 2009, 138, 822–829. [DOI] [PubMed] [Google Scholar]
- 15.Ma C, Fan R, Ahmad H, Shi Q, Comin-Anduix B, Chodon T, Koya RC, Liu CC, Kwong GA, Radu CG, Ribas A. & Heath JR A Clinical Microchip for Evaluation Of Single Immune Cells Reveals High Functional Heterogeneity in Phenotypically Similar T Cells. Nat Med. 2011, 17, 738–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamanaka YJ, Szeto GL, Gierahn TM, Forcier TL, Benedict KF, Brefo MS, Lauffenburger DA, Irvine DJ & Love JC Cellular Barcodes for Efficiently Profiling Single-Cell Secretory Responses by Microengraving. Anal Chem. 2012, 84, 10531–10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu Y, Chen JJ, Mu L, Xue Q, Wu Y, Wu PH, Li J, Vortmeyer AO, Miller-Jensen K, Wirtz D. & Fan R. High-Throughput Secretomic Analysis of Single Cells to Assess Functional Cellular Heterogeneity. Anal Chem. 2013, 85, 2548–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen YC, Cheng YH, Ingram P. & Yoon E. Single Cell Proteolytic Assays to Investigate Cancer Clonal Heterogeneity and Cell Dynamics Using an Efficient Cell Loading Scheme. Sci Rep. 2016, 6, 27154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Canavan HE, Cheng X, Graham DJ, Ratner BD & Castner DG Cell Sheet Detachment Affects the Extracellular Matrix: a Surface Science Study Comparing Thermal Liftoff, Enzymatic, and Mechanical Methods. J Biomed Mater Res A. 2005, 75, 1–13. [DOI] [PubMed] [Google Scholar]
- 20.Kudo LC, Vi N, Ma Z, Fields T, Avliyakulov NK, Haykinson MJ, Bragin A. & Karsten SL Novel Cell and Tissue Acquisition System (CTAS): Microdissection of Live and Frozen Brain Tissues. PlosOne. 2012, 7, e41564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guillaume-Gentil O, Zambelli, T. & Vorholt, J.A. Isolation of Single Mammalian Cells from Adherent Cultures by Fluidic Force Microscopy. Lab Chip. 2014, 14, 402–414. [DOI] [PubMed] [Google Scholar]
- 22.Sumaru K, Kikuchi K, Takagi T, Yamaguchi M, Satoh T, Morishita K. & Kanamori T. On-Demand Killing of Adherent Cells on Photo-Acid-Generating Culture Substrates. Biotechnol Bioeng. 2013, 110, 348–352. [DOI] [PubMed] [Google Scholar]
- 23.Gracz AD, Williamson IA, Roche KC, Johnston MJ, Wang F, Wang Y, Attayek PJ, Balowski J, Liu XF, Laurenza RJ, Gaynor LT, Sims CE, Galanko JA, Li L, Allbritton NL & Magness ST A High Throughput Platform for Stem Cell-Niche Co-Cultures and Downstream Gene Expression Analysis. Nat Cell Biol. 2015, 17, 340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sada T, Fujigaya T, Niidome Y, Nakazawa K. & Nakashima N. Near-IR Laser-Triggered Target Cell Collection Using a Carbon Nanotube-Based Cell-Cultured Substrate. ACS Nano. 2011, 5, 4414–4421. [DOI] [PubMed] [Google Scholar]
- 25.Baac HW, Ok JG, Maxwell A, Lee KT, Chen YC, Hart AJ, Xu Z, Yoon E, Guo LJ Carbon-Nanotube Optoacoustic Lens for Focused Ultrasound Generation and High-Precision Targeted Therapy. Sci Rep. 2012, 2, 989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baac HW, Lee T. & Guo LJ Micro-Ultrasonic Cleaving of Cell Clusters by Laser-Generated Focused Ultrasound and Its Mechanisms. Biomed. Opt. Express. 2013, 4, 1442–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen K, Wu M, Guo F, Li P, Chan CY, Mao Z, Li S, Ren L, Zhang R. & Huang TJ Rapid Formation of Size-Controllable Multicellular Spheroids Via 3D Acoustic Tweezers. Lab Chip. 2016, 16, 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baac HW, Ok JG, Lee T. & Guo LJ Nano-Structural Characteristics of Carbon Nanotube-Polymer Composite Films for High-Amplitude Optoacoustic Generation. Nanoscale. 2015, 7, 14460–14468. [DOI] [PubMed] [Google Scholar]
- 29.Noimark S, Colchester JR, Blackburn JB, Zhang ZE, Alles JE, Ourselin S, Beard CP, Papakonstantinou I, Parkin PI & Desjardins EA Carbon-Nanotube-PDMS Composite Coatings on Optical Fibers for All-Optical Ultrasound Imaging. Adv. Funct. Mater 2016, 26, 8390–8396. [Google Scholar]
- 30.Hong J, Choi HS, Lee KS & Shim SE Thermal Properties of Poly(Dimethylsiloxane) Nanocomposite Filled with Silicon Carbide and Multiwall Carbon Nanotubes. Polym. Int 2012, 61, 639–645. [Google Scholar]
- 31.Lukianova-Hleb EY, Anderson LJ, Lee S, Hafner JH & Lapotko DO Hot Plasmonic Interactions: a New Look at the Photothermal Efficacy of Goldnanoparticles. Phys Chem Chem Phys. 2010, 12, 12237–12244. [DOI] [PubMed] [Google Scholar]
- 32.Wu TH, Teslaa T, Kalim S, French CT, Moghadam S, Wall R, Miller JF, Witte ON, Teitell MA & Chiou PY Photothermal Nanoblade for Large Cargo Delivery into Mammalian Cells. Anal Chem. 2011, 83, 1321–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu H, Koo LY, Wang WM, Lauffenburger DA, Griffith LG & Jensen KF Microfluidic Shear Devices for Quantitative Analysis of Cell Adhesion. Anal Chem. 2004. 76(18), 5257–5264. [DOI] [PubMed] [Google Scholar]
- 34.Huang HL, Hsing HW, Lai TC, Chen YW, Lee TR, Chan HT, Lyu PC, Wu CL, Lu YC, Lin ST, Lin CW, Lai CH, Chang HT, Chou HC& Chan HL. Trypsin-Induced Proteome Alteration During Cell Subculture in Mammalian Cells. J Biomed Sci. 2010, 11, 17–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L, Cheng F, Liu T, Lu JR, Song K, Jiang L, Wu S. & Guo W. Comparison Of Mesenchymal Stem Cells Released from Poly(N-Isopropylacrylamide) Copolymer Film and by Trypsinization. Biomed Mater. 2012, 7, 035003. [DOI] [PubMed] [Google Scholar]
- 36.Kültz D. Molecular and Evolutionary Basis of the Cellular Stress Response. Annu Rev Physiol. 2005, 67, 225–257. [DOI] [PubMed] [Google Scholar]
- 37.Grudzien P, Lo S, Albain KS, Robinson P, Rajan P, Strack PR, Golde TE, Miele L. & Foreman KE Inhibition of Notch Signaling Reduces the Stem-Like Population of Breast Cancer Cells and Prevents Mammosphere Formation. Anticancer Res. 2010, 30, 3853–3868. [PubMed] [Google Scholar]
- 38.Cao L, Zhou Y, Zhai B, Liao J, Xu W, Zhang R, Li J, Zhang Y, Chen L, Qian H, Wu M. & Yin Z. Sphere-Forming Cell Subpopulations with Cancer Stem Cell Properties in Human Hepatoma Cell Lines. BMC Gastroenterol. 2011, 11, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, Hong S, Adams A, D’Angelo R, Ginestier C, Charafe-Jauffret E, Clouthier SG, Birnbaum D, Wong ST, Zhan M, Chang JC & Wicha MS Breast Cancer Stem Cells Transition Between Epithelial and Mesenchymal States Reflective of Their Normal Counterparts. Stem Cell Reports. 2014, 2, 78–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silver N, Best S, Jiang J. &Thein SL. Selection of Housekeeping Genes for Gene Expression Studies in Human Reticulocytes Using Real-Time PCR. BMC Mol Biol. 2006, 7, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo S, Liu, M. & Gonzalez-Perez, R.R. Role of Notch and Its Oncogenic Signaling Crosstalk in Breast Cancer. Biochim Biophys Acta. 2011, 1815, 197–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dawson SJ, Makretsov N, Blows FM, Driver KE, Provenzano E, Le Quesne J, Baglietto L, Severi G, Giles GG, McLean CA, Callagy G, Green AR, Ellis I, Gelmon K, Turashvili G, Leung S, Aparicio S, Huntsman D, Caldas C. & Pharoah P. BCL2 in Breast Cancer: a Favourable Prognostic Marker Across Molecular Subtypes and Independent of Adjuvant Therapy Received. Br J Cancer. 2010, 103, 668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ginestier C, Liu S, Diebel ME, Korkaya H, Luo M, Brown M, Wicinski J, Cabaud O, Charafe-Jauffret E, Birnbaum D, Guan JL, Dontu G. & Wicha MS CXCR1 Blockade Selectively Targets Human Breast Cancer Stem Cells in Vitro and in Xenografts. J Clin Invest. 2010, 120, 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ok JG, Tawfick HS, Juggernauth KA, Sun K, Zhang Y. & Hart AJ Electrically Addressable Hybrid Architectures of Zinc Oxide Nanowires Grown on Aligned Carbon Nanotubes. Adv. Funct. Mater 2010, 20, 2470–2480. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.