Abstract
It has recently been shown through DNA microarray analysis of Bacillus subtilis two-component regulatory systems (DegS-DegU, ComP-ComA, and PhoR-PhoP) that overproduction of a response regulator of the two-component systems in the background of a deficiency of its cognate sensor kinase affects the regulation of genes, including its target ones. The genome-wide effect on gene expression caused by the overproduction was revealed by DNA microarray analysis. In the present work, we newly analyzed 24 two-component systems by means of this strategy, leaving out 8 systems to which it was unlikely to be applicable. This analysis revealed various target gene candidates for these two-component systems. It is especially notable that interesting interactions appeared to take place between several two-component systems. Moreover, the probable functions of some unknown two-component systems were deduced from the list of their target gene candidates. This work is heuristic but provides valuable information for further study toward a comprehensive understanding of the B. subtilis two-component regulatory systems. The DNA microarray data obtained in this work are available at the KEGG Expression Database website (http://www.genome.ad.jp/kegg/expression).
Various organisms have developed sophisticated signaling systems for eliciting a variety of adaptive responses to their environment. Adaptive response systems of a class present in prokaryotes, lower eukaryotes, and plants each consist of at least two signal transduction proteins and are therefore referred to as two-component regulatory systems (12, 19, 20). A typical two-component regulatory system consists of two types of a sensor kinase and its cognate response regulator, which usually functions as a transcriptional factor. The sensor kinase receives some environmental signal, which induces autophosphorylation of a histidine residue. The phosphoryl group on the histidine residue is then transferred to a conserved aspartate residue on the cognate response regulator, resulting in modulation of the expression of its target genes in response to the environmental signal.
Genomic sequencing of various microorganisms has revealed the presence of many two-component regulatory systems in every species. In Bacillus subtilis, 36 sensor kinases and 35 response regulators have been found, among which each of 30 kinase-regulator pairs resides in an operon on the genome (4, 11). Of the B. subtilis two-component regulatory systems, approximately 10 have been characterized so far as to their roles (4, 14). Very recently, the DesK-DesR (formerly YocF-YocG) system was reported to be involved in thermosensing and signal transduction at low temperatures (1). However, the functions of the other systems remain unknown (4, 14). In a directed gene knockout study of the response regulators of unknown two-component systems, only the YycG-YycF system was found to be essential for growth (5, 8). The other response regulator null mutations did not noticeably affect colony morphology, growth, or sporulation on laboratory media (5; see also the websites of the Japan functional analysis network for B. subtilis [JAFAN {http://bacillus.genome.ad.jp/}] and the Microbial Advanced Database Organization [Micado {http://locus.jouy.inra.fr/micado}], which are provided by the Japanese and European consortia for Functional Analysis of the B. subtilis Genome, respectively). It is probably safe to conclude that most two-component regulation is used to enhance the versatility of the response of an organism to environmental stimuli through regulation of normally unexpressed genes.
Recently, DNA chip technology involving high-density arrays of open reading frame (ORF)-specific DNA fragments has been rapidly developed for transcriptome analysis of various microorganisms whose genome sequences have been determined. With respect to array analysis of B. subtilis two-component systems, the Spo0A and ResD regulons have been investigated (6, 25), and the respective global changes in gene expression in strains with disruptions in the spo0A and resD genes have been revealed.
The B. subtilis DegU, ComA, and PhoP regulons were recently analyzed using a DNA microarray (15). For the analysis, a strategy was used by which the response regulator genes were cloned downstream of the spac promoter (Pspac) in plasmid pDG148 (21) in Escherichia coli, and the constructed plasmids were then introduced into B. subtilis strains with disruptions in the cognate sensor kinase genes; this plasmid contains Pspac as well as lacI, E. coli, and B. subtilis replication origins derived from plasmids pS11 (24), pBR322, and pUB110, respectively (21). As the response regulator genes were placed under the control of Pspac and negatively regulated by LacI, the response regulators could be overproduced upon the addition of isopropyl-β-d-galactopyranoside (IPTG) to the medium. It was expected that this overproduction of the response regulators might result in altered expression of the target genes in the absence of the environmental signals responsible for their phosphorylation (15). Even if unknown signals that cause autophosphorylation of uncharacterized sensor kinases are present under the growth conditions employed, one can eliminate phosphorylation of the cognate response regulators by using the sensor kinase gene disruptants (15). Thus, one can examine alteration of the expression of the target genes of two-component regulatory systems only upon overproduction of the nonphosphorylated response regulators. Determination of the gene expression ratios of the fluorescence intensities obtained by DNA microarray analysis with RNA from cells grown with IPTG compared to those of cells grown without IPTG allowed for the detection of target gene candidates for the three response regulators, which included most of their known target genes as well as various unknown ones (15). Although some known target genes escaped detection (probably because they were also under the control of other regulatory mechanisms under the experimental conditions used), this strategy was certainly applicable to the detection of the target genes of many uncharacterized two-component systems of B. subtilis for which environmental signals for activation are unknown.
In this work, we comprehensively analyzed 24 B. subtilis two-component regulatory systems by means of the strategy described above. The target gene candidates for the 24 two-component regulatory systems detected in this work, which may be used to identify their target genes, are heuristic but provide valuable information for investigation of the functions of unknown two-component systems as well as further studies of the known ones.
MATERIALS AND METHODS
Bacterial strains and plasmids and their construction.
B. subtilis strains deficient in each of the sensor kinases are listed in Table 1. These mutants were constructed by means of integrational disruption of one of the pMUTIN plasmids (22) through a single-crossover event (26) at the 5′ site of each of the sensor kinase genes of B. subtilis strain 168 trpC2 (3) by the Japanese and European consortia for Functional Analysis of the B. subtilis Genome (see the websites of JAFAN and Micado for details of their construction). Derivatives of plasmid pDG148 (23) carrying the respective cognate response regulator genes (Table 1) were constructed as follows. DNA regions encompassing the structural genes for the response regulators and their Shine-Dalgarno (SD) sequences were amplified by PCR using the primer pairs listed in Table 1. The amplified DNA fragments were digested with HindIII (or SalI) and SalI (or SphI) and then cloned into plasmid pDG148, which had been digested with the same restriction enzymes, resulting in derivatives of plasmid pDG148 (Table 1). For this DNA manipulation, E. coli strain DH5α cells were grown in liquid or agar Luria-Bertani (LB) medium (16), and the transformants exhibiting ampicillin resistance (50 μg/ml) were selected. After the nucleotide sequences of the cloned DNA regions had been confirmed by sequence determination, B. subtilis strains with disruptions in the sensor kinase genes were transformed to have kanamycin resistance (10 μg/ml) by plasmids carrying the respective cognate response regulator genes.
TABLE 1.
B. subtilis strains deficient in each sensor kinase of the two-component regulatory systems, plasmids carrying their cognate response regulator genes, and primer pairs used for their construction
| System | Strain | Plasmid | Sequences of primer pair (forward, reverse) |
|---|---|---|---|
| CitS-CitT | CITSda | pDG148-citT | GATAAGCTTACTGTATTTATACCGAAGG, GATGTCGACCTAATCCGCCGCCAAATAATA |
| DesK-DesR | BFS687b | pDG148-yocG | AAACGCGTCGACGGGACCAAGCTTACCATGGC, GCAGCATGCAATTCTGGTTGCGACGAGG |
| LytS-LytT | LYTSda | pDG148-lytTc | GCGCAAGCTTGTCGACCCAATGCAACAGATGAAAGA, GCCGGGATCCGCATGCAAAGCGCTTACA AGATAACAT |
| YbdK-YbdJ | YBDKda | pDG148-ybdJ | GATGTCGACAAACTGTCCATGGAGTTTATG, GATGCATGCTCATTTCTTCACAACGGTATTCAGCC |
| YcbA-YcbB | YCBAda | pDG148-ycbB | GATGTCGACGATTCAGAAAGGGTGAACCAAAAGC, GATGCATGCTTATATCCTTCTTTGTTTCATCG |
| YcbM-YcbL | YCBMda | pDG148-ycbL | GATAAGCTTGTAAAGGTTAGGTGAGGAGATG, GATGTCGACCTAAAACTCTCCTAATTTATAACCG |
| YccG-YccH | YCCGda | pDG148-yccH | GATAAGCTTGATTCTATGTAACTAGGGGAA, GATGTCGACAAACTCCTCAAAAATAGTAG |
| YclK-YclJ | YCLKda | pDG148-yclJ | GATGTCGACAACCGGAGGATATAATGAAAATATT, GATGCATGCATATGGCTTAGCAGCAGCTGAT |
| YdbF-YdbG | YDBFda | pDG148-ydbG | GATGTCGACCCAATGAAGGGGGAGGAAGCA, GATGCATGCGGTCTCAGGTTTATCCTTTTAA |
| YdfH-YdfI | YDFHda | pDG148-ydfI | AAACGCGTCGACCAAATTGAGATCACTGTACC, GCAGCATGCTCTTTCTCATTGTAGTTGAC |
| YesM-YesN | BFS2288b | pDG148-yesN | AAGAAGCTTGGATTCATAGTGAGCATG, AAACGCGTCGACACACTCAAAAAACAGTGGCG |
| YfiJ-YfiK | YFIJda | pDG148-yfiK | GATCTCGAGTCGGGCCCGTGCAGCAAAAG, GATCTCGAGCGTCATATGTCAAAAGACA |
| yhcY-YhcZ | BFS1714b | pDG148-yhcZ | AAACGCGTCGACAGTCAGCGCTTGGAACTGG, GCAGCATGCTGGCGTGCCATTTATGAC |
| YkoH/ykoG | YKOHda | pDG148-ykoG | AAACGCGTCGACCATCATAATAGATGATAAGAATC, GCAGCATGCGAATCTTGGTCTTCAGCTTC |
| YrkQ-YrkP | YRKQda | pDG148-yrkP | GATAAGCTTAAGAATCATGAGGTGAGAGAGA, GATCTCGAGGCGTAAATTTAAGATGCGCC |
| YtsB-YtsA | BFS81b | pDG148-ytsA | AAACGCGTCGACGGCGTTTTGCTATACACTTG, GCAGCATGCAACAGCATCAAAGCCTGCTG |
| YufL-YufM | BFS1431b | pDG148-yufM | AAACGCGTCGACCCAAATGATCCTCACAAGCG, GCAGCATGCAACGCACTGCTCAAAGCAAG |
| YvcQ-YvcP | YVCQda | pDG148-yvcP | AAACGCGTCGACATTTACTATTCCGCACATGC, GCAGCATGCGGCTAAGCGGTCGATGAG |
| YvfT-YvfU | BFS1109b | pDG148-yvfU | AAACGCGTCGACCATAACGGAACGGTTGTGGC, GCAGCATGCCCGATCTCCCAGGAGTGCAG |
| YvqB-YvqA | BFS2462b | pDG148-yvqA | AAACGCGTCGACCTGCCGTATGTTAAAACTAG, GCAGCATGCCGGATATGACAACCCATATC |
| YvqE-YvqC | BFS2466b | pDG148-yvqC | AAACGCGTCGACAAGGCACTCAAATCGAAGTG, GCAGCATGCTGTCAGAGACTTCGGAACG |
| YvrG-YvrH | BFS2479b | pDG148-yvrH | AAACGCGTCGACGATAAGATGAAAAAAACAGTGACAG, GCAGCATGCTCACAATCAGCATCTGGC |
| YxdK-YxdJ | YXDKda | pDG148-yxdJ | AAACGCGTCGACTGCGTCCATACGCCCAAAACG, GCAGCATGCTCAGCACTGCATGAGACCGG |
| YxjM-YxjL | YXJMpa | pDG148-yxjL | AAACGCGTCGACTGCCGTACAGCCGAGCGGCC, GCAGCATGCTAGCATGGCGGGATGAGAG |
Construction of the mutants is described at the JAFAN website.
Mutants are described at the Micado website.
This plasmid was constructed by M. Serizawa in the laboratory of J. Sekiguchi (Shinshu University, Ueda, Japan).
Growth conditions and RNA isolation.
Cells of the strains deficient in sensor kinases, which carried the respective plasmids containing the cognate response regulator genes, were grown overnight in LB medium. After inoculation of the cells into 100 ml of Schaeffer medium (17) or LB medium in two 500-ml Erlenmeyer flasks, the cells were grown at 37°C to the early logarithmic phase. IPTG was added to one of the flasks at a concentration of 1 mM, and the cells were harvested after 2 h. The detailed growth conditions are available at the KEGG Expression Database website (http://www.genome.ad.jp/kegg/expression). Total RNA was isolated from the cells essentially as described previously (27).
Preparation of fluorescently labeled cDNA.
The fluorescently labeled cDNA probes used for hybridization to DNA microarrays were prepared by a two-step procedure, as described previously (15).
Hybridization and microarray analysis.
DNA microarrays were prepared as described previously (27). The microarrays that we used in this study contained 4,055 protein genes, 45 not being spotted due to a problem with DNA amplification by PCR, as well as 39 calf thymus DNA spots used as negative controls. The hybridization and microarray analysis were performed as described previously (15, 27). The mean Cy3 and Cy5 fluorescence intensities for each spot were calculated, the background being taken as the average of the intensities of the 39 calf thymus DNA spots. After subtracting the background from the intensities of each of the B. subtilis gene spots and normalizing their intensities using the total Cy3 and Cy5 intensities, we calculated the expression ratios (Table 2). To get reliable ratios, we ignored the spots with intensities used as numerators that were less than the background, and replaced the intensities used as denominators with the standard deviation of the average intensity of the negative controls if they were less than the standard deviation.
TABLE 2.
Target gene candidates for 24 B. subtilis two-component regulatory systemsa
| System | No. of genes | Upregulated genesb | Downregulated genes |
|---|---|---|---|
| CitS-CitT | 5 | (citS), citT, citM, yflN, yxiO | |
| DesK-DesR | 26 | des, (desK), desR, yorV, yvfU, yvfT, yvfS, yvfR, hisZ | ydbH, yesL, yesM, yfiI, yjdC, yjdD, yjdE, yjdF, yrxA, rbsR, rbsK, rbsD, rbsA, rbsC, rbsB, ywoG, licC |
| LytS-LytT | 6 | (ysbB), (ysbA), lytT, (lytS), ywjC | ywbH |
| YbdK-YbdJ | 9 | ybdJ, (ybdK), purC, purN, purH, ald | ykoM, ysfC, ysfD |
| YcbA-YcbB | 19 | ybgH, (ycbA), ycbB, lctE, sunA | ybbC, ndhF, ybcT, ybdB, ykzH, yoxA, lpd, bkdR, ysfC, ysfD, rbsB, hutH, hutU, hutI |
| YcbM-YcbL | 4 | ycbL, (ycbM), ysfC, ysfD | |
| YccG-YccH | 5 | (yccG), yccH, natA, natB, yccK | |
| YclK-YclJ | 30 | gerKB, yclH, yclI, yclJ, (yclK), yfiY, ykcB, ykcC, ykuO, pyrR, yngA, yngB, yngC, ysfC, ysfD, yukL, dhbB, dhbA, fhuD | acoA, acoB, acoC, acoL, yvdK, yvdH, atpD, atpA, atpE, qoxC, qoxB |
| YdbF-YdbG | 76 | ydbG, (ydbF), (ydbH), yhcP, yjcP, yqaS, mcpA, mcpB, flgK, flgM | feuC, ybbC, adaA, ndhF, ybcH, ybcL, ybdJ, ybdL, ybfF, ybfH, ycxB, ycxD, ycsO, ydaO, yesZ, yisZ, cotY, yjiA, yjiC, yjmG, xpf, xkdM, ykzH, pksI, ymaB, yngK, yoxD, yoaB, yoaJ, yoaV, yozH, yozJ, yobS, yobW, yqhB, yqfT, yqbE, yqbA, yqaL, yqaJ, yrkP, yrkO, yrdN, cypA, yrpE, sigZ, yraK, yrrD, sspA, yteQ, ytkD, ytaB, glgP, paiA, yurH, yvsH, yvaG, yvaP, opuCD, yvbG, araE, yvfA, yvoE, yvoA, ywrA, yycD |
| YdfH-YdfI | 29 | ydfE, (ydfH), ydfI, ydfJ, ydfT, ydgJ, ydjM, ykuD, pyrR, proJ, proH, yqjD, ysbA, rbsR, rbsK, rbsD, rbsA, rbsC, rbsB, licC, yydH | lctE, lctP, yqxL, ywcJ, ywbH, cydC, cydB, cydA |
| YesM-YesN | 21 | (yesM), yesN, (yesO), (yesP), (yesQ), (yesR), (yesS), (yesT), (yesU), (yesV), (yesW), (yesX), (yesY), (yesZ), (yetA), (lplA), (lplB), opuBD | yjdD, yjdE, licC |
| YfiJ-YfiK | 31 | (yfiJ), yfiK | ybdB, argC, argJ, argB, argD, carA, carB, argF, yjaZ, appF, appB, ykfA, yoeB, yqiZ, leuC, leuB, leuA, ilvN, ilvB, ytzD, argH, argG, yutL, thrB, hom, yxbC, yxbB, yxnB, yxaM |
| YhcY-YhcZ | 8 | (yhcY), yhcZ, yknU, bglS, licT, yxiP, yycE | yjdD |
| YkoH-YkoG | 26 | ybdJ, yclH, yclI, ykcB, ykcC, htrA, ykmA, ykoG, (ykoH), (ykoI), (ykoJ), ykoS, ykoT, yngA, yngB, yngC, yocH, yoqN, yqjD, ytlI, yvtB, yvtA, yvpB, yvpA, galE, yybQ | |
| YrkQ-YrkP | 11 | yjcE, ykcB, ykcC, yqaQ, yqaI, (yrkR), (yrkQ), yrkP, yrkO, yrkN, ywmG | |
| YtsB-YtsA | 9 | slp, yrkR, yrkQ, yrkP, (ytsD), (ytsC), (ytsB), ytsA, yvcR | |
| YufL-YufM | 98 | ybbC, ybbD, ybbE, ybbF, ybbH, ybbI, ybbJ, ybbK, ybfT, ycbP, nin, nucA, yckF, yckG, ycxB, ydfL, rapH, yfjU, ygxB, yhfS, yhfT, yhfU, comK, yhxD, yjgD, smf, yodF, yopY, yopX, yopW, yqjD, sinI, comGG, comGF, comGE, comGD, comGC, comGB, comGA, comEC, comEA, comER, comC, ysbA, ytxD, ccpA, (yufL), yufM, comFC, comFB, comFA, ywpJ, ywpH, ywkA, ywfM, yweA, licH, licC, licB, aldY, galE, yxiQ, bglH, yyaF | lctP, yhfB, pyrB, pyrC, pyrAA, pyrAB, pyrDII, pyrD, pyrE, yosO, yonB, yomU, yolF, yqjI, ythP, mcpB, yukL, yukM, dhbB, dhbC, dhbA, yvfC, yvfB, yvfA, yveR, yveQ, yveP, yveN, yveM, yveL, ywcJ, cydC, cydB, cydA |
| YvcQ-YvcP | 47 | yhaS, clpE, mreBH, yorK, yorI, yorG, yorF, yorE, yorA, yoqS, yoqL, yoqH, yoqD, yoqB, yoqA, yopZ, yopX, yonO, yonN, yonB, yomX, yomW, yomU, yomT, yomS, yomO, yomM, yomG, yomD, araM, araD, araB, araA, ytzE, ytsD, ytsC, yvqI, (yvcS), (yvcR), (yvcQ), yvcP, yvcB, yxdM, yxdL, yxaH | yukL, fliT |
| YvfT-YvfU | 10 | yocE, yvfU, (yvfT), yvfS, yvfR | ydaD, ydaS, yqgZ, yrpC, dhbB |
| YvqB-YvqA | 9 | ygxB, yjgD, htrA, yvtB, yvtA, yvqA | yjdC, yjdD, yjdE |
| YvqE-YvqC | 31 | yacD, cysK, ycsF, ycsG, ycsJ, ydhE, yhcY, yhcZ, yhdA, yhdB, yhdC, yjcL, ypuA, yqjD, gerAB, yvqC, (yvqE), yvqF, yvqG, yvqH, yvqI, yvrK, yvrL, yxjN, yxjM, yxjL, pepT, bglS, licT, yxiP, yycE | |
| YvrG-YvrH | 43 | ycbN, yfhI, yfhK, csbB, yfhO, yheN, wprA, yisO, yisW, ykcB, ykcC, ykfD, yocH, sunT, sunA, yokF, yokE, ypzC, yqjD, pstB2, pstA, (yvrG), yvrH, dltA, galE, yxiM, yxiL, yxiK, yxiJ, yxiI, yxzG, yxiH, yxiG, yxiF, yxxG, wapA, yxdL | ycdA, yorF, yrkO, yrhH, lytC, lytB |
| YxdK-YxdJ | 9 | ydbP, acoA, yqjD, yxdM, yxdL, (yxdK), yxdJ | glpT, yjdE |
| YxjM-YxjL | 28 | yhcY, yhcZ, yhdA, yhfO, yqjL, yqjD, yvqC, yvqE, yvqF, yvqG, yvqH, yvqI, yvrK, yxjN, yxjL, bglS, licT, yxiP, yycE | glpT, ycbK, yjdC, yjdE, pyrAB, pyrD, pyrE, yojI, ywhP |
RNA preparation and DNA microarray experiments were repeated until the two best quality hybridization profiles were obtained. The expression ratio for each gene was taken as the average of two ratios obtained from the hybridization profiles shown. The target gene candidates exhibited greater than fourfold upregulation or downregulation and are listed in clockwise order starting at the chromosomal location of oriC.
The genes in bold were cloned into plasmid pDG148 and overexpressed up on the addition of IPTG. The underlined genes are known target genes of the respective two-component systems. Transcription of the genes in parentheses can be induced by IPTG due to pMUTIN integration into sensor kinase genes without a response to overproduction of the cognate response regulator genes.
RESULTS
B. subtilis two-component regulatory systems subjected to DNA microarray analysis.
Out of the 35 response regulators found in the B. subtilis genome, three well-characterized regulons (DegU, ComA, and PhoP) were analyzed previously by means of the strategy described above (15). DNA microarray analysis of the DegU regulon revealed 116 target gene candidates whose expression was affected by DegU overproduction, including known target genes such as aprE, nprE, and ispA (15). Out of the target gene candidates, several (bpr, yukL, ycdA, and murD) were newly found to be under DegU regulation (15). Microarray analysis of the ComA and PhoP regulons revealed 33 and 23 target candidates, respectively. Target candidates for ComA included the known target genes srfAA, srfAB, srfAC, srfAD, and rapA, while those for PhoP included phoA, phoB, ydhF, phoD, tuaB, tuaC, tuaD, tuaE, tuaF, tuaG, tuaH, pstS, pstA, pstC, pstB1, pstB2, glpQ, and PhoR (15). Out of these target candidates, rapF was newly found to be under ComA regulation and yycP and yjdB were newly found to be under PhoR regulation. These results underscored the validity of our strategy of selecting target genes of the two-component systems even if their functions are not known (15).
Among the B. subtilis two-component regulatory systems, YycG-YycF has been reported to be essential for B. subtilis growth (5, 8), so we cannot apply this strategy involving disruption of the sensor kinase gene for its analysis. Furthermore, five response regulators (Spo0F, CheY, YneI, CheV, and CheB) do not possess a carboxy-terminal DNA-binding domain (4, 7, 10, 14, 18). These appear to play roles in protein-protein interactions rather than in direct regulation of genetic expression. To analyze B. subtilis two-component regulatory systems by means of this strategy, the overproduced response regulators should interact with the cis elements upstream of the genes to affect their expression. Therefore, we left out the regulons governed by these five response regulators from the present comprehensive analysis. In addition, we also left out the Spo0A and ResD regulons from the analysis, the rationale being as follows. First, macroarray and microarray analyses of the Spo0A and ResD regulons using spo0A and resD mutants have already been performed under the physiological conditions under which Spo0A and ResD operate, respectively (6, 25). Second, our current analysis involving overproduction of a response regulator upon the addition of IPTG is supposed to detect only the target genes solely under its control. Since many of the target genes of Spo0A and ResD are also known to be regulated by other regulatory proteins involved in sporulation and anaerobic respiration (6, 25), our analysis was considered not to be appropriate for the detection of their target genes.
To comprehensively analyze B. subtilis two-component regulatory systems by means of this strategy involving DNA microarray analysis, 24 other regulatory genes, including their SD sequences, were cloned into plasmid pDG148 (21), and then the constructed plasmids were transferred to B. subtilis mutants deficient in the cognate sensor kinases (Table 1). In the course of cloning the genes into plasmid pDG148, we found a sequencing error within the ORF of yvrH. The yvrH gene is reported to be 1,107 bp long (11), but our sequencing indicated that the 336th guanine should be deleted. This deletion introduces a frame shift, resulting in separation of the original yvrH coding region into two split ORFs. The latter ORF (237 codons), starting from the +397 nucleotide (+1 is the translation start nucleotide of the original yvrH sequence), exhibited greater similarity than the former ORF to various response regulators on a BLASTP search (2); its greatest similarity to any of the B. subtilis two-component regulatory proteins was to YyeF (E value [2], 4e−43). Thus, we considered this ORF to be yvrH.
Detection of target gene candidates by DNA microarray analysis.
To find target gene candidates for the 24 two-component regulatory systems, cells deficient in each of the sensor kinases, which carried a plasmid pDG148 derivative with its cognate response regulator gene, were grown with and without IPTG, and then their RNAs were subjected to DNA microarray analysis. The B. subtilis gene expression intensities obtained with RNA from cells grown with IPTG were divided by those obtained with RNA from cells grown without IPTG, which yielded their expression ratios. When RNAs were prepared from cells of strain 168 trpC2 bearing plasmid pDG148 that had been grown with and without IPTG, and gene expression profiles were obtained by means of microarray experiments involving them, none of the genes exhibited ratios greater than 1.8-fold upon the addition of IPTG. The DNA microarray data obtained on analysis of the response regulators are available at the KEGG Expression Database website.
In this study, we adopted the criterion of fourfold up- and downregulation upon the addition of IPTG to pick out the target gene candidates for the respective two-component systems. This cutoff was somewhat arbitrary, with a goal of covering most of the known target genes of the response regulators analyzed and possibly selecting direct target gene candidates for them. (See the KEGG website for both more relaxed and more rigorous criteria.) Table 2 is a list of the target gene candidates for the 24 newly analyzed two-component systems. As shown in Table 2, all of the response regulator genes overexpressed upon the addition of IPTG are included in the list of the upregulated genes. Some known members of the CitT (23) and DesR (1) regulons are also included in this list of target gene candidates. It is notable that the numbers of the target genes for the 24 two-component regulatory systems listed in Table 2 are greatly variable; the systems possessing large numbers of target gene candidates are likely to be more-global regulatory ones.
For analysis of the 24 two-component regulatory systems, mutant strains whose sensor kinase genes had been disrupted through integration of a pMUTIN series of plasmids (22) were utilized (Table 1). This integration caused truncated sensor kinase genes to be under the control of Pspac. Thus, IPTG addition can induce the transcription not only of a response regulator gene cloned into plasmid pDG148 but also of a truncated cognate sensor kinase gene lacking the SD sequence and the initiation codon, as well as of the downstream genes up to the transcription termination site. We therefore picked out the latter genes, indicated in parentheses in Table 2, by consulting the SubtiList web server (http://genolist.pasteur.fr/SubtiList /[13]) with respect to the putative transcription termination sites of the operons encoding the respective pairs of the two-component regulatory systems. Therefore, the operons encoding each pair of the two-component regulatory systems should be investigated further, especially with respect to autoregulation of their transcription.
DISCUSSION
Our strategy for detecting target gene candidates for known and unknown two-component regulatory systems by DNA microarray analysis was based on the assumption that overproduction of a nonphosphorylated response regulator in cells deficient in the cognate sensor kinase might affect the expression of its target genes (15). This strategy was previously applied to the three well-characterized systems (DegS-DegU, ComP-ComA, and PhoR-PhoP), and it appeared to work so well for the detection of their target genes (15) that further attempts were not immediately made to examine whether alternate strategies, such as one involving the use of the disruptant of the response regulator gene, also work for target gene detection. As all of the kinase-regulator gene pairs of the two-component systems examined in this study reside in the same operons on the B. subtilis genome (4, 11), our strategy is applicable to the analysis of all of them. We comprehensively analyzed 24 two-component systems by means of this strategy (Table 2). The expression levels of the target gene candidates for each of the two-component regulatory systems listed in Table 2 were affected by at least fourfold upon overexpression of the regulatory protein, providing valuable information for further investigation of the functions of the unknown two-component regulatory systems analyzed.
However, the following limitations of our present comprehensive analysis of the two-component systems should be kept in mind for any further investigation. (i) We were able to detect only the target genes that are solely regulated by each of the examined two-component systems. In other words, we could not detect the fraction of the target genes unexpressed under the present experimental conditions due to additional negative and/or positive regulation mediated by other regulatory proteins. (ii) We detected the target gene candidates for each of the two-component regulatory systems, the expression of which was affected upon overproduction of the nonphosphorylated response regulator. As discussed previously (15), it is conceivable that in some instances the up- and downregulation of gene expression might not reflect the in vivo regulation occurring when an actual signal that causes phosphorylation of the response regulator is transduced. Nevertheless, most known target genes for the three well-characterized systems were regulated in a manner similar to that which has been observed in physiological signal transduction (15). (iii) It is obvious that the target gene candidates listed in Table 2 include not only direct targets to which the response regulators bind, but also secondary target genes, the expression of which is a consequence of alteration of the expression of the direct target genes. To find only direct target genes for the two-component systems, further molecular genetic analyses of the interactions between the cis elements of the target gene candidates and the respective response regulators, involving such methods as gel retardation and DNA footprinting, are necessary. (iv) For the present analysis of two-component regulatory systems, we used the integrants of plasmid pMUTIN (22) into the sensor kinase genes, which carried plasmid pDG148 derivatives containing the cognate response regulator genes. Thus, IPTG addition induced the transcription not only of genes upregulated upon overproduction of the respective response regulators but also of the truncated sensor kinase genes themselves and their downstream genes, which reside in the same operons as those encoding the sensor-regulator pairs. (v) We have attempted to improve the DNA microarray analysis technique in order to reduce cross hybridization between paralogues, which affects the gene expression ratios (15, 27). However, these procedures are unlikely to eliminate this cross hybridization completely. In addition, our DNA microarrays are deficient in 45 genes which could not be amplified by PCR. Therefore, the potential problems of our microarray analysis itself must be taken into consideration upon interpretation of the target gene candidates listed in Table 2.
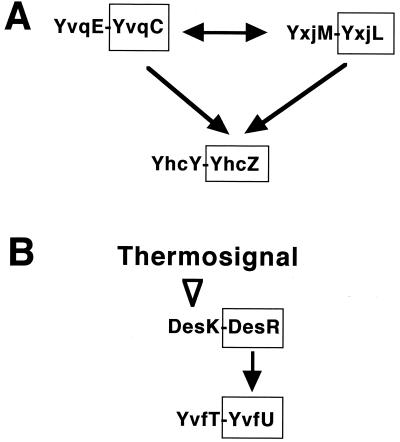
Some interesting features can be seen in the list of the target gene candidates for various unknown two-component regulatory systems (Table 2). There is a striking overlap between the respective target gene candidates of the YxjM-YxjL and YvqE-YvqC systems; out of 19 target gene candidates of YxjM-YxjL upregulated, 17 are included in those of YvqE-YvqC (Table 2). The target gene candidates for the DesK-DesR system also overlap those of YvfT-YvfU. The four response regulators (YxjL, YvqC, DesR, and YvfU) belong to the NarL family of response regulators (4). YxjL and DesR exhibited the highest similarity (e−43 and e−58) to YvqC and YvfU, respectively, among the 35 B. subtilis response regulators on a BLASTP search (2). Furthermore, the target gene candidates for the YvqE-YvqC system include yxjL as well as yhcZ, the response regulator of the YhcY-YhcZ system, while those of YxjM-YxjL include yvqC and yhcZ (Table 2). As shown in Fig. 1A, we can deduce an interaction between these two-component systems, which might explain this extensive overlapping of the target gene candidates for the YvqE-YvqC and YxiM-YxiL systems. Moreover, as shown in Fig. 1B, the target gene candidates of the DesK-DesR system include yvfU (Table 2), which likely explains the overlapping of the target gene candidates for the DesK-DesR and YvfT-YvfU systems. It would be quite interesting to determine how and when these two-component systems that interact with each other evolved from a common ancestor.
FIG. 1.
Probable interactions between some two-component regulatory systems. (A) Interaction between the YvqE-YvqC, YxjM-YxjL, and YhcY-YhcZ systems. (B) Regulation of the YvfT-YvfU system by the DesK-DesR system. DesK is the sensor of a thermosignal (1).
The functions of the target genes for three unknown two-component systems (YdbG-YdbF, YufL-YufM, and YvrG-YvrH) were examined by consulting JAFAN and SubtiList, the B. subtilis genome analysis websites. The target gene candidates for the YdbG-YdbF system included the mcpA, mcpB, flgK, and flgM genes (Table 2), suggesting that this system might be related to chemotaxis. The YufL-YufM system is likely to be involved in competence, because its target gene candidates include most members of the ComK regulon (9), such as comK, nucA, comGG, comGF, comGE, comGD, comGC, comGB, comGA, comEC, comEA, comER, comC, comFC, comFB, and comFA (Table 2). Furthermore, the YvrG-YvrH system appears to be related to cell membrane and cell wall function, because various genes encoding membrane proteins (YfhI, CsbB, YfhO, YkcB, YkcC, and YvrG), transporters (YcbN, YkfD, SunT, and YxdL), and wall-associated proteins (WprA, YocH, DltA, and WapA) are included among its target gene candidates. The target gene candidates for other unknown two-component systems also include more than a few genes whose functions are known or suggested (Table 2), providing valuable clues for unveiling the functions of these unknown two-component systems.
ACKNOWLEDGMENTS
We are grateful to M. Serizawa, H. Yamamoto, and J. Sekiguchi for construction of plasmid pDG148-lytT. We also thank H. Kitoh, T. Negishi, T. Saito, W. Shimizu, T. Takahashi, Y. Nakaura, and S. Tojo for their assistance.
This work was supported by a Grant-in-Aid for Scientific Research on Priority Areas (C) from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Aguilar P S, Hernandez-Arriaga A M, Cybulski L E, Erazo A C, de Mendora D. Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 2001;20:1681–1691. doi: 10.1093/emboj/20.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fabret C, Feher V A, Hoch J A. Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J Bacteriol. 1999;181:1975–1983. doi: 10.1128/jb.181.7.1975-1983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fabret C, Hoch J A. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol. 1998;180:6375–6383. doi: 10.1128/jb.180.23.6375-6383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fawcett P, Eichenberger P, Losick R, Youngman P. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 2000;97:8063–8068. doi: 10.1073/pnas.140209597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fredrick K L, Helmann J D. Dual chemotaxis signaling pathways in Bacillus subtilis: a ςD-dependent gene encodes a novel protein with both CheW and CheY homologous domains. J Bacteriol. 1994;176:2727–2735. doi: 10.1128/jb.176.9.2727-2735.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuchi K, Kasahara Y, Asai K, Kobayashi K, Moriya S, Ogasawara N. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis. Microbiology. 2000;146:1573–1583. doi: 10.1099/00221287-146-7-1573. [DOI] [PubMed] [Google Scholar]
- 9.Hamoen L W, Van Werkhoven A F, Bijlsma J J E, Dubnau D, Venema G. The competence transcription factor of Bacillus subtilis recognizes short A/T-rich sequences arranged in a unique, flexible pattern along the DNA helix. Genes Dev. 1998;12:1539–1550. doi: 10.1101/gad.12.10.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirsch M L, Peters P D, Hanlon D W, Kirby J R, Ordal G W. Chemotactic methylesterase promotes adaptation to high concentrations of attractant in Bacillus subtilis. J Biol Chem. 1993;268:18610–18616. [PubMed] [Google Scholar]
- 11.Kunst F, Ogasawara N, Moszer I, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 12.Loomis W F, Kuspa A, Shaulsky G. Two-component signal transduction systems in eukaryotic microorganisms. Curr Opin Microbiol. 1998;1:643–648. doi: 10.1016/s1369-5274(98)80109-4. [DOI] [PubMed] [Google Scholar]
- 13.Moszer I, Kunst F, Danchin A. The European Bacillus subtilis genome sequencing project: current status and accessibility of the data from a new world wide web site. Microbiology. 1996;142:2987–2991. doi: 10.1099/13500872-142-11-2987. [DOI] [PubMed] [Google Scholar]
- 14.Msadek T, Kunst F, Rapoport G. Two-component regulatory systems. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 729–745. [Google Scholar]
- 15.Ogura M, Yamaguchi H, Yoshida K, Fujita Y, Tanaka T. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of Bacillus subtilis two-component regulatory systems. Nucleic Acids Res. 2001;29:3804–3813. doi: 10.1093/nar/29.18.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Schaeffer P, Millet J, Aubert J P. Catabolite repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schiött T, von Wachenfeldt C, Hederstedt L. Identification and characterization of the ccdA gene, required for cytochrome c synthesis in Bacillus subtilis. J Bacteriol. 1997;179:1962–1973. doi: 10.1128/jb.179.6.1962-1973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stock J B, Surette M G, Levit M, Stock A M. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 25–51. [Google Scholar]
- 21.Stragier P, Bonamy C, Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 22.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 23.Yamamoto H, Murata M, Sekiguchi J. The CitST two-component system regulates the expression of the Mg-citrate transporter in Bacillus subtilis. Mol Microbiol. 2000;37:898–912. doi: 10.1046/j.1365-2958.2000.02055.x. [DOI] [PubMed] [Google Scholar]
- 24.Yansura D G, Henner D J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ye R W, Tao W, Bedzyk L, Young T, Chen M, Li L. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J Bacteriol. 2000;182:4458–4465. doi: 10.1128/jb.182.16.4458-4465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida K, Ishio I, Nagakawa E, Yamamoto Y, Yamamoto M, Fujita Y. Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome. Microbiology. 2000;146:573–579. doi: 10.1099/00221287-146-3-573. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida K, Kobayashi K, Miwa Y, Kang C-M, Matsunaga M, Yamaguchi H, Tojo S, Yamamoto M, Nishi R, Ogasawara N, Nakayama T, Fujita Y. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 2001;29:683–692. doi: 10.1093/nar/29.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]