Graphical abstract
Keywords: Wastewater, Environmental surveillance, WBE, Public health, Monkeypox virus
Abstract
As of 18 August 2022, 1087 confirmed cases of monkeypox are reported in the Netherlands. Monkeypox virus likely ends up in wastewater because i) skin flakes from areas affected by the typical rash and scabbing may wash off, and ii) monkeypox virus has been detected in animal and human feces. Here we describe a method to qualitatively detect monkeypox virus DNA in wastewater, that may prove a valuable surveillance tool for outbreaks.
1. Introduction
Since the beginning of May 2022, the zoonotic disease monkeypox is spreading outside of Africa, in non-endemic countries (Zumla et al., 2022). In contrast to previous outbreaks when the transmission route was mainly animal-to-human (Reynolds et al., 2006), human-to-human transmission is currently considered to be the main route of transmission (Bunge et al., 2022; WHO, 2022). The majority of monkeypox cases develop a (initially localized) rash across the body. In the current outbreak, cases of genital or peri-anal rash are reported (WHO, 2022). Virus concentrations are found to be particularly high in affected skin (Adler et al., 2022; Antinori et al., 2022). Over time, the rash typically turns into blisters that dry out and subsequently form crusts and scabs. Monkeypox virus (MPXV) from these crusts and scabs may be released in wastewater while rinsing, showering, or toilet usage (WHO, 2022). MPXV can also be detected in animal (Hutson et al., 2009) and human feces (Antinori et al., 2022), and human anal swabs are collected for diagnostic testing of infected individuals (personal communication; Centre for Infectious diseases research, Diagnostics and laboratory Surveillance (CIb, RIVM)). This indicates that feces either contain excreted MPXV, or become contaminated when defecating. Both skin and excretion processes are likely to result in viral particles ending up in wastewater. Here, we describe a method to detect MPXV DNA in wastewater, which may be a valuable surveillance tool to provide information about MPXV circulation in the human population.
The first case of monkeypox in the Netherlands was confirmed on 20 May 2022. As of 18 August 2022, the number of confirmed cases has further increased to 1087 (RIVM, 2022). The affected population is almost exclusively male. When patients are suspected of having monkeypox, diagnostic tests are performed. Municipal Public Health Services start source and contact tracing when cases are confirmed. Recently, on July 23, 2022, WHO declared monkeypox outbreak a Public Health Emergency of International Concern. Monkeypox was classified as an A-status notifiable disease in the Netherlands on May 21st, meaning that any suspected or confirmed case should be immediately reported to the public health services (Government of the Netherlands, 2022). Confirmed cases are reported throughout the country, the majority of which are currently located in Amsterdam (RIVM, 2022).
2. Materials and methods
2.1. Wastewater samples
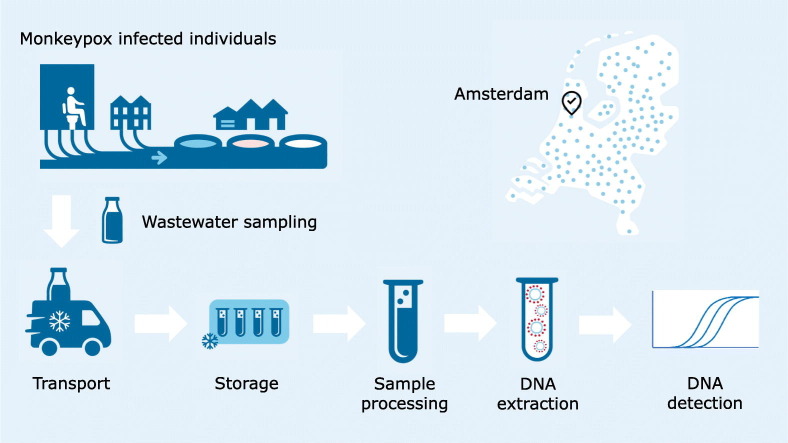
To monitor the spread of SARS-CoV-2 in the Netherlands a National Sewage Surveillance program has been established. As of September 2020, all wastewater treatment plants (WWTPs) send 500 mL of 24-h flow proportional wastewater samples to the RIVM. Additionally, wastewater samples were taken at the Schiphol Airport WWTP. Currently, each WWTP is sampled four times a week. As a pilot study, since the end of 2020 flow proportional samplers were placed in five city districts of Amsterdam to monitor the SARS-CoV-2 viral load at district level twice per week. In this study a selection of samples collected from the five Amsterdam city districts, the WWTP of Schiphol Airport and two WWTPs in Amsterdam, WWTP Amsterdam West and Amsterdam Westpoort (inhabitant equivalents shown in Table 1 ). The samples included in our study were collected in week 9 (28 February–6 March 2022) and weeks 20–26 (16 May 2022–3 July 2022).The February and March samples were included as negative control samples for our assay. All samples were stored in a refrigerator at 4 °C until further processed.
Table 1.
Monkeypox virus DNA detection in the wastewater samples taken at different locations from 16 May-3 July 2022 (n = 108). Shown are the average Ct values of the generic and the West-African specific assay, respectively.
| Date | WWTP Amsterdam Westpoort |
WWTP Amsterdam West |
Schiphol Airport |
||||||
|---|---|---|---|---|---|---|---|---|---|
| CD-1 64,000a | WWTP 310,762 | CD-2 16,000 | CD-3 49,000 | CD-4 118,000 | CD-5 121,000 | WWTP 648,560 | WWTP-c | ||
| Week 20 | 16 May | n.d. | n.d. | ||||||
| 17 May | n.d. | n.d. | n.d. | ||||||
| 18 May | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | |||
| 19 May | |||||||||
| 20 May | n.d. | n.d. | |||||||
| 21 May | |||||||||
| 22 May | 38.1/39.4 | ||||||||
| Week 21 | 23 May | n.d. | n.d. | n.d. | |||||
| 24 May | n.d. | ||||||||
| 25 May | n.d. | n.d. | n.d. | n.d. | n.d. | ||||
| 26 May | n.d. | ||||||||
| 27 May | n.d. | n.d. | |||||||
| 28 May | n.d. | ||||||||
| 29 May | 38.1/37.4 | ||||||||
| Week 22 | 30 May | 38.8/39.6 | |||||||
| 31 May | n.d. | n.d. | |||||||
| 1 Jun | n.d. | 41.8b/42.1b | n.d. | n.d. | n.d. | 42.9/42.4 | 39.0/38.0 | ||
| 2 Jun | |||||||||
| 3 Jun | 39.1/37.4 | 39.5/39.9 | |||||||
| 4 Jun | 39.3/39.2 | ||||||||
| 5 Jun | 40.2/39.3 | ||||||||
| Week 23 | 6 Jun | 38.2/37.4 | 38.7/38.3 | n.d. | |||||
| 7 Jun | n.d. | ||||||||
| 8 Jun | n.d. | 40.6/38.2 | n.d. | 39.8/39.2 | n.d. | n.d. | n.d. | ||
| 9 Jun | 38.6/37.6 | n.d. | |||||||
| 10 Jun | 40.3/38.6 | 39.0/39.2 | |||||||
| 11 Jun | 38.9/39.1 | n.d. | |||||||
| 12 Jun | |||||||||
| Week 24 | 13 Jun | 36.5/36.1 | n.d. | ||||||
| 14 Jun | 39.8/38.0 | 40.0/39.0 | n.d. | ||||||
| 15 Jun | n.d. | n.d. | 40.6/40.5 | 37.0/37.0 | 40.0/37.9 | 39.0/37.9 | 39.1/38.4 | n.d. | |
| 16 Jun | |||||||||
| 17 Jun | n.d. | n.d. | |||||||
| 18 Jun | |||||||||
| 19 Jun | 40.1/39.5 | ||||||||
| Week 25 | 20 Jun | 38.8/38.7 | n.d. | ||||||
| 21 Jun | 37.7/37.7 | 39.7/40.9 | n.d. | ||||||
| 22 Jun | 40.9/41.4 | 35.9/36.6 | n.d. | n.d. | 39.1/39.8 | 35.7/35.8 | 39.3/38.6 | ||
| 23 Jun | n.d. | ||||||||
| 24 Jun | n.d. | 42.1/41.4 | |||||||
| 25 Jun | n.d. | n.d. | |||||||
| 26 Jun | |||||||||
| Week 26 | 27 Jun | n.d. | 39.5/38.9 | ||||||
| 28 Jun | n.d. | 38.6/38.3 | 39.8/38.2 | ||||||
| 29 Jun | n.d. | n.d. | n.d. | n.d. | 39.4/38.8 | 39.6/39.6 | 41.0/40.5 | n.d. | |
| 30 Jun | |||||||||
| 1 Jul | 40.6/39.4 | 40.8/40.7 | |||||||
| 2 Jul | |||||||||
| 3 Jul | 37.9/38.4 | ||||||||
| Total | 7 | 26 | 7 | 7 | 7 | 7 | 24 | 23 | |
CD: Amsterdam city district; n.d.: sample tested, monkeypox virus DNA not detected.
The number of inhabitants connected to each sampling location.
Results from the 10-times diluted sample when the undiluted sample yielded no result.
No number of inhabitants is associated with the WWTP of Schiphol Airport since it has no permanent inhabitants.
2.2. Sample processing and DNA extraction
Twenty-five mL of homogenized wastewater was centrifuged for 10 min at 15,000 ×g at room temperature. A maximum of 2 mL of pellet and remaining wastewater was transferred to a 2-mL tube and centrifuged for 5 min at 20,000 ×g at room temperature. Subsequently supernatant was discarded resulting in a final volume of approximately 0.5 mL per sample.
The DNeasy blood and tissue kit (Qiagen, Hilden, Germany) was used for the extraction of nucleic acids from the pellet/wastewater samples. Briefly, 1:1 lysis buffer AL was added to the samples (for instance 0.5 mL sample + 0.5 mL lysis buffer) and thoroughly mixed by vortexing for 1 min and the samples were incubated in a thermomixer (Eppendorf) for 15 min at 56 °C (1250 rpm). The samples were centrifuged for 1 min at 10,000 ×g and the supernatant containing the lysis buffer with the nucleic acids was transferred to new 2-mL tubes. A volume of 96 % ethanol (EtOH) (1:2) was added (for instance 0.8 mL lysis buffer containing nucleic acids +0.4 mL 96 % EtOH) and mixed by vortexing. The samples were added to the spin columns and this process was repeated until the entire sample was processed. After two wash steps the DNA was eluted twice with 100 μL elution buffer, resulting in a DNA eluate volume of 200 μL. To remove any inhibitors present in the DNA eluates, 100 μL of each sample was processed with the Zymo OneStep™ PCR inhibitor Removal Kit (Zymo Research, Leiden, the Netherlands) and were either used directly in the qPCR or stored at −70 °C until further use.
2.3. Molecular detection
MPXV DNA was detected with a multiplex assay using primers and probes described by the CDC (Li et al., 2010), an assay for the generic MPXV detection and the specific detection of the West-African clade, with minor modifications, to better fit the currently circulating MPXV (modification is underlined in primer sequences). Briefly, a total volume of 20 μL reaction mixture contained 1× TaqMan® Universal PCR Master Mix (Applied Biosystems, Warrington, UK), 500 nM G2R_G forward (5′-GGA AAG TGT AAA GAC AAC GAA TAC AG-3′), 500 nM G2R_G reversed (5′-GCT ATC ACA TAA TCT GAA AGC GTA-3′), 250 nM G2R_G probe (5′-FAM-AAG CCG TAA TCT ATG TTG TCT ATC GTG TCC-BHQ1-3′), 500 nM G2R_WA forward (5′-CAC ACC GTC TCT TCC ACA GA-3′), 500 nM G2R_WA reversed (5′-GAT ACA GGT TAA TTT CCA CAT CG-3′), 250 nM G2R_WA probe (5′-Texas Red-AAC CCG TCG TAA CCA GCA ATA CAT TT-BHQ2-3′) and 5 μL of template DNA.
The qPCR was performed on a QiaQuant (Qiagen) Real-time PCR machine with cycling conditions of 50 °C for 1 min, 95 °C for 6 min followed by amplification consisting of 50 cycles of 95 °C for 15 s and at 60 °C for 45 s. In each qPCR run the extracted DNA was tested in duplicate on an undiluted and on a 10 times diluted DNA extract. In each of these runs a negative and positive control (containing MPXV DNA from a clinical sample, kindly provided by the Centre for Infectious diseases research, Diagnostics and laboratory Surveillance, CIb, RIVM) and plasmid pMP were included. Plasmid pMP contains the following sequence, which consists of several parts of the MPXV genome:
AGAGATTTAGCACCACATGCACCATCCAATGGAAAATGTAAAGACAACGAATACAGAAGCCGTAATCTATGTTGCTATCGTGTCCTCCGGGAACTTACGCTTCCAGATTATGTGATAGCAAGACTAATACACAATGTACGCCGTGTGGTCACACCGTCTCTTCCACAGATAAATGCGAACCCGTCGTAACCAGCAATACATTTAACTATATCGATGTGGAAATTAACCTGTATCCAGTCAACGACACATCGTGTACTCGGACGATGTCTACCTGGATACAGAAAGCAAAAAATGGGACCCATATATGCTAAATGTACCGGTACCGGATGGACACTCTTTAATCAATGTATTAAACGGAGATGCCCATCGCCTCGAGATATCGATAATGGCCAAC.
The sequence is synthesized and ligated into vector pMA-T by Invitrogen resulting in plasmid pMP. Plasmid pMP was diluted to a concentration of 0.01 pg/μL, and 5 μL was used as a positive control in the qPCR runs.
To confirm the specificity of the qPCR results, a selection of samples with Ct-values below 40 were used in a semi-nested PCR assay, combining the oligos from the generic and the West-African specific qPCR. Sanger sequencing was performed on the resulting 449 bp PCR products (Text S1).
3. Results
Since the majority of confirmed monkeypox cases were located in Amsterdam, we determined the presence of MPXV DNA in wastewater samples from five city districts in Amsterdam and two WWTPs, i.e. Amsterdam West and Amsterdam Westpoort. Additionally, wastewater samples from Schiphol Airport, a major travel hub, were included. DNA was extracted from (bio)solids present in wastewater, and the presence of MPXV DNA was determined using the generic and West-African specific qPCR assays.
A sample was considered positive when both the generic qPCR and the West-African clade qPCR yielded a signal resulting in a Ct-value. MPXV DNA was detected in 45/108 (42 %) wastewater samples collected between 16 May–3 July 2022: 11/35 (31 %) wastewater samples collected from the five city districts in Amsterdam, 28/50 (56 %) wastewater samples collected from the two WWTPs, and 6/23 (26 %) wastewater samples collected from the Schiphol Airport WWTP (Table 1). In week 20 and week 21, MPXV DNA could only be detected in 2 wastewater samples, both from Schiphol Airport. From week 22 onwards, each week approximately more than half of the samples were positive for the presence of MPXV DNA. Ct values ranged between 35 and 42, indicating relatively low DNA concentrations present in the tested samples. No MPXV DNA could be detected in samples taken in week 9 (28 February–6 March) (Table S1), corresponding to the absence of monkeypox cases in the Netherlands at that time.
In the qPCR that was performed on the DNA eluates directly after the DNeasy blood and tissue kit (Qiagen) we found high Ct-values (41–45) and variable results between the tested dilutions were obtained, which suggested the presence of inhibitory substances. After using the Zymo OneStep™ PCR inhibitor Removal Kit (Zymo Research) the qPCR data resulted in lower Ct-values (35–42) and improved S-curves in the qPCR data, in only one sample only the 10-times diluted samples yielded a qPCR signal. Although differences in Ct-values between samples were observed due to different compositions of the wastewater at different locations, the assays for the generic and West-African clade yield highly similar Ct-values within a sample (Table 1).
To confirm the specificity of the qPCR used in this study an additional conventional PCR was performed on a selection of samples with Ct values below 40. A PCR product was generated using the G2R_G forward primer and the G2R_WA reversed primer (475 bp), on which Sanger sequencing was performed. Sanger sequencing confirmed the amplification of MPXV DNA from wastewater (details described in Text S1).
4. Discussion
This study describes a method to detect MPXV DNA in (bio)solids in wastewater in a qualitative manner, meaning that when a sample is positive, the viral DNA is present. In contrast, when the virus is not detected this could either indicate that no individuals are excreting viral particles in that area or that the amount of DNA present in the sample is below the detection limit of the assay.
Direct nucleic acid extraction, as used for the SARS-CoV-2 analysis, was also tested in the MPXV qPCR but yielded no usable data. Furthermore, comparative experiments were performed by centrifuging 25–50 mL wastewater to separate the aqueous phase from the (bio)solids. The aqueous phase was then concentrated with centrifugal filters, and DNA was extracted from the concentrate and the (bio)solids. The DNA extraction on the (bio)solids yielded the best results and was therefore used throughout our study (Data not shown).
Currently, the process through which MPXV ends up in wastewater is unknown. Both excretion or secretion through feces is observed in infected animals and humans (Antinori et al., 2022; Hutson et al., 2009). Another plausible process is that skin flakes from affected areas of the body wash off into wastewater. The blisters and resulting scabs are reported to contain high virus concentrations (Adler et al., 2022; Antinori et al., 2022). Although the predominant origin of MPXV shedding is unknown, both pathways may lead to detectable DNA in wastewater. The locations that were selected in our study were based on the fact that the number of confirmed cases is relatively high in Amsterdam.
A caveat in terms of public health monitoring is that MPXV has been isolated from different rodents and other small mammals (Alakunle et al., 2020), suggesting that small mammals living in the sewerage, such as rats, could become a reservoir of MPXV. However, we believe our results show that MPXV shed by humans is currently being detected. Firstly because in the wastewater samples from Schiphol Airport, detection of MPXV DNA is intermittent, whereas it is stable in the WWTPs of Amsterdam where clinically confirmed cases are known. With a relatively small and highly specific catchment area, Schiphol Airport as a major travel hub (5.1 million passengers in May 2022) has an almost completely variable ‘human population’, whereas detection of MPXV deriving from animal reservoirs would be far more consistent over time. Secondly, as monkeypox is not shown or believed to be a currently endemic disease in animal populations, this provides further confidence that detection of the virus in the tested samples is currently the result of human shedding. Nonetheless, MPXV animal reservoirs remain possible, and could affect the applicability of wastewater monitoring in the future.
Future research will include testing of wastewater samples originating from WWTPs dispersed over the Netherlands, especially in regions where no or only a few confirmed monkeypox cases have been reported. This will provide additional results to assess the potential of wastewater-based surveillance and the possibility of an early-warning system for viruses such as MPXV. Asymptomatic and mild cases are inherently underrepresented in the group of confirmed cases, detection in wastewater may be an unbiased indicator. By performing this research, the potential of wastewater-based surveillance and especially an early-warning system for viruses, such as monkeypox, can be investigated further.
5. Conclusions
Our study shows a proof of principle of MPXV DNA detection in (bio)solids from wastewater. We present qualitative detections in various samples from five city districts, and two WWTPs in Amsterdam, chosen because of the relatively high number of confirmed cases. Additionally we present detections in samples from Schiphol Airport, an international transportation hub where infected patients had possibly travelled through. The occasional detections at Schiphol Airport and the increase in positive wastewater samples when confirmed cases increased in Amsterdam indicate that the detection of MPXV DNA in wastewater, besides clinical surveillance may be an additional, regionally applicable surveillance option to monitor the presence of monkeypox infected individuals.
The following is the supplementary data related to this article.
Monkeypox virus DNA detection in the wastewater samples taken at different locations from 28 February–6 March 2022 (n = 12).
CRediT authorship contribution statement
EFdJ and WJL designed the study and coordinated the activities. CMP and JMK performed the molecular analysis. EFdJ and WJL wrote the draft manuscript. EFdJ, AMRvdD, RFHJvdB, EN and WJL reviewed and edited the manuscript, and all authors approved the final version of the manuscript.
Funding statement
This research was financed by the Dutch Ministry of Health, Welfare and Sport.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to acknowledge the Dutch Water Authorities, water boards and water laboratories for their collaboration in the Dutch wastewater surveillance program (COVID). Also Evides Industriewater for collecting wastewater originating from Schiphol Airport. Furthermore the authors would like to thank our colleagues from the Centre for Infectious diseases research, Diagnostics and laboratory Surveillance (CIb, RIVM) for providing a MPXV DNA sample. We would also like to acknowledge Heike Schmitt and Partners4UrbanWater for their participation in the set-up and sampling of the wastewater originating from the Amsterdam city districts. Lastly, the authors would like to gratefully acknowledge the other colleagues from the National Sewage Surveillance department for their participation in the wastewater surveillance program.
Editor: Warish Ahmed
Data availability
All our data is presented in the results section.
References
- Adler H., Gould S., Hine P., Snell L.B., Wong W., Houlihan C.F., Osborne J.C., Rampling T., Beadsworth M.B., Duncan C.J., Dunning J., Fletcher T.E., Hunter C.J., Jacobs M., Khoo S.T., Newsholme W., Porter D., Porter R.J., Ratcliffe L., Schmid C.J., Semple M.G., Tunbridge A.J., Wingfield T., Price N.M., NHS England High Consequence Infectious Diseases (Airborne) Network Clinical features and management of human monkeypox: a retrospective observational study in the UK. Lancet Infect. Dis. 2022;22:1153–1162. doi: 10.1016/S1473-3099(22)00228-6. S1473-3099(22)00228-6. PMID: 35623380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alakunle E., Moens U., Nchinda G., Okeke M.I. Monkeypox virus in Nigeria: infection biology, epidemiology, and evolution. Viruses. 2020;12(11):1257. doi: 10.3390/v12111257. PMID: 33167496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinori A., Mazzotta V., Vita S., Carletti F., Tacconi D., Lapini L.E., D’Abramo A., Cicalini S., Lapa D., Pittalis S., Puro V., Rivano Capparuccia M., Giombini E., Gruber C.E.M., Garbuglia A.R., Marani A., Vairo F., Girardi E., Vaia F., Nicastri E., INMI Monkeypox Group Epidemiological, clinical and virological characteristics of four cases of monkeypox support transmission through sexual contact, Italy, May 2022. Euro Surveill. 2022;27(22) doi: 10.2807/1560-7917.ES.2022.27.22.2200421. PMID: 35656836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge E.M., Hoet B., Chen L., Lienert F., Weidenthaler H., Baer L.R., Steffen R. The changing epidemiology of human monkeypox—a potential threat? A systematic review. PLoS Negl. Trop. Dis. 2022;16(2) doi: 10.1371/journal.pntd.0010141. PMID: 35148313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Government.nl 2022. https://www.government.nl/latest/news/2022/05/24/monkeypox-cases-must-be-reported
- Hutson C.L., Olson V.A., Carroll D.S., Abel J.A., Hughes C.M., Braden Z.H., Weiss S., Self J., Osorio J.E., Hudson P.N., Dillon M., Karem K.L., Damon I.K., Regnery R.L. A prairie dog animal model of systemic orthopoxvirus disease using West African and Congo Basin strains of monkeypox virus. J. Gen. Virol. 2009;90(2):323–333. doi: 10.1099/vir.0.005108-0. PMID: 19141441. [DOI] [PubMed] [Google Scholar]
- Li Y., Zhao H., Wilkins K., Hughes C., Damon I.K. Real-time PCR assays for the specific detection of monkeypox virus West African and Congo Basin strain DNA. J. Virol. Methods. 2010;169(1):223–227. doi: 10.1016/j.jviromet.2010.07.012. PMID: 20643162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds M.G., Yorita K.L., Kuehnert M.J., Davidson W.B., Huhn G.D., Holman R.C., Damon I.K. Clinical manifestations of human monkeypox influenced by route of infection. J. Infect. Dis. 2006;194(6):773–780. doi: 10.1086/505880. [DOI] [PubMed] [Google Scholar]
- RIVM (National Institute for Public Health and the Environment) RIVM; Bilthoven: 2022. Monkeypox News.https://www.rivm.nl/en/monkeypox Available from. [Google Scholar]
- World Health Organization Disease Outbreak News; Multi-country monkeypox outbreak in non-endemic countries. 2022. https://www.who.int/emergencies/disease-outbreak-news/item/2022-DON390 [updated 04/06/2022]. [Accessed 4 Jul 2022]. Available from.
- Zumla A., Valdoleiros S.R., Haider N., Asogun D., Ntoumi F., Petersen E., Kock R. Monkeypox outbreaks outside endemic regions: scientific and social priorities. Lancet Infect. Dis. 2022;22(7):929–931. doi: 10.1016/S1473-3099(22)00354-1. PMID: 35636447. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Monkeypox virus DNA detection in the wastewater samples taken at different locations from 28 February–6 March 2022 (n = 12).
Data Availability Statement
All our data is presented in the results section.