Abstract
We have developed an antisense oligonucleotide microarray for the study of gene expression and regulation in Bacillus subtilis by using Affymetrix technology. Quality control tests of the B. subtilis GeneChip were performed to ascertain the quality of the array. These tests included optimization of the labeling and hybridization conditions, determination of the linear dynamic range of gene expression levels, and assessment of differential gene expression patterns of known vitamin biosynthetic genes. In minimal medium, we detected transcripts for approximately 70% of the known open reading frames (ORFs). In addition, we were able to monitor the transcript level of known biosynthetic genes regulated by riboflavin, biotin, or thiamine. Moreover, novel transcripts were also detected within intergenic regions and on the opposite coding strand of known ORFs. Several of these novel transcripts were subsequently correlated to new coding regions.
Gene expression in bacteria has been traditionally analyzed by transcriptional or translational fusions to promoterless “reporter” genes (e.g., lacZ, cat, and gus) or by direct detection of transcripts using Northern blotting or reverse transcription-PCR (RT-PCR). With the completion of many bacterial genomes and the development of large-scale analysis tools such as DNA genomic arrays, however, researchers have increasingly applied genomics tools in their research. Measurements of mRNA levels using genome arrays for Escherichia coli, Bacillus subtilis, Streptococcus pneumoniae, and Haemophilus influenzae (10, 11, 12, 17, 23, 24, 29, 32, 38, 39, 41) have been found to offer many advantages to traditional gene-monitoring methods. Since the structure of bacterial genomes is relatively simple, containing ca. 4,000 genes and few repetitive sequences, DNA arrays can monitor transcript levels of an entire genome in a single hybridization with high sensitivity. This can lead to the elucidation of complex interactions among genetic networks, which then can be coupled with results from other newer technologies that analyze global protein synthesis (proteome) and metabolite levels (metabolome) to provide a comprehensive picture of the physiology of the bacterium (13, 14, 18, 34, 35, 42).
Using the public B. subtilis genome sequence (20), we developed an oligonucleotide B. subtilis genome microarray using Affymetrix GeneChip technology (21, 40). This technology offers high sensitivity, high specificity, and excellent reproducibility (19). We show that the microarray can monitor gene expression changes in response to transition from the exponential to the stationary growth phases and exposure to three different vitamins that repress expression of biosynthetic genes. Moreover, we also present evidence that the microarray can be used to detect novel transcripts within intergenic regions and on the opposite strand of known genes, leading in some cases to the identification of previous unreported coding regions.
MATERIALS AND METHODS
Microarray design.
An “antisense” oligonucleotide array complementary to the Bacillus subtilis genome was custom designed by Affymetrix (Santa Clara, Calif.) using the published DNA sequence (GenBank accession no. NC_000964). A general description of Affymetrix GeneChip microarrays has been previously given (10, 11, 21, 30, 40). In the antisense format, the oligonucleotide sequence on the microarray is the same as the coding region sequence, and the labeled target sequence is complementary to the mRNA sequence. Each oligonucleotide probe is 25 nucleotides in length and is specifically selected using an Affymetrix proprietary algorithm. The probes are organized as probe pairs consisting of a perfect match probe (PM) and a mismatch probe (MM). Probe pairs are further organized into larger groups referred to as probe sets; one probe set is used to detect a single putative transcript. Probe sets were present for 4,112 predicted open reading frames (ORFs), as well as for 59 tRNA and 3 rRNA coding regions. More than 95% of these probe sets contained 20 probe pairs. Probe sets to most of the remaining genes contained 10 to 20 probe pairs; only a few had fewer than 10 probe pairs. A probe set to one gene (ypuE) could not be designed. Probe sets to 590 selected genes were also duplicated and placed at different sites on the microarray. Of these, 550 were also included in the sense format (probe sets complementary to the mRNA sequence). Probe sets specific to both strands of 603 intergenic or interoperon (“gap”) regions larger than 250 nucleotides are represented. In some cases, larger gap regions contained two or more probe sets. In a few cases, a probe set to only one strand was made. Also included were 50 putative ORFs with Bacillus-like ribosome binding sites (RBS); 15 of these were located on the noncoding strand of known genes, and the other 35 were located within intergenic regions.
Bacterial strains and growth conditions.
The B. subtilis strains used in this study were PY79 (SPβc prototroph) (43) and BS0011 (leuA8 metB5 polC::neo) (N. Mouncey and H.-P. Hohmann, unpublished data) and isogenic derivatives BI421 (3) containing a biotin (birA) deregulatory mutation and BS0012 (Mouncey and Hohmann, unpublished) containing a riboflavin (ribC) deregulatory mutation. Cells were grown overnight in either Luria-Bertani (LB) medium (without glucose) or Spizizen's minimal medium (SS) containing 0.04% sodium glutamate, 0.4% glucose, and trace amounts of micronutrients [CaCl2, FeSO4, MgSO4, ZnSO4, CoCl2, (NH4)6HMo7O24, AlCl3, and CuCl2] in the presence or absence of biotin (0.1 μg/ml), thiamine (0.34 μg/ml), or riboflavin (200 μg/ml). Overnight cultures were diluted 50-fold into fresh medium and grown to exponential growth phase (optical density at 600 nm = 0.7). Cells from half of the culture were pelleted, and the total RNA was immediately extracted. In some experiments, the remaining culture was allowed to grow to early stationary phase before RNA extraction. Early stationary phase was determined to be 30 min after glucose exhaustion.
RNA isolation and cDNA labeling.
Total RNA was extracted from cells as described by Wei et al. (38) except that the initial lysis buffer contained a mixture of glass beads (bead size, 106 μm), macaloid clay, phenol-chloroform, and sodium dodecyl sulfate (1). RNA isolation was performed at 4°C or on ice. Isolated RNA was treated with RNase-free DNase I to remove contaminating DNA and further purified by using the Qiagen RNeasy Midi kit according to the manufacturer's instructions. Purified total RNA was stored at −20°C. Preparation of cDNA targets was based on a previously described method (10). Random hexamer primers (9 μg [Gibco-BRL]) and total RNA (30 μg) were added to a mixture containing 6.0 μl of deoxynucleoside triphosphate mix (10 mM dCTP, dGTP, and dTTP; 4 mM dATP) and 30.0 μl of biotin-labeled Bio-14 dATP (1 mM [NEN Life Sciences Products]) in a total volume of 60 μl. The mixture was incubated at 70°C for 5 min to denature the primers and RNA. The mixture was cooled on ice, and 24 μl of 5× Strand buffer (250 mM Tris-HCl [pH 8.3], 375 mM KCl, 15 mM MgCl2), 12 μl of 0.1 M dithiothreitol, and 25 μl of Superscript II (200 U/μl; Gibco-BRL) was added to a final volume of 120 μl. cDNA synthesis was performed at room temperature for 10 min and then at 45°C for 2 to 4 h. The mixture was treated with NaOH to degrade the RNA strands, followed by neutralization with HCl and Tris-HCl (pH 7.0) (38). cDNA was further purified by two ethanol precipitations to remove unincorporated biotin-labeled dATP. Fragmentation was performed in One-Phor-All buffer (Amersham Pharmacia Biotech) containing 0.4 U of DNase I for 5 min at 37°C. DNase I was inactivated by heating at 99°C for 10 min. Fragmented cDNA was recovered after overnight ethanol precipitation at −20°C.
Genomic DNA labeling.
Genomic DNA was labeled by nick translation. Briefly, 5 to 10 μg of genomic DNA was added to a mixture of 50 μl of dCTP, dGTP, and dTTP (0.2 mM each), 30 μl of Bio-14 dATP (0.4 mM), and a combination of 15 U of DNA polymerase I and 12 μU of DNase I mix (Gibco-BRL). After incubation at 22°C for 2 h, the labeled DNA was then precipitated twice by ethanol to remove the unincorporated label.
Hybridization and staining procedures.
Microarrays were first prehybridized for 10 min at 43°C in a buffer consisting of 100 mM MES (morpholineethanesulfonic acid), 0.01% Tween 20, bovine serum albumin at 1.6 mg/ml, and fragmented yeast RNA at 0.4 mg/ml. Then, 240 μl of the hybridized mixture containing 1 μl of “Checkboard” (biotin-labeled oligonucleotide [Affymetrix] used to outline the microarray boarder) and 22.5 μg of labeled cDNA was added to the microarray, and the mixture was hybridized overnight at the same temperature in a rotisserie oven. Washing and staining with GeneChip were performed according to Affymetrix's standard protocol except that the stringent wash was performed at 45°C (9). No signal amplification was performed.
Data analysis.
Microarrays were scanned twice at 570 nm at a 3-μm resolution with an Affymetrix scanner and analyzed as previously described (11, 21) by using the Affymetrix gene expression analysis suite. Data were converted to a text format and normalized according to the mean of the sum of all of the comparable experiments. Gene mining software Genespring (Silicon Genetics, Inc.) and Spotfire (Spotfire, Inc.) were also used to further analyze the expression data.
Real-time RT-PCR.
Real-time RT-PCR using the SYBR green staining method was performed according to the protocol described by Wei et al. (39). PCR primer sequences (Gibco-BRL) to the following genes were as follows: bioA-forward (5′-CCGCGCTTTCCATTGAAT-3′), and bioA-reverse (5′-CAAATATCCTTCCGGCATCAC-3′), gap-forward (5′-CCTTGATCTTCCGCACAAAGA-3′) and gap-reverse (5′-GTTGATGTTGGGATGATGTTTTCA-3′), ribG-forward (5′-CGAAGGACAGACCGAATCCA) and ribG-reverse (5′-GACAATTTGTCCGTCCTTTACGA-3′), and thiA-forward (5′-CGTGAATGGATTATCCGCAATT-3′) and thiA-reverse (5′-TTTCAAGCGCCTGATAAATCG-3′).
RESULTS
Microarray quality.
Control experiments were carried out with total RNA isolated from a prototroph strain (PY79) grown to either late exponential or early stationary stage in minimal medium. By using biotin-labeled cDNA targets prepared from the RNA, the optimal hybridization temperature range was determined to be between 42 and 45°C, with 43°C used in all subsequent experiments. To determine the linear range of gene transcript levels, independent microarrays were hybridized with undiluted and diluted (1:2, 1:5, and 1:10) labeled targets. Comparison of the data sets indicated that the transcript levels were linear over an average intensity difference range of 50 to 20,000. An average intensity difference value of 20 corresponded to background noise. Measurement of transcripts from highly expressed genes (i.e., genes with an average difference of >20,000) was inconsistent. The presence of such highly abundant transcripts (e.g., rRNA) often resulted in significant hybridization to the mismatch probe pairs, causing the average difference metric calculations to indicate that transcript levels were low or nonexistent. In these instances, RT-PCR could be used to measure transcript levels.
Additional analysis of the microarray data revealed that >70% of the known B. subtilis ORFs produced a detectable RNA transcript. This indicated that the sensitivity of the system is sufficiently high to detect even low-abundance transcripts. Within the group that did not produce a detectable transcript, eight genes (cca, hisC, ynzH, yobE, yoaI, yorV, ypjA, and pyjC) did not hybridize to labeled genomic DNA isolated from B. subtilis strains PY79 and BS0011 combined. In addition, 15 probe sets from intergenic regions also failed to generate a detectable signal.
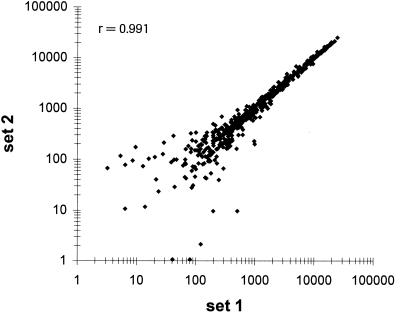
Comparison of the duplicated probes sets within a single hybridization experiment also showed good reproducibility of the fluorescent signals with a Pearson correlation coefficient (r) of >0.99 (Fig. 1). This result indicated uniform hybridization of the microarray and provided additional data sets for statistical analysis.
FIG. 1.
Comparison of transcript levels of duplicated probe sets within a single microarray. A scatter plot of average intensity difference values of duplicated probe sets from a single hybridization is shown. cDNA probes were prepared from total RNA isolated from a culture of B. subtilis PY79 grown in minimal medium to exponential growth phase. r is the Pearson correlation coefficient.
Differential expression of vitamin-regulated genes.
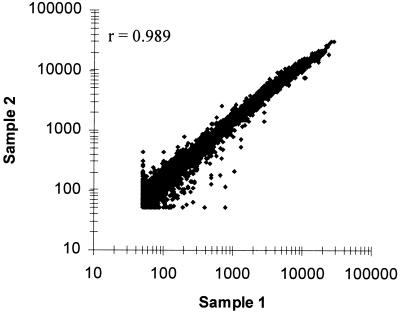
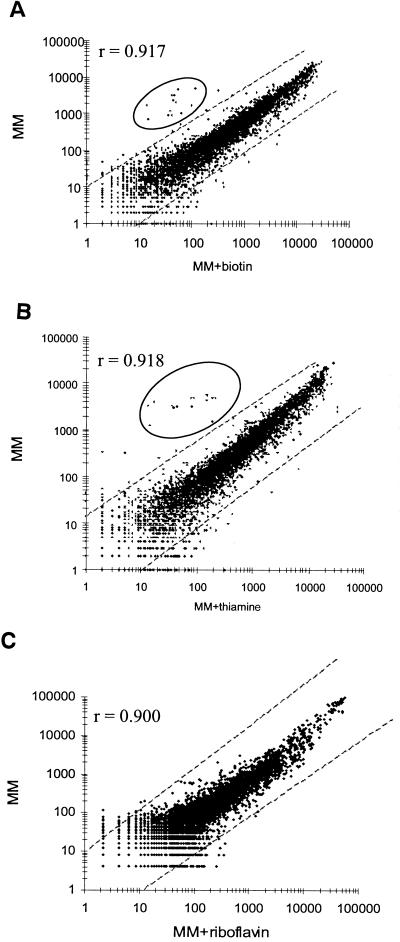
The Bacillus GeneChip microarray was also tested to detect differential gene expression patterns of known vitamin regulons for riboflavin, biotin, and thiamine. Control studies showed good reproducibility (r = 0.989) of transcript levels between duplicated bacterial cultures of B. subtilis PY79 grown to late exponential phase in minimal medium (Fig. 2). In contrast, significant and specific changes in transcript levels were detected when cells were grown with biotin (Fig. 3A), thiamine (Fig. 3B), or riboflavin (Fig. 3C) at levels known to repress transcription of their respective biosynthetic genes. Biotin biosynthetic genes bioWAFDBIorf2 exhibited a 30- to >100-fold repression in transcript levels by biotin (Table 1). As expected, genes outside this cluster (ytaP and ytcP) did not show any change in transcript levels. Moreover, in an isogenic strain (BI421) containing a B. subtilis birA mutation that derepresses the bio operon (3), constitutive high-level transcription of the biosynthetic genes was observed. Transcription of birA remained unchanged in both strains.
FIG. 2.
Comparison of transcript levels from duplicate bacterial cultures. A scatter plot of average intensity difference values from a hybridization experiment with cDNA prepared from duplicate cultures of B. subtilis PY79 grown to exponential growth phase in minimal medium is shown. Average intensity differences from 50 to >20,000 U are plotted. r is the Pearson correlation coefficient.
FIG. 3.
Identification of biotin-, thiamine-, and riboflavin-regulated genes. Scatter plots of average intensity difference values from hybridization experiments with cDNA prepared from cultures of B. subtilis PY79 grown to exponential growth phase in minimal medium in the presence (x axis) or absence (y axis) of biotin (0.1 μg/ml) (A), thiamine (0.34 μg/ml) (B), or riboflavin (200 μg/ml) (C) are shown. r is the Pearson correlation coefficient. The diagonal lines indicate a 10-fold change in transcript levels. Known and potential biotin- or thiamine-regulated genes are circled.
TABLE 1.
Transcript levels of genes involved in biosynthesis of riboflavin, biotin, and thiamine
| Biosynthesis type and genea | Enzyme and/or functiona | Ratiob
|
|
|---|---|---|---|
| wt−/wt+c | deg+/wt+ | ||
| Riboflavin | |||
| ribG | Riboflavin-specific deaminase or reductase | 2.0 | 12 |
| ribB | Riboflavin synthase (α subunit) | 2.5 | 28 |
| ribA | GTP cyclohydrolase IV 3,4-dihydroxy-2-butanone 4-phosphate synthase | 2.9 | 19 |
| ribH | Lumazine synthase (β subunit) | 2.7 | 13 |
| ribT | Unknown (not required for riboflavin synthesis) | NC | NC |
| ribC | Riboflavin kinase or FAD synthase | NC | NC |
| ribR | Cryptic riboflavin kinase | NT | NT |
| ypaA | Unknown transmembrane protein | NC | 11 |
| Biotin | |||
| bioW | Pimeloyl-CoA synthase | 89 | >100 |
| bioA | DAPA aminotransferase | >100 | >100 |
| bioF | 7-KAP synthase | 52 | 60 |
| bioD | DTB synthetase | 48 | 86 |
| bioB | Biotin synthase | 94 | >100 |
| bioI | Biotin cytochrome P-450 | 33 | 66 |
| ytbQ | Unknown and/or not required for biotin synthesis | 62 | 100 |
| birA | Biotin operon repressor/biotin-protein ligase | NC | NC |
| yuiG | Unknown function or homologous to B. sphaericus bioY | 21 | 17 |
| yhfU | Unknown function or homologous to B. sphaericus bioY | 11 | 16 |
| Thiamine | |||
| thiA | Biosynthesis of hydroxymethylpyrimidine phosphate | 62 | |
| thiK | Hydroxyethylthiazole kinase | NC | |
| thiC | Thiamine phosphate pyrophosphorylase | NC | |
| goxB | Glycine oxidase; possible thiO ortholog | 67 | |
| thiS (yjbS) | Biosynthesis of hydroxyethylthiazole phosphate | 84 | |
| thiG (yjbT) | Biosynthesis of hydroxyethylthiazole phosphate | 82 | |
| thiF (yjbU) | Biosynthesis of hydroxyethylthiazole phosphate | 90 | |
| thiD (yjbV) | Possible phosphomethylpyrimidine kinase | 32 | |
| ydiA | Unknown; possible thiL ortholog (TMP kinase) | NC | |
| ytbJ | Unknown; possible thiI ortholog (sulfur transferase) | NC | |
| dxs (yqiE) | 1-Deoxy-d-xylulose synthase | NC | |
Gene names and functions are as described in Perkins and Pero (27) and SubtiList (http://genolist.pasteur.fr/SubtiList). Abbreviations: CoA, coenzyme A; FAD, flavin adenine dinucleotide; DAPA, 7,8-diaminopelargonic acid; DTB, dethiobiotin; 7-KAP, 7-keto-8-aminopelargonic acid; TMP, thiamine monophosphate.
Transcript ratios were calculated by dividing the average difference values (after normalization) from hybridization experiments of wild-type cells grown to exponential phase in minimal medium without vitamin treatment by those with vitamin treatment (wt−/wt+) or from hybridization experiments of deregulated mutant cells grown to exponential phase in minimal medium with vitamin treatment by those of wild-type cells grown under the same condition (deg+/wt+). B. subtilis BI421 (birA) was used in the biotin treatment experiments, and B. subtilis BS0012 (ribC) was used in the riboflavin treatment experiments. For some genes (indicated in boldface), average difference values were obtained from duplicate probe sets per hybridization experiment. NC, no change in average difference values; NT, no transcript detected.
Average of two independent experiments. r values (vitamin-treated versus untreated datasets) for experiments 1 and 2: biotin, 0.91 and 0.89; thiamine, 0.94 and 0.95; and riboflavin, 0.90 and 0.90, respectively.
Similarly, a 30- to 90-fold thiamine-specific repression was observed with transcripts from two major thiamine biosynthetic operons, thiA and tenAI-goxB-yjbSTUV. However, transcript levels of a third operon containing thiK and thiC remained unchanged. Other putative biosynthetic genes, ydiA, ytbJ, and yqiE, also showed no regulation (Table 1).
Transcription of the riboflavin operon (ribGBAH) also exhibited riboflavin-specific repression, but the change in average difference levels was only threefold for genes ribA, ribB, and ribH and twofold for ribG (Table 1). Similar results were obtained when a related strain, B. subtilis BS0011, was grown under similar conditions (data not shown). Moreover, in the presence of the ribC mutation (in B. subtilis 1012), which has been shown to derepress the rib operon (6, 22), constitutive, high-level transcription of the biosynthetic genes was observed (Table 1). Transcripts levels of ribGBAH were increased by >10-fold in the ribC-containing strain B. subtilis BS0012 compared to the isogenic parent B. subtilis BS0011 when both were grown in the presence of riboflavin. Transcription of the ribC gene remained unchanged; transcripts of ribR, a ribC paralog (31), were not detected. Collectively, these results were in good agreement with past Northern blot and lacZ reporter gene fusion studies (1, 26, 29, 44, 45; K. Eichler, S. Taylor, C. Vockler, Y. Zhang, V. Delague, T. P. Begley, and A. P. G. M. van Loon, unpublished results).
Real-time RT-PCR experiments were also performed to assess the accuracy of the GeneChip microarray to detect changes in transcript levels. By using primers complementary to the 5′ and 3′ ends of the bioA, thiA, and ribG genes, transcript levels were increased 33-, 8-, and 3-fold, respectively, in bacteria grown in minimal medium compared to cells grown in the same medium supplemented with their respective vitamin (Table 2). In a parallel control experiment, the transcript level of a housekeeping gene, gap, showed no significant change under these growth conditions. Qualitatively, these results were in good agreement with microarray data, although in some cases the exact fold change in gene expression differed. Based on these results and standard deviation criteria, average intensity difference ratios of 3 or higher were considered significant.
TABLE 2.
Comparison of differential gene expression levels determined by real-time RT-PCR and microarray hybridization
| Expt and gene | Fold change in transcript levelsa as determined by:
|
|
|---|---|---|
| RT-PCR | Microarray | |
| +/− riboflavin treatment | ||
| ribG | 3 | 2.0 |
| thiA | NC | NC |
| bioA | NC | NC |
| gap | NC | NC |
| +/− biotin treatment | ||
| ribG | NC | NC |
| thiA | NC | NC |
| bioA | 8 | >100 |
| gap | NC | NC |
| +/− thiamine treatment | ||
| ribG | NC | NC |
| thiA | 33 | 62 |
| bioA | NC | NC |
| gap | NC | NC |
Triplet real-time RT-PCRs were performed on cDNA prepared from total RNA isolated from cells grown to exponential phase in minimal medium in the presence or absence of riboflavin (200 μg/ml), biotin (0.1 μg/ml), or thiamine (0.34 μg/ml) as described in Materials and Methods. Values presented were calculated by dividing the average fold change from untreated cells by those from vitamin-treated cells. The fold change values determined by microarray analysis were taken from Table 1 for comparison. NC, no change in real-time RT-PCR values.
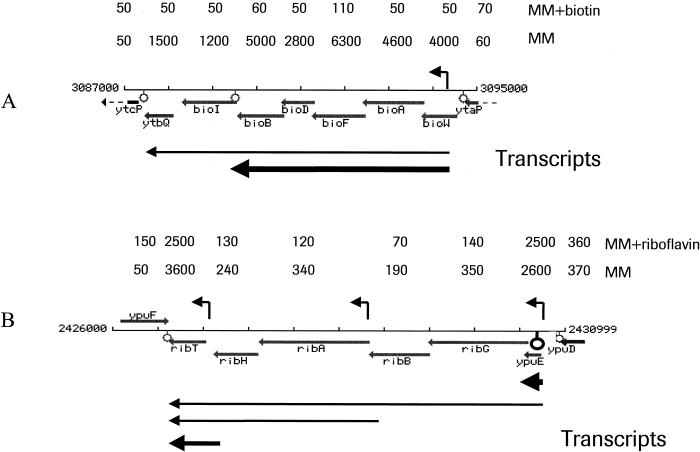
Analysis of the transcript levels of selected biosynthetic genes showed that the data conformed to known operon structures and regulation mechanisms. As examples, results with the biotin and riboflavin biosynthetic operons are illustrated in Fig. 4. Transcription of the biotin operon is tightly regulated by a repressor or operator mechanism involving biotin and the B. subtilis BirA-like repressor (3, 4). Location of a rho-independent transcription terminator between the fifth and sixth genes results in the synthesis of two polycistronic transcripts: a full-length transcript of 7.2 kb covering the entire operon (bioWAFDBI-ytbQ) and a shorter 5.2-kb mRNA species that cover the first five genes (bioWAFDB) (26). Northern blots indicated that abundance of the shorter transcript is ∼8-fold higher than the full-length transcript. Microarray data showed that, in cells grown in the absence of biotin, bioWAFDB transcript levels were threefold greater than those of bioI and ytbQ (Fig. 4). In the presence of biotin, however, no transcripts were detected. Interestingly, no readthrough transcription from the biotin operon into ytcP was detected, indicating strong transcription termination at the 3′ terminator.
FIG. 4.
Comparison of expression levels of individual genes within the biotin (A) and riboflavin (B) biosynthetic operons. Physical maps of the B. subtilis bio and rib operons are shown with alignment of the transcripts detected by Northern blotting (5, 26, 29). The relative steady-state abundance of individual transcripts is represented by the thickness of the arrows. Above each physical map are the average intensity difference values of individual genes or DNA segments determined from hybridization experiments described in Fig. 3. Symbols: angled arrow, ςA promoter sequence; lollipop, rho-independent transcription terminator. Drawings of the biotin and riboflavin operons are reproduced from the Subtilist website (24). Numbers at either end of the maps represent the B. subtilis genome location (in base pairs) of the operons.
The riboflavin operon is one of several well-studied operons that are regulated by a transcription termination-antitermination mechanism (27). The genes are transcribed from three vegetative promoters, two of which (ribP1 and ribP2) are regulated by FMN (flavin mononucleotide) and B. subtilis RibC (6, 22). A proximal rho-independent transcription terminator located between a vegetative promoter and the first structural gene is the key element in the regulatory mechanism that limits expression of the operon under excess FMN conditions. A probe set to the 5′ RFN leader region (15) easily detected the attenuator transcripts, which has been observed in previous Northern studies. These transcripts were approximately sevenfold higher than transcripts of the first four rib genes under derepressing growth conditions. Transcript levels of ribA and ribH were found to be similar to those of ribG and ribB, indicating that any increase in transcript levels contributed by the internal ribP2 promoter was not detected by the ribA and ribH probe sets. The fact that transcription from ribP2 is weaker than from ribP1 may account for this observation (28, 29). The last gene of the operon, ribT, was highly expressed under both growth conditions. Transcription from the unregulated ribP3 promoter might account for this result and “mask” transcripts from ribP1 and ribP2.
Interestingly, several new vitamin-regulated genes were identified by using an algorithm that groups genes according to their transcription patterns (GeneSpring). For example, transcription of ypaA, a putative transport gene shown to contain the riboflavin RFN regulatory region (15), was increased >10-fold in a ribC mutant compared to wild-type cells when both were grown in minimal medium containing riboflavin (Table 1). These results are in good agreement with recent ypaA-lacZ fusion studies (Mouncey and Hohmann, unpublished). Other examples included two biotin-regulated transcripts, yuiG and the yhfU. Both yuiG and yhfU encode proteins with strong similarity to the B. sphaericus bioY gene (16). The average difference levels of yuiG and the yhfU were 20- and 10-fold higher, respectively, in wild-type cells grown in minimal medium than in cells grown in the presence of biotin (Table 1). In addition, the transcription of both genes was derepressed in the B. subtilis birA mutant. Gene transcripts of three genes (yfhT, yfhS, and yfhR) adjacent to yfhU showed a similar biotin-regulated transcription pattern, suggesting that these four genes are organized as a single operon. Moreover, inspection of the predicted 5′ leader region of yuiG and yhfU revealed putative vegetative (ςA) promoter regions, followed by short DNA segments with strong sequence homology to the “regulatory” site of the B. subtilis bio operon (Fig. 5). The presence of these DNA elements is consistent with the structure of known biotin-regulated genes.
FIG. 5.
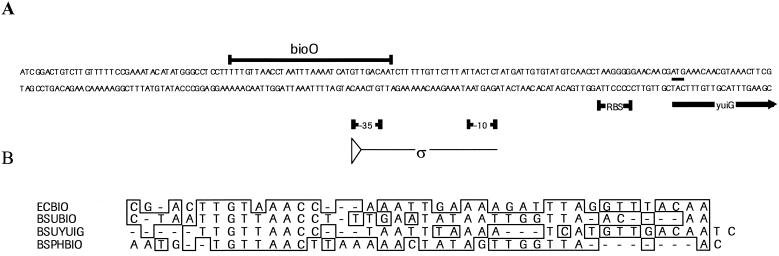
(A) Nucleotide sequence of the 5′ leader region of B. subtilis yuiG. Symbols: thin arrow, putative sigma A promoter sequence (ς); heavy underlines, −35 and −10 contact regions, the RBS, and the biotin regulatory region (bioO); heavy arrow, 5′ coding region of yuiG. (B) Consensus sequence of possible biotin regulatory regions. The 5′ leader regions of E. coli bioABFCD, B. subtilis bioWAFDBI-ytbQ (BSUBIO), B. subtilis yuiG (BSUYUIG), and B. sphaericus bioDAYB (BSPHBIO) were aligned by using the MegAlign algorithm of DNASTAR software. Conserved nucleotides are boxed.
Differential expression during transition from exponential to stationary growth.
Control studies initially showed poor reproducibility (r < 0.9) of transcript levels between duplicated bacterial cultures of PY79 when standard optical measurements were used to determine the onset of stationary phase. Reproducibility improved if a biochemical indicator was used to indicate the stationary phase. We arbitrarily chose 30 min after glucose exhaustion as an indicator of cells entering stationary phase. By this method, the transcript levels of 439 exponentially expressed genes decreased threefold or more when cells entered stationary phase. Conversely, transcription of 230 genes poorly or not expressed during exponential phase were increased threefold or more in stationary phase. Table 3 lists examples of the mostly highly expressed genes from both classes. Of notable interest, genes acoA, acoC, glvC, gapB, licB, licC, pckA, and tdh and the rbs gene cluster (rbsA and rbsD as examples) have been previously shown to be glucose repressed (22, 39). Others are involved in the cold shock response (cspB and cspD), carbohydrate (gap, kbl, lctE, pdhA, pdhB, yjdE, and yvdF) and lipid metabolisms (yusJ), transport (lctP), and ribosomal function (rplJ and rpsD).
TABLE 3.
Change in transcript levels of several highly expressed genes from exponential to stationary growth phase
| Gene | Enzyme and/or functionsa | Ratiob |
|---|---|---|
| acoA | Acetoin dehydrogenase E1 component (alpha subunit) | <0.01 |
| acoC | Acetoin dehydrogenase E2 component | 0.01 |
| gapB | Glyceraldehyde-3-phosphate dehydrogenase | 0.013 |
| glvC | PTS arbutin-like enzyme IIBC component | <0.01 |
| kbl | 2-Amino-3-ketobutyrate CoA ligase | 0.014 |
| licB | PTS lichenan-specific enzyme IIB component | 0.015 |
| licC | PTS lichenan-specific enzyme IIC component | 0.011 |
| pckA | Phosphoenolpyruvate carboxykinase | 0.04 |
| rbsA | Ribose ABC transporter (ATP-binding protein) | 0.016 |
| rbsD | Ribose ABC transporter (membrane protein) | 0.019 |
| tdh | Threonine 3-dehydrogenase | 0.038 |
| yjdE | Unknown; similar to mannose-6-phosphate isomerase | 0.023 |
| yusJ | Unknown; similar to butyryl-CoA dehydrogenase | 0.011 |
| yvdF | Unknown; similar to glucan 1,4-alpha-maltohydrolase | 0.011 |
| cspB | Cold shock protein | 13.8 |
| cspD | Cold shock protein | 9.7 |
| gap | Glyceraldehyde-3-phosphate dehydrogenase | 66.3 |
| lctE | l-Lactate dehydrogenase | 9.4 |
| lctP | l-Lactate permease | >100 |
| pdhA | Pyruvate dehydrogenase (E1 α subunit) | 14.3 |
| pdhB | Pyruvate dehydrogenase (E1 β subunit) | 18.3 |
| rplJ | Ribosomal protein L10 (BL5) | 91.5 |
| rspD | Ribosomal protein S4 (BS4) | 72.0 |
CoA, coenzyme A.
The repression or derepression ratios were calculated by dividing the average difference values (after normalization) from duplicate hybridization experiments of cells grown to exponential phase in minimal medium without vitamin treatment by those grown to stationary phase. See text for details. For most genes (indicated in boldface), average difference values were obtained from duplicate probe sets per microarray.
Identification of new transcripts.
Selinger et al. (30) have recently reported on the use of an E. coli oligonucleotide genome microarray to detect new transcripts within intergenic regions and located on the noncoding strand of known coding regions (i.e., antisense RNA). Inclusion of probe sets to the Bacillus microarray, which were complementary to intergenic regions of 250 bp and larger, allowed a similar screening for unidentified transcripts. Approximately 33% of these probe sets detected a transcript from cells grown in either minimal medium or LB broth. For many of these signals, we suspect that they represented either 3′ readthough transcripts or untranslated 5′ leader transcripts from existing genes. To determine whether any of these transcripts originated from coding regions, a subclass of transcripts were analyzed that were oriented in the opposite direction relative to transcripts from known flanking genes. All 35 such transcripts hybridized to a specific cluster of probe pairs, indicating the presence of a novel transcript. Subsequent placement of the oligonucleotide probe sequence onto the DNA sequence identified 20 ORFs with a B. subtilis-type RBS. In many cases, known Bacillus promoter sequences and rho-independent transcription termination sites could also be identified from the DNA sequence. Several examples of coding regions identified by this method are listed in Table 4. In most of these examples, changes in the transcript levels could be detected between exponential and stationary growth phases or between minimal medium- and LB medium-grown cells. Moreover, in all six examples tested (GAP74-R, GAP123-F, GAP163-2-R, GAP206A-F, GAP240-F, and GAP342-F), these gene expression changes could be confirmed qualitatively by real-time RT-PCR (data not shown). For two of the intergenic (GAP68D-R and GAP163-2-R) regions, two coding regions were identified. The recently updated Subtilist database now indicates that the GAP68D-R region contains a single gene, ygzB, which encompasses the two coding regions detected by the microarray. The deduced amino acid sequence of several coding regions showed significant homology to known B. subtilis genes (yobB) or to related species (e.g., Bacillus halodurans and Bacillus stearotheromophilus). In two cases, the detected genes have been recently identified and characterized (ynxB and sda). However, in coding regions from four intergenic regions, GAP96-R, GAP169A-R, GAP206A, and GAP342-F, no similarity was detected. Most of the coding regions listed in Table 4 are also present in a new public B. subtilis genome database developed by Integrated Genomics, Inc. (2, 25). Importantly, coding regions from four intergenic regions, GAP74-R, GAP169A-R, GAP206A, and GAP240-F, are not. Two of these did not show any homology to known proteins.
TABLE 4.
Locations and expression levels of several RNA transcripts within intergenic regions identified by microarray hybridization
| GAP region (flanking genes) | Coordinatesa (bp) | Positive probe pairs (position)b | Expression levelc in:
|
Homologyd | |||||
|---|---|---|---|---|---|---|---|---|---|
| MM
|
LB
|
||||||||
| Log | Stat | Fold change | Log | Stat | Fold change | ||||
| 68D-R (perR/ygxA) | 944257–944854 | 1–15 | 480 | 2,800 | 0.17 | 220 | 130 | 1.7 | B. subtilis ygzB, B. stearothermophilus ORF3663, and B. halodurans BH0952 |
| 74-R (yhbF/prkA) | 972141–972492 | 9–19 | 1,300 | 4,900 | 0.26 | 230 | 1,200 | 0.19 | B. halodurans BH1024 |
| 96-R (yjiC/yjjA) | 1293043–1293447 | 5–15 | 190 | 80 | 2.4 | <50 | <50 | NTe | None |
| 123-F (slp/cad) | 1533009–1533588 | 10–20 | <50 | 7,000 | 0.007 | 90 | 150 | 0.6 | B. stearothermophilus ORF4213 |
| 163-2-R (yoaM/yoaN) | 2036272–2036634 | 1–20 | 370 | 280 | 1.3 | 150 | <50 | 3.0 | B. subtilis ynxB and RBS04224 |
| 169A-R (xynA/yobD) | 2056788–2057011 | 3–13 | 260 | 100 | 2.6 | 90 | 80 | 1.1 | None |
| 172-1-F (yozJ/rapK) | 2059904–2060133 | 1–20 | 1,200 | 3,000 | 0.4 | 140 | <50 | 2.8 | B. subtilis yobB |
| 206A-F (ypjQ/ypiP) | 2298343–2298604 | 10–20 | 1,200 | 3,900 | 0.3 | 580 | 290 | 2.0 | None |
| 240-F (yqeN/comEC) | 2636363–2636768 | 9–20 | 2,600 | 4,600 | 0.56 | 880 | 110 | 8.0 | B. halodurans BH1336 |
| 241-F (yqeG/yqeF) | 2646336–2647145 | 4–20 | 570 | 290 | 2.0 | <50 | <50 | NT | B. subtilis sda and B. halodurans GH1321 |
| 342-F (yvcP/yvcN) | 3567166–3567559 | 2–19 | 200 | 90 | 2.2 | 150 | <50 | 3.0 | None |
Coordinates taken from SubtiList (http://genolist.pasteur.fr/SubtiList).
That is, the position of probe pairs within each probe set (20 probe pairs per probe set) that generated a positive hybridization signal according to Affymetrix hybridization metrics.
The average difference values (after normalization) of B. subtilis PY79 grown to exponential or stationary phase in minimal medium (MM) or LB broth (LB). Values were determined from either duplicate (MM) or single (LB) hybridization experiments. Fold change values indicated in boldface were confirmed by RT-PCR. Log, log phase; Stat, stationary phase.
Homology of putative coding regions that correspond to detected RNA transcripts.
NT, no transcript detected.
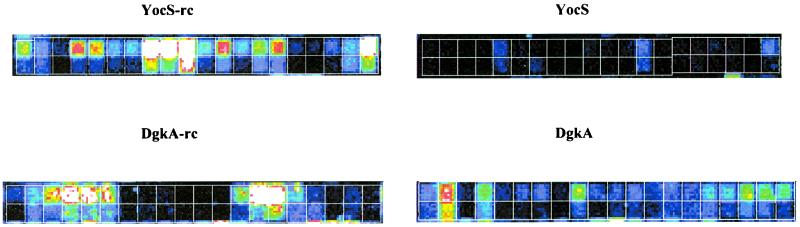
The presence of probe sets complementary to the noncoding strand of known coding regions also allowed a screening for “antisense” transcripts. In samples from cells grown in minimal or LB medium, hybridization signals exceeding noise by a factor of 5 or greater were observed for 18% (102 of 565) of the known genes tested (see Materials and Methods). In some cases, the transcript levels were observed to change under specific growth conditions or stages. The biological significance of these antisense transcripts is not known. The hybridization data of two such probe sets complementary to the noncoding strand of dgkA (diacyl glycerol kinase) and yocS (unknown function) are illustrated in Fig. 6. In minimal medium, the yocS antisense transcript was eightfold higher in the exponential stage than in the stationary phase, with a maximum average intensity difference of 3,600. This transcript could be the result of readthrough transcription from odhB, which is upstream from yocS and is oriented in the same direction as this antisense transcript. However, a putative rho-independent transcription terminator is located between odhB and yocC that could block readthrough transcription. Moreover, the expression profile of odhB is significantly different, showing high constitutive levels between exponential and stationary growth phases (average intensity difference of between 6,000 and 7,000). The dgkA antisense transcript, alternately, was observed to increase three- to fourfold as cells entered exponential growth phase, with a maximum average intensity difference of 1,600. This gene is located within the middle of a six-gene operon. An antisense transcript from the adjacent gene cdd was not detected, indicating that the dgkA antisense transcript was not caused by readthrough transcription.
FIG. 6.
Detection of antisense RNA transcripts. False-colored images of probe sets to the coding and noncoding (reverse complement) strand of B. subtilis yocS (top) and dgkA (bottom) are shown. The images were derived from hybridization experiments by using cDNA samples prepared from total RNA of B. subtilis PY79 grown in minimal medium to either the stationary (yocS/yocS-rc) or the exponential (dgkA/dgkA-rc) growth phase. The 20 probe pairs in these probe sets are outlined by a white grid, with the PM (perfect match) features on the top row and the MM (mismatch) features on the bottom row. Black indicates no hybridization.
DISCUSSION
Genome-wide profiling of B. subtilis has been performed previously with nylon filters (macroarrays) or microscope glass slides (microarrays) containing spotted PCR products that correspond to the annotated ORFs. Here we report on the development and testing of an oligonucleotide-based antisense microarray for B. subtilis using Affymetrix technology. The array was found to monitor gene expression changes in response to transition from exponential to stationary growth phase and exposure to three different vitamins that repress expression of biosynthetic genes. More importantly, these results were consistent with our current understanding of gene expression and regulation in B. subtilis. We anticipate that this array can be applied to assess other gene expression changes such as sporulation, heat shock, stress, competence, and chemotaxis with equal accuracy.
The Bacillus antisense microarray was found to detect transcripts for >70% of the known coding regions. This sensitivity level is in good agreement with gene detection levels reported for S. pneumoniae, H. influenzae, and E. coli antisense Affymetrix oligonucleotide microarrays (10, 17, 30). Caldwell et al. (R. Caldwell, R. Sapolsky, W. Weyler, R. R. Maile, S. Causey, and E. Ferrari, unpublished data) also describe an antisense B. subtilis oligonucleotide array that detects >70% of the known coding regions. In addition to antisense arrays, Affymetrix has commercialized a sense E. coli array. This array is reported to detect transcripts from only ca. 50% of the known coding regions (7). It is not clear why the sensitivity level of the sense microarray is lower than the antisense microarray. An underlying cause might be the fact that these microarray systems used two different targets during hybridization (cDNA for antisense arrays and total RNA for sense arrays). Differences in nucleic acid quantity, labeling efficiency of the targets, and hybridization conditions could also contribute to the performance differences of these microarrays.
Analysis of the transcript levels of three vitamin biosynthetic operons for riboflavin, biotin, and thiamine showed that the expression pattern of these operons was dramatically different. Whereas derepression of thiamine- and biotin-regulated biosynthetic genes in wild-type cells varied between 30- and 100-fold, only a 3-fold change was detected for riboflavin-regulated genes. However, a higher level of riboflavin derepression (10- to 30-fold) was observed in the ribC mutant strains. These differences could be related to the dissimilar regulatory mechanisms that these pathways exhibit. Biotin-regulated genes are reported to be regulated by a repressor-operator mechanism which controls the ability of the RNA polymerase to bind to the promoter. On the other hand, riboflavin biosynthetic genes are regulated by a termination-antitermination mechanism, which modulates the transcriptional flow from the promoter to the structural genes by means of an active or inactive transcription terminator. Thiamine-regulated genes have regulatory elements of both a highly conserved 39-bp sequence referred to as the thi box and a transcriptional terminator within the predicted 5′ leader region (27).
Transcription patterns that are common among a set of genes can be used to identify additional genes that are similarly regulated. Accordingly, we were able to use this fact to identify several new biotin-regulated genes by using a computer-based algorithm. As expected of biotin-regulated genes, the predicted 5′ leader regions of yuiG and yhfU contained sequences that were homologous to known bio operator sequences. However, it is unclear why the level of derepression was significantly lower than that observed for the biosynthetic genes. Further work is necessary to determine the function of these genes and whether they are involved in biotin biosynthesis.
One advantage of short single-stranded oligonucleotide arrays is the ability to monitor expression over a short DNA sequence or a specific region of a transcript without undue cross-hybridization (8, 9, 19, 30). We were able to make use of this property to detect transcripts from intergenic regions of the chromosome that were not previously annotated to contain coding regions. Although most of these represented either 3′ readthrough transcripts or untranslated 5′ leader transcripts from existing genes, analysis of a subclass of transcripts oriented in the opposite direction relative to transcripts from known flanking genes indicated that at least 20 transcripts were synthesized from specific coding regions. Many of these coding regions showed similarity to known genes from recently sequenced Bacillus species and from other microorganisms. However, several others did not. As the genomes of more bacilli and other microorganisms are sequenced, determination of the identity of these genes should be possible. In many cases, the expression level of these transcripts differed depending on the medium in which the cells were grown (Table 4). A systematic comparison of transcript levels from minimal and rich medium-grown samples should provide leads about the function and regulation of these transcripts. We have also begun to construct null mutations in several of these putative genes to ascertain their function in B. subtilis physiology. To date mutations within GAP123, GAP172, GAP206, and GAP240 have not resulted in a discernible phenotype when grown on complex or minimal medium. For several of these transcripts, a coding region could not be detected. It is possible that these regions encode small RNAs (sRNA) that have been shown to have a regulatory function (36). Alternatively, failure to identify an ORF might be caused by sequencing mistakes within the intergenic region that result in an incorrectly predicted ORF start or stop codon (30). In any event, analysis of the remaining intergenic regions is expected to yield more new transcripts. Additional work is necessary to determine their function and role in B. subtilis physiology. Recently, Wassarman et al. (37) reported on the detection of new sRNA in E. coli by a similar method, which also resulted in the detection of putative new short ORFs.
A smaller number of new transcripts detected by the microarray appeared to originate from the noncoding strand of the genome. Approximately 18% of 565 known genes generated an antisense transcript. This suggests that a significant portion of the noncoding strand of the genome is transcribed at a low level. This observation is consistent with a similar analysis of the E. coli genome with a nucleotide-based E. coli microarray (30). It remains to be determined, however, whether these “antisense” transcripts simply represent readthrough transcription from known coding regions or cryptic genetic elements or whether they are independent transcripts. Construction of gene fusions, as well as promoter and transcript mapping, should resolve the source of these transcripts. It is also not clear whether they have a regulatory function or indicate the presence of overlapping genes. However, analysis of several of these antisense transcripts did reveal the presence of possible ORFs. For example, a 156-bp ORF was located on the reverse complement of dgkA (Fig. 7). The TTG start of this putative ORF was preceded by a sequence (5′-GCAAGAGGGTG-3′) that strongly resembled a Bacillus-like RBS, with a calculated ΔG of −11 kcal/mol (33). The predicted amino acid sequence did not show significant homology to any known protein. In addition, the predicted amino acid sequences from several other ORFs also did not show significant homology to any known protein. Thus, it remains to be determined whether these putative ORFs encode protein products or are just artifacts. However, it is important to point out that the B. subtilis genome, like bacteriophage genomes, does contain genes encoded on the opposite strand of known genes. Integrated Genomics, Inc. has recently shown that ca. 50 overlapping genes have been identified in the B. subtilis genome (M. D'Souza, unpublished results). Further analysis of these transcripts is necessary to resolve these issues.
FIG. 7.
Nucleotide sequence of a putative ORF transcribed from the noncoding strand of B. subtilis dgkA. Nucleotide sequence from bp 2610711 to 2611170 of the B. subtilis genome is shown. Symbols: underline, deduced amino acid sequences; heavy underline, RBS.
ACKNOWLEDGMENTS
We gratefully acknowledge Detlef Wolf and Clemens Broger for their help with bioinformatics. We thank Antoine de Saizieu and Ulrich Certa for helpful discussions and critical reading of the manuscript. We also thank Integrated Genomics for providing unpublished genomics data.
REFERENCES
- 1.Azevedo V, Sorokin A, Ehrlich D, Serror P. The transcriptional organization of the Bacillus subtilis 168 chromosome region between the spoVAF and serA genetic loci. Mol Microbiol. 1993;10:397–405. doi: 10.1111/j.1365-2958.1993.tb02671.x. [DOI] [PubMed] [Google Scholar]
- 2.Bernal A, Ear U, Kyrpides N. Genomes online database (GOLD): a monitor of genome projects world-wide. Nucleic Acids Res. 2001;29:126–127. doi: 10.1093/nar/29.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bower S, Perkins J, Yocum R R, Serror P, Sorokin A, Rahaim P, Howitt C L, Prasad N, Ehrlich S D, Pero J. Cloning and characterization of the Bacillus subtilis birA gene encoding a repressor of the biotin operon. J Bacteriol. 1995;177:2572–2575. doi: 10.1128/jb.177.9.2572-2575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bower S, Perkins J B, Yocum R R, Howitt C L, Rahaim P, Pero J. Cloning, sequencing, and characterization of the Bacillus subtilis biotin biosynthetic operon. J Bacteriol. 1996;178:4122–4130. doi: 10.1128/jb.178.14.4122-4130.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bower, S. G., J. B. Perkins, R. R. Yocum, and J. Pero. May 2000. Biotin biosynthesis in Bacillus subtilis. U.S. patent 6,057,136.
- 6.Coquard D, Huecas H, Ott M, van Dijl J M, van Loon A P G M, Hohmann H-P. Molecular cloning and characterisation of the ribC gene from Bacillus subtilis: a point mutation in ribC results in riboflavin overproduction. Mol Gen Genet. 1997;254:81–84. doi: 10.1007/s004380050393. [DOI] [PubMed] [Google Scholar]
- 7.de Feo G. New human genome U95 set, design and performance characteristics and applications of the E. coli genome array. 3rd Annual Affymetrix User Group Meeting, Boston, Mass. Santa Clara, Calif: Affymetrix, Inc.; 2000. [Google Scholar]
- 8.DeRisi J, Penland L, Brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 9.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 10.de Saizieu A, Certa U, Warrington J, Gray C, Keck W, Mous J. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat Biotechnol. 1998;16:45–48. doi: 10.1038/nbt0198-45. [DOI] [PubMed] [Google Scholar]
- 11.de Saizieu A, Gardes C, Flint N, Wagner C, Kamber M, Mitchell T J, Keck W, Amrein K E, Lange R. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J Bacteriol. 2000;182:4696–4703. doi: 10.1128/jb.182.17.4696-4703.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fawcett P, Eichenberger P, Losick R, Youngman P. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc Natl Acad Sci USA. 2000;97:8063–8068. doi: 10.1073/pnas.140209597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey R N, Willmitzer L. Metabolic profiling for plant functional genomics. Nat Biotechnol. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- 14.Futcher B, Latter G I, Monardo P, McLaughlin C S, Garrels J I. A sampling of the yeast proteome. Mol Cell Biol. 1999;19:7357–7368. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gelfand M S, Mironov A A, Jomantas J, Kozlov Y I, Perumov D A. A conserved RNA structure element involved in regulation of bacterial riboflavin biosynthesis genes. Trends Genet. 1999;15:439–442. doi: 10.1016/s0168-9525(99)01856-9. [DOI] [PubMed] [Google Scholar]
- 16.Gloeckler R, Ohsawa I, Speck D, Ledoux C, Bernard S, Zinsius M, Villeval D, Kisou T, Kamogawa K, Lemoine Y. Cloning and characterization of the Bacillus sphaericus genes controlling the bioconversion of pimelate into dethiobiotin. Gene. 1990;87:63–70. doi: 10.1016/0378-1119(90)90496-e. [DOI] [PubMed] [Google Scholar]
- 17.Gmuender H, Kuratli K, Di Padova K, Gray C P, Keck W, Evers S. Gene expression changes triggered by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 2001;11:28–42. doi: 10.1101/gr.157701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gygi S P, Rochon Y, Franza B R, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kane M D, Jatkoe T, Stumpf C R, Lu J, Thomas J D, Madore S J. Assessment of the sensitivity and specificity of oligonucleotide (50mer) microarrays. Nucleic Acids Res. 2000;28:4552–4557. doi: 10.1093/nar/28.22.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Beverley S M, Bressieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 21.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 22.Mack M, van Loon A P G M, Hohmann H-P. Regulation of riboflavin biosynthesis in Bacillus subtilis is affected by the activity of the favokinase/flavin adenine dinucleotide synthetase encoded by ribC. J Bacteriol. 1998;180:950–955. doi: 10.1128/jb.180.4.950-955.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno M S, Schneider B L, Maile R R, Weyler W, Saier M H., Jr Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol Microbiol. 2001;39:1366–1381. doi: 10.1111/j.1365-2958.2001.02328.x. [DOI] [PubMed] [Google Scholar]
- 24.Moszer I. The complete genome of Bacillus subtilis: from sequence annotation to data management and analysis. FEBS Lett. 1998;430:28–36. doi: 10.1016/s0014-5793(98)00620-6. [DOI] [PubMed] [Google Scholar]
- 25.Overbeek R, Larsen N, Pusch G D, D'Souza M, Selkov E, Jr, Kyrpides N, Fonstein M, Maltsev N, Selkov E. WIT: integrated system for high-throughput genome sequence analysis and metabolic reconstruction. Nucleic Acids Res. 2000;28:123–125. doi: 10.1093/nar/28.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perkins J B, Bower S, Howitt C L, Yocum R R, Pero J. Identification and characterization of RNA transcripts from the biotin biosynthetic operon of Bacillus subtilis. J Bacteriol. 1996;178:6361–6365. doi: 10.1128/jb.178.21.6361-6365.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perkins J B, Pero J G. Vitamin biosynthesis. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and its relatives: from genes to cells. Washington, D.C.: American Society for Microbiology; 2001. pp. 279–293. [Google Scholar]
- 28.Perkins, J. B., J. G. Pero, and A. Sloma. November 1998. Riboflavin overproducing strains of bacteria. U.S. patent 5,837,528.
- 29.Perkins J B, Sloma A, Hermann T, Zachgo E, Erdenberger T, Hannett N, Chatterjee N P, Williams II V, Rufo G A, Jr, Pero J. Genetic engineering of Bacillus subtilis for the commercial production of riboflavin. J Ind Microbiol Biotechnol. 1999;22:8–18. [Google Scholar]
- 30.Selinger D W, Cheung K J, Mei R, Johansson E M, Richmond C S, Blattner F R, Lockhart D J, Church G M. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat Biotechnol. 2000;18:1262–1268. doi: 10.1038/82367. [DOI] [PubMed] [Google Scholar]
- 31.Solovieva I M, Kreneva R A, Leak D J, Perumov D A. The ribR gene encodes a monofunctional riboflavin kinase which is involved in regulation of the Bacillus subtilis riboflavin operon. Microbiology. 2001;145:67–73. doi: 10.1099/13500872-145-1-67. [DOI] [PubMed] [Google Scholar]
- 32.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tinoco I, Jr, Borer P N, Dengler B, Levine M D, Uhlenbeck O C, Crothers D M, Gralia J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 34.Tobisch S, Zühlke D, Bernhardt J, Stülke J, Hecker M. Role of CcpA in regulation of the central pathway of carbon catabolism in Bacillus subtilis. J Bacteriol. 1999;181:6994–7004. doi: 10.1128/jb.181.22.6996-7004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.VanBogelen R A, Greis K D, Blumenthal R M, Tani T H, Matthews R G. Mapping regulatory networks in microbial cells. Trends Microbiol. 1999;7:320–328. doi: 10.1016/s0966-842x(99)01540-1. [DOI] [PubMed] [Google Scholar]
- 36.Wassarman K M, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 37.Wassarman K M, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei Y, Lee J-M, Richmond C, Blattner F R, Rafalski J A, LaRossa R A. High-density microarray-mediated gene expression profiling of Escherichia coli. J Bacteriol. 2001;183:545–556. doi: 10.1128/JB.183.2.545-556.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei Y, Lee J-M, Smulski D R, LaRossa R A. Global impact of sdiA amplification revealed by comprehensive gene expression profiling of Escherichia coli. J Bacteriol. 2001;183:2265–2272. doi: 10.1128/JB.183.7.2265-2272.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wodicka L, Dong H, Mittmann M, Ho M-H, Lockhart D J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 41.Ye R W, Tao W, Bedzyk L, Young T, Chen M, Li L. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J Bacteriol. 2000;182:4458–4465. doi: 10.1128/jb.182.16.4458-4465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida K-I, Kobayashi K, Miwa Y, Kang C-M, Matsunaga M, Yamaguchi H, Tojo S, Yamamoto M, Nishi R, Ogasawara N, Nakayama T, Fujita Y. Combined transcriptome and proteome analysis as a powerful approach to study genes under glucose repression in Bacillus subtilis. Nucleic Acids Res. 2001;29:683–692. doi: 10.1093/nar/29.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Youngman P J, Perkins J B, Losick R. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, Begley T P. Cloning, sequencing, and regulation of thiA, a thiamin biosynthesis gene from Bacillus subtilis. Gene. 1997;198:73–82. doi: 10.1016/s0378-1119(97)00295-3. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y, T, Chiu H J, Begley T P. Characterization of the Bacillus subtilis thiC operon involved in thiamine biosynthesis. J Bacteriol. 1997;179:3030–3035. doi: 10.1128/jb.179.9.3030-3035.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]