Abstract
Down syndrome (DS) is the most common chromosomal condition associated with intellectual disability and is characterized by a variety of additional clinical findings. The pathogenesis of DS and the differences between the sexes are not clear. In order to identify differentially expressed proteins that might be employed as potential biological markers and elucidate the difference in pathogenesis between different genders of T21 fetuses, providing clues for individualized detection and treatment is essential. Amniocyte samples of T21 males, T21 females, CN males, and CN females were collected by amniocentesis. The quantitative value of the peptide corresponding to each sample was determined through quantitative analysis by mass spectrometry. We identified many differentially expressed proteins between T21 fetuses and CN fetuses/T21 males and CN males/T21 females and CN females/and T21 males and T21 females. These differential proteins are associated with many important biological processes and affect the development of multiple systems, including the heart, hematopoietic, immune, reproductive, and nervous systems. Our results show sex-specific modulation of protein expression and biological processes and provide new insights into sex-specific differences in the pathogenesis of DS.
Introduction
Down syndrome (DS) is the most common chromosomal condition associated with intellectual disability and is characterized by a variety of additional clinical findings. It occurs in approximately 1 of 800 births worldwide.1 A third copy of chromosome 21, trisomy 21, has long been recognized as the cause of DS. Life expectancy in children with DS has increased significantly over the past decade, but children with DS remain at a higher risk of neonatal and infant mortality than children without DS (1.65 vs 0.36% and 4 vs 0.48%).2
The incidence and presentation of DS vary depending on ethnic background and geographic region. The range and severity of DS phenotypic features vary from person to person. Some of the most noticeable characteristics of the DS phenotype include mental retardation and an increased incidence of congenital heart disease, hypothyroidism, diabetes, leukemia, and an increased risk of developing Alzheimer’s-like dementia by the age of 40.3,4 Furthermore, patients with DS show multiple defects in both numbers and functions of the B-cell compartment.5
Pregnancy progression and fetal development involve complex fetomaternal physiological processes that rely on intricate interactions between multiple genes and proteins.6 Therefore, multiple genes and other factors working in concert are expected to be responsible for the major DS phenotypes.3 Despite the high prevalence of DS and early identification of the cause, there are many studies on the pathogenesis of DS. However, its specific pathogenesis is still unclear, and specific treatments have consequently been practically unavailable.
In humans, differences in the pathogenesis and prevalence between males and females are being continuously identified in many diseases.7 Although sex disparities in brain function, cardiac homeostasis, heart disease, and humoral immune responses to immunization and infection are well documented, the science that explains these differences remains poorly understood.8 Very few studies have evaluated gender differences in DS, and differences in the pathogenesis and prevalence between T21 males and T21 females are also not clear.
Amniotic fluid cells are the most readily available fetal cells. Amniotic fluid can be divided into two major components: supernatant fluid and free-floating fetal cells called amniotic fluid cells (also known as amniocytes). Amniocytes are shed from all three germ layers of the fetus, and some of these cells that originate from embryonic and extra-embryonic tissues show stem cell-like properties.9 Proteomic analysis of amniotic fluid cells may be more responsive to consistent changes in multi-source cells than single-source cells.
Here, we utilized proteomics analysis to obtain a panel of proteins found to be differentially expressed in amniocytes between the following five groups: (1) trisomy 21 (T21) fetuses and chromosomal normal (CN) fetuses (containing male and female), (2) T21 males and CN males, (3) T21 females and CN females, (4) CN males and CN females, and (5) T21 males and T21 females.
This is the first study to report sex-specific proteomic changes in amniocytes of T21 fetuses to the best of our knowledge. The identified differentially expressed proteins could be used as potential biological markers to uncover differences in pathogenesis between T21 fetuses of different genders. Investigating the pathogenesis of DS is critical for developing more effective individualized screening, diagnosis, and treatment strategies.
Materials and Methods
Ethics Statement
Written informed consent was obtained from the patients and healthy control donors before the blood samples were drawn. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Amniocyte Culture
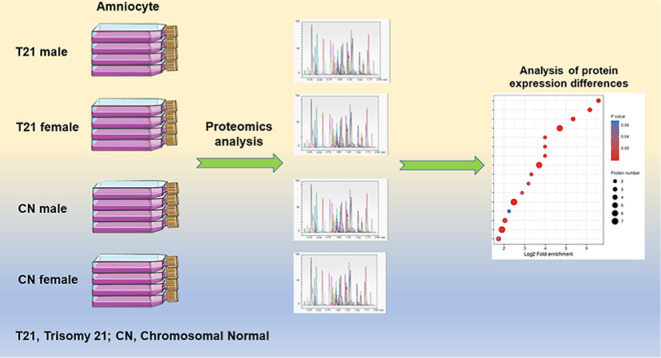
This study was performed at the Prenatal Diagnosis Center of West China Second University Hospital, Sichuan University, following a study protocol approved by the University Hospital. A total of 23 T21 (including 13 males and 10 females) and 29 CN (14 males and 15 females) amniocyte samples were collected by amniocentesis from women at 18–25 weeks of gestation, undergoing prenatal diagnosis. The amniotic fluid cells were a fraction of the cells obtained for cytogenetic analysis and chromosome microarray analysis. Before cytogenetic analysis and CMA, all samples were subjected to a quantitative fluorescent polymerase chain reaction to rapidly detect abnormal numbers of chromosomes 13/18/21 and sex chromosomes. 21-Trisomy cells and CN cells were grown in T-25 cm2 flasks for approximately 14–21 days in BIO-AMF TM-3 (complete culture medium for human amniotic fluid cells and chorionic villi samples (Biological Industries, ref 01-196-1B). The cells were then harvested.
Cell Lysis Protocol and Proteomics Analysis of Amniocytes
T21 male amniocyte samples (n = 13), T21 female amniocyte samples (n = 10), CN male amniocyte samples (n = 14), and CN female amniocyte samples (n = 15) were pooled together. Cell lysis and proteomics analyses were performed by PTM-Biolabs (HangZhou) Co., Ltd., and detailed materials and methods are shown in the Supporting Information Materials and Methods.
The quantitative value of the peptide corresponding to each sample was determined using mass spectrometry quantitative analysis, and each protein corresponded to multiple peptides. The p-value was calculated using the two-sample and two-tail t-test method after the quantitative value of the specific peptide corresponding to the protein in the two samples was calculated using log 2 (to make the data conform to normal distribution). When the p-value was <0.05, the change in the differential expression level was more than 1.3 as the threshold for significant upregulation and less than 1/1.3 as the threshold for significant downregulation.
Results
Identification and Quantification of Proteins
In this project experiment, 350,964.0 secondary spectra were obtained through mass spectrometry analysis. The number of available spectrograms was 103,763, and the utilization rate was 29.6%. A total of 53457.0 peptides were identified by spectrogram analysis, among which the specific peptide was 51684.0. A total of 6541.0 proteins were identified, of which 5543.0 were quantifiable (quantifiable proteins indicated that quantitative information was available in at least one comparison group). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository10 with the data set identifier PXD032883.
Identification of 105 Differentially Expressed Proteins with Important Biological Functions between T21 and CN Fetuses
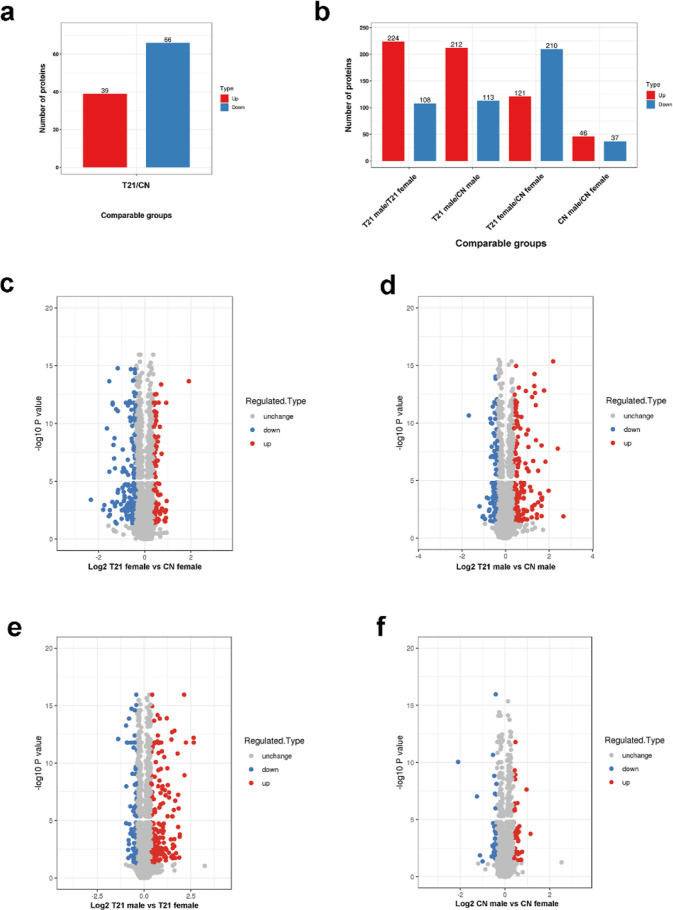
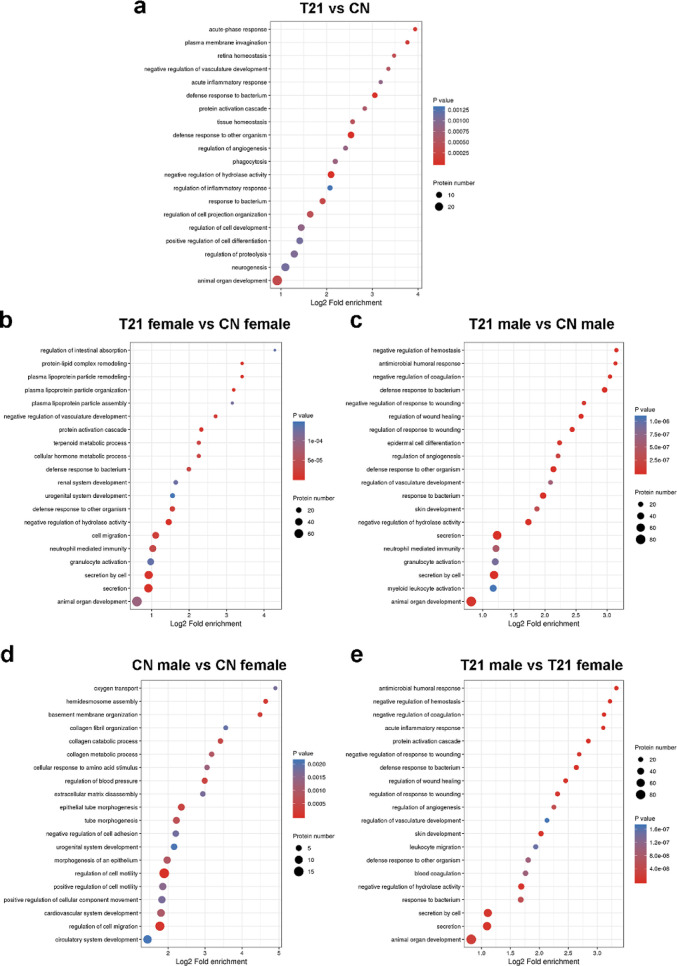
We identified 105 differentially expressed proteins between T21 fetuses and CN fetuses. In contrast to CN, there were 39 proteins with higher expression and 66 proteins with lower expression in the amniocytes of T21 fetuses (Figure 1a and Table S1). The differentially expressed proteins were CPA1, SHTN1, TTYH3, OLFM4, TCF25, PTTG1IP, IGKC, SFTPA1, FBN1, UMOD, IMUP, GC, AFP, SERPINA1, and others.
Figure 1.
Quantity distribution of differentially expressed proteins in different comparison groups. (a) Quantity distribution of differentially expressed proteins between T21 fetuses and CN fetuses (b) and between T21 female and CN female, T21 male and CN male, T21 male and T21 female, and CN male and CN female. (c) Quantitative volcanic map of differentially expressed proteins in T21 female and CN female group, (d) T21 male and CN male group, (e) T21 male and T21 female group, and (f) CN male and CN female group.
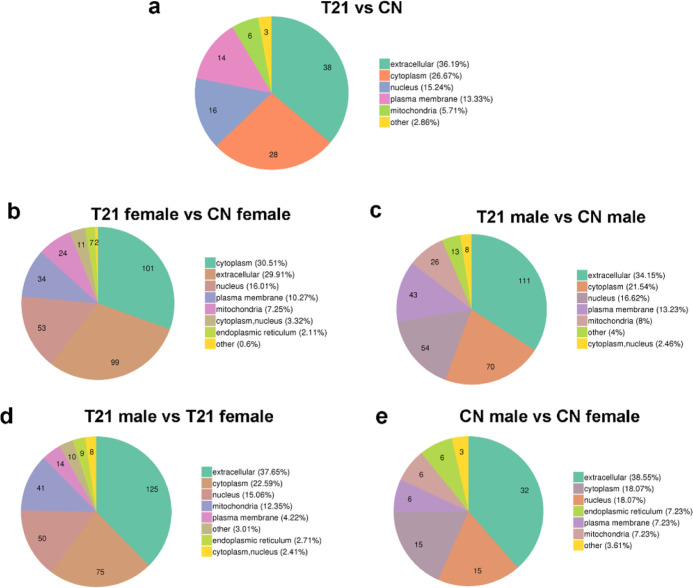
The subcellular localization of the differentially expressed proteins was mainly extracellular (Figure 3a). The cellular component categories of these proteins were cell, organelle, and extracellular region (Figure 2a), and the proteins mainly existed in vesicles, extracellular exosomes, extracellular organelles, and extracellular regions (Figure 6a).
Figure 3.
Subcellular structure and distribution of differentially expressed proteins. (a) T21 fetuses and CN fetuses group, (b) T21 female and CN female group, (c) T21 male and CN male group, (d) T21 male and T21 female group, and (e) CN male and CN female group.
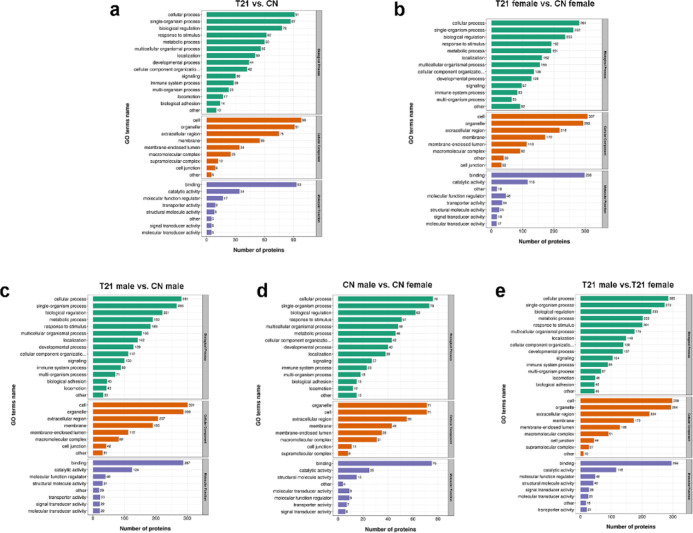
Figure 2.
Gene ontology analysis of the differential proteins. (a) Biological process, cellular component, and molecular function difference of the differential expressed proteins in T21 fetuses and CN fetuses group, (b) T21 female and CN female group, (c) T21 male and CN male group, (d) CN male and CN female group, and (e) T21 male and T21 female group.
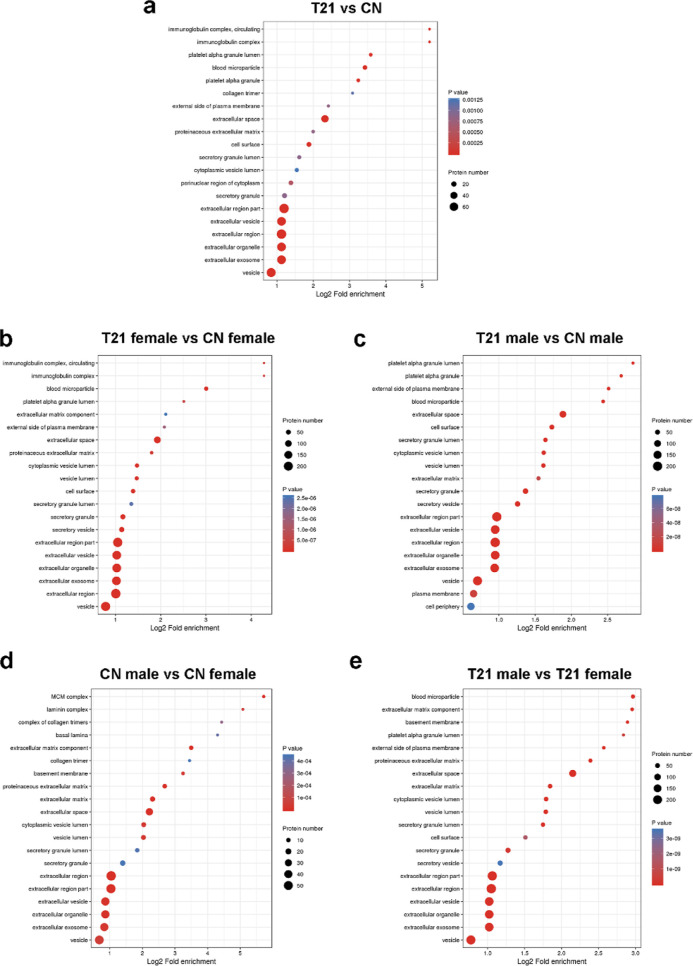
Figure 6.
Bubble diagram of enrichment and distribution of differentially expressed proteins in go functional classification—cellular component. (a) T21 fetuses and CN fetuses group, (b) T21 female and CN female group, (c) T21 male and CN male group, (d) CN male and CN female group, and (e) T21 male and T21 female group.
Dysregulated proteins are involved in many important biological processes, including cellular processes, single-organism processes, biological regulation, responses to stimuli, and metabolic processes (Figure 2a), and these differentially expressed proteins were mainly associated with animal organ development, neurogenesis, regulation of cell development, and positive regulation of cell differentiation (Figure 5a).
Figure 5.
Bubble diagram of enrichment and distribution of differentially expressed proteins in go functional classification—biological process. (a) T21 fetuses and CN fetuses group, (b) T21 female and CN female group, (c) T21 male and CN male group, (d) CN male and CN female group, and (e) T21 male and T21 female group.
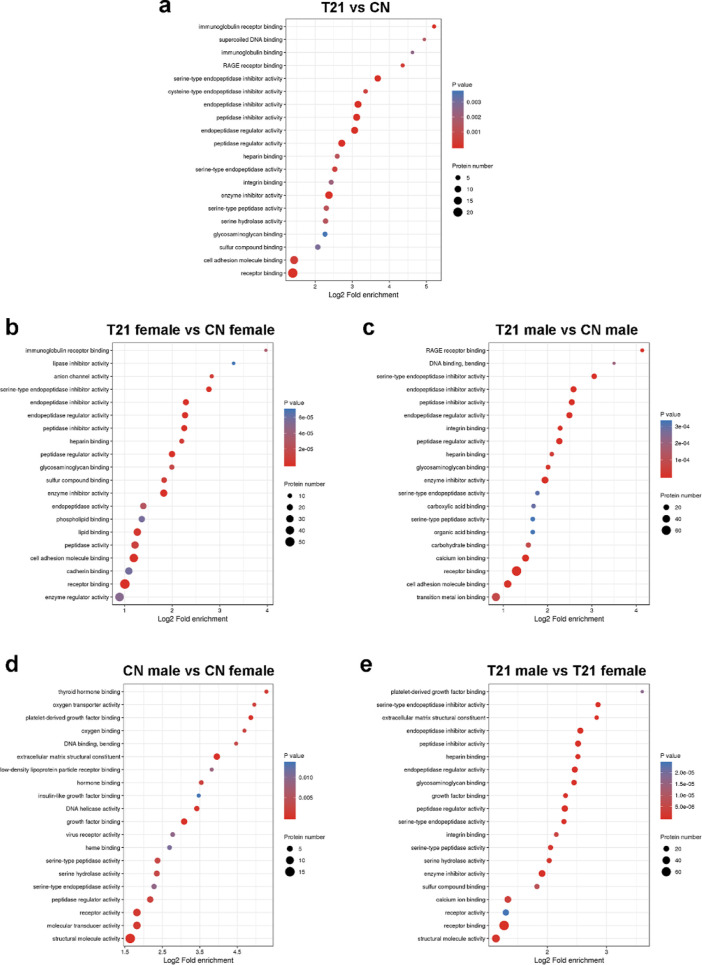
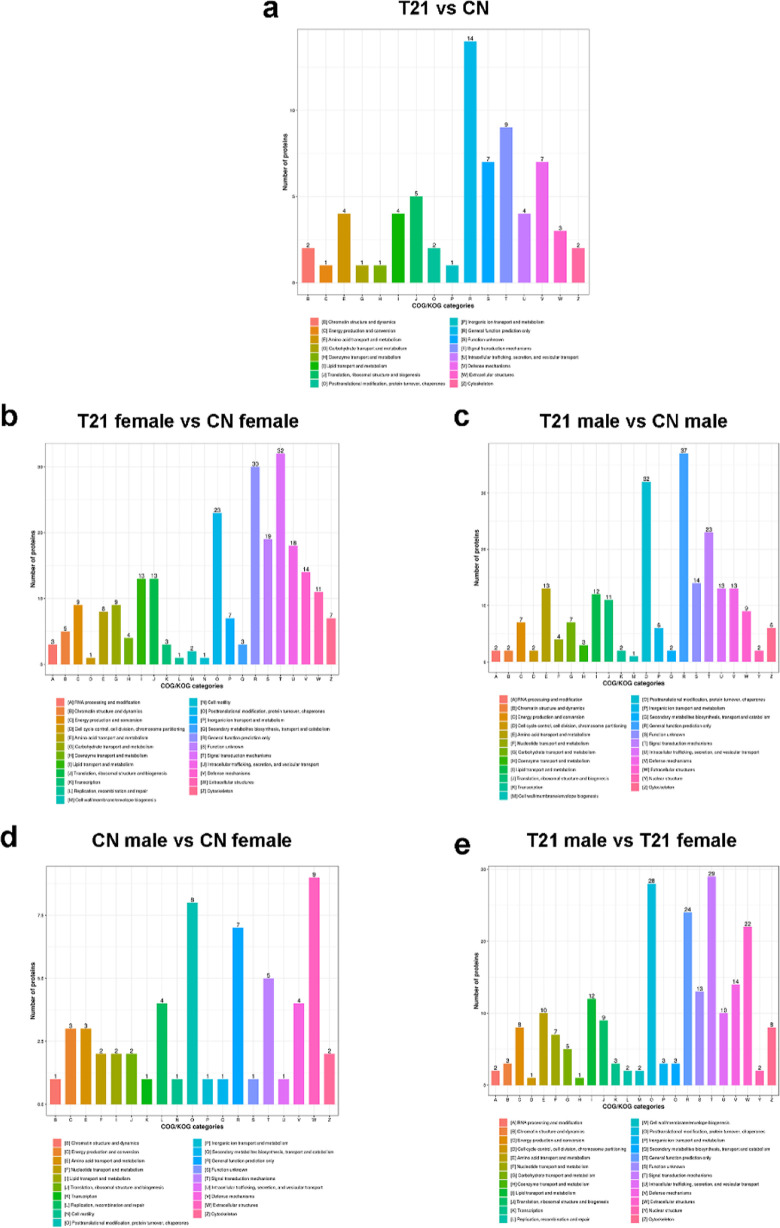
Molecular function-based enrichment results are shown in Figures 3a and 7a, where the differentially expressed proteins participated in binding [including receptor binding, cell adhesion molecule (CAM) binding, receptor for advanced glycated endproducts (RAGE) receptor binding], catalytic activity (enzyme inhibitor activity, serine hydrolase activity, peptidase regulator activity, and endopeptidase regulator activity), and molecular function regulator. The study of clusters of orthologous groups of proteins showed the enrichment of differentially expressed proteins in signal transduction mechanisms and defence mechanisms (Figure 4a).
Figure 7.
Bubble diagram of enrichment and distribution of differentially expressed proteins in go functional classification—molecule function. (a) T21 fetuses and CN fetuses group, (b) T21 female and CN female group, (c) T21 male and CN male group, (d) CN male and CN female group, and (e) T21 male and T21 female group.
Figure 4.
Clusters of orthologous groups of differentially expressed proteins. (a)T21 fetuses and CN fetuses group, (b) T21 female and CN female group, (c) T21 male and CN male group, (d) CN male and CN female group, and (e) T21 male and T21 female group.
In Addition to the Co-alternation Proteins, Identification of More Differentially Expressed Proteins with Important Biological Functions between T21 Males and CN Males Group/T21 Female and CN Females Group
Interestingly, in the T21 male versus CN male groups, except for the co-increased and co-decreased proteins shown above, the expression of other 222 proteins was also significantly different, including 173 proteins with increased expression and 49 proteins with decreased expression only in amniocytes of T21 male fetuses (Figure 1b,c, and Table S2), such as MARCKSL1 (0.749), HMGA2 (0.756), AKAP12 (0.756), HMGB3 (0.65), CRYAB (3.556), PLOD2 (1.876), UAP1 (1.482), AMIGO2 (1.309), CRNN (3.404), and SPRR2D (3.197). The other 230 proteins showed significant differences in levels between T21 females and CN females; the expression level of 84 of these proteins was increased and that of 146 proteins was decreased in amniocytes of T21 female fetuses (Figure 1b,d and Table S3), such as HPX (0.55), APOA1 (0.652), IGHG4 (0.288), PTGDS (0.341), TSPAN1 (1.889), TOP2A (1.888), and MFAP2 (1.806).
From our research, it is clear to see that the protein expression showed sex differences. The subcellular localization of the differentially expressed proteins in the female group was mainly in the cytoplasm, and some of the different proteins were in the endoplasmic reticulum, the differentially expressed proteins in the male group were mainly extracellular, and no proteins were located in the endoplasmic reticulum (Figure 3b,c).
The dysregulated proteins’ biological processes were enriched in cellular processes, single-organism processes, biological regulation, stimuli response, and so on. Additionally, in the T21 male versus CN male group, a few differentially expressed proteins were associated with biological adhesion and locomotion (Figure 2b,c). Meanwhile, differentially expressed proteins in the male group were involved in animal organ development, secretion, neutrophil-mediated immunity, hemostasis regulation, myeloid leukocyte activation, antimicrobial humoral response, skin development, and so on (Figure 5b). Differentially expressed proteins in the female group participated in animal organ development, secretion, neutrophil-mediated immunity, cell migration, and others (Figure 5c).
Molecular function-based enrichment analysis is shown in Figure 2b,c, where the differentially expressed proteins participated in binding, catalytic activity, and molecular function regulator. The specific molecular functions of the two groups were different; the proteins in the female group were associated with immunoglobulin receptor binding, anion channel activity, enzyme regulator activity, and others. The proteins in the male group were associated with RAGE receptor binding, DNA binding, bending, and carboxylic acid binding (Figure 7b,c).
In the cellular component of the category (Figure 2b,c), enrichment included cell, organelle, and extracellular regions. Except for vesicles, extracellular exosomes, extracellular organelles, and others, the cellular components of the differentially expressed proteins in the female group also participated in the immunoglobulin complex, extracellular matrix component, and proteinaceous extracellular matrix (Figure 6b). The cellular components of the differentially expressed proteins in the male group also participated in platelet alpha granules, extracellular matrix, plasma membrane, and cell periphery (Figure 6b).
The study of clusters of orthologous groups showed the enrichment of differentially expressed proteins between T21 females versus CN females and T21 males versus CN males in signal transduction mechanisms, general function prediction only, post-translational modification, protein turnover, and chaperones (Figure 4b,c).
Of note, these dysregulation proteins were involved in neurodevelopment (Figure S1), growth (Figure S2), heart development (Figure S3), hematopoietic (Figure S4), immunity (Figure S5), and reproduction (Figure S6).
More Differentially Expressed Proteins Identified in T21 Male versus T21 Female than CN Male versus CN Female Group
In the CN male versus CN female group, only 83 proteins showed highly significant differential expression. In contrast to CN females, there were 46 proteins with increased expression and 37 proteins with decreased expression in CN males (Figure 1b).
Compared with T21 females, the increased proteins were 121 in T21 males, and only 27 proteins were co-upregulated in both T21 males versus T21 females group and CN males versus CN females (Tables S4–S6). Meanwhile, the decreased proteins were 210 in T21 males, and only 16 proteins were co-downregulated in both T21 males versus T21 females group and CN males versus CN females (Tables S4–S6). The differential protein expression between males and females was more obvious in the T21 group (Figure 1e,f). In T21 male and CN male, compared with T21 female and CN female, the variation trend of some proteins is consistent, such as TACSTD2 (0.515/0.758), MISP (0.518/0.743), PALLD (0.53/0.752), AKR1C1 (1.998/2.216), ADIRF (1.547/1.398), and others. TMSB4X, which escapes X inactivation, is involved in cell proliferation, migration, and differentiation. The CN male/CN females ratio was 0.419, and the expression of TMSB4X in males was about half that in women, conforming to the escape of X chromosome inactivation. The expression of TMSB4X significantly decreased only in T21 females but not in T21 males.
The subcellular localization of the differentially expressed proteins in the two groups was mainly extracellular, and some of the different proteins were located in the endoplasmic reticulum only in the T21 group (Figure 3d,e).
The biological processes of the dysregulated proteins were investigated (Figure 2d,e), and they were found to be enriched in cellular processes, single-organism processes, biological regulation, and responses to stimuli in both groups. The biological processes involved in the differentially expressed proteins in the two groups were significantly different. In the CN group, the differentially expressed proteins participated in circulatory system development, regulation of cell migration, cardiovascular system development, regulation of cell motility, and so on. In the T21 group, the differentially expressed proteins participated in animal organ development, secretion, and response to bacteria (Figure 5d,e).
Molecular function-based enrichment results are shown in Figure 2d,e, where the differentially expressed proteins participated in binding, catalytic activity, and molecular function regulator. The differentially expressed proteins of the CN group were associated with structural molecule activity, molecular transducer activity, receptor activity, and others (Figure 7d). The differentially expressed proteins in the T21 group were associated with structural molecule activity, receptor binding, calcium ion binding, enzyme inhibitor activity, and others (Figure 7e).
In the cellular component category (Figure 2d,e), enrichment included cell, organelle, extracellular region, and others. Except for vesicles, extracellular exosomes, extracellular organelles, and others, the cellular components of the differentially expressed proteins in the CN group also participated in the minichromosome maintenance (MCM) complex, laminin complex, complex of collagen trimers, and others (Figure 6d). The cellular components of the differentially expressed proteins in the T21 group also participated in blood microparticles, basement membrane, platelet alpha granule lumen, and others (Figure 6e).
In the T21 male and T21 female groups, the study of clusters of orthologous groups of proteins showed the enrichment of differentially expressed proteins in post-translational modification, protein turnover, chaperones, signal transduction mechanisms, and general function prediction only (Figure 4e). The differentially expressed proteins were enriched in the extracellular structures (Figure 4d).
Human development-based enrichment analysis showed that these dysregulation proteins were involved in neurodevelopment (Figure S7), growth (Figure S8), heart development (Figure S9), hematopoietic (Figure S10), immunity (Figure S11), and reproduction (Figure S12).
Discussion
Several differentially expressed proteins between T21 female versus CN female, T21 male versus CN male, T21 female versus T21 male, and CN female versus CN male were identified in our study. The results suggested that the protein expression patterns of T21 males and T21 females were significantly different. The subcellular localization, biological processes, cellular components, and molecular functions of the proteins were different. These differential proteins are related to the processes of heart development, hematopoiesis, immunity, neural development, and reproduction.
Cho’s11 research revealed that over 900 proteins were dysregulated in amniocytes of T21. The changing trend of some proteins is consistent with that in our study, such as the upregulated proteins, CRYAB, PLOD2, UAP1, and AMIGO2, and the downregulated proteins, HPX, APOA1, MARCKSL1, HMGA2, and AKAP12. However in our study, HPX and APOA1 expression varied only in T21 females, while other differentially expressed proteins were only found in T21 males. Therefore, gender differences should be taken into account when studying differential protein expression. Our findings differ slightly from those of previous studies,11 which could be due to a variety of factors, the populations recruited, the experimental methods used, and the gender of the participants were all different.
Our study found that APP, a chromosome 21 gene that codes for amyloid precursor protein, was significantly upregulated in T21 male and T21 female fetuses. Virtually, all adults with DS show neuropathological changes in Alzheimer’s disease (AD) by the age of 40 years.12 This association is partially due to the overexpression of the amyloid precursor protein. Previous studies reported that increased expression of APP might drive the development of AD in individuals with DS by increasing the levels of amyloid-β (Aβ).
GC, which encodes a vitamin D binding protein, is downregulated in T21 male and T21 female amniocytes. The protein belongs to the albumin gene family. It is a multifunctional protein found in plasma, ascitic fluid, cerebrospinal fluid, and on the surface of many cell types. It binds to vitamin D and its plasma metabolites and transports them to the target tissues. DS patients may develop reduced bone mass accrual, predisposing them to fragility, fractures, and osteoporosis. Stagi et al. demonstrated a very high prevalence of vitamin D deficiency in different age groups of patients with DS.17 In DS individuals, vitamin D supplementation did not appear to be sufficient, even if 25(OH)D levels increased significantly after supplementation. In addition to abnormal bone development, vitamin D deficiency has been associated with immune system abnormalities and cardiovascular disease.13 The decreased expression of vitamin D-binding protein may be a critical reason for DS patients’ decreased vitamin D levels.
SHTN1, which encodes shootin-1, was downregulated in both T21 male and T21 female amniocytes. This protein is a key molecule involved in neuronal polarization and axon outgrowth, accumulates at the leading edge of axonal growth cones, and mediates the mechanical coupling between F-actin retrograde flow and the CAM L1-CAM.14 Hippocampal neurons display reduced axon length in Ts65Dn mouse brains.15 These results suggest that SHTN1 downregulation may be a critical reason for abnormal axon development.
A1BG was downregulated in both T21 males and T21 females, and previous studies have shown that loss of A1BG leads to cardiac defects in females but not in males. Congenital heart defects, particularly atrioventricular septal defects (AVSD), are more common in female patients with DS.16 These results suggest that A1BG is associated with heart disease in patients with DS.
TMSB4X has a homologue on chromosome Y and escapes X inactivation, according to GenBank. This gene escapes X inactivation and encodes an action sequestering protein that plays a role in regulating action polymerization. The protein is also involved in cell proliferation, migration, and differentiation. The CN male/CN females ratio was 0.419, and the expression of TMSB4X in males was about half that in women, conforming to the escape of X chromosome inactivation. The expression of TMSB4X significantly decreased only in T21 females but not in T21 males, implying that Trisomy 21 may affect the expression of genes that are not inactivated on the X chromosome.
Five candidate proteins (CEL, CPA1, MUC13, CLCA1, MUC5AC, and AFP) were significantly downregulated in the DS amniotic fluid samples. This trend is similar to what we found in amniotic fluid cells of both T21 males and T21 females, with the following ratios: CEL (0.518 and 0.422), CPA1 (0.549 and 0.298), CLCA1 (0.788 and 0.685), MUC5AC (0.653 and 0.635), and AFP (0.665 and 0.435). The extracellular protein AFP was used as a biomarker for DS serum screening, similar to AFP, and the other four proteins were extracellular proteins (Table S1). These proteins have potential as biomarkers for DS.
The ER is the largest organelle in the cell. It is an important protein synthesis and transport site, protein folding, lipid and steroid synthesis, carbohydrate metabolism, and calcium storage. The multifunctional nature of this organelle requires a myriad of proteins, unique physical structures, and coordination with and response to changes in the intracellular environment. The extra 21 chromosomes dysregulated the ER proteins in T21 females, suggesting that endoplasmic reticulum dysfunction may be associated with the onset of T21 females.
In addition to the above proteins, other proteins identified in our study may be involved in the occurrence of DS, and our results provide a possibility for further exploration of the pathogenesis of DS. Till date, we have found no proteomic study of DS amniotic fluid cells based on Gene Ontology (GO) analysis. Our study reveals a variety of biological processes involved in the pathogenesis of DS through proteomic GO analysis for the first time, providing rich information for future research on the molecular mechanism of the pathogenesis of DS. In the future, based on the information obtained from our GO analysis, it will be important and meaningful to study the specific biological processes involved in DS.
Conclusions
In summary, we report amniocyte proteomics in T21 fetuses. Our results showed sex-specific modulation of protein expression and biological processes. Comprehensive proteomic profiling analysis would provide new insights into sex-specific differences in the pathogenesis of DS. Our results suggest differences in clinical manifestations between T21 males and T21 females and provide clues for personalized diagnosis and treatment of DS.
Acknowledgments
This research was supported by grants from the Sichuan Science and Technology Program (nos. 2019YFS0350, 2020ZYD007, and 2021YFS0026); the National Natural Science Foundation of China (nos. 82271692, 81974226, 81974365, and 81670346); the Natural Science Foundation of Sichuan Province (2022NSFSC0782); and the Fundamental Research Funds for the Central Universities (SCU2022F4080).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c05152.
Differentially expressed proteins associated with neuro development in T21 male versus CN male group and T21 female versus CN female group; differentially expressed proteins associated with growth in T21 male versus CN male group and T21 female versus CN female group; differentially expressed proteins associated with cardiac development in T21 male versus CN male group and T21 female versus CN female group; differentially expressed proteins associated with hematopoietic in T21 male versus CN male group and T21 female versus CN female group; differentially expressed proteins associated with immune in T21 male versus CN male group and T21 female versus CN female group; differentially expressed proteins associated with reproduction in T21 male versus CN male group and T21 female versus CN female group; differentially expressed proteins associated with neuro development in T21 male versus T21 male group, CN male and CN female group; differentially expressed proteins associated with growth in T21 male versus T21 male group, CN male and CN female group; differentially expressed proteins associated with cardiac development in T21 male versus T21 male group, CN male and CN female group; differentially expressed proteins associated with hematopoietic in T21 male versus T21 male group, CN male and CN female group; differentially expressed proteins associated with immune in T21 male versus T21 male group, CN male and CN female group; and differentially expressed proteins associated with reproduction in T21 male versus T21 male group, CN male and CN female group (PDF)
Differential protein expression in amniocytes between T21 fetuses and CN fetuses; differential protein expression in amniocytes between T21 male and CN male fetuses; differential protein expression in amniocytes between T21 female and CN female fetuses; differential protein expression in amniocytes between T21 male versus T21female, CN male versus CN female; differential protein expression in amniocytes between CN male and CN female; differential protein expression in amniocytes between T21 male and T21 female; and differential expression pattern of genes on X chromosome (XLSX)
Author Contributions
H.S. and Y.L. conceived and designed the experiments; H.S., Y.L., and Z.L. performed the experiments; X.Z., H.Z., L.Z., and J.C. collected clinic samples; H.S., Y.L., H.W., and B.Z. analyzed the data; S.L. coordinated the project; and H.S. and Y.L. wrote the paper.
The authors declare no competing financial interest.
Supplementary Material
References
- Bull M. J. Down Syndrome. N. Engl. J. Med. 2020, 382, 2344–2352. 10.1056/NEJMra1706537. [DOI] [PubMed] [Google Scholar]
- Weijerman M. E.; van Furth A. M.; Vonk Noordegraaf A.; van Wouwe J. P.; Broers C. J.; Gemke R. J. Prevalence, neonatal characteristics, and first-year mortality of Down syndrome: a national study. J. Pediatr. 2008, 152, 15–19. 10.1016/j.jpeds.2007.09.045. [DOI] [PubMed] [Google Scholar]
- a Antonarakis S. E.; Lyle R.; Dermitzakis E. T.; Reymond A.; Deutsch S. Chromosome 21 and down syndrome: from genomics to pathophysiology. Nat. Rev. Genet. 2004, 5, 725–738. 10.1038/nrg1448.. [DOI] [PubMed] [Google Scholar]; b Lott I. T.; Head E. Alzheimer disease and Down syndrome: factors in pathogenesis. Neurobiol. Aging 2005, 26, 383–389. 10.1016/j.neurobiolaging.2004.08.005. [DOI] [PubMed] [Google Scholar]
- a Nistor M.; Don M.; Parekh M.; Sarsoza F.; Goodus M.; Lopez G. E.; Kawas C.; Leverenz J.; Doran E.; Lott I. T.; et al. Alpha- and beta-secretase activity as a function of age and beta-amyloid in Down syndrome and normal brain. Neurobiol. Aging 2007, 28, 1493–1506. 10.1016/j.neurobiolaging.2006.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Lai F.; Williams R. S. A prospective study of Alzheimer disease in Down syndrome. Arch. Neurol. 1989, 46, 849–853. 10.1001/archneur.1989.00520440031017. [DOI] [PubMed] [Google Scholar]; c Baban A.; Olivini N.; Cantarutti N.; Calì F.; Vitello C.; Valentini D.; Adorisio R.; Calcagni G.; Alesi V.; Di Mambro C.; Villani A.; Dallapiccola B.; Digilio M. C.; Marino B.; Carotti A.; Drago F.; et al. Differences in morbidity and mortality in Down syndrome are related to the type of congenital heart defect. Am. J. Med. Genet., Part A 2020, 182, 1342–1350. 10.1002/ajmg.a.61586. [DOI] [PubMed] [Google Scholar]; d Lanzillotta C.; Greco V.; Valentini D.; Villani A.; Folgiero V.; Caforio M.; Locatelli F.; Pagnotta S.; Barone E.; Urbani A.; et al. Proteomics Study of Peripheral Blood Mononuclear Cells in Down Syndrome Children. Antioxidants 2020, 9, 1112. 10.3390/antiox9111112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carsetti R.; Valentini D.; Marcellini V.; Scarsella M.; Marasco E.; Giustini F.; Bartuli A.; Villani A.; Ugazio A. G. Reduced numbers of switched memory B cells with high terminal differentiation potential in Down syndrome. Eur. J. Immunol. 2015, 45, 903–914. 10.1002/eji.201445049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel J. O.; Tirosh-Wagner T.; Urban A. E.; Chen X. N.; Kasowski M.; Dai L.; Grubert F.; Erdman C.; Gao M. C.; Lange K.; et al. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 12031–12036. 10.1073/pnas.0813248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. P.; Lai T. C.; Chern S. R.; Li S. H.; Chou H. C.; Chen Y. W.; Lin S. T.; Lu Y. C.; Wu C. L.; Li J. M.; et al. Proteome differences between male and female fetal cells in amniotic fluid. OMICS 2013, 17, 16–26. 10.1089/omi.2010.0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Shi W.; Sheng X.; Dorr K. M.; Hutton J. E.; Emerson J. I.; Davies H. A.; Andrade T. D.; Wasson L. K.; Greco T. M.; Hashimoto Y. Cardiac proteomics reveals sex chromosome-dependent differences between males and females that arise prior to gonad formation. Dev. Cell 2021, 56, 3019–−3034.e7. 10.1016/j.devcel.2021.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Zhao R.; Chen X.; Ma W.; Zhang J.; Guo J.; Zhong X.; Yao J.; Sun J.; Rubinfien J.; Zhou X.; et al. A GPR174-CCL21 module imparts sexual dimorphism to humoral immunity. Nature 2020, 577, 416–420. 10.1038/s41586-019-1873-0. [DOI] [PubMed] [Google Scholar]; c McCarthy M. M.; Arnold A. P.; Ball G. F.; Blaustein J. D.; De Vries G. J. Sex differences in the brain: the not so inconvenient truth. J. Neurosci. 2012, 32, 2241–2247. 10.1523/JNEUROSCI.5372-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Jazin E.; Cahill L. Sex differences in molecular neuroscience: from fruit flies to humans. Nat. Rev. Neurosci. 2010, 11, 9–17. 10.1038/nrn2754. [DOI] [PubMed] [Google Scholar]
- a Underwood M. A.; Gilbert W. M.; Sherman M. P. Amniotic fluid: not just fetal urine anymore. J. Perinatol. 2005, 25, 341–348. 10.1038/sj.jp.7211290. [DOI] [PubMed] [Google Scholar]; b Kim J.; Lee Y.; Kim H.; Hwang K. J.; Kwon H. C.; Kim S. K.; Cho D. J.; Kang S. G.; You J. Human amniotic fluid-derived stem cells have characteristics of multipotent stem cells. Cell Proliferation 2007, 40, 75–90. 10.1111/j.1365-2184.2007.00414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J.; Chen T.; Wu S.; Yang C.; Bai M.; Shu K.; Li K.; Zhang G.; Jin Z.; He F.; et al. iProX: an integrated proteome resource. Nucleic Acids Res. 2019, 47, D1211–D1217. 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C. K. J.; Drabovich A. P.; Karagiannis G. S.; Martínez-Morillo E.; Dason S.; Dimitromanolakis A.; Diamandis E. P. Quantitative proteomic analysis of amniocytes reveals potentially dysregulated molecular networks in Down syndrome. Clin. Proteomics 2013, 10, 2. 10.1186/1559-0275-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott I. T.; Head E. Dementia in Down syndrome: unique insights for Alzheimer disease research. Nat. Rev. Neurol. 2019, 15, 135–147. 10.1038/s41582-018-0132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagi S.; Lapi E.; Romano S.; Bargiacchi S.; Brambilla A.; Giglio S.; Seminara S.; de Martino M. Determinants of vitamin D levels in children and adolescents with Down syndrome. Int. J. Endocrinol. 2015, 2015, 896758. 10.1155/2015/896758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozmus D.; Ciesielska A.; Płomiński J.; Grzybowski R.; Fiedorowicz E.; Kordulewska N.; Savelkoul H.; Kostyra E.; Cieślińska A. Vitamin D Binding Protein (VDBP) and Its Gene Polymorphisms-The Risk of Malignant Tumors and Other Diseases. Int. J. Mol. Sci. 2020, 21, 7822. 10.3390/ijms21217822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo Y.; Baba K.; Toriyama M.; Minegishi T.; Sugiura T.; Kozawa S.; Ikeda K.; Inagaki N. Shootin1-cortactin interaction mediates signal-force transduction for axon outgrowth. J. Cell Biol. 2015, 210, 663–676. 10.1083/jcb.201505011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S.; Watts C. A.; Chung W. C. J.; Welshhans K. Neurodevelopmental wiring deficits in the Ts65Dn mouse model of Down syndrome. Neurosci. Lett. 2020, 714, 134569. 10.1016/j.neulet.2019.134569. [DOI] [PubMed] [Google Scholar]
- Diogenes T. C. P.; Mourato F. A.; de Lima Filho J. L.; Mattos S. D. S. Gender differences in the prevalence of congenital heart disease in Down’s syndrome: a brief meta-analysis. BMC Med. Genet. 2017, 18, 111. 10.1186/s12881-017-0475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.