Abstract
The Aurora kinases, including Aurora A, B and C, play critical roles in cell division. They have been found overexpressed in a number of types of cancer and may thus be potential targets in cancer therapy. Several Aurora kinase inhibitors have been identified and developed. Some of these have been used in clinical trials and have exhibited certain efficacy in cancer treatment. However, none of these has yet been applied clinically due to the poor outcomes. Oxostephanine is an aporphine alkaloid isolated from several plants of the genus Stephania. This compound has been reported to inhibit Aurora kinase activity in kinase assays and in cancer cells. The present study aimed to investigate the real-time effects of oxostephanine extracted from Stephania dielsiana Y.C. Wu leaves on the growth of an ovarian cancer cell line (OVCAR-8, human ovarian carcinoma); these effects were compared to those of the well-known Aurora kinase inhibitor, VX-680. The effects of oxostephanine on stromal cells, as well as endothelial cells were also examined. The results demonstrated that oxostephanine was an Aurora kinase inhibitor through the prevention of histone H3 phosphorylation at serine 10, the mislocalization of Aurora B and the induction of aneuploidy. Moreover, this substance was selectively cytotoxic to human umbilical vein endothelial cells (hUVECs), whereas it was less cytotoxic to human fibroblasts and umbilical cord-derived mesenchymal stem cells. In addition, this compound significantly attenuated the migration and tube formation ability of hUVECs. Taken together, the present study demonstrates that oxostephanine plays dual roles in inhibiting Aurora kinase activity and angiogenesis. Thus, it may have potential for use as a drug in cancer treatment.
Keywords: Aurora kinases, Aurora kinase inhibitor, ovary cancer cell line, angiogenesis, endothelial cells, growth factors
Introduction
The Aurora kinases, including Aurora A, B and C, are serine/threonine kinases that play a central role in regulating cell division and multiple signaling pathways. Aurora A functions in the formation of a typical bipolar spindle (1), the maturation of centrosomes, which is necessary for G2/M transition (2), and the formation and stimulation of the cyclin B-CDK1 complex (3). Moreover, Aurora A helps to increase both size and microtubule-nucleating capacity just before mitotic entry (3). Aurora B plays a function in the chromosome biorientation on the mitotic spindle. It mediates the attachment of the microtubule to the kinetochores and regulates the spindle assembly checkpoint (SAC) (4,5). The improper attachment of kinetochores promotes Aurora B to recruit and phosphorylate its substrates at the kinetochores to depolymerize the uncorrected attachment, allowing other microtubules to capture the unattached kinetochores. The inhibition of Aurora B can impair the chromosome arrangement at the mitotic spindle equator (6).
Furthermore, Aurora B phosphorylates histone H3 at the serine 10 (H3S10ph) residue at the beginning of the prophase and leads to a peak in H3S10ph at the prometaphase and metaphase. This phosphorylation contributes to the active chromosome conformation at the entry of mitosis (7). Other studies have reported that H3S10ph may involve chromosome condensation and Aurora B recruitment to the centromere (8,9). Most notably, Aurora B is the only enzymatic member of the chromosomal passenger protein complex (CPC). All members of CPC share the co-localization during mitosis: They concentrate in the kinetochore during the prophase, prometaphase and metaphase; transfer to the midzone with anaphase onset; and remain in the midbody in telophase and cytokinesis (10). The mislocalization of any CPC members, including Aurora B, can lead to a defection in mitosis and cytokinesis (10,11). Apart from the pivotal functions in cell division, Aurora A and B kinases are also involved in tumor angiogenesis. These enzymes phosphorylate MYCN, regulate vascular endothelial growth factor (VEGF) production, and inhibit the proliferation and tube formation of human endothelial cells (12-14). Aurora C kinase has been found in cells that undergo meiosis and has a unique physiological role in spermatogenesis (15). The limitation in understanding the role of Aurora C may stem from the high sequence homology between this kinase and Aurora B, leading to the overlapping in the function of these proteins (16). Aurora C can rescue the genetic stability of the cells in case Aurora B is absent (17). Previously, it was demonstrated that the overexpression of Aurora C induces abnormal cell division, resulting in centrosome amplification and multinucleation in cells (17).
The overexpression of Aurora kinases has been observed in a broad range of human solid tumors, such as gliomas, and colorectal, breast, ovarian and pancreatic cancer (18), as well as in liquid tumors such as diffuse large B-cell lymphoma (19). Moreover, Aurora kinases have been found to be associated with genetic instability and aneuploidy in tumors (20). Hence, it is not surprising that Aurora kinases have become attractive targets in cancer treatment. The development of Aurora inhibitors has drawn the attention of several scientists from academic institutes and pharmaceutical companies. Over the first two decades of the 21st century, a series of Aurora kinase inhibitors were produced, which were Aurora A- or B-selective, or pan inhibitors. Although these compounds exhibit preclinical and clinical efficacy, no Aurora kinase inhibitor has yet been approved for clinical use due to their poor outcomes (18). Thus, there is an urgent need for the identification of novel small molecule inhibitors.
Oxostephanine is a substance belonging to the group of aporphine alkaloids isolated from several plants of the genus Stephania. Previous studies have demonstrated that this substance exerts a potent cytotoxic effect on several cancer cell lines, such as KB (human epithelial carcinoma), HepG2 (human hepatocellular carcinoma), GLC4/Adr (human small cell lung adriamycin-resistant carcinoma), K562 (human chronic myelogenous leukemia) and K562/Adr (human chronic myelogenous leukemia resistant to adriamycin) (21), whereas it has a minimal toxic effect on normal cells (MRC-5; human fetal lung fibroblasts) (22). In addition, oxostephanine has been shown to exhibit potent activity against breast cancer cells and MOLT-3 acute lymphoblastic leukemia cells (21). Moreover, Knockleby et al (23) revealed that oxostephanine inhibited the activity of Aurora kinase A and B by the competition of ATP binding sites in an in vitro kinase assay.
The aim of the present study was to examined the effects of oxostephanine extracted from Vietnamese Stephania dielsiana Y.C. Wu (S. dielsiana) as a novel Aurora kinase inhibitor on an ovarian cancer cell line (OVCAR-8). As demonstrated herein, S. dielsiana may prove to be a potent Aurora kinase inhibitor, as well as an anti-angiogenic agent with potential to be developed into an anticancer drug.
Materials and methods
Compound preparation
The stems and leaves of S. dielsiana were collected in Ba Vi District, Hanoi, Vietnam in October, 2019 and identified by the Department of Botany, Hanoi University of Pharmacy, Hanoi, Vietnam. A voucher specimen (no. SD10/2019) has been deposited at the Department of Botany and Pharmacognosy, Vietnam University of Traditional Medicine, Hanoi, Vietnam. The process used for the isolation and characterization of oxostephanine from the leaves of S. dielsiana in Vietnam has been previously published (22,23). In brief, the leaves of S. dielsiana (7 kg) were extracted with 95% MeOH (3×15 liters, 3 days each) at room temperature. The extracts were concentrated in vacuo to yield a MeOH extract (680 g), which was suspended in H2O (2.5 liters) and adjusted to pH 3 with 10% HCl. The acidic aqueous phase was filtered off. The filtrate was loaded on ion-exchange resin, eluted with 20% MeOH until the eluate approached colorless to give the nonalkaloid parts, and then eluted with 2% NaOH in 65% MeOH solution (five-fold of retention volume) to yield the crude total alkaloids. The alkaloid-containing solution was acidified to pH 5 with 10% HCl and partitioned with EtOAc (3×2 liters) to yield the EtOAc extract (65 g).
The EtOAc-soluble portion was subjected to silica gel column chromatography eluted with gradient systems of CH2Cl2-MeOH (100:0, 100:10, 100:30 and 100:50, v/v). The eluted fractions were evaluated and pooled according to thin layer chromatography (TLC) analysis, resulting in six major fractions (SDE.1-SDE.6). The purification of SDE.6 over Sephadex LH-20 (100% MeOH) was performed using the same methodology, and subsequent preparative TLC, eluted with CH2Cl2-MeOH (20:1) yielded oxostephanine (8.6 mg). The purification of oxostephanine by repeating recrystallization in a mixture of methanol and ethanol yielded pure oxostephanine compound as an amorphous yellow-orange powder (purity 99.0% as a percentage of the peak area using a HPLC-DA system (Agilent 1260 Infinity II; Agilent Technologies, Inc.).
Cell lines and culture
OVCAR-8 (human ovarian carcinoma-8) and HeLa (Aurora B-GFP) cells were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.). Human dermal fibroblasts (hFBs) were cultured in DMEM/F12 medium (Gibco; Thermo Fisher Scientific, Inc.). The media were supplemented with 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc.), 100 units/ml penicillin and 100 µg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.). Human umbilical vein endothelial cells (hUVECs) were cultured in EBM-2 medium (Lonza Group, Ltd.). Umbilical cord-derived mesenchymal stem cells (UC-MSCs) were grown on the surface of culture flasks coated by CELLstart™ CTS™ (CELLstart) in StemMACS™ MSC Expansion medium (StemMACS) (Miltenyi Biotec). All the cells were cultured in an incubator at 37°C with 5% CO2. The hUVECs, hFBs and UC-MSCs were provided by Vinmec Research Institute of Stem cell and Gene Technology, and they were not immortalized cell lines. The protocols for cell isolation were approved by the Ethics Committee of Vinmec International Hospital (Document no. 40/2020/QD-Vinmec for hUVECs and UC-MSCs, signed and dated on December 24, 2020; Document no. 311/2018/QD-Vinmec for hFBs, signed and dated on September 11, 2018). The HeLa (Aurora B-GFP) cells were kindly provided as a gift from Professor Stefan Dimitrov at Institute Albert Bonniot (present name is Institute for Advanced Biosciences) (11,24).
Cell viability assay
Cell viability was assessed using sulforhodamine B (SRB) assay. The cells were seeded at a density of 3×103 cells/well in 96-well plates and incubated with oxostephanine for 24, 48 and 72 h at six concentrations differed by five from the highest of 25 to 5, 1, 0.2 and 0.04 µM. Subsequently, the medium was removed, and the cells were stained with 4% SRB (Millipore, Sigma) for 10 min at room temperature after fixing with 10% TCA (MilliporeSigma) for 1 h at 4°C. The absorbance was measured at 540 nm using a microplate reader (BioTech Power Wave XS; BioTek Instruments, Inc.).
Real-time analysis of cell proliferation using the xCELLigence system
The proliferation assay was performed using the xCelligence system (ACEA Biosciences; Agilent Technologies, Inc.). Media (100 µl/well) were added to each 96-well of an E-plate (ACEA Biosciences; Agilent Technologies, Inc.) to take the background reading for 15 min. In the meantime, the cells were resuspended in medium, and 80 µl cell suspension were added to yield a cell density of 3×103 cells/180 µl/well. Following incubation for 30 min at room temperature, the E-plate was placed into the RTCA SP station in an incubator. After 24 h, the cells were treated with oxostephanine (125, 25, 5, 1 and 0.2 µM) and VX-680 (Vertex and Merck; 25, 5, 1, 0.2 and 0.04 µM). Dynamic cell proliferation was monitored in 30-min intervals from the seeding point till the end of the experiment with a total of >200 h. The electrical impedance was measured using RTCA-integrated software of the xCEL-Ligence system as a dimensionless parameter termed cell index (CI). Normalized CI values were used to obtain the IC50 values, doubling times and other evaluations.
Immunofluorescence
The cells were grown on glass coverslips for 24 h before being treated with either oxostephanine (5 µM) or VX-680 (0.2 µM) with or without paclitaxel (0.035 µM; Millipore, Sigma) and incubated for 15 h in an incubator at 37°C with 5% CO2. Paclitaxel was used to synchronize the cells to the M phase in the cell cycle, in order to obtain dividing cells. The cells were then fixed with 4% paraformaldehyde and 2% sucrose for 15 min at 37°C, permeabilized with 0.2% Triton X-100 for 10 min, blocked with 5 mg/ml BSA, and incubated with primary antibodies for 2 h at room temperature. Phosphorylated histone H3 was detected using a polyclonal rabbit antibody (ab183626, Abcam), at a dilution of 1:500. Aurora B was detected using mouse monoclonal antibodies (36-5200, Invitrogen; Thermo Fisher Scientific, Inc.), at a dilution of 1:250. DNA was visualized with 5 µg/ml Hoechst 33342 (Invitrogen; Thermo Fisher Scientific, Inc.) or 2 µg/ml propidium iodide (PI; Thermo Fisher Scientific, Inc.). Images were collected using a ZEISS 510 Laser Scanning Confocal (LSM) microscope with 40X or 63X objectives (Carl Zeiss AG). For the HeLa (Aurora B-GFP), the cells were grown on a Lab-Tek chamber coverglass (Nalge Nunc International). Following 24 h of treatment with the compounds at concentrations of oxostephanine (5 µM) or VX-680 (0.2 µM), cells were observed without fixing.
As regards the cell nuclear morphological examination, the cells were incubated with either oxostephanine (5 µM) or VX-680 (0.2 µM) for 48 h. The cells were then fixed with 4% paraformaldehyde and 2% sucrose for 15 min at 37°C, permeabilized with 0.2% Triton X-100 for 10 min and stained with 5 µg/ml Hoechst 33342. Following incubation for 15 min, the cells were collected, washed with phosphate-buffered saline (PBS; Millipore, Sigma), and observed using a LSM microscope. Images were analyzed using LSM Image Browser (Carl Zeiss AG).
Apoptosis assay
Apoptosis assay was performed using the Alexa Fluor 488 Annexin V/dead cell apoptosis kit (Invitrogen; Thermo Fisher Scientific, Inc.). As mentioned in the kit, Annexin V is a phospholipid binding protein, and it specifically binds to negatively charged phosphatidylserine molecules exposure on the surface of apoptotic cells. Following treatment of the cells with either 0.5 µM oxostephanine or 0.2 µM VX-680 for 48 h, the cells were harvested and prepared for apoptosis analysis. Briefly, the cells were washed with PBS, then suspended in Annexin-binding buffer to obtain a density of 106 cells/ml. The cell solution was then incubated with 5 µl Alexa Fluor® 488-Annexin V and 100 µl PI working solution for 15 min at room temperature. Subsequently, 400 µl Annexin-binding buffer were gently mixed into the solution with and the cell solution was analyzed on a FACS Canto II System (BD Biosciences). For the visualization of apoptotic marker expression, following 24 h of treatment with the compounds, the cells were incubated with Alexa Fluor® 488-Annexin V for 30 min and observed under a LSM microscope.
Multicellular tumor spheroid assay
OVCAR-8 spheroids were created using the hanging drop method as previously described (25). A total of 15 µl of the medium that contained 5×103 cells were added to each circle on the inverted cover of a 96-well plate to create one spheroid. The cover was then placed upside down on the plate coated with sterile agarose 1.5% (w/v) containing 200 µl complete medium. Following 48 h of incubation in a humidified chamber with 5% CO2 at 37°C, spheroids were transferred from the cover into each well of the agarose-coated plate and further cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.). Spheroids were treated with oxostephanine under two conditions: i) The compound was added to the cell preparation before making the hanging drop; and ii) the compound was added after transferring the formed spheroids into the culture wells. Two concentrations at 5 and 1 µM of Oxostephanine were used in both conditions. Images were obtained using an Axiovert 40CFL microscope (Carl Zeiss AG) with Powershot G9 camera. These images were analyzed using Axio version 4.5 software (Carl Zeiss AG) to determine the spheroid diameter. The approximated volume (V) of each spheroid was calculated as follows: V= (4/3) x π x (D1/2) x (D2/2)2, where D1 and D2 were the longest and shortest diameters, respectively (26).
RNA extraction and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from the five cell lines using the RNeasy Mini kit (Qiagen GmbH) according to the manufacturer's instructions. A total of 1 µg total RNA from each sample was converted into cDNA using the M-MLV cDNA Synthesis kit (Enzynomics, Inc.). The reaction was performed at 25°C for 10 min, 42°C for 60 min, 95°C for 5 min, and held at 4°C on a SimpliAmp™ Thermal Cycler (Applied Biosystems; Thermo Fisher Scientific, Inc.). The cDNA products from each sample were used to perform qPCR. A total of 1 µl five-time diluted cDNA was used for qPCR, and reagents were mixed followed by PCR using the SensiFAST SYBR® Lo-ROX kit (Bioline Pty Ltd, Meridian Bioscience, Inc.). The primers used are listed in Table I. β-actin mRNA was used as an internal control gene to normalize the data. RT-qPCR was performed for the initial activation at 95°C for 20 sec, followed by 40 cycles at 95°C for 10 sec, 63°C for 30 sec, and 70°C for 1 sec. The melting curve was analyzed using the instrument default setting. The assays were performed in triplicate on a Light Cycle® 96 system (Roche Diagnostics). The DDCq method (31) was used for the quantification of mRNA expression.
Table I.
Sequences of specific primers used for RT-qPCR.
| Gene | Accession no. | Primer sequence | Amplicon size (bp) | (Refs.) |
|---|---|---|---|---|
| Aurora A | NM_198433.3 | Fw 5′-TTCCAGGAGGACCACTCTCTGT-3′ Rv 5′-TGCATCCGACCTTCAA TCATT-3′ |
69 | (27) |
| Aurora B | NM_001313950.2 | Fw 5′-CGCAGAGAGATCGAAATCCAG-3′ Rv 5′-AGATCCTCCTCCGGTCATAAAA-3′ |
85 | (28) |
| VEGF | NM_001025366.3 | Fw 5′-AGGAGGAGGGCAGAATCATCAC-3′; Rv 5′-ATGTCCACCAGGGTCTCGATTG-3′ |
90 | (29) |
| β-actin | NM_001101.5 | Fw 5′-ACAGAGCCTCGCCTTTG-3′ Rv 5′-CCTTGCACATGCCGGAG-3′ |
110 | (30) |
Fw, forward; Rv, reverse.
Wound healing assay
The hUVECs and hFBs were cultured in EGM-2 endothelial cell growth medium-2 Bulletkit (Lonza Bioscience) and DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS, respectively, to reach the completed confluence in 24-well plates. The cells were then supplemented with mitomycin C (5 µg/ml) to inhibit cell proliferation. Thereafter, the cells were cultured in serum-free medium for 24 h (hUVECs) and 48 h (hFBs). Scratches were created using cell scrapers SPLScar (SPL Life Sciences Co., Ltd.), and floating cells were removed by washing the wells twice with PBS. Oxostephanine was incubated with the cells at three concentrations of 25, 5 and 1 µM for 24 h (hUVECs) and 48 h (hFBs). Images were captured every 6 h (Olympus IX73 Inverted Microscope, Olympus Corporation) from the scars created. The cell migration ability was analyzed using ImageJ software (version 1.53e, National Institutes of Health).
Colony formation assay
The hUVECs and hFBs were seeded in a six-well plate at a density of 1×103 cells/well and treated with oxostephanine at four different concentrations (25, 5, 1 and 0.2 µM) for 24 h. The medium was refreshed, and the cells were then incubated in a humidified incubator with 5% CO2 at 37°C for a further 10 days. The cells were then stained with Giemsa (Millipore, Sigma) for 5 min at room temperature after fixing with 70% methanol for 10 min at room temperature. The formation of colony units of endothelial cells (CFU-ECs) and fibroblasts (CFU-Fs) was observed, photographed and counted using an Axiovert 40 Inverted Microscope (Carl Zeiss AG) (magnification, ×4). The number of colonies was determined per 1,000 cells at seeding.
Growth factor analysis using luminex assay
Growth factors, including VEGF-A, fibroblast growth factor-2 (FGF-2) and hepatocyte growth factor (HGF), were analyzed using Luminex assay with ProcartaPlexTM Multiplex Immunoassays (Human Custom ProcartaPlex 4-Plex kit; Thermo Fisher Scientific, Inc.). The conditioned media was prepared by culturing cells to 90% confluency in an appropriate medium without supplement or FBS for 48 h. The conditioned medium was then collected and kept on ice prior to use. Reagents and procedures were processed following the manufacturer's instructions. The luminescent signals of the growth factors were detected using a LuminexTM 100/200TM system equipped with the xPONENT 3.1 software (Luminex Co., Ltd).
Tube formation assay
The tube formation assay was performed using Angiogenesis Assay kit (ab204726, Abcam). Briefly, extracellular matrix solution (Matrigel, supplied with the kit, Abcam) was added to a 96-well plate and incubated for 1 h at 37°C to allow the solution to form a gel. hVUECs were seeded at 1.5×104 cells/well (three replicates per group) on the gel and incubated with oxostephanine at two concentrations of 5 and 1 µM. For the background control wells, no Matrigel was added. Suramin (supplied with the kit, Abcam) was used as an angiogenesis suppressor control. Following 8 h of incubation at 37°C in the incubator, the tube formation was examined using an inverted microscope. The total tube length, total branching points and mean tube length were analyzed using Wimasis software (Web-based version, wimasis.com).
Statistical analysis
All statistical analyses were performed using R software version 3.4.4. The differences between groups were assessed using an unpaired t-test, two-way ANOVA and Tukey's HSD tests. A P-value <0.05 was considered to indicate a statistically significant difference. All data are presented as the mean ± SD.
Results
Real-time analysis of the effects of oxostephanine on OVCAR-8 cancer cells
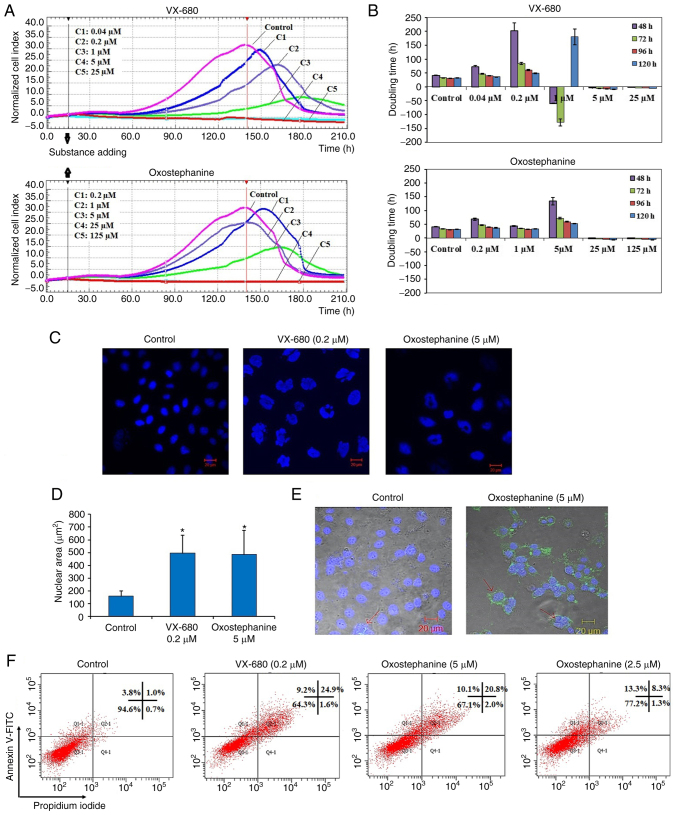
The present study performed a cytotoxicity analysis of oxostephanine using the OVCAR-8 cell line with the xCELLigence RTCA system. During >200 h of incubation, the viability, number, morphology and adherent ability of the cells were recorded and visualized as a graph (Fig. 1A). The utilities in the RTCA Control Unit software allowed for the creation of a dose-response curve and the calculation of the IC50 value of the drug at different time points. The results revealed that oxostephanine and VX-680 exerted a similar effect on cell proliferation; the higher concentrations of the compounds the greater the inhibitory effects on cell growth. In the control wells, the cell index values gradually increased and peaked at the time point of 140 h, with a CI of 32 (Fig. 1A). In the wells treated with the two highest concentrations of 125 and 25 µM oxostephanine, cell proliferation was entirely inhibited compared to the control with the CI values decreasing after 3 h of incubation, indicating that the cells could not grow, but were killed. At the oxostephanine concentration of 5 µM, the cell proliferation rate was approximately half that of the control, with the time to get the peak of CI values was 165 h. At the oxostephanine concentration of 1 µM, the peak was reached at the same time but with a smaller value equivalent to 78% of the control. At the smallest concentration of 0.2 µM oxostephanine, the cell proliferation was lower than that of the control. For the wells treated with VX-680, while all cells were killed at the two highest concentrations, the CI values at the peaks associated with the other concentrations were smaller and were observed at later time points than those of the control (Fig. 1A). Using RTCA software, the IC50 values at different time points of incubation from 24 to 120 h were calculated. The IC50 values were from 3.8-7.3 µM for oxostephanine and 0.2-0.6 µM for VX-680 (Table II).
Figure 1.
(A) Real-time analysis of OVCAR-8 cell proliferation following treatment with oxostephanine and VX-680. On the plot, the normalized cell index (CI) is shown at 15 h, which is the adding point of the substance. The horizontal axis of the graph was the time of the experiment. (B) Population doubling times of OVCAR-8 cells were calculated on RTCA system after 48, 72, 96 and 120 h of incubation with the two compounds at five concentrations, as indicated in the figures. Of note, the minus values of PDT indicated that the cells died when exposed to the compound at an early stage and no cell growth was counted. (C) Image of cell nuclei following incubation with oxostephanine and VX-680 for 48 h. (D) The average sizes of cell nuclear area were calculated and presented as the mean ± SD. Data were collected from three repeated experiments. (E) Oxostephanine induced the apoptosis of OVCAR-8 cancer cells. Immunofluorescence images of control and oxostephanine-treated cells stained with Annexin V-FITC indicated the higher expression of phosphatidylserine molecules on the cell surface in treated cells (green color). (F) Quantitative analysis of the percentage apoptosis in the oxostephanine- and VX-680-treated cells. *P<0.05, vs. control.
Table II.
IC50 values of oxostephanine and VX-680 in OVCAR-8 cancer cells with different incubation times.
| Compound | Incubation time (h)
|
||||
|---|---|---|---|---|---|
| 24 | 48 | 72 | 96 | 120 | |
| VX-680 (µM) | 0.3±0.02 | 0.6±0.06 | 0.2±0.04 | 0.2±0.07 | 0.2±0.05 |
| Oxostephanine (µM) | 7.3±1.5 | 6.6±0.9 | 5.6±0.7 | 4.6±0.8 | 3.8±0.5 |
The doubling time of the OVCAR-8 cells was also affected by these two compounds. Following treatment with VX-680, the cells did not grow and died rapidly following the addition of the substance expressed by the minus values of the doubling time at the three highest concentrations. In terms of oxostephanine, the majority of the doubling time was higher compared to the controls, indicating that the proliferation of cells was inhibited (Fig. 1B). Of note, a change in the size of the cells treated with oxostephanine and VX-680 at low concentrations was observed. The cells increased their size following the incubation time. Not only the cell size, but the immunostaining of these cells also indicated that there was a significant increase in the nuclei area (Fig. 1C). Additionally, the morphology of the cell nuclei was changed, with the nuclei becoming heterogeneous, multi-lobed and enlarged, that were not homogeneous or oval-shaped as in the controls (Fig. 1C). Using the LSM image browser software, the nuclear area was measured. The data indicated that the nucleic size of the cells treated with oxostephanine or VX-680 was three-fold larger than that in the control group (Fig. 1D). Taken together, these results demonstrated that oxostephanine inhibited the proliferation of OVCAR-8 cells in the micromolar range. The real-time effects of oxostephanine were comparable to those of VX-680, an Aurora kinase inhibitor.
Apoptosis induction is a characteristic of Aurora kinase inhibitors (18,23). Hence, the present study examined whether oxostephanine could induce the apoptosis of OVCAR-8 cancer cells. At the oxostephanine concentration of 5 µM, we observed the expression of phosphatidylserine molecule, an apoptosis marker, that binding to Annexin-V on the cell surface after 24 h of incubation (Fig. 1E). The rate of cells positive with Annexin-V was calculated from the sum of Q1-1 (early apoptosis) and Q2-1 (late apoptosis) quadrants in the flow cytometry plots (Fig. 1F). Accordingly, the percentage of oxostephanine (5 µM)-treated cells positive for Annexin-V was 30.4±6.8%, which was 7.4-fold higher than that of the control (4.1±0.8%). Moreover, a 33.7±5.1% cell population was positive for Annexin-V when treated with 0.2 µM VX-680 (Fig. 1F).
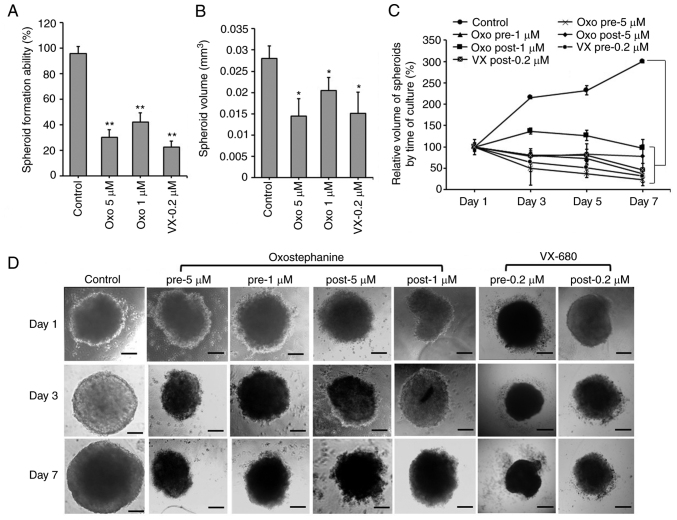
Oxostephanine inhibits the growth of OVCAR-8 spheroids
The effects of oxostephanine on the growth of OVCAR-8 cells in 3D culture were investigated. When adding the substance at the time of spheroid preparation, this compound prevented 70% spheroid formation at 5 µM and 58% spheroid formation at 1 µM. A similar result was obtained with VX-680; only 22.5% of spheroids could be formed at the concentration of 0.2 µM (Fig. 2A). Moreover, the volume of the formed spheroids was smaller than that of the control (Fig. 2B). After transferring the spheroids into agar plates, the growth was unaltered at the concentration of 1 µM, whereas this decreased at the concentration of 5 µM following the time of culture even with the absence of the compound (Fig. 2C). For the other treatments, oxostephanine was added and maintained in the medium after the spheroids were transferred into the agar plate. Under this condition, after 7 days, the substance inhibited the growth of spheroids, with the size decreasing 4.3-fold at 5 µM and 2.7-fold at 1 µM. The effect of oxostephanine on spheroid growth was even more prominent than that of VX-680 at 0.2 µM, with a decrease of 2.1-fold in the volume on day 7 of treatment. Moreover, the control increased the spheroid volume 3-fold on day 7 of culture on agar (Fig. 2C). Furthermore, the morphology of the treated spheroids was also changed into loose cell clusters with numerous cells separately surrounded, in contrast to the tight and impact control spheroids (Fig. 2D).
Figure 2.
Oxostephanine inhibits the formation and growth of OVCAR-8 spheroids. (A) Spheroid formation in the presence of the compounds. (B) The spheroid volume was reduced following incubation with the compounds. (C) The growth of tumor spheroids was prevented by the two types of treatment: The addition of the compound at the spheroid preparation (pre) and after spheroid formation (post). The days were counted from the time of transferring the spheroid from the hanging drop to the agar plates (day 1, etc.). (D) The morphology of spheroids of cells treated under the two conditions mentioned above. Pre, compounds were added at the time of spheroid preparation; post, compounds were added and maintained in the medium for spheroid growth in the agar plate. Scale bars, 100 µm. *P<0.05 and **P<0.01, vs. control. Oxo, oxostephanine; VX, VX-680.
Oxostephanine inhibits Aurora kinase expression and activity
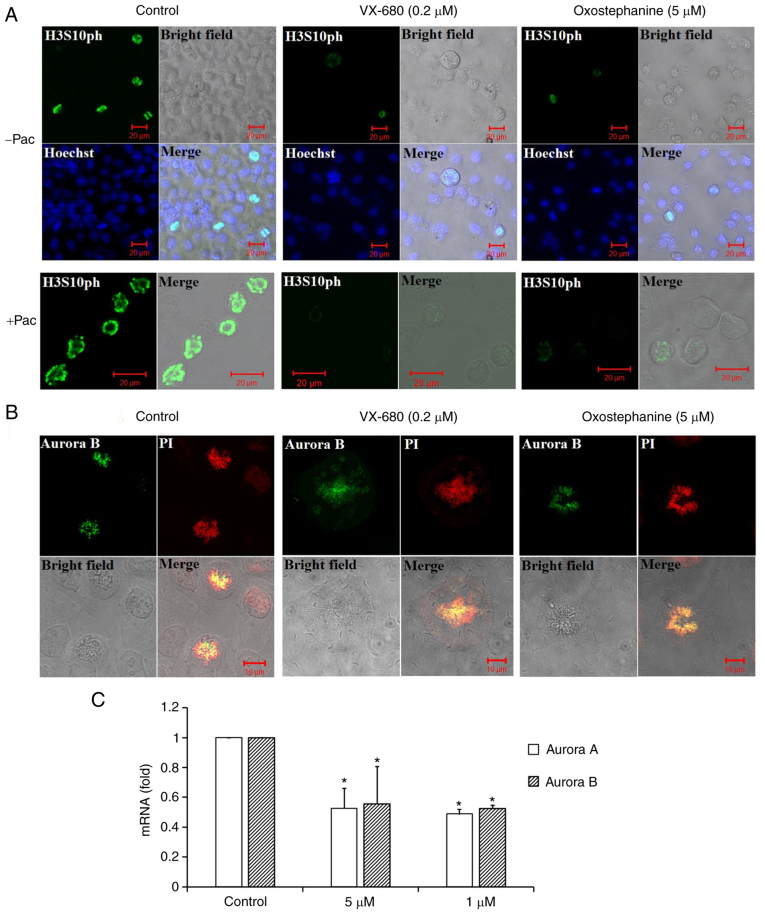
To characterize oxostephanine as a novel Aurora kinase inhibitor, the effect of this compound on the phosphorylation of H3S10ph was evaluated in OVCAR-8 cancer cells. To collect cells at the mitotic phase, the cell population was synchronized by the addition of paclitaxel followed by incubation with oxostephanine and VX-680 at concentrations of 5 and 0.2 µM, respectively. The images revealed that the fluorescence signal of H3S10ph was markedly decreased in mitotic cells incubated with oxostephanine and VX-680, even with or without paclitaxel (Fig. 3A).
Figure 3.
The phosphorylation of histone H3 at serine 10 and the localization of Aurora B kinase were disrupted in OVCAR-8 cancer cells treated with oxostephanine and VX-680. (A) H3S10ph (green) was suppressed in the presence of oxostephanine (5 µM) and VX-680 (0.2 µM) for 15 h. In the case of synchronization to the pro-metaphase, cells were pre-treated with paclitaxel (0.3 µg/ml) for 8 h, then incubated with the two substances as mentioned above. (B) Aurora B was deconcentrated on the chromosomal centromeres following treatment with the substances. (C) The expression of Aurora A and Aurora B was decreased at the mRNA level following treatment with oxostephanine and VX-680. *P<0.05, vs. control.
In addition, the distribution of Aurora B was affected by these compounds. In mitotic OVCAR-8 cells, this protein was not expressed at the centromere, but was diffused on the whole chromosomes at the metaphase. Moreover, Aurora B presented as bright dots in the centromere in the control cell group (Fig. 3B). Additionally, RT-qPCR revealed that the mRNA expression of Aurora B was decreased following incubation with oxostephanine in OVCAR-8 cells (Fig. 3C).
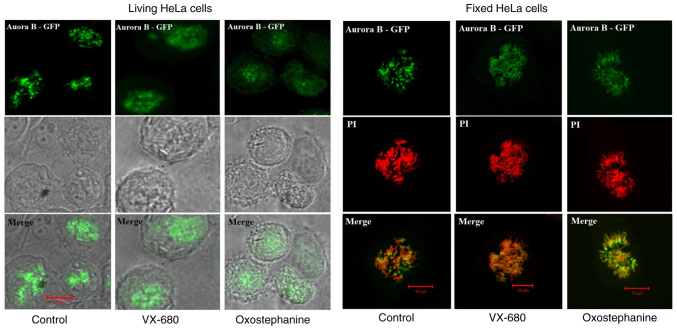
To determine the effects of oxostephanine on the localization of Aurora B kinase, HeLa cells stably expressing Aurora kinase B-GFP were used. Notably, the diffusion of Aurora B was observed in both living and fixed HeLa cells (Fig. 4). Furthermore, in mitotic cells treated with oxostephanine and VX-680, Aurora B-GFP was observed on the entire chromosomes when the cells were at metaphase.
Figure 4.
Effects of oxostephanine and VX-680 on the expression of Aurora kinase B in mitotic cells. The Aurora B distribution was determined on living HeLa cells stably expressing Aurora B-GFP and on fixed HeLa cells. Of note, in the control cells, this protein was located as bright dots on chromosomes at the metaphase; in treated cells, the protein was diffused in the whole chromosomes, particularly in VX-680-treated cells.
In summary, these data illustrated that the treatment of the cells with oxostephanine affected the behavior of Aurora B during the cell cycle in a similar manner to VX-680, but with a lower efficiency.
Oxostephanine is selectively cytotoxic on different cell types
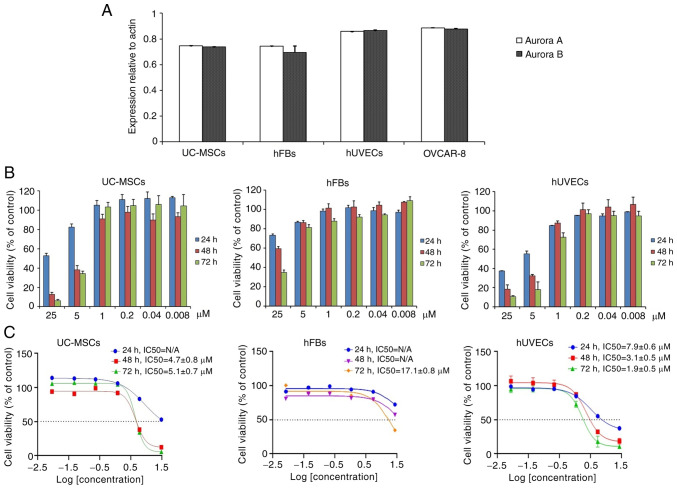
The present study also selected three cell lines, including human UC-MSCs, hUVECs and hFBs for the examination of oxostephanine cytotoxicity. Firstly, the expression of Aurora A and Aurora B kinase genes relative to the actin gene control was examined in normal and cancer cells. The results revealed that these genes were highly expressed at the mRNA level, with the highest levels observed in the hUVECs and OVCAR-8 cells, and the lowest in hFBs (Fig. 5A).
Figure 5.
Oxostephanine is selectively cytotoxic to different cell types. (A) mRNA expression of Aurora A and Aurora B kinase in normal and cancer cell lines. (B) Proliferation of hUVECs and hFBs treated with various concentrations of oxostephanine after 24, 48 and 72 h of incubation. (C) Dose-response growth inhibition curve for oxostephanine in the three cell types. hUVECs, human umbilical vein endothelial cells; hFB, human dermal fibroblasts.
Secondly, the cells were incubated with oxostephanine for the analysis of cell death. Following 24 h of incubation with oxostephanine, the death of hUVECs was observed at the two highest concentrations. After 48 and 72 h, the cell death number increased continuously in these wells containing hUVECs. Similar results were detected in UC-MSCs. On the other hand, in the wells of hFBs, no cell death was observed (Fig. 5B). Additionally, the IC50 values were consistent with these observations. The IC50 values from the hUVECs were 7.9±0.6, 3.1±0.5 and 1.9±0.5 µM after 24, 48 and 72 h of incubation, respectively. However, the IC50 values from the hFBs could not be determined after 24 and 48 h, but were 17.1±0.8 µM after 72 h of incubation. Notably, the cytotoxicity effect of oxostephanine on UC-MSCs was lower than that on hUVECs, but higher than that on hFBs, with IC50 values at 48 and 72 h were 4.7±0.8 and 5.1±0.7 µM, respectively (Fig. 5C). These data, as well as the results of the mRNA levels indicated that the oxostephanine may be more toxic to OVCAR-8 cancer cells and hUVECs, but less on hFBs and UC-MSCs.
Oxostephanine reduces colony formation and growth factor secretion by hUVECs and hFBs
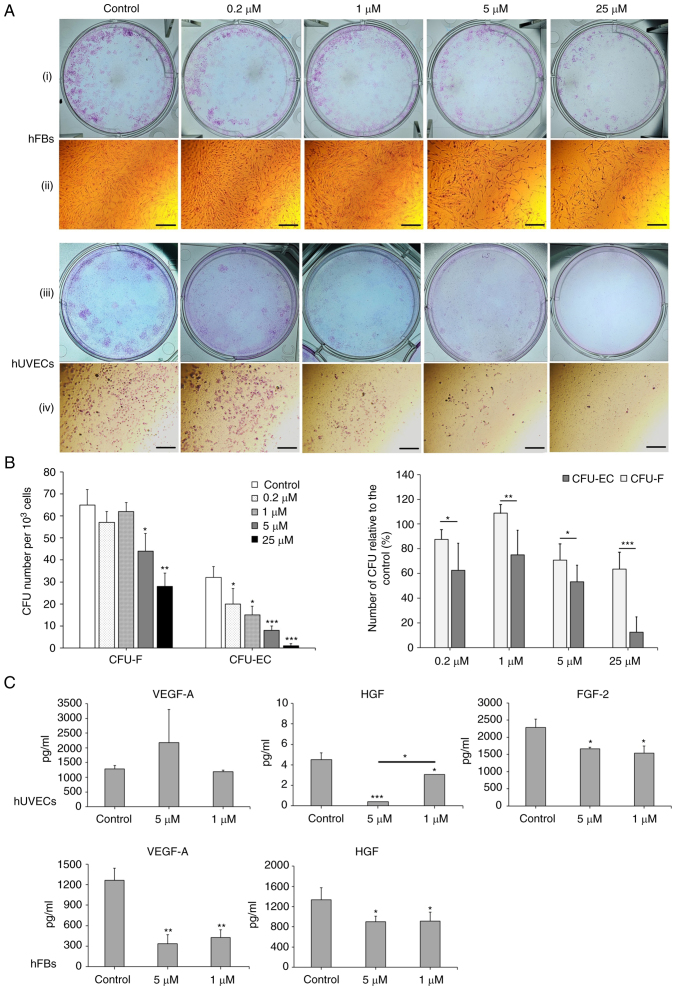
The effects of Oxostephanine on the capacity of endothelial progenitor cells and fibroblast precursor cells to form colonies were then examined. As shown in Fig. 6A, both the number of colonies and the density of cells/colonies were reduced in the treated wells compared to the controls. The numbers of CFU-ECs and CFU-Fs were significantly decreased with the two highest concentrations (25 and 5 µM) (P<0.01). Colony formation was also disrupted with the lower concentrations of oxostephanine, with a smaller number of colonies compared to the control in hUVECs (P<0.05) (Fig. 6B, left panel. In addition, the inhibitory effects of oxostephanine on colony formation were more prominent in hUVECs than in hFBs, with a smaller number of CFUs relative to the control (%) in the endothelial cells compared to that in the fibroblasts (P<0.05) (Fig. 6B, right panel).
Figure 6.
Oxostephanine reduces the colony formation and growth factor secretion by hUVECs and hFBs. (A) Images of CFU-Fs (hFBs) and CFU-ECs (hUVECs) and cell morphology in each type of CFU. CFUs were reduced in both the number of CFU and the number of cells per CFU. (i and iii) Macroscopic images of hFBs and hUVECs culture plates, respectively, following Giemsa staining; (ii and iv) microscopic of a single stained colony in hFBs and hUVECs, respectively. Scale bars, 200 µM. (B) The colony formation ability of hUVECs and hFBs treated with the indicated concentrations of oxostephanine. (C) The secretion of three types of growth factors (VEGF-A, HGF and FGF-2) in the presence of oxostephanine at various concentrations. *P<0.05, **P<0.01 and ***P<0.001, vs. control. hUVECs, human umbilical vein endothelial cells; hFB, human dermal fibroblasts; CFU, colony-forming units.
Three types of growth factors, including VEGF-A, FGF-2 and HGF, were measured in the cell culture medium after treating the cells with oxostephanine at 1 and 5 µM. The data indicated that the secretion of these proteins was differed between the cell types. In the controls, both the hUVECs and hFBs secreted HGF with values of 4.5, and 1,333±243.2 µg/ml, respectively (Fig. 6C). Additionally, both the hFbs and hUVECs produced VEGF-A into the medium at a concentration of around ~1,270 pg/ml. The hUVECs secreted a high amount of FGF-2 (2,285.8±240.1 pg/m). Following incubation with oxostephanine, the capacity of growth factor secretion by the cells was consistent with the control regarding the factor component that only hUVECs could secrete all three factors (VEGF-A, HGF and FGF-2) and hFBs secreted only VEGF-A and HGF. However, the amount of all tested growth factors decreased (P<0.05), apart from VEGF-A secreted by hUVECs treated with 5 µM oxostephanine (Fig. 6C). These results demonstrated that oxostephanine affected the secretion of growth factors by cells.
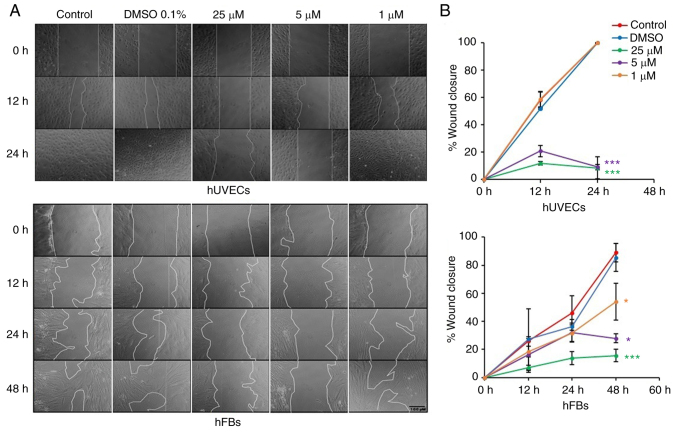
Oxostephanine inhibits the migration of hUVECs and hFBs
Fibroblast and endothelial cell migration is a critical step in the wound healing and angiogenesis processes (32). Thus, in the present study, a wound healing assay was performed to examine the capacity of oxostephanine to regulate the migration of endothelial cells and fibroblasts. In the control group, both hUVECs and hFBs expressed their ability to migrate to close the gap at a more rapid rate; the hUVECs exhibited a greater migratory ability (covering 100% of the wound after 24 h) compared to the hFBs (covering 46.1% of the wound after 24 h) (Fig. 7). When the cells were treated with oxostephanine, a significant decrease in the migration of hUVECs and hFBs were observed (P<0.05; Fig. 7). As regards the hUVECs, the percentage of the wound covered by cells treated with oxostephanine at the concentrations of 25 and 5 µM was ~11% compared to 100% of that in the control group after 24 h, which indicated that the compound inhibited the migration of hUVECs >10-fold (Fig. 7). This inhibitory effect was less prominent in hFBs at the two highest concentrations (5.7-fold decrease at 25 µM and 3.2-fold decrease at 5 µM at 48 h). However, at the concentration of 1 µM, the compound exerted more prominent inhibitory effects on the migration of hFBs than that of hUVECs. These results demonstrated that oxostephanine significantly inhibited the migration of hUVECs and hFBs.
Figure 7.
Oxostephanine inhibits the migration of hUVECs and hFBs. (A) Images of cell migration toward the gap in the presence of oxostephanine at the indicated concentrations. (B) Quantitative analysis of gap covering (%) after a time of cell migration. Experiments were repeated in triplicate and data are presented as the mean ± SD. *P<0.05 and ***P<0.001. hUVECs, human umbilical vein endothelial cells; hFB, human dermal fibroblasts.
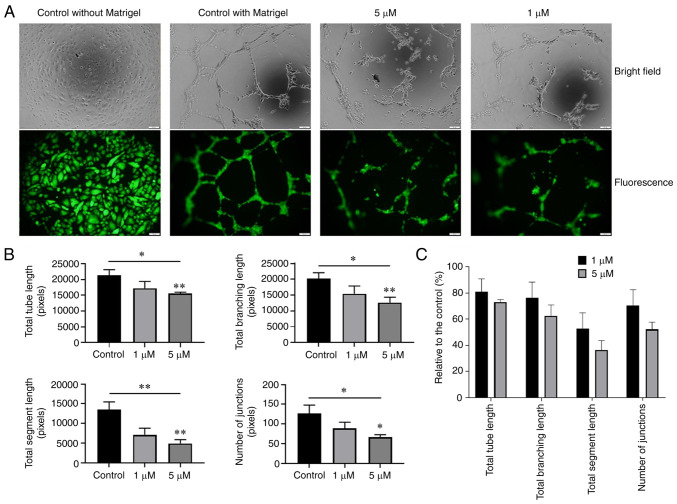
Oxostephanine suppresses angiogenesis in vitro
The effect of oxostephanine on the angiogenesis of hUVECs was examined using tube formation assay. As shown in Fig. 8A, the hUVECs formed a capillary-like network on the Matrigel, with the highest number of total tube lengths and tube branching points after 10 h. By contrast, the tube-formation capacity significantly decreased when the cells were treated with 5 µM oxostephanine (P<0.05) (Fig. 8B). The total tube length, tube branching, tube segments and the number of junctions were 72.9±2.1, 62.5±8.4, 36.4±7.2, and 52.1±5.6%, respectively, compared to the control group. The majority of hUVECs clustered, and very few tube-like structures were observed. When the cells were treated with 1 µM oxostephanine, the percentage of total tube length, branching, segments, and number of junctions reached 80.8±10, 76.2±12, 52.7±12.2, and 70.3±12.3%, respectively, compared to the control (Fig. 8C). These findings suggested that oxostephanine suppressed angiogenesis in vitro.
Figure 8.
Effect of osxostephanine on the tube formation assay of hUVECs. (A) Representative images and (B) quantification of the tube formation when seeding hUVECs on Matrigel in the presence of the test compounds. (C) Tube formation capacity relative to the control (%) of hUVECs incubated with oxostephanine. Experiments were repeated in triplicate and data are presented as the mean ± SD. *P<0.05 and **P<0.01.
Discussion
The crucial role of Aurora kinases, particularly Aurora A and B, in cell division, as well as the overexpression of these kinases in a wide range of cancers, renders them a potential target in cancer treatment (18). Oxostephanine extracted from the Stephania plant was first reported by Makarasen et al (21) for its activity in inhibiting the growth of a variety of cancer cell lines. The present study first aimed to characterize oxostephanine, extracted from S. dielsiana leaves in Vietnam, as a novel Aurora inhibitor by comparing the real-time effects of this substance on cancer cells to those of VX-680, a well-known Aurora kinase inhibitor (33). An ovarian cancer cell line (OVCAR-8), was used to examine the effects of oxostephanine, since Aurora kinase has been reported to be overexpressed in epithelial ovarian cancer, in addition to two recent clinical trials that have used Aurora kinase inhibitors to treat ovarian cancer (34-36). In the present study, the analysis using the xCelligence system revealed similar responses of the OVCAR-8 cells to both compounds (oxostephanine and VX-680) in real-time growth dose-response curves, cell population doubling time and cellular size change.
Of note, at low tested concentrations of oxostephanine (<5 µM) and VX-680 (1 µM), the cells became aneuploidy with an increase in their size, but not in their number. Previous research has indicated that when Aurora kinase activity is inhibited, the mitotic SAC is activated, which leads to mitotic arrest. However, this SAC could be overridden, which causes the mitotic slippage of cells in the presence of Aurora kinase inhibitors. This phenomenon eventually led to cells becoming aneuploidy or apoptotic (37). In the present study, OVCAR-8 cells treated with oxostephanine and VX-680 at low concentrations expressed enlarged and lobed nuclei. Moreover, as shown by immunofluorescence assay, both compounds downregulated the phosphorylation of protein histone H3 at serine 10 in cancer cells. These data are consistent with those of the study by Knockleby et al (23), demonstrating that oxostephanine inhibited H3S10ph in HeLa cells (23). As the phosphorylation of histone H3 at serine 10 is considered a marker of activated Aurora B kinase (7,8), hence oxostephanine could prevent the function of this kinase.
Previous studies have indicated that the activity of Aurora B is associated with auto-phosphorylation and centromere distribution (5,23). Under normal conditions, Aurora B must concentrate at the kinetochore to phosphorylate some proteins in the conserved outer kinetochore KNL1/Mis12 complex/Ndc80 complex (KMN) network, which plays a role in the kinetochore-microtubule attachment (4-6). The present study demonstrated that oxostephanine affected the normal localization of Aurora B kinase; thus, it may inhibit the auto-phosphorylation activity of this enzyme. In the presence of oxostephanine and VX-680, Aurora B diffused to all chromosome arms and in the cytoplasm. This phenomenon ocurred in all fixed and living OVCAR-8 and HeLa cells. By observing living HeLa cells that express Aurora B-GFP, it was noted that the cells that have chromosomes with diffused Aurora B remained longer in metaphase and eventually became aneuploidy. This phenomenon of Aurora B has been mentioned with the other inhibitors (24,25). This could be explained by the fact that Aurora B did not concentrate at the kinetochore, leading to an effect on the correct attachment of the chromosome to the microtubule and subsequently activating the SAC, consequently leading to mitotic slippage, as discussed above. Moreover, oxostephanine decreased the expression of both Aurora A and Aurora B at the mRNA level as did VX-680. The reduction in the levels of these proteins contributed to defects in cell division functions. Taken together, these data demonstrated that oxostephanine was an Aurora kinase inhibitor, and this compound was cytotoxic to OVCAR-8 cells in both monolayer culture and tumor spheroids. It is worth noting that Knockleby et al (23) indicated the effect of Oxostephanine on both Aurora A and Aurora B in the kinase assay. The present study first focused on Aurora B in OVCAR-8 cells. In future studies, the authors aim to continue to test the effects of oxostephanine on Aurora A kinase in cell culture.
Cancer-associated mesenchymal stem cells and fibroblasts have been proven to facilitate tumor progression. Recent research has revealed the function of mesenchymal stem cells in glioblastoma resistant to Aurora kinase inhibitor, leading to the recurrence of tumors (38). In acute myeloid leukemia (AML), one mechanism of mesenchymal stem cells used to protect leukemic cells from chemotherapeutic agents is activating Aurora A by increasing IL-6 secretion (39). In a co-culture system, fibroblasts have been shown to induce the upregulation of Aurora A in non-small cell lung carcinoma to protect the cancer cells from gefitinib treatment (40,41). Fibroblasts can be activated by Aurora B through Wilms tumor 1 signaling, leading to an induction of fibrogenesis (42). Moreover, the downregulation of Aurora B stimulates cellular senescence in hFBs (43). Aurora kinase and stromal cells exert synergistic effects on the development of cancer cells. Moreover, angiogenesis is necessary for the progression of tumors (44). Hence, in the present study examined the effects of oxostephanine on four cell types (UC-MSCs, hFBs, hUVECs and OVCAR-8). Firstly, it was found that all tested cells highly expressed Aurora A and B, with the highest expression level observed in OVCAR-8 cells and hUVECs, followed by UC-MSCs, and finally hFBs. Accordingly, the IC50 values of oxostephanine in these cell lines were the lowest in the OVCAR-8 cells and hUVECs, higher in the MSCs, and highest in the hFBs. Moreover, the reduction in the colony-forming units indicated that oxostephanine could inhibit the proliferation of endothelial progenitor cells and fibroblasts. One limitation of the present study was that the presentation of colonies needed improvement as the location of the closely clustered colonies could not be seen. However, the number of colonies could still be counted. At the concentration of 5 µM, oxostephanine significantly inhibited the colony formation of hUVECs; however, the colony-forming inhibitory effect was less prominent in hFBs (~30% CFUs). Additionally, in the wound healing assay, oxostephanine also exerted a more prominent inhibitory effect on the migration of hUVECs than hFBs. These results demonstrated the selective activity of oxostephanine toward hUVECs. The targeting of the compound to different cell types may result from the expression of Aurora kinase in these cells. Higher levels of Aurora kinase are associated with a more prominent effect of oxostephanine on the cells. Apart from cell growth, the function of hUVECs in angiogenesis was also disrupted by oxostephanine. These cells could not successfully form tubes in Matrigel in the presence of 5 µM oxostephanine. The anti-angiogenic effect of Aurora kinase inhibitors has been previously reported (13) through their involvement in a signaling pathway that enhances angiogenesis (45) and stabilizes N-Myc, which is a well-known oncogene (46,47). These results indicate that oxostephanine functions as a suppressor of angiogenesis.
Furthermore, the data indicated that oxostephanine decreased the production of VEGF-A, HGF and FGF-2, which functions in the proliferation, migration and tube formation processes (48-51), by both hUVECs and hFBs. Notably, in the present study, in hUVECs, the mRNA expression of VEGF-A in cells treated with oxostephanine was not considerably altered; however, the expression of FGF-2 was significantly decreased compared to the control. This activity of oxostephanine differed from VX-680, which has been shown to inhibit VEGF-A expression (13). Nonetheless, the decrease in the levels of FGF-2 and HFG was sufficient to inhibit the growth and function of hUVECs.
Of note, the effects of oxostephanine one growth factor secretion by cells have not yet been clarified. In addition, the involvement of Aurora kinases in angiogenesis have not yet been elucidated. However, it can be hypothesized that Aurora kinase inhibitors, such as oxostephanine, are cytotoxic toward ovarian cancer cells and endothelial cells, which leads to the inhibition of tumor angiogenesis. Furthermore, even though this compound was less cytotoxic to the other stromal cells such as hFBs and UC-MSCs, it prevented the cell functions that can result in stromal cells being inefficient in supporting tumor growth. This hypothesis was encouraged by a published study on the antitumor activity of the methanol fractional extraction from S. dielsiana roots on Swiss mice bearing Sarcoma-180 tumors, which reported a 4-fold decrease in tumor volume in the treated mine (52). It is necessary to examine the effects of oxostephanine in vivo using animal models transplanted with human tumor cells. The authors aim to perform such experiments in the future.
In conclusion, the findings of the present study indicate that oxostephanine is a potential Aurora kinase inhibitor. It inhibited the proliferation of ovarian cancer OVCAR-8 cells and multicellular tumor spheroids. Moreover, oxostephanine exhibited selective cytotoxicity to normal cells by inducing a high expression of Aurora kinase A and B. Furthermore, this compound downregulated the expression of growth factors, prevented the migration of hUVECs and hFBs, and reduced tube formation. However, further studies are required for oxostephanine to be developed as an anticancer drug. This compound needs to be tested on other ovarian cancer cell lines, particularly primary cell lines, to confirm its effects on ovarian cancer. In addition, the expression of Aurora A and B in different cell types needs to be quantified using effective methods, such as western blot analysis, in order to determine to the association of Aurora kinase expression and the effects of oxostephanine. More importantly, in the long term, experiments using in vivo tumor models need be performed to confirm the efficiency of oxostephanine.
Acknowledgments
Not applicable.
Funding Statement
The present study was funded by the Administration of Science Technology and Training-Ministry of Health-Vietnam (according to Decision no. 2721/QD-BYT, dated June 28, 2019, and Contract no. 09/HD-K2DT, dated September 18, 2019).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
THTT and LDBV were involved in the study experimental design and performance, data analysis and in the writing of the manuscript. XPTD, LDD and HBP were involved in performing the experiments and in data analysis. UTTT and THTP were involved in the guidance of the experimental design and in manuscript revision. KVTL and TPN were involved in the guidance of the experimental operations. MNTH and HQN were involved in the conceptualization of the study, in the provision of resources, in the experimental design, data analysis and in the writing and revision of the manuscript.
Ethics approval and consent to participate
The hUVECs, hFBs and UC-MSCs were provided by the Vinmec Research Institute of Stem cell and Gene Technology, and they were not immortalized cell lines. The protocols for cell isolation were approved by the Ethics Committee of Vinmec International Hospital (Document no. 40/2020/QD-Vinmec for hUVECs and UCMSCs, signed and dated on December 24, 2020; Document no. 311/2018/QD-Vinmec for hFBs, signed and dated on September 11, 2018).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cowley DO, Rivera-Pérez JA, Schliekelman M, He YJ, Oliver TG, Lu L, O'Quinn R, Salmon ED, Magnuson T, Van Dyke T. Aurora-A kinase is essential for bipolar spindle formation and early development. Mol Cell Biol. 2009;29:1059–1071. doi: 10.1128/MCB.01062-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barretta ML, Spano D, D'Ambrosio C, Cervigni RI, Scaloni A, Corda D, Colanzi A. Aurora-A recruitment and centrosomal maturation are regulated by a Golgi-activated pool of Src during G2. Nat Commun. 2016;7:11727. doi: 10.1038/ncomms11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmena M, Ruchaud S, Earnshaw WC. Making the Auroras glow: Regulation of Aurora A and B kinase function by inter-acting proteins. Curr Opin Cell Biol. 2009;21:796–805. doi: 10.1016/j.ceb.2009.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gurden MD, Anderhub SJ, Faisal A, Linardopoulos S. Aurora B prevents premature removal of spindle assembly checkpoint proteins from the kinetochore: A key role for Aurora B in mitosis. Oncotarget. 2016;9:19525–19542. doi: 10.18632/oncotarget.10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimada M, Goshima T, Matsuo H, Johmura Y, Haruta M, Murata K, Tanaka H, Ikawa M, Nakanishi K, Nakanishi M. Essential role of autoactivation circuitry on Aurora B-mediated H2AX-pS121 in mitosis. Nat Commun. 2016;7:12059. doi: 10.1038/ncomms12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lan W, Zhang X, Kline-Smith SL, Rosasco SE, Barrett-Wilt GA, Shabanowitz J, Hunt DF, Walczak CE, Stukenberg PT. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- 7.Mallm JP, Rippe K. Aurora kinase B regulates telomerase activity via a centromeric RNA in stem cells. Cell Rep. 2015;11:1667–1678. doi: 10.1016/j.celrep.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Rosasco-Nitcher SE, Lan W, Khorasanizadeh S, Stukenberg PT. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 2008;319:469–472. doi: 10.1126/science.1148980. [DOI] [PubMed] [Google Scholar]
- 9.Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, Gorbsky GJ, Higgins JMG. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330:231–235. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vader G, Medema RH, Lens SMA. The chromosomal passenger complex: Guiding Aurora-B through mitosis. J Cell Biol. 2006;173:833–837. doi: 10.1083/jcb.200604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delacour-Larose M, Thi MNH, Dimitrov S, Molla A. Role of survivin phosphorylation by aurora B in mitosis. Cell cycle. 2007;6:1878–1885. doi: 10.4161/cc.6.15.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otto T, Horn S, Brockmann M, Eilers U, Schüttrumpf L, Popov N, Kenney AM, Schulte JH, Beijersbergen R, Christiansen H, et al. Stabilization of N-myc is a critical function of aurora A in human neuroblastoma. Cancer Cell. 2009;15:67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Sun X, Niu S, Zhang Z, Wang A, Yang C, Guo Z, Hao Y, Li X, Wang X. Aurora kinase inhibitor VX-680 suppresses the proliferation and migration of HUVECs and angiogenesis. Mol Med Rep. 2019;19:3841–3847. doi: 10.3892/mmr.2019.9996. [DOI] [PubMed] [Google Scholar]
- 14.Romain CV, Paul P, Lee S, Qiao J, Chung DH. Targeting Aurora kinase A inhibits hypoxia-mediated neuroblastoma cell tumorigenesis. Anticancer Res. 2014;34:2269–2274. [PMC free article] [PubMed] [Google Scholar]
- 15.Tang CJC, Lin CY, Tang TK. Dynamic localization and functional implications of Aurora-C kinase during male mouse meiosis. Dev Biol. 2006;290:398–410. doi: 10.1016/j.ydbio.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 16.Balboula AZ, Schindler K. Selective disruption of aurora C kinase reveals distinct functions from aurora B kinase during meiosis in mouse oocytes. PLoS Genet. 2014;10:e1004194. doi: 10.1371/journal.pgen.1004194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quartuccio SM, Schindler K. Functions of Aurora kinase C in meiosis and cancer. Front Cell Dev Biol. 2015;3:50. doi: 10.3389/fcell.2015.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bavetsias V, Linardopoulos S. Aurora kinase inhibitors: Current status and outlook. Front Oncol. 2015;5:278. doi: 10.3389/fonc.2015.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inamdar KV, O'Brien S, Sen S, Keating M, Nguyen MH, Wang X, Fernandez M, Thomazy V, Medeiros LJ, Bueso-Ramos CE. Aurora-A kinase nuclear expression in chronic lymphocytic leukemia. Mod Pathol. 2008;21:1428–1435. doi: 10.1038/modpathol.2008.173. [DOI] [PubMed] [Google Scholar]
- 20.Giet R, Petretti C, Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 2005;15:241–250. doi: 10.1016/j.tcb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Makarasen A, Sirithana W, Mogkhuntod S, Khunnawutmanotham N, Chimnoi N, Techasakul S. Cytotoxic and antimicrobial activities of aporphine alkaloids isolated from stephania venosa (Blume) spreng. Planta Med. 2011;77:1519–1524. doi: 10.1055/s-0030-1270743. [DOI] [PubMed] [Google Scholar]
- 22.Thien DD, Thuy TT, Huy NQ, Van TH, Duong LTT, Tam NT. Cytotoxic alkaloids from stephania dielsiana. Chem Nat Compd. 2018;54:613–616. doi: 10.1007/s10600-018-2426-8. [DOI] [Google Scholar]
- 23.Knockleby J, Pradines B, Gendrot M, Mosnier J, Nguyen TT, Trinh TT, Lee H, Le PM. Cytotoxic and anti-plasmodial activities of stephania dielsiana Y.C. Wu extracts and the isolated compounds. Molecules. 2020;25:3755. doi: 10.3390/molecules25163755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoang TMN, Favier B, Valette A, Barette C, Nguyen CH, Lafanechère L, Grierson DS, Dimitrov S, Molla A. Benzo[e] pyridoindoles, novel inhibitors of the aurora kinases. Cell Cycle. 2009;8:765–772. doi: 10.4161/cc.8.5.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoang NTM, Phuong TT, Nguyen TTN, Tran YTH, Nguyen ATN, Nguyen TL, Bui KTV. In vitro characterization of derrone as an aurora kinase inhibitor. Biol Pharm Bull. 2016;39:935–945. doi: 10.1248/bpb.b15-00835. [DOI] [PubMed] [Google Scholar]
- 26.McMillan KS, McCluskey AG, Sorensen A, Boyd M, Zagnoni M. Emulsion technologies for multicellular tumour spheroid radiation assays. Analyst. 2016;141:100–110. doi: 10.1039/C5AN01382H. [DOI] [PubMed] [Google Scholar]
- 27.Lin YS, Su LJ, Yu CT, Wong FH, Yeh HH, Chen SL, Wu JC, Lin WJ, Shiue YL, Liu HS, et al. Gene expression profiles of the aurora family kinases. Gene Expr. 2006;13:15–26. doi: 10.3727/000000006783991962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He J, Qi Z, Zhang X, Yang Y, Liu F, Zhao G, Wang Z. Aurora kinase B inhibitor barasertib (AZD1152) inhibits glucose metabolism in gastric cancer cells. Anticancer Drugs. 2019;30:19–26. doi: 10.1097/CAD.0000000000000684. [DOI] [PubMed] [Google Scholar]
- 29.Romain C, Paul R, Kim KW, Lee S, Qiao J, Chung DH. Targeting Aurora kinase-A downregulates cell proliferation and angiogenesis in neuroblastoma. J Pediatr Surg. 2014;49:159–165. doi: 10.1016/j.jpedsurg.2013.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy JG, McElhaney JE, Verschoor CP. Reliable reference genes for the quantification of mRNA in human T-cells and PBMCs stimulated with live influenza virus. BMC Immunol. 2020;21:4. doi: 10.1186/s12865-020-0334-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Bi H, Li H, Zhang C, Mao Y, Nie F, Xing Y, Sha W, Wang X, Irwin DM, Tan H. Stromal vascular fraction promotes migration of fibroblasts and angiogenesis through regulation of extracellular matrix in the skin wound healing process. Stem Cell Res Ther. 2019;10:302. doi: 10.1186/s13287-019-1415-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Zhang ZF, Chen J, Huang D, Ding Y, Tan MH, Qian CN, Resau JH, Kim H, The BT. VX680/MK-0457, a potent and selective Aurora kinase inhibitor, targets both tumor and endothelial cells in clear cell renal cell carcinoma. Am J Transl Res. 2010;2:296–308. [PMC free article] [PubMed] [Google Scholar]
- 34.Pérez-Fidalgo JA, Gambardella V, Pineda B, Burgues O, Piñero O, Cervantes A. Aurora kinases in ovarian cancer. ESMO Open. 2010;5:e000718. doi: 10.1136/esmoopen-2020-000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cervantes A, Elez E, Roda D, Ecsedy J, Macarulla T, Venkatakrishnan K, Roselló S, Andreu J, Jung J, Sanchis-Garcia JM, et al. Phase I pharmacokinetic/pharmacodynamic study of MLN8237, an investigational, oral, selective Aurora a kinase inhibitor, in patients with advanced solid tumors. Clin Cancer Res. 2012;18:4764–4774. doi: 10.1158/1078-0432.CCR-12-0571. [DOI] [PubMed] [Google Scholar]
- 36.Falchook G, Coleman RL, Roszak A, Behbakht K, Matulonis U, Ray-Coquard I, Sawrycki P, Duska LR, Tew W, Ghamande S, et al. Alisertib in combination with weekly paclitaxel in patients with advanced breast cancer or recurrent ovarian cancer: A randomized clinical trial. JAMA Oncol. 2019;5:e183773. doi: 10.1001/jamaoncol.2018.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brito DA, Yang Z, Rieder CL. Microtubules do not promote mitotic slippage when the spindle assembly checkpoint cannot be satisfied. J Cell Biol. 2008;182:623–629. doi: 10.1083/jcb.200805072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willems E, Lombard A, Dedobbeleer M, Goffart N, Rogister B. The unexpected roles of aurora A kinase in gliobastoma recurrences. Target Oncol. 2017;12:11–18. doi: 10.1007/s11523-016-0457-2. [DOI] [PubMed] [Google Scholar]
- 39.Wang JD, Zhang W, Zhang JW, Zhang L, Wang LX, Zhou HS, Long L, Lu G, Liu Q, Long ZJ. A novel aurora kinase inhibitor attenuates leukemic cell proliferation induced by mesenchymal stem cells. Mol Ther Oncolytics. 2020;18:491–503. doi: 10.1016/j.omto.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu CC, Yu CTR, Chang GC, Lai JM, Hsu SL. Aurora-A promotes gefitinib resistance via a NF-κB signaling pathway in p53 knockdown lung cancer cells. Biochem Bioph Res Commun. 2011;405:168–172. doi: 10.1016/j.bbrc.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 41.Chen J, Lu H, Zhou W, Yin H, Zhu L, Liu C, Zhang P, Hu H, Yang Y, Han H. AURKA upregulation plays a role in fibroblast-reduced gefitinib sensitivity in the NSCLC cell line HCC827. Oncol Rep. 2015;33:1860–1866. doi: 10.3892/or.2015.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kasam RK, Ghandikota S, Soundararajan D, Reddy GB, Huang SK, Jegga AG, Madala SK. Inhibition of Aurora kinase B attenuates fibroblast activation and pulmonary fibrosis. EMBO Mol Med. 2020;12:e12131. doi: 10.15252/emmm.202012131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim HJ, Cho JH, Quan H, Kim JR. Down-regulation of Aurora B kinase induces cellular senescence in human fibro-blasts and endothelial cells through a p53-dependent pathway. FEBS Lett. 2011;585:3569–3576. doi: 10.1016/j.febslet.2011.10.022. [DOI] [PubMed] [Google Scholar]
- 44.Lugano R, Ramachandran M, Dimberg A. Tumor angiogenesis: Causes, consequences, challenges and opportunities. Cell Mol Life Sci. 2020;77:1745–1770. doi: 10.1007/s00018-019-03351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Zhao Y, An Z, Li W. Molecular links between angiogenesis and neuroendocrine phenotypes in prostate cancer progression. Front Oncol. 2020;9:1491. doi: 10.3389/fonc.2019.01491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villaume K, Blanc M, Gouysse G, Walter T, Couderc C, Nejjari M, Vercherat C, Cordier-Bussat M, Roche C, Scoazec JY. VEGF secretion by neuroendocrine yumor cells is inhibited by octreotide and by inhibitors of the PI3K/AKT/mTOR pathway. Neuroendocrinology. 2010;91:268–278. doi: 10.1159/000289569. [DOI] [PubMed] [Google Scholar]
- 47.Ton AT, Singh K, Morin H, Ban F, Leblanc E, Lee J, Lallous N, Cherkasov A. Dual-inhibitors of N-Myc and AURKA as potential therapy for neuroendocrine prostate cancer. Int J Mol Sci. 2020;21:8277. doi: 10.3390/ijms21218277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sedlář A, Trávníčková M, Matějka R, Pražák Š, Mészáros Z, Bojarová P, Bačáková L, Křen V, Slámová K. Growth factors VEGF-A165 and FGF-2 as multifunctional biomolecules governing cell adhesion and proliferation. Int J Mol Sci. 2021;22:1843. doi: 10.3390/ijms22041843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cao R, Eriksson A, Kubo H, Alitalo K, Cao Y, Thyberg J. Comparative evaluation of FGF-2-, VEGF-A-, and VEGF-C-induced angiogenesis, lymphangiogenesis, vascular fenestrations, and permeability. Circ Res. 2004;94:664–670. doi: 10.1161/01.RES.0000118600.91698.BB. [DOI] [PubMed] [Google Scholar]
- 50.Grugan KD, Miller CG, Yao Y, Michaylira CZ, Ohashi S, Klein-Szanto AJ, Diehl JA, Herlyn M, Han M, Nakagawa H, Rustgi AK. Fibroblast-secreted hepatocyte growth factor plays a functional role in esophageal squamous cell carcinoma invasion. Proc Natl Acad Sci USA. 2010;107:11026–11031. doi: 10.1073/pnas.0914295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sahni A, Francis CW. Stimulation of endothelial cell proliferation by FGF-2 in the presence of fibrinogen requires alphavbeta3. Blood. 2004;104:3635–3641. doi: 10.1182/blood-2004-04-1358. [DOI] [PubMed] [Google Scholar]
- 52.Huy NQ, Trang NTM. Evaluation the anti-tumor activity of SM2 fraction extracted from Stephania dielsiana Y.C.Wu on Swiss mice bearing S180 sarcoma tumor. Vietnam Pharm J. 2015;55:42–45. In Vietnamese. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.