ABSTRACT
Our previous studies have demonstrated that human papillomavirus (HPV)-16 E7 oncoprotein promoted epithelial-mesenchymal transition (EMT) in non-small cell lung cancer (NSCLC) cells. Moreover, recent studies have found that exosomes can mediate EMT of NSCLC cells and epidermal growth factor receptor (EGFR) is related to the progression of NSCLC. Here, we further investigated the role of exosomal EGFR in HPV-16 E7-induced EMT of NSCLC cells. Our results showed that the exosomes derived from the stable HPV-16 E7-overexpressing A549 and NCI-H460 NSCLC cells (E7 Exo) significantly increased migration, invasion, and proliferation abilities of NSCLC cells as compared with the exosomes derived from empty vector-infected NSCLC cells (ev Exo). Moreover, both in vitro and in vivo results demonstrated that E7 Exo dramatically enhanced EMT of NSCLC cells and promoted the growth of subcutaneous NSCLC xenografts. Additionally, HPV-16 E7 enhanced the expression of EGFR and p-EGFR in both NSCLC cells and exosomes. Furthermore, the inhibition of EGFR activation or exosome secretion suppressed E7 Exo-induced migration, invasion, and EMT of NSCLC. Moreover, 12 kinds of differentially expressed miRNAs between E7 Exo and ev Exo (fold change≥2, P ≤ .05) were screened out, of which 7 miRNAs were up-regulated while 5 miRNAs were down-regulated in A549 E7 Exo. Taken together, our findings suggest that exosomal EGFR is involved in HPV-16 E7-induced EMT of NSCLC cells, which may play a key role in the progression of HPV-related NSCLC.
KEYWORDS: Exosomes, EGFR, HPV-16 E7, EMT, NSCLC
Introduction
Lung cancer is the most common cancer with the highest rate of cancer-related deaths in both male and female populations, and has a relatively low 5-year survival rate.1,2 Smoking is the first cause of lung cancer,2 but nonsmokers account for a large proportion in the patients with lung cancer.3 In addition to smoking, air pollution, environmental exposure, genetics, and human papillomavirus (HPV) infection are also considered to be related to lung cancer.4
HPV infection is recognized as the main cause of cervical cancer in women. Recently, more and more studies also have shown that high-risk HPV infection may be associated with the occurrence and development of lung cancer.5–9 The multiple reports demonstrated that the positive rates of high-risk HPV DNA and oncoproteins (E6 and E7) in lung cancer were much higher than those in control tissues,5–8 and HPV-16 was found to be the most prevalent genotype in lung cancer tissues.5–7 Moreover, de Oliveira et al. demonstrated that the frequencies of HPV presence in non-small cell lung cancer (NSCLC) accounted for 81.82%.7 These reports indicate that HPV-16 may play an important role in the progression of lung cancer, especially in the progression of NSCLC. However, the underlying molecular mechanisms by which HPV-16 mediates the progression of NSCLC remain to be further systematically explored.
Exosomes, the extracellular vesicles with a diameter of 30 ~ 150 nm secreted by a variety of living cells, are the important bridges for cell-to-cell communication.10 Exosomes with the multiple compositions including proteins, lipids, mRNAs, microRNAs (miRNAs) et al play a crucial role in the progression of lung cancer and are regarded as the most promising biomarkers for early diagnosis, prognosis, and treatment of lung cancer.11–13 Accumulating evidence demonstrates that exosomes are of great significance in invasion,14 metastasis,15 and drug resistance of NSCLC.16–18 Especially, it has been found that exosomes also play a key role in epithelial-mesenchymal transition (EMT) of NSCLC cells.14,18,19 EMT, a process in which epithelial cells acquire mesenchymal characteristics,20 is involved in the progression of cancer including NSCLC.20,21 Moreover, a growing body of evidence demonstrates that EMT can promote the progression of NSCLC by mediating invasion,22 metastasis,22 and chemotherapy resistance.23–25 Interestingly, our previous studies have found that HPV-16 E7 oncoprotein enhanced EMT of NSCLC cells.26–28 However, whether exosomes are involved in EMT induced by HPV-16 E7 in NSCLC cells remains unclear.
Epidermal growth factor receptor (EGFR), a typical tyrosine kinase receptor, was highly expressed in over 60% of NSCLC patients and considered as an important target for the therapy of NSCLC.29 EGFR mutants were frequently found in NSCLC patients, triggering the resistance/insensitivity to epidermal growth factor receptor-tyrosine kinase inhibitors (EGFR-TKIs).30–32 Moreover, EMT was reported to mediate acquired EGFR-TKI resistance in NSCLC.23,33 Recently, exosomes secreted by lung cancer cells were found to inhibit immune checkpoint inhibitors by delivering EGFR into dendritic cells34 and exosomes were demonstrated to transmit T790M mutation-induced resistance in EGFR-mutant NSCLC via the activation of the PI3K/Akt signaling pathway.35 These reported indicated that exosomal EGFR may play a key role in the progression of lung cancer. In addition, among the 498 Asian NSCLC patients, the rate of EGFR mutation was found to be higher in HPV-positive patients than HPV-negative patients (52%:31%).36 Therefore, the role of exosomal EGFR in the progression of HPV infection-related NSCLC is worthy of further exploration.
In this study, we found for the first time, to the best of our knowledge, that exosomal EGFR is involved in HPV-16 E7-induced migration, invasion and EMT of NSCLC cells, indicating that exosomal EGFR may play a key role in the progression of HPV-16-related NSCLC.
Materials and methods
Reagents
Rat anti-human E-cadherin was purchased from Millipore (MA, USA). Rabbit anti-human N-cadherin, Vimentin, Snail1, Slug, Twist1, and β-actin monoclonal antibodies, and horseradish peroxidase (HRP)-conjugated secondary antibodies were from Cell Signaling Technology Inc. (Beverly, MA, USA). GW4869, an inhibitor of exosome release, and PKH26 were purchased from Sigma (St. Louis, MO, USA). PD168393, an irreversible EGFR inhibitor, was purchased from Beyotime Biotechnology Co. Ltd (Shanghai, China).
Cell lines and cell culture
The human NSCLC cell lines, A549 and NCI-H460, were respectively from American Type Culture Collection (Rockville, MD, USA) and Chinese Academy of Sciences Cell Bank of Type Culture Collection (Shanghai, China). The stable HPV-16 E7-overexpressing and empty vector-infected NSCLC cell lines (A549 and NCI-H460) were constructed by our laboratory.26 All NSCLC cells were cultured in RPMI-1640 media containing 10% fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 μg/mL) at 37°C in a humidified condition with 5% CO2.
Isolation of exosomes
Ultracentrifugation was used to isolate exosomes derived from the stable HPV-16 E7-overexpressing A549 and NCI-H460 NSCLC cells (E7 Exo) and empty vector-infected A549 and NCI-H460 NSCLC cells (ev Exo). All NSCLC cells were cultured in media with 10% exosome-free FBS. 48 h later, cell culture media were collected, and exosomes were isolated from the supernatant by consecutive centrifugation (300 g for 10 min, 2000 g for 30 min, and 12000 g for 30 min) at 4°C to remove the floating cells and cellular debris. Then, the supernatant was centrifuged at 120000 g for 90 min at 4°C. After centrifugation, the supernatant was discarded and the pellet was washed with PBS. The ultracentrifugation and wash were repeated one time again. The exosomes were collected at a ratio of 10 μL of PBS buffer per 10 mL of cell supernatant. After extraction, the exosomes were directly added into 6-well plates for co-culture or stored at −80°C for a short-term storage (no more than 1 week).
Transmission electron microscopy
10 μL of exosomes were put on the carbon-coated copper grid for 2 min and stained by 10 μL of 2% uranyl acetate for 2 min. After absorbing excess dye solution with dry filter paper, the copper meshes with stained exosomes were dried for 30 min at room temperature. Then, the dried copper meshes with stained exosomes were put under a transmission electron microscope (JEM-1400, JEOL, Japan) and the shapes of exosomes were observed at a working voltage of 80 KV.
Uptake of exosomes
100 μL of exosomes and 4 μL of PKH26 were respectively diluted with 200 μL of diluent (Sigma, St. Louis, MO, USA). The two diluted solutions were immediately mixed and incubated for 10 min at room temperature, and then an equal volume of serum was added to the mixture to stop the reaction. Afterward, the exosomes were re-extracted by centrifugation at 120000 g for 90 min at 4°C. The exosomes were added to the cells in 6-well plates and cultured for 4 h. The nucleus was stained by DAPI (Beyotime Biotechnology Co. Ltd., Shanghai, China) for 10 min and observed under a fluorescent microscope (Nikon, Tokyo, Japan).
Wound healing assay
5 × 105 cells/well were seeded into 6-well plates and cultured into monolayers. Wounds were generated in the cell monolayer by scratching with a sterile 10 µL pipette tip. Then, the cells in the plates were treated with the exosomes at 30 μg/well for 0, 12, 24, and 36 h, respectively, and wound healing was observed under a light microscope at different times.
Transwell migration and invasion assays
The assays were performed using 24-well Transwell chambers (Corning Costar, Corning, USA) containing 8 μm pore polycarbonate membranes with Matrigel (BD Biosciences, San Jose, CA) for invasion assay or without Matrigel for migration assay. The cells were starved overnight, and then inoculated into the upper chamber without serum. 24 h later, the passed cells were fixed with 4% formaldehyde, and then stained with 4% crystal violet for 20 min. Under a light microscope, 5 fields were randomly selected to calculate the number of the passed cells.
Colony formation assay
300 cells were seeded into 6-well plates and co-cultured with different exosomes or PBS (control). After the cells were attached to the plates, the exosomes at 30 μg/well were added to the medium every 2 days. 12 days later, the colonies in the plates were stained with 4% crystal violet for 10 min. The rate of clone formation was calculated according to the formula: the rate of clone formation (%) = the number of colonies/the number of plated cells × 100%.
Co-culture of cells
HPV-16 E7-overexpressing A549 and NCI-H460 NSCLC cells were pretreated for 24 h with 10 mmol/L of GW4869 to block the release of exosomes or 0.1 mg/L of PD168393 to inhibit the activation of EGFR. Afterward, HPV-16 E7-overexpressing NSCLC cells pretreated with GW4869 or PD168393 were plated into the upper chamber of a co-culture system with 0.4 μm pores (Corning, USA) at a density of 3 × 105 cells/well, and A549 and NCI-H460 NSCLC cells at 2 × 105 cells/well were put into the lower chamber. 24 h co-culture later, the cells in the lower chamber were collected for further experiments.
Western blot
The cells were lysed on ice for 1 h with lysis buffer (Beyotime Biotechnology Corporation, Shanghai, China) supplemented with complete protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA), and the supernatant was collected by centrifugation at 12000 × g for 10 min. The protein concentration was determined using BCA Kits according to the manufacturer’s instructions (Beyotime, Shanghai, China). The electrophoresis was performed on 8 ~ 10% SDS-PAGE gel and transferred to the polyvinylidene difluoride membrane. The membrane was blocked with 5% nonfat milk, and incubated with specific primary antibody (1:1000) and HRP-conjugated secondary antibody (1:2000), respectively. The signals were visualized using enhanced chemiluminescence. The analysis of β-actin protein expression served as an internal control.
RT-qPCR
Total RNA was extracted using a total RNA extraction kit (Tiangen Biotech Co. Ltd., Beijing, China) according to the manufacturer’s instructions. RNA samples were reverse-transcribed using a reverse transcription (RT) kit (Takara Biotechnology Co. Ltd., Dalian, China) and amplified by qPCR on ABI 7500 real-time system using SYBR Premix Ex Taq™ II (Takara Biotechnology Co. Ltd., Dalian, China). The sequences of the primers were listed in Table 1 and the primers were synthesized by Sangon biotech (shanghai) Co., Ltd. All relative mRNA levels were normalized to β-actin. The RT conditions were as follows: 37°C for 15 min and 85°C for 5 s. The qPCR conditions were as follows: 42°C for 5 min, 95°C for 10s, followed by 40 cycles at 95°C for 5 s and 60°C for 31s.
Table 1.
Sequences of the primers.
| Names | Sequences (5′→ 3′) | GenBank No. |
|---|---|---|
| E-cadherin | Forwards: GCTGGACCGAGAGAGTTTCC | NM_001317184.2 |
| Reverse: CAAAATCCAAGCCCGTGGTG | ||
| N-cadherin | Forwards: TTATCCTTGTGCTGATGTTTGTG | NM_001792.5 |
| Reverse: TCTTCTTCTCCTCCACCTTCTTC | ||
| Vimentin | Forwards: CGGGAGAAATTGCAGGAGGA | NM_003380.5 |
| Reverse: AAGGTCAAGACGTGCCAGAG | ||
| EGFR | Forwards: TGAGCTCTCTGAGTGCAACC | NM_001346897.1 |
| Reverse: CAGACAAGCCACTCACCAGG | ||
| β-actin | Forwards: TGGCACCCAGCACAATGAA | NM_001101.3 |
| Reverse: CTAAGTCATAGTCCGCCTAGAAGCA |
Screening of the differentially expressed miRNAs
Exosomes derived from HPV-16 E7-overexpressing A549 cells (A549 E7 Exo) and empty vector-infected A549 cells (A549 ev Exo) were respectively isolated by ultracentrifugation as described above, and the differentially expressed miRNAs between A549 E7 Exo and A549 ev Exo were screened by high-throughput sequencing analysis (Cloud-Seq Biotech Ltd. Co., Shanghai, China).
Animal experiments
The 4-week-old BALB/c nude mice were purchased from Guangzhou Dien Gene Technology Co., Ltd. (Guangzhou, China). All animal experiments were approved by Ethics Committee of Guangdong Medical University. NCI-H460 cells (2 × 106 cells/100 μL) were respectively injected subcutaneously into the two sides of dorsum of nude mice to establish the subcutaneous xenograft model. On day 7, when the xenograft nude mouse models were successfully established, the nude mice were classified into three groups, namely PBS negative control group, NCI-H460 ev Exo group, and NCI-H460 E7 Exo group (4 mice/group). The mice were respectively injected subcutaneously with 50 μL of PBS in negative control group, and the mice were respectively injected subcutaneously with the exosomes derived from empty vector-infected NCI-H460 cells in NCI-H460 ev Exo group and the exosomes derived from HPV-16 E7-overexpressing NCI-H460 cells in NCI-H460 E7 Exo group. The volume of subcutaneous tumors and the weight of nude mice were measured every two days. About 15 days, all the nude mice were sacrificed, and the subcutaneous tumors were taken out, weighed, and fixed in 10% formalin. Then, immunohistochemical staining was performed as described previously28,37 to analyze the expression of EMT markers including N-cadherin, E-cadherin, and vimentin in the tumors.
Statistical analysis
Each experiment was repeated three times. Statistical analysis was performed using GraphPad Prism 7.0 software. All data were expressed as mean ± SD. One-way ANOVA and t-test were used to determine the significant differences among the groups at P < .05.
Results
Uptake of exosomes derived from stable HPV-16 E7- overexpressing NSCLC cells and empty vector-infected NSCLC cells
The stable HPV-16 E7-overexpressing NSCLC cells and empty vector-infected NSCLC cells were cultured in media with exosome-free FBS. The isolated exosomes derived from these cells were observed under transmission electron microscope. The results showed that the isolated exosomes exhibited typical cup-shaped or spherical morphology with double-layer membrane (Figure 1a). Then, Western blot was performed to detect the exosome-related protein expression. Our results showed that the four transmembrane proteins including CD9 and CD81 and the nuclear protein TSG101 were expressed in all extracted exosomes, while the endoplasmic reticulum-related protein Grp94 was not found in these exosomes (Figure 1b), confirming that all extracts are exosomes not membrane structure or endosome. To further examine whether exosomes were taken up by A549 and NCI-H460 cells, the exosomes were stained with PKH26 dye and incubated with A549 and NCI-H460 cells for 4 h. The results showed that red fluorescence spots were found in A549 and NCI-H460 cells (Figure 1c), indicating that the exosomes derived from the stable HPV-16 E7-overexpressing NSCLC cells (E7 Exo) and empty vector-infected NSCLC cells (ev Exo) can be taken up by A549 and NCI-H460 cells.
Figure 1.
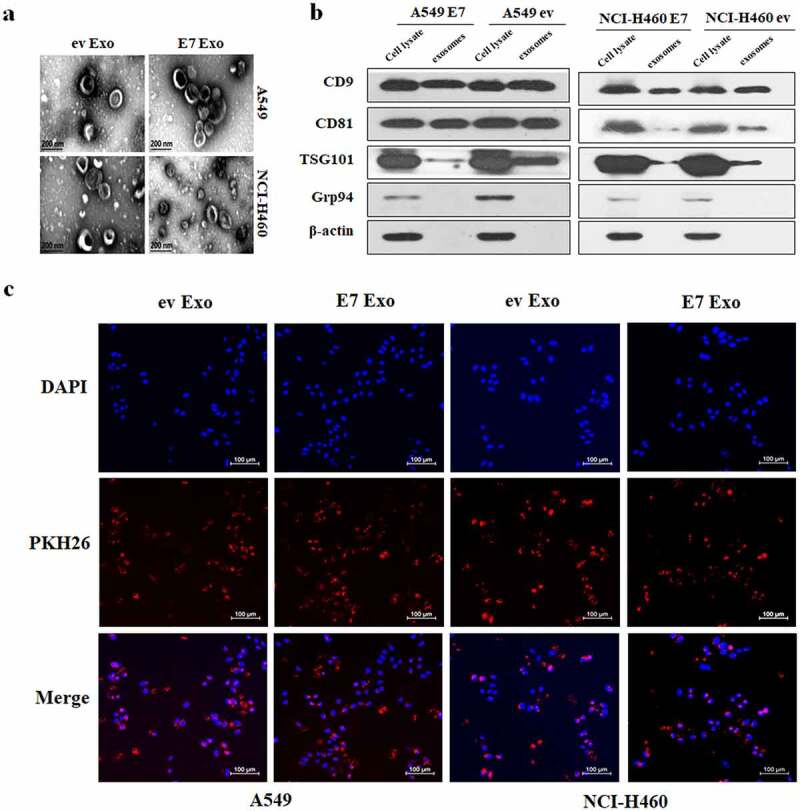
Identification and uptake of exosomes. (a) The images of transmission electron microscope of exosomes derived from the stable HPV-16 E7-overexpressing A549 and NCI-H460 cells (E7 Exo) and empty vector A549 and NCI-H460 cells (ev Exo), scale bar = 200 nm. (b) The expression of exosomal markers including CD9, CD81, and TSG101 and intracellular protein Grp94 (absence in exosomes) was analyzed by Western blot. (c) The fluorescent results of exosome uptake analysis, scale bar = 100 μm.
Exosomes derived from stable HPV-16 E7-overexpressing NSCLC cells enhanced migration, invasion, proliferation, and EMT of NSCLC cells
Our previous studies demonstrated that HPV-16 E7 enhanced migration and invasion of A549 and NCI-H460 cells.26–28 In order to investigate the role of exosomes in the migration and invasion of NSCLC cells induced by HPV-16 E7 oncoprotein, wound healing assay and Transwell assay were performed to observe the effect of exosomes on the migration and invasion abilities of NSCLC cells. The results showed that the exosomes derived from the stable HPV-16 E7-overexpressing A549 and NCI-H460 NSCLC cells (E7 Exo) significantly enhanced the migration and invasion abilities of NSCLC cells as compared with the exosomes derived from empty vector-infected cells (ev Exo) (P < .01, Figure 2a,b). Next, we further explored the effect of exosomes on the proliferation ability of NSCLC cells. Compared with the exosomes derived from empty vector-infected A549 cells, the exosomes derived from HPV-16 E7-overexpressing A549 cells significantly increased the rate of colony formation in A549 cells (Figure 2c).
Figure 2.
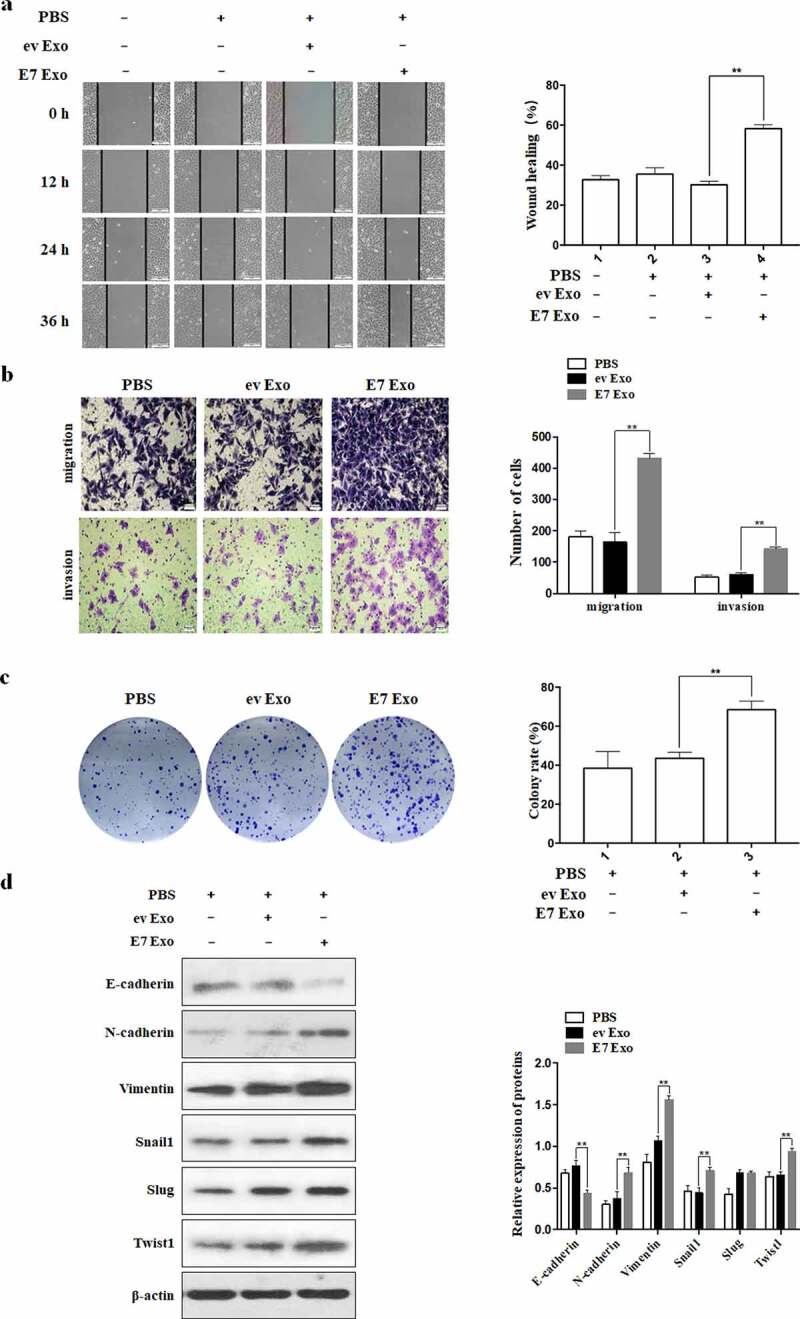
Exosomes derived from the stable HPV-16 E7-overexpressing A549 and NCI-H460 cells enhanced migration, invasion, proliferation, and EMT of NSCLC cells. (a) Wound healing results for A549 cells, scale bar = 500 μm. (b) Transwell results for the migration and invasion abilities of NCI-H460 cells, scale bar = 100 μm. (c) Colony formation results for the proliferation abilities of A549 cells. (d) Western blot analysis of expression of EMT related markers in A549 cells. E7 Exo: the cells treated with the exosomes derived from the stable HPV-16 E7-overexpressing cells; ev Exo: the cells treated with the exosomes derived from the empty vector-infected cells; PBS: the cells treated with PBS buffer. Left: one representative result of three independent experiments; right: the results for three independent experiments. All data are expressed as mean ± SD of three independent experiments. **P < .01.
Our previous studies demonstrated that HPV-16 E7 oncoprotein promoted the EMT of NSCLC cells.26–28 To further study the role of exosomes in the EMT of NSCLC cells induced by HPV-16 E7 oncoprotein, Western blot was performed to detect the expression of EMT-related epithelial marker (E-cadherin), mesenchymal markers (Vimentin and N-cadherin), and transcription factors (Snail1, Slug, and Twist1). Our results showed that the exosomes derived from the stable HPV-16 E7-overexpressing A549 and NCI-H460 NSCLC cells down-regulated the expression of E-cadherin while up-regulated the expression of Vimentin, N-cadherin, Snail1, and Twist1 (P < .01, Figure 2d), indicating that the exosomes derived from the stable HPV-16 E7-overexpressing NSCLC cells promoted EMT of NSCLC cells.
Collectively, the above results indicated that exosomes were involved in the migration, proliferation, and EMT of NSCLC cells induced by HPV-16 E7 oncoprotein.
Exosomes derived from stable HPV-16 E7-overexpressing NCI-H460 cells promoted tumor growth and EMT of NSCLC in vivo
To further confirm cell experiment results, we established the NCI-H460 NSCLC subcutaneous xenograft tumor model and observed the effect of the exosomes derived from HPV-16 E7-overexpressing NCI-H460 cells on the growth of NSCLC. Our results showed that the volume and the weight of the xenograft tumors in NCI-H460 E7 Exo group were significantly increased as compared with NCI-H460 ev Exo group (P < .05, Figure 3a-c). But the weight difference of nude mice among all groups was not obvious (Figure 3d). Moreover, immunohistochemistry results showed that the expression of E-cadherin was down-regulated but the expression of N-cadherin and Vimentin was up-regulated in NCI-H460 E7 Exo group as compared with NCI-H460 ev Exo group (P < .05, Figure 3e). Taken together, these results indicated that the exosomes derived from HPV-16 E7-overexpressing NCI-H460 cells significantly promoted the growth and EMT of NSCLC in vivo.
Figure 3.
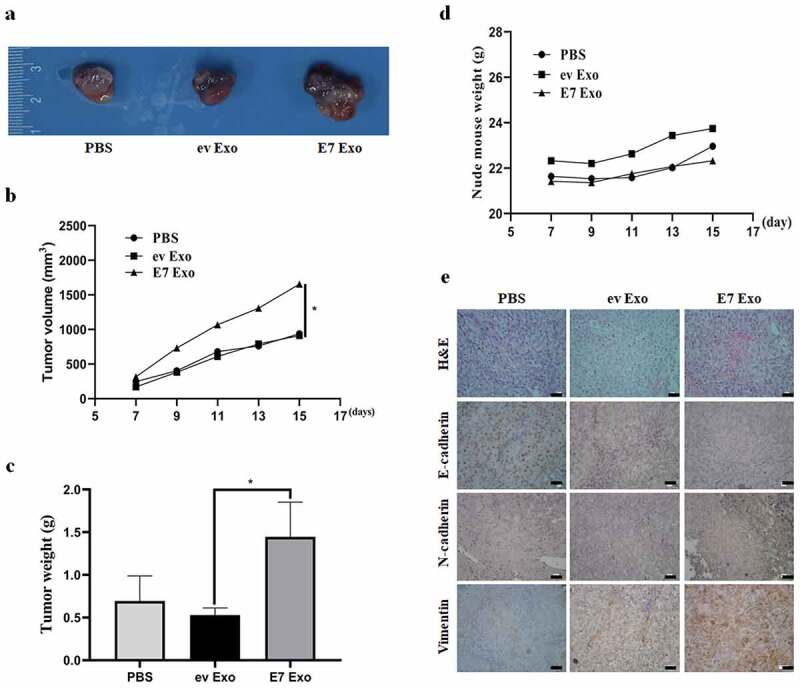
Exosomes derived from stable HPV-16 E7-overexpressing NCI-H460 cells promoted tumor growth and EMT of NSCLC in vivo. (a) The representative results of subcutaneous tumors. (b) Tumor volume (mm3). (c) Tumor weight (g). (d) Nude mouse weight (g). (e) The representative results of HE and immunohistochemical staining, scale bar = 100 μm.
EGFR induced by HPV-16 E7 oncoprotein can be delivered to NSCLC cells through exosomes
EGFR, a key carcinogen, is over-expressed in more than 60% of NSCLC patients.29 In this study, we demonstrated that HPV-16 E7 significantly enhanced the expression of EGFR and p-EGFR in A549 cells (P < .01, Figure 4a). Moreover, we further found that the exosomes derived from HPV-16 E7-overexpressing A549 and NCI-H460 cells (E7 Exo) also exhibited stronger EGFR expression than the exosomes derived from empty vector-infected cells (ev Exo) (P < .05, Figure 4b). Next, to determine whether the delivery of EGFR to A549 and NCI-H460 cells was dependent on exosomes, the stable HPV-16 E7-overexpressing A549 and NCI-H460 cells in the upper transwell chamber with 0.4 μm pores were respectively treated with GW4869 (an inhibitor of exosome release) and PD168393 (an irreversible EGFR inhibitor) for 24 h, followed by the analysis of EGFR expression in the lower transwell chamber NSCLC cells. The results showed that not only GW4869 but also PD168393 significantly inhibited HPV-16 E7-induced p-EGFR and EGFR protein expression at both A549 and NCI-H460 cells (P < .05, Figure 4c,d). Taken together, these findings suggested that the delivery of EGFR was related to exosomes.
Figure 4.
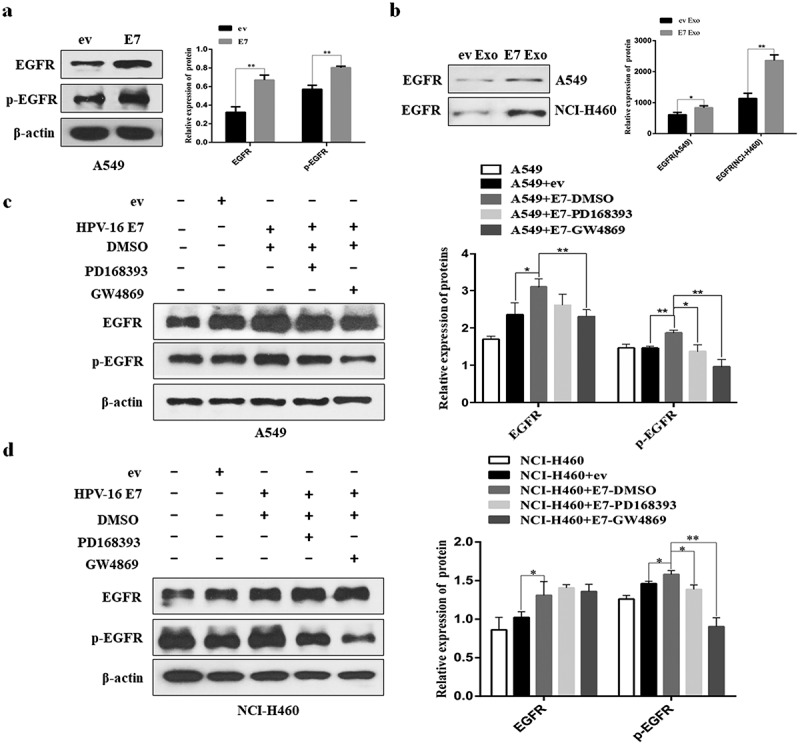
EGFR induced by HPV-16 E7 can be delivered to NSCLC cells through exosomes. (a) The expression of EGFR and p-EGFR in HPV-16 E7-overexpressing cells (E7 cells) and empty vector cells (ev cells). (b) The expression of EGFR in the exosomes derived from the stable HPV-16 E7-overexpressing cells (E7 Exo) and the exosomes derived from empty vector-infected cells (ev Exo). (c,d) The analysis of the protein levels of EGFR and p-EGFR in A549 (c) and NCI-H460 cells (d) co-cultured with ev cells and E7 cells treated with DMSO, PD168393, and GW4869, respectively. Left: one representative result of three independent experiments; right: the results of density for three independent experiments. All data are expressed as mean ± SD of three independent experiments. *P < .05, **P < .01.
Exosomal EGFR regulated HPV-16 E7-induced migration, invasion, and EMT in NSCLC cells
Our previous studies demonstrated that HPV-16 E7 promoted migration, invasion, and EMT in A549 and NCI-H460 cells.26–28 Moreover, in this study, we deeply confirmed that exosomes were involved in these processes (Figure 2). To further investigate the effect of exosomal EGFR on migration and invasion abilities promoted by HPV-16 E7, Transwell assay was used to analyze the migration and invasion abilities of NSCLC cells which were co-cultured with HPV-16 E7-overexpressing cells pretreated with different inhibitors. Our results showed that both exosome secretion inhibitor GW4869 and EGFR inhibitor PD168393 effectively inhibited the migration and invasion of NSCLC cells induced by HPV-16 E7 oncoprotein (P < .01, Figure 5a), indicating that exosomal EGFR was involved in migration and invasion of NSCLC cells induced by HPV-16 E7 oncoprotein.
Figure 5.
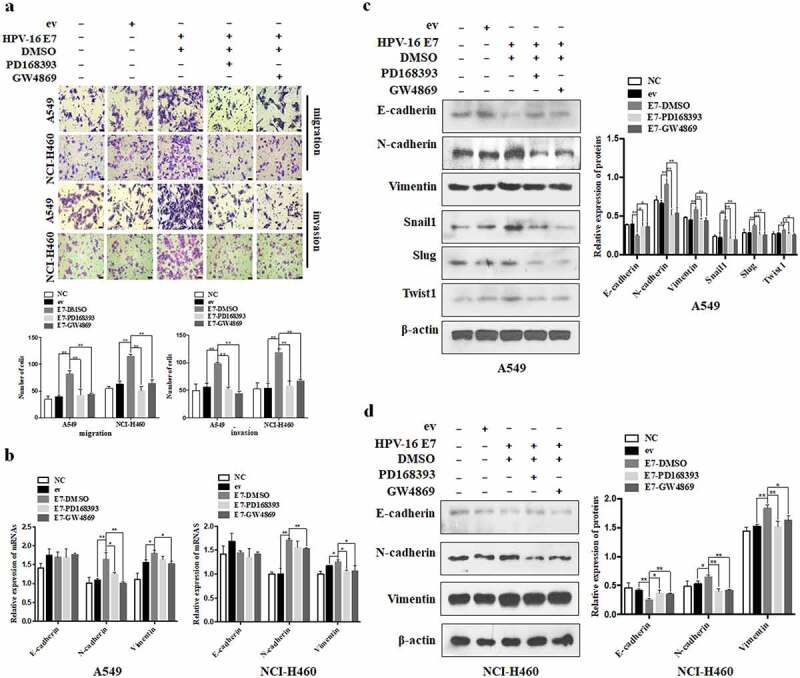
Exosomal EGFR was involved in migration, invasion, and EMT of NSCLC cells induced by HPV-16 E7. (a) Transwell assays were performed to analyze the migration and invasion abilities of A549 and NCI-H460 cells after co-cultured with HPV-16 E7-overexpressing cells (E7 cells) and empty vector cells (ev cells) treated with DMSO, PD168393, and GW4869, respectively; Up: one representative result of three independent experiments, scale bar = 100 μm; down: the number of migratory cells (left) and invasive cells (right) for three independent experiments. (b-d) EMT-related mRNA (b) and protein (c,d) levels were determined by RT-qPCR and Western blot after co-cultured with ev cells and E7 cells treated with DMSO, PD168393, and GW4869, respectively. The left of c (A549) and d (NCI-H460): one representative result of three independent experiments; the right of c (A549) and d (NCI-H460): the results of density for three independent experiments. All data are expressed as mean ± SD of three independent experiments. *P < .05, **P < .01.
Next, we analyzed the role of exosomal EGFR in HPV-16 E7-induced EMT of NSCLC cells. Our results demonstrated that HPV-16 E7 significantly up-regulated Vimentin and N-cadherin mRNA expression in both A549 and NCI-H460 cells (P < .05, Figure 5b). Moreover, HPV-16 E7 dramatically down-regulated E-cadherin expression while up-regulated N-cadherin and Vimentin expression in both A549 and NCI-H460 cells, and promoted Snail1, Slug, and Twist1 expression at protein levels in A549 cells (P < .05, Figure 5c,d). However, PD168393 and GW4869 remarkably reversed these effects (P < .05, Figure 5b-d). Collectively, these results indicated that exosomal EGFR mediated HPV-16 E7 oncoprotein-induced EMT in A549 and NCI-H460 cells.
Differential expression of miRNAs
In recent years, a large number of studies have shown that exosome-derived miRNAs play an important role in EMT of cancer cells.38–40 In order to further study whether exosome-derived miRNAs are involved in the EMT of NSCLC cells induced by HPV-16 E7 oncoprotein, the exosomes derived from the stable HPV-16 E7-overexpressing A549 cells and empty vector-infected A549 cells were analyzed by high-throughput sequencing. Our results showed that 12 kinds of differentially expressed miRNAs with fold change≥2 and P ≤ .05 were screened out, of which 7 kinds of miRNAs (let-7d-5p, miR-10b-5p, miR-221-3p, miR-30c-2-3p, miR-381-3p, miR-409-3p and miR-548o-3p) were up-regulated while 5 miRNAs (let-7d-3p, miR −101-3p, miR-125b-1-3p, miR-30c-1-3p and miR-30e-3p) were down-regulated in the exosomes derived from the stable HPV-16 E7-overexpressing A549 cells (Figure 6a-c). Moreover, the results of the signal pathway analysis of 12 differentially expressed miRNAs showed that the pathway in cancer was the most important, followed by the FoxO signaling pathway and the Hippo signaling pathway (Figure 6d).
Figure 6.
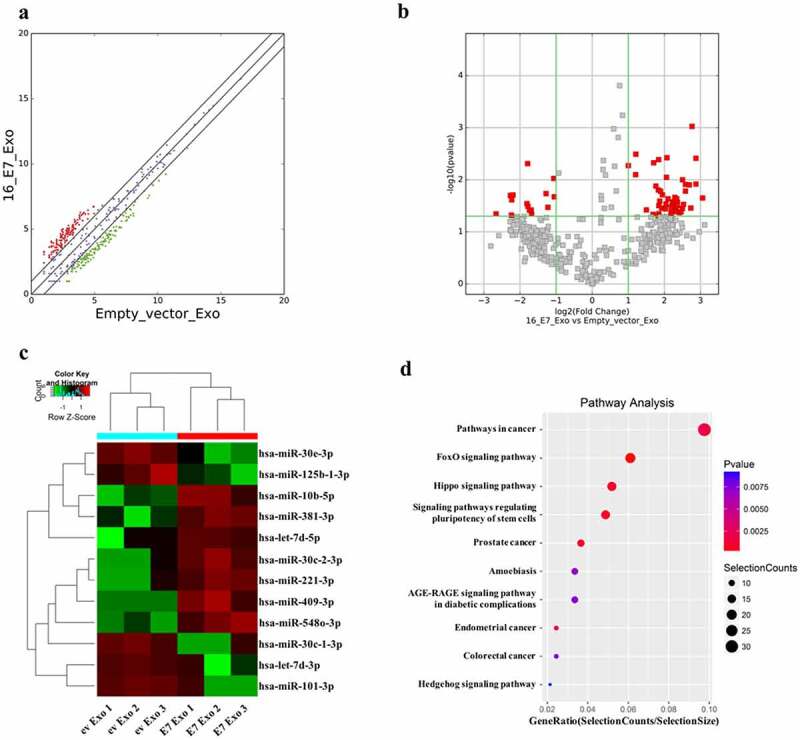
Screening of the differentially expressed miRNAs. The analysis of difference of miRNA expression between exosomes derived from the stable HPV-16 E7-overexpressing A549 cells (E7 Exo) and empty vector-infected A549 cells (ev Exo). (a) Scatter plots. (b) Volcano plots. (c) Clustering was performed on differentially expressed miRNAs between E7 Exo and ev Exo, fold change≥2 and P ≤ .05. (d) Pathway analysis of differentially expressed miRNAs in E7 Exo and ev Exo. All data are expressed as mean ± SD of three independent experiments.
Discussion
Exosomes are extracellular vesicles secreted by cells that mediate information exchange between the adjacent or distant cells. Accumulating evidence demonstrates that exosomes are involved in both physiological and pathological processes.41 Recently, exosomes have been found to play an important role in EMT of NSCLC cells.14,18,19 It was found that HPV-16 E7 can effectively promote EMT of NSCLC cells in our previous studies.26–28 However, the role of exosomes in HPV-16 E7-induced EMT has not been reported. In this study, we firstly demonstrated that the exosomes derived from the stable HPV-16 E7-overexpressing NSCLC cells significantly enhanced the proliferation, migration, and invasion of NSCLC cells as compared with the exosomes derived from empty vector-infected NSCLC cells (Figure 2a-c). EMT is the key step of migration and invasion, so we further analyzed the effect of the exosomes derived from the stable HPV-16 E7-overexpressing NSCLC cells on EMT of NSCLC cells. Our results showed that the exosomes derived from the stable HPV-16 E7-overexpressing NSCLC cells dramatically enhanced EMT in NSCLC cells (Figure 2d). Moreover, in vivo results further demonstrated the exosomes derived from the stable HPV-16 E7-overexpressing NSCLC cells promoted the growth and EMT of NSCLC (Figure 3). Therefore, our results indicate that exosomes can mediate HPV-16 E7-induced EMT in NSCLC cells.
Recent studies have shown that EGFR could be enriched in exosomes, and EGFR in exosomes was different from EGFR in cells in terms of quantity and binding strength with the targets.42 EGFR is a crucial target of NSCLC30–32 and exosomes may play a key role in the progression of EGFR-mutant NSCLC.34,35 Moreover, EMT was found to contribute to the resistance to EGFR-targeted therapies in NSCLC,43 and targeting the EMT transcription factor TWIST1 was demonstrated to overcome the resistance to EGFR inhibitors in EGFR-mutant NSCLC.44 However, the role of exosomal EGFR in HPV-16 E7-induced EMT of NSCLC cells is still unclear. In this study, we demonstrated that HPV-16 E7 promoted EGFR and p-EGFR protein expression in A549 cells (Figure 4a). Furthermore, EGFR protein expression was much higher in the exosomes derived from the stable HPV-16 E7-overexpressing NSCLC cells than that in the exosomes derived from empty vector-infected cells (Figure 4b). These results suggest that exosomes may be involved in the HPV-16 E7-induced EMT of NSCLC cells by delivering EGFR protein and exosomal EGFR may play a role in the progression of HPV-16-related NSCLC. In order to further confirm this proposal, in this study, NSCLC cells were pretreated with GW4869 (an inhibitor of exosome release) and PD168393 (an irreversible EGFR inhibitor), respectively, followed by the analysis of migration and invasion capabilities and the expression of EMT markers in NSCLC cells. Our results showed that both GW4869 and PD168393 inhibited HPV-16 E7-induced migration and invasion capabilities of NSCLC cells (Figure 5a). Furthermore, both GW4869 and PD168393 also suppressed HPV-16 E7-induced EMT of NSCLC cells (Figure 5b-d). Taken together, these findings suggest that exosomal EGFR mediated EMT of NSCLC cells induced by HPV-16 E7 oncoprotein.
Interestingly, in this study, we further found that EGFR inhibitor PD168393 exhibited less inhibitory effect on HPV-16 E7-induced EMT of NSCLC cells as compared with exosome release inhibitor GW4869 (Figure 5). We speculate that these novel phenomena may be because EGFR inhibitors can only inhibit the activation of EGFR in exosomes but have no effect on the delivery of other biomolecules by exosomes, exosomes can still carry other biomolecules besides EGFR protein into the recipient cells to produce biological effects.
Recent studies have shown that HPV-16 E7 can not only regulate the expression of cancer-related miRNAs in tumor cells,45,46 but also further affect miRNAs enrichment in exosomes.47 This is similar to our hypothesis that there are other biomolecules that can interact with exosomal EGFR. Therefore, we used high-throughput sequencing to analyze the differentially expressed miRNAs between the exosomes derived from the stable HPV-16 E7-overexpressing A549 cells and empty vector-infected A549 cells. The results showed that 7 kinds of miRNAs (let-7d-5p, miR-10b-5p, miR-221-3p, miR-30c-2-3p, miR-381-3p, miR-409-3p, miR-548o-3p) were up-regulated while 5 miRNAs (let-7d-3p, miR-101-3p, miR-125b-1-3p, miR-30c-1-3p and miR-30e-3p) were down-regulated in the exosomes derived from the stable HPV-16 E7-overexpressing A549 cells (Figure 6c). Previous studies showed that when the expression of HPV-18 E6/E7 in HeLa cells was inhibited, exosomal let-7d-5p was significantly reduced, confirming that the expression of exosomal let-7d-5p was E6/E7 dependent.47 Recently, it was reported that lung cancer cell derived exosomal let-7d-5p down-regulated OPRM1 to promote bone metastasis.48 HPV infection increased the expression of miR-221-3p,49 while exosomal miR-221-3p derived from cervical cancer cells promoted the migration, invasion, and angiogenesis of vascular endothelial cells by targeting thbs2, vash1 and mapk10, thereby promoting the progression of cervical cancer.50 Interestingly, miR-221-3p was highly expressed in lung cancer, and the inhibition of miR-221-3p expression reduced the growth and metastasis of lung cancer.51 Exosomal miR-10b-5p not only promoted the progression of glioma and gastric cancer,52,53 but also played an important role in the early diagnosis of liver cancer and NSCLC,54,55 especially lung adenocarcinoma.56All these previous studies suggest that the above miRNAs may be involved in exosomal EGFR-mediated EMT induced by HPV-16 E7 oncoprotein in NSCLC cells. Furthermore, we preliminarily found that FoxO signaling pathway and Hippo signaling pathway might be related to the regulation of 12 differentially expressed miRNAs (Figure 6d). Therefore, whether these signaling pathways can be involved in exosomal EGFR-mediated EMT of NSCLC cells induced by HPV-16 E7 oncoprotein via regulating the differential expression of these miRNAs and the underlying mechanisms need to be further explored
In conclusion, in this study, we firstly demonstrated that exosomal EGFR is involved in HPV-16 E7-induced EMT of NSCLC cells, suggesting that exosomes may play a key role in the progression of HPV-16-related NSCLC by delivering EGFR.
Biographies
Zhiyuan Zhou has got the degree of master from Guangdong Medical University. His research direction focuses on the relationship between exosomes and cancer, especially the role of exosomal proteins and miRNAs in NSCLC progression.
Xiaofeng Wu has got the degree of master from Guangdong Medical University. Currently, she is a senior MT in Affiliated Hospital of Guangdong Medical University. Her research direction focuses on the role of exosomes in NSCLC progression.
Riming Zhan has got the degree of master from Guangdong Medical University. Currently, he is a MT in Affiliated Hospital of Guangdong Medical University. His research direction focuses on the relationship between exosomes and cancer.
Xiangyong Li has got the degree of PhD from Wuhan University. Currently, he is a professor in Guangdong Medical University and mainly studies the role of small noncoding RNAs in tumorigenesis and development.
Dazhao Cheng is a graduate student of Guangdong Medical University. His research direction focuses on the role of exosomes in NSCLC progression.
Li Chen has got the degree of master from Guangdong Medical University. Her research direction focuses on the pathogenesis and treatment of NSCLC.
Tianyu Wang is a graduate student of Guangdong Medical University. Her research direction focuses on the mechanisms of NSCLC treatment.
Hua Yu is a graduate student of Guangdong Medical University. Her research direction focuses on the role of exosomes in NSCLC progression.
Guihong Zhang is a graduate student of Guangdong Medical University. His research direction focuses on the mechanisms of NSCLC treatment.
Xudong Tang has got the degree of PhD from Wuhan University. Currently, she is a professor and principal investigator in Guangdong Medical University. Her main research interests are the underlying molecular mechanisms of occurrence and development of NSCLC, especially the role of exosomes in NSCLC progression.
Funding Statement
This work was supported by the grants from National Natural Science Foundation of China, 81372511 (to X.T.); Guangdong Basic and Applied Basic Research Foundation, 2019A1515011081 (to X.T.); “Sail Plan” in Guangdong Province to Cultivate High-level Talents, 201635011 (to X.T.); and the Characteristic Innovation Project of Guangdong Province Ordinary University (Nature Science), 2022KTSCX048 (to X.T.).
Authors’ contributions
ZZ and XT designed the study and wrote the paper. ZZ, XW, TW, HY, and GZ performed cell experiments. RZ and DC performed animal experiments. ZZ, XW, RZ, and XT analyzed the data. ZZ, XL, and XT revised the paper. All authors contributed to the article and approved the submitted version.
Ethical Review Board approval
All experiments in this study have obtained the approval from Ethical Review Board of Guangdong Medical University.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F.. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Yuan J, Sun Y, Bu X, Ren H, Chen M.. Global, regional and national burden of lung cancer and its attributable risk factors in 204 countries and territories, 1990-2019. Eur J Cancer Prev. 2022;31(3):253–259. doi: 10.1097/CEJ.0000000000000687. [DOI] [PubMed] [Google Scholar]
- 3.Wu Z, Tan F, Yang Z, Wang F, Cao W, Qin C, Dong X, Zheng Y, Luo Z, Zhao L, et al. Sex disparity of lung cancer risk in non-smokers: a multicenter population-based prospective study based on China national lung cancer screening program. Chin Med J (Engl). 2022;135(11):1331–1339. doi: 10.1097/CM9.0000000000002161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corrales L, Rosell R, Cardona AF, Martín C, Zatarain-Barrón ZL, Arrieta O. Lung cancer in never smokers: the role of different risk factors other than tobacco smoking. Crit Rev Oncol Hematol. 2020;148:102895. doi: 10.1016/j.critrevonc.2020.102895. [DOI] [PubMed] [Google Scholar]
- 5.Karnosky J, Dietmaier W, Knuettel H, Freigang V, Koch M, Koll F, Zeman F, Schulz C. Schulz C. HPV and lung cancer: a systematic review and meta-analysis. Cancer Rep (Hoboken). 2021;4(4):e1350. doi: 10.1002/cnr2.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hussen BM, Ahmadi G, Marzban H, Fard Azar ME, Sorayyayi S, Karampour R, Nahand JS, Hidayat HJ, Moghoofei M. The role of HPV gene expression and selected cellular MiRNAs in lung cancer development. Microb Pathog. 2021;150:104692. doi: 10.1016/j.micpath.2020.104692. [DOI] [PubMed] [Google Scholar]
- 7.de Oliveira THA, do Amaral CM, de França São Marcos B, Nascimento KCG, de Miranda Rios AC, Quixabeira DCA, Muniz MTC, Silva Neto JDC, de Freitas AC. Presence and activity of HPV in primary lung cancer. J Cancer Res Clin Oncol. 2018;144(12):2367–2376. doi: 10.1007/s00432-018-2748-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang EY, Tang XD. Human papillomavirus type 16/18 oncoproteins: potential therapeutic targets in non-smoking associated lung cancer. Asian Pac J Cancer Prev. 2012;13(11):5363–5369. doi: 10.7314/apjcp.2012.13.11.5363. [DOI] [PubMed] [Google Scholar]
- 9.Huang JY, Lin C, Tsai SC, Lin FC. Human papillomavirus is associated with adenocarcinoma of lung: a population-based cohort study. Front Med (Lausanne). 2022;9:932196. doi: 10.3389/fmed.2022.932196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-Mediated metastasis: communication from a distance. Dev Cell. 2019;49(3):347–360. doi: 10.1016/j.devcel.2019.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Min L, Zhu T, Lv B, An T, Zhang Q, Shang Y, Yu Z, Zheng L, Wang Q. Exosomal LncRNA RP5-977B1 as a novel minimally invasive biomarker for diagnosis and prognosis in non- small cell lung cancer. Int J Clin Oncol. 2022;27(6):1013–1024. doi: 10.1007/s10147-022-02129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S, Lin Y, Wu Y, Chen H, Huang Z, Lin M, Dong J, Wang Y, Yang Z. The value of serum exosomal miR-184 in the diagnosis of NSCLC. J Healthc Eng. 2022;2022:9713218. doi: 10.1155/2022/9713218. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Xu K, Zhang C, Du T, Gabriel ANA, Wang X, Li X, Sun L, Wang N, Jiang X, Zhang Y. Progress of exosomes in the diagnosis and treatment of lung cancer. Biomed Pharmacother. 2021;134:111111. doi: 10.1016/j.biopha.2020.111111. [DOI] [PubMed] [Google Scholar]
- 14.Jouida A, O’Callaghan M, Mc Carthy C, Fabre A, Nadarajan P, Keane MP. Exosomes from EGFR-mutated adenocarcinoma induce a hybrid EMT and MMP9-dependant tumor invasion. Cancers (Basel). 2022;14(15):3776. doi: 10.3390/cancers14153776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang R, Liu H, Dong M, Huang D, Yi J. Exosomal hsa_circ_0000519 modulates the NSCLC cell growth and metastasis via miR-1258/RHOV axis. Open Med (Wars). 2022;17(1):826–840. doi: 10.1515/med-2022-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wan X, Xie B, Sun H, Gu W, Wang C, Deng Q, Zhou S. Exosomes derived from M2 type tumor-associated macrophages promote osimertinib resistance in non-small cell lung cancer through MSTRG.292666.16-miR-6836-5p-MAPK8IP3 axis. Cancer Cell Int. 2022;22(1):83. doi: 10.1186/s12935-022-02509-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi L, Zhu W, Huang Y, Zhuo L, Wang S, Chen S, Zhang B, Ke B. Cancer-associated fibroblast-derived exosomal microRNA- 20a suppresses the PTEN/PI3K-AKT pathway to promote the progression and chemoresistance of non-small cell lung cancer. Clin Transl Med. 2022;12(7):e989. doi: 10.1002/ctm2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hisakane K, Seike M, Sugano T, Yoshikawa A, Matsuda K, Takano N, Takahashi S, Noro R, Gemma A. Exosome-derived miR-210 involved in resistance to osimertinib and epithelial-mesenchymal transition in EGFR mutant non-small cell lung cancer cells. Thorac Cancer. 2021;12(11):1690–1698. doi: 10.1111/1759-7714.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.You J, Li M, Cao LM, Gu QH, Deng PB, Tan Y, Hu CP. Snail1-dependent cancer-associated fibroblasts induce epithelial-mesenchymal transition in lung cancer cells via exosomes. QJM. 2019;112(8):581–590. doi: 10.1093/qjmed/hcz093. [DOI] [PubMed] [Google Scholar]
- 20.Pastushenko I, Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29(3):212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Weinberg RA. Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front Med. 2018;12(4):361–373. doi: 10.1007/s11684-018-0656-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei L, Sun JJ, Cui YC, Jiang SL, Wang XW, Lv LY, Xie L, Song XR. Twist may be associated with invasion and metastasis of hypoxic NSCLC cells. Tumour Biol. 2016;37(7):9979–9987. doi: 10.1007/s13277-016-4896-2. [DOI] [PubMed] [Google Scholar]
- 23.Zhu X, Chen L, Liu L, Niu X. EMT-mediated acquired EGFR-TKI resistance in NSCLC: mechanisms and strategies. Front Oncol. 2019;9:1044. doi: 10.3389/fonc.2019.01044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adachi Y, Ito K, Hayashi Y, Kimura R, Tan TZ, Yamaguchi R, Ebi H. Epithelial-to-mesenchymal transition is a cause of both intrinsic and acquired resistance to KRAS G12C inhibitor in KRAS G12C-mutant non-small cell lung cancer. Clin Cancer Res. 2020;26(22):5962–5973. doi: 10.1158/1078-0432.ccr-20-2077. [DOI] [PubMed] [Google Scholar]
- 25.Nurwidya F, Takahashi F, Winardi W, Tajima K, Mitsuishi Y, Murakami A, Kobayashi I, Nara T, Hashimoto M, Kato M, et al. Zinc-finger E-box-binding homeobox 1 (ZEB1) plays a crucial role in the maintenance of lung cancer stem cells resistant to gefitinib. Thorac Cancer. 2021;12(10):1536–1548. doi: 10.1111/1759-7714.13937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, Wu X, Hu L, Ma Y, Xiu Z, Huang B, Feng Y, Tang X. Overexpression of human papillomavirus Type 16 oncoproteins enhances epithelial-mesenchymal transition via STAT3 signaling pathway in non-small cell lung cancer cells. Oncol Res. 2017;25(5):843–852. doi: 10.3727/096504016x14813880882288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Huang B, Xiu Z, Zhou Z, Liu J, Li X, Tang X. PI3K/Akt/HIF-1α signaling pathway mediates HPV-16 oncoprotein-induced expression of EMT-related transcription factors in non-small cell lung cancer cells. J Cancer. 2018;9(19):3456–3466. doi: 10.7150/jca.26112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang B, Zhou Z, Liu J, Wu X, Li X, He Q, Zhang P, Tang X. The role of monoamine oxidase A in HPV-16 E7-induced epithelial-mesenchymal transition and HIF-1α protein accumulation in non-small cell lung cancer cells. Int J Biol Sci. 2020;16(14):2692–2703. doi: 10.7150/ijbs.46966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.da Cunha Santos G, Shepherd FA, Tsao MS. EGFR mutations and lung cancer. Annu Rev Pathol. 2011;6(1):49–69. doi: 10.1146/annurev-pathol-011110-130206. [DOI] [PubMed] [Google Scholar]
- 30.Nishihara S, Yamaoka T, Ishikawa F, Ohmori T, Ando K, Kusumoto S, Kishino Y, Manabe R, Hasebe Y, Sagara H, et al. Diverse mechanisms of resistance against osimertinib, a third-generation EGFR-TKI, in lung adenocarcinoma cells with an EGFR-Activating mutation. Cells. 2022;11(14):2201. doi: 10.3390/cells11142201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren KH, Qin WW, Wang Y, Peng JC, Hu WX. Detection of an EML4-ALK fusion mutation secondary to epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI) therapy for lung cancer: a case report. Ann Palliat Med. 2022;11(7):2503–2509. doi: 10.21037/apm-22-744. [DOI] [PubMed] [Google Scholar]
- 32.Pacini L, AD Jenks, Vyse S, CP Wilding, Arthur A, PH Huang. Tackling drug resistance in EGFR Exon 20 insertion mutant lung cancer. Pharmgenomics Pers Med. 2021;14:301–317. doi: 10.2147/pgpm.s242045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MS Clement, KR Gammelgaard, AL Nielsen, BS Sorensen. Epithelial-to-mesenchymal transition is a resistance mechanism to sequential MET-TKI treatment of MET-amplified EGFR-TKI resistant non-small cell lung cancer cells. Transl Lung Cancer Res. 2020;9(5):1904–1914. doi: 10.21037/tlcr-20-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu S, Sha H, Qin X, Chen Y, Li X, Shi M, Feng J. EGFR E746-A750 deletion in lung cancer represses antitumor immunity through the exosome-mediated inhibition of dendritic cells. Oncogene. 2020;39(13):2643–2657. doi: 10.1038/s41388-020-1182-y. [DOI] [PubMed] [Google Scholar]
- 35.Liu X, Jiang T, Li X, Zhao C, Li J, Zhou F, Zhang L, Zhao S, Jia Y, Shi J, et al. Exosomes transmit T790M mutation-induced resistance in EGFR-mutant NSCLC by activating PI3K/AKT signalling pathway. J Cell Mol Med. 2020;24(2):1529–1540. doi: 10.1111/jcmm.14838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang H, Pan Z, Cai X, Wang W, Guo C, He J, Chen Y, Liu Z, Wang B, He J, et al. The association between human papillomavirus presence and epidermal growth factor receptor mutations in Asian patients with non-small cell lung cancer. Transl Lung Cancer Res. 2018;7(3):397–403. doi: 10.21037/tlcr.2018.03.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li G, He L, Zhang E, Shi J, Zhang Q, Le AD, Zhou K, Tang X. Overexpression of human papillomavirus (HPV) type 16 oncoproteins promotes angiogenesis via enhancing HIF-1α and VEGF expression in non-small cell lung cancer cells. Cancer Lett. 2011;311(2):160–170. doi: 10.1016/j.canlet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 38.Rezaei R, Baghaei K, Amani D, Piccin A, Hashemi SM, Asadzadeh Aghdaei H, Zali MR. Exosome-mediated delivery of functionally active miRNA-375-3p mimic regulate epithelial mesenchymal transition (EMT) of colon cancer cells. Life Sci. 2021;269:119035. doi: 10.1016/j.lfs.2021.119035. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Lai X, Yue Q, Cao F, Zhang Y, Sun Y, Tian J, Lu Y, He L, Bai J, et al. Bone marrow mesenchymal stem cells-derived exosomal microRNA-16-5p restrains epithelial-mesenchymal transition in breast cancer cells via EPHA1/NF-κB signaling axis. Genomics. 2022;114(3):110341. doi: 10.1016/j.ygeno.2022.110341 [DOI] [PubMed] [Google Scholar]
- 40.Wang S, Du P, Cao Y, Ma J, Yang X, Yu Z, Yang Y. Cancer associated fibroblasts secreted exosomal miR-1290 contributes to prostate cancer cell growth and metastasis via targeting GSK3β. Cell Death Discov. 2022;8(1):371. doi: 10.1038/s41420-022-01163-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He C, Zheng S, Luo Y, Wang B. Exosome theranostics: biology and translational medicine. Theranostics. 2018;8(1):237–255. doi: 10.7150/thno.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sung MS, Jung JH, Jeong C, Yoon TY, Park JH. Single-molecule co-immunoprecipitation reveals functional inheritance of EGFRs in extracellular vesicles. Small. 2018;14(42):e1802358. doi: 10.1002/smll.201802358. [DOI] [PubMed] [Google Scholar]
- 43.Tulchinsky E, Demidov O, Kriajevska M, Barlev NA, Imyanitov E. EMT:a mechanism for escape from EGFR-targeted therapy in lung cancer. Biochim Biophys Acta Rev Cancer. 2019;1871(1):29–39. doi: 10.1016/j.bbcan.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Yochum ZA, Cades J, Wang H, Chatterjee S, Simons BW, O’Brien JP, Khetarpal SK, Lemtiri-Chlieh G, Myers KV, Huang EH, et al. Targeting the EMT transcription factor TWIST1 overcomes resistance to EGFR inhibitors in EGFR-mutant non-small-cell lung cancer. Oncogene. 2019;38(5):656–670. doi: 10.1038/s41388-018-0482-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harden ME, Munger K. Human papillomavirus 16 E6 and E7 oncoprotein expression alters microRNA expression in extracellular vesicles. Virology. 2017;508:63–69. doi: 10.1016/j.virol.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiantore MV, Mangino G, Iuliano M, Zangrillo MS, De Lillis I, Vaccari G, Accardi R, Tommasino M, Columba Cabezas S, Federico M, et al. Human papillomavirus E6 and E7 oncoproteins affect the expression of cancer-related microRNAs: additional evidence in HPV-induced tumorigenesis. J Cancer Res Clin Oncol. 2016;142(8):1751–1763. doi: 10.1007/s00432-016-2189-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Honegger A, Schilling D, Bastian S, Sponagel J, Kuryshev V, Sültmann H, Scheffner M, Hoppe-Seyler K, Hoppe-Seyler F, Wells S. Dependence of intracellular and exosomal microRNAs on viral E6/E7 oncogene expression in HPV-positive tumor cells. PLoS Pathog. 2015;11(3):e1004712. doi: 10.1371/journal.ppat.1004712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X, Chen Y, Wang J, Jiang C, Huang Y. Lung cancer cell-derived exosomal let-7d-5p down-regulates OPRM1 to promote cancer-induced bone pain. Front Cell Dev Biol. 2021;9:666857. doi: 10.3389/fcell.2021.666857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Babion I, Miok V, Jaspers A, Huseinovic A, Steenbergen RDM, van Wieringen WN, Wilting SM. Identification of deregulated pathways, key regulators, and novel miRNA-mRNA interactions in HPV-mediated transformation. Cancers (Basel). 2020;12(3):700. doi: 10.3390/cancers12030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu XG, Zhou CF, Zhang YM, Yan RM, Wei WF, Chen XJ, Yi HY, Liang LJ, Fan LS, Liang L, et al. Cancer-derived exosomal miR-221-3p promotes angiogenesis by targeting THBS2 in cervical squamous cell carcinoma. Angiogenesis. 2019;22(3):397–410. doi: 10.1007/s10456-019-09665-1 [DOI] [PubMed] [Google Scholar]
- 51.Di Paolo D, Pontis F, Moro M, Centonze G, Bertolini G, Milione M, Mensah M, Segale M, Petraroia I, Borzi C, et al. Cotargeting of miR-126-3p and miR-221-3p inhibits PIK3R2 and PTEN, reducing lung cancer growth and metastasis by blocking AKT and CXCR4 signalling. Mol Oncol. 2021;15(11):2969–2988. doi: 10.1002/1878-0261.13036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qian M, Chen Z, Guo X, Wang S, Zhang Z, Qiu W, Qi Y, Zhang S, Xu J, Zhao R, et al. Exosomes derived from hypoxic glioma deliver miR-1246 and miR-10b-5p to normoxic glioma cells to promote migration and invasion. Lab Invest. 2021;101(5):612–624. doi: 10.1038/s41374-020-00522-0 [DOI] [PubMed] [Google Scholar]
- 53.Yan T, Wang X, Wei G, Li H, Hao L, Liu Y, Yu X, Zhu W, Liu P, Zhu Y, et al. Exosomal miR-10b-5p mediates cell communication of gastric cancer cells and fibroblasts and facilitates cell proliferation. J Cancer. 2021;12(7):2140–2150. doi: 10.7150/jca.47817.eCollection2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghosh S, Bhowmik S, Majumdar S, Goswami A, Chakraborty J, Gupta S, Aggarwal S, Ray S, Chatterjee R, Bhattacharyya S, et al. The exosome encapsulated microRNAs as circulating diagnostic marker for hepatocellular carcinoma with low alpha-fetoprotein. Int J Cancer. 2020;147(10):2934–2947. doi: 10.1002/ijc.33111 [DOI] [PubMed] [Google Scholar]
- 55.Jin X, Chen Y, Chen H, Fei S, Chen D, Cai X, Liu L, Lin B, Su H, Zhao L, et al. Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res. 2017;23(17):5311–5319. doi: 10.1158/1078-0432.CCR-17-0577 [DOI] [PubMed] [Google Scholar]
- 56.Shan X, Zhang L, Zhu DX, Zhou X, Zhang H, Liu QX, Tang JW, Wen W, Wang TS, Zhu W, et al. Serum microRNA expression profiling revealing potential diagnostic biomarkers for lung adenocarcinoma. Chin Med J (Engl). 2020;133(21):2532–2542. doi: 10.1097/CM9.0000000000001100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


