Abstract
Objective
We aimed to develop a deep learning (DL)–based algorithm for early glaucoma detection based on color fundus photographs that provides information on defects in the retinal nerve fiber layer (RNFL) and its thickness from the mapping and translating relations of spectral domain OCT (SD-OCT) thickness maps.
Design
Developing and evaluating an artificial intelligence detection tool.
Subjects
Pretraining paired data of color fundus photographs and SD-OCT images from 189 healthy participants and 371 patients with early glaucoma were used.
Methods
The variational autoencoder (VAE) network training architecture was used for training, and the correlation between the fundus photographs and RNFL thickness distribution was determined through the deep neural network. The reference standard was defined as a vertical cup-to-disc ratio of ≥0.7, other typical changes in glaucomatous optic neuropathy, and RNFL defects. Convergence indicates that the VAE has learned a distribution that would enable us to produce corresponding synthetic OCT scans.
Main Outcome Measures
Similarly to wide-field OCT scanning, the proposed model can extract the results of RNFL thickness analysis. The structural similarity index measure (SSIM) and peak signal-to-noise ratio (PSNR) were used to assess signal strength and the similarity in the structure of the color fundus images converted to an RNFL thickness distribution model. The differences between the model-generated images and original images were quantified.
Results
We developed and validated a novel DL-based algorithm to extract thickness information from the color space of fundus images similarly to that from OCT images and to use this information to regenerate RNFL thickness distribution images. The generated thickness map was sufficient for clinical glaucoma detection, and the generated images were similar to ground truth (PSNR: 19.31 decibels; SSIM: 0.44). The inference results were similar to the OCT-generated original images in terms of the ability to predict RNFL thickness distribution.
Conclusions
The proposed technique may aid clinicians in early glaucoma detection, especially when only color fundus photographs are available.
Keywords: Image-to-image translation, Glaucoma, Autoencoder, OCT, Retinal nerve fiber layer thickness
Abbreviations and Acronyms: DL, deep learning; DNN, deep neural network; GAN, generative adversarial network; PSNR, peak signal-to-noise ratio; RNFL, retinal nerve fiber layer; SD-OCT, spectral domain optical coherence tomography; SSIM, structural similarity index measure; VAE, variational autoencoder
Glaucoma, a multifactorial optic degenerative neuropathy, is the leading cause of irreversible blindness worldwide.1, 2, 3 It is characterized by a progressive loss of retinal ganglion cells and axons, followed by irreversible visual field deterioration.2,4,5 In 2013, approximately 64.3 million people (age: 40–80 years) had glaucoma,6 and an increase to 111.8 million is estimated by 2040. Approximately 60% of glaucoma cases are reported from Asia.6 However, effective glaucoma screening and detection are difficult because the condition is chronic and asymptomatic. In addition, glaucoma diagnosis requires high–diagnostic power equipment and professional evaluation by physicians.
Glaucoma is diagnosed on the basis of characteristic changes in the optic discs, defects in the retinal nerve fiber layer (RNFL), and the corresponding loss in the visual field.7,8 Diagnosis based on RNFL thickness, a major indicator of glaucoma, offers high accuracy.7 RNFL loss can be diffuse or local.9 Color fundus photography and spectral domain OCT (SD-OCT) are the most widely used clinical tools to noninvasively document and quantify retinal structures and can facilitate glaucoma diagnosis and screening.9,10 Fundus photography is the simplest method for assessing changes in the optic discs; however, interpretating the photographs is labor intensive and subjective. In addition, although OCT is highly accurate and reproducible, it is often unavailable in regions with few medical resources.
Artificial intelligence based on computer vision and deep learning (DL) has dominated medical care by revolutionizing diagnosis.10 Automated computer vision is used in glaucoma detection to identify the position of the optic cup and disc in a fundus image.11, 12, 13, 14, 15, 16, 17, 18 In addition to traditional computer vision analysis, DL performs favorably in glaucoma detection because trained algorithms can learn features from the data rather than from predefined rules.11 The DL algorithms developed by Ting et al to detect referable glaucoma exhibited high performance (area under the curve: 0.942).19,20 However, image-to-image translation entails visual and graphical problems, and the goal is to learn the mapping relations between the input images and output images. Image-to-image translation is mostly used for (1) noise reduction, (2) super resolution, (3) image synthesis, and (4) image reconstruction.21 These applications rely on 2 powerful deep neural networks (DNNs): autoencoders and generative adversarial networks (GANs).22, 23, 24, 25 An autoencoder’s network architecture consists of an encoder and a decoder. An autoencoder recodes features through compression and decompression and can be applied to the learning of correlations in input data and feature extraction, dimensionality reduction, generative models, and the pretraining of unsupervised learning models.26
This study identified the structure of and color mapping relations between fundus images and RNFL thickness by using the DL structure of an autoencoder.27 A medical image style-transfer technology was developed. This study used this technology to convert a fundus image into an RNFL thickness distribution map, similar to those created through OCT. The study created a model to assist clinicians with or without specialization in glaucoma for early glaucoma screening at clinics and medical institutions without OCT equipment.
Methods
DNN Architecture
U-Net is a U-shaped, symmetric convolutional network with a down-sampling contraction path and an up-sampling expansion path. It requires few training data and is particularly advantageous for image segmentation.28 This study adopted the U-Net as the core training network for the DNN to extract new images exhibiting the distribution of RNFL thickness from a fundus image database. The deep generative learning models were a variational autoencoder (VAE) and the GAN.29, 30, 31 The VAE enabled the model to encode and decode features under limited conditions. The model-generated vectors were forced to conform to a Gaussian distribution to control image generation.32 The GAN enabled the model to determine the distribution of the training data through competition between the generative and discriminative models. With the VAE and GAN, the model generated samples that conformed to this distribution and generation limitations. Thus, with the GAN as the core training network, the required sample size was large, and the model was unlikely to converge. Thus, in this study, the VAE-varied U-Net was used as the primary training network architecture. The paired fundus and RNFL thickness distribution images were used for model training (Fig 1). All paired data were resized to 256 × 256 × 3 and input into the model. Because the RNFL thickness distribution images were derived from OCT information across multiple hierarchies, the U-Net network design was advantageous. This decreased the probability of nonconvergence because of an insufficient number of training samples in the model.
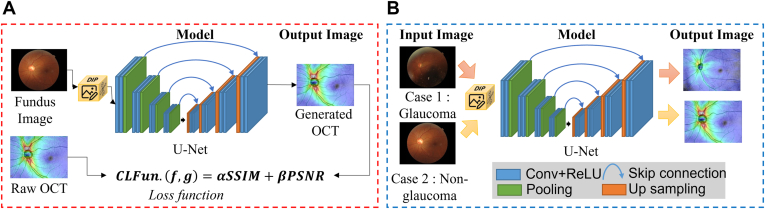
Figure 1.
Illustration of the pipelines in model training and model application for the conversion from fundus images to OCT thickness maps. A, Model training: Color fundus images were inputted into the U-Net for generating OCT thickness maps. In addition, model parameter correction was performed using raw OCT images and the compound loss function (CLFun.) proposed in this study. Images similar to the original OCT thickness maps were generated. B, Model application: Only the color fundus images of the patients were required to generate output OCT thickness maps, which can be used for early glaucoma diagnosis. Conv = convolution layer; PSNR = peak signal-to-noise ratio; ReLU = rectified linear unit; SSIM = structural similarity index measure.
Training Methods
To enable the model to efficiently identify mapping functions for image translation, images were normalized using a digital image-processing unit. During model training, all images were flipped and rotated before being input into the model, leading to random augmentation. To handle differences in brightness and contrast between images, adaptive histogram equalization was used for image preprocessing during model training; this enhanced the details of the images and suppressed background noise. The k-fold cross-validation technique is one of the most used approaches by practitioners for model selection and error estimation of classifiers.33 In this study, fivefold cross-validation was applied for training and validation with the original and augmented data sets. Retrospective samples were classified using random numbers. Among the samples, 80% were used for model training, and the remaining samples were reserved for the validation data set. The training samples were classified fivefold, with random numbers used in the model training process for model assessment. The validation samples were not used for model training and vice versa. The model could be assessed objectively when it received the test samples for the first time.
Table S1 presents the training process for pseudocode description. The U-Net was used as the DL training model, and the structural similarity index measure (SSIM) and peak signal-to-noise ratio (PSNR) were used as compound loss functions. Adam was used to optimize model parameter correction. Adam is simple and computationally efficient and requires little memory. It is widely used to train DNNs because of its adaptive learning rate and excellent performance.
Data Sets
To determine the feasibility of extracting RNFL thickness features from color fundus images, we paired fundus and OCT images. This retrospective case study was approved by the Human Experiment Ethics Committee of Taipei Chang Gung Memorial Hospital, Taiwan (IRB: 201900528B0C502). All participants underwent a comprehensive ophthalmologic examination, and their diagnoses were confirmed by glaucoma specialists on the basis of their history, the results of visual acuity measurement, slit lamp evaluation, intraocular pressure measurement, standard automated perimetry, fundus evaluation, and OCT scan images. Glaucoma was affirmed by the consensus of 2 glaucoma specialists, and a third glaucoma specialist was involved in case of disagreement. Only participants with open angles on gonioscopy were included in our study. Participants were excluded if they had any other ocular or systemic disease affecting the fundus structures or visual field. Color fundus photographs and OCT scan images were obtained from the Glaucoma Clinic in the Department of Ophthalmology of Chang Gung Memorial Hospital, Taipei, Taiwan. Low-quality fundus images with poor location were excluded from the training and validation data sets. Standard automated perimetry was performed using the Swedish interactive threshold algorithm with the central 24-2 program of the Humphrey Field Analyzer III (Carl Zeiss Meditec). The inclusion criteria were diagnosis of early glaucoma based on a repeatable Humphrey Field Analyzer 24-2 Swedish Interactive Thresholding Algorithm (SITA) standard visual field defect comparable with glaucoma, a mean deviation of at least −6 decibels (dB), and optic nerve head and RNFL damage confirmed through fundus photography and SD-OCT. The visual fields were considered reliable if the false-positive response rate was <15% and the fixation loss rate and false-negative response rate were <25%. Swept-source OCT (DRI OCT Triton, Topcon) was used for OCT detection. Traditional OCT machines have a small scanning range, whereas the swept-source OCT is a wide field (scanning area: 12 mm × 9 mm) and thus enables the effective assessment of early glaucoma. To increase the clinical diagnostic value of the conversion technology, the DRI OCT Triton was used to produce the OCT index.
Verification of the Proposed Model: Loss Function Design
The loss function was specially designed to ensure that the model achieved image generation control under limited conditions. The images displayed the anatomical structures in the fundus, such as the vessels (arteries and veins), optic disc, and macula, and a layer of jet-color heatmap to display the thickness distribution. An analysis of the quality of the training fundus images indicated that the luminance was low. In some samples, the pupils were small. Because incident was limited light, the entire fundus did not display completely. Therefore, a digital image-processing unit was used for image preprocessing with contrast-limited adaptive histogram equalization. Under contrast-limited conditions, the feature details in the bright and dark parts of the images improved.34
The SSIM, proposed by Wang et al,35 is an index used to assess the similarity of 2 images. It is highly correlated to the image quality of the human vision system. The SSIM uses the distortion characteristic function of images as a mathematical model. This model covers 3 image distortion assessment methods: loss of correlation, luminance distortion, and contrast distortion.36 Its formula is as follows:
| (1) |
where
| (2) |
The first part, l(f, g), compares luminance, with the average luminance of the 2 images being μf and μg. The second part, c(f, g), compares the contrast, and standard deviations σ of the images were calculated to determine whether they had similar contrast. The third part, s(f, g), is used to determine whether the images have similar structure. Image covariance σfg was calculated to assess the correlation coefficient of the images. The SSIM, obtained by multiplying the values of the 3 parts, was between 0 and 1. Structural similarity index measure values approaching 0 indicate no correlation between images, whereas those approaching 1 indicate identical images.
The PSNR was used to calculate the differences between the generated and reference images (ground truth) of the signal conversion, which indicated model performance. The PSNR, an objective index, is commonly used to assess the signal-to-noise ratios of images,37,38 such as image compression. According to its mathematical definition, when the mean square error of the color space of the assessed image and the ground truth approach 0, the 2 images are the same. Thus, the higher the PSNR is, the more similar are the signals of the 2 images.
The formula for PSNR is as follows:
| (3) |
where
| (4) |
where f is the reference image, g is the test image, MaxI is the maximum value of image pixels, and m and n represent image width and height, respectively.
The compound loss function is used to assess anatomical structure generation. The SSIM was used as the model performance assessment index to ensure the structural information obtained from the original fundus images.39 The PSNR was used to verify the signal conversion quality of the RNFL thickness mapping information (from the fundus images) extracted by the DNN. To ensure the optic disc and cup retained their structure after the conversion of the DNN, the SSIM and PSNR were used as the compound loss function of the model.
| (5) |
where
| (6) |
Results
This study used the data of 200 healthy participants and 550 patients with glaucoma. After the data were cleaned, the images from 560 participants were included (both eyes from 371 glaucoma patients and 189 nonglaucoma controls) for further analysis. Thus, the fundus images of 1120 eyes (742 in glaucoma and 378 in nonglaucoma) and their corresponding OCT thickness maps were included for model training and validation. Table 1 presents the demographic data of the participants. The average age was 51 years in the glaucoma group and 44 years in the nonglaucoma group. Among the participants in each group, 45% and 66% of them were female in glaucoma and nonglaucoma groups, respectively. As for myopia, 25% and 18% of the participants had myopia in glaucoma and nonglaucoma groups, respectively.
Table 1.
Demographic Data of the Study Participants
| Parameter | Glaucoma | Nonglaucoma |
|---|---|---|
| Number of subjects | 371 | 189 |
| Number of eyes | 742 | 378 |
| Age, years (SD) | 50.96 (11.96) | 44.41 (15.29) |
| Female (%) | 45.20 | 66.15 |
| Myopia (%) | 24.94 | 18.19 |
SD = standard deviation.
The hierarchy information of the color space in the retinal thickness distribution was analyzed. In addition to the image translation quality assessment, the performance of U-Net models with various depths was evaluated (Fig 2). As the number of layers in the network architecture increased, the pooling and sampling became deeper and more complex. Features such as color space and structural information extracted by the U-Net network were analyzed objectively, and the feature relations of the model in the RNFL mapping were identified. The model’s feasibility was determined through operational performance analysis.
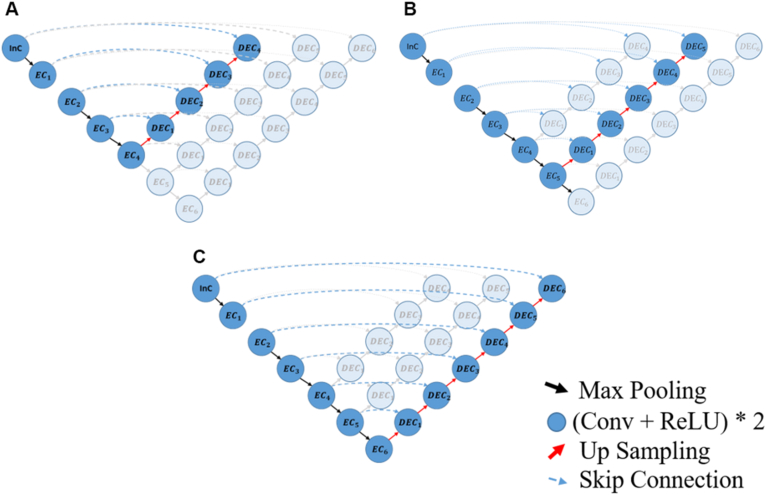
Figure 2.
U-Net architectures with different depths to compare image translation performances: A, 4 × 4 U-Net, B, 5 × 5 U-Net, and C, 6 × 6 U-Net. Conv = convolution layer; DEC = decoder convolution; EC = encoder convolution; InC = input convolution; ReLU = rectified linear unit.
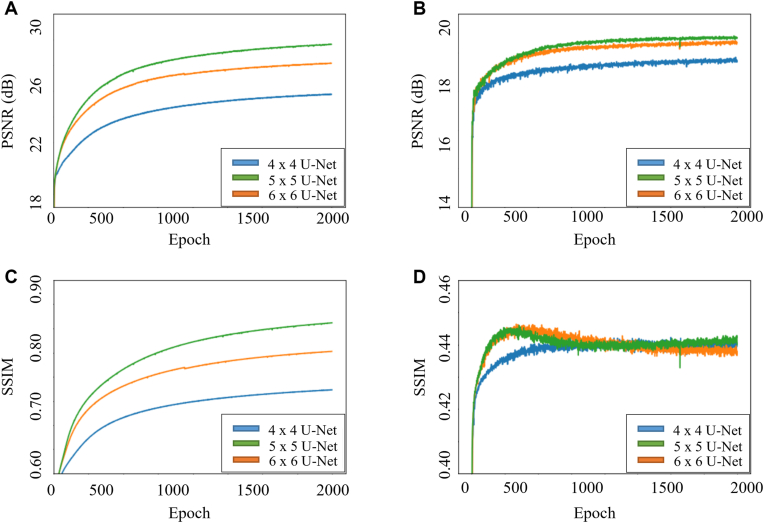
Figure 3 presents the learning curves of the networks during training and validation using different network architecture in the PSNR and SSIM. The performance of the training data set indicated that the network architecture depth led to significant differences in the PSNR and SSIM. The model performance index improved as the depth increased. For the validation set, the SSIM exhibited overfitting in the 5 × 5 and 6 × 6 architectures. For the 6 × 6 architecture, the SSIM decreased at approximately 300 epochs, whereas for the 5 × 5 architecture, it decreased at approximately 400 epochs. The performance of the model with different depths and parameters was also shown in Table 2. The results indicated that the PSNR increased with network architecture depth. The PSNR values were from 23.8 dB and 18.8 dB in 4 × 4 architecture to 27.6 dB and 19.4 dB in 6 × 6 architecture, respectively, during training and validation. However, the depth had little effect on the SSIM, which had the same value (0.44) in all architectures during validation.
Figure 3.
Learning curves of the model during training and validation with different network architectures in loss function measurement of peak signal-to-noise ratio (PSNR) and structural similarity index measure (SSIM). Learning curves of (A) training PSNR, (B) validation PSNR, (C) training SSIM, and (D) validation SSIM.
Table 2.
Performance of U-Net Networks and Models with Various Depths and Parameters
| Architecture | Parameters | Training |
Validation |
||
|---|---|---|---|---|---|
| PSNR (dB) | SSIM | PSNR (dB) | SSIM | ||
| 4 × 4 U-Net | 13.40 M | 23.76 ± 1.20 | 0.67 ± 0.03 | 18.84 ± 2.43 | 0.44 ± 0.09 |
| 5 × 5 U-Net | 53.52 M | 26.16 ± 1.10 | 0.75 ± 0.03 | 19.31 ± 3.06 | 0.44 ± 0.12 |
| 6 × 6 U-Net | 213.97 M | 27.63 ± 0.99 | 0.81 ± 0.12 | 19.42 ± 0.44 | 0.44 ± 0.02 |
dB = decibels; M = million; PSNR = peak signal-to-noise ratio; SSIM = structural similarity index measure.
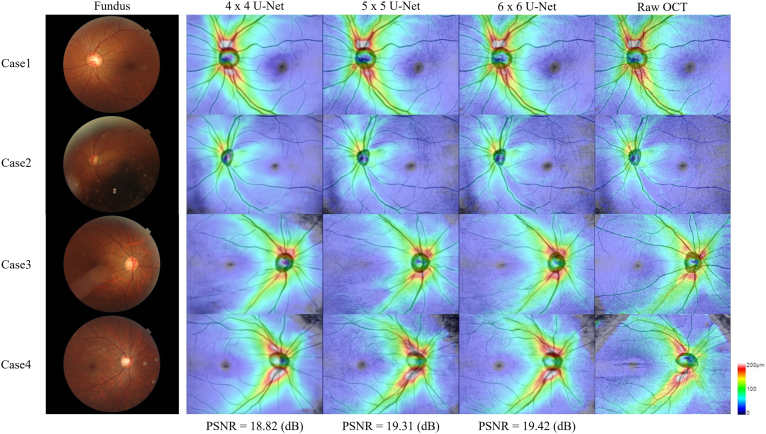
The 4 cases not included in model training were randomly selected as samples for demonstration of the results in different U-Net depths (Fig 4). The key feature details of the generated images were assessed. Minimal differences were noted in the translated and generated structures of the optic disc, optic cup, macula, and large blood vessels regarding to different network depths. Only peripheral blood vessels were unclear in the shallower networks (i.e., 4 × 4 U-Net). The RNFL thickness images presented in the heatmap graphs were similar to the raw OCT images.
Figure 4.
Four cases were used as examples to display the performance of a U-Net with various depths. The thickness information corresponding to the color is provided on the right side of the image. The cooler the color is, the thinner the relative thickness is. Comparing to the raw OCT, the 4 × 4 U-Net was not as ideal as the 5 × 5 and 6 × 6 U-Nets in the detailed translation of blood vessels. The 5 × 5 and 6 × 6 U-Nets differed little to the naked eye.
In addition to the translation performance of the models, the parameters and corresponding time required for the models with various depths were compared (Table S2). According to the GPU (Nvidia Geforce GTX2080Ti, 3,584 cuda cores and 11GB memory [GDDR5X]) operation time analysis, the 3 architectures had similar operating times (average: approximately 5.5 ms). However, the central processing unit operating time of the 6 × 6 architecture was > 0.5 sec, approximately twice that of the 4 × 4 architecture.
The characteristics of the color space on the ocular fundus include RNFL thickness information. Similarly to wide-field OCT scanning, the proposed model can extract the results of RNFL thickness analysis. Differences between the model-generated images and original images were quantified. The SSIM was used to verify the spatial structural consistency of the blood vessels and optic discs in the generated images. The generated thickness maps are suitable for glaucoma detection in clinics, and the generated images were similar to ground truth (PSNR: 19.31 dB; SSIM: 0.44).
Discussion
This study used DNNs to learn glaucoma classification features instead of using cup-to-disc ratios or fundus images as the model input for early glaucoma diagnosis. The advantages of using the model in clinical glaucoma diagnosis were achieved to overcome the problems of OCT availability and the difficulty in glaucoma screening. Model performance was assessed, and the results of RNFL thickness distribution prediction were similar to those obtained from the original OCT images.
OCT provides high-resolution cross- and longitudinal-sectional 3-dimensional images of various parts of the eye, enabling clinicians to accurately locate lesions.40, 41, 42 In clinical practices, fundus images and correlated RNFL thickness distribution in OCT scans were the basis for clinical diagnosis of glaucoma.43,44 However, OCT equipment is expensive and requires additional maintenance costs, so it is not widely used especially in an area with insufficient medical resources.45, 46, 47 This study determined the mapping relations of image translation between color fundus images and OCT thickness maps by the model using a VAE network. The results of our study can be applied to primary-care ophthalmology clinics, and doctors use our model to obtain information for diagnosis. In the proposed network, only color fundus photography was required to obtain RNFL thickness distribution images similar to those provided by high-level OCT medical imaging equipment. Such images can then be used for early glaucoma diagnosis and assessment.
The network depths were changed to improve the image translation performance of the models.48 Theoretically, deeper networks have superior feature extraction abilities. However, they often require more parameter numbers and higher costs.48 The number of parameters of the 6 × 6 architecture was almost 16 times that of the 4 × 4 architecture because of the difference in their depths. In our study, the PSNR of the OCT images generated by the 5 × 5 U-Net architecture was approximately 19.3 dB, which was acceptable for its quality. The color distribution was also nearly identical to the original OCT thickness maps, and the connected blood vessels were complete. Although the PSNR of the 6 × 6 architecture was higher (approximately 19.4 dB), it required > 4 times the parameters of the 5 × 5 architecture. The quality of the images generated by the 5 × 5 U-Net and 6 × 6 U-Net was similar, and the average processing time required for each fundus image was approximately 0.4 and 0.6 s, respectively. Under limited hardware conditions, the calculation time of the 6 × 6 U-Net may be even higher. With only 1 central processing unit, the 5 × 5 U-Net is a more suitable model. Comparing with the 4 × 4 U-Net, the PSNR of its image translation quality was lower (approximately 18.8 dB), though it has a lower cost than the 5 × 5 U-Net. Furthermore, the images generated by the 4 × 4 U-Net had less complete connected blood vessels because of the shallower network depth. Thus, the resources required for the 5 × 5 U-Net architecture and its model performance reached a balance, and this architecture was therefore more applicable to clinical settings.
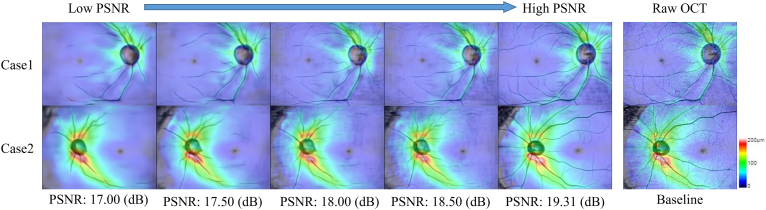
In our study, we used the PSNR to demonstrate the similarity in signal quality between the generated image and the original OCT RNFL thickness map. In traditional computer vision, the PSNR before and after image compression must be > 30 dB to be unnoticeable to the naked eye.49, 50, 51, 52 Image translation models with different PSNR performances were applied to compare 2 cases (Fig 5). The overall quality of the generated images could be divided into 2 parts, the connectivity of the retinal blood vessels and the distribution consistency of RNFL thickness. After the PSNR was > 17 dB, there was almost no significant difference in the distribution of RNFL thickness compared with the raw OCT images. However, the connectivity of the retinal blood vessels was not ideal until the PSNR reached 19.31 dB, when the generated images had a favorable overall quality. Therefore, this PSNR value of identifying correlation between fundus images and OCT thickness maps may be sufficient enough for clinical early glaucoma detection.
Figure 5.
Different peak signal-to-noise ratio (PSNR) indices used in validation and corresponding quality changes in the generated images. The higher PSNR value indicates the higher similarity of the generated images to the raw OCT.
The compound loss function index, containing the PSNR and SSIM,35,36 was proposed and used in our study to assess the model-generated images. The PSNR was used to assess the overall image generation and translation performance and heatmaps of RFNL thickness distribution, and the SSIM was used to assess structural features such as arteries, veins, capillaries, and optic discs in the generated images. This compound loss function index enabled non–GAN-based VAE DNNs to generate favorable image-generation results. The proposed loss function also solved problems in traditional GANs, namely, that they require large data sets for model training and have a low probability of convergence.
During the validation process, we noticed that overfitting started to present after 500 epochs in the SSIM (Fig 3D). However, the PSNR continued to converge without overfitting (Fig 3B). Comparing the training and validation data set, we found that the differences may be related to the overall brightness of the images. Higher the SSIM values were, which evaluated overall signal (brightness, contrast, and structure) variation,36 the higher the human eye perceived the similarity of the image. As for the PSNR index, we found that an increased PSNR was associated with better image details regarding RNFL thickness around the optic disc (Fig 5, case 1). It suggested that the PSNR index was more likely to reflect the details of generated RNFL thickness, especially around the optic disc. After reviewing the fundus images in the training and validation data sets, we observed the images of the validation data set were slightly darker, and the rendering of fundus vessels was less clear, especially on the vessels of the side branches, although the data set had been shuffled through the fivefold cross-validation technique. The image quality around the optic disc was comparable in both data sets. As a result, the overall brightness requirements of the model for the input image would be relatively higher during the validation process and caused an overfitting in the SSIM. In terms of actual clinical application, the impact may be ignorable. In clinical practice, physicians could still refer to the existing retinal fundus images and compare them to the RNFL thickness maps for glaucoma diagnosis at the same time.
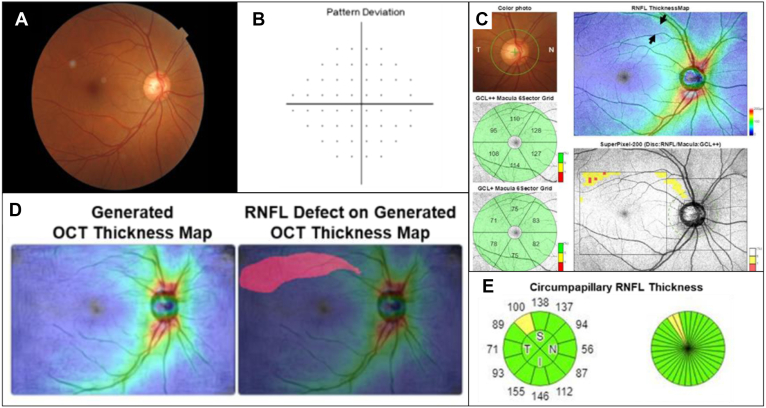
For demonstrating the early glaucoma detection in the clinical scenario, we introduced the proposed model to clinical practice, and one sample case was presented (Fig 6). In the fundus image from the right eye of a 50-year-old female patient, there was no obvious abnormality in the cup-to-disc ratio or significant RNFL defects. The fundus image was input to our model, and a predicting RNFL thickness map was generated. There was a distinct RNFL defect at the superior temporal retina in the generated thickness map. The patient also received a wide-field OCT examination, and an RNFL defect was confirmed in the same region as predicted by the model. This clinical sample could suggest the benefits of early glaucoma identification using the proposed model at the institutes without OCT devices.
Figure 6.
Example of early glaucoma detection by using our model to predict retinal nerve fiber layer (RNFL) thickness distribution on the retinal fundus image from the right eye of a 50-year-old female patient with low-grade myopia (axial length = 24.2 mm, spherical equivalent = −2.25 diopters). A, The conventional retinal fundus photography showed no visible RNFL defect. B, No scotoma was identified by using the 24-2 visual field test. C, The OCT thickness map automatically generated by the model showed a high probability of RNFL thinning in the superotemporal retina region (red color labeled area). D, Although the macular ganglion cell analysis revealed no abnormality, the thickness map from the wide-field swept-source OCT examination showed a wedge-shaped dark-blue area indicating RNFL thinning (black arrows). In addition, the OCT superpixel map revealed an arcuate pattern of contiguous abnormal yellow/red pixels over the corresponding macula area. The lesion identified by the OCT scan was highly correlated with the results generated by the model. E, Circumpapillary RNFL thickness analysis confirmed a borderline superotemporal RNFL thinning. GCL = ganglion cell layer.
There are some limitations in this study. The OCT images were produced based on the ocular fundus wide-field OCT scan by using Topcon’s swept-source OCT (DRI OCT Triton) only. The performance and results from other OCT devices may be different. If the input fundus images were taken by other fundus photography, the predicted RNFL thickness distribution images, particularly the structural features, would be affected. Furthermore, the results may be affected by the quality of the fundus images because the images were selected based on the quality for the model development in our study. Thus, the robustness of the overall model could be increased by including additional samples from different devices and by using data augmentation. Nevertheless, our study had some strengths. We used VAE U-Net DNNs for model training. The U-Net is often used for image extraction, such as segmenting the retinal vessels from color fundus images.53, 54, 55 It could ensure that the segmentation process combines a multiscale spatial context so that the network could be trained end to end on limited training data. In addition, for the loss function design of the model, unlike studies that have used solely the PSNR as the model performance quantification index, we used a compound index with the PSNR and SSIM. The differences between the model-generated images and original images could be quantified by the proposed compound loss function index.
Conclusion
This study constructed and validated a DL algorithm for extracting thickness information from the fundus images similarly to OCT measurement and for using this information to regenerate RNFL thickness distribution images. Detection and screening tools must be safe, simple, accurate, time saving, and cost effective. Despite some limitations, the automated DNN is a promising alternative for distinguishing referable glaucoma and for early glaucoma screening in clinical settings. This technique may aid clinicians in detecting early glaucoma, especially when an OCT scan is not accessible.
Acknowledgments
The authors thank their colleagues at Chang Gung Memorial Hospital for their assistance in collecting clinical data and providing professional clinical insight. They also thank their colleagues in the Industrial Technology Research Institute for their insight and expertise in data analysis and model building.
Manuscript no. XOPS-D-22-00039.
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Disclosure:
All authors have completed and submitted the ICMJE disclosures form.
The authors have no proprietary or commercial interest in any materials discussed in this article.
This research was supported by ITRI Research Projects J301ARB310, K301AR9410, and L301AR9510.
HUMAN SUBJECTS: This retrospective case study was approved by the Human Experiment Ethics Committee of Taipei Chang Gung Memorial Hospital, Taiwan (IRB: 201900528B0C502). The requirement for informed consent was waived because of the retrospective nature of the study.
No animal subjects were used in this study.
Author Contributions:
Research design: Henry Shen-Lih Chen, Jhen-Yang Syu, Eugene Yu-Chuan Kang
Data acquisition and/or research execution: Henry Shen-Lih Chen, Guan-An Chen, Lan-Hsin Chuang, Wei-Wen Su, Wei-Chi Wu, Eugene Yu-Chuan Kang
Data analysis and/or interpretation: Henry Shen-Lih Chen, Guan-An Chen, Jhen-Yang Syu, Jian-Hong Liu, Jian-Ren Chen, Su-Chen Huang, Eugene Yu-Chuan Kang.
Obtained funding: N/A
Manuscript preparation: Henry Shen-Lih Chen, Guan-An Chen, Eugene Yu-Chuan Kang
Contributor Information
Henry Shen-Lih Chen, Email: hslchen@yahoo.com.tw.
Eugene Yu-Chuan Kang, Email: yckang0321@gmail.com.
Supplementary Data
References
- 1.Davis B.M., Crawley L., Pahlitzsch M., et al. Glaucoma: the retina and beyond. Acta Neuropathol. 2016;132:807–826. doi: 10.1007/s00401-016-1609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kingman S. Glaucoma is second leading cause of blindness globally. Bull World Health Organ. 2004;82:887–888. [PMC free article] [PubMed] [Google Scholar]
- 3.Weinreb R.N., Aung T., Medeiros F.A. The pathophysiology and treatment of glaucoma: a review. JAMA. 2014;311:1901–1911. doi: 10.1001/jama.2014.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinreb R.N., Khaw P.T. Primary open-angle glaucoma. Lancet. 2004;363:1711–1720. doi: 10.1016/S0140-6736(04)16257-0. [DOI] [PubMed] [Google Scholar]
- 5.Quigley H.A., Broman A.T. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tham Y.C., Li X., Wong T.Y., et al. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121:2081–2090. doi: 10.1016/j.ophtha.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Sommer A., Miller N.R., Pollack I., et al. The nerve fiber layer in the diagnosis of glaucoma. Arch Ophthalmol. 1977;95:2149–2156. doi: 10.1001/archopht.1977.04450120055003. [DOI] [PubMed] [Google Scholar]
- 8.Hitchings R.A., Spaeth G.L. The optic disc in glaucoma. I: classification. Br J Ophthalmol. 1976;60:778–785. doi: 10.1136/bjo.60.11.778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muramatsu C., Hayashi Y., Sawada A., et al. Detection of retinal nerve fiber layer defects on retinal fundus images for early diagnosis of glaucoma. J Biomed Opt. 2010;15 doi: 10.1117/1.3322388. [DOI] [PubMed] [Google Scholar]
- 10.Esteva A., Chou K., Yeung S., et al. Deep learning-enabled medical computer vision. NPJ Digit Med. 2021;4:5. doi: 10.1038/s41746-020-00376-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson A.C., Jammal A.A., Medeiros F.A. A review of deep learning for screening, diagnosis, and detection of glaucoma progression. Transl Vis Sci Technol. 2020;9:42. doi: 10.1167/tvst.9.2.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinton G.E., Osindero S., Teh Y.W. A fast learning algorithm for deep belief nets. Neural Comput. 2006;18:1527–1554. doi: 10.1162/neco.2006.18.7.1527. [DOI] [PubMed] [Google Scholar]
- 13.Phene S., Dunn R.C., Hammel N., et al. Deep learning and glaucoma specialists: the relative importance of optic disc features to predict glaucoma referral in fundus photographs. Ophthalmology. 2019;126:1627–1639. doi: 10.1016/j.ophtha.2019.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Li Z., He Y., Keel S., et al. Efficacy of a deep learning system for detecting glaucomatous optic neuropathy based on color fundus photographs. Ophthalmology. 2018;125:1199–1206. doi: 10.1016/j.ophtha.2018.01.023. [DOI] [PubMed] [Google Scholar]
- 15.Son J., Shin J.Y., Kim H.D., et al. Development and validation of deep learning models for screening multiple abnormal findings in retinal fundus images. Ophthalmology. 2020;127:85–94. doi: 10.1016/j.ophtha.2019.05.029. [DOI] [PubMed] [Google Scholar]
- 16.Liu S., Graham S.L., Schulz A., et al. A deep learning-based algorithm identifies glaucomatous discs using monoscopic fundus photographs. Ophthalmol Glaucoma. 2018;1:15–22. doi: 10.1016/j.ogla.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 17.Thakur A., Goldbaum M., Yousefi S. Predicting glaucoma before onset using deep learning. Ophthalmol Glaucoma. 2020;3:262–268. doi: 10.1016/j.ogla.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shibata N., Tanito M., Mitsuhashi K., et al. Development of a deep residual learning algorithm to screen for glaucoma from fundus photography. Sci Rep. 2018;8 doi: 10.1038/s41598-018-33013-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bock R., Meier J., Nyúl L.G., et al. Glaucoma risk index: automated glaucoma detection from color fundus images. Med Image Anal. 2010;14:471–481. doi: 10.1016/j.media.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Ting D.S.W., Pasquale L.R., Peng L., et al. Artificial intelligence and deep learning in ophthalmology. Br J Ophthalmol. 2019;103:167–175. doi: 10.1136/bjophthalmol-2018-313173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaji S., Kida S. Overview of image-to-image translation by use of deep neural networks: denoising, super-resolution, modality conversion, and reconstruction in medical imaging. Radiol Phys Technol. 2019;12:235–248. doi: 10.1007/s12194-019-00520-y. [DOI] [PubMed] [Google Scholar]
- 22.Hinton G.E., Salakhutdinov R.R. Reducing the dimensionality of data with neural networks. Science. 2006;313:504–507. doi: 10.1126/science.1127647. [DOI] [PubMed] [Google Scholar]
- 23.Ting D.S.W., Cheung C.Y., Lim G., et al. Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes. JAMA. 2017;318:2211–2223. doi: 10.1001/jama.2017.18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yi X., Walia E., Babyn P. Generative adversarial network in medical imaging: a review. Med Image Anal. 2019;58 doi: 10.1016/j.media.2019.101552. [DOI] [PubMed] [Google Scholar]
- 25.Chen J.S., Coyner A.S., Chan R.V., et al. Deepfakes in ophthalmology applications and realism of synthetic retinal images from generative adversarial networks. Ophthalmol Sci. 2021;1 doi: 10.1016/j.xops.2021.100079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panda S.K., Cheong H., Tun T.A., et al. Describing the structural phenotype of the glaucomatous optic nerve head using artificial intelligence. Am J Ophthalmol. 2022;236:172–182. doi: 10.1016/j.ajo.2021.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Selvaraju R.R., Das A., Vedantam R., et al. Grad-CAM: why did you say that? Visual explanations from deep networks via gradient-based localization. arXiv. 2016 v. 2020. [Google Scholar]
- 28.Ronneberger O., Fischer P., Brox T. MICCAI. 2015. U-net: convolutional networks for biomedical image segmentation; pp. 234–241.https://arxiv.org/abs/1505.04597 [Google Scholar]
- 29.Wang X., Tan K., Du Q., et al. CVA2E: a conditional variational autoencoder with an adversarial training process for hyperspectral imagery classification. IEEE Trans Geosci Remote Sens. 2020;58:1–17. [Google Scholar]
- 30.Chen M., Xu Z., Weinberger K., Sha F. Proceedings of the 29th International Conference on Machine Learning. 2012. Marginalized denoising autoencoders for domain adaptation; pp. 1627–1634.https://arxiv.org/abs/1206.4683 [Google Scholar]
- 31.Kim T., Cha M., Kim H., et al. Proceedings of the 34th International Conference on Machine Learning. arxiv; 2017. Learning to discover cross-domain relations with generative adversarial networks.https://arxiv.org/abs/1703.05192 [Google Scholar]
- 32.Xu W., Keshmiri S., Wang W. Adversarially approximated autoencoder for image generation and manipulation. IEEE Trans Multimedia. 2019;39:355–368. [Google Scholar]
- 33.Anguita D., Ghelardoni L., Ghio A., et al. 20th ESANIN. i6doc.com; (online): 2012. The ‘K’ in K-fold cross validation; pp. 441–446. [Google Scholar]
- 34.Pizer S.M., Amburn E.P., Austin J.D., et al. Adaptive histogram equalization and its variations. Comput Vis Graph Image Process. 1987;39:355–368. [Google Scholar]
- 35.Wang Z., Bovik A., Sheikh H., Simoncelli E. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process. 2004;13:600–612. doi: 10.1109/tip.2003.819861. [DOI] [PubMed] [Google Scholar]
- 36.Horé A., Ziou D. The 20th International Conference on Pattern Recognition. IEEE; 2010. Image Quality Metrics: PSNR vs. SSIM; pp. 2366–2369.https://ieeexplore.ieee.org/document/5596999 [Google Scholar]
- 37.Kaur P., Singh J. A study on the effect of Gaussian noise on PSNR value for digital images. Int J Comput Electr Eng. 2011;3:319. [Google Scholar]
- 38.Bondzulic B.P., Pavlovic B.Z., Petrovic V.S., Andric M.S. Performance of peak signal-to-noise ratio quality assessment in video streaming with packet losses. Electron Lett. 2016;52:454–456. [Google Scholar]
- 39.Zhou C., Zhang X., Chen H. A new robust method for blood vessel segmentation in retinal fundus images based on weighted line detector and hidden Markov model. Comput Methods Programs Biomed. 2020;187 doi: 10.1016/j.cmpb.2019.105231. [DOI] [PubMed] [Google Scholar]
- 40.Wu Z., Weng D.S.D., Rajshekhar R., et al. Effectiveness of a qualitative approach toward evaluating OCT imaging for detecting glaucomatous damage. Transl Vis Sci Technol. 2018;7:7. doi: 10.1167/tvst.7.4.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hood D.C., De Cuir N., Blumberg D.M., et al. A single wide-field OCT protocol can provide compelling information for the diagnosis of early glaucoma. Transl Vis Sci Technol. 2016;5:4. doi: 10.1167/tvst.5.6.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hood D.C., Fortune B., Mavrommatis M.A., et al. Details of glaucomatous damage are better seen on OCT en face images than on OCT retinal nerve fiber layer thickness maps. Invest Ophthalmol Vis Sci. 2015;56:6208–6216. doi: 10.1167/iovs.15-17259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang P., Shen J., Chang R., et al. Machine learning models for diagnosing glaucoma from retinal nerve fiber layer thickness maps. Ophthalmol Glaucoma. 2019;2:422–428. doi: 10.1016/j.ogla.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim S.J., Cho K.J., Oh S. Development of machine learning models for diagnosis of glaucoma. PLoS One. 2017;12 doi: 10.1371/journal.pone.0177726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lisboa R., Leite M.T., Zangwill L.M., et al. Diagnosing preperimetric glaucoma with spectral domain optical coherence tomography. Ophthalmology. 2012;119:2261–2269. doi: 10.1016/j.ophtha.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bussel I.I., Wollstein G., Schuman J.S. OCT for glaucoma diagnosis, screening and detection of glaucoma progression. Br J Ophthalmol. 2014;98 Suppl 2(Suppl 2):ii15–ii19. doi: 10.1136/bjophthalmol-2013-304326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hood D.C. Improving our understanding, and detection, of glaucomatous damage: an approach based upon optical coherence tomography (OCT) Prog Retin Eye Res. 2017;57:46–75. doi: 10.1016/j.preteyeres.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan A., Sohail A., Zahoora U., Qureshi A.S. A survey of the recent architectures of deep convolutional neural networks. Artif Intell Rev. 2020;53:5455–5516. [Google Scholar]
- 49.Zhao R., Zhao J., Dai F., Zhao F. A new image secret sharing scheme to identify cheaters. Comput Stand Interfaces. 2009;31:252–257. [Google Scholar]
- 50.Hwang D.C., Park J.S., Kim S.C., et al. Magnification of 3D reconstructed images in integral imaging using an intermediate-view reconstruction technique. Appl Opt. 2006;45:4631–4637. doi: 10.1364/ao.45.004631. [DOI] [PubMed] [Google Scholar]
- 51.Kim H.J., Kim D.H., Lee C.L., et al. Comparison and evaluation of JPEG and JPEG2000 in medical images for CR (computed radiography) J Korean Phys Soc. 2010;56:856–862. [Google Scholar]
- 52.Kang S.J., Kim Y.H. Multi-histogram-based backlight dimming for low power liquid crystal displays. J Disp Technol. 2011;7:544–549. [Google Scholar]
- 53.Kumawat S., Raman S. ICASSP ICASSP 2019 - 2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) IEEE; 2019. Local phase U-net for fundus image segmentation; pp. 1209–1213.https://ieeexplore.ieee.org/document/8683390 [Google Scholar]
- 54.Yang D., Liu G., Ren M., et al. A multi-scale feature fusion method based on U-net for retinal vessel segmentation. Entropy. 2020;22:811. doi: 10.3390/e22080811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sathananthavathi V., Indumathi G. Encoder enhanced atrous (EEA) unet architecture for retinal blood vessel segmentation. Cogn Syst Res. 2021;67:84–95. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.