Abstract
Perception of non-self molecules known as microbe-associated molecular patterns (MAMPs) by host pattern recognition receptors (PRRs) activates plant pattern-triggered immunity (PTI). Pathogen infections often trigger the release of modified-self molecules, termed damage- or danger-associated molecular patterns (DAMPs), which modulate MAMP-triggered signaling to shape the frontline of plant immune responses against infections. In the context of advances in identifying MAMPs and DAMPs, cognate receptors, and their signaling, here, we focus on the most recent breakthroughs in understanding the perception and role of non-self and modified-self patterns. We highlight the commonalities and differences of MAMPs from diverse microbes, insects, and parasitic plants, as well as the production and perception of DAMPs upon infections. We discuss the interplay between MAMPs and DAMPs for emerging themes of the mutual potentiation and attenuation of PTI signaling upon MAMP and DAMP perception during infections.
Introduction
During the co-evolution with pathogens, sessile plants have developed plasma membrane (PM)-resident pattern recognition receptors (PRRs) to distinguish self from modified-self and non-self patterns and initiate pattern-triggered immunity (PTI) [1–3]. PTI provides plant basal immunity against a broad spectrum of pathogens. In the arms race, successful pathogens have evolved effector proteins, some of which could suppress PTI to promote pathogenicity [4]. To counteract, plants could activate effector-triggered immunity (ETI) mediated by intracellular nucleotide-binding domain leucine-rich repeat receptors (NLRs) [5], which mutually potentiates PTI to establish a unified defense system [6–10].
Pathogen infections often cause host cellular damages, resulting in the release of damage- or danger-associated molecular patterns (DAMPs), essentially as modified-self patterns [3,11]. DAMPs are classified into constitutive DAMPs (cDAMPs) and inducible DAMPs (iDAMPs) [3]. cDAMPs are ubiquitous molecules released upon cellular damage, while iDAMPs are endogenous peptides secreted upon infections and function as immunomodulatory signals, also known as phytocytokines [3,12,13]. Non-self patterns have been coined as pathogen- or microbe-associated molecular patterns (PAMPs or MAMPs) and are derived from bacteria, fungi, oomycetes, viruses, nematodes, insects, and parasitic plants [3,14–16].
Perception of MAMPs or DAMPs by PRRs often recruits co-receptors and triggers some overlapping signaling events, such as the increased cytosolic Ca2+ levels, the production of extracellular reactive oxygen species (ROS), the phosphorylation of receptor-like cytoplasmic kinases (RLCKs), the activation of mitogen-activated protein kinase (MAPK) cascades, transcriptional reprogramming of defense-associated genes, and callose deposition on the cell walls [1,13,17].
In the context of many knowledge advances in unveiling a plethora of MAMPs and DAMPs, cognate receptors, and PTI signaling events, instead of summarizing all the literature, we focus on the most recent breakthroughs in understanding the perception and mode-of-actions of modified self and non-self patterns. We will discuss the commonalities and differences of MAMPs from diverse microbes, insects, and parasitic plants. In parallel, the production and perception of DAMPs upon infections will be discussed. We finish with the complex interplay between MAMPs and DAMPs to piece together emerging themes of the mutual potentiation and/or attenuation of MAMP and DAMP signaling during infections.
Decoding non-self patterns from diverse pathogens, insects, and parasitic plants
In nature, plants constantly monitor their extracellular environment for the presence of external threats. Non-self patterns are generic molecules specifically present in invading microbes, insects, and parasitic plants but absent from hosts. Recognition of non-self patterns by PRRs is the frontline to trigger basal immune responses in plants. Plant PRRs are mainly composed of PM-resident receptor-like kinases (RLKs) and receptor-like proteins (RLPs). RLKs and RLPs possess diverse extracellular domains (ECDs), including leucine-rich repeats (LRRs), lysin motifs (LysM), lectin motifs, epidermal growth factor (EGF)-like repeats, and malectin-like motifs, which mainly mediate the pattern recognition [2,18].
MAMPs from bacterial pathogens
Diverse components, including flagellin, elongation factor Tu (EF-Tu), and peptidoglycans (PGNs) from bacteria, have been shown to bear immune-stimulating activities and function as MAMPs (Figure 1). Bacterial flagellin or a 22-amino acid synthetic peptide (flg22) corresponding to its N-terminus from both pathogenic and non-pathogenic bacteria is perceived by the LRR-RLK flagellin sensing 2 (FLS2) and BRI1-associated receptor kinase 1 (BAK1) receptor–coreceptor complex and triggers the immune response in Arabidopsis [19–21]. However, the immunogenic flagellin fragments recognized by FLS2 are buried in the flagellin polymer structure. How the immunogenic motifs like flg22 are accessible to PRRs during the natural infection remains elusive. Recently, the flg22 epitopes have been shown to be released from the glycosylated flagellin by plant-secreted apoplastic β-galactosidase 1 (BGAL1) and an unidentified protease [22]. Interestingly, some bacterial strains deploy BGAL1 inhibitors and glycan polymorphisms in flagellin to suppress BGAL1 function and escape recognition for microbial evasion of plant immunity during infections [22]. In addition, Ralstonia solanacearum bears a polymorphism in the flg22 sequence, avoiding the FLS2-BAK1 recognition in most plants except in soybean (Glycine max), which has co-evolved polymorphisms in the soybean flagellin receptors GmFLS2 and GmBAK1 in triggering the immune response [23]. Similarly, the polymorphic flg22 from Agrobacterium tumefaciens can be detected by an FLS2 allele in the riverbank grape (Vitis riparia) [24]. These results suggest the evolutionary flexibility and dynamics of the MAMP encoding and decoding process, in which MAMPs may mutate to escape the PRR detection while PRRs could co-evolve to recognize the polymorphic MAMPs.
Figure 1. Decoding MAMPs and DAMPs by the cognate receptors.
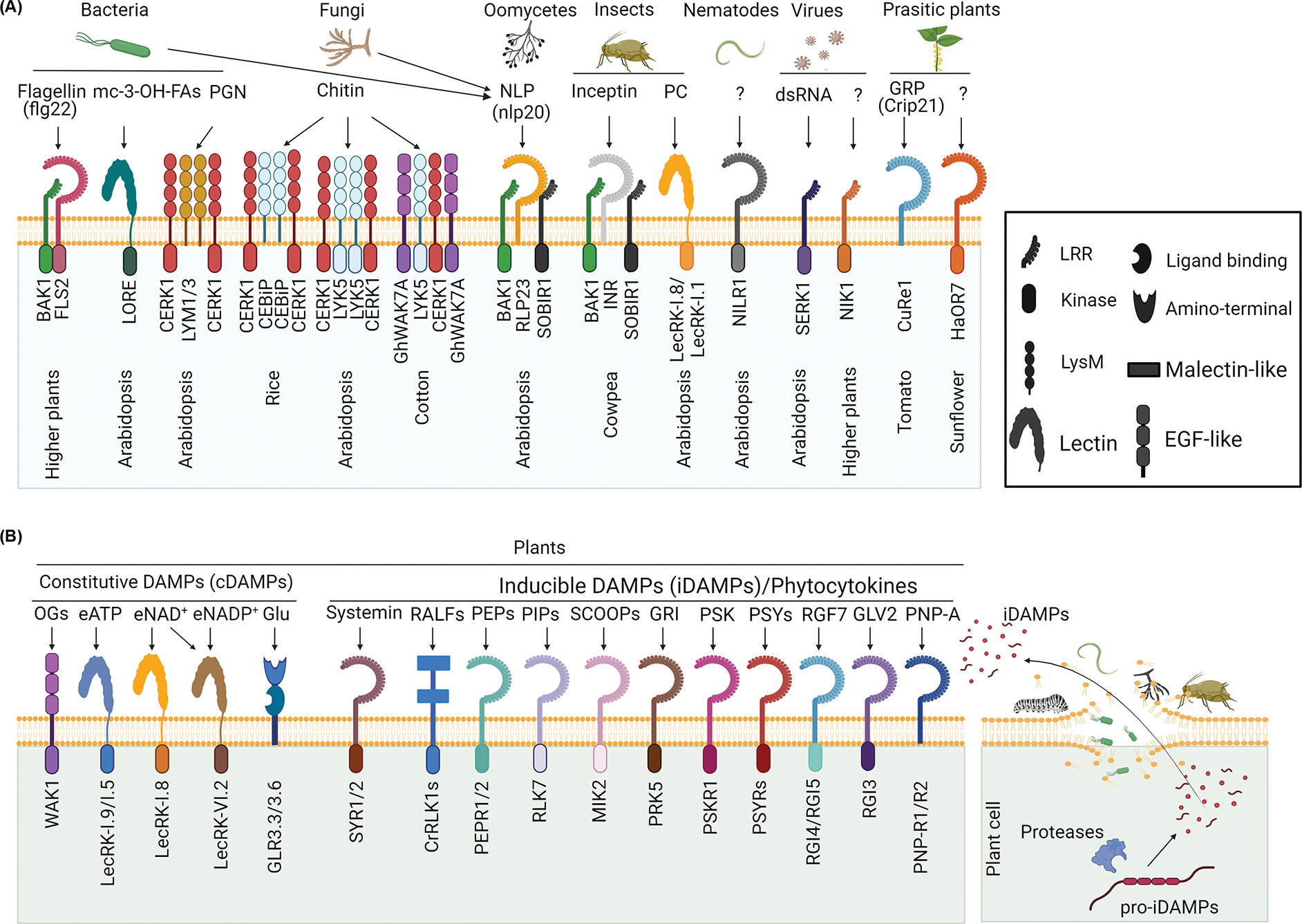
(A) Recognition of diverse MAMPs from bacteria, fungi, oomycetes, insects, nematodes, viruses, and parasitic plants by plant PM-resident PRRs. The bacterial flagellin or flg22 peptide is perceived by the LRR-RLKs FLS2 in complex with BAK1. Free mc-3-OH-FAs are recognized by LecRK LORE. Bacterial PGN is sensed by LysM-RLPs LYM1 and LYM3 with LysM-RLK CERK1. Fungal chitin is perceived by LysM-RLP OsCEBiP and OsCERK1 in rice, LysM-RLK LYK5 and CERK1 in Arabidopsis, and LysM-RLKs GhGLYK5 and GhCERK1 in cotton. Bacterial, fungal, and oomycete necrosis and ethylene-inducing peptide 1 (Nep1)-like protein (NLP) is recognized by RLP23 in complex with SOBIR1 and BAK1. Inceptin from herbivore oral secretions is recognized by LRR-RLP inceptin receptor (INR) in complex with SOBIR1 and SERKs. Insect egg-derived phosphatidylcholine (PC) is identified as an EAMP signaling through lectin receptor kinase-I.1 (LecRK-I.1) and LecRK-I.8. Nematode-induced LRR-RLK 1 (NILR1) is required for basal resistance to nematodes. Viral double-stranded RNA (dsRNA) stimulates PTI responses requiring SERK1, and LRR-RLK NIK1 positively regulates plant antiviral immunity. The parasitic plant Cuscuta reflexa cell wall protein CrGRP is sensed by tomato LRR-RLP CuRe1 by recognizing the epitope, Crip21. LRR-RLK HaOR7 from sunflower recognizes an unknown MAMP and confers resistance against parasitic plant Orobanche cumana. (B) Recognition of DAMPs by PRRs. cDAMPs are ubiquitous molecules with essential roles in plant homeostasis and released into the apoplasts upon infections. They include oligogalacturonide (OG), eATP, eNAD(P)+, and glutamate recognized by WAK1, LecRK-I.9, and LecRK-I.5, LecRK-I.8 and LecRK VI.2, and GLR3.3 and GLR3.6, respectively. iDAMPs, termed phytocytokines, are small secretory peptides processed from precursor proteins (pro-iDAMPs) by proteases upon infections and cell damages (right). Several well-studied iDAMPs and their receptors are shown on the left. Abbreviations: CERK1, chitin elicitor receptor kinase 1; CuRe1, Cuscuta receptor 1; eATP, extracellular adenosine 5′-triphosphate; eNAD(P)+, extracellular nicotinamide adenine dinucleotides in their phosphorylated or unphosphorylated forms; GLR, glutamate receptor-like; NIK1, NSP-interacting kinase 1; SOBIR1, suppressor of BIR1.
PGNs, major components of bacterial cell walls, are released by the plant chitinase lysozyme-like hydrolase 1 (LYS1) in triggering the immune response [25]. In Arabidopsis, PGN perception requires the heterologous complex consisting of the LysM-RLP LYM1, LYM3, and LysM-RLK chitin elicitor receptor kinase 1 (CERK1) [26], which show shared features with the fungal chitin perception complex in rice (Oryza sativa) [27]. In addition, bacterial medium-chain 3-hydroxy fatty acid (mc-3-OH-FA) metabolites, which are perceived by the lectin motif-RLK lipooligosaccharide-specific reduced elicitation (LORE), trigger immunity in Arabidopsis [28,29].
MAMPs from fungal and oomycete pathogens
Examples of MAMPs from fungi and oomycetes include the fungal cell wall structure building block chitin and polysaccharide β-glucan (Figure 1). The recognition systems and downstream signaling mediated by these MAMPs bear similarities and differences from those perceiving bacterial MAMPs.
Chitin, a major constituent of fungal cell walls, functions as an MAMP in a wide range of plants. Chitin perception in Arabidopsis genetically involves the multiple protein complexes with members of LysM-RLKs and LysM-RLPs [30,31]. Among them, LYK5 is the primary receptor for chitin, forming a chitin-inducible complex with CERK1/LYK1 to induce plant innate immunity [30]. Interestingly, chitin induces dynamic changes in the localization, association, or mobility of these receptors with only LYSM-containing GPI-anchored protein 2 (LYM2) and LYK4 being detected in the plasmodesmal PM, which is different from the nearby PM [31]. Chitin-LYM2-mediated signaling triggers callose deposition in plasmodesmata, thereby leading to plasmodesmata closure and cell isolation [31]. In rice, in addition to OsLYKs, LysM-RLP chitin elicitor-binding protein (OsCEBiP) was identified as the first chitin-binding PRR [32]. In cotton (Gossypium hirsutum), GhWAK7A, a wall-associated kinase phosphorylates GhLYK5 and promotes a complex formation of GhLYK5 with GhCERK1 in mediating chitin-induced responses [33]. Considering the involvement in both PGN and chitin-mediated responses, CERK1 likely functions as a coreceptor or signaling component in PRR complexes [26,27,30,33].
Necrosis and ethylene-inducing peptide 1 (Nep1)-like proteins (NLPs) are secreted cytotoxins present in bacteria, fungi, and oomycetes and trigger necrosis as well as defense responses on dicot plants through different mechanisms [34]. NLPs bind to plant sphingolipid glycosylinositol phosphorylceramides (GIPCs) to induce necrosis [34]. In contrast, a conserved 20-amino acid fragment (nlp20), which is uncoupled from the NLP-necrosis inducing region, is perceived by LRR-RLP RLP23 complexing with LRR-RLK suppressor of BIR1 (SOBIR1) and BAK1 in triggering PTI [35]. The data support the evolutionary diversification of MAMPs from the virulence motifs in pathogens.
MAMPs from commensal and symbiotic microbes
Considering that the commensal and symbiotic microbes also produce diverse MAMPs, an outstanding question is how MAMPs present in these beneficial, life-supporting microbes evade host recognition. A systemic investigation of the naturally occurring flg22 epitopes in commensal microbiota revealed substantial flagellin epitope variations, and most variants in the flg22 sequence from these commensal bacteria are non-immunogenic to the host [36]. Thus, to avoid plant immunity, commensal bacteria tend to lose the immunogenic activities of MAMPs. However, flagellins are the building blocks of the flagellum important for bacterial motility, an essential function for host colonization. Another question arises in terms of how such variations in flagellins affect bacterial motility and fitness. Mutagenesis analysis of flagellin unveiled that some variants showed altered host recognition and immunogenic activities but maintained normal motility [37], suggesting the evolutionary dynamics of immunogenic and motility functions in maximizing microbial fitness.
In addition, hosts also evolved strategies to counteract immunity triggered by MAMPs present in the symbiotic microbes. For example, a recent study showed that the rice OsMYR symbiotic receptor competed with the OsCEBiP MAMP receptor for the complex formation with OsCERK1 involved in the chitin-induced immune response [38]. By recognizing different lengths of chitooligosaccharides (COs), two distinct receptors discriminate the signals of friends and foes, and result in the opposing outcomes of symbiosis and defense.
MAMPs from insects and nematodes
Herbivores also release immunomodulatory molecules, which are termed, based on their origins, herbivore-associated molecular patterns (HAMPs) and egg-associated molecular patterns (EAMPs) [39]. Inceptin, a proteolytic fragment of plant chloroplast ATP synthase γ-subunit present in caterpillar oral secretions, is perceived as a HAMP by LRR-RLP Inceptin receptor (INR) complexing with SOBIR1 and somatic embryogenesis receptor kinases (SERKs) in cowpea (Vigna unguiculata) and common bean (Phaseolus vulgaris) [40]. However, it remains speculative for the precise processing and releasement of inceptin peptides from plant chloroplasts. Recently, egg-derived phosphatidylcholines (PCs) were identified as an EAMP that signals through lectin receptor kinase-I.1 (LecRK-I.1) and LecRK-I.8 [41,42]. Despite that it is yet to be determined whether PCs are bona fide ligands of the LecRK-I receptors, the data demonstrate the important function of plant RLKs in detecting non-self patterns from diverse invading structures, including insect eggs.
Plant-parasitic nematodes release the pheromone ascarosides, which trigger defense responses with the yet-identified receptor(s) [43]. Interestingly, nematode-induced LRR-RLK 1 (NILR1) and BAK1 are required for plant resistance against different nematodes [44]. It will be interesting to determine whether these RLKs perceive specific molecules from nematodes in mediating plant defense.
MAMPs from viral pathogens
Double-stranded RNA (dsRNA), an intermediate product during DNA and RNA viral replications, triggers RNA interference, a primary plant immune response against viral infections. Interestingly, dsRNA also stimulates PTI responses, which requires SERK1 but is independent of Dicer-like (DCL) proteins, critical components in binding and cleaving dsRNA in triggering RNA interference [45]. In addition, dsRNA induces callose deposition at the plasmodesmata, leading to plasmodesmata closure and restricting the viral movement in an RLCK BIK1-dependent but ROS-independent manner [46]. Additionally, geminiviral Nuclear shuttle protein (NSP) interacts with the LRR-RLK NSP-interacting kinase 1 (NIK1), which positively regulates plant antiviral immunity but negatively regulates plant antibacterial immunity [47]. Yet-identified viral MAMPs may induce NIK1 dimerization in suppressing viral RNA translation. Mechanisms underlying the opposite roles of NIK1 in antibacterial and antiviral immunity remain elusive.
MAMPs from parasitic plants
Like pathogens, parasitic plants constraint plant productivities. The resistance mechanism against parasitic plants could be attributed to the detection and activation of MAMP-mediating signaling [16,48]. For example, a Glycine-rich protein (GPR) from the cell wall of rootless parasite Cuscuta reflexa acts as an MAMP in tomato (Solanum lycopersicum) and is perceived by LRR-RLP Cuscuta receptor 1 (CuRe1), leading to defense response in resistant hosts [49]. Recently, LRR-RLK HaOR7 in sunflower (Helianthus annuus) was identified as a key genetic determinant in mediating resistance against parasitic plant sunflower broomrape (Orobanche cumana) [50]. Despite unclear modes-of-action, some unknown components or MAMPs from O. cumana may trigger the defense response blocking the connection of the sunflower root vascular system with sunflower broomrape [50]. These findings imply the convergent themes by which plants recognize and differentiate self from non-self patterns in parasitic plants, insects, and microbial pathogens.
Generating and recognizing modified-self patterns during infections
MAMP perceptions and pathogen infections induce the production of modified-self cDAMPs and iDAMPs [3]. cDAMPs are ubiquitous molecules released into certain cellular compartments upon cellular damages and infections. In contrast, iDAMPs, known as proteinaceous DAMPs or phytocytokines, are secretory peptides processed from precursor proteins upon infections.
cDAMPs
Various molecules, including oligosaccharides, nucleotides, and amino acids, can be released by enzymatic or physical degradation upon infections and act as the primary cDAMP signals [3].
Cell wall components
Oligogalacturonides (OGs) with varying degrees of polymerization are fragments of the plant cell wall and function as danger signals responding to changes in cell wall integrity, wounding, and pathogen infections [51,52]. Compared with long OGs (degree of polymerization from 10 to 15), trimeric OGs trigger more profound immune responses in plants [53]. Various plant cell wall-degrading enzymes secreted by necrotrophic bacteria and fungi contribute to the release of OGs [52]. The Arabidopsis Wall-associated kinase 1 (WAK1) recognizes long OGs and activates OG-mediated defense responses against fungal and bacterial pathogens [54].
Nucleotides
Adenosine 5′-triphosphate (ATP), a major biochemical energy source for all living cells, could be released into the apoplasts upon stresses, wounding, and pathogen infections. Extracellular ATP (eATP), released via the wounded cell membrane, exocytosis, ATP-binding ABC transporters, or nucleotide transporters, function as a cDAMP in animals and plants [55]. In Arabidopsis, the lectin-like RLK P2K1/DORN1/LecRK-I.9 receptor is essential for perception of eATP in response to wounding, herbivory, and pathogen [55,56]. A recent report shows that P2K2 (LecRK-I.5) is an alternative eATP receptor with a higher affinity than P2K1 [57].
Pathogen infections also induce the releasement of nicotinamide adenine dinucleotides either in their phosphorylated (NADP+) or unphosphorylated (NAD+) forms into the extracellular fluid, leading to plant immune activation [58]. A lectin-like RLK LecRK-VI.2 and BAK1 may function as a receptor complex for extracellular NAD+ and NADP+, which play a role in systemic acquired resistance (SAR) [58]. Furthermore, extracellular NAD(P)+ has been implicated in functioning as an SAR mobile signal in plants [58]. However, it awaits to be further illustrated whether and how endogenous eNAD(P)+ (extracellular nicotinamide adenine dinucleotides in their phosphorylated or unphosphorylated forms) moves during infections.
Amino acids
The amino acid glutamate is also an immunogenic signal in the wound- and insect-induced defenses [59,60]. Glutamate released from locally damaged tissues triggers long-distance defense signaling, which depends on PM-bound ion channels of Glutamate receptor-like (GLR) [60,61]. GLR3.3 and GLR3.6 sense the apoplastic glutamate and other amino acids and trigger Ca2+ influx, further propagating to distal leaves for systemic defense responses [60]. It remains elusive whether glutamates act in local tissues or are transmitted distantly.
iDAMPs or phytocytokines
iDAMPs function as secondary signals upon the perception of stimuli and modulate immunity, abiotic stresses, plant growth and development [3]. Phytocytokines usually contain a signal peptide at the N-terminus of the precursor proteins for secretion. However, there are some phytocytokines without a signal peptide [13,62]. Furthermore, phytocytokines undergo enzymatical and post-translational modifications to generate bioactive peptides [62,63].
Phytocytokines without a signal peptide
Despite the absence of a signal peptide, those phytocytokines are also released from the damaged cells into the apoplasts [62]. Systemin, an 18-amino acid peptide, is induced by wounding and herbivore attacks and triggers long-distance systemic defense responses in the Solanaceae family [64]. Two subtilisin-like proteases (phytaspases) mediate the prosystemin processing into Leu-systemin, which is less active than systemin [65]. How the fully bioactive systemin is generated and secreted remains elusive. Tomato LRR-RLK SYR1 and SYR2, as well as PORK1, are required for systemin-triggered responses and mediate plant defense against insect herbivory [66,67]. Plant elicitor peptides (Peps) are derived from their precursors PROPEPs with eight members in Arabidopsis, expression of which are induced by MAMPs, herbivores, and wounding [68,69]. Upon cellular damage, Ca2+-activated type-II metacaspases (MCs) cleave PROPEP1, leading to the releasement of Pep1, which functions in resistance to herbivores and pathogens across different plant species [70,71]. Peps are sensed by the LRR-RLKs Pep receptor 1 (PEPR1) and PEPR2 in complex with BAK1 [68,72]. How Peps are released into the apoplasts and whether they can trigger long-distance defense signaling remain a mystery. Maize (Zea mays) immune signaling peptide 1 (Zip1) is a maize-specific secreted peptide that activates the salicylic acid (SA)-mediated defense but exhibits no effect on the PTI signaling in maize [73]. Zip1 is released from its propeptide precursor by two Papain-like cysteine proteases, PLCP1 and PLCP2. It is not known how Zip1 is perceived by plant cells.
Phytocytokines with a signal peptide
Precursors of most secreted peptides harbor a signal peptide, which mediates their secretion and is subsequently cleaved [3]. Expression of these precursor genes is often induced by stimuli and bear cell- or tissue-type specificity [3]. Rapid alkalization factors (RALFs) are a large family of secreted cysteine-rich peptides forming disulfide bonds, which are implicated in diverse developmental processes as well as immunity [74,75]. Some RALFs are processed by subtilase site-1-protease (S1P) [76] and subsequently recognized by malectin-like receptor kinases (MLRs), also known as Catharanthus roseus receptor-like kinase-1-like proteins [77]. Interestingly, RALF17 positively but RALF23/RALF33 negatively regulate plant immunity in an MLR FER-dependent manner [76]. How different RALFs trigger opposing immune responses via FER awaits to be investigated. In addition, Serine-rich endogenous peptides (SCOOPs) represent a group of secreted peptides involved in plant development and immunity [78,79]. Precursors of several SCOOPs are induced upon pathogen infections and MAMP treatments [78,80]. LRR-RK male discoverer 1-interacting receptor-like kinase 2 (MIK2) in complex with BAK1 and SERK4 is required for sensing the conserved SCOOP signature motifs present in plant-derived SCOOPs as well as microbial SCOOP-like (SCOOPL) peptides [78,79].
In addition to proteolytic processing from precursors, several secreted peptides undergo post-translational modifications to release biologically active peptides [62,63]. Root meristem growth factors (RGFs), also known as GOLVEN (GLV), are a group of secreted and tyrosine-sulfated peptides and mainly expressed in root meristem with the RGF7 precursor-encoding gene (preRGF7) being up-regulated and preRGF6/9 down-regulated in leaves upon bacterial infections [81,82]. RGF7 is sensed by RGF1-insensitive (RGI) receptors RGI4 and RGI5 in complex with BAK1 and SERK4 in triggering plant immunity [81]. Like RGF7, RGF6/9 are processed via sulfation and cleaved by SUBTILASE 6.1 (SBT6.1) and SBT3.8 [83,84]. Interestingly, RGF6/9 perceived by RGI3 enhance the flg22-triggered immune response likely by forming a complex with FLS2 and increasing the FLS2 abundance [82].
Coordination of MAMPs and DAMPs in modulating plant immunity
DAMPs induced by MAMP perceptions and pathogen infections play essential roles in fortifying and/or amplifying the immune responses. To maintain the immune homeostasis without running amok from autoimmunity, DAMP-triggered signaling could also attenuate the immune outputs. The modes-of-action of DAMPs, including immunomodulatory phytocytokines, can act directly on the same target cell, adjacent cells, or distant cells, which resembles the autocrine, paracrine, or endocrine signaling in animals, respectively (Figure 2).
Figure 2. Mutual potentiation and/or attenuation of immunity by MAMPs and DAMPs and pathogen mimicry.
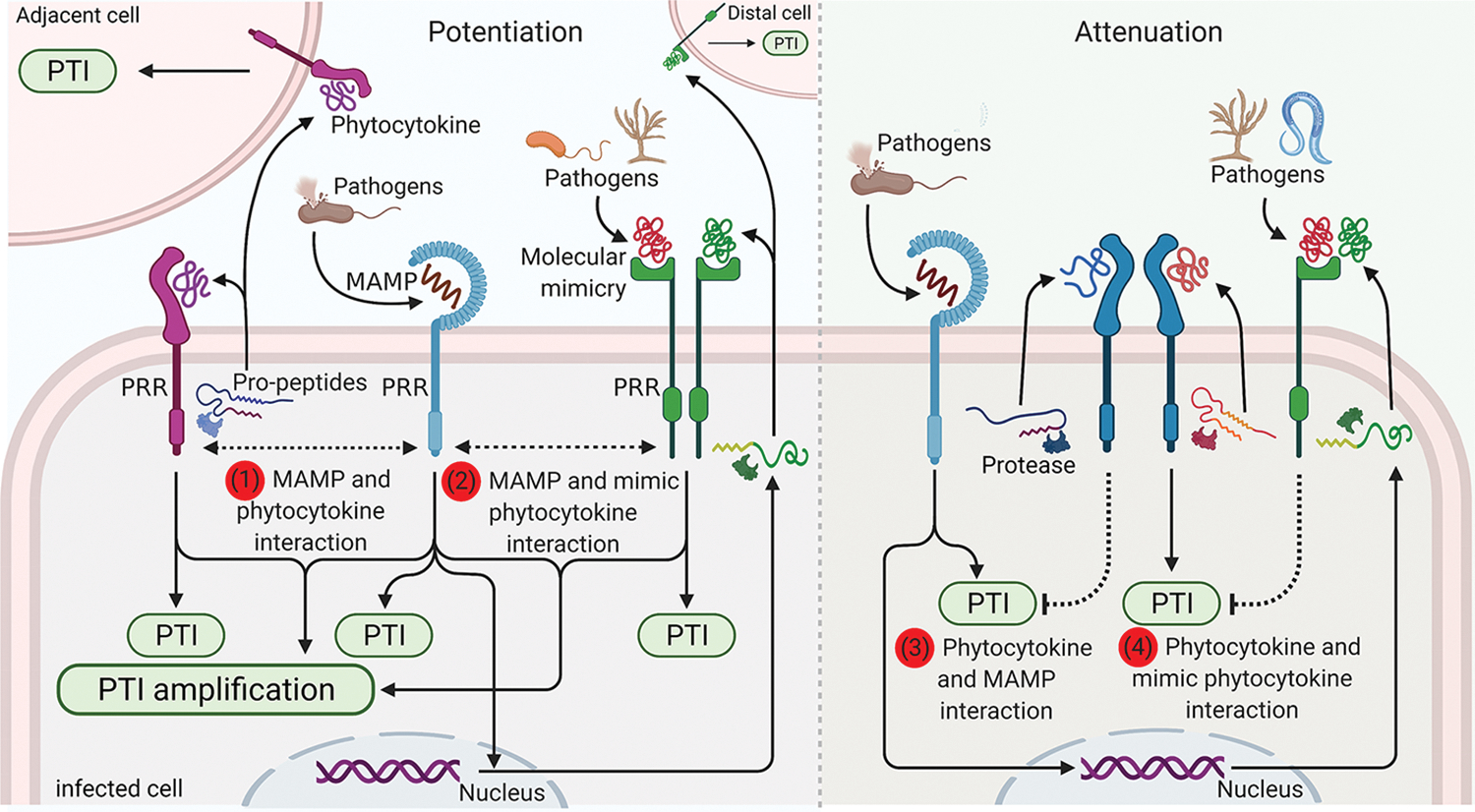
Perception of MAMPs, phytocytokines, and pathogen-derived phytocytokine-mimic peptides by PRRs induces the PTI signaling via acting on the same infected cell, adjacent cells, or distal cells. Pathogen infections or MAMP perceptions induce the production of DAMPs and phytocytokines, which further modulate the PTI signaling. Phytocytokines (1) and pathogen mimicries (2) mutually amplify MAMP-induced defense responses. Moreover, host-derived phytocytokine (3) or pathogen-derived phytocytokine-mimic peptides (4) can interfere with and attenuate the MAMP-or host phytocytokine-mediated immune signaling. All figures were generated using BioRender (biorender.com).
Mutual potentiation of immunity by MAMPs and DAMPs
MAMP treatment can induce the expression, maturation, production, and/or releasement of diverse DAMPs. cDAMPs, like OGs, eATP, eNAD(P)+, and glutamate, which are released into the apoplasts upon MAMP perceptions and elicit some overlapping immune responses with those triggered by MAMPs, including ROS burst and expression of defense-associated genes [3,85]. However, the timing, thresholds, and tissue specificity (local or distal) of the immune responses triggered by different MAMPs/DAMPs may vary. For example, unlike MAMPs and other DAMPs, eNAD(P)+ likely acts as a mobile signal to mediate LecRK-VI.2-dependent SAR and amplify the immune responses in distal tissues [58]. In general, the expressions of precursors of phytocytokines and their cognate receptors are induced by MAMPs. Those phytocytokines include RALFs, SCOOPs, RGFs, Peps, and PAMP-induced peptides (PIPs), which could potentiate MAMP-triggered immune responses. RLK7-mediated PIP1 signaling is required for flg22-induced defense responses cooperatively with Pep1 [86]. The observation that MIK2 perceives both plant- and microbe-derived SCOOPs suggests that plant-derived SCOOPs were evolved to amplify SCOOPL signaling [78,79]. Although the mechanism underlying DAMP-mediated amplification of MAMP-triggered signaling remains fragmented, members of RALFs and RGFs have been suggested to modulate the MAMP receptor abundance and membrane nanodomain assembly as scaffolds [82,87]. Grim reaper (GRI) peptides, which are proteolytically processed by metacaspase 9 (MC9) and recognized by LRR-RLK pollen-specific receptor-like kinase 5 (PRK5), induce oxidative stress, ROS-dependent cell death, and the defense hormone SA signaling [88]. Expression of GRI and MC9 is induced by oxidative stress and wound, suggesting ROS-mediated feedback regulation on the MC9-GRI-PRK5 signaling module. It remains unknown whether and how this signaling module mechanistically integrates with MAMP-mediated signaling since ROS burst is among essential early signaling events triggered by multiple MAMPs.
Attenuation of immunity by phytocytokines or microbial peptides mimicking host MAMPs
Some phytocytokines antagonistically modulate plant immunity to prevent excessive defense responses. For example, in contrast with RALF17, RALF23, 33, and 34 peptides attenuate MAMP-induced immune responses by complexing with FER, which scaffolds and promotes the MAMP-induced PRR complex formation [76]. Furthermore, plant natriuretic peptides (PNPs) also negatively regulate SA-mediated local and systemic resistance in an LRR-RLP PNP receptor (PNP-R1/2)-dependent manner [89]. Considering the important role of ROS and SA in plant PTI, these peptides may play a role in attenuating the immune signaling outputs.
Some pathogens also secrete phytocytokine-like peptides to interfere with the plant defense signaling and promote pathogenicity. For example, parasitic nematodes possess RALF-like proteins, which interact with plant FER receptors and dampen the plant immune responses to facilitate their parasitism [90]. In addition, Fusarium-RALF (F-RALF) peptides derived from Fusarium oxysporum trigger the plant apoplast alkalization in the FER-dependent manner, thereby enhancing the infectious opportunity and pathogenicity [91]. Furthermore, Xanthomonas axonopodis pv. citri-derived PNP-like peptide (XacPNP) induced upon infections is required for pathogenicity with an elusive mechanism [92]. These phytocytokine-mimics exemplify how microbial organisms exploit the host signaling machinery for survival and pathogenicity.
Conclusion and perspectives
The plant PM is decorated with an array of RLKs and RLPs with expanded family members (>600 RLKs and 57 RLPs in Arabidopsis), many of which play a fundamental role in sensing non-self patterns and triggering the immune response. Non-self patterns could be from pathogens, insects, and parasitic plants. In addition, plant genomes encode thousands of small peptides, some of which function as immunomodulatory phytocytokines. Their expression, maturation, and secretion could be modulated during pathogen infections. Moreover, cDAMPs, including eATP, eNAD(P)+, and glutamate, function as short- and long-distance signaling molecules in modulating plant immunity. Some phytocytokines cross-talk to each other and play roles in cell-to-cell communication in autocrine, endocrine, or paracrine manners [63]. Furthermore, phytocytokines coordinate with MAMPs to modulate defense responses either agonistically or antagonistically by different mechanisms. Apparently, pathogens also explore and modulate immune signaling by mimicking plant phytocytokines and vice versa. The signaling networks mediated by RLKs and RLPs sensing non-self and modified-self patterns shape the robust immunity against infections from a broad spectrum of pathogens, pests, and parasitic plants.
Despite a series of breakthroughs that have been made, particularly in the identification of MAMPs, DAMPs, and their cognate receptors, our knowledge on the concerted actions of MAMP- and DAMP-mediated immunity against natural infections is still at the tip of the iceberg, and outstanding questions await to be addressed. Although treatments of MAMPs and DAMPs trigger defense responses in plants, can all MAMPs and DAMPs be sensed by plants during natural infections? How are the immune-eliciting motifs of MAMPs and DAMPs accessible to the PM-resident PRRs enclosed by the cell wall? Do different cell types, for instance, leaf epidermis and mesophyll cells, bear similar or different sensitivities to MAMPs and DAMPs? Many RLKs, RLPs, and DAMPs are involved in plant growth, development, and abiotic stress responses, in addition to immunity. For instance, many phytocytokines induced by drought and salt stresses play essential roles in biotic and abiotic stresses [13]. Whether and how these processes are interconnected or uncoupled are not known in most cases. The importance of MAMPs and DAMPs in modulating plant immunity is mainly based on some classical and well-established PTI readouts. Whether they also explore other plants physiological and developmental processes to increase plant fitness as a defense strategy is unknown. For example, leaf shedding in cauline leaves mediated by inflorescence deficient in abscission (IDA) peptides perceived by LRR-RLK HAESA (HAE)/HSL2 in complex with BAK1/SERKs has been implicated as an antibacterial defense mechanism [93,94]. In addition, understanding how multiple MAMPs and DAMPs interplay with each other through their shared or non-shared downstream pathways and how host plants locally or systemically regulate MAMP- or DAMP-mediated responses will shed light on comprehension of their roles in coordinating stress responses and developmental regulation. In addition to efforts to discover and characterize known/unknown non-self and modified-self patterns, how they are translationally modified by certain processing enzymes and properly secreted with/without signal peptides are still far from clear. Rapid technological development, including single-cell sequencing and proteomics, single-molecule imaging, sensors for in vivo detection of small molecules and peptides, and tissue-/site-specific gene-editing, will undoubtedly further advance our understanding of the coordinated action of MAMPs and DAMPs in plant immunity in the context of growth, development, and coping with other environmental stresses.
Summary.
The perception of non-self and modified-self molecules by PRRs elicits PTI.
Non-self molecules, termed MAMPs, are generic molecules specifically present in invading microbes, insects, and parasitic plants but absent from hosts.
Modified-self molecules induced or released upon pathogen infections or MAMP perceptions are termed DAMPs.
Immunomodulatory phytocytokines are secretory peptides processed from precursor proteins upon infections.
Mutual potentiation and attenuation of MAMP- and DAMP-triggered signaling modulate plant immune homeostasis in the context of growth, development, and coping with other environmental stresses.
Plants and pathogens have co-evolved to possess phytocytokines and their mimics for defensive and pathogenicity adaptation.
Acknowledgements
We apologize to those whose work is not cited due to space limitations. We thank lab members and Dr. Ping He for insightful discussions and critical reading of the manuscript.
Funding
This work was supported by the National Institutes of Health (NIH) [grant number 1R35GM144275–01]; and the National Science Foundation (NSF) [grant number IOS-2049642 (to L.S.)].
Abbreviations
- ATP
adenosine 5′-triphosphate
- BAK1
BRI1-associated receptor kinase 1
- BGAL1
β-galactosidase 1
- cDAMP
constitutive DAMP
- CEBiP
chitin elicitor-binding protein
- CERK1
chitin elicitor receptor kinase 1
- DAMP
damage- or danger-associated molecular pattern
- dsRNA
double-stranded RNA
- eATP
extracellular adenosine 5′-triphosphate
- eNAD(P)+
extracellular nicotinamide adenine dinucleotides in their phosphorylated or unphosphorylated forms
- FLS2
flagellin sensing 2
- HAMP
herbivore-associated molecular pattern
- iDAMP
inducible DAMP
- LecRK-I.1
lectin receptor kinase-I.1
- LRR
leucine-rich repeat
- LysM
lysin motif
- MAMP
microbe-associated molecular pattern
- MC
metacaspase
- MIK2
male discoverer 1-interacting receptor-like kinase 2
- NIK1
NSP-interacting kinase 1
- NLP
necrosis and ethylene-inducing peptide 1 (Nep1)-like protein
- OG
oligogalacturonide
- PGN
peptidoglycan
- PIP
PAMP-induced peptide
- PM
plasma membrane
- PNP
plant natriuretic peptide
- PRK5
pollen-specific receptor-like kinase 5
- PRR
pattern recognition receptor
- PTI
pattern-triggered immunity
- RALF
rapid alkalization factor
- RGF
root meristem growth factor
- RLCK
receptor-like cytoplasmic kinase
- RLK
receptor-like kinase
- RLP
receptor-like protein
- ROS
reactive oxygen species
- SAR
systemic acquired resistance
- SCOOP
serine-rich endogenous peptide
- SCOOPL
SCOOP-like
- SOBIR1
suppressor of BIR1
- WAK1
wall-associated kinase 1
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.DeFalco TA and Zipfel C (2021) Molecular mechanisms of early plant pattern-triggered immune signaling. Mol. Cell, 10.1016/j.molcel.2021.09.028 [DOI] [PubMed] [Google Scholar]
- 2.Escocard de Azevedo Manhaes AM, Ortiz-Morea FA, He P and Shan L (2021) Plant plasma membrane-resident receptors: Surveillance for infections and coordination for growth and development. J. Integr. Plant Biol. 63, 79–101, 10.1111/jipb.13051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tanaka K and Heil M (2021) Damage-associated molecular patterns (DAMPs) in plant innate immunity: applying the danger model and evolutionary perspectives. Annu. Rev. Phytopathol. 59, 53–75, 10.1146/annurev-phyto-082718-100146 [DOI] [PubMed] [Google Scholar]
- 4.Dou D and Zhou JM (2012) Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12, 484–495, 10.1016/j.chom.2012.09.003 [DOI] [PubMed] [Google Scholar]
- 5.Yuan M, Ngou BPM, Ding P and Xin XF (2021) PTI-ETI crosstalk: an integrative view of plant immunity. Curr. Opin. Plant Biol. 62, 102030, 10.1016/j.pbi.2021.102030 [DOI] [PubMed] [Google Scholar]
- 6.Yuan M, Jiang Z, Bi G, Nomura K, Liu M, Wang Y et al. (2021) Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109, 10.1038/s41586-021-03316-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ngou BPM, Ahn HK, Ding PT and Jones JDG (2021) Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115, 10.1038/s41586-021-03315-7 [DOI] [PubMed] [Google Scholar]
- 8.Pruitt RN, Locci F, Wanke F, Zhang L, Saile SC, Joe A et al. (2021) The EDS1-PAD4-ADR1 node mediates Arabidopsis pattern-triggered immunity. Nature 598, 495–499, 10.1038/s41586-021-03829-0 [DOI] [PubMed] [Google Scholar]
- 9.Zhai K, Liang D, Li H, Jiao F, Yan B, Liu J et al. (2022) NLRs guard metabolism to coordinate pattern- and effector-triggered immunity. Nature 601, 245–251, 10.1038/s41586-021-04219-2 [DOI] [PubMed] [Google Scholar]
- 10.Tian H, Wu Z, Chen S, Ao K, Huang W, Yaghmaiean H et al. (2021) Activation of TIR signalling boosts pattern-triggered immunity. Nature 598, 500–503, 10.1038/s41586-021-03987-1 [DOI] [PubMed] [Google Scholar]
- 11.Boller T and Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406, 10.1146/annurev.arplant.57.032905.105346 [DOI] [PubMed] [Google Scholar]
- 12.Gust AA, Pruitt R and Nurnberger T (2017) Sensing danger: key to activating plant immunity. Trends Plant Sci. 22, 779–791, 10.1016/j.tplants.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 13.Hou S, Liu D and He P (2021) Phytocytokines function as immunological modulators of plant immunity. Stress Biol. 1, 8, 10.1007/s44154-021-00009-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou JM and Zhang Y (2020) Plant immunity: danger perception and signaling. Cell 181, 978–989, 10.1016/j.cell.2020.04.028 [DOI] [PubMed] [Google Scholar]
- 15.Niehl A and Heinlein M (2019) Perception of double-stranded RNA in plant antiviral immunity. Mol. Plant Pathol. 20, 1203–1210, 10.1111/mpp.12798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fishman MR and Shirasu K (2021) How to resist parasitic plants: pre- and post-attachment strategies. Curr. Opin. Plant Biol. 62, 102004, 10.1016/j.pbi.2021.102004 [DOI] [PubMed] [Google Scholar]
- 17.Yu X, Feng B, He P and Shan L (2017) From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 55, 109–137, 10.1146/annurev-phyto-080516-035649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kong L, Rodrigues B, Kim JH, He P and Shan L (2021) More than an on-and-off switch: Post-translational modifications of plant pattern recognition receptor complexes. Curr. Opin. Plant Biol. 63, 102051, 10.1016/j.pbi.2021.102051 [DOI] [PubMed] [Google Scholar]
- 19.Stringlis IA, Proietti S, Hickman R, Van Verk MC, Zamioudis C and Pieterse CMJ (2018) Root transcriptional dynamics induced by beneficial rhizobacteria and microbial immune elicitors reveal signatures of adaptation to mutualists. Plant J. 93, 166–180, 10.1111/tpj.13741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chinchilla D, Bauer Z, Regenass M, Boller T and Felix G (2006) The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18, 465–476, 10.1105/tpc.105.036574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C et al. (2013) Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science 342, 624–628, 10.1126/science.1243825 [DOI] [PubMed] [Google Scholar]
- 22.Buscaill P, Chandrasekar B, Sanguankiattichai N, Kourelis J, Kaschani F, Thomas EL et al. (2019) Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides. Science 364, eaav0748, 10.1126/science.aav0748 [DOI] [PubMed] [Google Scholar]
- 23.Wei Y, Balaceanu A, Rufian JS, Segonzac C, Zhao A, Morcillo RJL et al. (2020) An immune receptor complex evolved in soybean to perceive a polymorphic bacterial flagellin. Nat. Commun. 11, 3763, 10.1038/s41467-020-17573-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furst U, Zeng Y, Albert M, Witte AK, Fliegmann J and Felix G (2020) Perception of Agrobacterium tumefaciens flagellin by FLS2(XL) confers resistance to crown gall disease. Nat. Plants 6, 22–27, 10.1038/s41477-019-0578-6 [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Grabherr HM, Willmann R, Kolb D, Brunner F, Bertsche U et al. (2014) Host-induced bacterial cell wall decomposition mediates pattern-triggered immunity in Arabidopsis. eLife 3, 12, e01990, 10.7554/eLife.01990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willmann R, Lajunen HM, Erbs G, Newman MA, Kolb D, Tsuda K et al. (2011) Arabidopsis lysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 108, 19824–19829, 10.1073/pnas.1112862108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y et al. (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214, 10.1111/j.1365-313X.2010.04324.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kutschera A, Dawid C, Gisch N, Schmid C, Raasch L, Gerster T et al. (2019) Bacterial medium-chain 3-hydroxy fatty acid metabolites trigger immunity in Arabidopsis plants. Science 364, 178–181, 10.1126/science.aau1279 [DOI] [PubMed] [Google Scholar]
- 29.Luo X, Wu W, Liang Y, Xu N, Wang Z, Zou H et al. (2020) Tyrosine phosphorylation of the lectin receptor-like kinase LORE regulates plant immunity. EMBO J. 39, e102856, 10.15252/embj.2019102856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao YR, Liang Y, Tanaka K, Nguyen CT, Jedrzejczak RP, Joachimiak A et al. (2014) The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3, e03766, 10.7554/eLife.03766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheval C, Samwald S, Johnston MG, de Keijzer J, Breakspear A, Liu X et al. (2020) Chitin perception in plasmodesmata characterizes submembrane immune-signaling specificity in plants. Proc. Natl. Acad. Sci. U.S.A. 117, 9621–9629, 10.1073/pnas.1907799117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K et al. (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 11086–11091, 10.1073/pnas.0508882103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang P, Zhou L, Jamieson P, Zhang L, Zhao Z, Babilonia K et al. (2020) The cotton wall-associated kinase GhWAK7A mediates responses to fungal wilt pathogens by complexing with the chitin sensory receptors. Plant Cell 32, 3978–4001, 10.1105/tpc.19.00950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenarcic T, Pirc K, Hodnik V, Albert I, Borisek J, Magistrato A et al. (2019) Molecular basis for functional diversity among microbial Nep1-like proteins. PLoS Pathog. 15, e1007951, 10.1371/journal.ppat.1007951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Albert I, Bohm H, Albert M, Feiler CE, Imkampe J, Wallmeroth N et al. (2015) An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat. Plants 1, 15140, 10.1038/nplants.2015.140 [DOI] [PubMed] [Google Scholar]
- 36.Colaianni NR, Parys K, Lee HS, Conway JM, Kim NH, Edelbacher N et al. (2021) A complex immune response to flagellin epitope variation in commensal communities. Cell Host Microbe 29, 635.e9–649.e9, 10.1016/j.chom.2021.02.006 [DOI] [PubMed] [Google Scholar]
- 37.Parys K, Colaianni NR, Lee HS, Hohmann U, Edelbacher N, Trgovcevic A et al. (2021) Signatures of antagonistic pleiotropy in a bacterial flagellin epitope. Cell Host Microbe 29, 620.e9–634.e9, 10.1016/j.chom.2021.02.008 [DOI] [PubMed] [Google Scholar]
- 38.Zhang C, He J, Dai H, Wang G, Zhang X, Wang C et al. (2021) Discriminating symbiosis and immunity signals by receptor competition in rice. Proc. Natl. Acad. Sci. U.S.A. 118, 16, 10.1073/pnas.2023738118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reymond P (2021) Receptor kinases in plant responses to herbivory. Curr. Opin. Biotechnol. 70, 143–150, 10.1016/j.copbio.2021.04.004 [DOI] [PubMed] [Google Scholar]
- 40.Steinbrenner AD, Munoz-Amatriain M, Chaparro AF, Aguilar-Venegas JM, Lo S, Okuda S et al. (2020) A receptor-like protein mediates plant immune responses to herbivore-associated molecular patterns. Proc. Natl. Acad. Sci. U.S.A. 117, 31510–31518, 10.1073/pnas.2018415117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stahl E, Brillatz T, Ferreira Queiroz E, Marcourt L, Schmiesing A, Hilfiker O et al. (2020) Phosphatidylcholines from Pieris brassicae eggs activate an immune response in Arabidopsis. eLife 9, e60293, 10.7554/eLife.60293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Groux R, Stahl E, Gouhier-Darimont C, Kerdaffrec E, Jimenez-Sandoval P, Santiago J et al. (2021) Arabidopsis natural variation in insect egg-induced cell death reveals a role for LECTIN RECEPTOR KINASE-I.1. Plant Physiol. 185, 240–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manosalva P, Manohar M, von Reuss SH, Chen S, Koch A, Kaplan F et al. (2015) Conserved nematode signalling molecules elicit plant defenses and pathogen resistance. Nat. Commun. 6, 7795, 10.1038/ncomms8795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendy B, Wang’ombe MW, Radakovic ZS, Holbein J, Ilyas M, Chopra D et al. (2017) Arabidopsis leucine-rich repeat receptor-like kinase NILR1 is required for induction of innate immunity to parasitic nematodes. PLoS Pathog. 13, e1006284, 10.1371/journal.ppat.1006284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niehl A, Wyrsch I, Boller T and Heinlein M (2016) Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol. 211, 1008–1019, 10.1111/nph.13944 [DOI] [PubMed] [Google Scholar]
- 46.Huang C, Rocio Sede A, Mutterer J, Boutant E and Heinlein M (2021) Suppression of a dsRNA-induced plant immunity pathway by viral movement protein. bioRxiv, 10.1101/2021.10.30.466425 [DOI] [Google Scholar]
- 47.Li B, Ferreira MA, Huang M, Camargos LF, Yu X, Teixeira RM et al. (2019) The receptor-like kinase NIK1 targets FLS2/BAK1 immune complex and inversely modulates antiviral and antibacterial immunity. Nat. Commun. 10, 4996, 10.1038/s41467-019-12847-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Albert M, Axtell MJ and Timko MP (2021) Mechanisms of resistance and virulence in parasitic plant-host interactions. Plant Physiol. 185, 1282–1291, 10.1093/plphys/kiaa064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hegenauer V, Slaby P, Korner M, Bruckmuller JA, Burggraf R, Albert I et al. (2020) The tomato receptor CuRe1 senses a cell wall protein to identify Cuscuta as a pathogen. Nat. Commun. 11, 5299, 10.1038/s41467-020-19147-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duriez P, Vautrin S, Auriac MC, Bazerque J, Boniface MC, Callot C et al. (2019) A receptor-like kinase enhances sunflower resistance to Orobanche cumana. Nat. Plants 5, 1211–1215, 10.1038/s41477-019-0556-z [DOI] [PubMed] [Google Scholar]
- 51.Voxeur A, Habrylo O, Guenin S, Miart F, Soulie MC, Rihouey C et al. (2019) Oligogalacturonide production upon Arabidopsis thaliana-Botrytis cinerea interaction. Proc. Natl. Acad. Sci. U.S.A. 116, 19743–19752, 10.1073/pnas.1900317116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bacete L, Melida H, Miedes E and Molina A (2018) Plant cell wall-mediated immunity: cell wall changes trigger disease resistance responses. Plant J. 93, 614–636, 10.1111/tpj.13807 [DOI] [PubMed] [Google Scholar]
- 53.Davidsson P, Broberg M, Kariola T, Sipari N, Pirhonen M and Palva ET (2017) Short oligogalacturonides induce pathogen resistance-associated gene expression in Arabidopsis thaliana. BMC Plant Biol. 17, 19, 10.1186/s12870-016-0959-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brutus A, Sicilia F, Macone A, Cervone F and De Lorenzo G (2010) A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. U.S.A. 107, 9452–9457, 10.1073/pnas.1000675107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cho SH, Nguyen CT, Choi J and Stacey G (2017) Molecular mechanism of plant recognition of extracellular ATP. Adv. Exp. Med. Biol. 1051, 233–253 [DOI] [PubMed] [Google Scholar]
- 56.Li ZJ, Chakraborty S and Xu GZ (2016) X-ray crystallographic studies of the extracellular domain of the first plant ATP receptor, DORN1, and the orthologous protein from Camelina sativa. Acta Cryst. 72, 782–787, 10.1107/S2053230X16014278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pham AQ, Cho SH, Nguyen CT and Stacey G (2020) Arabidopsis lectin receptor kinase P2K2 is a second plant receptor for extracellular ATP and contributes to innate immunity. Plant Physiol. 183, 1364–1375, 10.1104/pp.19.01265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang C, Huang X, Li Q, Zhang Y, Li JL and Mou Z (2019) Extracellular pyridine nucleotides trigger plant systemic immunity through a lectin receptor kinase/BAK1 complex. Nat. Commun. 10, 4810, 10.1038/s41467-019-12781-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mousavi SA, Chauvin A, Pascaud F, Kellenberger S and Farmer EE (2013) GLUTAMATE RECEPTOR-LIKE genes mediate leaf-to-leaf wound signalling. Nature 500, 422–426, 10.1038/nature12478 [DOI] [PubMed] [Google Scholar]
- 60.Toyota M, Spencer D, Sawai-Toyota S, Wang JQ, Zhang T, Koo AJ et al. (2018) Glutamate triggers long-distance, calcium-based plant defense signaling. Science 361, 1112, 10.1126/science.aat7744 [DOI] [PubMed] [Google Scholar]
- 61.Alfieri A, Doccula FG, Pederzoli R, Grenzi M, Bonza MC, Luoni L et al. (2020) The structural bases for agonist diversity in an Arabidopsis thaliana glutamate receptor-like channel. Proc. Natl. Acad. Sci. U.S.A. 117, 752–760, 10.1073/pnas.1905142117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matsubayashi Y (2014) Posttranslationally modified small-peptide signals in plants. Annu. Rev. Plant Biol. 65, 385–413, 10.1146/annurev-arplant-050312-120122 [DOI] [PubMed] [Google Scholar]
- 63.Tavormina P, De Coninck B, Nikonorova N, De Smet I and Cammue BP (2015) The plant peptidome: an expanding repertoire of structural features and biological functions. Plant Cell 27, 2095–2118, 10.1105/tpc.15.00440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang H, Zhang H and Lin J (2020) Systemin-mediated long-distance systemic defense responses. New Phytol. 226, 1573–1582, 10.1111/nph.16495 [DOI] [PubMed] [Google Scholar]
- 65.Beloshistov RE, Dreizler K, Galiullina RA, Tuzhikov AI, Serebryakova MV, Reichardt S et al. (2018) Phytaspase-mediated precursor processing and maturation of the wound hormone systemin. New Phytol. 218, 1167–1178, 10.1111/nph.14568 [DOI] [PubMed] [Google Scholar]
- 66.Wang L, Einig E, Almeida-Trapp M, Albert M, Fliegmann J, Mithofer A et al. (2018) The systemin receptor SYR1 enhances resistance of tomato against herbivorous insects. Nat. Plants 4, 152–156, 10.1038/s41477-018-0106-0 [DOI] [PubMed] [Google Scholar]
- 67.Xu S, Liao CJ, Jaiswal N, Lee S, Yun DJ, Lee SY et al. (2018) Tomato PEPR1 ortholog receptor-like kinase1 regulates responses to systemin, necrotrophic fungi, and insect herbivory. Plant Cell 30, 2214–2229, 10.1105/tpc.17.00908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huffaker A (2015) Plant elicitor peptides in induced defense against insects. Curr. Opin. Insect Sci. 9, 44–50, 10.1016/j.cois.2015.06.003 [DOI] [PubMed] [Google Scholar]
- 69.Lori M, van Verk MC, Hander T, Schatowitz H, Klauser D, Flury P et al. (2015) Evolutionary divergence of the plant elicitor peptides (Peps) and their receptors: interfamily incompatibility of perception but compatibility of downstream signalling. J. Exp. Bot. 66, 5315–5325, 10.1093/jxb/erv236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hander T, Fernández-Fernández ÁD, Kumpf RP, Willems P, Schatowitz H, Rombaut D et al. (2019) Damage on plants activates Ca(2+)-dependent metacaspases for release of immunomodulatory peptides. Science 363, eaar7486, 10.1126/science.aar7486 [DOI] [PubMed] [Google Scholar]
- 71.Shen W, Liu J and Li JF (2019) Type-II metacaspases mediate the processing of plant elicitor peptides in Arabidopsis. Mol. Plant 12, 1524–1533, 10.1016/j.molp.2019.08.003 [DOI] [PubMed] [Google Scholar]
- 72.Krol E, Mentzel T, Chinchilla D, Boller T, Felix G, Kemmerling B et al. (2010) Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor at PEPR1 and its close homologue AtPEPR2. J. Biol. Chem. 285, 13471–13479, 10.1074/jbc.M109.097394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ziemann S, van der Linde K, Lahrmann U, Acar B, Kaschani F, Colby T et al. (2018) An apoplastic peptide activates salicylic acid signalling in maize. Nat. Plants 4, 172–180, 10.1038/s41477-018-0116-y [DOI] [PubMed] [Google Scholar]
- 74.Abarca A, Franck CM and Zipfel C (2021) Family-wide evaluation of RAPID ALKALINIZATION FACTOR peptides. Plant Physiol. 187, 996–1010, 10.1093/plphys/kiab308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blackburn MR, Haruta M and Moura DS (2020) Twenty years of progress in physiological and biochemical investigation of RALF peptides. Plant Physiol. 182, 1657–1666, 10.1104/pp.19.01310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N et al. (2017) The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355, 287–289, 10.1126/science.aal2541 [DOI] [PubMed] [Google Scholar]
- 77.Ortiz-Morea FA, Liu J, Shan L and He P (2022) Malectin-like receptor kinases as protector deities in plant immunity. Nat. Plants [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hou S, Liu D, Huang S, Luo D, Liu Z, Xiang Q et al. (2021) The Arabidopsis MIK2 receptor elicits immunity by sensing a conserved signature from phytocytokines and microbes. Nat. Commun. 12, 5494, 10.1038/s41467-021-25580-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rhodes J, Yang H, Moussu S, Boutrot F, Santiago J and Zipfel C (2021) Perception of a divergent family of phytocytokines by the Arabidopsis receptor kinase MIK2. Nat. Commun. 12, 705, 10.1038/s41467-021-20932-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gully K, Pelletier S, Guillou MC, Ferrand M, Aligon S, Pokotylo I et al. (2019) The SCOOP12 peptide regulates defense response and root elongation in Arabidopsis thaliana. J. Exp. Bot. 70, 1349–1365, 10.1093/jxb/ery454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang X, Zhang N, Zhang L, He Y, Cai C, Zhou J et al. (2021) Perception of the pathogen-induced peptide RGF7 by the receptor-like kinases RGI4 and RGI5 triggers innate immunity in Arabidopsis thaliana. New Phytol. 230, 1110–1125, 10.1111/nph.17197 [DOI] [PubMed] [Google Scholar]
- 82.Stegmann M, Zecua-Ramirez P, Ludwig C, Lee H-S, Peterson B, Nimchuk ZL et al. (2021) RGI-GOLVEN signalling promotes FLS2 abundance to regulate plant immunity. bioRxiv, 10.1101/2021.01.29.428839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ghorbani S, Hoogewijs K, Pečenková T, Fernandez A, Inzé A, Eeckhout D et al. (2016) The SBT6.1 subtilase processes the GOLVEN1 peptide controlling cell elongation. J. Exp. Bot. 67, 4877–4887, 10.1093/jxb/erw241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stührwohldt N, Scholl S, Lang L, Katzenberger J, Schumacher K and Schaller A (2020) The biogenesis of CLEL peptides involves several processing events in consecutive compartments of the secretory pathway. eLife 9, e55580, 10.7554/eLife.55580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hou S, Liu Z, Shen H and Wu D (2019) Damage-associated molecular pattern-triggered immunity in plants. Front. Plant Sci. 10, 646, 10.3389/fpls.2019.00646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hou S, Wang X, Chen D, Yang X, Wang M, Turrà D et al. (2014) The secreted peptide PIP1 amplifies immunity through receptor-like kinase 7. PLoS Pathog. 10, e1004331, 10.1371/journal.ppat.1004331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gronnier J, Franck CM, Stegmann M, DeFalco TA, Abarca A, von Arx M et al. (2022) Regulation of immune receptor kinase plasma membrane nanoscale organization by a plant peptide hormone and its receptors. eLife 11, e74162, 10.7554/eLife.74162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wrzaczek M, Vainonen JP, Stael S, Tsiatsiani L, Help-Rinta-Rahko H, Gauthier A et al. (2015) Grim reaper peptide binds to receptor kinase PRK5 to trigger cell death in Arabidopsis. EMBO J. 34, 55–66, 10.15252/embj.201488582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee KP, Liu K, Kim EY, Medina-Puche L, Dong H, Duan J et al. (2020) Plant natriuretic peptide A and its putative receptor PNP-R2 antagonize salicylic acid-mediated signaling and cell death. Plant Cell 32, 2237–2250, 10.1105/tpc.20.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang X, Peng H, Zhu S, Xing J, Li X, Zhu Z et al. (2020) Nematode-encoded RALF peptide mimics facilitate parasitism of plants through the FERONIA receptor kinase. Mol. Plant 13, 1434–1454, 10.1016/j.molp.2020.08.014 [DOI] [PubMed] [Google Scholar]
- 91.Masachis S, Segorbe D, Turra D, Leon-Ruiz M, Furst U, El Ghalid M et al. (2016) A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 1, 16043, 10.1038/nmicrobiol.2016.43 [DOI] [PubMed] [Google Scholar]
- 92.Gottig N, Garavaglia BS, Daurelio LD, Valentine A, Gehring C, Orellano EG et al. (2008) Xanthomonas axonopodis pv. citri uses a plant natriuretic peptide-like protein to modify host homeostasis. Proc. Natl. Acad. Sci. U.S.A. 105, 18631–18636, 10.1073/pnas.0810107105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Patharkar OR, Gassmann W and Walker JC (2017) Leaf shedding as an anti-bacterial defense in Arabidopsis cauline leaves. PLoS Genet. 13, e1007132, 10.1371/journal.pgen.1007132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meng X, Zhou J, Tang J, Li B, de Oliveira MVV, Chai J et al. (2016) Ligand-induced receptor-like kinase complex regulates floral organ abscission in Arabidopsis. Cell Rep. 14, 1330–1338, 10.1016/j.celrep.2016.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]


