Abstract
One objective of human genetics is to unveil the variants that contribute to human diseases. With the rapid development and wide use of next-generation sequencing (NGS), massive genomic sequence data have been created, making personal genetic information available. Conventional experimental evidence is critical in establishing the relationship between sequence variants and phenotype but with low efficiency. Due to the lack of comprehensive databases and resources which present clinical and experimental evidence on genotype-phenotype relationship, as well as accumulating variants found from NGS, different computational tools that can predict the impact of the variants on phenotype have been greatly developed to bridge the gap. In this review, we present a brief introduction and discussion about the computational approaches for variant impact prediction. Following an innovative manner, we mainly focus on approaches for non-synonymous variants (nsSNVs) impact prediction and categorize them into six classes. Their underlying rationale and constraints, together with the concerns and remedies raised from comparative studies are discussed. We also present how the predictive approaches employed in different research. Although diverse constraints exist, the computational predictive approaches are indispensable in exploring genotype-phenotype relationship.
Keywords: in silico prediction, human genetics, genotype-phenotype relationship, nonsynonymous variants, variant impact
1 Introduction
One of the primary goals of human genetics is to discover the genetic variants associated with the onset and progression of human disease. The challenge is a “a needle in haystack” problem: how to pinpoint the potential causative ones from millions of individual variants (Genomes Project et al., 2015) spreading over the newly assembled, non-gap 3.055 billion–base pair human genome sequence (Nurk et al., 2022). Efforts to achieve this goal, such as linkage analysis and genome-wide association studies, were inadequately effective in identifying causative candidates and had poor clinical predictive value (Tam et al., 2019).
Over the last decade, the next generation sequencing (NGS) has been extensively utilized in biomedical research as consequences of its substantially reduced cost and generation of large volume of data. According to the fact sheets on genomic cost provided by the National Human Genome Research Institute (NHGRI) (KA., 2021), NGS technology achieved one hundred-fold cost reduction compared to Sanger sequencing, and the price is currently less than $1,000 per human genome. Nowadays, NGS platforms can finish one run within 2 days producing billions of reads for up to 48 samples (Hu et al., 2021). With the raw NGS data, standard and well-recognized variant format files can be generated using upstream analysis pipeline (Kanzi et al., 2020). Whereas the downstream disease-causing variant fishing step among ∼50,000 variants from WES, or even millions of variants from WGS is the most challenge part (Eberle et al., 2017; Koboldt, 2020).
There are plenty of data resources storing evidenced genotype-phenotype relationship information. To a certain extent, clinicians and researchers are able to utilize these records to interpret the formation, progress, diagnosis and treatment of diseases from a genetic perspective. However, even the most well-recognized databases, such as ClinVar (Landrum et al., 2020), only contain around 14,000 of highly confident variants with evidence evaluated by genetic experts, which is a small fraction compared to the huge number of variants identified from NGS. This situation dramatically reduces clinical utility from genetics. In addition, it also poses great challenges for understanding differential actions of genes between/among individuals, populations and species, as well as deciphering the genotype-phenotype relationship (Orgogozo et al., 2015). To address these issues, computational tools for predicting variant impact have emerged which can help bridge the gap between vast amount of genomic data generated and limited known genetic evidence, and finally build up the potential genotype-phenotype relationship for the newly identified variants.
Variant call format (VCF) files store identified variants providing variant genomic position, nucleotide substitution, assessed quality score, genotype and other relevant information according to alignment and variant calling information (Danecek et al., 2011). Based on the specified information, variant annotation can locate them to specific genes or transcripts, classify them into different types and conclude on their impactable consequences (Wang et al., 2010; Cingolani et al., 2012; McLaren et al., 2016). Variants causing sequence alteration are mainly categorized into four types: insertion, deletion, single nucleotide variant (SNV) and other substitution, including multiple nucleotide variant (MNV) (Eilbeck et al., 2005). Among them, SNVs are the most frequently identified (Genomes Project et al., 2015; Lek et al., 2016) and annotated (Cunningham et al., 2015). SNVs are composed of non-synonymous SNVs (nsSNVs) and synonymous SNVs (sSNVs). Comparing to sSNVs, nsSNVs, which will cause amino acid change based on the protein translation codons, are estimated at higher frequency in individuals with excess deleteriousness (Genomes Project et al., 2012). Therefore, in this review, we focus on the computational approaches which are developed to infer the impact of nsSNVs in coding regions. The database resources that are utilized by majority of the predictive methods (we name them as predictors throughout this review) are firstly introduced. Following that, we discuss the underlying motivation and constraints of those predictors with which we group them into six categories in an innovative manner. We also present their corresponding predictive performance and concerns from assessment studies. Finally, we demonstrate the application performance of the predictors in large-scale studies, as well as their ability to reveal the genotype-phenotype associations.
2 Database resources for variant predictors
Models are not created out of thin air; rather, they are designed to identify hidden correlations in massive volumes of real data, allowing data to be interpreted and used to generate predictions. Since the deployment of the Human Genome Project in the 1990s, various relevant databases and knowledgebases have been established and maintained by academic institutions, organizations, consortia, and communities to collect, store, and retrieve records pertaining to genetic, clinical, and phenotypic information. They provide sufficient accessible evidences and facts to reliably demonstrate the genotype-phenotype association, which explains the functional and pathogenic importance of genetic variations (Johnston and Biesecker, 2013).
Databases can be categorized according to their scope, purpose, and scale. Several reviews (Thorisson et al., 2009; Brookes and Robinson, 2015; Zhang et al., 2019; Banck et al., 2021; Katsonis et al., 2022) provided comprehensive details of the content, usage, comparisons, and limitations for those databases. In this section, we briefly review the most frequently used databases (Table 1) containing sequence information, population-scale data, phenotype ontology, clinical and experimental evidence.
TABLE 1.
Summary of resources for human genotypes and phenotypes relationships.
| Type of data | Name | Full name | Techniques | Type of variants | Targeted diseases | Website | Containing entries (until writtern in June 2022) | Composition | First publication year | Last update (until writtern in June 2022) | Accessible | Publications |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein data | Uniprot | Universal protein resource | Curated | — | General | https://www.uniprot.org/ | 567,483 entries in Swiss-Prot and 231,354,261 entries in TrEMBL | UniProt Knowledgebase, UniProt Reference Clusters, and UniProt Archive | 1997 | 2 February 2021 | Free | UniProt, (2021) |
| Protein information | UniProtKB | Uniprot Knowledgebase | Curated | — | General | https://www.uniprot.org/uniprot/ | — | Swiss-Prot and TrEMBL | — | 22 November 2021 | Free | UniProt, (2021) |
| Protein sequences | UniRef | Uniprot Reference Clusters | Curated | — | General | https://www.uniprot.org/uniref/ | — | UniRef100, 90, 50 | — | 29 November 2021 | Free | UniProt, (2021) |
| Protein sequences | UniParc | Uniprot Archive | Curated | — | General | https://www.uniprot.org/uniparc/ | — | — | — | 24 March 2022 | Free | UniProt, (2021) |
| Protein, DNA and RNA structural data | PDB | Protein data bank | Structural data from X-ray, NMR, electron microscopy | — | General | https://www.rcsb.org/ | 191,565 Biological Macromolecular Structures | — | 1971 | 14 June 2022 | Free | Berman et al. (2000) |
| Protein data with themodynamic parameters | ProThermDB | Thermodynamic Database for Proteins and Mutants | Curated | — | General | https://web.iitm.ac.in/bioinfo2/prothermdb/index.html | ∼0.12 million thermodynamic data obtained for different organisms and cell lines, >32,000 entries, ∼20,000 mutations | — | 1999 | 22 September 2021 | Free | Nikam et al. (2021) |
| Protein data | ONGene | — | Curated | — | Cancer | https://ongene.bioinfo-minzhao.org/index.html | 803 oncogenes | — | 2016 | — | Free | Liu et al. (2017) |
| Protein data | TSGene2.0 | Tumor suppressor gene database | Curated | — | Cancer | https://bioinfo.uth.edu/TSGene/ | 1217 human tumor suppressor genes | — | 2012 | 4 January 2016 | Free | Zhao et al. (2016) |
| Population data | 1000 Genome Project | — | WGS | SNVs, indels | General | https://www.internationalgenome.org/ | Genotypes for 2,504 healthy donor samples from 26 populations | — | 2008 | 1 October 2015 | Free | Sudmant et al. (2015) |
| Population data | GnomAD (previously ExAC) | Genome aggregation database | WGS, WES | SNVs, indels | General | https://gnomad.broadinstitute.org/ | 76,156 genomes data of diverse ancestries in v3.1 and 141,456 individuals exomes or genomes data in v2 | — | 2014 | 21 January 2022 | Free | Karczewski et al. (2020) |
| Population data | ESP | The NHLBI exome sequencing project | WES | SNVs, indels | Disease-, phenotype-related | https://evs.gs.washington.edu/EVS/ | 6,503 unrelated individual exom data | — | 2011 | 23 April 2019 | Free | Fu et al. (2013) |
| Population data | UK Biobank | — | — | — | Disease-, phenotype-related | https://www.ukbiobank.ac.uk/ | 49,960 exome data | — | 2006 | 19 March 2019 | Registration fee needed | — |
| Population data | UK10K | — | WGS, WES | — | Healthy and disease-related cohorts | https://www.uk10k.org/ | Nearly 10,000 individuals in UK population | Whole genome, Neurodevelopment, Obesity, Rare Diseases Sample Sets | 2010 | 1 October 2015 | Access control | Consortium et al. (2015) |
| Phenotype and genotype data | OMIM | Online Mendelian Inheritance in Man | Classification | — | Disease-, phenotype-, gene-related | https://www.omim.org/ | 26,446 entries, including all known mendelian disorders and over 16,000 genes | — | 1960 | 27 May 2022 | Free | Amberger et al. (2019) |
| Phenotype and genotype data | Orphanet | The portal for rare diseases and orphan drugs | Classification | — | Disease-, phenotype-related | https://www.orpha.net/ | 6,172 disease, 5835 genes | — | 1997 | 31 May 2022 | Free | — |
| Ontology | HPO | Human phenotype ontology | Classification | — | Disease-, phenotype-, gene-related | https://hpo.jax.org/ | >13,000 terms, > 156,000 annotations | — | 2008 | 14 April 2022 | Free | Kohler et al. (2021) |
| Ontology | GO | Gene ontology | Classification | — | Gene-specific | http://geneontology.org/ | 7,510,543 annotations | Molecular Function, Cellular Component, and Biological Process | 2000 | 16 May 2022 | Free | (Ashburner et al., 2000; Gene Ontology, 2021) |
| Ontology | Mammalian Phenotype Ontology | — | Classification | — | Phenotype-related | https://bioportal.bioontology.org/ontologies/MP/?p=summary | 14,716 classes | — | 2005 | 14 June 2022 | Free | Smith et al. (2005) |
| Genomic data | HGMD | Human gene mutation database | Curated | SNVs, indels | Disease-, phenotype-related | http://www.hgmd.cf.ac.uk/ac/index.php | 352,731 mutation entries | 352,731 mutation entries | 1996 | 31 May 2022 | Registration needed | Maffucci et al. (2019) |
| Genomic data | VariBench | A benchmark database for variations | Curated | SNVs, indels | — | http://structure.bmc.lu.se/VariBench/index.php | — | VariBench datasets include disease-causing missense variations, neutral high frequency SNPs, protein stability affecting missense variations, variations affecting transcription factor binding sites, variations affecting splice sites | 2012 | — | Free | Sasidharan Nair and Vihinen, (2013) |
| Genomic data | VariSNP | — | Curated | SNVs, indels | — | http://structure.bmc.lu.se/VariSNP/index.php | 145,435,955 variants | Datasets selected from dbSNP which were filtered for disease-related variants found in ClinVar, Swiss-Prot and PhenCode | 2014 | 16 February 2017 | Free | Schaafsma and Vihinen, (2015) |
| Genomic data | dbSNP | Single nucleotide polymorphism database | Curated | SNVs, indels, retroposable element insertions and microsatellite repeat variations | General | https://www.ncbi.nlm.nih.gov/snp/ | 1,085,850,277 refSNP | — | 1999 | 26 May 2020 | Free | Sherry et al. (2001) |
| Genomic data | ClinVar | — | Curated | SNVs, indels | Disease-, phenotype-, gene-related | https://www.ncbi.nlm.nih.gov/clinvar/ | 1,540,318 unique variation records | — | 2013 | 5 May 2022 | Free | Landrum et al. (2020) |
| Genomic data | ClinGen | — | Curated | SNVs, indels | Disease-, phenotype-related | https://clinicalgenome.org/ | Unique 3692 variants in unique 2278 genes | — | 2013 | 1 April 2022 | Free | Rehm et al. (2015) |
| Genomic data | DoCM | Database of Curated Mutations | Curated | SNVs, indels | Cancer | http://www.docm.info/ | 1,364 variants among 122 disease type | — | 2014 | — | Free | Ainscough et al. (2016) |
| Genomic data | VKGL | Vereniging klinisch genetische | Curated | SNVs, indels | Disease-, phenotype-related | https://vkgl.molgeniscloud.org/ | 188,502 variants | — | 2018 | December 2021 | Free | Fokkema et al. (2019) |
| Genomic data | CIViC | Clinical interpretation of variants in cancer | Curated | SNVs, indels, SVs | Cancer | https://civicdb.org/welcome | 3165 variants, 470 genes with clinical interpretation | — | 2015 | 1 May 2022 | Free | Griffith et al. (2017) |
| Genomic data | COSMIC | Catalogue of somatic mutations in cancer | Curated | SNVs, indels | Cancer | https://cancer.sanger.ac.uk/cosmic | 29,399,170 variants, 1,207,190 CNVs, 19,422 fusions | — | 2004 | 31 May 2022 | Free | Tate et al. (2019) |
| Genomic data | LOVD3.0 | Leiden open variation database 3.0 | Curated | SNVs, indels | Disease-, phenotype-related | https://www.lovd.nl/3.0/home | 800,780 variants | — | 2002 | 17 August 2021 | Free | Fokkema et al. (2021) |
| Genomic data | InSight | The International Society for Gastrointestinal Hereditary Tumours | Curated | SNVs, indels | Gene-specific | http://insight-database.org/ | 35,644 variant entries from 9 genes related to gastrointestinal tumours | Variants are automatically sourced from LOVD3 | 2005 | — | Free | Fokkema et al. (2021) |
| Genomic data | HuVarBase | Human variants database | Curated | Missense, nonsense, insertion, deletion | Disease-, phenotype-related | https://www.iitm.ac.in/bioinfo/huvarbase/index.php | 774,863 variants from 18,318 proteins, including 702,048 disease-causing and 72,815 neutral variants | Sources from 1000 Genomes, ClinVar, COSMIC, Humsavar, SwissVar, MutHTP, PROXiMATE | 2018 | 15 October 2018 | Free | Ganesan et al. (2019) |
| Genomic data | DVD | Deafness variation database | Curated | SNVs, indels | Deafness-related | https://deafnessvariationdatabase.org/ | 223 genes | Sources from ClinVar, dbNSFP, gnomAD, VEP, CADD, dbSNP, Population Analysis and others | 2018 | 4 January 2021 | Free | Azaiez et al. (2018) |
| Genomic data | METABRIC | Molecular Taxonomy of Breast Cancer International Consortium | Targeted NGS | SNVs, indels | Breast cancer | Mutation details can be retrived from https://www.cbioportal.org/study/summary?id=brca_metabric | Mutation data in 173 genes from 2433 primary breast tumor samples and 650 normal controls | Genomic mutation data, copy number aberration (CNA), gene expression and long-term clinical follow-up data | 2012 | — | Free | (Curtis et al., 2012; Pereira et al., 2016) |
| Genomic data | TCGA-BRCA | — | WES | SNVs, indels | Breast cancer | https://portal.gdc.cancer.gov/projects/TCGA-BRCA | Mutation data from WES of 817 Breast Invasive Carcinoma tumor/normal pairs | Genomic mutation data, copy number aberration (CNA), gene expression and long-term clinical follow-up data | 2012 | 8 October 2015 | Free | Ciriello et al. (2015) |
| Genomic data | BRCA1 dataset | — | Saturation genome editing assays | SNVs | BRCA1 gene | https://sge.gs.washington.edu/BRCA1/ | 3,893 SNVs located within or near 13 exons that encode for the RING and BRCT domains of BRCA1 (exons 2–5 and 15–23, respectively) | — | 2018 | — | Free | Findlay et al. (2018) |
| Genomic data | VarCards | — | Curated | SNVs, indels | General | http://varcards.biols.ac.cn/ | 110,154,363 SNVs, and 1,223,370 indels in coding regions or splicing sites | Variant-level and gene-level resources | 2016 | 28 June 2020 | Free | Li et al. (2018) |
2.1 Sequence resources
GenBank (Sayers et al., 2022), hosted by National Institutes of Health (NIH), European Nucleotide Archive (ENA) (Baker et al., 2000), hosted by European Molecular Biology Laboratory’s European Bioinformatics Institute (EMBL-EBI), as well as the DNA Data Bank of Japan (DDBJ) (Okido et al., 2022) are the most widely used sequence databases, storing over 2.5 billion nucleotide sequences for over 504,000 formally described species. They serve as a basis for genetic analysis since aligning clean reads to the reference genome is an indispensable step in NGS analysis. As sequences of plethora species become accessible, protein sequences with 100%, 95%, and 50% identity are assembled to create clusters that are stored in informative databases such as UniProt Reference Clusters (UniRef) (Suzek et al., 2007), a branch of Universal Protein Resource (UniProt) (UniProt, 2021). These clusters are utilized to build multiple sequence alignment (MSA) sets, which form the basis of homology sequence-based approach.
2.2 Population resources
Several worldwide population projects exist, including the NCBI dbSNP (Smigielski et al., 2000), 1000 genome project (1KGP) (Sudmant et al., 2015), HapMap (International HapMap, 2003), UK10K (Consortium et al., 2015), Genome Aggregation Database (gnomAD) (Karczewski et al., 2020), and NHLBI GO Exome Sequencing Project (ESP) (Fu et al., 2013). With their progress and completion, their reports are now public and offer an exquisite view of the landscape of human genetic variants ranging from common to extremely rare ones. They also provide valuable information allowing the examination of variants between and within subpopulations with different ethnicities or disease status like heart, lung and blood disorders. Furthermore, minor allele frequency (MAF) from these databases is usually a useful indicator for prioritization or pertain as important feature for building prediction models.
2.3 Phenotype resources
Phenotype databases describe phenotypes and illnesses in conjunction with genetic information. The most widely known are OMIM (Online Mendelian Inheritance in Man) (Amberger et al., 2019) and Orphanet (Ayme et al., 1998). Their goal is to offer high-quality information on common and rare diseases or phenotypes in order to comprehensively review the genotype-phenotype association. To assist the investigation on connections between phenotypes and genes and to describe diseases in an algorithm-friendly data structure, ontology databases such as Human Phenotype Ontology (HPO) (Robinson et al., 2008), Mammalian Phenotype Ontology (Smith et al., 2005), and Gene Ontology (GO) (Ashburner et al., 2000) were developed. They are designed to annotate clinical phenotypes and genes with well-structured, computational-friendly, precise, and accurate terminology. Overall, these databases provide valuable insights for prioritization and interpretation of genetic data.
2.4 Clinical genetic resources
Several databases curated genetic data with clinical significance information. These databases are also known as Locus-Specific Databases (LSDB). Data and entries are often curated from literature and clinical trials. LSDBs range in scale from a single gene with roughly 4000 variants (Findlay et al., 2018) to hundreds of millions of variants (Schaafsma and Vihinen, 2015). The goal of LSDBs is to unambiguously and accurately define and categorize genotype-phenotype correlation, to understand gene functions and effects, to provide a map of genetic distribution across populations and diseases, and to assist clinicians/diagnostic laboratories in conducting further validation assays by providing detail molecule, pathogenicity, and effects of variants (Greenblatt et al., 2008). A well-curated and annotated LSDB is a valuable resource for constructing and evaluating prediction models. But note in mind that there would be overlapped variants in different LSDBs, even with contradictory classification of clinical impact due to inconsistent rules and subjective opinion of different curators. Phenotype-/disease- specific LSDBs are established, such as DVD (Deafness Variation Database) (Azaiez et al., 2018) for deafness, RAPID (Resource of Asian Primary Immunodeficiency Diseases) (Keerthikumar et al., 2009) for primary immunodeficiency disease, InSiGHT (The International Society for Gastrointestinal Hereditary Tumors) (Fokkema et al., 2021) for gastrointestinal tumors, fabry-database.org (Saito et al., 2011) for Fabry disease. Thanks to the effort of the Leiden Open source Variation Database (LOVD) (Fokkema et al., 2021) platform, a comprehensive list of public LSDBs are presented with details for researchers and clinicians to retrieve gene and mutation information from different resources.
3 Various variant predictors
Each predictor has a unique biochemical or biological basis. It is important to remember that the outcome of the predictor on different bases has different implications. The terms “dangerous,” “pathogenic,” “conservative,” and “damaging” do not necessarily denote causal of a specific phenotype or condition. Knowing the principles and drawbacks of each type of predictor aids in correctly interpreting the variants.
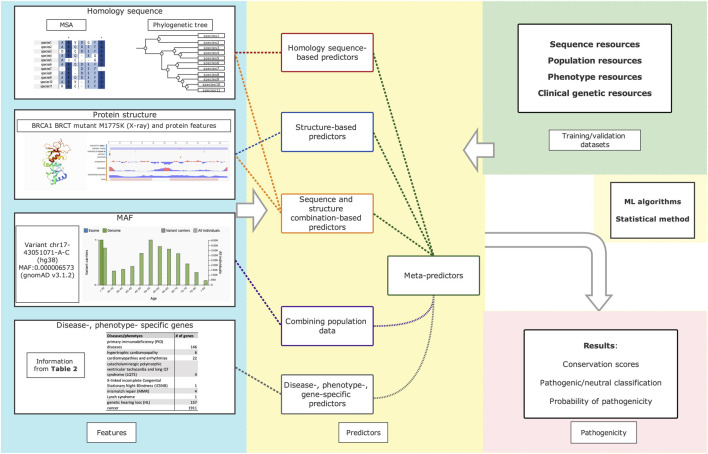
Variant impact predictors can be categorized in different ways: machine-learning (ML) and non-ML models based on the used algorithms; homology sequence-based and structural-based models regarding the features they used in prediction; supervised and unsupervised ML-models. Unlike the category of sequence-, structure- and meta-methodologies in other reviews (Hassan et al., 2019b; Yazar and Ozbek, 2021), we introduced an innovative category here based on the characteristics and included features of each type (Figure 1). We discuss these categories by outlining the rational reasoning behind the predictors and provide an overview of the constraints. Later, we discuss predictor performance evaluation and underline current concerns and remedies. Details for each tool are present in Supplementary Table S1 and Table 2.
FIGURE 1.
Summarized workflow of variants impact predictors. Protein structure and protein features of BRCA1 BRCT mutant M1775K are retrived from studies. (Birrane, 2006: Tischkowitz et al., 2008). The minor allele frequency (MAF) information of variant rs41293463 (chr17-43051071-A-C(GRCh38)) was retrived from gnomAD (Genome Aggregation Database).
TABLE 2.
Representative diseases-, phenotypes-, genes-specific variants impact predictors.
| Characteristic category | Name | Type of variants | Targeted disease/phenotype/gene | # of genes | Website | Distribution (web-server/stand-alone) | First publication | Programming language | Algorithm/model | Features | Dataset for modeling | Classification index | Classification | Additional data | Publication |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Meta-predictor | VIPPID (Variant Impact Predictor for PIDs) | Missense | Primary immunodeficiency (PID) diseases | 146 | https://mylab.shinyapps.io/VIPPID/ | Web and stand-alone | April 2022 | Perl, R | Conditional Inference Forest | 85 features including AA, exonic, protein structural, conservation, and 20 pre-existing prediction tools | 4,865 disease-associated variants from Asian Primary Immunodeficiency Diseases (RAPID) database, HGMD and ClinVar; 4,237 neutral variants from gnomAD | Classifier | Pathogenic/non-pathogenic | 26 reviewed P/LP variants of known PID pathogenic genes from 1318 patients cohort and 39 validated in-house variants | Fang et al. (2022) |
| Meta-predictor | CanPredict | Missense | Cancer | — | http://www.canpredict.org/or http://www.cgl.ucsf.edu/Research/genentech/canpredict/, both are not accessible | — | May 2007 | R | RF | SIFT, Pfam-based LogR.E-value and GO Similarity Score (GOSS) metrics | — | Classifier | Likely cancer/likely non-cancer/not determined | — | Kaminker et al. (2007) |
| Meta-predictor | PolyPhen-HCM | Missense | Hypertrophic cardiomyopathy | 6 | http://genetics.bwh.harvard.edu/hcm/ | Pre-computed results | February 2011 | — | Naïve bayes classifier | Prediction scores, protein structure comparison score | 74 curated variants from literitures and manually classified by Laboratory for Molecular Medicine standard variant-assessment pipeline (41 pathogenic, 26 benign) | Classifier | Pathogenic/benign/no call | — | Jordan et al. (2011) |
| Meta-predictor | Cadioboost | Missense | Cardiomyopathies and arrhythmias | 22 | https://www.cardiodb.org/cardioboost/ | Pre-computed results | October. 2020 | R | 2 Adaptive Boosting (Adaboost) classifiers | 76 functional features | CM datasets: 356 rare P/LP variants from 9,007 clinical CM patients, 302 rare missense variants in CM genes from 2,090 healthy controls. Inherited arrhythmia dataset: 252 P/LP in arrhythmia-associated genes from ClinVar, 237 rare missense variants in arrhythmia genes from 2,090 healthy controls | Pathogenicity score | Disease-causing/VUS/Benign | 4 datasets from ClinVar, HGMD, Oxford Medical Genetics Laboratory (OMGL), a large registry of HCM patients, SHaRe | Zhang et al. (2021) |
| Multiple features | GENESIS (GENe-specific EnSemble grId Search) | Variants of uncertain clinical significance | Catecholaminergic polymorphic ventricular tachycardia and long QT syndrome (LQTS) | 4 | https://github.com/rachellea/medgenetics | Stand-alone and pre-computed results | March 2022 | Python | Logistic regression and multilayer perceptron model | 8 kinds of features including AA features, domain, conservation, rate of evolution, signal-to-noise ratio, and a position-specific scoring matrix (PSSM) score | 717 pathogenic variants and 3,164 benign variants curated from literiture | Probabilities of pathogenicity | Pathogenic/VUS/benign | 925 VUS classified according to ACMG | Draelos et al. (2022) |
| Multiple features | CACNA1F-vp | Missense | X-linked incomplete Congenital Stationary Night Blindness (iCSNB) | 1 | https://github.com/shalawsallah/CACNA1F-variants-analysis | Stand-alone | April 2020 | Python | Logistic regression model | Variant-level features and structural features | 72 disease-implicated from HGMD or MGDL database, 322 benign variants from gnomAD | Probabilities of pathogenicity | Pathogenic/benign | - | Sallah et al. (2020) |
| Optimized PON-P2 | PON-MMR2 | AA substitution | Mismatch repair (MMR) | 4 | http://structure.bmc.lu.se/PON-MMR2/ | Web and stand-alone | September 2015 | R | RF | 5 features: sequence conservation, physical and biochemical properties of AA | 109 pathogenic, 99 neutral, 354 VUS from InSiGHT database and VariBench | Probabilities of pathogenicity | Pathogenic/VUS/benign | 354 VUS dataset | Niroula and Vihinen, (2015) |
| Optimized MAPP | CoDP (Combination of Different Properties of MSH6 protein) | Missense | Lynch syndrome (LS) | 1 | http://cib.cf.ocha.ac.jp/CoDP/ | Web | April 2013 | — | Logistic regression model | MSA, phylogenetic tree, structral properties, MAPP, SIFT, PolyPhen2 | 294 missense variants from InSiGHT, MMRUV, UniProt, dbSNP, ESP, HapMap Project, 1KGP and literature | Probabilities of pathogenicity | Likely LS/Unlikely LS | 260 unclassified variants dataset | Terui et al. (2013) |
| Meta-predictor with MAF as features | DvPred | nsSNVs | Genetic hearing loss (HL) | 157 | https://github.com/WCH-IRD/DVPred/tree/main/DVPred_score | Stand-alone and pre-computed results | February 2022 | Python | Gradient boosting decision tree (GBDT) | 65 features include conservation scores, prediction scores, MAF, gene intolerance scores and other features | 1,318 P/LP and 4,628 B/LB from China Deafness Genetics Consortium (CDGC), Deafness Variation Database (DVD), ClinVar, HGMD | DvPred score | Deleterious/neutral | 463 pathogenic and 454 benign variants from new version of CDGC and ClinVar | Bu et al. (2022) |
| Meta-predictor | NBDriver | Missense | Cancer | 58 | https://github.com/RamanLab/NBDriver | Stand-alone | May 2021 | Python | RF, extra tress (ET) classifier, generative KDE classifier | 3 types of features: one-hot encoding, overlapping k-mers, 27 genomic features | 5,265 disease-associated variants from five literatures | Classifier | — | — | Banerjee et al. (2021) |
| Combination of rule-based and meta-predictor | CancerVar | Exon variants, CNVs, indels | Cancer | 1911 | https://cancervar.wglab.org/index.php | Web, stand-alone and pre-computed results | May 2022 | Python | Semi-supervised generative adversarial network used in scoring method OPAI | 12 clinical evidence prediction scores and 23 precomputed scores by other computational tools | 13 million variants from 7 cancer knowledgebases | OPAI score | Oncogenic/benign | 4 datasets from OncoKB and CIViC, IARC and literatures | Li et al. (2022) |
*VUS, variant of uncertain significance.
3.1 Types of tools and their principles
3.1.1 Homologous sequence-based predictors
This class of predictors are derived from comparative genomics. The assumption is straightforward: under natural selection, amino acid changes in conservative sequences are more “deleterious” determined by homologous sequence searching across species, than that happened in other non-homologous positions which would be deemed as “tolerant” (Cooper and Shendure, 2011). Methodologically, these predictors firstly construct the multiple sequence alignment (MSA) either by grouping multiple protein sequences with a given similarity from BLAST alignment (Altschul et al., 1990), or just retrieval customed selective sequences from afroed-mentioned genomic databases (Section 2.1) for multiple alignment using MULTIZ (Blanchette et al., 2004), or MUSCLE (Edgar, 2004). Based on MSA, a position-specific scoring matrix (PSSM) (Gribskov et al., 1987) is computed to generate the prediction outcome with probability score (Ng and Henikoff, 2001), likelihood ratio (Chun and Fay, 2009), the average distance between targeted species and others in subfamilies (Choi et al., 2012), or the entropy difference (Reva et al., 2007; Hopf et al., 2017). The predictive outcomes are normally continuous values with the designer’s recommended threshold validated in mutation datasets.
Apart from computing scores using empirically rational equations, ML algorithms were commonly utilized as classifiers. Classical models include random forest (RF) (Capriotti et al., 2006), and hidden Markov Model (HMM) (Thomas et al., 2003; Siepel et al., 2005; Garber et al., 2009; Pollard et al., 2010; Shihab et al., 2013). Although they are both ML techniques, the attributes they employ are distinct. For example, PhD-SNP (Capriotti et al., 2006) converted MSA and mutation to a 40-feature variables in support vector machine (SVM). The 40 features are composed of two parts: the first 20 vectors explicitly define the mutation residues, with -1 for the wild-type residue, 1 for the mutation, and 0 for the others. The second set of 20 vectors represents the mutation sequence environment, which is the frequency of each 20 amino acid residue in a 20 amino acid length window centered on the targeted site. Unlike unweighted and balanced MSA, HMM is a probabilistic profile of MSA that captures position-specific information (Krogh et al., 1994). Two different configurations of HMM were observed. One assumed three hidden states: “match,” “insertion,” and “deletion” to build a profile-HMM MSA (Thomas et al., 2003; Shihab et al., 2013), while the other considered a two-hidden state as “conserved” and “non-conserved” according to the phylogenetic information from tree topologies (Siepel et al., 2005; Garber et al., 2009; Pollard et al., 2010).
More recently, a novel unsupervised ML model is utilized to discover patterns and correlations between absolute locations in the MSA, allowing direct observation of both conservation and coevolution (Riesselman et al., 2018; Frazer et al., 2021). This deep generative model captured the latent structure from MSA using Variational Autoencoders (VAEs), which was proved to be an outstanding model for separation of β-lactamase protein family, at the phyla level (Detlefsen et al., 2022). By assuming the observed data s are generated from latent variable z, the decode part of VAE consists of modeling the conditional probability. Hence, the encode part is the neural network modeling of approximate posterior distribution (Riesselman et al., 2018; Frazer et al., 2021).
ML models’ predictions were normally given as log odds ratio scores between the probabilities of “substitution” and “wild-type” or “conserved” and “non-conserved”. In other words, under wild-type or neutral model, higher scores represent higher probability of unexpected substitution, thus are more evolutionary constraint.
There are two considerations regarding homologous sequence-based predictions (Eilbeck et al., 2017). Firstly, many known disease-causing alleles reside in poorly or non-conserved regions will be false-negatively classified as neutral by predictors. Secondly, the tools are inadequate for predicting stop-gain and frameshift variations since they are not included in other organisms in the MSA (Eilbeck et al., 2017). The stop-gain and frameshift variants are rated as “HIGH” impact on biological sequence in annotation tools, e.g., VEP (McLaren et al., 2010) and SnpEff (Cingolani et al., 2012). But the impact on protein is not always concordant. The amino acid changes seem to be tolerant especially the ones located near C-terminal of protein (MacArthur et al., 2012). Some frameshift variants, even in homozygous state, were frequently observed among population suggesting limited impact on human health (Eilbeck et al., 2017). Thus, additional information such as protein structure might help improve the predictive power and efficiency of the predictors, which will be discussed in the following subsections in more detail.
3.1.2 Structure-based predictors
Apart from the primary structure of protein, the folding and stability are also essential for protein function normally. Early findings of variants that affect protein structure leading to aberrant phenotypes can be dated back to the 1950s, when the amino acid substitution in the half molecule of hemoglobin was discovered to cause sickle cell anemia (Ingram, 1957). Since then, thousands of mutations (Giardine et al., 2014) were described to impact on the function (increase (Jones et al., 1979) or decrease (Bonaventura and Riggs, 1968) oxygen affinity), stability (Martinez et al., 1977) and conformation (Moo-Penn et al., 1988) of hemoglobin. Indeed, missense variants also affect protein expression (Haraksingh and Snyder, 2013), post-translational modification (Kim et al., 2015) or binding affinity (Pires et al., 2015; Morningstar-Kywi et al., 2021).
An estimation of ∼75% disease-causing variants directly lead to protein destabilization, making protein stability the major contributor to disease pathology (Yue et al., 2005), whereas ∼7% variants in disease dataset also have functional role (Yue et al., 2014). The location of the mutation has a preference. In comparison to polymorphisms, disease-causing mutations predominantly impact the core of the protein, whereas ∼70% are found in structural and functionally essential regions (Sunyaev et al., 2000; de Beer et al., 2013). Protein-protein interfaces are hot spots for disease-causing nsSNVs (David et al., 2012; Petukh et al., 2015). Again, disease-causing variations were 49% more likely (interface core vs interface rim odds ratio (OR) 1.49, 95% CI 1.24–1.80, p < 0.00001) to be found in the interface core than in the rim, possibly due to their differences in energy contribution to protein stability, physicochemical and evolutionary properties (David and Sternberg, 2015).
Typically, nsSNVs impact on protein stability is estimated by computing the variation of Gibbs free energy change (∆∆Gf) resulting from an amino acid substitution. Physical effective energy function, statistical potential function, and empirical defined potential function are the three types of energy computing methodologies (Guerois et al., 2002). Because the first function is computationally intensive, the latter two are more frequently utilized. Structure-based predictors of protein stability mainly attribute to empirical potentials that integrate physical and statistical structure-related energy components (Guerois et al., 2002), and ML techniques (Dehouck et al., 2009; Laimer et al., 2015).
In theory, these approaches should potentially give greater insights into the mutation effect than the homologous sequence-based predictors since they are built on the direct impact of mutation on protein structure and function. However, the truth is that protein-based predictors are still limited because of the unbalance and intrinsic variability of the thermodynamic data and their prediction performance (Sanavia et al., 2020). On one hand, despite that the Protein Data Bank (PDB) (Berman et al., 2000) contains over 50,000 human protein records, many of them are redundant, covering only 70% of reference human proteome at a sequence identity level higher than 30% (Somody et al., 2017). The development of AlphaFold2 (Tunyasuvunakool et al., 2021), to an extent increases the protein structure coverage; but its capability to predict the impact of single mutation is questionable (Pak et al., 2021; Buel and Walters, 2022). On the other hand, sequence-based techniques, under certain circumstances, outperform structure-based stability prediction tools (Hoie et al., 2022). Thus, combining sequence with structural information may aid in improving prediction capacity of variant impact.
3.1.3 Sequence and structure combination-based predictors
The approaches of this category consider both the previously described homologous sequence and protein structure information. Predictions take benefit from the combination of homology sequence information (e.g., conservative scores), and the structure features, such as hydropathy, polarity, backbone angles and electrostatic interactions, supplemented with energy features and biochemical features such as solvent accessible surface area of the interface (Kulshreshtha et al., 2016). Those features are sometimes transformed or selected for model training to achieve high prediction efficiency. Sometimes hundreds of features might be incorporated into the final model (Niroula et al., 2015). Algorithmically, supervised ML approaches including SVM (Calabrese et al., 2009; Li et al., 2009), naïve bayes classifier (Adzhubei et al., 2010), neural network (NN) (Hecht et al., 2015), RF (Carter et al., 2013; Niroula et al., 2015) and boosted tree regression (Zhou et al., 2016) are commonly applied in the multiple features predictors.
3.1.4 Meta-predictors
Meta-predictors are tools that make predictions by integrating results of pre-existing predictors. The term “meta-” sometimes corresponds to the term “consensus” in other studies (Bendl et al., 2014). The basic idea behind meta-predictors is to leverage on potential complimentary performance of selected predictors in classifying variants.
There are mainly two improvements regarding meta-predictors comparing to aforementioned counterparts. First of all, meta-predictors give a comprehensive evaluation on the selected pre-existing tools. Each predictor has its own metric and scale making it difficult to compare across multiple predictors hindering the simultaneous usage. Meta-predictors have their own way to interpret scores from selected tools, by transforming to a comparable range as normalized scores (Bendl et al., 2014) or binary values (Gonzalez-Perez and Lopez-Bigas, 2011). In addition, meta-predictors are able to improve prediction performance by integrating prediction scores from different predictors, which allows the avoidance of bias and anti-generalization by single predictors (Kircher et al., 2014).
In terms of missing value, where partly pre-existing tools fail to predict, some meta-predictors impute them using deleterious/neutral threshold (Capriotti et al., 2013), average score (Kircher et al., 2014; Quang et al., 2015), fixed score (Quinodoz et al., 2022), the maximal pathogenic score (Jagadeesh et al., 2016), or a flexible imputation using average value of k-nearest neighbors (Ioannidis et al., 2016) and Bayesian principle component analysis (BPCA) (Dong et al., 2015). There is currently no gold standard for imputation. Although machine-learning imputation appears to be more accurate (Brock et al., 2008; Wei et al., 2018), meta-predictor builders revealed that missing values account for less than 10% of their training and testing datasets (Dong et al., 2015), making the imputation methods less significant difference.
While prediction performance studies suggested that meta-predictors surpassed other counterparts (Tian et al., 2019), concerns regarding circularity occurred, which will be discussed in Section 3.3.
3.1.5 Combining population data
A polymorphism is defined as an alteration in DNA sequence found in the general population at a MAF greater than 1%. According to The American College of Medical Genetics (ACMG) and the Association for Molecular Pathology (AMP) guidelines for clinical variant interpretation, a variant with >5% MAF is considered as a stand-alone support for benign interpretation for a rare Mendelian disorder (Richards et al., 2015). This is supported by the “neutral theory”, which defines neutral variants as the ones settled in the population through random genetic drift causing neither harmful nor beneficial effect to the survival of individual organisms (Kimura, 1979). When training and validating predictors, variants with higher than specified allele frequency (e.g., 5% or 1%) from population-scale databases were usually denoted as benign or neutral. However, predictors in predicting neutral variants differ greatly in capacity and specificity. For example, PON-P2 (Niroula et al., 2015) had a 95% specificity, while the poorest predictor incorrectly categorized more than one-third of polymorphisms as disease-causing (Niroula and Vihinen, 2019). Classifying the impact of variants according to their MAF were further argued by different hypothesis including “rare variant for Mendelian disease” (Pritchard and Cox, 2002), “Common disease, common variant” (CDCV) and “Common disease/rare variant” (CDRV).
Researchers now have access to an exquisitely detailed view of the landscape of common and rare human genetic variants. Another issue that predictors should be careful with when utilizing MAF is that MAF is largely dependent on the population size and varies among subpopulations leading to population stratification (Eilbeck et al., 2017). For example, rs79444516, which is common in African population (13%), exhibited its extreme rareness in European and Asian population, with MAF <0.05%. When estimated in the mixture population, the MAF is 1.2% which will cause confounding classification. Varied MAF for the same variant because of different scales of sample size could be largely mitigated with the completion of the huge population-scale projects.
To better utilize MAF in prediction, ClinPred (Alirezaie et al., 2018), a meta-predictor using ML approach, employs MAFs from diverse populations as part of their features, instead of classifying variants based on single arbitrary MAF cutoffs. Together with feature scores from 16 pre-existing tools, ClinPred trains on clinically curated pathogenic and benign datasets and outperforms other meta-predictors when applied to datasets of rare diseases and cancer (Alirezaie et al., 2018). Therefore, MAFs from population data is capable to enhance the prediction. Similarly, more and more tools (Chennen et al., 2020; do Nascimento et al., 2020; Lai et al., 2020; Li et al., 2020) integrated MAFs as predicting features and achieved competitive performance on pathogenicity prediction.
3.1.6 Disease-, phenotype-, gene-specific predictors
The ultimate goal of the variant prediction tools is to accelerate the development of precision medicine. Majority of the strategies discussed above aim to estimate disease occurrence based on the assumption that changes in protein function leads to a decrease in organismal fitness (Boucher et al., 2016). They are trained in a large-scale datasets in a genome-wide and pan-disease manner neglecting the complexity among different diseases and making the prediction results suboptimal (Dorfman et al., 2010). Therefore, with the necessity to precisely estimate the impact of variants on specific disease/phenotype, a class of disease-, gene-, phenotype-specific prediction has emerged.
The phenotype-targeting predictors range widely from common cardiac (Zhang et al., 2021), cancer (Kaminker et al., 2007), and neurodegenerative disease (Ahmed et al., 2015), to rare diseases, such as methylmalonic acidemia (Peng et al., 2019), X-linked incomplete Congenital Stationary Night Blindness (Sallah et al., 2020) and Pompe disease (Adhikari, 2019). More details are presented in Table 2. There are over 13,000 terms defined in HPO. While LSDBs provide benchmarked variant datasets, such as COSMIC, CIViC and OncoKB for cancer, which can be utilized for disease-specific predictors construction, limited datasets are available for majority of phenotypes. For a particular disease/phenotype, the training and validation datasets can be prepared by curation of variants and genes from literatures, or re-analysis of unpublished sequencing data of case-control cohorts, followed by manually classification using recognized guidelines such as ACMG/AMP. The scale of the curated databases for different diseases/phenotypes varies in gene (from one to hundreds) and variant number (from thousands to millions) which is largely dependent on the number of relevant publications.
With the curated databases, most of this category of predictors utilize ML methodology. They can be grouped into three classes. The first class overlaps with previous mentioned categories but has distinct characteristic. It includes sequence and structure combination-based (Sallah et al., 2020; Draelos et al., 2022), sharing the same strategy with previously mention predictors in Section 3.1.3, and meta-predictors (Jordan et al., 2011; Bu et al., 2022) similar to the ones discussed in Section 3.1.4. The distinct characteristic is the difference in training and validation datasets selection. Also, disease-related genes are known, making predictors capable of constructing sub-model for each gene, resulting in better prediction performance (Fang et al., 2022). The second class aims to optimize pre-existing predictor, usually sequence-based model, by re-constructing MSAs and phylogenetic tree of targeted gene(s) (Niroula and Vihinen, 2015; Fortuno et al., 2018). These predictors share the same strategy as their precursors, with distinct features selection. The third class predicts variants in a comprehensive and robust way, utilizing additional rule-based classification system. For example, CancerVar (Li et al., 2022), integrates rule-based categorization with ML-based meta-predictor scores to interpret the predicting clinical significance.
These well-calibrated and sculpted predictors demonstrate their capability in targeted sequencing disease-specific panels to the utmost (Peng et al., 2019). In contrast, their ability to generalize is then questioned. When utilizing these techniques, note in mind the key target phenotypes and genes.
3.2 Performance assessment of predictors
As dozens of predictors exist, choosing the appropriate one(s) becomes challenging for end users. Several assessment criteria, such as sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), accuracy, and Matthews correlation coefficient (MCC), are commonly used to demonstrate model performance (Vihinen, 2012). The values for sensitivity, specificity, PPV, NPV, and accuracy range from 0 to 1, with higher values indicating better performance. MCC benefits from true and false positives and negatives with values on a scale of -1 to 1, with values closer to 1 indicating perfect prediction. Furthermore, a visualization measurement, receiver operating characteristics (ROC) analysis is frequently used to intuitively compare the area under the ROC curve (AUC) of multiple predictors (Vihinen, 2012). For non-intersecting curves, the AUC value closer to 1 suggests better overall performance, while a value of 0.5 indicates random and useless classification.
Most predictors, when developed, would be assessed using respective training and validation datasets presenting supreme or acceptable performance. However, evaluation using consensus datasets would be more informative for tool selection.
There are dozens of comparison studies on the performance assessment of different selected tools using different benchmark datasets. When Performance evaluation of pathogenicity-computation methods for missense variants, meta-predictors such as REVEL, Meta-SNP, generally have better performance and stronger evidence in clinical interpretation (Accetturo et al., 2020; Cubuk et al., 2021; Anderson and Lassmann, 2022). In the assessment of 23 predictors, Li et al. (Li et al., 2018) revealed that meta-predictors achieved higher AUC than others of sequence-based and structure-based predictors using the ClinVar benchmark dataset, indicating better performance of meta-predictors. However, when regarding somatic variants and PPARG gene benchmark datasets, meta-predictors and structure-based predictors exhibited comparable performance (AUC>0.8) (Li et al., 2018), and were superior to homology sequence-based predictors (AUC>0.7). Hassan et al. (Hassan et al., 2019a) revealed that meta-predictor which integrated 4 pre-existing prediction scores, outperformed other 8 predictors achieving ∼10%, 20%, 15% improvement in specificity, sensitivity and AUC, respectively.
The performance of different categories are not always consistent, and sometimes are contradictory. Poon’s study (Poon, 2021) on BRCA1/2 datasets revealed that SIFT and PolyPhen2’s performance differed among genes. Meléndez-Aranda et al. (Melendez-Aranda et al., 2019) compared the performance of 6 in silico tools on 215 missense mutations in hemophilia B causative gene F9, and the results showed that the most popular tool, SIFT, was the most accurate. When applying to a somatic dataset containing 4319 somatic missense variants, the performance of SIFT was sub-optimal (Suybeng et al., 2020). As a result, it is critical to have pre-knowledge of your testing data and predictive goal when selecting appropriate tools.
In order to address the confounding situation and objectively determine the appropriate usage and accuracy of predictors, the Critical Assessment of Genome Interpretation (CAGI) (Andreoletti et al., 2019) community started their experiments in 2010. Until now, there are six editions with 63 challenges and over 50 articles released. Participants predict the phenotypic impact of unpublished genetic variants collected from experimental and clinical labs provided by CAGI. Later, independent assessors test the predictions against experimental characterized phenotypes, and the results will be presented at the CAGI conference and published in special journal issues. The challenges released include a wide range of topics, from nsSNVs to splicing variants, and from disease panels to databases including curated variants. However, the reality of the outcome is frequently far more complex than the challenges’ initial objective. Predictors with superior performance in one challenge, would fail to call the pathogenicity of variants in other datasets (Katsonis and Lichtarge, 2019; Savojardo et al., 2019). Complex gene datasets caused divergence predictions and confounding outcomes, raising concerns about the possibility of experimental mistakes as the basis of disagreement (Miller et al., 2019). All the above suggested the caution when interpreting the evaluation results.
3.3 Concerns of current predictors and remedies
Majority of predictors are trained, validated and tested using benchmarked sets of variants with explicit classification labels. When evaluated 10 predictors across major public databases, Grimm et al. (2015) raised concerns about “circularity” involving in the usage of predictors and conduction of comparative studies. The term “circularity” refers to the situation that same variants are recursively used in both training and evaluating models.
“Type 1 circularity” refers to the overlap between training and evaluation particularly for supervised ML-based predictors, resulting in poor generalization on new data (Grimm et al., 2015). Selecting predictions from unsupervised tools as features or filtering overlapping sets during training might assist to minimize the “type 1 circularity” during model construction (Alirezaie et al., 2018; Won et al., 2021). Furthermore, avoiding overuse of individual dataset (Vihinen, 2013; Weber et al., 2019) and choosing benchmark database which addressed overlapped issue (Sasidharan Nair and Vihinen, 2013; Sarkar et al., 2020) also helps when conducting comparative studies.
Grimm et al. (2015) observed that weighted FatHMM (Shihab et al., 2013) achieved outstanding performance in 2 datasets but severe drop in performance in subset from SwissVar. They found that the ratio of pathogenic and neutral variants in the same protein family was the key element for weighting scheme, leading to higher pathogenic score assigned to both neutral and pathogenic variants in the same gene with higher ratio (Grimm et al., 2015). This strategy made weighted FatHMM statistically successful in some datasets, but ultimately inappropriate. Therefore, they defined the “Type 2 circularity” as the circumstance in which all variants from the same gene are jointly labeled as pathogenic or neutral. To address this problem, it was suggested to use datasets with an appropriate pathogenic-to-neutral ratio and avoid genes with exclusive pathogenic or neutral variations when reporting performance (Bu et al., 2022; Quinodoz et al., 2022).
Another concern is about “collinearity,” which generally occurs with the regression models. ‘Collinearity’ refers to the circumstance in which significant correlation between two or more feature variables resulting in independent regression coefficients estimation problems and leading to redundancy in the set of variables (Bayman and Dexter, 2021). This situation might be mitigated via feature selection and estimator modification (Zheng et al., 2020; Chan et al., 2022). From another perspective, “collinearity” should not be a problem because more complicated machine learning algorithms including SVM, Random Forest, and Neural Network, can handle large-scale and multi-collinear datasets in a better way (Dong et al., 2015; Perez-Enciso and Zingaretti, 2019).
4 Application
In-silico approaches combined mathematical strategies with expert opinion allows researchers to analyze the biological meaning of genetic data efficiently and economically (Trisilowati and Mallet, 2012). In-silico predictors on variant effect aids in genome interpretation. The prediction-based categorization provides insight into variant characterization and prioritization.
Regards to large-scale population study, in silico predictors aid in variant classification for pattern overview and comparison at subpopulation level. For example, Palmer et al. (2022) subdivided missense variants by SIFT and PolyPhen2 prediction in research on bipolar disorder (BD) and revealed an obvious enrichment in ultra-rare harmful missense variation outside of confined missense areas, particularly in bipolar II disorder (BD2). This observation contrasted with the findings in schizophrenia cases (Singh et al., 2022) of enrichment within constrained missense regions. The authors speculated this signal may capture something distinct to mood disorders relative to psychotic disorders (Palmer et al., 2022).
For large-scale population, in silico predictors also facilitate the detection of variant-level signals under natural-selection for those living in extreme environments or with a diverse geographic distribution. Deng et al. (2019) ranked variants by calculating the functional importance score (FIS) from four in silico predictors. Based on the ranking of adaptive genetic variants, they revealed a seldom studied gene, TMEM247 with a missense variant rs116983452, to be the most-differentiated functional variant identified between Tibetan and non-Tibetan populations (Deng et al., 2019). When studying non-homogeneous Taiwanese Han population, integrated selection of allele favored by evolution (iSAFE) was incorporated with the CADD functional impact score to identify 16 natural-selection signals by geographic distribution that were unambiguously localized to 5 single genes (Lo et al., 2021). Meanwhile, in the western Roma population, Font-Porterias et al. (2021) categorized missense variants based on GERP, PolyPhen2 and CADD, revealing significant difference in common deleterious variant portion between Roma and non-Roma population. Furthermore, runs of homozygozity (ROH), which are continuous homozygous regions of the DNA sequence, exhibit ancestry-specific patterns of accumulation of deleterious homozygotes.
In addition to characterization for population-level study, predictors have also been widely used for prioritization of disease-causing candidates in case-control or pedigree studies, finally leading to the identification of genotype-phenotype association. There are commonly two strategies for variant prioritization in which predictors help. Several frameworks and platforms are listed in Table 3.
TABLE 3.
Representative prioritization frameworks and tools.
| Characteristic category | Name | Type of Targeted variants* | Website | Distribution (web-server/stand-alone) | First publication | Last update | Programming language | Algorithm/modules | Input type | Dataset for modeling | Publications |
|---|---|---|---|---|---|---|---|---|---|---|---|
| User-defined rule-based | VCF.Filter | SNVs, indels | https://biomedical-sequencing.at/VCFFilter/ | Web and stand-alone | July 2017 | — | Java | Filter cohort, prioritize on pedigree and search variant in cohort modules | VCF files, targeted regions, cohort allele frequencies, pedigree information | — | Muller et al. (2017) |
| User-defined rule-based | BiERapp | SNVs, indels, CNVs, MNVs, SVs | http://bioinfo.cipf.es/apps-beta/bierapp/2.0.0/#home | Web and stand-alone | April 2014 | — | HTML5 and JS | CellBase annotation, consecutive filtering strategy | Multi-sample VCF files | — | Aleman et al. (2014) |
| User-defined rule-based | KGGSeq | SNVs, indel, CNVs | http://pmglab.top/kggseq/ | Stand-alone | January. 2012 | 1 January 2022 | Java | 5 major modules: quality control, filtration, annotation, pathogenic prediction and statistic tests | VCF files, pedigree information | 7,296 disease-causing variants from OMIM and 48,089 neutral variants | Li et al. (2012); Li et al. (2017) |
| User-defined rule-based | VPOT (variant prioritization ordering tool) | SNVs, indel | https://github.com/VCCRI/VPOT/ | Stand-alone | November. 2019 | 27 October 2021 | Python | 2 steps: prioritization of variants based on user-defined parameters, post-processing of variant priority ordered list | ANNOVAR annotated VCF or TXT files, inheritance model | — | Ip et al. (2019) |
| ACMG guideline based | TAPES | SNVs, indel | https://github.com/a-xavier/tapes | Stand-alone | October. 2019 | — | Python | Bayesian classification framework | VCF files | — | Xavier et al. (2019) |
| ACMG guideline based | InterVar | SNVs, indel | https://github.com/WGLab/InterVar, http://wintervar.wglab.org/ | Web, stand-alone and pre-computed results | February 2017 | 13 June 2022 | Python | Automated or manually scoring system. Manual review and adjustment on specific criteria | Annotated or unannotated VCF files | — | Li and Wang, (2017) |
| ACMG guideline realted | VarFish | SNVs, indels | https://varfish-kiosk.bihealth.org/, https://github.com/bihealth/varfish-server | Web and stand-alone | July 2020 | June 2022 | Python | Quality control, database- and user-based annotation, filtering interface, joint filtering of multiple cases | VCF files, optional pedigree information | - | Holtgrewe et al. (2020) |
| Phenotype-driven | Exomiser | SNVs, indels | https://www.sanger.ac.uk/tool/exomiser/ | Stand-alone | November 2015 | November 2021 | Java | Filtering and Prioritization based on logistical regression model. Four prioritization method include PHIVE, PhenIX, ExomeWalker, hiPHIVE. | VCF files, HPO terms, optional pedigree information | — | Smedley et al. (2015) |
| Phenotype-driven | eXtasy | nsSNVs | https://extasy.esat.kuleuven.be/ | Web and stand-alone | September 2013 | — | Ruby | RF | VCF files, HPO terms | 24,454 disease-causing nsSNV from HGMD associated with 1,142 HPO terms. Control datasets: common polymophisms and rare variants from 1KGP, rare variants in in-house control samples | Sifrim et al. (2013) |
| Phenotype-driven | AMELIE (Automatic Mendelian Literature Evaluation) | Missense, stopgain, splicing, indels, duplication | https://amelie.stanford.edu/ | Web and stand-alone | May 2020 | May 2021 | — | Natural language processing (NLP) and logistic regression classifier | VCF files, HPO terms | A set of 681 simulated patients using data from OMIM, ClinVar and 1KGP | Birgmeier et al. (2020) |
| Phenotype-driven | Phen-Gen | Missense, nonsense, splice site and indels | https://github.com/pkuerten/phen-gen | Stand-alone | September 2014 | — | Perl | Random walk–with–restart algorithm, Bayesian framework based on genotype and phenotype data | VCF files, HPO terms | HGMD 2011.4 datasets | Javed et al. (2014) |
| Phenotype-driven | LIRICAL (LIkelihood Ratio Interpretation of Clinical AbnormaLities) | SNVs, indels | https://github.com/TheJacksonLaboratory/LIRICAL | Stand-alone | September 2020 | September 2021 | Java | Likelihood-ratio | VCF files, HPO terms | — | Robinson et al. (2020) |
| Phenotype only | Phrank (phenotype ranking) | — | https://bitbucket.org/bejerano/phrank/src/master/ | Stand-alone | February 2019 | — | Python | Boolean Bayesian network | HPO terms | Knowledgebase of gene-disease-phenotype relationships, HPO-A | Jagadeesh et al. (2019) |
| Phenotype only | PhenoRank | — | https://github.com/alexjcornish/PhenoRank | Stand-alone | June 2018 | — | Python | Phenotypic similarity measured by simGIC, gene scores calculation by random walk with restart (RWR) method | HPO terms | 5,685 unique associations between 4,729 diseases and 3,713 genes from ClinVar, OMIM and UniProtKB | Cornish et al. (2018) |
| Phenotype only | Phen2Gene | — | https://phen2gene.wglab.org/, https://github.com/WGLab/Phen2Gene | Web and stand-alone | June 2020 | March 2021 | Python | Weighting by skewness | HPO terms | HPO–gene annotation files downloaded from the Jackson Laboratory for Genomic Medicine; gene-disease databases OMIM, ClinVar, Orphanet, GeneReviews; gene-gene relationship databases HPRD, HGNC, Biosystem, HTRI | Zhao et al. (2020) |
First, empirical criteria are used to filter variations. With high quality variants, many studies (Ma et al., 2013; Blue et al., 2014) performed prioritization based on in silico predictions, MAF in population database and control groups, inheritance pattern, and functional effect. By this method, less than 10 variants are distilled out of hundreds of thousands obtained from WES analysis. Following the validation of orthogonal assays (e.g., Sanger sequencing), true positive causal candidates will be examined for the functional effect on protein in vitro and/or in vivo. The relationship between variants-phenotype is therefore thoroughly investigated. Several user-friendly rule-based frameworks (Coutant et al., 2012; Li et al., 2012; Aleman et al., 2014; Muller et al., 2017) have been built to make the filtering procedure easier to implement. Researchers can set their own criteria and get the findings in readable files with detailed annotation information. The prioritization can also be supplemented with adoption of consensus recommendations, such as ACMG/AMP standards and guidelines (Richards et al., 2015). The guideline includes a comprehensive set of definitions and criteria for variation interpretation, ranging from standardized nomenclature to evidence-based rating yielding a five-tier terminology system outcome. Results from in silico predictors are accounted as “supporting” evidence for benign or pathogenic classification. Some automatic tools (Li and Wang, 2017; Xavier et al., 2019) have also been developed for variant classification based on the guidelines, although the manual classification by professional geneticists would be deemed as the golden standard.
The second strategy refers to phenotype-driven frameworks, which combine phenotype and variants data for prioritization and interpretation. Clinical diagnosis would be straightforward when the disease is known. However, before the identification of candidate disease, the procedure to explain a set of clinical features is challenging due to the absence or presence of unrelated features and various degrees of specificity (Kohler et al., 2009). To extract standardized and normalized phenotypic terminologies from sparse clinical abnormalities in case studies, some tools like Phenomizer (Kohler et al., 2009) and Doc2HPO (Liu et al., 2019) are recommended to map the clinical symptoms to the list of known disorders and estimate the significance of each disease match. Prediction scores from in silico predictors are integrated in this kind of framework as “pathogenicity” or “deleteriousness” features. Most of phenotype-driven tools (Sifrim et al., 2013; Javed et al., 2014; Smedley et al., 2015; Birgmeier et al., 2020; Robinson et al., 2020) require variant files and HPO terms as input, while some tools (Cornish et al., 2018; Jagadeesh et al., 2019; Zhao et al., 2020) require only HPO terms. Yuan et al. (2022) investigated causal-gene prioritizing performance of both types on two benchmark datasets in a recent comparative study and revealed that former ones performed better overall than latter ones. Their results also indicated the complementarity of multiple phenotype-driven tools towards a viable integrated strategy may improve diagnostic efficiency (Yuan et al., 2022).
5 Discussion
In this review, we firstly summarized the database resources frequently used during predictor development. We then discussed the rational, necessity and limitations for the newly categorized predictors: homologous sequence-based, structural-based, combination of sequence and structural, meta-predictors, population-based, and gene-, phenotype-, disease-specific predictors. Predictor performance as well as their limitations and possible remedies were then outlined. The application of the predictors in real studies was finally presented demonstrating their efficient assistance in variant characterization and prioritization, as well as the discovery of genotype-phenotype association.
When building predictors, unambiguous labeled datasets are critical. Avoiding overlapping and contradicting data, as well as balancing the positive-negative ratio in training and validation datasets, will definitely minimize the negative influence of circularity. Further examination on the collinearity between/among feature variables will facilitate the optimization of prediction models, even though some algorithms are literally not affected.
Among the predictors, meta-predictors outperform others in general; however, their prediction performance is considerably discounted in some disease-specific datasets, raising concern about their applications especially in clinical settings (Schiemann and Stowell, 2016; Mahmood et al., 2017). Employment of disease-, gene-, phenotype-specific predictors can to an extent solve the above issue. When selecting predictors for a particular study, efforts should be given on screening whether the genes and phenotype predictor calibrated perfectly matching your research, and understanding the scope and predictive performance of each predictor. On the other hand, we look forward to more specialized predictors sculpted for a variety of phenotypes covering both common and rare diseases.
According to Variation Ontology (VariO) (Vihinen, 2014), variant impacts on protein level can be annotated with effects on function, structure and property. Variants impact on protein functional or property effects can be classified as follows: abundance, which includes gene dosage, expression, degradation and mis-localization; activity, which includes enzymatic, kinetic and regulation; enzymatic specificity, and molecular affinity (Vihinen, 2021). Most of above-mentioned predictors computed the possibility of pathogenic effect on protein function and structure in a broad range, rather the effects on protein abundance, activity or affinity properties separately. This may indicate a challenging future orientation of variant predictors development.
The correlation between variants pathogenic prediction on protein function or structure and abnormal clinical outcomes are validated by experimental facts at the current stage. For certain phenotypes, an evident enrichment of deleterious variants in a set of disease-related genes, such as the increased mutational burden in essential genes in autism spectrum disorder (Ji et al., 2016), WNT signaling genes in myelomeningocele (Hebert et al., 2020), a set of 5 genes in epilepsy (Leu et al., 2015). The gap between observed higher burden genes and clinical phenotype is then bridged by functional or mechanical experimental studies. For example, meiocytes with pathogenic mutation p.S167L in HSF2BP found in premature ovarian insufficiency (POI) patients from a family, showed a reduced number of foci formed by the recombinases RAD51/DMC1, leading to crossover defect, which provided an insight into the molecular mechanism of mutation in POI and subfertility (Felipe-Medina et al., 2020). Currently, variant impact predictors are insufficient for indicating molecular mechanism of pathogenicity. However, the advancement of protein structure prediction may assist the interpretation of pathogenic variants since structural information gives useful insights in evaluating variant impact on protein or biological systems (Diwan et al., 2021).
Impacts of mutations on protein synthesis includes transcriptional and translational influences. For SNVs, the impact on transcription involves in changes in transcript sequence and influence in gene regulation (Haraksingh and Snyder, 2013). Tools for predicting impact on gene regulation have been timely and systematically reviewed by other studies (Li et al., 2015; Ohno et al., 2018; Rojano et al., 2019; Canson et al., 2020). In terms of translation, SNVs-induced amino acid substitution causes protein structure and function abnormalities, and the prediction methods have been explored in this study. The deeper association between SNVs for protein folding and post-translation modification is still being investigated.
With the development of a cutting-edge structure prediction tool, AlphaFold2, the unstructured human protein narrowed down to less than 30% (Porta-Pardo et al., 2022). However, examples showed that AlphaFold2 was not capable for predicting protein structure modification caused by pathogenic mutations, particularly those having experimentally proven destabilizing effect (Buel and Walters, 2022). The reasons for this limitation may relate to the bioinformatics and physical methodologies utilized in modeling, as well as the resources from protein sequence and PDB structure data employed, instead of the fundamental driving forces of protein folding (Jumper et al., 2021; Buel and Walters, 2022). The AlphaFold team is presently considering solutions for new mutations, which may give better prediction on unfolding to folding state, based on protein physics instead of sequence evolutionary (Callaway, 2022). We anticipate that its success will usher in a new age of human genetic research, including the acceleration of in silico functional and mechanical genotype-phenotype association investigations.
Finally, although the variant effect predictors greatly help the genomic interpretation, end-users should keep in mind that the predictor’s role is only an assistance to clinical diagnosis, and merely a starting point (Eilbeck et al., 2017). The unequal relationship between predicted damaging effect and pathogenicity warns their usage. In addition, under some circumstances, the predicted scores overstating the effect of uncommon mutations, will cause inflated estimation affecting the specificity and sensitivity (Lanktree et al., 2018). Therefore, experimental validations, the golden standard in variant impact evaluation, are still indispensable.
Author contributions
YL and DC designed the structure of the review. YL wrote the draft manuscript, curated data, organized tables and figure. DC supervised the work and revised the manuscript. PC and WY supervised and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Shenzhen Fundamental Research Program, China (no. JCYJ20210324114807019).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.981005/full#supplementary-material
References
- Accetturo M., Bartolomeo N., Stella A. (2020). In-silico analysis of NF1 missense variants in ClinVar: Translating variant predictions into variant interpretation and classification. Int. J. Mol. Sci. 21 (3), E721. 10.3390/ijms21030721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari A. N. (2019). Gene-specific features enhance interpretation of mutational impact on acid alpha-glucosidase enzyme activity. Hum. Mutat. 40 (9), 1507–1518. 10.1002/humu.23846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., Bork P., et al. (2010). A method and server for predicting damaging missense mutations. Nat. Methods 7 (4), 248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed A. B., Znassi N., Chateau M. T., Kajava A. V. (2015). A structure-based approach to predict predisposition to amyloidosis. Alzheimers Dement. 11 (6), 681–690. 10.1016/j.jalz.2014.06.007 [DOI] [PubMed] [Google Scholar]
- Ainscough B. J., Griffith M., Coffman A. C., Wagner A. H., Kunisaki J., Choudhary M. N., et al. (2016). DoCM: A database of curated mutations in cancer. Nat. Methods 13(10), 806–807. 10.1038/nmeth.4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleman A., Garcia-Garcia F., Salavert F., Medina I., Dopazo J. (2014). A web-based interactive framework to assist in the prioritization of disease candidate genes in whole-exome sequencing studies. Nucleic Acids Res. 42, W88–W93. Web Server issue). 10.1093/nar/gku407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alirezaie N., Kernohan K. D., Hartley T., Majewski J., Hocking T. D. (2018). ClinPred: Prediction tool to identify disease-relevant nonsynonymous single-nucleotide variants. Am. J. Hum. Genet. 103 (4), 474–483. 10.1016/j.ajhg.2018.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 (3), 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Amberger J. S., Bocchini C. A., Scott A. F., Hamosh A. (2019). OMIM.org: Leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 47 (D1), D1038-D1043–D1043. 10.1093/nar/gky1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D., Lassmann T. (2022). An expanded phenotype centric benchmark of variant prioritisation tools. Hum. Mutat. 43 (5), 539–546. 10.1002/humu.24362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoletti G., Pal L. R., Moult J., Brenner S. E. (2019). Reports from the fifth edition of CAGI: The critical assessment of genome interpretation. Hum. Mutat. 40 (9), 1197–1201. 10.1002/humu.23876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., et al. (2000). Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25 (1), 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayme S., Urbero B., Oziel D., Lecouturier E., Biscarat A. C. (1998). Information on rare diseases: The Orphanet project. La Rev. Med. Interne 19 (3), 376S–377S. 10.1016/s0248-8663(98)90021-2 [DOI] [PubMed] [Google Scholar]
- Azaiez H., Booth K. T., Ephraim S. S., Crone B., Black-Ziegelbein E. A., Marini R. J., et al. (2018). Genomic landscape and mutational Signatures of deafness-associated genes. Am. J. Hum. Genet. 103 (4), 484–497. 10.1016/j.ajhg.2018.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker W., van den Broek A., Camon E., Hingamp P., Sterk P., Stoesser G., et al. (2000). The EMBL nucleotide sequence database. Nucleic Acids Res. 28 (1), 19–23. 10.1093/nar/28.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banck H., Dugas M., C M. U.-T., Sandmann S. (2021). Comparison of open-access databases for clinical variant interpretation in cancer: A case study of MDS/AML. Cancer Genomics Proteomics 18 (2), 157–166. 10.21873/cgp.20250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Raman K., Ravindran B. (2021). Sequence neighborhoods enable reliable prediction of pathogenic mutations in cancer genomes. Cancers (Basel) 13 (10), 2366. 10.3390/cancers13102366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayman E. O., Dexter F. (2021). Multicollinearity in logistic regression models. Anesth. Analg. 133 (2), 362–365. 10.1213/ANE.0000000000005593 [DOI] [PubMed] [Google Scholar]
- Bendl J., Stourac J., Salanda O., Pavelka A., Wieben E. D., Zendulka J., et al. (2014). PredictSNP: Robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput. Biol. 10 (1), e1003440. 10.1371/journal.pcbi.1003440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berliner N., Teyra J., Colak R., Garcia Lopez S., Kim P. M. (2014). Combining structural modeling with ensemble machine learning to accurately predict protein fold stability and binding affinity effects upon mutation. PLoS One 9 (9), e107353. 10.1371/journal.pone.0107353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., et al. (2000). The protein Data Bank. Nucleic Acids Res. 28 (1), 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgmeier J., Haeussler M., Deisseroth C. A., Steinberg E. H., Jagadeesh K. A., Ratner A. J., et al. (2020). AMELIE speeds Mendelian diagnosis by matching patient phenotype and genotype to primary literature. Sci. Transl. Med. 12 (544), eaau9113. 10.1126/scitranslmed.aau9113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrane G., Soni A., Ladias J. A. A. (2006). X-ray Structure of the BRCA1 BRCT mutant M1775K [Online]. Available at: https://gnomad.broadinstitute.org/variant/17-43051071-A-C?dataset=gnomad_r3 (Accessed September 13 2022) [abstract].
- Blanchette M., Kent W. J., Riemer C., Elnitski L., Smit A. F., Roskin K. M., et al. (2004). Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14 (4), 708–715. 10.1101/gr.1933104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blue G. M., Kirk E. P., Giannoulatou E., Dunwoodie S. L., Ho J. W., Hilton D. C., et al. (2014). Targeted next-generation sequencing identifies pathogenic variants in familial congenital heart disease. J. Am. Coll. Cardiol. 64 (23), 2498–2506. 10.1016/j.jacc.2014.09.048 [DOI] [PubMed] [Google Scholar]
- Bonaventura J., Riggs A. (1968). Hemoglobin Kansas, a human hemoglobin with a neutral amino acid substitution and an abnormal oxygen equilibrium. J. Biol. Chem. 243 (5), 980–991. 10.1016/s0021-9258(18)93612-4 [DOI] [PubMed] [Google Scholar]
- Boucher J. I., Bolon D. N., Tawfik D. S. (2016). Quantifying and understanding the fitness effects of protein mutations: Laboratory versus nature. Protein Sci. 25 (7), 1219–1226. 10.1002/pro.2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock G. N., Shaffer J. R., Blakesley R. E., Lotz M. J., Tseng G. C. (2008). Which missing value imputation method to use in expression profiles: A comparative study and two selection schemes. BMC Bioinforma. 9, 12. 10.1186/1471-2105-9-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg Y., Rost B. (2007). Snap: Predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 35 (11), 3823–3835. 10.1093/nar/gkm238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes A. J., Robinson P. N. (2015). Human genotype-phenotype databases: Aims, challenges and opportunities. Nat. Rev. Genet. 16 (12), 702–715. 10.1038/nrg3932 [DOI] [PubMed] [Google Scholar]
- Bu F., Zhong M., Chen Q., Wang Y., Zhao X., Zhang Q., et al. (2022). DVPred: A disease-specific prediction tool for variant pathogenicity classification for hearing loss. Hum. Genet. 141 (3-4), 401–411. 10.1007/s00439-022-02440-1 [DOI] [PubMed] [Google Scholar]
- Buel G. R., Walters K. J. (2022). Can AlphaFold2 predict the impact of missense mutations on structure? Nat. Struct. Mol. Biol. 29 (1), 1–2. 10.1038/s41594-021-00714-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese R., Capriotti E., Fariselli P., Martelli P. L., Casadio R. (2009). Functional annotations improve the predictive score of human disease-related mutations in proteins. Hum. Mutat. 30 (8), 1237–1244. 10.1002/humu.21047 [DOI] [PubMed] [Google Scholar]
- Callaway E. (2022). What's next for AlphaFold and the AI protein-folding revolution. Nature 604 (7905), 234–238. 10.1038/d41586-022-00997-5 [DOI] [PubMed] [Google Scholar]
- Canson D., Glubb D., Spurdle A. B. (2020). Variant effect on splicing regulatory elements, branchpoint usage, and pseudoexonization: Strategies to enhance bioinformatic prediction using hereditary cancer genes as exemplars. Hum. Mutat. 41 (10), 1705–1721. 10.1002/humu.24074 [DOI] [PubMed] [Google Scholar]
- Capriotti E., Altman R. B., Bromberg Y. (2013). Collective judgment predicts disease-associated single nucleotide variants. BMC Genomics 14 (3), S2. 10.1186/1471-2164-14-S3-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E., Calabrese R., Casadio R. (2006). Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 22 (22), 2729–2734. 10.1093/bioinformatics/btl423 [DOI] [PubMed] [Google Scholar]
- Carter H., Douville C., Stenson P. D., Cooper D. N., Karchin R. (2013). Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics 14, S3. 10.1186/1471-2164-14-S3-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. Y.-L., Leow S. M. H., Bea K. T., Cheng W. K., Phoong S. W., Hong Z.-W., et al. (2022). Mitigating the multicollinearity problem and its machine learning approach: A review. Mathematics 10 (8), 1283. 10.3390/math10081283 [DOI] [Google Scholar]
- Chennen K., Weber T., Lornage X., Kress A., Bohm J., Thompson J., et al. (2020). Mistic: A prediction tool to reveal disease-relevant deleterious missense variants. PLoS One 15 (7), e0236962. 10.1371/journal.pone.0236962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Chan A. P. (2015). PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31 (16), 2745–2747. 10.1093/bioinformatics/btv195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Sims G. E., Murphy S., Miller J. R., Chan A. P. (2012). Predicting the functional effect of amino acid substitutions and indels. PLoS One 7 (10), e46688. 10.1371/journal.pone.0046688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S., Fay J. C. (2009). Identification of deleterious mutations within three human genomes. Genome Res. 19 (9), 1553–1561. 10.1101/gr.092619.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P., Platts A., Wang le L., Coon M., Nguyen T., Wang L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. (Austin) 6 (2), 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G., Gatza M. L., Beck A. H., Wilkerson M. D., Rhie S. K., Pastore A., et al. (2015). Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 163 (2), 506–519. 10.1016/j.cell.2015.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium U. K., Walter K., Min J. L., Huang J., Crooks L., Memari Y., et al. (2015). The UK10K project identifies rare variants in health and disease. Nature 526 (7571), 82–90. 10.1038/nature14962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper G. M., Shendure J. (2011). Needles in stacks of needles: Finding disease-causal variants in a wealth of genomic data. Nat. Rev. Genet. 12 (9), 628–640. 10.1038/nrg3046 [DOI] [PubMed] [Google Scholar]
- Cooper G. M., Stone E. A., Asimenos G., Program N. C. S., Green E. D., Batzoglou S., et al. (2005). Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15 (7), 901–913. 10.1101/gr.3577405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish A. J., David A., Sternberg M. J. E. (2018). PhenoRank: Reducing study bias in gene prioritization through simulation. Bioinformatics 34 (12), 2087–2095. 10.1093/bioinformatics/bty028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutant S., Cabot C., Lefebvre A., Leonard M., Prieur-Gaston E., Campion D., et al. (2012). Eva: Exome Variation Analyzer, an efficient and versatile tool for filtering strategies in medical genomics. BMC Bioinforma. 13, S9. Suppl 14. 10.1186/1471-2105-13-S14-S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubuk C., Garrett A., Choi S., King L., Loveday C., Torr B., et al. (2021). Clinical likelihood ratios and balanced accuracy for 44 in silico tools against multiple large-scale functional assays of cancer susceptibility genes. Genet. Med. 23 (11), 2096–2104. 10.1038/s41436-021-01265-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F., Moore B., Ruiz-Schultz N., Ritchie G. R., Eilbeck K. (2015). Improving the Sequence Ontology terminology for genomic variant annotation. J. Biomed. Semant. 6, 32. 10.1186/s13326-015-0030-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C., Shah S. P., Chin S. F., Turashvili G., Rueda O. M., Dunning M. J., et al. (2012). The genomic and transcriptomic architecture of 2, 000 breast tumours reveals novel subgroups. Nature 486 (7403), 346–352. 10.1038/nature10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27 (15), 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David A., Razali R., Wass M. N., Sternberg M. J. (2012). Protein-protein interaction sites are hot spots for disease-associated nonsynonymous SNPs. Hum. Mutat. 33 (2), 359–363. 10.1002/humu.21656 [DOI] [PubMed] [Google Scholar]
- David A., Sternberg M. J. (2015). The contribution of missense mutations in core and rim residues of protein-protein interfaces to human disease. J. Mol. Biol. 427 (17), 2886–2898. 10.1016/j.jmb.2015.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov E. V., Goode D. L., Sirota M., Cooper G. M., Sidow A., Batzoglou S. (2010). Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 6 (12), e1001025. 10.1371/journal.pcbi.1001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer T. A., Laskowski R. A., Parks S. L., Sipos B., Goldman N., Thornton J. M. (2013). Amino acid changes in disease-associated variants differ radically from variants observed in the 1000 genomes project dataset. PLoS Comput. Biol. 9 (12), e1003382. 10.1371/journal.pcbi.1003382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehouck Y., Grosfils A., Folch B., Gilis D., Bogaerts P., Rooman M. (2009). Fast and accurate predictions of protein stability changes upon mutations using statistical potentials and neural networks: PoPMuSiC-2.0. Bioinformatics 25 (19), 2537–2543. 10.1093/bioinformatics/btp445 [DOI] [PubMed] [Google Scholar]
- Deng L., Zhang C., Yuan K., Gao Y., Pan Y., Ge X., et al. (2019). Prioritizing natural-selection signals from the deep-sequencing genomic data suggests multi-variant adaptation in Tibetan highlanders. Natl. Sci. Rev. 6 (6), 1201–1222. 10.1093/nsr/nwz108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detlefsen N. S., Hauberg S., Boomsma W. (2022). Learning meaningful representations of protein sequences. Nat. Commun. 13 (1), 1914. 10.1038/s41467-022-29443-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diwan G. D., Gonzalez-Sanchez J. C., Apic G., Russell R. B. (2021). Next generation protein structure predictions and genetic variant interpretation. J. Mol. Biol. 433 (20), 167180. 10.1016/j.jmb.2021.167180 [DOI] [PubMed] [Google Scholar]
- do Nascimento P. M., Medeiros I. G., Falcao R. M., Stransky B., de Souza J. E. S. (2020). A decision tree to improve identification of pathogenic mutations in clinical practice. BMC Med. Inf. Decis. Mak. 20 (1), 52. 10.1186/s12911-020-1060-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., et al. (2015). Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 24 (8), 2125–2137. 10.1093/hmg/ddu733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman R., Nalpathamkalam T., Taylor C., Gonska T., Keenan K., Yuan X. W., et al. (2010). Do common in silico tools predict the clinical consequences of amino-acid substitutions in the CFTR gene? Clin. Genet. 77 (5), 464–473. 10.1111/j.1399-0004.2009.01351.x [DOI] [PubMed] [Google Scholar]
- Draelos R. L., Ezekian J. E., Zhuang F., Moya-Mendez M. E., Zhang Z., Rosamilia M. B., et al. (2022). Genesis: Gene-specific machine learning models for variants of uncertain significance found in catecholaminergic polymorphic ventricular Tachycardia and Long QT syndrome-associated genes. Circ. Arrhythm. Electrophysiol. 15 (4), e010326. 10.1161/CIRCEP.121.010326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle M. A., Fritzilas E., Krusche P., Kallberg M., Moore B. L., Bekritsky M. A., et al. (2017). A reference data set of 5.4 million phased human variants validated by genetic inheritance from sequencing a three-generation 17-member pedigree. Genome Res. 27 (1), 157–164. 10.1101/gr.210500.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32 (5), 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilbeck K., Lewis S. E., Mungall C. J., Yandell M., Stein L., Durbin R., et al. (2005). The sequence ontology: A tool for the unification of genome annotations. Genome Biol. 6 (5), R44. 10.1186/gb-2005-6-5-r44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilbeck K., Quinlan A., Yandell M. (2017). Settling the score: Variant prioritization and mendelian disease. Nat. Rev. Genet. 18 (10), 599–612. 10.1038/nrg.2017.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M., Su Z., Abolhassani H., Itan Y., Jin X., Hammarstrom L. (2022). Vippid: A gene-specific single nucleotide variant pathogenicity prediction tool for primary immunodeficiency diseases. Brief. Bioinform., bbac176. 10.1093/bib/bbac176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felipe-Medina N., Caburet S., Sanchez-Saez F., Condezo Y. B., de Rooij D. G., Gomez H. L., et al. (2020). A missense in HSF2BP causing primary ovarian insufficiency affects meiotic recombination by its novel interactor C19ORF57/BRME1. Elife 9, e56996. 10.7554/eLife.56996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B. J. (2017). Perch: A unified framework for disease gene prioritization. Hum. Mutat. 38 (3), 243–251. 10.1002/humu.23158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay G. M., Daza R. M., Martin B., Zhang M. D., Leith A. P., Gasperini M., et al. (2018). Accurate classification of BRCA1 variants with saturation genome editing. Nature 562 (7726), 217–222. 10.1038/s41586-018-0461-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkema I., Kroon M., Lopez Hernandez J. A., Asscheman D., Lugtenburg I., Hoogenboom J., et al. (2021). The LOVD3 platform: Efficient genome-wide sharing of genetic variants. Eur. J. Hum. Genet. 29 (12), 1796–1803. 10.1038/s41431-021-00959-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokkema I., van der Velde K. J., Slofstra M. K., Ruivenkamp C. A. L., Vogel M. J., Pfundt R., et al. (2019). Dutch genome diagnostic laboratories accelerated and improved variant interpretation and increased accuracy by sharing data. Hum. Mutat. 40 (12), 2230–2238. 10.1002/humu.23896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Font-Porterias N., Caro-Consuegra R., Lucas-Sanchez M., Lopez M., Gimenez A., Carballo-Mesa A., et al. (2021). The counteracting effects of demography on functional genomic variation: The roma paradigm. Mol. Biol. Evol. 38 (7), 2804–2817. 10.1093/molbev/msab070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortuno C., James P. A., Young E. L., Feng B., Olivier M., Pesaran T., et al. (2018). Improved, ACMG-compliant, in silico prediction of pathogenicity for missense substitutions encoded by TP53 variants. Hum. Mutat. 39 (8), 1061–1069. 10.1002/humu.23553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer J., Notin P., Dias M., Gomez A., Min J. K., Brock K., et al. (2021). Disease variant prediction with deep generative models of evolutionary data. Nature 599 (7883), 91–95. 10.1038/s41586-021-04043-8 [DOI] [PubMed] [Google Scholar]
- Frederic M. Y., Lalande M., Boileau C., Hamroun D., Claustres M., Beroud C., et al. (2009). UMD-Predictor, a new prediction tool for nucleotide substitution pathogenicity -- application to four genes: FBN1, FBN2, TGFBR1, and TGFBR2. Hum. Mutat. 30 (6), 952–959. 10.1002/humu.20970 [DOI] [PubMed] [Google Scholar]
- Fu W., O'Connor T. D., Jun G., Kang H. M., Abecasis G., Leal S. M., et al. (2013). Analysis of 6, 515 exomes reveals the recent origin of most human protein-coding variants. Nature 493 (7431), 216–220. 10.1038/nature11690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan K., Kulandaisamy A., Binny Priya S., Gromiha M. M. (2019). HuVarBase: A human variant database with comprehensive information at gene and protein levels. PLoS One 14 (1), e0210475. 10.1371/journal.pone.0210475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M., Guttman M., Clamp M., Zody M. C., Friedman N., Xie X. (2009). Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics 25 (12), i54–62. 10.1093/bioinformatics/btp190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology C. (2021). The gene ontology resource: Enriching a GOld mine. Nucleic Acids Res. 49 (D1), D325–D334. 10.1093/nar/gkaa1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C., Abecasis G. R., Auton A., Brooks L. D., DePristo M. A., Durbin R. M., et al. (2012). An integrated map of genetic variation from 1, 092 human genomes. Nature 491 (7422), 56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project C., Auton A., Brooks L. D., Durbin R. M., Garrison E. P., Kang H. M., et al. (2015). A global reference for human genetic variation. Nature 526 (7571), 68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardine B., Borg J., Viennas E., Pavlidis C., Moradkhani K., Joly P., et al. (2014). Updates of the HbVar database of human hemoglobin variants and thalassemia mutations. Nucleic Acids Res. 42, D1063–D1069. 10.1093/nar/gkt911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Perez A., Lopez-Bigas N. (2011). Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am. J. Hum. Genet. 88 (4), 440–449. 10.1016/j.ajhg.2011.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt M. S., Brody L. C., Foulkes W. D., Genuardi M., Hofstra R. M., Olivier M., et al. (2008). Locus-specific databases and recommendations to strengthen their contribution to the classification of variants in cancer susceptibility genes. Hum. Mutat. 29 (11), 1273–1281. 10.1002/humu.20889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., McLachlan A. D., Eisenberg D. (1987). Profile analysis: Detection of distantly related proteins. Proc. Natl. Acad. Sci. U. S. A. 84 (13), 4355–4358. 10.1073/pnas.84.13.4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith M., Spies N. C., Krysiak K., McMichael J. F., Coffman A. C., Danos A. M., et al. (2017). CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat. Genet. 49 (2), 170–174. 10.1038/ng.3774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D. G., Azencott C. A., Aicheler F., Gieraths U., MacArthur D. G., Samocha K. E., et al. (2015). The evaluation of tools used to predict the impact of missense variants is hindered by two types of circularity. Hum. Mutat. 36 (5), 513–523. 10.1002/humu.22768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerois R., Nielsen J. E., Serrano L. (2002). Predicting changes in the stability of proteins and protein complexes: A study of more than 1000 mutations. J. Mol. Biol. 320 (2), 369–387. 10.1016/S0022-2836(02)00442-4 [DOI] [PubMed] [Google Scholar]
- Haraksingh R. R., Snyder M. P. (2013). Impacts of variation in the human genome on gene regulation. J. Mol. Biol. 425 (21), 3970–3977. 10.1016/j.jmb.2013.07.015 [DOI] [PubMed] [Google Scholar]
- Hassan M. S., Shaalan A. A., Dessouky M. I., Abdelnaiem A. E., ElHefnawi M. (2019b). A review study: Computational techniques for expecting the impact of non-synonymous single nucleotide variants in human diseases. Gene 680, 20–33. 10.1016/j.gene.2018.09.028 [DOI] [PubMed] [Google Scholar]
- Hassan M. S., Shaalan A. A., Dessouky M. I., Abdelnaiem A. E., ElHefnawi M. (2019a). Evaluation of computational techniques for predicting non-synonymous single nucleotide variants pathogenicity. Genomics 111 (4), 869–882. 10.1016/j.ygeno.2018.05.013 [DOI] [PubMed] [Google Scholar]
- Hebert L., Hillman P., Baker C., Brown M., Ashley-Koch A., Hixson J. E., et al. (2020). Burden of rare deleterious variants in WNT signaling genes among 511 myelomeningocele patients. PLoS One 15 (9), e0239083. 10.1371/journal.pone.0239083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht M., Bromberg Y., Rost B. (2015). Better prediction of functional effects for sequence variants. BMC Genomics 16 (8), S1. 10.1186/1471-2164-16-S8-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoie M. H., Cagiada M., Beck Frederiksen A. H., Stein A., Lindorff-Larsen K. (2022). Predicting and interpreting large-scale mutagenesis data using analyses of protein stability and conservation. Cell. Rep. 38 (2), 110207. 10.1016/j.celrep.2021.110207 [DOI] [PubMed] [Google Scholar]
- Holtgrewe M., Stolpe O., Nieminen M., Mundlos S., Knaus A., Kornak U., et al. (2020). VarFish: Comprehensive DNA variant analysis for diagnostics and research. Nucleic Acids Res. 48 (W1), W162-W169–W169. 10.1093/nar/gkaa241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf T. A., Ingraham J. B., Poelwijk F. J., Scharfe C. P., Springer M., Sander C., et al. (2017). Mutation effects predicted from sequence co-variation. Nat. Biotechnol. 35 (2), 128–135. 10.1038/nbt.3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T., Chitnis N., Monos D., Dinh A. (2021). Next-generation sequencing technologies: An overview. Hum. Immunol. 82 (11), 801–811. 10.1016/j.humimm.2021.02.012 [DOI] [PubMed] [Google Scholar]
- Ingram V. M. (1957). Gene mutations in human haemoglobin: The chemical difference between normal and sickle cell haemoglobin. Nature 180 (4581), 326–328. 10.1038/180326a0 [DOI] [PubMed] [Google Scholar]
- International HapMap C. (2003). The international HapMap project. Nature 426 (6968), 789–796. 10.1038/nature02168 [DOI] [PubMed] [Google Scholar]
- Ioannidis N. M., Rothstein J. H., Pejaver V., Middha S., McDonnell S. K., Baheti S., et al. (2016). Revel: An ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 99 (4), 877–885. 10.1016/j.ajhg.2016.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip E., Chapman G., Winlaw D., Dunwoodie S. L., Giannoulatou E. (2019). Vpot: A customizable variant prioritization ordering tool for annotated variants. Genomics Proteomics Bioinforma. 17 (5), 540–545. 10.1016/j.gpb.2019.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagadeesh K. A., Birgmeier J., Guturu H., Deisseroth C. A., Wenger A. M., Bernstein J. A., et al. (2019). Phrank measures phenotype sets similarity to greatly improve Mendelian diagnostic disease prioritization. Genet. Med. 21 (2), 464–470. 10.1038/s41436-018-0072-y [DOI] [PubMed] [Google Scholar]
- Jagadeesh K. A., Wenger A. M., Berger M. J., Guturu H., Stenson P. D., Cooper D. N., et al. (2016). M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 48 (12), 1581–1586. 10.1038/ng.3703 [DOI] [PubMed] [Google Scholar]
- Javed A., Agrawal S., Ng P. C. (2014). Phen-gen: Combining phenotype and genotype to analyze rare disorders. Nat. Methods 11 (9), 935–937. 10.1038/nmeth.3046 [DOI] [PubMed] [Google Scholar]
- Ji X., Kember R. L., Brown C. D., Bucan M. (2016). Increased burden of deleterious variants in essential genes in autism spectrum disorder. Proc. Natl. Acad. Sci. U. S. A. 113 (52), 15054–15059. 10.1073/pnas.1613195113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston J. J., Biesecker L. G. (2013). Databases of genomic variation and phenotypes: Existing resources and future needs. Hum. Mol. Genet. 22 (R1), R27–R31. 10.1093/hmg/ddt384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. M., Charache S., Hathaway P. J. (1979). The effect of hemoglobin F-Chesapeake (alpha 2 92 Arg. leads to Leu gamma 2) on fetal oxygen affinity and erythropoiesis. Pediatr. Res. 13 (7), 851–853. 10.1203/00006450-197907000-00011 [DOI] [PubMed] [Google Scholar]
- Jordan D. M., Kiezun A., Baxter S. M., Agarwala V., Green R. C., Murray M. F., et al. (2011). Development and validation of a computational method for assessment of missense variants in hypertrophic cardiomyopathy. Am. J. Hum. Genet. 88 (2), 183–192. 10.1016/j.ajhg.2011.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596 (7873), 583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ka W. (2021). DNA sequencing costs: Data from the NHGRI genome sequencing Program (GSP). [Online]. Available: www.genome.gov/sequencingcostsdata (Accessed June 1, 2022 2022). [Google Scholar]
- Kaminker J. S., Zhang Y., Watanabe C., Zhang Z. (2007). CanPredict: A computational tool for predicting cancer-associated missense mutations. Nucleic Acids Res. 35, W595–W598. 10.1093/nar/gkm405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanzi A. M., San J. E., Chimukangara B., Wilkinson E., Fish M., Ramsuran V., et al. (2020). Next generation sequencing and bioinformatics analysis of family genetic inheritance. Front. Genet. 11, 544162. 10.3389/fgene.2020.544162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski K. J., Francioli L. C., Tiao G., Cummings B. B., Alföldi J., Wang Q., et al. (2020). The mutational constraint spectrum quantified from variation in 141, 456 humans. Nature 581 (7809), 434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski K. J., Francioli L. C., Tiao G., Cummings B. B., Alfoldi J., Wang Q., et al. (2020). The mutational constraint spectrum quantified from variation in 141, 456 humans. Nature 581 (7809), 434–443. 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsonis P., Lichtarge O. (2019). CAGI5: Objective performance assessments of predictions based on the Evolutionary Action equation. Hum. Mutat. 40 (9), 1436–1454. 10.1002/humu.23873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsonis P., Wilhelm K., Williams A., Lichtarge O. (2022). Genome interpretation using in silico predictors of variant impact. Hum. Genet. 10.1007/s00439-022-02457-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keerthikumar S., Raju R., Kandasamy K., Hijikata A., Ramabadran S., Balakrishnan L., et al. (2009). Rapid: Resource of asian primary immunodeficiency diseases. Nucleic Acids Res. 37, D863–D867. 10.1093/nar/gkn682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Kang C., Min B., Yi G. S. (2015). Detection and analysis of disease-associated single nucleotide polymorphism influencing post-translational modification. BMC Med. Genomics 8 (2), S7. 10.1186/1755-8794-8-S2-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. (1979). The neutral theory of molecular evolution. Sci. Am. 241 (5), 98–100. 10.1038/scientificamerican1179-98 [DOI] [PubMed] [Google Scholar]
- Kircher M., Witten D. M., Jain P., O'Roak B. J., Cooper G. M., Shendure J. (2014). A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46 (3), 310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt D. C. (2020). Best practices for variant calling in clinical sequencing. Genome Med. 12 (1), 91. 10.1186/s13073-020-00791-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S., Gargano M., Matentzoglu N., Carmody L. C., Lewis-Smith D., Vasilevsky N. A., et al. (2021). The human phenotype ontology in 2021. Nucleic Acids Res. 49 (D1), D1207–D1217. 10.1093/nar/gkaa1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler S., Schulz M. H., Krawitz P., Bauer S., Dolken S., Ott C. E., et al. (2009). Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am. J. Hum. Genet. 85 (4), 457–464. 10.1016/j.ajhg.2009.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Brown M., Mian I. S., Sjolander K., Haussler D. (1994). Hidden Markov models in computational biology. Applications to protein modeling. J. Mol. Biol. 235 (5), 1501–1531. 10.1006/jmbi.1994.1104 [DOI] [PubMed] [Google Scholar]
- Kulshreshtha S., Chaudhary V., Goswami G. K., Mathur N. (2016). Computational approaches for predicting mutant protein stability. J. Comput. Aided. Mol. Des. 30 (5), 401–412. 10.1007/s10822-016-9914-3 [DOI] [PubMed] [Google Scholar]
- Lai C., Zimmer A. D., O'Connor R., Kim S., Chan R., van den Akker J., et al. (2020). Leap: Using machine learning to support variant classification in a clinical setting. Hum. Mutat. 41 (6), 1079–1090. 10.1002/humu.24011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laimer J., Hofer H., Fritz M., Wegenkittl S., Lackner P. (2015). MAESTRO--multi agent stability prediction upon point mutations. BMC Bioinforma. 16, 116. 10.1186/s12859-015-0548-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum M. J., Chitipiralla S., Brown G. R., Chen C., Gu B., Hart J., et al. (2020). ClinVar: Improvements to accessing data. Nucleic Acids Res. 48 (D1), D835-D844–D844. 10.1093/nar/gkz972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanktree M. B., Haghighi A., Guiard E., Iliuta I. A., Song X., Harris P. C., et al. (2018). Prevalence estimates of polycystic kidney and liver disease by population sequencing. J. Am. Soc. Nephrol. 29 (10), 2593–2600. 10.1681/ASN.2018050493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M., Karczewski K. J., Minikel E. V., Samocha K. E., Banks E., Fennell T., et al. (2016). Analysis of protein-coding genetic variation in 60, 706 humans. Nature 536 (7616), 285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu C., Balestrini S., Maher B., Hernandez-Hernandez L., Gormley P., Hamalainen E., et al. (2015). Genome-wide polygenic burden of rare deleterious variants in sudden unexpected death in epilepsy. EBioMedicine 2 (9), 1063–1070. 10.1016/j.ebiom.2015.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Krishnan V. G., Mort M. E., Xin F., Kamati K. K., Cooper D. N., et al. (2009). Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 25 (21), 2744–2750. 10.1093/bioinformatics/btp528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Shi L., Zhang K., Zhang Y., Hu S., Zhao T., et al. (2018). VarCards: An integrated genetic and clinical database for coding variants in the human genome. Nucleic Acids Res. 46 (D1), D1039-D1048–D1048. 10.1093/nar/gkx1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhao T., Zhang Y., Zhang K., Shi L., Chen Y., et al. (2018). Performance evaluation of pathogenicity-computation methods for missense variants. Nucleic Acids Res. 46 (15), 7793–7804. 10.1093/nar/gky678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Li J., Li M. J., Pan Z., Hsu J. S., Liu D. J., et al. (2017). Robust and rapid algorithms facilitate large-scale whole genome sequencing downstream analysis in an integrative framework. Nucleic Acids Res. 45 (9), e75. 10.1093/nar/gkx019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. X., Gui H. S., Kwan J. S., Bao S. Y., Sham P. C. (2012). A comprehensive framework for prioritizing variants in exome sequencing studies of Mendelian diseases. Nucleic Acids Res. 40 (7), e53. 10.1093/nar/gkr1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Ren Z., Cao K., Li M. M., Wang K., Zhou Y. (2022). CancerVar: An artificial intelligence-empowered platform for clinical interpretation of somatic mutations in cancer. Sci. Adv. 8 (18), eabj1624. 10.1126/sciadv.abj1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wang K. (2017). InterVar: Clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am. J. Hum. Genet. 100 (2), 267–280. 10.1016/j.ajhg.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., van der Velde K. J., de Ridder D., van Dijk A. D. J., Soudis D., Zwerwer L. R., et al. (2020). Capice: A computational method for consequence-agnostic pathogenicity interpretation of clinical exome variations. Genome Med. 12 (1), 75. 10.1186/s13073-020-00775-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chen C. Y., Kaye A. M., Wasserman W. W. (2015). The identification of cis-regulatory elements: A review from a machine learning perspective. Biosystems. 138, 6–17. 10.1016/j.biosystems.2015.10.002 [DOI] [PubMed] [Google Scholar]
- Liu C., Peres Kury F. S., Li Z., Ta C., Wang K., Weng C. (2019). Doc2Hpo: A web application for efficient and accurate HPO concept curation. Nucleic Acids Res. 47 (W1), W566-W570–W570. 10.1093/nar/gkz386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Sun J., Zhao M. (2017). ONGene: A literature-based database for human oncogenes. J. Genet. Genomics 44 (2), 119–121. 10.1016/j.jgg.2016.12.004 [DOI] [PubMed] [Google Scholar]
- Lo Y. H., Cheng H. C., Hsiung C. N., Yang S. L., Wang H. Y., Peng C. W., et al. (2021). Detecting genetic ancestry and adaptation in the Taiwanese han people. Mol. Biol. Evol. 38 (10), 4149–4165. 10.1093/molbev/msaa276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Roman-Campos D., Austin E. D., Eyries M., Sampson K. S., Soubrier F., et al. (2013). A novel channelopathy in pulmonary arterial hypertension. N. Engl. J. Med. 369 (4), 351–361. 10.1056/NEJMoa1211097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur D. G., Balasubramanian S., Frankish A., Huang N., Morris J., Walter K., et al. (2012). A systematic survey of loss-of-function variants in human protein-coding genes. Science 335 (6070), 823–828. 10.1126/science.1215040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffucci P., Bigio B., Rapaport F., Cobat A., Borghesi A., Lopez M., et al. (2019). Blacklisting variants common in private cohorts but not in public databases optimizes human exome analysis. Proc. Natl. Acad. Sci. U. S. A. 116 (3), 950–959. 10.1073/pnas.1808403116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood K., Jung C. H., Philip G., Georgeson P., Chung J., Pope B. J., et al. (2017). Variant effect prediction tools assessed using independent, functional assay-based datasets: Implications for discovery and diagnostics. Hum. Genomics 11 (1), 10. 10.1186/s40246-017-0104-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G., Lima F., Colombo B. (1977). Haemoglobin J Guantanamo (alpha 2 beta 2 128 (H6) Ala replaced by Asp). A new fast unstable haemoglobin found in a Cuban family. Biochim. Biophys. Acta 491 (1), 1–6. 10.1016/0005-2795(77)90034-4 [DOI] [PubMed] [Google Scholar]
- McLaren W., Gil L., Hunt S. E., Riat H. S., Ritchie G. R., Thormann A., et al. (2016). The ensembl variant effect predictor. Genome Biol. 17 (1), 122. 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. (2010). Deriving the consequences of genomic variants with the ensembl API and SNP effect predictor. Bioinformatics 26 (16), 2069–2070. 10.1093/bioinformatics/btq330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendez-Aranda L., Jaloma-Cruz A. R., Pastor N., Romero-Prado M. M. J. (2019). In silico analysis of missense mutations in exons 1-5 of the F9 gene that cause hemophilia B. BMC Bioinforma. 20 (1), 363. 10.1186/s12859-019-2919-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Ebert D., Muruganujan A., Mills C., Albou L. P., Mushayamaha T., et al. (2021). PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 49 (D1), D394–D403. 10.1093/nar/gkaa1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M., Wang Y., Bromberg Y. (2019). What went wrong with variant effect predictor performance for the PCM1 challenge. Hum. Mutat. 40 (9), 1486–1494. 10.1002/humu.23832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moo-Penn W. F., Jue D. L., Johnson M. H., Olsen K. W., Shih D., Jones R. T., et al. (1988). Hemoglobin brockton [beta 138 (H16) ala----pro]: An unstable variant near the C-terminus of the beta-subunits with normal oxygen-binding properties. Biochemistry 27 (20), 7614–7619. 10.1021/bi00420a007 [DOI] [PubMed] [Google Scholar]
- Morningstar-Kywi N., Haworth I. S., Mosley S. A. (2021). Ligand-specific pharmacogenetic effects of nonsynonymous mutations. Pharmacogenet. Genomics 31 (4), 75–82. 10.1097/FPC.0000000000000424 [DOI] [PubMed] [Google Scholar]
- Muller H., Jimenez-Heredia R., Krolo A., Hirschmugl T., Dmytrus J., Boztug K., et al. (2017). VCF.Filter: Interactive prioritization of disease-linked genetic variants from sequencing data. Nucleic Acids Res. 45 (W1), W567-W572–W572. 10.1093/nar/gkx425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P. C., Henikoff S. (2001). Predicting deleterious amino acid substitutions. Genome Res. 11 (5), 863–874. 10.1101/gr.176601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P. C., Henikoff S. (2003). Sift: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31 (13), 3812–3814. 10.1093/nar/gkg509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikam R., Kulandaisamy A., Harini K., Sharma D., Gromiha M. M. (2021). ProThermDB: Thermodynamic database for proteins and mutants revisited after 15 years. Nucleic Acids Res. 49 (D1), D420–D424. 10.1093/nar/gkaa1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niroula A., Urolagin S., Vihinen M. (2015). PON-P2: Prediction method for fast and reliable identification of harmful variants. PLoS One 10 (2), e0117380. 10.1371/journal.pone.0117380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niroula A., Vihinen M. (2015). Classification of amino acid substitutions in Mismatch Repair proteins using PON-MMR2. Hum. Mutat. 36 (12), 1128–1134. 10.1002/humu.22900 [DOI] [PubMed] [Google Scholar]
- Niroula A., Vihinen M. (2019). How good are pathogenicity predictors in detecting benign variants? PLoS Comput. Biol. 15 (2), e1006481. 10.1371/journal.pcbi.1006481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S., Koren S., Rhie A., Rautiainen M., Bzikadze A. V., Mikheenko A., et al. (2022). The complete sequence of a human genome. Science 376 (6588), 44–53. 10.1126/science.abj6987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno K., Takeda J. I., Masuda A. (2018). Rules and tools to predict the splicing effects of exonic and intronic mutations. WIREs RNA 9 (1). 10.1002/wrna.1451 [DOI] [PubMed] [Google Scholar]
- Okido T., Kodama Y., Mashima J., Kosuge T., Fujisawa T., Ogasawara O. (2022). DNA Data Bank of Japan (DDBJ) update report 2021. Nucleic Acids Res. 50 (D1), D102–D105. 10.1093/nar/gkab995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgogozo V., Morizot B., Martin A. (2015). The differential view of genotype-phenotype relationships. Front. Genet. 6, 179. 10.3389/fgene.2015.00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak M. A., Markhieva K. A., Novikova M. S., Petrov D. S., Vorobyev I. S., Maksimova E. S., et al. (2021). Using AlphaFold to predict the impact of single mutations on protein stability and function. bioRxiv [Preprint]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer D. S., Howrigan D. P., Chapman S. B., Adolfsson R., Bass N., Blackwood D., et al. (2022). Exome sequencing in bipolar disorder identifies AKAP11 as a risk gene shared with schizophrenia. Nat. Genet. 54 (5), 541–547. 10.1038/s41588-022-01034-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandurangan A. P., Ochoa-Montano B., Ascher D. B., Blundell T. L. (2017). Sdm: A server for predicting effects of mutations on protein stability. Nucleic Acids Res. 45 (W1), W229-W235–W235. 10.1093/nar/gkx439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejaver V., Urresti J., Lugo-Martinez J., Pagel K. A., Lin G. N., Nam H. J., et al. (2020). Inferring the molecular and phenotypic impact of amino acid variants with MutPred2. Nat. Commun. 11 (1), 5918. 10.1038/s41467-020-19669-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Shen P., Gandotra N., Le A., Fung E., Jelliffe-Pawlowski L., et al. (2019). Combining newborn metabolic and DNA analysis for second-tier testing of methylmalonic acidemia. Genet. Med. 21 (4), 896–903. 10.1038/s41436-018-0272-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira B., Chin S. F., Rueda O. M., Vollan H. K., Provenzano E., Bardwell H. A., et al. (2016). The somatic mutation profiles of 2, 433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 7, 11479. 10.1038/ncomms11479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Enciso M., Zingaretti L. M. (2019). A guide for using deep learning for complex trait genomic prediction. Genes. (Basel) 10 (7), E553. 10.3390/genes10070553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukh M., Kucukkal T. G., Alexov E. (2015). On human disease-causing amino acid variants: Statistical study of sequence and structural patterns. Hum. Mutat. 36 (5), 524–534. 10.1002/humu.22770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires D. E., Blundell T. L., Ascher D. B. (2015). Platinum: A database of experimentally measured effects of mutations on structurally defined protein-ligand complexes. Nucleic Acids Res. 43, D387–D391. 10.1093/nar/gku966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K. S., Hubisz M. J., Rosenbloom K. R., Siepel A. (2010). Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20 (1), 110–121. 10.1101/gr.097857.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon K. S. (2021). In silico analysis of BRCA1 and BRCA2 missense variants and the relevance in molecular genetic testing. Sci. Rep. 11 (1), 11114. 10.1038/s41598-021-88586-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porta-Pardo E., Ruiz-Serra V., Valentini S., Valencia A. (2022). The structural coverage of the human proteome before and after AlphaFold. PLoS Comput. Biol. 18 (1), e1009818. 10.1371/journal.pcbi.1009818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Cox N. J. (2002). The allelic architecture of human disease genes: Common disease-common variant.or not? Hum. Mol. Genet. 11 (20), 2417–2423. 10.1093/hmg/11.20.2417 [DOI] [PubMed] [Google Scholar]
- Quang D., Chen Y., Xie X. (2015). Dann: A deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics 31 (5), 761–763. 10.1093/bioinformatics/btu703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinodoz M., Peter V. G., Cisarova K., Royer-Bertrand B., Stenson P. D., Cooper D. N., et al. (2022). Analysis of missense variants in the human genome reveals widespread gene-specific clustering and improves prediction of pathogenicity. Am. J. Hum. Genet. 109 (3), 457–470. 10.1016/j.ajhg.2022.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm H. L., Berg J. S., Brooks L. D., Bustamante C. D., Evans J. P., Landrum M. J., et al. (2015). ClinGen--the clinical genome resource. N. Engl. J. Med. 372 (23), 2235–2242. 10.1056/NEJMsr1406261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva B., Antipin Y., Sander C. (2007). Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol. 8 (11), R232. 10.1186/gb-2007-8-11-r232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva B., Antipin Y., Sander C. (2011). Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 39 (17), e118. 10.1093/nar/gkr407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., et al. (2015). Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet. Med. 17 (5), 405–424. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesselman A. J., Ingraham J. B., Marks D. S. (2018). Deep generative models of genetic variation capture the effects of mutations. Nat. Methods 15 (10), 816–822. 10.1038/s41592-018-0138-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson P. N., Kohler S., Bauer S., Seelow D., Horn D., Mundlos S. (2008). The human phenotype ontology: A tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 83 (5), 610–615. 10.1016/j.ajhg.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson P. N., Ravanmehr V., Jacobsen J. O. B., Danis D., Zhang X. A., Carmody L. C., et al. (2020). Interpretable clinical genomics with a likelihood ratio paradigm. Am. J. Hum. Genet. 107 (3), 403–417. 10.1016/j.ajhg.2020.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojano E., Seoane P., Ranea J. A. G., Perkins J. R. (2019). Regulatory variants: From detection to predicting impact. Brief. Bioinform. 20 (5), 1639–1654. 10.1093/bib/bby039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Ohno K., Sakuraba H. (2011). Fabry-database.org: Database of the clinical phenotypes, genotypes and mutant alpha-galactosidase A structures in Fabry disease. J. Hum. Genet. 56 (6), 467–468. 10.1038/jhg.2011.31 [DOI] [PubMed] [Google Scholar]
- Salgado D., Desvignes J. P., Rai G., Blanchard A., Miltgen M., Pinard A., et al. (2016). UMD-predictor: A high-throughput sequencing compliant system for pathogenicity prediction of any human cDNA substitution. Hum. Mutat. 37 (5), 439–446. 10.1002/humu.22965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallah S. R., Sergouniotis P. I., Barton S., Ramsden S., Taylor R. L., Safadi A., et al. (2020). Using an integrative machine learning approach utilising homology modelling to clinically interpret genetic variants: CACNA1F as an exemplar. Eur. J. Hum. Genet. 28 (9), 1274–1282. 10.1038/s41431-020-0623-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanavia T., Birolo G., Montanucci L., Turina P., Capriotti E., Fariselli P. (2020). Limitations and challenges in protein stability prediction upon genome variations: Towards future applications in precision medicine. Comput. Struct. Biotechnol. J. 18, 1968–1979. 10.1016/j.csbj.2020.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A., Yang Y., Vihinen M. (2020). Variation benchmark datasets: Update, criteria, quality and applications. Oxford: Database. 10.1093/database/baz117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasidharan Nair P., Vihinen M. (2013). VariBench: A benchmark database for variations. Hum. Mutat. 34 (1), 42–49. 10.1002/humu.22204 [DOI] [PubMed] [Google Scholar]
- Savojardo C., Babbi G., Bovo S., Capriotti E., Martelli P. L., Casadio R. (2019). Are machine learning based methods suited to address complex biological problems? Lessons from CAGI-5 challenges. Hum. Mutat. 40 (9), 1455–1462. 10.1002/humu.23784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E. W., Cavanaugh M., Clark K., Pruitt K. D., Schoch C. L., Sherry S. T., et al. (2022). GenBank. GenBank. Nucleic Acids Res. 50 (D1), D161–D164. 10.1093/nar/gkab1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaafsma G. C., Vihinen M. (2015). VariSNP, a benchmark database for variations from dbSNP. Hum. Mutat. 36 (2), 161–166. 10.1002/humu.22727 [DOI] [PubMed] [Google Scholar]
- Schiemann A. H., Stowell K. M. (2016). Comparison of pathogenicity prediction tools on missense variants in RYR1 and CACNA1S associated with malignant hyperthermia. Br. J. Anaesth. 117 (1), 124–128. 10.1093/bja/aew065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz J. M., Cooper D. N., Schuelke M., Seelow D. (2014). MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 11 (4), 361–362. 10.1038/nmeth.2890 [DOI] [PubMed] [Google Scholar]
- Schwarz J. M., Rodelsperger C., Schuelke M., Seelow D. (2010). MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods 7 (8), 575–576. 10.1038/nmeth0810-575 [DOI] [PubMed] [Google Scholar]
- Sherry S. T., Ward M. H., Kholodov M., Baker J., Phan L., Smigielski E. M., et al. (2001). dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29 (1), 308–311. 10.1093/nar/29.1.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab H. A., Gough J., Cooper D. N., Day I. N., Gaunt T. R. (2013a). Predicting the functional consequences of cancer-associated amino acid substitutions. Bioinformatics 29 (12), 1504–1510. 10.1093/bioinformatics/btt182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab H. A., Gough J., Cooper D. N., Stenson P. D., Barker G. L., Edwards K. J., et al. (2013b). Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 34 (1), 57–65. 10.1002/humu.22225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab H. A., Gough J., Cooper D. N., Stenson P. D., Barker G. L., Edwards K. J., et al. (2013). Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum. Mutat. 34 (1), 57–65. 10.1002/humu.22225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab H. A., Gough J., Mort M., Cooper D. N., Day I. N., Gaunt T. R. (2014). Ranking non-synonymous single nucleotide polymorphisms based on disease concepts. Hum. Genomics 8, 11. 10.1186/1479-7364-8-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J. S., Hinrichs A. S., Hou M., Rosenbloom K., et al. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15 (8), 1034–1050. 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifrim A., Popovic D., Tranchevent L. C., Ardeshirdavani A., Sakai R., Konings P., et al. (2013). eXtasy: variant prioritization by genomic data fusion. Nat. Methods 10 (11), 1083–1084. 10.1038/nmeth.2656 [DOI] [PubMed] [Google Scholar]
- Sim N. L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P. C. (2012). SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 40, W452–W457. Web Server issue). 10.1093/nar/gks539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh T., Poterba T., Curtis D., Akil H., Al Eissa M., Barchas J. D., et al. (2022). Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature 604 (7906), 509–516. 10.1038/s41586-022-04556-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley D., Jacobsen J. O., Jager M., Kohler S., Holtgrewe M., Schubach M., et al. (2015). Next-generation diagnostics and disease-gene discovery with the Exomiser. Nat. Protoc. 10 (12), 2004–2015. 10.1038/nprot.2015.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smigielski E. M., Sirotkin K., Ward M., Sherry S. T. (2000). dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 28 (1), 352–355. 10.1093/nar/28.1.352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Goldsmith C. A., Eppig J. T. (2005). The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 6 (1), R7. 10.1186/gb-2004-6-1-r7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somody J. C., MacKinnon S. S., Windemuth A. (2017). Structural coverage of the proteome for pharmaceutical applications. Drug Discov. Today 22 (12), 1792–1799. 10.1016/j.drudis.2017.08.004 [DOI] [PubMed] [Google Scholar]
- Steinhaus R., Proft S., Schuelke M., Cooper D. N., Schwarz J. M., Seelow D. (2021). MutationTaster2021. Nucleic Acids Res. 49 (W1), W446–W451. 10.1093/nar/gkab266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E. A., Sidow A. (2005). Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 15 (7), 978–986. 10.1101/gr.3804205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant P. H., Rausch T., Gardner E. J., Handsaker R. E., Abyzov A., Huddleston J., et al. (2015). An integrated map of structural variation in 2, 504 human genomes. Nature 526 (7571), 75–81. 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunyaev S., Ramensky V., Bork P. (2000). Towards a structural basis of human non-synonymous single nucleotide polymorphisms. Trends Genet. 16 (5), 198–200. 10.1016/s0168-9525(00)01988-0 [DOI] [PubMed] [Google Scholar]
- Suybeng V., Koeppel F., Harle A., Rouleau E. (2020). Comparison of pathogenicity prediction tools on somatic variants. J. Mol. Diagn. 22 (12), 1383–1392. 10.1016/j.jmoldx.2020.08.007 [DOI] [PubMed] [Google Scholar]
- Suzek B. E., Huang H., McGarvey P., Mazumder R., Wu C. H. (2007). UniRef: Comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23 (10), 1282–1288. 10.1093/bioinformatics/btm098 [DOI] [PubMed] [Google Scholar]
- Tam V., Patel N., Turcotte M., Bosse Y., Pare G., Meyre D. (2019). Benefits and limitations of genome-wide association studies. Nat. Rev. Genet. 20 (8), 467–484. 10.1038/s41576-019-0127-1 [DOI] [PubMed] [Google Scholar]
- Tate J. G., Bamford S., Jubb H. C., Sondka Z., Beare D. M., Bindal N., et al. (2019). Cosmic: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 47 (D1), D941-D947–D947. 10.1093/nar/gky1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terui H., Akagi K., Kawame H., Yura K. (2013). CoDP: Predicting the impact of unclassified genetic variants in MSH6 by the combination of different properties of the protein. J. Biomed. Sci. 20, 25. 10.1186/1423-0127-20-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. D., Campbell M. J., Kejariwal A., Mi H., Karlak B., Daverman R., et al. (2003). Panther: A library of protein families and subfamilies indexed by function. Genome Res. 13 (9), 2129–2141. 10.1101/gr.772403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. D., Kejariwal A. (2004). Coding single-nucleotide polymorphisms associated with complex vs. Mendelian disease: Evolutionary evidence for differences in molecular effects. Proc. Natl. Acad. Sci. U. S. A. 101 (43), 15398–15403. 10.1073/pnas.0404380101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. D., Kejariwal A., Guo N., Mi H., Campbell M. J., Muruganujan A., et al. (2006). Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 34, W645–W650. Web Server issue). 10.1093/nar/gkl229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorisson G. A., Muilu J., Brookes A. J. (2009). Genotype-phenotype databases: Challenges and solutions for the post-genomic era. Nat. Rev. Genet. 10 (1), 9–18. 10.1038/nrg2483 [DOI] [PubMed] [Google Scholar]
- Tian Y., Pesaran T., Chamberlin A., Fenwick R. B., Li S., Gau C. L., et al. (2019). REVEL and BayesDel outperform other in silico meta-predictors for clinical variant classification. Sci. Rep. 9 (1), 12752. 10.1038/s41598-019-49224-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischkowitz M., Hamel M. A, Carvalho M., Birrane G., Soni A., van Beers E. H. (2008). Pathogenicity of the BRCA1 missense variant M1775K is determined by the disruption of the BRCT phosphopeptide-binding pocket: a multi-modal approach. Eur J Hum Genet 16 (7), 820–832. 10.1038/ejhg.2008.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trisilowati, , Mallet D. G. (2012). In silico experimental modeling of cancer treatment. ISRN Oncol. 2012, 828701. 10.5402/2012/828701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Zidek A., et al. (2021). Highly accurate protein structure prediction for the human proteome. Nature 596 (7873), 590–596. 10.1038/s41586-021-03828-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt C. (2021). UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 49 (D1), D480–D489. 10.1093/nar/gkaa1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R., Adusumalli S., Leng S. N., Sikic M., Ng P. C. (2016). SIFT missense predictions for genomes. Nat. Protoc. 11 (1), 1–9. 10.1038/nprot.2015.123 [DOI] [PubMed] [Google Scholar]
- Vihinen M. (2021). Functional effects of protein variants. Biochimie 180, 104–120. 10.1016/j.biochi.2020.10.009 [DOI] [PubMed] [Google Scholar]
- Vihinen M. (2013). Guidelines for reporting and using prediction tools for genetic variation analysis. Hum. Mutat. 34 (2), 275–282. 10.1002/humu.22253 [DOI] [PubMed] [Google Scholar]
- Vihinen M. (2012). How to evaluate performance of prediction methods? Measures and their interpretation in variation effect analysis. BMC Genomics 13 (4), S2. 10.1186/1471-2164-13-S4-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihinen M. (2014). Variation Ontology for annotation of variation effects and mechanisms. Genome Res. 24 (2), 356–364. 10.1101/gr.157495.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M., Hakonarson H. (2010). Annovar: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38 (16), e164. 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber L. M., Saelens W., Cannoodt R., Soneson C., Hapfelmeier A., Gardner P. P., et al. (2019). Essential guidelines for computational method benchmarking. Genome Biol. 20 (1), 125. 10.1186/s13059-019-1738-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R., Wang J., Su M., Jia E., Chen S., Chen T., et al. (2018). Missing value imputation approach for mass spectrometry-based metabolomics data. Sci. Rep. 8 (1), 663. 10.1038/s41598-017-19120-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witvliet D. K., Strokach A., Giraldo-Forero A. F., Teyra J., Colak R., Kim P. M. (2016). ELASPIC web-server: Proteome-wide structure-based prediction of mutation effects on protein stability and binding affinity. Bioinformatics 32 (10), 1589–1591. 10.1093/bioinformatics/btw031 [DOI] [PubMed] [Google Scholar]
- Won D. G., Kim D. W., Woo J., Lee K. (2021). 3Cnet: Pathogenicity prediction of human variants using multitask learning with evolutionary constraints. Bioinformatics 37, 4626–4634. 10.1093/bioinformatics/btab529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth C. L., Preissner R., Blundell T. L. (2011). SDM--a server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 39, W215–W222. Web Server issue). 10.1093/nar/gkr363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier A., Scott R. J., Talseth-Palmer B. A. (2019). TAPES: A tool for assessment and prioritisation in exome studies. PLoS Comput. Biol. 15 (10), e1007453. 10.1371/journal.pcbi.1007453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazar M., Ozbek P. (2021). In silico tools and approaches for the prediction of functional and structural effects of single-nucleotide polymorphisms on proteins: An expert review. OMICS 25 (1), 23–37. 10.1089/omi.2020.0141 [DOI] [PubMed] [Google Scholar]
- Yuan X., Wang J., Dai B., Sun Y., Zhang K., Chen F., et al. (2022). Evaluation of phenotype-driven gene prioritization methods for Mendelian diseases. Brief. Bioinform. 23 (2), bbac019. 10.1093/bib/bbac019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P., Li Z., Moult J. (2005). Loss of protein structure stability as a major causative factor in monogenic disease. J. Mol. Biol. 353 (2), 459–473. 10.1016/j.jmb.2005.08.020 [DOI] [PubMed] [Google Scholar]
- Yue P., Melamud E., Moult J. (2006). SNPs3D: Candidate gene and SNP selection for association studies. BMC Bioinforma. 7, 166. 10.1186/1471-2105-7-166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue W. W., Froese D. S., Brennan P. E. (2014). The role of protein structural analysis in the next generation sequencing era. Top. Curr. Chem. 336, 67–98. 10.1007/128_2012_326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Zhang H., Yang H., Li M., Xie Z., Li W. (2019). Computational resources associating diseases with genotypes, phenotypes and exposures. Brief. Bioinform. 20 (6), 2098–2115. 10.1093/bib/bby071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Walsh R., Whiffin N., Buchan R., Midwinter W., Wilk A., et al. (2021). Disease-specific variant pathogenicity prediction significantly improves variant interpretation in inherited cardiac conditions. Genet. Med. 23 (1), 69–79. 10.1038/s41436-020-00972-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Havrilla J. M., Fang L., Chen Y., Peng J., Liu C., et al. (2020). Phen2Gene: Rapid phenotype-driven gene prioritization for rare diseases. Nar. Genom. Bioinform. 2, lqaa032. 10.1093/nargab/lqaa032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., Kim P., Mitra R., Zhao J., Zhao Z. (2016). TSGene 2.0: An updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 44 (D1), D1023–D1031. 10.1093/nar/gkv1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Amos C. I., Frost H. R. (2020). Cancer prognosis prediction using somatic point mutation and copy number variation data: A comparison of gene-level and pathway-based models. BMC Bioinforma. 21 (1), 467. 10.1186/s12859-020-03791-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Gao M., Skolnick J. (2016). Entprise: An algorithm for predicting human disease-associated amino acid substitutions from sequence entropy and predicted protein structures. PLoS One 11 (3), e0150965. 10.1371/journal.pone.0150965 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.