Abstract
Purpose
To investigate atrophy expansion rate (ER) using ultra-widefield (UWF) fundus autofluorescence (FAF) in Stargardt disease (STGD1).
Design
Retrospective, longitudinal study.
Participants
Patients with biallelic ABCA4 mutations who were evaluated with UWF FAF and Heidelberg 30° × 30° and 55° × 55° FAF imaging.
Methods
Patients with atrophy secondary to STGD1 were classified into genotype groups: group A, biallelic severe or null-like variants with early-onset disease; group B, 1 intermediate variant in trans with severe or null-like variant; and group C, 1 mild variant in trans with severe or null-like variant or late-onset disease. The boundaries of definitely decreased autofluorescence (DDAF) were outlined manually and areas (in square millimeters) were recorded at baseline and follow-up. Bland-Altman analysis was conducted to examine agreement between observers and devices. Linear mixed modeling was used to evaluate predictors of ER in DDAF area and square root area (SRA).
Main Outcome Measures
Patient and ocular predictors of DDAF area ER and DDAF SRA ER included age at onset, duration of symptoms, genotype group, baseline visual acuity, and baseline atrophy size.
Results
A total of 138 eyes from 69 patients (33 men [47%]; mean age ± standard deviation, 41 ± 20 years; range, 10–83 years) carrying 61 unique ABCA4 variants were recruited. Ultra-widefield FAF measurements were equivalent to Heidelberg 30° × 30° imaging. Baseline DDAF area was the only significant predictor of DDAF area ER (P < 0.001). Age at baseline and genotype group were predictors for DDAF SRA ER. Definitely decreased autofluorescence area ER ranged from 4.65 mm2/year (group A) to 0.62 mm2/year (group C).
Conclusions
Ultra-widefield FAF is a feasible and reliable method for assessing atrophy ER in STGD1. The value of ABCA4 mutation severity in predicting atrophy ER warrants further investigation.
Keywords: ABCA4-associated retinopathy, Clinical trial end point, Inherited retinal disease, Macular dystrophy, Retinal dystrophy, Retinal imaging
Abbreviations and Acronyms: DDAF, definitely decreased autofluorescence; ER, expansion rate; ETDRS, Early Treatment Diabetic Retinopathy Study; FAF, fundus autofluorescence; NIR AF, near-infrared autofluorescence; SD, standard deviation; SRA, square root area; STGD1, Stargardt disease; SW AF, short-wavelength autofluorescence; UWF, ultra-widefield; VA, visual acuity
Stargardt disease (STGD1; Online Mendelian Inheritance in Man identifier, 248200), caused by biallelic mutations in the ATP-binding cassette transporter subfamily A4 (ABCA4) gene, is the most common of all inherited retinal diseases.1, 2, 3, 4 The formation and expansion of retinal pigment epithelium atrophy in the macular region is a hallmark of STGD1 progression.5 Fundus autofluorescence (FAF) imaging allows reliable quantification of the expansion rate (ER) in retinal pigment epithelium atrophy as defined by the area of definitely decreased autofluorescence (DDAF).6 Consequently, DDAF area has been proposed as a trial end point in the treatment of STG1.7 To date, studies have limited their evaluation of DDAF area to the central 30° × 30° field of view, although expansion beyond this region frequently occurs in STGD1.8,9 Notwithstanding the emergence of ABCA4 mutation-specific therapies, a paucity of data is available on genotype-specific ER in DDAF area.10, 11, 12
Single-center case series and multicenter studies have used the 30° × 30° lens on the Heidelberg scanning laser ophthalmoscope device to capture and measure DDAF area.7,10, 11, 12, 13 Some research groups have enabled the use of the widefield 55° × 55° lens, on the assumption that the DDAF area is equivalent to the 30° × 30° view.7,13,14 However, this assumption has not been examined. Most studies reported the baseline DDAF area as the most significant factor in predicting ER, although the actual mean baseline DDAF area was relatively small (0.74–3.93 mm2).7,10,11,13 To eliminate the dependence of ER on the baseline DDAF area, 2 studies examined the square root area (SRA) ER.12,14 Lindner et al14 reported an ER of 0.23 mm/year in SRA by using both 30° × 30° and 55° × 55° lenses, whereas Muller et al12 found an ER of 0.20 mm/year in a study using only the 30° × 30° lens. Both of these studies reported small baseline lesion sizes of 6.2 mm2 and 1.58 mm2, respectively.12,14 More recently, Chen et al8 used the ultra-widefield (UWF) Optos device to examine lesion extent in STGD1. They found only 26% of patients with STGD1 harbored lesions confined to the 30° × 30° field. Hence, an unmet clinical need exists to examine the ER of DDAF that extends beyond the posterior pole and the dependence of ER on baseline lesion sizes that are much larger. Several studies have investigated the relationship between genotype and DDAF area ER.10, 11, 12 However, their classification system was limited to pathogenicity and single variants, rather than the combined severity of biallelic ABCA4 variants. This study aimed to evaluate the agreement in DDAF area and SRA between the Heidelberg 30° × 30° and 55° × 55° FAF images and the Optos UWF FAF images and investigate the impact of various genotypes classified by variant severity on ER of DDAF area and SRA in a large, genetically defined STGD1 cohort.
Methods
Study Design and Population
This was a longitudinal analysis of data that were collected retrospectively and prospectively at the Lions Eye Institute, Perth, Australia, from June 2011 through December 2020. The study protocol adhered to the tenets of the Declaration of Helsinki, and ethics approval was obtained from the Human Ethics Office of Research Enterprise, the University of Western Australia (identifier, RA/4/1/7916), and Sir Charles Gairdner Hospital Human Research Ethics Committee (identifier, 2001-053). Informed consent was obtained from all study participants.
All patients with both a clinical and genetic molecular diagnosis of STGD1 who had undergone UWF FAF imaging were eligible for inclusion. The clinical diagnosis was established by the senior author (F.K.C.) based on the presence of central vision loss resulting from macular atrophy with or without surrounding flecks on fundus examination. Patient DNA was collected through the Australian Inherited Retinal Disease Registry and DNA Bank,15 and genetic molecular diagnosis was confirmed by Casey Eye Institute or Molecular Vision Laboratory through the detection of biphasic, suspected disease-causing variants in ABCA4. Patients with variants shown to be in trans configuration were included, and those with only 1 variant in the ABCA4 gene or with 2 or more variants without evidence of biphasic status were excluded.
Patients were enrolled into 1 of the 3 genotype groups according to the combined severities of the 2 ABCA4 alleles. Group A showed biallelic null or severe variants or extensive panretinal degeneration with onset of symptoms before 14 years of age. Group B showed a known intermediate variant in trans with a null or severe variant, that is, in a hemizygous-like state. Group C showed a known mild or hypomorphic variant in a hemizygous-like state or demonstrated clinical features of localized foveal lesion or foveal-sparing macular lesion with late-onset disease. Variants were considered null-like if they were a stop mutation or a frame-shift mutation resulting in a premature stop codon predicted to undergo nonsense-mediated decay. Missense and splice-site mutations were assigned null-like or severe status based on published clinical data or in vitro assays.16, 17, 18 Patients were recruited consecutively as they were referred to our center for assessment of suspected STGD1. Clinical data including age, gender, symptom onset, and best-corrected visual acuity as measured on the Early Treatment Diabetic Retinopathy Study (ETDRS) chart were recorded.
Procedures and Outcomes
Ultra-widefield FAF images were obtained using the Optomap or the California device (Optos PLC), which captures retinal features spanning 200° of the internal eye angle from the center of the globe (approximately 135° field angle, covering 892 mm2) using a green excitation laser at 532 nm.19 The total area of the retina has been estimated to be 1081.57 mm2 based on the calculation by Atkinson and Mazo.19 Hence, the maximum retinal area imaged by the Optos device was approximately 82.5% of the total retina. Short-wave (excitation λ, 488 nm; barrier filter transmitting λ, 500–680 nm) fundus autofluorescence (HRA2; Heidelberg Engineering) with 30° × 30° and 55° × 55° lenses also were acquired with special care to ensure that all atrophic lesions were captured if possible. All images were graded by 2 expert image graders (R.C.H.J. and F.K.C.) using the OptosAdvance and Heidelberg Explorer software. The outermost boundary of DDAF was outlined manually and the area in planimetric square millimeters was recorded (Fig 1). Areas of DDAF that extended outside the posterior pole were included only if they were contiguous with or within 3 disc diameters of the main central DDAF lesion. Areas of reduced autofluorescence resulting from masking by pigment plaques were not included in the DDAF marking. Subsequently, small DDAF lesions discontinuous from the primary lesion and pigment plaque masking of autofluorescence in the extreme periphery were excluded.
Figure 1.
Images showing the outermost boundary of definitely decreased autofluorescence outlined manually and the area in planimetric square millimeters (mm2) recorded for (A1, B1, C1, D1) Heidelberg 30° × 30° fundus autofluorescence (FAF), (A2, B2, C2, D2) Heidelberg 55° × 55° FAF, and (A3, B3, C3, D3) ultra-widefield FAF imaging. Patient shown in C (ID:22) had lesions that spread outside the 30° × 30° field of view. Patient shown in (D) (patient 12) demonstrated widespread lesions that were outside the 30° × 30° and 55° × 55° field of view.
Genetic Analysis
Genomic DNA was analyzed using various disease-specific next-generation sequencing SmartPanels, which evolved throughout the study (Table S1, available at www.ophthalmologyscience.org).20 Identified candidate ABCA4 mutations were confirmed by Sanger sequencing (genetic testing performed by Casey Eye Institute Molecular Diagnostics Laboratory, Portland, OR). Phase segregation was performed for all families. Variant nomenclature was described in relationship to ABCA4 coding DNA reference sequence NM_000350.2 and was reported in accordance with the recommendations of the Human Genome Variation Society.21 Pathogenicity was assessed as described previously22 and was interpreted according to the joint guidelines of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology23 and associated literature.24
Statistical Analysis
Visual acuity of counting fingers, hand movements, and light perception were assigned –15, –30, and –45 letter scores, respectively, based on the mean relative logarithm of minimum angle of resolution values (2.0, 2.3, and 2.6, respectively) assigned to these off-chart measurements using the Freiburgh test.25,26 Categorical variables were summarized by frequencies and proportions. Continuous variables were described by mean ± standard deviation (SD). Baseline features were compared across genotype groups using a 1-way analysis of variance test.
Bland-Altman analysis was performed to compare the DDAF area and SRA measurements by the 2 graders (F.K.C. and R.C.H.J.). Mean and 95% limits of agreement were calculated. The DDAF area and SRA derived from the UWF FAF image (Optos200) and the 30° × 30° and 55° × 55° Heidelberg systems were compared using the Bland-Altman analysis. Only pairs of FAF images in which all of the atrophic lesions were captured by both imaging methods were used for Bland-Altman analysis to prevent bias generated by truncation of an atrophic lesion related to the small field of view. The SRA was determined to eliminate the dependence of ER on the baseline DDAF area. Interocular symmetry in DDAF area and SRA were assessed using the Bland-Altman analysis. Paired sample Student t tests were performed to compare the means to determine the significance of bias.
The ER of DDAF area was calculated using the formula:
Square root transformation of DDAF area was performed before calculation of ER SRA to adjust for baseline area using the formula:
Linear mixed modeling was used to assess patient and ocular predictors of DDAF area ER and DDAF SRA ER. Right and left eye measurements were treated as repeated measures. Patient factors recorded included gender, age at symptom onset (years), age at baseline DDAF assessment (years), and genotype group (A, B, or C). Ocular measurements included visual acuity (ETDRS letters), DDAF area (square millimeters), and DDAF SRA (millimeters) recorded at baseline. Preliminary analyses were conducted to identify and remove outliers to ensure no violation of the assumptions of normality. Linearity, multicollinearity, and homoscedasticity were examined. Data were analyzed using IBM SPSS Statistics for Windows software version 26 (IBM Corp). P values for post hoc testing were adjusted for false discovery rate (FDR). Statistical tests were considered significant at P < 0.05.
Results
Baseline Demographics
A total of 69 patients (33 men [47%]; mean ± SD age, 41 ± 20 years; range, 10–83 years) carrying 61 unique ABCA4 variants were recruited from 52 families (Table S2, available at www.ophthalmologyscience.org). The pathogenicity assessment and severity for each ABCA4 variant are shown in Table S3 (available at www.ophthalmologyscience.org). Overall, the mean ± SD age at symptom onset and duration of disease were 24.5 ± 22.6 years and 16.3 ± 14.0 years (including 6 patients who were asymptomatic at baseline), respectively. The mean ± SD best-corrected visual acuities were 38 ± 29 ETDRS letters (Snellen equivalent, 20/174) and 37 ± 31 ETDRS letters (Snellen equivalent, 20/182) in the right and left eyes, respectively. A significant difference was found in baseline features across the 3 genotype groups for all variables, including age at onset, age at baseline examination, duration of disease, and baseline visual acuity (Table 1).
Table 1.
Baseline Demographics for Each Genotype Group
| Group A (n = 24) | Group B (n = 21) | Group C (n = 24) | P Value∗ | |
|---|---|---|---|---|
| Age at symptom onset (yrs) | 8.9 ± 1.9 | 20.3 ± 14.5 | 44.8 ± 25.6 | < 0.001 |
| Age at baseline assessment (yrs) | 28.5 ± 16.1 | 40.6 ± 13.8 | 53.2 ± 20.7 | < 0.001 |
| Disease duration (yrs) | 19.6 ± 16.0 | 20.5 ± 13.9 | 9.2 ± 8.9 | 0.008 |
| Baseline VA right eye (ETDRS letters) | 18 ± 23 | 37 ± 28 | 59 ± 20 | < 0.001 |
| Baseline DDAF area in right eye (mm2) | 73 ±152 | 18 ± 25 | 2.9 ± 5.1 | 0.024 |
| Baseline DDAF square-root-area in right eye (mm) | 5.6 ± 6.6 | 3.5 ± 2.6 | 1.2 ± 1.2 | 0.003 |
| Patients with follow-up data | 16 (67) | 20 (95) | 15 (63) | N/A |
DDAF = definitely decreased autofluorescence; ETDRS = Early Treatment Diabetic Retinopathy Study; N/A = not applicable; VA = visual acuity.
Data are presented as mean±standard deviation or no. (%).
One-way analysis of variance.
Eight of the 69 patients (12%) showed no DDAF in either eye over a mean ± SD follow-up of 1.9 ± 1.8 years. Of the remaining 61 patients with DDAF at baseline, 2 eyes from 2 patients had ungradable images because of extension of the DDAF beyond the field of view on UWF FAF, leaving 120 eyes from 61 patients (59 with bilateral disease and 2 with unilateral disease) with gradable images for measuring baseline DDAF areas. Of these, 19 eyes of 10 patients did not have follow-up data, leaving 101 eyes of 51 patients (50 with bilateral disease and 1 with unilateral disease) with paired baseline and follow-up DDAF areas for calculating ER (Fig 2). The mean ± SD DDAF area and SRA at baseline (n = 59 pairs of eyes) were 23 ± 57 mm2 and 3.3 ± 3.5 mm in the right eye and 25 ± 69 mm2 and 3.5 ± 3.7 mm in the left eye, respectively.
Figure 2.
Flow chart showing those patients who were excluded, leaving 101 eyes of 51 patients (50 with bilateral disease and 1 with unilateral disease) with paired baseline and follow-up definitely decreased autofluorescence (DDAF) areas. FAF = fundus autofluorescence; STGD1 = Stargardt disease; UWF = ultra-widefield.
Interobserver, Interdevice, and Interocular Agreements
A total of 221 baseline and follow-up Optos UWF FAF images were marked by the 2 graders. No significant difference was found in baseline DDAF area and DDAF SRA between the 2 image graders (Table 2; Figure S1 (available at www.ophthalmologyscience.org). Given the agreement between graders, the averaged DDAF areas were used for comparison with Heidelberg DDAF and for calculation of ER in all subsequent analyses.
Table 2.
Interobserver, Interdevice, and Interocular Agreements in Definitely Decreased Autofluorescence Area and Square Root Area
| Comparisons | Sample Size | Mean Difference | Limits of Agreement∗ | P Value† |
|---|---|---|---|---|
| DDAF area (mm2) | ||||
| Interobserver | ||||
| F.K.C. vs. R.C.H.J.‡ baseline RE | 61 | –0.12 | –3.13 to +2.87 | 0.528 |
| F.K.C. vs. R.C.H.J.‡ baseline LE | 59 | –0.46 | –4.68 to +3.73 | 0.104 |
| Interdevice | ||||
| O200 vs. H30 RE only | 45 | –0.19 | –2.38 to +1.99 | 0.266 |
| H55 vs. O200 RE only | 51 | +2.79 | –6.84 to +12.37 | < 0.001 |
| H55 vs. H30 RE only | 44 | +1.20 | –2.26 to +4.65 | < 0.001 |
| Interocular | ||||
| RE vs. LE baseline | 59 | –2.24 | –30.41 to +25.79 | 0.236 |
| RE vs. LE final | 50 | –2.03 | –22.75 to +18.58 | 0.180 |
| DDAF SRA (mm) | ||||
| Interobserver | ||||
| F.K.C. vs. R.C.H.J.‡ baseline RE | 61 | –0.02 | –0.20 to +0.17 | 0.168 |
| F.K.C. vs. R.C.H.J.‡ baseline LE | 59 | –0.02 | –0.31 to +0.26 | 0.197 |
| Interdevice | ||||
| O200 vs. H30 RE only | 45 | –0.03 | –0.37 to +0.30 | 0.221 |
| H55 vs. O200 RE only | 51 | +0.27 | –0.41 to +0.91 | < 0.001 |
| H55 vs. H30 RE only | 44 | +0.16 | –0.17 to +0.49 | < 0.001 |
| Interocular | ||||
| RE vs. LE baseline | 59 | –0.11 | –1.82 to +1.60 | 0.355 |
| RE vs. LE final | 50 | –0.13 | –2.03 to +1.75 | 0.330 |
DDAF = definitely decreased autofluorescence; LE = left eye; H30 = Heidelberg 30° × 30° lens; H55 = Heidelberg 55° × 55° lens; O200 = Optos200 device; RE = right eye; SRA = square root area.
Bland-Altman 95% limits of agreement defined by mean ± 1.95 × standard deviation.
Paired sample t test.
Author initials.
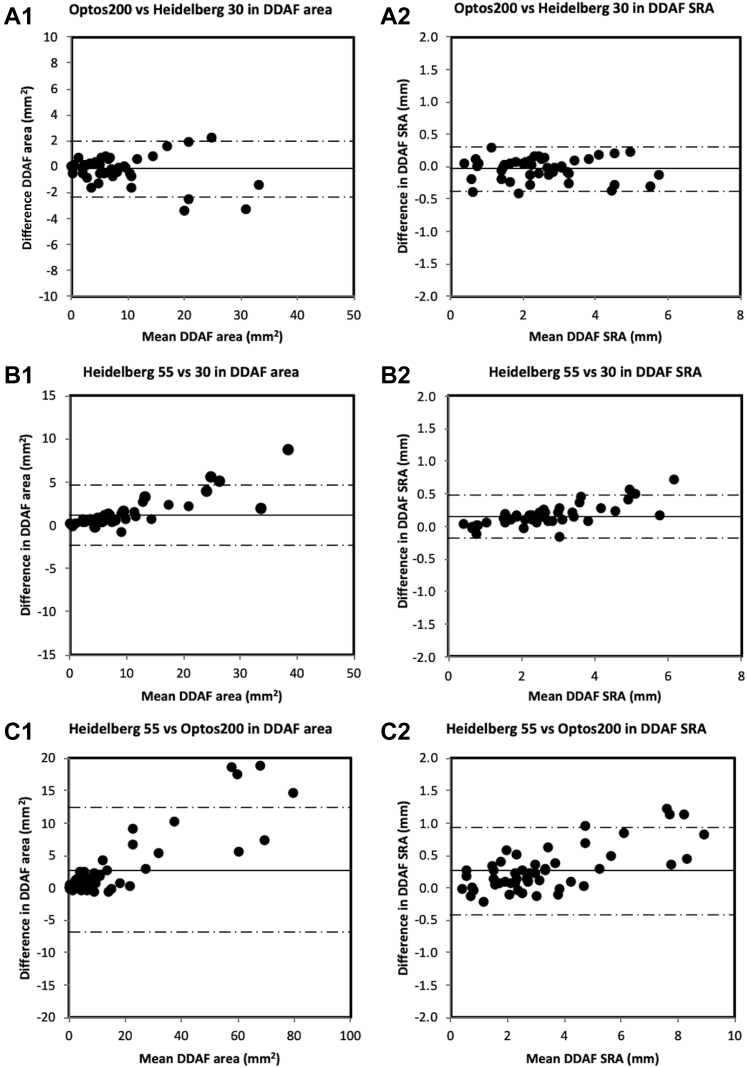
For interdevice agreement validation, the graders marked the DDAF boundaries of 51 55° × 55° and 45 30° × 30° FAF images from the right eyes of patients with DDAF visualized entirely within the respective fields of view. The interdevice difference in DDAF area between UWF FAF and Heidelberg 30° × 30° FAF images increased with larger lesions (Table 2; Fig 3). However, this relationship was not evident after square root transformation. Heidelberg 55° × 55° FAF imaging tended to overestimate DDAF area when compared with UWF FAF and Heidelberg 30° × 30° imaging systems, respectively (Table 2).
Figure 3.
Bland-Altman plots demonstrating interdevice and intermethod agreement between the Optos200 and Heidelberg systems and between 30° × 30° and 55° × 55° imaging methods on the Heidelberg system (Optos200 vs. Heidelberg 30° × 30°, Heidelberg 55° × 55° vs. Heidelberg 30° × 30°, and Heidelberg 55° × 55° vs. Optos200) for (A1, B1, C1) definitely decreased autofluorescence (DDAF) area as well as (A2, B2, C2) DDAF square root area (SRA). The solid line indicates the mean difference and the dashed lines indicate the 95% limits of agreement. Overall, Optos200 and Heidelberg 30° × 30° DDAF SRA showed similar results.
The 2 eyes had similar DDAF area and SRA, but the limits of agreement were wider than interobserver or interdevice comparisons (Table 2; Fig S2, available at www.ophthalmologyscience.org).
Predictors of Definitely Decreased Autofluorescence Expansion Rates
Preliminary analysis identified the patient with a DDAF area ER of 32.29 mm2/year as an outlier, and the individual subsequently was removed from the analysis. Data for 2 other patients also were removed because the baseline DDAF areas were more than 390 mm2, whereas all other patients included in the linear mixed modeling showed DDAF areas of less than 220 mm2. Definitely decreased autofluorescence area was the only significant predictor of DDAF area ER (F = 20.516; P < 0.001; Table 3). In contrast, increasing patient baseline age (F = 7.477; P = 0.008) was associated with greater DDAF SRA ER, whereas increasing baseline DDAF SRA was associated with reduced DDAF SRA ER (F = 5.640; P = 0.020). Furthermore, significant differences in DDAF SRA ER were observed among the genotypes (F = 3.298; P = 0.046) whereby the mean ER for genotype C was significantly lower than both genotype group A (P = 0.041, FDR) and group B (P = 0.041, FDR). No significant difference was found in DDAF SRA ER between genotype groups A and B (P = 0.547, FDR; Table 3).
Table 3.
Linear Mixed Modeling for Predictors of Expansion Rates in Definitely Decreased Autofluorescence Area and Square Root Area
| Predictor | Category | Data∗ | Definitely Decreased Autofluorescence Area Expansion Rate (mm2/yr) |
Definitely Decreased Autofluorescence Square Root Area Expansion Rate (mm/yr) |
||||
|---|---|---|---|---|---|---|---|---|
| Model Estimate ± SE | F(v1, v2)† | P Value | Model Estimate ± SE | F(v1, v2)† | P Value | |||
| Gender | Female | 24 (49) | –0.188 ± 0.341 | F(1, 41.5) = 0.304 | 0.584 | –0.042 ± 0.038 | F(1, 43.1) = 1.225 | 0.274 |
| Male | 25 (51) | 1.00 (reference)‡ | 1.00 (reference)‡ | |||||
| Eye | Left | 49 (50) | 0.046 ± 0.205 | F(1, 47.6) = 0.050 | 0.824 | 0.001 ± 0.016 | F(1, 47.6) = 0.006 | 0.939 |
| Right | 49 (50) | 1.00 (reference)‡ | 1.00 (reference)‡ | |||||
| Genotype group | A | 14 (28.6) | 0.25 ± 0.67 | F(2, 42.7) = 1.549 | 0.224 | 0.169 ± 0.074 | F(2, 46.2) = 3.298 | 0.046 |
| B | 20 (40.8) | 0.757 ± 0.502 | 0.137 ± 0.056 | |||||
| C | 15 (30.6) | 1.00 (reference)‡ | 1.00 (reference)‡ | |||||
| Age at baseline assessment (yrs) | 41.3 ± 20.2 | 0.004 ± 0.023 | F(1, 47.4) = 0.025 | 0.876 | 0.007 ± 0.003 | F(1, 67.8) = 7.477 | 0.008 | |
| Age at symptom onset (yrs) | 26 ± 23.2 | 0.008 ± 0.021 | F(1, 49.6) = 0.129 | 0.721 | –0.004 ± 0.002 | F(1, 63.6) = 2.466 | 0.121 | |
| Baseline visual acuity | 39.9 ± 29.9 | –0.012 ± 0.009 | F(1, 77) = 1.807 | 0.183 | –0.0005 ± 0.001 | F(1, 88.3) = 0.293 | 0.590 | |
| Baseline DDAF area/SRA§ | 16.4 ± 28.3 | 0.044 ± 0.01 | F(1, 52.4) = 20.516 | < 0.001 | –0.026 ± 0.011 | F(1, 87.1) = 5.640 | 0.020 | |
DDAF = definitely decreased autofluorescence; SE = standard error; SRA = square root area.
Presented as no. (%) or mean ± standard deviation for continuous variables.
Observed F statistic with v1 and v2 degrees of freedom.
The reference level in the categorical predictor is defined as 1.00 (reference).
Definitely decreased autofluorescence area was used as the predictor for area expansion rate, whereas DDAF SRA was the predictor for SRA expansion rate.
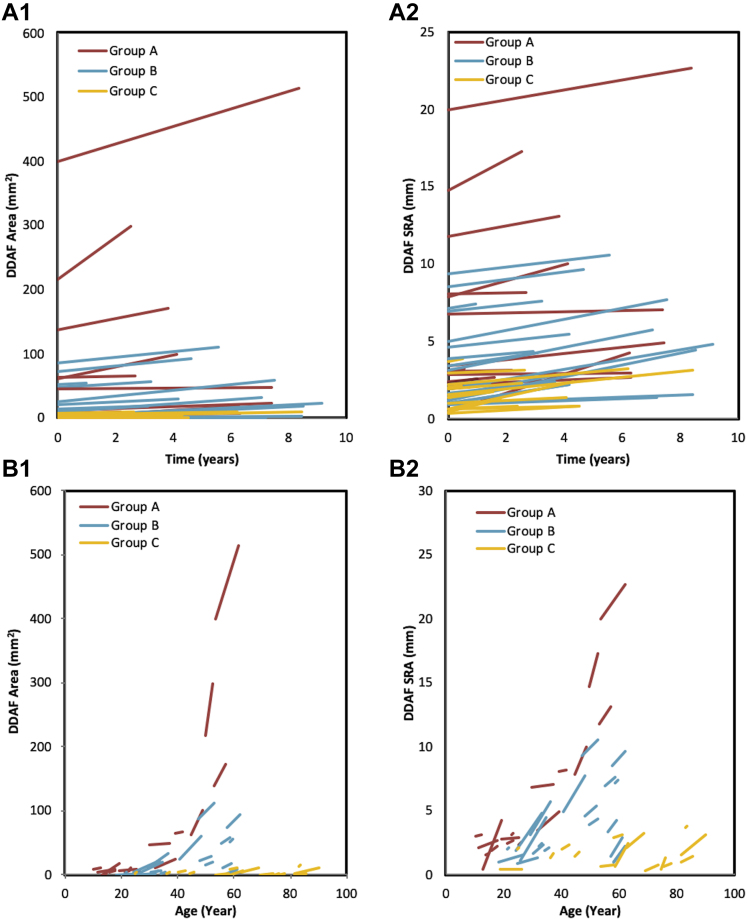
The mean ER in DDAF area ranged from 0.62 mm2/year in the mildest genotype group (group C) to 4.65 mm2/year in the most severe genotype group (group A). Similarly, the ER in DDAF SRA ranged from 0.18 mm/year in group C to 0.25 mm/year in group A (Table 4). Figure S3 (available at www.ophthalmologyscience.org) illustrates the UWF DDAF baseline and follow-up calculations for each genotype group. A striking contrast exists between the trajectories of DDAF area or SRA increase for each patient among the 3 genotype groups, as illustrated by Figure 4.
Table 4.
Expansion Rate in Definitely Decreased Autofluorescence Area and Square Root Area for Each Genotype Group
| Group A (n = 16) | Group B (n = 20) | Group C (n = 15) | |
|---|---|---|---|
| Annual DDAF area expansion rate (mm2)∗ | 4.65 ± 8.43 (0.06–32.3) | 2.00 ± 1.41 (0.14–4.58) | 0.62 ± 0.52 (0.00–1.90) |
| Annual DDAF SRA expansion rate (mm)∗ | 0.25 ± 0.27 (0.01–1.01) | 0.25 ± 0.12 (0.06–0.57) | 0.18 ± 0.14 (0.00–0.47) |
DDAF = definitely decreased autofluorescence; SRA = square root area.
Data are presented as mean ± standard deviation (range).
Right eye only.
Figure 4.
Graphs showing (A1, B1) definitely decreased autofluorescence (DDAF) area or (A2, B2) square root area (SRA) expansion for each patient according to the 3 genotype groups. Group A (red) showed biallelic severe or null-like variants with early-onset disease, group B (blue) showed an intermediate variant in trans with a severe or null-like variant, and group C (orange) carried a mild variant in trans with a severe null-like variant or late-onset disease.
Discussion
This study raises significant questions regarding the validity of using Heidelberg 30° × 30° and 55° × 55° viewing systems interchangeably in DDAF area as reported in previous studies.7,13,14 Furthermore, we demonstrated the feasibility and usefulness of UWF FAF for measuring DDAF area expansion across a wide spectrum of STGD1. In addition to the established predictive factor of DDAF area ER, our finding of the association between genotype group and DDAF SRA ER has significant implications for patient selection and clinical trial designs.
To our knowledge, the feasibility and repeatability of UWF FAF in the longitudinal assessment of DDAF has not been reported previously. In our study, we selected UWF FAF as the primary imaging method to estimate atrophy enlargement rate in STGD1. To date, short-wavelength (SW) FAF-based DDAF area measurement using the central 30° × 30° or 55° × 55° field has been the main outcome measure in published studies, including the ProgStar study,9,13 whereas Optos-derived UWF FAF imaging has not been explored as an end point in this large multicenter trial. The use of UWF FAF has advantages, most notably for pediatric populations or patients with impaired mobility as well as for those with disease extending outside of the central 30° × 30° or 55° × 55° field. In addition, Chen et al8 found that UWF FAF images can have reduced uniformity, as compared with SW AF, most pronounced in the superior and inferior fundus. The reduced visibility was compensated partially by imaging through a dilated pupil, but the entire DDAF atrophic lesion was still not visible with UWF FAF in 2 of 24 patients in group A. Fernanda Ablem et al27 showed that UWF FAF imaging in STGD1 was correlated with electroretinography and kinetic perimetry, thus supporting the use of this more accessible clinical tool for evaluating both central and peripheral retinal involvement. Given the ease of use (single flash compared with prolonged averaging), reduced potential for toxicity, wider availability, and greater field of view of UWF FAF, we propose that UWF FAF may be more appropriate for evaluation of larger lesions and should be included in the assessment of all patients with STGD1. In further support of UWF FAF, Chen et al8 also reported that foveal-sparing lesions were more readily demonstrable by green-wavelength FAF imaging. Müller et al6 also found that green FAF-based quantification of lesion size provided similar results to SW FAF. Although we provided evidence, for the first time, that the Heidelberg 30° × 30° imaging systems is equivalent to Optos UWF FAF imaging with respect to DDAF area, we also found that Heidelberg 55° × 55° FAF imaging tended to overestimate DDAF area when compared with 30° × 30° imaging and UWF FAF imaging. Chen et al8 compared the different imaging methods near-infrared autofluorescence, SW AF, and green UWF FAF in 34 patients with STGD1 and found that 18 of 34 patients (53%) harbored lesions extending outside the 55° × 55° field. Similarly, Klufus et al9 found that most patients with STGD1 demonstrate changes in the peripheral retina. While comparing the 3 viewing systems, we also found 26% and 16% of patients showed DDAF boundaries beyond the 30° × 30° and 55° × 55° fields, respectively. Similar to Müller et al,6 who reported high interobserver agreement with the 30° × 30° imaging system, we also observed good interobserver agreement in DDAF boundary demarcation for area and SRA measurements using Optos UWF FAF imaging.
After demonstrating the feasibility of UWF FAF for measuring DDAF area, we calculated the annual ER of DDAF area and SRA in STGD1 to investigate predictive factors. This DDAF area ER was significantly greater than that of previous reports because we did not exclude atrophic lesions that extended beyond the limits of the 30° × 30° or 55° × 55° viewing systems. The mean annual DDAF area ER was 2.4 mm2/year (range, 0.06–32.3 mm2/year) as compared with less than 1 mm2/year reported by all previous studies that also reported varied annual ER because of differences in baseline atrophy sizes.6, 7, 8,12,13 Consistent with previous literature, we also found that baseline DDAF area was a significant predictor of DDAF area ER.6,12,13 Interestingly, baseline age was associated positively with DDAF SRA ER, whereas baseline DDAF SRA was associated negatively with DDAF SRA ER. In a retrospective study, Strauss et al7 reported a mean annual DDAF progression rate of 0.51 mm2 (95% confidence interval, 0.42–0.61 mm2) with a mean ± SD baseline lesion size of 2.2 ± 2.7 mm2 where the rate of progression depended on the baseline lesion size. In their subsequent prospective study, the annual DDAF progression increased to 0.76 mm2 (95% confidence interval, 0.54–0.97 mm2) as the baseline lesion size also increased to 3.9 ± 4.4 mm2.13 Within the lesion size category, Strauss et al7,13 found that the growth rates were age dependent where a linear mixed model stratified by initial lesion size provided the best fit in estimating annual progression rates. Müller et al6 also reported a low mean annual DDAF progression rate of 0.89 ± 0.13 mm2. They found that the number of DDAF lesions, full-field electroretinographic findings, and age at onset were the most significant predictors for future growth. Fujinami et al2 reported DDAF progression in 67 patients with STGD1, of whom only 35 patients underwent mutation screening. They reported a median annual DDAF progression rate of only 0.45 mm2 and found the age and size of atrophy at baseline to be significant predictors. Cicinelli et al11 also found multifocal disease, larger baseline lesion, and worse visual acuity to be associated with a faster rate of progression. Lindner et al14 reported an annual DDAF SRA progression rate of 0.23 mm, which is identical to our mean ER of 0.23 ± 0.18 mm/year. However, their study only included 66 eyes from 38 patients 45 years of age or older, in whom the mean ± SD age was 63.4 ± 9.9 years, in contrast to 24.5 ± 22.6 years in our cohort and in the Strauss et al13 and Muller et al12 series, where the mean age was 21.9 ± 13.3 years and 33.6 ± 17.2 years, respectively. Although baseline atrophy size has been identified as the key predictor of ER, less is known about the effect of genotype, especially classified by the combined severity of biallelic ABCA4 mutations. Given the importance of genotype in predicting age at onset and extent of lesion area, we added genotype group to the linear mixed model for ER.
A key novel finding in this study is the effect of genotype on DDAF SRA ER, whereby the mean ER for genotype group C was significantly lower than that of both genotype groups A and B. Fujinami et al2 assessed the effect of specific STGD1 genotypes on DDAF progression rates and found an annual median rate of atrophy expansion of 0.45 mm2 and 0.39 mm2 in patients harboring c.5461–10T→C and c.6079C→T variants, respectively. In contrast, those with the c.5882G→A variant showed an expansion rate of only 0.20 mm2/year, suggesting a potential association between patients carrying 1 truncating or severe variant and more progressive FAF patterns. Di Iorio et al28 also found a significant association between severe STGD1 phenotypes and faster progression on OCT. However, both these studies considered only 1 allele, although the phenotype in STGD1 is dependent on the combined effect of both paternal and maternal variants. In contrast, our study takes into consideration both alleles, with known severities, to determine the effect of genotype on DDAF progression rates. In a retrospective cohort study of 28 patients with STGD1, Cicinelli et al11 found that genetic variants were not associated with the rate of DDAF progression. However, their patients were divided into 2 simplistic genotype groups of (1) patients with 1 or more null variant or (2) patients with 2 or more missense variants, which does not take into account the severity of the impact of these variants on ABCA4 function. Our results have implications on clinical trials design regarding inclusion criteria and stratification based on baseline DDAF area as well as genotype characteristics of the study cohort.
One limitation of our study is the small sample size in the nullizygous and the hemizygous-like genotype groups. The images also were evaluated by 2 unmasked investigators (R.C.H.J. and F.K.C.) to ensure that all atrophic lesions were marked. Significantly lower numbers of follow-up data were available for some patients, which limited our investigation of the actual lesion area growth rate for specific mutations. Although UWF FAF imaging exhibits less shadowing with greater penetration of 532-nm light through the macular pigment, direct correlation in lesion size between Heidelberg Spectralis SW AF 30° × 30° or 55° × 55° fields and UWF FAF imaging has been hindered by optical distortions inherent to the UWF FAF platform as well as no real-time averaging to reduce background noise.8 Ultra-widefield FAF image analysis depends on high-quality, well-centered images that can be affected by several artefacts, potentially influencing our results and area calculations for DDAF size. In addition, some of our patients had difficulty fixating on a central target, and consequently some of the images obtained may have centration issues and may have deviated from the standard retinal region typically captured by Optos.
In summary, we demonstrated the feasibility and reliability of using UWF FAF to detect atrophy ER in STGD1. Ultra-widefield FAF offers many advantages, including better visualization of the perifoveal and equatorial retina, ability to capture (duration, 250 ms) retinal lesions rapidly in undilated children, and a more comfortable photography experience (λ, 532 nm, green flash) for the patients. We demonstrated genotype-dependent variations in ER and found that patients belonging to genotype group C showed a significantly slower DDAF SRA ER. Further multicenter studies of UWF FAF DDAF area expansion and its correlation with other pathogenic ABCA4 variants are warranted.
Acknowledgments
The authors thank Amanda Scurry and Jayme Glynn for their assistance in organizing the patient appointments and Maryam Saadat for performing retinal imaging. The Australian Inherited Retinal Disease Registry and DNA Bank acknowledges the assistance of Ling Hoffman and Isabella Urwin from the Department of Medical Technology and Physics at Sir Charles Gairdner Hospital.
Manuscript no. D-21-00006.
Footnotes
Supplemental material available atwww.ophthalmologyscience.org/.
Disclosure(s): All authors have completed and submitted the ICMJE disclosures form.
The author(s) have made the following disclosure(s): I.L.M.: Board membership – Novartis, Bayer
F.K.C.: Board membership – Roche; Consultant – Bayer, Novartis, Allergan, Heidelberg Engineering, Alcon, Pfizer; Financial support – Novartis, Bayer
Supported by the National Health & Medical Research Council, Australia; (project and fellowship grant nos.: GNT116360 [F.K.C.], GNT1188694 [F.K.C.], GNT1054712 [F.K.C.], and MRF1142962 [F.K.C.]); the McCusker Foundation (F.K.C.); the Miocevich Retina Fellowship (R.C.H.J.); and Retina Australia (J.A.T., T.L., J.N.D.R., T.L.M.), an independent medical research institute and not-for-profit company.
HUMAN SUBJECTS: Human subjects were included in this study. The human ethics committees at the University of Western Australia and Sir Charles Gairdner Hospital approved the study. All research adhered to the tenets of the Declaration of Helsinki. All participants provided informed consent.
No animal subjects were included in this study.
Author Contributions:
Conception and design: Heath Jeffery, Chen
Analysis and interpretation: Heath Jeffery, Thompson, Lo, Lamey, McLaren, De Roach, Chen
Data collection: Heath Jeffery, Thompson, Lamey, McLaren, McAllister, Mackey, Constable, De Roach, Chen
Obtained funding: Study was performed as part of regular employment duties at Lions Eye Institute, The University of Western Australia, Royal Perth Hospital, Sir Charles Gairdner Hospital, and Edith Cowan University. No additional funding was provided.
Overall responsibility: Heath Jeffery, Thompson, Lo, Lamey, McLaren, McAllister, Mackey, Constable, De Roach, Chen
Supplementary Data
References
- 1.Michaelides M., Hunt D.M., Moore A.T. The genetics of inherited macular dystrophies. J Med Genet. 2003;40:641–650. doi: 10.1136/jmg.40.9.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujinami K., Zernant J., Chana R.K., et al. Clinical and molecular characteristics of childhood-onset Stargardt disease. Ophthalmology. 2015;122:326–334. doi: 10.1016/j.ophtha.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Roach J.N., McLaren T.L., Thompson J.A., et al. The Australian Inherited Retinal Disease Registry and DNA Bank. Tasman Med J. 2020;2:60–67. [Google Scholar]
- 4.Westeneng-van Haaften S.C., Boon C.J., Cremers F.P., et al. Clinical and genetic characteristics of late-onset Stargardt’s disease. Ophthalmology. 2012;119:1199–1210. doi: 10.1016/j.ophtha.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 5.Chen B., Tosha C., Gorin M.B., Nusinowitz S. Analysis of autofluorescent retinal images and measurement of atrophic lesion growth in Stargardt disease. Exp Eye Res. 2010;91:143–152. doi: 10.1016/j.exer.2010.03.021. [DOI] [PubMed] [Google Scholar]
- 6.Müller P.L., Pfau M., Mauschitz M.M., et al. Comparison of green versus blue fundus autofluorescence in ABCA4-related retinopathy. Transl Vis Sci Technol. 2018;7:13. doi: 10.1167/tvst.7.5.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strauss R.W., Muoz B., Ho A., et al. Progression of Stargardt disease as determined by fundus autofluorescence in the retrospective progression of Stargardt disease study (ProgStar report no. 9) JAMA Ophthalmol. 2017;135:1232–1241. doi: 10.1001/jamaophthalmol.2017.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L., Lee W., de Carvalho J.R.L., et al. Multi-platform imaging in ABCA4-associated disease. Sci Rep. 2019;9:6436. doi: 10.1038/s41598-019-42772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klufas M.A., Tsui I., Sadda S.R., et al. Ultrawidefield autofluorescence in ABCA4 Stargardt disease. Retina. 2018;38:403–415. doi: 10.1097/IAE.0000000000001567. [DOI] [PubMed] [Google Scholar]
- 10.Fujinami K., Lois N., Mukherjee R., et al. A longitudinal study of Stargardt disease: quantitative assessment of fundus autofluorescence, progression, and genotype correlations. Invest Ophthalmol Vis Sci. 2013;54:8181–8190. doi: 10.1167/iovs.13-12104. [DOI] [PubMed] [Google Scholar]
- 11.Cicinelli M.V., Rabiolo A., Brambati M., et al. Factors influencing retinal pigment epithelium-atrophy progression rate in Stargardt disease. Transl Vis Sci Technol. 2020;9:33. doi: 10.1167/tvst.9.7.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muller P.L., Gliem M., McGuinnes M., et al. Quantitative fundus autofluorescence in ABCA4-related retinopathy: functional relevance and genotype-phenotype correlation. Am J Ophthalmol. 2020;222:340–350. doi: 10.1016/j.ajo.2020.08.042. [DOI] [PubMed] [Google Scholar]
- 13.Strauss R.W., Kong X., Ho A., et al. Progression of Stargardt disease as determined by fundus autofluorescence over a 12-month period: ProgStar report no. 11. JAMA Ophthalmol. 2019;137:1134–1145. doi: 10.1001/jamaophthalmol.2019.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindner M., Lambertus S., Mauschitz M.M., et al. Differential disease progression in atrophic age-related macular degeneration and late-onset Stargardt disease. Invest Ophthalmol Vis Sci. 2017;58:1001–1007. doi: 10.1167/iovs.16-20980. [DOI] [PubMed] [Google Scholar]
- 15.De Roach J.N., McLaren T.L., Paterson R.L., et al. Establishment and evolution of the Australian Inherited Retinal Disease Register and DNA Bank. Clin Exp Ophthalmol. 2013;41:476–483. doi: 10.1111/ceo.12020. [DOI] [PubMed] [Google Scholar]
- 16.Cremers F.P.M., Lee W., Collin R.W.J., Allikmets R. Clinical spectrum, genetic complexity and therapeutic approaches for retinal disease caused by ABCA4 mutations. Prog Retin Eye Res. 2020;79:100861. doi: 10.1016/j.preteyeres.2020.100861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Curtis S.B., Molday L.L., Garces F.A., Molday R.S. Functional analysis and classification of homozygous and hypomorphic ABCA4 variants associated with Stargardt macular degeneration. Hum Mutat. 2020;41:1944–1956. doi: 10.1002/humu.24100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fakin A., Robson A.G., Chiang J., et al. The effect on retinal structure and function of 15 specific ABCA4 mutations: a detailed examination of 82 hemizygous patients. Invest Ophthalmol Vis Sci. 2016;57:5963–5973. doi: 10.1167/iovs.16-20446. [DOI] [PubMed] [Google Scholar]
- 19.Atkinson A., Mazo C. Imaged area of the retina. www.optos.com/global/documents/casestudies_imagedareaoftheretina.pdf Available at: Accessed 12.19.20.
- 20.Chiang J.P., Lamey T., McLaren T., et al. Progress and prospects of next-generation sequencing testing for inherited retinal dystrophy. Expert Rev Mol Diagn. 2015;15:1269–1275. doi: 10.1586/14737159.2015.1081057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.den Dunnen J.T., Dalgleish R., Maglott D.R., et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37:564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 22.Thompson J.A., De Roach J.N., McLaren T.L., et al. The genetic profile of Leber congenital amaurosis in an Australian cohort. Mol Genet Genomic Med. 2017;5:652–667. doi: 10.1002/mgg3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards S., Nazneen A., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarvik G.P., Browning B.L. Consideration of cosegregation in the pathogenicity classification of genomic variants. Am J Hum Genet. 2016;98:1077–1081. doi: 10.1016/j.ajhg.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulze-Bonsel K., Feltgen N., Curau H., et al. Visual acuities “hand motion” and “counting fingers” can be quantified with the Freiburg visual acuity test. Invest Ophthalmol Vis Sci. 2006;47:1236–1240. doi: 10.1167/iovs.05-0981. [DOI] [PubMed] [Google Scholar]
- 26.Lange C., Feltgen N., Junker B., et al. Resolving the clinical acuity categories “hand motion” and “count fingers” using the Freiburg visual acuity test (FrACT) Graefes Arch Clin Exp Ophthalmol. 2009;247:137–142. doi: 10.1007/s00417-008-0926-0. [DOI] [PubMed] [Google Scholar]
- 27.Fernanda Abalem M., Otte B., Andrews C., et al. Peripheral visual fields in ABCA4 Stargardt disease and correlation with disease extent on ultra-widefield fundus autofluorescence. Am J Ophthalmol. 2017;184:181–188. doi: 10.1016/j.ajo.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Di Iorio V., Orrico A., Esposito G., et al. Disease progression in Italian Stargardt patients; a retrospective natural history study. Retina. 2019;39:1399–1409. doi: 10.1097/IAE.0000000000002151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.