Abstract
Aims
While electrocardiogram (ECG) characteristics have been associated with life-threatening ventricular arrhythmias (LTVA) in dilated cardiomyopathy (DCM), they typically rely on human-derived parameters. Deep neural networks (DNNs) can discover complex ECG patterns, but the interpretation is hampered by their ‘black-box’ characteristics. We aimed to detect DCM patients at risk of LTVA using an inherently explainable DNN.
Methods and results
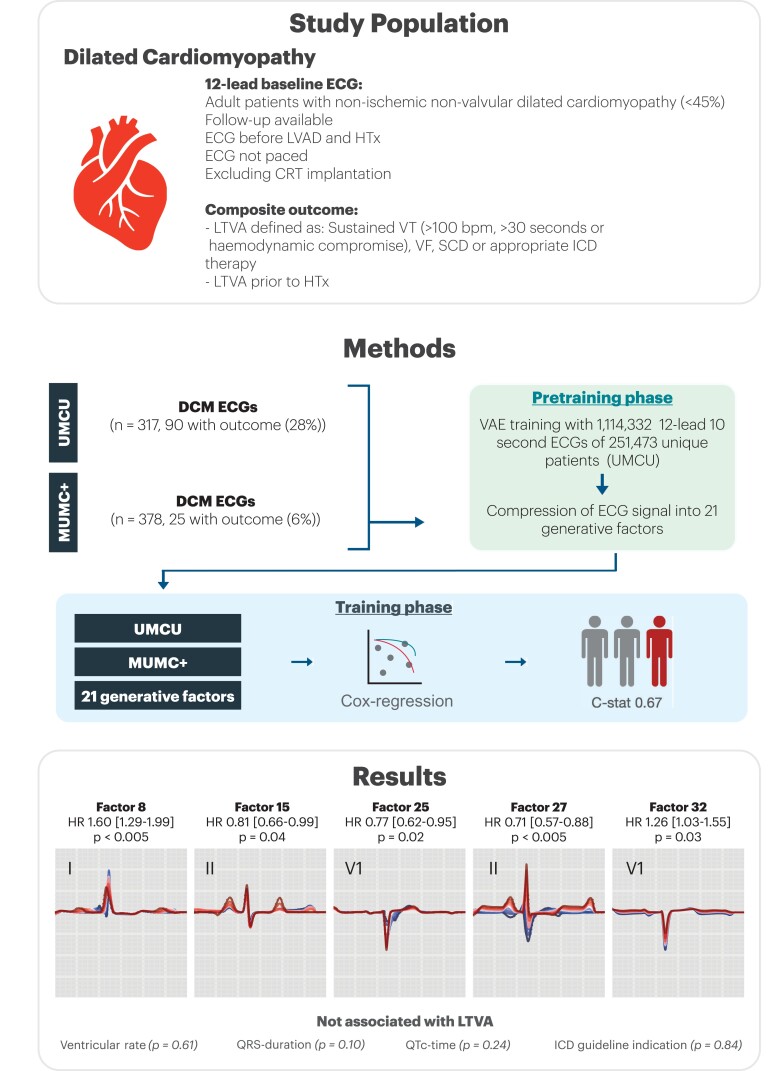
In this two-phase study, we first developed a variational autoencoder DNN on more than 1 million 12-lead median beat ECGs, compressing the ECG into 21 different factors (F): FactorECG. Next, we used two cohorts with a combined total of 695 DCM patients and entered these factors in a Cox regression for the composite LTVA outcome, which was defined as sudden cardiac arrest, spontaneous sustained ventricular tachycardia, or implantable cardioverter-defibrillator treated ventricular arrhythmia. Most patients were male (n = 442, 64%) with a median age of 54 years [interquartile range (IQR) 44–62], and median left ventricular ejection fraction of 30% (IQR 23–39). A total of 115 patients (16.5%) reached the study outcome. Factors F8 (prolonged PR-interval and P-wave duration, P < 0.005), F15 (reduced P-wave height, P = 0.04), F25 (increased right bundle branch delay, P = 0.02), F27 (P-wave axis P < 0.005), and F32 (reduced QRS-T voltages P = 0.03) were significantly associated with LTVA.
Conclusion
Inherently explainable DNNs can detect patients at risk of LTVA which is mainly driven by P-wave abnormalities.
Keywords: Dilated cardiomyopathy, Deep neural network, Prognosis, Sudden cardiac death, Implantable cardioverter-defibrillator
What’s new?
This is the first study to use an inherently interpretable deep neural network (variational autoencoder, VAE) for the prediction of life-threatening arrhythmias in patients with dilated cardiomyopathy using raw electrocardiogram (ECG) signals.
The FactorECG summarizes the median beat ECG, including most of its features, into 21 generative factors of variation [see https://dcm.ecgx.ai/]. This novel strategy allows to simultaneously evaluate most characteristics that make up an ECG automatically, rather than using selected and human-derived ECG features.
While the VAE encompasses complete electrocardiograms, visualisation of pivotal ECG features showed that network predictions were mainly driven by P-wave abnormalities.
These P-wave abnormalities did not correlate with their anatomical analogues, such as left atrial dimensions, suggesting an electrophysiological substrate.
Introduction
Patients with non-ischaemic dilated cardiomyopathy (DCM) have an estimated annual risk of life-threatening ventricular arrhythmias (LVTAs) of 4.5% and may potentially benefit from implantable cardioverter-defibrillator (ICD) implantation.1,2 A novel risk model (DCM-SVA risk) for predicting LTVA was recently published and includes easily accessible clinical parameters, such as the history of non-sustained ventricular tachycardia (VT), QRS duration, and left ventricular ejection fraction (LVEF).3 More complex electrocardiogram (ECG) characteristics such as fragmented QRS waves, heart rate variability, and T-wave alternans have also been associated with LTVA, but rely on manually derived ECG parameters that remain difficult to standardize, hampering their integration into daily clinical practice.1 By using raw ECG signals and machine learning techniques, manual feature extraction is not necessary. Moreover, novel and more subtle parameters may be detected.4
Deep neural networks (DNN) have proven to be potent machine learning algorithms for diagnostic classification tasks using raw ECGs signals. Previous studies using DNNs on raw ECG signals in cardiomyopathies report high performance in disease classification and triaging.5,6 However, because of the inherent lack of ‘explainability’ of DNNs, clinical implementation remains limited.7 Different techniques may assist in interpreting DNNs. A recently introduced pipeline for fully explainable DNNs for ECG analysis uses variational autoencoders (VAEs),8 that can compress the ECG into a lower number of explanatory and independent generative factors (FactorECG), which can subsequently be used in interpretable algorithms (such as Cox regression).9
In this study, we aimed (i) use an inherently interpretable DNN for predicting potentially LTVA based on ECGs in patients with non-ischemic DCM, assess its added value above conventional ECG parameters and current guidelines and (ii) interpret the model by visualizing pivotal ECG features.
Methods
Study participants
In this retrospective cohort study, we included consecutive adult patients with DCM as defined by the European Society of Cardiology (ESC) guidelines were included from the UMCU and MUMC+.2 Only patients with a baseline non-paced 12-lead ECG acquired before left ventricular assist device (LVAD) implantation or heart transplantation (HTx) were eligible. Patients with a cardiac resynchronization therapy (CRT) were excluded, as it positively affects reverse remodelling which may reduce arrhythmias.10 This study was conducted in accordance with the principles laid out in the Declaration of Helsinki and in line with guidelines provided by ethics committees and the national GDPR legislature. The participants from the UMCU cohort were included using the opt-out procedure. The UMCU cohort was exempt from the Medical Research Involving Human Subjects Act (WMO) as per the judgement of the Medical Ethics Committee (18/446 and 19/222 UMCU, The Netherlands) including the requirement for informed consent. The participants of the Maastricht cohort signed informed consent at enrolment.
Data acquisition
For all subjects, the ECG closest to the date of the first presentation was obtained which was considered ‘baseline” for the purpose of this study. The median time between diagnosis and ECG was 0 (IQR 0–28) days. All ECGs were exported from the MUSE ECG system (version 8; GE Healthcare, Chicago, IL, USA) in raw voltage format. The recordings were made using a General Electric MAC V, 5000 or 5500 device and acquired at either 250 or 500 Hz. Resampling to 500 Hz was performed via linear interpolation and transformation into 1.2-s median beats was achieved by aligning all QRS-complexes of the same shape (e.g. excluding premature ventricular complexes) and taking the median voltage to generate a representative P-QRS-T complex. Echocardiographic measurements were extracted from the electronic health record using methods described before.11
Pre-training and explainability of the variational autoencoder
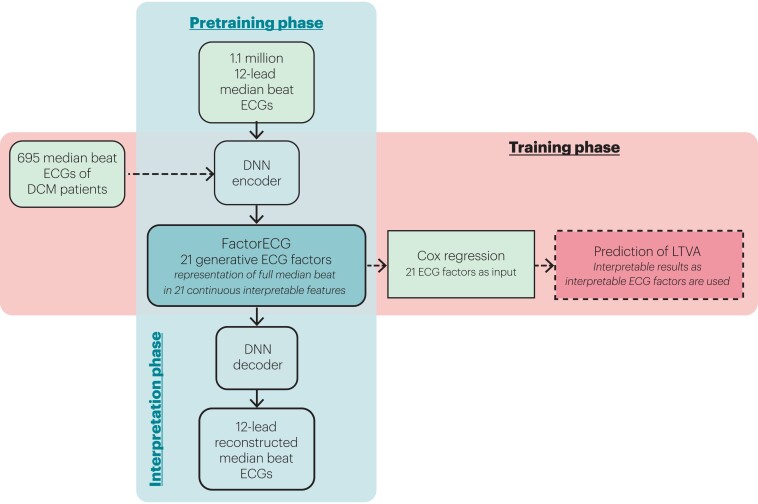
A two-phase approach was used in this study, where a VAE was first pre-trained on the complete UMCU ECG dataset, and them used in the training step to find associations with LTVA (Figure 1). VAEs are unsupervised deep learning encoder–decoder convolutional neural networks that are optimized to reconstruct their training data with a lower-dimensional representation (i.e. using less data) than the original training data (in this case ECGs). The current VAE network is enforced with a specific function to reach maximum disentanglement of lower-dimensional representation (i.e. to produce generative factors in the ECG that operate independently: the FactorECG).12 Resting 12-lead 10-s ECGs of 251 473 unique patients (1 114 331 ECGs) were exported from the UMCU ECG system and used for pre-training of the VAE. In a prior study, the optimal number of dimensions was found to be 21, considering the trade-off of good reconstruction disentanglement and encoding for visible ECG abnormalities.8
Figure 1.
Overview of the pre-training and training phases of the FactorECG algorithm. During the pre-training phase, 1.1 million 12-lead median beat ECGs were included for the training of the VAE. The VAE was trained to compress all 12-lead median beat ECGs into 21 continuous factors of variation (the FactorECG), that can subsequently be used to reconstruct the median beat ECG. The VAE is explainable by visualizing the influence of the individual ECG factors on the ECG morphology using the decoder. In the training phase, for each of the 695 DCM patients, median beat ECGs were encoded into 21 generative factors using the pre-trained encoder. These 21 generative ECG factors were used as an input in a Cox regression model to predict life-threatening ventricular arrhythmias. The importance of each ECG factor was then determined by investigating the hazard ratios of the standardized ECG factors. DCM, dilated cardiomyopathy; DNN, deep neural network; LTVA, life-threatening ventricular arrhythmia.
Explainability of the individual factors was obtained on a model level using factor traversals. Starting with a mean FactorECG for this population (i.e. the mean value of the 21 ECG factors), a median beat ECG is reconstructed using the decoder. Subsequently, for each individual ECG factor, values between −4 and 4 are added and ECGs are reconstructed for every value. Meanwhile, values for the other factors are kept constant. This way we are able to visualize the effect of a single ECG factor on the median beat ECG morphology in this cohort (Figure 2). On the individual patient level, explainability is obtained by investigating the FactorECG values of that specific ECG. A tool to visualize the factors interactively can be found at https://dcm.ecgx.ai. The architecture and model training process were implemented using PyTorch (version 1.7.0 + cu110) in Python (version 3.6.7).
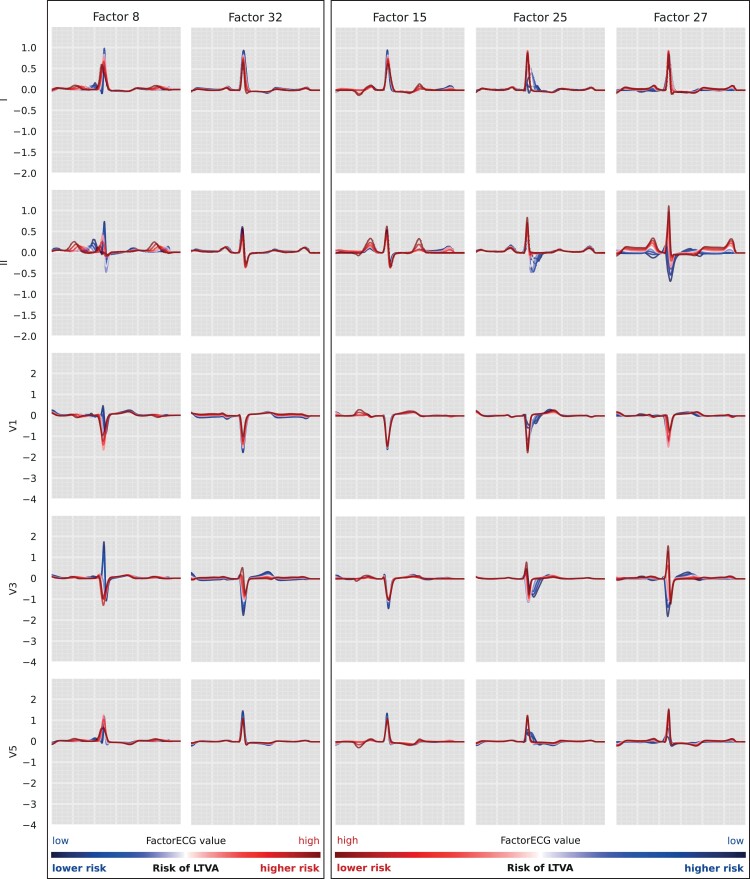
Figure 2.
Factor traversals for the ECG factors that were associated with LTVA in the DCM cohort. We start with a mean FactorECG for this population (i.e. the mean value of the 21 ECG factors) and reconstruct an ECG using the decoder (white). Subsequently, for each individual ECG factor, values between −4 (blue) and 4 (red) are added and ECGs are reconstructed for every value. Meanwhile, values for the other factors are kept constant. This way we are able to visualize the effect of a single ECG factor on the median beat ECG morphology in this cohort. For factors 8 and 32, high values of the factors were associated with a higher risk of LTVA (left). For factors 15, 25, and 27, conversely, low values of the factors were associated with a higher risk of LTVA (right). The FactorECG decoder reconstructs the full 12-lead median beat ECG, a selection of leads is shown in this figure. DCM, dilated cardiomyopathy; LTVA, life-threatening ventricular arrhythmia.
Outcome definitions
The primary study outcome was LTVA, defined as the composite outcome of sustained ventricular tachycardia (VT) >100 b.p.m. lasting >30 s or with hemodynamic compromise, ventricular fibrillation (VF), sudden cardiac death (SCD, defined as the death of cardiac origin that occurred unexpectedly within 1 h after the onset of new symptoms) or appropriate ICD therapy (defined as any ICD therapy delivered by the device in response to VT or VF according to stored intracardiac electrograms).3
Statistical analyses
For the baseline table, mean ± SD or median (interquartile range) were used where appropriate. Missingness in baseline data was not addressed. Each baseline ECG’s generative factors (the FactorECGs, as computed by the VAE encoder) were included in a Cox proportional hazards model (Figure 1). All patients had a digitalized ECG available. The proportional hazards assumption was tested. Hazard ratios (HR) were reported, and 95% confidence intervals (CIs) were computed using 2000 bootstrap samples. To rule out that the VAE model was solely considering already established ECG characteristics (ventricular rate, PR-interval, QRS-duration, and Bazett corrected QT-interval), a Cox proportional hazard model was also fitted using these variables in a complete case analysis. The correlations of the significant ECG factors were plotted against the left atrial (LA) dimension and left atrial volume index (LAVI) measured on standard care clinical echocardiography using both the first (closest to baseline) and last (closest to follow-up) available measurements.11 Additionally a Kaplan–Meier curve was plotted for one of the significant VAE generative factors. All analyses were performed using Python (version 3.8.5).
Results
Patient characteristics
Baseline characteristics stratified by centre and outcome are depicted in Table 1. A total of 695 patients were included from the UMCU and MUMC+, which were predominantly male (n = 442, 64%) with a median age of 54 years [interquartile range (IQR) 44–62] and median LVEF of 30% (IQR 23–39%). A total of 115 (17%) reached the study outcome in both centres combined during a median follow-up of 4.3 years (IQR 2.0–7.5). In summary, patients from the MUMC+ cohort had less severe symptoms at baseline with primarily New York Heart Association classes I and II as opposed to the UMCU cohort with primarily II and III, and a median LVEF of 33% (IQR 25–40). A lower proportion of MUMC+ patients (25, 6%) reached the study outcome of LTVA compared with 90 (28%) UMCU patients.
Table 1.
Patient characteristics at baseline (first evaluation) stratified by centre and outcome
| UMCU all (n = 317) | UMCU without LTVA (n = 227) | UMCU with LTVA (n = 90) | MUMC all (n = 378) | MUMC without LTVA (n = 353) | MUMC with LTVA (n = 25) | |
|---|---|---|---|---|---|---|
| Age (years), median (Q1–Q3) | 52 (42–61) | 51 (41–60) | 52 (42–62) | 55 (47–63) | 56 (47–63) | 54 (49–63) |
| Male sex | 195 (62%) | 129 (57%) | 66 (74%) | 247 (65%) | 228 (65%) | 19 (76%) |
| NYHA-class | ||||||
| I | 53 (20%)* | 36 (18%)* | 17 (23%)* | 158 (42%) | 150 (43%) | 8 (32%) |
| II | 102 (39%)* | 71 (36%)* | 31 (41%)* | 175 (46%) | 163 (47%) | 12 (48%) |
| III | 79 (30%)* | 56 (28%)* | 23 (32%)* | 37 (10%) | 32 (9%) | 5 (20%) |
| IV | 27 (10%)* | 36 (18%)* | 4 (5%)* | 8 (2%) | 8 (2%) | 0 (0)% |
| Diabetes mellitus | 42 (13%) | 31 (13%) | 11 (12%) | 52 (14%) | 50 (14%) | 2 (8%) |
| Hypercholesterolemia | 37 (13%) | 26 (13%) | 11 (13%) | 41 (11%) | 38 (11%) | 3 (12%) |
| (Ever) smoked | 203 (64%) | 145 (64%) | 57 (63%) | 77 (20%) | 72 (20%) | 5 (20%) |
| History of LTVA | 42 (13%) | 17 (7%) | 25 (27%) | 8 (2%) | 7 (2%) | 1 (4%) |
| Family history of DCM | 133 (42%) | 97 (43%) | 36 (40%) | 47 (14%) | 39 (11%) | 8 (32%) |
| ICD implantation | 233 (74%) | 145 (63%) | 88 (97%) | 0 (0%) | 0 (0%) | 0 (0%) |
| LVEF (%), median (Q1–Q3) | 25 (20–33) | 25 (20–33) | 25 (19–32) | 33 (25–40) | 28 (22–37) | 33 (25–41) |
| MRI LGE | 84 (56%**) | 60 (51%*) | 24 (71%**) | n/a | n/a | n/a |
Baseline characteristics of the included cohorts. NYHA, New York Heart Association; LTVA, life-threatening ventricular arrhythmia; ICD, implantable cardioverter-defibrillator; LVEF, left ventricular ejection fraction; *, of valid, in patients for which a NYHA class was noted in the electronic health record; MRI LGE, magnetic resonance imaging late gadolinium enhancement; **, of valid, in patients with cardiac MRIs.
Prediction of LTVA with established ECG variables
Established ECG variables (such as ventricular rate, PR-interval, QRS-duration, and QTc-time) were entered in a ‘baseline’ Cox regression model controlled for guideline indication [complete case analysis with n = 577, excluding patients without a measurable PR-interval (n = 118, due to atrial fibrillation/flutter)]. This baseline model had a C-statistic of 0.58 (95% CI 0.52–0.64) and no significant effects of: ventricular rate [HR 0.94 per 10 beat/s increase (95% CI 0.81–1.09), P = 0.41], QRS duration [HR 1.08 per 10 ms increase (95% CI 0.98–1.19), P = 0.13] and QTc-time [HR 0.95 per 10 ms increase (95% CI 0.88–1.03), P = 0.19]. The PR-interval was, however, significantly associated with LTVA [HR 1.06 per 10 ms increase (95% CI 1.00–1.13), P = 0.04]. The results of this model are depicted in Supplementary material online, Table S1.
Prediction of LTVA with FactorECG
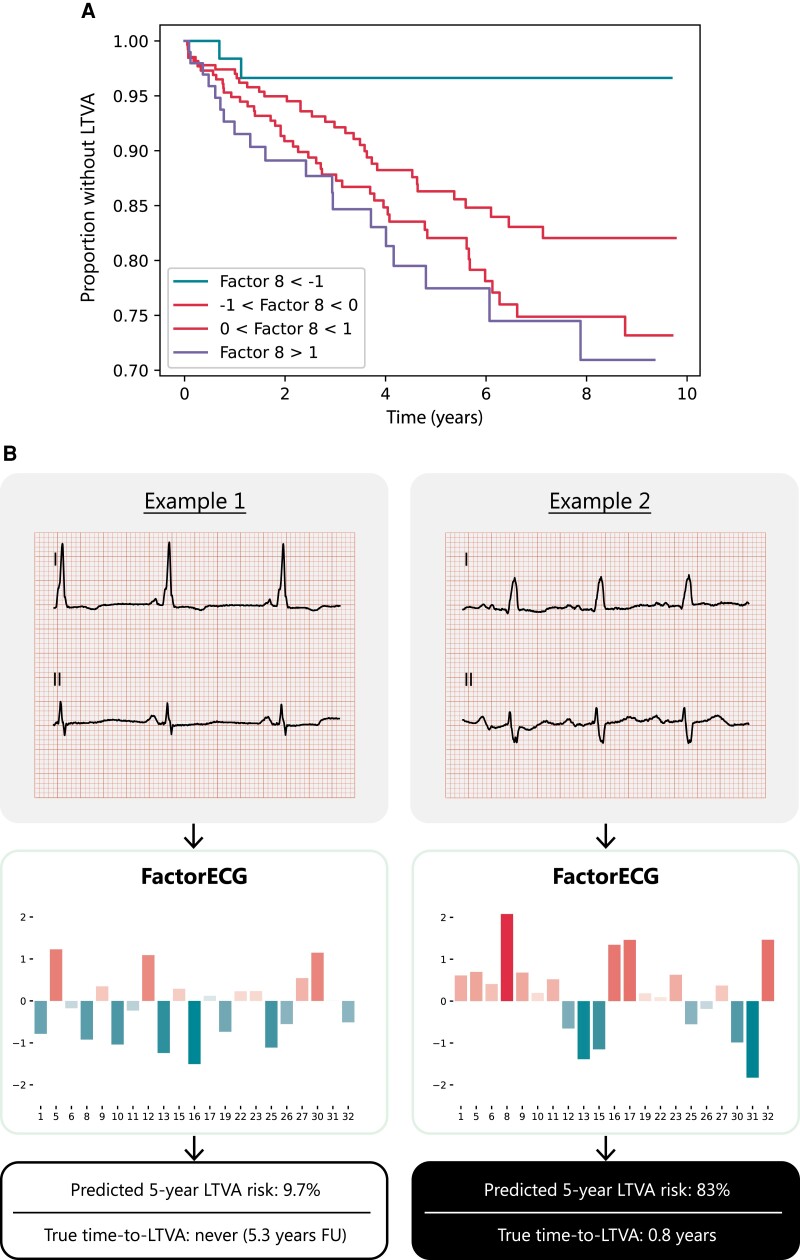
The VAE compressed the ECG data into 21 different ECG factors and their factor traversals are available in Supplementary material online, Figure S1. In Cox regression, F8 [HR 1.60; 95% CI (1.29–1.99), P < 0.005], F15 [HR 0.81; 95% CI (0.66–0.99), P = 0.04], F25 [HR 0.77 95% CI (0.62–0.95), P = 0.02], F27 [HR 0.71, 95% CI (0.57–0.88), P < 0.005], and F32 [HR 1.26, 95% CI (1.03–1.55), P = 0.03] were significantly associated with the outcome after correcting for guideline indication (NYHA II/III and LVEF < 35%, P = 0.84). C-statistic for the model was 0.67 (95% CI 0.62–0.72). A reconstruction of the significant generative factors (F8, F15, F25, F27, and F32) has been illustrated in Figure 2. F8 encodes for PR-interval and P-wave morphology, where high values increase PR-interval and broaden the P-wave. F15 encodes for P-wave height and P/T-overlap, where low values are correlated with atrial fibrillation and third-degree AV-block. F25 encodes for conduction delays in the right bundle (right bundle branch block), where low values increase the block. F27 encodes for P- and R-axis deviation, where low values flatten out the P-wave. F32 encodes for QRS-T amplitudes, with low values reconstruct QRS-T microvoltages. Results of the Cox regression model and the descriptions of the generative factors are present in Table 2 and Supplementary material online, Table S2. The partial effects on outcome per significant factor have been plotted in Supplementary material online, Figure S2. As an example, the ECGs and their corresponding values of the generative factors of two patients were plotted in Figure 3. A summary figure of this study was depicted in Figure 4. To address the possible effect of cardiac memory after pacing, a subgroup analysis was run excluding patients with a pacemaker (n = 32) which showed similar factors to be important (Supplementary material online, Table S3).
Table 2.
Cox proportional hazards model of generative factors in both cohorts
| Factors | Factor descriptions | Hazard ratio | 95% CI | P-value |
|---|---|---|---|---|
| F1 | Inferolateral ST deviation | 0.91 | 0.72–1.14 | 0.39 |
| F5 | Inferolateral T-wave height and orientation | 1.17 | 0.92–1.48 | 0.19 |
| F6 | P-wave height and/or shape | 1.14 | 0.90–1.44 | 0.27 |
| F8a | PR-interval (high values associated with first degree AV-block and reduced LVEF) | 1.61 | 1.29–1.99 | <0.005 |
| F9 | T-wave height and orientation | 1.12 | 0.89–1.39 | 0.34 |
| F10 | Ventricular rate | 0.93 | 0.76–1.13 | 0.46 |
| F11 | Subtle P- and T-wave changes | 1.00 | 0.83–1.21 | 0.97 |
| F12 | Onset of depolarization | 1.08 | 0.86–1.36 | 0.50 |
| F13 | Anterior ST deviation | 0.85 | 0.68–1.06 | 0.14 |
| F15a | P-wave height and P/T-overlap (low values associated with third-degree AV-block and junctional tachycardia) | 0.81 | 0.66–0.99 | 0.04 |
| F16 | T-wave morphology | 1.14 | 0.94–1.40 | 0.19 |
| F17 | Lateral ST deviation | 1.08 | 0.85–1.38 | 0.51 |
| F19 | Precordial R-wave progression and combined P-QRS-T-amplitude | 1.07 | 0.87–1.33 | 0.51 |
| F22 | Subtle T-wave changes | 1.02 | 0.83–1.25 | 0.85 |
| F23 | P-wave height and/or shape | 1.13 | 0.93–1.37 | 0.21 |
| F25a | Right bundle branch delay (low values associated with ventricular tachycardia, RBBB, and reduced LVEF) | 0.77 | 0.62–0.95 | 0.02 |
| F26 | Left bundle branch delay | 1.02 | 0.81–1.29 | 0.85 |
| F27a | P- and R-axis deviation (low values associated with AF, junctional bradycardia, ventricular tachycardia, and left axis deviation) | 0.71 | 0.57–0.88 | <0.005 |
| F30 | QR interval | 0.92 | 0.74–1.16 | 0.48 |
| F31 | QRS-T amplitudes | 0.86 | 0.71–1.05 | 0.15 |
| F32a | QRS-T amplitudes (high values associated with microvoltages) | 1.26 | 1.02–1.55 | 0.03 |
Results of Cox regression and explanation of (significant) factors including their association with known electrocardiographic and echocardiographic pathologies as described in Van de Leur and Bos et al.8
Significant.
Figure 3.
Kaplan–Meier curve and examples for the predictive value of factor 8. A Kaplan–Meier (A) for factor 8 and (B) two ECGs of patients with a low and high predicted 5-year LTVA risk are depicted. The values for each factor are depicted below the ECG, along with the outcomes of the patient. The ECG on the left had a low value for factor 8, corresponding to a short PR and P-wave duration: this patient had a low predicted risk of LTVA and did not reach the endpoint. The ECG on the right had a high value for factor 8, corresponding to a broad P-wave with a long PR-interval: this patient had a high predicted risk of LTVA and reached the outcome.
Figure 4.
Study summary figure, including the methods and results. The study population were patients with dilated cardiomyopathy, in which an explainable pre-trained deep neural network (FactorECG) was trained for the outcome of life-threatening ventricular arrhythmias. This network encoded the median beat ECG into 21 factors to generate an ECG using only these factors, allowing to evaluate most characteristics that make up an ECG automatically, in a relatively small dataset. LVAD, left ventricular assist device. HTx, heart transplantation; CRT, cardiac resynchronization therapy; ECG, electrocardiogram; VT, ventricular tachycardia; VF, ventricular fibrillation; SCD, sudden cardiac death; ICD, implantable cardioverter-defibrillator; HR, hazard ratio; UMCU, University Medical Centre Utrecht; MUMC+ = Maastricht University Medical Centre.
LA dimensions
To investigate the possibility that the identified factors were an effect of anatomical substrates of P-wave abnormalities, such as atrial remodelling, first and last LAVI and LA dimensions (by outcome) of complete UMCU cases (n = 219) were plotted (Supplementary material online, Figures S4–S7, respectively). LA’s were significantly larger in the last echocardiography, compared with the first (P = 0.02, Supplementary material online, Figure S6). Next, the relationship of LA dimensions with the significant factors was plotted (Supplementary material online, Figure S8) which showed no association between F8, F15, F25, F27, and F32 and LA dimensions.
Discussion
This is the first study to use an explainable DNNs trained with (baseline) ECGs for LTVA prediction in DCM patients on a multicentre dataset. By using an inherently explainable DNN architecture, we were able to distinguish patients at risk for LTVA whilst allowing interpretation and visualisation of pivotal ECG features.7 The model was able to identify patients at the highest risk with a predominant network focus on P-wave abnormalities. Furthermore, these identified P-wave abnormalities did not correlate to their anatomical analogues (LA dimension/LAVI), suggesting an electrophysiological substrate.
FactorECG findings in relation to prior studies
The FactorECG encompasses the median beat ECG, including most of its features, into 21 generative factors of variation (https://dcm.ecgx.ai/). This novel strategy allows to simultaneously evaluate most characteristics that make up an ECG automatically in much smaller datasets, rather than using selected and human-derived ECG features. Overall, the factors that were most predictive for LTVA primarily encoded for several P-wave characteristics, such as PR-duration, P-wave morphology, and P-wave axis (Figure 2). The combination of reconstructed ECGs together with the hazard ratios allows for a novel in-depth interpretation of a DNN’s features. A high value in F8 for instance, leads to PR-prolongation with a broadened P-wave, whereas a low value in F27 leads to removal of the P-wave, which is associated with atrial fibrillation, a known clinical risk factor for LTVA in DCM.3 Because the baseline model using established ECG variables performed poorly, this indicated that the VAE generative factors are more complex than solely the standard ECG intervals. The combination of the 21 generative factors as well as their interpretation allows for LTVA prediction and feature detection (Figure 3).
The fact that atrial (i.e. P-wave) abnormalities predict ventricular events (i.e. LTVA) may be considered remarkable. However, this association has been described before, and has been thought to be due to shared mechanistic pathologies between atria and ventricles, such as ion-channel abnormalities, or atrioventricular fibrosis due to atrial remodelling.1,3,13 In a recently published population study of 13 580 participants, abnormal P-wave indices were independently associated with LTVA, after adjustment for age, sex, race, and study centre.14 As it is likely that these P-wave indices are caused by atrial remodelling, we investigated the association of anatomical LA characteristics and our identified ECG factors. As expected, LA dimensions increased significantly over time, indicating disease progression. However, we did not find any association to the significant ECG factors, suggesting an exclusive electrophysiological substrate. This is in line with other reports, in which individual ECG P-wave changes were not reliable predictors of anatomic atrial enlargement.15,16
Myocardial fibrosis is often seen in patients with DCM and may cause zones of slow conduction in the myocardium, resulting in zigzag pathways that are prone to causing ventricular tachycardias.17 These cellular mechanisms may be visible on the ECG as increased QRS duration and bundle branch block or low voltages. In this study, F25 reflects increased QRS duration in case of right bundle branch blocks and was associated with LTVA as well. Left bundle branch block (F26), however, was not associated with LTVA. As these patients generally are CRT recipients which were excluded from our analyses, this may have caused an underestimation of the effect of left bundle branch blocks in our model. Lower voltages seem to be reflected in the FactorECG in F32, which is also associated with LTVA. More complex electrocardiographic markers, such as QRS fractionation, T-wave alternans, and QRS-T angle, have also been proposed.1 QRS fractionation and T-wave abnormalities, however, did not appear as an explanatory ECG variable in our model. T-wave alternans (defined as changing T-wave morphology, occurring in each alternant beat) has been repeatedly associated with LTVA in DCM, but cannot be measured in the single ECG median beat that is used in the current research. All these ECG markers are limited by standardization difficulties, which may be decreased by (automatic) interpretation using DNNs.1,4,18,19 As these networks are generally ‘black-box’ algorithms that need very large datasets for training, a strategy of reducing the ECG into its generative factors was used. These interpretable factors were then used in a common statistical model (Cox regression), that allowed for pivotal ECG features to be visualized.
Future studies are warranted to prospectively validate the identified ECG abnormalities and their electrophysiological substrate for LTVA prediction in DCM, including a comparison with accepted risk factors for LTVA. Since longer PR-interval and wide QRS duration were associated with LTVA, assessment of the value of hemiblocks may also be considered. Importantly, the addition of prolonged measurements (such as exercise tests or Holter for T-wave alternans) in DNNs remains to be investigated.
Genotype–phenotype associations
DCM has a genetic basis in 30–50% of cases and specific genotype–phenotype associations are known to lead to arrhythmogenic phenotypes. One study analysed over 75.000 ECGs from the UK Biobank and established several genetic ECG signatures. A polygenic effect on PR-interval for instance, was identified, as well as genetic variants related to the Q-wave in DCM. The strongest Q-wave locus was discovered in BAG3: a gene in which pathogenic variants have been described for DCM with high penetrance and a high risk of progressive heart failure.20 As our VAE model assessed the entire ECG, an interesting significant factor included QRS-T voltages (F32), with high values in this factor associated with microvoltages. These microvoltages are an established ECG characteristic for phospholamban cardiomyopathy, which can lead to both a highly arrhythmogenic DCM phenotype and arrhythmogenic cardiomyopathy.2 Integrating genome and phenome provides unique opportunities to study ECG biology in relation to genetic risk which can be explored by future studies using DNNs.20–22 Furthermore, these studies may pave the way for using artificial intelligence models for risk prediction in DCM patients to estimate an individual’s lifetime (genetic) risk of developing a specific arrhythmogenic DCM phenotype.
Limitations
This study has several limitations to address. Given the nature of retrospective cohorts, data may contain missingness not at random and bias may be present requiring prospective evaluation of the findings. As the UMCU is a heart transplantation centre, this may have caused a selection bias. To account for this, an external cohort was added from the MUMC+ (non-heart transplantation centre) of which the patients logically presented with less severe phenotypes (Table 1). Unfortunately, the characteristics of the implanted ICDs in this population were not available, which may have biased our findings. More importantly, since ICD shocks are not a true surrogate for sudden cardiac death in patients with DCM, the results need confirmation in a study population with fewer ICD carriers or considering only fast events (i.e. >250/min).23 Because DCM is relatively rare, the results may be due to sample size and require confirmation in larger (prospective) studies.
Supplementary Material
Contributor Information
Arjan Sammani, Department of Cardiology, University Medical Centre Utrecht, University of Utrecht, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands.
Rutger R van de Leur, Department of Cardiology, University Medical Centre Utrecht, University of Utrecht, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands.
Michiel T H M Henkens, Department of Cardiology, CARIM, Maastricht University Medical Centre, Maastricht, The Netherlands; Netherlands Heart Institute (NLHI), Utrecht, The Netherlands.
Mathias Meine, Department of Cardiology, University Medical Centre Utrecht, University of Utrecht, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands.
Peter Loh, Department of Cardiology, University Medical Centre Utrecht, University of Utrecht, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands.
Rutger J Hassink, Department of Cardiology, University Medical Centre Utrecht, University of Utrecht, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands.
Daniel L Oberski, Department of Cardiology, University Medical Centre Utrecht, University of Utrecht, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands; Department of Methodology and Statistics, Faculty of Social Sciences, Utrecht University and University Medical Centre Utrecht, Utrecht, The Netherlands.
Stephane R B Heymans, Department of Cardiology, CARIM, Maastricht University Medical Centre, Maastricht, The Netherlands; Netherlands Heart Institute (NLHI), Utrecht, The Netherlands; Department of Cardiovascular Research, University of Leuven, Leuven, Belgium.
Pieter A Doevendans, Department of Cardiology, University Medical Centre Utrecht, University of Utrecht, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands; Netherlands Heart Institute (NLHI), Utrecht, The Netherlands; Central Military Hospital, Utrecht, The Netherlands.
Folkert W Asselbergs, Department of Cardiology, University Medical Centre Utrecht, University of Utrecht, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands; Institute of Cardiovascular Science and Institute of Health Informatics, Faculty of Population Health Sciences, University College London, London, UK.
Anneline S J M te Riele, Department of Cardiology, University Medical Centre Utrecht, University of Utrecht, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands.
René van Es, Department of Cardiology, University Medical Centre Utrecht, University of Utrecht, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands.
Conclusion
To the best of our knowledge, this study is the first to use interpretable DNNs trained with ECGs for LTVA prediction in DCM patients. We observed that the VAE network combined with an interpretable Cox regression can distinguish patients at risk of LTVA. The use of this inherently explainable DNN pipeline allowed interpretation and visualisation of pivotal ECG features.7 While the VAE network encompasses the complete ECG, predictions were mainly driven by P-wave abnormalities that did not correlate with LA dimensions, suggesting an electrophysiological substrate. Future studies are warranted to validate these findings and elucidate their electrophysiological substrate for LTVA prediction in DCM.
Supplementary material
Supplementary material is available at Europace online.
Funding
This study was financed by The Netherlands Organisation for Health Research and Development (ZonMw) with grant number 104021004 and the Dutch Heart Foundation with grant number 2019B011. A.S. is supported by the Alexandre Suerman Stipendium. F.W.A. is supported by UCL Hospitals NIHR Biomedical Research Centre and CVON-AI: 2018B017. P.A.D. is supported by the Fondation Leducq CURE-PLaN. A.S.J.M. t.R. is supported by the Netherlands Heart Foundation (2015T058), the UMC Utrecht Fellowship Clinical Research Talent, and CVON eDETECT. This work is further supported by the Netherlands Cardiovascular Research Initiative, with the support of the Dutch Heart Foundation (CVON 2015-12 eDETECT) and by the focus area of Applied Data Science at Utrecht University, The Netherlands.
Data Availability
The dataset is not available due to patient privacy restrictions.
References
- 1. Sammani A, Kayvanpour E, Bosman LP, Sedaghat-Hamedani F, Proctor T, Gi W, et al. Predicting sustained ventricular arrhythmias in dilated cardiomyopathy: a meta-analysis and systematic review. ESC Heart Fail 2020;7:1430–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McDonagh TA, Metra M, Adamo M, Gardner RS, Baumbach A, Böhm M, et al. 2021 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure: developed by the Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC) With the special contribution of the Heart Failure Association (HFA) of the ESC. Eur Heart J 2021;42:ehab368. [DOI] [PubMed] [Google Scholar]
- 3. Kayvanpour E, Sammani A, Sedaghat-Hamedani F, Lehmann DH, Broezel A, Koelemenoglu J, et al. A novel risk model for predicting potentially life-threatening arrhythmias in non-ischemic dilated cardiomyopathy (DCM-SVA risk). Int J Cardiol 2021;339:75–82. [DOI] [PubMed] [Google Scholar]
- 4. van de Leur RR, Boonstra MJ, Bagheri A, Roudijk RW, Sammani A, Taha K, et al. Big data and artificial intelligence: opportunities and threats in electrophysiology. Arrhythmia Electrophysiol Rev 2020;9:146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ko W-Y, Siontis KC, Attia ZI, Carter RE, Kapa S, Ommen SR, et al. Detection of hypertrophic cardiomyopathy using a convolutional neural network-enabled electrocardiogram. J Am Coll Cardiol 2020;75:722–733. [DOI] [PubMed] [Google Scholar]
- 6. van de Leur RR, Blom LJ, Gavves E, Hof IE, van der Heijden JF, Clappers NC, et al. Automatic triage of 12-lead ECGs using deep convolutional neural networks. J Am Heart Assoc 2020;9:e015138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rudin C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat Mach Intell 2019;1:206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van de Leur RR, Bos MN, Taha K, Sammani A, van Duijvenboden S, Lambiase PD, et al. Inherently explainable deep neural network-based interpretation of electrocardiograms using variational auto-encoders. medRxiv 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van de Leur R, Taha K, Bos MN, van der Heijden JF, Gupta D, Cramer MJ, et al. Discovering and visualizing disease-specific electrocardiogram features using deep learning: proof-of-concept in phospholamban gene mutation carriers. Circ Arrhythm Electrophysiol 2021;14:e009056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sapp JL, Parkash R, Wells GA, Yetisir E, Gardner MJ, Healey JS, et al. Cardiac resynchronization therapy reduces ventricular arrhythmias in primary but not secondary prophylactic implantable cardioverter defibrillator patients: insight from the resynchronization in ambulatory heart failure trial. Circ Arrhythm Electrophysiol 2017;10:e004875. [DOI] [PubMed] [Google Scholar]
- 11. Sammani A, Jansen M, Linschoten M, Bagheri A, de Jonge N, Kirkels H, et al. UNRAVEL: big data analytics research data platform to improve care of patients with cardiomyopathies using routine electronic health records and standardised biobanking. Neth Heart J 2019;27:426–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Higgins I, Matthey L, Pal A, Burgess C, Glorot X, Botvinick M, et al. Beta-VAE: learning basic visual concepts with a constrained variational framework. Conference Track Proceedins 5th International Conference on Learning Representations; 2018.
- 13. Spezzacatene A, Sinagra G, Merlo M, Barbati G, Graw SL, Brun F, et al. Arrhythmogenic phenotype in dilated cardiomyopathy: natural history and predictors of life-threatening arrhythmias. J Am Heart Assoc 2015;4:e002149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maheshwari A, Norby FL, Soliman EZ, Alonso A, Sotoodehnia N, Chen LY. Association of P-wave abnormalities with sudden cardiac and cardiovascular death: the ARIC study. Circ Arrhythm Electrophysiol 2021;14:e009314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsao CW, Josephson ME, Hauser TH, O’Halloran TD, Agarwal A, Manning WJ, et al. Accuracy of electrocardiographic criteria for atrial enlargement: validation with cardiovascular magnetic resonance. J Cardiovasc Magn Reson 2008;10:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Truong QA, Charipar EM, Ptaszek LM, Taylor C, Fontes JD, Kriegel M, et al. Usefulness of electrocardiographic parameters as compared with computed tomography measures of left atrial volume enlargement: from the ROMICAT trial. J Electrocardiol 2011;44:257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de Bakker JM, van Capelle FJ, Janse MJ, Tasseron S, Vermeulen JT, de Jonge N, et al. Slow conduction in the infarcted human heart. “Zigzag” course of activation. Circulation 2018;88:915–926. [DOI] [PubMed] [Google Scholar]
- 18. Pei J, Li N, Gao Y, Wang Z, Li X, Zhang Y, et al. The J wave and fragmented QRS complexes in inferior leads associated with sudden cardiac death in patients with chronic heart failure. Europace 2012;14:1180–1187. [DOI] [PubMed] [Google Scholar]
- 19. Vandenberk B, Robyns T, Goovaerts G, Claeys M, Helsen F, Soest SV, et al. Inter- and intra-observer variability of visual fragmented QRS scoring in ischemic and non-ischemic cardiomyopathy. J Electrocardiol 2018;51:549–554. [DOI] [PubMed] [Google Scholar]
- 20. Verweij N, Benjamins J-W, Morley MP, van de Vegte YJ, Teumer A, Trenkwalder T, et al. The genetic makeup of the electrocardiogram. Cell Syst 2020;11:229–238.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Meder B, Rühle F, Weis T, Homuth G, Keller A, Franke J, et al. A genome-wide association study identifies 6p21 as novel risk locus for dilated cardiomyopathy. Eur Heart J 2014;35:1069–1077. [DOI] [PubMed] [Google Scholar]
- 22. Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med 2020;12:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ellenbogen KA, Levine JH, Berger RD, Daubert JP, Winters SL, Greenstein E, et al. Are implantable cardioverter defibrillator shocks a surrogate for sudden cardiac death in patients with nonischemic cardiomyopathy? Circulation 2006;113:776–782. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset is not available due to patient privacy restrictions.