Abstract
The goals of this review paper on deep learning (DL) in medical imaging and radiation therapy are to (a) summarize what has been achieved to date; (b) identify common and unique challenges, and strategies that researchers have taken to address these challenges; and (c) identify some of the promising avenues for the future both in terms of applications as well as technical innovations. We introduce the general principles of DL and convolutional neural networks, survey five major areas of application of DL in medical imaging and radiation therapy, identify common themes, discuss methods for dataset expansion, and conclude by summarizing lessons learned, remaining challenges, and future directions.
Keywords: computer‐aided detection/characterization; deep learning, machine learning; reconstruction; segmentation; treatment
1. Introduction
In the last few years, artificial intelligence (AI) has been rapidly expanding and permeating both industry and academia. Many applications such as object classification, natural language processing, and speech recognition, which until recently seemed to be many years away from being able to achieve human levels of performance, have suddenly become viable.1, 2, 3 Every week, there is a news story about an AI system that has surpassed humans at various tasks ranging from playing board games4 to flying autonomous drones.5 One report shows that revenues from AI will increase by around 55% annually in the 2016–2020 time period from roughly $8 billion to $47 billion.6 Together with breakthroughs in other areas such as biotechnology and nanotechnology, the advances in AI are leading to what the World Economic Forum refers to as the fourth industrial revolution.7 The disruptive changes associated with AI and automation are already being seriously discussed among economists and other experts as both having the potential to positively improve our everyday lives, for example, by reducing healthcare costs, as well as to negatively affect society, for example, by causing large‐scale unemployment and rising income inequality8, 9 (according to one estimate, half of all working activities can be automated by existing technologies10). The advances in AI discussed above have been almost entirely based on the groundbreaking performance of systems that are based on deep learning (DL). We now use DL‐based systems on a daily basis when we use search engines to find images on the web or talk to digital assistants on smart phones and home entertainment systems. Given its widespread success in various computer vision applications (among other areas), DL is now poised to dominate medical image analysis and has already transformed the field in terms of performance levels that have been achieved across various tasks as well as its application areas.
1.A. Deep learning, history, and techniques
Deep learning is a subfield of machine learning, which in turn is a field within AI. In general, DL consists of massive multilayer networks of artificial neurons that can automatically discover useful features, that is, representations of input data (in our case images) needed for tasks such as detection and classification, given large amounts of unlabeled or labeled data.11, 12
Traditional applications of machine learning using techniques such as support vector machines (SVMs) or random forests (RF) took as input handcrafted features, which are often developed with a reliance on domain expertise, for each separate application such as object classification or speech recognition. In imaging, handcrafted features are extracted from the image input data and reduce the dimensionality by summarizing the input into what is deemed to be the most relevant information that helps with distinguishing one class of input data from another Using the image pixels as the input, the image data can be flattened into a high‐dimensional vector; for example, in mammographic mass classification, a 500 × 500 pixel region of interest will result in a vector with 250,000 elements. Given all the possible variations of a mass's appearance due to differences in breast type, dose, type and size of a mass, etc., finding the hyperplane that separates the high‐dimensional vectors of malignant and benign masses would require a very large number of examples if the original pixel values are used. However, each image can be summarized into a vector consisting of a few dozen or a few hundred elements (as opposed to over a million elements in the original format) by extracting specialized features that for instance describe the shape of the mass. This lower dimensional representation is more easily separable using fewer examples if the features are relevant. A key problem with this general approach is that useful features are difficult to design, often taking the collective efforts of many researchers over years or even decades to optimize. The other issue is that the features are domain or problem specific. One would not generally expect that features developed for image recognition should be relevant for speech recognition, but even within image recognition, different types of problems such as lesion classification and texture identification require separate sets of features. The impact of these limitations has been well demonstrated in experiments that show the performance of top machine learning algorithms to be very similar when they are used to perform the same task using the same set of input features.13 In other words, traditional machine learning algorithms were heavily dependent on having access to good feature representations; otherwise, it was very difficult to improve the state‐of‐the‐art results on a given dataset.
The key difference between DL and traditional machine learning techniques is that the former can automatically learn useful representations of the data, thereby eliminating the need for handcrafted features. What is more interesting is that the representations learned from one dataset can be useful even when they are applied to a different set of data. This property, referred to as transfer learning14, 15, is not unique to DL, but the large training data requirements of DL make it particularly useful in cases where relevant data for a particular task are scarce. For instance, in medical imaging, a DL system can be trained on a large number of natural images or those in a different modality to learn proper feature representations that allow it to “see.” The pretrained system can subsequently use these representations to produce an encoding of a medical image that is used for classification.16, 17, 18 Systems using transfer learning often outperform the state‐of‐the‐art methods based on traditional handcrafted features that were developed over many years with a great deal of expertise.
The success of DL compared to traditional machine learning methods is primarily based on two interrelated factors: depth and compositionality.11, 12, 19 A function is said to have a compact expression if it has few computational elements used to represent it (“few” here is a relative term that depends on the complexity of the function). An architecture with sufficient depth can produce a compact representation, whereas an insufficiently deep one may require an exponentially larger architecture (in terms of the number of computational elements that need to be learned) to represent the same function. A compact representation requires fewer training examples to tune the parameters and produces better generalization to unseen examples. This is critically important in complex tasks such as computer vision where each object class can exhibit many variations in appearance which would potentially require several examples per type of variation in the training set if a compact representation is not used. The second advantage of deep architectures has to do with how successive layers of the network can utilize the representations from previous layers to compose more complex representations that better capture critical characteristics of the input data and suppress the irrelevant variations (for instance, simple translations of an object in the image should result in the same classification). In image recognition, deep networks have been shown to capture simple information such as the presence or absence of edges at different locations and orientations in the first layer. Successive layers of the network assemble the edges into compound edges and corners of shapes, and then into more and more complex shapes that resemble object parts. Hierarchical representation learning is very useful in complicated tasks such as computer vision where adjacent pixels and object parts are correlated with each other and their relative locations provide clues about each class of object, or speech recognition and natural language processing where the sequence of words follow contextual and grammatical rules that can be learned from the data. This distributed hierarchical representation has similarities with the function of the visual and auditory cortexes in the human brain where basic features are integrated into more complex representations that are used for perception.20, 21
As discussed earlier, DL is not a completely new concept, but rather mostly an extension of previously existing forms of artificial neural networks (ANNs) to larger number of hidden layers and nodes in each layer. In the late 1990s until early 2000s, ANNs started to lose popularity in favor of SVMs and decision‐tree‐based methods such as random forests and gradient boosting trees that seemed to be more consistently outperforming other learning methods.22 The reason for this was that ANNs were found to be both slow and difficult to train aside from shallow networks with one to two hidden layers as well as prone to getting stuck in local minima. However, starting around 2006, a combination of several factors led to faster and more reliable training of deep networks. One of the first influential papers was a method for efficient unsupervised (i.e., using unlabeled data, as opposed to supervised training that uses data labeled based on the ground truth) layer by layer training of deep restricted Boltzmann machines.23 As larger datasets became more commonplace, and with availability of commercial gaming graphical processing units (GPUs), it became possible to explore training of larger deeper architectures faster. At the same time, several innovations and best practices in network architecture and training led to faster training of deep networks with excellent generalization performance using stochastic gradient descent. Some examples include improved methods for network initialization and weight updates,24 new neuron activation functions,25 randomly cutting connections or zeroing of weights during training,26, 27 and data augmentation strategies that render the network invariant to simple transformations of the input data. Attention to these improvements was still mostly concentrated within the machine learning community and not being seriously considered in other fields such as computer vision. This changed in 2012 in the ImageNet28 competition in which more than a million training images with 1000 different object classes were made available to the challenge participants. A DL architecture that has since been dubbed AlexNet outperformed the state‐of‐the‐art results from the computer vision community by a large margin and convinced the general community that traditional methods were on their way out.29
The most successful and popular DL architecture in imaging is the convolutional neural network (CNN).30 Nearby pixels in an image are correlated with one another both in areas that exhibit local smoothness and areas consisting of structures (e.g., edges of objects or textured regions). These correlations typically manifest themselves in different parts of the same image. Accordingly, instead of having a fully connected network where every pixel is processed by a different weight, every location can be processed using the same set of weights to extract various repeating patterns across the entire image. These sets of trainable weights, referred to as kernels or filters, are applied to the image using a dot product or convolution and then processed by a nonlinearity (e.g., a sigmoid or tanh function). Each of these convolution layers can consist of many such filters resulting in the extraction of multiple sets of patterns at each layer. A pooling layer (e.g., max pooling where the output is the maximum value within a window) often follows each convolution layer to both reduce the dimensionality and impose translation invariance so that the network becomes immune to small shifts in location of patterns in the input image. These convolution and pooling layers can be stacked to form a multilayer network often ending in one or more fully connected layers as shown in Fig. 1, followed by a softmax layer. The same concepts can be applied in one‐dimensional and three‐dimensional (3D) to accommodate time series and volumetric data, respectively. Compared to a fully connected network, CNNs contain far fewer trainable parameters and therefore require less training time and fewer training examples. Moreover, since their architecture is specifically designed to take advantage of the presence of local structures in images, they are a natural choice for imaging applications and a regular winner of various imaging challenges.
Figure 1.
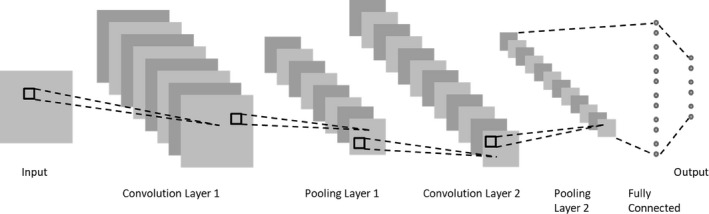
CNN with two convolution layers each followed by a pooling layer and one fully connected layer.
Another very interesting type of network is the recurrent neural network (RNN) which is ideal for analyzing sequential data (e.g., text or speech) due to having an internal memory state that can store information about previous data points. A variant of RNNs, referred to as long short‐term memory (LSTM),31 has improved memory retention compared to a regular RNN and has demonstrated great success across a range of tasks from image captioning32, 33 to speech recognition1, 34 and machine translation.35
Generative adversarial networks (GANs) and its different variants (e.g., WGAN36, CycleGAN37, etc.) are another promising class of DL architectures that consist of two networks: a generator and a discriminator.38 The generator network produces new data instances that try to mimic the data used in training, while the discriminator network tries to determine the probability of whether the generated candidates belong to the training samples or not. The two networks are trained jointly with backpropagation, with the generative network becoming better at generating more realistic samples and the discriminator becoming better at detecting artificially generated samples. GANs have recently demonstrated great potential in medical imaging applications such as image reconstruction for compressed sensing in magnetic resonance imaging (MRI).39
1.B. Deep learning in medical imaging
In medical imaging, machine learning algorithms have been used for decades, starting with algorithms to analyze or help interpret radiographic images in the mid‐1960s.40, 41, 42 Computer‐aided detection/diagnosis (CAD) algorithms started to make advances in the mid 1980s, first with algorithms dedicated to cancer detection and diagnosis on chest radiographs and mammograms,43, 44 and then widening in scope to other modalities such as computed tomography (CT) and ultrasound.45, 46 CAD algorithms in the early days predominantly used a data‐driven approach as most DL algorithms do today. However, unlike most DL algorithms, most of these early CAD methods heavily depended on feature engineering. A typical workflow for developing an algorithm for a new task consisted of understanding what types of imaging and clinical evidence clinicians use for the interpretation task, translating that knowledge into computer code to automatically extract relevant features, and then using machine learning algorithms to combine the features into a computer score. There were, however, some notable exceptions. Inspired by the neocognitron architecture,47 a number of researchers investigated the use of CNNs48, 49, 50, 51 or shift‐invariant ANNs52, 53 in the early and mid‐1990s, and massively trained artificial neural networks (MTANNs)54, 55 in the 2000s for detection and characterization tasks in medical imaging. These methods all shared common properties with current deep CNNs (DCNNs): Data propagated through the networks via convolutions, the networks learned filter kernels, and the methods did not require feature engineering, that is, the inputs into the networks were image pixel values. However, severely restricted by computational requirements of the time, most of these networks were not deep, that is, they mostly consisted of only one or two hidden layers. In addition, they were trained using much smaller datasets compared to a number of high‐profile DCNNs that were trained using millions of natural images. Concepts such as transfer learning,14 residual learning,56 and fully convolutional networks with skip connections57 were generally not well developed. Thus, these earlier CNNs in medical imaging, as competitive as they were compared to other methods, did not result in a massive transformation in machine learning for medical imaging.
With the advent of DL, applications of machine learning in medical imaging have dramatically increased, paralleling other scientific domains such as natural image and speech processing. Investigations accelerated not only in traditional machine learning topics such as segmentation, lesion detection, and classification58 but also in other areas such as image reconstruction and artifact reduction that were previously not considered as data‐driven topics of investigation. Figure 2 shows the number of peer‐reviewed publications in the last 6 yr in the areas of focus for this paper, DL for radiological images, and shows a very strong trend: For example, in the first 3 months of 2018, more papers were published on this topic than the whole year of 2016.
Figure 2.
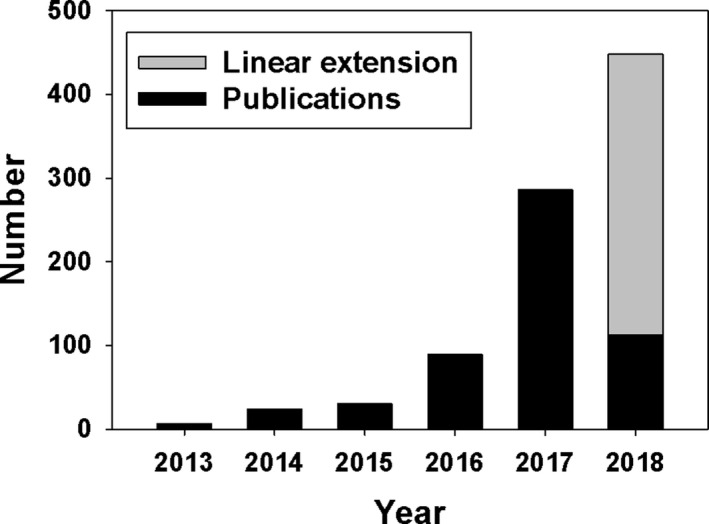
Number of peer‐reviewed publications in radiologic medical imaging that involved DL. Peer‐reviewed publications were searched on PubMed using the criteria (“deep learning” OR “deep neural network” OR deep convolution OR deep convolutional OR convolution neural network OR “shift‐invariant artificial neural network” OR MTANN) AND (radiography OR x‐ray OR mammography OR CT OR MRI OR PET OR ultrasound OR therapy OR radiology OR MR OR mammogram OR SPECT). The search only covered the first 3 months of 2018 and the result was linearly extended to the rest of 2018.
Using DL involves making a very large number of design decisions such as number of layers, number of nodes in each layer (or number and size of kernels in the case of CNNs), type of activation function, type and level of regularization, type of network initialization, whether to include pooling layers and if so what type of pooling, type of loss function, and so on. One way to avoid using trial and error for devising the best architecture is to follow the same exact architectures that have shown to be successful in natural image analysis such as AlexNet,29 VGGNet,59 ResNet,56 DenseNet,60 Xception,61 or Inception V3.62 These networks can be trained from scratch for the new task.63, 64, 65, 66, 67 Alternatively, they can be pretrained on natural images that are more plentiful compared to medical images so that the weights in the feature extraction layers are properly set during training (see Section 3.B for more details). The weights only in the last fully connected layer or last few layers (including some of the convolutional layers) can then be retrained using medical images to learn the class associations for the desired task.
1.C. Existing platforms and resources
A large number of training examples are required to estimate the large number of parameters of a DL system. One needs to perform backpropagation throughout many iterations using stochastic gradient descent over minibatches consisting of a small subset of samples at any given time to train hundreds of thousands to hundreds of millions or even billions of parameters. A single or multicore central processing unit (CPU) or a cluster of CPU nodes in a high‐performance computing (HPC) environment could be used for training, but the former approach would take an extremely long amount of time while the latter requires access to costly infrastructure.
Fortunately, in the last 10 yr gaming, GPUs have become cheaper, increasingly powerful, and easier to program. This has resulted in simultaneously far cheaper hardware requirements for running DL (compared to HPC solutions) and training times that are several orders of magnitude shorter compared to a solution run on a CPU.27, 68 The most common setup for training DL models is therefore to train networks on a desktop workstation containing one or more powerful gaming GPUs that can be easily configured for a reasonable price. There are also several cloud‐based solutions including Amazon Web Services (AWS)69 and Nvidia GPU cloud70 that allow users to train or deploy their models remotely. Recently, Google has developed application‐specific integrated circuit (ASIC) for neural networks to run its wide variety of applications that utilize DL. These accelerators, referred to as tensor processing units (TPUs), are several times faster than CPU or GPU solutions and have recently been made available to general users via Google Cloud.71
In line with the rapid improvements in performance of GPUs, several open‐source DL libraries have been developed and made public that free the user from directly programming GPUs. These frameworks allow the users to focus on how to setup a particular network and explore different training strategies. The most popular DL libraries are TensorFlow,72 Caffe,73 Torch,74 and Theano.75 They all have application programming interfaces (APIs) in different programming languages, with the most popular language being Python.
1.D. Organization of the paper
Throughout the paper, we strived to refer to published journal articles as much as we could. However, DL is a very fast‐changing field, and reports of many excellent and new studies either appear as a conference proceeding paper only, or as a preprint in online resources such as arxiv. We did not refrain from citing articles from these resources whenever necessary. In sections other than Section 2, to better summarize the state‐of‐the‐art, we have included publications from many different medical imaging and natural imaging. However, to keep the length of the paper reasonable, in Section 2, we focused only on applications in radiological imaging and radiation therapy, although there are other areas in medical imaging that have seen influx of DL applications, such as digital pathology and optical imaging. This paper is organized as follows. In Section 2, we summarize applications of DL to radiological imaging and radiation therapy. In Section 3, we describe some of the common themes among DL applications, which include training and testing with small dataset sizes, pretraining and fine tuning, combining DL with radiomics applications, and different types of training, such as supervised, unsupervised, and weakly supervised. Since dataset size is a major bottleneck for DL applications in medical imaging, we have devoted Section 4 to special methods for dataset expansion. In Section 5, we summarize some of the perceived challenges, lessons learned, and possible trends for the future of DL in medical imaging and radiation therapy.
2. Application areas in radiological imaging and radiation therapy
2.A. Image segmentation
DL has been used to segment many different organs in different imaging modalities, including single‐view radiographic images, CT, MR, and ultrasound images.
Image segmentation in medical imaging based on DL generally uses two different input methods: (a) patches of an input image and (b) the entire image. Both methods generate an output map that provides the likelihood that a given region is part of the object being segmented. While patch‐based segmentation methods were initially used, most recent studies use the entire input image to give contextual information and reduce redundant calculations. Multiple works subsequently refine these likelihood maps using classic segmentation methods, such as level sets,76, 77, 78, 79, graph cuts,80 and model‐based methods,81, 82 to achieve a more accurate segmentation than using the likelihood maps alone. Popular DL frameworks used for segmentation tasks include Caffe, Matlab™, and cuda‐convnet.
2.A.1. Organ and substructure segmentation
Segmentation of organs and their substructures may be used to calculate clinical parameters such as volume, as well as to define the search region for computer‐aided detection tasks to improve their performance. Patch‐based segmentation methods, with refinements using traditional segmentation methods, have been shown to perform well for different segmentation tasks.76, 83 Table 1 briefly summarizes published performance of DL methods in organ and substructure segmentation tasks using either Dice coefficient or Jaccard index, if given, as the performance metric.
Table 1.
Organ and substructure segmentation summary and performance using DL
| Region | Segmentation object | Network input | Network architecture basis | Dataset (train/test) | Dice coefficient on test set |
|---|---|---|---|---|---|
| Abdomen | Skeletal muscle89 | Whole image | FCN | 250/150 patients | 0.93 |
| Subcutaneous and visceral fat areas90 | Image patch | Custom | 20/20 patients | 0.92–0.98 | |
| Liver, spleen, kidneys91 | Whole image | Custom | 140 scans fivefold CV | 0.94–0.96 | |
| Bladder | Bladder76 | Image patch | CifarNet | 81/93 patients | 0.86 |
| Brain | Anterior visual pathway92 | Whole image | AE | 165 patients LOO CV | 0.78 |
| Bones86 | Whole image | U‐net | 16 patients LOO CV | 0.94 | |
| Striatum93 | Whole image | Custom | 15/18 patients | 0.83 | |
| Substructures94 | Image patch | Custom | 15/20 patients | 0.86–0.95 | |
| Substructures95 | Image patch | Custom | 20/10 patients | 0.92 | |
| Substructures96 | Image patch | Deep Residual Network92 | 18 patients sixfold CV | 0.69–0.83 | |
| Substructures97 | Whole image | FCN | 150/947 patients | 0.86–0.92 | |
| Breast | Dense tissue and fat98 | Image patch | Custom | 493 images fivefold CV | 0.63–0.95 |
| Breast and fibroglandular tissue85 | Whole image | U‐net | 66 patients threefold CV | 0.85–0.94 | |
| Head and neck | Organs‐at‐risk83 | Image patch | Custom | 50 patients fivefold CV | 0.37–0.90 |
| Heart | Left ventricle79 | Whole image | AE | 15/15 patients | 0.93 |
| Left ventricle82 | Whole image | AE | 15/15 patients | 0.94 | |
| Left ventricle99 | Image patch | Custom | 100/100 patients | 0.86 | |
| Left ventricle100 | Image Patch | Custom | 100/100 patients | 0.88 | |
| Fetal left ventricle101 | Image patch | Custom | 10/41 patients | 0.95 | |
| Right ventricle78 | Whole image | AE | 16/16 patients | 0.82 | |
| Kidney | Kidney102 | Whole image | Custom | 2000/400 patients | 0.97 |
| Kidney103 | Whole image | FCN | 165/79 patients | 0.86 | |
| Knee | Femur, femoral cartilage, tibia, tibial cartilage81 | Whole image | Custom | 60/40 images | – |
| Liver | Liver80 | Image patch | Custom | 78/40 patients | – |
| Liver104 | Image patch | Custom | 109/32 patients | 0.97 | |
| Portal vein83 | Image Patch | Custom | 72 scans eightfold CV | 0.70 | |
| Lung | Lung105 | Whole image | HNN | 62 slices/31 patients | 0.96–0.97 |
| Pancreas | Pancreas106 | Image patch | Custom | 80 patients sixfold CV | 0.71 |
| Pancreas107 | Image patch | Custom | 82 patients fourfold CV | 0.72 | |
| Prostate | Prostate108 | Image patch | AE | 66 patients twofold CV | 0.87 |
| Prostate109 | Image patch | Custom | 30 patients LOO CV | 0.87 | |
| Prostate110 | Whole image | FCN | 41/99 patients | 0.85 | |
| Prostate87 | Whole image | HNN | 250 patients fivefold CV | 0.90 | |
| Rectum | Organs‐at‐risk111 | Whole Image | VGG‐16 | 218/60 patients | 0.88–0.93 |
| Spine | Intervertebral disk112 | Image Patch | Custom | 18/6 scans | 0.91 |
| Whole body | Multiple organs113 | Whole Image | FCN | 228/12 scans | – |
| Multiple organs | Liver and heart (blood pool, myocardium)114 | Whole Image | Custom |
Liver: 20/10 patients Heart: 10/10 patients |
0.74–0.93 |
A “–” on the performance metrics means that the authors report different segmentation accuracy metrics. AE, autoencoder; FCN, fully convolutional network; HNN, holistically nested network; LOO, leave‐one‐out; CV, cross‐validation.
A popular network architecture for segmentation is the U‐net.84 It was originally developed for segmentation of neuronal structures in electron microscope stacks. U‐nets consist of several convolution layers, followed by deconvolution layers, with connections between the opposing convolution and deconvolution layers (skip connections), which allow for the network to analyze the entire image during training, and allow for obtaining segmentation likelihood maps directly, unlike the patch‐based methods. Derivatives of U‐net have been used for multiple tasks, including segmenting breast and fibroglandular tissue85 and craniomaxillofacial bony structures.86
Another DL structure that is being used for segmentation of organs is holistically nested networks (HNN). HNN uses side outputs of the convolutional layers, which are multiscale and multilevel, and produce a corresponding edge map at different scale levels. A weighted average of the side outputs is used to generate the final output, and the weights for the average are learned during the training of the network. HNN has been successfully implemented in segmentation of the prostate87 and brain tumors.88
2.A.2. Lesion segmentation
Lesion segmentation is a similar task to organ segmentation; however, lesion segmentation is generally more difficult than organ segmentation, as the object being segmented can have varying shapes and sizes. Multiple papers covering many different lesion types have been published for DL lesion segmentation (Table 2). A common task is the segmentation of brain tumors, which could be attributed to the availability of a public database with dedicated training and test sets for use with the brain tumor segmentation challenge held by the Medical Image Computing and Computer Assisted Intervention (MICCAI) conference from 2014 to 2016, and continuing in 2017 and 2018. Methods evaluated on this dataset include patch‐based autoencoders,115, 116 U‐net‐based structures117 as well as HNN.88
Table 2.
Lesion segmentation summary and performance using DL
| Region | Segmentation object | Network input | Network architecture basis | Dataset (train/test) | Dice coefficient on test set |
|---|---|---|---|---|---|
| Bladder | Bladder lesion77 | Image patch | CifarNet | 62 patients LOO CV | 0.51 |
| Breast | Breast lesion118 | Image patch | Custom | 107 patients fourfold CV | 0.93 |
| Bone | Osteosarcoma119 | Whole image | ResNet‐50 | 15/8 patients | 0.89 |
| Osteosarcoma120 | Whole image | FCN | 1900/405 images from 23 patients | 0.90 | |
| Brain | Brain lesion121 | Image patch | Custom | 61 patients fivefold CV | 0.65 |
| Brain metastases122 | Image patch | Custom | 225 patients fivefold CV | 0.67 | |
| Brain tumor115 | Image patch | AE |
HGG: 150/69 patients, LGG: 20/23 patients |
HGG: 0.86 LGG: 0.82 |
|
| Brain tumor117 | Image patch | Custom |
HGG: 220, LGG: 54, fivefold CV |
HGG: 0.85–0.91 LGG: 0.83–0.86 |
|
| Brain tumor123 | Whole image | Custom | 30/25 patients | 0.88 | |
| Brain tumor124 | Whole image | FCN | 274/110 patients | 0.82 | |
| Brain tumor88 | Whole image | HNN | 20/10 patients | 0.83 | |
| Ischemic lesions125 | Whole image | DeConvNet | 380/381 patients | 0.88 | |
| Multiple sclerosis lesion126 | Whole image | Custom | 250/77 patients | 0.64 | |
| White matter hyperintensities116 | Image patch | AE | 100/135 patients | 0.88 | |
| White matter hyperintensities127 | Image patch | Custom | 378/50 patients | 0.79 | |
| Head and neck | Nasopharyngeal cancer128 | Whole image | VGG‐16 | 184/46 patients | 0.81–0.83 |
| Thyroid nodule129 | Image patch | HNN | 250 patients fivefold CV | 0.92 | |
| Liver | Liver lesion130 | Image patch | Custom | 26 patients LOO CV | 0.80 |
| Lung | Lung nodule131 | Image patch | Custom | 350/493 nodules | 0.82 |
| Lymph nodes | Lymph nodes132 | Whole image | HNN | 171 patients fourfold CV | 0.82 |
| Rectum | Rectal cancer133 | Image patch | Custom | 70/70 patients | 0.68 |
| Skin | Melanoma134 | Image patch | Custom | 126 images fourfold CV | – |
A “–” on the performance metrics means that the authors report different segmentation accuracy metrics. AE, autoencoder; FCN, fully convolutional network; HNN, holistically nested network; LOO, leave‐one‐out; CV, cross‐validation; HGG, high‐grade glioma; LGG, low‐grade glioma.
2.B. Detection
2.B.1. Organ detection
Anatomical structure detection is a fundamental task in medical image analysis, which involves computing the location information of organs and landmarks in 2D or 3D image data. Localized anatomical information can guide more advanced analysis of specific body parts or pathologies present in the images, that is, organ segmentation, lesion detection, and radiotherapy planning. In a similar fashion to counterparts using traditional machine learning techniques, DL‐based organ/landmark detection approaches can be mainly divided into two groups, that is, classification‐based methods and regression‐based ones. While classification‐based methods focus on discriminating body parts/organs on the image or patch level, regression‐based methods target at recovering more detailed location information, for example, coordinates of landmarks. Table 3 illustrates a list of the DL‐based anatomical structure detection methods together with their performance on different evaluation settings.
Table 3.
Organ and anatomical structure detection summary and performance
| Organ | Detection object | Network input | Network architecture basis | Dataset (train/test) | Error (mean ± SD) |
|---|---|---|---|---|---|
| Bone | 37 hand landmarks147 | X‐ray images | Custom CNN | 895 images threefold CV | 1.19 ± 1.14 mm |
| Femur bone135 | MR 2.5D image patches | Custom 3D CNN | 40/10 volumes | 4.53 ± 2.31 mm | |
| Vertebrae148 | MR/CT image patches | Custom CNN | 1150 patches/110 images | 3.81 ± 2.98 mm | |
| Vertebrae149 | US/x‐ray images | U‐Net | 22/19 patients | F1:0.90 | |
| Vessel | Carotid artery150 | CT 3D image patches | Custom 3D CNN | 455 patients fourfold CV | 2.64 ± 4.98 mm |
| Ascending aorta139 | 3D US | Custom CNN | 719/150 patients | 1.04 ± 0.50 mm | |
| Fetal anatomy | Abdominal standard scan plane136, 151 | US image patches | Custom CNN | 11942/8718 images | F1:0.71136, 0.75151 |
| 12 standard scan planes137 | US images | Custom CNN | 800/200 images | F1:0.42–0.93 | |
| 13 standard scan planes138 | US images | AlexNet | 5229/2339 images | Acc: 0.10–0.94 | |
| Body | Body parts152 | CT images | AlexNet + FCN | 450/49 patients | 3.9 ± 4.7 voxels |
| Body parts153 | CT images | AlexNet | 3438/860 images | AUC: 0.998 | |
| Multiple Organ154 | 3D CT images | Custom CNN | 200/200 scans | F1:0.97 | |
| Body parts141, 142 | CT images | LeNet | 2413/4043 images | F1:0.92 | |
| Brain | Brain landmarks155 | MR images | FCN | 350/350 images | 2.94 ± 1.58 mm |
| Lung | Pathologic Lung156 | CT images | FCN | 929 scans fivefold CV | 0.76 ± 0.53 mm |
| Extremities | Thigh muscle157 | MR images | FCN | 15/10 patients | 1.4 ± 0.8 mm |
| Heart | Ventricle landmarks143, 144, 145 | MRI images | Custom CNN + RL | 801/90 images | 2.9 ± 2.4 mm |
FCN, fully convolutional network; RL, reinforcement learning; F1, harmonic average of the precision (positive predictive value) and recall (sensitivity); AUC, area under the receiver operating characteristic curve; CV, cross‐validation.
Early classification‐based approaches often utilized off‐the‐shelf CNN features to classify image or image patches that contain anatomical structures. Yang et al.135 adopted a CNN classifier to locate two‐dimensional (2D) image patches (extracted from 3D MR volumes) that contain possible landmarks as an initialization of the follow‐up segmentation process for the femur bone. Chen et al.136 adopted an ImageNet pretrained model and fine‐tuned the model using fetal ultrasound frames from recorded scan videos to classify the fetal abdominal standard plane images.
A variety of information in addition to original images could also be included to help the detection task. For the same standard plane detection task in fetal ultrasound, Baumgartner et al.137 proposed a joint CNN framework to classify 12 standard scan planes and also localize the fetal anatomy using a series of ultrasound fetal midpregnancy scans. The final bounding boxes were generated based on the saliency maps computed as the visualization of network activation for each plane class.
Improvements were also achieved by adapting the CNN network with more advanced architecture and components. Kumar et al.138 composed a two‐path CNN network with features computed from both original images and pregenerated saliency maps in each path. The final standard plane classification was performed using SVM on a set of selected features.
Another category of methods tackle the anatomy detection problems with regression analysis techniques. Ghesu et al.139 formulated the 3D heart detection task as a regression problem, targeting at the 3D bounding box coordinates and affine transform parameters in transesophageal echocardiogram images. This approach integrated marginal space learning into the DL framework and learned sparse sampling to reduce computational cost in the volumetric data setting.140
Yan et al.141, 142 formulated body part localization using DL. The system was developed using an unsupervised learning method with two intersample CNN loss functions. The unsupervised body part regression built a coordinate system for the body and output a continuous score for each axial slice, representing the normalized position of the body part in the slice.
Besides the two common categories of methods discussed above, modern techniques (e.g., reinforcement learning) are also adopted to tackle the problem from a different direction. Ghesu et al.143 present a good example of combining reinforcement learning and DL in anatomical detection task. With the application in multiple image datasets across a number of different modalities, the method could search the optimal paths from a random starting point to the predefined anatomical landmark via reinforcement learning with the help of effective hierarchical features extracted via DCNN models. Furthermore, the system was further extended to search 3D landmark positions with 3D volumetric CNN features.144, 145 Later on, Xu et al.146 further extended this approach by turning the optimal action path searching problem into an image partitioning problem, in which a global action map across the whole image was constructed and learned by a DCNN network to guide the searching action.
2.B.2. Lesion detection
Detection of abnormalities (including tumors and other suspicious growths) in medical images is a common but costly and time‐consuming part of the daily routine of physicians, especially radiologists and pathologists. Given that the location is often not known a priori, the physician should search across the 2D image or 3D volume to find deviations compared to surrounding tissue and then to determine whether that deviation constitutes an abnormality that requires follow‐up procedures or something that can be dismissed from further investigation. This is often a difficult task that can lead to errors in many situations either due to the vast amount of data that needs to be searched to find the abnormality (e.g., in the case of volumetric data or whole‐slide images) or because of the visual similarity of the abnormal tissue with normal tissue (e.g., in the case of low‐contrast lesions in mammography). Automated computer detection algorithms have therefore been of great interest in the research community for many years due to their potential for reducing reading costs, shortening reading times and thereby streamlining the clinical workflow, and providing quality care for those living in remote areas who have limited access to specialists.
Traditional lesion detection systems often consist of long processing pipelines with many different steps.158, 159 Some of the typical steps include preprocessing the input data, for example, by rescaling the pixel values or removing irrelevant parts of the image, identification of locations in the image that are similar to the object of interest according to rule‐based methods, extraction of handcrafted features, and classification of the candidate locations using a classifier such as SVM or RF. In comparison, DL approaches for lesion detection are able to avoid the time‐consuming pipeline design approach. Table 4 presents a list of studies that used DL for lesion detection, along with some details about the DL architecture.
Table 4.
Lesion detection using DL
| Detection organ | Lesion type | Dataset (train/test) | Network input | Network architecture basis |
|---|---|---|---|---|
| Lung and thorax | Pulmonary nodule | 888 patients fivefold CV168 | Image patch168, 169, 173, 174, 175, 176, 177Whole image178, 179, 180 | CNN168, 169, 173, 175, 176, 177, 178, 179, 180SDAE/CNN174 |
| 888 patients tenfold CV169 | ||||
| 303 patients tenfold CV173 | ||||
| 2400 images tenfold CV174 | ||||
| 104 patients fivefold CV175 | ||||
| 1006 patients tenfold CV176 | ||||
| Multiple pathologies | 35,038/2,443 radiographs178 | |||
| 76,000/22,000 chest x rays180 | ||||
| ImageNet Pretraining, 433 patients LOO CV181 | ||||
| Tuberculosis | 685/151 chest radiographs179 | |||
| Brain | Cerebral aneurism | 300/100 magnetic resonance angiography images182 | Image patch182Whole image170, 172 | CNN182FCN/CNN170, 172 |
| Cerebral microbleed | 230/50 brain MR scans172 | |||
| Lacune | 868/111 brain MR scans170 | |||
| Breast | Solid cancer | 40,000/18,000 mammographic images64 | Image patch17, 64, 183Whole image66, 161 | CNN17, 64, 66, 183FCN/CNN161 |
| 161/160 breast MR images183 | ||||
| Mass | Pretraining on ~2300 mammography images, 277/47 DBT cases17 | |||
| ImageNet pretraining, 306/163 breast ultrasounds images161 | ||||
| Malignant mass and mircocalcification | ImageNet Pretraining, 3476/115 FFDM images66 | |||
| Colon | Polyp | 394/792 CT colonography cases166 | Whole image,184Image patch166, 185 | CNN166, 184, 185 |
| 101 CT colonography cases; tenfold CV185 | ||||
| Colitis | ImageNet Pretraining, 160 abdominal CT cases; fourfold CV184 | |||
| Multiple | Lymph node | ImageNet Pretraining, 176 CT cases; threefold CV160 | Image patch160, 166, 186 | CNN160, 166, 186 |
| 69/17 abdominal CT cases166 | ||||
| 176 abdominal CT cases; threefold CV186 | ||||
| Liver | Tumor | NA/37187 | Image patch187 | CNN187 |
| Thyroid | Nodule | 21,523 ultrasound images; tenfold CV188 | Image patch188 | CNN188 |
| Prostate | Cancer | 196 MR cases; tenfold CV189 | Whole image189 | FCN189 |
| Pericardium | Effusion | 20/5 CT cases190 | Whole image190 | FCN190 |
| Vascular | Calcification | ImageNet pretraining; 84/28191 | Image patch191 | FCN191 |
SDAE, stacked denoising autoencoder; FCN, fully convolutional network; LOO, leave‐one‐out; CV, cross‐validation.
Many of the papers focused on detection tasks use transfer learning with architectures from computer vision.160 Examples of this approach can be found in many publications, including those for lesion detection in breast ultrasound,161 for the detection of bowel obstructions in radiography,162 and for the detection of the third lumbar vertebra slice in a CT scan.163 Usage of CNNs in lesion detection is not limited to architectures taken directly from computer vision but also includes some applications where custom architectures are used.164, 165, 166, 167
Most of the early applications used 2D CNNs, even if the data were 3D. Due to prior experience with 2D architectures, limitations in the amount of available memory of GPUs, and higher number of samples needed for training the larger number of parameters in a 3D architecture, many DL systems used multiview 2D CNNs for analysis of CT and MRI datasets in what is referred to as 2.5D analysis. In these methods, orthogonal views of a lesion or multiple views at different angles through the lesion were used to train an ensemble of 2D CNNs whose scores would be merged together to obtain the final classification score.166, 168 More recently, 3D CNNs that use 3D convolution kernels are successfully replacing 2D CNNs for volumetric data. A common approach to deal with the small number of available cases is to train the 3D CNNs on 3D patches extracted from each case. This way, each case can be used to extract hundreds or thousands of 3D patches. Combined with various data augmentation methods, it is possible to generate sufficient number of samples to train 3D CNNs. Examples of using 3D patches can be found for the detection of pulmonary nodules in chest CT169 and for the detection of lacunar strokes in brain MRI.170 Due to the large size of volumetric data, it would be very inefficient to apply the CNN in a sliding window fashion across the entire volume. Instead, once the model is trained on patches, the entire network can be converted into a fully convolutional network171 so that the whole network acts as a convolution kernel that can be efficiently applied to an input of arbitrary size. Since convolution operations are highly optimized, this results in fast processing of the entire volume when using a 3D CNN on volumetric data.172
2.C. Characterization
Over the past decades, characterization of diseases has been attempted with machine learning leading to computer‐aided diagnosis (CADx) systems. Radiomics, the –omics of images, is an expansion of CADx to other tasks such as prognosis and cancer subtyping. Radiomic features can be described as (a) “hand‐crafted”/“engineered”/“intuitive” features or (b) deep‐learned features. Characterization of disease types will depend on the specific disease types and the clinical question. With handcrafted radiomic features, the features are devised based on imaging characteristics typically used by radiologists in their interpretation of a medical image. Such features might include tumor size, shape, texture, and/or kinetics (for dynamic contrast‐enhanced imaging). Various review papers have already been written about these handcrafted radiomic features that are merged with classifiers to output estimates of, for example, the likelihood of malignancy, tumor aggressiveness, or risk of developing cancer in the future.158, 159
DL characterization methods may take as input a region of the image around the potential disease site, such as a region of interest (ROI) around a suspect lesion. How that ROI is determined will likely affect the training and performance of the DL. Thinking of how a radiologist is trained during residency will lend understanding of how a DL system needs to be trained. For example, an ROI that is cropped tightly around a tumor will provide different information to a DL system than an ROI that is much larger than the encompassing tumor since with the latter, more anatomical background is also included in the ROI.
More and more DL imaging papers are published each year, but there are still only a few methods that are able to characterize among the vast range of radiological presentations across subtle disease states. Table 5 presents a list of published DL characterization studies in radiological imaging.
Table 5.
Characterization using DL
| Anatomic site | Object or task | Network input | Network architecture | Dataset (train/test) |
|---|---|---|---|---|
| Breast | Cancer risk assessment192 | Mammograms | Pretrained Alexnet followed by SVM | 456 patients LOO CV |
| Cancer risk assessment193 | Mammograms | Modified AlexNet | 14,000/1850 images randomly selected 20 times | |
| Cancer risk assessment194 | Mammograms | Custom DCNN | 478/183 mammograms | |
| Cancer risk assessment195 | Mammograms | Fine‐tuned a pretrained VGG16Net | 513/91 women | |
| Diagnosis196 | Mammograms | Pretrained AlexNet followed by SVM | 607 cases fivefold CV | |
| Diagnosis197 | Mammograms, MRI, US | Pretrained VGG19Net followed by SVM | 690 MRI, 245 FFDM 1125 US, LOO CV | |
| Diagnosis198 | Breast tomosynthesis | Pretrained Alexnet followed by evolutionary pruning | 2682/89 masses | |
| Diagnosis199 | Mammograms | Pretrained AlexNet | 1545/909 masses | |
| Diagnosis200 | MRI MIP | Pretrained VGG19Net followed by SVM | 690 cases with fivefold CV | |
| Diagnosis201 | DCE‐MRI | LSTM | 562/141 cases | |
| Solitary cyst diagnosis202 | Mammograms | Modified VGG Net | 1600 lesions eightfold CV | |
| Prognosis203 | Mammograms | VGG16Net followed by logistic regression classifier | 79/20 cases randomly selected 100 times | |
| Chest — lung | Pulmonary nodule classification204 | CT patches | ResNet | 665/166 nodules |
| Tissue classification205 | CT patches | Restricted Boltzmann machines | Training 50/100/150/200; testing 20,000/1000/20,000/20,000 image patches | |
| Interstitial disease206 | CT patches | Modified AlexNet | 100/20 patients | |
| Interstitial disease207 | CT patches | Modified VGG |
Public: 71/23 scans Local: 20/6 scans |
|
| Interstitial disease208 | CT patches | Custom | 480/(120 and 240) | |
| Interstitial disease209 | CT patches | Custom | 36,106/1050 patches | |
| Pulmonary nodule staging210 | CT | DFCNet | 11/7 patients | |
| Prognosis211 | CT | Custom | 7983/(1000 and 2164) subjects | |
| Chest — cardiac | Calcium scoring212 | CT | Custom | 1181/506 scans |
| Ventricle quantification213 | MR | Custom (CNN + RNN + Bayesian multitask) | 145 cases, fivefold CV | |
| Abdomen | Tissue classification214 | Ultrasound | CaffeNet and VGGNet | 136/49 Studies |
| Liver tumor classification215 | Portal Phase 2D CT | GAN | 182 cases, threefold CV | |
| Liver Fibrosis216 | DCE‐CT | Custom CNN | 460/100 scans | |
| Fatty liver disease217 | US | Invariant scattering convolution network | 650 patients, five‐ and tenfold CV | |
| Brain | Survival218 | Multiparametric MR | Transfer learning as feature extractor, CNN‐S | 75/37 patients |
| Skeletal | Maturity219 | Hand radiographs | Deep residual network | 14,036/(200 and 913) examinations |
FCN, fully convolutional network; LOO, leave‐one‐out; CV, cross‐validation.
2.C.1. Lesion characterization
When it comes to computer algorithms and specific radiological interpretation tasks, there is no one‐size‐fits‐all for either conventional radiomic machine learning methods or DL approaches. Each computerized image analysis method requires customizations specific to the task as well as the imaging modality.
Lesion characterization is mainly being conducted using conventional CAD/radiomics computer algorithms, especially when the need is to characterize (i.e., describe) a lesion rather than conduct further machine learning for disease assessment. For example, characterization of lung nodules and characterization of the change in lung nodules over time are used to track the growth of lung nodules in order to eliminate false‐positive diagnoses of lung cancer.
Other examples involving computer characterization of tumors occur in research in imaging genomics. Here, radiomic features of tumors are used as image‐based phenotypes for correlative and association analysis with genomics as well as histopathology. A well‐documented, multiinstitutional collaboration on such was conducted through the TCGA/TCIA Breast Phenotype Group.220, 221, 222, 223, 224
Use of DL methods as feature extractors can lend itself to tumor characterization; however, the extracted descriptors (e.g., CNN‐based features) are not intuitive. Similar to “conventional” methods that use handcrafted features, DL‐extracted features could characterize a tumor relative to some known trait — such as receptor status — during supervised training, and that subsequent output could be used in imaging genomics discovery studies.
Additional preprocessing and data use methods can further improve characterization such as in the past use of unlabeled data with conventional features to enhance the machine learning training.225, 226 Here, the overall system can learn aspects of the data structure without the knowledge of the disease state, leaving the labeled information for the final classification step.
2.C.2. Tissue characterization
Tissue characterization is sought when specific tumor regions are not relevant. Here, we focus on the analysis of nondiseased tissue to predict a future disease state (such as texture analysis on mammograms in order to assess the parenchyma with the goal to predict breast cancer risk159) and the characterization of tissue that includes diffuse disease, such as in various types of interstitial lung disease and liver disease.227, 228
In breast cancer risk assessment, computer‐extracted characteristics of breast density and/or breast parenchymal patterns are computed and related to breast cancer risk factors. Using radiomic texture analysis, Li et al. have found that women at high risk for breast cancer have dense breasts with parenchymal patterns that are coarse and low in contrast.229 DL is now being used to assess breast density.194, 195 In addition, parenchymal characterization is being conducted using DL, in which the parenchymal patterns are related through the CNN architecture to groups of women using surrogate markers of risk. One example is shown by Li et al. assessing the performance of DL in the distinction between women at normal risk of breast cancer and those at high risk based on their BRCA1/2 status.192
Lung tissue has been analyzed with conventional texture analysis and DL for a variety of diseases. Here, characterization of the lung pattern lends itself to DL as patches of the lung may be informative of the underlying disease, commonly interpreted by the radiologist's eye–brain system. Various investigators have developed CNNs, including those to classify interstitial lung diseases characterized by inflammation of the lung tissue.207, 208, 209 These disease characterizations can include healthy tissue, ground glass opacity, micronodules, consolidation, reticulation, and honeycombing patterns.179
Assessing liver tissue lends itself to DCNNs in the task of staging liver fibrosis on MRI by Yasaka et al.216 and ultrasonic fatty liver disease characterization by Bharath et al.217
2.C.3. Diagnosis
Computer‐aided diagnosis (CADx) involves the characterization of a region or tumor, initially indicated by either a radiologist or a computer, after which the computer characterizes the suspicious region or lesion and/or estimates the likelihood of being diseased (e.g., cancerous) or nondiseased (e.g., non‐cancerous), leaving the patient management to the physician.158, 159 Note that CADx is not a localization task but rather a characterization/classification task. The subtle difference between this section and the preceding two sections is that here the output of the machine learning system is related to the likelihood of disease and not just a characteristic feature of the disease presentation.
Many review papers have been written over the past two decades on CADx, radiomic features, and machine learning,158, 159 and thus, details will not be presented in this paper.
An active area of DL is CADx of breast cancer. Training CNNs “from scratch” is often not possible for CAD and other medical image interpretation tasks, and thus, methods to use CNNs trained on other data (transfer learning) are considered. Given the initial limited datasets and variations in tumor presentations, investigators explored the use of transfer learning to extract tumor characteristics using CNNs trained on nonmedical tasks. The outputs from layers can be considered as characteristic features of the lesion and serve as input to classifiers, such as linear discriminant analysis and SVMs. Figure 3(a) shows an example in which AlexNet is used as a feature extractor for an SVM, and Fig. 3(b) shows the performance of the SVM based on features from each layer of AlexNet.
Figure 3.
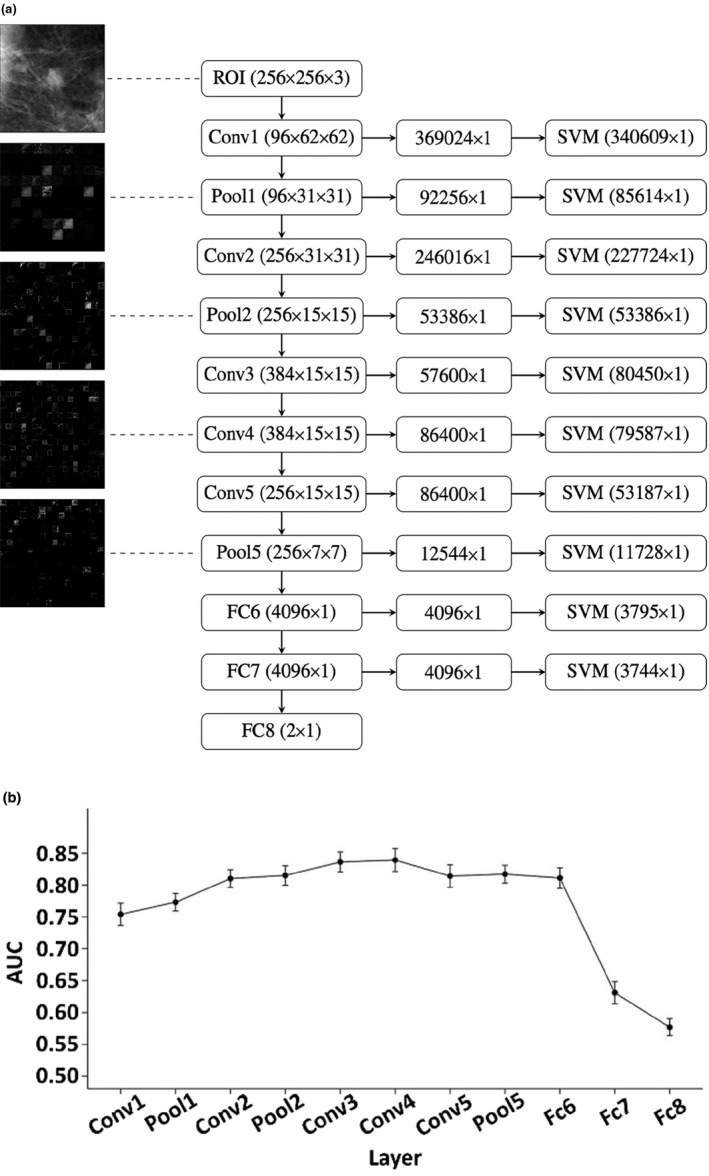
Use of CNN as a feature extractor.196 (a) Each ROI is sent through AlexNet and the outputs from each layer are preprocessed to be used as sets of features for an SVM. The filtered image outputs from some of the layers can be seen in the left column. The numbers in parentheses for the center column denote the dimensionality of the outputs from each layer. The numbers in parentheses for the right column denote the length of the feature vector per ROI used as an input for the SVM after zero‐variance removal. (b) Performance in terms of area under the receiver operating characteristic curve for classifiers based on features from each layer of AlexNet in the task of distinguishing between malignant and benign breast tumors.
Researchers have found that performance of the conventional radiomics CADx and that of the CNN‐based CADx yielded similar levels of diagnostic performance in the task of distinguishing between malignant and benign breast lesions, and thus when combined, via a deep feature fusion methodology, gave a statistically significant level of performance.196, 197 Figure 4 shows one possible method for combining CNN‐extracted and conventional radiomics features.
Figure 4.
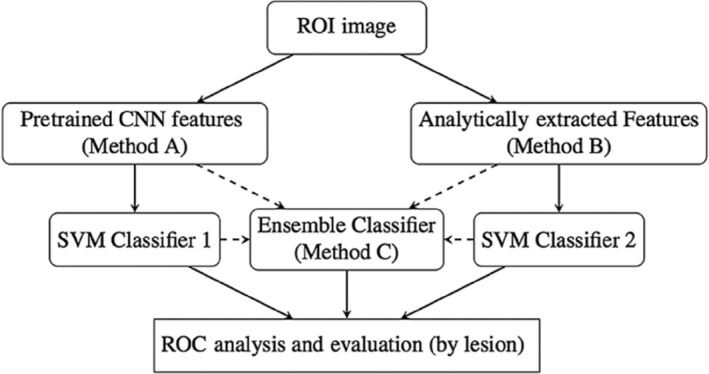
CNN‐extracted and conventional features can be combined in a number of ways, including a traditional classifier such as an SVM.196
In an effort to augment, under limited dataset constraints, CNN performance with dynamic contrast‐enhanced MRI, investigators have looked to vary the image types input to the CNN. For example, instead of replicating a single image region to the three RGB channels of VGG19Net, investigators have used the temporal images obtained from DCE‐MRI, inputting the precontrast, the first postcontrast, and the second postcontrast MR images to the RGB channels, respectively. In addition, to exploit the four‐dimensional nature of DCE‐MRI (3D and temporal), Antropova et al. have input MIP (maximum intensity projections) images to the CNN.200 Incorporation of temporal information into the DL efforts has resulted in the use of RNN, such as LSTM recurrent networks.201, 230
Instead of using transfer learning for feature extraction, investigators have used transfer learning for fine tuning by either (a) freezing the earlier layers of a pretrained CNN and training the later layers, that is, fine tuning or (b) training on one modality, such as digitized screen/film mammography (dSFM), for use on a related modality, such as full‐field digital mammography (FFDM). The latter has been shown by Samala et al.199 to be useful in the training of CNN‐based CADx for lesion diagnosis on FFDMs.
Investigations on DL for CADx are continuing across other cancers, that is, lung cancer, and other disease types, and similar methods can be used.204, 205, 206, 207, 208, 209, 210, 211, 212, 213, 214, 215, 216, 217, 218, 219 The comparison to more conventional radiomics‐based CADx is also demonstrated further, which is potentially useful for both understanding the CNN outputs and providing additional decision support.
2.C.4. Prognosis and staging
Once a cancer is identified, further workup through biopsies gives information on stage, molecular subtype, and/or genomics to yield information on prognosis and potential treatment options. Cancers are spatially heterogeneous, and therefore, investigators are interested whether imaging can provide information on that spatial variation. Currently, many imaging biomarkers of cancerous tumors include only size and simple enhancement measures (if dynamic imaging is employed), and thus, there is interest in expanding, through radiomic features, the knowledge that can be obtained from images. Various investigators have used radiomics and machine learning in assessing the stage and prognosis of cancerous tumors.220, 231 Now, those analyses are being investigated further with DL. It is important to note that when using DL to assess prognosis, one can analyze the tumor from medical imaging, such as MRI or ultrasound, or from pathological images. Also, in the evaluation, one needs to determine the appropriate comparison — a radiologist, a pathologist, or some other histopathological/genomics test.
The goal is to better understand the imaging presentation of cancer, that is, to obtain prognostic biomarkers from image‐based phenotypes, including size, shape, margin morphology, enhancement texture, kinetics, and variance kinetic phenotypes. For example, enhancement texture phenotypes can characterize the tumor texture pattern of contrast‐enhanced tumors on DCE‐MRI though an analysis of the first postcontrast images, and thus quantitatively characterize the heterogeneous nature of contrast uptake within the breast tumor.220 Here, the larger the enhancement texture entropy, the more heterogeneous is the vascular uptake pattern within the tumor, which potentially reflects the heterogeneous nature of angiogenesis and treatment susceptibility, and serves as a location‐specific “virtual digital biopsy.” Understanding the relationships between image‐based phenotypes and the corresponding biopsy information could potentially lead to discoveries useful for assessing images obtained during screening as well as during treatment follow‐up, that is, when an actual biopsy is not practical.
Shi et al.203 demonstrated the prediction of prognostic markers using DL on mammography in distinguishing between DCIS with occult invasion from pure DCIS. Staging on thoracic CTs is being investigated by Masood et al. through DL by relating CNN output to metastasis information for pulmonary nodules.210 In addition, Gonzalez et al. evaluated DL on thoracic CTs in the detection and staging of chronic obstructive pulmonary disease and acute respiratory disease.211 While the use of DL in the evaluation of thoracic CTs is promising, more development is needed to reach clinical applicability.
2.C.5. Quantification
Use of DL in quantification requires a CNN output that correlates significantly with a known quantitative medical measurement. For example, DL has been used in automatic calcium scoring in low‐dose CTs by Lessmann et al.212 and in cardiac left ventricle quantification by Xue et al.213 Similar to cancer workup, in cardiovascular imaging, use of DL is expected to augment clinical assessment of cardiac defect/function or uncover new clinical insights.232 Larson et al. turned to DL to assess skeletal maturity on pediatric hand radiographs with performance levels rivaling that of an expert radiologist.219 DL has been used to predict growth rates for pancreatic neuroendocrine tumors233 on PET‐CT scans.
2.D. Processing and reconstruction
In the previous parts of this section, we focused on applications in which image pixels or ROIs are classified into multiple classes (e.g., segmentation, lesion detection, and characterization), the subject is classified into multiple classes (e.g., prognosis, staging), or a feature in the image (or the ROI) is quantified. In this part, we focus on applications in which the output of the machine learning algorithm is also an image (or a transformation) that potentially has a quantifiable advantage over no processing or traditional processing methods.Table 6 presents a list of studies that used DL for image processing or reconstruction, and that produced an image as the DL algorithm output.
Table 6.
Image processing and reconstruction with DL
| Task | Imaging modality | Performance measure | Network output | Network architecture basis |
|---|---|---|---|---|
| Filtering | CT,234Chest x ray,235x ray fluoro236 | MSE234, CAD performance,234PSNR,235, 236 SSIM,235, 236Runtime236 | Likelihood of nodule,234Bone image,235CLAHE filtering236 | Custom CNN,234, 235Residual CNN,236Residual AE236 |
| Noise reduction | CT,237, 238, 239, 240PET241 | PSNR,237, 238, 239, 240, 241 RMSE,237, 238 SSIM,237, 238, 240 NRMSE,239 NMSE241 | Noise‐reduced image237, 238, 239, 240, 241 | Custom CNN,237, 238, 239Residual AE,237, 238Concatenated CNNs,241U‐net240 |
| Artifact reduction | CT,242, 243MRI244 | SNR,242, 243 NMSE,244 Qualitative,243 Runtime244 | Sparse‐view recon,242, 244 Metal artifact reduced image243 | U‐net,242, 244Custom CNN243 |
| Recons | MRI245, 246, 247, 248 | RMSE,245, 248 runtime,245 MSE,246, 247 NRMSE,246 SSIM,246 SNR248 | Image of scalar measures,245 MR reconstruction246, 247, 248 | Custom CNN,245, 248Custom NN,246Cascade of CNNs247 |
| Registration | MRI249, 250, 251, 252x‐ray to 3D253, 254 | DICE,249, 250 Runtime,250 Target Overlap,251 SNR,252 TRE,254 Image and vessel sharpness,252 mTREproj253 | Deformable registration,249, 250, 251, 252 Rigid body 3D transformation253, 254 | Custom CNN,249, 251, 252, 253, 254SAE250 |
| Synthesis of one modality from another | CT from MRI,255, 256, 257, 258, 259MRI from PET,260PET from CT261 | MAE,255, 256 PSNR,255, 259 ME,256 MSE,256 Pearson correl,256 PET image Quality,257, 258 SSIM,260 SUVR of MR‐less methods,260 Tumor detection by radiologist261 | Synthetic CT,255, 256, 257, 258Synthetic MRI,260Synthetic PET261 | Custom 3D FCN,255GAN,259, 260, 261U‐net,256, 257AE258 |
| Image quality assessment |
US,262 MRI265 |
AUC,262, 264 IOU,262 Correlation between TRE estimation and ground truth,263 Concordance with readers265 | ROI localization and classification,262 TRE estimation,263 Estimate of image diagnostic value264, 265 | Custom CNN,262, 265Custom NN,263VGG19264 |
MSE, mean‐squared error; RMSE, Root MSE; NSME, normalized MSE; NRMSE, normalized RMSE; SNR, signal‐to‐noise ratio; PSNR, peak SNR; SSIM, structural similarity; DICE, segmentation overlap index; TRE, target registration error; mTREproj, mean TRE in projection direction; MAE, mean absolute error; ME, mean error; SUVR, standardized uptake value ratio; AUC, area under the receiver operating characteristic curve; IOU, intersection over union; CLAHE, contrast‐limited adaptive histogram equalization.
2.D.1. Filtering, noise/artifact reduction, and reconstruction
Filtering
Going back to the early days of application of CNNs to medical images, one can find the examples of CNNs that produced output images for further processing. Zhang et al.52 trained a shift‐invariant ANN that aimed at having a high or low pixel value in an output image depending on whether the pixel was determined to be the center of a microcalcification by an expert mammographer. Suzuki et al.234 trained an MTANN as a supervised filter for the enhancement lung nodules on thoracic CT scans. More recently, Yang et al.235 used a cascade of CNNs for bone suppression in chest radiography. Using ground truth images extracted from dual‐energy subtraction chest x rays, the authors trained a set of multiscale networks to predict bone gradients at different scales and fuse these results to obtain a bone image from a standard chest x ray. Another advantage of CNNs for image filtering is speed; Mori236 investigated several types of residual convolutional autoencoders and residual CNNs for contrast‐limited adaptive histogram equalization filtering and denoising of x‐ray fluoroscopic imaging during treatment, without specialized hardware.
Noise reduction
The past couple of years have seen a proliferation of applications of DL to improve the noise quality of reconstruction medical images. One application area is low‐dose image reconstruction. This is important in modalities with ionizing radiation such as CT or PET for limiting patient dose,237, 238, 239, 241 or for limiting damage to samples in synchrotron‐based x‐ray CT.240 Chen et al.237 designed a DL algorithm for noise reduction in reconstructed CT images. They used the mean‐squared pixelwise error between the ideal image and the denoised image as the loss function, and synthesized noisy projections based on patient images to generate training data.238 They later combined a residual autoencoder with a CNN in an architecture called the RED‐CNN,238 which has a stack of encoders and a symmetrical stack of decoders that are connected with shortcuts for the matching layers. Kang et al.239 applied a DCNN to the wavelet transform coefficients of low‐dose CT images and used a residual learning architecture for faster network training and better performance. Their method won the second best place at the 2016 “Low‐Dose CT Grand Challenge.266 Xiang et al. used low‐dose PET images combined with T1‐weighted images acquired on a PET/MRI scanner to obtain standard acquisition quality PET images. In comparison to the papers above that started denoising with reconstructed images, Yang et al. aimed at improving the quality of recorded projections. They used a CNN‐based approach for learning the mapping between a number of pairs of low‐ and high‐dose projections. After training with a limited number of high‐dose training examples, they used the trained network to predict high‐dose projections from low‐dose projections and then used the predicted projections for reconstruction.
Artifact reduction
Techniques similar to those described for denoising have been applied to artifact reduction. Jin et al.242 described a general framework for the utilization of CNNs for inverse problems, applied the framework to reduce streaking artifacts in sparse‐view reconstruction on parallel beam CT, and compared their approach to filtered back projection (FBP) and total variation (TV) techniques. Han et al.244 used DL to reduce streak artifacts resulting from limited number of radial lines in radial k‐space sampling in MRI. Zhang et al.243 used a CNN‐based approach to reduce metal artifacts on CT images. They combined the original uncorrected image with images corrected with the linear interpolation and beam hardening correction methods to obtain a three‐channel input. This input was fed into a CNN, whose output was further processed to obtain “replacement” projections for the metal‐affected projections.
Reconstruction
Several studies indicated that DL may be useful in directly attacking the image reconstruction problem. In one of the early publications in this area, Golkov et al.245 applied a DL approach to diffusion‐weighted MR images (DWI) to derive rotationally invariant scalar measures for each pixel. Hammernik et al.246 designed a variational network to learn a complete reconstruction procedure for multichannel MR data, including all free parameters which would otherwise have to be set empirically. To obtain a reconstruction, the undersampled k‐space data, coil sensitivity maps, and the zero‐filling solution are fed into the network. Schlemper et al.247 evaluated the applicability of CNNs for reconstructing undersampled dynamic cardiac MR data. Zhu et al.248 introduced an automated transform by manifold approximation approach to replace the conventional image reconstruction with a unified image reconstruction framework that learns the reconstruction relationship between sensor and image domain without expert knowledge. They showed examples in which their approach resulted in superior immunity to noise and a reduction in reconstruction artifacts compared with conventional reconstruction methods.
2.D.2. Image registration
To establish accurate anatomical correspondences between two medical images, both handcrafted features and features selected based on a supervised method are frequently employed in deformable image registration. However, both types of features have drawbacks.249 Wu et al.249 designed an unsupervised DL approach to directly learn the basis filters that can effectively represent all observed image patches and used the coefficients by these filters for correspondence detection during image registration. They subsequently further refined the registration performance by using a more advanced convolutional stacked autoencoder and comprehensively evaluated the registration results with respect to current state‐of‐the‐art deformable registration methods.250 A deep encoder–decoder network was used for predictions for the large deformation diffeomorphic metric mapping model by Yang et al.251 for fast deformable image registration. In a feasibility study, Lv et al.252 trained a CNN for respiratory motion correction for free‐breathing 3D abdominal MRI. For the problem of 2D–3D registration, Miao et al.253 used a supervised CNN regression approach to find a rigid transformation from the object coordinate system to the x‐ray imaging coordinate system. The CNNs were trained using synthetic data only. The authors compared their method with for intensity‐based 2D–3D registration methods and a linear regression‐based method and showed that their approach achieved higher robustness and larger capture range as well as higher computational efficiency. A later study by the same research group identified a performance gap when the model trained with synthetic data is tested on clinical data.254 To narrow the gap, the authors proposed a domain adaptation method by learning domain invariant features with only a few paired real and synthetic data.
2.D.3. Synthesis of one modality from another
A number of studies have recently investigated using DL to generate synthetic CT (sCT) images from MRI. This is important for at least two applications. First, for accurate PET image reconstruction and uptake quantification, tissue attenuation coefficients can be readily estimated from CT images. Thus, estimation of sCT from MRI in PET/MRI imaging is desirable. Second, there is an interest in replacing CT with MRI in the treatment planning process mainly because MRI is free of ionizing radiation. Nie et al.255 used a 3D CNN to learn an end‐to‐end nonlinear mapping from an MR image to a CT image. The same research group in their later research added a context‐aware GAN for improved results.259 Han et al.256 adopted and modified the U‐net architecture for sCT generation from MRI. Current commercially available MR attenuation correction (MRAC) methods for body PET imaging use a fat/water map derived from a two‐echo Dixon MRI sequence. Leynes et al.257 used multiparametric MRI consisting of Dixon MRI and proton‐density‐weighted zero (ZTE) echo‐time MRI to generate sCT images with the use of a DL model that also adopted the U‐net architecture.267 Liu et al.258 trained a deep network (deep MRAC) to generate sCT from T1‐weighted MRI and compared deep MRAC with Dixon MRAC. Their results showed that significantly lower PET reconstruction errors were realized with deep MRAC. Choi et al.260 investigated a different type of synthetic image generation. They noted that although PET combined with MRI is useful for precise quantitative analysis, not all subjects have both PET and MR images in the clinical setting and used a GAN‐based method to generate realistic structural MR images from amyloid PET images. Ben‐Cohen et al.261 aimed at developing a system that can generate PET images from CT, to be used in applications such as evaluation of drug therapies and detection of malignant tumors that require PET imaging, and found that a conditional GAN is able to create realistic looking PET images from CT.
2.D.4. Quality assessment
In addition to traditional characterization tasks in medical imaging, such as classification of ROIs as normal or abnormal, DL has been applied to image quality assessment. Wu et al.262 proposed a DCNN for computerized fetal US image quality assessment to assist the implementation of US image quality control in the clinical obstetric examination. The proposed system has two components: the L‐CNN that locates the ROI of the fetal abdominal region in the US image and the C‐CNN that evaluates the image quality by assessing the goodness of depiction for the key structures of stomach bubble and umbilical vein. Neylon et al.263 used a deep neural network as an alternative to image similarity metrics to quantify deformable image registration performance.
Since the image quality strongly depends on both the characteristics of the patient and the imager, both of which are highly variable, using simplistic parameters like noise to determine the quality threshold is challenging. Lee et al.264 showed that DL using fine‐tuning of a pretrained VGG19 CNN was able to predict whether CT scans meet the minimal image quality threshold for diagnosis, as deemed by a chest radiologist.
Esses et al.265 used a DCNN for automated task‐based image quality evaluation of T2‐weighted liver MRI acquisition and compared this automated approach to image quality evaluation by two radiologists. Both the CNN and the readers classified a set of test images as diagnostic or nondiagnostic. The concordance between the CNN and reader 1 was 0.79, that between the CNN and reader 2 was 0.73, and that between the two readers was 0.88. The relatively lower concordance of the CNN with the readers was mostly due to cases that the readers agreed to be diagnostic, but the CNN did not agree with readers. The authors concluded that although the accuracy of the algorithm needs to be improved, the algorithm could be utilized to flag cases as low‐quality images for technologist review.
2.E. Tasks involving imaging and treatment
Radiotherapy and assessment of response to treatment are not areas that are traditionally addressed using neural networks or data‐driven approaches. However, these areas have recently seen a strong increase in the application of DL techniques. Table 7 summarizes studies in this fast‐developing DL application area.
Table 7.
Radiotherapy and assessment of response to treatment with DL
| Anatomic site | Object or task | Network input | Network architecture | Dataset (train/test) |
|---|---|---|---|---|
| Bladder | Treatment response assessment268 | CT | CifarNet | 82/41 patients |
| Brain | Glioblastoma multiforme treatment options and survival prediction218 | MRI | Custom | 75/37 patients |
| Assessment of treatment effect in acute ischemic stroke269 | MRI | CNN based on SegNet | 158/29 patients | |
| Breast | Response to neoadjuvant chemotherapy270 | MRI | Pretrained VGGNet followed by LDA | 561 examinations from 64 subjects LOO CV |
| Response to neoadjuvant chemotherapy271 | MRI | Custom | 133/33 patients | |
| Segmentation of clinical target volume272 | CT | Deep dilated residual network | 800 patients fivefold CV | |
| Cancer cell lines | Prediction of drug effectiveness in cancer cell lines273 | Multiple omics data from cancer cells (gene expression data, copy number variation data, mutation data, and cell line annotations) | Deep autoencoder | 520/104 cell lines |
| Head and Neck | Organ segmentation274 | CT | U‐Net based with shape retention model | 22/10 scans |
| Kidney | Renal segmentation275 | CT | Custom | 89/24 patients |
| Early detection of acute renal transplant rejection276 | DWI‐MRI | Stacked autoencoders | 100 patients fourfold, tenfold and LOO CV | |
| Liver | Hepatobiliary toxicity prediction after liver SBRT277 | CT and patient demographics, clinical information | Custom CNN trained on other organs, fine‐tuned on liver SBRT | 125 patients 20‐fold CV |
| Lung | Estimation of dose protocols in Radiotherapy278 | FDG‐PET/CT, clinical, genetic, imaging radiomics features, tumor and lung dosimetric variables, treatment plans | Deep Q‐Network | 114 real train/4000 synthesized test cases |
| Dynamic tracking during therapy279 | DRRs from 4D CT | DenseNet | 1/9 volumes | |
| Prostate | Prediction of dose from patient image contours280 | IMRT | U‐net | 80/8 patients |
| Prediction of dosimetric eligibility of prostate cancer patients undergoing IMRT281 | CT | Fine‐tuned AlexNet | 60 patients fivefold CV | |
| Pelvis | Generating synthetic CTs from MR‐only radiotherapy282 | MRI | CGAN | 123/59 patients |
| Assessment of toxicity to normal organs and tissue283 | Rectum surface dose maps | Fine‐tuned VGG‐16 | 42 patients tenfold and LOO CV | |
| Rectum | Segmentation of rectal tumors on T2‐MRI and clinical target volume segmentation on CT272 | T2‐MRI or CT | Novel CNN involving cascaded atrous convolution and spatial pyramid pooling | 70 T2‐MR and 100 CT fivefold CV |
| Prediction of pathologic complete response after chemoradiation284 | CT | DNN classifier custom estimator | 95 patients fivefold CV |
IMRT, intensity‐modulated radiation therapy; SBRT, stereotactic body radiotherapy; DWI, diffusion‐weighted MRI; DRR, digitally reconstructed radiographs; LDA, linear discriminant analysis; LOO, leave‐one‐out; CV, cross‐validation.
2.E.1. Discovery: imaging genomics (radiogenomics)
A major need in breast cancer research is the elucidation of the relationship between the macroscopic image‐based presentation of the tumor and its environment and cancer biology indicators of risk, diagnosis, prognosis, or treatment response. Imaging genomics, that is, “radiogenomics,” aims to find these relationships between imaging data and clinical data, molecular data, genomic data, and outcome data.222, 224 Of interest is whether DL can provide sufficient detailed information to relate to genetic data as have handcrafted radiomic phenotypes.285
2.E.2. Radiotherapy
The goals of DL in radiation oncology are to assist in treatment planning, assess response to therapy, and provide automated adaptation in treatments over time. Deep reinforcement learning using both prior treatment plans and methods for assessing tumor local control was used to automatically estimate dose protocols.278 Such adaptive radiotherapy methods may provide clinical decision support for dose adaptation.
Much of the needs in treatment planning relate to the segmentation of organs (discussed earlier) and in the prediction of dose distributions from contours. Nguyen et al.280 used a U‐net to predict dose from patient image contours on prostate intensity‐modulated radiation therapy (IMRT) patients and demonstrated desired radiation dose distributions. Foote et al.279 combined a DCNN with motion tracking to recover anatomical positions from a single projection radiographic image in real time in order to achieve dynamic tracking of a lung tumor volume.
As discussed earlier, DL can be used to convert between modalities (Section 2.D.3), which can benefit both diagnosis and therapy. Maspero et al.282 have developed a DL method for creating synthetic CTs from MR‐only radiotherapy, leading to online adaptive replanning. Such methods, in order to allow for real‐time changes, need to rapidly generate synthetic CTs, thus modeling the radiation attenuation and dose calculations.
While DL methods are being developed to plan and predict radiation therapy to specific tumor sites, they are also being investigated to assess toxicity to normal organs and tissue. Zhen et al.283 used a transfer learning strategy to predict rectum dose toxicity for cervical cancer radiotherapy.
Segmentation methods to aid in the assessment of treatment plans have been developed as well; Tong et al. developed a CNN‐based method for multiorgan segmentation for use in head and neck cancer radiotherapy274, Men et al. developed a target tumor volume segmentation for rectal cancer272 and breast cancer,286 while Jackson et al. focused on renal segmentation for automated radiation dose estimation.275 Dose estimation was also the aim of Kajikawa et al. who investigated the feasibility of DL in the automated determination of dosimetric eligibility of prostate cancer patients undergoing intensity‐modulated radiation therapy.281
Just as with imaging genomics, as discussed earlier, incorporation of both image‐based phenotypes and genomics in treatment planning and response assessment may yield new relationships and improved therapeutics.273
Overall, however, the use of DL in radiation planning is still at a very early stage in development.
2.E.3. Response to treatment
Just as DL is used to extract tumor characteristics for diagnosis and prognosis, it can also be used in decision‐making for assessing response to therapy. In machine learning, various classifiers can be used to merge the tumor image‐based phenotypes into a response prediction. Thus, DL can also be used to analyze medical image(s) over time to predict response. For example, CNNs were used with breast DCE‐MRI to assess response to neoadjuvant chemotherapy, where the inputs varied over contrast time points as well as treatment examination times.270
Cha et al.268 have explored the feasibility of DL through CNNs on pre‐ and posttreatment CT of bladder cancer patients to assist in the assessment of treatment response. In addition, assessing prognosis of a tumor contributes to decision‐making on treatment options and predicting survival. Lao et al.218 investigated MRI radiomic features and DL as a means to predict survival in glioblastoma multiforme. Bibault et al. used DL to predict pathologic complete response after chemoradiation in locally advanced rectal cancer,284 while Ibramov et al. predicted hepatobiliary toxicity after liver stereotactic body radiotherapy.277 In research unrelated to oncology, the interest in using DL to assess response to treatment has increased as well. Shehata et al.276 used autoencoders for early detection/prediction of acute renal rejection after kidney transplant. Nielsen et al. used DL to predict outcome and to assess the effect of treatment with recombinant tissue‐type plasminogen activator in ischemic stroke patients.269
3. Common themes
3.A. Training and testing with size‐limited datasets
The rapid and immense success of DCNNs in many challenging computer vision problems is achieved through accessibility to large‐scale well‐annotated datasets, for example, PASCAL VOC,287 ImageNet,28 and MS COCO.288 ImageNet pretrained DCNN models29, 73 serve as the foundation in many higher level tasks, that is, image captioning,289 visual question answering,290 and instance relationship extraction.291 Compared to natural image datasets, existing medical image datasets are typically smaller in size. This is because the collection of medical image datasets is often a challenging, time‐consuming process, which involves multiple steps, such as searching in large hospital PACS systems with moderately structured clinical information, selection of a relatively small number of useful clinical cases, and further data annotation by expert physicians. In this subsection, we explore some of the challenges for applying DL on relatively small datasets. The concepts and principles discussed below, such as overfitting, the need for independent training and test datasets, and dependence of performance on training dataset size, apply to most machine learning algorithms, including traditional (shallow) neural networks. However, some aspects may be exacerbated due to the large number of tunable parameters in DL networks.
3.A.1. Overfitting
It has long been recognized that training a complex classifier with a small dataset invites the risk of overfitting (also termed overtraining). According to the Oxford English dictionary, overfitting is “the production of an analysis that corresponds too closely or exactly to a particular set of data, and may therefore fail to fit additional data or predict future observations reliably.” In other words, overfitting occurs when a classifier models the training data too well, resulting in it failing to generalize and performing poorly on new unseen data. John von Neumann famously said “With four parameters I can fit an elephant, and with five I can make him wiggle his trunk.”292 Both shallow neural networks and DL exhibit overtraining. Surprisingly, compared to the huge number of tunable parameters in DL networks, they may exhibit a more limited amount of overfitting compared to a shallow network designed to achieve the same functionality. One possible explanation for this, as discussed in the introduction, is that DL learns a hierarchical representation that matches the composition of the individual components that the data consist of.293 Another possible explanation, using concepts from information theory, contends that a deep network helps better to compress the irrelevant information in the input data and thus can achieve better generalization.294
A number of ways have been suggested in the literature to reduce overfitting, including regularization,295 early stopping,296 and dropout.11, 26 Regularization involves the addition of an extra term to the loss function during training akin to the use of a Lagrange multiplier to satisfy certain boundary conditions. The regularization term is typically chosen to penalize overly complex solutions and for example imposes rules for the smoothness of the solution. Early stopping can be seen as regularization in time. The longer a network is trained, the more complex its solutions become, so by regularizing on time (through early stopping), the complexity will be reduced and generalizability improved. When to stop training is usually determined by monitoring the loss on a validation set (see next paragraph). Dropout is another very efficient way to prevent overfitting and the term “dropout” refers to dropping out units in a neural network.
3.A.2. Training, validation, and testing
Ideally, one has access to three large independent datasets to serve as training, validation, and test set for the training and evaluation of any machine learning approach. Although the terms “validation set” and “test set” may not be defined consistently among all communities, here we use the term “validation set” for the set used for fine‐tuning as part of training and “test set” for the set used for final performance evaluation. Figure 5 shows how the training, validation, and test sets can be used in a supervised machine learning system in an ideal scenario with a large number of available cases. However, when the total number of available cases is small, such a scenario may be inadequate to make full use of the limited‐size dataset. For example, if a total of a hundred cases are available, then it may not be reasonable to randomly assign 20% as a test set and divide the remaining 80 cases into training and validation. The statistical variability in the classification performance for 20 cases will typically be large, limiting the usefulness of the reported performance. Instead, it may be clinically more useful to use a cross‐validation approach (with multiple training/validation and testing data splits) for obtaining a more realistic performance estimate. Using a cross‐validation training/validation and testing approach is a way to obtain a realistic performance estimate for the entire dataset when done correctly but does not result in a single model. Care must be taken to perform all training and validation steps only within the training fold of the cross‐validation, so that there is no leakage of information from the different folds into each other that might bias the cross‐validation performance estimate. In Section 4, methods to help overcome problems related to training DL on a small dataset are discussed, but one should keep in mind that these methods do not overcome the most important limitation of having a small dataset, that is, the potential inadequacy of a small sample to accurately represent the population of interest.
Figure 5.
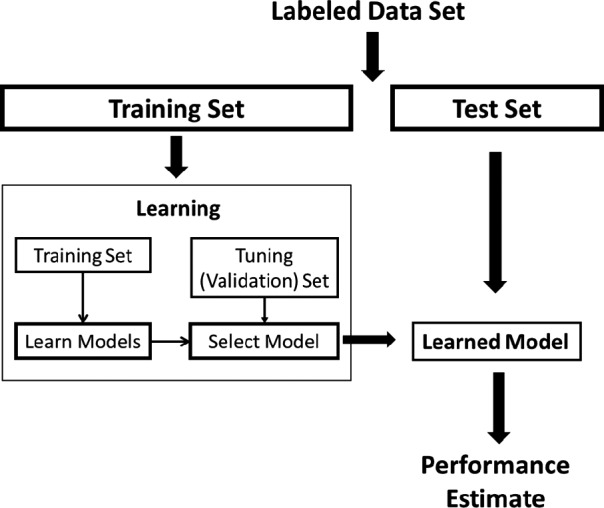
The use of training, validation, and test sets for the design and performance evaluation of a supervised machine learning algorithm.
3.A.3. Dependence of test performance on training set size
A number of studies in the literature have investigated the effect of training size on the performance of the machine learning system.297, 298, 299, 300, 301 The general trend is that as the number of training cases increases, overtraining decreases and the performance on the targeted population improves. There is also a number beyond which increasing the training set size only marginally improves the test performance. However, this number is believed to be a function of the machine learning system architecture, the task, and the system inputs. A few papers studied the effect of varying the training set size on the performance of their DL network.16, 63, 193, 302, 303 Mohamed et al.193 found that for breast density classification, there is a small increase in test performance (the area under the receiver operating characteristic curve increases from 0.95 to 0.99, P < 0.001) when their training set size increased from 2000 images to 6000 images. Azizi et al.16 also found that increasing the training dataset increased the performance of a DL model used for prostate cancer detection in ultrasound. Gulshan et al.63 showed that for their detection algorithm of diabetic retinopathy in retinal fundus photographs, the relative specificity at a given sensitivity on their validation set consistently increased as the number of training samples increased from around 200 samples to around 60,000 samples, at which point the performance plateaued. Using natural images datasets, where the available labeled data are much more abundant compared to medical images, Sun et al.303 demonstrated that the test performance of the DL network continued to increase when going from 10 million training samples up to 300 million training samples for both object detection and semantic segmentation tasks. While it is difficult to obtain datasets of annotated medical images similar in size to datasets for natural images, the trend that increasing the training dataset size increases the performance of the DL network on a target population still applies.
3.B. Transfer learning and fine tuning
Transfer learning is a technique in which a DL network trained on a large dataset from one domain is used to retrain or fine‐tune the DL network with a smaller dataset associated with another domain.160 The limited size of the annotated medical image datasets and the current trend of using deeper and larger structures increase the risk of overtraining and make transfer learning more appealing in medical imaging.
Transfer learning in medical imaging commonly starts with a CNN that was already trained on natural images, that is, a pretrained model. The limited medical image dataset is then used to fine‐tune the pretrained model or, in some applications, no fine‐tuning is performed at all. During fine‐tuning, the DL architecture typically remains fixed, and only a subset of the weights may be retrained.
A commonly used dataset for pretraining of DL structures is ImageNet28 composed of natural scene images. It has been used in more than 75% of the reported transfer learning studies. Different datasets also used for pretraining include CIFAR‐10,204 Places205,304 and texture datasets, such as ALOT, DTD, FMD, and KTH‐TIPS‐2b, as discussed in the literature.209
Transfer learning within the same domain of the target task has also been performed. Kooi et al.202 pretrained DCNN on a large mammogram data set and then retrained the DCNN on a different smaller mammogram dataset for the task of discriminating benign solitary cysts from malignant masses in digital mammography. Samala et al. first pretrained a DCNN on ImageNet198 or a larger mammogram dataset17 and then fine‐tuned on a digital breast tomosynthesis (DBT) dataset for the classification and detection of masses on DBT. Zheng et al.254 pretrained on synthetic data and retrained on clinical data for 2D–3D registration of preoperative 3D image data. Azizi et al.16 used radiofrequency ultrasound images as a source domain to pretrain the DCNN and fine‐tuned it on B‐mode images as a target domain for prostate cancer detection.
A number of studies used pretrained CNNs for extracting features, which are sometimes referred to as the off‐the‐shelf CNN features.305 A relatively small labeled dataset can then be used to train a classifier such as an SVM for the problem at hand. A number of studies173, 181, 192, 196, 306, 307, 308 extracted the outputs of the fully connected layers of a DL network that has been pretrained ImageNet, and used those features as input to SVMs to build classification models, which suggests that a network pretrained on natural images is useful for extracting features for medical image analysis purposes.
Many of the studies that use transfer learning fine‐tune their models by performing additional training on all the network layers, thus using transfer learning like a weight initialization step. With the assumption that the earlier layers perform more common filtering tasks and later layers (usually fully connected layers) focus more on semantic and high‐level features for specific purposes, others have fine‐tuned only a few of the last layers within the network.110 Samala et al.199 studied the effects of fine‐tuning different layers of the AlexNet architecture and found that fine‐tuning different layer combinations resulted in different performance. For their task, they found that freezing the weights of just the first convolution layer achieved higher performance compared to freezing additional layers or fine‐tuning all the convolution layers. Similar trends were observed by Lee et al.309. However, the dataset size for the fine‐tuning may also need to be taken into consideration when using transfer learning, as Samala et al.310 saw a trend where the performance of the fine‐tuned network increased with increasing dataset size of the target task domain used for fine‐tuning.
3.C. Combining deep learning with radiomics approaches
Before DL was applied to medical imaging, handcrafted features‐based approaches were generally used to analyze the images. By using DL, it is expected that given enough data, the network will learn image descriptors useful for analysis. However, it is possible to combine the outputs of DL methods with the knowledge the field of medical imaging analysis has accumulated with computer‐extracted, handcrafted features.166 Several works, including Antropova et al.,197 Li et al.,192 Huynh et al.,196 and Ben‐Cohen et al.307 combined features extracted from the fully connected layers of a DL architecture, with traditional handcrafted features (morphology, intensity, texture). Feature selection was performed to reduce the number of features; then, a machine learning classifier, such as SVM or RF, was used to generate a model using the extracted features. These studies suggest that supplementing DL with information already known to be useful, may improve the performance of these DL models.
3.D. Supervised/weakly supervised/unsupervised learning
The majority of the DL applications utilize supervised learning; there is ground truth or labels that the system is trying to match. However, there are also unsupervised methods that attempt to draw inferences from unlabeled data, that is, without the help of a supervisor (or label) that provides a degree of error for each observation, and weakly supervised methods, that use noisy labels, or images labeled as positive or negative, without localization information, to train for a specific task.
Unsupervised learning in DL is generally performed by autoencoders or independent subspace analysis (ISA).249, 250, 311 The outputs of these networks may be further processed in a supervised manner, by extracting the features from the network and applying a machine learning classifier. In weakly supervised learning, the reference standard used to train does not contain the full information.311, 312 For example, Feng et al.313 trained a system for lung nodule segmentation with a binary label if a nodule was present for a given image slice. Yang et al.167 used a weakly supervised network in a system that aimed to generate a cancer response map with each pixel indicating the likelihood to be cancerous. Both methods refined the initial results with additional DL networks. There are also methods that use a combination of weakly supervised and supervised methods.180, 314 Wang et al.180 and Rajpurkar et al.314 used supervised learning to label chest x rays with one or multiple specific lung diseases and used weakly supervised learning to localize the region with the disease.
4. Expanding datasets for deep learning
As discussed above, DL performs significantly better than previous shallow learning methods and handcrafted image features. However, this comes at the cost of requiring greater amounts of training data compared to previous methods. In the medical domain, publicly available large‐scale image datasets that contain images from tens of thousands of patients are not available (except the recently released ChestX‐ray14 dataset.180). Although vast amounts of clinical images/annotations/reports are stored in many hospitals’ digital warehouse, for example, picture archiving and communication systems (PACS) and oncology information system (OIS), obtaining semantic labels on a large‐scale medical image database is another bottleneck to train highly effective DL models for image analysis.
It is difficult to directly borrow conventional means of collecting image annotations that are used for annotating natural scene images (e.g., Google image search uses terms from NEIL knowledge315 base followed by crowdsourcing28) and apply them in medical images. Medical annotations are difficult to obtain from clinically untrained annotators. Using well‐trained radiologists is expensive. Moreover, the task of “assigning labels to images” is not aligned with their regular clinical routine, which can cause drastic interobserver variations or inconsistency. There is a lot of definition ambiguity to assign image labels based on visible anatomic structures, pathological findings, or using both cues. In addition, a high quality or large capacity medical image search engine is a prerequisite to locate relevant image studies. For example, the radiological data stored in the PACS server are only indexed with dates, patient names, and scan protocols, and it often takes extra effort to find all the cases with a disease pattern of interest. Natural language processing‐based systems that text mine radiology reports are just beginning to become available.316
A wide variety of techniques have been developed for tackling the data shortage problem for both the general computer vision and medical image analysis domain. Data augmentation is the most straightforward way to increase the size of a dataset for training purposes. It has been proved to be extremely effective for currently existing datasets,160 which often contain a small number (hundreds of cases) of hand‐labeled data. Others believe that DL and humans‐in‐the‐loop inspection may have to be interleaved and integrated to construct labels for a large‐scale image database, rather than being employed as two independent labeling processes. It can involve selectively labeling critical samples via active learning. A few recent works focus on transferring the tremendous number of imaging studies accompanied by radiological reports (i.e., loosely labeled samples) into machine trainable data format. Both image and textual features could be utilized for this retrospective and cost‐effective process. In addition to using hand‐labeled ground truth, others317, 318 utilize the algorithm‐generated ground truth of existing image data for training the CNN models. They assume that the model can learn from these less accurate examples and produce refined results in an iterative training process. Furthermore, approaches based on generative adversarial networks38 (GAN) can create image samples for training, either from random initialization or from more advanced clues for image generation. Recent results have shown examples of its promising and useful outcomes. In the following sections, we will summarize these techniques individually.
4.A. Data augmentation
Data augmentation creates new samples based on existing samples in a dataset or according to a generative model. These new samples can then be combined with the original samples to increase the variability in data points in a dataset. This class of techniques has become a common practice in DL‐based applications since it has been shown to be extremely effective for increasing the size of training sets, reducing the chance of overfitting and eliminating the unbalance issue in multiclass datasets, which is critical for achieving generalizable models and testing results.
Common data augmentation techniques adopted in medical image analysis applications84, 107, 319 include cropping, translation, rotation, flipping, and scaling of images. Instead of augmenting whole images, Gao et al.206 randomly jittered and cropped subimages as patches from each original CT slice to generate more samples for classifying interstitial lung diseases. Pezeshk et al.320 introduced an image blending tool that can seamlessly embed a lesion patch into a CT scan or mammography. Furthermore, the lesion patches could be inserted with various types of transformations to the lesion shape and characteristics. Improved classification performances were presented even for small training datasets. Zhang et al.321 intended to tackle the unbalanced data issue for common medical image classification tasks. They proposed a new data augmentation method called unified learning of feature representation and similarity matrix. A single DCNN was trained on the seed‐labeled dataset to obtain image feature representations and a similarity matrix simultaneously, which could be used for searching more similar images to each class of colonoscopy and upper endoscopy images.
Another type of data augmentation involves synthesizing images or data using an object model and physics principles of image formation. Depending on the ultimate purpose of the DL algorithm, the degree of sophistication for the models and image formation approximations can vary.322 Yang et al.240 created a synthetic CT dataset through the use of the Radon transform for a known object and modeled different exposure conditions through adding noise to the data, for training a CNN to estimate high‐dose projections from low‐dose ones. Cui et al.323 simulated dynamic PET emission data in order to train a stacked sparse autoencoder‐based reconstruction framework for dynamic PET imaging. Chen et al.237 synthesized noisy projections based on patient images to generate training data for developing a DL algorithm for noise reduction in reconstructed CT images. Miao et al.253 used synthetic data only to train a CNN for 2D–3D image registration.
4.B. Data annotation via mining text reports
Over the decades, large amounts of radiological data (e.g., images, clinical annotations, and radiological reports) have accumulated in many hospitals’ PACS. How to transform those retrospective radiological data into a machine learnable format has become a big challenge in the DL era. A radiological report could contain many types of information. Generally speaking, it is a free‐text summary of all the clinical findings and impressions determined during examination of a radiological image study. It can contain richer information than just the description of disease findings but also may consist of negation and uncertainty statements. In the “findings” section, a list of normal and abnormal observations is listed for each part of the body examined in the image. Attributes of the disease patterns, for example, specific location and severity, are also noted. Furthermore, critical diagnosis information is often presented in the “impression” section by considering all findings, patient history, and previous studies. Additional or follow‐up imaging studies are recommended if suspicious findings are located. As such, reports consist of a challenging mixture of information. A key for machine learning is extracting the relevant parts for particular applications.324
Schlegl et al.325 relied on existing optical coherence tomography (OCT) volume data and corresponding diagnostic reports to correlate image content and geometry with semantic concepts described in the reports. Increasing classification accuracy for intraretinal cystoid fluid, subretinal fluid, and normal retinal tissue was demonstrated while mining the voxel‐level annotation of class labels.
Following an initial work using MeSH (medical subject headings) manual annotations on chest radiographs,326 Shin et al.33 extracted sentences from the original radiology reports describing key images (images identified during clinical image interpretation as having important findings). The authors used natural language processing (NLP) to analyze about 780,000 patients’ radiology reports and found 215,786 key images mentioned in the reports from scans of 61,845 unique patients. The key images were then extracted from their institution's PACS. Corresponding image labels were then mined via unsupervised hierarchical Bayesian document clustering, that is, generative latent Dirichlet allocation topic modeling, to form 80 classes at the first level of hierarchy. Zech et al.316 applied a similar methodology to a set of 96,303 head computed tomography reports. While mining topic labels in a fully unsupervised manner,33 they adopted latent Dirichlet allocation together with bag of words to compute the feature representation of corpuses. Then, a regression model was trained using a small subset (1004) of annotated reports to initialize the clustering of those unlabeled text reports.
The purely text‐computed information offers some coarse level of radiology semantics but is often limited and disconnected from the associated image. First, the classes could be highly unbalanced, which means that one dominating category may contain many more images while other classes may contain few. Furthermore, the images in a class assigned purely by text analysis may not be visually coherent since the image appearance is not considered in the clustering process. Wang et al.327 exploited a combination of image features and textual information extracted from reports to label groups of images to alleviate these limitations. Figure 6 shows the flowchart of the framework. A CNN‐based joint mining framework was developed to iteratively improve the extracted CNN image features and clustering labels. Consequently, NLP‐mined disease keywords were assigned to each image cluster.
Figure 6.
A disease image categorization framework using both images and texts.327
More advanced NLP techniques have demonstrated better performances in extracted disease keywords for image‐labeling task in recent studies. Wang et al.180 introduced a two‐stage pathology extraction approach by first detecting all disease keywords mentioned in the report using ontology‐based tools and then building negation and uncertainty elimination rules on the dependency graph of sentences. Figure 7 shows sample disease categories mined from the retrospective data. The authors publicly released their dataset of 112,120 frontal view chest x‐ray images of 30,805 unique patients along with image annotations of 14 disease categories. Subsequent research led to a 6% average improvement in the area under the receiver operating characteristic curve through the use of a multilevel attention model in a DL pipeline that included both CNNs and RNNs.328
Figure 7.

Eight sample disease keywords and images mined from PACS.180
Chen et al.329 applied a CNN‐based textual classification framework to find the presence, chronicity, and location of pulmonary embolism in CT examination reports. A human‐in‐the‐loop NLP annotation strategy was adopted to reduce the labeling cost for CNN training. The final CNN model was trained using a total of 2512 radiologist‐annotated CT reports.
Yan et al.330, 331 mined radiology reports and images to extract lesion measurements. The lesion measurements were made in the course of routine clinical interpretation of CT scans. They were bidimensional measurements performed for RECIST (Response Evaluation Criteria in Solid Tumors) assessment, many as part of oncology clinical trials. Their dataset, named “DeepLesion,” consisted of 32,120 axial CT slices, each containing a measured lesion, from 10,594 CT imaging studies of 4459 unique patients. The dataset consists of a large variety of lesion types, including those involving lung, liver, kidney, pancreas, and lymph nodes. The authors’ DL algorithm, which used a triple network and ImageNet pretrained weights, was able to retrieve images of specified type, location, and size with an average accuracy of 90.5%.
Possibilities for text mining do not need to be limited to radiology reports but extend to other clinical reports. The presence of electronic health records (EHR) yields the potential to collect both imaging and clinical/pathology data in order to input to DL to predict diagnosis, outcome, and guide treatments within a clinical workflow.332 Dai et al.333 proposed a clinical report guided CNN which leverages a small amount of supervised information in clinical reports to identify the potential microaneurysms in fundus images. During training, both fundus images and clinical reports are presented to the network. In the testing stage, the input is a fundus image only, and the output is a probabilistic map of the lesion types in the image. Zhang et al.334 proposed a multimodal network that jointly learns from medical images and their diagnostic reports, in which semantic information interacts with visual information to improve the image understanding ability by teaching the network to distill informative features. Applied to bladder cancer images and the corresponding diagnostic reports, the network demonstrated improved performance compared to baseline CNN that only use image information for training.
4.C. Data annotation via active learning
Another approach for assembling large datasets for DL is to try to increase the efficiency of collecting hand‐labeled data to minimize the annotation cost. Active learning is one group of methods for increasing number of annotated data points by including human annotators in the loop of incremental learning and performance improvement. Two key aspects are usually considered for selecting the candidate data for the expensive annotation process: Uncertainty and representativeness of the candidate data.
Different types of information could be utilized to measure the uncertainty and representativeness in order to select samples. Top et al.335 computed the uncertainty values of radius bone regions in the image for segmentation by considering boundary, regional, smoothness, and entropy energies of those image regions. Annotators were then required to label those regions in a CT plane with maximum uncertainty. Zhu et al.336 leveraged the structured information (e.g., data from individual patients) when selecting batch of candidate unlabeled samples. The proposed learning framework enforced a set of specifically designed diversity constraints for the histopathological image annotation task. The visual saliency of objects337 inside an image was considered as a measure for selecting samples. The similarities between labeled and unlabeled data were computed and encoded in a graph. Then, random walks were adopted for searching the most informative node (with largest classification uncertainty and minimum overlap with labeled data). Lee et al.338 believe that the most informative instances (hard examples) are those closest to the SVM hyperplane. Together with balanced sampling, their proposed learning framework was able to achieve a more than 40% classification performance increase on the testing set.
A batch mode‐based active learning339 method was proposed and applied to medical image classification applications. The Fisher information matrix was adopted to select informative unlabeled samples in a groupwise manner. The framework developed an efficient greedy searching algorithm to find a subset of the unlabeled data that can minimize the Fisher information of remaining unlabeled set. The experiments demonstrated the effectiveness of this batch‐mode‐based active learning approach. Konyushkova et al.340 trained a segmentation classifier to decide if a set of supervoxels was most in need to be annotated in 3D image volumes. Geometric priors were utilized in this process to compute geometric uncertainty for each voxel, indicating whether a clear boundary was present. For segmenting electron microscopy images, the model trained using 100 selected pixels with annotations (less than 0.03% of the total training set) achieved even higher classification performance than the one trained with all available labeled training pixels.
Recent approaches further utilized DCNN features to compute and representativeness criteria. Yang et al.341 presented a deep fully convolutional network‐based active learning framework to reduce annotation effort in image that contain multiple instances, for example, pathological images. The uncertainty and similarity information computed from network activations are utilized to select the most cost‐effective annotation areas. Zhou et al.342 measured the uncertainty and diversity of candidate image samples using the CNN classification prediction values computed for all the image patches extracted from the candidate image. In comparison to previous methods, this method has the advantage that no seed‐labeled sample is required. A newly annotated sample will further improve the candidate selection process after CNN mode is fine‐tuned again based on the new training set. They demonstrated that the CNN's classification performance could be incrementally enhanced by continuously fine‐tuning the CNN in an iterative manner.
There are other methods that do not require even a small number of initial hand‐labeled data. Gaur et al.343 started the selection process with a deep model trained on a similar domain. Then, they interpreted the active learning problem of increasing the size of limited labeled dataset as an optimization problem by maximizing both the uncertainty and abundancy. Only a minimum number of data fulfilling both criterions were selected and annotated by a human expert. Mosinska et al.344 tailored the uncertainty sampling‐based active learning approach for the delineation of complex linear structures problem, which significantly reduced the size (up to 80%) of training dataset while achieving equivalent performance. Multiple samples inside the same image were simultaneously presented to the annotator while the interactive annotation framework kept the selected samples informative, representative, and diverse.
4.D. Expanding the training dataset via domain adaptation
Instead of manually annotating selective number of data, another strategy for training data hungry DL paradigms is to leverage labeled data from a different domain, for example, ImageNet database of natural images, and then fine‐tune based on the pretrained CNN parameters in the target domain via transfer learning, as discussed in Section 3.B. The assumption is that the essential pattern learned and recorded in CNN weights, especially in the earlier layers, to some extent are shared by different kinds of images from different domains. Under this assumption, transfer learning using a pretrained model is rather straightforward, but the underlying differences in structures and features in data cross domains is overlooked. In contrast to this straightforward application of pretraining, domain adaptation attempts to alter a source domain to bring the distribution of the source closer to that of the target. In‐depth analyses have been conducted to measure the distribution difference or nonlinear mapping of features between source and target domains for domain adaptation.
Heimann et al.345 employed a discriminative learning‐based approach to localize the transesophageal echocardiography transducer in x‐ray images. Instance weighting was applied on unlabeled fluoroscopy image samples to estimate the differences in feature space density and correct covariate shift to align the data distribution cross domains. Wachinger et al.346 employed a similar instance weighting strategy in a supervised domain adaptation problem with a small training set as supervision from the target domain. Conjeti et al.347 computed tissue‐specific backscattering signal statistics for calcified, lipidic, and fibrotic arterial plaques and used decision forest‐based method to align the distribution shift of signal statistics between in vitro and in vivo image domains.
Schlegl et al.205 trained a CNN in an unsupervised manner for learning more general low‐level image features for images from multiple sites (as domains). Then, another CNN model was fine‐tuned based on the previous CNN model (with domain information injected) to classify lung tissue in high‐resolution CT data using a small set of annotated data from on‐site. Improved classification performance was demonstrated by adopting unsupervised pretraining with data cross domains.
Different acquisition and staining processes can cause large variability in microscopic brain images even on the same part of brain.348 Normalized cross correlation was introduced to locate image patches in the images from target domain, which shared the similar selected features with an image patch from the source domain. Those located image patches will also share the same label as their counterpart from the annotated source domain. Then, a multiple instance learning‐based classification framework was used to utilize those newly labeled (and also possibly noisy) patches for the image classification task. For the same problem, Becker et al.349 proposed to learn a nonlinear mapping of the data features between two domains (acquisitions in this case), together with decision boundary for the regression‐based classification.
Azizi et al.16 applied an unsupervised domain adaptation method based on DL for the prostate cancer detection problem. A deep belief network was trained using both B‐mode (target domain) and radiofrequency (source domain) ultrasound images to effectively align features from two domains in a common latent feature space. The alignment was achieved by minimizing the divergence between the source and target distributions through the training. Similar ideas were presented for multiple sclerosis lesion segmentation in MR images using fully convolutional networks.350 A modified U‐net architecture was designed to take both labeled (source domain) and unlabeled (target domain) data and simultaneously minimize both the segmentation loss and the discrepancy between embedded features from two domains.
4.E. Data synthesis via generative adversarial networks
Generative adversarial networks have attracted tremendous attentions and have grown into a big family of methods in the past two years, from the original GAN framework38 to recent CycleGAN.37 The quality of synthesized images also evolved rapidly from 32 × 32 snapshots to high‐resolution CT/MR images. There have been quite a few successful applications of GANs in the medical imaging domain. Compared to the conventional generative model‐based methods, for example, characteristic modeling,351 random walk sampling,352 and image decomposition,353 GANs intend to produce better images from an image appearance perspective. However, these images are often less meaningful from a clinical point of view since the image intensity on each pixel in a real clinical image has semantic meanings, for example, high values in PET image usually represent high take‐up tumor regions. To overcome such limitations, a variety of constraints and additional information need to be included to help produce more clinically meaningful medical images.
Calimeri et al.354 cascaded the GAN models as a multiscale pyramid based refinement framework with different size image inputs at each level so that a high‐resolution MR image could be synthesized and then improved from coarse to fine. Frid‐Adar et al.215 started with standard data augmentation methods to create a larger dataset that could be used to train a deep convolutional GAN. The synthetic data samples created for each lesion class, that is, cysts, metastases, and hemangiomas, by the GAN were then inputted to the training process of the final lesion classifier together with the enlarged training set from previous data augmentation. Lahiri et al.355 extended the discriminator for classifying patches from multiple categories in addition to answering the fake or real binary question. This design has proven to be more data efficient for adversarial training. Zhang et al.356 applied the same strategy on the semantic segmentation task, where the discriminator not only evaluated the segmentation results itself but also tried to differentiate the labeled and unlabeled data. The segmentation results from unlabeled data were weighted less (compared to the counterpart from labeled data) in the adversarial training procedure to produce more accurate results for the next iteration.
Generating realistic images from scratch (initialized with noise vectors from the latent space) is extremely challenging, especially for medical images. However, more meaningful images could be synthesized if some prior knowledge was provided, for example, an image similar to the target one but in different modality.357 Costa et al.358 proposed to generate retinal images by using corresponding vessel tree images. Different from the standard pairwise GAN generative framework, an autoencoder was first trained to learn the distribution of realistic retinal vessel trees and the retinal images were generated from the representations learned via the autoencoder.
Instead of using paired images for training, Chartsias et al.359 adopted the CycleGAN framework in synthesizing cardiac MR images and masks from view‐aligned CT ones in a loosely supervised manner. The pairwise constraints (e.g., paired images with similar anatomical structure) were eliminated in this case. A 15% increase in segmentation accuracy was demonstrated by using both real and synthetic data compared to using real data alone. The application of CycleGAN in the unpaired MRI to CT image synthesis was also demonstrated.360
Although it is still in its early stage, GAN‐based medical image generation has provided a promising alternative to other data augmentation approaches. Chuquicusma et al.361 reported a visual Turing test that involved two radiologists (with different years of experience) to evaluate the quality of the synthesized nodules. A mixed set of (benign or malignant) nodule patches was shown to the radiologists individually for determining whether they were real or generated. The results showed that the majority (67% and 100%, respectively) of the generated nodules were recognized as real by the two radiologists.
5. Challenges, lessons learned, and the future
As discussed in previous sections, recent advances in DL show that computers can extract more information from images, more reliably, and more accurately than ever before. However, further developing and optimizing DL techniques for the characteristics of medical images and medical data remain an important and relevant research challenge.
5.A. Evaluation and robustness
As discussed previously, data augmentation is often used to alleviate the problem of limited dataset sizes. Data augmentation is powerful, but must be used correctly. One cannot train a network on a set of images pertaining to a given case and then test this trained network on a different set of images pertaining to that same case. Similarly, when dealing with 3D images, it might be tempting to treat every image slice as an independent entity. This would be incorrect, however, since slices of the same case are correlated and slices of a given case either need to be all in the training/validation set or all in the test set. If not done correctly, the performance will be substantially overestimated and not be generalizable. It is also important to keep in mind that performance needs to be evaluated “by case,” whether a “case” is a lesion, patient, or whatever is relevant to the clinical task at hand. No matter how one slices and dices the data, if there are 100 patients, there really are only 100 patients, and evaluation needs to be done accordingly.
When DL is used as a feature extractor, even in transfer learning when a completely trained deep net is applied to new images, the sheer number of extracted features poses a challenge. With the use of data augmentation, one would hope that the number of features will not exceed the number of data points so that dimension reduction or feature selection is possible in a meaningful way before further classification with a different classifier such as a shallow neural net or SVM. Feature selection, however, is likely to be a rather unstable undertaking with different features being selected depending on how the dataset is partitioned. Additionally, it is common practice to use P‐values to choose which of numerous features should be used, but P‐values themselves are highly variable.362, 363 P‐values are data‐dependent statistics that vary from sample to sample even when underlying effects, population, and sampling are the same.364 Hence, utmost care needs to be taken when using DL methods as feature extractors.
Robustness and repeatability are concerns with any machine learning approach,365 and even more so with DL. Since medical image datasets are so difficult to come by compared to those of natural images and generally are of limited size, researchers like to reuse the same data for different tasks. Hence, correction for multiple comparisons366, 367 is crucial in the statistical evaluation of performance. The requirement that datasets need to be of sufficient size and quality is not unique to DL or medical imaging. It is, for example, reminiscent of issues observed in genomics where lack of reproducibility was observed when looking for predictive gene lists in small datasets (~100s of cases).368, 369 There, thousands of samples are needed to generate a robust gene list to predict the outcome in cancer.369 A 2012 study of 53 landmark papers in basic cancer research was able to replicate the original results of just six of these studies.370 Moreover, a study reviewing radiomics using texture features, that is, “conventional” radiomics, for the prediction of survival, found that all of the results of nine published studies failed to reach statistical significance after properly correcting P‐values for multiple comparisons and the use of an optimal cutoff (if applicable) in Kaplan–Meier analysis.371 Results of DL‐based methods, if analysis is not performed correctly, may be even less likely to hold up to scrutiny.
5.B. Datasets and curation
Perhaps the most important challenge when it comes to medical imaging data sets is to obtain data of a sufficiently large number of properly annotated cases. The bottleneck is not necessarily obtaining the images, but obtaining annotations and reference standards. For segmentation tasks, for example, the reference standard or “truth” would be the manual outline of one, or preferably more, expert radiologists. For cancer classification tasks, for example, the reference standard would be the pathological truth as determined by biopsy or surgery which needs to be extracted from pathology reports. The reference standard has to be of high quality, especially when used for training but also for performance evaluation. Obtaining high‐quality image data, annotations, and reference standards are expensive and time‐consuming. Patient privacy laws, while absolutely necessary, further complicate data collection because all protected health information needs to be removed from image data and corresponding radiology, pathology, and other reports. Moreover, relevant information needs to be extracted from the radiology, pathology, and other text reports which are time‐consuming and potentially error prone when done manually and not trivial when performed automatically (Section 4.B). There is immense value in sharing annotated image data and anonymized publicly accessible databases such as provided by the Cancer Imaging Archive (www.cancerimagingarchive.net/).
Another challenge for medical image datasets is that imaging devices are not measurement devices. Unlike a ruler or a Volt meter, which are calibrated and expected to give consistent and correct results within the calibration accuracy, imaging devices generate images through often proprietary image processing techniques. Images are usually not quantitative and primarily designed to be interpretable by humans, not by computers. Robustness of “conventional” and DL‐based methods with respect to image manufacturer or image preprocessing methods needs to be investigated. There has been effort investigating robustness of “conventional” methods with respect to manufacturer for breast cancer diagnosis on ultrasound,372, 373 the assessment of risk of future breast cancer on digital mammography,374 and lung nodule features.375 Work has also been done toward the harmonization of image data with respect to different CT scanners.376 One of the advantages of DL‐based methods, however, is that they may be less sensitive than “conventional” methods to differences in images due to the use of imaging equipment of different manufacturers. Having been designed for natural images in which, for example, a dog in the shade is still a dog, may make them better able to deal with differences in image appearance and quality.
Class imbalance is another challenge related to many medical imaging datasets, not only to DL‐based methods but also to “conventional” methods as well. In screening mammography, for example, the cancer prevalence is so low that developing a method to detect cancer without causing undue false positives is a formidable task. One approach to alleviate the problem of class imbalance in the training of DL methods is to use data augmentation of the underrepresented class only in classifier training as explained in more detail in Section 4.
5.C. Interpretability
When a deep neural net is used as a feature extractor thousands of features are extracted. Unlike engineered handcrafted features, these features do not directly relate to something radiologists can easily interpret. Engineered features often describe something directly related to characteristics radiologists use in their clinical assessment, such as lesion size or shape. Such characteristics can be described by multiple mathematical descriptors, that is, engineered features. For example, the “simplest” feature of maximum linear dimension is both used by a radiologist and can be automatically calculated by a radiomics method. It is then easy for a radiologist to assess whether to trust the radiomics output. But even for “traditional” approaches, this direct interpretability diminishes for more “complicated” features such as for the many that describe texture. For features extracted from deep neural nets, this interpretability is almost completely lost. Radiologists may not care about all the DL parameters and how an application works, however, and it may be more a matter of human trust in the capabilities of the proverbial DL “black box.” The “believability” of DL approaches — both as classifiers and as feature extractors — then, relies on past performance reported for large independent test sets. For example, in diagnosis of breast cancer, the believability of the probability of malignancy output by a DL method relies on the knowledge of past performance on independent test data. Acceptance of DL in medical imaging may benefit from success of DL in other applications such as self‐driving cars and robotics. There may be legal implications to using DL in medical imaging applications since it will be more difficult than for “conventional” applications to pinpoint exactly what went wrong if the output is incorrect (potentially negatively impacting patient care).
Recently, there has been increasing interest in making AI methods (including those involving DL) transparent, interpretable, and explainable.377 This, in part, has been driven by the European general data protection regulation that will go into effect in May 2018 and will make “black‐box” approaches difficult to use in business. These new rules require it to be at least possible to trace results on demand.377 Although traditional approaches tend to be at least interpretable in the sense that users can understand the underlying math of an algorithm, until recently, DL systems tended to be more opaque offering little or no insight into their inner workings. However, there has been increasing effort in making DL methods more transparent and methods have been proposed to assess the sensitivity of the prediction with respect to changes in the input or to decompose the decision in terms of the input variables.378
It is possible to provide visual “explanations,” for example, to show heat maps visualizing the importance of each pixel for the prediction. These visualization techniques could help to further optimize a CNN training approach and ensure that the CNN is “paying attention” to the correct regions of an image in analysis. For example, if a CNN were to be trained to detect pneumothorax on chest x rays, it would be important to know whether the CNN correctly “looked at” the pneumothorax region of images or instead focused on chest tubes that are often present in patients with pneumothorax. Most popular visualization techniques are either perturbation based or backpropagation based. Perturbation‐based methods modify parts of the image and study the effect on the CNN output.379, 380 Backpropagation‐based methods propagate either the output probability score, or the gradient of the output with respect to the input in order to construct heatmaps. Some of the most popular backpropagation‐based methods include the saliency map,381 the class activation map,382 and the gradient‐weighted class activation map.383 Backpropagation‐based methods are computationally cheaper because they use the fundamental property of propagating signals through convolutions, instead of propagating each modification through the network as in done in perturbation‐based methods.
5.D. Competitive challenges
There have been a number of competitive challenges in the field of medical image analysis (https://grand-challenge.org/all_challenges/). The prevalence of DL‐based methods has clearly increased over the last couple of years and DL methods have become top performers in medical image analysis competitions. They often, but not always, perform as well as or better than “conventional” methods. In a literature review on DL, Litjens et al.384 noted that the exact DL architecture does not seem to be the most important determinant in getting a good solution. For example, in the Kaggle Diabetic Retinopathy Challenge (https://www.kaggle.com/c/diabetic-retinopathy-detection), many researchers used the exact same architectures, the same type of networks, but obtained widely varying results. Data augmentation methods and preprocessing techniques seem to contribute substantially to good performance and robustness. It remains an open question how the results from these competitive challenges can be leveraged to benefit the medical image analysis research community at large.
5.E. Lessons learned
Looking back into the history of medical image analysis, it appears that popularity of certain methods fluctuated in time. For example, ANNs gathered a lot of attention in the early 90s, were replaced by SVMs in many applications in late 1990s and early 2000s, only to make a comeback in the form of DL in the 2010s. Likewise, the popularity of wavelet methods and feature extraction techniques such as SIFT evolved in time. The successes already achieved by DL methods, many of them discussed above, are undeniable and well established. We believe that the application areas of DL will evolve in time like other methods, and will likely be supplemented and complemented by newer methods. However, one important lesson learned that will likely be maintained into the future is one about data quality, or the “garbage‐in garbage‐out” principle. Quality of the image data and annotations is crucial and analysis needs to be carried out correctly. Another important lesson is the difference between statistical significance and clinical significance/relevance. Although establishing statistical significance is a very important step in research and publications, we should never lose sight of what the clinically relevant questions are, and just because there is a newer more complicated CNN, does not necessarily mean that it will better help (or replace) radiologists. Expert knowledge about the clinical task can provide advantages that go beyond adding more layers to a CNN, and incorporating expert medical knowledge to optimize methods, for example, through novel data preprocessing or augmentation techniques, for a specific clinical task is often crucial in obtaining good performance.
Plenty of challenges remain for “conventional” medical image analysis and DL‐based methods, including computational and statistical aspects. We need to investigate and improve image data harmonization, develop standards for reporting as well as experiments, and have better access to annotated image data such as publicly available datasets to serve as independent benchmarks.
5.F. Future of deep learning in imaging and therapy
Machine learning, including DL, is a fast‐moving research field that has great promise for future applications in imaging and therapy. It is evident that DL has already pervaded almost every aspect of medical image analysis. “Conventional” image analysis methods were never intended to replace radiologists but rather to serve as a second opinion. Likewise, DL‐based methods are unlikely to replace human experts any time soon. The performance of DL has equaled or surpassed human performance for some nonmedical tasks such as playing computer games385 and, as illustrated by the many cited publications in this paper, DL has also been quite successful in a variety of medical imaging applications. However, most medical imaging tasks are far from solved386 and the optimal DL method and architecture for each individual task and application area have not yet been established. Moreover, the integration of medical image analysis methods and other patient data — such as patient history, age, and demographics — also remains an area of active research that could further improve performance of clinical decision‐making aids.
Three aspects that will drive the DL revolution are availability of big data, advances in DL algorithms, and processing power. As discussed above, there is abundant new research aimed at alleviating the limited dataset size problem in medical imaging, and some of the custom DL architectures and algorithms specifically designed for medical imaging have shown great promise. There has been an explosion of research papers published on DL in medical imaging, most within the past couple of years, and this trend is expected continue. The emergence of conferences solely dedicated to DL in medical imaging (such as the “Medical Imaging with Deep Learning Conference” to be held in July 2018, https://midl.amsterdam/) is very telling. The potential of DL in medical imaging has also not gone unnoticed by the healthcare industry. Companies, both big and small, are taking big steps in developing and commercializing new applications that are based on DL, and large medical imaging vendors have already made significant investments. DL is here to stay, and its future in medical imaging and radiation therapy seems bright.
Conflicts of interest
MLG is a stockholder in R2/Hologic, scientific advisor, cofounder, and equity holder in Quantitative Insights, makers of QuantX, shareholder in Qview, and receives royalties from Hologic, GE Medical Systems, MEDIAN Technologies, Riverain Medical, Mitsubishi, and Toshiba. KD receives royalties from Hologic. RMS receives royalties from iCAD, Inc., Koninklijke Philips NV, ScanMed, LLC, PingAn, and receives research support from Ping An Insurance Company of China, Ltd., Carestream Health, Inc. and NVIDIA Corporation.
Acknowledgments
MLG and KD were supported in part by NIH grants CA 195564, CA 166945, and CA 189240; and The University of Chicago CTSA UL1 TR000430 pilot awards. KHC was supported in part through a Critical Path grant from the U.S. Food and Drug Administration, and by an appointment to the Research Participation Program at the Center for Devices and Radiological Health administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration. XW and RMS were supported by the Intramural Research Programs of the NIH Clinical Center. The mention of commercial products, their sources, or their use in connection with material reported herein is not to be construed as either an actual or implied endorsement of such products by the Department of Health and Human Services.
References
- 1. Amodei D, Ananthanarayanan S, Anubhai R, et al. Deep speech 2: end‐to‐end speech recognition in English and Mandarin. In: International Conference on Machine Learning; 2016:173–182.
- 2. Peters ME, Neumann M, Iyyer M, et al. Deep contextualized word representations; 2018. arXiv preprint arXiv:1802.05365.
- 3. Zoph B, Vasudevan V, Shlens J, Le QV. Learning transferable architectures for scalable image recognition, 2; 2017. arXiv preprint arXiv:1707.07012.
- 4. Silver D, Huang A, Maddison CJ, et al. Mastering the game of go with deep neural networks and tree search. Nature. 2016;529:484–489. [DOI] [PubMed] [Google Scholar]
- 5. Gandhi D, Pinto L, Gupta A. Learning to fly by crashing; 2017. arXiv:1704.05588.
- 6. Worldwide Semiannual Cognitive and Artificial Intelligence Systems Spending Guide; 2016. https://www.idc.com/getdoc.jsp?containerId=prUS41878616.
- 7. The Fourth Industrial Revolution: what it means, how to respond; 2016. https://www.weforum.org/agenda/2016/01/the-fourth-industrial-revolution-what-it-means-and-how-to-respond/.
- 8. Brynjolfsson E, McAfee A. The Second Machine Age: Work, Progress, and Prosperity in a Time of Brilliant Technologies. New York, NY: WW Norton & Company; 2014. [Google Scholar]
- 9. Schwab K. The Fourth Industrial Revolution. New York, NY: Crown Business; 2017. [Google Scholar]
- 10. Harnessing automation for a future that works; 2017. https://www.mckinsey.com/global-themes/digital-disruption/harnessing-automation-for-a-future-that-works.
- 11. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. [DOI] [PubMed] [Google Scholar]
- 12. Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. 2015;61:85–117. [DOI] [PubMed] [Google Scholar]
- 13. Caruana R, Niculescu‐Mizil A. An empirical comparison of supervised learning algorithms. In: Proceedings of the 23rd International Conference on Machine Learning. Pittsburgh, PA, USA: ACM; 2006:161–168. [Google Scholar]
- 14. Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2010;22:1345–1359. [Google Scholar]
- 15. Weiss K, Khoshgoftaar TM, Wang D. A survey of transfer learning. J Big Data. 2016;3:9. [Google Scholar]
- 16. Azizi S, Mousavi P, Yan P, et al. Transfer learning from RF to B‐mode temporal enhanced ultrasound features for prostate cancer detection. Int J Comput Assist Radiol Surg. 2017;12:1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Samala RK, Chan HP, Hadjiiski L, Helvie MA, Wei J, Cha K. Mass detection in digital breast tomosynthesis: deep convolutional neural network with transfer learning from mammography. Med Phys. 2016;43:6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kumar A, Kim J, Lyndon D, Fulham M, Feng D. An ensemble of fine‐tuned convolutional neural networks for medical image classification. IEEE J Biomed Health Inf. 2017;21:31–40. [DOI] [PubMed] [Google Scholar]
- 19. Bengio Y. Learning deep architectures for AI. Found Trends Mach Learn. 2009;2:1–127. [Google Scholar]
- 20. Felleman DJ, Van Essen DC. Distributed hierarchical processing in the primate cerebral cortex. Cereb Cortex. 1991;1:1–47. [DOI] [PubMed] [Google Scholar]
- 21. Wessinger CM, Van Meter J, Tian B, Van Lare J, Pekar J, Rauschecker JP. Hierarchical organization of the human auditory cortex revealed by functional magnetic resonance imaging. J Cogn Neurosci. 2001;13:1–7. [DOI] [PubMed] [Google Scholar]
- 22. Caruana R, Karampatziakis N, Yessenalina A. An empirical evaluation of supervised learning in high dimensions. In: Proceedings of the 25th International Conference on Machine Learning. Helsinki, Finland: ACM; 2008:96–103. [Google Scholar]
- 23. Hinton GE, Osindero S, Teh YW. A fast learning algorithm for deep belief nets. Neural Comput. 2006;18:1527–1554. [DOI] [PubMed] [Google Scholar]
- 24. Sutskever I, Martens J, Dahl G, Hinton G. On the importance of initialization and momentum in deep learning. In: Sanjoy D, David M, eds. Proceedings of the 30th International Conference on Machine Learning (PMLR, Proceedings of Machine Learning Research), Vol. 28; 2013:1139–1147.
- 25. Glorot X, Bordes A, Bengio Y. Deep sparse rectifier neural networks. In: Geoffrey G, David D, Miroslav D, eds. Proceedings of the Fourteenth International Conference on Artificial Intelligence and Statistics (PMLR, Proceedings of Machine Learning Research), Vol. 15; 2011:315–323.
- 26. Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15:1929–1958. [Google Scholar]
- 27. Wan L, Zeiler M, Zhang S, Cun YL, Fergus R. Regularization of neural networks using DropConnect. In: Sanjoy D, David M, eds. Proceedings of the 30th International Conference on Machine Learning (PMLR, Proceedings of Machine Learning Research), Vol. 28; 2013:1058–1066.
- 28. Deng J, Dong W, Socher R, Li L‐J, Li K, Fei‐Fei L. Imagenet: a large‐scale hierarchical image database. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2009:248–255. [Google Scholar]
- 29. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ, eds. Advances in Neural Information Processing Systems, Vol. 25. New York, NY: Curran Associates, Inc.; 2012:1097–1105. [Google Scholar]
- 30. LeCun Y, Kavukcuoglu K, Farabet C. Convolutional networks and applications in vision. In: IEEE International Symposium on Circuits and Systems; 2010;253–256.
- 31. Hochreiter S, Schmidhuber R. Long short‐term memory. Neural Comput. 1997;9:1735–1780. [DOI] [PubMed] [Google Scholar]
- 32. Vinyals O, Toshev A, Bengio S, Erhan D. Show and tell: a neural image caption generator In: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Boston, MA, USA: IEEE. 2015:3156–3164. [Google Scholar]
- 33. Shin H‐C, Lu L, Kim L, Seff A, Yao J, Summers R. Interleaved text/image deep mining on a large‐scale radiology database for automated image interpretation. J Mach Learn Res. 2016;17:2. [Google Scholar]
- 34. Graves A, Mohamed A‐R, Hinton G. Speech recognition with deep recurrent neural networks. In: 2013 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). Vancouver, BC Canada: IEEE; 2013:6645–6649. [Google Scholar]
- 35. Sutskever I, Vinyals O, Le QV. Sequence to sequence learning with neural networks. In: Advances in Neural Information Processing Systems. Montreal, QC Canada: NIPS; 2014:3104–3112. [Google Scholar]
- 36. Arjovsky M, Chintala S, Bottou L. Wasserstein Gan; 2017. arXiv preprint arXiv:1701.07875.
- 37. Zhu JY, Park T, Isola P, Efros AA. Unpaired image‐to‐image translation using cycle‐consistent adversarial networks. In: IEEE International Conference on Computer Vision; 2017.
- 38. Goodfellow I, Pouget‐Abadie J, Mirza M, et al. Generative adversarial nets. Advances in Neural Information Processing Systems. Montreal, QC Canada: NIPS. 2014;2672–2680. [Google Scholar]
- 39. Mardani M, Gong E, Cheng JY, et al. Deep generative adversarial networks for compressed sensing automates MRI; 2017. arXiv:1706.00051.
- 40. Meyers PH, Nice CM, Becker HC, Nettleton WJ, Sweeney JW, Meckstroth GR. Automated computer analysis of radiographic images. Radiology. 1964;83:1029–1034. [DOI] [PubMed] [Google Scholar]
- 41. Becker HC, Nettleton WJ, Meyers PH, Sweeney JW, Nice CM. Digital computer determination of a medical diagnostic index directly from chest X‐ray images. IEEE Trans Biomed Eng. 1964;11:67–72. [DOI] [PubMed] [Google Scholar]
- 42. Lodwick GS, Keats TE, Dorst JP. The coding of roentgen images for computer analysis as applied to lung cancer. Radiology. 1963;81:185–200. [DOI] [PubMed] [Google Scholar]
- 43. Chan H‐P, Doi K, Galhotra S, Vyborny CJ, MacMahon H, Jokich PM. Image feature analysis and computer‐aided diagnosis in digital radiography. 1. Automated detection of microcalcifications in mammography. Med Phys. 1987;14:538–548. [DOI] [PubMed] [Google Scholar]
- 44. Giger ML, Doi K, MacMahon H. Image feature analysis and computer aided diagnosis in digital radiography. 3. Automated detection of nodules in peripheral lung fields. Med Phys. 1988; 15:158–166. [DOI] [PubMed] [Google Scholar]
- 45. Kanazawa K, Kawata Y, Niki N, et al. Computer‐aided diagnosis for pulmonary nodules based on helical CT images. Comput Med Imaging Graph. 1998;22:157–167. [DOI] [PubMed] [Google Scholar]
- 46. Abe C, Kahn CE, Doi K, Katsuragawa S. Computer‐aided detection of diffuse liver‐disease in ultrasound images. Invest Radiol. 1992; 27:71–77. [DOI] [PubMed] [Google Scholar]
- 47. Fukushima K, Miyake S, Ito T. Neocognitron: a neural network model for a mechanism of visual pattern recognition. IEEE Trans Syst Man Cybern. 1983;SMC‐13:826–834. [Google Scholar]
- 48. Lin JS, Ligomenides PA, Freedman MT, Mun SK. Application of artificial neural networks for reduction of false‐positive detections in digital chest radiographs. In: Proceedings of the Symposium on Computer Applications in Medical Care; 1993:434–438. [PMC free article] [PubMed]
- 49. Lo SC, Lou SL, Lin JS, Freedman MT, Mun SK. Artificial convolution neural network techniques and applications to lung nodule detection. IEEE Trans Med Imaging. 1995;14:711–718. [DOI] [PubMed] [Google Scholar]
- 50. Chan H‐P, Lo SCB, Sahiner B, Lam KL, Helvie MA. Computer‐aided detection of mammographic microcalcifications: pattern recognition with an artificial neural network. Med Phys. 1995;22:1555–1567. [DOI] [PubMed] [Google Scholar]
- 51. Sahiner B, Heang‐Ping C, Petrick N, et al. Classification of mass and normal breast tissue: a convolution neural network classifier with spatial domain and texture images. IEEE Trans Med Imaging. 1996;15:598–610. [DOI] [PubMed] [Google Scholar]
- 52. Zhang W, Doi K, Giger ML, Wu Y, Nishikawa RM, Schmidt RA. Computerized detection of clustered microcalcifications in digital mammograms using a shift‐invariant artificial neural network. Med Phys. 1994;21:517–524. [DOI] [PubMed] [Google Scholar]
- 53. Zhang W, Doi K, Giger ML, Nishikawa RM, Schmidt RA. An improved shift‐invariant artificial neural network for computerized detection of clustered microcalcifications in digital mammograms. Med Phys. 1996;23:595–601. [DOI] [PubMed] [Google Scholar]
- 54. Suzuki K, Armato SG, Li F, Sone S, Doi K. Massive training artificial neural network (MTANN) for reduction of false positives in computerized detection of lung nodules in low‐dose computed tomography. Med Phys. 2003;30:1602–1617. [DOI] [PubMed] [Google Scholar]
- 55. Suzuki K, Li F, Sone S, Doi K. Computer‐aided diagnostic scheme for distinction between benign and malignant nodules in thoracic low‐dose CT by use of massive training artificial neural network. IEEE Trans Med Imaging. 2005;24:1138–1150. [DOI] [PubMed] [Google Scholar]
- 56. He KM, Zhang XY, Ren SQ, Sun J. Deep residual learning for image recognition. In 2016 IEEE Conference on Computer Vision and Pattern Recognition; 2016:770–778.
- 57. Drozdzal M, Vorontsov E, Chartrand G, Kadoury S, Pal C. The importance of skip connections in biomedical image segmentation. In: Carneiro G, Mateus D, Peter L, Bradley A, Tavares J, Belagiannis V, Papa JP, Nascimento JC, Loog M, Lu Z, Cardoso JS, Cornebise J, eds. Deep Learning and Data Labeling for Medical Applications, Vol. 10008. Berlin: Springer; 2016:179–187. [Google Scholar]
- 58. Greenspan H, van Ginneken B, Summers RM. Guest editorial deep learning in medical imaging: overview and future promise of an exciting new technique. IEEE Trans Med Imaging. 2016;35:1153–1159. [Google Scholar]
- 59. Simonyan K, Zisserman A. Very deep convolutional networks for large‐scale image recognition; 2014. arXiv:1409.1556.
- 60. Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In: CVPR, 3; 2017.
- 61. Chollet F. Xception: deep learning with depthwise separable convolutions. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017:1800–1807.
- 62. Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016:2818–2826.
- 63. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. [DOI] [PubMed] [Google Scholar]
- 64. Kooi T, Litjens G, van Ginneken B, et al. Large scale deep learning for computer aided detection of mammographic lesions. Med Image Anal. 2017;35:303–312. [DOI] [PubMed] [Google Scholar]
- 65. Liu Y, Gadepalli K, Norouzi M, et al. Detecting cancer metastases on gigapixel pathology images; 2017. arXiv:1703.02442.
- 66. Ribli D, Horváth A, Unger Z, Pollner P, Csabai I. Detecting and classifying lesions in mammograms with deep learning. Sci Rep. 2018;8:4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tajbakhsh N, Shin JY, Gurudu SR, et al. Convolutional neural networks for medical image analysis: full training or fine tuning? IEEE Trans Med Imaging. 2016;35:1299–1312. [DOI] [PubMed] [Google Scholar]
- 68. Coates A, Huval B, Wang T, Wu D, Catanzaro B, Andrew N. Deep learning with COTS HPC systems. Int Conf Mach Learn. 2013;1337–1345. [Google Scholar]
- 69. Amazon Web Services. https://aws.amazon.com/.
- 70. NVidia GPU Cloud. https://www.nvidia.com/en-us/gpu-cloud/.
- 71. Google Cloud TPU. https://cloud.google.com/tpu/.
- 72. Abadi M, Barham P, Chen J, et al. TensorFlow: a system for large‐scale machine learning. OSDI. 2016;265–283.
- 73. Jia Y, Shelhamer E, Donahue J, et al. Caffe: convolutional architecture for fast feature embedding. In: Proceedings of the 22nd ACM international conference on Multimedia; 2014:675–678.
- 74. Collobert R, Kavukcuoglu K, Farabet C. Torch7: a matlab‐like environment for machine learning. In: BigLearn, NIPS Workshop; 2011.
- 75. Bastien F, Lamblin P, Pascanu R, et al. Theano: new features and speed improvements; 2012. arXiv:1211.5590.
- 76. Cha KH, Hadjiiski L, Samala RK, Chan HP, Caoili EM, Cohan RH. Urinary bladder segmentation in CT urography using deep‐learning convolutional neural network and level sets. Med Phys. 2016;43:1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Cha KH, Hadjiiski LM, Samala RK, et al. Bladder cancer segmentation in CT for treatment response assessment: application of deep‐learning convolution neural network – a pilot study. Tomography. 2016;2:421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Avendi MR, Kheradvar A, Jafarkhani H. Automatic segmentation of the right ventricle from cardiac MRI using a learning‐based approach. Magn Reson Med. 2017;78:2439–2448. [DOI] [PubMed] [Google Scholar]
- 79. Ngo TA, Lu Z, Carneiro G. Combining deep learning and level set for the automated segmentation of the left ventricle of the heart from cardiac cine magnetic resonance. Med Image Anal. 2017;35:159–171. [DOI] [PubMed] [Google Scholar]
- 80. Lu F, Wu F, Hu P, Peng Z, Kong D. Automatic 3D liver location and segmentation via convolutional neural network and graph cut. Int J Comput Assist Radiol Surg. 2017;12:171–182. [DOI] [PubMed] [Google Scholar]
- 81. Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med. 2018;79:2379–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Avendi MR, Kheradvar A, Jafarkhani H. A combined deep‐learning and deformable‐model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal. 2016;30:108–119. [DOI] [PubMed] [Google Scholar]
- 83. Ibragimov B, Xing L. Segmentation of organs‐at‐risks in head and neck CT images using convolutional neural networks. Med Phys. 2017;44:547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ronneberger O, Fischer P, Brox T. U‐Net: convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI), Cham; 2015:234–241.
- 85. Dalmis MU, Litjens G, Holland K, et al. Using deep learning to segment breast and fibroglandular tissue in MRI volumes. Med Phys. 2017;44:533–546. [DOI] [PubMed] [Google Scholar]
- 86. Nie D, Wang L, Trullo R, et al. Segmentation of craniomaxillofacial bony structures from MRI with a 3D deep‐learning based cascade framework. Mach Learn Med Imaging. 2017;10541:266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cheng R, Roth HR, Lay N, et al. Automatic magnetic resonance prostate segmentation by deep learning with holistically nested networks. J Med Imaging (Bellingham). 2017;4:041302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zhuge Y, Krauze AV, Ning H, et al. Brain tumor segmentation using holistically nested neural networks in MRI images. Med Phys. 2017;44:5234–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lee H, Troschel FM, Tajmir S, et al. Pixel‐level deep segmentation: artificial intelligence quantifies muscle on computed tomography for body morphometric analysis. J Digit Imaging. 2017;30:487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wang Y, Qiu Y, Thai T, Moore K, Liu H, Zheng B. A two‐step convolutional neural network based computer‐aided detection scheme for automatically segmenting adipose tissue volume depicting on CT images. Comput Methods Programs Biomed. 2017;144:97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Hu P, Wu F, Peng J, Bao Y, Chen F, Kong D. Automatic abdominal multi‐organ segmentation using deep convolutional neural network and time‐implicit level sets. Int J Comput Assist Radiol Surg. 2017;12:399–411. [DOI] [PubMed] [Google Scholar]
- 92. Mansoor A, Cerrolaza JJ, Idrees R, et al. Deep learning guided partitioned shape model for anterior visual pathway segmentation. IEEE Trans Med Imaging. 2016;35:1856–1865. [DOI] [PubMed] [Google Scholar]
- 93. Choi H, Jin KH. Fast and robust segmentation of the striatum using deep convolutional neural networks. J Neurosci Methods. 2016;274:146–153. [DOI] [PubMed] [Google Scholar]
- 94. Moeskops P, Viergever MA, Mendrik AM, de Vries LS, Benders MJ, Isgum I. Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans Med Imaging. 2016;35:1252–1261. [DOI] [PubMed] [Google Scholar]
- 95. Wachinger C, Reuter M, Klein T. DeepNAT: deep convolutional neural network for segmenting neuroanatomy. NeuroImage. 2018;170:434–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Chen H, Dou Q, Yu L, Qin J, Heng PA. VoxResNet: deep voxelwise residual networks for brain segmentation from 3D MR images. NeuroImage. 2018;170:446–455. [DOI] [PubMed] [Google Scholar]
- 97. Dolz J, Desrosiers C, Ben I. Ayed, “3D fully convolutional networks for subcortical segmentation in MRI: a large‐scale study”. NeuroImage. 2018;170:456–470. [DOI] [PubMed] [Google Scholar]
- 98. Kallenberg M, Petersen K, Nielsen M, et al. Unsupervised deep learning applied to breast density segmentation and mammographic risk scoring. IEEE Trans Med Imaging. 2016;35:1322–1331. [DOI] [PubMed] [Google Scholar]
- 99. Tan LK, McLaughlin RA, Lim E, Abdul Aziz YF, Liew YM. Fully automated segmentation of the left ventricle in cine cardiac MRI using neural network regression. J Magn Reson Imaging. 2018;48:140–452. [DOI] [PubMed] [Google Scholar]
- 100. Tan LK, Liew YM, Lim E, McLaughlin RA. Convolutional neural network regression for short‐axis left ventricle segmentation in cardiac cine MR sequences. Med Image Anal. 2017;39:78–86. [DOI] [PubMed] [Google Scholar]
- 101. Yu L, Guo Y, Wang Y, Yu J, Chen P. Segmentation of fetal left ventricle in echocardiographic sequences based on dynamic convolutional neural networks. IEEE Trans Biomed Eng. 2017;64:1886–1895. [DOI] [PubMed] [Google Scholar]
- 102. Kline TL, Korfiatis P, Edwards ME, et al. Performance of an artificial multi‐observer deep neural network for fully automated segmentation of polycystic kidneys. J Digit Imaging. 2017;30:442–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Sharma K, Rupprecht C, Caroli A, et al. Automatic segmentation of kidneys using deep learning for total kidney volume quantification in autosomal dominant polycystic kidney disease. Sci Rep. 2017;7:2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hu P, Wu F, Peng J, Liang P, Kong D. Automatic 3D liver segmentation based on deep learning and globally optimized surface evolution. Phys Med Biol. 2016;61:8676–8698. [DOI] [PubMed] [Google Scholar]
- 105. Kovacs W, Hsieh N, Roth H, et al. Holistic segmentation of the lung in cine MRI. J Med Imaging (Bellingham). 2017;4:041310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Farag A, Lu L, Roth HR, Liu J, Turkbey E, Summers RM. A bottom‐up approach for pancreas segmentation using cascaded superpixels and (Deep) image patch labeling. In: IEEE Transactions on Image Processing; 2016. [DOI] [PubMed]
- 107. Roth HR, Lu L, Farag A, et al. Deeporgan: multi‐level deep convolutional networks for automated pancreas segmentation. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2015:556–564.
- 108. Guo Y, Gao Y, Shen D. Deformable MR prostate segmentation via deep feature learning and sparse patch matching. IEEE Trans Med Imaging. 2016;35:1077–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Liao S, Gao Y, Oto A, Shen D. Representation learning: a unified deep learning framework for automatic prostate MR segmentation. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2013;254–261. [DOI] [PMC free article] [PubMed]
- 110. Tian Z, Liu L, Zhang Z, Fei B. PSNet: prostate segmentation on MRI based on a convolutional neural network. J Med Imaging (Bellingham). 2008;5:021208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Men K, Dai J, Li Y. Automatic segmentation of the clinical target volume and organs at risk in the planning CT for rectal cancer using deep dilated convolutional neural networks. Med Phys. 2017;44:6377. [DOI] [PubMed] [Google Scholar]
- 112. Li X, Dou Q, Chen H, et al. 3D multi‐scale FCN with random modality voxel dropout learning for Intervertebral disc localization and segmentation from multi‐modality MR images. Med Image Anal. 2018;45:41–54. [DOI] [PubMed] [Google Scholar]
- 113. Zhou X, Takayama R, Wang S, Hara T, Fujita H. Deep learning of the sectional appearances of 3D CT images for anatomical structure segmentation based on an FCN voting method. Med Phys. 2017;44:5221–5233. [DOI] [PubMed] [Google Scholar]
- 114. Dou Q, Yu L, Chen H, et al. 3D deeply supervised network for automated segmentation of volumetric medical images. Med Image Anal. 2017;41:40–54. [DOI] [PubMed] [Google Scholar]
- 115. Alex V, Vaidhya K, Thirunavukkarasu S, Kesavadas C, Krishnamurthi G. Semisupervised learning using denoising autoencoders for brain lesion detection and segmentation. J Med Imaging (Bellingham). 2017;4:041311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Korfiatis P, Kline TL, Erickson BJ. Automated segmentation of hyperintense regions in FLAIR MRI using deep learning. Tomography. 2016;2:334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Iqbal S, Ghani MU, Saba T, Rehman A. Brain tumor segmentation in multi‐spectral MRI using convolutional neural networks (CNN). Microsc Res Tech. 2018;81:419–427. [DOI] [PubMed] [Google Scholar]
- 118. Al‐antari MA, Al‐masni MA, Choi M‐T, Han S‐M, Kim T‐S. A fully integrated computer‐aided diagnosis system for digital X‐ray mammograms via deep learning detection, segmentation, and classification. Int J Med Inform. 2018;117:44–54. [DOI] [PubMed] [Google Scholar]
- 119. Zhang R, Huang L, Xia W, Zhang B, Qiu B, Gao X. Multiple supervised residual network for osteosarcoma segmentation in CT images. Comput Med Imaging Graph. 2018;63:1–8. [DOI] [PubMed] [Google Scholar]
- 120. Huang L, Xia W, Zhang B, Qiu B, Gao X. MSFCN‐multiple supervised fully convolutional networks for the osteosarcoma segmentation of CT images. Comput Methods Programs Biomed. 2017;143:67–74. [DOI] [PubMed] [Google Scholar]
- 121. Kamnitsas K, Ledig C, Newcombe VFJ, et al. Efficient multi‐scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal. 2017;36:61–78. [DOI] [PubMed] [Google Scholar]
- 122. Liu Y, Stojadinovic S, Hrycushko B, et al. A deep convolutional neural network‐based automatic delineation strategy for multiple brain metastases stereotactic radiosurgery. PLoS ONE. 2017;12:e0185844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Havaei M, Davy A, Warde‐Farley D, et al. Brain tumor segmentation with deep neural networks. Med Image Anal. 2017;35:18–31. [DOI] [PubMed] [Google Scholar]
- 124. Zhao X, Wu Y, Song G, Li Z, Zhang Y, Fan Y. A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Med Image Anal. 2018;43:98–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Chen L, Bentley P, Rueckert D. Fully automatic acute ischemic lesion segmentation in DWI using convolutional neural networks. Neuroimage Clin. 2017;15:633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Brosch T, Tang LY, Youngjin Y, Li DK, Traboulsee A, Tam R. Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans Med Imaging. 2016;35:1229–1239. [DOI] [PubMed] [Google Scholar]
- 127. Ghafoorian M, Karssemeijer N, Heskes T, et al. Location sensitive deep convolutional neural networks for segmentation of white matter hyperintensities. Sci Rep. 2017;7:5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Men K, Chen X, Zhang Y, et al. Deep deconvolutional neural network for target segmentation of nasopharyngeal cancer in planning computed tomography images. Front Oncol. 2017;7:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Ma J, Wu F, Jiang T, Zhao Q, Kong D. Ultrasound image‐based thyroid nodule automatic segmentation using convolutional neural networks. Int J Comput Assist Radiol Surg. 2017;12:1895–1910. [DOI] [PubMed] [Google Scholar]
- 130. Li W, Jia F, Hu Q. Automatic segmentation of liver tumor in CT images with deep convolutional neural networks. J Comput Commun. 2015;3:146. [Google Scholar]
- 131. Wang S, Zhou M, Liu Z, et al. Central focused convolutional neural networks: developing a data‐driven model for lung nodule segmentation. Med Image Anal. 2017;40:172–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Nogues I, Lu L, Wang X, et al. Automatic lymph node cluster segmentation using holistically‐nested neural networks and structured optimization in CT images. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2016:388–397.
- 133. Trebeschi S, van Griethuysen JJM, Lambregts DMJ, et al. Deep learning for fully‐automated localization and segmentation of rectal cancer on multiparametric MR. Sci Rep. 2017;7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Jafari MH, Nasr‐Esfahani E, Karimi N, Soroushmehr SMR, Samavi S, Najarian K. Extraction of skin lesions from non‐dermoscopic images for surgical excision of melanoma. Int J Comput Assist Radiol Surg. 2017;12:1021–1030. [DOI] [PubMed] [Google Scholar]
- 135. Yang D, Zhang S, Yan Z, Tan C, Li K, Metaxas D. Automated anatomical landmark detection on distal femur surface using convolutional neural network. In: 2015 IEEE 12th International Symposium on Biomedical Imaging; 2005:17–21.
- 136. Chen H, Ni D, Qin J, et al. Standard plane localization in fetal ultrasound via domain transferred deep neural networks. IEEE J Biomed Health Inform. 2015;19:1627–1636. [DOI] [PubMed] [Google Scholar]
- 137. Baumgartner CF, Kamnitsas K, Matthew J, Smith S, Kainz B, Rueckert D. Real‐time standard scan plane detection and localisation in fetal ultrasound using fully convolutional neural networks. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2016:203–211.
- 138. Kumar A, Sridar P, Quinton A, et al. Plane identification in fetal ultrasound images using saliency maps and convolutional neural networks. In: 2016 IEEE 13th International Symposium on Biomedical Imaging; 2016:791–794.
- 139. Ghesu FC, Krubasik E, Georgescu B, et al. Marginal space deep learning: efficient architecture for volumetric image parsing. IEEE Trans Med Imaging. 2016;35:1217–1228. [DOI] [PubMed] [Google Scholar]
- 140. Wu H, Bailey C, Rasoulinejad P, Li S. Automatic landmark estimation for adolescent idiopathic scoliosis assessment using BoostNet. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2017:127–135.
- 141. Yan K, Lu L, Summers RM. Unsupervised body part regression using convolutional neural network with self‐organization; 2017. arXiv:1707.03891.
- 142. Yan Z, Zhan Y, Peng Z, et al. Multi‐instance deep learning: discover discriminative local anatomies for bodypart recognition. IEEE Trans Med Imaging. 2016;35:1332–1343. [DOI] [PubMed] [Google Scholar]
- 143. Ghesu FC, Georgescu B, Mansi T, Neumann D, Hornegger J, Comaniciu D. An artificial agent for anatomical landmark detection in medical images. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2016:229–237.
- 144. Ghesu FC, Georgescu B, Zheng Y, et al. Multi‐scale deep reinforcement learning for real‐time 3D‐landmark detection in CT scans. IEEE Trans Patt Anal Mach Intell. 2017. [Epub ahead of print]. 10.1109/TPAMI.2017.2782687 [DOI] [PubMed] [Google Scholar]
- 145. Ghesu FC, Georgescu B, Grbic S, Maier AK, Hornegger J, Comaniciu D. Robust multi‐scale anatomical landmark detection in incomplete 3D‐CT data. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2017:194–202.
- 146. Xu Z, Huang Q, Park J, et al. Supervised action classifier: approaching landmark detection as image partitioning. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2017:338–346.
- 147. Payer C, Štern D, Bischof H, Urschler M. Regressing heatmaps for multiple landmark localization using CNNs. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2016:230–238.
- 148. Cai Y, Landis M, Laidley DT, Kornecki A, Lum A, Li S. Multi‐modal vertebrae recognition using transformed deep convolution network. Comput Med Imaging Graph. 2016;51:11–19. [DOI] [PubMed] [Google Scholar]
- 149. Baka N, Leenstra S, Walsum TV. Ultrasound aided vertebral level localization for lumbar surgery. IEEE Trans Med Imaging. 2017;36:2138–2147. [DOI] [PubMed] [Google Scholar]
- 150. Zheng Y, Liu D, Georgescu B, Nguyen H, Comaniciu D. 3D Deep learning for efficient and robust landmark detection in volumetric data. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2015:565–572.
- 151. Chen H, Dou Q, Ni D, et al. Automatic fetal ultrasound standard plane detection using knowledge transferred recurrent neural networks. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2015:507–514.
- 152. Lu X, Xu D, Liu D. Robust 3D organ localization with dual learning architectures and fusion. In: Deep Learning and Data Labeling for Medical Applications. Berlin: Springer. 2016:12–20. [Google Scholar]
- 153. Roth HR, Lee CT, Shin H‐C, et al. Anatomy‐specific classification of medical images using deep convolutional nets. In: 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI); 2015:101–104.
- 154. de Vos BD, Wolterink JM, de Jong PA, Leiner T, Viergever MA, Išgum I. ConvNet‐based localization of anatomical structures in 3‐D medical images. IEEE Trans Med Imaging. 2017;36:1470–1481. [DOI] [PubMed] [Google Scholar]
- 155. Zhang J, Liu M, Shen D. Detecting anatomical landmarks from limited medical imaging data using two‐stage task‐oriented deep neural networks. IEEE Trans Image Process. 2017;26:4753–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Harrison AP, Xu Z, George K, Lu L, Summers RM, Mollura DJ. Progressive and multi‐path holistically nested neural networks for pathological lung segmentation from CT images. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention; 2017:621–629.
- 157. Yao J, Kovacs W, Hsieh N, Liu C‐Y, Summers RM. Holistic segmentation of intermuscular adipose tissues on thigh MRI. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention; 2017:737–745.
- 158. Giger ML, Chan HP, Boone J. Anniversary paper: history and status of CAD and quantitative image analysis: the role of medical physics and AAPM. Med Phys. 2008;35:5799–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Giger, M. L. , Karssemeijer, N. , & Schnabel, J. A. (2013). Breast image analysis for risk assessment, detection, diagnosis, and treatment of cancer. Annual review of biomedical engineering., 15, 327‐357. [DOI] [PubMed] [Google Scholar]
- 160. Shin H‐C, Roth HR, Gao M, et al. Deep convolutional neural networks for computer‐aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35:1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Yap MH, Pons G, Martí J, et al. Automated breast ultrasound lesions detection using convolutional neural networks.IEEE J Biomed Health Inform. 2017:22;1218–1226. [DOI] [PubMed] [Google Scholar]
- 162. Cheng PM, Tejura TK, Tran KN, Whang G. Detection of high‐grade small bowel obstruction on conventional radiography with convolutional neural networks. Abdom Radiol. 2017; 43:1120–1127. [DOI] [PubMed] [Google Scholar]
- 163. Belharbi S, Chatelain C, Hérault R, et al. Spotting L3 slice in CT scans using deep convolutional network and transfer learning. Comput Biol Med. 2017;87:95–103. [DOI] [PubMed] [Google Scholar]
- 164. Albarqouni S, Baur C, Achilles F, Belagiannis V, Demirci S, Navab N. Aggnet: deep learning from crowds for mitosis detection in breast cancer histology images. IEEE Trans Med Imaging. 2016;35:1313–1321. [DOI] [PubMed] [Google Scholar]
- 165. Orlando JI, Prokofyeva E, del Fresno M, Blaschko MB. An ensemble deep learning based approach for red lesion detection in fundus images. Comput Methods Programs Biomed. 2018;153:115–127. [DOI] [PubMed] [Google Scholar]
- 166. Roth HR, Lu L, Liu J, et al. Improving computer‐aided detection using convolutional neural networks and random view aggregation. IEEE Trans Med Imaging. 2016;35:1170–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Yang X, Liu C, Wang Z, et al. Co‐trained convolutional neural networks for automated detection of prostate cancer in multi‐parametric MRI. Med Image Anal. 2017;42:212–227. [DOI] [PubMed] [Google Scholar]
- 168. Setio AA, Ciompi F, Litjens G, et al. Pulmonary nodule detection in CT images: false positive reduction using multi‐view convolutional networks. IEEE Trans Med Imaging. 2016;35:1160–1169. [DOI] [PubMed] [Google Scholar]
- 169. Dou Q, Chen H, Yu L, Qin J, Heng PA. Multilevel contextual 3‐D CNNs for false positive reduction in pulmonary nodule detection. IEEE Trans Biomed Eng. 2017;64:1558–1567. [DOI] [PubMed] [Google Scholar]
- 170. Ghafoorian M, Karssemeijer N, Heskes T, et al. Deep multi‐scale location‐aware 3D convolutional neural networks for automated detection of lacunes of presumed vascular origin. Neuroimage Clin. 2017;14:391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2015:3431–3440. [DOI] [PubMed]
- 172. Qi D, Hao C, Lequan Y, et al. Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans Med Imaging. 2016;35:1182–1195. [DOI] [PubMed] [Google Scholar]
- 173. Ciompi F, de Hoop B, van Riel SJ, et al. Automatic classification of pulmonary peri‐fissural nodules in computed tomography using an ensemble of 2D views and a convolutional neural network out‐of‐the‐box. Med Image Anal. 2015;26:195–202. [DOI] [PubMed] [Google Scholar]
- 174. Chen S, Qin J, Ji X, et al. Automatic scoring of multiple semantic attributes with multi‐task feature leverage: a study on pulmonary nodules in CT images. IEEE Trans Med Imaging. 2017;36:802–814. [DOI] [PubMed] [Google Scholar]
- 175. Teramoto A, Fujita H, Yamamuro O, Tamaki T. Automated detection of pulmonary nodules in PET/CT images: ensemble false‐positive reduction using a convolutional neural network technique. Med Phys. 2016;43:2821–2827. [DOI] [PubMed] [Google Scholar]
- 176. Jiang H, Ma H, Qian W, Gao M, Li Y. An automatic detection system of lung nodule based on multi‐group patch‐based deep learning network. IEEE J Biomed Health Inform. 2017;22:1227–1237. [DOI] [PubMed] [Google Scholar]
- 177. Bar Y, Diamant I, Wolf L, Lieberman S, Konen E, Greenspan H. Chest pathology detection using deep learning with non‐medical training. In: 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI); 2015:294–297.
- 178. Cicero M, Bilbily A, Colak E, et al. Training and validating a deep convolutional neural network for computer‐aided detection and classification of abnormalities on frontal chest radiographs. Invest Radiol. 2017;52:281–287. [DOI] [PubMed] [Google Scholar]
- 179. Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284:574–582. [DOI] [PubMed] [Google Scholar]
- 180. Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. Chestx‐ray8: hospital‐scale chest x‐ray database and benchmarks on weakly‐supervised classification and localization of common thorax diseases. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017:3462–3471.
- 181. Bar Y, Diamant I, Wolf L, Greenspan H. Deep learning with non‐medical training used for chest pathology identification. In: Hadjiiski LM, Tourassi GD, eds. Proc. SPIE Medical Imaging, Vol. 9414; 2015:94140V.
- 182. Nakao T, Hanaoka S, Nomura Y, et al. Deep neural network‐based computer‐assisted detection of cerebral aneurysms in MR angiography. J Magn Reson Imaging. 2018;47:948–953. [DOI] [PubMed] [Google Scholar]
- 183. Dalmış MU, Vreemann S, Kooi T, Mann RM, Karssemeijer N, Gubern‐Mérida A. Fully automated detection of breast cancer in screening MRI using convolutional neural networks. J Med Imaging. 2018;5:014502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Liu J, Wang D, Lu L, et al. Detection and diagnosis of colitis on computed tomography using deep convolutional neural networks. Med Phys. 2017;44:4630–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Nappi JJ, Pickhardt P, Kim DH, Hironaka T, Yoshida H. Deep learning of contrast‐coated serrated polyps for computer‐aided detection in CT colonography. In: Armato SG, Petrick NA, eds. Medical Imaging 2017: Computer‐Aided Diagnosis, Vol. 10134; 2017.
- 186. Roth HR, Lu L, Seff A, et al. A new 2.5D representation for lymph node detection using random sets of deep convolutional neural network observations. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2014:520–527. [DOI] [PMC free article] [PubMed]
- 187. Vivanti R, Szeskin A, Lev‐Cohain N, Sosna J, Joskowicz L. Automatic detection of new tumors and tumor burden evaluation in longitudinal liver CT scan studies. Int J Comput Assist Radiol Surg. 2017;12:1945–1957. [DOI] [PubMed] [Google Scholar]
- 188. Ma J, Wu F, Jiang TA, Zhu J, Kong D. Cascade convolutional neural networks for automatic detection of thyroid nodules in ultrasound images. Med Phys. 2017;44:1678–1691. [DOI] [PubMed] [Google Scholar]
- 189. Tsehay Y, Lay N, Wang X, et al. Biopsy‐guided learning with deep convolutional neural networks for prostate cancer detection on multiparametric MRI. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017); 2017:642–645.
- 190. Liu J, Chellamuthu K, Lu L, Bagheri M, Summers RM. A coarse‐to‐fine approach for pericardial effusion localization and segmentation in chest CT scans. In: Petrick N, Mori K, eds. Proc. SPIE Medical Imaging, Vol. 10575; 2018:105753B.
- 191. Chellamuthu K, Liu J, Yao J, et al. Atherosclerotic vascular calcification detection and segmentation on low dose computed tomography scans using convolutional neural networks. In: 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017); 2017:388–391.
- 192. Li H, Giger ML, Huynh BQ, Antropova NO. Deep learning in breast cancer risk assessment: evaluation of convolutional neural networks on a clinical dataset of full‐field digital mammograms. J Med Imaging (Bellingham). 2017;4:041304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Mohamed AA, Berg WA, Peng H, Luo Y, Jankowitz RC, Wu S. A deep learning method for classifying mammographic breast density categories. Med Phys. 2018;45:314–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Li SF, Wei J, Chan HP, et al. Computer‐aided assessment of breast density: comparison of supervised deep learning and feature‐based statistical learning. Phys Med Biol. 2018;63:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Lee J, Nishikawa RM. Automated mammographic breast density estimation using a fully convolutional network. Med Phys. 2018;45:1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Huynh BQ, Li H, Giger ML. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J Med Imaging (Bellingham). 2016;3:034501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Antropova N, Huynh BQ, Giger ML. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med Phys. 2017;44:5162–5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Samala RK, Chan HP, Hadjiiski LM, Helvie MA, Richter C, Cha K. Evolutionary pruning of transfer learned deep convolutional neural network for breast cancer diagnosis in digital breast tomosynthesis. Phys Med Biol. 2018;63:095005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Samala RK, Chan HP, Hadjiiski LM, Helvie MA, Cha KH, Richter CD. Multi‐task transfer learning deep convolutional neural network: application to computer‐aided diagnosis of breast cancer on mammograms. Phys Med Biol. 2017;62:8894–8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Antropova N, Abe H, Giger ML. Use of clinical MRI maximum intensity projections for improved breast lesion classification with deep convolutional neural networks. J Med Imaging (Bellingham). 2018;5:014503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Antropova N, Huynh B, Giger ML. Long short‐term memory networks for efficient breast DCE‐MRI classification. In: NIPS: Neural Information Processing Systems, Medical Imaging Meets NIPS; 2017.
- 202. Kooi T, van Ginneken B, Karssemeijer N, den Heeten A. Discriminating solitary cysts from soft tissue lesions in mammography using a pretrained deep convolutional neural network. Med Phys. 2017;44:1017–1027. [DOI] [PubMed] [Google Scholar]
- 203. Shi BB, Grimm LJ, Mazurowski MA, et al. Prediction of occult invasive disease in ductal carcinoma in situ using deep learning features. J Am Coll Radiol. 2018;15:527–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Nibali A, He Z, Wollersheim D. Pulmonary nodule classification with deep residual networks. Int J Comput Assist Radiol Surg. 2017;12:1799–1808. [DOI] [PubMed] [Google Scholar]
- 205. Schlegl T, Ofner J, Langs G. Unsupervised pre‐training across image domains improves lung tissue classification. In: Medical Computer Vision: Algorithms for Big Data; 2014:82–93.
- 206. Gao M, Bagci U, Lu L, et al. Holistic classification of CT attenuation patterns for interstitial lung diseases via deep convolutional neural networks. Comput Methods Biomech Biomed Eng. 2018;6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Anthimopoulos M, Christodoulidis S, Ebner L, Christe A, Mougiakakou S. Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans Med Imaging. 2016;35:1207–1216. [DOI] [PubMed] [Google Scholar]
- 208. Kim GB, Jung KH, Lee Y, et al. Comparison of shallow and deep learning methods on classifying the regional pattern of diffuse lung disease. J Digit Imaging. 2017;31:415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Christodoulidis S, Anthimopoulos M, Ebner L, Christe A, Mougiakakou S. Multisource transfer learning with convolutional neural networks for lung pattern analysis. IEEE J Biomed Health Inform. 2017;21:76–84. [DOI] [PubMed] [Google Scholar]
- 210. Masood A, Sheng B, Li P, et al. Computer‐assisted decision support system in pulmonary cancer detection and stage classification on CT images. J Biomed Inform. 2018;79:117–128. [DOI] [PubMed] [Google Scholar]
- 211. Gonzalez G, Ash SY, Vegas‐Sanchez‐Ferrero G, et al. Disease staging and prognosis in smokers using deep learning in chest computed tomography. Am J Respir Crit Care Med. 2018;197:193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Lessmann N, van Ginneken B, Zreik M, et al. Automatic calcium scoring in low‐dose chest CT using deep neural networks with dilated convolutions. IEEE Trans Med Imaging. 2018;37:615–625. [DOI] [PubMed] [Google Scholar]
- 213. Xue WF, Brahm G, Pandey S, Leung S, Li S. Full left ventricle quantification via deep multitask relationships learning. Med Image Anal. 2018;43:54–65. [DOI] [PubMed] [Google Scholar]
- 214. Cheng PM, Malhi HS. Transfer learning with convolutional neural networks for classification of abdominal ultrasound images. J Digit Imaging. 2017;30:234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Frid‐Adar M, Klang E, Amitai M, Goldberger J, Greenspan H. Synthetic data augmentation using GAN for improved liver lesion classification. In: IEEE International Symposium on Biomedical Imaging; 2018.
- 216. Yasaka K, Akai H, Abe O, Kiryu S. Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast‐enhanced CT: a preliminary study. Radiology. 2018;286:899–908. [DOI] [PubMed] [Google Scholar]
- 217. Bharath R, Rajalakshmi P. Deep scattering convolution network based features for ultrasonic fatty liver tissue characterization. In: 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society; 2017:1982–1985. [DOI] [PubMed]
- 218. Lao J, Chen Y, Li ZC, et al. A deep learning‐based radiomics model for prediction of survival in glioblastoma multiforme. Sci Rep. 2017;7:10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Larson DB, Chen MC, Lungren MP, Halabi SS, Stence NV, Langlotz CP. Performance of a deep‐learning neural network model in assessing skeletal maturity on pediatric hand radiographs. Radiology. 2018;287:313–322. [DOI] [PubMed] [Google Scholar]
- 220. Li H, Zhu Y, Burnside ES, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer. 2016;2:16012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Burnside ES, Drukker K, Li H, et al. Using computer‐extracted image phenotypes from tumors on breast magnetic resonance imaging to predict breast cancer pathologic stage. Cancer. 2016;122:748–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Zhu YT, Li H, Guo WT, et al. Deciphering genomic underpinnings of quantitative MRI‐based radiomic phenotypes of invasive breast carcinoma. Sci Rep. 2015;5:17787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Li H, Zhu YT, Burnside ES, et al. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology. 2016;281:382–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Guo W, Li H, Zhu Y, et al. Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. J Med Imaging (Bellingham). 2015;2:041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Sun WQ, Tseng TL, Zhang JY, Qian W. Enhancing deep convolutional neural network scheme for breast cancer diagnosis with unlabeled data. Comput Med Imaging Graph. 2017;57:4–9. [DOI] [PubMed] [Google Scholar]
- 226. Jamieson AR, Giger ML, Drukker K, Pesce LL. Enhancement of breast CADx with unlabeled data. Med Phys. 2010;37:4155–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Katsuragawa S, Doi K, MacMahon H, MonnierCholley L, Ishida T, Kobayashi T. Classification of normal and abnormal lungs with interstitial diseases by rule‐based method and artificial neural networks. J Digit Imaging. 1997;10:108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. van Ginneken B, Katsuragawa S, Romeny BMT, Doi K, Viergever MA. Automatic detection of abnormalities in chest radiographs using local texture analysis. IEEE Trans Med Imaging. 2002;21:139–149. [DOI] [PubMed] [Google Scholar]
- 229. Li H, Giger ML, Lan L, et al. Computerized analysis of mammographic parenchymal patterns on a large clinical dataset of full‐field digital mammograms: robustness study with two high‐risk datasets. J Digit Imaging. 2015;25:591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. He H, Yang X, Wu L, et al. Dual long short‐term memory networks for sub‐character representation learning; 2018. arXiv:1712.08841.
- 231. Garapati SS, Hadjiiski L, Cha KH, et al. Urinary bladder cancer staging in CT urography using machine learning. Med Phys. 2017;44:5814–5823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232. Henglin M, Stein G, Hushcha PV, Snoek J, Wiltschko AB, Cheng S. Machine learning approaches in cardiovascular imaging. Circ Cardiovasc Imaging. 2017;10:e005614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Zhang L, Lu L, Summers RM, Kebebew E, Yao J. Convolutional invasion and expansion networks for tumor growth prediction. IEEE Trans Med Imaging. 2018;37:638–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Suzuki K. A supervised ‘lesion‐enhancement’ filter by use of a massive‐training artificial neural network (MTANN) in computer‐aided diagnosis (CAD). Phys Med Biol. 2009;54:S31–S45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235. Yang W, Chen YY, Liu YB, et al. Cascade of multi‐scale convolutional neural networks for bone suppression of chest radiographs in gradient domain. Med Image Anal. 2017;35:421–433. [DOI] [PubMed] [Google Scholar]
- 236. Mori S. Deep architecture neural network‐based real‐time image processing for image‐guided radiotherapy. Phys Med. 2017;40:79–87. [DOI] [PubMed] [Google Scholar]
- 237. Chen H, Zhang Y, Zhang W, et al. Low‐dose CT via convolutional neural network. Biomed Opt Express. 2017;8:679–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Chen H, Zhang Y, Kalra MK, et al. Low‐dose CT with a residual encoder‐decoder convolutional neural network. IEEE Trans Med Imaging. 2017;36:2524–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Kang E, Min J, Ye JC. A deep convolutional neural network using directional wavelets for low‐dose X‐ray CT reconstruction. Med Phys. 2017;44:e360–e375. [DOI] [PubMed] [Google Scholar]
- 240. Yang X, De Andrade V, Scullin W, et al. Low‐dose x‐ray tomography through a deep convolutional neural network. Sci Rep. 2018;8:2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241. Xiang L, Qiao Y, Nie D, An L, Wang Q, Shen D. Deep auto‐context convolutional neural networks for standard‐dose PET image estimation from low‐dose PET/MRI. Neurocomputing. 2017;267:406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Jin KH, McCann MT, Froustey E, Unser M. Deep convolutional neural network for inverse problems in imaging. IEEE Trans Image Process. 2017;26:4509–4522. [DOI] [PubMed] [Google Scholar]
- 243. Zhang YB, Chu Y, Yu HY. Reduction of metal artifacts in x‐ray CT images using a convolutional neural network. In: Muller B, Wang G, eds. Proc. SPIE Developments in X‐Ray Tomography XI, Vol. 10391. 2017:103910V. [Google Scholar]
- 244. Han Y, Yoo J, Kim HH, Shin HJ, Sung K, Ye JC. Deep learning with domain adaptation for accelerated projection‐reconstruction MR. Magn Reson Med. 2018;80:1189–1205. [DOI] [PubMed] [Google Scholar]
- 245. Golkov V, Dosovitskiy A, Sper JI, et al. q‐space deep learning: twelve‐fold shorter and model‐free diffusion MRI scans. IEEE Trans Med Imaging. 2016;35:1344–1351. [DOI] [PubMed] [Google Scholar]
- 246. Hammernik K, Klatzer T, Kobler E, et al. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med. 2018;79:3055–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Schlemper J, Caballero J, Hajnal JV, Price AN, Rueckert D. A deep cascade of convolutional neural networks for dynamic MR image reconstruction. IEEE Trans Med Imaging. 2018;37:491–503. [DOI] [PubMed] [Google Scholar]
- 248. Zhu B, Liu JZ, Cauley SF, Rosen BR, Rosen MS. Image reconstruction by domain‐transform manifold learning. Nature. 2018;555:487–492. [DOI] [PubMed] [Google Scholar]
- 249. Wu G, Kim M, Wang Q, Gao Y, Liao S, Shen D. Unsupervised deep feature learning for deformable registration of MR brain images. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2013:649–656. [DOI] [PMC free article] [PubMed]
- 250. Wu G, Kim M, Wang Q, Munsell BC, Shen D. Scalable high‐performance image registration framework by unsupervised deep feature representations learning. IEEE Trans Biomed Eng. 2016;63:1505–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Yang X, Kwitt R, Styner M, Niethammer M. Quicksilver: fast predictive image registration – a deep learning approach. NeuroImage. 2017;158:378–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Lv J, Yang M, Zhang J, Wang XY. Respiratory motion correction for free‐breathing 3D abdominal MRI using CNN‐based image registration: a feasibility study. Br J Radiol. 2018;91:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Miao S, Wang ZJ, Liao R. A CNN regression approach for real‐time 2D/3D registration. IEEE Trans Med Imaging. 2016;35:1352–1363. [DOI] [PubMed] [Google Scholar]
- 254. Zheng J, Miao S, Jane Wang Z, Liao R. Pairwise domain adaptation module for CNN‐based 2‐D/3‐D registration. J Med Imaging (Bellingham). 2018;5:021204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Nie D, Cao X, Gao Y, Wang L, Shen D. Estimating CT image from MRI data using 3D fully convolutional networks. Deep Learn Data Label Med Appl. 2016;2016:170–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256. Han X. MR‐based synthetic CT generation using a deep convolutional neural network method. Med Phys. 2017;44:1408–1419. [DOI] [PubMed] [Google Scholar]
- 257. Leynes AP, Yang J, Wiesinger F, et al. Direct PseudoCT generation for pelvis PET/MRI attenuation correction using deep convolutional neural networks with multi‐parametric MRI: zero echo‐time and dixon deep pseudoCT (ZeDD‐CT). J Nucl Med. 2017;59:852–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB. Deep learning MR imaging‐based attenuation correction for PET/MR imaging. Radiology. 2018;286:676–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Nie D, Trullo R, Lian J, et al. Medical image synthesis with context‐aware generative adversarial networks. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2017: 417–425. [DOI] [PMC free article] [PubMed]
- 260. Choi H, Lee DS. Generation of structural MR images from amyloid PET: application to MR‐less quantification. J Nucl Med. 2017;59:1111–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Ben‐Cohen A, Klang E, Raskin SP, Amitai MM, Greenspan H. Virtual PET images from CT data using deep convolutional networks: initial results. In: International Workshop on Simulation and Synthesis in Medical Imaging; 2017;49–57.
- 262. Wu L, Cheng JZ, Li S, Lei B, Wang T, Ni D. FUIQA: fetal ultrasound image quality assessment with deep convolutional networks. IEEE Trans Cybern. 2017;47:1336–1349. [DOI] [PubMed] [Google Scholar]
- 263. Neylon J, Min YG, Low DA, Santhanam A. A neural network approach for fast, automated quantification of DIR performance. Med Phys. 2017;44:4126–4138. [DOI] [PubMed] [Google Scholar]
- 264. Lee JH, Grant BR, Chung JH, Reiser I, Giger ML. Assessment of diagnostic image quality of computed tomography (CT) images of the lung using deep learning. In: Proc. SPIE Medical Imaging, Vol. 10573; 2018:105731M
- 265. Esses SJ, Lu X, Zhao T, et al. Automated image quality evaluation of T2‐weighted liver MRI utilizing deep learning architecture. J Magn Reson Imaging. 2018;47:723–728. [DOI] [PubMed] [Google Scholar]
- 266. McCollough C. Low dose CT grand challenge, the Mayo Clinic, the American Association of Physicists in Medicine, and grants EB017095 and grants EB017185 from the National Institute of Biomedical Imaging and Bioengineering; 2016.
- 267. Delso G, Wiesinger F, Sacolick LI, et al. Clinical evaluation of zero‐echo‐time MR imaging for the segmentation of the skull. J Nucl Med. 2015;56:417–422. [DOI] [PubMed] [Google Scholar]
- 268. Cha KH, Hadjiiski L, Chan HP, et al. Bladder cancer treatment response assessment in CT using radiomics with deep‐learning. Sci Rep. 2017;7:8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Nielsen A, Hansen MB, Tietze A, Mouridsen K. Prediction of tissue outcome and assessment of treatment effect in acute ischemic stroke using deep learning. Stroke. 2018;49:1394–1401. [DOI] [PubMed] [Google Scholar]
- 270. Huynh BQ, Antropova N, Giger ML. Comparison of breast DCE‐MRI contrast time points for predicting response to neoadjuvant chemotherapy using deep convolutional neural network features with transfer learning. In: Armato SG, Petrick NA, eds. Proc. SPIE Medical Imaging, Vol. 10134; 2017:101340U.
- 271. Ravichandran K, Braman N, Janowczyk A, Madabhushi A. A deep learning classifier for prediction of pathological complete response to neoadjuvant chemotherapy from baseline breast DCE‐MRI. In: Petrick N, Mori K, eds. Proc. SPIE Medical Imaging, Vol. 10575; 2018: 105750C. [Google Scholar]
- 272. Men K, Boimel P, Janopaul‐Naylor J, et al. Cascaded atrous convolution and spatial pyramid pooling for more accurate tumor target segmentation for rectal cancer radiotherapy. Phys Med Biol. 2018;63:185016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Ding MQ, Chen LJ, Cooper GF, Young JD, Lu XH. Precision oncology beyond targeted therapy: combining omics data with machine learning matches the majority of cancer cells to effective therapeutics. Mol Cancer Res. 2018;16:269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Tong N, Gou S, Yang S, Ruan D, Sheng K. Fully automatic multi‐organ segmentation for head and neck cancer radiotherapy using shape representation model constrained fully convolutional neural networks. Med Phys. 2018;45:4558–4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Jackson P, Hardcastle N, Dawe N, Kron T, Hofman MS, Hicks RJ. Deep learning renal segmentation for fully automated radiation dose estimation in unsealed source therapy. Front Oncol. 2018;8:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Shehata M, Khalifa F, Soliman A, et al. Computer‐aided diagnostic system for early detection of acute renal transplant rejection using diffusion‐weighted MRI. IEEE Trans Biomed Eng. 2018; [Epub ahead of print]. 10.1109/TBME.2018.2849987 [DOI] [PubMed] [Google Scholar]
- 277. Ibragimov B, Toesca D, Chang D, Yuan Y, Koong A, Xing L. Development of deep neural network for individualized hepatobiliary toxicity prediction after liver SBRT. Med Phys. 2018;45:4763–4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Tseng HH, Luo Y, Cui S, Chien JT, Ten Haken RK, El Naqa I. Deep reinforcement learning for automated radiation adaptation in lung cancer. Med Phys. 2017;44:6690–6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Foote MD, Zimmerman B, Sawant A, Joshi S. Real‐time patient‐specific lung radiotherapy targeting using deep learning. In: International Conference on Medical Imaging with Deep Learning (MIDL); 2018.
- 280. Nguyen D, Long T, Jia X, et al. Dose prediction with U‐Net: a feasibility study for predicting dose distributions from contours using deep learning on prostate IMRT patients. 2017; arXiv:1709.09233.
- 281. Kajikawa T, Kadoya N, Ito K, et al. Automated prediction of dosimetric eligibility of patients with prostate cancer undergoing intensity‐modulated radiation therapy using a convolutional neural network. Radiol Phys Technol. 2018;11:320–327. [DOI] [PubMed] [Google Scholar]
- 282. Maspero M, Savenije MHG, Dinkla AM, et al. Fast synthetic CT generation with deep learning for general pelvis MR‐only radiotherapy; 2018. arXiv:1802.06468. [DOI] [PubMed]
- 283. Zhen X, Chen JW, Zhong ZC, et al. Deep convolutional neural network with transfer learning for rectum toxicity prediction in cervical cancer radiotherapy: a feasibility study. Phys Med Biol. 2017;62:8246–8263. [DOI] [PubMed] [Google Scholar]
- 284. Bibault JE, Giraud P, Durdux C, et al. Deep learning and radiomics predict complete response after neo‐adjuvant chemoradiation for locally advanced rectal cancer. Sci Rep. 2018;8:12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285. Giger ML. Machine learning in medical imaging. J Am Coll Radiol. 2018;15:512–520. [DOI] [PubMed] [Google Scholar]
- 286. Men K, Zhang T, Chen X, et al. Fully automatic and robust segmentation of the clinical target volume for radiotherapy of breast cancer using big data and deep learning. Phys Med. 2018;50:13–19. [DOI] [PubMed] [Google Scholar]
- 287. Everingham M, Eslami SMA, Van Gool L, Williams C, Winn J, Zisserman A. The Pascal visual object classes challenge: a retrospective. Int J Comput Vis. 2015;111:98–136. [Google Scholar]
- 288. Lin T‐Y, Maire M, Belongie S, et al. Microsoft coco: common objects in context. Eur Conf Comput Vis. 2014;740–755. [Google Scholar]
- 289. Karpathy A, Fei‐Fei L. Deep visual‐semantic alignments for generating image descriptions. IEEE Trans Pattern Anal Mach Intell. 2017;39:664–676. [DOI] [PubMed] [Google Scholar]
- 290. Nam H, Ha J‐W, Kim J. Dual attention networks for multimodal reasoning and matching. In: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017:299–307.
- 291. Dai B, Zhang Y, Lin D. Detecting visual relationships with deep relational networks. In: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2014:3076–3086.
- 292. Dyson F. A meeting with Enrico Fermi. Nature. 2004;427:297. [DOI] [PubMed] [Google Scholar]
- 293. Mhaskar H, Liao Q, Poggio TA. When and why are deep networks better than shallow ones? In: Proceedings of the Thirty‐First AAAI Conference on Artificial Intelligence. San Francisco, CA; 2017: 2343–2349.
- 294. Schwartz‐Ziv R, Tishby N. Opening the black box of deep neural networks via information; 2017. arXiv:1703.00810.
- 295. Goodfellow I, Bengio Y, Courville A. Regularization for deep learning. In: Deep Learning. Cambridge: MIT Press; 2016:221–265. [Google Scholar]
- 296. Prechelt J. Early Stopping – But When? Berlin, Heidelberg: Springer; 2012. [Google Scholar]
- 297. Fukunaga K, Hayes RR. Effects of sample size in classifier design. IEEE Trans Pattern Anal Mach Intell. 1989;11:873–885. [Google Scholar]
- 298. Wagner RF, Chan H‐P, Sahiner B, Petrick N, Mossoba JT. Finite‐sample effects and resampling plans: applications to linear classifiers in computer‐aided diagnosis. In: Medical Imaging 1997: Image Processing; 1997:467–478.
- 299. Chan HP, Sahiner B, Wagner RF, Petrick N. Classifier design for computer‐aided diagnosis: effects of finite sample size on the mean performance of classical and neural network classifiers. Med Phys. 1999;26:2654–2668. [DOI] [PubMed] [Google Scholar]
- 300. Sahiner B, Chan HP, Petrick N, Wagner RF, Hadjiiski L. Feature selection and classifier performance in computer‐aided diagnosis: the effect of finite sample size. Med Phys. 2000;27:1509–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. Kupinski MA, Edwards DC, Giger ML, Metz CE. Ideal observer approximation using Bayesian classification neural networks. IEEE Trans Med Imaging. 2001;20:886–899. [DOI] [PubMed] [Google Scholar]
- 302. Cho J, Lee K, Shin E, Choy G, Do S. How much data is needed to train a medical image deep learning system to achieve necessary high accuracy?; 2015. arXiv:1511.06348.
- 303. Sun C, Shrivastava A, Singh S, Gupta A. Revisiting unreasonable effectiveness of data in deep learning era. In: 2017 IEEE International Conference on Computer Vision (ICCV); 2017:843–852.
- 304. Zhang R, Zheng Y, Mak TWC, et al. Automatic detection and classification of colorectal polyps by transferring low‐level CNN features from nonmedical domain. IEEE J Biomed Health Inf. 2017;21:41–47. [DOI] [PubMed] [Google Scholar]
- 305. van Ginneken B, Setio AAA, Jacobs C, Ciompi F. Off‐the‐shelf convolutional neural network features for pulmonary nodule detection in computed tomography scans. In: 2015 IEEE 12th International Symposium on Biomedical Imaging; 2015:286–289.
- 306. Lopes UK, Valiati JF. Pre‐trained convolutional neural networks as feature extractors for tuberculosis detection. Comput Biol Med. 2017;89:135–143. [DOI] [PubMed] [Google Scholar]
- 307. Ben‐Cohen A, Klang E, Diamant I, et al. CT image‐based decision support system for categorization of liver metastases into primary cancer sites: initial results. Acad Radiol. 2017;24:1501–1509. [DOI] [PubMed] [Google Scholar]
- 308. Abidin AZ, Deng BT, Dsouza AM, Nagarajan MB, Coan P, Wismuller A. Deep transfer learning for characterizing chondrocyte patterns in phase contrast X‐ray computed tomography images of the human patellar cartilage. Comput Biol Med. 2018;95:24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Lee H, Tajmir S, Lee J, et al. Fully automated deep learning system for bone age assessment. J Digit Imaging. 2017;30:427–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Samala RK, Chan H‐P, Hadjiiski L, Helvie MA, Richter C, Cha K. Cross‐domain and multi‐task transfer learning of deep convolutional neural network for breast cancer diagnosis in digital breast tomosynthesis. In: Medical Imaging 2018: Computer‐Aided Diagnosis; 2018:105750Q.
- 311. Shin HC, Orton MR, Collins DJ, Doran SJ, Leach MO. Stacked autoencoders for unsupervised feature learning and multiple organ detection in a pilot study using 4D patient data. IEEE Trans Pattern Anal Mach Intell. 2013;35:1930–1943. [DOI] [PubMed] [Google Scholar]
- 312. Wang S, Cong Y, Fan H, et al. Computer‐aided endoscopic diagnosis without human‐specific labeling. IEEE Trans Biomed Eng. 2016;63:2347–2358. [DOI] [PubMed] [Google Scholar]
- 313. Feng X, Yang J, Laine AF, Angelini ED. Discriminative localization in CNNs for weakly‐supervised segmentation of pulmonary nodules. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI), Vol. 10435; 2017:568–576. [DOI] [PMC free article] [PubMed]
- 314. Rajpurkar P, Irvin J, Zhu K, et al. CheXNet: radiologist‐level pneumonia detection on chest x‐rays with deep learning; 2017. arXiv:1711.05225.
- 315. Chen X, Shrivastava A, Gupta A. NEIL: extracting visual knowledge from web data. In: Proc. of ICCV; 2013.
- 316. Zech J, Pain M, Titano J, et al. Natural language‐based machine learning models for the annotation of clinical radiology reports. Radiology. 2018;287:570–580. [DOI] [PubMed] [Google Scholar]
- 317. Ghafoorian M, Teuwen J, Manniesing R, et al. Student beats the teacher: deep neural networks for lateral ventricles segmentation in brain MR. In: Angelini ED, Landman BA, eds. Proc. SPIE Medical Imaging, Vol. 10574; 2018:105742U.
- 318. Zhang L, Gopalakrishnan V, Lu L, Summers RM, Moss J, Yao J. Self‐learning to detect and segment cysts in lung CT images without manual annotation. In: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018); 2018:1100–1103.
- 319. Asperti A, Mastronardo C. The effectiveness of data augmentation for detection of gastrointestinal diseases from endoscopical images. In: The 5th International Conference on Bioimaging; 2017.
- 320. Pezeshk A, Petrick N, Chen W, Sahiner B. seamless lesion insertion for data augmentation in CAD training. IEEE Trans Med Imaging. 2017;36:1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Zhang C, Tavanapong W, Wong J, de Groen PC, Oh J. Real data augmentation for medical image classification. In: Cardoso MJ, Arbel T, Lee S‐L, Cheplygina V, Balocco S, Mateus D, et al., eds. Intravascular Imaging and Computer Assisted Stenting and Large‐Scale Annotation of Biomedical Data and Expert Label Synthesis, Springer International Publishing;2017:67–76. [Google Scholar]
- 322. Badano A, Badal A, Glick S, et al. In silico imaging clinical trials for regulatory evaluation: initial considerations for VICTRE, a demonstration study. In: Flohr TG, Lo JY, eds. Proc. SPIE Medical Imaging, Vol. 10132, Schmidt TG; 2017:1013220.
- 323. Cui J, Liu X, Wang Y, Liu H. Deep reconstruction model for dynamic PET images. PLoS ONE. 2017;12:e0184667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Shin H‐C, Lu L, Summers RM. Natural language processing for large‐scale medical image analysis using deep learning. In: Deep Learning for Medical Image Analysis. San Diego, CA, USA: Elsevier; 2017:405–421. [Google Scholar]
- 325. Schlegl T, Waldstein SM, Vogl W‐D, Schmidt‐Erfurth U, Langs G. Predicting semantic descriptions from medical images with convolutional neural networks. In: Information Processing in Medical Imaging; 2015:437–448. [DOI] [PubMed]
- 326. Shin H‐C, Roberts K, Lu L, Demner‐Fushman D, Yao J, Summers RM. Learning to read chest x‐rays: recurrent neural cascade model for automated image annotation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2016:2497–2506.
- 327. Wang X, Lu L, Shin H‐C, et al. Unsupervised joint mining of deep features and image labels for large‐scale radiology image categorization and scene recognition. In: 2017 IEEE Winter Conference on Applications of Computer Vision (WACV); 2017:998–1007.
- 328. Wang X, Peng Y, Lu L, Lu Z, Summers RM. TieNet: Text‐image embedding network for common thorax disease classification and reporting in chest X‐rays. In: International Conference of Computer Vision and Pattern Recognition; 2018.
- 329. Chen MC, Ball RL, Yang L, et al. Deep learning to classify radiology free‐text reports. Radiology. 2018;286:845–852. [DOI] [PubMed] [Google Scholar]
- 330. Yan K, Wang X, Lu L, Summers RM. DeepLesion: automated deep mining, categorization and detection of significant radiology image findings using large‐scale clinical lesion annotations; 2017. arXiv preprint arXiv:1710.01766.
- 331. Yan K, Wang X, Lu L, et al. Deep lesion graphs in the wild: relationship learning and organization of significant radiology image findings in a diverse large‐scale lesion database. In: International Conference of Computer Vision and Pattern Recognition; 2018.
- 332. Bibault JE, Giraud P, Burgun A. Big Data and machine learning in radiation oncology: state of the art and future prospects. Cancer Lett. 2016;382:110–117. [DOI] [PubMed] [Google Scholar]
- 333. Dai L, Fang R, Li H, et al. Clinical report guided retinal microaneurysm detection with multi‐sieving deep learning. IEEE Trans Med Imaging. 2018;37:1149–1161. [DOI] [PubMed] [Google Scholar]
- 334. Zhang Z, Chen P, Sapkota M, Yang L. TandemNet: distilling knowledge from medical images using diagnostic reports as optional semantic references. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2017:320–328.
- 335. Top A, Hamarneh G, Abugharbieh R. Active learning for interactive 3D image segmentation. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI), Berlin, Heidelberg; 2011:603–610. [DOI] [PubMed]
- 336. Zhu Y, Zhang S, Liu W, Metaxas DN. Scalable histopathological image analysis via active learning. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2014:369–376. [DOI] [PubMed]
- 337. Dwarikanath Mahapatra JMB. Visual saliency‐based active learning for prostate magnetic resonance imaging segmentation. J Med Imaging. 2016;3:014003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Lee J, Wu Y, Kim H. Unbalanced data classification using support vector machines with active learning on scleroderma lung disease patterns. J Appl Stat. 2015;42:676–689. [Google Scholar]
- 339. Hoi SC, Jin R, Zhu J, Lyu MR. Batch mode active learning and its application to medical image classification. In: Proceedings of the 23rd International Conference on Machine learning; 2006:417–424.
- 340. Konyushkova K, Sznitman R, Fua P. Introducing geometry in active learning for image segmentation. In: Proceedings of the IEEE International Conference on Computer Vision ; 2015:2974–2982.
- 341. Yang L, Zhang Y, Chen J, Zhang S, Chen DZ. Suggestive annotation: a deep active learning framework for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2017:399–407.
- 342. Zhou Z, Shin J, Zhang L, Gurudu S, Gotway M, Liang J. Fine‐tuning convolutional neural networks for biomedical image analysis: actively and incrementally. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017: 4761–4772. [DOI] [PMC free article] [PubMed]
- 343. Gaur U, Kourakis M, Newman‐Smith E, Smith W, Manjunath BS. Membrane segmentation via active learning with deep networks. In: 2016 IEEE International Conference on Image Processing (ICIP); 2016:1943–1947.
- 344. Mosinska‐Domanska A, Sznitman R, Glowacki P, Fua P. Active learning for delineation of curvilinear structures. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016:5231–5239.
- 345. Heimann T, Mountney P, John M, Ionasec R. Learning without labeling: domain adaptation for ultrasound transducer localization. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2013: 49–56. [DOI] [PubMed]
- 346. Wachinger C, Reuter M. Domain adaptation for Alzheimer's disease diagnostics. NeuroImage. 2016;139:470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347. Conjeti S, Katouzian A, Roy AG, et al. Supervised domain adaptation of decision forests: Transfer of models trained in vitro for in vivo intravascular ultrasound tissue characterization. Med Image Anal. 2016;32:1–17. [DOI] [PubMed] [Google Scholar]
- 348. Bermúdez‐Chacón R, Becker C, Salzmann M, Fua P. Scalable unsupervised domain adaptation for electron microscopy. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2016:326–334.
- 349. Becker C, Christoudias CM, Fua P. Domain adaptation for microscopy imaging. IEEE Trans Med Imaging. 2015;34:1125–1139. [DOI] [PubMed] [Google Scholar]
- 350. Baur C, Albarqouni S, Navab N. Semi‐supervised deep learning for fully convolutional networks. In: International Conference on Medical Image Computing and Computer‐Assisted Intervention (MICCAI); 2017:311–319.
- 351. Saunders R, Samei E, Baker J, Delong D. Simulation of mammographic lesions. Acad Radiol. 2006;13:860–870. [DOI] [PubMed] [Google Scholar]
- 352. Rashidnasab A, Elangovan P, Yip M, et al. Simulation and assessment of realistic breast lesions using fractal growth model. Phys Med Biol. 2013;58:5613–5627. [DOI] [PubMed] [Google Scholar]
- 353. Martinez‐Murcia FJ, Górriz JM, Ramírez J, et al. Functional Brain Imaging Synthesis Based on Image Decomposition and Kernel Modeling: Application to Neurodegenerative Diseases. Front Neuroinform. 2017;11:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354. Calimeri F, Marzullo A, Stamile C, Terracina G. Biomedical data augmentation using generative adversarial neural networks. In: Artificial Neural Networks and Machine Learning (ICANN); 2017:626–634.
- 355. Lahiri A, Ayush K, Biswas PK, Mitra P. Generative adversarial learning for reducing manual annotation in semantic segmentation on large scale miscroscopy images: automated vessel segmentation in retinal fundus image as test case. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops; 2017:42–48.
- 356. Zhang L, Gooya A, Frangi AF. Semi‐supervised assessment of incomplete LV coverage in cardiac MRI using generative adversarial nets. In: International Workshop on Simulation and Synthesis in Medical Imaging; 2017:61–68.
- 357. Bayramoglu N, Kaakinen M, Eklund L, Heikkila J. Towards virtual H&E staining of hyperspectral lung histology images using conditional generative adversarial networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 2017:64–71.
- 358. Costa P, Galdran A, Meyer MI, et al. End‐to‐end adversarial retinal image synthesis. In: IEEE Transactions on Medical Imaging; 2017. [DOI] [PubMed]
- 359. Chartsias A, Joyce T, Dharmakumar R, Tsaftaris SA. Adversarial image synthesis for unpaired multi‐modal cardiac data. In: International Workshop on Simulation and Synthesis in Medical Imaging; 2017:3–13.
- 360. Wolterink JM, Dinkla AM, Savenije MH, Seevinck PR, van den Berg CA, Išgum I. Deep MR to CT synthesis using unpaired data. In: International Workshop on Simulation and Synthesis in Medical Imaging; 2017:14–23.
- 361. Chuquicusma MJ, Hussein S, Burt J, Bagci U. How to fool radiologists with generative adversarial networks? A visual turing test for lung cancer diagnosis. In: IEEE International Symposium on Biomedical Imaging; 2018.
- 362. Obuchowski NA, Barnhart HX, Buckler AJ, et al. Statistical issues in the comparison of quantitative imaging biomarker algorithms using pulmonary nodule volume as an example. Stat Methods Med Res. 2015;24:107–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363. Obuchowski NA, Reeves AP, Huang EP, et al. Quantitative imaging biomarkers: a review of statistical methods for computer algorithm comparisons. Stat Methods Med Res. 2015;24:68–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364. Lazzeroni LC, Lu Y, Belitskaya‐Levy I. P‐values in genomics: apparent precision masks high uncertainty. Mol Psychiatry. 2014;19:1336–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365. Drukker K, Pesce L, Giger M. Repeatability in computer‐aided diagnosis: application to breast cancer diagnosis on sonography. Med Phys. 2010;37:2659–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366. Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 367. Curran‐Everett D. Multiple comparisons: philosophies and illustrations. Am J Physiol Regul Integr Comp Physiol. 2000;279:R1–R8. [DOI] [PubMed] [Google Scholar]
- 368. Ein‐Dor L, Kela I, Getz G, Givol D, Domany E. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics. 2005;21:171–178. [DOI] [PubMed] [Google Scholar]
- 369. Ein‐Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci USA. 2006;103:5923–5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370. Begley CG, Ellis LM. Raise standards for preclinical cancer research. Nature. 2012;483:531–533. [DOI] [PubMed] [Google Scholar]
- 371. Chalkidou A, O'Doherty MJ, Marsden PK. False discovery rates in PET and CT studies with texture features: a systematic review. PLoS ONE. 2015;10:e0124165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372. Gruszauskas NP, Drukker K, Giger ML, Sennett CA, Pesce LL. Performance of breast ultrasound computer‐aided diagnosis: dependence on image selection. Acad Radiol. 2008;15:1234–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373. Gruszauskas NP, Drukker K, Giger ML, et al. Breast US computer‐aided diagnosis system: robustness across urban populations in South Korea and the United States. Radiology. 2009;253:661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374. Mendel KR, Li H, Lan L, et al. Quantitative texture analysis: robustness of radiomics across two digital mammography manufacturers’ systems. J Med Imaging. 2018;5:011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375. Kalpathy‐Cramer J, Mamomov A, Zhao BS, et al. Radiomics of lung nodules: a multi‐institutional study of robustness and agreement of quantitative imaging features. Tomography. 2016;2:430–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376. Court LE. Harmonization & robustness in radiomics. Med Phys. 2016;43:3695–3696. [Google Scholar]
- 377. Holzinger A, Biemann C, Pattichis CS, Kell DB. What do we need to build explainable AI systems for the medical domain?; 2017. arXiv:1712.09923.
- 378. Samek W, Wiegand T, Müller K‐R. Explainable artificial intelligence: understanding, visualizing and interpreting deep learning models; 2017. arXiv:1708.08296.
- 379. Zeiler MD, Fergus R. Visualizing and understanding convolutional networks; 2014. arXiv:1311.2901.
- 380. Ribeiro MT, Singh S, Guestrin C. Why Should I Trust You? Explaining the predictions of any classifier; 2016. arXiv:1602.04938.
- 381. Simonyan K, Vedaldi A, Zisserman A. Deep inside convolutional networks: visualising image classification models and saliency maps; 2014. arXiv:1312.6034.
- 382. Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning deep features for discriminative localization; 2015. arXiv:1512.04150.
- 383. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad‐CAM: visual explanations from deep networks via gradient‐based localization; 2017. arXiv:1610.02391.
- 384. Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. [DOI] [PubMed] [Google Scholar]
- 385. Adil K, Jiang F, Liu SH, Grigorev A, Gupta BB, Rho S. Training an agent for FPS doom game using visual reinforcement learning and VizDoom. Int J Adv Comput Sci Appl. 2017;8:32–41. [Google Scholar]
- 386. Summers RM. Are we at a crossroads or a plateau? Radiomics and machine learning in abdominal oncology imaging”. Abdom Radiol. 2018;1–5. [Epub ahead of print]. 10.1007/s00261-018-1613-1 [DOI] [PubMed] [Google Scholar]


