Abstract
Purpose
Develop computer vision models for image-based differentiation of bacterial and fungal corneal ulcers and compare their performance against human experts.
Design
Cross-sectional comparison of diagnostic performance.
Participants
Patients with acute, culture-proven bacterial or fungal keratitis from 4 centers in South India.
Methods
Five convolutional neural networks (CNNs) were trained using images from handheld cameras collected from patients with culture-proven corneal ulcers in South India recruited as part of clinical trials conducted between 2006 and 2015. Their performance was evaluated on 2 hold-out test sets (1 single center and 1 multicenter) from South India. Twelve local expert cornea specialists performed remote interpretation of the images in the multicenter test set to enable direct comparison against CNN performance.
Main Outcome Measures
Area under the receiver operating characteristic curve (AUC) individually and for each group collectively (i.e., CNN ensemble and human ensemble).
Results
The best-performing CNN architecture was MobileNet, which attained an AUC of 0.86 on the single-center test set (other CNNs range, 0.68–0.84) and 0.83 on the multicenter test set (other CNNs range, 0.75–0.83). Expert human AUCs on the multicenter test set ranged from 0.42 to 0.79. The CNN ensemble achieved a statistically significantly higher AUC (0.84) than the human ensemble (0.76; P < 0.01). CNNs showed relatively higher accuracy for fungal (81%) versus bacterial (75%) ulcers, whereas humans showed relatively higher accuracy for bacterial (88%) versus fungal (56%) ulcers. An ensemble of the best-performing CNN and best-performing human achieved the highest AUC of 0.87, although this was not statistically significantly higher than the best CNN (0.83; P = 0.17) or best human (0.79; P = 0.09).
Conclusions
Computer vision models achieved superhuman performance in identifying the underlying infectious cause of corneal ulcers compared with cornea specialists. The best-performing model, MobileNet, attained an AUC of 0.83 to 0.86 without any additional clinical or historical information. These findings suggest the potential for future implementation of these models to enable earlier directed antimicrobial therapy in the management of infectious keratitis, which may improve visual outcomes. Additional studies are ongoing to incorporate clinical history and expert opinion into predictive models.
Keywords: Artificial intelligence, Bacterial keratitis, Computer vision, Convolutional neural networks, Corneal ulcer, Deep learning, Fungal keratitis, Infectious keratitis
Abbreviations and Acronyms: AUC, area under the receiver operating characteristic curve; CI, confidence interval; CNN, convolutional neural network; MUTT, Mycotic Ulcer Treatment Trials; SCUT, Steroids for Corneal Ulcers Trial
Corneal opacification is the fifth leading cause of blindness worldwide, with most cases attributable to infection.1, 2, 3 Prompt identification of the cause of infectious keratitis is important to guide antimicrobial therapy. Of particular importance is the expeditious differentiation between bacterial and fungal keratitis, which together account for more than 95% of corneal ulcers.4, 5, 6 Cultures of corneal scrapings are the current gold standard for determining the causative pathogen of corneal ulcers, but show false-negative results in approximately half of cases.4,7 Even when culture results are positive, the results typically are not available for several days, which may result in a delay in effective antimicrobial therapy and worse visual outcomes. In the absence of microbiologic data, empiric therapy must be selected based on the clinical impression of the cause of infection, which is unreliable even among cornea specialists.8, 9, 10 Applying artificial intelligence using deep learning with convolutional neural networks (CNNs) for image-based diagnosis of infectious keratitis may minimize the delay in initiating targeted antimicrobial therapy.
In the past 2 decades, deep CNNs have achieved unprecedented performance in computer vision applications, particularly using supervised learning for image classification.11 State-of-the-art model architectures leverage the exponential gains in modern computer processing speeds to abstract complex features and develop highly nonlinear decision boundaries in multidimensional feature space. Transfer learning has enabled machine learning engineers to repurpose models initially trained on very large image sets to perform medically relevant image classification tasks using the relatively small training databases available in health care.12 This has resulted in human-level or even superhuman performance in image-based diagnosis of a variety of eye diseases, including glaucoma, macular degeneration, diabetic retinopathy, and retinopathy of prematurity, among others.13, 14, 15, 16 Several prior studies have described deep learning models for image-based differentiation of bacterial and fungal keratitis using photographs from slit-lamp cameras with some success.17, 18, 19, 20 However, infrastructure limitations in low- and middle-income countries where the burden of disease is highest result in relatively limited potential for implementation of models that require slit-lamp–mounted cameras. Telemedicine applications using less expensive and more portable imaging methods may significantly increase the potential public health impact of this technology. Herein, we describe the development and evaluation of deep CNN models for automated image-based differentiation of bacterial and fungal ulcers using images from handheld portable cameras.
Methods
Image Sets
Several large clinical trials evaluating management options for bacterial and fungal keratitis were conducted at the Aravind Eye Care System in South India between 2006 and 2015, including the Steroids for Corneal Ulcers Trial (SCUT) and Mycotic Ulcer Treatment Trials (MUTT) I and II.21, 22, 23 Each corneal ulcer in these trials was microbiologically proven to be either bacterial or filamentous fungal keratitis, and each patient underwent corneal photography at initial presentation using a handheld Nikon D-series digital single-lens reflex camera according to a standardized lighting and photography protocol. From these trials, we collated a database of 980 high-quality images from 980 patients with corneal ulcers photographed at the initial presentation, including 500 fungal ulcers and 480 bacterial ulcers. Only 1 image per ulcer was included. All images were obtained from 1 of 4 study sites within the Aravind Eye Care System in South India (Madurai, Coimbatore, Pondicherry, or Tirunelveli). No culture-negative or polymicrobial infections were included.
The 2 largest-volume sites (Madurai and Coimbatore) participated in both bacterial and fungal ulcer trials during this period, whereas the smaller sites participated only in the fungal ulcer trials (Pondicherry) or only the bacterial ulcer trial (Tirunelveli). Initial iterations of CNN model development using a randomly selected training set comprising 90% of the above image database demonstrated that deep CNNs were leveraging this fact by learning to identify the study site (likely using subtle differences in room lighting, flash used, etc.) to predict whether an ulcer was bacterial or fungal, a phenomenon known as “label leakage.”24 This allowed the models to perform reasonably well on a multicenter test set comprising the remaining 10% of images, but provided little generalizable predictive usefulness when evaluated on a holdout test set from a single site. To obviate this issue, we restricted the training set to only a single location (Madurai), resulting in 396 training cases comprising 215 bacterial ulcers (54%) and 181 fungal ulcers. This forced the model to identify more generalizable features to distinguish between bacterial and fungal ulcers. We then tuned model hyperparameters according to performance on a separate validation set of 50 ulcers from a different study site (Coimbatore) consisting of 25 bacterial ulcers (50%) and 25 fungal ulcers to assess generalizability. Finally, we evaluated model performance using 2 holdout test sets: the first consisting of 100 images from Coimbatore selected using stratified random sampling to contain 50% bacterial ulcers and 50% fungal ulcers, and the second comprising 80 images randomly selected from the multicenter image database containing images from all 4 study sites (48 from bacterial ulcers [60%] and 32 from fungal ulcers). Figure S1 depicts the partitioning of the image database into training, validation, and testing sets.
Deep Convolutional Neural Network Model Development
During CNN model training, image augmentation was performed to reduce overfitting using random flipping along the vertical axis and random rotation of up to 20° in either direction.25 Images were then resized and preprocessed according to the standard required for each model architecture. No manual annotation or additional image preprocessing was performed.
We trained and evaluated 5 deep CNN architectures: MobileNetV2,26 DenseNet201,27 ResNet152V2,28 VGG19,29 and Xception.30 Each model was pretrained on the ImageNet imaging database consisting of more than 10 million images with 10000 class labels and was repurposed to this classification task using a transfer learning approach.12,31 Specifically, the learned parameters from the ImageNet training set were used as the initialization values for subsequent training on this image set. The final layer of each model (consisting of 10 000 densely connected nodes with a softmax activation function) was removed and replaced with a global average pooling layer, a dropout layer for regularization effect, and a final layer consisting of a single L2-regularized node with a sigmoid activation function. This resulted in an output value ranging from 0 to 1, which in this binary classification task could be interpreted as an estimated probability of fungal etiology [P(fungal)] and estimated probability of bacterial etiology [1 – P(fungal)]. Optimization of model parameters was performed using minibatch gradient descent with RMSprop minimizing the binary cross-entropy loss function.
Each model was first trained for several epochs with all layers frozen except for the final layer, allowing the model to essentially act as a feature extractor using the learned features from ImageNet. We subsequently fine-tuned each model by unfreezing the parameters of deeper layers (with the learning rate reduced by an order of magnitude) to allow the models to learn complex features more specific to corneal ulcer images. The number of frozen layers and number of epochs during the feature extractor and fine-tuning phases were treated as hyperparameters and were adjusted according to model performance on the validation set. The primary outcome measure for model tuning and evaluation was the area under the receiver operating characteristic curve (AUC).
All model training was conducted in Python 3 using Tensorflow 2 with the Keras application programming interface on an AWS EC2 remote instance (Amazon, Inc) with a Tesla V100 GPU (NVIDIA Corp).32 All source code used to develop the optimal model is publicly available on GitHub (https://github.com/tkredd2/MobileNet_corneal_ulcers).
Human Grading
A detailed description of the quantification of human performance in image-based differentiation of bacterial and fungal keratitis using this multicenter test set was previously published.10 Briefly, an international cohort of 66 expert cornea specialists individually interpreted these images via a web-based portal. Participants provided their estimated probability that an ulcer image represented bacterial or fungal infection. This allowed direct comparison against the CNN model outputs. Graders were informed that each image represented either a culture-proven bacterial ulcer or a culture-proven fungal ulcer, but no additional information regarding clinical history or examination findings was provided. Each human grader thus had access to the same amount of information as the CNNs to formulate their predictions. This prior study determined that experts practicing in India demonstrated statistically significantly better performance than their counterparts practicing outside India in classifying these ulcer images from South India.10 As a result, in the current analysis, we restricted the human comparison group to include only these 12 Indian cornea specialists to ensure the most rigorous standard of comparison against CNN model performance.
Statistical Analysis
Because each of the 2 classes (bacterial ulcers and fungal ulcers) are of equal importance in determining appropriate directed antimicrobial therapy and our test sets were well balanced with respect to the 2 classes, we elected to use the AUC as the primary evaluation metric of CNN and human grader performance. The 95% confidence interval (CI) of the AUC was computed using bootstrapping with 2000 replications.33 To summarize overall human and CNN performance, we generated a CNN ensemble prediction and a human ensemble prediction by determining the group mean predicted probability for each image. We compared CNN and human ensembles using the DeLong method for nonparametric statistical comparisons of correlated receiver operating characteristic curves.34 The ensemble predictions were further analyzed categorically using confusion matrices to determine their accuracy for bacterial ulcers and for fungal ulcers specifically. Because the outputs of both human and CNN predictions were a probability value on a continuous scale ranging from 0 to 1, this required identifying a threshold value to determine whether to classify a prediction as bacterial or fungal. The probability cutoff value that maximized both sensitivity and specificity (Youden’s index) was assigned as this threshold value for each of the 2 ensembles and was used to determine the categorical prediction for each image.35 If the estimated P(fungal) was more than this threshold, the prediction was labeled as fungal; if the estimated P(fungal) was less than the threshold, the prediction was labeled bacterial. Of note, these threshold values were used only to facilitate categorical analysis of these binary classification models. Final threshold values for production models will be determined using prospectively collected, population-based samples based on sensitivity, specificity, and positive and negative predictive values.
Gradient class activation heatmaps were used to qualitatively assess which image regions exerted the greatest influence on the best-performing CNN’s prediction.36 Twenty heatmaps were generated in total; 10 visualized the images on which the CNN performed best (i.e., images on which the difference between model prediction and ground truth were smallest) and 10 depicted images on which the CNN performed worst. All statistical analyses were performed in R software version 4.0.5 (R Foundation for Statistical Computing, Vienna, Austria).
This study adhered to the tenets of the Declaration of Helsinki and was approved by the internal review board at Oregon Health & Science University. Informed consent was obtained from all human expert participants.
Results
The final hyperparameters used to train each model are reported in Table S1.
Performance on the Single-Center External Test Set
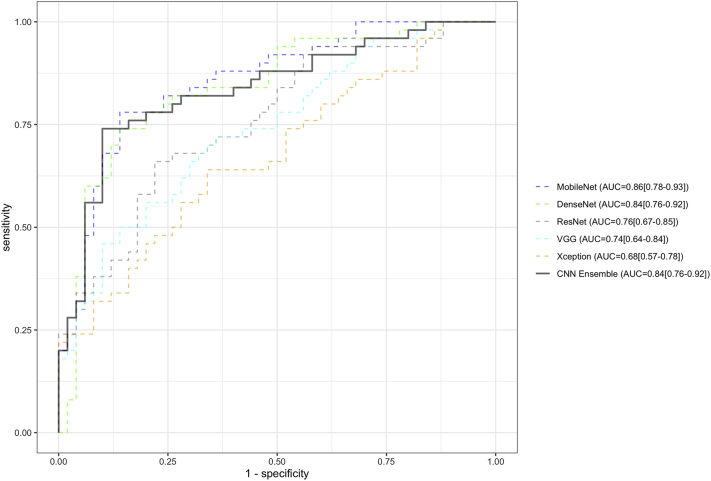
Figure 1 depicts receiver operating characteristic curves for each of the 5 CNN architectures (trained only on images from Madurai) on the single-center external testing set comprising 100 images (50 bacterial and 50 fungal) exclusively from Coimbatore. MobileNet achieved the highest performance with an AUC of 0.86 (95% CI, 0.78–0.93), followed by DenseNet (AUC, 0.84; 95% CI, 0.76–0.92), ResNet (AUC, 0.76; 95% CI, 0.67–0.85), VGG (AUC, 0.74; 95% CI, 0.64–0.84), and Xception (AUC, 0.68; 95% CI, 0.57–0.78). The ensemble of all 5 CNNs reached an AUC of 0.84 (95% CI, 0.76–0.92).
Figure 1.
Receiver operating characteristic curves of 5 deep convolutional neural network (CNN) models on a single-center external testing set consisting of 100 corneal ulcer images (50% fungal, 50% bacterial) from Coimbatore. The performance of the CNN ensemble is also depicted, representing the average output of all 5 models for each image. MobileNet demonstrated the highest performance among the model architectures tested. Ninety-five percent confidence intervals are depicted next to each area under the receiver operating characteristic curve (AUC) estimate.
Performance on Multicenter Test Set
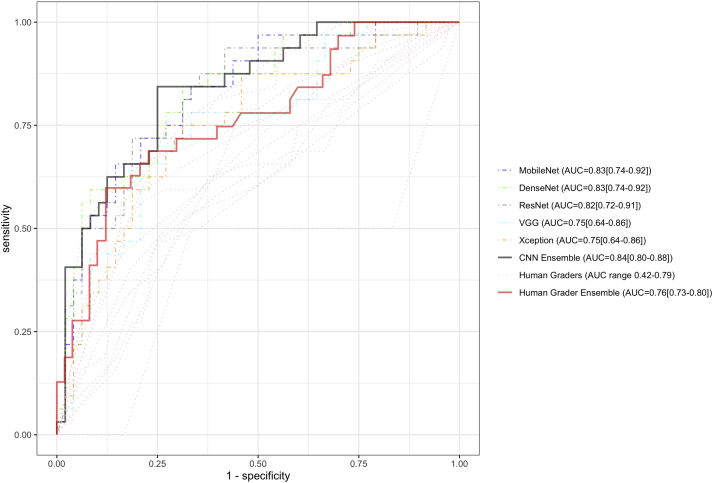
Figure 2 depicts the receiver operating characteristic curves of all 5 CNNs and the 12 human graders on the multicenter holdout test set of 80 images randomly selected from the image database collated from all 4 study sites. MobileNet and DenseNet again achieved the highest performance (AUC, 0.83 for both), followed by ResNet (AUC, 0.82), VGG (AUC, 0.75), and Xception (AUC, 0.75; Table 1). The AUC among individual human graders ranged from 0.42 to 0.79. The CNN ensemble achieved an AUC of 0.84, which was statistically significantly higher than the AUC of the human ensemble (AUC, 0.76; P < 0.01; Fig 2).
Figure 2.
Receiver operating characteristic curves of the 5 convolutional neural network (CNN) models and 12 human graders on a multicenter testing set consisting of 80 corneal ulcer images (48 bacterial, 32 fungal). The CNN ensemble demonstrated a statistically significantly higher area under the receiver operating characteristic curve (AUC; 0.84) than the human ensemble (AUC, 0.76; P < 0.01). Ninety-five percent confidence intervals are depicted next to each AUC estimate.
Table 1.
Convolutional Neural Network and Human Grader Performance on External Test Sets
| Single Center Test Set (Coimbatore) | Multicenter Test Set | |
|---|---|---|
| Classifier | AUC (95% CI) | AUC (95% CI) |
| CNNs | ||
| MobileNet | 0.86 (0.78-0.93) | 0.83 (0.74-0.92) |
| DenseNet | 0.84 (0.76-0.92) | 0.83 (0.74-0.92) |
| ResNet | 0.76 (0.67-0.85) | 0.82 (0.72-0.91) |
| VGG | 0.74 (0.64-0.84) | 0.75 (0.64-0.86) |
| Xception | 0.68 (0.57-0.78) | 0.75 (0.64-0.86) |
| CNN Ensemble | 0.84 (0.76-0.92) | 0.84 (0.80-0.88)∗ |
| Human Graders | ||
| Grader 1 | - | 0.79 (0.69-0.89) |
| Grader 2 | - | 0.78 (0.68-0.88) |
| Grader 3 | - | 0.76 (0.65-0.87) |
| Grader 4 | - | 0.73 (0.61-0.83) |
| Grader 5 | - | 0.70 (0.58-0.81) |
| Grader 6 | - | 0.69 (0.57-0.81) |
| Grader 7 | - | 0.67 (0.20-1.00) |
| Grader 8 | - | 0.65 (0.52-0.77) |
| Grader 9 | - | 0.64 (0.41-0.87) |
| Grader 10 | - | 0.61 (0.48-0.73) |
| Grader 11 | - | 0.57 (0.45-0.69) |
| Grader 12 | - | 0.42 (0.05-0.95) |
| Human Grader Ensemble | - | 0.76 (0.73-0.80)∗ |
CNN = convolutional neural network; — = not available.
Boldface values indicate ensemble rather than individual performance. Data are presented as area under the receiver operating curve (95% confidence interval).
P < 0.01 (DeLong method).
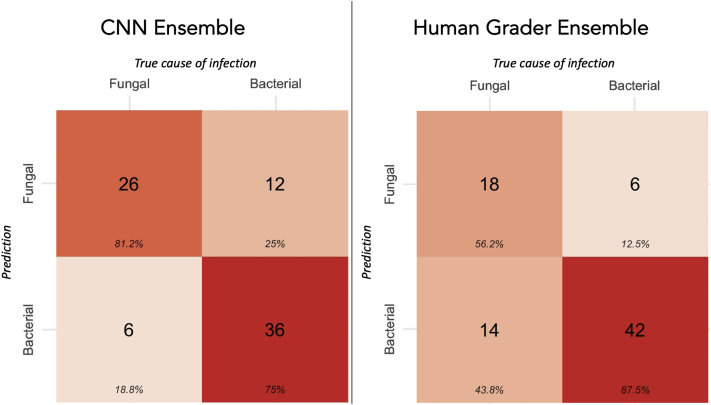
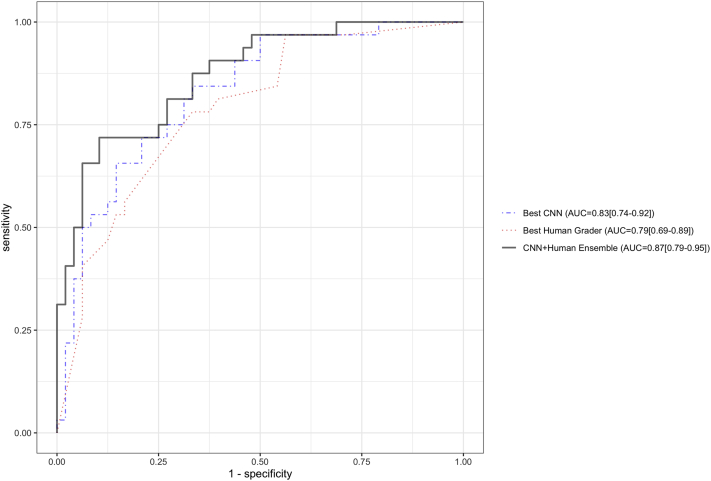
The ideal threshold value (Youden’s index) for the CNN ensemble prediction was 0.36. Using this cut point, the CNN ensemble achieved 81% accuracy for identifying fungal ulcers and 75% accuracy for identifying bacterial ulcers (Fig 3). The ideal threshold for the human grader ensemble was 0.58. The human grader ensemble demonstrated higher accuracy for identifying bacterial ulcers (88%), but lower accuracy for fungal ulcers (56%). This suggested that the 2 classifiers may be complementary to each other when combining their outputs into a single prediction. An ensemble (arithmetic mean of the 2 outputs: the CNN plus the human ensemble) of the best-performing CNN (MobileNet) and the best-performing human grader (grader 1) attained an AUC of 0.87 (95% CI, 0.79–0.95). This was higher than the AUC of grader 1 (AUC, 0.79) and MobileNet (AUC, 0.83) individually, but the differences were not statistically significantly (P = 0.09 and P = 0.17, respectively; Fig 4). The optimal threshold for the CNN plus the human ensemble was 0.51, which demonstrated 72% accuracy for fungal ulcers and 90% accuracy for bacterial ulcers. Similarly, an ensemble of the outputs of the CNN ensemble and the human grader ensemble (an ensemble of ensembles) attained an AUC of 0.87 (95% CI, 0.79–0.95).
Figure 3.
Confusion matrices of the convolutional neural network (CNN) and human ensembles, with prediction categories (bacterial or fungal) assigned according to the threshold defined by Youden’s index. Percent values indicate column proportions. For example, the CNN ensemble showed 81% accuracy for identifying fungal infections and 75% accuracy for bacterial infections.
Figure 4.
Receiver operating characteristic curves of the best-performing convolutional neural network (CNN) (MobileNet), best-performing human (grader 1), and the ensemble of the 2 (CNN plus human ensemble). The area under the receiver operating characteristic curve (AUC) of the ensemble was 0.87, compared with an AUC of 0.79 for grader 1 (P = 0.09) and AUC of 0.83 for MobileNet (P = 0.17). Ninety-five percent confidence intervals are depicted next to each AUC estimate.
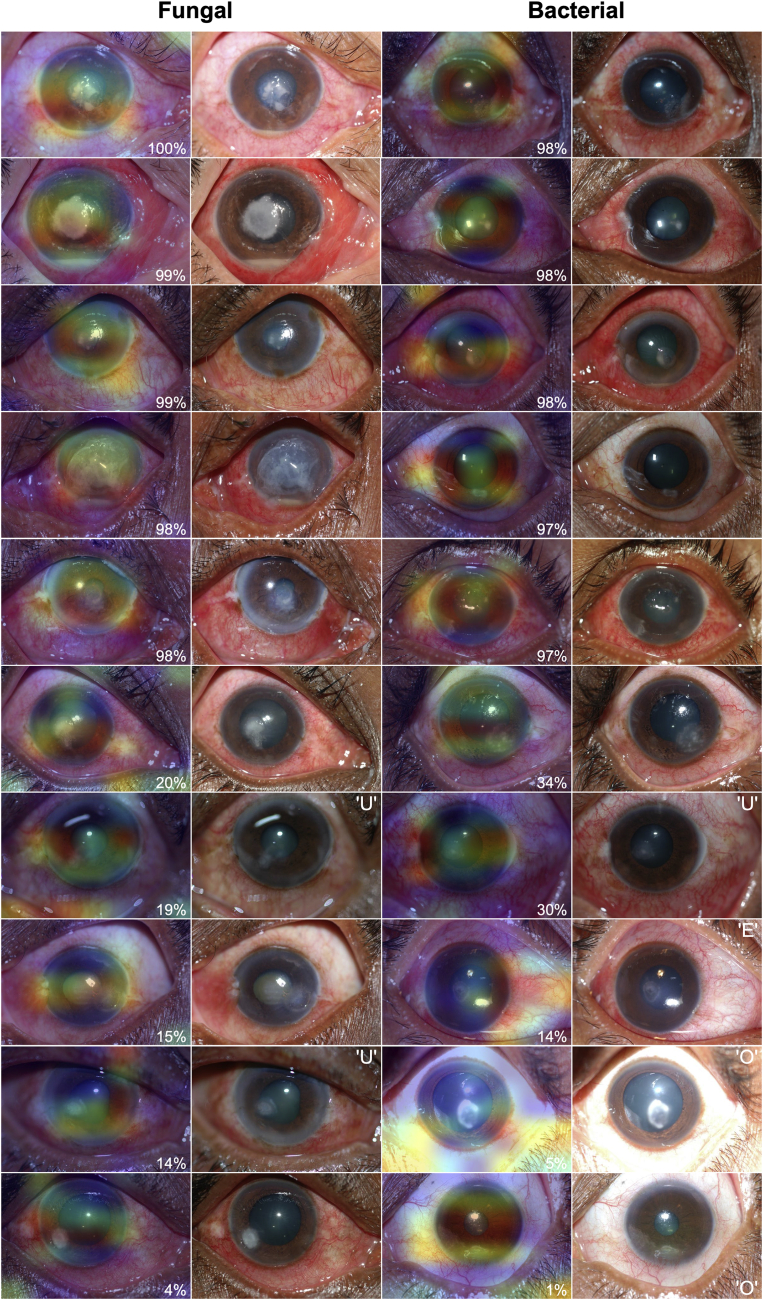
Figure 5 demonstrates gradient class activation heatmaps for the MobileNet model including the 10 images for which the model performed best and the 10 images demonstrating worst performance. Although this is a limited sample and subject to confirmation bias, qualitative review of the heatmaps suggests that the model seems to identify the cornea (specifically the corneal infiltrate) consistently as a region of interest without any human annotation or a priori indication that the model should pay attention to this area. These examples also emphasize the impact of image quality on model performance; 6 of the 10 images on which the model performed worst demonstrated relatively limited quality, including underexposure, overexposure, and eccentric gaze.
Figure 5.
Representative gradient class activation heatmaps of the images on which the best-performing convolutional neural network model (MobileNet) achieved highest agreement (top 5 fungal and top 5 bacterial) between model prediction and ground truth and lowest agreement (bottom 5 fungal and bottom 5 bacterial). Red coloration indicates regions of the input image that conferred the highest influence on the model’s prediction. Superimposed on each image is the percent agreement between the model’s prediction and the ground truth (for fungal images, P(fungal) × 100%; for bacterial images: 1 – P(fungal) × 100%). Adjacent to each heatmap is the raw image input to the model. The model tended to perform well when focusing attention on the corneal infiltrate, with relatively worse performance when areas of attention strayed from the cornea or when tested on images with quality limitations including overexposure (O), underexposure (U), or eccentric fixation (E).
Discussion
These results demonstrate that deep learning computer vision models can achieve superhuman performance in image-based differentiation of bacterial and fungal keratitis, surpassing even expert cornea specialists. Our MobileNet model achieved an AUC of 0.83 interpreting ulcer images from handheld cameras, which may enable greater portability and lower cost compared to slit-lamp image classifiers. Combining human and AI predictions seemed to further increase predictive value on post hoc analysis, but additional studies are required to investigate this possibility. These results emphasize the potential of computer vision applications for automated image interpretation in the etiologic evaluation of infectious keratitis.
Several prior studies have investigated computer vision applications in the differentiation of bacterial and fungal corneal ulcers using slit-lamp cameras. Kuo et al17 described a DenseNet model with a small training set that achieved an AUC of only 0.65. Ghosh et al18 reported an ensemble of 3 CNN architectures that achieved a reasonably high F1 score of 0.83 (the harmonic mean of precision [positive predictive value] and recall [sensitivity]), but was only evaluated on a very small test set of < 20 patients. Hung et al19 described a DenseNet161 model that attained nearly identical performance to our best-performing model (AUC, 0.85), but required the addition of a preceding U2 segmentation model. Xu et al37 developed a deep sequential feature learning model to differentiate bacterial and fungal keratitis that also attained similar accuracy to our own (84% for fungal keratitis, 65% for bacterial keratitis), but required manual annotation of images and a complicated and computationally expensive pipeline of models. In addition to providing improved potential for telemedicine implementation, our MobileNet model has several advantageous features within the context of these prior studies: (1) every image in our database was obtained from a culture-proven bacterial or fungal ulcer, resulting in a robust ground truth label for training and evaluation; (2) we used only 1 image from each participant in our training, validation, and testing sets, which reduces the risk of label leakage; (3) we evaluated model performance on both single-center and multicenter testing sets; and (4) we directly compared CNN performance against expert cornea specialists.
The CNNs in our study attained a statistically significantly higher AUC than expert humans with experience examining this population of corneal ulcer patients. Specifically, CNNs tended to perform well identifying fungal ulcers, whereas humans showed high accuracy for bacterial ulcers. As a result of this finding, we evaluated an ensemble model combining the predictions of humans and CNNs that achieved an even higher AUC (0.87). This suggests the possibility that future prediction models may benefit from incorporating multiple sources of input, including computer vision model predictions and expert opinion. Several structured data elements have also been shown to provide predictive information relating to the cause of infection, including aspects of the clinical history (onset of symptoms, contact lens wear, trauma, etc.) and slit-lamp examination findings (including the elevation and texture of corneal infiltrates).38, 39, 40, 41 In much the same way that an expert clinician typically gathers information from multiple sources before formulating a diagnosis, predictive models for identifying the underlying cause of infection will likely benefit from accessing this additional contextual information in addition to imaging data. These models may also benefit from adopting a Bayesian decision framework accounting for the pretest probability of different infectious agents based on local epidemiologic factors, which demonstrates marked geographic variability.5,42 Theoretically, future iterations of these prediction models may enable users to take a picture, enter their own expert prediction of the causative organism, input basic information from the clinical history and slit-lamp examination, and receive a unified output from the prediction model detailing the suspected cause of infection based on all of this information. Efforts are ongoing to develop the prospectively collected training databases necessary to develop such models and to allow determination of their diagnostic performance including sensitivity, specificity, and positive and negative predictive values.
A major advantage of our MobileNet model is its portability and potential for telemedicine implementation, making it a particularly attractive option considering the high burden of morbidity resulting from corneal ulceration in low- and middle-income countries.43 In addition to being the best-performing model in our study, MobileNet is also the smallest in terms of memory requirements (22 MB) and requires no manual image annotation or complicated computational pipeline, making it a strong candidate for potential implementation in portable imaging devices. This model interprets images from handheld cameras, which are less expensive and more portable than slit-lamp–mounted cameras and may provide better substrate for computer vision models compared with high-magnification slit-lamp images because of their greater depth of field. Difficulty maintaining focus on the entire cornea with slit-lamp imaging was identified as a potential source of error in the model developed by Kuo et al.17 For similar reasons, the advent of ubiquitous smartphone cameras makes this imaging method a particularly appealing candidate for future model implementation. Additional imaging databases consisting of smartphone imaging of corneal ulcers will be required to determine the feasibility of this possibility.
Broad geographic generalizability of computer vision models for etiologic diagnosis of infectious keratitis is complicated by the significant geographic variability in ulcer epidemiology.5,42 During the early stages of model development using a multicenter training set, we identified significant label leakage that markedly impaired model performance. This occurred because of differential participation in the SCUT, MUTT I, and MUTT II trials among different study sites. Although this would not necessarily be the case with a prospectively collected multicenter sample of corneal ulcers, differences in local epidemiologic features among sites may still result in some degree of problematic label leakage when used as a multicenter batch of training images. A federated learning approach may help to address this, but will require a data-centric mentality with care to avoid this label leakage issue.44,45 In this case, our final models were trained on data from a single site, but were evaluated on both a multicenter test set and an external single-center testing set. The single-center test set confirmed that the model was not learning site-specific features to perform this task, whereas the multicenter test set demonstrated that its performance generalized well in several centers throughout South India.
Several limitations should be considered in interpreting these results. First, image quality is of crucial importance to both AI and human performance in image classification, as demonstrated by the gradient class activation heatmaps (Fig 5). However, in the randomized trials from which this database was derived, image collection was repeated until at least 1 high-quality photograph was obtained for each ulcer, which was achieved in nearly every case (e.g., only 2 of the 500 ulcers photographed in SCUT did not produce acceptable images).21 Nonetheless, we are currently investigating the impact of image quality on model performance and training additional classifiers to interpret image quality automatically, which may be enable real-time feedback for future users of these models. Second, we used only bacterial and fungal ulcers to train and evaluate these models, but a small minority of ulcers are caused by other pathogens, including parasitic organisms. It is also possible that these models may not generalize to culture-negative infections. Data collection efforts are ongoing to train multiclass, multilabel classification models to address these limitations and to evaluate performance on all types of ulcers, including culture-negative and polymicrobial infections. Third, we demonstrated generalizability to 4 sites in India, but this does not imply that the model will generalize successfully to other geographic regions. Additional testing and model adaptation among geographically diverse populations will be required before implementation. Fourth, we trained and evaluated these models on acute, severe corneal ulcers, but both duration of infection and ulcer severity likely impact model performance. These aspects will need to be evaluated further before implementation. Fifth, we compared human experts and CNNs using the multicenter test set, but not the single-center test set from Coimbatore. However, the multicenter test set is a better representation of the potential for generalizability, and thus a better measure of classification performance. Finally, images were obtained from distinct clinical trials, and thus do not necessarily represent a cohesive cohort. However, the photography protocols for SCUT, MUTT I, and MUTT II were identical, and images for each trial were obtained with the same equipment, so this would not be expected to introduce label leakage.
Based on these findings, we conclude that sufficient information is contained within 2-dimensional corneal ulcer images obtained using handheld cameras to predict whether an ulcer is bacterial or fungal with an AUC of 0.87. Nearly all of this predictive information was identified by state-of-the-art deep CNN models, which demonstrated superhuman performance on this task compared with local cornea experts who previously were shown to outperform an international cohort of cornea specialists. Further work is underway to generate models capable of identifying less common causes of corneal infection, to incorporate structured clinical and historical data as well as human input, and to evaluate additional imaging methods, including smartphone cameras. Future implementation of this technology may allow earlier initiation of directed antimicrobial therapy and better visual outcomes for patients with infectious keratitis.
Manuscript no. D-21-00230
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Disclosure(s):
All authors have completed and submitted the ICMJE disclosures form.
The author(s) have no proprietary or commercial interest in any materials discussed in this article.
Supported by the National Institutes of Health, Bethesda, Maryland (grant nos.: K12EY027720, P30EY10572, U10EY015114, and U10EY018573); and Research to Prevent Blindness, Inc., New York, New York (unrestricted departmental funding). The funding organizations had no role in the design or conduct of this research.
HUMAN SUBJECTS: Human subjects were included in this study. The IRB at Oregon Health & Science University approved the study. All research adhered to the tenets of the Declaration of Helsinki. All participants provided informed consent.
No animal subjects were included in this study.
Author Contributions:
Conception and design: Redd, Seitzman, Lietman, Keenan, Campbell, Song
Analysis and interpretation: Redd, Lietman, Keenan
Data collection: Redd, Prajna, Srinivasan, Lalitha, Krishnan, Rajaraman, Venugopal, Acharya, Lietman, Keenan
Obtained funding: Redd
Overall responsibility: Redd, Prajna, Srinivasan, Lalitha, Krishnan, Rajaraman, Venugopal, Acharya, Seitzman, Lietman, Keenan, Campbell, Song
Supplementary Data
References
- 1.Flaxman S.R., Bourne R.R.A., Resnikoff S., et al. Global causes of blindness and distance vision impairment 1990–2020: a systematic review and meta-analysis. Lancet Global Health. 2017;5(12):e1221–e1234. doi: 10.1016/S2214-109X(17)30393-5. [DOI] [PubMed] [Google Scholar]
- 2.Whitcher J.P., Srinivasan M., Upadhyay M.P. Corneal blindness: a global perspective. Bull World Health Organ. 2001;79:214–221. [PMC free article] [PubMed] [Google Scholar]
- 3.Pascolini D., Mariotti S.P. Global estimates of visual impairment: 2010. Br J Ophthalmol. 2012;96(5):614–618. doi: 10.1136/bjophthalmol-2011-300539. [DOI] [PubMed] [Google Scholar]
- 4.Varaprasathan G., Miller K., Lietman T., et al. Trends in the etiology of infectious corneal ulcers at the F. I. Proctor Foundation. Cornea. 2004;23(4):360–364. doi: 10.1097/00003226-200405000-00009. [DOI] [PubMed] [Google Scholar]
- 5.Srinivasan M., Gonzales C.A., George C., et al. Epidemiology and aetiological diagnosis of corneal ulceration in Madurai, south India. Br J Ophthalmol. 1997;81(11):965–971. doi: 10.1136/bjo.81.11.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bharathi M.J., Ramakrishnan R., Meenakshi R., et al. Microbial keratitis in South India: influence of risk factors, climate, and geographical variation. Ophthalmic Epidemiol. 2007;14(2):61–69. doi: 10.1080/09286580601001347. [DOI] [PubMed] [Google Scholar]
- 7.Mcleod S.D., Kolahdouz-isfahani A., Rostamian K., et al. The role of smears, cultures, and antibiotic sensitivity testing in the management of suspected infectious keratitis. Ophthalmology. 1996;103:23–28. doi: 10.1016/s0161-6420(96)30738-0. [DOI] [PubMed] [Google Scholar]
- 8.Dalmon C., Porco T.C., Lietman T.M., et al. The clinical differentiation of bacterial and fungal keratitis: a photographic survey. Invest Ophthalmol Vis Sci. 2012;53(4):1787–1791. doi: 10.1167/iovs.11-8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahlgren M.A., Lingappan A., Wilhelmus K.R. The clinical diagnosis of microbial keratitis. Am J Ophthalmol. 2007;143(6):940–944. doi: 10.1016/j.ajo.2007.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redd T.K., Prajna N.V., Srinivasan M., Al E. Expert performance in visual differentiation of bacterial and fungal keratitis. Ophthalmology. 2022;129(2):227–230. doi: 10.1016/j.ophtha.2021.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lecun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 12.Pan S.J., Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2010;22(10):1345–1359. [Google Scholar]
- 13.Asaoka R., Murata H., Iwase A., Araie M. Detecting preperimetric glaucoma with standard automated perimetry using a deep learning classifier. Ophthalmology. 2016;123(9):1974–1980. doi: 10.1016/j.ophtha.2016.05.029. [DOI] [PubMed] [Google Scholar]
- 14.Gulshan V., Peng L., Coram M., et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316(22):2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 15.Lee C.S., Baughman D.M., Lee A.Y. Deep learning is effective for classifying normal versus age-related macular degeneration OCT images. Ophthalmol Retina. 2017;1:322–327. doi: 10.1016/j.oret.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown J.M., Campbell J.P., Beers A., et al. Automated diagnosis of plus disease in retinopathy of prematurity using deep convolutional neural networks. JAMA Ophthalmol. 2018;136:803–810. doi: 10.1001/jamaophthalmol.2018.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo M.T., Hsu B.W.Y., Yin Y.K., et al. A deep learning approach in diagnosing fungal keratitis based on corneal photographs. Sci Rep. 2020;10:14424. doi: 10.1038/s41598-020-71425-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh AK, Thammasudjarit R, Jongkhajornpong P, et al. Deep learning for discrimination between fungal keratitis and bacterial keratitis: DeepKeratitis. Cornea. 2021 Sep 29. doi: 10.1097/ICO.0000000000002830. Online ahead of print. [DOI] [PMC free article] [PubMed]
- 19.Hung N., Shih A.K.Y., Lin C., et al. Using slit-lamp images for deep learning-based identification of bacterial and fungal keratitis: model development and validation with different convolutional neural networks. Diagnostics. 2021;11(7):1246. doi: 10.3390/diagnostics11071246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu F., Qin Y., He W., et al. A deep transfer learning framework for the automated assessment of corneal inflammation on in vivo confocal microscopy images. PloS One. 2021;16(6) doi: 10.1371/journal.pone.0252653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srinivasan M., Mascarenhas J., Rajaraman R., et al. Corticosteroids for bacterial keratitis: the Steroids for Corneal Ulcers Trial (SCUT) Arch Ophthalmol. 2012;130(2):143–150. doi: 10.1001/archophthalmol.2011.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prajna N.V., Krishnan T., Rajaraman R., et al. Effect of oral voriconazole on fungal keratitis in the Mycotic Ulcer Treatment Trial II (MUTT II): a randomized clinical trial. JAMA Ophthalmol. 2016;134(12):1365–1372. doi: 10.1001/jamaophthalmol.2016.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prajna N.V., Krishnan T., Mascarenhas J., et al. The Mycotic Ulcer Treatment Trial. JAMA Ophthalmol. 2013;131(4):422–429. doi: 10.1001/jamaophthalmol.2013.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaufman S., Rosset S., Perlich C., Stitelman O. Leakage in data mining. ACM Trans Knowl Discov Data. 2012;6(4):1–21. [Google Scholar]
- 25.Shorten C., Khoshgoftaar T.M. A survey on image data augmentation for deep learning. J Big Data. 2019;6(1):1–48. doi: 10.1186/s40537-021-00492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sandler M., Howard A., Zhu M., et al. MobileNetV2: inverted residuals and linear bottlenecks. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit. 2018:4510–4520. [Google Scholar]
- 27.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. IEEE Conf Comput Vis Pattern Recognit; 2017. Densely connected convolutional networks. 2017; pp. 2261–2269. [Google Scholar]
- 28.He K., Zhang X., Ren S., Sun J. Deep residual learning for image recognition. 2016. IEEE Conf Comput Vis Pattern Recognit. 2016:770–778. [Google Scholar]
- 29.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 3rd International Conference on Learning Representations (ICLR). 2015:1–14.
- 30.Chollet F. Xception: deep learning with depthwise separable convolutions. 2017 IEEE Conf Comput Vis Pattern Recognit. 2017:1800–1807.
- 31.ImageNet. ImageNet Large Scale Visual Recognition Challenge (ILSVRC). Available at: https://image-net.org/challenges/LSVRC/. Accessed 21.06.21.
- 32.Abadi M, Agarwal A, Barham P, et al. TensorFlow: large-scale machine learning on heterogenous systems. arXiv. 2016:1603.04467.1-13.
- 33.Robin X., Turck N., Hainard A., et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837. [PubMed] [Google Scholar]
- 35.Youden W. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 36.Selvaraju R.R., Cogswell M., Das A., et al. Grad-CAM: visual explanations from deep networks via gradient-based localization. Int J Comput Vis. 2016;128(2):336–359. [Google Scholar]
- 37.Xu Y., Kong M., Xie W., et al. Deep sequential feature learning in clinical image classification of infectious keratitis. Engineering. 2021;7(7):1002–1010. [Google Scholar]
- 38.Thomas P.A., Leck A.K., Myatt M. Characteristic clinical features as an aid to the diagnosis of suppurative keratitis caused by filamentous fungi. Br J Ophthalmol. 2005;89(12):1554–1558. doi: 10.1136/bjo.2005.076315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leck A., Burton M. Distinguishing fungal and bacterial keratitis on clinical signs. Community Eye Health. 2015;28(89):6. [PMC free article] [PubMed] [Google Scholar]
- 40.Srinivasan M. Fungal keratitis. Curr Opin Ophthalmol. 2004;15(4):321–327. doi: 10.1097/00055735-200408000-00008. [DOI] [PubMed] [Google Scholar]
- 41.Saini J.S., Jain A.K., Kumar S., et al. Neural network approach to classify infective keratitis. Curr Eye Res. 2003;27(2):111–116. doi: 10.1076/ceyr.27.2.111.15949. [DOI] [PubMed] [Google Scholar]
- 42.Jeng B.H., Gritz D.C., Kumar A.B., et al. Epidemiology of ulcerative keratitis in Northern California. Arch Ophthalmol. 2010;128(8):1022–1028. doi: 10.1001/archophthalmol.2010.144. [DOI] [PubMed] [Google Scholar]
- 43.Ung L., Acharya N.R., Agarwal T., et al. Infectious corneal ulceration: a proposal for neglected tropical disease status. Bull World Health Org. 2019;97(12):854–856. doi: 10.2471/BLT.19.232660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaissis G.A., Makowski M.R., Rückert D., Braren R.F. Secure, privacy-preserving and federated machine learning in medical imaging. Nature Machine Intelligence. 2020;2:305–311. [Google Scholar]
- 45.Miranda L.J. Towards data-centric machine learning: a short review; 2021. 2021. https://ljvmiranda921.github.io/notebook/2021/07/30/data-centric-ml/ Available at: Accessed 04.10.21.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.