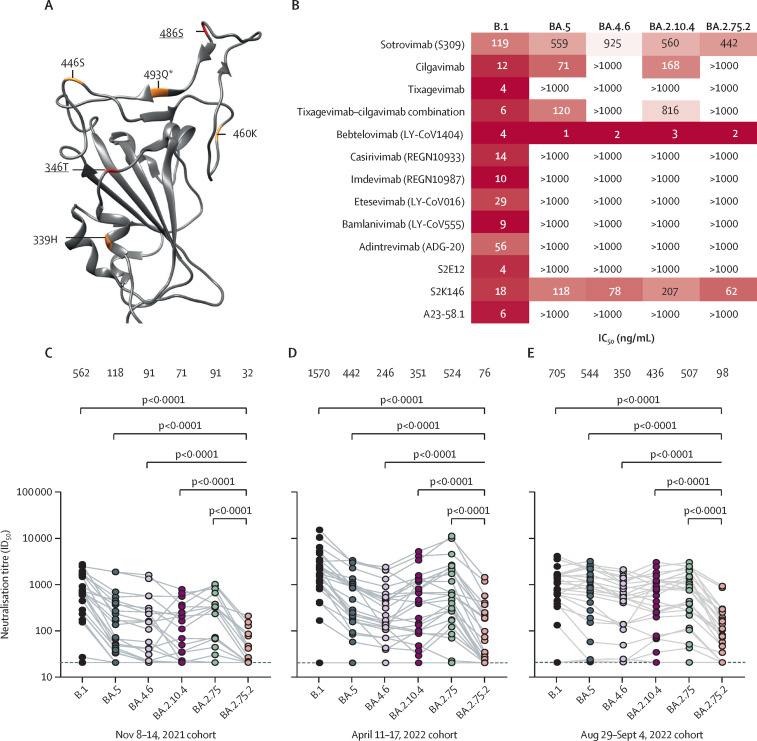
SARS-CoV-2 omicron sublineage BA.2.75 expanded rapidly in parts of the world, but it has so far not outcompeted BA.5 globally. Despite similar geometric mean neutralising titres (GMT) to BA.5, BA.2.75 remained sensitive to classes of antibodies that BA.5 had escaped,1, 2 suggesting scope for antibody evasion. The emergence of a sublineage of BA.2.75 carrying additional mutations R346T, F486S, and D1199N (BA.2.75.2; figure A ; appendix p 1), growing rapidly (appendix p 2), suggested more extensive escape from neutralising antibodies.
Figure.
BA.2.75.2 escapes neutralising antibodies
(A) Differences from BA.2 in BA.2.75 (orange), and BA.2.75.2 (red, underlined), depicted upon the SARS-CoV-2 BA.2 receptor binding domain (pdb:7UB0). Sensitivity of SARS-CoV-2 omicron sublineages and ancestral B.1 (D614G) to neutralisation by monoclonal antibodies (B), and randomly sampled sera from blood donated in Stockholm, Sweden, between Nov 8–14, 2021 (n=25; C), April 11–17, 2022 (n=25; D), and Aug 29 to Sept 4, 2022 (n=25; E). Sera with neutralisation less than 50% at the lowest dilution tested (20) are plotted as 20 (dotted line). IC50=50% inhibitory concentration. ID50=50% inhibitory dilution. *Indicates reversion.
F486V contributed to escape in BA.5,3 mutations at spike residue 346 have occurred in earlier variants,4, 5 and have been specifically highlighted for their capacity to confer additional in escape in omicron.6 Numerous sublineages of the omicron variant have been detected carrying convergent mutations at position 346. R346S is present in BA.4.7, and R346I in BA.5.9.7 BA.4.6, carrying R346T and N658S, is currently the dominant 346T-carrying lineage, detected across a wide geographic distribution. Similarly, several lineages are emerging carrying mutations at residue 486, including BA.2.10.4 harbouring a highly mutated spike, including F486P.
Here we report the sensitivity of emerging omicron sublineages BA.2.75.2, BA.4.6, and BA.2.10.4 to neutralisation by a panel of clinically relevant and preclinical monoclonal antibodies and by serum from blood donated in Stockholm, Sweden. BA.2.75.2 and BA.4.6 both show complete escape from cilgavimab and a combination of cilgavimab and tixagevimab, whereas BA.2.10.4 retains some sensitivity to cilgavimab (figure B). Sotrovimab exhibits similarly low potency against BA.5, BA.2.75.2, and BA.2.10.4, with some reduction against BA.4.6. Bebtelovimab still potently neutralises all variants we tested.
To characterise the evolving resistance to immunity at the population level, we evaluated the sensitivity to neutralisation by serum from random blood donors in Stockholm, including a cohort sampled Nov 8–14, 2021 (November 2021 cohort; figure C), before the emergence of the omicron variant; a cohort sampled April 11–17, 2022 (April 2022 cohort; figure D), after both a large wave of infections (driven by BA.1 then BA.2) and the rollout of third vaccine doses; and a cohort sampled Aug 29 to Sept 4, 2022 (September 2022 cohort; figure E), after the spread of BA.5.
In only the September 2022 cohort, ancestral B.1 titres were at a similar level to those against omicron variants (figure E), potentially as a result of omicron infections. There was a significant difference between the neutralisation of BA.5 and B.1 in the November 2021 and April 2022 cohorts (p<0·0001); however, neutralisation of the two variants was not statistically different in the September 2022 cohort. BA.4.6 (GMT 350 [95% CI 195–629]) and BA.2.10.4 (GMT 436 [248–767]) were moderately more resistant to neutralisation than BA.5 (GMT 544 [287–1033]).
Across all three timepoints, neutralisation of BA.2.75.2 by serum antibodies was significantly lower than all other variants tested (figure C, D, and E). Both R346T and F486S mutations contributed to the significantly enhanced resistance of BA.2.75.2 compared with BA.2.75 (appendix p 3). 14 (56%) of 25 samples from the November 2021 cohort had ID50 titres against BA.2.75.2 that were below the limit of detection (<20). In samples from April 2022 and September 2022 cohorts, BA.2.75.2 was neutralised with a GMT approximately 6·5-times lower than that of the currently dominant BA.5 sublineage, representing the most resistant variant characterised at the time of writing (Sept 19, 2022). Taken together, these data identify profound antibody escape by the emerging omicron sublineage BA.2.75.2, suggesting that it effectively evades current humoral immunity in the population.
STR is a cofounder of and held shares in deepCDR Biologics, which has been acquired by Alloy Therapeutics. DJS, GBKH, and BM have intellectual property rights associated with antibodies that neutralise omicron variants. All other authors declare no competing interests.
Supplementary Material
References
- 1.Cao Y, Song W, Wang L, et al. Characterization of the enhanced infectivity and antibody evasion of Omicron BA.2.75. Cell Host Microbe. 2022 doi: 10.1016/j.chom.2022.09.018. published online Sept 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheward DJ, Kim C, Fischbach J, et al. Evasion of neutralising antibodies by omicron sublineage BA.2.75. Lancet Infect Dis. 2022 doi: 10.1016/S1473-3099(22)00524-2. published online Sept 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao Y, Yisimayi A, Jian F, et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608:593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halfmann PJ, Kuroda M, Armbrust T, et al. Characterization of the SARS-CoV-2 B.1.621 (Mu) variant. Sci Transl Med. 2022;14 doi: 10.1126/scitranslmed.abm4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cele S, Jackson L, Khoury DS, et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2021;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greaney AJ, Starr TN, Bloom JD. An antibody-escape estimator for mutations to the SARS-CoV-2 receptor-binding domain. Virus Evol. 2022;8 doi: 10.1093/ve/veac021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jian F, Yu Y, Song W, et al. Further humoral immunity evasion of emerging SARS-CoV-2 BA.4 and BA.5 subvariants. Lancet Infect Dis. 2022 doi: 10.1016/S1473-3099(22)00642-9. published online Sept 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.