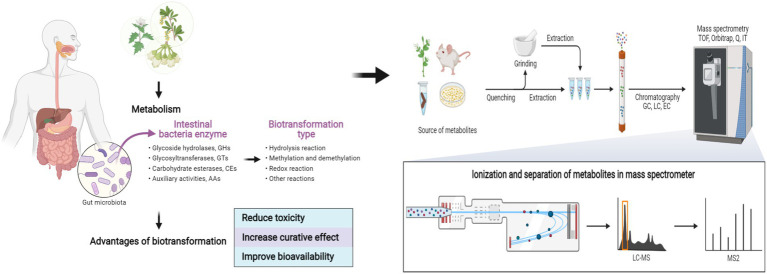
Abstract
Natural products have been extensively applied in clinical practice, characterized by multi-component and multi-target, many pharmacodynamic substances, complex action mechanisms, and various physiological activities. For the oral administration of natural products, the gut microbiota and clinical efficacy are closely related, but this relationship remains unclear. Gut microbes play an important role in the transformation and utilization of natural products caused by the diversity of enzyme systems. Effective components such as flavonoids, alkaloids, lignans, and phenols cannot be metabolized directly through human digestive enzymes but can be transformed by enzymes produced by gut microorganisms and then utilized. Therefore, the focus is paid to the metabolism of natural products through the gut microbiota. In the present study, we systematically reviewed the studies about gut microbiota and their effect on the biotransformation of various components of natural products and highlighted the involved common bacteria, reaction types, pharmacological actions, and research methods. This study aims to provide theoretical support for the clinical application in the prevention and treatment of diseases and provide new ideas for studying natural products based on gut biotransformation.
Keywords: natural products, gut microbes, enzyme system, biotransformation, bioavailability
Graphical Abstract.
Biotransformation and metabolism of natural products based on gut microbes. Created with BioRender.com.
Introduction
The gut microbiota is composed of 1,000–1,250 kinds of bacteria that interact with humans in various forms, such as symbiosis and parasitism, and this interaction greatly affects human health via microbial metabolites as signal molecules (Liu et al., 2017; de Vos et al., 2022). The gut microbes constitute a dynamic and diversified micro-ecosystem, which is a natural barrier to resisting pathogenic bacteria (Chopyk and Grakoui, 2020; Zhao and Maynard, 2022). Gut microbes have abundant enzyme systems, including glucosidase, reductase, lyase, transferase, etc., and greatly expand the metabolic response pool in the human body (Wilson and Nicholson, 2017; Fushinobu and Abou Hachem, 2021).
Natural products are small molecules produced naturally by any organism including primary and secondary metabolites.1 This article mainly describes natural products of plant origin, including nutrients and drugs. They easily interact with gut microbiota because of their complex components and long residence time in the gut. Generally, the residence time for exogenous substances is 1–6 h in the small intestine and 1–3 days in the colon (Chu and Traverso, 2022). Specific gut microbes decompose and transform natural products to produce rich metabolites and functional compounds with physiological activities that cannot be synthesized by the host itself (Koppel et al., 2017; Xie et al., 2020). Microbial transformation in natural products usually refers to the chemical reactions that are used to modify the structure of natural products substrates, such as hydrolysis, methylation, demethylation, redox, and cyclization reaction (Morgan et al., 2022; Rocchetti et al., 2022). Gut microbiota remarkably affects the chemical modification, pharmacological activity, and metabolic mechanism of natural products. The potential utility of gut microbes for large-scale synthesis of active metabolites and production of compounds has not been investigated. Studying these gut microbes, metabolites, and the reactions involved in the interactions between natural products and gut microbiota is of great significance in the exploration of the pharmacological mechanisms and utilization of natural products. In this review, we introduce the resident gut microbes that contribute to the transformation of natural products and summarize the transformation pathways between natural products and specific microbes classified by the reactions. Moreover, the advantages, research methods, and future directions of gut microbial in the conversion of natural products are discussed to provide a theoretical basis for the modern application of natural products and the precise treatment through gut microbiota.
Key gut microbes in the biotransformation of natural products
Oral administration is the preferred route for drug delivery, and oral drugs account for 84% of the top 50 best-selling drugs in the US and European markets (Lennernäs and Abrahamsson, 2005; Vinarov et al., 2021). In recent years, the influence of gut microbiota on the stability of oral administration of natural products has received much attention. The intestinal tract has abundant bacteria that help with normal digestive function, in which about 98% of gut microbes in healthy subjects can be classified into four phyla, Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria (Manor et al., 2020; Ye et al., 2021; de Vos et al., 2022). Some gut microbes such as Escherichia coli, Bifidobacterium, Eubacterium, Lactobacillus, Bacteroides, and Streptococcus participate in the biotransformation of natural products, and part of their metabolites are conducive to intestinal absorption and play a notable pharmacological role (Al-Ishaq et al., 2021; Augusti et al., 2021; Figure 1).
Figure 1.
Biotransformation of natural products by key gut microbes. (A) β-glucuronidase of E. coli HGU-3 catalyzes hydrolysis of baicalin to yield baicalein (Han et al., 2016; Li D. et al., 2019). (B) Feruloyl esterases from B. animalis, L. reuteri, L. helveticus, and L. fermentum catalyzes hydrolysis of chlorogenic acid into caffeic acid (Raimondi et al., 2015; Pang et al., 2016; Aguirre Santos et al., 2018). (C) β-glucosidase of Eubacterium L-8 and Streptococcus LJ-22 catalyzes hydrolysis of glycyrrhizin to 18β-glycyrrhetinic acid (Kim et al., 1999; Chen B. et al., 2021). (D) α-L-rhamnosidase from Bacteroides sp. 45 catalyzes hydrolysis of rutin to quercetin 3-O-glucoside (Riva et al., 2020). (E) O-Methyltransferase from Blautia sp. MRG-PMF1 catalyzes the demethylation of curcumin to demethoxycurcumin (Burapan et al., 2017a,b). Created with BioRender.com.
Escherichia coli
Escherichia coli is a Gram-negative, spore-free, facultative anaerobic bacterium, which mainly inhabits the intestines of vertebrates (Foster-Nyarko and Pallen, 2022). Part of E. coli can produce glycosidase to participate in the transformation of exogenous substances, resulting in its beneficial role (Rodríguez-Daza et al., 2021; Candeliere et al., 2022). For example, E. coli HGU-3 produces β-glucuronidase to hydrolyze the O-glycosidic bond in baicalin to produce baicalein (Kim et al., 1996, 1998; Han et al., 2016). Baicalein depresses histamine-induced scratching behavior more effectively than baicalin at the same dose and presents anti-inflammatory and anti-oxidant effects by inhibiting Nrf2-ARE and NF-κB signaling pathway (Chen et al., 2000; Trinh et al., 2010; Ye et al., 2014). Some E. coli strains have high specific activity for curcumin conversion. Curcumin is reduced to dihydrocurcumin (DHC) and tetrahydrocurcumin (THC) by the highly expressed NADPH-dependent curcumin/dihydrocurcumin reductase (CurA) of E. coli DH10B, whose whole-genome sequence had already been determined (Hassaninasab et al., 2011; Tan et al., 2014). DHC and THC (20 μM) reduce triglyceride levels in OA-induced L02 and HepG2 cells by regulating mRNA and protein expression levels of SREBP-1C and PPARα and attenuate OA-induced liver adipogenesis in an AMPK-dependent manner; DHC and THC have novel therapeutic benefits over curcumin in hepatic steatosis (Chen et al., 2018; Yu et al., 2018). E. coli Nu, E. coli MC, and E. coli WC-1 have cinnamyl esterase activity, which can release hydroxycinnamic acids through the hydrolysis of conjugated hydroxycinnamates and free hydroxycinnamates exhibit antioxidant and anticancer properties both in vitro and in vivo (Couteau et al., 2001). At present, a good understanding of the genetic and biochemical characteristics of E. coli may contribute to the synthesis of natural product derivatives with various health activities in vitro.
Bifidobacterium
Bifidobacterium is a widespread and abundant genus belonging to the phylum Actinobacteria and is among the first colonizers of gut microbiota for humans (Satti et al., 2021; He et al., 2022). The most common Bifidobacterium in the human gut include B. adolescentis, B. angulatum, B. bifidum, B. breve, B. catenulatum, B. dentium, B. longum, B. pseudocatenulatum, and B. pseudolongum (Turroni et al., 2009; Hidalgo-Cantabrana et al., 2017), accounting for <10% of the adult human microbiome, but they are linked to host health (Turroni et al., 2008). Certain species of Bifidobacterium can generate phenolic acids by expressing feruloyl esterase. For example, the feruloyl esterase of B. animalis can hydrolyze chlorogenic acid (CHA) into caffeic acid (CAA; Raimondi et al., 2015). CAA (10–30 mg/kg) can prevent acetaminophen-induced acute liver injury in mice by increasing Nrf2 transcription (Raimondi et al., 2015; Pang et al., 2016). The participation of partial Bifidobacterium promotes the metabolism of flavanones, glycosides, and saponins in the gut. β-glucosidase and demethylase in B. longum R0175 promote 3-(3′-hydroxyphenyl) propionic acid and 3-(phenyl) propionic acid production from hesperidin through ring-cleavage and demethylation (Pereira-Caro et al., 2018). B. longum SBT2928 hydrolyzes six major human and two animal bile salts (Tanaka et al., 2000). Thus, Bifidobacterium may regulate bile acid metabolism and reduce cholesterol levels in vivo. In addition, B. breve ATCC 15700 produces β-glucosidase to cleave glycoside at the C-3 and C-20 positions of ginsenoside Rd. to generate deglycosylated ginsenoside compound K (Zhong et al., 2016; Zhang R. et al., 2019). These metabolic characteristics make Bifidobacterium a prime candidate for the development of symbiosis to make natural products potentially beneficial.
Eubacterium
The genus of Eubacterium strains is Gram-positive, which forms one of the core genera of the human gut microbiota and shows widespread colonization of the human gut (Mukherjee et al., 2020). Some Eubacterium species produce glycosidase, reductase, etc., and participate in the metabolism of exogenous substances (Zhang J. et al., 2019; Ellenbogen et al., 2021). E. ramulus is one of the most widely studied flavonoid-degrading gut bacteria, and it is prevalent in the human intestine. Chalcone isomerase and flavanone-/flavanonol-cleaving reductase from E. ramulus degrade certain flavonoids to produce chalcone, and dihydrochalcone (Gall et al., 2014). Dihydrochalcone and its metabolites have anti-inflammatory and antioxidant effects, which can down-regulate the secretion of pro-inflammatory cytokines in RAW 264.7 and rescue LPS-induced oxidative phosphorylation (Choi et al., 2021). Braune et al. investigated the degradation of flavonol quercetin and flavone luteolin by E. ramulus strain wK1 and found that resting cells and enzyme preparations convert these flavonoids into 3, 4-dihydroxyphenylacetic acid, and 3-(3, 4-dihydroxyphenyl) propionic acid via the reduction of 2, 3-position double bonds and subsequent ring fission (Braune et al., 2001). Phloretin hydrolase from E. ramulus strain wK1 hydrolytically cleaves the C-C bond, which is adjacent to the aromatic A-ring of phloretin to 3-(4-hydroxyphenyl)-propionic acid and phloroglucinol (Schoefer et al., 2004; Braune et al., 2019). E. cellulosolvens ATCC 43171T may contribute to the deglycosylation of flavonoid O- and C-glucosides (luteolin 6-C-glucoside and apigenin 6-C-glucoside) through the fermentation of the liberated glucose portion. The deglycosylation of C-glucosides is exclusively catalyzed by bacterial enzymes (Braune and Blaut, 2012; Braune et al., 2016). Eubacterium L-8 hydrolyzed terpenoid glycyrrhizin (GL) to 18β-glycyrrhetinic acid (18β-GA; Kim et al., 2000). 18β-GA prevents OVA-induced airway allergic inflammation by inhibiting NF-κB phosphorylation and enhancing the Nrf2/HO-1 pathway (Liu et al., 2022). These metabolic transformations provide more information about the diverse array of benefits that humans derive from Eubacterium spp. However, further in vivo studies are necessary to maximize the potential benefits the Eubacterium genus has to offer.
Lactobacillus
The genus Lactobacillus belongs to the phylum Firmicutes, which can balance the micro-community and protect gastrointestinal mucosa (Dempsey and Corr, 2022). Some Lactobacillus species are rich in metabolic enzymes, such as α-rhamnosidases, tannase, gallate decarboxylases, etc. and they transform exogenous substances (Reverón et al., 2017; Li B.C. et al., 2019; Ferreira-Lazarte et al., 2021). L. rhamnosus NCTC 10302, which has both β-glucosidase and α-rhamnosidase activities, converts hesperetin-7-O-rutinoside and naringenin-7-O-rutinoside to their respective aglycones and 3-(phenyl) propionic acid by hydrolysis, ring fission, and dehydroxylation (Pereira-Caro et al., 2018). L. plantarum expresses tannase to hydrolyze gallate, protocatechuate esters with a short aliphatic alcohol substituent, and complex gallic tannins to produce gallic acid (Jiménez et al., 2014). Gallic acid (11.5–46 μg/ml) plays a protective role in LPS-induced inflammation and oxidative stress by inhibiting the MAPK/NF-κB pathway and activating the Akt/AMPK/Nrf2 pathway (Tanaka et al., 2018). Fang et al. observed that gallic acid and pyrogallol are produced by the degradation of gallotannins by gallotannin-metabolizing enzymes in L. plantarum WCFS1. This study implies the potential role of prebiotic-probiotic interactions in the prevention of diet-induced metabolic disorders (Reverón et al., 2015; Fang et al., 2019). Daidzein is reduced to dihydrodaidzein by Lactobacillus sp. Niu-O16 with daidzein reductase activity (Wang et al., 2007; Heng et al., 2019). Dihydrodaidzein (2.5–5 μM) inhibits NF-κB activation and MAPK phosphorylation, thereby improving osteoporosis (Kim et al., 2019). L. casei, L. plantarum, and L. acidophilus highly influence the deglycosylation of piceid to resveratrol (Basholli-Salihu et al., 2016). This conversion is important for increasing the bioavailability and bioactivity of piceid. Feruloyl esterases from L. reuteri, L. helveticus, and L. fermentum hydrolyze chlorogenic acid to release caffeic acid (Aguirre Santos et al., 2018). These findings open a new perspective on the role of Lactobacillus in health-promoting pharmaceutical and food product applications. However, the underlying transformation mechanism deserves further study.
Bacteroides
Members of the genus Bacteroides are Gram-negative obligate anaerobes, which account for 25% of the total bacteria in the colon and play multiple roles in the human gut bacteriome (Zafar and Saier, 2021). Bacteroides species such as B. fragilis, B. distasonis, B. ovatus, and B. thetaiotaomicron are commonly detected in the clinic (Wexler, 2007). Bacteroidetes spp. possesses a series of hydrolases and participates in inter-species cross-feeding relationships with their microbial neighbors by converting foreign substances (Sonnenburg et al., 2004; Zafar and Saier, 2021). In vitro co-incubation experiments showed that certain Bacteroides species are involved in the biotransformation of flavonoids. Bacteroides sp. 45 expresses α-L-rhamnosidase and β-rutinosidase for the hydrolysis of rutin into quercetin 3-O-glucoside, quercetin, and leucocyanidin (Yang et al., 2012; Riva et al., 2020; Ferreira-Lazarte et al., 2021). Quercetin 3-O-glucoside is better absorbed than other forms of quercetin and can suppress the inflammatory response in mice with TNBS-induced colitis via the inhibition of the NF-κB and MAPK signaling pathways (Zhang D. et al., 2019). Bacteroides sp. 54 metabolizes quercitrin to hydroxyquercitrin and desmethylquercitrin. Quercitrin is also degraded to quercetin by α-L-rhamnosidase and undergoes further ring-cleavage to yield 3,4-dihydroxybenzoic acid by Bacteroides sp. 45 (Jiang et al., 2014). β-glucuronidase, which is expressed by Bacteroidetes J-37, metabolizes GL to 18β-GA (Kim et al., 1999; Guo et al., 2018). Based on the review of existing studies, natural products are biotransformed under the action of Bacteroidetes to produce metabolites with different functional activities. It is important to understand the whole process of natural products occurring in the body to assess the effect on human health.
Streptococcus
The Streptococcus species are Gram-positive, spherical, or ovoid cells, which are usually arranged in chains or pairs and widely exist in human feces and nasopharynx (Lannes-Costa et al., 2021). Meta-transcriptomic analysis indicates that the phosphotransferase system is majority expressed by Streptococcus, suggesting that these bacteria are the main utilizers of the available carbohydrates in the small intestinal (Zoetendal et al., 2012). Streptococcus LJ-22 expresses β-glucuronidase to metabolize GL to 18β-glycyrrhetinic acid-3-O-β-D-glucuronic acid (GAMG; Kim et al., 2000; Park et al., 2004; Guo et al., 2018). GAMG has anti-allergic activity against LPS-induced RAW264.7 cells with IC50 value of 0.28 mM (Park et al., 2004). In addition, tannic acid is degraded by tannase of Streptococcus gallolyticus subsp. Gallolyticus (SGG) to produce pyrogallol. SGG may contribute to the development of colorectal cancer by eliminating the toxicity of tannic acid to tumor cells (Oehmcke-Hecht et al., 2020). Therefore, further in vivo studies are necessary to determine whether the elimination of these tannic acid-degrading microbes can support the effective treatment of colorectal cancer. S. thermophilus GIM 1.321 has a high production capacity of β-glucosidase for the degradation of fructus anthocyanins into CHA, CAA, and ferulic acid (Cheng J.R. et al., 2016). The administration of CAA and CHA (10/15 mg/kg/day) can lower blood pressure and exert an anti-oxidant effect (Agunloye et al., 2019). Streptococcus strains might be a commensal, pathogenic, and opportunistic pathogen in the gut, and more information is needed about its effect on human health. A better understanding of how Streptococcus metabolizes natural products may allow the regulation of the gut microbiome to improve therapeutic efficacy.
Blautia
Blautia species are strictly anaerobic, nonmotile, usually spherical or oval, and widely found in the gut and feces of mammals (Liu X. et al., 2021). There is increasing evidence for the probiotic properties of Blautia on the biotransformation of natural products (Tremaroli and Bäckhed, 2012). In the course of flavonoid biotransformation, the reactions catalyzed by Blautia include demethylation, O-and C- deglycosylation, and C-ring cleavage (Braune and Blaut, 2016), which may be catalyzed by the corresponding enzymes, such as O-glycosidase and β-glucosidases (Braune et al., 2016). Research indicates that the strain Blautia sp. MRG-PMF1 has a hydrolytic ability on aryl methyl ether functional groups by converting 5,7-dimethoxyflavone and 5,7,4-trimethoxyflavone into bioactive chrysin and apigenin, respectively. Blautia sp. MRG-PMF1 also possesses deglycosylation activity, and various isoflavones, flavones, and flavones were found to be metabolized into the corresponding aglycones (Kim et al., 2014). Besides, under anaerobic conditions, Blautia sp. MRG-PMF1 strain metabolizes icariin further to desmethylicaritin with estrogenic effects (Wu et al., 2016). The strain can also catalyze curcumin to produce demethoxycurcumin with anti-inflammatory and anti-cancer properties (Burapan et al., 2017a; Hatamipour et al., 2019). In addition, Blautia sp. AUH-JLD56 is capable of solely biotransforming arctiin or arctigenin into demethylated products with better antioxidant capacity (Liu et al., 2013). Recently, a growing academic interest has been witnessed in the biotransformation and metabolism of herbal plants and functional foods by Blautia. Exploring the biotransformation of Blautia is of great significance for the development of new enzymes and bioactive metabolites (Meng et al., 2020).
Key transformation types involved in natural products microbial metabolism
Complex microbial enzymes catalyze the metabolism of natural products in the gut, resulting in lipophilic and low-molecule-weight metabolites conducive to host utilization/excretion (Weersma et al., 2020). Unlike human genetics, the gut microbiome is modifiable in terms of characteristics, making it a potential therapeutic target to optimize therapy. After oral natural products enter the digestive tract, they will first come into contact with a large number of gut microbes and the active enzymes produced by them. Therefore, natural products’ gut biotransformation may occur before the first-pass effect through the liver (Xie et al., 2020). Natural products can be modified/deconjugated by the gut microbiome, and can also be transported to the liver to modify/bind and then excreted into the gut to react with gut microbes to form a series of metabolites (Koppel et al., 2017). The metabolites transformed by the host-microbial co-metabolic system may be functionally novel and not clearly defined. Therefore, the combination of specific strains, specific metabolic pathways, and specific enzymes associated with health/disease is important for the determination of the effect of gut microbes on the host.
Hydrolysis
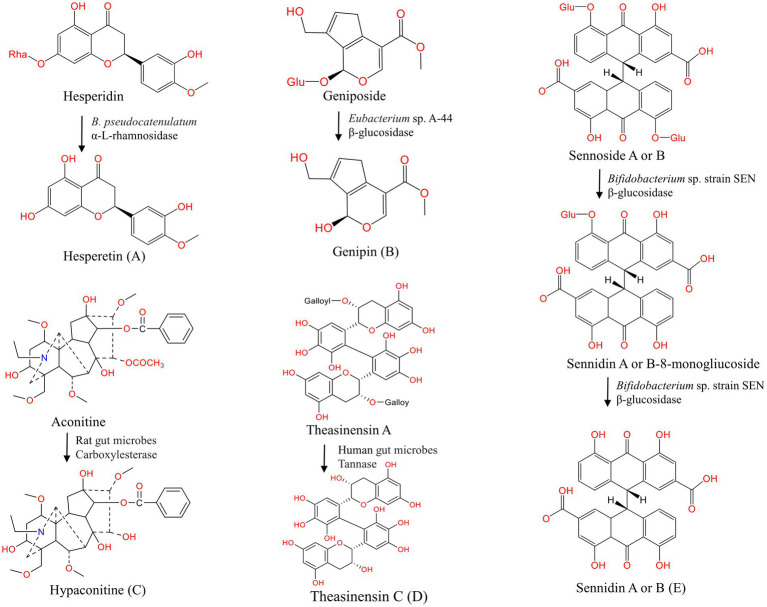
Certain natural products have high molecular weight and low lipid solubility, and they are difficult to be absorbed by the body in the intestine and have low bioavailability (Hostetler et al., 2017). Through gut microbes-mediated hydrolysis, their physical properties are changed, and their biological activity and bioavailability are greatly improved (Wu and Tan, 2019). Slámová et al. indicated that most glycosides have low activity and are considered “natural prodrugs” (Slámová et al., 2018). After interacting with gut microbes, the sugar groups of glycosides are removed, and then, the aglycone portion is absorbed by intestinal cells to exert physiological effects (Wilson and Nicholson, 2017; Murota et al., 2018). The hydrolysis reaction is required for further transformation, and the products (e.g., sugars) participate in promoting the growth and survival of gut microorganisms (Theilmann et al., 2017). Figure 2 shows the hydrolysis reaction of partial natural products under the action of gut microbes.
Figure 2.
Hydrolysis of natural products under the action of gut microbes. (A–E) Hydrolysis of hesperidin (Mas-Capdevila et al., 2020), geniposide (Jiang et al., 2016), aconitine (Zhang et al., 2015), theasinensin A (Liu Z. et al., 2021), sennoside A or B (Matsumoto et al., 2012).
Flavonoids
Flavonoids are natural phenolic compounds found abundantly in fruits and vegetables. Gut microbes may be partly responsible for the efficacy of flavonoids (glycoside forms), which have low bioavailability because of the presence of water-soluble sugar components (Murota et al., 2018; Al-Ishaq et al., 2021). Flavanols with 3-hydroxyflavone base (3-hydroxy-2-phenylchromen-4-one) and planar ring system constitute a significant class of flavonoids. In the study of Du et al., isorhamnetin-3-O-neohesperidoside was first deglycosylated to isorhamnetin-3-O-glucoside and subsequently to the aglycone isorhamnetin by Escherichia sp. 23 (Du et al., 2017). The gut microbes and derived enzymes (lactase phlorizin hydrolase) jointly controlled the metabolism of epimedium koreanum nakai-prenylated flavonoids as determined by in vitro assays. In the present study, gut enzymes metabolized flavonoids faster than gut microbes (Zhou et al., 2013). Wu et al. found that α-L-rhamnosidase from Bacteroides thetaiotaomicron VPI-5482 could hydrolyze the α-1,2 glycosidic bond of epimedin C to produce icariside (Wu et al., 2018). β-xylosidase Dt-2,286, which is derived from Dictyoglomus turgidum, is highly active in hydrolyzing xylose and glucose groups in epimedium B to obtain baohuoside I and sagittatoside B (Tong et al., 2021). Flavanones have a 2,3-dihydro-2-phenylchromen-4-one structure. Hesperidin is converted to its active form hesperetin by α-L-rhamnosidase, which is expressed by B. pseudocatenulatum (Mas-Capdevila et al., 2020). Isoflavones are mainly found in legumes. B. breve MCC1274 possesses the highest β-glucosidase activity for the conversion of daidzin to daidzein (Yao et al., 2019). The anthocyanin cyanidin 3-glucoside is converted to cyanidin by E. ramulus and Clostridium saccharogumia (Hanske et al., 2013). Human gut enzymes such as β-glucuronidase play a key role in the hydrolysis of wogonoside to its aglycone form wogonin (Xing et al., 2014). Theasinensin A, a bioactive catechin dimer found in black tea, is degalloylated to yield theasinensin C by human fecal microbiota (Liu Z. et al., 2021). In the present study, we observed the metabolic differences in flavane-3-ols, and the results suggest that steric hindrance may limit the degradation of partial flavane-3-ols C-ring by bacterial enzymes during gut microbial fermentation. Many other flavonoids can also undergo hydrolysis reactions under the action of gut microbes, as shown in Table 1. Notably, considering the structural differences of flavonoids, the degree of degradation of flavonoids by gut microbes varies greatly, thus affecting their bioaccessibility. Further efforts are required to investigate the role of gut metabolism in the bioavailability and absorption of flavonoids and the possible bacteria-flavonoid interaction ctivities.
Table 1.
Hydrolysis reaction of gut bacteria to natural products.
| Classification | Gut microbiota | Enzyme | Substrate | End-product | Changes | Ref. |
|---|---|---|---|---|---|---|
| Flavonoid glycosides |
E. coli HGU-3; L. brevis RO1 |
β-glucuronidase | Baicalin | Baicalein; oroxylin A |
Bioavailability↑ anti-inflammation↑ |
Yim et al. (2004), Trinh et al. (2010) and Han et al. (2016) |
| E. cellulosolvens ATCC 43171T | β-glucosidase | Luteolin 7-O-glucoside; apigenin 7-O-glucoside |
Luteolin; apigenin |
Bioavailability↑ | Braune and Blaut (2012) and Braune et al. (2016) | |
| E. cellulosolvens ATCC 43171T | NA | Luteolin 6-C-glucoside; apigenin 6-C-glucoside |
Luteolin; apigenin |
Bioavailability↑ | Braune and Blaut (2012) and Braune et al. (2016) | |
| Human gut microbes | β-glucuronidase | Wogonoside | Wogonin | Anti-inflammation↑ | Xing et al. (2014) | |
|
Bacteroides JY-6; Fusobacterium K-60 |
β-glucosidase α-L-rhamnosidase β-rutinosidase |
Rutin | Quercetin-3-O-glucoside; quercetin; leucocyanidin |
Bioavailability↑ anti-oxidant↑ |
Riva et al. (2020) and Ferreira-Lazarte et al. (2021) | |
| Escherichia sp. 23 | β-glucosidase | Isorhamnetin-3-O-neohesperidoside | Isorhamnetin-3-O-glucoside; isorhamnetin |
Bioavailability↑ anti-inflammation↑ |
Du et al. (2017) | |
| Rat gut microbes; B. thetaiotaomicron VPI-5482 |
β-glucosidase α-L-rhamnosidase |
Epimedin A, B, C | Icariin II; icariin A, B |
Anti-osteoporosis↑ | Cui et al. (2013), Cui et al. (2014) and Wu et al. (2018) | |
| Dictyoglomus turgidum | β-xylosidase Dt-2,286 | Epimedium B | Baohuoside I; sagittatoside B |
Anti-osteoporosis↑ | Tong et al. (2021) | |
| B. animalis subsp. lactis AD011 | β-glucosidase | Quercetin 3-O-glucoside isorhamnetin 3-O-glucoside |
Quercetin; isorhamnetin |
Anti-tumor↑ anti-inflammatory↑ |
Youn et al. (2012) | |
| Lactobacillus paracasei A221 | β-glucosidase | Kaempferol-3-O-sophoroside | Kaempferol | Anti-aging↑ | Shimojo et al. (2018) | |
| Enterococcus. sp. 8B, 8-2,9-2 | β-glucosidase | Astilbin | Taxifolin | Cardiovascular protection↑ anti-tumor↑ anti-inflammatory↑ |
Zhao et al. (2014) and Zhao et al. (2021) | |
| Human gut microbes; B. pseudocatenulatum |
α-L-rhamnosidase; β-glucosidase |
Hesperidin | Hesperetin | Anti-oxidant ↑ anti-inflammatory ↑ |
Mas-Capdevila et al. (2020) | |
| Rat gut microbes | β-glucosidase | Calycosin-7-O-β-D-glucoside | Calycosin | Neuroprotection↑ anti-oxidant ↑ |
Ruan et al. (2015) | |
|
E. ramulus; B. breve MCC1274 |
β-glucosidase | Daidzin | Daidzein | Neuroprotection↑ | Mace et al. (2019) and Yao et al. (2019) | |
| Dorea species PUE | C-deglycosylation enzymes (DgpB-C) | 3″-oxo-puerarin | Daidzein | Bioavailability↑ | Nakamura et al. (2020) | |
|
E. ramulus; Clostridium saccharogumia |
β-glucosidase | Cyanidin 3-glucoside | Cyanidin | Bioavailability↑ | Hanske et al. (2013) | |
| Human gut microbes | Tannase | Theasinensins A | Theasinensins C | Bioavailability↑ | Liu Z. et al. (2021) | |
|
Bacillus sp. KM7-1; Bacteroides sp. MANG |
C-C glucosyl-cleaving enzyme | Mangiferin | Norathyriol | Anti-cancer↑anti-diabetes↑ | Hasanah et al. (2021) | |
| Strain CG19-1 | NA | Mangiferin | Norathyriol | Braune and Blaut (2011) | ||
| Terpenoids | Eubacterium. sp. A-44 | β-glucosidase, carboxylesterases | Geniposide | Genipin; geniposidic acid |
Bile secretion↑ anti-hepatitis↑ |
Akao et al. (1994), Jiang et al. (2016) and Tian et al. (2013) |
|
B. fragilis; L. brevis; rat gut microbes |
β-glucosidase | Paeoniflorin | PM-I; albiflorin and its aglycone; Deacyl-paeonifloridin |
Anti-convulsant↑ Bioavailability↑ |
He et al. (2007) and Ke et al. (2016) | |
| Rat gut microbes | Glycoside hydrolases | Asiaticoside | Corresponding aglycones | Bioavailability↑ | Weng et al. (2006) | |
| Eubacterium sp. A-44 | β-glucosidase; β-D-fucosidase |
Saikosaponin B1 | Prosaikogenin; saikogenin A | Anti-inflammatory↑ Anti-oxidant↑ |
Kida et al. (1998) | |
|
Eubacterium L-8; Bacteroidetes J-37; Streptococcus LJ-22 |
β-glucuronidase | GL | 18β-GA; GAMG |
Anti-platelet aggregation↑ anti-allergic↑ anti-tumor↑ anti-bacterial↑ |
Kim et al. (1999), Kim et al. (2000), Park et al. (2004) and Guo et al. (2018) | |
|
Eubacterium sp. A-44; B. breve ATCC 15700 |
β-glucosidase | Ginsenoside Rh2 | Ginsenoside F2; compound K |
Bioavailability↑ | Zhong et al. (2016), Zhang R. et al. (2019) and Kim (2018) | |
|
B. breve; B. longum |
Esterases | Albiflorin | Benzoic acid | Anti-depression↑ | Zhao et al. (2018) and Peng et al. (2022) | |
| Human/rat gut microbes | β-glucosidase; α-L-rhamnosidase |
Ardipusillosides I | Deglycosylated product | Bioavailability↑ | Cao et al. (2015) | |
| Human gut microbes | NA | Mogroside III | Mogroside II mogrol |
Bioavailability↑ | Yang et al. (2007) | |
|
B. adolescentis; B. breve |
NA | Pedunculoside | Deglycosylated products | Bioavailability↑ | Wu et al. (2019) | |
| Rat gut microbes | NA | Capilliposide C | Deglycosylated products esterolysis products |
Cheng Z. et al. (2016) | ||
| Anthraquinones | Bifidobacterium sp. strain SEN | β-glucosidase | Sennoside A and B | Sennidin A/B-8-monoglucoside | Purgation↑ | Matsumoto et al. (2012) |
| Alkaloids Phenols |
Human gut microbes | CEs | DDAs | MDAs | Toxicity↓ | Zhang et al. (2015) |
| Rat gut microbes | NA | Scopolamine | Scopine | Anti-tumor↑ anti-inflammatory↑ |
Dey (2019) | |
| L. plantarum | Tannase | Gallic tannins | Gallic acid | Anti-oxidant↑ anti-inflammatory↑ |
Jiménez et al. (2014) | |
| Akkermansia muciniphila | Tannase | Ellagitannins | Ellagic acid | Neuroprotection↑ | Luca et al. (2020) | |
| Rat gut microbes | β-glucosidase | Amygdalin | Mandelonitrile; prunasin; phenylacetonitrile; hydrogen cyanide |
Toxicity↑ | Kim et al. (2008) and Qin et al. (2021) | |
| B. animalis | Feruloyl esterase | CHA | CAA | Anti-oxidant↑ | Raimondi et al. (2015) | |
|
L. plantarum; L. johnsonii; L. acidophilus |
Feruloyl esterases | CAA; p-coumaric acids |
ferulic acid | Anti-oxidant↑ | Fritsch et al. (2017) | |
|
E. coli Nu; E. coli MC; E. coli WC-1 |
Cinnamyl esterase | Conjugated hydroxycinnamates | Free hydroxycinnamates | Anti-oxidant↑ anti-cancer↑ |
Couteau et al. (2001) | |
| Steroids | Human gut microbes | NA | Pulsatilla saponin D | Corresponding deglycosylation products | Bioavailability↑ | Yan et al. (2018) |
| Other | Mouse gut microbes | β-glucosidase | Cycasin | Diazomethane | Toxicity↑ | Goldin (1990) |
Terpenoids
Terpenoids are the largest class of natural products with anti-cancer, anti-inflammatory, and neuroprotective effects (Agatonovic-Kustrin et al., 2020; El-Baba et al., 2021). Part of terpenoids can also be hydrolyzed by gut microbes. Geniposide produces genipin with the action of β-glucosidase expressed by Eubacterium sp. A-44 (Akao et al., 1994; Jiang et al., 2016). Paeoniflorin is transformed into PM-I under the action of β-glucosidase, which is expressed by L. brevis and B. fragilis (Abdel-Hafez et al., 1999; He et al., 2007). By incubating with rat anaerobic gut microbiota, paeoniflorin is also deglucosed and dephenyled into albiflorin and acyl albiflorin with a small molecular weight (Ke et al., 2016). Peng et al. demonstrated that several Bifidobacteria species with esterase can hydrolyze albiflorin to benzoic acid in vitro (Peng et al., 2022). In vitro study shows that asiaticoside is gradually deglycosylated by glycoside bond hydrolase and produces corresponding aglycones (Weng et al., 2006). Saikosaponin B1 is gradually hydrolyzed to prosaikogenin and saikogenin A under the action of β-glucosidase and β-D-focusidase, which are expressed by Eubacterium sp. A-44 (Kida et al., 1998). Except for the compounds mentioned above, terpenoids ginsenoside Rh2 (Guo et al., 2019), ardipusillosides I (Cao et al., 2015), mogroside III (Yang et al., 2007), and pedunculoside (Wu et al., 2019) can also undergo hydrolysis reactions under the action of gut microbes (Table 1). Therefore, gut microbes play an important role in terpenoid metabolism, and the effects of their metabolites on gut microbiome and human health need to be further studied.
Other compounds
Ellagitannins, which have a very low bioavailability perform a pharmacological role only when it is hydrolyzed into derivatives such as ellagic acid and uroliths under the action of tannase from Gordonibacter urolithinfaciens, Gordonibacter pamelaeae, and Ellagibacter isourolithinifaciens (Beltrán et al., 2018; García-Villalba et al., 2020; Tang et al., 2021). The anthraquinone glycosides extracted from rhubarb are hydrolyzed into anthraquinone aglycones by gut microbes (Li Q. et al., 2020). Sennoside A, a major component of rhubarb extract, is metabolized into rhein anthrone by β-glucosidase of Bifidobacterium sp. strain SEN (Matsumoto et al., 2012; Kon et al., 2014). Under the action of carboxylesterase (CEs), which are expressed by gut microbes, diester diterpenoid alkaloids (DDAs, such as aconitine) hydrolyze the ester bonds of C-8 and C-14 to produce monoester diterpene alkaloids (MDAs, such as hypaconitine), which are less toxic (Zhang et al., 2015). Pulsatilla Chinensis is commonly used in Asia, and its major saponin anemoside B4 can be degraded by gut microbes to produce deglycosylation products (Wan et al., 2017). Table 1 shows that the alkaloids scopolamine (Wu et al., 2019), steroid compound pulsatilla saponin D (Yan et al., 2018), and cycasin (Goldin, 1990) undergo hydrolysis reactions under the action of gut microbes. The hydrolysis reaction is an important step in the metabolism of natural products by gut microbes and is required for the expression of biological activity and further biotransformation. The specific microorganisms and enzymes involved in this reaction should be focused on to fully understand the ultimate fate of natural products and their impact on human health and provide a basis for personalized treatment.
Methylation and demethylation
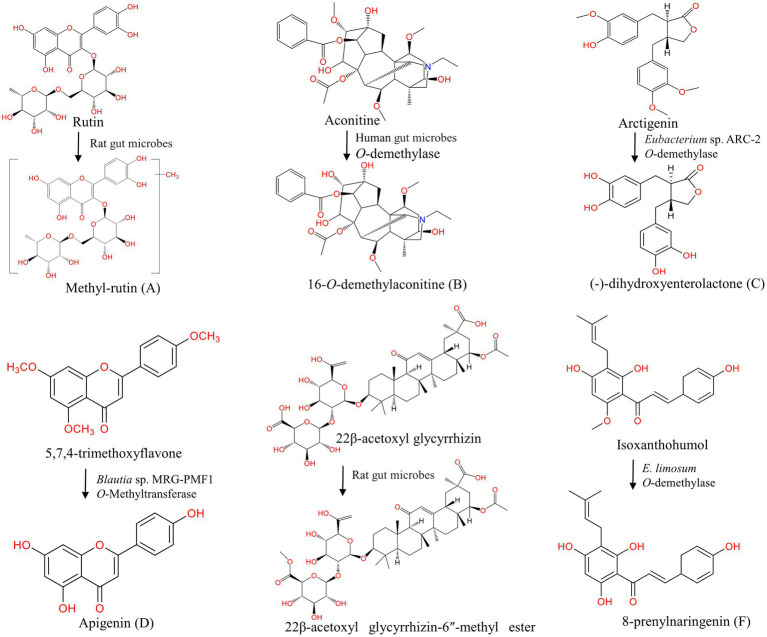
Gut microbes can express transferases and move functional groups between the two substrates through nucleophilic substitution reactions (Koppel et al., 2017). The addition of methyl to exogenous substances by gut microbes requires chemically activated co-substrates, such as acetyl coenzyme A, adenosine triphosphate, or S-adenosylmethionine, while demethylation requires cofactors that can undergo nucleophilic catalysis, such as COB (I) alamin, and tetrahydrofolate (Kumano et al., 2016). Methylation modification can optimize the physiological activity of natural products, and demethylation can release polar groups for further binding and excretion from the body, and provide a carbon source for the growth of gut microbes (Ticak et al., 2014). Figure 3 shows the methylation and demethylation of natural products under the action of gut microbes.
Figure 3.
Methylation and demethylation of natural products under the action of gut microbes. (A) Methylation of rutin (Wu et al., 2017); (B–F) Demethylation of aconitine (Zhang et al., 2017), arctigenin (Jin et al., 2013), 5,7,4-trimethoxyflavone (Kim et al., 2014), 22β-acetoxyl glycyrrhizin (Wang Q. et al., 2015), isoxanthohumol (Paraiso et al., 2019).
Flavonoids
The methylation modification can be carried out at the C-2, C-3, C-4, C-5, C-6, C-7, and C-8 positions in the structure of flavonoids, and the bioavailability of methylated flavonoids is greatly improved (Wen and Walle, 2006). Bernini et al. found that O-methylated flavonoids have remarkable anti-cancer activity and resistance to hepatic metabolism (Bernini et al., 2011; Choi, 2019). After oral administration of rutin in rats, many methylated metabolites, such as methylrutin, methylisoquercetin, and methylquercetin sulfate, are detected in fecal samples (Yang et al., 2012; Wu et al., 2017; Riva et al., 2020). Methoxylated isoflavonoids formononetin and biochanin A undergo demethylation to produce daidzein and genistein under the action of E. limosum ATCC 8486 (Hur and Rafii, 2000). Isoxanthohumol yields demethylation products 8-prenylnaringenin by E. limosum (Paraiso et al., 2019). Hesperidin, hesperetin (Pereira-Caro et al., 2018; Jiao et al., 2020), 5,7-dimethoxyflavone, xanthohumol (Paraiso et al., 2019), and 5,7,4′-trimethoxyflavone (Kim et al., 2014) can also undergo demethylation reactions under the action of gut microbes (Table 2).
Table 2.
Methylation and demethylation reaction of gut microbes to natural products.
| Classification | Gut microbiota | Enzyme | Substrate | End-product | Changes | Ref. |
|---|---|---|---|---|---|---|
| Flavonoids | Rat gut microbes | Methyltransferase | Rutin | Methylrutin; Methyl-isoquercetin; methylquercetin sulfate |
Bioavailability↑ | Wu et al. (2017) |
| Mice gut microbes | NA | Myricetin | Mono- and di-methylated myricetin | Toxicity↓ | Zhang et al. (2020) | |
| Rat gut microbes | NA | Hesperidin; hesperetin |
Demethylated products | Bioavailability↑ | Pereira-Caro et al. (2018) and Jiao et al. (2020) | |
| E. limosum | O-demethylase | Formononetin; biochanin A |
Daidzein; genistein |
Estrogen effect↑ | Hur and Rafii (2000) | |
| Blautia sp. MRG-PMF1 | Methyltransferase | 5,7-dimethoxyflavone; 5,7,4′-trimethoxyflavone |
Chrysin; apigenin |
Anti-oxidant↑ anti-inflammatory↑ anti-cancer↑ |
Kim et al. (2014) | |
| Blautia sp. MRG-PMF1 | Methyltransferase | Icariin | Desmethylicaritin | Estrogenic effects↑ | Wu et al. (2016) | |
| E. limosum | O-demethylase | Isoxanthohumol | 8-prenylnaringenin | Anti-androgen↑ anti-osteoporosis↑ |
Paraiso et al. (2019) | |
| Alkaloids | Human gut microbes | Methyltransferase | Quassic ketone | Quassic alkali butyl | Chen F.Z. et al. (2021) | |
| Rat gut microbes | Methyltransferase | Palmatine | Columbamine; Jatrorrhizine; demethyleneberberine |
Bioavailability↑ | He et al. (2017) and Liao et al. (2021) | |
| Human gut microbes | O-demethylase | Aconitine | 16-O-demethylaconitine | Toxicity↓ | Zhao et al. (2008) and Zhang et al. (2017) | |
| Lignans |
Eubacterium sp. ARC-2; Blautia sp. AUH-JLD56 |
O-demethylase | Arctiin; arctigenin |
DHENL; 3′-DMAG |
Anti-oxidant↑ estrogen effect↑ |
Jin et al. (2007), Jin et al. (2013), Liu et al. (2013) and Seyed Hameed et al. (2020) |
|
Blautia producta DSM3507; Gordonibacter strains 3C and 28C; Lactonifactor longoviformis DSM17459T |
Guaiacol lignan methyltransferase; catechol lignan dehydroxylase; enterodiol lactonizing enzyme |
Secoisolariciresinol | Enterolactone; enterodiol |
Estrogen effect↑ | Bess et al. (2020) and Tse et al. (2022) | |
| Human gut microbes | SesA | Sesamin | Enterolactone; enterodiol |
Estrogen effect↑ | Peñalvo et al. (2005) | |
| Rat gut microbes | O-demethylase | Matairesinol | 2,3-bis(3,4-dihydroxybenzyl) butyrolactone; enterolactone |
Anti-inflammatory↑ estrogen effect↑ |
Clavel et al. (2006), Yamawaki et al. (2011) and Michalak et al. (2018) | |
| Human/rat gut microbes | O-demethylase | Phillygenin | Enterolactone | Anti-inflammatory↑ estrogen effect↑ |
Michalak et al. (2018) | |
|
E. limosum ZL-II; human gut microbes |
O-demethylase | Silybin A and B | Demethylsilybin A; demethylsilybin B |
Anti-Alzheimer’s disease↑ | Zhang et al. (2014) and Valentová et al. (2020) | |
| Diketones | Blautia sp. MRG-PMF1 | Co O-Methyltransferase | Curcumin | DMC; bDMC |
Anti-tumor↑ anti-inflammatory↑ |
Burapan et al. (2017a,b) |
| Phenols | Rat gut microbes | NA | Danshensu | 3-(3-O-methyl-4-hydroxyphenyl)-2-hydroxypropanoic acid | Bioavailability↑ | Gu et al. (2014) |
| Rat gut microbes | O-demethylase | Dihydro-isoferulic acid | Dihydrocaffeic acid | Anti-oxidant↑ anti-apoptosis↑ |
Kay et al. (2017) | |
| Terpenoids | Rat gut microbes | NA | 22β-acetoxyl glycyrrhizin | 22β-acetoxyl glycyrrhizin-6″-methyl ester | Bioavailability↑ | Wang Q. et al. (2015) |
| Eubacterium sp. A-44 | NA | Genipin | Geniposidic acid | Anti-oxidant↑ | Akao et al. (1994) | |
| Stilbenoids | Human gut microbes | O-demethylase | Thunalbene | Isoresveratrol | Anti-oxidant↑ | Jarosova et al. (2019) |
| Steroids | Human gut microbes | NA | Pulsatilla saponin B3 | Corresponding Deglycosylation products | Bioavailability↑ | Liu et al. (2015) |
Alkaloids
Alkaloids are nitrogen-containing compounds, which are biosynthesized by both marine and terrestrial organisms, and they have anti-cancer (Tse et al., 2022) and anti-viral activity (Abookleesh et al., 2022). Under the action of enzymes expressed by gut microbes, quassic ketone, the main alkaloid component in bitter wood, is methylated into quassic alkali butyl (Fan et al., 2013; Chen et al., 2021). Isoquinoline alkaloid palmatine yields demethylation products such as columbamine, jatrorrhizine, demethyleneberberine, and demethyleneberberine via in vitro anaerobic incubation (He et al., 2017; Liao et al., 2021). The demethylation of aconitine by gut microbes is demonstrated by ion trap electrospray ionization tandem mass spectrometry, and 16-O-demethylaconitine is produced (Zhao et al., 2008; Zhang et al., 2017).
Lignans
Dietary lignans are phytoestrogens that are mostly found in seeds, nuts, legumes, and vegetables. Arctiin can be demethylated to (−)-dihydroxyenterolactone (DHENL) and other products by Eubacterium sp. ARC-2 strain (Jin et al., 2007, 2013; Seyed Hameed et al., 2020). Liu et al. isolated a bacterium named Blautia sp. AUH-JLD56 from human fecal bacteria, and this species could efficiently transform arctiin or arctigenin into a demethylation metabolite 3′-desmethylarctigenin (3′-DMAG; Liu et al., 2013). Secoisolariciresinol, which is one of the most common lignans found in flaxseed, can be demethylated in the presence of Blautia producta, Gordonibacter and Lactonifactor longoviformis to form enterolactone and enterodiol (Bess et al., 2020; Tse et al., 2022). Sesamin is metabolized into mammalian lignan enterolactone and enterodiol through methylation, demethylation, and other reactions by gut microbes (Peñalvo et al., 2005). Matairesinol and phillygenin can also be demethylated to produce enterolactone (Clavel et al., 2006; Yamawaki et al., 2011; Michalak et al., 2018). Silybin A and B are demethylated into demethylsilybin A and demethylsilybin B by human fecal microbiota (Zhang et al., 2014; Valentová et al., 2020).
Other compounds
Polyphenol compound curcumin is demethylated by Blautia sp. MRG-PMF1 to produce metabolites demethoxycurcumin (DMC) and bis-demethoxycurcumin (bDMC; Burapan et al., 2017a,b). The demethylated products of dihydro-isoferulic acid, such as dihydrocaffeic acid, are also obtained in fecal metabolites (Kay et al., 2017). Wang et al. found that the methylation reaction occurs at the internal and external glucuronic acid residues of the licorice saponins 22β-acetoxyl glycyrrhizin sugar chain, yielding 22β-acetoxyl glycyrrhizin-6″-methyl ester (Wang et al., 2015). Compounds such as polyphenols danshensu (Gu et al., 2014), terpenoids genipin (Akao et al., 1994), stilbenoids thunalbene (Jarosova et al., 2019), and steroids pulsatilla saponin B3 (Liu et al., 2015) undergo methylation and demethylation under the action of gut microbes, as shown in Table 2. Methylation and demethylation reactions are important pathways of gut microbial metabolism, and have been confirmed in many studies. However, the genes/enzymes that mediate this reaction have not been fully characterized.
Redox reaction
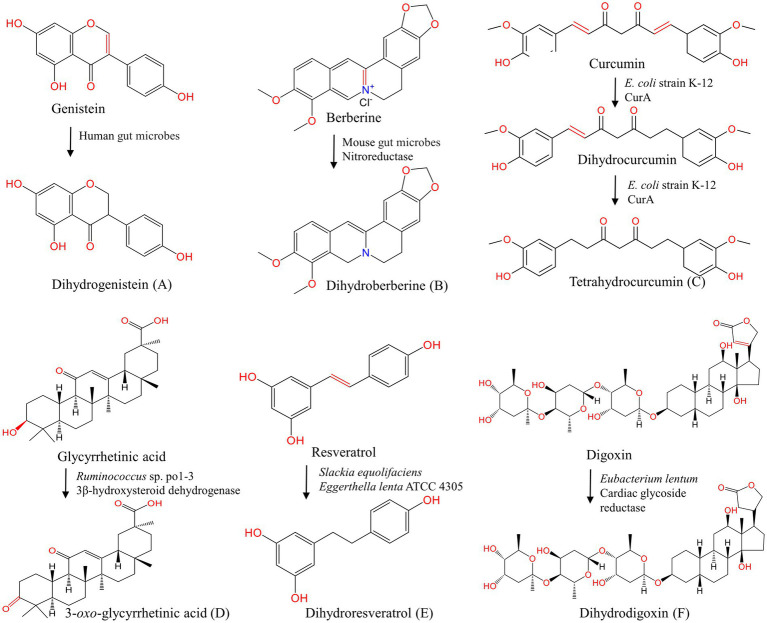
Gut microbes can express many oxidoreductases and transform natural compounds by adjusting various functional groups, such as olefins, carboxylic acid derivatives, nitro, N-oxides, and a, b-unsaturated carboxylic acid derivatives, which influence the activity of natural products in vivo (Lavrijsen et al., 1995; Haiser et al., 2013; Abookleesh et al., 2022). Various cofactors such as NADH, NADPH, flavin, Fe/S cluster, heme, and molybdenum cofactor are involved in the mediation of the transfer of electron or hydride equivalent (H+, 2e−) to the substrate (Vanoni, 2021; Lubner et al., 2022). Figure 4 shows the oxidation and reduction reactions of natural products under the action of gut microbes.
Figure 4.
Oxidation and reduction of natural products under the action of gut microbes. (A–C, E, F) Reduction of genistein (Mace et al., 2019), berberine (Feng et al., 2015), curcumin (Hassaninasab et al., 2011), resveratrol (Pallauf et al., 2019), digoxin (Kumar et al., 2018); (D) glycyrrhetinic acid.
Flavonoids
Daidzein is reduced to dihydrodaidzein and further tetrahydrodaidzein under the action of Clostridium sp. strain HGH6 and Lactobacillus. sp. Niu-O16 (Zhao et al., 2011; Heng et al., 2019). The reduced product dihydrogenistein is produced by genistein under the action of human fecal bacteria (Mace et al., 2019). By using UPLC-ESI-Q-TOF-MS/MS analysis, compounds such as the deoxidized metabolites kaempferol and the C2-C3 double bond hydrogenation reduction product taxifolin were identified in the culture solution of rat gut fluid by incubation with quercetin under anaerobic conditions (Qin et al., 2017). Yang et al. discovered a flavone reductase from Flavonifractor plautii ATCC 49531, and this enzyme specifically catalyzes the hydrogenation of the C2-C3 double bound of flavones/flavanols C-ring and acts during the initial step of the entire biodegradation pathway of flavonoid (Goris et al., 2021; Yang et al., 2021). O-desmethylxanthohumol, a chalcone compound, is reduced to O-desmethyl-α, β-dihydroxanthohumol by E. ramulus (Paraiso et al., 2019).
Alkaloids
Nitroreductase, which is produced by gut microbes, catalyzes ether and coordination bond reactions in alkaloids. Berberine (BBR), as the main component of Coptis Chinensis, can be reduced to dihydroberberine (dhBBR) by nitroreductase expressed by gut microbes, and this reduction product has high polarity. dhBBR could be absorbed in the intestine and then oxidized into the prototype BBR into the blood. The absorption rate of dhBBR in the intestine is five times that of BBR (Feng et al., 2015). Li et al. found that the gut microbes could transform BBR into oxyberberine via oxidation (Li et al., 2020). Oxyberberine, a novel metabolite of BBR, may be a promising bioactive agent worthy to be explored. Coptisine is a natural protoberberine alkaloid with the same maternal structure as BBR. After oral administration of coptisine, the C-O bond is opened and cracked, followed by a reduction reaction to produce hydrogenated BBR (Cui et al., 2018). Avenanthramide-C is reduced by mice and the human gut microbiota into dihydroavenanthramide-C (Wang et al., 2015).
Phenylpropanoids
Caffeic acid (CAA), as the main dietary polyphenol in food and beverage, can easily enter the colon and react with gut microbiota after esterification. CAA is transformed to 3-hydroxyphenylpropionic acid through C4 double bond reduction and dehydroxylation, and then rapidly converted to 3-phenyl propionic acid via the β-oxidation of gut microbes in vitro (Gonthier et al., 2006). CAA can also be dehydroxylated to m-coumaric acid or hydrogenated to dihydrocaffeic acid (García-Villalba et al., 2020). Danshensu, the major monomer phenolic acid of Salvia Miltiorrhiza, undergo dehydrogenation and deoxygenation by gut microbiota to produce 3-phenyl-2-hydroxy propionic acid, 3-(3,4-dihydroxy phenyl) 2-acrylic acid (caffeic acid), and 3-(3,4-dihydroxy phenyl)-propionate (Gu et al., 2014).
Other compounds
Glycyrrhetic acid generates 3-oxo-glycyrrhetinic acid by 3β-hydroxysteroid dehydrogenase of Ruminococcus sp. po1-3 in the cecum. Sennosides, a class of natural anthraquinone derivative and dimeric glycosides, are first hydrolyzed by β-glucosidase to produce sennoside-8-O-monoglycoside, and then reduced to rhubaranthrone with purgative effect by Streptococcus in vivo (Hattori et al., 1988). Stilbenoids resveratrol is reduced to dihydroresveratrol by Slackia equolifaciens and Eggerthella lenta ATCC 4305 (Bode et al., 2013; Pallauf et al., 2019) Moreover, diketones curcumin (Hassaninasab et al., 2011; Tan et al., 2014), steroid compounds digoxin (Kumar et al., 2018) and other compounds aristolochic acid (Feng et al., 2019) can also be reduced in the presence of gut microbes (Table 3). Gut microbial flavone reductase and nitroreductase have special catalytic selectivity, filling key gaps in gut microbial transformation pathways. However, the specific genes and enzymes that mediate gut microbial reduction have not been fully determined.
Table 3.
Reduction and oxidation reaction of gut microbes to natural products.
| Classification | Gut microbiota | Enterobacterial metabolic enzyme | Substrate | End-product | Changes | Ref. |
|---|---|---|---|---|---|---|
| Flavonoids |
Clostridium sp. strain HGH6; Lactobacillus sp. Niu-O16 |
Dihydrodaidzein reductase; tetrahydrodaidzein reductase |
Daidzein | Dihydrodaidzein; tetrahydrodaidzein |
Anti-osteoporosis↑ | Wang et al. (2007) and Heng et al. (2019) |
| Aeroto Niu-O16 | NA | Genistein | Dihydrogenistein | Bioavailability↑ | Mace et al. (2019) | |
| Rat gut microbes E. ramulus |
Flavone reductase | Quercetin | Kaempferol; taxifolin |
Bioavailability↑ | Qin et al. (2017) | |
| E. ramulus | Flavanone-/flavanonol-cleaving reductase | Xanthohumol; O-desmethylxanthohumol |
α, β-dihydroxanthohumol; O-desmethyl-α, β-dihydroxanthohumol |
Anti-bacterial↑ | Paraiso et al. (2019) | |
| Alkaloids | Mouse gut microbes | Nitroreductase | BBR; coptisine |
dhBBR; hydrogenated berberine |
Bioavailability↑ anti-inflammatory↑ |
Feng et al. (2015) and Cui et al. (2018) |
| Mouse gut microbes | NA | BBR | Oxyberberine | Anti-fungal↑ | Li C. et al. (2020) | |
| Mouse/human gut microbes | NA | Avenanthramide-C | Dihydroavenanthramide-C | Anti-inflammation↑ anti-atherogenesis↑ |
Wang P. et al. (2015) | |
| Phenolic acids | Human gut microbes | NA | CAA | Dihydrocaffeic acid | Bioavailability↑ | Gonthier et al. (2006) and García-Villalba et al. (2020) |
| Rat gut microbes | NA | Isoferulic acid | Dihydrocaffeic acid | Anti-oxidant↑ anti-apoptosis↑ |
Kay et al. (2017) | |
| Rat gut microbes | NA | Dansensu | 3-phenyl-2-hydroxy propionic acid; 3-(3,4-dihydroxy phenyl) 2-acrylic acid; 3-(3,4-dihydroxy phenyl)-propionate |
Bioavailability↑ | Gu et al. (2014) | |
| Gordonibacter urolithinfaciens | Catechol-dehydroxylase | Chlorogenic acid; rosmarinic acid |
Dihydro-chlorogenic acid; dihydro-rosmarinic acid |
Bioavailability↑ | García-Villalba et al. (2020) | |
| Terpenoids | Ruminococcus sp. po1-3 | 3β-hydroxysteroid dehydrogenase | Glycyrrhetinic acid | 3-oxo-glycyrrhetinic acid | Anti-inflammatory↑ | |
| Anthraquinone | Human gut microbes; Streptococcus spp. |
NA | Sennoside-8-O-monoglycoside | Rhubaranthrone | Purgation↑ | Hattori et al. (1988) and Matsumoto et al. (2012) |
| Stilbenes | Slackia equolifaciens; Eggerthella lenta ATCC 4305 | NA | Resveratrol | Dihydroresveratrol | Anti-oxidant↑ | Jung et al. (2009), Bode et al. (2013) and Pallauf et al. (2019) |
| Diketones |
E. coli strain K-12; E. fergusonii ATCC 35469; E. coli strains ATCC 8739 and DH10B |
CurA | Curcumin | DHC; THC |
Anti-oxidant↑ lipid-lowering↑ |
Hassaninasab et al. (2011) and Tan et al. (2014) |
| Steroids | Eubacterium lenta | Cardiac glycoside reductase | Digoxin | Dihydrodigoxin | Bioavailability↓ | Kumar et al. (2018) |
| Other classes | Human gut microbes | NA | Aristolochic acid | Aristololactams | Anti-cancer↑ | Feng et al. (2019) |
Other reactions
As shown in Table 4, natural products are also transformed by gut microbes through ring fission, sulfuration, aromatization, and other reactions. Gentiopicroside, a natural iridoid glycoside, can be hydrolyzed to gentianaldehyde by gut microbial β-glucosidase, and then to nitrogen-containing compounds via N-heterocyclic reaction (el-Sedawy et al., 1989). The partial ring-opening of genipin acetone alcohol results in the formation of dialdehyde by gut microbes (Kang et al., 2012). Quinic acid can be aromatized to hippuric acid in the presence of gut microbes (Pero and Lund, 2011). Maren et al. incubated kaempferol-O-glycosides and apigenin-C-glycosides with human fecal samples to generate 3-(4-hydroxyphenyl) propionic acid, 3-phenyl propionic acid, and phenylacetic acid through deglycosylation, ring fission and other reactions (Vollmer et al., 2018). The main hydrolytic and ring-cleaved metabolites, namely, benzoic acid, 2-(3,4-dihydroxy phenyl) acetic acid, and 5-(3,4-dihydroxy phenyl)-γ-valerolactone were obtained via in vitro fermentation of flavan-3-ols procyanidin B2 and A2 with human gut microbes (Stoupi et al., 2010; Ou et al., 2014; Le Bourvellec et al., 2019). Sulfated and hydrogen-reduced metabolites have been detected in the fecal samples of rats after oral administration of luteolin (Li et al., 2017; Káňová et al., 2020). The conversion of daidzein to equol, which is facilitated by gut microbes is another interesting example (Li et al., 2000; Hur et al., 2002; Mayo et al., 2019). Eggerthella lenta and Flavonifractor plautii reductively cleaved the heterocyclic C-ring of both (−)-epicatechin and (+)-catechin giving rise to 1-(3,4-dihydroxyphenyl)-3-(2,4,6-trihydroxyphenyl) propan-2-ol, δ-(31,41-dihydroxyphenyl)-γ-valerolactone, and δ-(31,41-dihydroxyphenyl)-γ-valeric acid (Ozdal et al., 2016). Tea polyphenols are metabolized by gut microbiota (Liu et al., 2018). Tea polyphenols first undergo structural modifications such as methylation and sulfation in the small intestine and then enter the colon to be cleaved into small phenolic acids, which is conducive to absorption (Cheng et al., 2018). SesA, a sesamin-metabolizing enzyme from Sinomonas sp. no. 22, catalyzes the methylene group transfer from sesamin or sesamin monocatechol to tetrahydrofolate with ring cleavage, yielding sesamin mono- or di-catechol and 5,10-methylenetetrahydrofolate (Kumano et al., 2016). The terpenoids astragaloside A (He et al., 2019), flavonoids quercitrin (Jiang et al., 2014) and myricetin (Zhang et al., 2019), the phenol anthocyanidin (Aura et al., 2005), the alkaloid strychnine N-oxide (el-Mekkawy et al., 1993) and the aliphatic myristic acid (Du et al., 2014) can all undergo biotransformation reactions to generate active metabolites under the action of gut microbes. These studies demonstrate the enormous metabolic potential of various gut microbiomes. The gut microbial metabolism of natural products and their role in host health should be the focus of future research.
Table 4.
Other reactions of gut microbes to natural products.
| Classification | Gut microbiota | Biotransformation | Enterobacterial metabolic enzyme | Substrate | End-product | Changes | Ref. | |
|---|---|---|---|---|---|---|---|---|
| Terpenoids | Human gut microbes | Cyclization | β-glucosidase | Gentiopicroside | Gentisaldehyde; nitrogen-containing compounds |
Anti-inflammatory↑ | el-Sedawy et al. (1989) | |
| Human gut microbes | Cyclization | NA | Geniposide | Nitrogen-containing compounds | Bioavailability↑ | Kawata et al. (1991) | ||
| Human gut microbes | Deglycosylation; deacetylation; dehydrogenation |
NA | Astragaloside A | Cycloastragenol | Bioavailability↑ | He et al. (2019) | ||
| Phenolic acids | Rat gut microbes | Aromatization | NA | Quinic acid | Hippuric acid | Anti-cancer↑ anti-bacterial↑ anti-viral↑ |
Pero and Lund (2011) | |
| Egerthella lenta | Dehydroxylation | Catechol dehydroxylases | Dihydrocaffeic acid | 3-(3-hydroxyphenyl) propionic acid | Bioavailability↑ | Maini Rekdal et al. (2020) | ||
| Human gut microbes | Ring cleavage; sulfation methylation |
NA | Tea polyphenols |
Phenolic acids | Bioavailability↑ | Cheng et al. (2018) | ||
| L. plantarum WCFS1 | Ring fission; hydrolysis |
Tannase; gallate decarboxylase |
Gallotannins | Gallic acid; pyrogallol |
Anti-oxidant↑ anti-inflammatory↑ |
Reverón et al. (2015) and Fang et al. (2019) | ||
| SGG | Ring fission; hydrolysis |
Tannase; gallate decarboxylase |
Gallotannins | Gallic acid; pyrogallol |
Anti-cancer↓ | Oehmcke-Hecht et al. (2020) | ||
|
Gordonibacter urolithinfaciens; Goronibacter pamelaeae; Ellagibacter isourolithinifacens |
Decarboxylation; lactone-ring cleavage; dehydroxylation |
NA | Ellagic acid | Urolithins | Anti-cancer↑ anti-oxidant↑ anti-inflammatory↑ |
Beltrán et al. (2018), García-Villalba et al. (2020) and Tang et al. (2021) | ||
| Flavonoids | Rat gut microbes | Sulfation | Aryl sulfotransferase | Luteolin | Luteolin-3′-O-sulfate; luteolin-4′-O-sulfate |
Li et al. (2017) and Káňová et al. (2020) | ||
| Clostridium sp. strain HGH136 | C-ring fission | 2-dehydro-O-demethylangolensin | Daidzein | O-desmethylangolensin | Anti-cancer↑ | Hur et al. (2002) | ||
|
Eggerthella sp. strain YY7918; B. breve ATCC 15700 T; B. longum BB536; L. paracasei CS2 |
Ring-fission | Dihydrodaidzein racemase | Dihydrodaidzein | S-equol | Estroge effect↑ | Yokoyama and Suzuki (2008) and Mayo et al. (2019) | ||
| E. ramulus | Ring-fission; reduction |
Chalcone isomerase; flavanone-/flavanonol-cleaving reductase |
Naringenin; eriodictyol |
Naringenin chalcone; phloretin; 3-hydroxyphloretin |
Bioavailability↑ anti-inflammatory↑ |
Gall et al. (2014) and Braune et al. (2019) | ||
| E. ramulus strain wK1 | Ring-fission | Phloretin hydrolase | Phloretin | 3-(4-hydroxyphenyl)-propionic acid; phloroglucinol |
Bioavailability↑ | Schoefer et al. (2004) and Braune et al. (2019) | ||
|
Bacteroides sp. 45; B. fragilis; E. ramulus |
Ring-fission | Chalcone isomerase; phloretin hydrolase | Quercetin; luteolin |
4-hydroxybenzoic acid 3,4-dihydroxyphenylacetic acid; 3,4-dihydroxybenzoic; 3-(3-hydroxyphenyl) propionic acid |
Anti-platelet aggregation↑anti-tumor↑ | Jiang et al. (2014) and Braune et al. (2001) | ||
| Rat gut microbes | Ring-fission sulfation |
Chalcone isomerase; phloretin hydrolase | Myricetin | 3,4,5-trihydroxyphenylacetic acid; myricetin-3′-O-sulfate |
Anti-inflammatory↑ | Zhang S. et al. (2019) and Káňová et al. (2020) | ||
| B. longum R0175 | Ring-cleavage; demethylation |
Phloretin hydrolase; demethylase |
Hesperidin | 3-(3′-hydroxyphenyl) propionic acid; 3-(phenyl) propionic acid |
Bioavailability↑ | Pereira-Caro et al. (2018) | ||
|
Eggerthella lenta;
Flavonifractor plautii |
C-ring cleavage | NA | (−)-epicatechin; (+)-catechin |
1-(3,4-dihydroxyphenyl)-3-(2,4,6-trihydroxyphenyl) propan-2-ol; δ-(31,41-dihydroxyphenyl)-γ-valerolactone; δ-(31,41-dihydroxyphenyl)-γ-valeric acid |
Bioavailability↑ | Ozdal et al. (2016) | ||
| Human gut microbes | C-ring cleavage A-ring fission dehydroxylation |
NA | Anthocyanidin | Protocatechuic acid; syringic acid; vanillic acid; phloroglucinol aldehyde |
Bioavailability↑ | Aura et al. (2005) | ||
| Human gut microbes | C-ring cleavage A-ring fission dehydroxylation, etc |
Tannase | Procyanidin B2 and A2 | 2-(3,4-dihydroxyphenyl) acetic acid; 5-(3,4-dihydroxyphenyl)-γ-valerolactone; benzoic acid |
Anti-oxidant↑ | Stoupi et al. (2010), Ou et al. (2014) and Le Bourvellec et al. (2019) | ||
| Alkaloids | Human gut microbes | Ring fission | NA | Strychnine N-oxide | Strychnine; 16-hydroxystrychnine |
Toxicity↓ | el-Mekkawy et al. (1993) | |
| Lignins | Eggerthella lenta | Ring cleavage | Benzyl ether reductase | Pinoresinol | Lariciresinol; secoisolariciresinol |
Anti-apoptosis↑ | Bess et al. (2020)s and Xiao et al. (2021) | |
| Aliphatics | Human gut microbes | Dehydroxylation | NA | Myristic acid | Quercetin-3-O-rhamnoside; quercetin; aglycone myricetin |
Anti-oxidant↑ | Du et al. (2014) |
This section summarizes the biotransformation of gut microbiota-mediated natural products from a single reaction. However, some limitations are observed. Firstly, considering the complexity of gut microbes and the diversity of gut microbial enzymes, natural products undergo complex transformations in the intestinal tract. A single reaction can only describe a certain process of metabolism. Therapy can be optimized by activating/inhibiting this process. In addition, considering that gut microbes contain various potentially multifunctional enzymes, more biotransformation reactions underplayed by natural products can be expected from gut microbes. To elucidate how gut microbial metabolism affects human health, researchers should link the functions of interest to genes and enzymes. A deep understanding of the gene sequences of functional enzymes allows organisms with similar sequences to be assigned the same biological activity. Moreover, in addition to the regulation of gut microbes on the disposal of natural products, the regulation of natural products on gut microecology is important as a potential mechanism of efficacy.
Biotransformation contributions to mining the active substance and mechanism
The increasing research about gut microbiota gradually reveals the relationship between high pharmacological action and low oral availability of most natural products. Most glycosides have complex parent structures and are difficult to be absorbed by the intestine cells, thus limiting their tissue-specific bio-accessibility. These compounds are transformed into small molecule metabolites/unique metabolites through degradation reactions that are dependent on microbial/gut microbial enzymes and thus have a wide range of effects on the host (Wardman et al., 2022). Gut microbes also act on dietary phenolics to produce functional metabolites that contribute to host health (Loo et al., 2020).
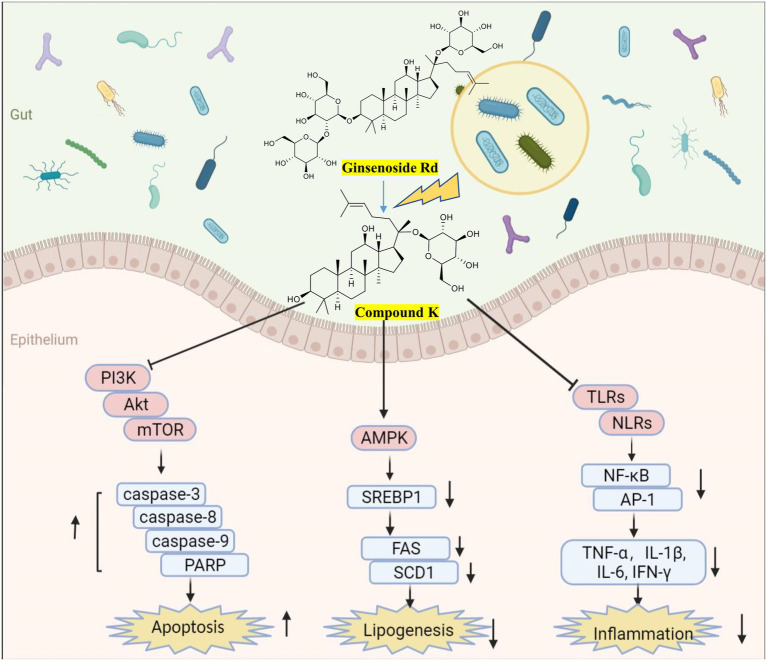
Importantly, the biotransformation by gut microbes facilitates the therapeutic effects of natural products. The typical metabolization model of ginsenosides to compound K (CK) has been widely reported (Figure 5), with enhanced anti-tumor, anti-inflammatory, and lipid-lowering effects (Kim et al., 2013; Kim, 2018). At 50 μM, CK inhibits the growth of glioblastoma cells by upregulating caspase-3-, caspase-8-, caspase-9- and cAMP-dependent protein kinases (Lee et al., 2017); At 20 μM, CK reduces hepatic lipid accumulation in human hepatocellular carcinoma cells by activating AMPK (Zhang et al., 2022); CK attenuates macrophage inflammation and foam cell formation via autophagy induction and by modulating NF-κB, p38 and JNK/MAPK signaling (Lu et al., 2020). The bioavailability of curcumin metabolites is dependent on the microbiota dependent (Hassaninasab et al., 2011). For instance, DMC increases PPARγ expression, resulting in autophagy and NF-κB inhibition and subsequently inhibiting LPS-induced inflammation (Tang et al., 2021). DMC mitigates inflammatory responses in vivo and in vitro by inhibiting the secretion of inflammatory factors and activation of MAPK and NF-κB pathways (Lu et al., 2022). The chemical stability of DMC increases because of the absence of the methoxyl group in their prototype benzene ring structure, thus explaining the strong beneficial effects of curcumin (Burapan et al., 2017a). Notably, urolithin A (UA), a natural compound that is produced by gut microbes from ingested ellagitannins and ellagic acid, has significant anti-inflammatory and neuroprotective effects. At 1 μM, UA is sufficient for the decreased production of TNF-α and MCP-1 and the inactivation of TLR3/TRIF signaling in poly (I:C)-induced RAW264.7 cells (Huang et al., 2022). UA improves systemic insulin sensitivity and reduces liver IL-1β levels in high-fat diet mice (Toney et al., 2019). UA ameliorates cognitive impairment in APP/PS1 mice and inhibits neuroinflammation by decreasing the levels of IL-6, IL-1β, and TNF-α in the cortex and hippocampus (Gong et al., 2019). These studies highlight the importance of identifying natural products-microbial metabolism. Moreover, many in vitro pharmacological activity measurements should be performed in conjunction with microbial metabolites, which actually interact with biochemical receptors in vivo.
Figure 5.
Biotransformation of ginsenosides (Kim et al., 2008) and efficacy of metabolite compound K. Created with BioRender.com.
The composition, structure, function, and metabolites of gut microbes have become potential targets for natural products to exert beneficial effects and reduce toxicity as well. For instance, gut microbes can catalyze the ester bond hydrolysis of C-8 and C-14 of DDAs through CEs or catalyze the ester exchange of C-8 to produce less toxic MDAs (Zhang et al., 2015; Ding et al., 2019). The digoxin-reducing type strains of E. lenta contain cardiac glycoside reductase that can reduce the α and β-unsaturated lactone on the digoxin ring and metabolize it into dihydrodigoxin with less activity, thereby inhibiting its possible cardiotoxicity (Kumar et al., 2018). However, this ability is limited, and 50% of digoxin can be inactivated by gut microbial transformation (Lu et al., 2014). Cardiac glycoside reductase may be an effective biomarker for digoxin inactivation, and its expression can be inhibited by arginine (Haiser et al., 2013). Therefore, diet could explain the inter-individual variations in digoxin reduction and may modulate microbial metabolic activity in vivo. By contrast, toxic compounds can be produced by gut microbes. Cycasin is hydrolyzed into carcinogenicity diazomethane under the action of β-glucosidase from gut microbes (Goldin, 1990). Therefore, small molecule inhibitors of microbial gut enzymes should be developed to play a regulatory role in specific transformation in this complex habitat. The toxicity difference between metabolites transformed by gut microbiota and precursor substances is worthy of further study. Moreover, excessive drugs may cause imbalance and adverse reactions in gut microbes (Lindell et al., 2022), and the effects of different doses of natural products on gut microbes and metabolism need further investigation.
Multivariate technologies for studying biotransformation
Considering that gut microbes can increase the host’s complex and variable response to drugs/natural products, this process is of great interest to researchers. Research on biotransformation is mainly conducted via in vitro approach (Sousa et al., 2008) as follows: (1) Intestinal fluid transformation. The large-scale preparation of transformed products can be realized by intestinal fluid biotransformation; (2) Incubation with a sample of the host microbiota. The type and quantity of prototype drugs and metabolites can be detected using the method. It has the advantage of accurate representation of the entire gut microbiome of the individual; (3) Incubation of representative strains. This method affords high-throughput potential, which is valuable for large-scale drug studies and contributes to the industrial production of beneficial metabolites. In addition, organ-on-a-chip microphysiological systems (Ashammakhi et al., 2020), gastrointestinal organoids (Singh et al., 2020), and various predictive/computational tools (Machado et al., 2018; Chowdhury and Fong, 2020) may help improve our understanding of microbial metabolism in the future.
In addition, the relationships between natural product metabolism and gut microbes have been studied in animal models, and the results can be used to investigate the distribution and form of metabolites (Yoshisue et al., 2000). Germ-free/antibiotic-treated animals with conventional animals have been compared to prove the key roles of gut microbes on natural product metabolism. The limitation of this method is that inherent gastrointestinal and microbiological differences exist between humans and rodents (Nguyen et al., 2015). Detailed microbiota and metabolite analysis of feces collected from subjects in clinical trials can comprehensively reflect the metabolic process of natural products in vivo and be used to explain individual differences. In addition, the application of sequencing technology needs to be increased to study the microbial transcriptional activity and metabolic profile. By using the single-cell method, the physiological structure of gut microbes can be characterized to determine their metabolic activity (Zheng et al., 2022). Metatranscriptomics (RNA-Seq) allows the direct analysis of gene expression profiles of microorganisms with strong metabolic activity in the human gut (Berlinberg et al., 2022). The combination of single-cell methods, metatranscriptomics, and metagenomics has been used to identify and characterize the active subsets of gut microbiota and determine their metabolic responses to natural products.
Conclusions and future remarks
The gut microbiota is a reservoir of genes that encode various metabolic enzymes (Flint et al., 2012). The activation of biological activities and potential health benefits of most natural products (e.g., flavonoids, alkaloids, and lignin) are extremely dependent on gut microbes as a substrate-machining factory (Braune and Blaut, 2016; Seyed Hameed et al., 2020; Plamada and Vodnar, 2021). Much research effort has been devoted to understanding how microbes uniquely modify natural products and the effects of these metabolites on host health (Luca et al., 2020; Shabbir et al., 2021). The following conclusions have been made: (1) gut microbes can transform natural products (Xie et al., 2020); (2) natural products can regulate the composition and abundance of gut microbes (Saccon et al., 2021); and (3) gut microbes can mediate the multi-component synergy of natural products (Feng et al., 2019). Although high-throughput methods are being developed to help people understand the importance of the gut microbiome in the metabolism of natural products, microbial metabolism-based screening has not been adopted as part of the drug development process, because its mechanism remains unclear (Zimmermann et al., 2019). Moreover, the great plasticity and interindividual differences of gut microbes are notable (Vujkovic-Cvijin et al., 2020). Therefore, researchers need to improve the understanding of the physiological, chemical, and microbial contributions of gut microbes to the metabolism of natural products to help in explaining the individual differences in natural product responses and provide support for personalized treatment (Kolodziejczyk et al., 2019; Javdan et al., 2020). Most of the data in the present study were obtained independently of the clinic, but clinical trials are already underway, and the results will influence clinical practice in the foreseeable future.
Increasing studies on the mechanism of how to exert the curative effect, the application of fecal transplantation, specific bacterial transplantation, and animal models will help in clarifying the role of gut microbes. Nevertheless, standardization of operation, reproducibility of experimental results, and variation between species and individuals greatly reduce the authenticity and stability of the research, and a standard and scientific operating procedure remain to be put forward. Thus, confirming the symbolic functional extremely involved in biotransformation and its material basis will help in exploring the mechanism of natural products in the treatment of diseases and explaining the treatment mode of indirect interaction between natural products with low bioavailability and gut microbiota.
Author contributions
YZ contributed to the data collection and preparation of the original draft. XZ, JY, and CS provided brief article ideas and language modifications. XZ and XW supervised and revised the manuscripts. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China under grant no. 81873104, 81830112, and 82192914.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
Abbreviations
- DHC
dihydrocurcumin
- THC
tetrahydrocurcumin
- CurA
NADPH-dependent curcumin/dihydrocurcumin reductase
- CHA
chlorogenic acid
- CA
caffeic acid
- GL
glycyrrhizin
- 18β-GA
18β-glycyrrhetinic acid
- PM-I
paeonimetabolin-I
- GAMG
18β-glycyrrhetinic acid-3-O-β-D-glucuronic acid
- SGG
Streptococcus gallolyticus subsp. Gallolyticus
- DDAs
diester diterpenoid alkaloids
- MDAs
monoester diterpene alkaloids
- DHENL
(−)-dihydroxyenterolactone
- 3′-DMAG
3′-desmethylarctigenin
- BBR
Berberine
- dhBBR
dihydroberberine
- DMC
demethoxycurcumin
- bDMC
bis-demethoxycurcumin
- CK
compound K
- UA
Urolitin A
Footnotes
References
- Abdel-Hafez A. A., Meselhy M. R., Nakamura N., Hattori M., Watanabe H., Murakami Y., et al. (1999). Anticonvulsant activity of paeonimetabolin-I adducts obtained by incubation of paeoniflorin and thiol compounds with lactobacillus brevis. Biol. Pharm. Bull. 22, 491–497. doi: 10.1248/bpb.22.491, PMID: [DOI] [PubMed] [Google Scholar]
- Abookleesh F. L., Al-Anzi B. S., Ullah A. (2022). Potential antiviral action of alkaloids. Molecules 27:903. doi: 10.3390/molecules27030903, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agatonovic-Kustrin S., Kustrin E., Gegechkori V., Morton D. W. (2020). Anxiolytic terpenoids and aromatherapy for anxiety and depression. Adv. Exp. Med. Biol. 1260, 283–296. doi: 10.1007/978-3-030-42667-5_11, PMID: [DOI] [PubMed] [Google Scholar]
- Aguirre Santos E. A., Schieber A., Weber F. (2018). Site-specific hydrolysis of chlorogenic acids by selected lactobacillus species. Food Res. Int. 109, 426–432. doi: 10.1016/j.foodres.2018.04.052, PMID: [DOI] [PubMed] [Google Scholar]
- Agunloye O. M., Oboh G., Ademiluyi A. O., Ademosun A. O., Akindahunsi A. A., Oyagbemi A. A., et al. (2019). Cardio-protective and antioxidant properties of caffeic acid and chlorogenic acid: mechanistic role of angiotensin converting enzyme, cholinesterase and arginase activities in cyclosporine induced hypertensive rats. Biomed. Pharmacother. 109, 450–458. doi: 10.1016/j.biopha.2018.10.044, PMID: [DOI] [PubMed] [Google Scholar]
- Akao T., Kida H., Kanaoka M., Hattori M., Kobashi K. (1998). Intestinal bacterial hydrolysis is required for the appearance of compound K in rat plasma after oral administration of ginsenoside Rb1 from Panax ginseng. J. Pharm. Pharmacol. 50, 1155–1160. doi: 10.1111/j.2042-7158.1998.tb03327.x, PMID: [DOI] [PubMed] [Google Scholar]
- Akao T., Kobashi K., Aburada M. (1994). Enzymic studies on the animal and intestinal bacterial metabolism of geniposide. Biol. Pharm. Bull. 17, 1573–1576. doi: 10.1248/bpb.17.1573, PMID: [DOI] [PubMed] [Google Scholar]
- Al-Ishaq R. K., Liskova A., Kubatka P., Büsselberg D. (2021). Enzymatic metabolism of flavonoids by gut microbiota and its impact on gastrointestinal cancer. Cancers 13:3934. doi: 10.3390/cancers13163934, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashammakhi N., Nasiri R., Barros N. R., Tebon P., Thakor J., Goudie M., et al. (2020). Gut-on-a-chip: current progress and future opportunities. Biomaterials 255:120196. doi: 10.1016/j.biomaterials.2020.120196, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augusti P. R., Conterato G. M. M., Denardin C. C., Prazeres I. D., Serra A. T., Bronze M. R., et al. (2021). Bioactivity, bioavailability, and gut microbiota transformations of dietary phenolic compounds: implications for COVID-19. J. Nutr. Biochem. 97:108787. doi: 10.1016/j.jnutbio.2021.108787, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aura A. M., Martin-Lopez P., O'Leary K. A., Williamson G., Oksman-Caldentey K. M., Poutanen K., et al. (2005). In vitro metabolism of anthocyanins by human gut microflora. Eur. J. Nutr. 44, 133–142. doi: 10.1007/s00394-004-0502-2, PMID: [DOI] [PubMed] [Google Scholar]
- Basholli-Salihu M., Schuster R., Mulla D., Praznik W., Viernstein H., Mueller M. (2016). Bioconversion of piceid to resveratrol by selected probiotic cell extracts. Bioprocess Biosyst. Eng. 39, 1879–1885. doi: 10.1007/s00449-016-1662-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrán D., Romo-Vaquero M., Espín J. C., Tomás-Barberán F. A., Selma M. V. (2018). Ellagibacter isourolithinifaciens gen. Nov., sp. nov., a new member of the family Eggerthellaceae, isolated from human gut. Int. J. Syst. Evol. Microbiol. 68, 1707–1712. doi: 10.1099/ijsem.0.002735, PMID: [DOI] [PubMed] [Google Scholar]
- Berlinberg A. J., Brar A., Stahly A., Gerich M. E., Fennimore B. P., Scott F. I., et al. (2022). A novel approach toward less invasive multiomics gut analyses: a pilot study. Microbiol. Spectr. 10:e0244621. doi: 10.1128/spectrum.02446-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernini R., Crisante F., Ginnasi M. C. (2011). A convenient and safe O-methylation of flavonoids with dimethyl carbonate (DMC). Molecules 16, 1418–1425. doi: 10.3390/molecules16021418, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bess E. N., Bisanz J. E., Yarza F., Bustion A., Rich B. E., Li X., et al. (2020). Genetic basis for the cooperative bioactivation of plant lignans by Eggerthella lenta and other human gut bacteria. Nat. Microbiol. 5, 56–66. doi: 10.1038/s41564-019-0596-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode L. M., Bunzel D., Huch M., Cho G. S., Ruhland D., Bunzel M., et al. (2013). In vivo and in vitro metabolism of trans-resveratrol by human gut microbiota. Am. J. Clin. Nutr. 97, 295–309. doi: 10.3945/ajcn.112.049379, PMID: [DOI] [PubMed] [Google Scholar]
- Braune A., Blaut M. (2011). Deglycosylation of puerarin and other aromatic C-glucosides by a newly isolated human intestinal bacterium. Environ. Microbiol. 13, 482–494. doi: 10.1111/j.1462-2920.2010.02352.x, PMID: [DOI] [PubMed] [Google Scholar]
- Braune A., Blaut M. (2012). Intestinal bacterium Eubacterium cellulosolvens deglycosylates flavonoid C- and O-glucosides. Appl. Environ. Microbiol. 78, 8151–8153. doi: 10.1128/aem.02115-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braune A., Blaut M. (2016). Bacterial species involved in the conversion of dietary flavonoids in the human gut. Gut Microbes 7, 216–234. doi: 10.1080/19490976.2016.1158395, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braune A., Engst W., Blaut M. (2016). Identification and functional expression of genes encoding flavonoid O- and C-glycosidases in intestinal bacteria. Environ. Microbiol. 18, 2117–2129. doi: 10.1111/1462-2920.12864, PMID: [DOI] [PubMed] [Google Scholar]
- Braune A., Gütschow M., Blaut M. (2019). An NADH-dependent reductase from Eubacterium ramulus catalyzes the stereospecific heteroring cleavage of flavanones and flavanonols. Appl. Environ. Microbiol. 85:e01233-19. doi: 10.1128/aem.01233-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braune A., Gütschow M., Engst W., Blaut M. (2001). Degradation of quercetin and luteolin by Eubacterium ramulus. Appl. Environ. Microbiol. 67, 5558–5567. doi: 10.1128/aem.67.12.5558-5567.2001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burapan S., Kim M., Han J. (2017a). Curcuminoid demethylation as an alternative metabolism by human intestinal microbiota. J. Agric. Food Chem. 65, 3305–3310. doi: 10.1021/acs.jafc.7b00943, PMID: [DOI] [PubMed] [Google Scholar]
- Burapan S., Kim M., Han J. (2017b). Demethylation of polymethoxyflavones by human gut bacterium, Blautia sp. MRG-PMF1. J. Agric. Food Chem. 65, 1620–1629. doi: 10.1021/acs.jafc.7b00408, PMID: [DOI] [PubMed] [Google Scholar]
- Candeliere F., Raimondi S., Ranieri R., Musmeci E., Zambon A., Amaretti A., et al. (2022). β-Glucuronidase pattern predicted from gut metagenomes indicates potentially diversified pharmacomicrobiomics. Front. Microbiol. 13:826994. doi: 10.3389/fmicb.2022.826994, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W. Y., Wang Y. N., Wang P. Y., Lei W., Feng B., Wang X. J. (2015). Ardipusilloside-I metabolites from human intestinal bacteria and their antitumor activity. Molecules 20, 20569–20581. doi: 10.3390/molecules201119719, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. W., Kong Z. L., Tsai M. L., Lo C. Y., Ho C. T., Lai C. S. (2018). Tetrahydrocurcumin ameliorates free fatty acid-induced hepatic steatosis and improves insulin resistance in HepG2 cells. J. Food Drug Anal. 26, 1075–1085. doi: 10.1016/j.jfda.2018.01.005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. Z., Long J., Sun G. F., Tian C., Li S. H. (2021). Isolation of main alkaloids and transformation of intestinal microorganisms from Sophora alba (in Chinese). J. Leshan Norm. Univ. 36, 14–19. doi: 10.16069/j.cnki.51-1610/g4.2021.08.003 [DOI] [Google Scholar]
- Chen Y., Yang L., Lee T. J. (2000). Oroxylin A inhibition of lipopolysaccharide-induced iNOS and COX-2 gene expression via suppression of nuclear factor-kappaB activation. Biochem. Pharmacol. 59, 1445–1457. doi: 10.1016/s0006-2952(00)00255-0, PMID: [DOI] [PubMed] [Google Scholar]
- Chen B., Zhu D., Xie C., Shi Y., Ni L., Zhang H., et al. (2021). 18β-Glycyrrhetinic acid inhibits IL-1β-induced inflammatory response in mouse chondrocytes and prevents osteoarthritic progression by activating Nrf2. Food Funct. 12, 8399–8410. doi: 10.1039/d1fo01379c, PMID: [DOI] [PubMed] [Google Scholar]
- Cheng Z., Huang M., Chen G., Yang G., Zhou X., Chen C., et al. (2016). Cell-based assays in combination with ultra-high performance liquid chromatography-quadrupole time of flight tandem mass spectrometry for screening bioactive capilliposide C metabolites generated by rat intestinal microflora. J. Pharm. Biomed. Anal. 119, 130–138. doi: 10.1016/j.jpba.2015.11.029, PMID: [DOI] [PubMed] [Google Scholar]
- Cheng J. R., Liu X. M., Chen Z. Y., Zhang Y. S., Zhang Y. H. (2016). Mulberry anthocyanin biotransformation by intestinal probiotics. Food Chem. 213, 721–727. doi: 10.1016/j.foodchem.2016.07.032, PMID: [DOI] [PubMed] [Google Scholar]
- Cheng M., Zhang X., Zhu J., Cheng L., Cao J., Wu Z., et al. (2018). A metagenomics approach to the intestinal microbiome structure and function in high fat diet-induced obesity mice fed with oolong tea polyphenols. Food Funct. 9, 1079–1087. doi: 10.1039/c7fo01570d, PMID: [DOI] [PubMed] [Google Scholar]
- Choi Y. H. (2019). Isorhamnetin induces ROS-dependent cycle arrest at G2/M phase and apoptosis in human hepatocarcinoma Hep3B cells. Gen. Physiol. Biophys. 38, 473–484. doi: 10.4149/gpb_2019038, PMID: [DOI] [PubMed] [Google Scholar]
- Choi S., Yu S., Lee J., Kim W. (2021). Effects of neohesperidin dihydrochalcone (NHDC) on oxidative phosphorylation, cytokine production, and lipid deposition. Foods 10:1408. doi: 10.3390/foods10061408, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopyk D. M., Grakoui A. (2020). Contribution of the intestinal microbiome and gut barrier to hepatic disorders. Gastroenterology 159, 849–863. doi: 10.1053/j.gastro.2020.04.077, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury S., Fong S. S. (2020). Leveraging genome-scale metabolic models for human health applications. Curr. Opin. Biotechnol. 66, 267–276. doi: 10.1016/j.copbio.2020.08.017, PMID: [DOI] [PubMed] [Google Scholar]
- Chu J. N., Traverso G. (2022). Foundations of gastrointestinal-based drug delivery and future developments. Nat. Rev. Gastroenterol. Hepatol. 19, 219–238. doi: 10.1038/s41575-021-00539-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel T., Borrmann D., Braune A., Doré J., Blaut M. (2006). Occurrence and activity of human intestinal bacteria involved in the conversion of dietary lignans. Anaerobe 12, 140–147. doi: 10.1016/j.anaerobe.2005.11.002, PMID: [DOI] [PubMed] [Google Scholar]
- Couteau D., McCartney A. L., Gibson G. R., Williamson G., Faulds C. B. (2001). Isolation and characterization of human colonic bacteria able to hydrolyse chlorogenic acid. J. Appl. Microbiol. 90, 873–881. doi: 10.1046/j.1365-2672.2001.01316.x, PMID: [DOI] [PubMed] [Google Scholar]
- Cui L., Sun E., Zhang Z., Qian Q., Tan X., Xu F., et al. (2013). Metabolite profiles of epimedin B in rats by ultraperformance liquid chromatography/quadrupole-time-of-flight mass spectrometry. J. Agric. Food Chem. 61, 3589–3599. doi: 10.1021/jf304625x, PMID: [DOI] [PubMed] [Google Scholar]
- Cui L., Sun E., Zhang Z., Tan X., Xu F., Jia X. (2014). Metabolite profiles of epimedin C in rat plasma and bile by ultra-performance liquid chromatography coupled with quadrupole-TOF-MS. Biomed. Chromatogr. 28, 1306–1312. doi: 10.1002/bmc.3174, PMID: [DOI] [PubMed] [Google Scholar]
- Cui X., Tao J. H., Jiang S., Wei X. Y., Xu J., Qian D. W., et al. (2018). Study on the interaction between rhizoma coptis extract and intestinal microflora (in Chinese). Chin. Herb. Med. 49, 2103–2107. [Google Scholar]
- de Vos W. M., Tilg H., Van Hul M., Cani P. D. (2022). Gut microbiome and health: mechanistic insights. Gut 71, 1020–1032. doi: 10.1136/gutjnl-2021-326789, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey E., Corr S. C. (2022). Lactobacillus spp. for gastrointestinal health: current and future perspectives. Front. Immunol. 13:840245. doi: 10.3389/fimmu.2022.840245, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey P. (2019). Gut microbiota in phytopharmacology: a comprehensive overview of concepts, reciprocal interactions, biotransformations and mode of actions. Pharmacol. Res. 147:104367. doi: 10.1016/j.phrs.2019.104367, PMID: [DOI] [PubMed] [Google Scholar]
- Ding J. F., Song X. H., Ji Y., Gu Y. Q. (2019). Research progress on metabolism mechanism of aconitum alkaloids in vivo (in Chinese). Chin. J. Forensic Med. 34, 375–378. doi: 10.13618/j.issn.1001-5728.2019.04.012 [DOI] [Google Scholar]
- Du L. Y., Zhao M., Tao J. H., Qian D. W., Jiang S., Shang E. X., et al. (2017). The metabolic profiling of isorhamnetin-3-O-neohesperidoside produced by human intestinal flora employing UPLC-Q-TOF/MS. J. Chromatogr. Sci. 55, 243–250. doi: 10.1093/chromsci/bmw176, PMID: [DOI] [PubMed] [Google Scholar]
- Du L. Y., Zhao M., Xu J., Qian D. W., Jiang S., Shang E. X., et al. (2014). Identification of the metabolites of myricitrin produced by human intestinal bacteria in vitro using ultra-performance liquid chromatography/quadrupole time-of-flight mass spectrometry. Expert Opin. Drug Metab. Toxicol. 10, 921–931. doi: 10.1517/17425255.2014.918954, PMID: [DOI] [PubMed] [Google Scholar]
- El-Baba C., Baassiri A., Kiriako G., Dia B., Fadlallah S., Moodad S., et al. (2021). Terpenoids' anti-cancer effects: focus on autophagy. Apoptosis 26, 491–511. doi: 10.1007/s10495-021-01684-y, PMID: [DOI] [PubMed] [Google Scholar]
- Ellenbogen J. B., Jiang R., Kountz D. J., Zhang L., Krzycki J. A. (2021). The MttB superfamily member MtyB from the human gut symbiont Eubacterium limosum is a cobalamin-dependent γ-butyrobetaine methyltransferase. J. Biol. Chem. 297:101327. doi: 10.1016/j.jbc.2021.101327, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Mekkawy S., Meselhy M. R., Kawata Y., Kadota S., Hattori M., Namba T. (1993). Metabolism of strychnine N-oxide and brucine N-oxide by human intestinal bacteria. Planta Med. 59, 347–350. doi: 10.1055/s-2006-959698 [DOI] [PubMed] [Google Scholar]
- el-Sedawy A. I., Hattori M., Kobashi K., Namba T. (1989). Metabolism of gentiopicroside (gentiopicrin) by human intestinal bacteria. Chem. Pharm. Bull. 37, 2435–2437. doi: 10.1248/cpb.37.2435, PMID: [DOI] [PubMed] [Google Scholar]
- Fan H., Qi D., Yang M., Fang H., Liu K., Zhao F. (2013). In vitro and in vivo anti-inflammatory effects of 4-methoxy-5-hydroxycanthin-6-one, a natural alkaloid from Picrasma quassioides. Phytomedicine 20, 319–323. doi: 10.1016/j.phymed.2012.11.016, PMID: [DOI] [PubMed] [Google Scholar]
- Fang C., Kim H., Yanagisawa L., Bennett W., Sirven M. A., Alaniz R. C., et al. (2019). Gallotannins and lactobacillus plantarum WCFS1 mitigate high-fat diet-induced inflammation and induce biomarkers for thermogenesis in adipose tissue in gnotobiotic mice. Mol. Nutr. Food Res. 63:e1800937. doi: 10.1002/mnfr.201800937, PMID: [DOI] [PubMed] [Google Scholar]
- Feng W., Ao H., Peng C., Yan D. (2019). Gut microbiota, a new frontier to understand traditional Chinese medicines. Pharmacol. Res. 142, 176–191. doi: 10.1016/j.phrs.2019.02.024, PMID: [DOI] [PubMed] [Google Scholar]
- Feng R., Shou J. W., Zhao Z. X., He C. Y., Ma C., Huang M., et al. (2015). Transforming berberine into its intestine-absorbable form by the gut microbiota. Sci. Rep. 5:12155. doi: 10.1038/srep12155, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Lazarte A., Plaza-Vinuesa L., de Las Rivas B., Villamiel M., Muñoz R., Moreno F. J. (2021). Production of α-rhamnosidases from lactobacillus plantarum WCFS1 and their role in deglycosylation of dietary flavonoids naringin and rutin. Int. J. Biol. Macromol. 193, 1093–1102. doi: 10.1016/j.ijbiomac.2021.11.053, PMID: [DOI] [PubMed] [Google Scholar]
- Flint H. J., Scott K. P., Duncan S. H., Louis P., Forano E. (2012). Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3, 289–306. doi: 10.4161/gmic.19897, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster-Nyarko E., Pallen M. J. (2022). The microbial ecology of Escherichia coli in the vertebrate gut. FEMS Microbiol. Rev. 46:fuac008. doi: 10.1093/femsre/fuac008, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch C., Jänsch A., Ehrmann M. A., Toelstede S., Vogel R. F. (2017). Characterization of cinnamoyl esterases from different lactobacilli and Bifidobacteria. Curr. Microbiol. 74, 247–256. doi: 10.1007/s00284-016-1182-x, PMID: [DOI] [PubMed] [Google Scholar]
- Fushinobu S., Abou Hachem M. (2021). Structure and evolution of the bifidobacterial carbohydrate metabolism proteins and enzymes. Biochem. Soc. Trans. 49, 563–578. doi: 10.1042/bst20200163, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall M., Thomsen M., Peters C., Pavlidis I. V., Jonczyk P., Grünert P. P., et al. (2014). Enzymatic conversion of flavonoids using bacterial chalcone isomerase and enoate reductase. Angew. Chem. Int. Ed. Engl. 53, 1439–1442. doi: 10.1002/anie.201306952, PMID: [DOI] [PubMed] [Google Scholar]
- García-Villalba R., Beltrán D., Frutos M. D., Selma M. V., Espín J. C., Tomás-Barberán F. A. (2020). Metabolism of different dietary phenolic compounds by the urolithin-producing human-gut bacteria Gordonibacter urolithinfaciens and Ellagibacter isourolithinifaciens. Food Funct. 11, 7012–7022. doi: 10.1039/d0fo01649g, PMID: [DOI] [PubMed] [Google Scholar]
- Goldin B. R. (1990). Intestinal microflora: metabolism of drugs and carcinogens. Ann. Med. 22, 43–48. doi: 10.3109/07853899009147240, PMID: [DOI] [PubMed] [Google Scholar]
- Gong Z., Huang J., Xu B., Ou Z., Zhang L., Lin X., et al. (2019). Urolithin A attenuates memory impairment and neuroinflammation in APP/PS1 mice. J. Neuroinflammation 16:62. doi: 10.1186/s12974-019-1450-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonthier M. P., Remesy C., Scalbert A., Cheynier V., Souquet J. M., Poutanen K., et al. (2006). Microbial metabolism of caffeic acid and its esters chlorogenic and caftaric acids by human faecal microbiota in vitro. Biomed. Pharmacother. 60, 536–540. doi: 10.1016/j.biopha.2006.07.084, PMID: [DOI] [PubMed] [Google Scholar]
- Goris T., Cuadrat R. R. C., Braune A. (2021). Flavonoid-modifying capabilities of the human gut microbiome-an in silico study. Nutrients 13:2688. doi: 10.3390/nu13082688, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J. F., Feng L., Zhang M. H., Qin D., Jiang J., Cheng X. D., et al. (2014). New metabolite profiles of Danshensu in rats by ultraperformance liquid chromatography/quadrupole-time-of-flight mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 955-956, 20–25. doi: 10.1016/j.jchromb.2014.02.010, PMID: [DOI] [PubMed] [Google Scholar]
- Guo Y. P., Chen M. Y., Shao L., Zhang W., Rao T., Zhou H. H., et al. (2019). Quantification of Panax notoginseng saponins metabolites in rat plasma with in vivo gut microbiota-mediated biotransformation by HPLC-MS/MS. Chin. J. Nat. Med. 17, 231–240. doi: 10.1016/s1875-5364(19)30026-3, PMID: [DOI] [PubMed] [Google Scholar]
- Guo L., Katiyo W., Lu L., Zhang X., Wang M., Yan J., et al. (2018). Glycyrrhetic acid 3-O-mono-β-d-glucuronide (GAMG): an innovative high-potency sweetener with improved biological activities. Compr. Rev. Food Sci. Food Saf. 17, 905–919. doi: 10.1111/1541-4337.12353, PMID: [DOI] [PubMed] [Google Scholar]
- Haiser H. J., Gootenberg D. B., Chatman K., Sirasani G., Balskus E. P., Turnbaugh P. J. (2013). Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science 341, 295–298. doi: 10.1126/science.1235872, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han D. H., Lee Y., Ahn J. H. (2016). Biological synthesis of baicalein derivatives using Escherichia coli. J. Microbiol. Biotechnol. 26, 1918–1923. doi: 10.4014/jmb.1605.05050, PMID: [DOI] [PubMed] [Google Scholar]
- Hanske L., Engst W., Loh G., Sczesny S., Blaut M., Braune A. (2013). Contribution of gut bacteria to the metabolism of cyanidin 3-glucoside in human microbiota-associated rats. Br. J. Nutr. 109, 1433–1441. doi: 10.1017/s0007114512003376, PMID: [DOI] [PubMed] [Google Scholar]
- Hasanah U., Miki K., Nitoda T., Kanzaki H. (2021). Aerobic bioconversion of C-glycoside mangiferin into its aglycone norathyriol by an isolated mouse intestinal bacterium. Biosci. Biotechnol. Biochem. 85, 989–997. doi: 10.1093/bbb/zbaa121, PMID: [DOI] [PubMed] [Google Scholar]
- Hassaninasab A., Hashimoto Y., Tomita-Yokotani K., Kobayashi M. (2011). Discovery of the curcumin metabolic pathway involving a unique enzyme in an intestinal microorganism. Proc. Natl. Acad. Sci. U. S. A. 108, 6615–6620. doi: 10.1073/pnas.1016217108, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatamipour M., Ramezani M., Tabassi S. A. S., Johnston T. P., Sahebkar A. (2019). Demethoxycurcumin: a naturally occurring curcumin analogue for treating non-cancerous diseases. J. Cell. Physiol. 234, 19320–19330. doi: 10.1002/jcp.28626, PMID: [DOI] [PubMed] [Google Scholar]
- Hattori M., Namba T., Akao T., Kobashi K. (1988). Metabolism of sennosides by human intestinal bacteria. Pharmacology 36, 172–179. doi: 10.1159/000138437, PMID: [DOI] [PubMed] [Google Scholar]
- He C. Y., Fu J., Shou J. W., Zhao Z. X., Ren L., Wang Y., et al. (2017). In vitro study of the metabolic characteristics of eight isoquinoline alkaloids from natural plants in rat gut microbiota. Molecules 22:932. doi: 10.3390/molecules22060932, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J. X., Goto E., Akao T., Tani T. (2007). Interaction between Shaoyao-Gancao-Tang and a laxative with respect to alteration of paeoniflorin metabolism by intestinal bacteria in rats. Phytomedicine 14, 452–459. doi: 10.1016/j.phymed.2006.09.014, PMID: [DOI] [PubMed] [Google Scholar]
- He Y., Hu Z., Li A., Zhu Z., Yang N., Ying Z., et al. (2019). Recent advances in biotransformation of saponins. Molecules 24:2365. doi: 10.3390/molecules24132365, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B. L., Xiong Y., Hu T. G., Zong M. H., Wu H. (2022). Bifidobacterium spp. as functional foods: a review of current status, challenges, and strategies. Crit. Rev. Food Sci. Nutr. 23, 1–18. doi: 10.1080/10408398.2022.2054934, PMID: [DOI] [PubMed] [Google Scholar]
- Heng Y., Kim M. J., Yang H. J., Kang S., Park S. (2019). Lactobacillus intestinalis efficiently produces equol from daidzein and chungkookjang, short-term fermented soybeans. Arch. Microbiol. 201, 1009–1017. doi: 10.1007/s00203-019-01665-5, PMID: [DOI] [PubMed] [Google Scholar]
- Hidalgo-Cantabrana C., Delgado S., Ruiz L., Ruas-Madiedo P., Sánchez B., Margolles A. (2017). Bifidobacteria and their health-promoting effects. Microbiol. Spectr. 5. doi: 10.1128/microbiolspec.BAD-0010-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostetler G. L., Ralston R. A., Schwartz S. J. (2017). Flavones: food sources, bioavailability, metabolism, and bioactivity. Adv. Nutr. 8, 423–435. doi: 10.3945/an.116.012948, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W. C., Liou C. J., Shen S. C., Hu S., Chao J. C., Hsiao C. Y., et al. (2022). Urolithin A inactivation of TLR3/TRIF signaling to block the NF-κB/STAT1 axis reduces inflammation and enhances antioxidant defense in poly(I:C)-induced RAW264.7 cells. Int. J. Mol. Sci. 23:4697. doi: 10.3390/ijms23094697, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur H. G., Beger R. D., Heinze T. M., Lay J. O., Jr., Freeman J. P., Dore J., et al. (2002). Isolation of an anaerobic intestinal bacterium capable of cleaving the C-ring of the isoflavonoid daidzein. Arch. Microbiol. 178, 8–12. doi: 10.1007/s00203-002-0414-6, PMID: [DOI] [PubMed] [Google Scholar]
- Hur H., Rafii F. (2000). Biotransformation of the isoflavonoids biochanin A, formononetin, and glycitein by Eubacterium limosum. FEMS Microbiol. Lett. 192, 21–25. doi: 10.1111/j.1574-6968.2000.tb09353.x, PMID: [DOI] [PubMed] [Google Scholar]
- Jarosova V., Vesely O., Marsik P., Jaimes J. D., Smejkal K., Kloucek P., et al. (2019). Metabolism of stilbenoids by human faecal microbiota. Molecules 24:1155. doi: 10.3390/molecules24061155, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javdan B., Lopez J. G., Chankhamjon P., Lee Y. J., Hull R., Wu Q., et al. (2020). Personalized mapping of drug metabolism by the human gut microbiome. Cells 181, 1661–1679.e22. doi: 10.1016/j.cell.2020.05.001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Ma Y., Gao Y., Li Z., Lian S., Xu Z., et al. (2016). Comprehensive evaluation of the metabolism of genipin-1-β-d-gentiobioside in vitro and in vivo by using HPLC-Q-TOF. J. Agric. Food Chem. 64, 5490–5498. doi: 10.1021/acs.jafc.6b01835, PMID: [DOI] [PubMed] [Google Scholar]
- Jiang S., Yang J., Qian D., Guo J., Shang E. X., Duan J. A., et al. (2014). Rapid screening and identification of metabolites of quercitrin produced by the human intestinal bacteria using ultra performance liquid chromatography/quadrupole-time-of-flight mass spectrometry. Arch. Pharm. Res. 37, 204–213. doi: 10.1007/s12272-013-0172-9, PMID: [DOI] [PubMed] [Google Scholar]
- Jiao Q., Xu L., Jiang L., Jiang Y., Zhang J., Liu B. (2020). Metabolism study of hesperetin and hesperidin in rats by UHPLC-LTQ-Orbitrap MS (n). Xenobiotica 50, 1311–1322. doi: 10.1080/00498254.2019.1567956, PMID: [DOI] [PubMed] [Google Scholar]
- Jiménez N., Esteban-Torres M., Mancheño J. M., de Las Rivas B., Muñoz R. (2014). Tannin degradation by a novel tannase enzyme present in some lactobacillus plantarum strains. Appl. Environ. Microbiol. 80, 2991–2997. doi: 10.1128/aem.00324-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J. S., Lee J. H., Hattori M. (2013). Ligand binding affinities of arctigenin and its demethylated metabolites to estrogen receptor alpha. Molecules 18, 1122–1127. doi: 10.3390/molecules18011122, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J. S., Zhao Y. F., Nakamura N., Akao T., Kakiuchi N., Min B. S., et al. (2007). Enantioselective dehydroxylation of enterodiol and enterolactone precursors by human intestinal bacteria. Biol. Pharm. Bull. 30, 2113–2119. doi: 10.1248/bpb.30.2113, PMID: [DOI] [PubMed] [Google Scholar]
- Jung C. M., Heinze T. M., Schnackenberg L. K., Mullis L. B., Elkins S. A., Elkins C. A., et al. (2009). Interaction of dietary resveratrol with animal-associated bacteria. FEMS Microbiol. Lett. 297, 266–273. doi: 10.1111/j.1574-6968.2009.01691.x, PMID: [DOI] [PubMed] [Google Scholar]
- Kang M. J., Khanal T., Kim H. G., Lee D. H., Yeo H. K., Lee Y. S., et al. (2012). Role of metabolism by human intestinal microflora in geniposide-induced toxicity in HepG2 cells. Arch. Pharm. Res. 35, 733–738. doi: 10.1007/s12272-012-0418-y, PMID: [DOI] [PubMed] [Google Scholar]
- Káňová K., Petrásková L., Pelantová H., Rybková Z., Malachová K., Cvačka J., et al. (2020). Sulfated metabolites of luteolin, myricetin, and ampelopsin: chemoenzymatic preparation and biophysical properties. J. Agric. Food Chem. 68, 11197–11206. doi: 10.1021/acs.jafc.0c03997, PMID: [DOI] [PubMed] [Google Scholar]
- Kawata Y., Hattori M., Akao T., Kobashi K., Namba T. (1991). Formation of nitrogen-containing metabolites from geniposide and gardenoside by human intestinal bacteria. Planta Med. 57, 536–542. doi: 10.1055/s-2006-960201, PMID: [DOI] [PubMed] [Google Scholar]
- Kay C. D., Pereira-Caro G., Ludwig I. A., Clifford M. N., Crozier A. (2017). Anthocyanins and flavanones are more bioavailable than previously perceived: A review of recent evidence. Annu. Rev. Food Sci. Technol. 8, 155–180. doi: 10.1146/annurev-food-030216-025636, PMID: [DOI] [PubMed] [Google Scholar]
- Ke Z. C., Yang L., Hou X. F., Wang A. D., Feng L., Jia X. B. (2016). Metabolism of paeoniflorin by rat intestinal flora in vitro (in Chinese). Zhongguo Zhong Yao Za Zhi 41, 3839–3845. doi: 10.4268/cjcmm20162021, PMID: [DOI] [PubMed] [Google Scholar]
- Kida H., Akao T., Meselhy M. R., Hattori M. (1998). Metabolism and pharmacokinetics of orally administered saikosaponin b1 in conventional, germ-free and Eubacterium sp. A-44-infected gnotobiote rats. Biol. Pharm. Bull. 21, 588–593. doi: 10.1248/bpb.21.588, PMID: [DOI] [PubMed] [Google Scholar]
- Kim D. H. (2018). Gut microbiota-mediated pharmacokinetics of ginseng saponins. J. Ginseng Res. 42, 255–263. doi: 10.1016/j.jgr.2017.04.011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Hong S. W., Kim B. T., Bae E. A., Park H. Y., Han M. J. (2000). Biotransformation of glycyrrhizin by human intestinal bacteria and its relation to biological activities. Arch. Pharm. Res. 23, 172–177. doi: 10.1007/bf02975509, PMID: [DOI] [PubMed] [Google Scholar]
- Kim D.-H., Jang I.-S., Lee H.-K., Jung E.-A., Lee K.-Y. (1996). Metabolism of glycyrrhizin and baicalin by human intestinal bacteria. Arch. Pharm. Res. 19, 292–296. doi: 10.1007/BF02976243 [DOI] [Google Scholar]
- Kim K. A., Jung I. H., Park S. H., Ahn Y. T., Huh C. S., Kim D. H. (2013). Comparative analysis of the gut microbiota in people with different levels of ginsenoside Rb1 degradation to compound K. PLoS One 8:e62409. doi: 10.1371/journal.pone.0062409, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Jung E. A., Sohng I. S., Han J. A., Kim T. H., Han M. J. (1998). Intestinal bacterial metabolism of flavonoids and its relation to some biological activities. Arch. Pharm. Res. 21, 17–23. doi: 10.1007/bf03216747, PMID: [DOI] [PubMed] [Google Scholar]
- Kim Y. S., Kim J. J., Cho K. H., Jung W. S., Moon S. K., Park E. K., et al. (2008). Biotransformation of ginsenoside Rb1, crocin, amygdalin, geniposide, puerarin, ginsenoside re, hesperidin, poncirin, glycyrrhizin, and baicalin by human fecal microflora and its relation to cytotoxicity against tumor cells. J. Microbiol. Biotechnol. 18, 1109–1114. doi: 10.4014/jmb.1511.11047, PMID: [DOI] [PubMed] [Google Scholar]
- Kim M., Kim N., Han J. (2014). Metabolism of kaempferia parviflora polymethoxyflavones by human intestinal bacterium Bautia sp. MRG-PMF1. J. Agric. Food Chem. 62, 12377–12383. doi: 10.1021/jf504074n, PMID: [DOI] [PubMed] [Google Scholar]
- Kim D. H., Lee S. W., Han M. J. (1999). Biotransformation of glycyrrhizin to 18beta-glycyrrhetinic acid-3-O-beta-D-glucuronide by streptococcus LJ-22, a human intestinal bacterium. Biol. Pharm. Bull. 22, 320–322. doi: 10.1248/bpb.22.320, PMID: [DOI] [PubMed] [Google Scholar]
- Kim J. S., Lee H., Nirmala F. S., Jung C. H., Kim M. J., Jang Y. J., et al. (2019). Dihydrodaidzein and 6-hydroxydaidzein mediate the fermentation-induced increase of antiosteoporotic effect of soybeans in ovariectomized mice. FASEB J. 33, 3252–3263. doi: 10.1096/fj.201800953R, PMID: [DOI] [PubMed] [Google Scholar]
- Kolodziejczyk A. A., Zheng D., Elinav E. (2019). Diet-microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 17, 742–753. doi: 10.1038/s41579-019-0256-8, PMID: [DOI] [PubMed] [Google Scholar]
- Kon R., Ikarashi N., Nagoya C., Takayama T., Kusunoki Y., Ishii M., et al. (2014). Rheinanthrone, a metabolite of sennoside A, triggers macrophage activation to decrease aquaporin-3 expression in the colon, causing the laxative effect of rhubarb extract. J. Ethnopharmacol. 152, 190–200. doi: 10.1016/j.jep.2013.12.055, PMID: [DOI] [PubMed] [Google Scholar]
- Koppel N., Maini Rekdal V., Balskus E. P. (2017). Chemical transformation of xenobiotics by the human gut microbiota. Science 356:aag2770. doi: 10.1126/science.aag2770, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano T., Fujiki E., Hashimoto Y., Kobayashi M. (2016). Discovery of a sesamin-metabolizing microorganism and a new enzyme. Proc. Natl. Acad. Sci. U. S. A. 113, 9087–9092. doi: 10.1073/pnas.1605050113, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar K., Jaiswal S. K., Dhoke G. V., Srivastava G. N., Sharma A. K., Sharma V. K. (2018). Mechanistic and structural insight into promiscuity based metabolism of cardiac drug digoxin by gut microbial enzyme. J. Cell. Biochem. 119, 5287–5296. doi: 10.1002/jcb.26638, PMID: [DOI] [PubMed] [Google Scholar]
- Lannes-Costa P. S., de Oliveira J. S. S., da Silva Santos G., Nagao P. E. (2021). A current review of pathogenicity determinants of streptococcus sp. J. Appl. Microbiol. 131, 1600–1620. doi: 10.1111/jam.15090 [DOI] [PubMed] [Google Scholar]
- Lavrijsen K., van Dyck D., van Houdt J., Hendrickx J., Monbaliu J., Woestenborghs R., et al. (1995). Reduction of the prodrug loperamide oxide to its active drug loperamide in the gut of rats, dogs, and humans. Drug Metab. Dispos. 23, 354–362. PMID: [PubMed] [Google Scholar]
- Le Bourvellec C., Bagano Vilas Boas P., Lepercq P., Comtet-Marre S., Auffret P., Ruiz P., et al. (2019). Procyanidin-cell wall interactions within apple matrices decrease the metabolization of procyanidins by the human gut microbiota and the anti-inflammatory effect of the resulting microbial metabolome in vitro. Nutrients 11:664. doi: 10.3390/nu11030664, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Kwon M. C., Jang J. P., Sohng J. K., Jung H. J. (2017). The ginsenoside metabolite compound K inhibits growth, migration and stemness of glioblastoma cells. Int. J. Oncol. 51, 414–424. doi: 10.3892/ijo.2017.4054, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennernäs H., Abrahamsson B. (2005). The use of biopharmaceutic classification of drugs in drug discovery and development: current status and future extension. J. Pharm. Pharmacol. 57, 273–285. doi: 10.1211/0022357055263, PMID: [DOI] [PubMed] [Google Scholar]
- Li C., Ai G., Wang Y., Lu Q., Luo C., Tan L., et al. (2020). Oxyberberine, a novel gut microbiota-mediated metabolite of berberine, possesses superior anti-colitis effect: impact on intestinal epithelial barrier, gut microbiota profile and TLR4-MyD88-NF-κB pathway. Pharmacol. Res. 152:104603. doi: 10.1016/j.phrs.2019.104603, PMID: [DOI] [PubMed] [Google Scholar]
- Li Q., Guo Y., Yu X., Liu W., Zhou L. (2020). Protective mechanism of rhubarb anthraquinone glycosides in rats with cerebral ischaemia-reperfusion injury: interactions between medicine and intestinal flora. Chin. Med. 15:60. doi: 10.1186/s13020-020-00341-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. M., He M. Z., Feng Y. L., Li T. E., Li Y., Wu B., et al. (2017). Study on metabolism of luteolin and luteolin in rats (in Chinese). Tradit. Chin. Drug Res. Pharmacol. 28, 61–68. doi: 10.19378/j.issn.1003-9783.2017.01.13 [DOI] [Google Scholar]
- Li C., Lee M. J., Sheng S., Meng X., Prabhu S., Winnik B., et al. (2000). Structural identification of two metabolites of catechins and their kinetics in human urine and blood after tea ingestion. Chem. Res. Toxicol. 13, 177–184. doi: 10.1021/tx9901837, PMID: [DOI] [PubMed] [Google Scholar]
- Li D., Shi G., Wang J., Zhang D., Pan Y., Dou H., et al. (2019). Baicalein ameliorates pristane-induced lupus nephritis via activating Nrf2/HO-1 in myeloid-derived suppressor cells. Arthritis Res. Ther. 21:105. doi: 10.1186/s13075-019-1876-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. C., Zhang T., Li Y. Q., Ding G. B. (2019). Target discovery of novel α-L-rhamnosidases from human fecal metagenome and application for biotransformation of natural flavonoid glycosides. Appl. Biochem. Biotechnol. 189, 1245–1261. doi: 10.1007/s12010-019-03063-5, PMID: [DOI] [PubMed] [Google Scholar]
- Liao C. P., Liu X. C., Dong S. Q., An M., Zhao L., Zhang A. J., et al. (2021). Investigation of the metabolites of five major constituents from Berberis amurensis in normal and pseudo germ-free rats. Chin. J. Nat. Med. 19, 758–771. doi: 10.1016/s1875-5364(21)60082-1, PMID: [DOI] [PubMed] [Google Scholar]
- Lindell A. E., Zimmermann-Kogadeeva M., Patil K. R. (2022). Multimodal interactions of drugs, natural compounds and pollutants with the gut microbiota. Nat. Rev. Microbiol. 20, 431–443. doi: 10.1038/s41579-022-00681-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., de Bruijn W. J. C., Sanders M. G., Wang S., Bruins M. E., Vincken J. P. (2021). Insights in the recalcitrance of theasinensin A to human gut microbial degradation. J. Agric. Food Chem. 69, 2477–2484. doi: 10.1021/acs.jafc.1c00727, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Hong J., Xu X., Feng Q., Zhang D., Gu Y., et al. (2017). Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 23, 859–868. doi: 10.1038/nm.4358, PMID: [DOI] [PubMed] [Google Scholar]
- Liu M. Y., Li M., Wang X. L., Liu P., Hao Q. H., Yu X. M. (2013). Study on human intestinal bacterium Blautia sp. AUH-JLD56 for the conversion of arctigenin to (−)-3′-desmethylarctigenin. J. Agric. Food Chem. 61, 12060–12065. doi: 10.1021/jf403924c, PMID: [DOI] [PubMed] [Google Scholar]
- Liu X., Mao B., Gu J., Wu J., Cui S., Wang G., et al. (2021). Blautia-a new functional genus with potential probiotic properties? Gut Microbes 13, 1–21. doi: 10.1080/19490976.2021.1875796, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. L., Song Y. G., Wei S. F., Zhang L., Yang S. L., Wang M., et al. (2015). The metabolites of Pulsatilum pulsatilum saponin B_3 in rat intestinal flora were analyzed by UPLC-ESI-Q-TOF-MS/MS (in Chinese). Chin. J. Exp. Formulae 21, 72–76. doi: 10.13422/j.cnki.syfjx.2015230072 [DOI] [Google Scholar]
- Liu A. B., Tao S., Lee M. J., Hu Q., Meng X., Lin Y., et al. (2018). Effects of gut microbiota and time of treatment on tissue levels of green tea polyphenols in mice. Biofactors 44, 348–360. doi: 10.1002/biof.1430, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Xu Y., Yan M., Yu Y., Guo Y. (2022). 18β-Glycyrrhetinic acid suppresses allergic airway inflammation through NF-κB and Nrf2/HO-1 signaling pathways in asthma mice. Sci. Rep. 12:3121. doi: 10.1038/s41598-022-06455-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo Y. T., Howell K., Chan M., Zhang P., Ng K. (2020). Modulation of the human gut microbiota by phenolics and phenolic fiber-rich foods. Compr. Rev. Food Sci. Food Saf. 19, 1268–1298. doi: 10.1111/1541-4337.12563, PMID: [DOI] [PubMed] [Google Scholar]
- Lu B., Chen X., Chen H., Li Q., Li H., Xu Y., et al. (2022). Demethoxycurcumin mitigates inflammatory responses in lumbar disc herniation via MAPK and NF-κB pathways in vivo and in vitro. Int. Immunopharmacol. 108:108914. doi: 10.1016/j.intimp.2022.108914, PMID: [DOI] [PubMed] [Google Scholar]
- Lu S., Luo Y., Sun G., Sun X. (2020). Ginsenoside ompound K attenuates ox-ldl-mediated macrophage inflammation and foam cell formation via autophagy induction and modulating NF-κB, p38, and JNK MAPK signaling. Front. Pharmacol. 11:567238. doi: 10.3389/fphar.2020.567238, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Wu Y., Zuo L., Luo X., Large P. J. (2014). Intestinal microbiome and digoxin inactivation: meal plan for digoxin users? World J. Microbiol. Biotechnol. 30, 791–799. doi: 10.1007/s11274-013-1507-x, PMID: [DOI] [PubMed] [Google Scholar]
- Lubner C. E., Artz J. H., Mulder D. W., Oza A., Ward R. J., Williams S. G., et al. (2022). A site-differentiated [4Fe-4S] cluster controls electron transfer reactivity of clostridium acetobutylicum [FeFe]-hydrogenase I. Chem. Sci. 13, 4581–4588. doi: 10.1039/d1sc07120c, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca S. V., Macovei I., Bujor A., Miron A., Skalicka-Woźniak K., Aprotosoaie A. C., et al. (2020). Bioactivity of dietary polyphenols: the role of metabolites. Crit. Rev. Food Sci. Nutr. 60, 626–659. doi: 10.1080/10408398.2018.1546669, PMID: [DOI] [PubMed] [Google Scholar]
- Mace T. A., Ware M. B., King S. A., Loftus S., Farren M. R., McMichael E., et al. (2019). Soy isoflavones and their metabolites modulate cytokine-induced natural killer cell function. Sci. Rep. 9:5068. doi: 10.1038/s41598-019-41687-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado D., Andrejev S., Tramontano M., Patil K. R. (2018). Fast automated reconstruction of genome-scale metabolic models for microbial species and communities. Nucleic Acids Res. 46, 7542–7553. doi: 10.1093/nar/gky537, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maini Rekdal V., Nol Bernadino P., Luescher M. U., Kiamehr S., Le C., Bisanz J. E., et al. (2020). A widely distributed metalloenzyme class enables gut microbial metabolism of host- and diet-derived catechols. Elife 9:e50845. doi: 10.7554/eLife.50845, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor O., Dai C. L., Kornilov S. A., Smith B., Price N. D., Lovejoy J. C., et al. (2020). Health and disease markers correlate with gut microbiome composition across thousands of people. Nat. Commun. 11:5206. doi: 10.1038/s41467-020-18871-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas-Capdevila A., Teichenne J., Domenech-Coca C., Caimari A., Del Bas J. M., Escoté X., et al. (2020). Effect of hesperidin on cardiovascular disease risk factors: the role of intestinal microbiota on hesperidin bioavailability. Nutrients 12:1488. doi: 10.3390/nu12051488, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M., Ishige A., Yazawa Y., Kondo M., Muramatsu K., Watanabe K. (2012). Promotion of intestinal peristalsis by Bifidobacterium spp. capable of hydrolysing sennosides in mice. PLoS One 7:e31700. doi: 10.1371/journal.pone.0031700, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayo B., Vázquez L., Flórez A. B. (2019). Equol: a bacterial metabolite from the daidzein isoflavone and its presumed beneficial health effects. Nutrients 11:2231. doi: 10.3390/nu11092231, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X., Zhang G., Cao H., Yu D., Fang X., de Vos W. M., et al. (2020). Gut dysbacteriosis and intestinal disease: mechanism and treatment. J. Appl. Microbiol. 129, 787–805. doi: 10.1111/jam.14661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak B., Filipek A., Chomicki P., Pyza M., Woźniak M., Żyżyńska-Granica B., et al. (2018). Lignans from forsythia x intermedia leaves and flowers attenuate the pro-inflammatory function of leukocytes and their interaction with endothelial cells. Front. Pharmacol. 9:401. doi: 10.3389/fphar.2018.00401, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan E. W., Perdew G. H., Patterson A. D. (2022). Multi-omics strategies for investigating the microbiome in toxicology research. Toxicol. Sci. 187, 189–213. doi: 10.1093/toxsci/kfac029, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Lordan C., Ross R. P., Cotter P. D. (2020). Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health. Gut Microbes 12:1802866. doi: 10.1080/19490976.2020.1802866, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murota K., Nakamura Y., Uehara M. (2018). Flavonoid metabolism: the interaction of metabolites and gut microbiota. Biosci. Biotechnol. Biochem. 82, 600–610. doi: 10.1080/09168451.2018.1444467 [DOI] [PubMed] [Google Scholar]
- Nakamura K., Zhu S., Komatsu K., Hattori M., Iwashima M., Zhou N.-Y. (2020). Deglycosylation of the isoflavone C-glucoside puerarin by a combination of two recombinant bacterial enzymes and 3-oxo-glucose. Appl. Environ. Microbiol. 86, e00607–e00620. doi: 10.1128/AEM.00607-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T. L., Vieira-Silva S., Liston A., Raes J. (2015). How informative is the mouse for human gut microbiota research? Dis. Model. Mech. 8, 1–16. doi: 10.1242/dmm.017400, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oehmcke-Hecht S., Mandl V., Naatz L. T., Dühring L., Köhler J., Kreikemeyer B., et al. (2020). Streptococcus gallolyticus abrogates anti-carcinogenic properties of tannic acid on low-passage colorectal carcinomas. Sci. Rep. 10:4714. doi: 10.1038/s41598-020-61458-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou K., Sarnoski P., Schneider K. R., Song K., Khoo C., Gu L. (2014). Microbial catabolism of procyanidins by human gut microbiota. Mol. Nutr. Food Res. 58, 2196–2205. doi: 10.1002/mnfr.201400243, PMID: [DOI] [PubMed] [Google Scholar]
- Ozdal T., Sela D. A., Xiao J., Boyacioglu D., Chen F., Capanoglu E. (2016). The reciprocal interactions between polyphenols and gut microbiota and effects on bioaccessibility. Nutrients 8:78. doi: 10.3390/nu8020078, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallauf K., Chin D., Günther I., Birringer M., Lüersen K., Schultheiß G., et al. (2019). Resveratrol, lunularin and dihydroresveratrol do not act as caloric restriction mimetics when administered intraperitoneally in mice. Sci. Rep. 9:4445. doi: 10.1038/s41598-019-41050-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang C., Zheng Z., Shi L., Sheng Y., Wei H., Wang Z., et al. (2016). Caffeic acid prevents acetaminophen-induced liver injury by activating the Keap1-Nrf2 antioxidative defense system. Free Radic. Biol. Med. 91, 236–246. doi: 10.1016/j.freeradbiomed.2015.12.024, PMID: [DOI] [PubMed] [Google Scholar]
- Paraiso I. L., Plagmann L. S., Yang L., Zielke R., Gombart A. F., Maier C. S., et al. (2019). Reductive metabolism of xanthohumol and 8-prenylnaringenin by the intestinal bacterium Eubacterium ramulus. Mol. Nutr. Food Res. 63:e1800923. doi: 10.1002/mnfr.201800923, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H. Y., Park S. H., Yoon H. K., Han M. J., Kim D. H. (2004). Anti-allergic activity of 18beta-glycyrrhetinic acid-3-O-beta-D-glucuronide. Arch. Pharm. Res. 27, 57–60. doi: 10.1007/bf02980047, PMID: [DOI] [PubMed] [Google Scholar]
- Peñalvo J. L., Heinonen S. M., Aura A. M., Adlercreutz H. (2005). Dietary sesamin is converted to enterolactone in humans. J. Nutr. 135, 1056–1062. doi: 10.1093/jn/135.5.1056, PMID: [DOI] [PubMed] [Google Scholar]
- Peng R., Han P., Fu J., Zhang Z. W., Ma S. R., Pan L. B., et al. (2022). Esterases from Bifidobacteria exhibit the conversion of albiflorin in gut microbiota. Front. Microbiol. 13:880118. doi: 10.3389/fmicb.2022.880118, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Caro G., Fernández-Quirós B., Ludwig I. A., Pradas I., Crozier A., Moreno-Rojas J. M. (2018). Catabolism of citrus flavanones by the probiotics Bifidobacterium longum and lactobacillus rhamnosus. Eur. J. Nutr. 57, 231–242. doi: 10.1007/s00394-016-1312-z, PMID: [DOI] [PubMed] [Google Scholar]
- Pero R. W., Lund H. (2011). Dietary quinic acid supplied as the nutritional supplement AIO + AC-11® leads to induction of micromolar levels of nicotinamide and tryptophan in the urine. Phytother. Res. 25, 851–857. doi: 10.1002/ptr.3348, PMID: [DOI] [PubMed] [Google Scholar]
- Plamada D., Vodnar D. C. (2021). Polyphenols-gut microbiota interrelationship: a transition to a new generation of prebiotics. Nutrients 14:137. doi: 10.3390/nu14010137, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X. L., Sun H. Y., Yang W., Li Y. J., Zheng L., Liu T., et al. (2017). The metabolism of quercetin in intestinal flora of rats was analyzed by UPLC-ESI-Q-TOF-MS/MS (in Chinese). China J. Chin. Materia Medica 42, 357–362. doi: 10.19540/j.cnki.cjcmm.20161222.018 [DOI] [PubMed] [Google Scholar]
- Qin Y., Wang S., Wen Q., Xia Q., Wang S., Chen G., et al. (2021). Corrigendum: interactions between ephedra sinica and prunus armeniaca: from stereoselectivity to deamination as a metabolic detoxification mechanism of amygdalin. Front. Pharmacol. 12:831921. doi: 10.3389/fphar.2021.831921, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi S., Anighoro A., Quartieri A., Amaretti A., Tomás-Barberán F. A., Rastelli G., et al. (2015). Role of Bifidobacteria in the hydrolysis of chlorogenic acid. Microbiology 4, 41–52. doi: 10.1002/mbo3.219, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverón I., de las Rivas B., Matesanz R., Muñoz R., López de Felipe F. (2015). Molecular adaptation of lactobacillus plantarum WCFS1 to gallic acid revealed by genome-scale transcriptomic signature and physiological analysis. Microb. Cell Factories 14:160. doi: 10.1186/s12934-015-0345-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverón I., Jiménez N., Curiel J. A., Peñas E., López de Felipe F., de Las Rivas B., et al. (2017). Differential gene expression by lactobacillus plantarum WCFS1 in response to phenolic compounds reveals new genes involved in tannin degradation. Appl. Environ. Microbiol. 83:e03387-16. doi: 10.1128/aem.03387-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riva A., Kolimár D., Spittler A., Wisgrill L., Herbold C. W., Abrankó L., et al. (2020). Conversion of rutin, a prevalent dietary flavonol, by the human gut microbiota. Front. Microbiol. 11:585428. doi: 10.3389/fmicb.2020.585428, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocchetti G., Gregorio R. P., Lorenzo J. M., Barba F. J., Oliveira P. G., Prieto M. A., et al. (2022). Functional implications of bound phenolic compounds and phenolics-food interaction: a review. Compr. Rev. Food Sci. Food Saf. 21, 811–842. doi: 10.1111/1541-4337.12921, PMID: [DOI] [PubMed] [Google Scholar]
- Rodríguez-Daza M. C., Pulido-Mateos E. C., Lupien-Meilleur J., Guyonnet D., Desjardins Y., Roy D. (2021). Polyphenol-mediated gut microbiota modulation: toward prebiotics and further. Front. Nutr. 8:689456. doi: 10.3389/fnut.2021.689456, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan J. Q., Li S., Li Y. P., Wu W. J., Lee S. M., Yan R. (2015). The presystemic interplay between gut microbiota and orally administered Calycosin-7-O-β-D-glucoside. Drug Metab. Dispos. 43, 1601–1611. doi: 10.1124/dmd.115.065094, PMID: [DOI] [PubMed] [Google Scholar]
- Saccon T. D., Nagpal R., Yadav H., Cavalcante M. B., Nunes A. D. C., Schneider A., et al. (2021). Senolytic combination of dasatinib and quercetin alleviates intestinal senescence and inflammation and modulates the gut microbiome in aged mice. J. Gerontol. A Biol. Sci. Med. Sci. 76, 1895–1905. doi: 10.1093/gerona/glab002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satti M., Modesto M., Endo A., Kawashima T., Mattarelli P., Arita M. (2021). Host-diet effect on the metabolism of Bifidobacterium. Genes 12:609. doi: 10.3390/genes12040609, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoefer L., Braune A., Blaut M. (2004). Cloning and expression of a phloretin hydrolase gene from Eubacterium ramulus and characterization of the recombinant enzyme. Appl. Environ. Microbiol. 70, 6131–6137. doi: 10.1128/aem.70.10.6131-6137.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyed Hameed A. S., Rawat P. S., Meng X., Liu W. (2020). Biotransformation of dietary phytoestrogens by gut microbes: a review on bidirectional interaction between phytoestrogen metabolism and gut microbiota. Biotechnol. Adv. 43:107576. doi: 10.1016/j.biotechadv.2020.107576, PMID: [DOI] [PubMed] [Google Scholar]
- Shabbir U., Rubab M., Daliri E. B., Chelliah R., Javed A., Oh D. H. (2021). Curcumin, quercetin, catechins and metabolic diseases: the role of gut microbiota. Nutrients 13:206. doi: 10.3390/nu13010206, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo Y., Ozawa Y., Toda T., Igami K., Shimizu T. (2018). Probiotic lactobacillus paracasei A221 improves the functionality and bioavailability of kaempferol-glucoside in kale by its glucosidase activity. Sci. Rep. 8:9239. doi: 10.1038/s41598-018-27532-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Poling H. M., Spence J. R., Wells J. M., Helmrath M. A. (2020). Gastrointestinal organoids: a next-generation tool for modeling human development. Am. J. Physiol. Gastrointest. Liver Physiol. 319, G375–g381. doi: 10.1152/ajpgi.00199.2020, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slámová K., Kapešová J., Valentová K. (2018). "sweet flavonoids": glycosidase-catalyzed modifications. Int. J. Mol. Sci. 19:2126. doi: 10.3390/ijms19072126, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg J. L., Angenent L. T., Gordon J. I. (2004). Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 5, 569–573. doi: 10.1038/ni1079, PMID: [DOI] [PubMed] [Google Scholar]
- Sousa T., Paterson R., Moore V., Carlsson A., Abrahamsson B., Basit A. W. (2008). The gastrointestinal microbiota as a site for the biotransformation of drugs. Int. J. Pharm. 363, 1–25. doi: 10.1016/j.ijpharm.2008.07.009 [DOI] [PubMed] [Google Scholar]
- Stoupi S., Williamson G., Viton F., Barron D., King L. J., Brown J. E., et al. (2010). In vivo bioavailability, absorption, excretion, and pharmacokinetics of [14C]procyanidin B2 in male rats. Drug Metab. Dispos. 38, 287–291. doi: 10.1124/dmd.109.030304, PMID: [DOI] [PubMed] [Google Scholar]
- Tan S., Rupasinghe T. W., Tull D. L., Boughton B., Oliver C., McSweeny C., et al. (2014). Degradation of curcuminoids by in vitro pure culture fermentation. J. Agric. Food Chem. 62, 11005–11015. doi: 10.1021/jf5031168, PMID: [DOI] [PubMed] [Google Scholar]
- Tanaka H., Hashiba H., Kok J., Mierau I. (2000). Bile salt hydrolase of Bifidobacterium longum-biochemical and genetic characterization. Appl. Environ. Microbiol. 66, 2502–2512. doi: 10.1128/aem.66.6.2502-2512.2000, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Kishimoto Y., Sasaki M., Sato A., Kamiya T., Kondo K., et al. (2018). Terminalia bellirica (Gaertn.) Roxb. Extract and gallic acid attenuate LPS-induced inflammation and oxidative stress via MAPK/NF-κB and Akt/AMPK/Nrf2 pathways. Oxid. Med. Cell. Longev. 2018:9364364. doi: 10.1155/2018/9364364, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Tan X., Huang X., Zhang J., Chen L., Li A., et al. (2021). Dual targeting of autophagy and NF-κB pathway by PPARγ contributes to the inhibitory effect of demethoxycurcumin on NLRP3 inflammasome priming. Curr. Mol. Pharmacol. 14, 914–921. doi: 10.2174/1874467214666210301121020, PMID: [DOI] [PubMed] [Google Scholar]
- Theilmann M. C., Goh Y. J., Nielsen K. F., Klaenhammer T. R., Barrangou R., Abou Hachem M. (2017). Lactobacillus acidophilus metabolizes dietary plant glucosides and externalizes Ttheir bioactive phytochemicals. MBio 8:e01421-17. doi: 10.1128/mBio.01421-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J. S., Shi B. Y., Xiang H., Gao S., Qin X. M., Du G. H. (2013). 1H-NMR-based metabonomic studies on the anti-depressant effect of genipin in the chronic unpredictable mild stress rat model. PLoS One 8:e75721. doi: 10.1371/journal.pone.0075721, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticak T., Kountz D. J., Girosky K. E., Krzycki J. A., Ferguson D. J., Jr. (2014). A nonpyrrolysine member of the widely distributed trimethylamine methyltransferase family is a glycine betaine methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 111, E4668–E4676. doi: 10.1073/pnas.1409642111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toney A. M., Fan R., Xian Y., Chaidez V., Ramer-Tait A. E., Chung S. (2019). Urolithin A, a gut metabolite, improves insulin sensitivity through augmentation of mitochondrial function and biogenesis. Obesity (Silver Spring) 27, 612–620. doi: 10.1002/oby.22404, PMID: [DOI] [PubMed] [Google Scholar]
- Tong X., Qi Z., Zheng D., Pei J., Li Q., Zhao L. (2021). High-level expression of a novel multifunctional GH3 family β-xylosidase/α-arabinosidase/β-glucosidase from Dictyoglomus turgidum in Escherichia coli. Bioorg. Chem. 111:104906. doi: 10.1016/j.bioorg.2021.104906, PMID: [DOI] [PubMed] [Google Scholar]
- Tremaroli V., Bäckhed F. (2012). Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–249. doi: 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- Trinh H. T., Joh E. H., Kwak H. Y., Baek N. I., Kim D. H. (2010). Anti-pruritic effect of baicalin and its metabolites, baicalein and oroxylin A, in mice. Acta Pharmacol. Sin. 31, 718–724. doi: 10.1038/aps.2010.42, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse T. J., Guo Y., Shim Y. Y., Purdy S. K., Kim J. H., Cho J. Y., et al. (2022). Availability of bioactive flax lignan from foods and supplements. Crit. Rev. Food Sci. Nutr., 1–16. doi: 10.1080/10408398.2022.2072807, PMID: [DOI] [PubMed] [Google Scholar]
- Turroni F., Marchesi J. R., Foroni E., Gueimonde M., Shanahan F., Margolles A., et al. (2009). Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J. 3, 745–751. doi: 10.1038/ismej.2009.19, PMID: [DOI] [PubMed] [Google Scholar]
- Turroni F., Ribbera A., Foroni E., van Sinderen D., Ventura M. (2008). Human gut microbiota and bifidobacteria: from composition to functionality. Antonie Van Leeuwenhoek 94, 35–50. doi: 10.1007/s10482-008-9232-4, PMID: [DOI] [PubMed] [Google Scholar]
- Valentová K., Havlík J., Kosina P., Papoušková B., Jaimes J. D., Káňová K., et al. (2020). Biotransformation of silymarin flavonolignans by human fecal microbiota. Meta 10:29. doi: 10.3390/metabo10010029, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoni M. A. (2021). Iron-sulfur flavoenzymes: the added value of making the most ancient redox cofactors and the versatile flavins work together. Open Biol. 11:210010. doi: 10.1098/rsob.210010, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinarov Z., Abrahamsson B., Artursson P., Batchelor H., Berben P., Bernkop-Schnürch A., et al. (2021). Current challenges and future perspectives in oral absorption research: an opinion of the UNGAP network. Adv. Drug Deliv. Rev. 171, 289–331. doi: 10.1016/j.addr.2021.02.001, PMID: [DOI] [PubMed] [Google Scholar]
- Vollmer M., Esders S., Farquharson F. M., Neugart S., Duncan S. H., Schreiner M., et al. (2018). Mutual interaction of phenolic compounds and microbiota: metabolism of complex phenolic apigenin-C- and kaempferol-O-derivatives by human fecal samples. J. Agric. Food Chem. 66, 485–497. doi: 10.1021/acs.jafc.7b04842, PMID: [DOI] [PubMed] [Google Scholar]
- Vujkovic-Cvijin I., Sklar J., Jiang L., Natarajan L., Knight R., Belkaid Y. (2020). Host variables confound gut microbiota studies of human disease. Nature 587, 448–454. doi: 10.1038/s41586-020-2881-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J. Y., Zhang Y. Z., Yuan J. B., Yang F. Q., Chen Y., Zhou L. D., et al. (2017). Biotransformation and metabolic profile of anemoside B4 with rat small and large intestine microflora by ultra-performance liquid chromatography-quadrupole time-of-flight tandem mass spectrometry. Biomed. Chromatogr. 31. doi: 10.1002/bmc.3873, PMID: [DOI] [PubMed] [Google Scholar]
- Wang P., Chen H., Zhu Y., McBride J., Fu J., Sang S. (2015). Oat avenanthramide-C (2c) is biotransformed by mice and the human microbiota into bioactive metabolites. J. Nutr. 145, 239–245. doi: 10.3945/jn.114.206508, PMID: [DOI] [PubMed] [Google Scholar]
- Wang X. L., Kim H. J., Kang S. I., Kim S. I., Hur H. G. (2007). Production of phytoestrogen S-equol from daidzein in mixed culture of two anaerobic bacteria. Arch. Microbiol. 187, 155–160. doi: 10.1007/s00203-006-0183-8, PMID: [DOI] [PubMed] [Google Scholar]
- Wang Q., Qian Y., Wang Q., Yang Y. F., Ji S., Song W., et al. (2015). Metabolites identification of bioactive licorice compounds in rats. J. Pharm. Biomed. Anal. 115, 515–522. doi: 10.1016/j.jpba.2015.08.013, PMID: [DOI] [PubMed] [Google Scholar]
- Wardman J. F., Bains R. K., Rahfeld P., Withers S. G. (2022). Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat. Rev. Microbiol. 20, 542–556. doi: 10.1038/s41579-022-00712-1, PMID: [DOI] [PubMed] [Google Scholar]
- Weersma R. K., Zhernakova A., Fu J. (2020). Interaction between drugs and the gut microbiome. Gut 69, 1510–1519. doi: 10.1136/gutjnl-2019-320204, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X., Walle T. (2006). Methylated flavonoids have greatly improved intestinal absorption and metabolic stability. Drug Metab. Dispos. 34, 1786–1792. doi: 10.1124/dmd.106.011122, PMID: [DOI] [PubMed] [Google Scholar]
- Weng J., Lv Q. T., Tian Y., Zhang M., Bian G. X., Wen L. Q. (2006). Study on metabolism and transformation of Asiaticoside by intestinal flora (in Chinese). Chin. Herb. Med. 07, 1008–1011. [Google Scholar]
- Wexler H. M. (2007). Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621. doi: 10.1128/cmr.00008-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I. D., Nicholson J. K. (2017). Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl. Res. 179, 204–222. doi: 10.1016/j.trsl.2016.08.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Kang A., Shan C., Chai C., Zhou Z., Lin Y., et al. (2019). LC-Q-TOF/MS-oriented systemic metabolism study of pedunculoside with in vitro and in vivo biotransformation. J. Pharm. Biomed. Anal. 175:112762. doi: 10.1016/j.jpba.2019.07.010, PMID: [DOI] [PubMed] [Google Scholar]
- Wu H., Kim M., Han J. (2016). Icariin metabolism by human intestinal microflora. Molecules 21:1158. doi: 10.3390/molecules21091158, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Pei J., Ge L., Wang Z., Ding G., Xiao W., et al. (2018). Characterization of a α-l-rhamnosidase from Bacteroides thetaiotaomicron with high catalytic efficiency of epimedin C. Bioorg. Chem. 81, 461–467. doi: 10.1016/j.bioorg.2018.08.004, PMID: [DOI] [PubMed] [Google Scholar]
- Wu X. M., Tan R. X. (2019). Interaction between gut microbiota and ethnomedicine constituents. Nat. Prod. Rep. 36, 788–809. doi: 10.1039/c8np00041g, PMID: [DOI] [PubMed] [Google Scholar]
- Wu M. J., Wu X. L., Zhang D. Q., Ding L. Q., Qiu F., Zhang H. M. (2017). Identification of metabolites of rutin in rats by UPLC-Q-TOF/MS (in Chinese). Chin. J. Exp. Formulae 23, 91–97. doi: 10.13422/j.cnki.syfjx.2017170091 [DOI] [Google Scholar]
- Xiao Y., Shao K., Zhou J., Wang L., Ma X., Wu D., et al. (2021). Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases. Nat. Commun. 12:2828. doi: 10.1038/s41467-021-23095-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Hu F., Xiang D., Lu H., Li W., Zhao A., et al. (2020). The metabolic effect of gut microbiota on drugs. Drug Metab. Rev. 52, 139–156. doi: 10.1080/03602532.2020.1718691 [DOI] [PubMed] [Google Scholar]
- Xing S., Wang M., Peng Y., Chen D., Li X. (2014). Simulated gastrointestinal tract metabolism and pharmacological activities of water extract of Scutellaria baicalensis roots. J. Ethnopharmacol. 152, 183–189. doi: 10.1016/j.jep.2013.12.056, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki M., Nishi K., Nishimoto S., Yamauchi S., Akiyama K., Kishida T., et al. (2011). Immunomodulatory effect of (−-)-matairesinol in vivo and ex vivo. Biosci. Biotechnol. Biochem. 75, 859–863. doi: 10.1271/bbb.100781, PMID: [DOI] [PubMed] [Google Scholar]
- Yan T., Yi T., Zheng T. T. (2018). Effects of human intestinal flora on the metabolic transformation of ginsenoside Rg_3 and Ginsenoside D of Pulsatilla chinensis (in Chinese). Proprietary Chin. Med. 40, 1902–1909. [Google Scholar]
- Yang G., Hong S., Yang P., Sun Y., Wang Y., Zhang P., et al. (2021). Discovery of an ene-reductase for initiating flavone and flavonol catabolism in gut bacteria. Nat. Commun. 12:790. doi: 10.1038/s41467-021-20974-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Qian D., Jiang S., Shang E. X., Guo J., Duan J. A. (2012). Identification of rutin deglycosylated metabolites produced by human intestinal bacteria using UPLC-Q-TOF/MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 898, 95–100. doi: 10.1016/j.jchromb.2012.04.024, PMID: [DOI] [PubMed] [Google Scholar]
- Yang X. W., Zhang J. Y., Xu W. (2007). Biotransformation of mogroside III by human intestinal bacteria. Beijing Da Xue Xue Bao 39, 657–662. PMID: [PubMed] [Google Scholar]
- Yao R., Wong C. B., Nakamura K., Mitsuyama E., Tanaka A., Kuhara T., et al. (2019). Bifidobacterium breve MCC1274 with glycosidic activity enhances in vivo isoflavone bioavailability. Benef Microbes 10, 521–531. doi: 10.3920/bm2018.0179, PMID: [DOI] [PubMed] [Google Scholar]
- Ye M., Wang Q., Zhang W., Li Z., Wang Y., Hu R. (2014). Oroxylin A exerts anti-inflammatory activity on lipopolysaccharide-induced mouse macrophage via Nrf2/ARE activation. Biochem. Cell Biol. 92, 337–348. doi: 10.1139/bcb-2014-0030, PMID: [DOI] [PubMed] [Google Scholar]
- Ye M., Yu J., Shi X., Zhu J., Gao X., Liu W. (2021). Polysaccharides catabolism by the human gut bacterium-Bacteroides thetaiotaomicron: advances and perspectives. Crit. Rev. Food Sci. Nutr. 61, 3569–3588. doi: 10.1080/10408398.2020.1803198, PMID: [DOI] [PubMed] [Google Scholar]
- Yim J. S., Kim Y. S., Moon S. K., Cho K. H., Bae H. S., Kim J. J., et al. (2004). Metabolic activities of ginsenoside Rb1, baicalin, glycyrrhizin and geniposide to their bioactive compounds by human intestinal microflora. Biol. Pharm. Bull. 27, 1580–1583. doi: 10.1248/bpb.27.1580, PMID: [DOI] [PubMed] [Google Scholar]
- Yokoyama S., Suzuki T. (2008). Isolation and characterization of a novel equol-producing bacterium from human feces. Biosci. Biotechnol. Biochem. 72, 2660–2666. doi: 10.1271/bbb.80329, PMID: [DOI] [PubMed] [Google Scholar]
- Yoshisue K., Masuda H., Matsushima E., Ikeda K., Nagayama S., Kawaguchi Y. (2000). Tissue distribution and biotransformation of potassium oxonate after oral administration of a novel antitumor agent (drug combination of tegafur, 5-chloro-2,4-dihydroxypyridine, and potassium oxonate) to rats. Drug Metab. Dispos. 28, 1162–1167. PMID: [PubMed] [Google Scholar]
- Youn S. Y., Park M. S., Ji G. E. (2012). Identification of the beta-glucosidase gene from Bifidobacterium animalis subsp. lactis and its expression in B. bifidum BGN4. J. Microbiol. Biotechnol. 22, 1714–1723. doi: 10.4014/jmb.1208.08028, PMID: [DOI] [PubMed] [Google Scholar]
- Yu Q., Liu Y., Wu Y., Chen Y. (2018). Dihydrocurcumin ameliorates the lipid accumulation, oxidative stress and insulin resistance in oleic acid-induced L02 and HepG2 cells. Biomed. Pharmacother. 103, 1327–1336. doi: 10.1016/j.biopha.2018.04.143, PMID: [DOI] [PubMed] [Google Scholar]
- Zafar H., Saier M. H., Jr. (2021). Gut Bacteroides species in health and disease. Gut Microbes 13, 1–20. doi: 10.1080/19490976.2020.1848158, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Huang X. M., Yan H. J., Liu X. Y., Zhou Q., Luo Z. Y., et al. (2019). Highly selective production of compound K from Ginsenoside Rd by hydrolyzing glucose at C-3 glycoside using β-glucosidase of Bifidobacterium breve ATCC 15700. J. Microbiol. Biotechnol. 29, 410–418. doi: 10.4014/jmb.1808.08059, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang J., Lacroix C., Wortmann E., Ruscheweyh H. J., Sunagawa S., Sturla S. J., et al. (2019). Gut microbial beta-glucuronidase and glycerol/diol dehydratase activity contribute to dietary heterocyclic amine biotransformation. BMC Microbiol. 19:99. doi: 10.1186/s12866-019-1483-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ma X., Fan D. (2022). Ginsenoside CK ameliorates hepatic lipid accumulation via activating the LKB1/AMPK pathway in vitro and in vivo. Food Funct. 13, 1153–1167. doi: 10.1039/d1fo03026d, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang M., Peng C. S., Li X. B. (2015). In vivo and in vitro metabolites from the main diester and monoester diterpenoid alkaloids in a traditional Chinese herb, the aconitum species. Evid. Based Complement. Alternat. Med. 2015:252434. doi: 10.1155/2015/252434, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Peng C. S., Li X. B. (2017). Human intestine and liver microsomal metabolic differences between C19-diester and monoester diterpenoid alkaloids from the roots of Aconitum carmichaelii Debx. Toxicol. In Vitro 45, 318–333. doi: 10.1016/j.tiv.2017.09.011, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang S., Wang R., Zhao Y., Tareq F. S., Sang S. (2019). Biotransformation of myricetin: a novel metabolic pathway to produce aminated products in mice. Mol. Nutr. Food Res. 63:e1900203. doi: 10.1002/mnfr.201900203, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang S., Xiao L., Lv L., Sang S. (2020). Trapping methylglyoxal by myricetin and its metabolites in mice. J. Agric. Food Chem. 68, 9408–9414. doi: 10.1021/acs.jafc.0c03471, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang Y., Yang D. H., Zhang Y. T., Chen X. M., Li L. L., Cai S. Q. (2014). Biotransformation on the flavonolignan constituents of Silybi Fructus by an intestinal bacterial strain Eubacterium limosum ZL-II. Fitoterapia 92, 61–71. doi: 10.1016/j.fitote.2013.10.001, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang D., Zhu P., Liu Y., Shu Y., Zhou J. Y., Jiang F., et al. (2019). Total flavone of Abelmoschus manihot ameliorates Crohn's disease by regulating the NF-κB and MAPK signaling pathways. Int. J. Mol. Med. 44, 324–334. doi: 10.3892/ijmm.2019.4180, PMID: [DOI] [PubMed] [Google Scholar]
- Zhao Z. X., Fu J., Ma S. R., Peng R., Yu J. B., Cong L., et al. (2018). Gut-brain axis metabolic pathway regulates antidepressant efficacy of albiflorin. Theranostics 8, 5945–5959. doi: 10.7150/thno.28068, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q., Maynard C. L. (2022). Mucus, commensals, and the immune system. Gut Microbes 14:2041342. doi: 10.1080/19490976.2022.2041342, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Qi Z., Yi L., Li J., Cui Y., Ur Rehman F., et al. (2021). The interaction between gut microbiota and flavonoid extract from Smilax glabra Roxb. and its potent alleviation of fatty liver. Food Funct. 12, 7836–7850. doi: 10.1039/d1fo00727k, PMID: [DOI] [PubMed] [Google Scholar]
- Zhao Y. F., Song F. R., Wang X. Y., Guo X. H., Liu Z. Q., Liu S. Y. (2008). Study on biotransformation of 16-O-demethylaconitine and electrospray ionization mass spectrometry (in Chinese). J. Chem. 05, 525–530. [Google Scholar]
- Zhao H., Wang X. L., Zhang H. L., Li C. D., Wang S. Y. (2011). Production of dihydrodaidzein and dihydrogenistein by a novel oxygen-tolerant bovine rumen bacterium in the presence of atmospheric oxygen. Appl. Microbiol. Biotechnol. 92, 803–813. doi: 10.1007/s00253-011-3278-3, PMID: [DOI] [PubMed] [Google Scholar]
- Zhao M., Xu J., Qian D., Guo J., Jiang S., Shang E. X., et al. (2014). Identification of astilbin metabolites produced by human intestinal bacteria using UPLC-Q-TOF/MS. Biomed. Chromatogr. 28, 1024–1029. doi: 10.1002/bmc.3111, PMID: [DOI] [PubMed] [Google Scholar]
- Zheng W., Zhao S., Yin Y., Zhang H., Needham D. M., Evans E. D., et al. (2022). High-throughput, single-microbe genomics with strain resolution, applied to a human gut microbiome. Science 376:eabm1483. doi: 10.1126/science.abm1483, PMID: [DOI] [PubMed] [Google Scholar]
- Zhong F. L., Ma R., Jiang M., Dong W. W., Jiang J., Wu S., et al. (2016). Cloning and characterization of ginsenoside-hydrolyzing β-glucosidase from lactobacillus brevis that transforms ginsenosides Rb1 and F2 into ginsenoside Rd and compound K. J. Microbiol. Biotechnol. 26, 1661–1667. doi: 10.4014/jmb.1605.05052, PMID: [DOI] [PubMed] [Google Scholar]
- Zhou J., Chen Y., Wang Y., Gao X., Qu D., Liu C. (2013). A comparative study on the metabolism of Epimedium koreanum Nakai-prenylated flavonoids in rats by an intestinal enzyme (lactase phlorizin hydrolase) and intestinal flora. Molecules 19, 177–203. doi: 10.3390/molecules19010177, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M., Zimmermann-Kogadeeva M., Wegmann R., Goodman A. L. (2019). Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570, 462–467. doi: 10.1038/s41586-019-1291-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal E. G., Raes J., van den Bogert B., Arumugam M., Booijink C. C., Troost F. J., et al. (2012). The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 6, 1415–1426. doi: 10.1038/ismej.2011.212, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]