Abstract
The control of infectious diseases is seriously threatened by the increase in the number of microorganisms resistant to antimicrobial agents. Antibiotic-resistant bacteria have also been identified in the water environment. A field study was performed sampling drinking water sources in seven districts of southern Ghana targeting boreholes, dams, hand-dug wells, and streams during baseflow conditions. Bacteria were isolated (N = 110) from a total of 67 water samples to investigate their antimicrobial susceptibility and to determine their carriage of select antibiotic resistance genes. Bacterial identification was performed using conventional selective media methods and the analytical profile index (API) method. Antibiotic susceptibility tests were carried out using the Kirby–Bauer method. Results indicated that all water sources tested were of poor quality based on the presence of fecal indicator organisms. The most commonly occurring bacterium isolated from water was Klebsiella spp. (N = 24, 21.8%), followed by E. coli (N = 23, 20.9%). Gram-negative bacteria isolates were most commonly resistant to cefuroxime (24.5%), while the Gram-positives were most commonly resistant to meropenem (21.3%). The highest rates of bacterial resistances to more than one antibiotic were observed in Klebsiella spp. (30.0%) followed by E. coli (27.8%). PCR was used to detect the presence of a select antibiotic resistance genes in the Gram-negative isolates. The presence of blaNDM-1, sull, tet(O), and tet(W) were observed in isolates from all water sources. In contrast, ermF was not detected in any of the Gram-negative isolates from any water source. Most (28.7%) of the resistance genes were observed in E. coli isolates. Reducing microbial contamination of the various water sources is needed to protect public health and to ensure the sustainability of this resource. This further calls for education of the citizenry.
1. Introduction
Good quality water is vital for human health, which directly relates to the socioeconomic progress of a country [8, 78]. It is also critical towards the attainment of UN Sustainable Development Goal (SDG) number six, which is aimed at ensuring the availability and sustainable management of water and sanitation for all. However, clean fresh water today is a scarce resource, particularly in the developing world. Globally, 663 million people do not have access to safe water [23]. Rural communities in Sub-Saharan Africa account for more than 50% of those people [27, 57]. Most of these communities, therefore, rely on untreated sources such as streams, dams, boreholes, wells, and rivers to meet fundamental needs such as drinking, sanitation, cooking, and for their sustainable development [54, 58]. Ghana missed the United Nations Millennium Development target on sanitation [11], which has a direct impact on food safety and security as well [40].
The global burden of waterborne disease is further complicated by climate change altering the patterns of disease, and more importantly by the increasing occurrences of antibiotic resistance observed both in the clinic and in the water environment (Gimelli, et al., 2018). Antibiotic-resistant genes (ARGs) conferring resistance to a wide variety of antibiotics have been identified in a large range of water environments, including drinking water in both developed and developing countries (e.g., [42, 46]. The major risk for public health is that resistance genes can be transferred from environmental bacteria to human pathogens. With this, antimicrobial resistance has become an important public health issue globally [14, 64]. Antimicrobial-resistant bacteria (ARB) and ARGs have therefore been considered environmental contaminants with widespread distribution in various environments, including water sources and drinking water systems [14, 77]. Importantly, the rapid and widespread increase of new ARB and ARGs all over the world has accelerated in recent years often associated with an increase in the discharge of antibiotics and other pollutants into the environment [64]; Schafhauser et al., 2015). In light of this, antimicrobial resistance has become an important theme in environmental and health science.
Several studies in Sub-Saharan African countries have reported the presence of antibiotic-resistant strains of bacteria, all showing high levels of resistance to antimicrobial agents [25, 39, 72]. In Ghana specifically, there is some evidence of increasing bacteria resistance to antibiotics. However, these data are skewed towards clinical isolates [28, 48, 79]. Thus, there is a paucity of data on environmental antibiotic resistance. It is worth noting that in Ghana, antibiotics are readily available over the counter without a doctor's prescription. Antibiotics are widely used in agriculture, particular for livestock rearing [79].
Unfortunately, environmental monitoring in Ghana is hardly part of the treatment and advocacy process for these diseases, nor for water quality. This surveillance is critical to public health and safety, as it contributes and supports improvements in water quality and antimicrobial resistance control (Hope et al., 2020; Gara, et al., 2018.) Furthermore, antimicrobial resistance (AMR) data could inform decisions and raise awareness among stakeholders and policymakers. Surveillance of AMR under a ‘One Health' framework is thus needed to provide data for awareness and decision making and to enhance understanding of links between environmental and clinical AMR. Therefore, the objectives of this study were as follows: (1) determine the occurrence and identity of bacteria in Ghanaian drinking water sources, (2) determine antibiotic susceptibility profiles of the bacterial isolates, and (3) determine the resistance genes associated with the bacteria.
2. Material and Methods
2.1. Sample Collection Sites
Seven communities were then identified and selected for sampling after several preliminary visits were made to communities across the study area (Figure 1). The sampling sites in each community comprised of four different water sources as follows: boreholes (typical depths > 5 m to 50m), dams, hand-dug wells (typical depths < 1 m to 3 m), and streams. Table 1 shows details of the water sampled.
Figure 1.
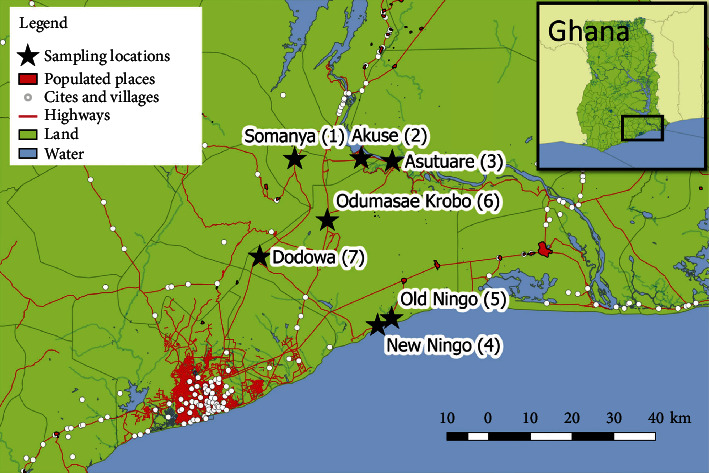
Map of sampling communities and sampling location number (L number). Insert map shows location of the study area within Ghana.
Table 1.
Composition of sampling locations and frequency.
| Water source | Sampling locations | |||||||
|---|---|---|---|---|---|---|---|---|
| L1 | L2 | L3 | L4 | L5 | L6 | L7 | Total | |
| Borehole | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 14 |
| Dam | 2 | 2 | 2 | 2 | 2 | 3 | 3 | 16 |
| Hand-dug well | 3 | 2 | 2 | 3 | 2 | 3 | 3 | 18 |
| Streams | 2 | 2 | 3 | 3 | 3 | 3 | 3 | 19 |
| Total | 9 | 8 | 9 | 10 | 9 | 11 | 11 | 67 |
L1: Somanya; L2: Akuse; L3: Asutsuare; L4: New Ningo; L5: Old Ningo; L6: Odumase Krobo; L7: Dodowa.
Sample sites were chosen to be representative of community water sources based primarily on factors such as popular water locations, extent of usage, and level of patronage of water from these sources. Prior to water sampling, observations were made around the sampling sites. These observations included the sanitary conditions, as well as possible sources of contamination which could influence water quality from the sources sampled. Field records for environmental factors, presence of animals, and fecal accidents, among others, were noted.
2.2. Sample Size and Sampling Frequency
A total of sixty-seven composite water samples were collected for assessment. One liter of water was collected in each sample. Water samples were taken in duplicates from each sample site to form the composite sample for analysis. All water sampling and preservation procedures were performed according to Standard Methods for the Examination of Water and Wastewater [9, 10] and WHO guidelines for drinking water quality [16, 33]. Sampling for bacteriological analysis was performed aseptically. Field blanks consisted of autoclaved distilled water, carried into the field, and analyzed to ensure that the samples were not contaminated during the sampling process. All samples were transported on ice to the laboratory within 2 hours.
2.3. Bacterial Isolation and Identification
Isolates from overnight cultures of the water samples were further characterized by streaking on MacConkey agar and incubated overnight at 37°C. This was done to obtain pure isolates prior to identification. All Gram-positive bacteria were identified by conventional methods including Gram stain, positive catalase, tube coagulase, and deoxyribonucleases (DNAse) test [32]. An API 20E kit was used to identify and differentiate the Gram-negative bacteria of the family Enterobacteriaceae following the manufacturer's instructions.
2.4. Antibacterial Susceptibility Testing
Each of the bacterial isolates was subjected to antibiotic susceptibility testing using the Kirby–Bauer method that has been standardized and evaluated by the methods of Clinical and Laboratory Standards Institute (CLSI) [30]. Isolates grown overnight on nutrient agar were suspended in sterile normal saline (0.9% w/v NaCl) using a sterile wire loop until the turbidity was equal to 0.5 Mcfarland standards. Sterile nontoxic cotton swabs dipped into the standardized inoculum were used to streak the entire surface of Mueller–Hinton agar plates. Gram-positive bacteria were tested against 12 antibiotics as follows: ampicillin (10 μg), cloxacillin (10 μg), erythromycin (15 μg), tetracycline (30 μg), cotrimoxazole (25 μg), cefuroxime (30 μg), gentamicin (10 μg), penicillin (10 IU), ciprofloxacin (5 μg), augmentin (30 μg), vancomycin (30 μg), and meropenem (25 μg). Gram-negative bacteria were tested against 11 antibiotics as follows: ampicillin (10 μg), tetracycline (30 μg), cotrimoxazole (25 μg), cefuroxime (30 μg), chloramphenicol (30 μg), ceftriaxone (25 μg), cefotaxime (30 μg), ciprofloxacin (5 μg), amikacin (30 μg), vancomycin (30 μg), and meropenem (25 μg). Antibiotic disks were aseptically placed using sterile forceps, and all plates were incubated at 37°C for 24 hrs. The results were interpreted using CLSI [76]. The susceptibility testing was repeated for each isolate to ensure that the results obtained were consistent.
2.5. DNA Extraction and PCR
Bacterial cultures stored Mueller–Hinton broth were extracted for identification of ARGs. To extract DNA, 1 mL of the suspension was transferred into a test tube containing 1 mL of sterile molecular biology grade deionized water (ddH2O) and heat shocked for 10 minutes at 95°C in a water bath. A negative control of ddH2O was simultaneously processed and tested to assess possible contamination during the DNA extraction. The solution was then centrifuged for five minutes at 14,000 × g. The supernatant containing bacterial DNA was transferred to a new 2 mL tube and stored at -20°C for downstream molecular analysis.
Polymerase chain reaction (PCR) targeting the bacterial 16S rRNA gene (Table 2) was used to confirm the DNA extraction method. PCR targeting eight different ARGs (ermF, mexB, blandm1, sul1, sul2, tet(G), tet(O), and tet(W)) was used to determine the presence or absence of each gene in all Gram-negative bacterial isolates. ermF encodes for macrolide resistance, mexB for a multidrug efflux pump, blandm1 encodes for a metallobetalactamase, the sul genes for sulfonamide resistance, and the tet genes for resistance to tetracycline antibiotics. These genes were chosen to represent a range of resistance types including some that are commonly observed (i.e., sul and tet genes) in environmental matrices and some of high medical relevance (i.e., blandm1). The PCR mixture for each reaction contained 12.5 μL of 2 × Taq PCR Master Mix ((0.1 U Taq polymerase/μL, 0.5 mM dNTP, and 3 MgCl2,), 0.5 μL of each primer (1 μM), 1 μL of template DNA, and ddH2O to a final volume of 25 μL. No template controls were performed during each PCR reaction. Positive controls were comprised of 1 μL containing 105 copies of DNA standards for each ARG, quantified with gel electrophoresis. PCR thermocycler condition and primer sequences for each ARG and the 16S rRNA gene are summarized in Table 2. The presence or absence of a given target gene was assessed through observation of the expected amplicon length (determined using a DNA ladder) via gel electrophoresis.
Table 2.
Primer sequences, PCR conditions, amplicon lengths for each ARG, and 16S rRNA gene.
| Gene | Primer sequence (5′-3′) | PCR conditions | Amplicon length (bp) | Source |
|---|---|---|---|---|
| sul1 | CGCACCGGAAACATCGCTGCAC | 95°C for 2 m (98°C for 5 s, 69.9°C for 5 s) ×40 cycles | 163 | 8 |
| TGAAGTTCCGCCGCAAGGCTCG | ||||
| sul2 | TCC GGT GGA GGC CGG TAT CTG G | 95°C for 2 m (98°C for 5 s, 65°C for 5 s) ×40 cycles | 191 | 8 |
| CGG GAA TGC CAT CTG CCT TGA G | ||||
| tet(G) | GCAGAGCAGGTCGCTGG | 98°C for 2 m (98°C for 5 s, 64°C for 5 s) ×40 cycles | 134 | 9 |
| CCYGCAAGAGAAGCCAGAAG | ||||
| ermF | CGACACAGCTTTGGTTGAAC GGACCTACCTCATAGACAAG |
95°C for 4 m (94°C for 30 s, 56°C for 30 s and 72°C for 30 s) ×40 cycles | 309 | 10 |
| tet(O) | ACGGARAGTTTATTGTATACC TGGCGTATCTATAATGTTGAC |
98°C for 2 m (98°C for 5 s, 50°C for 5 s) ×40 cycles | 171 | 12 |
| tet(W) | GAGAGCCTGCTATATGCCAGC GGGCGTATCCACAATGTTAAC |
98°C for 2 m (98°C for 5 s, 60°C for 5 s) ×40 cycles | 168 | 12 |
| blaNDM-1 | TTTCAGTCCGACACAACGCG CAGCCACCAAAAGCGATGTC 6-FAM-CAACCGCGCCCAACTTTGGC-TAMRA |
98°C for 15 m (98°C for 30 s, 59°C for 1 m) ×40 cycles | 155 | 6 |
| 16S rRNA | CCTACGGGAGGCAGCAG ATTACCGCGGCTGCTGG |
95°C for 10 m (95°C for 15 s, 60°C for 1 m) ×40 cycles | 202 | 14 |
2.6. Statistical Analysis
All the statistical analyzes were performed in R (R Team, 2018). To compare the genomic antibiotic resistance profile as a factor water sources, a value of one was assigned to the detected genes and a value of zero was assigned to the nondetected genes to create a presence/absence antibiotic resistance profile for each isolate. Statistical differences were determined through a permutational multivariate analysis of variance (PERMANOVA) and a post hoc pairwise PERMANOVA with a Bonferroni p adjustment: Pairwise Adonis package version 0.3 [12, 34]. Differences in multiple antibiotic resistance (MAR) indices as a factor of water source were evaluated using a Kruskal–Wallis test with a post hoc pairwise t-test with a Bonferroni correction for multiple comparisons. Nonnormality of the data was confirmed by a Shapiro-Wilk test.
3. Results
A total of 110 bacteria isolates were obtained across all of the water sources sampled during the period of study (Table 3). The most commonly occurring bacterium isolated from the water samples was Klebsiella spp. with 24 isolates (21.8% of total study isolates). The second most commonly observed bacteria was E. coli. with 23 isolates (20.9% of total study isolates). The highest number of bacterial isolates were obtained from stream water sources with 42 isolates obtained (38.2% of total study isolates), while the least were isolated from borehole water sources with nine isolates (8.2% of the total study isolates).
Table 3.
Occurrence and distribution of bacteria isolated from the drinking water sources.
| Bacteria | Boreholes | Dams | Hand-dug wells | Streams | Total | |
|---|---|---|---|---|---|---|
| No. | (%) | |||||
| Acinetobacter spp. | 0 | 2 | 1 | 3 | 6 | 5.5 |
| Bacillus spp. | 0 | 0 | 1 | 0 | 1 | 0.9 |
| Citrobacter freundii | 0 | 1 | 0 | 1 | 2 | 1.8 |
| Enterobacter spp. | 1 | 3 | 2 | 5 | 11 | 10.0 |
| Enterococcus spp. | 0 | 1 | 0 | 0 | 1 | 0.9 |
| Escherichia coli | 3 | 7 | 4 | 9 | 23 | 20.9 |
| Klebsiella spp. | 1 | 9 | 2 | 12 | 24 | 21.8 |
| Proteus vulgaris | 0 | 1 | 0 | 1 | 2 | 1.8 |
| Providencia spp. | 0 | 1 | 0 | 1 | 2 | 1.8 |
| Pseudomonas aeruginosa | 0 | 2 | 1 | 2 | 5 | 4.5 |
| Salmonella spp. | 0 | 0 | 0 | 1 | 1 | 0.9 |
| Staphylococcus aureus | 3 | 4 | 5 | 4 | 16 | 14.5 |
| Staphylococcus epidermidis | 0 | 2 | 3 | 1 | 6 | 5.5 |
| Streptococcus agalactiae | 0 | 2 | 0 | 1 | 3 | 2.7 |
| Vibrio spp. | 1 | 2 | 3 | 1 | 7 | 6.4 |
| Total | 9 (8.2)∗ | 37 (33.6) | 22 (20.0) | 42 (38.2) | 110 | 100.0 |
∗Number in parentheses represents the percent of the total isolates obtained.
The antibiotic resistance profiles observed are presented in Tables 4 and 5 for Gram-negative and Gram-positive bacteria isolated, respectively. Similar MAR values were detected among the isolated bacterial taxa with N = 3 or higher (p > 0.14, pairwise t-test) The Gram-negative bacterial isolates were most commonly resistant to CRX (cefuroxime) (24.5%) followed by cefotaxime and MEM (meropenem) with each exhibiting 21.3% resistance. Klebsiella spp. isolates had phenotypic resistance most often (30.0%) followed by Escherichia coli (27.8%). The Gram-positive bacterial isolates were commonly resistant to MEM (21.3%) followed by VAN and AUG (both 17%). Among these isolates, multidrug resistance was most common among S. aureus (59.6%) and S. epidermis (36.2%).
Table 4.
Antibiotic resistance patterns of Gram-negative bacteria isolated from the water sources.
| Isolate | Pattern of antibiotic resistance: (number of resistant strains per antibiotic) | Multiple resistances | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | TET | COT | CRX | CHL | CTR | CTX | CIP | AMK | VAN | MEM | No. | % | MAR | |
| Acinetobacter spp. (n = 6) | 0 | 0 | 0 | 3 | 0 | 2 | 2 | 0 | 0 | 2 | 5 | 14 | 6.5 | 1.3 |
| Citrobacter freundii (n = 2) | 1 | 0 | 1 | 2 | 0 | 1 | 1 | 0 | 0 | 1 | 2 | 9 | 4.2 | 0.8 |
| Enterobacter spp. (n = 11) | 2 | 2 | 1 | 7 | 1 | 2 | 6 | 0 | 0 | 2 | 4 | 27 | 12.5 | 2.5 |
| Escherichia coli (n = 23) | 5 | 3 | 4 | 12 | 0 | 4 | 14 | 0 | 0 | 4 | 14 | 60 | 27.8 | 5.5 |
| Klebsiella spp. (n = 24) | 1 | 1 | 5 | 16 | 0 | 4 | 17 | 0 | 0 | 7 | 14 | 65 | 30.1 | 5.9 |
| Proteus vulgaris (n = 2) | 0 | 0 | 0 | 2 | 0 | 1 | 2 | 0 | 0 | 0 | 1 | 6 | 2.8 | 0.5 |
| Providencia spp. (n = 2) | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0.9 | 0.2 |
| Pseudomonas aeruginosa (n = 5) | 2 | 1 | 1 | 5 | 0 | 2 | 2 | 0 | 0 | 2 | 4 | 19 | 8.8 | 1.7 |
| Salmonella spp. (n = 1) | 0 | 1 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 6 | 2.8 | 0.5 |
| Vibro spp. (n = 7) | 1 | 0 | 1 | 4 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 8 | 3.7 | 0.7 |
| Total | 12 (5.6) | 9 (4.2) | 13 (6.0) | 53 (24.5) | 1 (0.5) | 17 (7.9) | 46 (21.3) | 0 (0.0) | 0 (0.0) | 19 (8.8) | 46 (21.3) | 216 | 100 | |
Key: AMP: ampicillin; TET: tetracycline; CRX: cefuroxime; CHL: chloramphenicol; CTR: ceftriaxone; CTX: cefotaxime; CIP: ciprofloxacin; AMK: amikacin; VAN: vancomycin; MEM: meropenem.
Table 5.
Antibiotic resistance patterns of Gram-positive bacteria isolated from the water sources.
| Isolate | Pattern of antibiotic resistance: (number of resistant strains per antibiotic) | Multiple resistances | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP | COX | ERY | TET | COT | CRX | GEN | PEN | CIP | AUG | VAN | MEM | No. | % | MAR | |
| Bacillus spp. (n = 1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 | 0.0 |
| Enterococcus spp. (n = 1) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0 | 0.0 |
| Staphylococcus aureus (n = 16) | 1 | 4 | 1 | 1 | 1 | 2 | 0 | 1 | 1 | 5 | 5 | 6 | 28 | 59.6 | 2.3 |
| Staphylococcus epidermidis (n = 6) | 0 | 3 | 0 | 1 | 0 | 3 | 0 | 0 | 0 | 3 | 3 | 4 | 17 | 36.2 | 1.4 |
| Streptococcus agalactiae (n = 3) | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 4.3 | 0.2 |
| Total | 1 (2.1) | 7 (14.9) | 1 (2.1) | 2 (4.3) | 1 (2.1) | 7 (14.9) | 0 (0.0) | 1 (2.1) | 1 (2.1) | 8 (17.0) | 8 (17.0) | 10 (21.3) | 47 | 100 | |
Key: AMP: ampicillin; COX: cloxacillin; ERY: erythromycin; TET: tetracycline; COT: cotrimoxazole; CRX: cefuroxime; GEN: gentamicin; PEN: penicillin; CHL: chloramphenicol; CTR: ceftriaxone; CTX: cefotaxime; CIP: ciprofloxacin; AUG: augmentin; VAN: vancomycin; MEM: meropenem.
MAR indices of the bacterial isolates were determined for the various water sources (Table 6). The multiple antibiotic resistance (MAR) index is defined as a/b, where a represents the number of antibiotics to which the isolate was resistant and b represents the number of antibiotics to which the isolate was subjected. The aggregate MAR index for a sampling sources (MAR q) is defined as the ratio between the number of resistant tests at the sampling sources and the total number of tests performed at the sampling source. Stream water sources recorded the highest MAR q values of 0.9. This was followed by a MAR q for the dam water sources which was 0.8 and hand-dug well water sources with a recorded value of 0.6. Stream water sources resulted in a significantly higher MAR value than hand-dug well (p = 0.01, pairwise t-test); however, no other differences were detected as in MAR values as a factor of source.
Table 6.
Multiple antibiotic-resistant indexes of bacteria isolate at various water sources.
| Water source | Total numbers of test (isolates) | No. of resistant test (resistant isolates) | MAR q | ||||
|---|---|---|---|---|---|---|---|
| Gram negative | Gram positive | Total | Gram negative | Gram positive | Total | ||
| Borehole | 6 | 3 | 9 | 5 | 1 | 6 | 0.7 |
| Dam | 29 | 8 | 37 | 26 | 3 | 29 | 0.8 |
| Hand-dug well | 13 | 9 | 22 | 12 | 2 | 14 | 0.6 |
| Streams | 36 | 6 | 42 | 31 | 5 | 36 | 0.9 |
| Total | 84 | 26 | 110 | 74 | 11 | 85 | |
MAR q: MAR index per sampling source.
The presence of eight (8) different antibiotic resistance genes were tested for each Gram-negative bacterium isolated from the samples using PCR amplification (Table 7). blaNDM-1, sul1, tet(O), and tet(W) resistance genes were detected in isolates collected from all water sources, while ermF was not detected in isolates from any of the water sources. The number of genes amplified from isolates from each water source is presented in Table 7. All water sources resulted in a similar antibiotic resistance profile (p = 1; pairwise PERMANOVA) except for dam compared to the hand-dug well which resulted in a significantly different profile (p < 0.006; pairwise PERMANOVA). The MAR index was similar among all water sources (p > 0.16; pairwise t-test).
Table 7.
PCR detection of antibiotic resistance genes in DNA extracted from bacteria isolates at different water sources.
| Water source | No. of test isolates | ARGs | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ermF | mexB | bla NDM-1 | sul1 | sul2 | tet(G) | tet(O) | tet(W) | Total | |||
| No. | % | ||||||||||
| Borehole | 6 | 0 | 0 | 5 | 6 | 0 | 0 | 1 | 1 | 13 | 7.47 |
| Dam | 29 | 0 | 2 | 16 | 28 | 2 | 2 | 1 | 2 | 53 | 30.46 |
| Hand-dug well | 13 | 0 | 0 | 9 | 13 | 0 | 1 | 1 | 1 | 25 | 14.37 |
| Streams | 36 | 0 | 7 | 31 | 33 | 5 | 3 | 2 | 2 | 83 | 47.70 |
| Total | 84 | 0 (0.0) | 9 (5.17) | 61 (35.06) | 80 (45.98) | 7 (4.02) | 6 (3.45) | 5 (2.87) | 6 (3.45) | 174 | 100 |
An inventory of ARGs identified in the bacterial isolates is presented in Table 8. Note that most (28.7%) of the resistance genes were obtained from Escherichia coli (E. coli) isolates. This was followed by Klebsiella spp. with 27.6%. Enterobacter spp. accounted for 12.6% of the resistance genes observed, representing the third highest. The most frequently detected ARG was sul1, which was observed in 46% of isolates tested. This was followed by blaNDM-1, identified in 35.1% of isolates tested.
Table 8.
Inventory of antibiotic resistance genes identified in each bacteria isolate.
| Isolate | ARGs | Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ermF | mexB | bla NDM-1 | sul1 | sul2 | tet(G) | tet(O) | tet(W) | No. | % | |
| Acinetobacter spp. | 0 | 0 | 5 | 6 | 0 | 0 | 1 | 0 | 12 | 6.9 |
| Citrobacter freundii | 0 | 1 | 1 | 2 | 0 | 1 | 0 | 0 | 5 | 2.9 |
| Enterobacter spp. | 0 | 1 | 9 | 11 | 0 | 1 | 0 | 0 | 22 | 12.6 |
| Enterococcus spp. | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0.6 |
| Escherichia coli | 0 | 3 | 17 | 23 | 2 | 1 | 1 | 3 | 50 | 28.7 |
| Klebsiella spp. | 0 | 2 | 17 | 22 | 1 | 3 | 2 | 1 | 48 | 27.6 |
| Proteus vulgaris | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 3 | 1.7 |
| Providencia spp. | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 4 | 2.3 |
| Pseudomonas aeruginosa | 0 | 1 | 4 | 5 | 0 | 0 | 1 | 1 | 12 | 6.9 |
| Salmonella spp. | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 2 | 1.1 |
| Vibrio spp. | 0 | 0 | 6 | 5 | 4 | 0 | 0 | 0 | 15 | 8.6 |
| Total | 0 | 9 | 61 | 80 | 7 | 6 | 5 | 6 | 174 | 100.0 |
4. Discussion
4.1. Isolate Observations and Implications for Microbial Water Quality
In the present study, several bacteria of public health importance were identified in Ghanaian water sources: Salmonella spp. (typhoid fever and acute diarrheal infection), Vibrio spp. (cholera), and Klebsiella spp. (pneumonia and urinary/lower biliary tract disease [62]). Vibrio spp. and Klebiella spp. were isolated from each water source type in the present study while Salmonella spp. were isolated from the streams. Similar reports of the isolation of these organisms were made by Moges et al. [47] and also by Shahina et al. [69], from an assessment of ground and surface water sources in India.
The most commonly isolated bacteria were E. coli which were isolated from each type of water source. While the aim of this study was not quantifying E. coli in the water sources, it is worth noting that detection of one E. coli CFU per 100 mL is in exceedance of the World Health Organization's guidance values for water intended for drinking [73]. E. coli is widespread in the environment but elevated levels are indicative of fecal pollution [29] and the prevalence of water-related gastroenteritis [61]. While most E. coli strains are not pathogenic, given that some strains are pathogenic they have been used for disease risk assessment [53]. Recent studies have shown that rural water sources in Ghana have high occurrences of coliforms [49, 51, 55], another fecal indicating organism. Similarly, Nogueira et al. [50] reported that untreated water sources were more deeply contaminated with fecal coliforms than treated water sources.
4.2. Antimicrobial Resistance among Isolates
The current study also evaluated both phenotypic and genotypic antibiotic resistance among the waterborne isolates. Phenotypically, we observed multiple antibiotic resistance in both Gram-positive and Gram-negative isolates to commonly used antibiotics in the study area [4, 38]. These antibiotics include ampicillin, cloxacillin, erythromycin, tetracycline, cotrimoxazole, cefuroxime, gentamicin, penicillin, ciprofloxacin, augmentin, vancomycin, meropenem, chloramphenicol, ceftriaxone, amikacin, and meropenem [38]. Data from hospital surveys [38] in rural Ghana indicate that two of the antibiotics tested here were among top five most frequently prescribed (i.e., ceftriaxone and cefuroxime). Despite legal COT sales over-the-counter in Ghana, COT resistance was observed at a lower prevalence than several other antibiotics tested. The fact that illegal sales of other antibiotics from Licensed Chemical Sellers is known [4] may explain why the most available antibiotic was not associated with the most commonly observed phenotypic resistance.
In terms of Gram-positive isolates, S. aureus accounted for 59.6% of all multidrug resistances observed in the Gram-positive isolates. S. aureus also had a high MAR value of 2.3. S. aureus isolates were resistant to 12 of the antibiotics it was tested against, all except gentamicin. Multidrug-resistant S. aureus occurs commonly and has been observed in several diverse environments, including drinking water and food, indicating an important public health concern [2].
Multiple drug resistance was also commonly observed in Gram-negative bacteria. Klebsiella spp. and Escherichia coli showed a high prevalence of resistance to cefuroxime and cefotaxime. Likewise, a high prevalence of resistance to cefuroxime and cefotaxime has been recorded from clinical isolates in Ghana [5, 37]: [56]. We observed low resistance to ampicillin in contrast to findings from Moges et al. [47] who observed all isolates of Klebsiella spp. and Escherichia coli were resistant to ampicillin. Interestingly, we did not observe any bacterial resistance to ciprofloxacin. This observation is in contrast to another study done in Bangladesh, where 100% of waterborne Gram-negative bacteria isolates were resistant to ciprofloxacin [31], and in a similar study in Nigeria, where 54.7% of Gram-negative isolates obtained from water were found to resistant to ciprofloxacin (Ojayi and Ojo 2018).
In this current study, we also investigated the aggregate MAR index for sampling sources (MAR q). A number of factors could be responsible for the resistance observed at the sampling sites. For example, we made an interesting observation of the presences of two bacteria of public health importance Shigella spp. and Salmonella typhi, known to cause dysentery and typhoid fever/acute diarrheal infection, respectively [63] from ground water sources (boreholes and hand-dug wells). After a careful assessment of location, we discovered that the boreholes and hand-dug wells in question were likely contaminated with these enteric bacteria from the rural public ground toilet systems that were situated at an average of 50 meters from the location of those ground water sources sampled.
The MAR indices of isolates from surface water sources were comparable with those of previous studies [7, 17, 60]. Similar to the results presented here, Tambekar et al. [70] reported high MAR indices due to human and nonhuman fecal contamination of surface, ground, and public supply water sites in Akola and Buldhana of Vidarbha district. Likewise, a similar study by Chatterjee et al. [19] noted that drinking water sources of Uttarakhand region were contaminated with high MAR index E. coli originating from potential risk sources.
4.3. Antimicrobial Resistance among Isolates
PCR was performed to detect the presence of eight ARGs. The genes were determined for each Gram-negative bacterium isolated and for each sampled water source (Table 6). sul1 and blaNDM-1 resistance genes was found to be prevalent in isolates (46% and 31%, respectively) from all sampled water sources. The high prevalence of sul1 is not surprising; this gene is frequently observed in bulk environmental samples thus a suggested target for monitoring efforts (Vikesland et al., 2017). Tetracycline-resistant genes tet(O) and tet(W) were infrequently observed in isolates across water sources, which is contrast with similar studies by Chee-Sanford et al. [20] where tet(O) and tet(Q) were found to be regularly present in resistance isolates from water sources. However, in another study by Adesoji et al. [3], tet(O) was not detected in any of the isolates from water sources. Of particular interest is the frequent observation of the blaNDM-1 gene (35.1% of tested isolates). The blaNDM-1 gene is known to be transferred between bacterial genera ([36, 80] and has been reported in sewage and surface water isolates [36].
5. Conclusions
Bacteria associated with fecal contamination were observed in various water sources in the study communities. The study demonstrated multiple drug resistance to the commonly used antibiotics is high in rural communities in Ghana. Antibiotic-resistant bacteria were found to carry several antimicrobial-resistant genes. High MAR index values recorded in the study indicate a potential hazard associated with the sampled water sources, particularly the surface waters studied, potentially due poor sanitation contaminating the surface waters. This observation raises concern about water quality in rural Ghana and indicates a need to understand the sources of fecal and other contamination (i.e., human sewage) and opportunities for water treatment. Further investigation into the sources of the fecal microbes observed and measurements of resistance genes across in the source water microbiome could help provide further insight into the overall hazard posed by the waters tested and opportunities for mitigation. Future studies should include collection of other water quality parameters to understand any relationships between the water chemistry and ARGs or high MAR indices. Finally, the deployment of tools such as microbial source tracking for periodic monitoring of antibiotic sensitivity of the water sources is of importance to detect any changing patterns that may arise in the future. Further work in these areas may allow for the creation of more curative measures towards better management of water resources.
Acknowledgments
Funding for this study was provided by a Fulbright Fellowship to S.T.O. This work was also supported by a grant from the National Science Foundation 1510461 to support WRMM's time and 1846815 for supplies.
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Abdelgader S. A., Shi D., Chen M., et al. Antibiotics Resistance Genes Screening and Comparative Genomics Analysis of Commensal _Escherichia coli_ Isolated from Poultry Farms between China and Sudan. BioMed research international . 2018;2018:9. doi: 10.1155/2018/5327450.5327450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abulreesh H. H. Multidrug-resistant staphylococci in the environment. In International Conference on Biotechnology and Environment Management; 2011; pp. 1–6. [Google Scholar]
- 3.Adesoji A. T., Ogunjobi A. A., Olatoye I. O., Douglas D. R. Prevalence of tetracycline resistance genes among multi-drug resistant bacteria from selected water distribution systems in southwestern Nigeria. Annals of Clinical Microbiology and Antimicrobials . 2015;14(1):1–8. doi: 10.1186/s12941-015-0093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afari-Asiedu S., Kinsman J., Boamah-Kaali E., et al. To sell or not to sell; the differences between regulatory and community demands regarding access to antibiotics in rural Ghana. Journal of Pharmaceutical Policy and Practice . 2018;11(1):1–10. doi: 10.1186/s40545-018-0158-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agyepong N., Govinden U., Owusu-Ofori A., Essack S. Y. Multidrug-resistant gram-negative bacterial infections in a teaching hospital in Ghana. Antimicrobial Resistance & Infection Control . 2018;7(1):p. 37. doi: 10.1186/s13756-018-0324-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ojayi A. O., Ojo B. O. Antibiotics susceptibility profile of microorganisms encountered in riverine areas of Ondo state, Nigeria. Journal of Disease and Global Health . 2016;7(2):100–107. [Google Scholar]
- 7.Akturk S., Dincer S., Toroglu S. Determination of microbial quality and plasmid mediated multidrug resistant bacteria in fountain drinking water sources in Turkey. Journal of Environmental Biology . 2012;33:1127–1136. [PubMed] [Google Scholar]
- 8.Anthonj C., Diekkrüger B., Borgemeister C., Kistemann T. Health risk perceptions and local knowledge of water-related infectious disease exposure among Kenyan wetland communities. International Journal of Hygiene and Environmental Health . 2019;222(1):34–48. doi: 10.1016/j.ijheh.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Apha. Standard Methods for the Examination of Water and Wastewater . 19th edition. Washington, DC, USA: American Public Health Association, American Water Works Association and Water Environment Federation; 1995. [Google Scholar]
- 10.Apha. Standard Methods for the Examination of Water and Wastewater . 20th edition. Baltimore, MA, USA: United Book Press; 1998. [Google Scholar]
- 11.Appiah-Effah E., Duku G. A., Azangbego N. Y., Aggrey R. K. A., Gyapong-Korsah B., Nyarko K. B. Ghana's post-MDGs sanitation situation: an overview. Journal of Water, Sanitation and Hygiene for Development . 2019;9(3):397–415. doi: 10.2166/washdev.2019.031. [DOI] [Google Scholar]
- 12.Arbizu P. M. pairwiseAdonis: pairwise multilevel comparison using adonis . 2019.
- 13.Bengtsson-Palme J., Larsson D. J. Concentrations of antibiotics predicted to select for resistant bacteria: proposed limits for environmental regulation. Environment International . 2016;86:140–149. doi: 10.1016/j.envint.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 14.Berendonk T. U., Manaia C. M., Merlin C., et al. Tackling antibiotic resistance: the environmental framework. Nature Reviews Microbiology . 2015;13(5):310–317. doi: 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- 15.Bergeron S., Raj B., Nathaniel R., Corbin A., LaFleur G. Presence of antibiotic resistance genes in raw source water of a drinking water treatment plant in a rural community of USA. International Biodeterioration & Biodegradation . 2017;124:3–9. doi: 10.1016/j.ibiod.2017.05.024. [DOI] [Google Scholar]
- 16.Bürgmann H., Frigon D., Gaze W. H., et al. Water and sanitation: an essential battlefront in the war on antimicrobial resistance. FEMS microbiology ecology . 2018;94(9) doi: 10.1093/femsec/fiy101. [DOI] [PubMed] [Google Scholar]
- 17.Chandran A., Hatha A. A. M., Varghese S., Sheeja K. M. Prevalence of multiple drug resistant Escherichia coli serotypes in a tropical estuary, India. Microbes and Environments . 2008;23(2):153–158. doi: 10.1264/jsme2.23.153. [DOI] [PubMed] [Google Scholar]
- 18.Chapman D. Water Quality Assessments. A Guide to Use of Biota, Sediments and Water in Environmental Monitoring . 2nd. 1996. E&FN Spon: 417.
- 19.Chatterjee R., Sinha S., Aggarwal S., et al. Studies on susceptibility and resistance patterns of various E. coli isolated from different water samples against clinically significant antibiotics. Int. J Bioassays . 2012;1:156–161. [Google Scholar]
- 20.Chee-Sanford J. C., Aminov R. I., Krapac I. J., Garrigues-Jeanjean N., Mackie R. I. Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Applied and Environmental Microbiology . 2001;67(4):1494–1502. doi: 10.1128/AEM.67.4.1494-1502.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J., Li W., Zhang J., et al. Prevalence of antibiotic resistance genes in drinking water and biofilms: the correlation with the microbial community and opportunistic pathogens. Chemosphere . 2020;259:p. 127483. doi: 10.1016/j.chemosphere.2020.127483. [DOI] [PubMed] [Google Scholar]
- 22.Chitanand M. P., Kadam T. A., Gyananath G., Totewad N. D., Balhal D. K. Multiple antibiotic resistance indexing of coliforms to identify high risk contamination sites in aquatic environment. Indian Journal of Microbiology . 2010;50(2):216–220. doi: 10.1007/s12088-010-0042-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chitonge H., Mokoena A., Kongo M. In Africa and the Sustainable Development Goals . Cham: Springer; 2020. Water and Sanitation Inequality in Africa: Challenges for SDG 6; pp. 207–218. [DOI] [Google Scholar]
- 24.Das B. K., Behera B. K., Chakraborty H. J., et al. Metagenomic study focusing on antibiotic resistance genes from the sediments of River Yamuna. Gene . 2020;758:p. 144951. doi: 10.1016/j.gene.2020.144951. [DOI] [PubMed] [Google Scholar]
- 25.Elton L., Thomason M. J., Tembo J., et al. Antimicrobial resistance preparedness in sub-Saharan African countries. Antimicrobial Resistance & Infection Control . 2020;9(1):1–11. doi: 10.1186/s13756-020-00800-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrer N., Folch A., Masó G., Sanchez S., Sanchez-Vila X. What are the main factors influencing the presence of faecal bacteria pollution in groundwater systems in developing countries? Journal of Contaminant Hydrology . 2020;228:p. 103556. doi: 10.1016/j.jconhyd.2019.103556. [DOI] [PubMed] [Google Scholar]
- 27.Furtatova A., Kamenik L. Modeling features of sustainable urban development in modern conditions of water supply. In MATEC Web of Conferences . 2018;170:p. 04002. doi: 10.1051/matecconf/201817004002. [DOI] [Google Scholar]
- 28.García-Vello P., González-Zorn B., Saba C. K. S. Antibiotic resistance patterns in human, animal, food and environmental isolates in Ghana: a review. The Pan African Medical Journal . 2020;35:p. 37. doi: 10.11604/pamj.2020.35.37.18323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hachich E. M., Di Bari M., Christ A. P. G., Lamparelli C. C., Ramos S. S., Sato M. I. Z. Comparison of thermotolerant coliforms and Escherichia coli densities in freshwater bodies. Brazilian Journal of Microbiology . 2012;43(2):675–681. doi: 10.1590/S1517-83822012000200032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hudzicki J. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol . 2009.
- 31.Islam M. J., Uddin M. S., Hakim M. A., Das K. K., Hasan M. N. Role of untreated liquid hospital waste to the development of antibiotic resistant bacteria. J Innov Dev Strategy . 2008;2(2):17–21. [Google Scholar]
- 32.Ito T., Sekizuka T., Kishi N., Yamashita A., Kuroda M. Conventional culture methods with commercially available media unveil the presence of novel culturable bacteria. Gut Microbes . 2019;10(1):77–91. doi: 10.1080/19490976.2018.1491265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jang J., Hur H. G., Sadowsky M. J., Byappanahalli M. N., Yan T., Ishii S. Environmental Escherichia coli: ecology and public health implications-a review. Journal of Applied Microbiology . 2017;123(3):570–581. doi: 10.1111/jam.13468. [DOI] [PubMed] [Google Scholar]
- 34.Jari Oksanen F. G. B., Friendly M., Kindt R., et al. Community ecology package. R package version 2.5-6 . 2019. https://CRAN.R-project.org/package=vegan .
- 35.Krumperman P. H. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Applied and Environmental Microbiology . 1983;46(1):165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumarasamy K. K., Toleman M. A., Walsh T. R., et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. The Lancet Infectious Diseases . 2010;10(9):597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Labi A. K., Bjerrum S., Enweronu-Laryea C. C., Ayibor P. K., Nielsen K. L., Marvig R. L. High carriage rates of multidrug-resistant gram-negative bacteria in neonatal intensive care units from Ghana . US: Oxford University Press; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Labi A. K., Obeng-Nkrumah N., Nartey E. T., et al. Antibiotic use in a tertiary healthcare facility in Ghana: a point prevalence survey. Antimicrobial Resistance & Infection Control . 2018;7(1):p. 15. doi: 10.1186/s13756-018-0299-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leopold S. J., van Leth F., Tarekegn H., Schultsz C. Antimicrobial drug resistance among clinically relevant bacterial isolates in sub-Saharan Africa: a systematic review. Journal of Antimicrobial Chemotherapy . 2014;69(9):2337–2353. doi: 10.1093/jac/dku176. [DOI] [PubMed] [Google Scholar]
- 40.Lu Y., Song S., Wang R., et al. Impacts of soil and water pollution on food safety and health risks in China. Environment International . 2015;77:5–15. doi: 10.1016/j.envint.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 41.Mahvi A. H., Karyab H. Risk assessment for microbial pollution in drinking water in small community and relation to diarrhoea disease. American-Eurasian Journal of Agricultural and Environmental Science . 2007;2(4):404–406. [Google Scholar]
- 42.Marathe N. P., Pal C., Gaikwad S. S., Jonsson V., Kristiansson E., Larsson D. J. Untreated urban waste contaminates Indian river sediments with resistance genes to last resort antibiotics. Water Research . 2017;124:388–397. doi: 10.1016/j.watres.2017.07.060. [DOI] [PubMed] [Google Scholar]
- 43.Martineau F., Picard F. J., Lansac N., et al. Correlation between the resistance genotype determined by multiplex PCR assays and the antibiotic susceptibility patterns of Staphylococcus aureus and Staphylococcus epidermidis. Antimicrobial Agents and Chemotherapy . 2000;44(2):231–238. doi: 10.1128/AAC.44.2.231-238.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLellan S. L., Eren A. M. Discovering new indicators of fecal pollution. Trends in Microbiology . 2014;22(12):697–706. doi: 10.1016/j.tim.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McLellan S. L., Daniels A. D., Salmore A. K. Clonal populations of thermotolerant Enterobacteriaceae in recreational water and their potential interference with fecal Escherichia coli counts. Applied and Environmental Microbiology . 2001;67(10):4934–4938. doi: 10.1128/AEM.67.10.4934-4938.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mezrioui N., Baleux B. Resistance patterns of E. coli strains isolated from domestic sewage before and after treatment in both aerobic lagoon and activated sludge. Water Research . 1994;28(11):2399–2406. doi: 10.1016/0043-1354(94)90056-6. [DOI] [Google Scholar]
- 47.Moges F., Endris M., Belyhun Y., Worku W. Isolation and characterization of multiple drug resistance bacterial pathogens from waste water in hospital and non-hospital environments, Northwest Ethiopia. BMC Research Notes . 2014;7(1):p. 215. doi: 10.1186/1756-0500-7-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newman M. J., Frimpong E., Donkor E. S., Opintan J. A., Asamoah-Adu A. Resistance to antimicrobial drugs in Ghana. Infection and drug resistance . 2011;4 doi: 10.2147/IDR.S21769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nkansah M. A., Boadi N. O., Badu M. Assessment of the quality of water from hand-dug wells in Ghana. Environmental Health Insights . 2010;4(2):p. EHI.S3149. doi: 10.4137/EHI.S3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nogueira G., Nakamura C. V., Tognim M. C. B., Filho B. A. A., Filho B. P. D. Microbiological quality of drinking water of urban and rural communities, Brazil. Rev Saúde Pública . 2003;37(2):232–236. doi: 10.1590/S0034-89102003000200011. [DOI] [PubMed] [Google Scholar]
- 51.Obiri-Danso C. A., Weobong C. A., Jones K. Aspects of health-related microbiology of the Subin, an urban river in Kumasi, Ghana. Journal of Water and Health . 2005;3(1):69–76. doi: 10.2166/wh.2005.0007. [DOI] [PubMed] [Google Scholar]
- 52.Odonkor S. T., Ampofo J. K. Escherichia coli as an indicator of bacteriological quality of water: an overview. Microbiology Research . 2013;4(1):2–e2. doi: 10.4081/mr.2013.e2. [DOI] [Google Scholar]
- 53.Odonkor S. T., Mahami T. _Escherichia coli_ as a Tool for Disease Risk Assessment of Drinking Water Sources. International Journal of Microbiology . 2020;2020:7. doi: 10.1155/2020/2534130.25434130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Odonkor S. T., Addo K. K. Prevalence of multidrug-resistant Escherichia coli isolated from drinking water sources. International journal of microbiology . 2018;2018:7. doi: 10.1155/2018/7204013.7204013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Omari S., Yeboah-Manu D. The study of bacterial contamination of drinking water sources: a case study of Mpraeso, Ghana. The Internet Journal of Microbiology . 2012;10(1):6–11. [Google Scholar]
- 56.Opintan J. A., Newman M. J. Prevalence of antimicrobial resistant pathogens from blood cultures: results from a laboratory based nationwide surveillance in Ghana. Antimicrobial Resistance & Infection Control . 2017;6(1):p. 64. doi: 10.1186/s13756-017-0221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osunla C., Okoh A. Vibrio pathogens: a public health concern in rural water resources in sub-Saharan Africa. International Journal of Environmental Research and Public Health . 2017;14(10):p. 1188. doi: 10.3390/ijerph14101188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Owusu P. A., Asumadu-Sarkodie S., Ameyo P. A review of Ghana’s water resource management and the future prospect. Cogent Engineering . 2016;3(1):p. 1164275. doi: 10.1080/23311916.2016.1164275. [DOI] [Google Scholar]
- 59.Pan M., Chu L. M. Occurrence of antibiotics and antibiotic resistance genes in soils from wastewater irrigation areas in the Pearl River Delta region, southern China. Science of the Total Environment . 2018;624:145–152. doi: 10.1016/j.scitotenv.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 60.Parveen S., Murphee R. L., Edmiton L., Kaspar C. W., Portier K. M., Tamplin M. L. Association of multiple-antibiotic resistance profiles with point and nonpoint sources of Escherichia coli in Apalachicola Bay. Applied and Environmental Microbiology . 1997;63(7):2607–2612. doi: 10.1128/aem.63.7.2607-2612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petit F., Clermont O., Delannoy S., et al. Change in the structure of Escherichia coli population and the pattern of virulence genes along a rural aquatic continuum. Frontiers in Microbiology . 2017;8:p. 609. doi: 10.3389/fmicb.2017.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Podschun R., Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clinical Microbiology Reviews . 1998;11(4):589–603. doi: 10.1128/CMR.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prabhu D., Pandian R. S., Vasan P. T. Pathogenicity, antibiotic susceptibility and genetic similarity of environmental and clinical isolates of Vibrio cholerae . 2007. [PubMed]
- 64.Qiao M., Ying G. G., Singer A. C., Zhu Y. G. Review of antibiotic resistance in China and its environment. Environment International . 2018;110:160–172. doi: 10.1016/j.envint.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 65.Quiroz K. L., Rodriguez N. G., Murinda S., Ibekwe M. Determination of the Water Quality of a Constructed Wetland Monitoring Fecal Indicator Bacteria . 2018.
- 66.Resende A. C. B., de Bastos Ascenso Soares R., Santos D. B., Montalvão E. R., do Carmo Filho J. R. Detection of antimicrobial-resistant Gram-negative bacteria in hospital effluents and in the sewage treatment station of Goiânia Brazil. O Mundo da Saúde . 2009;33(4):385–391. doi: 10.15343/0104-7809.20094385391. [DOI] [Google Scholar]
- 67.Rodrigues C., Cunha M. Â. Assessment of the microbiological quality of recreational waters: indicators and methods. Euro-Mediterranean Journal for Environmental Integration . 2017;2(1):p. 25. doi: 10.1007/s41207-017-0035-8. [DOI] [Google Scholar]
- 68.Schafhauser B. H., Kristofco L. A., de Oliveira C. M. R., Brooks B. W. Global review and analysis of erythromycin in the environment: occurrence, bioaccumulation and antibiotic resistance hazards. Environmental Pollution . 2018;238:440–451. doi: 10.1016/j.envpol.2018.03.052. [DOI] [PubMed] [Google Scholar]
- 69.Shahina J., Sandhiya D., Rafiq S. Bacteriological quality assessment of groundwater and surface water in Chennai. Nature Environment & Pollution Technology . 2020;19(1) [Google Scholar]
- 70.Tambekar D. H., Dhanorkar D. V., Gulhane S. R., Khandelwal V. K., Dudhane M. N. Antibacterial susceptibility of some urinary tract pathogens to commonly used antibiotics. African Journal of Biotechnology . 2006;5(17) [Google Scholar]
- 71.Team R. C. R: A Language and Environment for Statistical Computing . Vienna, Austria: R Foundation for Statistical Computing; 2018. [Google Scholar]
- 72.Wangai F. K., Masika M. M., Lule G. N., et al. Bridging antimicrobial resistance knowledge gaps: the East African perspective on a global problem. PLoS One . 2019;14(2, article e0212131) doi: 10.1371/journal.pone.0212131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.World Health Organization. Guidelines for Drinking-Water Quality: Volume 2: Surveillance and Control of Community Supplies . 1997.
- 74.Who. Guidelines for Drinking Water Quality: Recommendations . Vol. 1. Geneva: World Health Organization; 2004. [Google Scholar]
- 75.Who. Guidelines for Drinking- Water Quality: Incorporating First Addendum . 3rd. Vol. 1. Recommendations; 2006. [Google Scholar]
- 76.Wikler M. A. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: approved standard. CLSI (NCCLS) . 2006;26:M7–A7. [Google Scholar]
- 77.Xu L., Ouyang W., Qian Y., Su C., Su J., Chen H. High-throughput profiling of antibiotic resistance genes in drinking water treatment plants and distribution systems. Environmental Pollution . 2016;213:119–126. doi: 10.1016/j.envpol.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 78.Xue X., Cashman S., Gaglione A., et al. Holistic analysis of urban water systems in the Greater Cincinnati region:(1) life cycle assessment and cost implications. Water research X . 2019;2:p. 100015. doi: 10.1016/j.wroa.2018.100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yevutsey S. K., Buabeng K. O., Aikins M., et al. Situational analysis of antibiotic use and resistance in Ghana: policy and regulation. BMC Public Health . 2017;17(1):p. 896. doi: 10.1186/s12889-017-4910-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang C., Qiu S., Wang Y., et al. Higher isolation of NDM-1 producing Acinetobacter baumannii from the sewage of the hospitals in Beijing. PLoS One . 2013;8(6, article e64857) doi: 10.1371/journal.pone.0064857. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included within the article.


